Abstract
Glycoproteins M (gM), E (gE), and I (gI) of pseudorabies virus (PrV) are required for efficient formation of mature virions. The simultaneous absence of gM and the gE/gI complex results in severe deficiencies in virion morphogenesis and cell-to-cell spread, leading to drastically decreased virus titers and a small-plaque phenotype (A. Brack, J. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter, J. Virol. 73:5364–5372, 1999). Serial passaging in noncomplementing cells of a virus mutant unable to express gM, gE, and gI resulted in a reversion of the small-plaque phenotype and restoration of infectious virus formation to the level of a gM− mutant. Genetic analyses showed that reversion of the phenotype was accompanied by a genomic rearrangement which led to the fusion of a portion of the gE gene encoding the cytoplasmic domain to the 3′ end of the glycoprotein D gene, resulting in expression of a chimeric gD-gE protein. Since this indicated that the intracytoplasmic domain of gE was responsible for the observed phenotypic alterations, the UL10 (gM) gene was deleted in a PrV mutant, PrV-107, which specifically lacked the cytoplasmic tail of gE. Regarding one-step growth, plaque size, and virion formation as observed under the electron microscope, the mutant lacking gM and the gE cytoplasmic tail proved to be very similar to the gE/I/M triple mutant. Thus, our data indicate that it is the cytoplasmic tail of gE which is responsible for the observed phenotypic effects in conjunction with deletion of gM. We hypothesize that the cytoplasmic domain of gE specifically interacts with components of the capsid and/or tegument, leading to efficient secondary envelopment of intracytoplasmic capsids.
Herpes virions consist of four morphologically differentiable structures. The nucleoprotein core contains the linear double-stranded genomic DNA associated with proteins which is contained within an icosahedral capsid consisting of 162 capsomers. Capsids are assembled and DNA is packaged in the nucleus of infected cells. During virion formation, a proteinaceous layer of electron-dense material, designated the tegument, is added to capsids, which, in turn, is enclosed in a lipid bilayer envelope containing virus-encoded (glyco)proteins (35, 47). How and where herpes virions gain the tegument and final envelope is still a matter of debate. It has been proposed for herpes simplex virus type 1 (HSV-1) that intranuclear capsids bud through the inner nuclear membrane and retain this first envelope during passage through the endoplasmic reticulum and the secretory pathway (7, 24). On the other hand, for the alphaherpesviruses varicella-zoster virus (19, 59) and pseudorabies virus (PrV) (21, 57) and for the betaherpesvirus human cytomegalovirus (46), it is proposed that capsids which acquired a primary envelope by budding through the inner nuclear membrane subsequently fuse with the outer nuclear membrane, leading to release of capsids into the cytoplasm. The final envelope containing mature glycoproteins is then acquired by budding of intracytoplasmic capsids into vesicles of the trans-Golgi network. This model is based on data obtained from electron microscopic or confocal laser scan analysis, in part after treatment of infected cells with drugs such as brefeldin A. Recently, elegant mutational analysis uncovered results which favor this de-/reenvelopment model for HSV-1 also (6, 58).
In the past, we identified several gene products of PrV which play a role in virion morphogenesis. In the absence of the UL3.5 protein, capsids are detected adjacent to vesicles but budding does not occur (18). Lack of the UL20 gene product results in accumulation of enveloped virions within huge vesicles (17). Deletion of the gK (UL53) gene led to release of enveloped virions, which, however, appear to immediately fuse with the cell they left (28). Any one of these gene products is required for virus replication, and respective viral mutants could only be obtained and propagated on trans-complementing cell lines. Recently, we described another egress phenotype after deletion of glycoproteins gM, gE, and gI from PrV (4). Glycoproteins gE and gI form a noncovalently linked complex which is assumed to represent the functional unit (22, 61). Mutants with either gM or gE/I deleted can be isolated on normal cells, indicating that these proteins are not essential for viral replication (9, 36). Deletion of gM results in a decrease in virus titers of ca. 50- to 100-fold (9), whereas deletion of gE/I has only a slight effect on virus titers in vitro (this report). In the absence of either gM (10) or gE/I (36, 60), plaque size is slightly diminished, which testifies for a somewhat impaired capability for direct cell-to-cell spread. However, simultaneous deletion of gM and gE/I is strongly detrimental to the virus, and virus titers drop to low levels. Electron microscopy showed that in the absence of these three glycoproteins, numerous large intracytoplasmic inclusions containing capsids associated with amorphous material, presumably tegument, were formed. There also was a lack of secondary envelopment and, consequently, an absence of extracellular enveloped virus particles (4).
Whereas deletion of gE/I has no dramatic effect on virus replication in tissue culture, it is one of the prime determinants for neuroinvasion and neurovirulence of PrV in model systems such as mice and rats and in the natural host, pigs (2, 8, 15, 30, 32, 38, 39, 56). Recent studies indicated that a gE lacking the cytoplasmic domain is still capable of complexing with gI and of proper maturation and transport (54). However, a mutant virus expressing tailless gE exhibited neurovirulence properties different from those of wild-type viruses. In the absence of the gE cytoplasmic tail, virulence in rats is reduced to that of a gE− mutant, although viral spread in the rodent nervous system still occurs quite efficiently (54). Interestingly, the gE proteins of varicella-zoster virus and PrV have been shown to contain endocytosis signals in their cytoplasmic domains which result in retrieval of the membrane-bound protein from the plasma membrane into vesicles (1, 41, 51, 52). However, the role of gE endocytosis in the viral life cycle is unclear, and all endocytosis is shut off by 6 h after infection (51).
The HSV-1 gE/I complex (23) has also been shown to be involved in neurovirulence of the virus (14). Moreover, it localizes to components of cell junctions in polarized cells in culture (13), which led to the hypothesis that interaction of gE/I with cell junctions is responsible for transfer of HSV-1 across cell borders by direct cell-to-cell spread (12). However, up till now it is unclear how cell-to-cell spread is effected at the molecular level.
Deletion of gM in HSV-1, PrV, and equine herpesvirus type 1 (EHV-1) resulted in a decrease in virus titers of 10- to 100-fold and slightly reduced plaque size (9, 10, 33, 42). Interestingly, deletion mutants were attenuated in both model hosts (HSV-1 and EHV-1) and the natural host (PrV), presumably due to a defect in neuroinvasion and/or neuronal spread (11, 33, 40). However, the molecular basis for these defects is not known.
In this report, we further define the function of gM and uncover an important role for the gE cytoplasmic tail in the formation of mature virions.
MATERIALS AND METHODS
Viruses and cells.
The viruses used in this study included wild-type PrV strain Kaplan (PrV-Ka) (25) and isogenic mutants unable to express gM (PrV-gM−) (11), gE/I (PrV-gE/I−) (36), or gM and gE/I (PrV-gE/I/M−) (4). In PrV-gE/I− and PrV-gE/I/M−, the first 322 nucleotides of the gI gene are still present and fused out of frame to the last 312 nucleotides of the gE gene (see Fig. 4). Thus, these mutants still carry sequences encoding the gE cytoplasmic tail but are unable to express it. PrV-1112, which carries a β-galactosidase expression cassette in the nonessential gG gene, exhibits growth properties similar to those of the parental strain PrV-Ka (37). PrV-107 is derived from wild-type PrV strain Becker (PrV-Be) (3) and contains a stop codon introduced after the sequences coding for the gE transmembrane domain. Therefore, this virus expresses a membrane-anchored gE which lacks the cytoplasmic domain (52). Viruses were propagated on rabbit (RK13), porcine (PSEK), bovine (MDBK), or African green monkey kidney (Vero) cells or on trans-complementing cell lines. Rabbit kidney cells expressing gM (RK13-gM) or gE/I (RK13-gE/I) have been described (4). Cells were cultured in Eagle's minimum essential medium supplemented with 5% (10% for RK13 cells) fetal calf serum. Cotransfections were performed by calcium phosphate coprecipitation (20).
