Abstract
Cytotoxic T lymphocytes (CTLs) play a central role in the control of persistent human cytomegalovirus (HCMV) infection in healthy virus carriers. Previous analyses of the specificity of HCMV-reactive CD8+ CTLs drawn from in vitro models in which antigen-presenting cells were autologous fibroblasts infected with laboratory HCMV strains have shown focusing of CTL responses against the major tegument protein, pp65. By contrast, the 72-kDa major immediate-early protein (IE1) was identified as a minor target for this response. Here we have studied the fine specificity and T-cell-receptor features of T-cell clones generated against autologous B lymphoblastoid cell lines stably transfected with HCMV cDNA coding for either pp65 or a natural variant of IE1. This strategy allowed efficient generation of T-cell clones against IE1 and pp65 and led to the identification of several new IE1 and pp65 epitopes, including some located in polymorphic regions of IE1. Such an approach may provide relevant information about the characteristics of the CTL response to IE1 and the effect of viral polymorphism on the immune response against HCMV.
Human cytomegalovirus (HCMV), the largest of the betaherpesviruses, infects between 40 and 60% of individuals in developed countries and nearly 100% in developing countries. After primary infection, which is asymptomatic in immunocompetent hosts, the virus persists lifelong under the control of the immune system in latent sites, including cells of the myeloid lineage (20, 32, 34, 35). The maintenance of this latent state depends on the cellular immune response, and cytotoxic T lymphocytes (CTLs) have been shown to play a major role in the control of HCMV reactivation and replication (25, 29, 30, 38).
Due to inhibition of the major histocompatibility complex (MHC) class I antigen presentation pathway by a set of viral proteins acting in a cooperative and multistep process during the HCMV replication cycle (1, 3, 13, 15–18, 39, 41, 42), the CTL response against HCMV appears to be targeted against a small set of proteins (for a review, see reference 5). Among these have been identified, on the one hand, structural proteins which are delivered to the cytosol by the infecting virion and rapidly presented to CD8+ CTL by MHC class I before blockade of their processing and, on the other hand, the immediate-early proteins, the presentation of which depends on de novo synthesis very early after virus infection (15).
Early analyses of the CTL response in seropositive individuals have suggested that the 72-kDa immediate-early protein IE1 was a dominant target for CD8+ CTLs (4). Nevertheless, more recent observations, drawn from an in vitro experimental model with autologous fibroblasts infected with the cytopathic AD169 viral strain as a stimulator to propagate HCMV-specific CD8+ CTLs, have designated the major tegument protein pp65 as a dominant target and have shown a highly focused peptide-specific memory CTL response against few dominant epitopes (21, 28, 40, 43).
In the present study, we have developed a novel strategy allowing analysis of the composition and fine specificity of the CTL response of HCMV-seropositive healthy donors against selected immunogenic HCMV proteins. B lymphoblastoid cells (BLCs) were derived by Epstein-Barr virus (EBV) transformation from peripheral blood mononuclear cells (PBMCs) and stably transfected with plasmidic vectors coding for either pp65 or IE1 proteins. These cells were then used as targets to stimulate a specific autologous T-lymphocyte response and to amplify reactive T-cell subsets. After cloning, the cells recognizing the recombinant HCMV protein targets were selected, the characteristics of their TCR repertoire was studied by sequencing their T-cell-receptor β-chain junctions, and their fine specificity and HLA restriction were determined. This study led to the identification of several new class I MHC-restricted CTL epitopes against which were focused the anti-pp65 and -IE1 responses. Since pp65 and IE1 cDNA used in this study were cloned from a clinical strain, the IE1-expressed recombinant protein was a genetic variant which diverged by several amino acids from corresponding products of the laboratory strains Towne and AD169, classically used for analysis of memory CTL response to HCMV infection (4, 21, 28, 40, 43). Interestingly, several polymorphic residues were found within three of the four epitopes recognized by HCMV-specific T-cell clones generated against the clinical IE1 variant from a single donor (i.e., 22 of 22 clones restricted by HLA-A2 molecules and 23 of 25 characterized in the HLA-B18 context). Thus, the present experimental system represents a powerful tool for characterizing new target epitopes of the anti-HCMV T-cell response and should allow in-depth analysis of the effect of viral polymorphism on the modulation of the host repertoire.
MATERIALS AND METHODS
Cells.
PBMCs were prepared from fresh heparinized peripheral blood of three CMV+ healthy donors expressing known HLA class I haplotypes (donor A, A1A1 B8B35 Cw4Cw7; donors B and C, A2A25 B18B44 Cw5Cw12) by Ficoll-Hypaque (Eurobio, les Ulis, France) density gradient centrifugation. HLA class I genotyping was established in the HLA typing Laboratory of Centre Régional de Transfusion Sanguine de Nantes by PCR amplification techniques (6, 36). There was no parental link between donors B and C. All donors carried antibodies against EBV. BLC lines were established from PBMCs by EBV transformation as previously described (24).
