Abstract
Mouse hepatitis virus (MHV)-infected cells contain full-length and subgenomic-length positive- and negative-strand RNAs. The origin and function of the subgenomic negative-strand RNAs is controversial. In this report we demonstrate that the synthesis and molar ratios of subgenomic negative strands are similar in alternative host cells, suggesting that these RNAs function as important mediators of positive-strand synthesis. Using kinetic labeling experiments, we show that the full-length and subgenomic-length replicative form RNAs rapidly accumulate and then saturate with label, suggesting that the subgenomic-length negative strands are the principal mediators of positive-strand synthesis. Using cycloheximide, which preferentially inhibits negative-strand and to a lesser extent positive-strand synthesis, we demonstrate that cycloheximide treatment equally inhibits full-length and subgenomic-length negative-strand synthesis. Importantly, following treatment, previously transcribed negative strands remain in transcriptionally active complexes even in the absence of new negative-strand synthesis. These findings indicate that the subgenomic-length negative strands are the principal templates of positive-strand synthesis during MHV infection.
Mouse hepatitis virus (MHV), a coronavirus in the Nidovirales order, contains a ∼32-kb linear, single-stranded, positive-polarity RNA genome (8, 21, 28). Upon entry into the cell, the viral genome is transcribed into seven to eight subgenomic mRNAs ranging in size from ∼1.0 to 32.0 kb (21, 32). The positive-strand mRNAs are arranged in a 3′ coterminal nested set, and each contains a 5′-end ∼72-nucleotide leader RNA sequence which is derived from the 5′ end of the genome (20, 37). Leader RNA sequences are joined to body sequences of each subgenomic-length mRNA at highly conserved intergenic (IG) sites located just upstream from the coding sequences of each viral gene (7, 14, 27, 32). In addition to the viral mRNAs, both full-length as well as subgenomic-length negative-strand RNAs and replicative form (RF) RNAs have been detected in porcine transmissible gastroenteritis virus-, bovine coronavirus-, and MHV-infected cells (13, 30, 32–35). The subgenomic-length negative-strand RNAs contain antileader RNA sequences (31, 35). While MHV negative-strand RNA synthesis is rapidly inhibited by inhibitors of protein synthesis, positive-strand synthesis is significantly more resistant, suggesting that continued protein synthesis is essential for MHV negative-strand synthesis (29).
Several discontinuous transcription models have been proposed to explain the presence of leader RNA sequences on positive-strand RNAs and antileader RNA sequences on full-length and subgenomic-length negative-strand RNAs (3, 10, 30, 31, 34, 35). It is generally agreed that these smaller RNAs do not originate from larger precursors (3, 15, 31). The leader-primed transcription model proposed that a free leader RNA was synthesized from the 3′ end of the full-length minus strand. In trans, the free leader RNA binds with highly conserved internal IG elements in a full-length minus-strand template to prime transcription of each of the subgenomic mRNAs (1, 3, 6, 19, 25). More recently, it was suggested that leader body fusion might result from quasi-continuous synthesis across looped-out regions of the template which are brought together by protein-protein interactions (22, 23). Following primary transcription of the subgenomic mRNA, these mRNAs may act as templates for the synthesis of subgenomic-length negative strands containing antileader RNA (34). Nascent labeling experiments and temperature shift experiments with a temperature-sensitive (ts) mutant defective in negative-strand synthesis strongly suggest that the subgenomic-length negative-strand RNAs are transcriptionally active after 6 h postinfection (30, 33). Data from other groups have suggested that the subgenomic negative strands may represent dead-end transcriptional products involved in the synthesis of a single mRNA (16, 24, 40).
It has also been proposed that the subgenomic-length negative strands may be synthesized directly from the incoming genomic RNA, either by trans splicing of full-length negative strands or by transcription attenuation within the IG elements (3, 22, 23, 30). While trans splicing is less attractive since full-length replicative intermediates (RIs) cannot be degraded into subgenomic-length RF RNA (3, 30), the transcription attenuation model readily accounts for most of the observations central to coronavirus discontinuous transcription (30). This model proposes that subgenomic-length negative strands are synthesized directly from full-length genomic RNA by transcription attenuation within the IG elements. The incomplete negative strands then act in trans to prime the synthesis of antileader RNA sequences located at the 5′ end of the genome. Protein protein interactions may also mediate this discontinuous transcription event to acquire antileader RNA sequences on the subgenomic negative strand (31). Subgenomic negative strands containing antileader RNAs then serve as the predominant templates for the synthesis of each equivalently sized mRNA.
In this study we have used kinetic radiolabeling experiments and cycloheximide treatment to demonstrate that the subgenomic-length negative-strand RNAs function as the principal templates for mRNA synthesis during MHV infection. The implications of these findings for coronavirus transcription are discussed.
MATERIALS AND METHODS
Virus and cell lines.
The A59 strain of MHV (MHV-A59) and the host range variant MHV-H2 were used throughout these studies. MHV-A59 stocks were propagated in 150-cm2 flasks containing murine astrocytoma (DBT) cells and were titered by plaque assay in DBT cells at 37°C (12). The MHV-H2 variant was selected for growth in Syrian baby hamster kidney cells (BHK) as previously described by our laboratory (5). MHV-H2 stocks were grown in 150-cm2 flasks of BHK cells for 40 to 48 h at 37°C and titered by plaque assay in DBT cells.
DBT cells were maintained at 37°C in Eagle's modified essential medium (MEM) containing 6% fetal clone II (HyClone Laboratories, Logan, Utah), 5% tryptose phosphate broth, gentamicin (0.05 μg/ml), and kanamycin (0.25 μg/ml) (GIBCO). BHK cells were kindly provided by Robert E. Johnston (University of North Carolina) and propagated at 37°C in αMEM containing 10% fetal calf serum (FCS), 5% tryptose phosphate broth, gentamicin (0.05 μg/ml), and kanamycin (0.25 μg/ml). DDT-1 cells, a Syrian hamster smooth muscle cell line, were propagated at 37°C in MEM containing 10% FCS, 5% tryptose phosphate broth, gentamicin (0.05 μg/ml), and kanamycin (0.25 μg/ml). Chinese hamster ovary cells (CHO) were maintained at 37°C in MEM containing 10% FCS, 5% tryptose phosphate broth, gentamicin (0.05 μg/ml), and kanamycin (0.25 μg/ml). The 17CL1 cells (kindly provided by Stanley Sawicki) were maintained in MEM containing 6% FCS, 5% tryptose phosphate broth, and gentamicin (0.05 μg/ml), and kanamycin (0.25 μg/ml). To reduce virus-induced fusion in the 17CL1 cells, pH 6.8 medium was used as previously described (30).
Radiolabeling and isolation of viral RNA.