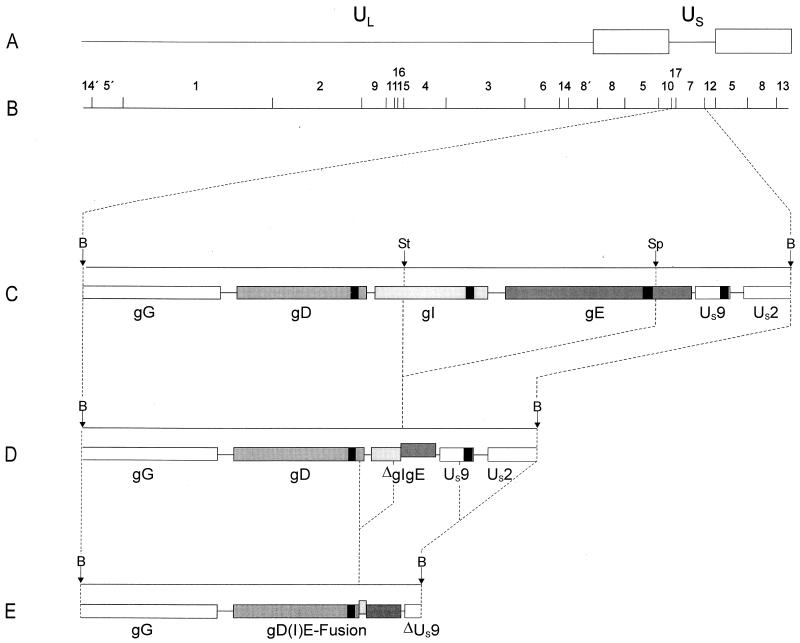
FIG. 4.
Genomic arrangement in the US region of PrV-gE/I/M−Pass. (A) Schematic diagram of the PrV genome. (B) BamHI restriction fragment map. UL, unique long region; US, unique short region. Open boxes, inverted repeat sequences which bracket the US region. (C) Enlargement of BamHI fragment 7 located in the US region, with relevant cleavage sites (B, BamHI; St, StuI; Sp, SphI). (D) Genomic arrangement in the US regions of PrV-gE/I− and PrV-gE/I/M−. It is evident that by deletion of gI sequences from the StuI site and gE sequences up to the SphI site (36), the 5′ terminus of the gI gene had been fused to the 3′ terminus of the gE gene. Note that the gE sequences in the ΔgI/gE fusion gene are out of frame with the gI sequences. (E) Deletion events that occurred during passaging were mapped by sequencing. One deletion resulted in the formation of a chimeric gD(I)E hybrid gene, in which the gE sequences are in frame with the gD sequences and the gI remnant is out of frame. Another rearrangement led to truncation of the US9 gene and at least partial deletion of the US2 gene. The locations of the gene sequences encoding the transmembrane portions of the gD, gI, gE, and US9 proteins are indicated by black boxes.
Construction of mutant PrV-107-gM−.
For isolation of PrV-107-gM−, plasmid ΔUL10β (11) was cotransfected with DNA from PrV-107 (52) into RK13-gM cells. Recombinant viruses were identified by their blue-plaque phenotype under an agarose overlay containing 300 μg of Bluo-Gal (Life Technologies, Eggenstein, Germany) per ml. Blue plaques were picked by aspiration and purified until all plaques stained blue. One plaque isolate, designated PrV-107-gM−, was chosen for further analysis. Correct recombination was verified by Southern blot analysis of mutant virus DNA.
Passaging of PrV-gE/I/M−.
Confluent Vero cells in a 75-cm2 tissue culture flask were inoculated at a multiplicity of infection (MOI) of 0.01 with PrV-gE/I/M−. Cells were repeatedly split 1:1 until a complete cytopathic effect (CPE) was observed. After development of complete CPE, the supernatant was removed, and residual adherent cells were trypsinized and reseeded with ca. 107 noninfected Vero cells in 15 ml of medium. This procedure was performed 20 times, and one plaque isolate of the 20th passage, designated PrV-gE/I/M−Pass, was further characterized.
Southern blot analysis.
Southern blot analysis of BamHI-restricted viral DNA was performed by standard procedures (31, 48).
Determination of plaque size.
Plaque size was measured after titration of wild-type PrV and the different mutants on the various cell lines. After 2 days of incubation at 37°C under methylcellulose medium, cells were fixed and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as described previously (4). Cells infected with β-galactosidase-negative PrV-Be, PrV-107, or PrV-gE/I− were fixed with 5% formaldehyde and stained with crystal violet. Thirty plaques per virus strain and cell line were measured microscopically, and average plaque sizes were determined.
One-step growth analysis.
For analysis of growth characteristics, RK13 cells were infected with PrV-1112, PrV-gM−, PrV-gE/I−, PrV-gE/I/M−Pass, PrV-Be, PrV-107, or phenotypically gM-complemented PrV-gE/I/M− or PrV-107-gM− at an MOI of 5. After incubation for 1 h at 4°C, the inoculum was replaced with warmed medium, and cells were further incubated for 90 min at 37°C to allow virus penetration. Thereafter, remaining extracellular virus was inactivated by low-pH treatment (34). Supernatants and cells were harvested separately immediately thereafter (0 h) and after 4, 8, 12, 24, and 36 h of incubation at 37°C. Titers of progeny virus were determined by plaque assays on RK13 cells. Since no significant difference in the ratio of intra- and extracellular infectious virus was observed between the different viruses, the titers were added, and the average values and standard deviations of two independent experiments were calculated.
Electron microscopy.
RK13 cells were infected with virus at an MOI of 1, fixed 16 h after infection, and prepared for ultrathin sectioning as described before (4, 21). Sections were examined with an electron microscope (EM400T; Philips, Eindhoven, The Netherlands). For intracellular labeling of viral proteins, RK13 cells were infected with PrV-gE/I/M− and analyzed 16 h after infection (4). Cells were fixed with 0.5% glutaraldehyde in phosphate-buffered saline (PBS) (pH 7.2) for 30 min, embedded in low-melting-point agarose (Biozym), and postfixed for another 30 min. Thereafter, samples were incubated in 0.5 M NH4Cl in PBS for 60 min, washed with PBS, stained in 0.5% aqueous uranyl acetate for 15 min, dehydrated in ethanol with progressive lowering of the temperature, embedded in the acrylic resin Lowicryl K4M (Lowi, Waldkraiburg, Germany) at −35°C, and polymerized with UV light (λ = 360 nm). Postembedding labeling of ultrathin sections was performed after surfaces were blocked with 1% coldwater fish gelatin–0.02 M glycine–1% bovine serum albumin fraction V (Sigma, Deisenhofen, Germany) in PBS, by either overnight incubation at 4°C or 2 h of incubation at room temperature with a rabbit antiserum directed against a glutathione-S-transferase–UL49 fusion protein, diluted in PBS-bovine serum albumin. Gold-tagged antispecies antibodies (GAR15; Biocell International, Cardiff, United Kingdom) were added for 60 min at room temperature, and excess conjugate was removed by washing. The specificity of the reaction was controlled by incubation of noninfected cells, by using gold conjugate without primary antibody, and by using antisera directed against non-PrV proteins (anti-Newcastle disease virus and anti-rabbit hemorrhagic disease virus), as well as by incubation with normal rabbit serum.
Sequence determination.
Sequencing was performed by the method of Sanger et al. (49) on double-stranded DNA with pUC19-specific primers (New England Biolabs) and PrV-gD-, PrV-gI-, and PrV-gE-specific primers (Life Technologies), which were derived from the available sequence information (43–45). For sequence determination, the 3.3-kb BamHI fragment 7 of PrV-gE/I/M−Pass was isolated and cloned into vector pUC19 (New England Biolabs). The sequenced portion of the fragment extended from the start of the gD gene to the BamHI site in the US2 gene. The gD-, gI-, and gE-specific sequences of PrV-gE/I/M−Pass were compared with the respective sequences of wild-type PrV strain Ka determined by us (accession no. AJ271967 and AJ271966).
Western blot analyses of purified virions.
Confluently grown MDBK cells in 150-cm2 tissue culture flasks were infected at an MOI of 0.1. After a complete CPE was observed, cells were lysed by freezing (−70°C) and thawing (37°C), and cellular debris was removed by low-speed centrifugation. For reduction of volume, the virus-containing supernatant was centrifuged for 1 h at 22,000 rpm. The pellet was resuspended in 10 ml of TBSal (10 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1.8 mM CaCl2, 20 mM MgCl2) and ultracentrifuged for 1 h at 20,000 rpm through a sucrose cushion (30% sucrose in TBSal). Virions were resuspended in TBSal and lysed in sample buffer. A total of 10 μg of protein per lane was electrophoresed through discontinuous sodium dodecyl sulfate (SDS)–10 or 13% polyacrylamide gels. Western blotting was performed essentially as described previously (27, 55) using monoclonal antibodies (MAbs) directed against PrV gE and gC (B. G. Klupp and E. Weiland, unpublished data) or antisera directed against PrV gD (26), gI (A. R. Brack, B. G. Klupp, and T. C. Mettenleiter, unpublished data), gM (9), the gE cytoplasmic tail (53), or the US9 protein (5) at the indicated dilutions.