Expression vectors.
IE1 and pp65 full-length coding sequences were amplified by reverse transcription (RT)-PCR from MRC-5 fibroblasts infected with a CMV strain selected from a clinical isolate, cloned in KS+ vectors (Stratagene, La Jolla, Calif.) and sequenced. The IE1 coding sequence used subsequently for expression exhibited nucleotidic variation when compared to the corresponding nucleotidic sequence of AD169 and Towne laboratory strains. All of these mutations were also found in two other IE1 PCR products independently amplified from the same clinical strain and then were due to natural variation. Such a polymorphism was not found in the pp65 coding sequence. IE1 and pp65 cDNAs were excised from KS+ and subcloned in pRc/CMV expression vector (Invitrogen Corp., San Diego, Calif.) under the control of CMV promoter.
Expression vectors containing various genomic DNA or cDNA encoding HLA class I alleles were used to determine the HLA restriction of IE1 or pp65 peptide recognition. The genomic HLA-A*0101 was kindly provided by J. Girdlestone (Medical Research Council, Cambridge, United Kingdom). HLA-B*0801 and -B*3501, a gift of L. Statz (Laboratory of Immunogenetics, Hospital de Clinicas Jose de San Martin, Buenos Aires, Argentina), and HLA-B18, a gift of E. Scotet (INSERM U463, Institut de Biologie, Nantes, France), were previously cloned into pcDNA3 (31). Genomic HLA-A*0201 cloned into pSV2 vector and HLA-B*4402 cDNA cloned into pcDNA3 were kindly supplied by T. Boon and P. Coulie (Ludwig Institute, Brussels, Belgium). The cDNA HLA-Cw4 cloned into pBK-RSV and the genomic DNA HLA-Cw7 cloned into pSV2neo were kindly furnished by M. Colonna (Basel Institute for Immunology, Basel, Switzerland).
Transfection of BLCs.
Five million actively dividing BLCs were resuspended in 0.8 ml of complete medium, mixed with 15 μg of vector, incubated for 1 min at room temperature, and electroporated in an Easyject Plus apparatus (Eurogentec, Seraing, Belgium). One pulse of 250 V/cm using a 1,500-μF capacitor was delivered. The cells were immediately plated with 5 ml of fresh medium. The same protocol was repeated three times to obtain 1.5 × 107 electroporated BLCs for each transfection assay. Selection with G418 (0.8 mg/ml) started 2 days after electroporation. After 1 week, dead cells and debris were eliminated by centrifugation through a Ficoll-Hypaque gradient, and the cells from the same three electroporations were pooled.
Generation of specific T lymphocytes.
Anti-pp65 and anti-IE1-specific T lymphocytes were amplified against BLCs expressing stably these viral recombinant proteins using the following protocol: donor PBMCs were plated at 2 × 106 cells per well in 24-well plates containing RPMI 1640 supplemented with 10% pooled human serum and 1 mM l-glutamine and then stimulated with 5 × 104 irradiated (30 Gy) autologous BLCs expressing either pp65 or IE1. After 9 days of coculture, 5 × 105 cells per well were restimulated with 1.25 × 105 autologous recombinant irradiated BLCs. Nine days later, recombinant interleukin-2 was added to the culture (20 IU/ml). Twenty days after the second stimulation, cells were cloned from the amplified lines by limiting dilution as previously described (37) at 0.3 cells per well. Growing colonies with a probability of monoclonality of >95% were kept for further analysis and expanded in vivo using standard conditions (9). Cells were maintained for 3 to 4 weeks without restimulation prior to functional analysis in order to decrease their spontaneous tumor necrosis factor (TNF) release and their nonspecific cytotoxic activity.
Flow cytometry analysis and functional assays.
Expression of pp65 and IE1 recombinant proteins was checked by flow cytometry in stably transfected BLCs permeabilized and then fixed as described previously (14). (1C3 plus AYM-1)-fluorescein isothiocyanate (FITC) monoclonal antibodies (MAbs) (Argene) were used to detect pp65, and E13-FITC MAb (Biosys) was used to detect IE1.
T cells were phenotyped by indirect fluorescence as previously described (37). Anti-coreceptor and -TCR MAb were purchased from Immunotech-Coulter Corp. (Marseille, France).
The proliferative and cytotoxic activity of T-cell clones against autologous irradiated BLCs stably transfected with pp65, IE1, or mock vector were estimated as previously described (8, 37).
HLA restriction characterization.
