Abstract
Bacteriophage P2 requires several host proteins for lytic replication, including helicase DnaB but not the helicase loader, DnaC. Some genetic studies have suggested that the loading is done by a phage-encoded protein, P2 B. However, a P2 minichromosome containing only the P2 initiator gene A and a marker gene can be established as a plasmid without requiring the P2 B gene. Here we demonstrate that P2 B associates with DnaB. This was done by using the yeast two-hybrid system in vivo and was confirmed in vitro, where 35S-labeled P2 B bound specifically to DnaB adsorbed to Q Sepharose beads and monoclonal antibodies directed against the His-tagged P2 B protein were shown to coprecipitate the DnaB protein. Finally, P2 B was shown to stabilize the opening of a reporter origin, a reaction that is facilitated by the inactivation of DnaB. In this respect, P2 B was comparable to λ P protein, which is known to be capable of binding and inactivating the helicase while acting as a helicase loader. Even though P2 B has little similarity to other known or predicted helicase loaders, we suggest that P2 B is required for efficient loading of DnaB and that this role, although dispensable for P2 plasmid replication, becomes essential for P2 lytic replication.
Many phages depend on the host DNA replication machinery, but they usually code for one or two proteins that direct the host machinery to their own genomes. For example, phage λ codes for λ O, an origin-binding protein, and λ P, a protein that interacts with λ O as well as with Escherichia coli DnaB helicase (36). The P protein can thus direct the helicase to the phage origin and is considered an analogue of E. coli DnaC protein, which loads the helicase onto the bacterial origin (51).
Bacteriophage P2 is a temperate coliphage that replicates via a modified rolling-circle mechanism generating monomeric double-stranded circles (7). P2 also utilizes several host components for its replication, like DNA polymerase III, primase, DnaB, and the Rep helicase (9, 11), but encodes at least two proteins for its own replication. One is protein A, which initiates replication by introducing a sequence-specific, single-stranded cut at the origin of replication (ori) (33). The second P2 protein required for phage replication is the B protein (31, 32). It is believed to be required for lagging-strand synthesis, since the displaced strand during rolling-circle replication remains single stranded in the absence of B (20). Since lagging-strand synthesis requires loading of the helicase (DnaB)-primase (DnaG) complex, a defect in helicase loading could account for the result. The requirement for the E. coli DnaB protein in P2 DNA replication is known (9). Moreover, a P2 rlb1 mutation, within the coding part of the B gene, has been shown to partially suppress a dnaB(Ts) mutation (49), suggesting that P2 B interacts directly with DnaB. These results led to the hypothesis that it is a DnaC analogue (23). However, the B protein was not required for replication of a P2 minichromosome containing only the P2 A gene and a marker gene, indicating that the function of the B protein is not essential for the loading of the helicase to the P2 ori (34). In this report we have provided in vivo and in vitro evidence for a physical association between P2 B and E. coli DnaB in support of the view that the phage protein is a helicase loader.
MATERIALS AND METHODS
Biological materials.
The bacteria, yeast, and plasmids used are listed in Table 1.
TABLE 1.
Bacteria, yeast, and plasmids used
| Strain or plasmid | Pertinent features | Reference or source |
|---|---|---|
| E. coli | ||
| BL21(DE3) | E. coli B strain carrying the T7 RNA polymerase gene under the control of lac promoter | 48 |
| DH5Δlac | recA | M. L. Berman |
| PC2 | dnaC2(Ts) | 12 |
| S. cerevisiae EGY48 | Matα trp1 his3 ura3 lexAops-LEU2 | 22 |
| Plasmids | ||
| pEE853 | pEG202 derivative expressing LexA DNA binding domain-DnaB fusion protein | This work |
| pEE854 | pJG4-51 derivative expressing B42 activation domain-P2 B fusion protein | This work |
| pEE855 | pJG4-51 derivative expressing B42 activation domain–P2 B-rlb1 fusion protein | This work |
| pEG202 | lexA DNA binding domain fusion plasmid | 22 |
| pET16b | pET plasmid containing the T7 promoter and a His tag | 48; Novagen |
| pJG4-51 | B42 activation domain fusion plasmid | 22 |
| pRLM109 | pBR322 derivative containing the λ P gene under the control of λPL promoter and cI857 repressor | R. McMacken |
| pSH18.34 | lacZ reporter plasmid. Contains eight lexA operators that control lacZ expression | 22 |
| pSH17-4 | Expresses LexA DNA binding domain-GAL4 activation domain fusion protein | 22 |
| pSP102 | Mini-P1 plasmid containing P1 ori + initiator gene (P1 coordinates −228 to 1565) | 41 |
| pST1 | pET16b derivative expressing P2 His-B fusion protein under the control of T7 promoter | This work |
| pRLM105 | pBR322 derivative containing the dnaB gene under the control of λPL promoter and cI857 repressor | R. McMacken |
Plasmid construction.
All constructions were performed according to standard procedures (45). All constructions were verified by automated DNA sequencing using an ABI Prism dye terminator cycle-sequencing ready-reaction kit (Perkin-Elmer) in an ABI Prism 377 DNA sequencer (Perkin-Elmer). The synthetic oligonucleotides used were obtained from DNA Technology, Aarhus, Denmark.
(i) pEE853.
The E. coli dnaB gene was amplified from plasmid pRM105 by PCR with primers DnaB-R (5′-GCAGGAAATAAACCCTTCAA) and DnaB-L (5′-GCCGCTTGCATTTGTGTTCC). After purification and phosphorylation, the PCR fragment was inserted into the filled-in and dephosphorylated BamHI site of pEG202. Strain HB101 was used as the initial recipient of the construct.
(ii) pEE854.
The P2 B gene was amplified by PCR with P2 DNA as a template and 79.5R (5′-GACAGTGATGACGCTCAATC) and 80.4L (see below) as primers. After purification and phosphorylation, the PCR fragment was inserted into the filled-in and dephosphorylated EcoRI site of pJG4-51. Strain HB101 was used as the initial recipient of the construct.
