Abstract
Ufmylation is implicated in multiple cellular processes, but little is known about its functions and regulation in protein trafficking. Here, we demonstrate that the genetic depletion of core components of the ufmylation cascade, including ubiquitin-fold modifier 1 (UFM1), UFM1 activation enzyme 5, UFM1-specific ligase 1 (UFL1), UFM1-specific protease 2, and UFM1-binding protein 1 (UFBP1) each markedly inhibits the endoplasmic reticulum (ER)–Golgi transport, surface delivery, and recruitment to COPII vesicles of a subset of G protein–coupled receptors (GPCRs) and UFBP1’s function partially relies on UFM1 conjugation. We also show that UFBP1 and UFL1 interact with GPCRs and UFBP1 localizes at COPII vesicles coated with specific Sec24 isoforms. Furthermore, the UFBP1/UFL1-binding domain identified in the receptors effectively converts non-GPCR protein transport into the ufmylation-dependent pathway. Collectively, these data reveal important functions for the ufmylation system in GPCR recruitment to COPII vesicles, biosynthetic transport, and sorting at ER via UFBP1 ufmylation and interaction directly.
Ufmylation, a posttranslational modification, regulates the forward delivery and segregation of G protein–coupled receptors.
INTRODUCTION
Ufmylation is a ubiquitination-like posttranslational modification in which ubiquitin-fold modifier 1 (UFM1) is conjugated to the targeted proteins. UFM1 consists of 85 amino acid residues and is structurally related to ubiquitin. Similar to ubiquitin and many other ubiquitin-like molecules, UFM1 conjugation is carried out by a three-enzyme cascade involving the E1 UFM1 activation enzyme 5 (UBA5), the E2 UFM1-conjugating enzyme 1 (UFC1), and the E3 UFM1-specific ligase 1 (UFL1). UBA5 activates UFM1 through adenylation and thioesterification reactions and transfers UFM1 to UFC1, which subsequently works together with UFL1 to deliver UFM1 to substrates. UFM1 on the ufmylated proteins can be removed by UFM1-specific proteases 1 and 2 (UFSP1 and UFSP2) (1–5). Ufmylation has been known for two decades with only a few known UFM1 substrates (2, 6–15). Recent studies have shown that ufmylation is implicated in human diseases, such as hematopoietic diseases (16–18), heart diseases (19), diabetes (20), intestinal exocrine diseases (21), neuronal diseases (22–25), and cancer (9, 12). At the cellular level, ufmylation regulates endoplasmic reticulum (ER) stress (11, 26, 27), ER-phagy (6, 28, 29), ribosome- and translocation-associated quality control (14, 30–33), genomic integrity (8, 15), and cell development and death (15, 34–36). However, virtually nothing is known about its functions and regulation in cellular protein trafficking.
Coat protein complex II (COPII) vesicles exclusively mediate cargo export from the ER. The formation of COPII vesicles is initiated by the activation of Sar1 by the transmembrane guanine nucleotide exchange factor Sec12 on ER exit sites (ERES). Activated Sar1 interacts with Sec23 to recruit heterodimeric Sec23-Sec24 complex that further recruits the Sec13-Sec31 complex, leading to the formation of COPII vesicles. Selective cargo recruitment to COPII is directed by export motifs in cargoes, which directly interact with Sec24, or by cargo receptors that interact with both Sec24 and cargoes. Four Sec24 isoforms (Sec24A/B/C/D) exist in human cells and they have multiple, distinct cargo-binding sites, allowing for efficient accommodation of diverse cargo molecules (37, 38). Recent studies have demonstrated that COPII coat proteins regulate the formation of a tubular network for ER-Golgi transport (39) and function as gatekeepers at the boundary between the ER and the ERES in selecting and concentrating cargo molecules (40).
G protein–coupled receptors (GPCRs) represent the largest superfamily of cell surface signaling proteins that regulate a wide variety of cell functions under physiological and pathological conditions (41–45). It has been known that GPCR export from the ER controls their maturation kinetics, the number of the receptors at the functional destinations, and the magnitude and duration of receptor-elicited cellular responses (46, 47), and its dysregulation is directly associated with the pathogenesis of human diseases (48). In the past decades, a number of regulatory proteins, such as Golgi-localized, γ-adaptin ear-containing, ARF-binding proteins (49), Rab guanosine triphosphatases (50, 51), Tre2-Bub2-Cdc16 domain-containing proteins (52), class II phosphoinositide-3 kinase α (53), Yip1 interacting factor homolog B (54), C1orf27 (55, 56), and coiled-coil α-helical rod protein 1 (57), have been demonstrated to regulate the biosynthetic transport of some GPCRs. Several motifs, such as di-leucine (58, 59), di-acidic (60, 61), and positively charged motifs (62, 63), have also been identified to control the COPII recruitment and export of GPCRs. These data demonstrate that, although GPCRs share a common structural topology, their anterograde delivery to the cell surface after synthesis in the ER may be mediated through distinct mechanisms.
Here, we have investigated the function of the ufmylation system in the transport of plasma membrane (PM) proteins along the biosynthetic pathway using knockout (KO) cells depleting UFM1, UBA5, UFL1, UFSP2, or UFM1-binding protein 1 (UFBP1). We have demonstrated that ufmylation is required for the COPII vesicle recruitment and subsequent delivery to the Golgi and the cell surface of a group of GPCRs. Our data have also shown that direct interactions between the receptors and ufmylation components dictate receptor trafficking and sorting. These data reveal important roles and mechanisms for the ufmylation system in the COPII-mediated export and segregation of nascent GPCRs at the ER and provide important insights into the regulation of general membrane trafficking.
RESULTS
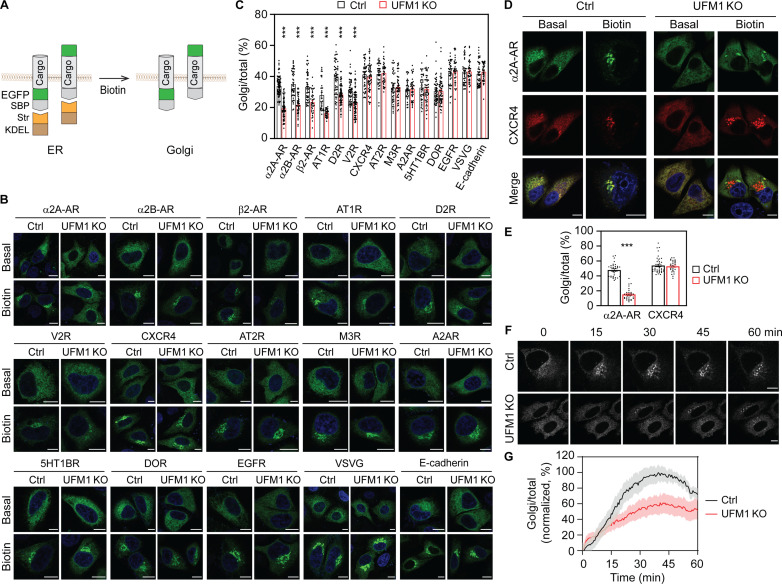
UFM1 knockout inhibits the ER-Golgi transport of some GPCRs
As an initial approach to explore the functions of ufmylation in protein trafficking, we used the retention using the selective hooks (RUSH) assays (55, 61, 64) to determine the effects of UFM1 KO by CRISPR-Cas9 on the ER-to-Golgi transport of newly synthesized PM proteins, including 12 GPCRs [α2A-, α2B-, and β2-adrenergic receptors (ARs); angiotensin II type 1 and type 2 receptors (AT1R and AT2R); dopamine D2 receptor (D2R); vasopressin V2 receptor (V2R); chemokine receptor CXCR4; muscarinic acetylcholine receptor type 3 (M3R); adenosine A2A receptor (A2AR); 5-hydroxytryptamine receptor 1B (5HT1BR); and δ-opioid receptor (DOR)] and three non-GPCR proteins [epidermal growth factor receptor (EGFR), vesicular stomatitis virus glycoprotein (VSVG), and E-cadherin] in HeLa cells. In RUSH assays, the ER retention signal Lys-Asp-Glu-Leu (KDEL) fused to streptavidin (Str-KDEL) was used as a hook and individual cargoes were conjugated with a green fluorescent protein (GFP) at either terminus and a streptavidin-binding peptide (SBP) at the N terminus (NT) (Fig. 1A). The ER-Golgi transport was quantified by measuring the Golgi expression relative to the total expression after biotin induction to disrupt the SBP-streptavidin interaction. Because these PM proteins have been studied previously in RUSH assays (55, 61, 64) and because α2A-AR and AT2R as examples were extensively colocalized with the ER marker Sec61 before biotin induction (fig. S1A) and biotin incubation for 30 min induced their colocalization with the ERGIC marker p58 (fig. S1B) and the Golgi markers giantin (fig. S1C) and β1,4-galactosyltransferase 1 (Gal T) (fig. S1D), the concentration of individual cargoes in the juxtanuclear region after biotin induction was considered to be transported to the Golgi, without using Golgi markers.
Fig. 1. Screening for UFM1-regulated PM proteins in RUSH assays.
(A) The RUSH system to measure the ER-Golgi transport of PM proteins tagged with GFP at the NT or the CT. (B) Representative images showing the ER-Golgi transport of PM proteins in control and UFM1 KO cells in RUSH assays. HeLa cells were transfected with individual RUSH plasmids for 20 hours and then incubated with biotin for 10 min (VSVG), 15 min (α2A-AR, α2B-AR, β2-AR, AT1R, D2R, CXCR4, AT2R, EGFR, and E-cadherin), or 30 min (V2R, M3R, A2AR, 5HT1BR, and DOR). (C) Quantitative data shown in (B). The ER-Golgi transport was measured as the ratio of the Golgi expression to the total expression. (D) Effects of UFM1 KO on the ER-Golgi transport of GFP–α2A-AR and mCherry-CXCR4 expressed in same cells after biotin induction for 15 min. (E) Quantitative data shown in (D). (F) Live cell imaging showing the ER-Golgi export of α2A-AR in control and UFM1 KO cells. (G) The ER-Golgi transport kinetics of α2A-AR in control and UFM1 KO cells. HeLa cells were transfected with α2A-AR RUSH plasmids and the ER export was induced by the addition of biotin at 0 min. Images were captured at an interval of 30 s and the Golgi/total ratio at each time was normalized to the highest ratio after subtraction from the ratio at time 0 in individual cells. The quantitative data are means ± SE; n = 30 to 80 cells in (C), 30 to 39 cells in (E), and 8 cells in (G), each from at least three separate experiments. ***P < 0.001 versus control. Scale bars, 10 μm.
Among the 15 PM proteins studied, the ER-Golgi transport of six GPCRs (α2A-AR, α2B-AR, β2-AR, AT1R, D2R, and V2R) was markedly attenuated in UFM1 KO HeLa cells as compared with that in control cells. In contrast, the ER-Golgi transport of the other nine PM proteins was very much the same in control and UFM1 KO cells (Fig. 1, B and C, and fig. S2A). UFM1 KO similarly inhibited the ER-Golgi transport of α2A-AR at 15 and 30 min after biotin induction in HeLa (fig. S2, B and C) and human embryonic kidney (HEK) 293 cells (fig. S2, D and E). To confirm the differential effects of UFM1 KO on different GPCRs, α2A-AR and CXCR4 were coexpressed and their transport was compared in cells expressing both receptors. After biotin induction, α2A-AR and CXCR4 were similarly transported and extensively colocalized in control cells, whereas the ER-Golgi export of α2A-AR, but not CXCR4, was attenuated in UFM1 KO cells (Fig. 1, D and E).
We next characterized the effect of UFM1 KO on the export kinetics of GPCRs using α2A-AR as a representative in live cell RUSH assays. The time course curve of α2A-AR transport to the Golgi was delayed and the maximal Golgi expression was reduced by approximately 40% in UFM1 KO cells as compared with those in control cells (Fig. 1, F and G, and movies S1 and S2). These data demonstrate that the normal function of UFM1 is required for the ER export of some, but not all, nascent PM proteins.