Radiolabeling of viral RNAs was performed in 17CL1, BHK, CHO, or DDT-1 cells. Cultures of cells were seeded at densities approaching 5.0 × 105 cells/35-mm2 dish and maintained in phosphate-free MEM (ΔMEM) containing 6% fetal clone II, gentamicin (0.05 μg/ml), and kanamycin (0.25 μg/ml) for 12 to 16 h prior to infection (overnight). After infection with MHV-A59 or MHV-H2 at a multiplicity of infection (MOI) of 10 for 1 h at room temperature, the virus inocula were removed and the cultures were overlaid with ΔMEM (pH 6.8) containing 2% fetal clone II, gentamicin (0.05 μg/ml), and kanamycin (0.25 μg/ml). The cultures were maintained at 37°C. At least 30 min prior to the addition of radioisotope, the medium was removed and 100% ΔMEM containing actinomycin D (AMD) (Sigma) at a concentration of 10 μg/ml was added to the culture. At the indicated times, cultures were then radiolabeled with 32Pi (300 μCi/ml) for 1 h to radiolabel virus-specific mRNA and RI RNAs. The RNA was isolated as described previously (33).
Kinetic labeling experiments.
Cultures of murine 17CL1 cells were seeded at densities of 5.0 × 105 cells per dish and grown in complete ΔMEM for 12 to 16 h (overnight). The cultures were then infected with MHV-A59 at an MOI of 10 for 1 h, virus inocula were removed, and the cultures were incubated in ΔMEM containing 2% fetal clone II, gentamicin (0.05 μg/ml), and kanamycin (0.25 μg/ml). At 5 h postinfection, ΔMEM containing 20 μg of AMD per ml was added to the cultures. Higher concentrations of AMD were used to reduce background labeling of cellular RNAs and DNAs associated with the higher concentrations of radioisotope. At 6.0 h postinfection, cultures were radiolabeled with 32Pi (1,000 μCi/ml) for 5, 15, 30, 45, and 60 min. After radiolabeling, the monolayers were washed with cold phosphate-buffered saline and the cells were lysed in LET buffer (0.1 M LiCl, 0.01 M Tris-HCl [pH 7.4], 0.002 M EDTA [pH 8.0]) containing 50 mg of dodecyl lithium sulfate (LDS) (Fluka) per ml (30, 33). The lysate was passed through a 26-gauge needle to shear DNA and incubated for 15 min at 37°C in the presence of proteinase K (200 μg/ml). The RNA was extracted with an equal volume of low-pH (pH 4.3) phenol (Amresco, Solon, Ohio) by vortexing for 5 min. After centrifugation for 4 to 5 min in an Eppendorf centrifuge, the supernatant was extracted twice with an equal volume of low-pH phenol-chloroform-isoamyl alcohol (25:24:1) (Amresco) and once with an equal volume of chloroform. After vortexing for 4 to 5 min, the samples were centrifuged in an Eppendorf centrifuge for 4 to 5 min at 12,000 rpm. After the final extraction, the supernatant was precipitated with 3 volumes of 100% ethanol. The RNA was collected by centrifugation, dried, and resuspended in sterile, deionized, diethylpyrocarbonate-treated water for mRNA and RF RNA analysis.
Cycloheximide treatment.
Cycloheximide experiments were performed as described by Sawicki and Sawicki (29). Briefly, cultures of 17CL1 cells were seeded at densities of 5.0 × 105 cells/35-mm2 dish and maintained in ΔMEM containing 6% fetal clone II, gentamicin (0.05 μg/ml), and kanamycin (0.25 μg/ml). The cultures were infected with MHV-A59 at an MOI of 10 for 1 h at room temperature, virus inocula were removed, and the cultures were incubated in ΔMEM containing 2% fetal clone II, gentamicin (0.05 μg/ml), and kanamycin (0.25 μg/ml). At 4 h postinfection, the cultures were treated with AMD (10 μg/ml in ΔMEM). Half of the cultures were then treated with cycloheximide (100 μg/ml; Sigma) for 30 min at 4.5 h postinfection. Cultures were then radiolabeled with 32Pi (300 μCi/ml) for 1 h from 4.5 to 5.5, 6.0 to 7.0, and 7.0 to 8.0 h postinfection. Following radiolabeling, intracellular RNA was isolated as previously described, precipitated with ethanol, and resuspended in sterile, diethylpyrocarbonate-treated deionized H2O (33).
Analysis of viral RNA.
Viral mRNA and RF RNA were analyzed as described by Sawicki and Sawicki (30). Briefly, the RNA (in a total volume of 15 μl) was treated with 0.16 U of DNase I (Fluka) in 1× DNase buffer (5× DNase buffer is 500 mM NaCl, 50 mM Tris [pH 7.8], 10 mM CaCl2, and 10 mM MgCl2) for 15 min at 30°C. One-sixth of the total RNA was set aside to be heat denatured at 90°C and analyzed as mRNA. The remaining five-sixths of the sample (in a volume of 21 μl) was digested with 1.0 ng of RNase A (Worthington) in 1× RNase buffer (3× RNase buffer is 700 mM NaCl, 10 mM Tris [pH 7.4], and 30 mM EDTA [pH 8.0]) for an additional 15 min at 30°C for RF RNA analysis. Following enzymatic digestion, the RNA was mixed with 2 to 4 μl of 10% LDS containing 5 mg of proteinase K per ml for 15 min at 30°C and electrophoresed in 1× TBE (Tris-borate-EDTA)-0.8% agarose gels. The gels were dried under vacuum before exposure to Hyperfilm-MP (Amersham) at −70°C. Dried gels containing 32Pi-labeled viral RNAs were scanned for 8 to 12 h by an AMBIS radioanalytic imaging system (RIS) (Ambis, San Diego, Calif.), and the relative percent molar ratio of each RNA was calculated from the percentage of total radiolabel. To calculate the percent molar ratios of mRNA and RF RNA, the percent counts per minute in each viral RNA was divided by the molecular weight of the respective mRNA/RF RNA and normalized to 100% (33).
RESULTS
MHV subgenomic RI RNAs are transcriptionally active in alternative hosts.
MHV is generally species specific in vivo and in vitro, and limited replication with MHV-JHM (∼104 PFU/ml) has been observed in some human cell lines (5, 17). Species specificity appears mediated at entry since MHV genomic RNA is infectious and MHV replicates efficiently in nonpermissive hosts that express the MHV receptor for entry (5, 17). Using a model system that may be reflective of conditions present in heavily immunosuppressed xenograph recipients, we previously isolated an MHV variant (MHV-H2) that replicated efficiency (107 to 108 PFU/ml) in mouse, hamster, and primate cell lines (5). It has been suggested that the subgenomic negative strands represent dead-end products of transcription (16, 24, 40). Accordingly, efficient virus replication in alternative host species may alter the molar ratio or, perhaps, not require the synthesis of subgenomic-length negative-strand RNAs. To test this hypothesis, we characterized the abundance, synthesis, and regulation of the positive- and negative-strand templates during MHV-H2 infection in alternative hosts species.