(iii) pEE855.
The construction of pEE855 was identical to that of pEE854, except that P2 rlb1 DNA (49) was used as a template for the PCR.
(iv) pST1.
The P2 B gene was amplified by PCR with primers 79.4R (5′-TGACAGTGATGACGCTCAAT) and 80.4L (5′-AGAAGCCCCGCACAATTAAG). After purification and phosphorylation, the PCR fragment was inserted into the filled-in and dephosphorylated NdeI site of plasmid pET16b. Strain C-1a was used as the initial recipient for the construct.
β-Galactosidase assays in the yeast two-hybrid system.
The two-hybrid system was used as described previously (22). All assays were performed with Saccharomyces cerevisiae strain EGY48. The cells were transformed by the one-step method (14) with the reporter plasmid pSH18-34, the DNA binding domain fusion plasmid, and/or the transcriptional activation domain fusion plasmid. The activation of the lacZ reporter gene was determined by a liquid assay (10). The X-Gal (5-bromo-4-chloro-3-indolyl-β-d-thiogalactopyranoside) overlay method was described previously (18).
Purification of DnaB protein.
For DnaB purification, a scaled-down version of the procedure of Arai et al. (4) was followed except that a HighTrap Q column (Amersham Pharmacia Biotech) was used instead of a DEAE-cellulose column. DnaB eluted at 0.37 M KCl. Purified DnaB (>95% pure) was stored in buffer D (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 1 mM dithiothreitol, 25% glycerol, and 1 mM ATP) at a final concentration of 800 ng/μl.
In vitro coupled transcription-translation.
[35S]methionine (15 mCi/ml; Amersham Pharmacia Biotech)-labeled P2 His-B was produced using an E. coli T7 S30 extract system (Promega) with plasmid pST1 as a template, and labeled DnaB was produced using an E. coli S30 extract system (Promega) with pRLM105 as a template, following the instructions of the manufacturer. Labeled PinPoint-CAT fusion protein and β-lactamase (Promega) were obtained similarly. The products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on the PhastSystem (Amersham Pharmacia Biotech), followed by autoradiography.
Anion-exchange chromatography.
DnaB (6 μg) in 250 μl of buffer D, supplemented with 30 mM KCl and 0.5% Tween 20, was mixed with 40 μl of Q Sepharose Fast Flow beads and incubated at room temperature for 20 min. The mixture was centrifuged to remove unbound DnaB. Five microliters of the labeled proteins was diluted in 250 μl of buffer D, supplemented with 0.15 M KCl and 0.5% Tween 20, and allowed to bind to DnaB immobilized on Q Sepharose beads for 45 min at room temperature. The supernatants were then recovered to measure the amount of unbound protein. The beads were washed with 250 μl of buffer D with 30 mM KCl and 0.5% Tween 20, and DnaB was eluted with buffer D with 0.4 M KCl. After centrifugation, the eluted proteins in the supernatants were acetone precipitated. One milliliter of acetone was added to 250 μl of the supernatant, and the mixture was left on ice for 15 min. After centrifugation, the pellet was dried under vacuum for 15 min and finally dissolved in SDS-urea loading buffer and analyzed by SDS-PAGE on the PhastSystem, followed by autoradiography.
Immunoprecipitation.
Ten microliters of monoclonal His antibodies (Santa Cruz Biotechnology) and 20 μl of protein G PLUS-agarose (Santa Cruz Biotechnology) were incubated in 240 μl of IP buffer (0.15 M NaCl, 9.1 mM Na2HPO4, 1.7 mM NaH2PO4, 1 mM ATP, and 5 mM MgCl2) for 3 h at 4°C. The antibody-agarose complex was washed twice with IP buffer and recovered by centrifugation. Ten microliters of labeled P2 His-B and 240 μl of IP buffer were added to the complex and incubated for 2 h at 4°C. The supernatant was retained to measure the amount of unbound P2 His-B. Ten microliters of labeled DnaB or PinPoint control and 240 μl of IP buffer were added, and the mixture was incubated for another 3 h at 4°C. The multiprotein-antibody-agarose complexes were washed twice with IP buffer and recovered by centrifugation. The supernatants were retained to measure the amount of unbound protein and acetone precipitated as described above. The precipitates were collected by centrifugation and were dissolved in SDS-urea loading buffer and analyzed by SDS-PAGE on the PhastSystem, followed by autoradiography.
Potassium permanganate probing of origin opening in vivo.
Bacterial cultures were grown in M9 medium (45) supplemented with 0.2% Casamino Acids, 0.002% thymine, and 0.001% vitamin B1. When appropriate, 100 μg of ampicillin/ml and/or 20 μg of chloramphenicol/ml were added to the medium. Fresh overnight cultures of BL21(DE3) carrying different plasmids were diluted 100-fold and grown at 37°C to an optical density at 600 nm of ∼0.2. The culture was distributed in 10-ml aliquots, and IPTG (isopropyl-β-d-thiogalactopyranoside) was added to the final concentrations of 0, 50, 100, and 500 mM. Incubation was continued for another hour for induction of the P2 B protein. Potassium permanganate was added to a final concentration of 3 mM and the mixture was incubated for 1 min at 37°C. The reaction was terminated by mixing the culture with 10 ml of ice-cold STE buffer (100 mM NaCl, 10 mM Tris, pH 8.0, and 1 mM EDTA) with 5 mM dithiothreitol and chilling the mixture on ice.
For PC2 or DH5Δlac/pRLM109, the cultures were grown at 30°C to an optical density at 600 nm of ∼0.2 and divided into two 10-ml aliquots. One set was maintained at 30°C as a control, while the other set was shifted to 42°C for inactivation of DnaC in the case of PC2 or for thermal induction of λ P protein from pRLM109. Incubation at either temperature was continued for another hour. Subsequently, reaction with potassium permanganate was performed at 42°C for 1 min; otherwise, the conditions were identical to those described for BL21(DE3).