HLA restriction of T cells was characterized, as previously described (31), by testing their TNF content when added to COS cells cotransfected with an expression vector coding for their target viral protein and an expression vector coding for one of the HLA.
Characterization of target peptides.
Synthetic peptides (23-mer overlapping by 12 residues) encompassing the whole primary amino acid sequence of pp65 and IE1 were obtained from Chiron Technologies (Pty Ltd., Clayton, Victoria, Australia). The pp65 peptides are numbered 1 to 50, and IE1 peptides are numbered 1 to 44 from the N terminus to the C terminus. Minimal 9- or 10-mer peptides corresponding to proposed MHC-restricted epitopes were obtained from Genosys (Pampisford, United Kingdom).
Peptides recognized by T-cell clones directed against pp65 and IE1 were characterized by determining the TNF release in an autopresentation assay. Ten-thousand T cells were incubated for 2 h with 10 μM pp65 or IE1 individual synthetic peptides. The quantification of TNF release was determined as described earlier (31).
Analysis of TCR β transcripts.
T-cell-receptor β (TCR β) cDNAs were amplified by RT-PCR from total RNA of clones cDNAs as previously described (31) and then directly sequenced.
RESULTS
Amplification and cloning of anti-pp65 or IE1 CTLs.
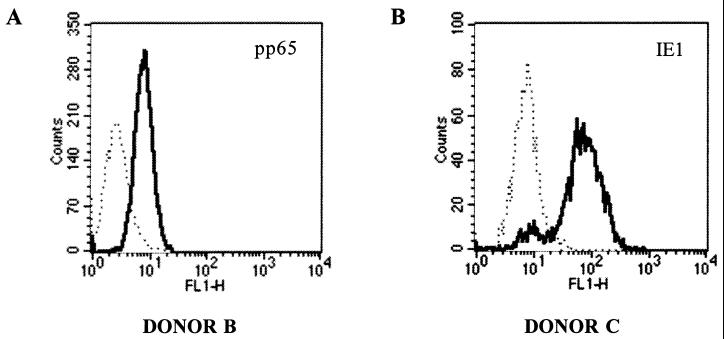
BLCs expressing stably pp65 were generated from donors A and B. BLCs expressing stably IE1 were generated from donor C (see, for example, Fig. 1). They were subsequently used as stimulators to amplify specific T cells from autologous PBMCs. The presence of specific CTLs in bulk cultures was checked using a technique in which their TNF release against COS cells cotransfected with pRc/CMV-pp65 or -IE1, and the DNA coding for individual HLA class I alleles was measured. Cells amplified from donors A and B were reactive against COS cells coexpressing pp65 and HLA-A1 or -A2 molecules, respectively. Furthermore, the bulk CTL line from donor C recognized COS cells expressing IE1 and either HLA-A2 or HLA-B18. Cell lines were cloned by limiting dilution, and colonies were tested for their reactivity against target cells expressing or not expressing recombinant pp65 or IE1 proteins.
FIG. 1.
Flow cytometry analysis of pp65 expression in BLCs of donor B stably transfected with pRc/CMV-pp65 ([MAb 1C3 plus AYM-1]-FITC) (A) and IE1 expression in BLCs of donor C stably transfected with pRc/CMV-IE1 (MAb E13-FITC) (B). The BLC lines transfected with pRc/CMV mock were used as negative controls (dotted lines).
Selection of T-lymphocyte clones reactive against either pp65 or IE1 and characterization of their HLA-restricting allele.
Two methods were used to screen for the specificity of T-cell clones derived from bulk cultures. Clones obtained from PBMCs stimulated against autologous BLCs expressing pp65 (donors A and B) were selected on the basis of their differential proliferative activities against autologous BLCs stably transfected with either pRc/CMV-pp65 or the empty pRc/CMV control. CD8+ CTL clones obtained from PBMCs stimulated against autologous BLCs expressing IE1 (donor C) were selected using a COS cell cotransfection assay which allowed us to characterize at the same time the HLA molecules restricting the presentation of HCMV peptides.
From 70 T-cell clones generated from donor A, a single clone (A18) proliferating against autologous targets expressing pp65 was isolated. Clone A18 recognized specifically pp65 when expressed in COS cells together with the HLA-A1 allele. From 75 clones generated from donor B, 2 clones (B3 and B20) were shown to recognize pp65-transfected BLCs. These two clones recognized pp65 in the HLA-A2 context, as indicated by COS screening assays. Of the 75 T-cell clones generated from donor C, 22 were reactive against IE1 in the HLA-A2 context and 26 were reactive in HLA-B18 context (Table 1).
TABLE 1.
