Abstract
Foamy viruses (FVs) are complex retroviruses which have been isolated from different animal species including nonhuman primates, cattle, and cats. Here, we report the isolation and characterization of a new FV isolated from blood samples of horses. Similar to other FVs, the equine foamy virus (EFV) exhibits a highly characteristic ultrastructure and induces syncytium formation and subsequent cell lysis on a large number of cell lines. Molecular cloning of EFV reveals that the general organization is that of other known FVs, whereas sequence similarity with its bovine FV counterpart is only 40%. Interestingly, EFV buds exclusively from the plasma membrane and not from the endoplasmic reticulum (ER), as previously shown for other FVs. The absence of the ER retrieval dilysine motif in EFV Env is likely responsible for this unexpected sorting pathway.
Foamy viruses (FVs), also known as spumaviruses, are widespread complex retroviruses which have been isolated from nonhuman primates, cattle, and cats. Although highly lytic in vitro, these viruses, which are innocuous in vivo, are known to induce a persistent infection in their hosts (20). All FVs characterized to date have very large genomes (between 12 to 13 kb) with the classical gag, pol, and env structural genes and additional regulatory open reading frames (ORFs) located at the 3′ end of the env gene which are under the control of both the 5′ long terminal repeat (LTR) and an internal promoter (IP) (14). In the case of the human foamy virus (HFV), the prototype FV, two accessory proteins, Tas and Bet, have been described. While Tas (originally called Bel1) is the potent DNA binding transactivator of viral gene expression of both the LTR and the IP, Bet has been shown to play a key role in the establishment and control of viral persistence in vitro (1, 19).
The molecular biology of retroviruses was highly focused on human T-cell leukemia virus (HTLV) and human immunodeficiency virus (HIV), clearly associated with human pathologies. However, recent findings regarding FVs raise important issues of general interest. Indeed, some of their features, such as the formation of a specific pol mRNA and the infectivity of the incoming viral DNA contained in extracellular virions (26, 28), set FVs apart from all other retroviruses. By virtue of these two features, FVs resemble pararetroviruses.
By analogy with lentiviruses, which were isolated from cattle, cats, primates, goats, and horses, we decided to look for the presence of an FV in horses. Here, we report the isolation of a new nonprimate FV from blood samples of naturally infected horses. The equine foamy virus (EFV) has been characterized by molecular cloning and nucleotide sequence analysis. The ultrastructure of the extracellular virion and the genomic organization were investigated and compared to those of other cloned FVs. Although displaying limited sequence similarities with primate FVs, EFV is phylogenetically closer to nonprimate FVs, especially to the bovine foamy virus (BFV). Interestingly, in contrast to other known FVs, EFV buds exclusively from the plasma membrane.
MATERIALS AND METHODS
Cell cultures.
Human neural U373-MG, simian COS-6, rabbit RK13, and hamster BHK-21 cells were maintained in Dulbecco modified Eagle medium supplemented with sodium pyruvate and 10% fetal calf serum (FCS). ED, a horse fibroblast cell line, was propagated in RPMI medium with 8% FCS. Blood samples from domestic horses were collected in heparin tubes, and lymphocytes were isolated on Ficoll-Hypaque gradients. Peripheral blood lymphocytes (PBLs) were cultured in RPMI medium supplemented with sodium pyruvate and 20% FCS. T cells were stimulated with phytohemagglutinin P (PHA-P; Sigma) at 3 μg/ml. COS-6 or ED cells were transfected with the Lipofectin reagent (Gibco BRL) according to the manufacturer's instructions, and luciferase expression was monitored by a LUMAT LB 9501 luminometer (Berthold).
Protein analysis.
For radioimmunoprecipitation (RIP) assays, HFV acutely infected cells (107 cells) were labeled with [35S]methionine-cysteine (75 μCi/ml; specific activity, 1,245 Ci/mmol; Dupont NEN) for 18 h in minimal essential medium lacking methionine and cysteine and supplemented with 5% FCS. Cells were lysed in 50 mM Tris-HCl (pH 7.4)–100 mM NaCl–5 mM MgCl2–1% Triton X-100–0.5% sodium deoxycholate–0.05% sodium dodecyl sulfate–1 mM phenylmethylsulfonyl fluoride for 30 min at 4°C. After centrifugation, the supernatant was collected and immunoprecipitated with a rabbit anti-whole virus antiserum for the positive control and with horse sera as described elsewhere (3).
Molecular cloning.
Linear unintegrated EFV viral DNA was cloned in λEMBL3 after addition of BamHI/XmnI adapters, using a Gigapack III Gold cloning kit from Stratagene. Positive plaques were identified in situ with the 5′-TGGCGCCCAACGTGGGGC-3′ oligonucleotide corresponding to the primer binding sequence (PBS). Lambda recombinant DNA was purified using a Wizard Lambda Preps kit from Promega.
Both strands of the EFV DNA were automatically sequenced, and determination of sequence similarities and alignments were performed with ALIGNn (http://www.infobiogen.fr/services/analyseq/cgi-bin/alignn_in.pl), ALIGNp (http://www.infobiogen.fr/services/analyseq/cgi-bin/alignp_in.pl), and CLUSTALW (http://www.infobiogen.fr/services/analyseq/cgi-bin/clustalw_in.pl) programs, respectively.