Plasmid DNA was isolated and analyzed by primer extension exactly as described previously (42).
Database analysis.
Predicted secondary structures were obtained from the Jpred server (http://circinus.ebi.ac.uk:8081). The secondary structures presented are consensus predictions based upon the algorithms DSC, MUL, NNSSP, PHD, PRED, and ZPRED (15). A sequence similarity search was performed using ψ-BLAST (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-psi_blast) (3).
Autoradiography quantification.
Protein bands on films were quantified using the program NIH Image version 1.62 (http://rsb.info.nih.gov/nih-image/).
RESULTS
In vivo interaction between P2 B and E. coli DnaB in the yeast two-hybrid system.
We used the yeast two-hybrid system to show a possible interaction between P2 B and DnaB (19). In this system, one of the proteins is fused to a DNA binding domain and the other is fused to a transcriptional activation domain. Interactions between the two fusion proteins allow transcriptional activation of one or more reporter genes. In the present study, the E. coli DnaB protein was fused to the DNA binding domain of LexA and the P2 B protein was fused to the N-terminal B42 transactivation domain. The plasmids expressing the fusion proteins were introduced into the yeast strain EGY48, which contains two reporter systems. One is the chromosomal leu2 gene, which has its upstream activating region replaced by lexA operators, and the other is a reporter plasmid, pSH18-34, which contains the lacZ gene under the control of the lexA operator.
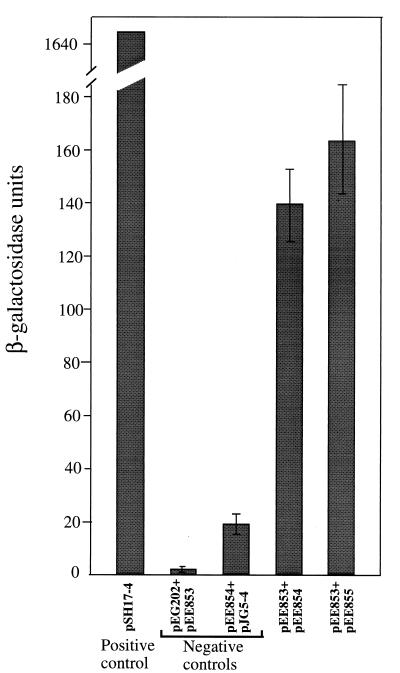
The capacity of the transformed EGY48 strain to grow on complete minimal medium lacking leucine was first analyzed. Cells transformed with plasmids expressing both the B and the DnaB fusion proteins grew as well as those transformed with the positive control, pSH17-4. Cells transformed with plasmids lacking inserts or one of the fusion genes did not grow on plates lacking leucine. Next, the expression of the lacZ gene was analyzed by plating the transformed yeast strain on complete minimal medium, and after exposure to chloroform, by the addition of X-Gal. The colors of the colonies were recorded after incubation at 30°C for 1 h. These were dark blue for cells with the positive control plasmid, blue for cells expressing both fusion proteins, and white for all negative controls (data not shown). These results confirm those obtained with the leu2 reporter. In order to quantify the level of lacZ expression, β-galactosidase activity was determined in a liquid assay. As shown in Fig. 1, cells containing plasmids expressing both fusion proteins showed a significant increase in enzyme activity, even though it was about 1/10 the activity obtained with cells containing the positive control plasmid pSS17-4 (1.646 U). One explanation of this difference could be that in one case activation depended upon transport into the nucleus of two fusion proteins and interaction between them in trans whereas in the other case transport and activation required one protein that has the DNA binding domain (LexA) and the activation domain (GAL4) in the same protein.
FIG. 1.
Comparison of β-galactosidase levels in extracts of yeast strain EGY48 transformed with different plasmids. The positive-control plasmid, pSH17-4, expresses a LexA DNA binding domain-GAL4 activation domain fusion protein. pEG202 expresses only the LexA DNA binding domain, pJG4-51 expresses only a B42 activation domain, pEE853 expresses a LexA DNA binding domain-DnaB fusion protein, pEE854 expresses a B42 activation domain-P2 B fusion protein, and pEE855 expresses a B42 activation domain–P2 B-rlb1 mutant fusion protein. The error bars indicate standard deviations.
Wild-type P2 is unable to plaque on E. coli dnaB(Ts) mutants at 37°C, in contrast to P2 rlb1 mutants, which can plaque under these conditions (49). Therefore, it was of interest to see if the rlb1 mutation caused a stronger interaction between B and DnaB. The mutated B gene was cloned in frame with the gene encoding the B42 activation domain, and the capacity of the mutated B protein to interact with DnaB fused to LexA was determined as described above. As can be seen in Fig. 1, a small increase in the mean β-lactamase activity was obtained, but the difference may not be significant, as it could be accounted for by experimental variations. The mutated protein, therefore, may not interact significantly better with the wild-type DnaB under the conditions of the present experiment.
In vitro interaction between P2 B and E. coli DnaB.
Having obtained an indication of a P2 B-DnaB interaction in vivo using the yeast two-hybrid system, we wished to confirm the interaction in vitro. Our initial approach was to use purified B and DnaB proteins, but purification of P2 B turned out to be problematic. P2 B was cloned into plasmid pET16b, which resulted in the addition of an in-frame N-terminal His tag under the control of the T7 promoter. The His-tagged B protein (His-B) fully complemented P2 amB116, indicating that the tag did not significantly affect the biological activity of the B protein. Upon induction of T7 polymerase, the His-B protein formed inclusion bodies, which could be solubilized in 6 M guanidine hydrochloride. However, after purification using a Ni column, and refolding by dialysis, the His-B protein was found to precipitate at concentrations above 30 ng/μl. Overexpression of DnaB did not result in inclusion bodies, and fractionation on a Q Sepharose column gave a preparation of DnaB that was >95% pure and was soluble at a protein concentration of about 800 ng/ml.