Amino acid sequence of TCR β-chain junction of T-cell clones derived from three HCMV+ donors and recognizing either pp65 or IE1a
| Donor | HLA restriction | Amino acid position in:
|
TCR β-chain sequence
|
No. of clones | |||||
|---|---|---|---|---|---|---|---|---|---|
| IE1 | pp65 | Vβ | CDR3 | Jβ | |||||
| A | A1 | 353–375 | Vβ1S1 | CAS | SPRGFDAGGYWLD | LRF | Jβ1.2 | 1 | |
| Total | 1 | ||||||||
| B | A2 | 485–507 | Vβ6S4A1 | CAS | SYGQNPLNTE | AFF | Jβ1.1 | 1 | |
| B | A2 | 485–507 | Vβ14S1 | CAS | SLEGYTE | AFF | Jβ1.1 | 1 | |
| Total | 2 | ||||||||
| C | A2 | 309–331 | Vβ1S1 | CAS | SVDGTGGALGNT | IYF | Jβ1.3 | 1 | |
| C | A2 | 309–331 | Vβ4S1 | CSV | EXSPPGEEWWLT | LRF | Jβ1.6 | 1 | |
| C | A2 | 309–331 | Vβ210S1 | CAW | SWGDSTNSP | LHF | Jβ1.6 | 19 | |
| C | A2 | 309–331 | Vβ21S3 | CAS | SLVGGTGSVSY | QFF | Jβ2.1 | 1 | |
| Total | 22 | ||||||||
| C | B18 | 188–210, 199–221 | Vβ3S1 | CAS | SLLMATNEK | LFF | Jβ1.4 | 1 | |
| C | B18 | 188–210, 199–221 | Vβ6S5 | CAS | SYRANYE | QYF | Jβ2.7 | 2 | |
| C | B18 | 188–210, 199–221 | Vβ9S2 | CAS | SQAVASGGGRSE | QFF | Jβ2.1 | 1 | |
| C | B18 | 188–210, 199–221 | Vβ22S1 | CAS | VTGSTDT | QYF | Jβ2.3 | 7 | |
| C | B18 | 188–210, 199–221 | Vβ22S1 | CAS | SDGTAYE | QYF | Jβ2.7 | 10 | |
| C | B18 | 265–287 | Vβ7S1 | CAS | SLARNQP | QHF | Jβ1.5 | 1 | |
| C | B18 | 265–287 | Vβ17S1 | CAS | GGADMKTE | AFF | Jβ1.1 | 1 | |
| C | B18 | 375–397 | Vβ2S1 | CSA | NPDWGLQLWLH | LRF | Jβ2.1 | 1 | |
| C | B18 | 375–397 | Vβ6S4 | CAS | SPGGGKET | QYF | Jβ2.5 | 1 | |
| C | B18 | ND | Vβ6S4 | CAS | RLRGSSSYNE | QFF | Jβ2.1 | 1 | |
| Total | 26 | ||||||||
The recognized epitope and restricting HLA molecule are given for each clone. The total number of anti-pp65 or IE1 T-cell clones restricted by a given HLA context for each donor is in boldface. ND, not determined.
All three anti-pp65 clones and the 48 anti-IE1 selected T-cell clones specifically lysed pp65- or IE1-transfected BLCs, respectively, but not the mock-transfected autologous BLCs. As expected, when analyzed by flow cytometry, all clones selected from the three donors were TCR αβ+ and CD8+.
Analysis of the fine epitope specificity of pp65- and IE1-reactive CTL clones.
The fine specificity of HCMV-reactive T-cell clones was studied by screening their reactivity to synthetic peptides encompassing the whole IE1 and pp65 proteins. The results from such screenings are summarized in Table 1. The donor A-derived clone A18 recognized the pp65353–375 peptide in the HLA-A1 context. The two clones B3 and B20 were specific for the HLA-A2-restricted pp65485–507 peptide. The anti-IE1 response of donor C-derived T cells was focused against the HLA-A2-restricted IE1309–331 peptide and a dominant B18-restricted peptide located within the IE1188–210-IE1199–221 overlapping peptidic region, which was recognized by 21 of 26 T-cell clones. Nine-mer peptides recognized in the HLA-A2 and HLA-B18 contexts by T-cell clones from donor C were further characterized through screening of an additional set of peptides (Table 2).
TABLE 2.
Nine-mer peptides recognized by the anti-IE1 clones generated from donor C
| Amino acid sequencea | Amino acid position in IE1 | Restricting HLA allele | No. of clones |
|---|---|---|---|
| YILEETSVM | 315–323 | A2 | 22 |
| ELKRKMIYM | 199–207 | B18 | 21 |
| CVETMCNEY | 279–287 | B18 | 2 |
| DEEDAIAAY | 379–387 | B18 | 2 |
Target residues for natural polymorphism are indicated in boldface.