Construction of EFV LTR, IP, and U3R reporter plasmids and EFV Tas expression vector.
LTR, IP, and U3R reporter plasmids directing the expression of the firefly luciferase gene were constructed using the promoterless pGL3basic plasmid (Promega). EFV inserts were PCR cloned using the following primers: LTR, 5′ GGGGTACCTGTCATGGAATGAGGATCCAG (KpnI)/3′ GAAGATCTATTGTCGCGGTATCTCCTTAA (BglII); IP, 5′ GGGGTACCATATGAACCTGTACTAGTAACA (KpnI)/3′ GAAGATCTGCAGGCCAGATGTCTTCTA (BglII); and U3R, 5′ GTGGATTAATGTCATGGAATGAGGATCCAGAGA (VspI)/3′ CATGCCATGGCCAGTCTGTGGCCTGCTCGAC (NcoI). The PCR products were first inserted into the pGEM-T vector (Promega) and then subsequently subcloned into the pGL3basic after KpnI and BglII digestion, except for the NcoI U3R insert, which was subcloned into the SmaI site after Klenow treatment. The resulting clones were named pEFV-LTR, pEFV-IP, and pEFV-U3R. The pRL-CMV vector, directing the expression of the renilla luciferase under the control of the cytomegalovirus immediate-early promoter, was used for normalization (Dual-Luciferase Reporter Assay System; Promega).
EFV Tas was amplified with the Tfl polymerase (Promega), using the primer 5′ GGAATTCAGGATATTATCATGGCTAGCA (EcoRI)/3′ CCCAAGCTTATGGTTCTCGAATAAAGCGGT (HindIII). The PCR product was first inserted in the pGEM-T vector and then subcloned into the pSG5-M eukaryotic expression vector at the EcoRI and HindIII sites. Integrity of the tas gene was confirmed by DNA sequencing. The corresponding DNA clone was designated pEFV-Tas.
Southern blotting.
DNA was extracted either by the Hirt method to obtain extrachromosomal DNA or by the technique described by Saïb et al. (19) to isolate high-molecular-weight DNA.
For single-stranded DNA analysis, Hirt supernatant DNA was denatured with glyoxal and electrophoretically separated in a 1.1% agarose gel as already described (23).
Hybridization was carried out in 50% formamide at 42°C, using specific probes. The DNA fragment corresponding to EFV LTR was labeled with [α-32P]dCTP by using the Prime-a-Gene labeling system from Promega. The 18-nucleotide (nt) PBS(+) and PBS(−) probes were end labeled with [γ-32P]ATP and T4 polynucleotide kinase.
Electron microscopy.
Cocultures of horse PBLs and human U373-MG cells were examined by electron microscopy. Monolayers were fixed in situ with 1.6% glutaraldehyde (Taab Laboratory Equipment Ltd., Reading, United Kingdom) in 0.1 M Sörensen phosphate buffer (pH 7.3 to 7.4) for 1 h at 4°C. Cells were scraped from the plastic substratum and centrifuged. The resulting pellets were successively postfixed with 2% aqueous osmium tetroxide for 1 h at room temperature, dehydrated in ethanol, and embedded in Epon. Ultrathin sections were collected on 200-mesh copper grids coated with Formvar and carbon and stained with uranyl acetate and lead citrate prior to be observed with a Philips 400 transmission electron microscope (80 kV) at a magnification of ×28,000 to ×36,000.
Nucleotide sequence accession number.
The complete EFV DNA sequence can be obtained from GenBank with accession no. AF201902.
RESULTS
Immunological reactivity of horse sera against HFV antigens.
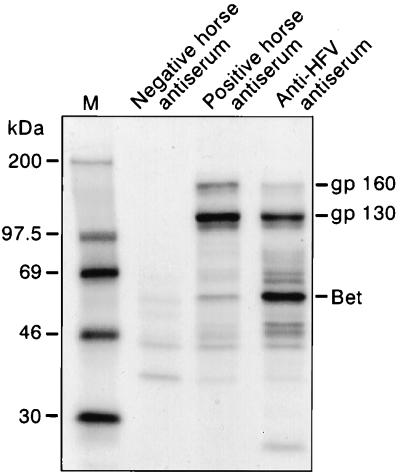
While several distinct primate FV isolates have been characterized, reflecting their widespread distribution among primate species, only BFV and the recently cloned feline foamy virus (FeFV) are available as nonprimate FVs (12). In an attempt to isolate new nonprimate FVs, sera from horses were tested for the presence of anti-FV antibodies by indirect immunofluorescence assay (IFA) on HFV-infected U373-MG cells. A significant fluorescent staining was detected with 9 sera among 36 tested, even at a 1/100 dilution, whereas mock-infected cells were completely negative in these conditions (data not shown). All horse sera were further screened by RIP on radiolabeled protein extracts from HFV- or mock-infected U373-MG cells. Eleven sera (including all nine previous identified by IFA) were found positive at a 1/25 dilution. All of these sera recognized in HFV-infected cells mainly the HFV 130-kDa Env precursor as well as a higher-molecular-weight band recently defined as the 160-kDa Env-Bet fusion glycoprotein (Fig. 1) (4, 11).
FIG. 1.