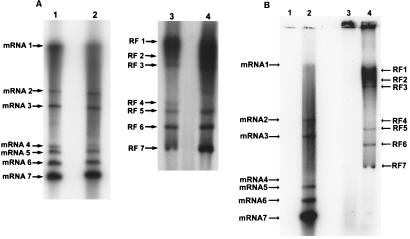
Cultures of 17CL1 cells were maintained in ΔMEM overnight and infected with MHV-A59 or MHV-H2. The cultures were treated with AMD (10 μg/ml) at 5 h postinfection and radiolabeled with 300 μCi of 32Pi from 6 to 7 h postinfection. Intracellular RNA was isolated for mRNA and RF RNA analysis. As shown in Fig. 1A, both the MHV-H2 variant and the parental virus synthesized six to seven subgenomic mRNAs in murine cell lines. As previously reported, the MHV-H2 mRNAs 2 and 3 were somewhat smaller than those in MHV-A59, reflecting the presence of a deletion in the MHV-H2 S glycoprotein gene (5). Both viruses synthesized full-length and subgenomic-length RI RNAs, as evidenced by the presence of full-length and subgenomic-length RF RNAs in mouse cells. Following MHV-A59 infection in BHK cell lines, no viral mRNAs or RF RNAs were observed, consistent with the nonpermissive phenotype of these host cells for wild-type MHV strains (Fig. 1B). In contrast, seven viral mRNAs and RF RNAs were detected in MHV-H2-infected BHK cells, effectively linking productive viral infection with the synthesis of transcriptionally active subgenomic-length negative strands. Importantly, MHV-H2 mRNA and negative-strand synthesis were tightly regulated, as the relative percent molar ratio of each RF RNA generally reflected the abundance of its mRNA in both 17CL1 and BHK cell lines (Table 1).
FIG. 1.
MHV-H2 RNA synthesis in murine and hamster cell lines. Cultures of murine 17CL1 (A) cells and syrian hamster BHK (B) cells were infected with MHV-A59 or MHV-H2 at an MOI of 10 for 1 h. At 6 and 17 h postinfection respectively, cultures of 17CL1 cells and BHK cells were labeled with 300 μCi of 32Pi as described in Materials and Methods. Intracellular RNA was isolated from infected cells and analyzed for the presence of virus-specific mRNAs and RF RNAs. Lanes: 1, MHV-A59 mRNAs; 2, MHV-H2 mRNAs; 3, MHV-A59 RF RNAs; 4, MHV-H2 RF RNAs.
TABLE 1.
Relative percent molar ratios of MHV-H2 mRNA and RF RNA in BHK, CHO, Vero, and 17Cl-1 cells
| RNA species | Relative % molar ratio
|
|||||||
|---|---|---|---|---|---|---|---|---|
| mRNA
|
RF RNA
|
|||||||
| Vero | CHO | 17Cl-1 | BHK | Vero | CHO | 17Cl-1 | BHK | |
| 1 | 1.3 | 0.7 | 4.5 | 1.7 | 4.4 | 5.2 | 5.8 | 3.2 |
| 2 | 6.3 | 3.4 | 8.6 | 5.8 | 8.5 | 8.5 | 11.3 | 5.9 |
| 3 | 7.8 | 4.3 | 6.1 | 7.1 | 9.1 | 6.6 | 11.2 | 6.4 |
| 4 | 9.3 | 7.9 | 6.4 | 11.2 | 11.3 | 10.6 | 7.6 | 12.8 |
| 5 | 10.9 | 9.2 | 11.5 | 12.6 | 13.8 | 15.8 | 11.2 | 18.0 |
| 6 | 16.0 | 15.7 | 18.9 | 16.5 | 21.3 | 21.5 | 17.4 | 23.8 |
| 7 | 48.7 | 58.8 | 44.1 | 45.1 | 30.3 | 32.4 | 31.9 | 30.0 |
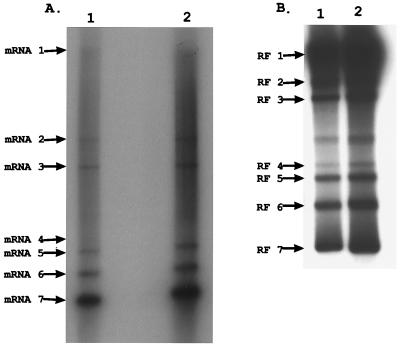
To further demonstrate the linkage between efficient MHV-H2 replication and the synthesis of subgenomic negative-strand RNAs, transcription was studied in cell lines that were not used in the selection of the variant. Cultures of CHO and DDT-1 cells were infected and radiolabeled with 32Pi (300 μCi/ml) between 8 and 9 h postinfection in CHO cells and 17 to 18 h postinfection in DDT-1 cells. The different labeling times were chosen to reflect peak levels of viral replication and to accommodate the delay in MHV-H2 RNA transcription noted in Syrian hamster DDT-1 cells (5). While MHV-A59 replication was completely restricted in these cell lines, productive MHV-H2 replication was always associated with the synthesis of full-length and subgenomic-length mRNA and RF RNA (Fig. 2). Similar findings were noted in primate (Vero) and human (HRT) cell lines (data not shown). Although the molar ratios of the RF RNAs generally reflected the abundance of each viral mRNAs, a 1-to-1 correlation was often not noted between a given RF RNA and its mRNA. This variance is likely due to the times required to synthesize different-sized mRNA from subgenomic-length templates, i.e., 1 to 2 min to synthesize the ∼1.7-kb mRNA 7 versus 10 to 20 min to synthesize the 32-kb genomic RNA (33). Concentrations of the smaller subgenomic mRNAs (mRNA 7 especially) accumulate more rapidly than genomic RNA and the other larger mRNAs, thereby altering the relative percent molar ratios of each mRNA. Consequently, percent molar ratios of each positive-strand RNA generally reflected the relative abundance of each corresponding subgenomic-length negative-strand RNA and the time required to transcribe the different-sized mRNA products from these templates.
FIG. 2.
MHV-H2 mRNA and RF RNA synthesis in CHO and DDT-1 cells. Cultures of CHO and DDT-1 Syrian smooth muscle cell lines were infected with MHV-H2 at an MOI of 10 for 1 h. At 8 and 17 h postinfection, respectively, the cultures were labeled with 300 μCi of 32Pi for 1 h. Intracellular RNA was isolated and treated as described in Materials and Methods and then separated in 0.8% agarose gels for mRNA (A) and RF RNA (B) analysis. Lanes: 1, CHO cells; 2, DDT-1 cells.
Kinetics of MHV full-length and subgenomic-length RI RNA synthesis.