We also measured the effect of UFM1 KO on α2A-AR transport from the Golgi to the PM in RUSH assays in combination with the temperature-induced block of cargo export from the Golgi. The cells transfected with α2A-AR RUSH plasmids were incubated with biotin at 20°C to release the receptors from the ER but block them in the Golgi, and receptor export from the Golgi to the PM was synchronized by incubation at 37°C (fig. S2F). In both control and UFM1 KO cells, about 60% of α2A-AR transported to the Golgi after biotin induction for 3 hours at 20°C and about 75% of the Golgi-accumulated α2A-AR exported to the cell surface after incubation at 37°C for 30 min (fig. S2, G and H). These data suggest that UFM1 conjugation is unlikely involved in the post-Golgi transport of α2A-AR.
Depletion of ufmylation and de-ufmylation enzymes impedes the ER-Golgi transport of GPCRs
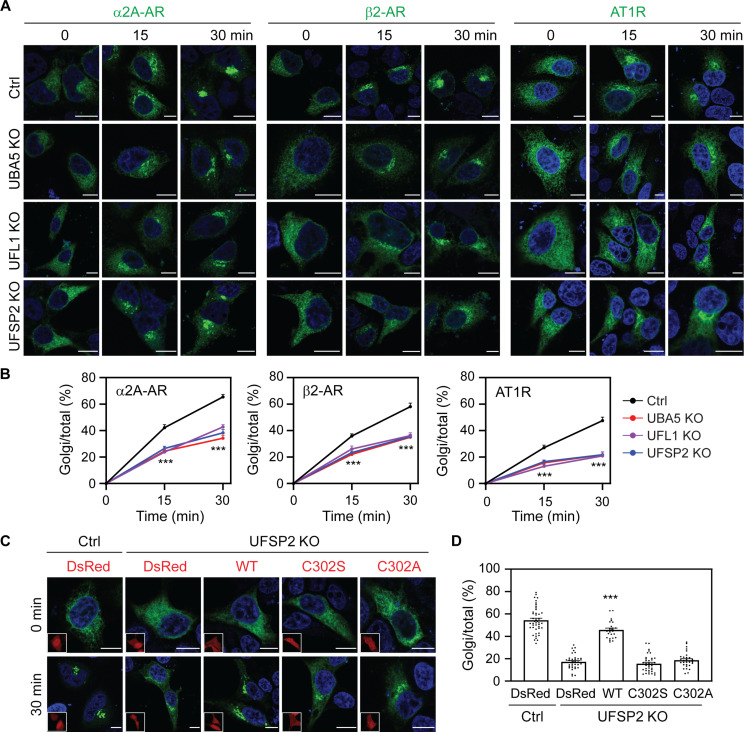
To further study the role of ufmylation in protein transport, we generated KO cells lacking the ufmylation ligases UBA5 and UFL1 and the de-ufmylation protease UFSP2 by CRISPR-Cas9 in HeLa and HEK293 cells (fig. S3, A and B). Individual KO of UFM1, UBA5, UFL1, and UFSP2 had no apparent effects on the expression of other components, except that UFSP2 KO reduced the level of free UFM1 which is consistent with previous studies (fig. S3B) (4, 14), suggestive of enhanced protein ufmylation in UFSP2 KO cells. To study cargo transport in these KO cells, we chose three GPCRs, α2A-AR, β2-AR, and AT1R, whose transport was impaired in UFM1 KO cells. Similar to the results observed in UFM1 KO cells, depletion of UBA5, UFL1, and UFSP2 significantly reduced the ER-Golgi transport of all three receptors in RUSH assays in HeLa cells (Fig. 2, A and B). UBA5, UFL1, and UFSP2 KO also inhibited the ER-Golgi transport of α2A-AR in HEK293 cells (fig. S3, C and D). These data demonstrate that both ufmylation and de-ufmylation enzymes are equally important for the normal ER-Golgi export of some GPCRs.
Fig. 2. Depletion of UBA5, UFL1, and UFSP2 impedes the ER-Golgi transport of α2A-AR, β2-AR, and AT1R.
(A) Representative images showing the effect of UBA5, UFL1, and UFSP2 KO on the ER-Golgi transport of three GPCRs in RUSH assays. HeLa cells were transfected with individual GPCRs in RUSH plasmids for 20 hours and then incubated with biotin for 15 and 30 min. (B) Quantitative data shown in (A). (C) Rescue of α2A-AR transport in UFSP2 KO cells by UFSP2 and its mutants. HeLa cells were transfected with α2A-AR RUSH plasmids together with CRISPR-Cas9–resistant DsRed-UFSP2 or its mutants and then incubated with biotin for 30 min. Insets show expression of DsRed or DsRed-UFSP2. (D) Quantitative data shown in (C). The quantitative data are the Golgi/total ratio and expressed as means ± SE; n = 55 to 80 cells in (B) and 25 to 44 cells in (D), each from at least three repeats. ***P < 0.001 between individual KO and control in (B), and ***P < 0.001 versus DsRed, C302S, or C302A in UFSP2 KO cells in (D). Scale bars, 10 μm.
We next measured the abilities of UFSP2 and its catalytically inactive mutants to rescue the defective transport of α2A-AR in UFSP2 KO cells. The expression of CRISPR-Cas9–resistant wild-type (WT) UFSP2 strongly enhanced the ER-Golgi transport of α2A-AR after biotin incubation for 30 min in UFSP2 KO cells, whereas the expression of two UFSP2 inactive mutants, C302S and C302A, was ineffective (Fig. 2, C and D). These data suggest that the action of UFSP2 on GPCR transport relies on its enzymatic activity in de-ufmylation.
Distinct ufmylation substrates differentially regulate the ER-Golgi traffic of GPCRs
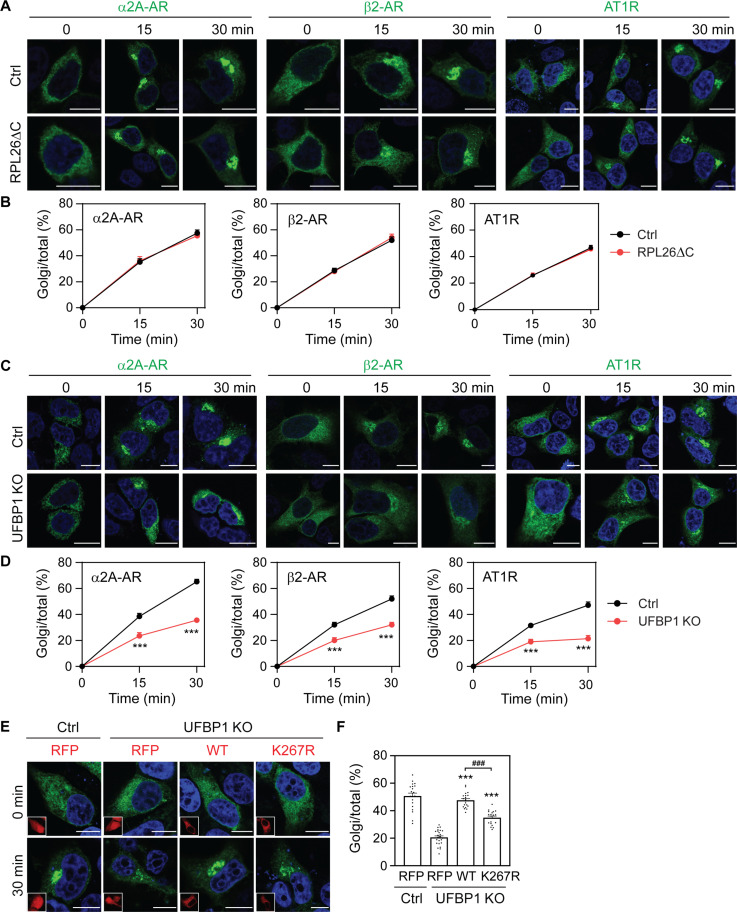
We next sought to define whether the ufmylation of substrates was able to affect the ER-Golgi transport of GPCRs by focusing on ribosomal protein L26 (RPL26) and UFBP1 (also known as DDRGK1, dashurin, and C20orf116). RPL26 is a component of the large ribosomal subunit involved in protein synthesis; it has been identified as the principle ufmylation substrate (14) and its ufmylation is involved in the regulation of ribosome-associated quality control at the ER (30–32). UFBP1 is an ER-anchored protein (65) and is the first identified ufmylation target (2). Recent studies suggest that UFBP1 is an important regulator in the ER localization and function of UFL1 (66). To study the function of RPL26 ufmylation, we made use of previously characterized RPL26ΔC knock-in HEK293 cells that lack RPL26 ufmylation sites (30). The ER-Golgi transport of α2A-AR, β2-AR, and AT1R was almost identical in WT control and RPL26ΔC cells after biotin induction in RUSH assays (Fig. 3, A and B).
Fig. 3. Distinct functions of RPL26 and UFBP1 ufmylation in the ER-Golgi transport of GPCRs.
(A) Representative images showing the ER-Golgi transport of α2A-AR, β2-AR, and AT1R in WT and RPL26ΔC HEK293 cells in RUSH assays. (B) Quantitative data shown in (A). (C) The ER-Golgi transport of α2A-AR, β2-AR, and AT1R in control and UFBP1 KO HEK293 cells. (D) Quantitative data shown in (C). (E) Rescue of α2A-AR transport in UFBP1 KO cells by UFBP1 and its mutant K267R. The cells were transfected with α2A-AR RUSH plasmids together with CRISPR-Cas9–resistant UFBP1-red fluorescent protein (RFP) or K267R and then incubated with biotin for 30 min. Insets show expression of RFP or UFBP1-RFP. (F) Quantitative data shown in (E). The quantitative data are the Golgi/total ratio and expressed as means ± SE; n = 45 to 70 cells in (B), 45 to 72 cells in (D), and 45 to 80 cells in (F), each from at least three experiments. ***P < 0.001 versus RFP in UFBP1 KO cells, and ###P < 0.001 between WT and K267R. Scale bars, 10 μm.
By contrast, the ER-Golgi export of all three receptors was suppressed in UFBP1 KO cells as compared with that in control cells (Fig. 3, C and D, and fig. S3E). In rescue experiments, the expression of CRISPR-Cas9–resistant WT UFBP1 and its ufmylation-deficient K267R enhanced α2A-AR transport in UFBP1 KO cells (Fig. 3, E and F). However, α2A-AR transport was still significantly lower in cells expressing the mutant K267R than in cells expressing WT UFBP1 (P < 0.001; Fig. 3, E and F). These data imply that the ufmylation of UFBP1, but not RPL26, is important in the ER-Golgi transport of GPCRs and that the function of UFBP1 is mediated through multiple mechanisms.
Ufmylation controls the recruitment of GPCRs to COPII vesicles
Previous studies have demonstrated that the ufmylation system plays an important role in the ribosome-associated quality control (14, 30–33). To elucidate the possible mechanisms underlying the function of ufmylation in the ER-Golgi transport of GPCRs, we sought to define whether UFM1 KO could cause the misfolding of GPCRs, leading to their defective ER export, which in turn induced the unfolded protein response (UPR). Treatment with alprenolol, a nonselective, membrane-permeant β-AR antagonist that is known to function as a pharmacological chaperone to enhance the export of misfolded β-ARs from the ER (67, 68), did not rescue the defective ER-Golgi transport of β2-AR in UFM1 KO cells (fig. S4, A and B). α2A-AR was similarly degraded over time in control and UFM1 KO cells as measured in cycloheximide (CHX) chase assays (fig. S4, C and D). Although the expression levels of GRP78 and activating transcription factor 4 (ATF4), two UPR markers, were increased by 10 to 16% in UFM1 KO cells as compared with those in control cells (P > 0.05), they were not altered by α2A-AR transfection in both control and UFM1 KO cells (fig. S4, E and F).
