Abstract
In a study of the evolution and distribution of avian retroviruses, we found avian sarcoma and leukosis virus (ASLV) gag genes in 26 species of galliform birds from North America, Central America, eastern Europe, Asia, and Africa. Nineteen of the 26 host species from whom ASLVs were sequenced were not previously known to contain ASLVs. We assessed congruence between ASLV phylogenies based on a total of 110 gag gene sequences and ASLV-host phylogenies based on mitochondrial 12S ribosomal DNA and ND2 sequences to infer coevolutionary history for ASLVs and their hosts. Widespread distribution of ASLVs among diverse, endemic galliform host species suggests an ancient association. Congruent ASLV and host phylogenies for two species of Perdix, two species of Gallus, and Lagopus lagopus and L. mutus also indicate an old association with vertical transmission and cospeciation for these ASLVs and hosts. An inference of horizontal transmission of ASLVs among some members of the Tetraoninae subfamily (grouse and ptarmigan) is supported by ASLV monophyletic groups reflecting geographic distribution and proximity of hosts rather than host species phylogeny. We provide a preliminary phylogenetic taxonomy for the new ASLVs, in which named taxa denote monophyletic groups.
Most work on avian retroviruses has focused on the avian type C retrovirus group also known as alpharetroviruses or avian sarcoma and leukosis viruses (ASLVs). ASLVs are known predominantly from the domestic chicken (Gallus gallus) and include both endogenous and exogenous forms. Based on envelope properties and env gene sequence similarities, ASLVs have been placed in nine different subgroups (13, 24). Variable presence or absence of one of the endogenous ASLVs, Rous-associated virus-0 (RAV-0), was examined in seven species from the avian order Galliformes (including the chicken), leading to the conclusion that RAV-0-related viruses have infected the germ line of galliform birds on multiple independent occasions relatively recently (10, 25). ASLV-related retroviruses that infect birds include the endogenous avian retroviruses (EAVs), the E51 group, and avian retrotransposons from chickens (ART-CHs). In contrast to RAV-0, EAVs seem to have infected an ancestral Gallus lineage prior to speciation and to have subsequently cospeciated with their hosts (2). The phylogenetic distribution of ART-CHs remains to be determined. Other retroviruses infecting birds appear to be only distantly related to ASLVs, and these include the reticuloendotheliosis viruses, which are more closely related to the mammalian type C retroviruses, and disparate retroviruses recently sequenced for reverse transcriptase and protease genes from representative members of the Passeriformes, Anseriformes, Columbiformes, and Tinamiformes (11, 17, 18).
Relatively little is known about avian retrovirus diversity, evolution, and host species range outside of a few such studies focusing on domesticated species. It is increasingly evident, however, that an understanding of retroviral origins, life histories, and mechanisms of transmission requires a greater knowledge of the diversity and distribution of retroviruses in nondomesticated host taxa. This includes a need for well-corroborated retrovirus and host species phylogenies, because an assessment of their phylogenetic congruence can aid in the discovery of the relative frequency of horizontal retrovirus transmission among host species compared to vertical transmission and cospeciation of retroviruses and host taxa.
In this study, we report on new ASLVs discovered in 26 species of birds in the order Galliformes, representing three families and 14 genera. Galliformes are medium- to large-sized birds, including commercially important members such as the domestic chicken and game birds such as grouse and pheasants; the order consists of about 280 species with worldwide distribution. We compared ASLV phylogenies based on gag gene sequences and host phylogenies based on mitochondrial 12S and ND2 sequences and found that ASLV dispersal among hosts, ancient infection followed by cospeciation, as well as duplication of ASLV elements within host species all played a role in ASLV and host coevolution. We also provide a preliminary phylogenetic taxonomy for the new ASLVs, in which named taxa denote monophyletic groups.
MATERIALS AND METHODS
DNA preparation.
Combined nuclear and mitochondrial genomic DNA was extracted from tissue (muscle, liver, and heart) using a QIAamp Tissue Kit (Qiagen) and the manufacturer's recommended protocols for a total of 60 species representing 19 avian orders. Positive results were only found in the order Galliformes, which is the focus of this report. We sampled 31 species or subspecies representing each of the five recognized families in the order Galliformes (Phasianidae, Numididae, Cracidae, Odontophoridae, and Megapodiidae). A total of one individual from 26 species, two individuals from four species, and four individuals from one species were sampled (Table 1).
TABLE 1.
Host species, genetic distances, and geographic information for ASLV gag genes
| Avian host species (family or subfamily) | Clone abbreviationc | Common name | No. of clones sequenced | Mean % pairwise distance between clones | Geographic distribution of host | Collection locality (state or country) |
|---|---|---|---|---|---|---|
| Phasianidae | ||||||
| Bambusicola thoracica | BATH | Chinese bamboo-partridge | 5 | 3.01 | China | Japan (introduced) |
| Gallus gallusa (2)b | GAGA | Domestic chicken | 8 | 2.21 | Southeast Asia | Captive |
| Gallus variusa (2) | GAVA | Green jungle fowl | 7 | 0.55 | Java | Texas (captive) |
| Perdix dauurica | PEDA | Daurian partridge | 2 | 2.12 | Palearctic | Buraytiya, Russia |
| Perdix perdixa | PEPE | Grey partridge | 3 | 2.02 | Palearctic | Astrakhan, Russia |
| Phasianus colchicus strauchia (2) | PHCO | Ring-necked pheasant | 8 | 4.17 | China | China (captive) |
| Coturnix ypsilophora australis | Brown quail | Australia | Australia | |||
| Coturnix coturnix | Common quail | South Africa | South Africa | |||
| Francolinus swainsonii | FRSW | Swainson's spurfowl | NCd | South Africa | South Africa | |
| Tetraoninae | ||||||
| Bonasa bonasia | BOBO | Hazel grouse | 5 | 1.85 | Palearctic | Sakhalin, Russia |
| Bonasa sewerzowi | BOSE | Chinese grouse | 4 | 0.47 | Palearctic | China |
| Bonasa umbellus (4) | BOUM | Ruffed grouse | 10 | 2.48 | Neartic | Michigan |
| Centrocercus urophasianus | CEUR | Sage grouse | 9 | 5.15 | Neartic | Colorado |
| Dendragapus canadensis franklinii | DEFR | Franklin's (spruce) grouse | 3 | 4.34 | Neartic | Washington |
| Dendragapus canadensis | DECA | Spruce grouse | 2 | 0.31 | Neartic | Alaska |
| Dendragapus falcipennis | DEFA | Siberian grouse | 2 | 2.79 | Palearctic | Khabarovsk, Russia |
| Dendragapus obscurus | DEOB | Blue grouse | 4 | 5.00 | Neartic | Colorado |
| Lagopus lagopusa | LALA | Willow ptarmigan | 4 | 9.13 | Holarctic | Sakhalin, Russia |
| Lagopus leucurus | LALE | White-tailed ptarmigan | 4 | 5.31 | Nearctic | Washington |
| Lagopus mutus (2) | LAMU | Rock ptarmigan | 7 | 6.59 | Holarctic | Magadan, Russia; Alaska |
| Tetrao parvirostris | TEPA | Black-billed capercaillie | 3 | 1.03 | Palearctic | Magadan, Russia |
| Tetrao tetrix | TETE | Black grouse | NCd | Palearctic | Moscow, Russia | |
| Tympanuchus cupido attwateri | TYCU | Greater prairie chicken | 6 | 3.50 | Nearctic | Texas |
| Tympanuchus pallidicinctus | TYPA | Lesser prairie chicken | NCd | Nearctic | New Mexico | |
| Tympanuchus phasianellus | TYPH | Sharp-tailed grouse | NCd | Nearctic | Idaho | |
| Odontophoridae | ||||||
| Colinus cristatusa | COCR | Crested bobwhite | 2 | 2.82 | South America | Nicaragua |
| Colinus virginianus | COVI | Northern bobwhite | 4 | 1.75 | Nearctic | Nebraska |
| Callipepla californica | CACA | California quail | NCd | California | California | |
| Megapodiidae | ||||||
| Leipoa ocellata | Malleefowl | Australia | Australia | |||
| Numididae | ||||||
| Numida meleagrisa | NUME | Helmeted guinea fowl | NCd | South Africa | South Africa | |
| Cracidae | ||||||
| Ortalis cinereiceps | Grey-headed chachalaca | Central America | Nicaragua |
Numbers in parentheses indicate number of individuals sampled if greater than one.