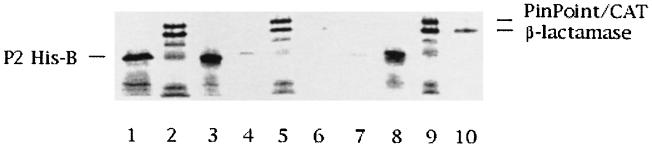
Due to the problem of keeping the P2 His-B protein soluble, we decided to use 35S-labeled His-B protein synthesized in vitro by coupled transcription-translation. Furthermore, since DnaB was found to elute at a very narrow range of salt concentrations with Q Sepharose, we found it appropriate to immobilize DnaB to Q Sepharose beads to demonstrate interaction between His-B and DnaB. Two negative-control proteins, PinPoint-CAT fusion and β-lactamase, were labeled, as was His-B in vitro. Purified DnaB was adsorbed to Q Sepharose beads. Equal amounts of labeled negative-control proteins and His-B were added separately to either Q Sepharose or DnaB-loaded Q Sepharose. Neither His-B nor the control proteins showed affinity for Q Sepharose, since most of the proteins remained in the supernatant after centrifugation (Fig. 2, lanes 3 and 5). Quantifications of the nonspecific binding of His-B and the controls, PinPoint-CAT fusion and β-lactamase, were 8, 4, and 9%, respectively. However, when labeled proteins were added to DnaB-loaded Q Sepharose, only His-B was found to bind specifically; 81% of the loaded His-B protein was bound, whereas 88% of the PinPoint-CAT fusion and 75% of β-lactamase remained in the supernatant (Fig. 2, lanes 7 to 10). Thus, the anion-exchange chromatography confirmed the in vivo results showing that P2 B specifically interacts with DnaB helicase. As can be seen in Fig. 2, the coupled transcription-translation system gave rise not only to the expected protein bands but also to some shorter polypeptides, which we believe consist of truncated forms of the proteins caused by premature termination during translation.
FIG. 2.
Autoradiogram of 12.5% acrylamide-SDS PhastGels showing interaction between DnaB and P2 His-B. Lanes 1 and 2, input of in vitro-synthesized 35S-labeled P2 His-B (lane 1) and control proteins, PinPoint-CAT fusion protein and β-lactamase (called C) (lane 2). Lanes 3 to 6, amounts of P2 His-B and C proteins that do not (lanes 3 and 5) and do (lanes 4 and 6) bind to Q Sepharose beads. Only trace amounts of the proteins are bound to the Q Sepharose. Lanes 7 to 10, Amounts of P2 His-B and C proteins that do not (lanes 7 and 9) and do (lanes 8 and 10) bind to DnaB immobilized on Q Sepharose beads. Almost all of the applied P2 His-B (81%) is present in the bound fraction, while almost all of the applied C proteins (88 and 75%) is found in the nonbound fraction.
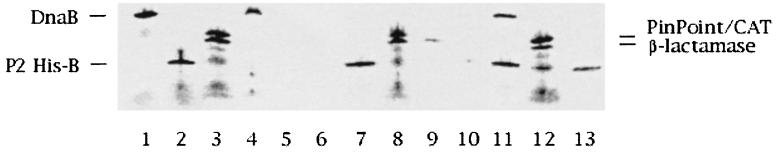
To further analyze the in vitro interaction, another approach was also undertaken. Since the B protein contains a His tag at the N terminus, we used monoclonal antibodies directed against the His tag to analyze whether the DnaB protein would coprecipitate with His-B upon addition of His antibodies and protein-G bound to agarose. As can be seen in Fig. 3, DnaB and the control proteins, PinPoint-CAT fusion and β-lactamase (lanes 1 and 5 and lanes and 3 and 6), did not bind to any large extent to protein-G–agarose in the presence of His antibodies: the background levels were 14, 12, and 29%, respectively. In contrast, 89% of P2 His-B was found to bind to G-agarose upon addition of antibodies (lanes 2 and 7). When DnaB was added to the agarose-antibody complex already loaded with P2 His-B, DnaB bound with a 78% efficiency (lanes 10 and 11), while the control proteins, PinPoint-CAT fusion and β-lactamase (lanes 12 and 13), showed reduced background levels: 11 and 14%, respectively. Thus, in the presence of P2 His-B, DnaB binding increased fivefold while the control proteins showed decreased background levels. These results are consistent with the data from the anion-exchange chromatography showing that P2 B and DnaB interact specifically in vitro.
FIG. 3.
Autoradiogram of 10 to 15% acrylamide gradient-SDS PhastGels showing interaction between in vitro-synthesized 35S-labeled P2 His-B and DnaB. Lanes 1 to 3, input of in vitro-synthesized 35S-labeled DnaB (lane 1), in vitro-synthesized 35S-labeled P2 His-B (lane 2), and control proteins, PinPoint-CAT fusion protein and β-lactamase (called C) (lane 3). Lanes 4 to 9, amounts of DnaB (lanes 4 and 5), P2 His-B (lanes 6 and 7), and C proteins (lanes 8 and 9) that do not and that do bind to protein-G–agarose saturated with monoclonal His antibodies. Lanes 10 to 13, amounts of DnaB (lanes 10 and 11) and C (lanes 12 and 13) proteins that do not and that do bind to P2 His-B immobilized on protein-G–agarose saturated with monoclonal His antibodies.
Stabilization of origin opening of plasmid P1 by P2 B protein.