We next determined the effect of KO of ufmylation components on the formation of COPII vesicles using Sec24D as a marker. The average numbers of the punctate structures containing GFP-Sec24D were largely similar in KO cells depleting UFM1, UBA5, UFL1, UFSP2, and UFBP1 as compared with those in respective control cells (fig. S5, A and B). Consistent with our previous studies (61), the expression of Sar1B GDP-bound G37A and GTP-bound H79G mutants strongly inhibited the ER-Golgi transport of α2A-AR and AT1R (fig. S5, C and D), indicative of COPII-mediated ER export.
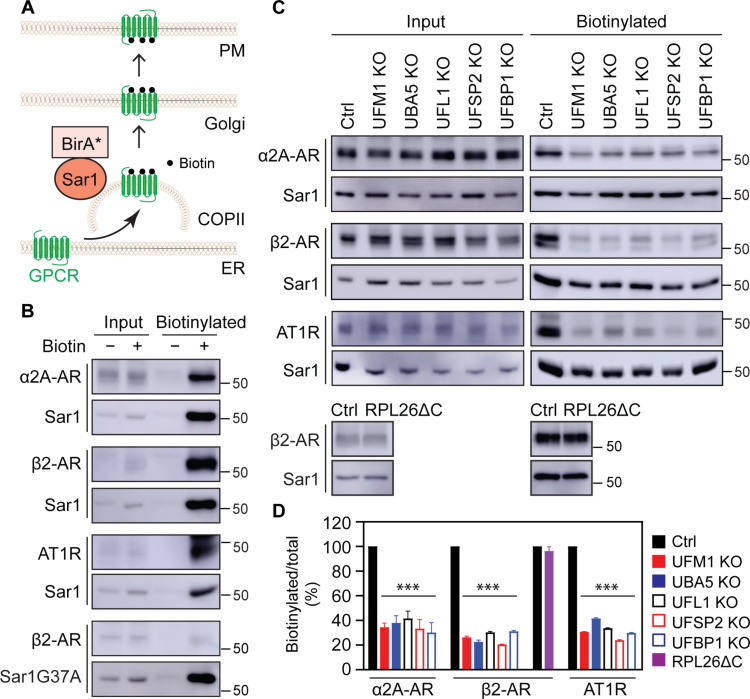
We then studied if ufmylation regulated GPCR recruitment to COPII vesicles in proximity-dependent biotinylation assays in which Sar1B was conjugated to the promiscuous biotin ligase BirA R118G (Sar1B-BirA*) that catalyzes biotinylation of neighboring proteins (69). This method has been used to identify cargoes, cargo receptors, and components of transport machinery in COPII-mediated secretion (69–71). For this purpose, cells were transfected with individual receptors together with Sar1B-BirA* for 6 hours and then treated with biotin for 24 hours (Fig. 4A). α2A-AR, β2-AR, and AT1R all were strongly biotinylated upon addition of biotin (Fig. 4B). In marked contrast, β2-AR biotinylation by Sar1BG37A-BirA* (Fig. 4B) and the biotinylation of the β2-AR mutant L64A, an ER export deficient (72), by Sar1B-BirA* (fig. S5E) were barely detected. These data suggest that GPCR biotinylation by Sar1-BirA* is likely specific that occurs when they are exported from the ER via COPII vesicles. The biotinylation of α2A-AR, β2-AR, and AT1R was markedly inhibited by 60 to 80% in UFM1, UBA5, UFL1, UFSP2, and UFBP1 KO cells as compared with that in control cells, whereas β2-AR biotinylation was very much the same in WT control and RPL26ΔC cells (Fig. 4, C and D). These data strongly demonstrate that ufmylation controls the recruitment of nascent GPCRs to COPII vesicles, without affecting vesicle formation at the ER membrane and receptor folding.
Fig. 4. Depleting ufmylation components inhibits the recruitment of GPCRs to COPII vesicles.
(A) Cartoon showing the biotinylation of GPCRs by Sar1B-BirA* in COPII vesicles. (B) The biotinylation of GPCRs by Sar1B-BirA* or Sar1BG37A-BirA*. HEK293 cells were transfected with individual HA-tagged GPCRs plus Myc-tagged Sar1B-BirA* or Sar1BG37A-BirA*, and then incubated with (+) or without (−) biotin. After pulldown with streptavidin-conjugated beads, receptors and Sar1 were measured by immunoblotting using HA and Myc antibodies, respectively. Similar results were obtained in at least three repeats. (C) Effects of depleting ufmylation components on Sar1B-BirA*–mediated biotinylation of GPCRs. Control, KO, and RPL26ΔC HEK293 cells were transfected with HA-receptors and Myc-Sar1B-BirA* and then incubated with biotin. (D) Quantitative data shown in (C). Input contains 3% of the total proteins used in the experiments. The quantitative data are means ± SE; n = 3. ***P < 0.001 between individual KO and control.
Ufmylation regulates the surface delivery and signaling of GPCRs
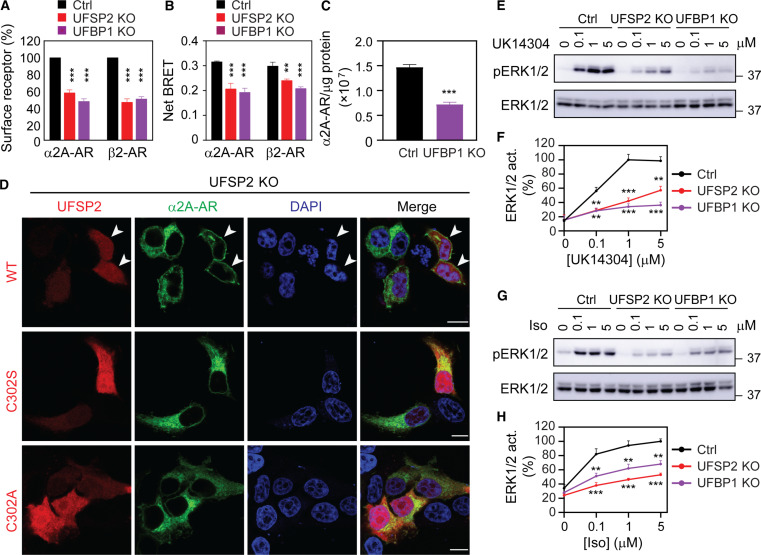
We next investigated whether ufmylation could affect the steady-state expression of GPCRs at the cell surface which is the functional destination for most GPCRs, by studying two KO cells (UFSP2 and UFBP1) and two GPCRs (α2A-AR and β2-AR). The receptors were transiently expressed in cells for at least 24 hours and their surface expression was quantified in intact live cells. The surface expression of both receptors was significantly attenuated in UFSP2 and UFBP1 KO cells as compared with that in control cells measured in radioligand binding (Fig. 5A) and bioluminescence resonance energy transfer (BRET) assays (Fig. 5B).
Fig. 5. UFSP2 and UFBP1 KO attenuate the surface expression and signaling of α2A-AR and β2-AR.
(A) Surface expression of α2A-AR and β2-AR in control and KO cells as measured by radioligand binding of intact live cells using [3H]-RX821002 and [3H]-CGP12177, respectively. HEK293 cells transfected with individual receptors were incubated with the radioligand at 20 nM for 90 min. (B) Surface expression of α2A-AR and β2-AR as measured by live cell BRET assays. The cells were transfected with α2A-AR–Rluc8 or β2-AR–Rluc8 together with Venus-kRas or pcDNA3.1. (C) Effect of UFBP1 KO on the surface expression of endogenous α2A-AR in HT29 cells as measured by radioligand binding of membrane preparations using [3H]-RX-821002 at 2 nM. The data expressed are the numbers of α2A-AR per microgram of membrane protein. (D) Rescue of α2A-AR surface expression in UFSP2 KO cells. HEK293 cells were transfected with α2A-AR–GFP together with CRISPR-Cas9–resistant DsRed-UFSP2 or its inactive mutants for 24 hours. Arrows in the top panel indicate the cell surface expression of α2A-AR in cells expressing WT UFSP2. (E and G) Effect of UFSP2 and UFBP1 KO on ERK1/2 activation by α2A-AR (E) and β2-AR (G). HEK293 cells transfected with α2A-AR and β2-AR were treated with UK14304 and Iso, respectively, for 5 min. (F and H) Quantitative data shown in (E) and (G). The quantitative data are means ± SE; n = 3. **P < 0.01 and ***P < 0.001 versus respective control. Scale bars, 10 μm.
To study the effects of ufmylation on the cell surface expression of endogenous GPCRs, we used HT29 cells that express endogenously α2A-AR. Depletion of UFBP1 (fig. S6A) attenuated the surface expression of endogenous α2A-AR by approximately 50% in HT29 cells as measured in radioligand binding assays (Fig. 5C). In addition, UFBP1 KO also inhibited the ER-Golgi transport of α2A-AR, β2-AR, and AT1R in HT29 cells in RUSH assays (fig. S6, B and C).
In rescue experiments, transient expression of WT UFSP2 for 24 hours restored the surface presentation of α2A-AR in UFSP2 KO cells (Fig. 5D, top), whereas the receptors were localized in perinuclear regions in cells without UFSP2 transfection or in cells expressing UFSP2 mutants, C302S and C302A (Fig. 5D).
We next determined if UFSP2 and UFBP1 KO could affect the concomitant function of α2A-AR and β2-AR by measuring activation of the mitogen-activated protein kinases ERK1/2. Consistent with the reduced cell surface expression of α2A-AR and β2-AR, ERK1/2 activation after stimulation with the α2-AR agonist UK14304 and the β2-AR agonist isoproterenol (Iso) was markedly attenuated in UFSP2 and UFBP1 KO cells as compared with that in control cells (Fig. 5, E to H). These data demonstrate that ufmylation controls not only GPCR transport from the ER to the Golgi but also their delivery to the cell surface and function.
UFBP1 and UFL1 interact with GPCRs at the ER and UFBP1 localizes at COPII vesicles
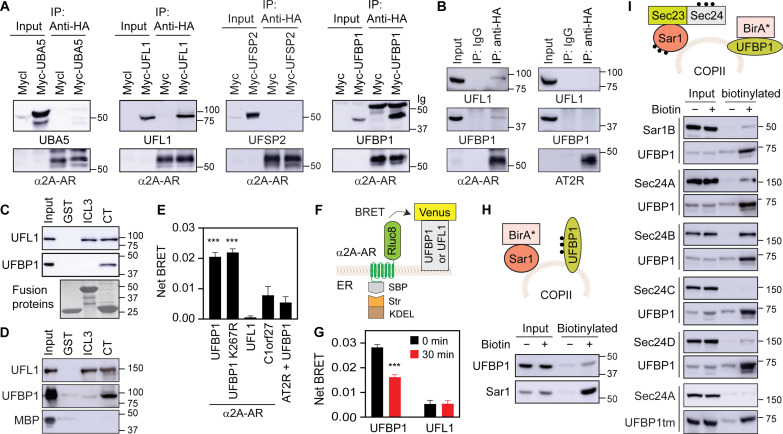
To further delineate the mechanisms of how ufmylation regulates GPCR export from the ER, we measured the interactions of GPCRs with ufmylation components. In coimmunoprecipitation (co-IP) assays using cell lysates expressing α2A-AR together with individual ufmylation components. α2A-AR was found to form complexes with UFL1 and UFBP1, but not UBA5 and UFSP2 (Fig. 6A). α2A-AR, but not AT2R (a UFM1-independent receptor), pulled down endogenous UFL1 and UFBP1 in co-IP assays (Fig. 6B). To identify the binding domains of α2A-AR, its third intracellular loop (ICL3) and C terminus (CT) known to mediate receptor interaction with a number of regulatory proteins involved in signaling and trafficking (52, 73) were generated as glutathione S-transferase (GST) fusion proteins and incubated with cell lysates expressing Myc-tagged UFL1 or UFBP1. We found that the CT strongly bound both UFL1 and UFBP1, whereas the ICL3 bound UFL1, but not UFBP1 (Fig. 6C). To further study the direct interaction between α2A-AR and UFL1 or UFBP1, GST fusion proteins containing the ICL3 or the CT were incubated with purified UFL1 and UFBP1 tagged with maltose-binding protein (MBP). The CT interacted with MBP-UFL1 and MBP-UFBP1 but not MBP alone, whereas the ICL3 interacted with MBP-UFL1 but not MBP-UFBP1 and MBP (Fig. 6D).