No abbreviation indicates that the individuals tested are negative for retroviral sequences using our primers. Additional birds testing negative for gag are: Struthioniformes: Rhea americana, Struthio camelus, and Dromaius novaehollandiae; Tinamiformes: Eudromia elegans; Procellariiformes: Diomedea nigripes; Sphenisciformes: Aptenodytes patagonicus; Gaviiformes: Gavia immer and Gavia pacifica; Pelecaniformes: Phalacrocorax pelagicus and Phaethon rubricauda; Ciconiiformes: Mycteria americana, Nyctanassa violacea, Eudocimus albus, and Scopus umbretta; Falconiformes: Falco peregrinus and Accipiter cooperii; Anseriformes: Aythya americana; Gruiformes: Grus canadensis, Fulica americana, and Cepphus columba; Columbiformes: Zenaida macroura; Psittaciformes: Cacatua goffini; Cuculiformes: Tauraco hartlaubi, Coccyzus erythropthalmus, and Coccyzus americanus; Caprimulgiformes: Chordeiles minor; Apodiformes: Aeronautes saxatalis; Coliiformes: Colius striatus; Trogoniformes: Trogon curucui; Piciformes: Sphyrapicus varius.
Sequenced to identify ASLVs but not cloned.
Retrovirus PCR and direct sequencing.
PCR was performed on avian genomic DNA by using two sets of primers designed to match conserved regions of various published Gallus gallus retrovirus gag sequences. Primer locations within gag are illustrated in Fig. 1 and have the following sequences: GAG.F1, 5′-GCCGTCATAAAGGTGATTTCGTC-3′; GAG.R1, 5′-TCAATTTTGGCTCCAGAGGGGTC-3′; GAG.F2, 5′-TGACTGGGCRAGGRTYAGGG-3′; and GAG.R2, 5′-AAGGACTCAGATGGTCCCTG-3′. GAG.R2 was designed based on the major homology region of gag which appears to be conserved across all retroviruses except spumaviruses. In most cases, GAG.F1 was paired with GAG.R2 to amplify a predicted 1.2-kb fragment. gag was chosen for its apparent intermediate rate of sequence evolution (19), being conserved enough to yield information from sampling of diverse host species and variable enough to provide sequence differences among conspecific hosts. We also chose to work with gag because some replication-defective ASLVs are known to lack the pol gene (e.g., Fujinami sarcoma virus (FUSV) and avian myelocytomatosis virus-29) and we wanted to be able to place our findings in a phylogenetic context that included diverse, published ASLV sequences.
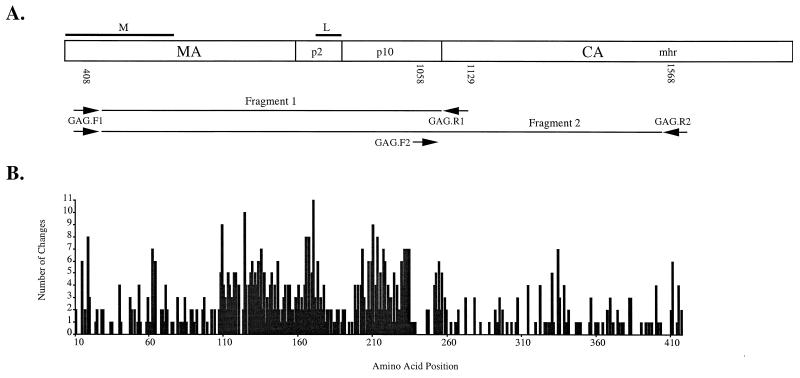
FIG. 1.
Primer locations within avian retrovirus gag genes sequenced and number of amino acid changes relative to the Gag polyprotein. Nucleotide and amino acid position numbers are relative to those of the published RSV genome (27). The known functional domains of assembly (L) and membrane binding (M) are indicated above the gag gene diagram. (A) Fragment 1 indicates the region sequenced for 102 retrovirus clones, and fragment 2 indicates the region sequenced for 40 retrovirus clones. Numbers below the diagram are nucleotide positions, starting at the 3′ base of primers. (B) Graphical representation of the number of amino acid changes over the region sequenced. Amino acid changes were determined from phylogenetic tree branch lengths as described in the text.
The 50-μl PCRs were performed using standard buffer and MgCl2 concentrations, 0.2 mM each deoxynucleoside triphosphate, 0.4 μM each primer, 1.5 U of Taq polymerase, and ≈100 ng of genomic DNA. Thermocycler profiles were: 94°C for 2 min and then 35 cycles of 94°C for 15 s, 55°C for 20 s, and 72°C for 1 min 20 s, with a final extension of 10 min at 72°C. We determined size and gel-purified PCR products by using 1.5% low-melting-point agarose, excised target bands from the gel, and recovered DNA using a gel extraction kit (Qiagen). Sequence reactions were performed using PCR primers with Taq DNA polymerase FS and either dRhodamine or Big Dye chemistry (Applied Biosystems). Reaction products were sequenced with an ABI 377 automated DNA sequencer.
Subcloning of retrovirus PCR products.
Multiple gag gene copies were detected in all PCR products sequenced, indicated by double peaks on sequence chromatograms and insertion/deletion events in alternative gene copies. To determine the sequence from single gag genes, PCR products were subcloned using a TA cloning kit (Invitrogen). We selected 5 to 10 individual colonies for sequencing analysis for each PCR-positive individual. Individual bacterial clones were lysed at 96°C for 10 min and then placed directly into a PCR mix containing the standard reagents listed above and universal M13 forward and reverse primers or gag-specific primers. We obtained 2 to 10 gag sequences from each positive individual and used these sequences for further analyses.
Host gene sequencing.
Mitochondrial 12S and ND2 gene sequences for galliform host taxa were obtained by using thermocycler profiles, purification of PCR products, and the sequencing protocols described above. Primer sequences for the 12S and ND2 genes are given by Sorenson et al. (30). The same individual that was gag positive was used for host gene sequencing except where multiple individuals from one species were tested and in the case where we have host sequence from Francolinus africanus but gag sequence from Francolinus swainsonii.
Sequence alignment and phylogenetic analyses.