For positive-strand RNA viruses, it is understood that viral mRNA is transcribed from a complementary negative-strand RNA. Nascent viral RNAs are rapidly radiolabeled and located in a partially single-stranded, partially double-stranded RI RNA (2, 36). Following RNase digestion, much of the recently transcribed RNA remains affiliated with double-stranded RF RNA cores (2, 36). Previous studies by Sethna et al. (34, 35) used probe hybridization to identify the presence of full-length and subgenomic-length negative-strand RNAs during transmissible gastroenteritis virus infection. These experiments could not address whether these negative strands were templates for mRNA synthesis. Sawicki and Sawicki used short-pulse-labeling experiments to demonstrate that full-length and subgenomic-length negative-strand RNAs were actively transcribing nascent plus strands (31). These experiments, however, were criticized because the subgenomic-length negative-strand RNAs could be dead-end products of transcription engaged in the synthesis of a single positive-strand RNA (16, 33). If negative strands were actively engaged in continual nascent plus-strand synthesis, both full-length and subgenomic-length RI RNAs should rapidly incorporate radiolabel, then saturate as the nascent positive strands become maximally radiolabeled, and then dissociate from the RI complex (36). In contrast, mRNA levels should increase steadily until degradation and transcription rates become equal.
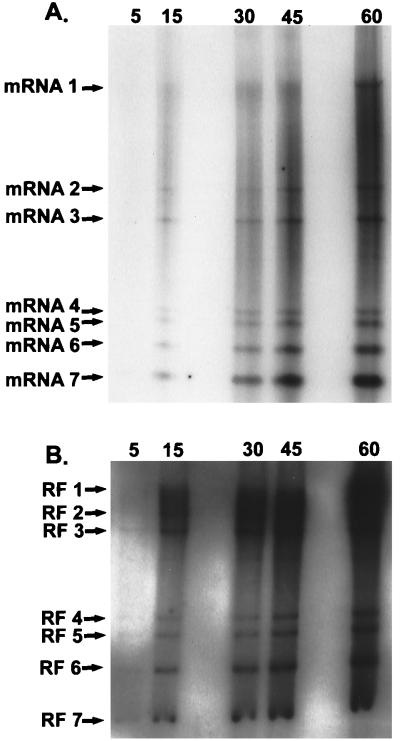
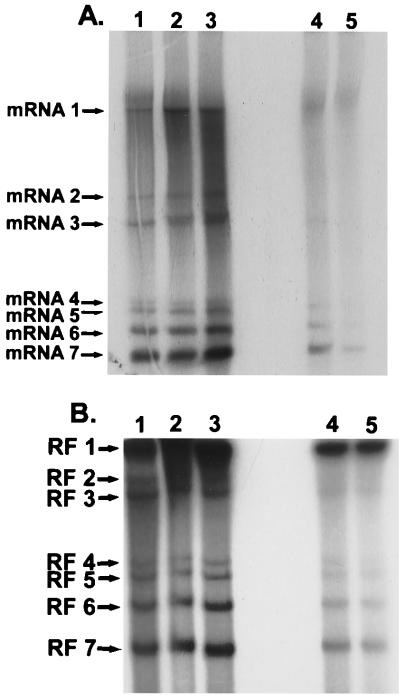
To study the saturation kinetics of full-length and subgenomic-length RI RNAs, cultures of 17CL1 cells were infected with MHV-A59 at an MOI of 10. The cultures were treated with AMD (20 μg/ml) at 5 h postinfection for 1 h. At 6 h postinfection, the cultures were radiolabeled with 32Pi (1,000 μCi/ml) for 5, 15, 30, 45, and 60 min. Under these conditions, nascent positive strands are preferentially labeled as rates of negative-strand synthesis are reduced at later times postinfection (29). The intracellular RNAs were isolated and analyzed for mRNA and RF RNA. Similar to findings reported by Sawicki and Sawicki (30), increasing amounts of radiolabel were incorporated into all viral mRNAs following longer labeling periods (Fig. 3A). Five-sixths of the intracellular RNA sample from each labeling period was also treated with DNase and RNase for RF RNA analysis. In agreement with previous reports (30, 33), full-length and subgenomic-length RF RNAs were rapidly labeled after a 5-min pulse, demonstrating that both full-length and subgenomic-length negative strands were actively engaged in mRNA synthesis. Increasing amounts of radiolabel were evident in the RF RNAs with longer labeling periods (15 to 30 min), after which time the amount of radiolabel slowly increased in the RF RNAs (Fig. 3B). Such findings were consistent with the hypothesis that the nascent plus strands on full-length and subgenomic-length RI RNAs rapidly saturated with label and that all negative strands remained actively engaged in the synthesis of many new mRNAs.
FIG. 3.
MHV mRNA and RF RNA synthesis during MHV infection. Cultures of 17CL1 cells were infected with MHV-A59 at an MOI of 10 for 1 h. The cultures were treated with AMD and radiolabeled with 1,000 μCi of 32Pi for 5, 15, 30, 45, and 60 min at 6.0 h postinfection. Intracellular RNA was isolated and treated as described in Materials and Methods for mRNA (A) and RF RNA (B) analysis in 0.8% agarose gels.
Saturation kinetics of full-length and subgenomic-length RI RNAs.
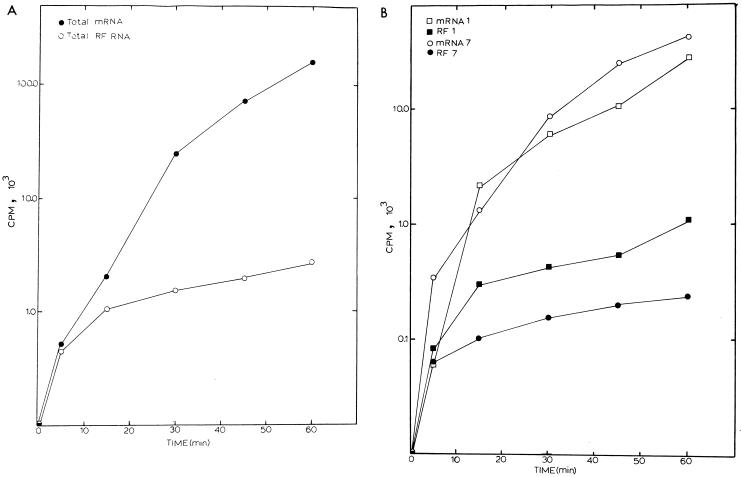
If subgenomic-length negative-strand RNAs were dead-end products of transcription, a linear increase in the amount of RF RNA should be evident over time as each newly synthesized negative strand synthesized a single positive-strand RNA and then “burned out” as a double-stranded RF RNA core. In contrast, if the subgenomic-length negative-strand RNAs function in transcriptionally active RI RNAs synthesizing many new nascent positive-strand RNAs, then full-length and subgenomic-length RI RNAs should generally saturate with label over time. This is because nascent strands will become fully labeled and then dissociate from the template as complete mRNAs. The 32Pi-labeled gels in Fig. 3 were scanned by AMBIS RIS for 8 to 12 h, and the total amounts of radiolabel in each mRNA and RF RNA were quantified. Total amounts of viral mRNA increased steadily over the labeling period (Fig. 4A). In contrast rapid increases in total RF RNA synthesis were evident only during the first 15- to 30-min pulse-label, after which a gradual linear increase in total RF RNA levels was evident. The gradual increase in total RF RNA after 30 min was most likely due to the presence of hotter pools of nucleotide precursors and continued negative-strand synthesis, which occurs at low levels throughout MHV infection (29–31).