RESULTS
Reversion analysis of PrV-gE/I/M−.
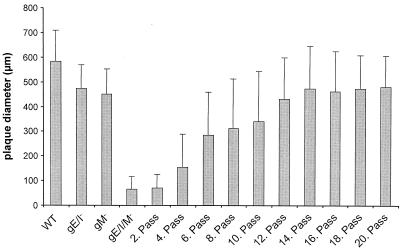
We previously showed in two instances that PrV can overcome supposedly lethal defects in virus entry by compensatory mutations which were acquired by copassaging virus-infected cells with noninfected cells (29, 50). A prerequisite for this approach is a minimal capacity of the virus mutant to perform direct cell-to-cell spread. PrV-gE/I/M− has this capacity (4). Therefore, Vero cells infected with PrV-gE/I/M− were copassaged with noninfected cells as described before (29, 50). For each passage, one parallel plate was used for determination of plaque size. As early as passage 4, larger plaques appeared among the progeny viruses and the average plaque size of the virus population started to increase, reaching the level of either gM or gE/I-deleted virus mutants after passage 14 (Fig. 1). Thus, the defect in plaque formation appears to have been overcome by a compensatory mutation(s).
FIG. 1.
Reversion of small-plaque phenotype by passaging of PrV-gE/I/M− in Vero cells. Vero cells were infected with PrV-gE/I/M−. After development of CPE, cells were trypsinized and reseeded with uninfected cells. For each second passage, a parallel plate was fixed, stained with X-Gal, and used for determination of plaque size. The average diameters of plaques are shown. Bars indicate standard deviation. A total of 30 plaques were measured for each assay.
Identification of genomic rearrangement correlating with phenotypic reversion.
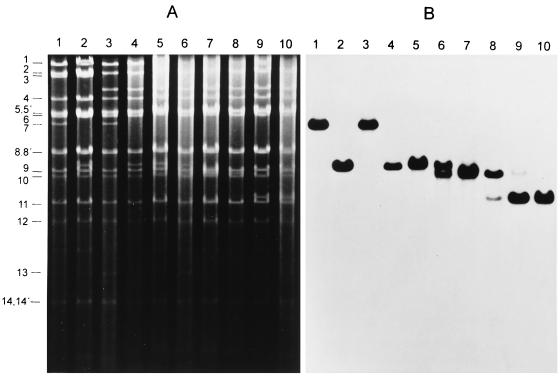
To identify the compensatory mutation(s), Southern blot hybridizations were performed on the passaged virus stock. As shown in Fig. 2, there appear to be at least two distinct mutational events resulting in alteration of BamHI fragment 7 which correlate with the increase in plaque size. A first deletion of about 300 bp was observed as early as passage 4 (Fig. 2, lanes 6), resulting in a loss of the parental truncated BamHI fragment 7 as present in PrV-gE/I/M− (Fig. 2, lanes 4). After passage 6, the virus population contained only the smaller fragment (Fig. 2, lanes 7). However, at passage 10, another even smaller fragment was detected resulting from deletion of approximately 700 bp (Fig. 2, lanes 8), which was the predominant fragment found after 20 passages (Fig. 2, lanes 9). From the virus population at passage 20, one plaque isolate possessing the short BamHI fragment 7 (Fig. 2, lanes 10) was designated PrV-gE/I/M−Pass and characterized further.
FIG. 2.
Southern blot analysis of PrV-gE/I/M−Pass. Southern blot analysis was performed with DNAs from wild-type PrV-Ka (lanes 1), PrV-gE/I− (lanes 2), PrV-gM− (lanes 3), PrV-gE/I/M− (lanes 4), and PrV-gE/I/M− passaged 2 (lanes 5), 4 (lanes 6), 6 (lanes 7), 10 (lanes 8), and 20 times (lanes 9) as well the single-plaque isolate PrV-gE/I/M−Pass (lanes 10) after cleavage with BamHI. (A) Ethidium bromide-stained gel. (B) Hybridization with genomic BamHI fragment 7 derived from the US region (see Fig. 4 for location). The locations of the BamHI fragments of PrV wild-type DNA are on the left.
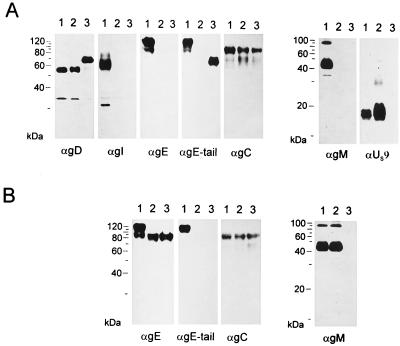
To determine the effect of these mutations on the expression of proteins from this genomic region, purified virions of either wild-type PrV (Fig. 3A, lanes 1), PrV-gE/I/M− (Fig. 3A, lanes 2) or PrV-gE/I/M−Pass (Fig. 3A, lanes 3) were analyzed by Western blotting for the presence of viral proteins gD, gI, gE, gC, gM, US9, and the gE cytoplasmic tail. As expected, neither PrV-gE/I/M− or PrV-gE/I/M−Pass reacted with the anti-gE MAb, the anti-gI serum, or the anti-gM serum. Thus, reversion of the phenotype was not due to inadvertent rescue of either of these genes or contamination of the mutant virus stock with wild-type PrV. Surprisingly, whereas gC was expressed similarly by all viruses, there was a distinct difference in the appearance of gD. The gD of PrV-gE/I/M−Pass migrated with an apparent molecular mass about 15 kDa higher than those for the gD proteins of wild-type PrV-Ka and PrV-gE/I/M−. A protein of similar size was also recognized in PrV-gE/I/M−Pass virions by an antiserum specific for the gE cytoplasmic tail (53), whereas in PrV-Ka, a protein of the size of mature gE was detected. In addition, expression of the US9 protein was no longer detectable in PrV-gE/I/M−Pass.
FIG. 3.
Western blot analysis. Purified virions of PrV-Ka (A, lanes 1), PrV-gE/I/M− (A, lanes 2), PrV-gE/I/M−Pass (A, lanes 3), PrV-Be (B, lanes 1), PrV-107 (B, lanes 2), and PrV-107-gM− (B, lanes 3) were lysed, and proteins were separated in an SDS–10 or 13% polyacrylamide gel. After electrophoretic transfer, nitrocellulose membranes were probed with antibodies against gD, gI, gE, the gE cytoplasmic tail, gC, gM, or the US9 protein. After incubation with peroxidase-conjugated secondary antibody, bound antibody was visualized by chemiluminescence recorded on X-ray films.
Phenotypic reversion is associated with creation of a gD-gE hybrid gene.
To identify the mutational events at the DNA level, a 3.3-kb fragment of PrV-gE/I/M−Pass was isolated and partially sequenced. As summarized in Fig. 4, two deletion events must have occurred during passaging. A 309-bp deletion removed the last 42 nucleotides of the gD gene, resulting in deletion of the carboxy-terminal 14 codons extending into the remnant of the gI gene, thereby eliminating the first 244 nucleotides encoding the amino-terminal 82 amino acids (aa). By this fusion event, the remaining sequences of the gE gene, which comprise 312 nucleotides encoding the carboxy-terminal 104 aa, are fused in-frame behind the transmembrane domain of gD with a spacer of 26 aa derived from the gI gene, which, however, are not related to gI sequences due to a frameshift in the remaining gI gene. The resulting chimeric protein thus consists of the first 386 aa of gD fused to 26 nonsense amino acids derived from the gI gene and the carboxy-terminal 104 aa of gE. The predicted increase in molecular mass for the gD(I)E hybrid protein of about 13 kDa matched well with the apparent molecular mass determined from gel electrophoresis (see above).
A second deletion removed at least 740 bp from the 3′ end of the US9 gene, extending into the 5′ part of the US2 gene.
Role of the gE cytoplasmic tail in plaque formation.