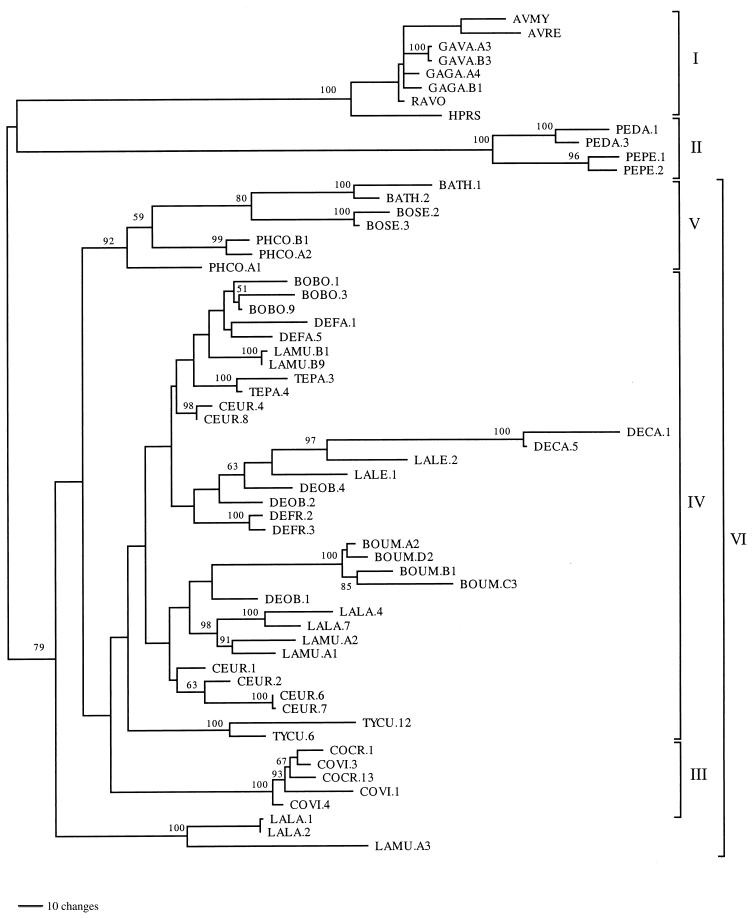
ASLV gag and avian mitochondrial DNA sequence alignments were based on the alignment of inferred amino acid sequences using Clustal X (32) and adjusted by eye to minimize mismatches. Avian mitochondrial 12S ribosomal DNA (rDNA) sequences were also adjusted to maintain alignment of conserved secondary structure features across species (21). Some gag and mitochondrial 12S rDNA sequences were too varied across taxa for unambiguous alignment and were excluded from phylogenetic analyses. We used the alignments to calculate pairwise genetic distances using the Kimura two-parameter model and accounting for unequal frequency of transitions and transversions (15). We performed phylogenetic analyses using maximum parsimony (MP), maximum likelihood (ML), and neighbor-joining (NJ) methods as implemented in PAUP* (31). Heuristic MP analyses for amino acids and nucleotides were conducted with 100 replicate searches with random addition of taxa. The PROTPARS weight matrix was used to assign greater weight to amino acid substitutions requiring more nucleotide changes. ML analyses were performed using either the Hasegawa-Kishino-Yano (HKY) or a general time reversible (GTR) model accommodating unequal base composition and evolutionary rate heterogeneity across sites with a discrete approximation of the gamma distribution. NJ analyses were based on pairwise distances, also using the Kimura two-parameter model. To perform bootstrap analyses, we used 100 replicates for MP and NJ trees. For MP analyses of host taxa and fragment 2 sequences (Fig. 1), we calculated decay indices which indicate the number of additional steps needed to invalidate specific nodes (3). These are calculated by inputting a constraint tree into PAUP* and finding the shortest tree that lacks particular nodes found originally in the most parsimonious tree. To investigate levels of amino acid conservation across gag gene regions, the numbers of amino acid changes were plotted on one of the most parsimonious trees (see Fig. 4) based on fragment 2 (Fig. 1) nucleotide sequence data and counted using MacClade (16). To assess potential selective effects, the proportion of synonymous and nonsynonymous nucleotide changes per synonymous and nonsynonymous site was calculated for fragment 2 alignments using the Synonymous Nonsynonymous Analysis Program (22). A ratio of synonymous to nonsynonymous substitution greater than one suggests purifying selection as amino acid replacement changes are selected against, while a ratio less than one suggests positive selection for amino acid replacements.
FIG. 4.
Strict consensus of 16 equally parsimonious trees for ASLV gag gene fragment 2 (Fig. 1, GAG.F1-GAG.R2) amplified from 16 species and one subspecies of galliform birds. Of 1,537 nucleotide characters, 557 are parsimony informative. Bootstraps followed by decay indices are indicated on branches, the latter denoting the number of additional steps required to collapse a particular node. Nodes with bootstrap values less than 50 are indicated by a dash. Biogeographic labels are given where ASLV relationships reflect geographic proximity of host species rather than host phylogeny to emphasize inferred independent colonizations (Table 2). An inferred duplication event is also indicated. Roman numerals for clades correspond to those in Fig. 2 and 3.
Sequences from databases and nucleotide sequence accession numbers.
We used published ASLV and mitochondrial sequences from GenBank as follows: ASLVs; M379890, Z46390, AF033809, X13744, J02342, D10652, AF033810, L10922, L10924.1, V01170.1, M10455.1, M30517, M73497, U83740, and U83742; 12S, NC_001323, X57245, and NC_000877; ND2, NC_001323, X57246, and NC_000877. New sequences reported and used here have accession numbers as follows: 12S rDNA, AF222570 to AF222590, AF222592 to AF222593, and AF222596 to AF222598; ND2, AF222538 to AF222555, AF222557 to AF222561, AF222563 to AF222564, and AF222567 to AF222569; gag, AF225298 to AF225399.
RESULTS
PCR and sequencing.
We found retroviral gag sequences in 26 species from three families of galliform birds from captive or natural populations in North America, Central America, eastern Europe, Asia, and Africa (Table 1). Nineteen of the 26 host species whose ASLVs were sequenced had not been known previously to contain retroviral elements. ASLV infection has been reported in seven of the species examined here (9, 10, 25); however, gag gene sequence has only been reported from one, Gallus gallus (27). All phasianids examined were positive for gag except for two species in the genus Coturnix. However, Coturnix species were found to be positive for an ASLV-related retrovirus in previous studies using Southern blot analysis (25). The two earliest diverging galliform families, the Cracidae and Megapodiidae, were negative for gag using our primers. We tested an additional 30 avian species representing 18 orders, and all were PCR negative (Table 1). This included two individuals from the sister group to the Galliformes, the Anseriformes.
Including multiple clones sequenced from individual hosts, we obtained a total of 102 fragment 1 gag sequences from 20 species of Galliformes and an additional 40 sequences spanning fragment 2 from 16 species of Galliformes (Fig. 1). These 16 species are a subset of the 20 species used to obtain fragment 1 sequences. Only three fragment 1 sequences were interrupted by stop codons. Two stop codons were located in the same position of gag from each of two Bonasa umbellus individuals, and the third was found in a different location in sequence from Bambusicola thoracia. Four fragment 2 sequences contained stop codons: one in each of two Lagopus mutus clones and one each from Bonasa sewerzowi and Colinus virginianus clones.
The gag fragment 1 alignment and the fragment 2 alignments were 1,125 and 1,564 characters long, respectively, including gaps. Based on 2 to 10 sequenced gag fragment 1 elements from each host, the mean pairwise distance within individuals ranged from 0.31% in Dendragapus canadensis to 9.13% in Lagopus lagopus (Table 1). A pairwise distance of 10.06% was found for all clones isolated from different host species within the subfamily Tetraoninae. The mean pairwise distance among all 110 sequences from fragment 1, which includes eight published ASLV sequences, was 17.5%. This compares to 18.0% when fragment 2 sequences are compared. When published Rous sarcoma virus (RSV) sequence was compared to that of one representative clone from each host species for fragment 2, we found a mean pairwise distance of 24.3%. The greatest pairwise distance was between Perdix perdix ASLV and RSV, with a genetic distance of 33.5%. These genetic distances are comparable to genetic distances found between sequences comprising other retroviral genera (35). Limited similarity was found at the amino acid level between recently sequenced gag from ev/J and our newly sequenced retroviruses. For example, ev/J Gag is 42% and 41% identical to Colinus virginianus and Lagopus leucurus Gag, respectively. This is comparable to the reported 46% identity between ev/J and RSV Gag (26). We conducted preliminary phylogenetic analyses which placed ev/J between a tetraonine clade and a Gallus clade (not shown). Even less similarity was observed between our Gag and those from ART-CH and lymphoproliferative disease virus of turkeys, such that only short stretches of amino acids could be reliably aligned.