TCR β-chain features of IE1- and pp65-reactive T-cell clones.
TCR Vβ gene usage by anti-pp65 and -IE1 CTLs generated from the three HCMV+ donors was first studied by flow cytometry using a panel of 22 TCR Vβ-specific MAbs (results are summarized in Table 1). The clone generated from donor A against pp65 expressed the Vβ1 segment. Of the two clones generated from donor B against pp65, one expressed the Vβ6 segment and the other expressed the Vβ14 segment. Of the 48 clones generated from donor C, 19 of 22 clones (i.e., 86%) recognizing the 9-mer IE1315–323 peptide in the HLA-A2 context expressed the TCR Vβ20 gene segment and 17 of 26 clones (i.e., 63%) recognizing IE1 in the HLA-B18 context expressed the Vβ22 gene segment. All of these Vβ22+ T-cell clones recognized the IE1199–207 epitope.
TCR β-chain sequencing of anti-pp65 and -IE1 CTL clones is presented in Table 1. All Vβ20 T-cell clones reactive against the IE1–HLA-A2 epitope exhibited identical junctional characteristics (i.e., Jβ1.6 segment and identical CDR3 sequence), suggesting that they were the progeny of a single clone. Similarly, of 17 CTL clones restricted by the HLA-B18 molecule and expressing the Vβ22 gene segment, 10 exhibited the Jβ2.7 segment and identical junctional features. The seven other clones used the Jβ2.3 segment and showed identical junctional sequences. All of these 17 clones exhibited conserved length of their CDR3 loop and a shared NDN-encoded Gly at a conserved position within the CDR3 loop.
DISCUSSION
In this report, we present data on TCR β-chain usage and the fine specificity of anti-HCMV CD8+ CTL clones obtained through stimulation of PBMCs from HCMV-seropositive individuals against pp65- and IE1-BLC transfectants.
Although recombinant proteins were similarly expressed in the stably transfected BLC stimulator cells, generation of T-cell clones reactive against pp65 was less efficient than against IE1. Such a result is in disagreement with those obtained by other authors who used a fibroblast infection model to generate anti-CMV CTLs (21, 28, 40, 43), but it is consistent with more recent data (19, 31). In an infection model, (i) the substantial level of dense bodies that carry only tegument proteins in laboratory strains (for a review, see reference 23), (ii) blockade of IE1 processing and presentation due to its phosphorylation (12), and (iii) downregulation of MHC class I expression (1, 3, 13, 15–18, 39, 41, 42) may together lead to an underestimation of the importance of CTL response against IE1. However, we cannot exclude that, in our expression model, we underestimated the CTL response against pp65.
The TCR repertoire of IE1-specific T-cell clones identified in donor C was highly heterogeneous, showing that the response to CMV proteins is more diversified, as has already been suggested by others (40, 43). In our experiments, we only stimulated donors A and B with pp65 and donor C with IE1, but conclusive information should be obtained from further investigations by using our antigenic presentation strategy to generate CTLs against both IE1 and pp65 from PBMCs of an HCMV-seropositive individual.
The clones generated from donors A and B against autologous BLCs expressing pp65 that were not specific of this HCMV product proliferated mainly against autologous untransfected BLCs and were then probably directed against EBV antigens (data not shown). Such a result is consistent with a very recent report showing that BLCs transduced with a recombinant retrovirus encoding pp65 can be used as antigen-presenting cells to stimulate expansion of EBV- and CMV-specific CTLs simultaneously (33).
Amplification of anti-EBV CD4+ and CD8+ CTLs against BLC targets has been already described (reference 22 and our own results). Therefore, CD4+ anti-HCMV clones should be theoretically generated against BLCs expressing recombinant HCMV proteins. Other reports have shown a specific CD4+ T-cell response against IE1 in HCMV-seropositive individuals (2, 4, 10). Further investigations are necessary to obtain conclusive data regarding the efficiency of CD4+ T-lymphocyte-specific amplifications when the antigens are nuclear proteins synthesized by presenting BLCs.
Of the six epitopes characterized in this study, four were new. The pp65485–507 peptide recognized in the HLA-A2 context contained the 495-NLVPMVATV-503 9-mer epitope, which had been identified in previous studies as an immunodominant epitope (40, 43). This minimal epitope was recognized by the two clones generated from donor B against pp65 (data not shown). Concerning IE1, the 9-mer peptide 379-DEEDAIAAY-387, recognized in the B18 restricted context, was internal to a 12-mer epitope 378-SDEEEAIVAYTL-389 previously characterized in the same HLA context but which differed from its sequence by two residues (11).