Reactivity of horse sera on HFV antigens tested by immunoprecipitation on protein extracts from HFV-infected U373-MG cells. The pattern obtained for one positive and one negative horse antiserum is shown in parallel with one characteristic profile visualized with anti-HFV antibodies. The 130-kDa Env precursor, the 160-kDa Env-Bet HFV glycoprotein, and the Bet protein are indicated.
Isolation of a new FV from blood samples from horses.
Blood samples were collected from two horses which were seropositive by IFA and RIP. PBLs isolated by centrifugation over a Ficoll-Hypaque gradient were cultured for 2 days with the mitogenic lectin PHA-P. These lymphocytes were then cocultivated with the highly sensitive human U373-MG or hamster BHK-21 monolayer cells. Four weeks later, a characteristic cytopathic effect (CPE) with giant multinucleated cells presenting numerous vacuoles was observed in cell cultures, in both U373-MG and BHK-21 cells for both horses and not in controls (Fig. 2A). Later, this effect was expanded to the entire cell monolayer and followed by a drastic cell lysis. These effects are generally the hallmarks of an infection with an FV and hence have facilitated the isolation of numerous FVs in the past. In addition, a manganese-dependent reverse transcriptase (RT) activity was detected in supernatants from cultures presenting this CPE, highly suggestive for the presence of a foamy retrovirus (data not shown).
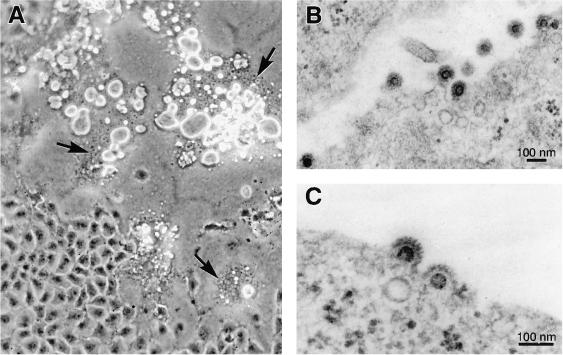
FIG. 2.
(A) Cytopathic effect in vitro on human U373-MG cells. Note the formation of syncytia (arrows) presenting numerous vacuoles. (B and C) Electron microscopy of ultrathin sections from infected cells. The viral particle has the typical FV appearance, enveloped particles surrounded by spikes and a clear central core.
Observations by electron microscopy of ultrathin sections of cells presenting this characteristic CPE revealed the presence of viral particles of about 100 nm in diameter, coated with multiple spikes and harboring a clear central core. As shown in Fig. 2B and C, these virions present all ultrastructural characteristics of FVs, consistent with the hypothesis that this is a new FV. Interestingly, virions seem to bud exclusively from the plasma membrane, in contrast to what has been described for all FVs.
Analysis of DNA from infected cells.
In a preliminary experiment, native Hirt supernatant DNA extracted from infected cells was analyzed by Southern blotting. Hybridization at low stringency with a full-length HFV probe revealed a weak band at about 12 kb, whereas no signal was visualized in noninfected cells (data not shown). In all FVs sequenced so far, the PBS is complementary to the 3′ end of the cellular tRNA1,2Lys used as primer for minus-strand DNA synthesis. Therefore, using the PBS probe, a strong and specific signal was detected at 12 kb in DNA from infected cells (data not shown).
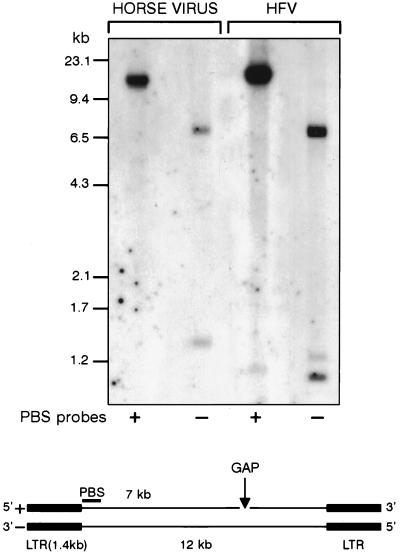
One important feature of FVs (shared with lentiviruses) is the dual initiation of plus-strand DNA synthesis being primed at the conventional 3′ LTR polypurine tract (PPT) and also at an internal PPT site located at the 3′ end of pol (10). This mode of replication results in the formation of gapped linear DNA duplex intermediates. To determine the structure of the viral genome, Hirt supernatant DNA extracted from infected U373-MG cells was denatured with glyoxal and analyzed by Southern blotting. Hybridization was performed with two strand-specific oligonucleotide probes of 18-nt PBS sequence. For a control, HFV DNA was run in parallel. As shown in Fig. 3, profiles are similar in both samples: the PBS(+) probe revealed a single band at 12 kb; with the PBS(−) probe, two bands of 7 and 1.4 kb were detected. By analogy to HFV, these three bands correspond to the full-length strand, the 5′ half part of the viral genome, and the LTR, respectively. These results indicate that the positive strand of the unintegrated EFV genome is gapped as previously shown for HFV (10).
FIG. 3.
Analysis of denatured viral DNA by Southern blotting. The conserved PBS(+) and PBS(−) sequences were used as probes and revealed that the viral genome is gapped.
Altogether, immunological cross-reactivity with HFV, electron microscopy observations, and structural analysis of the viral genome demonstrated that the virus isolated from seropositive horses belongs to the Spumavirinae subfamily and was therefore named equine foamy virus.