Phylogenetic analyses of galliform ASLV gag sequences.
Analyses for gag sequences from galliform bird hosts were either unrooted or midpoint rooted along the longest internode within the unrooted tree, because we do not have gag sequence from an appropriate outgroup for the ASLVs. Using NJ analysis for 110 fragment 1 sequences, we found that 10 clones from four Bonasa umbellus individuals formed a monophyletic group (not shown), suggesting a single infectious event with subsequent duplication and diversification. Monophyly was observed in 10 additional cases in which virus sequences from individual species grouped together (Fig. 2 and 3). However, monophyly for sequence fragments from single individual hosts was not observed for Centrocercus urophasianus, Dendragapus obscurus, Lagopus lagopus, and Lagopus mutus. Similarly, gag sequences from two different Lagopus mutus individuals did not form a clade. We excluded some of the retroviral elements that were clearly monophyletic with others from the same individual hosts; this reduced computing time, but did not affect the relationships among ASLVs from different host species.
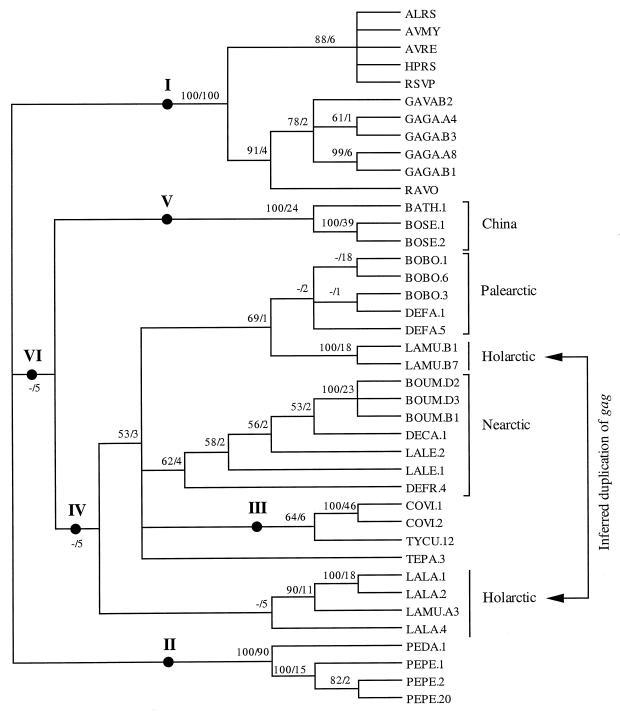
FIG. 2.
Unrooted NJ tree constructed using Kimura's two-parameter corrected distances, showing the relationship of 85 retroviral sequences based on 720 nucleotide sites from fragment 1 (Fig. 1A) of the gag gene. Bootstrap values are presented for the earliest divergences. An NJ tree using all 112 retroviral sequences has essentially the same topology; we removed 27 taxa representing multiple clones from single individuals from the analysis shown for clarity. Viral sequences are named for their host species (see Table 1 for abbreviations) and a number that identifies clones from the same host individual. When clones were sequenced from multiple individuals from a single species, a letter identifies the individual.
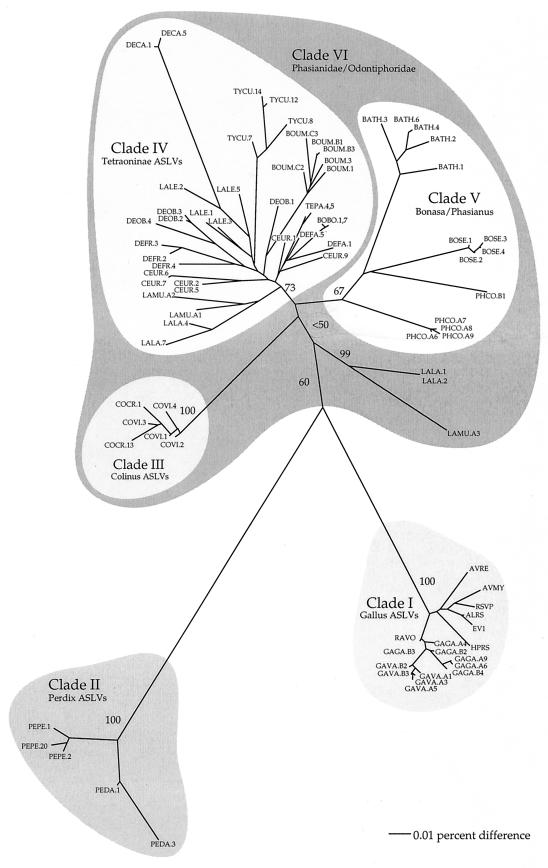
FIG. 3.
MP analysis of 61 gag retroviral sequences for fragment 1 isolated from 20 species of galliform birds (see Table 1 for sequence abbreviations and host species). This phylogeny is one of 24 equally parsimonious trees and is based on 654 characters, of which 378 are parsimony informative. Variation in the 24 trees was limited to the relative position of clones from the same individual within groups that remain monophyletic. Bootstrap values greater than 50 are shown on branches (100 replicates). This tree is midpoint rooted.
The unrooted NJ tree (Fig. 2) and the midpoint-rooted MP tree (Fig. 3) show the same major clades (monophyletic groups) of ASLVs, which we identify with Roman numerals. Clade I (Fig. 2) includes elements isolated from Gallus gallus and G. varius as well as existing, published Gallus gallus ASLVs. Thus, all published and newly sequenced Gallus gag sequences were more closely related to each other than they were to gag sequences isolated from other avian host species, indicating the close relationships between the new and published ASLVs. Clades II to V (Fig. 2) represent newly discovered lineages of ASLVs and show the same trend, in which retroviral sequences tend to group with their host taxa (species, genus, and family). The clades found were fairly well supported as indicated by bootstrap values. Clades I to III had bootstrap values of 100 in both NJ and MP analyses (Fig. 2 and 3). Clade V represents ASLVs isolated from species with natural (noncaptive or nonintroduced) distributions in China. We found retrovirus sequences from Bonasa sewerzowi, a tetraonine, placed within clade V, not in clade IV with other ASLVs from tetraonines and other species of Bonasa. Although the major clades were well supported, the internal nodes joining these clades were short and had relatively low support. The sister clade to the tetraonine clade (IV) varied, depending on the type of analysis done. Using MP, the Colinus clade representing the family Odontophoridae (clade III) was sister to clade IV, while NJ analyses indicated the Bonasa/Phasianidae clade (V) as sister to clade IV.
We attempted to better resolve relationships among ASLV clades I to V by sequencing an additional 409 bases (fragment 2, Fig. 1) from 16 species and one subspecies. MP analysis (consensus tree in Fig. 4) of nucleotides supported the same five ASLV clades found with fragment 1 (Fig. 2 and 3). Our analyses of amino acid residues using either PROTPARS or equal weights resulted in a phylogeny that was essentially congruent with the consensus tree in Fig. 4, except that fewer nodes were resolved (not shown). Although relationships among the five major clades still had low bootstrap and decay indices with this expanded set of sequence characters, relationships common to those shown in Fig. 2 to 4 included close relationships between clades I and II and among III to V.