Opening of the strands of the P1 plasmid origin, as assayed by reactivity to KMnO4 in vivo, was found to be greatly stimulated when the loading of DnaB to the plasmid origin was blocked (42). This was achieved in one of two ways: by inactivation of DnaC using a dnaC(Ts) host at the nonpermissive temperature or by supplying λ P, which inactivates DnaB by forming λ P-DnaB complexes (36). DnaC is essential for P1 plasmid replication in vitro, most likely for the loading of DnaB (5).
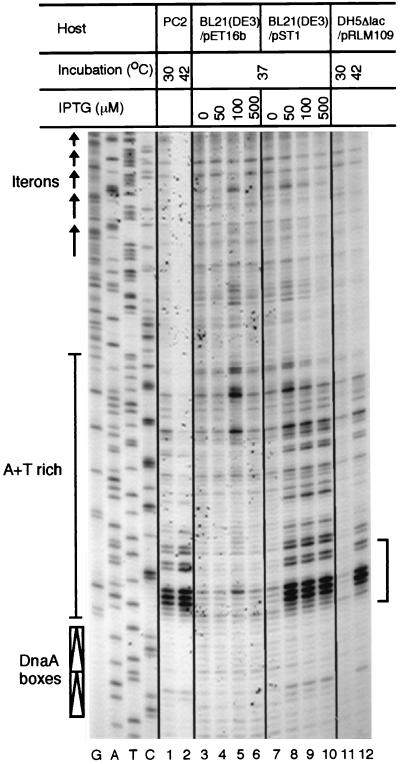
The results of origin opening of the mini-P1 plasmid (pSP102) in a dnaC(Ts) host, PC2, is shown in Fig. 4, lanes 1 and 2. As previously observed (42), an A+T-rich sequence spanning about 15 bp adjacent to the DnaA boxes became hypersensitive to the KMnO4 reaction at the nonpermissive temperature, whereas reactivity to the surrounding sequences became less sensitive, indicating that the absence of DnaC makes the opening more localized. The results were similar when, in a dnaC+ host, the P2 B protein was induced with IPTG (Fig. 4, lanes 8 to 10) or λ P was induced by thermal inactivation of λ cI857 repressor (Fig. 4, lane 12). Although not as dramatic as that in lane 2, some suppression of background reactivity was also seen as the concentration of IPTG was increased. From the overall similarity of the results of P2 B and those of λ P induction and DnaC inactivation, we conclude that P2 B can sequester DnaB from DnaC and thereby prevent DnaC-mediated loading of DnaB onto the P1 ori.
FIG. 4.
KMnO4 reactivity of the origin region of a P1-derived plasmid, pSP102. The origin region is bounded by DnaA boxes and the five iterons and contains an internal A+T-rich region. The reactivity of the bottom strand is shown in four different hosts: PC2, BL21(DE3)/pET16b, BL21(DE3)/pST1, and DH5Δlac/pRLM109. The KMnO4-reacted bases were visualized by primer extension. Localized increase of reactivity (bracket) is seen when the temperature of a dnaC(Ts) host (PC2) is raised to 42°C (lane 2), when P2 B is induced with IPTG (lanes 8 to 10), or when λ P protein is induced by temperature inactivation of cI857(Ts) repressor (lane 12). The first four lanes (G, A, T, and C) show the dideoxy sequencing reaction products of the P1 origin region.
DISCUSSION
Bacteriophage P2 requires the E. coli DnaB helicase for lagging-strand DNA synthesis, but not the helicase loader, DnaC (9, 20). In this work, we have demonstrated a physical interaction between P2 B and DnaB proteins in vivo as well as in vitro, supporting the hypothesis that P2 B is analogous to DnaC (23). P2 B was also comparable to the λ P protein, as they both appeared capable of competing with DnaC for binding to DnaB (Fig. 4).
The helicases unwind duplex DNA and generate transient single-stranded templates for new DNA synthesis. Since the helicases do not bind DNA in a sequence-specific manner, they have to be recruited to the origin by other origin-binding proteins. At the E. coli origin, the DnaA protein serves that role and recruits the DnaB-DnaC complex by a direct interaction with DnaB (37). DnaB forms a hexameric ring, which binds six DnaC monomers, and in this complex the helicase (and the ATPase) activity of DnaB is blocked (28, 30, 51, 52). To activate the DnaB helicase, DnaC must be released from the complex, a process that is believed to be associated with ATP hydrolysis by DnaC (8, 51). During lagging-strand synthesis, the E. coli DnaG primase has been shown to interact directly with the DnaB helicase, and this interaction is required for optimal primer synthesis (35). Some phages, like the well-studied phage λ, encode their own origin-binding proteins but utilize the host helicase(s) to separate the strands. The first step in λ replication is the specific binding of λ O protein to the phage origin, forming a complex called the O-some (17, 50, 54, 55). Next, the λ P protein, which is analogous to E. coli DnaC, binds to the DnaB hexamer, and the P-DnaB complex is recruited to the O-some through interactions between the λ O and λ P proteins (1, 21, 53, 55). The activation of DnaB requires partial disassembly of the complex by the DnaJ, DnaK, and GrpE chaperone system that depends upon ATP hydrolysis (2, 56). Compared to DnaC, λ P binds to DnaB more strongly, which enables the phage to compete with the host chromosome for the helicase (36). The situation appears to be similar for P2 replication, since P2 B binding to the DnaB helicase appears to be strong enough to prevent DnaC binding to DnaB for loading onto the P1 ori. It is not known whether the P2 A protein is involved in the recruitment of the B-DnaB complex to the origin or whether helicase activation requires the E. coli chaperone system. P2 does not plaque on dnaJ null mutants (our unpublished results), but it is not known what stage of the growth cycle is affected.