Molecular cloning and analysis of the EFV sequence.
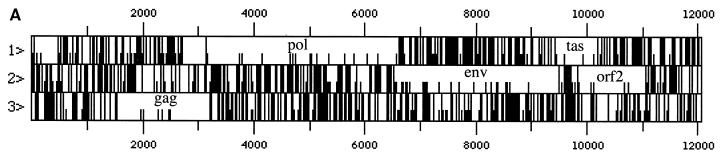
To get insights into the genomic structure of EFV, viral DNA from Hirt supernatants of acutely infected cells was directly cloned in λEMBL3. Ten positive clones were isolated from a total of approximately 10,000 phage plaques. To identify full-length clones, we looked for the presence of a 7-kb BamHI restriction fragment detected in the provirus from EFV Hirt extracts. Clones 1 and 4 contain apparently a full-length viral genome with a 12-kb insert and the specific 7-kb BamHI restriction fragment. Clone 1 was entirely sequenced on both strands, and critical regions were sequenced on the two strands of clone 4 (Fig. 4A).
FIG. 4.
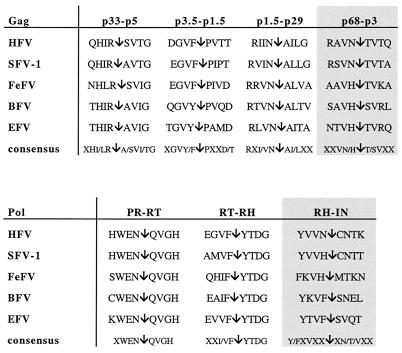
Schematic representation of predicted ORFs of EFV (A) and major hallmarks of the EFV Gag (B) and Env (C) gene products revealed by sequence comparison with other cloned FVs. Conserved residues among FVs are marked by asterisks, and arrows represent cleavage sites. (D) Amino acid sequence of EFV Tas.
Sequence analysis led to the observation that EFV harbored most of the FVs features while presenting some important distinctions.
(i) Noncoding regions.
The LTR of EFV is 1,449 bp long, flanked at the 3′ end of the 5′ LTR by the highly conserved 18-bp PBS sequence which is complementary to the 3′ end of the eukaryotic tRNA1,2Lys. By analogy with other cloned FV LTRs, the putative TATA box can be located at position 1105, while the R region starts at position 1125. As found for other FVs, the leader sequence is rather short (61 bp) and the first splice donor site is located at position 1186. At the 3′ LTR, the predicted polyadenylation signal, which harbors the consensus AATAAA sequence, is located at position 11861. Between the PBS and the first Met residue of Gag, the SI region as well as the palindromic SII sequence ([A/T]TCCCTAGGG[T/A]), which have been implicated in HFV RNA dimerization in vitro, are completely conserved in EFV, demonstrating their importance among this viral family. A sequence located at the 3′ end of pol (nt 7004 to 7013) consists of a duplication of the 3′ PPT (AAGGAGAGGG, nt 10577 to 10586) and may represent the central PPT used for positive-strand DNA synthesis (9).
(ii) gag gene.
The predicted Gag protein of EFV consists of 559 amino acids (aa) and starts at the Met residue at position 1527. Although EFV Gag is the largest among nonprimate FV Gag proteins, it is smaller than its primate FV Gag counterparts. Among the specific features setting FVs apart from other retroviruses (12), one is the peculiar proteolytic processing of the Gag precursors during the virus cycle. Indeed, mature infectious virus particles do not contain processed Gag, whereas partial cleavages occur only during the early steps of infection (3). Recently, four cleavage sites giving rise to mature products were delineated and shown to be present in all FV Gag proteins (16). These domains are also found in the EFV Gag sequence as shown in Fig. 5. Sequence identity of EFV Gag varies from 30% (simian FV1 [SFV1]) to 45% (BFV) compared to other FV Gag polypeptides.
FIG. 5.
By analogy with other known FVs, Gag and Pol putative cleavage sites of EFV were defined. RNase H (RH)-IN and p68-p3 cleavages occur at high frequency in infected cells and mature virions, whereas other cleavages are not well documented.
The sequence of amino acids GXWGX3RX7L(Q/V)D found in the N-terminal part of all FV Gag precursors, which resembles the morphogenetic signal of the Mason-Pfizer monkey virus, is conserved in EFV (24). At the C-terminal end of Gag, three Gly-Arg-rich sequences (GRI, GRII, and GRIII boxes) have been described for primate FVs (21). In EFV, only GRI and GRII, which have been shown to be implicated in viral nucleic acid binding (GRI) and nuclear localization of Gag (GRII) (27), are present (Fig. 4B).
(iii) pol gene.
The ORF of the pol gene, which harbors the protease (Pro), RT, RNase H and integrase (IN) domains, begins at position 3157 and consists of 1,154 residues. Interestingly, while other FV proteases display either a DSGA (HFV, BFV, SFV1, SFV3) or DSQA (FeFV) motif sequence as the active center of the enzyme, in EFV this domain consists of DTGA, similar to the one harbored by HIV and HTLV proteases (17, 18). However, other consensus motifs which have been shown to be conserved among FVs, such as the YVDD catalytic site of the RT enzyme or the GRK motif found just upstream the active center of Pro, are found in EFV Pol. As with all FVs, the canonical HH-CC zinc finger motif is present in the IN domain (24). The EFV Pol shows 60% identity with other FV Pol proteins.
