Abstract
Influenza virus RNA polymerase with the subunit composition PB1-PB2-PA is a multifunctional enzyme with the activities of both synthesis and cleavage of RNA and is involved in both transcription and replication of the viral genome. In order to produce large amounts of the functional viral RNA polymerase sufficient for analysis of its structure-function relationships, the cDNAs for RNA segments 1, 2, and 3 of influenza virus A/PR/8, each under independent control of the alcohol oxidase gene promoter, were integrated into the chromosome of the methylotrophic yeast Pichia pastoris. Simultaneous expression of all three P proteins in the yeast P. pastoris was achieved by the addition of methanol. To purify the P protein complexes, a sequence coding for a histidine tag was added to the PB2 protein gene at its N terminus. Starting from the induced P. pastoris cell lysate, we partially purified a 3P complex by Ni2+-agarose affinity column chromatography. The 3P complex showed influenza virus model RNA-directed and ApG-primed RNA synthesis in vitro but was virtually inactive without addition of template or primer. The kinetic properties of model template-directed RNA synthesis and the requirements for template sequence were analyzed using the 3P complex. Furthermore, the 3P complex showed capped RNA-primed RNA synthesis. Thus, we conclude that functional influenza virus RNA polymerase with the catalytic properties of a transcriptase is formed in the methylotrophic yeast P. pastoris.
Influenza A virus contains eight different RNA segments of negative polarity in its genome, each encoding one or two unique viral proteins. The viral RNA polymerase is associated with each RNA segment as a viral component and in infected cells, responsible for both transcription and replication of the viral genome (for reviews, see references 7, 13, and 21). The pathway for the synthesis of viral mRNA involves multiple-step reactions, consisting of endonucleolytic cleavage of host cell mRNA, capped oligonucleotide-primed transcription of viral RNA (vRNA), and the addition of a poly(A) tail to the nascent mRNA. On the other hand, replication takes place in two steps, vRNA-directed synthesis of full-length complementary RNA (cRNA) without any modification at both the 5′ and 3′ termini and cRNA-directed reproduction of vRNA.
Starting from the viral ribonucleoprotein (RNP), we isolated an RNA-3P (PB1, PB2, and PA) protein complex without NP by equilibrium centrifugation in cesium sulfate or cesium chloride (15, 16). The RNA-3P complex is enzymatically active in the in vitro synthesis of short attenuated RNA chains (but NP is needed for RNA chain elongation [12]), indicating that the three P proteins participate in the catalytic function of RNA polymerization. The vRNA-free 3P complex, consisting of one molecule each of the three P proteins, was isolated from the RNA-3P complex by centrifugation in cesium chloride or cesium trifluoroacetate (11). The 3P complexes exhibited RNA synthesis only when a model vRNA template with 5′- and 3′-terminal conserved vRNA sequences was added (31).
The molecular composition of influenza virus RNA polymerase was confirmed after establishment of the in vitro reconstitution system using the three P proteins which were individually expressed in insect cells after infection with recombinant baculoviruses (20, 39), but the reconstitution of the functional RNA polymerase from the three overexpressed P proteins was not high, mainly due to insufficient refolding of the P proteins from inclusion bodies in expressed cell lysates. Recently, we have succeeded in expressing each P protein in Escherichia coli by changing the nucleotide sequence (without changing the amino acid sequence) near the translation initiation site so as to adjust the codon usage to the E. coli pattern (Y. Asano and A. Ishihama, unpublished data), but the E. coli system has the same drawbacks as the baculovirus expression system. On the other hand, an enzymatically active RNP was also reconstituted from a mixture of three P proteins and NP, which were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), electroblotted onto a membrane, recovered from the membrane, and refolded with the help of E. coli thioredoxin (43), but this system is not practical for large-scale preparation of the functional RNA polymerase. At present, the use of in vitro reconstitution systems for large-scale production of the RNA polymerase is costly, time-consuming, and technically inconvenient.
In order to meet the demand for a large amount of functional influenza virus RNA polymerase in template-free form, an alternative approach has since been employed, in which all three P proteins were expressed in the same cells transiently after infection of recombinant vaccinia viruses (25, 41, 48) or permanently after integration of all three P protein cDNAs into the chromosome (18, 29). The expression levels of P proteins in these coexpression systems were, however, not sufficient for large-scale purification of functional RNA polymerase. In order to improve the expression levels, we used the methylotrophic yeast Pichia pastoris system as well as the recombinant baculovirus system. Here we describe the first successful expression of negative-strand viral RNA polymerase in a yeast.