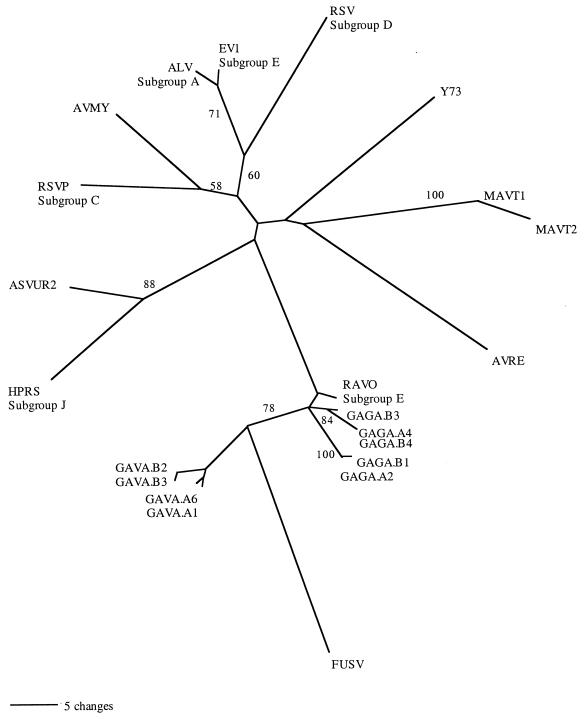
MP and ML analyses restricted to a set of new and existing Gallus ASLVs yielded essentially the same topology (Fig. 5). Existing Gallus ASLVs in the analyses based on gag sequences represent both endogenous and exogenous viruses and include ASLV subgroups A, C, D, E, and J, as well as replication-defective viruses that lack portions of the full-length retroviral genome. Our new sequences appeared most closely related to RAV-0, and we found that FUSV falls within this clade of newly sequenced gag fragments. HPRS, an exogenous virus of meat-type chickens (23), was closely related to other exogenous Gallus ASLVs. HPRS is thought to be a product of recombination, with its env gene most closely related to endogenous EAV-HP elements and the remaining genes arising from exogenous ASLVs (29). The gag gene from the newly described ev/J (1) has 46% amino acid sequence identity to those of published exogenous ASLVs, suggesting that ev/J arose from a virus only distantly related to published ASLVs (26). All ASLV sequences that we amplified had a higher amino acid similarity to those of published ASLVs than to ev/J.
FIG. 5.
Unrooted MP analysis of ASLV gag DNA sequences isolated from birds in the genus Gallus. This phylogeny is based on 1,165 characters (fragment 2) of which 95 are parsimony informative. This is one of two equally parsimonious hypotheses. Bootstrap values above 50 are shown on branches (100 replicates). ML analysis using the GTR model yields the same topology. Sequences GAGA and GAVA are newly sequenced elements from two individuals (A and B) of Gallus gallus and Gallus varius, respectively. gag sequences for the following viruses were obtained from databases: avian leukemia virus, subgroup A (ALV); exogenous avian leukosis virus, subgroup J (HPRS-103); avian myelocytomatosis virus (AVMY); avian retrovirus IC10 (AVRE); RSV (Prague), subgroup C (RSVP); RSV (Schmidt-Ruppin), subgroup D (RSV); FUSV; myeloblastosis-associated virus 1 (MAVT1); myeloblastosis-associated virus 2 (MAVT2); avian sarcoma virus Y73 (Y73); avian sarcoma virus UR2 (ASVUR2); Rous sarcoma-defective endogenous virus, subgroup E (EV1); chicken provirus RAV-0, subgroup E (RAV0) (accession numbers are listed in Materials and Methods).
Phylogenetic analyses of galliform birds.
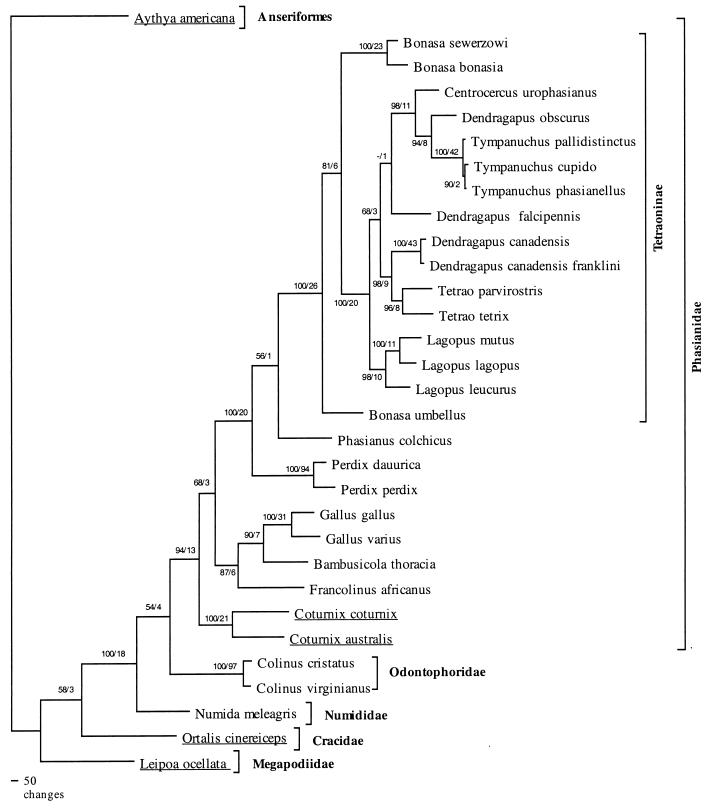
We obtained 1,041 bp of mitochondrial ND2 and 1,034 bp of mitochondrial 12S rDNA for 30 species in the avian order Galliformes. MP analyses yielded a single optimal tree (Fig. 6). Placement of Megapodiidae and Cracidae as diverging prior to the other three families agrees with results of previous studies, including those based on DNA-DNA hybridization (28), although our findings differed in not placing those two families as sisters. Monophyly for a Numididae, Odontophoridae, and Phasianidae clade was well supported. Monophyly of the family Phasianidae was well supported, with a bootstrap value of 94 and a decay index of 13, as was monophyly of the grouse and ptarmigan subfamily (Tetraoninae), with a bootstrap value of 100 and a decay index of 26. Within the Tetraoninae, our analyses indicated nonmonophyly for the genera Dendragapus and Bonasa. Nonmonophyly for these two genera is supported by analyses of mitochondrial cytB sequences as well (8). We also found close relationships between Gallus, Bambusicola, and Francolinus, indicating that the traditional groups pheasants and partridges were not phylogenetically meaningful, in agreement with the findings of Kimball et al. (14).
FIG. 6.
Inferred phylogeny for ASLV hosts in the avian order Galliformes based on MP analyses of 2,075 characters (703 informative sites) from the mitochondrial 12S rDNA and ND2 genes, with a waterfowl species (Aythya americana, redhead) as the outgroup. See Table 1 for bird species common names and corresponding ASLV names. Numbers on branches are bootstrap values based on 100 replicate searches and decay indices, respectively. Underlined host taxa were negative for retrovirus infection using our PCR primers.
Relative conservation of gag gene sequences.