Three-dimensional reconstruction of cryo-electron microscopic images of the DnaB-DnaC complex indicates that each monomer of DnaC interacts with two neighboring monomers of DnaB and vice versa and that the protein-protein interactions occur over an extensive contact surface (46). A computer search with ψ-BLAST for proteins homologous to DnaC detected a hypothetical protein orf9 in Salmonella with 38% identity (N. Figueroa-Bossi and L. Bossi, unpublished data). The predicted secondary structures of DnaC and orf9 are shown in Fig. 5A. The structures appear identical. The λ P protein, on the other hand, is nonhomologous to DnaC, in amino acid sequence as well as in predicted secondary structure. The Rep14 protein of the lambdoid phage Φ80, however, shows 42% identity to λ P over the whole protein, but the region of identity is mainly located in the N-terminal part. The first 110 amino acids show 65% identity (40). Since λ P and Rep14 have different C-terminal sequences, it has been suggested that these parts are involved in binding to the λ O protein and the Φ80 gene 15 protein, respectively, whereas the N termini are believed to interact with some host protein (40). The P2 B protein is shorter than the other proven or presumptive DnaB loaders and shares only some homology (and little identity) in the N-terminal part with the other loaders (Fig. 5B). One region is located at the predicted α-helix 1 that includes the amino acid change due to the rlb1 mutation, believed to be involved in the interaction between B and DnaB. In addition, there is an MRRI motif in both λ P and P2 B at the end of α-helix 1. Some homology can also be found in α-helix 2. These observations support the hypothesis that the N-terminal parts of the proteins interact with DnaB (Fig. 5B). DnaC and orf9 of Salmonella contain ATP-binding motifs (29), and they are located in regions nonhomologous to the phage helicase loaders. This is consistent with the fact that disassociation of the DnaB-DnaC complexes proceeds without the help of auxiliary proteins whereas λ P-DnaB, and probably P2 B-DnaB, requires the ATPase activity of the chaperone system to release the helicase from the helicase loader (47).
FIG. 5.
(A) Predicted secondary structure for putative DnaB loaders. The structures were obtained from the Jpred server as described in Materials and Methods. The tubes represent α-helices, and the arrows represent β-strands, and they are numbered sequentially, increasing from N to C terminal. ATP motifs and possible protein-protein interaction domains are indicated. Predicted helices with four or fewer amino acids (aa) were omitted for clarity. (B) Amino acid alignments of P2 B, DnaC, and λ P. Alignments i and iii show similarity in α-helix 1, whereas alignment ii shows similarity in α-helix 2. Amino acid identity is indicated by I, and similarity is indicated by ∶. The His→Arg change in P2 B (i) corresponds to the P2 rlb1 mutation.
In contrast to λ dv, which requires the P protein to be maintained as a plasmid (6, 38, 39), the phage P2 minichromosome does not require the P2 B protein (34). However, the P2 minichromosome still requires the E. coli Rep function, which is believed to be required to displace the parental strand to allow leading-strand synthesis during rolling-circle replication (11, 13). Thus, during replication of the minichromosome, the P2 replicon seems to be able to recruit the DnaB-DnaC complex to the origin for lagging-strand synthesis, perhaps involving the A protein or some other origin-binding proteins. This loading activity is apparently not efficient enough to support the level of replication required for plaque formation. It has been found that the interfaces of permanent protein-protein complexes have in general a higher frequency of certain hydrophobic residues than do those on the protein surfaces, and among nonobligate protein-protein complexes some polar residues are also common (26, 27). We have noticed a high frequency of those hydrophobic and polar residues in α-helix 4 and β-sheet 5 of P2 B and in α-helix 8 and β-sheet 5 of DnaC, making them possible candidates for residues of protein interacting domains. Even though these domains do not show any amino acid sequence similarity and only resemble each other in predicted secondary structures, it is tempting to visualize these domains as P2 A interacting domains. If DnaC could interact with P2 A, DnaB loading and thus P2 minichromosome replication would be possible in the absence of P2 B (34). It is known that several plasmid replication initiators can interact with DnaB directly in order to recruit the helicase to plasmid origins (16, 43). Also worth noting is the finding that a P2-related phage, 186, does not code for a DnaC analogue and consequently does require DnaC for replication, which suggests that the 186 A protein is able to recruit the DnaB-DnaC complex to the phage origin (25). However, in contrast to P2 (32), 186 has a gene, dhr, that encodes a protein that can depress host replication, which should make more of the DnaB-DnaC complex available for phage replication (44). Since dhr is nonessential, and an N-terminal deletion of dhr reduces the burst size to only 30% of the wild-type level, the 186 A protein or some other origin-binding protein must bind the DnaB-DnaC complex. In conclusion, the similarity of biological properties and some parts of the structure of P2 B to known helicase loaders makes the protein a strong candidate for a DnaB helicase loader during lytic replication. It is also clear that different pathways of helicase loading can exist in P2 and in other phages and plasmids.
ACKNOWLEDGMENTS
We thank Rich Calendar for suggesting the P1 origin-opening experiment and Roger MacMacken for providing the plasmid pRLM109. The in vivo and in vitro protein interaction experiments were performed at Stockholm University, and the origin-opening experiments were performed at NIH.
This work was supported in part by grant 72 from the Swedish Medical Research Council.