Putative cleavage sites of Gag and Pol are depicted in Fig. 5.
(iv) env gene.
The env gene encodes a predicted 987-residue Env precursor glycoprotein which starts at position 6536. By analogy with other known FV Env proteins, a putative subtilisin-like protease cleavage site between SU (surface) and TM (transmembrane) domains could be located between residues 559 and 564 (GRRK→RG), dividing Env into the SU (562 aa) and TM (425 aa) mature products. Similarity of the SU domain with other FV SU proteins varies from 39% (SFV1) to 45% (BFV), while the TM protein is 49% (SFV1) to 62% (BFV) identical to the other FV TMs. The TMpred program revealed that two regions from aa 64 to 86 and 935 to 965 represent the signal peptide and the hydrophobic membrane-spanning domain (MSD), respectively. The putative fusion peptide is predicted to lie near the TM protein N terminus, and the typical LX2SX3MX2AIX2LX2IS pattern which presents an amphipatic α helix structure can be found between aa 570 and 588. The GOR secondary structure prediction program predicted that the EFV TM protein presents the same specific pattern already defined in FV envelopes, characterized by three domains, two α-helix regions (downstream of the fusion peptide and upstream of the MSD), and a central region consisting of β sheets and loops, which represents a specific feature of FV TMs (24). As for all FVs, the cytoplasmic tail of the EFV Env protein is short (21 aa for EFV, the largest among FVs). The cytoplasmic domain of TM from known FVs is also characterized by the presence of a dilysine motif, conferring to the envelope the particularity of budding from membranes of the endoplasmic reticulum (ER) (6, 7). Consistent with what has been observed by electron microscopy, this motif is absent in EFV TM cytoplasmic tail. In addition, a conserved tryptophan residue found in FV Env between the MSD and the ER retrieval signal is present in EFV (Fig. 4C) (24).
(v) Regulatory region.
In addition to the gag, pol, and env genes, EFV, like all complex retroviruses, codes for other gene products from the 3′ end of the viral genome. The first ORF (ORF1) consists of 249 aa and is located downstream of a TATA box motif (nt 9172) from the presumed IP. In that sense, the nucleotide sequence GAGCTA, located at position 9202, might represent a putative transcriptional start site in EFV since it resembles the one found in the BFV IP. The ProfileScan program predicts that the EFV Tas protein, which is similar in size to the BFV transactivator, will be nuclear since it presents, as is the case for HFV Tas/Bel1, a bipartite nuclear localization signal with the consensus sequence KRIASYQMQGSGGKRRAT. The sequence YXCXXCX35–37R/KH, of unknown function and repeatedly found in FV Tas proteins, is also present in the EFV transactivator (25). As for BFV and FeFV Tas proteins, a region in the N-terminal part of the primate Tas which has been shown to be nonessential for transactivation is absent in the EFV Tas, explaining the smaller size of nonprimate FV transactivators (Fig. 4D) (25).
ORF2 starts at position 10076 and consists of 329 aa. As already described, sequence similarities between ORF2 from FVs are very weak. In the case of EFV ORF2, we find between 18% (SFV1) and 29% (BFV) identity with other FVs. Putative splice sites generating the Bet fusion protein are conserved in EFV (A. Saïb, unpublished data). Note the absence of a third ORF. Compared to other genes, accessory EFV ORFs present limited sequence similarities with other FVs, suggesting that secondary structure rather than peptide sequence is a major determinant of their function. Table 1 summarizes the sizes of viral gene products from EFV compared to those from primate and nonprimate FVs.
TABLE 1.
Sizes of predicted EFV gene products and LTR compared to those of primate and nonprimate FVsa
| Virus (GenBank accession no.) | Size
|
|||||
|---|---|---|---|---|---|---|
| LTR (bp) | Gag (aa) | Pol (aa) | Env (aa) | Tas (aa) | ORF2 (aa) | |
| EFV | 1,449 | 559 | 1,154 | 987 | 249 | 329 |
| BFV (U94514) | 1,305 | 544 | 1,220 | 990 | 249 | 335 |
| FeFV (Y08851) | 1,355 | 574 | 1,156 | 982 | 209 | 313 |
| SFV1 (X54482) | 1,623 | 647 | 1,158 | 985 | 308 | 403 |
| SFVcpz (U04327) | 1,769 | 653 | 1,146 | 988 | 300 | 364 |
| HFV13 (U21247) | 1,123 | 648 | 1,143 | 989 | 300 | 356 |
Southern blot analysis of proviral EFV DNA.