A protein-coding reading frame was conserved across all but seven of the ASLVs sequenced, in which either point substitutions or insertions led to a premature stop codon. To visualize variation in the level of amino acid conservation across the sequenced region, the inferred number of amino acid substitutions for each position in the amino acid sequence (Fig. 1B) was plotted onto one of the most parsimonious trees (Fig. 4). The gag L domain (PPPPYV in the chicken), which is an assembly domain required for efficient budding of virus-like particles (37, 38), was highly conserved in all ASLVs sequenced. In contrast, regions flanking the L domain were more variable. Most ASLVs, except the Gallus ASLVs, shared a deletion of one proline in the L domain. Perdix ASLVs were the most divergent in overall amino acid sequence, and they have a PPPTY motif in the L domain. The N-terminal and C-terminal residues were also well conserved across ASLV taxa. The M domain, located in the first 85 residues of the matrix protein, functions in membrane binding and particle formation (34) (Fig. 1B) and was also highly conserved across all ASLVs sequenced. To investigate the possibility that purifying selection is acting on these sequences, we calculated pairwise proportions of synonymous and nonsynonymous substitutions per synonymous and nonsynonymous site for all fragment 2 sequences. Using the average for all pairwise comparisons, we found a ratio of 5.57 to 1 synonymous to nonsynonymous substitutions, consistent with the operation of purifying selection.
DISCUSSION
Inferences of cospeciation and vertical transmission.
We found ASLVs in 26 species of galliform birds collected from natural populations throughout the world, including 19 species not previously known to have ASLVs. Our phylogenetic analyses of ASLVs and their hosts suggest a long association of ASLVs and galliform birds that includes vertical transmission and cospeciation with host lineages, as well as more recent horizontal transmission among host species (Table 2). Given the endemic nature of many of the galliform species surveyed and their various distributions within North America, Central America, South America, Russia, China, Asia, and South Africa, the presence of ASLVs in all but two of the Phasianidae species surveyed suggests an early ASLV and galliform association. Based on our current understanding of ASLV distribution within galliform birds, this association could date back to the divergence of the Phasianidae, Numididae, and Odontophoridae from the other galliform families, possibly as many as 50 million years ago (28, 33, 36). A representative of the family Numididae (Numida meleagris) was also found to contain ASLVs; however, sequence suitable for phylogenetic analyses from a single clone has not yet been determined (Table 1). We did not find ASLVs within the single Cracidae and Megapodiidae species examined. We were also unable to amplify ASLV gag sequences with our primers from representatives of 18 other avian orders (Table 1), including two individuals of the Anseriformes, the sister order to the Galliformes (4, 28, 20). We note, however, that repeated negative PCR results do not establish ASLV absence and that further analyses, including assessment of additional species, are needed.
TABLE 2.
Summary of inferred host-virus interactions
| Clade and/or taxon | Inferencea | Evidence | Figure no. |
|---|---|---|---|
| Clade II, Perdix | Vertical transmission/cospeciation | Phylogenetic congruenceb | 2, 3, 4 |
| Clade I, Gallus | Vertical transmission/cospeciation | Phylogenetic congruence | 2, 5 |
| L. lagopus/L. mutus | Vertical transmission/cospeciation | Phylogenetic congruence | 2, 3 |
| L. lagopus/L. mutus | Duplication | gag copies from the same individual are not sister taxa | 2, 3, 4 |
| Clade V, Bambusicola, Phasianus, and B. sewerzowi | Horizontal transmission | Phylogenetic incongruence, geographic affinityc | 2, 3 |
| Clade IV, L. leucurus, T. cupido, and D. canadensis | Horizontal transmission | Phylogenetic incongruence | 2 |
| Clade IV, D. obscurus, B. umbellus, and Centrocercus | Horizontal transmission | Phylogenetic incongruence | 2 |
| Clade IV, Tetraoninae | Horizontal transmission | Phylogenetic incongruence, geographic affinity | 3, 4 |
| Clade IV, C. urophasianus | Duplication | gag copies from the same individual are not sister taxa | 2, 3 |
Vertical transmission denotes germ line transmission from host parent to offspring.
Congruence of ASLV and host phylogenetic hypotheses.
ASLV phylogeny reflects similar geographic distribution of host species rather than phylogenetic relationships of host species.
Instances of congruence across ASLV and host phylogenies include Fig. 2 clade II, with distinct clades for ASLVs from Perdix perdix and P. dauurica, suggesting cospeciation of Perdix and ASLV taxa. Similarly, Fig. 2 clade I includes distinct sister clades for new ASLVs from Gallus gallus and G. varius. Lagopus lagopus and L. mutus ASLVs (Fig. 2 clade IV) also reflect host phylogeny; however, they show evidence of a duplication with subsequent cospeciation in the separate L. lagopus and L. mutus ASLV clade in Fig. 2 and 3 outside of clade IV (Table 2). Our evidence indicates the ASLV duplication event also occurred prior to the divergence of L. lagopus and L. mutus.
The history of domestication for Gallus gallus includes frequent long-distance transport by humans and the potential for horizontal transmission among host species not in geographic proximity prior to domestication. Earliest references in art and texts point to domestication dates of more than 4,000 years ago for apparent representatives of Gallus gallus (12). Our finding of monophyly for all Gallus ASLVs, however, does not provide evidence in support of such transmission events involving domesticated Gallus.
Inferences of horizontal transmission and dispersal.
ASLV and host phylogenies show more instances of incongruence than congruence, suggesting a greater frequency of ASLV dispersal and horizontal transmission across host species. It is not possible to infer the direction of transmission between any two host species (donor versus recipient) from phylogenies which show relationships among ASLV descendants and not those among descendants and ancestors. However, it is possible to note multiple host species involved in ASLV transmission based on phylogenetic analyses. For example, incongruence between ASLV and host phylogenies indicates likely horizontal transmission among the following host species sets: (i) Bambusicola, Phasianus, and Bonasa sewerzowi (Fig. 2 clade V); (ii) Lagopus leucurus, Tympanuchus cupido, and Dendragapus canadensis (Fig. 2 clade IV); and (iii) Dendragapus obscurus, Bonasa umbellus, and Centrocercus (Fig. 2 clade IV). Horizontal transmission is also indicated by noting monophyly in the host phylogeny but not in ASLV trees for Bonasa, Dendragapus, and Lagopus gag sequences.
There is a logical geographic pattern for the horizontal transmission inferences in which we found monophyly for ASLVs from disparate hosts with overlapping ranges. For example, clade V of Fig. 2 and 3 includes ASLVs from hosts having an eastern Palearctic distribution (Bambusicola thoracica, Bonasa sewerzowi, and Phasianus colchicus) but not ASLVs from species in one of the same host genera found in the Nearctic (Bonasa umbellus). In turn, Bonasa umbellus ASLVs were more closely related to ASLVs from other North American host species like Dendragapus canadensis and Lagopus leucurus (Table 2). Similarly, ASLVs from the two Lagopus hosts collected in Russia were sister taxa (Fig. 2 clades IV and VI and Fig. 3), whereas ASLVs from Lagopus leucurus collected in Washington state were more closely related to ASLVs from other North American hosts. For the Tetraoninae subfamily (grouse and ptarmigan) in Fig. 4 we denote three monophyletic ASLV groups which reflect host geographic distribution rather than phylogeny. These examples provide evidence for horizontal transmission of ASLVs facilitated by geographic proximity.
Overlaid on ancient associations, potentially dating back to galliform family divergences, is a more recent history of, at least occasional, horizontal transmission of ASLVs among the Tetraoninae species (grouse and ptarmigan). This is indicated by the Tetraoninae ASLV clades in Fig. 4, reflecting geographic distribution and proximity of hosts rather than host species phylogeny. The modern tetraonine lineages may have arisen during the mid-Pleistocene (0.8 to 0.4 million years ago) on the basis of fossils assigned to tetraonines (12, 33). Thus, apparent horizontal transmission among host species, for example Bonasa umbellus, Dendragapus canadensis, and Lagopus leucurus, likely postdate the mid-Pleistocene. Perhaps the best case for horizontal transmission involves the Chinese species, Bonasa sewerzowi. gag ASLV sequences from B. sewerzowi fall within clade V of Fig. 2, which includes other Chinese host species with overlapping ranges but does not include the other Bonasa species in our sample. These relationships are found in all phylogenetic analyses performed and have strong bootstrap support. These results suggest B. sewerzowi acquired gag through infection by a Chinese phasianid and not through vertical transmission. The possibility that all the galliform ASLVs arose and spread through the host species during the past few thousand years cannot be ruled out but seems unlikely due to the endemic and isolated nature of some of the host species ranges and habitats and due to the instances of congruence between ASLV and host trees.