REFERENCES
- 1.Alfano C, McMacken R. Ordered assembly of nucleoprotein structures at the bacteriophage lambda replication origin during the initiation of DNA replication. J Biol Chem. 1989;264:10699–10708. [PubMed] [Google Scholar]
- 2.Alfano C, McMacken R. Heat shock protein-mediated disassembly of nucleoprotein structures is required for the initiation of bacteriophage lambda replication. J Biol Chem. 1989;25:10709–10718. [PubMed] [Google Scholar]
- 3.Altschul S F, Schaffer A A, Zhang J, Zhang Z, Miller W, Lippman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arai K, Yasuda S, Kornberg A. Mechanism of DnaB protein action. J Biol Chem. 1981;256:5247–5252. [PubMed] [Google Scholar]
- 5.Baker T A, Wickner S H. Genetics and enzymology of DNA replication in Escherichia coli. Annu Rev Genet. 1992;26:447–477. doi: 10.1146/annurev.ge.26.120192.002311. [DOI] [PubMed] [Google Scholar]
- 6.Berg D E. Genes of lambda essential for lambda dv plasmids. Virology. 1974;62:224–233. doi: 10.1016/0042-6822(74)90317-1. [DOI] [PubMed] [Google Scholar]
- 7.Bertani L E, Six E W. The P2-like phages and their parasite, P4. In: Calendar R, editor. The bacteriophages. Vol. 2. New York, N.Y: Plenum; 1988. pp. 73–143. [Google Scholar]
- 8.Biswas S B, Biswas E E. Regulation of dnaB function in DNA replication in Escherichia coli by dnaC and lambda P gene products. J Biol Chem. 1987;262:7831–7838. [PubMed] [Google Scholar]
- 9.Bowden D, Twersky R S, Calendar R. Escherichia coli deoxyribonucleic acid synthesis mutants: their effect upon bacteriophage P2 and satellite bacteriophage P4 deoxyribonucleic acid synthesis. J Bacteriol. 1975;124:167–175. doi: 10.1128/jb.124.1.167-175.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brent R, Ptashne M. An eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985;43:729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- 11.Calendar R, Lindqvist B, Sironi G, Clark A J. Characterization of REP− mutants and their interaction with P2 phage. Virology. 1970;40:72–83. doi: 10.1016/0042-6822(70)90380-6. [DOI] [PubMed] [Google Scholar]
- 12.Carl P L. Escherichia coli mutants with temperature-sensitive synthesis of DNA. Mol Gen Genet. 1970;109:107–122. doi: 10.1007/BF00269647. [DOI] [PubMed] [Google Scholar]
- 13.Chattoraj D K. Strand-specific break near the origin of bacteriophage P2 DNA replication. Proc Natl Acad Sci USA. 1978;75:1685–1689. doi: 10.1073/pnas.75.4.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen D C, Yang B C, Kuo T T. One-step transformation of yeast in stationary phase. Curr Genet. 1992;21:83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- 15.Cuff J A, Barton G J. Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. Proteins. 1999;34:508–519. doi: 10.1002/(sici)1097-0134(19990301)34:4<508::aid-prot10>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 16.Datta H J, Khatri G S, Bastia D. Mechanism of recruitment of DnaB helicase to the replication origin of the plasmid pSC101. Proc Natl Acad Sci USA. 1999;96:73–78. doi: 10.1073/pnas.96.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dodson M, Roberts J, McMacken R, Echols H. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: complexes with lambda O protein and with lambda O, lambda P, and Escherichia coli DnaB proteins. Proc Natl Acad Sci USA. 1985;82:4678–4682. doi: 10.1073/pnas.82.14.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duttweiler H M. A highly sensitive and non-lethal β-galactosidase plate assay for yeast. Trends Genet. 1996;12:340–341. doi: 10.1016/s0168-9525(96)80008-4. [DOI] [PubMed] [Google Scholar]
- 19.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 20.Funnell B E, Inman R B. Bacteriophage P2 DNA replication. Characterization of the requirement of the gene B protein in vivo. J Mol Biol. 1983;167:311–334. doi: 10.1016/s0022-2836(83)80338-6. [DOI] [PubMed] [Google Scholar]
- 21.Furth M E, McLeester C, Dove W F. Specificity determinants for bacteriophage lambda DNA replication. I. A chain of interactions that controls the initiation. J Mol Biol. 1978;126:195–225. doi: 10.1016/0022-2836(78)90359-5. [DOI] [PubMed] [Google Scholar]
- 22.Golemis E A, Gyuris J, Brent R. Two hybrid systems/interaction traps. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1994. pp. 13.14.1–13.14.17. [Google Scholar]
- 23.Haggård-Ljungquist E, Kockum K, Bertani L E. DNA sequence of bacteriophage P2 early genes cox and B and their regulatory sites. Mol Gen Genet. 1987;208:52–56. doi: 10.1007/BF00330421. [DOI] [PubMed] [Google Scholar]
- 24.Hay J, Cohen G. Requirement of E. coli DNA synthesis functions for the lytic replication of bacteriophage P1. Virology. 1983;131:193–206. doi: 10.1016/0042-6822(83)90545-7. [DOI] [PubMed] [Google Scholar]
- 25.Hooper I, Egan B J. Coliphage 186 infection requires host initiation functions dnaA and dnaC. J Virol. 1981;40:599–601. doi: 10.1128/jvi.40.2.599-601.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones S, van Heynigen P, Berman H M, Thornton J M. Protein-DNA interactions: a structural analysis. J Mol Biol. 1999;287:877–896. doi: 10.1006/jmbi.1999.2659. [DOI] [PubMed] [Google Scholar]
- 27.Jones S, Thornton J M. Principles of protein-protein interactions. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobori J A, Kornberg A. The Escherichia coli dnaC gene product. III. Properties of the DnaB-DnaC protein complex. J Biol Chem. 1982;257:13770–13775. [PubMed] [Google Scholar]