Fig. 6. UFBP1 and UFL1 interactions with α2A-AR and UFBP1 localization at COPII vesicles.
(A) Co-IP of α2A-AR and individual ufmylation components. HEK293 cells were transfected with HA–α2A-AR together with Myc-tagged UBA5, UFL1, UFSP2, or UFBP1 and subjected to IP with HA antibodies. Antibodies used were anti-UBA5 for UBA5, anti-Myc for UFL1 and UFBP1, anti-UFSP2 for UFSP2, and anti-HA for α2A-AR. (B) Co-IP of α2A-AR or AT2R and endogenous UFL1 or UFBP1. HEK293 cells were transfected with HA–α2A-AR (left) or HA-AT2R (right) and subjected to IP with IgG or HA antibodies. (C) Interactions of the third ICL3 and CT of α2A-AR with UFL1 and UFBP1 using cell lysates expressing Myc-tagged UFL1 or UFBP1 in GST fusion protein pulldown assays. (D) Interactions of the ICL3 and CT of α2A-AR with purified MBP-UFL1, MBP-UFBP1, and MBP. (E) Live cell BRET. HEK293 cells were transfected with Rluc8-tagged α2A-AR or AT2R together with Venus-tagged UFBP1, the UFBP1 mutant K267R, UFL1, or C1orf27. (F) Cartoon of RUSH-based BRET assays. (G) RUSH-based BRET between α2A-AR and UFBP1 or UFL1. HEK293 cells were transfected with α2A-AR–Rluc8 in RUSH plasmids together with Venus-tagged UFBP1 or UFL1 and then treated with biotin for 30 min. (H) UFBP1 biotinylation by Sar1B-BirA*. HEK293 cells were transfected with Sar1B-BirA*-HA and Myc-UFBP1 and then incubated with (+) or without (−) biotin. (I) Biotinylation of Sar1B and Sec24A/B/C/D by UFBP1-BirA* or UFBP1tm-BirA*. HEK293 cells were transfected with UFBP1-BirA*-HA or UFBP1tm-BirA*-HA and GFP-tagged Sec24 or Sar1B. In each panel, similar results were obtained in at least three experiments. Input contains 3% of the total proteins used in the experiments, except in (D) with 10%. The quantitative data are means ± SE; n = 3 to 5. ***P < 0.001 versus UFL1, C1orf27, and AT2R plus UFBP1 in (E) or versus 0 min in (G).
We next used two different BRET assays to characterize the interactions of UFBP1 and UFL1 with α2A-AR in live cells. In regular BRET assays, very similar net BRET was observed between α2A-AR and UFBP1 and between α2A-AR and UFBP1 ufmylation mutant K267R, both of which were much greater than those between α2A-AR and C1orf27 and between AT2R and UFBP1 (Fig. 6E). C1orf27 is an ER protein and was recently shown to interact with α2A-AR (55). These data suggest that UFBP1 interaction with α2A-AR is independent of its ufmylation. In RUSH-based BRET assays in which α2A-AR–Rluc8 in RUSH plasmids and UFBP1-Venus were coexpressed (Fig. 6F), net BRET between α2A-AR and UFBP1 was clearly detected in the absence of biotin which was significantly reduced after biotin incubation to induce α2A-AR export from the ER for 30 min (Fig. 6G). These data suggest that UFBP1 interaction with α2A-AR occurs at the ER. However, net BRET between α2A-AR and UFL1 was very weak as measured in both BRET assays in live cells (Fig. 6, E and G).
We then determined if UFBP1 was able to localize at COPII vesicles in proximity biotinylation assays. To eliminate that UFBP1 was in close proximity with COPII components during its export via COPII vesicles, HEK293 cells were transfected with UFBP1 plus Sar1B-BirA* (Fig. 6H) or with UFBP1-BirA* plus individual Sec24 isoforms or Sar1B (Fig. 6I) for 24 hours before treatment with CHX and biotin for 8 hours. UFBP1 was biotinylated in cells expressing Sar1B-BirA* (Fig. 6H), and Sar1B was biotinylated in cells expressing UFBP1-BirA* (Fig. 6I). Sec24A, Sec24B, and Sec24D, but not Sec24C, were biotinylated in cells expressing UFBP1-BirA* (Fig. 6I). In addition, Sec24A biotinylation was undetectable in cells expressing the UFBP1 mutant containing only transmembrane domain (UFBP1tm) fused to BirA* (Fig. 6I, bottom two panels). Sec24B biotinylation was very much the same in cells expressing UFBP1-BirA* and UFBP1K267R-BirA* (fig. S7A). Furthermore, image analysis showed that UFBP1 was partially colocalized with Sec24A (fig. S7B). However, UFBP1 was found not to interact with Sec24A, Sec24D, and Sar1B in co-IP assays (fig. S7C). These data demonstrate that UFBP1 localizes at COPII vesicles, particularly those coated with Sec24A/B/D isoforms, in a UFM1 conjugation-independent fashion, but it does not physically associate with COPII components.
The ufmylation system has the sorting function in GPCR export from the ER
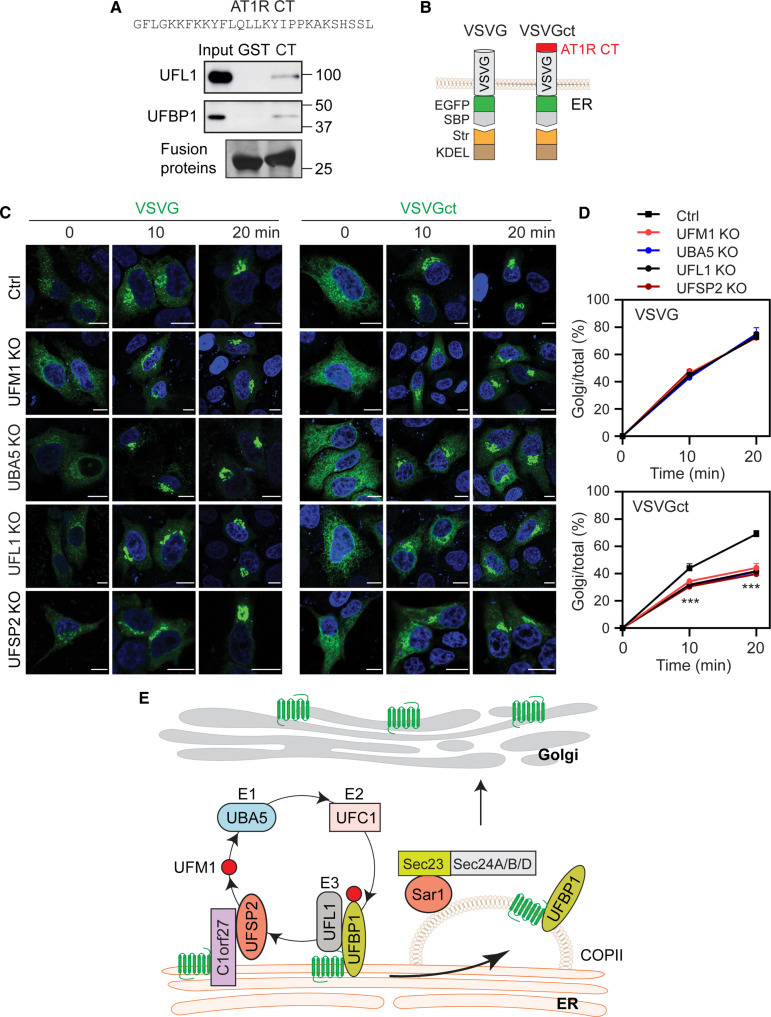
We next investigated the possible function of ufmylation in the sorting of PM proteins at the ER. For this purpose, we generated a chimeric protein in which VSVG was fused at its CT with the α2A-AR CT that binds UFBP1 and UFL1. Unfortunately, this chimeric VSVG was unable to export from the ER after biotin induction, most likely due to misfolding. As the AT1R CT also bound both UFBP1 and UFL1 in GST fusion protein pulldown assays (Fig. 7A), we generated the chimera VSVGct in which VSVG was fused to the AT1R CT (Fig. 7B). VSVG and VSVGct had similar degradation kinetics as measured in CHX chase experiments (fig. S8, A and B). Similar to WT VSVG, VSVGct was transported to the Golgi after biotin addition (Fig. 7, C and D). The ER-Golgi transport of VSVG was essentially identical within 20 min after biotin addition in UFM1, UBA5, UFL1, UFSP2, and UFBP1 KO cells and RPL26ΔC knock-in cells as compared with that in respective control cells (Fig. 7, C and D, and fig. S8, C and D). In marked contrast, the transport of VSVGct from the ER to the Golgi after biotin induction at both 10 and 20 min was much less in all KO cells studied than that in control cells (Fig. 7, C and D).
Fig. 7. Effect of the UFBP1/UFL1-binding domain identified in the AT1R CT on the ER-Golgi transport of VSVG.
(A) Interactions of the AT1R CT with UFBP1 and UFL1 in GST fusion protein pulldown assays. GST fusion proteins containing the AT1R CT were incubated with cell lysates prepared from cells transfected with Myc-tagged UFL1 or UFBP1. UFBP1 and UFL1 were measured by using Myc antibodies. Similar results were obtained in three experiments. (B) Cartoon showing generation of the VSVG chimera containing the AT1R CT (VSVGct). (C) Representative images showing the ER-Golgi transport of VSVG and VSVGct in control and KO cells. HeLa cells were transfected with VSVG or VSVGct in RUSH plasmids and then incubated with biotin for 10 and 20 min. (D) Quantitative data shown in (C). (E) A model depicting the functions and mechanisms of the ufmylation system in the COPII recruitment and ER-Golgi transport of nascent GPCRs (see text for detail). Input contains 3% of the total proteins used in the experiments. The quantitative data are means ± SE; n = 34 to 44 cells from at least three separate experiments. ***P < 0.001 between individual KO and control. Scale bars, 10 μm.
To further compare the ER-Golgi transport of VSVG and VSVGct, we used the temperature-sensitive mutant VSVGtsO45 which was misfolded and retained within the ER at a restrictive temperature and correctly delivered to the Golgi at permissive temperature (74). Both VSVG and VSVGct were retained in the ER in control and individual KO cells cultured at 40°C. After the shift to 32°C for 30 min, VSVG transport to the Golgi was comparable in control and KO cells, whereas VSVGct transport to the Golgi was significantly lower in each KO cell than in control cells (fig. S8, E and F). These data indicate that the UFL1/UFBP1-binding domain identified in the AT1R CT is sufficient to dictate VSVG transport to be dependent on ufmylation.
DISCUSSION
In this study, we have revealed an important function for the ufmylation system in the anterograde trafficking of nascent PM proteins, specifically a subset of GPCRs, after synthesis in the ER (Fig. 7E). This function was first identified in a screening assay in which UFM1 KO markedly inhibited the ER-Golgi transport of 6 GPCRs, out of the 15 PM proteins tested. This was further supported by studying the role of ufmylation and de-ufmylation enzymes and targets, demonstrating that depletion of UBA5, UFL1, UFSP2, and UFBP1 similarly disrupted the ER-Golgi export of α2A-AR, β2-AR, and AT1R. Both activity-dependent rescue by UFSP2 and ufmylation-dependent rescue by UFBP1 of α2A-AR transport provided another strong evidence indicating a role of protein ufmylation and de-ufmylation in GPCR transport. In addition, depleting ufmylation components reduced the surface delivery and function of GPCRs. These data together indicate that ufmylation plays a critical role in the maturation processing of some GPCRs. However, there is controversy over the function of UFSP2 in GPCR transport with one report suggesting that ODR8, Caenorhabditis elegans homolog of UFSP2, modulates the surface localization of some olfactory receptors in chemosensory neurons in an activity- and UFM1-independent manner (75). Furthermore, we cannot exclude the possibility that the depletion of ufmylation components may disrupt the trafficking of signaling molecules involved in the activation of the ERK1/2 pathway, leading to abnormal GPCR signaling to ERK1/2.