Horizontal transmission for ASLVs has been postulated by Frisby et al. (10) who found similar RAV-0 sequence fragments, based on DNA hybridization experiments, in Gallus gallus and two species of Phasianus without finding similar fragments in other species of Gallus or other pheasant species more closely related to Phasianus. This is a reasonable interpretation; however, the possibility of a single early infectious event with subsequent loss of RAV-0 elements in some host species cannot be ruled out. Relatively ancient infection, predating species divergences within the genus Gallus, followed by vertical transmission and cospeciation for EAVs and their hosts has been indicated by Resnick et al. (25) and Boyce-Jacino et al. (2).
gag gene duplications.
One of the most difficult issues to deal with in phylogenetic analyses of endogenous retroviruses is the potential for both gene duplications and multiple infectious events. Phylogenetic analyses should be based on orthologous genes, that is, the same gene in different hosts, rather than paralogous genes, which are alternative copies of a gene arising from gene duplications within a host individual. Mixing comparisons of orthologous and paralogous retrovirus genes can potentially confound inference of phylogenetic history for the retroviruses. Given the appearance of numerous, similar copies of most endogenous retroviruses within host individuals, as we have found with our ASLVs (see Materials and Methods), the assumption of orthology for all sequences compared among host species is unwarranted. We can, however, evaluate the possibility of multiple independent infectious events within species by assessing monophyly for multiple clones from individual hosts (Table 1).
Monophyly for all clones from a particular host individual suggests a single infectious event with subsequent sequence duplication and divergence. Nonmonophyly would indicate either an additional infectious event or a disparate sampling of divergent paralogs across host individuals, in which the phylogenetic signal is obscured due to convergent similarity in paralogous gene sequences. The problem of obscured phylogenetic signal can be addressed through increased sampling and phylogenetic analyses considering variable rates of evolution across taxa and sequence characters, as we have attempted. Our finding of duplicated sister relationships for Lagopus lagopus and L. mutus ASLVs based on different sets of ASLV clones (Fig. 2 and 3) suggests two independent infectious events in their common ancestor, with subsequent vertical transmission and cospeciation of the ASLVs with the hosts. The possibility that we are being misled in this view by variable sampling of disparate paralogs within the two host species is less likely given the levels of bootstrap support (99 and 73 in Fig. 2; 100 and 98 in Fig. 3) and congruent topologies in NJ, MP, and ML analyses using alternative models of molecular evolution. Nonmonophyly for ASLV clones from individual Centrocercus urophasianus and Dendragapus canadensis individuals (Fig. 2 and 3) also suggests multiple independent infectious events within those host lineages.
Phylogenetic taxonomy for new ASLVs.
To facilitate discussion of our findings we provide a preliminary phylogenetic taxonomy (Table 3). A phylogenetic taxonomy provides a classification and set of taxon names seeking to convey information about evolutionary relationships (7). The six new names in Table 3 are based on the monophyletic groups (clades) seen in Fig. 2 and 3.
TABLE 3.
| Category | Taxon name |
|---|---|
| Family | Retroviridae |
| Genus | Alpharetrovirus or ASLVs |
| Subgenus 1 | Gallus ASLV (includes taxa in Fig. 2 clade I) |
| Perdix ASLV (includes taxa in Fig. 2 clade II) | |
| Phasianidae/Odontophoridae ASLV (includes taxa in Fig. 2 clade VI) | |
| Subgenus 2 | Colinus ASLVs (includes taxa in Fig. 2 clade III) |
| Tetraonine ASLVs (includes taxa in Fig. 2 clade IV) | |
| Bonasa/Phasianus ASLVs (includes taxa in Fig. 2 clade V) |
The taxon names Retroviridae and ‘Alpharetrovirus’ follow the ICTV convention. The term ASLV follows the usage of Payne et al. (24). Alpharetroviruses have also been known as avian type C retroviruses. We suggest that the genus ASLV includes other avian retroviruses, including lymphoproliferative disease virus, EAV-E51, ev/J, and ART-CH, based on limited sequence comparisons.
We refer to the elements newly described here as ASLVs because of their close relationship to published ASLV sequences. Our Gallus gag sequences are more closely related to RAV-0 and FUSV (Fig. 2 and 5) published ASLVs than to the other gag sequences from galliform hosts amplified with the same set of primers, indicating that our primers are amplifying ASLVs. Further, our gag sequences are not readily alignable with those of any previously known retroviruses except the ASLVs and have an average 76% identity at the nucleotide level compared to that of published RSV. There are no published gag sequences for EAVs for comparison.
ASLV is often used synonymously with avian type C retrovirus as well as the genus Alpharetrovirus currently recognized by the ICTV. We follow that convention here and suggest inclusion of other avian retroviruses such as EAVs, LDVs, ev/J, and ART-CHs within the ASLV genus (Table 3) based on limited sequence comparisons indicating relatedness, so that these groups will not be without a genus placement. The names used in Table 3 consist of the host taxon, to the extent currently known, as a prefix to ASLV.
Conservation and purifying selection of gag gene sequences.
It is unclear why endogenous retrovirus sequences are maintained in host genomes. If endogenous ASLV sequences have no function, they could be expected to change rapidly, corresponding to the mutation rate, with ensuing loss of reading frame and coding function. As mentioned above, however, we find conservation of reading frame across all but seven of the ASLVs sequenced. Purifying selection maintaining the reading frame and protein function is indicated by the ratio of synonymous to nonsynonymous nucleotide substitutions being greater than one. This sequence conservation is consistent with either (i) recent horizontal transmission or (ii) ancient infection with subsequent vertical transmission accompanied by purifying selection to maintain gag gene function. Both of these scenarios are plausible explanations for various ASLV lineages, based on our results. One scenario that can be ruled out, however, is relatively old infectious events with subsequent vertical transmission not accompanied by purifying selection. Thus, those instances of ASLV and host tree congruence indicating ancient infection with subsequent vertical transmission, such as those within the Gallus and Perdix clades (Fig. 2 clades I and II), suggest that there has been purifying selection dating at least from the host speciation events. There has been frequent speculation that endogenous retroelements such as ASLVs may be conserved over time as a means for training the host's immune system and stimulating antibody production prior to exogenous infection (1, 5, 6), and although we cannot address this directly, the hypothesis appears consistent with some of our findings.
ACKNOWLEDGMENTS
This work was supported by University of Michigan graduate student Block Grant funds to D.E.D. and M.K. and by NSF grants to D.P.M.
We thank L. N. Payne for helpful comments on an earlier draft of this report. We thank Michael Sorenson for excellent technical assistance. Tissue samples were kindly provided by the University of Michigan Museum of Zoology, the University of Washington Burke Museum, and Thomas Quinn.