The idea of using a yeast as a host for the growth of animal viruses originated from the finding that the budding yeast Saccharomyces cerevisiae can be a host for the replication of the genome of a plant virus, brome mosaic virus (14). P. pastoris is able to utilize methanol as its sole carbon source and has been developed as a host for the expression of heterologous proteins. The major advantages of this expression system include (i) a strong, tightly regulated alcohol oxidase (AOX) promoter, 5′AOX1, is available; (ii) large-scale protein production can be achieved in a large-volume fermentor culture; (iii) a secretary pathway allows the product to be secreted into the medium, separating the foreign protein from most of the host proteins; (iv) the expression system can be easily set up; and (v) the cost is as low as that of the E. coli expression system (2, 4, 6, 23). Two membrane proteins of influenza virions, neuraminidase (NA) and hemagglutinin (HA), were successfully expressed in P. pastoris and produced as secreted forms in the culture medium (24, 37). These influenza virus proteins served as recombinant vaccines that elicit partial or fully protective antibodies in mice.
The main concern of this study is to express and purify template-free influenza virus RNA polymerase using P. pastoris as a host strain and subsequently to examine its catalytic properties. The results indicate that (i) an expression system producing reasonable amounts of the three P proteins in P. pastoris was established after a search for optimum induction times and the optimum concentration of methanol to give the maximum level of induction; (ii) by adding a histidine tag to the PB2 protein, the 3P protein complex was isolated from the cell lysates after Ni2+-nitrilotriacetic acid (NTA)-agarose affinity chromatography; (iii) by quantitative Western blotting, all three P proteins in the isolated 3P complex were detected in stoichiometric molar ratios; (iv) the catalytic activity of RNA synthesis was detected for the 3P complex only when model vRNA or cRNA templates with conserved terminal sequences were added; and (v) both synthetic dinucleotides and globin mRNA served as primers, but the 3P protein complex was virtually inactive in the absence of primers. Template recognition specificity was also examined by using various kinds of model vRNA or cRNA with and without the terminal conserved sequences. Taking all the results together, we conclude that functional influenza virus RNA polymerase can be produced in the methylotrophic yeast P. pastoris in amounts sufficient for detailed functional analysis and structural studies.
MATERIALS AND METHODS
Construction of plasmids and transformation into P. pastoris.
The cDNAs for the PB1, PB2, and PA proteins of influenza virus were prepared as described previously (20). The PB1 cDNA was inserted into plasmid pPic9 (Invitrogen) at the BamHI site between the promoter and the terminator of the AOX gene to construct pPic9-PB1 (Fig. 1). The PA cDNA was integrated into the pPic9 vector at the BamHI site to generate pPic9-PA, into which the HaeII fragment of pPicZαA containing the Zeor coding sequence was inserted to generate pPic9Z-PA. The PB2 cDNA was first inserted into pEH31 between the SacI and XhoI sites to generate pEH-PB2, from which the PB2 cDNA was PCR amplified with a 5′ primer containing a BamHI site and a histidine tag sequence and a 3′ primer containing an XhoI site. The N-terminal leader sequence, MSHTHEHLHHHEL, of the Klebsiella nitrile hydratase α subunit fused to the factor Xa recognition sequence IEGR was used as the histidine tag sequence (27) and was added at the N terminus of the PB2 gene (this sequence is hereafter referred to as Hx in this paper). The PCR-amplified HxPB2 sequence was inserted into pPic9 between the BamHI and XhoI sites to construct pPic-HxPB2. A HaeII fragment of pPicZαA containing the Zeor coding sequence was inserted into pPic-HxPB2 to produce pPicZ-HxPB2 (Fig. 1). All three P protein genes are under the independent control of the AOX promoter (Fig. 1). The construction of all expression plasmids was confirmed by DNA sequencing.