FIG. 4.
Labeling kinetics of full-length and subgenomic-length RNAs during MHV infection. The gels shown in Fig. 3 were scanned by AMBIS RIS for 12 h, and radioactivity in each mRNA and RF RNA was determined and plotted as a function of time. (A) Incorporation of label into all seven viral mRNAs and RF RNAs; (B) radiolabeling kinetics of mRNA 1, mRNA 7, RF RNA 1, and RF RNA 7.
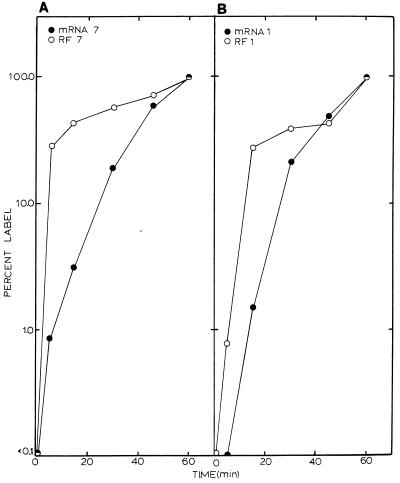
Labeling kinetics of individual MHV full-length and subgenomic-length RF RNAs reflected the kinetics of total RF RNA labeling. Rapid increases in the levels of RF RNA 1 and RF RNA 7 were noted during the first 30-min pulse-labeling period, after which total counts in these RF RNAs became relatively constant. In contrast, total counts in mRNAs 1 and 7 increased steadily over the same labeling period (Fig. 4B). To provide additional evidence that the full-length and subgenomic-length negative-strand RNAs were in transcriptionally active RI structures, we compared the saturation kinetics of mRNAs and RF RNAs 1 and 7 over the 1-h time period (Fig. 5). Importantly, while steady increases in mRNAs 1 and 7 were noted throughout the labeling period, their RF RNAs rapidly saturated with label. By comparing the percent label incorporated as a function of total mRNA or RF RNA, saturation kinetics revealed that over 70% of the radiolabel was incorporated into full-length and subgenomic-length RF RNAs within 30 min of the addition of label (Fig. 5). Only slight increases in the amount of RF RNA 1 or 7 were detected after this time. In contrast, under identical conditions, only about 15% of the total label was incorporated into mRNA 1 or 7 within the first 30 min. These data were consistent with the hypothesis that both full-length and subgenomic-length negative strands remained in transcription-active RI complexes engaged in the continual synthesis of numerous positive-strand RNAs.
FIG. 5.
Saturation kinetics of full-length and subgenomic-length RF RNAs. Radiolabeling kinetics of the MHV viral mRNAs and RF RNAs 1 and 7 were plotted as a function of percent label in a 1-h labeling period. (A) mRNA and RF RNA 7; (B) mRNA and RF RNA 1.
While the slight increase in total levels of the RF RNAs noted after 30 min was most likely due to new negative-strand synthesis and hotter pools of nucleotide precursors, this could also represent RF RNA structures which had burned out and were accumulating during infection. If this were the case, then the relative percent molar ratios of the transcriptionally active full-length RF RNA should decrease over time in proportion to the low rate of increase in the molar ratios of the slowly accumulating burned out subgenomic RF RNAs. To address this possibility, we calculated the percent molar ratio of each RF RNA and mRNA during the labeling period. Relative percent molar ratios of the viral mRNAs reflected the relative abundance of each RF RNA and remained relatively constant throughout the labeling period as well (Tables 2 and 3). Although insufficient counts were present in some of the larger mRNAs after the 5-min labeling period, molar ratios of the mRNAs remained remarkably constant from 15 to 60 min. The relative percent molar ratio of each RF RNA also remained nearly constant between 5 and 60 min. It seems likely that the full-length and subgenomic-length negative-strand RNAs exist in transcriptionally active RI structures involved in the synthesis of many new nascent positive strands and do not accumulate as dead-end products of transcription (Table 3) (10, 30, 33–35).
TABLE 2.
Relative percent molar ratios of MHV-A59 mRNAs over time
| mRNA species | Relative % molar ratio
|
||||
|---|---|---|---|---|---|
| Time (min)
|
Avg ± SD | ||||
| 15 | 30 | 45 | 60 | ||
| 1 | 2.2 | 1.3 | 0.8 | 1.3 | 1.4 ± 0.6 |
| 2 | 6.7 | 5.9 | 6.0 | 6.2 | 6.2 ± 0.4 |
| 3 | 8.2 | 7.1 | 7.5 | 7.9 | 7.7 ± 0.5 |
| 4 | 6.9 | 13.0 | 11.4 | 11.8 | 10.8 ± 2.7 |
| 5 | 9.7 | 16.4 | 14.1 | 14.7 | 13.7 ± 2.9 |
| 6 | 13.5 | 19.5 | 19.1 | 18.8 | 17.7 ± 2.8 |
| 7 | 52.8 | 36.9 | 41.1 | 39.0 | 42.5 ± 7.1 |
TABLE 3.
Relative percent molar ratio of MHV-A59 RF RNAs over time
| RF RNA species | Relative % molar ratio
|
|||||
|---|---|---|---|---|---|---|
| Time (min)
|
Avg ± SD | |||||
| 5 | 15 | 30 | 45 | 60 | ||
| 1 | 2.3 | 3.5 | 4.3 | 5.4 | 6.0 | 4.3 ± 1.5 |
| 2 | 5.9 | 9.5 | 11.6 | 12.6 | 14.3 | 10.8 ± 3.2 |
| 3 | 5.8 | 9.4 | 10.3 | 10.6 | 11.7 | 9.6 ± 2.3 |
| 4 | 10.2 | 12.0 | 10.1 | 10.9 | 9.5 | 10.5 ± 0.9 |
| 5 | 15.3 | 14.7 | 13.9 | 13.8 | 11.7 | 13.9 ± 1.4 |
| 6 | 23.3 | 21.3 | 21.9 | 20.5 | 20.9 | 21.6 ± 1.1 |
| 7 | 37.3 | 29.9 | 27.6 | 27.4 | 26.1 | 29.7 ± 4.5 |
Subgenomic negative strands remain transcriptionally active following cycloheximide treatment.
Studies using a ts mutant defective in negative-strand RNA synthesis have demonstrated that previously transcribed full-length and subgenomic-length negative strands remained in transcriptionally active RI structures engaged in the synthesis of many new nascent positive strands (33). These findings indicate that subgenomic negative strands were transcriptionally active after 6 h postinfection. To determine if the subgenomic negative strands functioned as the principal templates for mRNA synthesis, an alternative approach was developed to assess the function of the preexisting subgenomic negative strands in the absence of new negative-strand RNA synthesis. Previous studies by Sawicki and Sawicki (29) demonstrated that cycloheximide treatment rapidly inhibited the synthesis of well over 90% of the new negative-strand RNAs almost immediately after the addition of the drug to MHV-infected cultures. It was not clear whether full-length and subgenomic-length negative strands were equally inhibited by treatment. Positive-strand synthesis was much more stable and declined slowly over the next few hours (29).