REFERENCES
- 1.Best S, Le Tissier P, Towers G, Stoye J P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 2.Boyce-Jacino M T, O'Donoghue K, Faras A J. Multiple complex families of endogenous retroviruses are highly conserved in the genus Gallus. J Virol. 1992;66:4919–4929. doi: 10.1128/jvi.66.8.4919-4929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bremer K. Branch support and tree stability. Cladistics. 1994;10:295–304. [Google Scholar]
- 4.Cracraft J, Mindell D P. The early history of modern birds: a comparison of molecular and morphological evidence. In: Fernholm B, Bremer K, Jörnvall H, editors. The hierarchy of life: molecules and morphology in phylogenetic analysis. Amsterdam, The Netherlands: Elsevier Science Publishers; 1989. pp. 389–403. [Google Scholar]
- 5.Crittenden L B, McMahon S, Halpern M S, Fadly A M. Embryonic infection with the endogenous avian leukosis virus Rous-associated virus-0 alters responses to exogenous avian leukosis virus infection. J Virol. 1987;61:722–725. doi: 10.1128/jvi.61.3.722-725.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crittenden L B, Witter R L, Fadly A M. Low incidence of lymphoid tumors in chickens continuously producing endogenous virus. Avian Dis. 1979;23:646–653. [PubMed] [Google Scholar]
- 7.DeQueiroz K, Gauthier J. Phylogenetic taxonomy. Ann Rev Ecol Systemat. 1992;23:449–480. [Google Scholar]
- 8.Ellsworth D L, Honeycutt R L, Silvy N J. Systematics of grouse and ptarmigan determined by nucleotide sequences of the mitochondrial cytochrome-B gene. Auk. 1996;113:811–822. [Google Scholar]
- 9.Frisby D, MacCormick R, Weiss R, editors. Origin of RAV-0. The endogenous retrovirus of chickens. Vol. 7. New York, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 10.Frisby D P, Weiss R A, Roussel M, Stehelin D. The distribution of endogenous chicken retrovirus sequences in the DNA of galliform birds does not coincide with avian phylogenetic relationships. Cell. 1979;17:623–634. doi: 10.1016/0092-8674(79)90270-8. [DOI] [PubMed] [Google Scholar]
- 11.Herniou E, Martin J, Miller K, Cook J, Wilkinson M, Tristem M. Retroviral diversity and distribution in vertebrates. J Virol. 1998;72:5955–5966. doi: 10.1128/jvi.72.7.5955-5966.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoyo J D, Elliott A, Sargatal J, Cabot J. Handbook of the birds of the world. Barcelona, Spain: Lynx Edicions; 1992. [Google Scholar]
- 13.Hunter E. Viral entry and receptors. In: Coffin J M, Hughes S H, Varmus H, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 71–119. [PubMed] [Google Scholar]
- 14.Kimball R T, Braun E L, Zwartjes P W, Crowe T M, Ligon J D. A molecular phylogeny of the pheasants and partridges suggests that these lineages are not monophyletic. Mol Phylogenet Evol. 1999;11:38–54. doi: 10.1006/mpev.1998.0562. [DOI] [PubMed] [Google Scholar]
- 15.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 16.Maddison W P, Maddison D R. MacClade, 3.01 ed. Sunderland, Mass: Sinauer Assoc., Inc.; 1992. [Google Scholar]
- 17.Martin J, Herniou E, Cook J, O'Neill R W, Tristem M. Interclass transmission and phyletic host tracking in murine leukemia virus-related retroviruses. J Virol. 1999;73:2442–2449. doi: 10.1128/jvi.73.3.2442-2449.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin J, Herniou E, Cook J, O'Neill R W, Tristem M. Human endogenous retrovirus type I-related viruses have an apparently widespread distribution within vertebrates. J Virol. 1997;71:437–443. doi: 10.1128/jvi.71.1.437-443.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClure M A, Johnson M S, Feng D F, Doolittle R F. Sequence comparisons of retroviral proteins: relative rates of change and general phylogeny. Proc Natl Acad Sci USA. 1988;85:2469–2473. doi: 10.1073/pnas.85.8.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mindell D P, Sorenson M D, Dimcheff D E, Hasegawa M, Ast J C, Yuri T. Interordinal relationships of birds and other reptiles based on whole mitochondrial genomes. Syst Biol. 1999;48:138–152. doi: 10.1080/106351599260490. [DOI] [PubMed] [Google Scholar]
- 21.Mindell D P, Sorenson M D, Huddleston C J, Miranda H, Knight A, Sawchuk S J, Yuri T. Phylogenetic relationships among and within select avian orders based on mitochondrial DNA. In: Mindell D P, editor. Avian molecular evolution and systematics. New York, N.Y: Academic Press; 1997. pp. 211–245. [Google Scholar]
- 22.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 23.Payne L N, Brown S R, Bumstead N, Howes K, Frazier J A, Thouless M E. A novel subgroup of exogenous avian leukosis virus in chickens. J Gen Virol. 1991;72:801–807. doi: 10.1099/0022-1317-72-4-801. [DOI] [PubMed] [Google Scholar]
- 24.Payne L N, Howes K, Gillespie A M, Smith L M. Host range of Rous sarcoma virus pseudotype RSV(HPRS-103) in 12 avian species: support for a new avian retrovirus envelope subgroup, designated J. J Gen Virol. 1992;73:2995–2997. doi: 10.1099/0022-1317-73-11-2995. [DOI] [PubMed] [Google Scholar]
- 25.Resnick R M, Boyce-Jacino M T, Fu Q, Faras A J. Phylogenetic distribution of the novel avian endogenous provirus family EAV-0. J Virol. 1990;64:4640–4653. doi: 10.1128/jvi.64.10.4640-4653.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruis B L, Benson S J, Conklin K F. Genome structure and expression of the ev/J family of avian endogenous viruses. J Virol. 1999;73:5345–5355. doi: 10.1128/jvi.73.7.5345-5355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwartz D E, Tizard R, Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983;32:853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- 28.Sibley C G, Ahlquist J E. Phylogeny and classification of birds: a study in molecular evolution. New Haven, Conn: Yale University Press; 1990. [Google Scholar]
- 29.Smith L M, Toye A A, Howes K, Bumstead N, Payne L N, Venugopal K. Novel endogenous retroviral sequences in the chicken genome closely related to HPRS-103 (subgroup J) avian leukosis virus. J Gen Virol. 1999;80:261–268. doi: 10.1099/0022-1317-80-1-261. [DOI] [PubMed] [Google Scholar]
- 30.Sorenson M D, Ast J C, Dimcheff D E, Yuri T, Mindell D P. Primers for a PCR-based approach to mitochondrial genome sequencing in birds and other vertebrates. Mol Phylogenet Evol. 1999;12:105–114. doi: 10.1006/mpev.1998.0602. [DOI] [PubMed] [Google Scholar]
- 31.Swofford D L. PAUP*: phylogenetic analysis using parsimony (*and other methods), 4.0 ed. Sunderland, Mass: Sinauer; 1999. [Google Scholar]
- 32.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Unwin D M. The fossil record 2. In: Benton M J, editor. Aves. New York, N.Y: Chapman and Hall; 1993. pp. 717–737. [Google Scholar]
- 34.Verderame M F, Nelle T D, Wills J W. The membrane-binding domain of the Rous sarcoma virus Gag protein. J Virol. 1996;70:2664–2668. doi: 10.1128/jvi.70.4.2664-2668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogt V M. Retroviral Virions and Genomes. In: Coffin J M, Hughes S H, Varmus H, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 27–70. [PubMed] [Google Scholar]
- 36.Waddell P J, Cao Y, Hasegawa M, Mindell D P. Assessing the cretaceous superordinal divergence times within birds and placental mammals by using whole mitochondrial protein sequences and an extended statistical framework. Syst Biol. 1999;48:119–137. doi: 10.1080/106351599260481. [DOI] [PubMed] [Google Scholar]
- 37.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang Y, Cameron C E, Wills J W, Leis J. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]