Our studies have demonstrated that the function of ufmylation in GPCR export is likely mediated through controlling their recruitment to COPII vesicles (Fig. 7E). This became evident as depletion of ufmylation components markedly blocked Sar1B-BirA*–mediated biotinylation of all GPCRs studied, including α2A-AR, β2-AR, and AT1R. These results are consistent with our data showing that the ER export of these receptors is mediated through COPII vesicles (61). We have found that UFBP1 was in close vicinity to the COPII components Sar1, Sec24A, Sec24B, and Sec24D, but not Sec24C, as measured in proximity biotinylation assays using Sar1B-BirA* and UFBP1-BirA*, demonstrating that UFBP1 localizes on the cytoplasmic face of COPII vesicles, particularly those coated with Sec24A/B/D (Fig. 7E). Confocal imaging revealed partial localization of UFBP1 in COPII vesicles using Sec24A as a marker. COPII localization of UFBP1 is also supported by a recent study in which UFBP1 was identified in vesicles by proteomics (76). As an expression of UFBP1-BirA* and K267R-BirA* similarly induced Sec24B biotinylation, UFBP1 localization at COPII does not rely on its UFM1 conjugation. These data, together with our data showing that UFBP1 interacts with GPCRs on the ER as discussed below, strongly suggest that UFBP1 and GPCRs are recruited to COPII as a complex (Fig. 7E). However, we cannot completely rule out the possibility that UFBP1-BirA* expressed at the ER may induce the biotinylation of Sec24 and Sar1 in COPII vesicles. Several pieces of evidence, including alprenolol was unable to rescue the defective export of β2-AR in UFM1 KO cells, UFM1 KO did not alter α2A-AR degradation kinetics, the α2A-AR expression did not induce UPR, and depleting any ufmylation components had no clear effects on the formation of COPII vesicles, imply that the defective COPII recruitment and ER export of GPCRs in KO cells are not due to misfolding of the receptors and disruption of the assembly of COPII proteins to form the vesicles, which is consistent with the fact that KO of the ufmylation cascade did not globally disrupt the ER export of all cargoes studied.
We have also demonstrated that GPCRs and ufmylation components form multi-protein complexes on the ER membrane that likely contributes to COPII recruitment of the receptors (Fig. 7E). In particular, UFL1 and UFBP1 that are known to form a complex interacted with α2A-AR and AT1R and the UFBP1–α2A-AR interaction occurred spatially on the ER as measured in live cell RUSH-based BRET assays. In contrast, both proteins did not interact with AT2R, a UFM1-independent GPCR, as measured in co-IP and BRET assays. However, the sequence alignments of the CT and ICL3 of GPCRs did not reveal any specific sequences or motifs that may be responsible for ufmylation regulation (fig. S9). In particular, the membrane-proximal C-terminal portions of GPCRs are well conserved, containing highly hydrophobic and positively charged residues and structurally form the amphipathic helix 8 which may be anchored to the membrane by the cysteine residue. Among the UFM1-regulated GPCRs, D2R has the shortest CT that contains helix 8 only. In helix 8, the di-leucine motif (L/I-I/L) known to be important for ER export of GPCRs (58, 59) is found in all UFM1-regulated GPCRs but also exists in some UFM1-independent GPCRs (fig. S9, A and B). As UFBP1 did not interact with Sec24 as measured in co-IP assays, it is unlikely to function as a receptor or an adaptor for the GPCRs it regulates. Previous studies have shown that the ER-anchored protein C1orf27 binds UFSP2 and recruits it to the ER (56, 75, 77), which, together with UFL1 recruitment to the ER via UFBP1 (66), likely anchors the ufmylation system at the ER (Fig. 7E). Similar to UFSP2, C1orf27 KO enhanced protein ufmylation in cell (77). C1orf27 was also shown to interact with and regulate the ER export of some, but not all, GPCRs, including α2A-AR (55, 56, 75). Together, these data suggest two ternary complexes, GPCR/C1orf27/UFSP2 and GPCR/UFBP1/UFL1, formed on the ER membrane that may regulate GPCR recruitment to COPII vesicles and subsequent export from the ER (Fig. 7E). A recent study has shown that the Epstein-Barr virus–encoded, constitutively active GPCR BILF1 binds UFL1 to modulate mitochondrial membrane cargo turnover (78), further implicating the importance of protein-protein interactions in mediating the functions of ufmylation.
Another important finding presented here is the sorting function of ufmylation in the ER export of PM proteins or GPCRs. A fundamental but poorly addressed question in studying the trafficking of the GPCR superfamily with more than 800 members is how they are sorted from one another and from non-GPCR PM proteins. Previous studies have identified several proteins that are capable of sorting GPCRs in their biosynthetic transport via interaction (49–51). The fact that the transport of only some, but not all, GPCRs or PM proteins are affected by depleting ufmylation components suggests not only that the observed effects are likely specific but also that the ufmylation system can segregate different GPCRs and PM proteins at the ER level. Our finding that after fusion with the AT1R CT that binds UFL1 and UFBP1, VSVG changed its responsiveness to manipulation of ufmylation as measured in both RUSH and temperature-sensitive transport assays provides the direct evidence indicating the role of these protein-protein interactions in cargo separation. Hence, physical associations between cargoes (e.g., GPCRs) and ufmylation components may control both cargo recruitment to COPII and cargo selection to be regulated by ufmylation. It is also interesting to note that a recent study has demonstrated that the ufmylation of α-synuclein enhances its transport via the unconventional protein secretion pathway (79).
Our recent studies have shown that GPCR targeting to COPII vesicles may be mediated through the default bulk flow and active capture mechanisms (61). It is interesting to note that all six UFM1-dependent GPCRs use bulk flow for recruitment to COPII vesicles. However, the other four GPCRs (M3R, A2AR, 5HT1BR, and DOR) that use bulk flow for COPII recruitment are not subjected to ufmylation regulation. Nevertheless, these data suggest that protein ufmylation may provide an important mechanism for the separation of different GPCRs during their COPII recruitment which is the first step in their intracellular trafficking. The vesicles carrying distinct GPCRs may be further sorted into different transport pathways. These data, together with previous studies demonstrating that different GPCRs are regulated by distinct proteins (49, 50, 54, 56, 57), indicate multiple sorting mechanisms and different transport routes to dictate the cell surface delivery of nascent GPCRs.
As compared with ubiquitination, the physiological and pathological significance of ufmylation is relatively much less well defined. Emerging evidence suggests that it has multiple functions at the ER. As demonstrated here, the ufmylation system can control the COPII-mediated transport of GPCRs at the ER, in which UFBP1 plays a crucial role and its function is mediated through multiple mechanisms. In addition to being a UFM1 target, UFBP1 directly interacts with the cargo GPCRs and localizes to COPII vesicles in a ufmylation-independent manner. It is also worth mentioning that, in addition to UFBP1, the ufmylation of other targets and the overall ufmylation/de-ufmylation homeostasis of the ER may contribute to the observed effects in cells depleting ufmylation components. As dysregulation of GPCR trafficking and signaling and protein ufmylation are directly associated with the pathogenesis of human diseases, a thorough elucidation of the functions and underlying mechanisms of ufmylation in the biosynthetic transport of GPCRs may provide a foundation for therapeutic innovation. In particular, since the genetic variants of the ufmylation pathway have been identified in humans with neurodevelopmental diseases (5) and most UFM1-regulated GPCRs are functionally important neuronal receptors, the regulation of GPCR trafficking by ufmylation in neurons and its contribution to the development of neurological disorders merit further investigation.
In summary, our present study has identified crucial functions of the ufmylation system in the recruitment to COPII vesicles, anterograde ER-Golgi-surface trafficking, and sorting of a group of nascent GPCRs, which are mediated by virtue of its abilities to ufmylate specific targeted proteins and to interact with the receptors directly. These data also provide important insights into COPII-mediated membrane trafficking.
MATERIALS AND METHODS
Materials
Antibodies against UFM1 were from Abcam; antibodies against UBA5, UFSP2, and UFBP1 were from Proteintech; antibodies against UFL1 were from Bethyl Laboratories; antibodies against Myc, MBP, GFP, β-actin, and phospho-ERK1/2 were from Santa Cruz Biotechnology; antibodies against ERK1/2 and rabbit host antibodies against HA epitope tag were from Cell Signaling Technology; mouse host antibodies against HA were from Roche. Lipofectamine 2000, d-biotin, CHX, Dynabeads MyOne streptavidin C1 beads, and Dynabeads Protein G were from Thermo Fisher Scientific. UK14304, Iso, and alprenolol were obtained from Sigma-Aldrich. Linear polyethyleneimine (PEI; MW 25000) was from Polysciences. MagneGST glutathione particles were from Promega. [3H]-CGP12177 (specific activity = 51 Ci/mmol) and [3H]-RX821002 (50 Ci/mmol) were purchased from PerkinElmer. All other materials were obtained as described elsewhere (55, 61).
Plasmids and constructions
RUSH plasmids expressing α2A-AR, α2B-AR, AT1R, AT2R, D2R, V2R, CXCR4, A2AR, 5HT1BR, DOR, EGFR, VSVG, and E-cadherin in which GFP was tagged at the NT; RUSH plasmids expressing β2-AR and M3R tagged with GFP at the CT, GFP-tagged Sec24 and Sar1B; and GST fusion protein plasmids expressing the ICL3 and the CT of α2A-AR were generated as described previously (55, 57, 61). RUSH plasmids expressing α2A-AR in which mCherry was tagged at the NT were generated as described (57).