The sequences of IE1 have long been assumed to be well preserved from genetic variation. Nevertheless, we and others recently described polymorphic regions in the coding sequence of this viral protein (7, 27, 44) which could interfere with presentation by MHC molecules and/or recognition by the immune system. Our data suggest that a large part of the memory CD8 CTL response against IE1 protein cloned from a clinical strain is directed against polymorphic epitopes restricted by two HLA molecules. A single residue exchange within a viral CTL epitope could later recognition or destruction by specific proteasomal cleavage, resulting in a lack of antigen presentation (26). Important information on the role possibly played by the IE1 natural polymorphism on the modulation of the anti-HCMV memory repertoire should be obtained from analysis of the T-cell-clone reactivity against target cells expressing IE1 proteins derived from various clinical strains or loaded with modified peptides.
In summary, the results presented here show that IE1-specific CTL clones can be efficiently generated through stimulation of PBMCs against autologous IE1 BLC transfectants. Study of the TCR and antigenic characteristics of CD8+ CTL clones obtained through this technique should help us analyze the role of IE1 in the control of viral reactivation and infection in HCMV-seropositive individuals with various HLA genotypes and to assess the impact of its peptidic polymorphism on the modulation of the host response against numerous HCMV isolates.
ACKNOWLEDGMENTS
This work was supported by the Institute de la Santé et de la Recherche Médicale and by the Association pour la Recherche sur le Cancer.
REFERENCES
- 1.Ahn K, Früh K. Human cytomegalovirus inhibits antigen presentation by a sequential multistep process. Proc Natl Acad Sci USA. 1996;93:10990–10995. doi: 10.1073/pnas.93.20.10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alp N J, Allport T D, Van Zanten J, Rodgers B, Sissons J G P, Borysiewicz L K. Fine specificity of cellular immune responses in humans to human cytomegalovirus immediate-early 1 protein. J Virol. 1991;65:4812–4820. doi: 10.1128/jvi.65.9.4812-4820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beersma M F C, Bijlmakers M J E, Ploegh H L. Human cytomegalovirus down-regulates HLA class I expression by reducing the stability of class I H chains. J Immunol. 1993;151:4455–4464. [PubMed] [Google Scholar]
- 4.Borysiewicz L K, Hickling J K, Graham S, Sinclair J, Cranage M P, Smith G L, Sissons J G P. Human cytomegalovirus-specific cytotoxic T cells. Relative frequency of stage-specific CTL recognizing the 72-kD immediate early protein and glycoprotein B expressed by recombinant vaccinia viruses. J Exp Med. 1988;168:919–931. doi: 10.1084/jem.168.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Britt W J, Alford C A. Cytomegalovirus. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2493–2522. [Google Scholar]
- 6.Cereb N, Maye P, Lee S, Kong Y, Yang S Y. Locus-specific amplification of HLA class I genes from genomic DNA: locus-specific sequences in the first and third introns of HLA-A, -B and -C alleles. Tissue Antigens. 1995;45:1–11. doi: 10.1111/j.1399-0039.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- 7.Chou S. Effect of interstrain variation on diagnostic DNA amplification of the cytomegalovirus major immediate-early gene region. J Clin Microbiol. 1992;30:2307–2310. doi: 10.1128/jcm.30.9.2307-2310.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davodeau F, Peyrat M A, Hallet M M, Gaschet J, Houde I, Vivien R, Vié H, Bonneville M. Close correlation between Daudi and mycobacterial antigen recognition by human γδ T cells and expression of V9JPC1γ/V2DJCδ-encoded T cell receptors. J Immunol. 1993;151:1214–1223. [PubMed] [Google Scholar]
- 9.Davodeau F, Peyrat M A, Gaschet J, Hallet M M, Triebel F, Vié H, Kabelitz D, Bonneville M. Surface expression of functional T cell receptor chains formed by interlocus recombination on human T lymphocytes. J Exp Med. 1994;180:1685–1691. doi: 10.1084/jem.180.5.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gautier N, Chavant E, Prieur E, Monsarrat B, Mazarguil H, Davrinche C, Gairin J E, Davignon J L. Characterization of an epitope of the human cytomegalovirus protein IE1 recognized by a CD4+ T cell clone. Eur J Immunol. 1996;26:1110–1117. doi: 10.1002/eji.1830260523. [DOI] [PubMed] [Google Scholar]