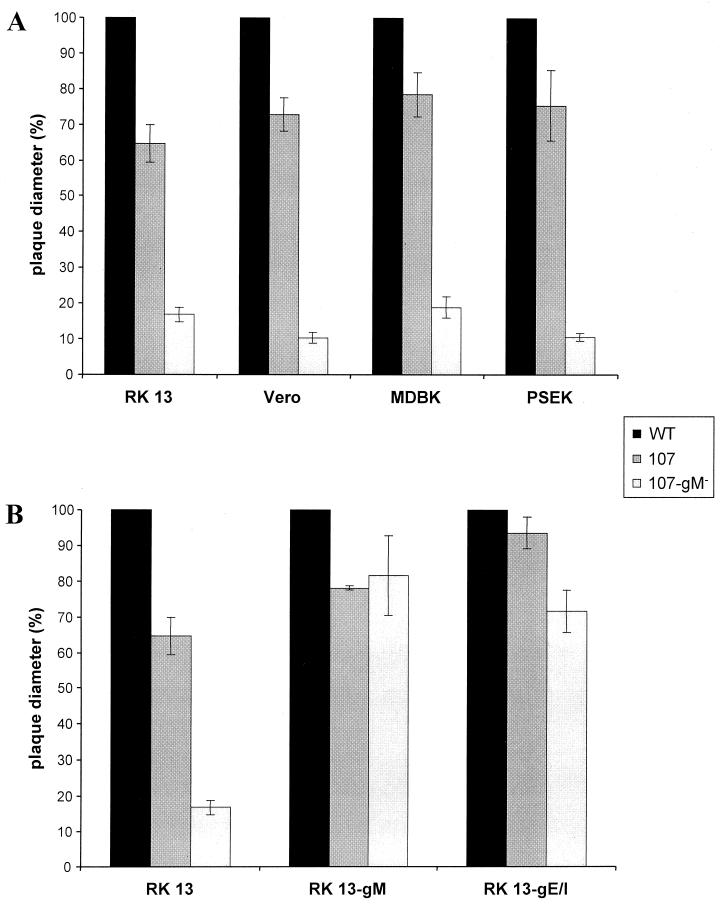
Since these data implied a role for the gE cytoplasmic tail in the phenotypic reversion of plaque size, we analyzed whether deletion of gM in a PrV mutant which expressed a C-terminally truncated gE protein resulted in the same phenotype as after complete deletion of gE/I. To this end, DNA from PrV-107, which contains a stop codon engineered behind the coding sequences for the gE transmembrane domain (52), was cotransfected with plasmid ΔUL10β (11) into gM-expressing cells. Blue plaques were purified from the transfection progeny and analyzed by Southern and Western blotting. One single plaque isolate, PrV-107-gM−, which contained the correct deletion in the UL10 gene encoding gM, expressed β-galactosidase and the truncated gE (Fig. 3B), and lacked gM in the Western blot (Fig. 3B) was further analyzed. As shown in Fig. 5A, PrV-107 produced plaques which were about 30% smaller than isogenic wild-type PrV-Be plaques on RK13, Vero, MDBK, and PSEK cells. However, deletion of gM drastically reduced plaque size to approximately that observed after deletion of gE/I and gM (4). This defect in PrV-107-gM− could be rescued on cells expressing either gM or gE/I (Fig. 5B), which was again similar to the situation with PrV-gE/I/M−. Thus, the absence of only the cytoplasmic tail of gE has the same consequences in combination with deletion of gM as elimination of gE and gI.
FIG. 5.
Plaque sizes of PrV-107 and PrV-107-gM−. RK13, Vero, MDBK, and PSEK cells (A), and RK13, RK13-gM, and RK13-gE/I cells (B) were infected under plaque assay conditions with PrV-107 and PrV-107-gM−. Two days after infection, plaque diameters were measured microscopically and compared with the average diameter of plaques induced by parental PrV-Be (WT, solid bars), which was set at 100%. Average values and standard deviations after measurement of at least 30 plaques in three independent experiments each are indicated.
One-step growth analyses.
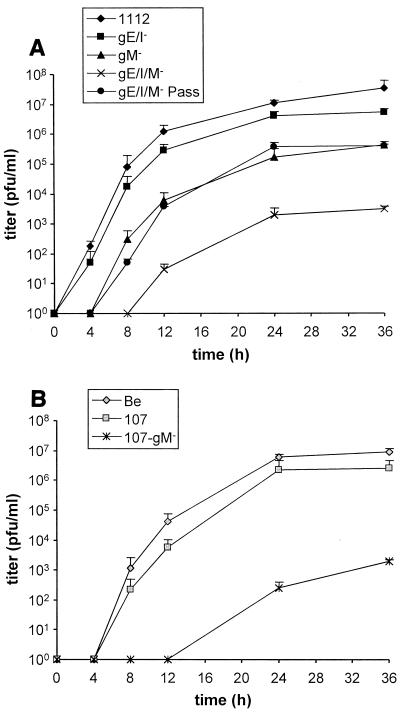
Mutant viruses were analyzed for their replication in noncomplementing RK13 cells by establishing one-step growth kinetics. As shown in Fig. 6A, PrV-Ka and PrV-gE/I− exhibited similar growth properties. In contrast, as described before (4, 9), PrV-gM− replicated to ∼100-fold-lower titers. The titers of the triple mutant PrV-gE/I/M− were again about 100-fold lower than those of PrV-gM−, with final titers reaching only 103 PFU/ml. PrV-gE/I/M−Pass exhibited a growth performance similar to that of PrV-gM−, indicating that the compensatory mutation(s) present in PrV-gE/I/M−Pass rescued the gE/I defect but not the gM-associated defect. The role of the cytoplasmic gE tail in the growth deficiency of PrV in the absence of gM was also analyzed. The data in Fig. 6B demonstrated that the absence of the gE tail has no significant effect on the growth properties of PrV-Be. In contrast, additional deletion of gM resulted in a growth curve which was very similar to that seen for PrV-gE/I/M−. Thus, the absence of the gE cytoplasmic tail yielded the same results as the absence of the whole gE/I complex.
FIG. 6.
One-step growth analysis. RK13 cells were infected at an MOI of 5 with PrV-1112, PrV-gE/I−, PrV-gM−, PrV-gE/I/M−, and PrV-gE/I/M−Pass (A) or PrV-Be, PrV-107, and PrV-107-gM− (B). At the indicated times after infection, supernatant and cells were harvested, titers were determined on RK13 cells, and the titers were added. Average values and standard deviations of two independent experiments are shown.