To generate the RUSH plasmid Str-KDEL_SBP-mCherry-CXCR4, CXCR4 was amplified by polymerase chain reaction (PCR) using primers (forward, 5′-GATCGGCCGGCCAATGGAGGGGATATC-3′ and reverse, 5′-GATCTCTAGATTAAGAGCTATGAAATGAGCTGG-3′), digested with Fsel and Xbal and then ligated to the plasmid Str-KDEL_SBP-mCherry-Ecadherin (Addgene, plasmid #65287) which was digested with the same enzymes to release E-cadherin. To generate the RUSH plasmid Str-KDEL_SBP-EGFP-VSVGct, the AT1R CT fragment 303-330 was first amplified by PCR using primers (forward, 5′-GATCGAATTCCGGCTTTCTGGGGAAG-3′ and reverse, 5′-GATCTCTAGATTACAGGCTTGAGTGGGAC-3′) and inserted into the VSVG CT in the Str-Ii_VSVG-SBP-EGFP plasmid (Addgene, plasmid #65300). VSVGct without signal peptide was then amplified by PCR using primers (forward, 5′-GATCGGCCGGCCAAAGTTCACCATAGTTTTTC-3′ and reverse, 5′-GATCTCTAGATTACAGGCTTGAGTGGGAC-3′), digested with Fsel and Xbal, and ligated to the plasmid Str-KDEL_SBP-EGFP-Ecadherin (Addgene, plasmid #65286) which was digested with the same enzymes. To generate AT2R-Rluc8, AT2R was amplified by PCR using primers (forward, 5′-GATCCTCGAGATGAAGGGCAACTCCAC-3′ and reverse, 5′-GATCGGATCCCGAGACACAAAGGTCTCC-3′) and cloned into the Rluc8-N1 vector at Xhol and BamHl sites. To generate Venus-tagged UFL1, UFL1 was amplified by using primers (forward, 5′-GATCGAGCTCATGGCGGACGCCTGG-3′ and reverse, 5′-GATCGGATCCCGCTCTTCCGTCACAGATG-3′) and cloned into the Venus-N1 vector at Sac1 and BamHl sites. To generate Venus-C1orf27, C1orf27 was amplified by using primers (forward, 5′-CCGCTCGAGAAATGGGAAGAACCTACATTG-3′ and reverse, 5′-GGGGTACCCCCTACTAATCACTGAAGTAATGAA-3′) and cloned into the Venus-C1 vector at Xhol and Kpnl sits. GST fusion protein plasmids expressing the AT1R CT were generated as described (51). α2A-AR, β2-AR, and AT1R tagged with HA or Rluc8 were generated as described (55). UBA5, UFL1, and UFBP1 tagged with Myc were generated as described (65). UFSP2 tagged with Myc was generated by using primers (forward, 5′-GATCGAATTCGGATGGTGATTTCAGAAAGTATG-3′ and reverse, 5′-GGGGTACCCCTTAAATCATATTTGGTCGCTG-3′) and inserted into the pCMV-Myc vector after digestion with EcoRl and Kpnl. UFSP2 and its mutants tagged with DsRed were generated by PCR using primers (forward, 5′-CGGAATTCTATGGTGATTTCAGAAAGTATG-3′ and reverse, 5′-CGGGATCCCGTTAAATCATATTTGGTCG-3′) and cloned into the pDsRed-Monomer-C1 vector at EcoRl and BamHl restriction sites. UFBP1 and its mutant K267R tagged with red fluorescent protein (RFP) in the pTagRFP-N vector or tagged with Venus in the Venus-N1 vector were generated by PCR using primers (forward, 5′-GATCAAGCTTATGGTGGCGCCTGTGTGGTAC-3′ and reverse, 5′-GATCGGATCCCGGGCTGGGGCTTGG-3′) and cloned into Hindlll and BamHl sites. To generate Sar1B-BirA*-HA, UFBP1-BirA*-HA, and UFBP1tm-BirA*-HA, Sar1B, UFBP1, and UFBPtm were amplified by using primers (Sar1B: forward, 5′-GATCACCGGTATGTCCTTCATATTTGACTGG-3′ and reverse, 5′-GATCGAATTCATCGATGTACTGTGCCATC-3′; UFBP1: forward, 5′-GATCACCGGTATGGTGGCGCCTGTG-3′ and reverse, 5′-GATCGAATTCGGCTGGGGCTTGGGC-3′; UFBP1tm: forward, 5′-GATCACCGGTATGGTGGCGCCTGTG-3′ and reverse, 5′-GATCGAATTCTGATGCCGCCCGGCC-3′) and cloned into the pcDNA3.1 MCS-BirA(R118G)-HA vector (Addgene, plasmid #36047) at BshTI and EcoRI sites. To generate Myc-Sar1B-BirA*, Sar1B-BirA* was amplified using Sar1B-BirA*-HA as template and primers (forward, 5′-GATCGTCGACCATGTCCTTCATATTTGACTGG-3′ and reverse, 5′-GATCGGTACCCTACTTCTCTGCGCTTCTC-3′) and inserted into the pCMV-Myc vector after digestion with SalI and KpnI. Myc-Sar1BG37A-BirA* and GFP-Sar1BG37A were generated by mutagenesis using primers (forward, 5′-CTTGGATTGGATAATGCAGCGAAAACAACGTTGCTACAC-3′ and reverse, 5′-GTGTAGCAACGTTGTTTTCGCTGCATTATCCAATCCAAG-3′). CRISPR-Cas9–resistant constructs of UFSP2 and UFBP1 were generated by mutagenesis using primers (UFSP2: forward, 5′-GAAAGTATGGATATACTATTTCGCATCCGCGGAGGCCTTGATTTG-3′ and reverse, 5′-CAAATCAAGGCCTCCGCGGATGCGAAATAGTATATCCATACTTTC-3′; UFBP1: forward, 5′-CAGCCGCCTACAGGCACAGCGTCGAGCCCAGC-3′ and reverse, 5′-GCTGGGCTCGACGCTGTGCCTGTAGGCGGCTG-3′). All mutations were generated by using the QuikChange site-directed mutagenesis kit (Agilent). All constructs used in the present study were verified by nucleotide sequence analysis.
Cell culture and transfection
HeLa, HEK293, and HT29 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml). Transient transfection of cells was carried out by using PEI or Lipofectamine 2000.
Generation of KO cell lines using the CRISPR-Cas9 genome editing technology
The sgRNA sequences are 5′-TCACGCTGACGTCGGACCCA-3′ for UFM1, 5′-TCCCGAGGAGCGGCGACGGA-3′ for UBA5, 5′-CCAGCGGGCGCAGTTCGCCG-3′ for UFL1, 5′-ATACTCTTCAGAATAAGAGG-3′ for UFSP2, and 5′-CCACCCGCTGGGCTCGACGC-3′ for UFBP1. The sgRNAs were constructed into the pLentiCRISPR v2 vector (Addgene, plasmid #52961). Lentiviral particles expressing individual sgRNA and Cas-9 were generated by standard HEK293T cell transfection. After infection, cell clones were selected by puromycin and limiting dilution. KO of the targeted proteins was determined by immunoblotting.
RUSH assays
RUSH assays were essentially carried out as described (55, 61, 64). Briefly, to measure ER-Golgi transport, cells were transfected with RUSH plasmids for 20 hours and then incubated with biotin at a final concentration of 40 μM plus CHX (400 μg/ml) for different time periods as indicated in figure legends. To measure Golgi-PM transport, cells were transfected and incubated with biotin plus CHX at 20°C for 3 hours. After replacing the cell culture medium with DMEM without biotin, the cells were incubated at 37°C for 30 min. Protein expression in the Golgi and the whole cell was quantified by measuring the fluorescence intensities using National Institutes of Health (NIH) ImageJ. The ER-Golgi and Golgi-PM transport were expressed as the Golgi/total expression ratio. In live cell RUSH assays, the Golgi/total ratio at each time point was subtracted from the ratio at time 0 and then normalized to the highest ratio in control cells which was defined as 100%.
Fluorescence microscopy
To measure α2A-AR transport in live cells, images were captured using LAS X software at an interval of 30 s with a 63× objective on a Leica Stellaris 5 confocal microscope equipped with an Okolab UNO stage top incubator as described previously (55, 61). To measure protein transport in fixed cells, cells were fixed in a mixture of 4% paraformaldehyde and 4% sucrose for 15 min before imaging.
Proximity-dependent biotinylation and streptavidin pulldown assays
To study COPII-mediated GPCR transport, HEK293 cells seeded on 6-cm dishes were transfected with Myc-Sar1B-BirA* or Myc-Sar1BG37A-BirA* and individual HA-tagged GPCRs (2 μg plasmids each) for 6 hours and then treated with or without 15 μM biotin for additional 24 hours. To study the proximity of UFBP1 with COPll components, HEK293 cells were transfected with Sar1B-BirA*-HA together with Myc-UFBP1 or transfected with UFBP1-BirA*-HA or UFBP1tm-BirA*-HA together with GFP-tagged Sec24 or Sar1B for 24 hours and then treated with biotin (15 μM) plus CHX (400 μg/ml) for 8 hours. The cells were harvested and lysed with 300 μl of radioimmunoprecipitation assay (RIPA) buffer [50 mM tris-HCl (pH 7.4), 150 mM NaCl, 0.25% deoxycholic acid, 1% NP-40, and 1 mM EDTA] with protease inhibitors. After centrifugation, the supernatants were incubated with 20 μl of Dynabeads MyOne streptavidin C1 beads overnight at 4°C. The beads were washed three times with RIPA buffer and biotinylated proteins were eluted with SDS loading buffer and detected by immunoblotting.
CHX chase assays
The degradation of α2A-AR, VSVG, and VSVGct was analyzed in CHX chase assays. Cells were transfected with individual cargo proteins tagged with GFP in RUSH plasmids for 24 hours and then treated with CHX at a concentration of 200 μg/ml for 8, 16, and 24 hours. The expression levels of cargo proteins were measured by immunoblotting using GFP antibodies.
Radioligand binding
The cell surface expression of α2A-AR and β2-AR was measured by ligand binding of intact live cells using [3H]-RX821002 and [3H]-CGP12177, respectively, as described previously (80). Briefly, cells were transfected with individual receptors and then incubated with the radioligand at a concentration of 20 nM for 90 min. The nonspecific binding was determined in the presence of rauwolscine (20 μM for α2A-AR) and alprenolol (20 μM for β2-AR). The PM expression of endogenous α2A-AR in HT29 cells was measured by radioligand binding of membrane preparations essentially as described (59). Briefly, the membrane suspension containing 250 μg of protein was incubated with [3H]-RX-821002 at 2 nM in a total volume of 100 μl for 1 hour and the reaction was terminated by vacuum filtration. The radioactivity was measured by liquid scintillation spectrometry in the AccuFLEX LSC-8000 counter (Hitachi Aloka).
BRET assays
Live cell–based BRET assays were used to measure the cell surface receptor expression and protein-protein interaction as described previously (55, 73). Briefly, to measure the surface receptor expression, cells were cultured on six-well plates and transfected with 250 ng of α2A-AR–Rluc8 or β2-AR–Rluc8 together with 750 ng of Venus-kRas or pcDNA3.1. To measure GPCR interaction, cells were seeded on six-well plates and transfected with 250 ng of Rluc8-tagged α2A-AR or AT2R and 750 ng of Venus-tagged UFBP1, UFL1, or C1orf27 for 24 hours. To measure α2A-AR interaction with UFBP1 or UFL1 in RUSH-based BRET assays, cells were transfected with 250 ng of Str-KDEL_SBP–α2A-AR–Rluc8 together with 750 ng of Venus-tagged UFBP1 or UFL1 for 24 h and then incubated with biotin at 40 μM for 30 min. The cells were harvested and split onto black 96-well plates. After the addition of coelenterazine h (5 mM), luminescence was immediately measured using a Mithras LB940 photon-counting plate reader (Berthold Technologies GmbH, Bad Wildbad, Germany). The BRET signals were calculated by dividing the emission intensity at 520 to 545 nm by the emission intensity at 475 to 495 nm. Net BRET was this ratio minus the same ratio measured from cells expressing Rluc8-tagged receptors and pcDNA3.1.
Measurement of ERK1/2 activation
ERK1/2 activation by α2A-AR and β2-AR was determined by measuring their phosphorylation by immunoblotting as described previously (51). Briefly, cells were transfected with α2A-AR or β2-AR for 24 hours and then starved for 8 hours before stimulation with different concentrations of UK14304 or Iso for 5 min.
Co-IP
Co-IP assays were carried out as described previously (51). Briefly, HEK293 cells were cultured on 10-cm dishes and transfected with HA–α2A-AR together with Myc-tagged UBA5, UFL1, UFSP2, or UFBP1 (10 μg plasmids each) for 24 hours. The cells were harvested and lysed with 500 μl of RIPA buffer containing protease inhibitors. After centrifugation, the supernatants were incubated with 2 μg of HA antibodies (Roche, mouse host) overnight at 4°C, followed by incubation with 30 μl of Dynabeads Protein G for 1 hour at 4°C. To measure receptor interaction with endogenous proteins, HEK293 cells were transfected with HA–α2A-AR or HA-AT2R for 24 hours and subjected to IP with IgG or HA antibodies. To measure UFBP1 interaction with Sec24 and Sar1B, HEK293 cells were transfected with Myc-UFBP1 together with GFP-tagged Sec24 or Sar1B and subjected to IP with Myc antibodies. The beads were collected and washed with lysis buffer. Immunoprecipitated proteins were detected by immunoblotting.
GST fusion protein pulldown assays
GST fusion protein pulldown assays were carried out using the MagneGST pulldown system (Promega) as described previously (51, 73). MBP and MBP-tagged UFL1 and UFBP1 in the pMAL-c5X vector were expressed in bacteria, purified by amylose-conjugated beads, and eluted by incubation with maltose. GST fusion proteins were incubated with HEK293 cell homogenates expressing Myc-tagged proteins or purified MBP-tagged proteins (5 μg) in a total volume of 400 μl of binding buffer containing 20 mM Tris-HCl (pH 7.4), 140 mM NaCl, 1% NP-40, and 10% glycerol overnight at 4°C. After washing three times with the binding buffer, the bound proteins were detected by immunoblotting.