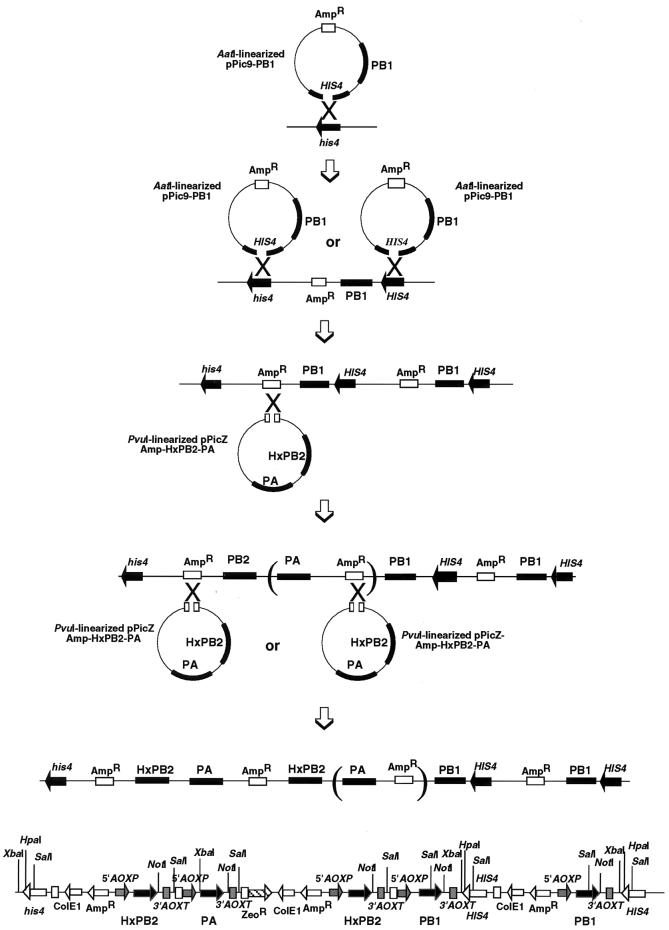
FIG. 1.
Construction of P. pastoris F3P carrying the 3P subunit genes for influenza virus RNA polymerase. Transformation of the cDNA for each of the three P proteins of influenza virus was carried out in two steps. In the first-step transformation, plasmid pPic9-PB1 carrying cDNA for the PB1 protein was linearized by treatment with AatI within the HIS4 gene and transformed into P. pastoris KM71 by the LiCl method, and the transformants were screened for His+. Since Southern blot analysis indicated integration of two copies of the PB1 cDNA (Fig. 2), the crossing-over must have taken place twice to yield P. pastoris FPB1. In the second-step transformation, plasmid pPicZ-Amp-HxPB2-PA carrying the cDNAs for both PB2 and PA was linearized by treatment with PvuI and transformed into P. pastoris FPB1, and cultures were screened for zeocin-resistant transformants. Southern blot analysis indicated the integration of two copies of the PB2 gene and one copy of the PA gene in the transformant P. pastoris F3P used for expression of the influenza virus RNA polymerase. The most probable pathway involves two consecutive cross-overs of the plasmid followed by deletion of the PA gene after the integration into the Pichia chromosome. An alternative mechanism involves the integration of two different sequences into the Pichia chromosome, one complete copy of the PB2-PA plasmid and the other incomplete copy of only the PA gene. The F3P genome, as estimated from the Southern blot patterns, contains two PB1, two HxPB2 (Hx tag conjugated at the N terminus of PB2), and one PA cDNAs. The promoter (5′AOX1) and terminator (3′AOXTT) of the alcohol oxidase gene are located upstream and downstream, respectively, of each P gene.
Transformation into P. pastoris KM71 mutS was carried out in two steps. First, pPic9-PB1 was transformed, after linearization by treatment with AatI, and His+ colonies were selected (FPB1 strain). In the second step, pPicZ-Amp-HxPB2-PA was transformed, after treatment with PvuI, into strain FPB1 to select zeocin-resistant transformants. Transformation was performed by the standard LiCl method (Invitrogen). The F3P strain carrying the genes for the 3P proteins was isolated after confirmation of the respective genes by Southern blot analysis.
Expression of the 3P proteins in P. pastoris.