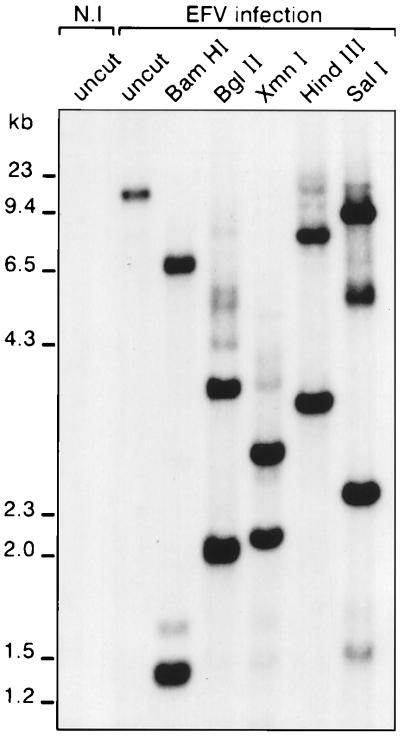
To confirm that EFV is not an endogenous virus, total DNA from mock-infected ED horse cells and Hirt supernatant DNA from EFV-infected ED cells digested with several enzymes were analyzed by Southern blotting. No signal was detected in genomic DNA from uninfected ED cells, whereas enzymatic restrictions of DNA from EFV-infected cells gave rise to distinct fragments which sizes are consistent to sequence analysis predictions. These results demonstrate that EFV is an exogenous FV (Fig. 6).
FIG. 6.
Southern blot analysis of total DNA or Hirt supernatant DNA from mock-infected (N.I) or EFV-infected horse ED cells. The EFV LTR, used as a probe, revealed that the viral genome is absent from uninfected horse cells, thus demonstrating that EFV is an exogenous virus.
Transactivation and cross-reactivity experiments.
To determine whether EFV encodes a viral transactivator as do other known FVs, the U3R sequence, the entire LTR, and the region between nt 8972 and 9290 (encompassing the putative IP) were cloned upstream of the luciferase gene, leading to reporter plasmids pEFV-U3R, pEFV-LTR, and pEFV-IP. To determine whether the viral transactivator mapped to the ORF1 gene, this ORF was cloned into the eukaryotic pSG5M vector and used in cotransfection experiments with simian COS-6 or horse ED cells. In the absence of ORF1, luciferase expression in ED cells from pEFV-LTR, pEFV-U3R, and pEFV-IP is low. However, the IP presents a higher basal activity (data not shown), consistent with what has been described for other FVs (13, 15). Coexpression of the ORF1 dramatically increased luciferase activity both in horse ED and in simian COS-6 cells (Table 2). Transactivation from the entire LTR is reduced compared to the U3R construct, reflecting the presence of negative regulatory elements in U5, as already found for other FVs (Table 2). These results demonstrated that ORF1 is the viral transactivator of EFV and therefore may be called Tas.
TABLE 2.
Transactivation of EFV promoters upon ORF1 expression and cross-reactivity between HFV and EFV
| Construct | Fold increasea
|
|||
|---|---|---|---|---|
| COS cells
|
ED cells
|
|||
| EFV Tas | HFV Tas | EFV Tas | HFV Tas | |
| pGL3basic | 2 | 0.5 | 2 | 4 |
| EFV LTR | 1,058 | 1 | 260 | 2 |
| EFV IP | 184 | ND | 123 | ND |
| EFV U3R | 4,015 | ND | 1,141 | ND |
| HFV LTR | 2 | 40 | 2 | 2 |
Values are from three independent transfection experiments performed in duplicate. ND, not determined.
FV Tas proteins were constantly shown to present weak or even no cross-reactivity on heterologous FV promoters. To see whether EFV Tas was able to transactivate the LTR from HFV, or reciprocally whether HFV Tas could activate gene expression from the two EFV promoters, cotransfection experiments were performed. As shown in Table 2, while homologous transactivation leads to a strong luciferase expression, no cross-activation was detected between EFV and HFV. Surprisingly, we were unable to detect any transactivation of the HFV LTR with its homologous Tas in ED horse cells, although these cells were permissive to HFV.
DISCUSSION
Discovered 50 years ago, FVs were isolated mainly from nonhuman primates (20). However, two nonprimate FV isolates have been described: BFV and the recently cloned FeFV. Availability of FV isolates of diverse origins is needed to determine conserved features which will reflect their importance in the biology of these retroviruses.
At the immunological level, sera from domestic horses reacted against viral antigens from the virus prototype HFV, especially the Env and Env-Bet glycoproteins. This result is consistent with reports showing a high degree of conservation of the env gene among FVs (reference 23 and this report). Similar homology in the env gene is shared by the murine leukemia viruses and viruses from the HTLV group but not by lentiviruses (24). Besides antigenic and structural characteristics, both viral particles and genome structure confirmed the existence of this new FV, EFV. Like other FVs, EFV exhibits a very broad cell tropism, as it can productively infect cell lines from different species (hamster BHK-21, rabbit RK13, simian COS-6, and human U373-MG cells), likely reflecting the ubiquity of the viral receptor (8). Preliminary studies suggest that EFV-infected horses do not exhibit any obvious pathological conditions. Whether EFV could be a positive cofactor in equine lentivirus infections remains to be investigated.
Sequence analysis of two full-length clones confirmed that EFV is a complex FV which harbors the hallmarks of this viral family, although some specific features clearly distinguish EFV from other members of the Spumavirinae.
The gag gene harbors most of the motifs found in other FV Gag proteins. Glycine-arginine-rich boxes implicated in nucleic acids binding (GRI) and nuclear localization (GRII) of FV Gag are present in EFV, whereas a GRIII box, the function of which is unknown in the viral cycle, is absent, similar to what has been described for FeFV Gag (25). Surprisingly, despite the strong similarity between FV Gag proteins, no cross-reactivity was observed between positive horse sera and HFV Gag precursors in Western blot or immunoprecipitation assay. Although cleavage sites recently described in FV Gag and Pol proteins exist in EFV, further analysis of FV proteins is needed to confirm these predictions.