Measurement of VSVG transport from the ER to the Golgi
In addition to RUSH assays, VSVG transport from the ER to the Golgi was also measured by using its temperature-sensitive mutant VSVGtsO45 (74). HEK293 cells grown on coverslips in 12-well plates were transfected with 0.25 μg of VSVG-GFP or VSVGct-GFP constructs and cultured for 24 hours at 40°C to induce the accumulation of VSVG in the ER. The cells were then transferred to 32°C for 30 min to allow VSVG to transport to the Golgi. VSVG expression in the Golgi and the whole cell was quantified by measuring the fluorescence intensities using NIH ImageJ and its ER-Golgi transport was expressed as the Golgi/total expression ratio.
Statistical analysis
Statistical differences were analyzed by using one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons posttest for more than two groups or by unpaired two-tailed t test for two groups. All data were expressed as means ± SE as indicated in each figure. Significance levels are *P < 0.05, **P < 0.01, and ***P < 0.001.
Acknowledgments
We thank Y. Ye (National Institute of Diabetes and Digestive and Kidney Diseases, NIH) for sharing RPL26 knock-in cells.
Funding: This work was supported by NIH grant R35GM136397 (to G.W.)
Author contributions: Conceptualization: X.X., Z.D., H.L., and G.W. Methodology: X.X., H.L., and G.W. Investigation: X.X., W.H., C.N.B., H.L., and G.W. Validation: X.X., W.H., H.L., and G.W. Formal analysis: X.X., W.H., C.N.B., and G.W. Data curation: X.X. and G.W. Resources: X.X., H.L., and G.W. Visualization: X.X., W.H., and G.W. Writing—original draft: X.X., W.H., H.L., and G.W. Writing—review and editing: X.X., W.H., Z.D., H.L., and G.W. Supervision: G.W. Project administration: G.W. Funding acquisition: H.L. and G.W.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Legends for movies S1 and S2
Uncropped gels
Other Supplementary Material for this manuscript includes the following:
Movies S1 and S2
REFERENCES AND NOTES
- 1.Komatsu M., Chiba T., Tatsumi K., Iemura S. I., Tanida I., Okazaki N., Ueno T., Kominami E., Natsume T., Tanaka K., A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J. 23, 1977–1986 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tatsumi K., Sou Y. S., Tada N., Nakamura E., Iemura S., Natsume T., Kang S. H., Chung C. H., Kasahara M., Kominami E., Yamamoto M., Tanaka K., Komatsu M., A novel type of E3 ligase for the Ufm1 conjugation system. J. Biol. Chem. 285, 5417–5427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang S. H., Kim G. R., Seong M., Baek S. H., Seol J. H., Bang O. S., Ovaa H., Tatsumi K., Komatsu M., Tanaka K., Chung C. H., Two novel ubiquitin-fold modifier 1 (Ufm1)-specific proteases, UfSP1 and UfSP2. J. Biol. Chem. 282, 5256–5262 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Millrine D., Cummings T., Matthews S. P., Peter J. J., Magnussen H. M., Lange S. M., Macartney T., Lamoliatte F., Knebel A., Kulathu Y., Human UFSP1 is an active protease that regulates UFM1 maturation and UFMylation. Cell Rep. 40, 111168 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerakis Y., Quintero M., Li H., Hetz C., The UFMylation system in proteostasis and beyond. Trends Cell Biol. 29, 974–986 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishimura R., El-Gowily A. H., Noshiro D., Komatsu-Hirota S., Ono Y., Shindo M., Hatta T., Abe M., Uemura T., Lee-Okada H. C., Mohamed T. M., Yokomizo T., Ueno T., Sakimura K., Natsume T., Sorimachi H., Inada T., Waguri S., Noda N. N., Komatsu M., The UFM1 system regulates ER-phagy through the ufmylation of CYB5R3. Nat. Commun. 13, 7857 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou J., Ma X., He X., Chen B., Yuan J., Jin Z., Li L., Wang Z., Xiao Q., Cai Y., Zou Y., Dysregulation of PD-L1 by UFMylation imparts tumor immune evasion and identified as a potential therapeutic target. Proc. Natl. Acad. Sci. U.S.A. 120, e2215732120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin B., Yu J., Nowsheen S., Wang M., Tu X., Liu T., Li H., Wang L., Lou Z., UFL1 promotes histone H4 ufmylation and ATM activation. Nat. Commun. 10, 1242 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoo H. M., Kang S. H., Kim J. Y., Lee J. E., Seong M. W., Lee S. W., Ka S. H., Sou Y. S., Komatsu M., Tanaka K., Lee S. T., Noh D. Y., Baek S. H., Jeon Y. J., Chung C. H., Modification of ASC1 by UFM1 is crucial for ERα transactivation and breast cancer development. Mol. Cell 56, 261–274 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Wang Z., Gong Y., Peng B., Shi R., Fan D., Zhao H., Zhu M., Zhang H., Lou Z., Zhou J., Zhu W. G., Cong Y. S., Xu X., MRE11 UFMylation promotes ATM activation. Nucleic Acids Res. 47, 4124–4135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo H., Jiao Q.-B., Shen C.-B., Gong W.-Y., Yuan J.-H., Liu Y.-Y., Chen Z., Liu J., Xu X.-L., Cong Y.-S., Zhang X.-W., UFMylation of HRD1 regulates endoplasmic reticulum homeostasis. FASEB J. 37, e23221 (2023). [DOI] [PubMed] [Google Scholar]
- 12.Wang K., Chen S., Wu Y., Wang Y., Lu Y., Sun Y., Chen Y., The ufmylation modification of ribosomal protein L10 in the development of pancreatic adenocarcinoma. Cell Death Dis. 14, 350 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komatsu M., Inada T., Noda N. N., The UFM1 system: Working principles, cellular functions, and pathophysiology. Mol. Cell 84, 156–169 (2024). [DOI] [PubMed] [Google Scholar]
- 14.Walczak C. P., Leto D. E., Zhang L., Riepe C., Muller R. Y., DaRosa P. A., Ingolia N. T., Elias J. E., Kopito R. R., Ribosomal protein RPL26 is the principal target of UFMylation. Proc. Natl. Acad. Sci. U.S.A. 116, 1299–1308 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee L., Perez Oliva A. B., Martinez-Balsalobre E., Churikov D., Peter J., Rahmouni D., Audoly G., Azzoni V., Audebert S., Camoin L., Mulero V., Cayuela M. L., Kulathu Y., Geli V., Lachaud C., UFMylation of MRE11 is essential for telomere length maintenance and hematopoietic stem cell survival. Sci. Adv. 7, eabc7371 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai Y., Pi W., Sivaprakasam S., Zhu X., Zhang M., Chen J., Makala L., Lu C., Wu J., Teng Y., Pace B., Tuan D., Singh N., Li H., UFBP1, a key component of the Ufm1 conjugation system, Is essential for ufmylation-mediated regulation of erythroid development. PLOS Genet. 11, e1005643 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M., Zhu X., Zhang Y., Cai Y., Chen J., Sivaprakasam S., Gurav A., Pi W., Makala L., Wu J., Pace B., Tuan-Lo D., Ganapathy V., Singh N., Li H., RCAD/Ufl1, a Ufm1 E3 ligase, is essential for hematopoietic stem cell function and murine hematopoiesis. Cell Death Differ. 22, 1922–1934 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tatsumi K., Yamamoto-Mukai H., Shimizu R., Waguri S., Sou Y. S., Sakamoto A., Taya C., Shitara H., Hara T., Chung C. H., Tanaka K., Yamamoto M., Komatsu M., The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nat. Commun. 2, 181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J., Yue G., Ma W., Zhang A., Zou J., Cai Y., Tang X., Wang J., Liu J., Li H., Su H., Ufm1-specific ligase Ufl1 regulates endoplasmic reticulum homeostasis and protects against heart failure. Circ. Heart Fail. 11, e004917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemaire K., Moura R. F., Granvik M., Igoillo-Esteve M., Hohmeier H. E., Hendrickx N., Newgard C. B., Waelkens E., Cnop M., Schuit F., Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PLOS ONE 6, e18517 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai Y., Zhu G., Liu S., Pan Z., Quintero M., Poole C. J., Lu C., Zhu H., Islam B., Riggelen J. V., Browning D., Liu K., Blumberg R., Singh N., Li H., Indispensable role of the Ubiquitin-fold modifier 1-specific E3 ligase in maintaining intestinal homeostasis and controlling gut inflammation. Cell Discov. 5, 7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubio M. D., Wood K., Haroutunian V., Meador-Woodruff J. H., Dysfunction of the ubiquitin proteasome and ubiquitin-like systems in schizophrenia. Neuropsychopharmacology 38, 1910–1920 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J., Zhu H., Liu S., Quintero M., Zhu T., Xu R., Cai Y., Han Y., Li H., Deficiency of murine UFM1-specific E3 ligase causes microcephaly and inflammation. Mol. Neurobiol. 59, 6363–6372 (2022). [DOI] [PubMed] [Google Scholar]
- 24.Nahorski M. S., Maddirevula S., Ishimura R., Alsahli S., Brady A. F., Begemann A., Mizushima T., Guzmán-Vega F. J., Obata M., Ichimura Y., Alsaif H. S., Anazi S., Ibrahim N., Abdulwahab F., Hashem M., Monies D., Abouelhoda M., Meyer B. F., Alfadhel M., Eyaid W., Zweier M., Steindl K., Rauch A., Arold S. T., Woods C. G., Komatsu M., Alkuraya F. S., Biallelic UFM1 and UFC1 mutations expand the essential role of ufmylation in brain development. Brain 141, 1934–1945 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Endo R., Chen Y. K., Burke J., Takashima N., Suryawanshi N., Hui K. K., Miyazaki T., Tanaka M., Dysregulation of ribosome-associated quality control elicits cognitive disorders via overaccumulation of TTC3. Proc. Natl. Acad. Sci. U.S.A. 120, e2211522120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao Z., Ma X., Jing Y., Shen M., Ma X., Zhu J., Liu H., Zhang G., Chen F., Ufmylation on UFBP1 alleviates non-alcoholic fatty liver disease by modulating hepatic endoplasmic reticulum stress. Cell Death Dis. 14, 584 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Zhang M., Wu J., Lei G., Li H., Transcriptional regulation of the Ufm1 conjugation system in response to disturbance of the endoplasmic reticulum homeostasis and inhibition of vesicle trafficking. PLOS ONE 7, e48587 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang J. R., Lingeman E., Luong T., Ahmed S., Muhar M., Nguyen T., Olzmann J. A., Corn J. E., A genome-wide ER-phagy screen highlights key roles of mitochondrial metabolism and ER-resident UFMylation. Cell 180, 1160–1177.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huber J., Obata M., Gruber J., Akutsu M., Lohr F., Rogova N., Guntert P., Dikic I., Kirkin V., Komatsu M., Dotsch V., Rogov V. V., An atypical LIR motif within UBA5 (ubiquitin like modifier activating enzyme 5) interacts with GABARAP proteins and mediates membrane localization of UBA5. Autophagy 16, 256–270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L., Xu Y., Rogers H., Saidi L., Noguchi C. T., Li H., Yewdell J. W., Guydosh N. R., Ye Y., UFMylation of RPL26 links translocation-associated quality control to endoplasmic reticulum protein homeostasis. Cell Res. 30, 5–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scavone F., Gumbin S. C., Da Rosa P. A., Kopito R. R., RPL26/uL24 UFMylation is essential for ribosome-associated quality control at the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 120, e2220340120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L., Xu Y., Yun S., Yuan Q., Satpute-Krishnan P., Ye Y., SAYSD1 senses UFMylated ribosome to safeguard co-translational protein translocation at the endoplasmic reticulum. Cell Rep. 42, 112028 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishimura R., Ito S., Mao G., Komatsu-Hirota S., Inada T., Noda N. N., Komatsu M., Mechanistic insights into the roles of the UFM1 E3 ligase complex in ufmylation and ribosome-associated protein quality control. Sci. Adv. 9, eadh3635 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang R., Wang H., Kang B., Chen B., Shi Y., Yang S., Sun L., Liu Y., Xiao W., Zhang T., Yang J., Zhang Y., Zhu M., Xu P., Chang Y., Jia Y., Huang Y., CDK5RAP3, a UFL1 substrate adaptor, is crucial for liver development. Development 146, dev169235 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Zhu H., Bhatt B., Sivaprakasam S., Cai Y., Liu S., Kodeboyina S. K., Patel N., Savage N. M., Sharma A., Kaufman R. J., Li H., Singh N., Ufbp1 promotes plasma cell development and ER expansion by modulating distinct branches of UPR. Nat. Commun. 