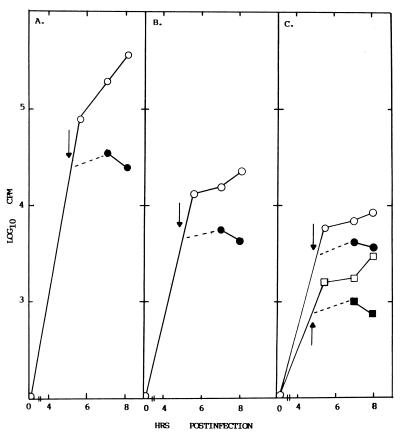
Cultures of 17CL1 cells were infected with MHV-A59, and one half was treated with cycloheximide (100 μg/ml) at 4.5 h postinfection. Cultures were then radiolabeled with 32Pi (300 μCi/ml) from 4.5 to 5.5, 6 to 7, and 7 to 8 h postinfection. Intracellular RNA was isolated, and the viral mRNAs and RF RNAs were separated in 0.8% agarose gels. Consistent with previous finding (29), positive-strand RNA synthesis continued for at least 3 h after the addition of drug (Fig. 6A). Importantly, transcriptionally active full-length and subgenomic-length RF RNAs were evident throughout the labeling period (Fig. 6B). The gels were scanned by AMBIS RIS for determination of radioactivity in each mRNA and RF RNA (Fig. 7A and B). Although cycloheximide treatment rapidly prevented additional increases of radiolabel into the RF RNAs probably by blocking new negative-strand RNA synthesis and to a lesser extent the rate of positive-strand RNA synthesis (29), previously transcribed negative-strand RNAs remained in transcriptionally active RI RNAs. Cycloheximide treatment also appeared to equally affect the synthesis of new full-length and subgenomic-length negative strands, as evidenced by reduced levels of RF RNAs (Fig. 7C). These data indicate that the subgenomic negative strands remain actively engaged in the synthesis of many new positive-strand RNAs and likely function as the principal templates for mRNA synthesis during MHV infection.
FIG. 6.
Effect of cycloheximide treatment on full-length and subgenomic-length mRNA and RF RNA synthesis. Cultures of 17Cl1 cells were infected with MHV-A59 at an MOI of 10 for 1 h. At 4.5 h postinfection, cycloheximide was added to one half of the cultures for 30 min; the cultures were radiolabeled with 32Pi (300 μCi/ml) for 1 h at 4.5 to 5.5, 6 to 7, and 7 to 8 h postinfection. Intracellular RNAs were isolated and treated as described in Materials and Methods and separated in 0.8% agarose gels. The gels were dried and exposed to X-ray film. (A) mRNA synthesis; (B) RF RNA synthesis. Lanes: 1, 4.5 to 5.5 h postinfection without cycloheximide treatment; 2, 6 to 7 h postinfection without cycloheximide treatment; 3, 7 to 8 h postinfection without cycloheximide treatment; 4, 6 to 7 h postinfection after cyclohixamide treatment; 5, 7 to 8 h postinfection after cycloheximide treatment.
FIG. 7.
Subgenomic-length negative strands remain in transcriptionally active complexes after cycloheximide treatment. The gels in Fig. 6 were scanned by AMBIS RIS for 12 h, and radioactivity in each mRNA and RF RNA was quantified by counting. (A) Total mRNA synthesis before (○) and after (●) cycloheximide treatment; (B) total RF RNA synthesis before (○) and after (●) cycloheximide treatment; (C) effect of cycloheximide treatment on mRNA 1 (○), RF RNA 1 (●), mRNA 7 (□), and RF RNA 7 (■) synthesis.
DISCUSSION
Previous studies have clearly demonstrated that a discontinuous transcription event is needed to join leader RNA sequences located at the 5′ end of the MHV genome to the body sequences of each mRNA (21). Since the seminal observation that subgenomic-length negative strands were also present in coronavirus-infected cells, it is less clear whether discontinuous transcription occurs during positive- or negative-strand RNA synthesis (13, 34, 35). Subgenomic negative strands may originate from each corresponding mRNA or from the genome-length RNA. The function of the subgenomic negative strands is also unclear, with some laboratories suggesting that they function as the principal templates for mRNA synthesis (30, 33–35) or represent dead-end products of transcription (16, 40). In this report, we provide additional evidence demonstrating that the subgenomic-length negative-strand RNAs function as the principal templates for positive-strand RNA synthesis during MHV infection.
The xenotropic MHV-H2 host range variant was isolated from mixed cell cultures of murine and hamster cell lines and efficiently replicates in murine, hamster, human, and primate cell lines. Although host range expansion was likely mediated at the level of entry (5), MHV transcription probably requires the presence of specific host factors which may regulate the efficiency of subgenomic RNA synthesis (22, 23, 43). We have demonstrated that the MHV-H2 variant transcribes both full-length and subgenomic-length negative-stranded RNAs in murine, hamster, and human cell lines. The relative percent molar ratio of each RF RNAs and each corresponding mRNA are stable and similar in alternative hosts, suggesting that the ratio of genomic RNA to subgenomic mRNAs is determined by the relative abundance of the full-length and subgenomic-length RI RNAs and the time required to synthesize different-sized mRNAs from these templates. These data also suggest that the proportion of negative- to positive-strand RNA is tightly regulated by virally encoded factors. Although these data only indirectly support a role for the subgenomic negative strands in MHV replication, they confirms earlier findings that the ratio of genome-length and subgenomic-length negative-strand RNAs directly influences the relative abundance of each corresponding positive-strand RNA (30, 31, 33).
RI RNAs likely contain many actively transcribed nascent plus strands associated with a single negative-strand template (2, 3, 36). Studies with Sindbis virus have demonstrated that RIs rapidly incorporate label and then saturate over time (2, 36). In this report, we demonstrate that the full-length and subgenomic-length MHV RI RNAs display similar labeling kinetics and saturate with label as has been described for alphavirus RI RNAs (2, 36). The relative percent molar ratio of each RF RNA also remained remarkably constant over the 1-h labeling period, consistent with the hypothesis that both full-length and subgenomic-length negative strands were actively engaged in the synthesis of many new mRNAs. Labeling kinetics were not significantly different between genome-length and subgenomic-length RI RNAs, refuting the hypothesis that the subgenomic-length negative strands represented dead-end products of transcription engaged in the synthesis of a single mRNA. Importantly, the relative abundance of the subgenomic negative strands reflected the relative molar ratio of each corresponding mRNA throughout the 1-h labeling period. Together the data argue that the subgenomic negative strands function as the principal templates for mRNA synthesis during MHV infection (30, 33–35).