Concerning Pol, a conserved ORF is present upstream of the initiator ATG of pol. Since this ORF is conserved in many FVs, it may have a yet to be defined function (Fig. 4A). Instead of a DSGA or DSQA motif present in cloned FVs, the active center of the Pro enzyme consists of a DTGA motif, as described for HIV and HTLV proteases. It will be interesting to determine whether antiprotease molecules which block the catalytic site of immunodeficiency viruses can be active on EFV replication.
As with all FVs, EFV possesses an IP located upstream of the viral transactivator encoded by ORF1 (14). In HFV, the IP is essential as it directs early expression of regulatory genes, the tas gene in particular, which in turn will transactivate viral gene expression from the IP and the 5′ LTR (13). In the presence of EFV Tas, levels of LTR- and IP-driven expression are greatly enhanced. The fact that EFV Tas failed to transactivate the HFV LTR, and reciprocally HFV Tas does not transactivate the EFV LTR, is consistent with previous reports of cross-reactivity experiments between FVs. The dramatic induction of EFV promoters with its own Tas warrants future studies on the mechanism involved in this transcriptional activation. Similarly, the absence of HFV transactivation in horse cells is surprising and suggests the involvement of specific cellular cofactors.
The strategy employed by FVs for capsid assembly resembles that of type B-D retroviruses and takes place in the cytoplasm of infected cells. However, unlike other retroviruses, FVs bud mainly in the ER (6). This is due to the presence of an ER retrieval motif consisting of two lysines at positions −3 and either −4 or −5 from the carboxyl terminus of TM and identified in the primate and feline FV isolates for which sequence is available (7). In contrast to other FVs, electron microscopy observations of EFV-infected cells revealed that virions bud exclusively from the plasma membrane, demonstrating that this new FV does not follow the same sorting pathway. In that sense, sequence analysis of the EFV env gene revealed that the dilysine motif is absent from the cytoplasmic tail of TM. For HFV, this motif was shown to be nonessential for efficient production of extracellular viral particles or for Gag-Env interactions since a mutated ΔKK HFV provirus is still infectious (5). The nonconservation of this motif in EFV confirm its dispensability in viral replication. It will be interesting to study the budding of the bovine isolate since, similar to EFV, it lacks the dilysine motif in the cytoplasmic tail of TM (Fig. 4B).
Recently, due to their wide cellular tropism and large genomes, FVs were considered interesting tools for gene therapy. The ability of EFV to bud at the cell surface rather than intracellularly could make this virus an attractive backbone for the design of a new generation of FV-based vectors, avoiding artificial disruption of internal cellular membranes to obtain higher viral titers.
ACKNOWLEDGMENTS
We thank Marc Foursin for providing horse blood samples and the Laboratoire de Photographie de l'Institut d'Hématologie for photographic work.
REFERENCES
- 1.Bock M, Heinkelein M, Lindemann D, Rethwilm A. Cells expressing the human foamy virus (HFV) accessory Bet protein are resistant to productive HFV superinfection. Virology. 1998;250:194–204. doi: 10.1006/viro.1998.9362. [DOI] [PubMed] [Google Scholar]
- 2.De Celis J, Tobaly-Tapiero J, Hampe A, Emanoil-Ravier R. Structure and function of the long terminal repeat of the chimpanzee foamy virus isolates (SFV-6) Arch Virol. 1994;138:345–355. doi: 10.1007/BF01379137. [DOI] [PubMed] [Google Scholar]
- 3.Giron M L, Colas S, Wybier J, Rozain F, Emanoil-Ravier R. Expression and maturation of human foamy virus Gag precursor polypeptides. J Virol. 1997;71:1635–1639. doi: 10.1128/jvi.71.2.1635-1639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giron M L, de Thé H, Saïb A. An evolutionarily conserved splice generates a secreted Env-Bet fusion protein during human foamy virus infection. J Virol. 1998;72:4906–10. doi: 10.1128/jvi.72.6.4906-4910.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goepfert P A, Shaw K, Wang G, Bansal A, Edwards B H, Mulligan M J. An endoplasmic reticulum retrieval signal partitions human foamy virus maturation to intracytoplasmic membranes. J Virol. 1999;73:7210–7217. doi: 10.1128/jvi.73.9.7210-7217.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goepfert P A, Shaw K L, Ritter G, Jr, Mulligan M J. A sorting motif localizes the foamy virus glycoprotein to the endoplasmic reticulum. J Virol. 1997;71:778–784. doi: 10.1128/jvi.71.1.778-784.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goepfert P A, Wang G, Mulligan M J. Identification of an ER retrieval signal in a retroviral glycoprotein. Cell. 1995;82:543–544. doi: 10.1016/0092-8674(95)90026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill C L, Bieniasz P D, McClure M O. Properties of human foamy virus relevant to its development as a vector for gene therapy. J Gen Virol. 1999;80:2003–2009. doi: 10.1099/0022-1317-80-8-2003. [DOI] [PubMed] [Google Scholar]