- 11.Gavin M A, Gilbert M J, Riddell S R, Greenberg P D, Bevan M J. Alkali hydrolysis of recombinant proteins allows for the rapid identification of class I MHC-restricted CTL epitopes. J Immunol. 1993;151:3971–3980. [PubMed] [Google Scholar]
- 12.Gilbert M J, Riddell S R, Platcher B, Greenberg P D. Cytomegalovirus selectively blocks antigen processing and presentation of its immediate-early gene product. Nature. 1996;383:720–722. doi: 10.1038/383720a0. [DOI] [PubMed] [Google Scholar]
- 13.Hengel H, Monburg F. A viral ER-resident glycoprotein inactivates the MHC-encoded peptide transporter. Immunity. 1997;6:623–632. doi: 10.1016/s1074-7613(00)80350-7. [DOI] [PubMed] [Google Scholar]
- 14.Imbert-Marcille B M, Robillard N, Poirier A S, Coste-Burel M, Cantarovich D, Milpied N, Billaudel S. Development of a method for direct quantification of cytomegalovirus antigenemia by flow cytometry. J Clin Microbiol. 1997;35:2665–2669. doi: 10.1128/jcm.35.10.2665-2669.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson D C, Hill A B. Herpesvirus evasion of the immune system. Curr Top Microbiol Immunol. 1998;232:149–177. doi: 10.1007/978-3-642-72045-1_8. [DOI] [PubMed] [Google Scholar]
- 16.Jones T R. Multiple independent loci within the HCMV unique short region downregulate expression of major histocompatibility complex class I heavy chains. J Virol. 1995;69:4830–4841. doi: 10.1128/jvi.69.8.4830-4841.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones T R, Ploegh H L. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc Natl Acad Sci USA. 1996;93:11327–11333. doi: 10.1073/pnas.93.21.11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones R, Sun L. Human CMV US2 destabilizes major histocompatibility complex class I heavy chains. J Virol. 1997;71:2970–2979. doi: 10.1128/jvi.71.4.2970-2979.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kern F, Surel I P, Faulhaber N, Frömmel C, Schneider-Mergener J, Schönemann C, Reinke P, Volk H-D. Target structures of the CD8+ T-cell response to human cytomegalovirus: the 72-kilodalton major immediate-early protein revisited. J Virol. 1999;73:8179–8184. doi: 10.1128/jvi.73.10.8179-8184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo K, Kaneshima H, Mocarski E S. HCMV latent infection of granulocyte-macrophage progenitors. Proc Natl Acad Sci USA. 1994;91:11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLaughlin-Taylor E, Pande H, Forman S J, Tanamachi B, Li C R, Zaia J A, Greenberg P D, Riddell S R. Identification of the major late human cytomegalovirus matrix protein pp65 as a target antigen for CD8+ virus-specific cytotoxic T lymphocytes. J Med Virol. 1994;43:103–110. doi: 10.1002/jmv.1890430119. [DOI] [PubMed] [Google Scholar]
- 22.Misko I S, Sculley T B, Schmidt C, Moss D J, Soszynski T, Burman K. Composite response of naive T cells to stimulation with the autologous lymphoblastoid cell line is mediated by CD4 cytotoxic T cell clones and induces an Epstein-Barr virus-specific component. Cell Immunol. 1991;132:295–307. doi: 10.1016/0008-8749(91)90029-b. [DOI] [PubMed] [Google Scholar]
- 23.Mocarski E S. Cytomegaloviruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2447–2492. [Google Scholar]
- 24.Moreau J F, Bonneville M, Peyrat M A, Godard A, Jacques Y, Desgranges C, Soulillou J P. T lymphocytes cloning from rejected human kidney allografts. J Clin Investig. 1986;78:874–879. doi: 10.1172/JCI112674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinnan G V, Burns W H, Kirmani N, Rook A H, Manischewitz J, Jackson L, Santos G W, Saral R. HLA-restricted cytotoxic T lymphocytes are an early immune response and important defense mechanism in cytomegalovirus infections. Rev Infect Dis. 1984;6:156–162. doi: 10.1093/clinids/6.2.156. [DOI] [PubMed] [Google Scholar]
- 26.Ossendorp F, Eggers M, Neisig A, Ruppert T, Groettrup M, Sijts A, Mengedé E, Kloetzel P-M, Neefjes J, Koszinowski U, Melief C. a single residue exchange within a viral CTL epitope alters proteasome-mediated degradation resulting in lack of antigen presentation. Immunity. 1996;5:115–124. doi: 10.1016/s1074-7613(00)80488-4. [DOI] [PubMed] [Google Scholar]
- 27.Retière C, Imbert B M, David G, Courcoux P, Hallet M M. A polymorphism in the major immediate-early gene delineates groups among cytomegalovirus clinical isolates. Virus Res. 1998;57:43–51. doi: 10.1016/s0168-1702(98)00083-5. [DOI] [PubMed] [Google Scholar]