Although previous studies demonstrated that MHV negative-strand synthesis required continual protein synthesis, it was not clear whether cycloheximide treatment equally inhibited the synthesis of both full-length and subgenomic-length negative strands (29). This is important, as an equine arteritis virus (EAV) replicase mutant which actively transcribes full-length, but ∼100-fold less subgenomic-length, RNAs (plus and minus) than the wild type has been described (38). These findings suggest that different viral factors may regulate transcription of full-length and subgenomic-length RNAs in Nidovirales. Our experiments demonstrate that cycloheximide treatment rapidly inhibited an increase in both full-length and subgenomic-length RI RNA, suggesting that continued protein synthesis was critical for all MHV negative-strand synthesis. If the MHV negative-strand polymerase is like the EAV enzyme, then the Nidovirales negative-strand polymerase may consist of a cycloheximide-sensitive protein factor and one or more additional proteins that regulate full- and subgenomic-length negative-strand synthesis. Importantly, in the absence of new negative-strand synthesis, these data also show that previously transcribed full-length and subgenomic-length negative strands remain actively engaged in the transcription of new mRNAs. As cycloheximide treatment rapidly inhibits the synthesis of new negative strands, these data support earlier findings with MHV ts mutants that demonstrated that preexisting negative strands remain actively engaged in the synthesis of multiple new mRNAs (33). Our results clearly support earlier findings by Sethna et al. (34, 35), Sawicki and Sawicki (30, 31), and Schaad and Baric (33) and indicate that the subgenomic negative strands function as the predominant templates for mRNA synthesis.
The finding of transcriptionally active subgenomic negative strands does not directly address the mechanism by which leader RNA sequences are joined to the body sequences of each subgenomic RNA. However, the leader-primed transcription model was heavily based on the presence of full-length negative strands in infected cells and the presence of subgenomic nascent RNAs in the full-length RIs (3, 6, 19). The former observation is clearly incorrect. The latter findings are more difficult to interpret in light of recent findings of high-frequency template switching between nascent RNAs in subgenomic-length and full-length RI RNAs during infection (3, 11, 14). Coronavirus-infected cells clearly contain negative-strand copies of subgenomic mRNA that contain antileader RNA sequences at their 3′ end (35; R. S. Baric et al., unpublished data). The finding of an EAV replicase mutant deficient in subgenomic negative-strand synthesis further supports the role of these RNA templates in mRNA synthesis (38). The origin of these negative-strand RNAs is less clear. Although transfected mRNAs can function in recombination and repair of defective MHV genomes, infected cells do not appear to efficiently replicate these exogenous mRNAs (14, 38; Baric et al., unpublished data). However, it remains possible that these mRNAs are not presented in the appropriate context or subcellular compartment to interact with the replication complex of the virus.
Alternatively, most of the available data supporting the original leader-primed transcription model are compatible with the transcription attenuation model proposed by Sawicki and Sawicki (30, 31). In this model, the insertion sequence elements in the genome-length template act as discontinuous elongation sites that allow for the polymerase and nascent minus strand to detach and reinitiate transcription (in cis or in trans) at the 5′ end of the genome. This model predicts that all subgenomic negative strands originate from the genomic RNA directly, rather than from mRNA. In support of this hypothesis, tandem placement of a coronavirus promoter resulted in enhanced mRNA synthesis from the downstream-most initiation sites (18). During EAV transcription, the IG element in subgenomic mRNA is derived from the IG sequences encoded in the genomic RNA during negative-strand synthesis rather than from leader sequences encoded at the 5′ end of the genome. These studies support a mechanism of discontinuous negative-strand synthesis reminiscent of the process of copy choice RNA recombination (39). Clearly, transcription attenuation is an attractive hypothesis which deserves serious attention, as it is consistent with the available data presented in this and other reports and represents a more direct mechanism to regulate subgenomic RNA synthesis (9, 14, 18, 25, 38). Recent UV transcription-mapping studies with mRNA transcribed from DI RNAs, however, have suggested that the subgenomic mRNAs directly originate from genome-length templates (16, 24). Unfortunately, these studies did not directly determine whether full-length or subgenomic-length templates were functioning during infection.
Additional studies must be designed to directly address the mechanism by which leader RNA sequences are joined to the body sequences of subgenomic RNA and determine whether this occurs from genome-length positive- or negative-strand RNAs. The presence of transcriptionally active subgenomic-length negative-strand RNAs provides viruses within the Nidovirales order not only a unique mechanism to regulate expression of individual viral genes but also ample opportunity for rapid evolution by recombination-mediated processes in the structural genes (4, 11, 26, 41).
ACKNOWLEDGMENTS
This research was supported by grant AI23946 from the National Institutes of Health.
We thank Sheila Peel and Wan Chen for encouraging suggestions and technical assistance.
REFERENCES
- 1.Baker S C, Lai M M C. An in vitro system for the leader-primed transcription of coronavirus mRNAs. EMBO J. 1990;9:4173–4179. doi: 10.1002/j.1460-2075.1990.tb07641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baric R S, Lineberger D W, Johnston R E. Reduced synthesis of Sindbis virus negative-strand RNA in cultures treated with host transcription inhibitors. J Virol. 1983;47:46–54. doi: 10.1128/jvi.47.1.46-54.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baric R S, Stohlman S A, Lai M M C. Characterization of replicative intermediate RNA of mouse hepatitis virus: presence of leader RNA sequences on nascent chains. J Virol. 1983;48:633–640. doi: 10.1128/jvi.48.3.633-640.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baric R S, Fu K S, Schaad M C, Stohlman S A. Establishing a genetic recombination map for MHV-A59 complementation groups. Virology. 1990;177:646–656. doi: 10.1016/0042-6822(90)90530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baric R S, Yount B, Hensley L, Peel S A, Chen W. Episodic evolution mediates interspecies transfer of a murine coronavirus. J Virol. 1997;71:1946–1955. doi: 10.1128/jvi.71.3.1946-1955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brayton P R, Stohlman S A, Lai M C C. Further characterization of mouse hepatitis virus RNA-dependent RNA polymerases. Virology. 1984;133:197–201. doi: 10.1016/0042-6822(84)90439-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budzilowicz C J, Wilczynski S P, Weiss S R. Three intergenic regions of coronavirus mouse hepatitis virus strain A59 genome RNA contain a common nucleotide sequence that is homologous to the 3′ end of the viral mRNA leader sequence. J Virol. 1985;53:834–840. doi: 10.1128/jvi.53.3.834-840.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavanagh D. Nidovirales: a new order comprising coronaviridae and arterioviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 9.Chang R Y, Krishnan R, Brian D A. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J Virol. 1996;70:2720–2729. doi: 10.1128/jvi.70.5.2720-2729.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.den Boon J A, Spaan W J M, Snijder E J. Equine arteritis virus subgenomic mRNA synthesis: analysis of leader-body junctions and replicative form RNAs. J Virol. 1996;70:4291–4298. doi: 10.1128/jvi.70.7.4291-4298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu K, Baric R S. Evidence for variable rates of recombination in the MHV genome. Virology. 1992;189:88–102. doi: 10.1016/0042-6822(92)90684-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirano N, Fujiwara K, Hino S, Matsumoto M. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch Gesamte Virusforsch. 1974;44:298–302. doi: 10.1007/BF01240618. [DOI] [PubMed] [Google Scholar]