P. pastoris F3P, which carries the PB1, PB2, and PA cDNAs integrated in the chromosome, was cultured overnight at 30°C in YPD medium supplemented with zeocin (final concentration, 0.1 mg/ml). The preculture was inoculated into 20 ml of fresh MG medium (1.34% yeast nitrogen base, 0.00004% biotin, and 1% [vol/vol] glycerol) containing zeocin (0.1 mg/ml). Cells were grown with vigorous shaking at 30°C until the cell density reached an A600 of 1.5 or 2.2.
For induction of 3P protein expression, the cells were harvested by centrifugation, washed once with MM medium (1.34% nitrogen base and 0.00004% biotin) and resuspended in MM medium containing zeocin (0.1 mg/ml) and 0.5% (vol/vol) methanol. During the incubation, methanol and zeocin were added to the culture at 24-h intervals, each time to give final concentrations of 0.5% and 0.1 mg/ml, respectively. At 24, 48, 72, and 96 h after induction, cells were harvested by centrifugation, washed once with breaking buffer (50 mM sodium phosphate [pH 7.4], 5% [vol/vol] glycerol, and 1 mM phenylmethylsulfonyl fluoride), and stored at −80°C until use.
Purification of the 3P protein complex.
The frozen cells were resuspended in breaking buffer, and an equal volume of acid-washed glass beads was added to the cell suspension. After cell breakage by vigorous vortexing at 4°C, the disrupted cell suspension was centrifuged at 12,000 rpm for 15 min. The remaining cell pellet was resuspended in an equal volume of breaking buffer and centrifuged. The combined supernatants were used as the cell lysate.
For isolation of the 3P proteins, 4.05 g of (NH4)2SO4 powder was added slowly to 10 ml of cell lysate to 60% saturation and mixed for at least 30 min at 4°C. After centrifugation at 12,000 rpm for 30 min at 4°C, the supernatant was pooled, and 1.7 g of solid (NH4)2SO4 was added to give a saturation of 77%. After centrifugation at 12,000 rpm for 30 min, the pellet was stored and dissolved in 2 ml of 1× Ni2+-NTA affinity binding buffer (20 mM Tris-HCl [pH 7.9], 0.5 M NaCl, 5 mM imidazole) and then dialyzed against 2 liters of the same buffer overnight at 4°C. The dialysate was loaded onto a 2.5-ml Ni2+-NTA-agarose column, and the column was washed once with 10 volumes of 1× binding buffer and once with 6 volumes of 1× washing buffer (20 mM Tris-HCl [pH 7.6], 0.5 M NaCl, 25 mM imidazole) and finally eluted with 6 volumes of 1× elution buffer (40 mM Tris-HCl [pH 7.9], 0.5 M NaCl, 60 mM imidazole). Fractions containing the 3P proteins were pooled into a small beaker and precipitated by the addition of (NH4)2SO4. After centrifugation at 15,000 rpm for 20 min, the precipitates were dissolved in 1× storage buffer (50 mM Tris-HCl [pH 7.6], 100 mM NaCl, 10 mM MgCl2, 2 mM dithiothreitol [DTT], 50% glycerol). Finally, the crude 3P fraction was dialyzed against the same storage buffer overnight at 4°C and subsequently stored at −80°C.
PAGE and Western blotting.
Samples of either the crude cell extract or the Ni2+-agarose column chromatography eluates were separated by SDS–10% PAGE, and the proteins were visualized by staining with Coomassie brilliant blue. For Western blotting, the proteins in the gel were transferred to a polyvinylidine difluoride membrane (Pall Gelman Laboratories) using semidry apparatus in transfer buffer (39 mM glycine, 48 mM Tris base, 0.037% SDS [electrophoresis grade], 20% [vol/vol] methanol). After blocking with bovine serum albumin, the membrane was incubated with rabbit polyclonal antibodies against PB1, PB2, or PA (1:1,000 dilution). The membrane was washed and then reacted with the secondary antibody, goat anti-rabbit immunoglobulin G conjugated to horseradish peroxides (1:1,000 dilution) (Promega). Finally, the membrane was washed and developed with 0.05% diaminobenzidine tetrahydrochloride in the presence of H2O2.
Polyclonal antibodies against each of the 3P proteins were raised in rabbits using the 3P proteins which were expressed in E. coli and purified to apparent homogeneity.
Isolation of genomic DNA from P. pastoris F3P.