Role of the gE cytoplasmic tail in formation of infectious virus.
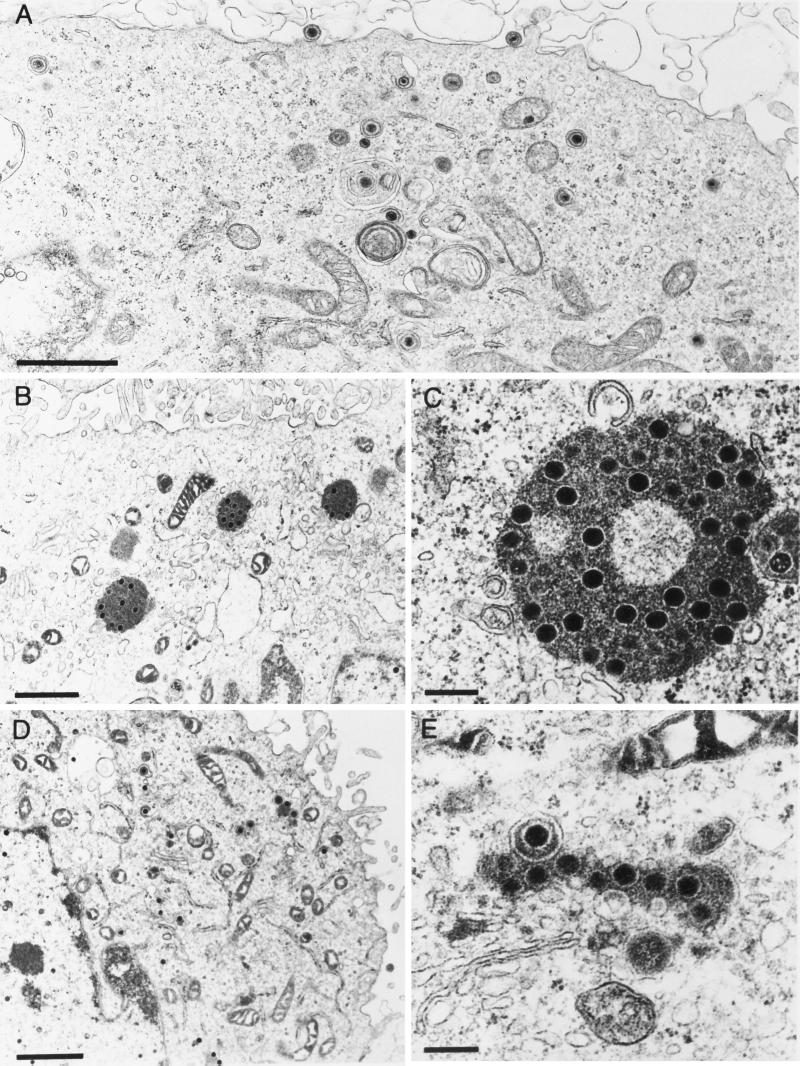
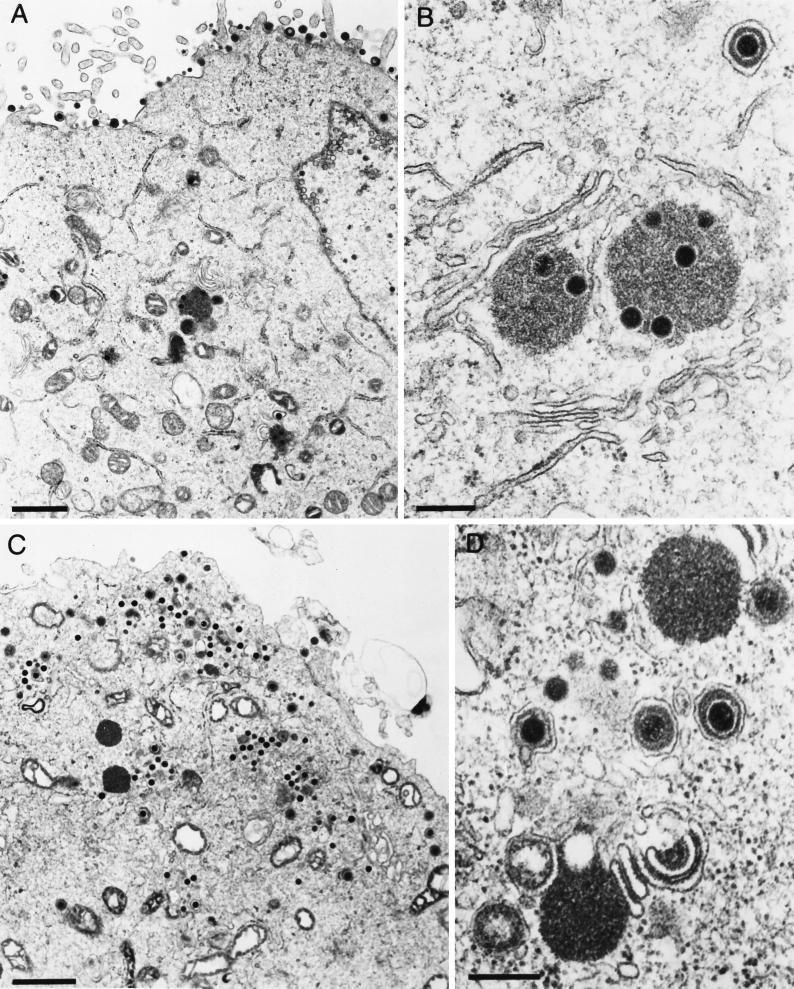
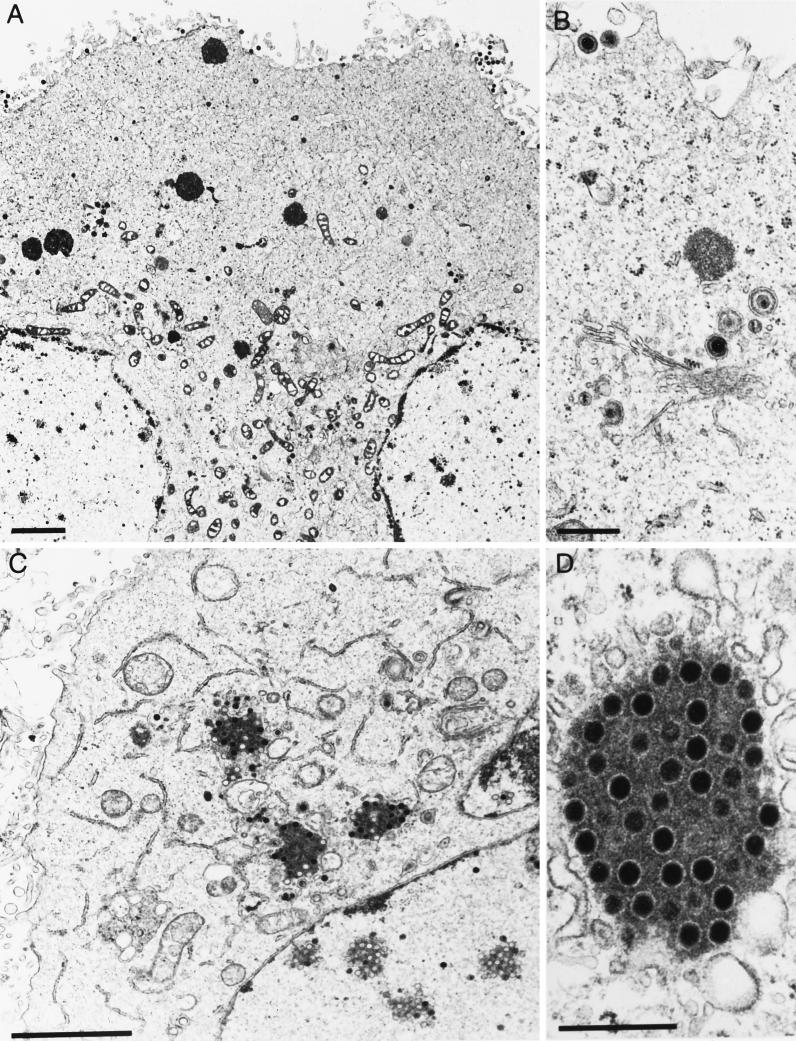
Ultrastructural studies revealed that in the absence of gE/I and gM, large intracytoplasmic inclusions were formed which contained electron-dense material, presumably tegument components, associated with capsids. In addition, secondary envelopment and release of enveloped virions were not observed. This correlated with dramatically decreased titers of PrV-gE/I/M− (4). To analyze the contribution of gE/I, gM, and the gE cytoplasmic tail to this phenotype, several isogenic virus mutants were analyzed under the electron microscope. These included PrV-gE/I/M−, PrV-gE/I/M−Pass, PrV-gE/I−, and PrV-gM−, which were all derived from wild-type PrV-Ka. PrV-107 and PrV-107-gM−, derived from PrV-Be, were also assayed. As already reported (4), in PrV-gE/I/M−-infected cells, capsids accumulated in large intracytoplasmic inclusions (Fig. 7B and C) which were absent from wild-type-virus-infected cells (Fig. 7A). In addition, neither secondary envelopment nor extracellular virions were detected. In PrV-gE/I/M−Pass-infected cells, enveloped particles were easily detected intra- and extracellularly, and only small accumulations of capsids and amorphous protein material were observed (Fig. 7D and E). Surprisingly, and in contrast to our previous observations, inclusions containing capsids, although less numerous and generally smaller, were also present in cells infected with PrV-gM− (Fig. 8A and B). Thus, it appears as if the formation of these inclusions is primarily due to the absence of gM. However, despite the formation of inclusion bodies, secondary envelopment was observed and extracellular enveloped virions were detected (Fig. 8A). In the absence of gE and gI in PrV-gE/I−, accumulations of electron-dense material in the cytoplasm were occasionally observed, but they did not contain capsids. Moreover, virion morphogenesis appeared to be uninhibited by these inclusions, since secondary envelopment and extracellular virions were readily observed (Fig. 8C and D). Interestingly, cells infected with PrV-107, which expresses a C-terminally truncated gE, gave a picture similar to that with PrV-gE/I−-infected cells, i.e., occasional intracytoplasmic accumulations of electron-dense material devoid of capsids as well as secondary envelopment and extracellular virions (Fig. 9A and B). Deletion of gM resulted in the formation of intracytoplasmic inclusions containing capsids, absence of secondary envelopment, and failure to detect extracellular virions (Fig. 9C and D). In conclusion, the absence of the cytoplasmic tail of gE in conjunction with the deletion of gM resulted in the same inhibition of formation of enveloped infectious virus as did the concomitant absence of gE/I and gM.
FIG. 7.
Electron microscopy of RK13 cells infected with (A) PrV-Ka, (B and C) PrV-gE/I/M−, and (D and E) PrV-gE/I/M−Pass, analyzed 16 h after infection. Bars: (A, B, and D) 1 μm; (C and E) 250 nm.
FIG. 8.
Electron microscopy of RK13 cells infected with (A and B) PrV-gM− and (C and D) PrV-gE/I−, analyzed 16 h after infection. Bars: (A and C) 1 μm; (B and D) 250 nm.
FIG. 9.