- 28.Riddell S R, Rabin M, Geballe A P, Britt W J, Greenberg P D. Class I MHC-restricted cytotoxic T lymphocyte recognition of cells infected with human cytomegalovirus does not require endogenous viral gene expression. J Immunol. 1991;146:2795–2804. [PubMed] [Google Scholar]
- 29.Riddell S R, Watanabe K S, Goodrich J M, Li C R, Agha E, Greenberg P D. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science. 1992;257:238–241. doi: 10.1126/science.1352912. [DOI] [PubMed] [Google Scholar]
- 30.Riddell S R, Greenberg P D. Therapeutic reconstitution of human viral immunity by adoptive transfer of cytotoxic T lymphocyte clones. Curr Top Microbiol Immunol. 1994;189:9–34. doi: 10.1007/978-3-642-78530-6_2. [DOI] [PubMed] [Google Scholar]
- 31.Scotet E, Peyrat M A, Saulquin X, Retière C, Couedel C, Davodeau F, Dulphy N, Toubert A, Bignon J D, Lim A, Vié H, Hallet M M, Liblau R, Weber M, Berthelot J M, Houssaint E, Bonneville M. Frequent enrichment for CD8 T cells reactive against common herpes viruses in chronic inflammatory lesions: toward a reassessment of the physiopathological significance of T cell clonal expansions found in autoimmune inflammatory processes. Eur J Immunol. 1999;29:973–985. doi: 10.1002/(SICI)1521-4141(199903)29:03<973::AID-IMMU973>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 32.Söderberg-Nauclér C, Fish K N, Nelson J A. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell. 1997;91:119–126. doi: 10.1016/s0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 33.Sun Q, Pollock K E, Burton R L, Dai L J, Britt W, Emmanuel D J, Lucas K G. Simultaneous ex vivo expansion of cytomegalovirus and Epstein-Barr virus-specific cytotoxic T lymphocytes using B-lymphoblastoid cell lines expressing cytomegalovirus pp65. Blood. 1999;94:3242–3250. [PubMed] [Google Scholar]
- 34.Taylor-Wiedeman J, Sinclair J H. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J Gen Virol. 1991;72:2059–2064. doi: 10.1099/0022-1317-72-9-2059. [DOI] [PubMed] [Google Scholar]
- 35.Taylor-Wiedeman J, Sissons P, Sinclair J. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J Virol. 1994;68:1597–1604. doi: 10.1128/jvi.68.3.1597-1604.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tonks S, Marsh S G E, Bunce M, Moses J H, Krausa P, Slader A M, Petronzelli F, Bodmer J G. HLA class I DNA typing study. In: Charron D, editor. Genetic diversity of HLA: functional and medical implication. Paris, France: EDK; 1997. pp. 199–215. [Google Scholar]
- 37.Vié H, Chevalier S, Garand R, Moisan J P, Praloran V, Devilder M C, Moreau J F, Soulillou J P. Clonal expansion of lymphocytes bearing the gamma/delta receptor in a patient with a large granular lymphocyte disorder. Blood. 1989;74:285–290. [PubMed] [Google Scholar]
- 38.Walter E A, Greenberg P D, Gilbert M J, Finch R J, Watanabase K S, Thomas E D, Ridell S R. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N Engl J Med. 1995;333:1038–1044. doi: 10.1056/NEJM199510193331603. [DOI] [PubMed] [Google Scholar]
- 39.Warren A P, Ducroq D H, Lehner P J, Borysiewicz L K. Human cytomegalovirus-infected cells have unstable assembly of major histocompatibility complex class I complexes and are resistant to lysis by cytotoxic T lymphocytes. J Virol. 1994;68:2822–2829. doi: 10.1128/jvi.68.5.2822-2829.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weekes M P, Wills M R, Mynard K, Carmichael A J, Sissons J G P. The memory cytotoxic T-lymphocyte (CTL) response to human cytomegalovirus infection contains individual peptide-specific CTL clones that have undergone extensive expansion in vivo. J Virol. 1999;73:2099–2108. doi: 10.1128/jvi.73.3.2099-2108.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiertz E J H J, Ploegh H L. The human CMV US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 42.Wiertz E J H J, Ploegh H L. Sec-61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 43.Wills M R, Carmichael A J, Mynard K, Jin X, Weekes M P, Platcher B, Sissons J G P. The human cytotoxic T-lymphocyte (CTL) response to CMV is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J Virol. 1996;70:7569–7579. doi: 10.1128/jvi.70.11.7569-7579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wirgart B Z, Brytting M, Linde A, Wahren B, Grillner L. Sequence variation within three important cytomegalovirus gene regions in isolates from four different patient populations. J Clin Microbiol. 1998;36:3662–3669. doi: 10.1128/jcm.36.12.3662-3669.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]