- 9.Kupiec J J, Sonigo P. Reverse transcriptase jumps and gaps. J Gen Virol. 1996;77:1987–1991. doi: 10.1099/0022-1317-77-9-1987. [DOI] [PubMed] [Google Scholar]
- 10.Kupiec J J, Tobaly-Tapiero J, Canivet M, Santillana-Hayat M, Flügel R M, Périès J, Emanoil-Ravier R. Evidence for a gapped linear duplex DNA intermediate in the replicative cycle of human and simian spumaviruses. Nucleic Acids Res. 1988;16:9557–9565. doi: 10.1093/nar/16.20.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindemann D, Rethwilm A. Characterization of a human foamy virus 170-kilodalton Env-Bet fusion protein generated by alternative splicing. J Virol. 1998;72:4088–4094. doi: 10.1128/jvi.72.5.4088-4094.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linial M L. Foamy viruses are unconventional retroviruses. J Virol. 1999;73:1747–1755. doi: 10.1128/jvi.73.3.1747-1755.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Löchelt M, Flügel R M, Aboud M. The human foamy virus internal promoter directs the expression of the functional Bel 1 transactivator and Bet protein early after infection. J Virol. 1994;68:638–645. doi: 10.1128/jvi.68.2.638-645.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Löchelt M, Muranyi W, Flügel R M. Human foamy virus genome possesses an internal, Bel-1-dependent and functional promoter. Proc Natl Acad Sci USA. 1993;90:7317–7321. doi: 10.1073/pnas.90.15.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Löchelt M, Yu S F, Linial M L, Flügel R M. The human foamy virus internal promoter is required for efficient gene expression and infectivity. Virology. 1995;206:601–610. doi: 10.1016/s0042-6822(95)80077-8. [DOI] [PubMed] [Google Scholar]
- 16.Pfrepper K I, Löchelt M, Rackwitz H R, Schnolzer M, Heid H, Flügel R M. Molecular characterization of proteolytic processing of the Gag proteins of human spumavirus. J Virol. 1999;73:7907–7911. doi: 10.1128/jvi.73.9.7907-7911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfrepper K I, Löchelt M, Schnolzer M, Flügel R M. Expression and molecular characterization of an enzymatically active recombinant human spumaretrovirus protease. Biochem Biophys Res Commun. 1997;237:548–553. doi: 10.1006/bbrc.1997.7187. [DOI] [PubMed] [Google Scholar]
- 18.Pfrepper K I, Rackwitz H R, Schnolzer M, Heid H, Löchelt M, Flügel R M. Molecular characterization of proteolytic processing of the Pol proteins of human foamy virus reveals novel features of the viral protease. J Virol. 1998;72:7648–7652. doi: 10.1128/jvi.72.9.7648-7652.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saïb A, Koken M H, van der Spek P, Périès J, de Thé H. Involvement of a spliced and defective human foamy virus in the establishment of chronic infection. J Virol. 1995;69:5261–5268. doi: 10.1128/jvi.69.9.5261-5268.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saïb A, Périès J, de Thé H. Recent insights into the biology of the human foamy virus. Trends Microbiol. 1995;3:173–178. doi: 10.1016/s0966-842x(00)88916-7. [DOI] [PubMed] [Google Scholar]
- 21.Schliephake A W, Rethwilm A. Nuclear localization of foamy virus Gag precursor protein. J Virol. 1994;68:4946–4954. doi: 10.1128/jvi.68.8.4946-4954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt M, Herchenröder O, Heeney J, Rethwilm A. Long terminal repeat U3 length polymorphism of human foamy virus. Virology. 1997;230:167–178. doi: 10.1006/viro.1997.8463. [DOI] [PubMed] [Google Scholar]
- 23.Tobaly-Tapiero J, Kupiec J J, Santillana-Hayat M, Canivet M, Périès J, Emanoil-Ravier R. Further characterization of the gapped DNA intermediates of human spumavirus: evidence for a dual initiation of plus-strand DNA synthesis. J Gen Virol. 1991;72:605–608. doi: 10.1099/0022-1317-72-3-605. [DOI] [PubMed] [Google Scholar]
- 24.Wang G, Mulligan M J. Comparative sequence analysis and predictions for the envelope glycoproteins of foamy viruses. J Gen Virol. 1999;80:245–254. doi: 10.1099/0022-1317-80-1-245. [DOI] [PubMed] [Google Scholar]
- 25.Winkler I, Bodem J, Haas L, Zemba M, Delius H, Flower R, Flügel R M, Löchelt M. Characterization of the genome of feline foamy virus and its proteins shows distinct features different from those of primate spumaviruses. J Virol. 1997;71:6727–6741. doi: 10.1128/jvi.71.9.6727-6741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu S F, Baldwin D N, Gwynn S R, Yendapalli S, Linial M L. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science. 1996;271:1579–1582. doi: 10.1126/science.271.5255.1579. [DOI] [PubMed] [Google Scholar]
- 27.Yu S F, Edelmann K, Strong R K, Moebes A, Rethwilm A, Linial M L. The carboxyl terminus of the human foamy virus Gag protein contains separable nucleic acid binding and nuclear transport domains. J Virol. 1996;70:8255–8262. doi: 10.1128/jvi.70.12.8255-8262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu S F, Sullivan M D, Linial M L. Evidence that the human foamy virus genome is DNA. J Virol. 1999;73:1565–1572. doi: 10.1128/jvi.73.2.1565-1572.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]