Electron microscopy of RK13 cells infected with (A and B) PrV-107 and (C and D) PrV-107-gM−, analyzed 16 h after infection. Bars: (A and D) 2 μm; (B and C) 500 nm.
Inclusion bodies contain tegument protein.
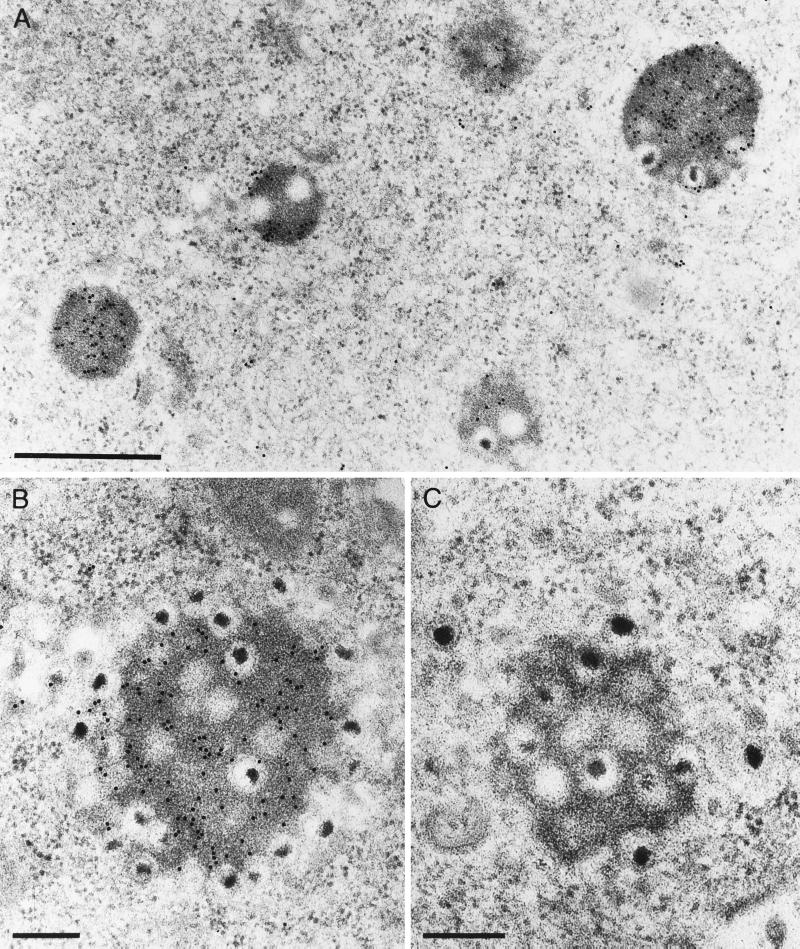
We hypothesized that the electron-dense material surrounding the capsids in the intracytoplasmic inclusions may consist of tegument proteins. To prove this assumption, we prepared an antiserum against the PrV homolog of the HSV-1 tegument protein VP22, the product of the UL49 gene, and used it in immunoelectron microscopic analyses of thin sections of RK13 cells infected with PrV-gE/I/M−. As shown in Fig. 10A and B, the anti-PrV UL49 antiserum specifically labeled the amorphous material in the inclusion bodies, whereas a control antiserum did not (Fig. 10C). Thus, the inclusion bodies contain at least one prominent tegument protein, which substantiates our hypothesis that they indeed consist of tegument material.
FIG. 10.
Immunoelectron microscopy of PrV-gE/I/M−-infected RK13 cells analyzed 16 h after infection. Thin sections were incubated with either a monospecific anti-PrV UL49 antiserum (A and B) or a control antiserum directed against Newcastle disease virus (C). Bars: (A) 750 nm; (B and C) 250 nm.
DISCUSSION
The formation of infectious PrV virions involves budding of capsids through the nuclear membrane, acquisition of tegument, secondary envelopment by budding into trans-Golgi vesicles, and release of intravesicular enveloped virus at the cell surface (21, 57). However, so far it is unclear how the different components find and interact with each other. Our observation that combined deletion of the gE/I complex and gM resulted in a severe block in the formation of infectious virus, a phenotype which has not been observed after deletion of either gE/I or gM, for the first time pointed to a role of these “nonessential” glycoproteins in an important step of virion morphogenesis which precedes secondary envelopment (4).
The data presented in this report show that deletion of the cytoplasmic domain of gE resulted in a deficiency in the formation of infectious virus in the absence of gM similar to that caused by deletion of the gE/I complex. Thus, the cytoplasmic tail of gE is responsible for gE function in virion morphogenesis. This has been shown by deletion of gM in a genetically engineered PrV mutant expressing a tailless gE and by the spontaneous creation in a phenotypic revertant of a hybrid gene in which sequences encoding the complete extracellular and transmembrane portions of gD have been fused to the gE cytoplasmic tail. The latter result was obtained after passaging of a gE/I/M-negative PrV mutant in cell culture, a procedure which we have used with success in isolation of infectious revertants of otherwise lethal mutations (29, 50).
Surprisingly, although parallel passage experiments were performed on Vero and RK13 cells, restoration of the phenotype occurred only in the virus population passaged in Vero cells. However, the resulting revertant virus exhibited a large-plaque phenotype on all cell types studied, including primate, swine, bovine, and rabbit kidney cells (data not shown). This can be explained by the infrequent and chance event of recombination occurring so that the functionally important gE tail is hooked onto a different glycoprotein, processed and transported properly, and presented in such a way that virion morphogenesis can occur. However, we cannot exclude a cell type-specific effect, as already observed for PrV-gE/I/M−, which exhibited larger foci (or even small plaques) on MDBK cells than on Vero, PSEK, or RK13 cells.
Several functions have been attributed to the gE cytoplasmic tail. It contains two YXXL endocytosis motifs, of which the first is responsible for retrieval of gE/gI from the cytoplasmic membrane (52). However, the role of endocytosis in viral replication is largely unclear. A defect in endocytosis did not affect the incorporation of gE into virions, gE-mediated virulence, or virus spread in the central nervous system of the rat (51). However, there was a correlation between the endocytosis defect and a moderate reduction in plaque size. Whether this phenotype is connected to the inhibition of virion morphogenesis described in this report remains to be analyzed. In addition, the gE cytoplasmic tail appears to be involved in the antibody-induced redistribution of viral glycoproteins in the cytoplasmic membrane (16). The biological relevance of this phenomenon is unclear, however.
The role of the cytoplasmic tail of gE in virulence is unclear. An attenuated mutant virus initially thought to specifically lack the gE tail proved to contain a second mutation resulting in a frameshift and translation of a novel cytoplasmic tail (54). Analysis of a clean tailless gE mutant, PrV-107 (52), showed that in the absence of the cytoplasmic tail, gE is still incorporated into virions and is defective in endocytosis, and the virus is still capable of infecting and spreading in the rat central nervous system. However, the animals survived longer and showed markedly less severe symptoms than those infected with wild-type virus. We report here that the absence of the gE cytoplasmic tail only slightly affects the plaque size of PrV, similar to the deletion of gE/I. However, additional deletion of gM severely impairs viral replication, and the mutant virus exhibits the same phenotype as the triply deleted PrV-gE/I/M−. This also correlates with electron microscopic observations: the prominent intracytoplasmic inclusions containing capsids associated with electron-dense material were regularly observed in large numbers in PrV-gE/I/M−- and PrV-107-gM−-infected cells. The reactivity of the amorphous material surrounding the capsids in the inclusion bodies with the anti-UL49 serum substantiates our hypothesis that it contains tegument proteins.
In contrast to our previous observations, similar inclusions were also observed in PrV-gM−-infected cells, although they are less numerous and generally smaller. Thus, the absence of gM appears to be predominantly responsible for the formation of these aggregates. However, in the absence of gM and continuous presence of gE/I, secondary envelopment and extracellular virions are still observed, which correlates with the decrease in titer in PrV-gM− compared with wild-type PrV or PrV-gE/I−. In the additional absence of gE/I or the gE cytoplasmic tail, secondary envelopment and extracellular virions could no longer be detected. This again matches the results from growth analyses which showed only very little infectious virus being formed in these cells. It is surprising in this context that deletion of gE/I or the gE tail alone had only a slight effect on virus titers. However, cells infected with PrV-gE/I− or PrV-107 occasionally contained intracytoplasmic aggregates of amorphous material which resembled those detected in PrV-gM−- or PrV-gE/I/M−-infected cells but did not contain any capsids.