- 29.Koonin E V. DnaC protein contains a modified ATP-binding motif and belongs to a novel family of ATPases including also DnaA. Nucleic Acids Res. 1992;20:1997. doi: 10.1093/nar/20.8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanka E, Schuster H. The dnaC protein of Escherichia coli. Purification, physical properties and interaction with dnaB protein. Nucleic Acids Res. 1983;11:987–997. doi: 10.1093/nar/11.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindahl G. Genetic map of bacteriophage P2. Virology. 1969;39:839–860. doi: 10.1016/0042-6822(69)90021-x. [DOI] [PubMed] [Google Scholar]
- 32.Lindqvist B H. Vegetative DNA of temperate coliphage P2. Mol Gen Genet. 1971;110:178–196. doi: 10.1007/BF00332647. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Haggård-Ljungquist E. Studies of bacteriophage P2 DNA replication: localization of the cleavage site of the A protein. Nucleic Acids Res. 1994;22:5204–5210. doi: 10.1093/nar/22.24.5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Saha S, Haggård-Ljungquist E. Studies of bacteriophage P2 DNA replication. The DNA sequence of the cis-acting gene A and ori region and construction of a P2 mini-chromosome. J Mol Biol. 1993;231:361–374. doi: 10.1006/jmbi.1993.1288. [DOI] [PubMed] [Google Scholar]
- 35.Lu Y-B, Ratnakar P V A L, Mohanty B K, Bastia D. Direct physical interaction between DnaG primase and DnaB helicase of Escherichia coli is necessary for optimal synthesis of primer RNA. Proc Natl Acad Sci USA. 1996;93:12902–12907. doi: 10.1073/pnas.93.23.12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mallory J B, Alfano C, McMacken R. Host virus interactions in the initiation of bacteriophage lambda DNA replication. Recruitment of Escherichia coli DnaB helicase by lambda P replication protein. J Biol Chem. 1990;265:13297–13307. [PubMed] [Google Scholar]
- 37.Marszalek J, Kaguni J M. DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J Biol Chem. 1994;269:4883–4890. [PubMed] [Google Scholar]
- 38.Matsubara K. Genetic structure and regulation of a replicon of plasmid lambda dv. J Mol Biol. 1976;102:427–439. doi: 10.1016/0022-2836(76)90325-9. [DOI] [PubMed] [Google Scholar]
- 39.Matsubara K, Kaiser A D. Lambda dv: an autonomously replicating DNA fragment. Cold Spring Harbor Symp Quant Biol. 1968;33:769–775. doi: 10.1101/sqb.1968.033.01.088. [DOI] [PubMed] [Google Scholar]
- 40.Ogawa T, Ogawa H, Tomizawa J-I. Organization of the early region of bacteriophage Φ80. J Mol Biol. 1988;202:537–550. doi: 10.1016/0022-2836(88)90284-7. [DOI] [PubMed] [Google Scholar]
- 41.Pal S K, Mason R J, Chattoraj D K. P1 plasmid replication. Role of initiator titration in copy number control. J Mol Biol. 1986;192:275–285. doi: 10.1016/0022-2836(86)90364-5. [DOI] [PubMed] [Google Scholar]
- 42.Park K, Mukhopadhyay S, Chattoraj D K. Requirements for and regulation of origin opening of plasmid P1. J Biol Chem. 1998;273:24906–24911. doi: 10.1074/jbc.273.38.24906. [DOI] [PubMed] [Google Scholar]
- 43.Ratnakar P V A L, Mohanty B K, Lobert M, Bastia D. The replication initiator protein π of the plasmid R6K specifically interacts with the host-encoded helicase DnaB. Proc Natl Acad Sci USA. 1996;93:5522–5526. doi: 10.1073/pnas.93.11.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richardson H, Egan J B. DNA replication studies with coliphage 186. II. Depression of host replication by a 186 gene. J Mol Biol. 1989;206:59–68. doi: 10.1016/0022-2836(89)90523-8. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 46.San Martin C, Radermacher M, Wolpensinger B, Engel A, Miles C S, Dixon N E, Carazo J-M. Three-dimensional reconstructions from cryoelectron microscopy images reveal an intimate complex between helicase DnaB and its loading partner DnaC. Structure. 1998;6:501–509. doi: 10.1016/s0969-2126(98)00051-3. [DOI] [PubMed] [Google Scholar]
- 47.Stephens K M, McMacken R. Functional properties of replication fork assemblies established by the bacteriophage λ O and P replication proteins. J Biol Chem. 1997;272:28800–28813. doi: 10.1074/jbc.272.45.28800. [DOI] [PubMed] [Google Scholar]
- 48.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 49.Sunshine M, Usher D, Calendar R. Interaction of P2 bacteriophage with the dnaB gene of Escherichia coli. J Virol. 1975;16:284–289. doi: 10.1128/jvi.16.2.284-289.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsurimoto T, Matsubara K. Replication of lambda dv plasmid in vitro promoted by purified lambda O and P proteins. Proc Natl Acad Sci USA. 1982;79:7639–7643. doi: 10.1073/pnas.79.24.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wahle E, Lasken R S, Kornberg A. The DnaB-DnaC replication protein complex of Escherichia coli. II. Role of the complex in mobilizing DnaB function. J Biol Chem. 1989;264:2463–2468. [PubMed] [Google Scholar]
- 52.Wickner S, Hurwitz J. Interaction of Escherichia coli dnaB and dnaC(D) gene products in vitro. Proc Natl Acad Sci USA. 1975;7:921–925. doi: 10.1073/pnas.72.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wickner S H, Zahn K. Characterization of the DNA binding domain of bacteriophage lambda O protein. J Biol Chem. 1986;261:7537–7543. [PubMed] [Google Scholar]
- 54.Zahn B, Blattner F R. Binding and bending of the lambda replication origin by the phage O protein. EMBO J. 1985;4:3605–3616. doi: 10.1002/j.1460-2075.1985.tb04124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zylicz M, Gorska L, Taylor K, Georgopoulus C. Bacteriophage lambda replication proteins: formation of a mixed oligomer and binding to the origin lambda DNA. Mol Gen Genet. 1984;196:401–406. doi: 10.1007/BF00436186. [DOI] [PubMed] [Google Scholar]
- 56.Zylicz M, Ang D, Liberek K, Georgopoulus C. Initiation of lambda DNA replication with purified host- and bacteriophage-encoded proteins: the role of the DnaK, DnaJ, and GrpE heat shock proteins. EMBO J. 1989;8:1601–1608. doi: 10.1002/j.1460-2075.1989.tb03544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]