10, 1084 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuehnle N., Osborne S. M., Liang Z., Manzano M., Gottwein E., CRISPR screens identify novel regulators of cFLIP dependency and ligand-independent, TRAIL-R1-mediated cell death. Cell Death Differ. 30, 1221–1234 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller E. A., Beilharz T. H., Malkus P. N., Lee M. C. S., Hamamoto S., Orci L., Schekman R., Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell 114, 497–509 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Chatterjee S., Choi A. J., Frankel G., A systematic review of Sec24 cargo interactome. Traffic 22, 412–424 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Weigel A. V., Chang C. L., Shtengel G., Xu C. S., Hoffman D. P., Freeman M., Iyer N., Aaron J., Khuon S., Bogovic J., Qiu W., Hess H. F., Lippincott-Schwartz J., ER-to-Golgi protein delivery through an interwoven, tubular network extending from ER. Cell 184, 2412–2429 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Shomron O., Nevo-Yassaf I., Aviad T., Yaffe Y., Zahavi E. E., Dukhovny A., Perlson E., Brodsky I., Yeheskel A., Pasmanik-Chor M., Mironov A., Beznoussenko G. V., Mironov A. A., Sklan E. H., Patterson G. H., Yonemura Y., Sannai M., Kaether C., Hirschberg K., COPII collar defines the boundary between ER and ER exit site and does not coat cargo containers. J. Cell Biol. 220, e201907224 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Zastrow M., Sorkin A., Mechanisms for regulating and organizing receptor signaling by endocytosis. Annu. Rev. Biochem. 90, 709–737 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu X., Wu G., Non-canonical Golgi-compartmentalized Gβγ signaling: Mechanisms, functions, and therapeutic targets. Trends Pharmacol. Sci. 44, 98–111 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morales Rodríguez L. M., Crilly S. E., Rowe J. B., Isom D. G., Puthenveedu M. A., Location-biased activation of the proton-sensor GPR65 is uncoupled from receptor trafficking. Proc. Natl. Acad. Sci. U.S.A. 120, e2302823120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ippolito M., De Pascali F., Hopfinger N., Komolov K. E., Laurinavichyute D., Reddy P. A. N., Sakkal L. A., Rajkowski K. Z., Nayak A. P., Lee J., Lee J., Cao G., Donover P. S., Reichman M., An S. S., Salvino J. M., Penn R. B., Armen R. S., Scott C. P., Benovic J. L., Identification of a β-arrestin-biased negative allosteric modulator for the β2-adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 120, e2302668120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grimsey N. J., Narala R., Rada C. C., Mehta S., Stephens B. S., Kufareva I., Lapek J., Gonzalez D. J., Handel T. M., Zhang J., Trejo J., A tyrosine switch on NEDD4-2 E3 ligase transmits GPCR inflammatory signaling. Cell Rep. 24, 3312–3323.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang G., Wu G., Small GTPase regulation of GPCR anterograde trafficking. Trends Pharmacol. Sci. 33, 28–34 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang M., Wu G., Mechanisms of the anterograde trafficking of GPCRs: Regulation of AT1R transport by interacting proteins and motifs. Traffic 20, 110–120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conn P. M., Ulloa-Aguirre A., Ito J., Janovick J. A., G protein-coupled receptor trafficking in health and disease: Lessons learned to prepare for therapeutic mutant rescue in vivo. Pharmacol. Rev. 59, 225–250 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Zhang M., Davis J. E., Li C., Gao J., Huang W., Lambert N. A., Terry A. V. Jr., Wu G., GGA3 interacts with a G protein-coupled receptor and modulates its cell surface export. Mol. Cell. Biol. 36, 1152–1163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei Z., Xu X., Fang Y., Khater M., Naughton S. X., Hu G., Terry A. V. Jr., Wu G., Rab43 GTPase directs postsynaptic trafficking and neuron-specific sorting of G protein-coupled receptors. J. Biol. Chem. 296, 100517 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li C., Wei Z., Fan Y., Huang W., Su Y., Li H., Dong Z., Fukuda M., Khater M., Wu G., The GTPase Rab43 controls the anterograde ER-Golgi trafficking and sorting of GPCRs. Cell Rep. 21, 1089–1101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei Z., Zhang M., Li C., Huang W., Fan Y., Guo J., Khater M., Fukuda M., Dong Z., Hu G., Wu G., Specific TBC domain-containing proteins control the ER-Golgi-plasma membrane trafficking of GPCRs. Cell Rep. 28, 554–566 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shiwarski D. J., Darr M., Telmer C. A., Bruchez M. P., Puthenveedu M. A., PI3K class II α regulates δ-opioid receptor export from the trans-Golgi network. Mol. Biol. Cell 28, 2202–2219 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carrel D., Masson J., Al Awabdh S., Capra C. B., Lenkei Z., Hamon M., Emerit M. B., Darmon M., Targeting of the 5-HT1A serotonin receptor to neuronal dendrites is mediated by Yif1B. J. Neurosci. 28, 8063–8073 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu X., Wu G., Human C1orf27 protein interacts with α2A-adrenergic receptor and regulates its anterograde transport. J. Biol. Chem. 298, 102021 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dwyer N. D., Troemel E. R., Sengupta P., Bargmann C. I., Odorant receptor localization to olfactory cilia is mediated by ODR-4, a novel membrane-associated protein. Cell 93, 455–466 (1998). [DOI] [PubMed] [Google Scholar]
- 57.Xu X., Qiu L., Zhang M., Wu G., Segregation of nascent GPCRs in the ER-to-Golgi transport by CCHCR1 via direct interaction. J. Cell Sci. 137, jcs261685 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duvernay M. T., Dong C., Zhang X., Zhou F., Nichols C. D., Wu G., Anterograde trafficking of G protein-coupled receptors: Function of the C-terminal F(X)6LL motif in export from the endoplasmic reticulum. Mol. Pharmacol. 75, 751–761 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duvernay M. T., Zhou F., Wu G., A conserved motif for the transport of G protein-coupled receptors from the endoplasmic reticulum to the cell surface. J. Biol. Chem. 279, 30741–30750 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Zhang X., Dong C., Wu Q. J., Balch W. E., Wu G., Di-acidic motifs in the membrane-distal C termini modulate the transport of angiotensin II receptors from the endoplasmic reticulum to the cell surface. J. Biol. Chem. 286, 20525–20535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu X., Lambert N. A., Wu G., Sequence-directed concentration of G protein-coupled receptors in COPII vesicles. iScience 26, 107969 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dong C., Nichols C. D., Guo J., Huang W., Lambert N. A., Wu G., A triple Arg motif mediates α2B-adrenergic receptor interaction with Sec24C/D and export. Traffic 13, 857–868 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shiwarski D. J., Crilly S. E., Dates A., Puthenveedu M. A., Dual RXR motifs regulate nerve growth factor-mediated intracellular retention of the delta opioid receptor. Mol. Biol. Cell 30, 680–690 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boncompain G., Divoux S., Gareil N., de Forges H., Lescure A., Latreche L., Mercanti V., Jollivet F., Raposo G., Perez F., Synchronization of secretory protein traffic in populations of cells. Nat. Methods 9, 493–498 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Wu J., Lei G., Mei M., Tang Y., Li H., A novel C53/LZAP-interacting protein regulates stability of C53/LZAP and DDRGK domain-containing Protein 1 (DDRGK1) and modulates NF-κB signaling. J. Biol. Chem. 285, 15126–15136 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peter J. J., Magnussen H. M., DaRosa P. A., Millrine D., Matthews S. P., Lamoliatte F., Sundaramoorthy R., Kopito R. R., Kulathu Y., A non-canonical scaffold-type E3 ligase complex mediates protein UFMylation. EMBO J. 41, e111015 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lan T. H., Liu Q., Li C., Wu G., Lambert N. A., Sensitive and high resolution localization and tracking of membrane proteins in live cells with BRET. Traffic 13, 1450–1456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kobayashi H., Ogawa K., Yao R., Lichtarge O., Bouvier M., Functional rescue of beta-adrenoceptor dimerization and trafficking by pharmacological chaperones. Traffic 10, 1019–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nie C., Wang H., Wang R., Ginsburg D., Chen X. W., Dimeric sorting code for concentrative cargo selection by the COPII coat. Proc. Natl. Acad. Sci. U.S.A. 115, E3155–E3162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Emmer B. T., Hesketh G. G., Kotnik E., Tang V. T., Lascuna P. J., Xiang J., Gingras A.-C., Chen X.-W., Ginsburg D., The cargo receptor SURF4 promotes the efficient cellular secretion of PCSK9. eLife 7, e38839 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qiu Z. P., Lin Z. C., Hu A., Liu Y. B., Zeng W. E., Zhao X., Shi X. J., Luo J., Song B. L., GRAMD1/ASTER-mediated cholesterol transport promotes Smoothened cholesterylation at the endoplasmic reticulum. EMBO J. 42, e111513 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duvernay M. T., Dong C., Zhang X., Robitaille M., Hébert T. E., Wu G., A single conserved leucine residue on the first intracellular loop regulates ER export of G protein-coupled receptors. Traffic 10, 552–566 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li C., Fan Y., Lan T. H., Lambert N. A., Wu G., Rab26 modulates the cell surface transport of α2-adrenergic receptors from the Golgi. J. Biol. Chem. 287, 42784–42794 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Presley J. F., Cole N. B., Schroer T. A., Hirschberg K., Zaal K. J. M., Lippincott-Schwartz J., ER-to-Golgi transport visualized in living cells. Nature 389, 81–85 (1997). [DOI] [PubMed] [Google Scholar]
- 75.Chen C., Itakura E., Weber K. P., Hegde R. S., de Bono M., An ER complex of ODR-4 and ODR-8/Ufm1 specific protease 2 promotes GPCR maturation by a Ufm1-independent mechanism. PLOS Genet. 10, e1004082 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang Y., Yin H., Li B., Wu Q., Liu Y., Poljak K., Maldutyte J., Tang X., Wang M., Wu Z., Miller E. A., Jiang L., Yao Z. P., Guo Y., An in vitro vesicle formation assay reveals cargo clients and factors that mediate vesicular trafficking. Proc. Natl. Acad. Sci. U.S.A. 118, e2101287118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang T., Yu H., Hughes N. W., Liu B., Kendirli A., Klein K., Chen W. W., Lander E. S., Sabatini D. M., Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell 168, 890–903.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yiu S. P. T., Zerbe C., Vanderwall D., Huttlin E. L., Weekes M. P., Gewurz B. E., An Epstein-Barr virus protein interaction map reveals NLRP3 inflammasome evasion via MAVS UFMylation. Mol. Cell 83, 2367–2386.e15 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang L., Xu Y., Fukushige T., Saidi L., Wang X., Yu C., Lee J. G., Krause M., Huang L., Ye Y., Mono-UFMylation promotes misfolding-associated secretion of α-synuclein. Sci. Adv. 10, eadk2542 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu X., Wei Z., Wu G., Specific motifs mediate post-synaptic and surface transport of G protein-coupled receptors. iScience 25, 103643 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Legends for movies S1 and S2
Uncropped gels
Movies S1 and S2