- 13.Hofmann M A, Sethna P B, Brian D A. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J Virol. 1990;64:4108–4114. doi: 10.1128/jvi.64.9.4108-4114.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsue B, Masters P S. Insertion of a new transcriptional unit into the genome of mouse hepatitis virus. J Virol. 1999;73:6128–6135. doi: 10.1128/jvi.73.7.6128-6135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobs L, Spaan W J M, Horzinek M C, van der Zeijst B A M. Synthesis of subgenomic mRNAs of mouse hepatitis virus is initiated independently: evidence from UV transcriptional mapping. J Virol. 1981;39:401–406. doi: 10.1128/jvi.39.2.401-406.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong Y S, Makino S. Mechanism of coronavirus transcription: duration of primary transcription initiation activity and effects of subgenomic RNA transcription on RNA replication. J Virol. 1992;66:3339–3346. doi: 10.1128/jvi.66.6.3339-3346.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koetters P J, Hassanieh L, Stohlman S A, Gallagher T, Lai M M C. Mouse hepatitis virus strain JHM infects a human hepatocellular carcinoma cell lines. Virology. 1999;264:398–409. doi: 10.1006/viro.1999.9984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krishnan R, Chang R-Y, Brian D A. Tandem placement of a coronavirus promoter results in enhanced mRNA synthesis from the downstream-most initiation site. Virology. 1996;218:400–405. doi: 10.1006/viro.1996.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai M M C, Patton C D, Stohlman S A. Replication of mouse hepatitis virus: negative-stranded RNA and replicative form RNA are of genome length. J Virol. 1982;44:487–492. doi: 10.1128/jvi.44.2.487-492.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai M M C, Baric R S, Brayton P R, Stohlman S A. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic virus. Proc Natl Acad Sci USA. 1984;81:3626–3630. doi: 10.1073/pnas.81.12.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai M M C, Liano C-L, Lin Y-J, Zhang X. Coronavirus, how a large viral RNA genome is replicated and transcribed. Infect Agents Dis. 1994;3:98–105. [PubMed] [Google Scholar]
- 22.Li H-P, Zhang X M, Duncan R, Comai L, Lai M M C. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc Natl Acad Sci USA. 1997;94:9544–9549. doi: 10.1073/pnas.94.18.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H-P, Huang P, Park S, Lai M M C. Polypyrimidine tract-binding protein binds to the leader RNA of mouse hepatitis virus and serves as a regulator of viral transcription. J Virol. 1999;73:772–777. doi: 10.1128/jvi.73.1.772-777.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maeda A, An S, Makino S. Importance of coronavirus negative-strand genomic RNA synthesis prior to subgenomic RNA transcription. Virus Res. 1998;57:35–42. doi: 10.1016/S0168-1702(98)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makino S, Stohlman S A, Lai M M C. Leader sequences of murine coronavirus mRNAs can be freely reassorted: evidence for the role of free leader RNA in transcription. Proc Natl Acad Sci USA. 1986;83:4204–4208. doi: 10.1073/pnas.83.12.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makino S, Keck J G, Stohlman S A, Lai M M C. High-frequency RNA recombination of murine coronaviruses. J Virol. 1986;57:729–737. doi: 10.1128/jvi.57.3.729-737.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makino S, Joo M, Makino J. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J Virol. 1991;65:6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pachuk C J, Bredenbeck D J, Zoltick P W, Spaan W J M, Weiss S R. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus strain A59. Virology. 1989;171:141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawicki S G, Sawicki D L. Coronavirus minus-strand synthesis and effect of cycloheximide on coronavirus RNA synthesis. J Virol. 1986;57:328–334. doi: 10.1128/jvi.57.1.328-334.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawicki S G, Sawicki D L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawicki S G, Sawicki D L. A new model for coronavirus transcription. Adv Exp Biol Med. 1998;280:215–218. doi: 10.1007/978-1-4615-5331-1_26. [DOI] [PubMed] [Google Scholar]
- 32.Schaad M C, Baric R S. Evidence for new transcriptional units encoded at the 3′ end of the mouse hepatitis virus genome. Virology. 1993;196:190–198. doi: 10.1006/viro.1993.1467. [DOI] [PubMed] [Google Scholar]
- 33.Schaad M C, Baric R S. Genetics of mouse hepatitis virus transcription: evidence that subgenomic negative strands are functional templates. J Virol. 1994;68:8169–8197. doi: 10.1128/jvi.68.12.8169-8179.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sethna P B, Hung S-L, Brian D A. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc Natl Acad Sci USA. 1989;86:5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sethna P B, Hoffman M A, Brian D A. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J Virol. 1991;65:320–325. doi: 10.1128/jvi.65.1.320-325.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simmons D T, Strauss J H. Replication of Sindbis virus. II. Multiple forms of double-stranded RNA isolated from infected cells. J Mol Biol. 1972;71:615–631. doi: 10.1016/s0022-2836(72)80027-5. [DOI] [PubMed] [Google Scholar]
- 37.Spaan W, Delius H, Skinner M, Armstrong J, Rottier P, Smeekens S, van der Zeijst B A M, Siddell S G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Marle G, van Dinten L C, Spaan W J M, Luytjes W, Snijder E J. Characterization of an equine arteritis virus replicase mutant defective in subgenomic mRNA synthesis. J Virol. 1999;71:5274–5281. doi: 10.1128/jvi.73.7.5274-5281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Marle G, Dobbe J C, Gultyaev A P, Luytjes W, Spaan W J M, Snijder E J. Arteriovirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription regulating sequences. Proc Natl Acad Sci USA. 1999;96:12056–12061. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yokomori K, Banner L R, Lai M M C. Coronavirus mRNA transcription: UV light transcriptional mapping studies suggest an early requirement for a genomic-length template. J Virol. 1992;66:4671–4678. doi: 10.1128/jvi.66.8.4671-4678.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan S, Nelsen C J, Murtaugh M P, Schmitt B J, Faabeg K S. Recombination between North American strains of porcine reproductive and respiratory syndrome virus. Virus Res. 1999;61:87–98. doi: 10.1016/S0168-1702(99)00029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, Liano C-L, Lai M M C. Coronavirus leader RNA regulates and initiates subgenomic mRNA transcription both in trans and in cis. J Virol. 1994;68:4738–4746. doi: 10.1128/jvi.68.8.4738-4746.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X M, Lai M M C. Interactions between the cytoplasmic proteins and the intergenic (promoter) sequence of murine hepatitis virus RNAs: correlation with the amounts of subgenomic mRNA transcribed. J Virol. 1995;69:1637–1644. doi: 10.1128/jvi.69.3.1637-1644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]