During passaging of PrV-gE/I/M−, two deletion events appeared to have occurred in BamHI fragment 7. An early deletion of 309 bp which correlated with a first increase in plaque size resulted in the creation of a fusion gene containing the gE cytoplasmic tail fused in frame to the gD transmembrane region, with intervening nonsense amino acids originating from the remnant of the gI gene. It might be argued that PrV-gE/I−, which contains a remnant of the 5′ portion of the gI gene fused out of frame to the 3′ portion of the gE gene, is not a “clean” deletion mutant. This is certainly correct, although expression of the hypothetical chimeric protein has never been detected. However, it was fortunate for the reversion analysis that the mutant still contained sequences encoding the cytoplasmic tail of gE. Thus, the spontaneous formation of the gD(I)E hybrid gene led us to the role of the gE tail in virion morphogenesis. The second deletion of at least 740 bp extending beyond the terminal BamHI site was again accompanied by an increase in plaque size. Its 3′ boundary has not yet been determined exactly, but portions of the US9 and US2 genes have been removed (Fig. 4). What role this second deletion plays in increasing plaque size is unclear. It may be that either one of these proteins is directly involved in direct viral cell-to-cell spread. Alternatively, the second deletion may have increased expression of the chimeric gD(I)E gene, which in turn further increased plaque size. However, simultaneous deletion of the US9 and UL10 (gM) genes did not result in a small-plaque phenotype (Brack et al., unpublished data).
Taken together, our data show that the absence of gM results in accumulation of capsids with tegument material in the cytoplasm but that virion morphogenesis still proceeds to produce infectious virus, albeit at a lower level. In contrast, the absence of gE/I or the gE tail does not markedly impair the production of infectious virus, although infected cells were observed to occasionally contain intracytoplasmic aggregations of amorphous material devoid of capsids. Deletion of gM and gE/I or of gM and the gE cytoplasmic tail drastically impairs the formation of infectious virus. We previously speculated that gM and gE/I act at a similar step in virion morphogenesis in a synergistic fashion, either simultaneously or sequentially. From the data presented here, we further hypothesize that gM is primarily involved in directing tegumented capsids to the budding site for secondary envelopment, presumably by its presence in the vesicle membrane and interaction with tegument components. The gE cytoplasmic tail may also be involved in this process but, in the presence of gM, plays a subordinate role. However, if gM is missing, gE/I is the only functional unit left which can provide the required signals, albeit less efficiently than gM. Thus, gE/I function is of paramount importance in the absence of gM. The relative contribution of gE/I to virion morphogenesis may well be a cell type-specific effect and thus may explain the striking phenotypes of gE-deleted PrV in vivo compared with their only moderate alteration in cell culture.
ACKNOWLEDGMENTS
This study was supported by grant DFG Me 854/4-1 from the Deutsche Forschungsgemeinschaft to T.C.M. and by National Institute of Neurological Disorders and Stroke grant 1R0133506 to L.W.E.
REFERENCES
- 1.Alconada A, Bauer U, Sodeik B, Hoflack B. Intracellular traffic of herpes simplex virus glycoprotein gE: characterization of the sorting signals required for its trans-Golgi network localization. J Virol. 1999;73:377–387. doi: 10.1128/jvi.73.1.377-387.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babic N, Klupp B G, Brack A, Mettenleiter T C, Ugolini G, Flamand A. Deletion of glycoprotein gE reduces the propagation of pseudorabies virus in the nervous system of the mouse after intranasal inoculation. Virology. 1996;219:279–284. doi: 10.1006/viro.1996.0247. [DOI] [PubMed] [Google Scholar]
- 3.Becker C H. Zur primären Schädigung vegetativer Ganglien nach Infektion mit dem Herpes suis Virus bei verschiedenen Tierarten. Experientia. 1967;23:209–217. doi: 10.1007/BF02136291. [DOI] [PubMed] [Google Scholar]
- 4.Brack A R, Dijkstra J, Granzow H, Klupp B G, Mettenleiter T C. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J Virol. 1999;73:5364–5372. doi: 10.1128/jvi.73.7.5364-5372.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brideau A D, Banfield B W, Enquist L W. The Us9 gene product of pseudorabies virus, an alphaherpesvirus, is a phosphorylated, tail-anchored type II membrane protein. J Virol. 1998;72:4560–4570. doi: 10.1128/jvi.72.6.4560-4570.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browne H, Bell S, Minson T, Wilson D W. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J Virol. 1996;70:4311–4316. doi: 10.1128/jvi.70.7.4311-4316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campadelli-Fiume G, Farabegoli F, DiGaeta S, Roizman B. Origin of unenveloped capsids in the cytoplasm of cells infected with herpes simplex virus 1. J Virol. 1991;65:1589–1595. doi: 10.1128/jvi.65.3.1589-1595.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Card J P, Whealy M E, Robbins A K, Enquist L W. Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat visual system. J Virol. 1992;66:3032–3041. doi: 10.1128/jvi.66.5.3032-3041.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dijkstra J, Visser N, Mettenleiter T C, Klupp B G. Identification and characterization of pseudorabies virus glycoprotein gM as a nonessential virion component. J Virol. 1996;70:5684–5688. doi: 10.1128/jvi.70.8.5684-5688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dijkstra J, Mettenleiter T C, Klupp B G. Intracellular processing of pseudorabies virus glycoprotein M (gM): gM of strain Bartha lacks N-glycosylation. Virology. 1997;237:113–122. doi: 10.1006/viro.1997.8766. [DOI] [PubMed] [Google Scholar]
- 11.Dijkstra J, Gerdts V, Klupp B G, Mettenleiter T C. Deletion of glycoprotein gM of pseudorabies virus results in attenuation for the natural host. J Gen Virol. 1997;78:2147–2151. doi: 10.1099/0022-1317-78-9-2147. [DOI] [PubMed] [Google Scholar]
- 12.Dingwell K, Johnson D C. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J Virol. 1998;72:8933–8942. doi: 10.1128/jvi.72.11.8933-8942.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dingwell K, Brunetti C, Hendricks R, Tang O, Tang M, Rainbow A, Johnson D C. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J Virol. 1994;68:834–845. doi: 10.1128/jvi.68.2.834-845.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dingwell K, Doering L, Johnson D C. Glycoproteins E and I facilitate neuron-to-neuron spread of herpes simplex virus. J Virol. 1995;69:7087–7098. doi: 10.1128/jvi.69.11.7087-7098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enquist L W, Husak P, Banfield B W, Smith G A. Infection and spread of alphaherpesviruses in the nervous system. Adv Virus Res. 1999;51:237–347. doi: 10.1016/s0065-3527(08)60787-3. [DOI] [PubMed] [Google Scholar]
- 16.Favoreel H, Nauwynck H, Pensaert M. Role of the cytoplasmic tail of gE in antibody-induced redistribution of viral glycoproteins expressed on pseudorabies-virus-infected cells. Virology. 1999;259:141–147. doi: 10.1006/viro.1999.9749. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs W, Klupp B G, Granzow H, Mettenleiter T C. The UL20 gene product of pseudorabies virus functions in virus egress. J Virol. 1997;71:5639–5646. doi: 10.1128/jvi.71.7.5639-5646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs W, Klupp B G, Rziha H-J, Mettenleiter T C. Identification and characterization of the pseudorabies virus UL3.5 protein, which is involved in virus egress. J Virol. 1996;70:3517–3527. doi: 10.1128/jvi.70.6.3517-3527.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gershon A A, Sherman D L, Zhu Z, Gabel C A, Ambron R T, Gershon M D. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J Virol. 1994;68:6372–6390. doi: 10.1128/jvi.68.10.6372-6390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 21.Granzow H, Weiland F, Jöns A, Klupp B, Karger A, Mettenleiter T C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs L. Glycoprotein E of pseudorabies virus and homologous proteins in other Alphaherpesvirinae. Arch Virol. 1994;137:209–228. doi: 10.1007/BF01309470. [DOI] [PubMed] [Google Scholar]
- 23.Johnson D C, Feenstra V. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J Virol. 1987;61:2208–2216. doi: 10.1128/jvi.61.7.2208-2216.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson D C, Spear P G. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J Virol. 1982;43:1102–1112. doi: 10.1128/jvi.43.3.1102-1112.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaplan A S, Vatter A E. A comparison of herpes simplex and pseudorabies viruses. Virology. 1959;7:394–407. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- 26.Klupp B G, Baumeister J, Karger A, Visser N, Mettenleiter T C. Identification and characterization of a novel structural glycoprotein in pseudorabies virus, gL. J Virol. 1994;68:3868–3878. doi: 10.1128/jvi.68.6.3868-3878.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klupp B G, Fuchs W, Weiland E, Mettenleiter T C. Pseudorabies virus glycoprotein L is necessary for virus infectivity but dispensable for virion localization of glycoprotein H. J Virol. 1997;71:7687–7695. doi: 10.1128/jvi.71.10.7687-7695.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klupp B G, Baumeister J, Dietz P, Granzow H, Mettenleiter T C. Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but is not required for entry. J Virol. 1998;72:1949–1958. doi: 10.1128/jvi.72.3.1949-1958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klupp B G, Mettenleiter T C. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J Virol. 1999;73:3014–3022. doi: 10.1128/jvi.73.4.3014-3022.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knapp A C, Husak P J, Enquist L W. The gE and gI homologs from two alphaherpesviruses have conserved and divergent neuroinvasive properties. J Virol. 1997;71:5820–5827. doi: 10.1128/jvi.71.8.5820-5827.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kopp A, Mettenleiter T C. Stable rescue of a glycoprotein gII deletion mutant of pseudorabies virus by glycoprotein gI of bovine herpesvirus 1. J Virol. 1992;66:2754–2762. doi: 10.1128/jvi.66.5.2754-2762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kritas S K, Pensaert M, Mettenleiter T C. Role of gI, gp63 and gIII in the invasion and spread of Aujeszky's disease virus (ADV) in the olfactory nervous pathway of pigs. J Gen Virol. 1994;75:2319–2327. doi: 10.1099/0022-1317-75-9-2319. [DOI] [PubMed] [Google Scholar]
- 33.MacLean C A, Robertson L M, Jamieson F E. Characterization of the UL10 gene product of herpes simplex virus type 1 and investigation of its role in vivo. J Gen Virol. 1993;74:975–983. doi: 10.1099/0022-1317-74-6-975. [DOI] [PubMed] [Google Scholar]
- 34.Mettenleiter T C. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology. 1989;171:623–625. doi: 10.1016/0042-6822(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 35.Mettenleiter T C. Pseudorabies (Aujeszky's disease) virus: state of the art. Acta Vet Hung. 1994;42:153–177. [PubMed] [Google Scholar]
- 36.Mettenleiter T C, Schreurs C, Zuckermann F, Ben-Porat T. Role of pseudorabies virus glycoprotein gI in virus release from infected cells. J Virol. 1987;61:2764–2769. doi: 10.1128/jvi.61.9.2764-2769.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mettenleiter T C, Rauh I. A glycoprotein gX-βgalactosidase fusion gene as insertional marker for rapid identification of pseudorabies virus mutants. J Virol Methods. 1990;30:55–66. doi: 10.1016/0166-0934(90)90043-f. [DOI] [PubMed] [Google Scholar]
- 38.Mettenleiter T C, Zsak L, Kaplan A S, Ben-Porat T, Lomniczi B. Role of a structural glycoprotein of pseudorabies in virus virulence. J Virol. 1987;61:4030–4032. doi: 10.1128/jvi.61.12.4030-4032.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mulder W, Jacobs L, Priem J, Kok G, Wagenaar F, Kimman T, Pol J. Glycoprotein gE-negative pseudorabies virus has a reduced capability to infect second- and third-order neurons of the olfactory and trigeminal routes in the porcine central nervous system. J Gen Virol. 1994;75:3095–3106. doi: 10.1099/0022-1317-75-11-3095. [DOI] [PubMed] [Google Scholar]
- 40.Neubauer A, Beer M, Brandmüller C, Kaaden O R, Osterrieder N. Equine herpesvirus 1 mutants devoid of glycoprotein B or M are apathogenic for mice but induce protection against challenge infection. Virology. 1997;239:36–45. doi: 10.1006/viro.1997.8857. [DOI] [PubMed] [Google Scholar]
- 41.Olson J K, Grose C. Endocytosis and recycling of varicella-zoster virus Fc receptor glycoprotein gE: internalization mediated by a YXXL motif in the cytoplasmic tail. J Virol. 1997;71:4042–4054. doi: 10.1128/jvi.71.5.4042-4054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osterrieder N, Neubauer A, Brandmüller C, Braun B, Kaaden O-R, Baines J. The equine herpesvirus 1 glycoprotein gp21/22a, the herpes simplex virus type 1 gM homolog, is involved in virus penetration and cell-to-cell spread of virions. J Virol. 1996;70:4110–4115. doi: 10.1128/jvi.70.6.4110-4115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrovskis E, Timmins J, Giermann T, Post L E. Deletions in vaccine strains of pseudorabies virus and their effect on synthesis of glycoprotein gp63. J Virol. 1986;60:1166–1169. doi: 10.1128/jvi.60.3.1166-1169.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petrovskis E, Timmins J G, Armentrout M A, Marchioli C C, Yancy R J, Post L E. DNA sequence of the gene for pseudorabies virus gp50, a glycoprotein without N-linked glycosylation. J Virol. 1986;59:216–223. doi: 10.1128/jvi.59.2.216-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petrovskis E A, Timmins J G, Post L E. Use of λgt11 to isolate genes for two pseudorabies virus glycoproteins with homology to herpes simplex virus and varicella-zoster virus glycoproteins. J Virol. 1986;60:185–193. doi: 10.1128/jvi.60.1.185-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radsak K, Eickmann M, Mockenhaupt T, Bogner E, Kern H, Eis-Huebinger A, Reschke M. Retrieval of human cytomegalovirus glycoprotein B from the infected cell surface for virus envelopment. Arch Virol. 1996;141:557–572. doi: 10.1007/BF01718317. [DOI] [PubMed] [Google Scholar]
- 47.Roizman B, Desrosiers R, Fleckenstein B, Lopez C, Minson A C, Studdert M J. The family Herpesviridae: an update. Arch Virol. 1992;123:425–449. doi: 10.1007/BF01317276. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 49.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmidt J, Klupp B G, Karger A, Mettenleiter T C. Adaptability in herpesviruses: glycoprotein D-independent infectivity of pseudorabies virus. J Virol. 1997;71:17–24. doi: 10.1128/jvi.71.1.17-24.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tirabassi R S, Enquist L W. Role of envelope protein gE endocytosis in the pseudorabies virus life cycle. J Virol. 1998;72:4571–4579. doi: 10.1128/jvi.72.6.4571-4579.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tirabassi R S, Enquist L W. Mutation of the YXXL endocytosis motif in the cytoplasmic tail of pseudorabies virus gE. J Virol. 1999;73:2717–2728. doi: 10.1128/jvi.73.4.2717-2728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tirabassi R S, Enquist L W. Role of the pseudorabies virus gI cytoplasmic domain in neuroinvasion, virulence, and posttranslational N-linked glycosylation. J Virol. 2000;74:3505–3516. doi: 10.1128/jvi.74.8.3505-3516.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tirabassi R S, Townley R A, Eldridge M G, Enquist L W. Characterization of pseudorabies virus mutants expressing carboxy-terminal truncations of gE: evidence for envelope incorporation, virulence, and neurotropism domains. J Virol. 1997;71:6455–6464. doi: 10.1128/jvi.71.9.6455-6464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whealy M, Card J P, Robbins A K, Dubin J R, Rziha H-J, Enquist L W. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J Virol. 1993;67:3786–3797. doi: 10.1128/jvi.67.7.3786-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whealy M E, Card J P, Meade R P, Robbins A K, Enquist L W. Effect of brefeldin A on alphaherpesvirus membrane protein glycosylation and virus egress. J Virol. 1991;65:1066–1081. doi: 10.1128/jvi.65.3.1066-1081.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whiteley A, Bruun B, Minson T, Browne H. Effects of targeting herpes simplex virus type 1 gD to the endoplasmic reticulum and trans-Golgi network. J Virol. 1999;73:9515–9520. doi: 10.1128/jvi.73.11.9515-9520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu Z, Gershon M D, Hao Y, Ambron R T, Gabel C A, Gershon A A. Envelopment of varicella-zoster virus: targeting of viral glycoproteins to the trans-Golgi network. J Virol. 1995;69:7951–7959. doi: 10.1128/jvi.69.12.7951-7959.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zsak L, Zuckermann F, Sugg N, Ben-Porat T. Glycoprotein gI of pseudorabies virus promotes cell fusion and virus spread via direct cell-to-cell transmission. J Virol. 1992;66:2316–2325. doi: 10.1128/jvi.66.4.2316-2325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuckermann F, Mettenleiter T C, Schreurs C, Sugg N, Ben-Porat T. Complex between glycoproteins gI and gp63 of pseudorabies virus: its effect on virus replication. J Virol. 1988;62:4622–4626. doi: 10.1128/jvi.62.12.4622-4626.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]