A single colony of P. pastoris F3P was inoculated into 10 ml of YPD containing Zeocin (0.1 mg/ml) and cultured at 30°C overnight. A 0.05-ml aliquot of the preculture was inoculated into 10 ml of minimal medium MD (1.34% YNB, 0.00004% biotin, 2% glucose) containing zeocin (0.1 mg/ml) and grown for at least for 20 h at 30°C with shaking at 300 rpm. Cells were harvested by centrifugation, washed twice with distilled water, and resuspended in 2 ml of SCED buffer (1 M sorbitol, 10 mM sodium citrate [pH 7.5], 10 mM EDTA, 10 mM DTT). To the cell suspension, 0.3 mg of Zymolyase (Seikagaku Kogyo, Tokyo, Japan) was added, and the mixture was incubated at 37°C for 1 h with continuous shaking. For isolation of DNA, 2 ml of 1% SDS was added, and the cell suspension was set on ice for 5 min. After adding 1.5 ml of 5 M potassium acetate (pH 8.9), the cell lysate was centrifuged to remove the cell debris. To the supernatant, 90 μl of 10-mg/ml RNase A was added, and the mixture was incubated at 37°C for 1 h. DNA was precipitated by adding 2 volumes of ethanol. The DNA pellet was dissolved in 1 ml of distilled water and purified several times by treatment with phenol-chloroform (1:1, vol/vol).
Southern blot analysis.
DNA probes for Southern blotting were generated by PCR using the genomic DNA from P. pastoris F3P as a template (Table 1). The PCR mixture contained, in 50 μl, 25 mM N-Tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid (TAPS) buffer (pH 9.3); 50 mM KCl; 3.25 mM MgCl2; 1 mM 2-mercaptoethanol; 20 ng of purified genomic DNA; 200 ng each of the forward and reverse primers; 0.5 mM each dATP, dGTP, and dTTP; 0.05 mM dCTP; 0.67 μM [α-32P]dCTP; and 2.5 U of Taq polymerase (Takara, Otsu, Japan). PCR products were purified by using the SUPREC-02 cartridge (Takara).
TABLE 1.
PCR primers used for preparation of Southern blot probes
| Primer | Sequence | Position on vRNA |
|---|---|---|
| PB1 forward | 5′-ATGGATGTCAATCCGACCTTACTTTTCTTA-3′ | 25→54 |
| PB1 backward | 5′-CGTTTCAATACACGAGTTTTC-3′ | 354←334 |
| PB2 forward | 5′-ATGGAAAGAATAAAAGAACTAAGAAATCTA-3′ | 28→57 |
| PB2 backward | 5′-TATTTGTTATTGGTCCATTCC-3′ | 349←329 |
| PA forward | 5′-ATGGAAGATTTTGTGCGACAATGCTTCAAT-3′ | 25→54 |
| PA backward | 5′-CCCTGTAGTGTTGCAAATACT-3′ | 321←301 |
Southern blotting was performed according to the standard procedure. In brief, genomic DNA from P. pastoris F3P was digested with NotI, SalI, XbaI, or HpaI at 37°C overnight. DNA fragments were separated by electrophoresis on a SeaKem GTG agarose gel and visualized by staining with ethidium bromide. The agarose gel was treated for 10 min with depurination solution (250 mM HCl), for 25 min with denaturation solution (1.5 M NaCl, 0.5 M NaOH), and then for 25 min with neutralization solution (0.5 M Tris-HCl [pH 7.5], 1.5 M NaCl), with each step done at room temperature with gentle agitation. DNA fragments were allowed to transfer to Hybond N+ (Amersham) membranes overnight in transfer buffer, 20× SSC (3 M NaCl, 0.3 M sodium citrate [pH 7]). The DNA was fixed to the membrane by UV cross-linking. Before hybridization, the membrane was soaked in 5× SSC and then incubated in hybridization buffer (5× SSC, 0.1% SDS, 5% dextran sulfate, 20-fold-diluted Liquid block [Amersham]) for 30 min at 60°C. Approximately 5 × 105 cpm of DNA per ml was added to the hybridization buffer, and the membrane was further incubated at 60°C for 18 h. The 32P-labeled DNA probe was heated at 95°C for 10 min and then immediately cooled on ice before being added to the hybridization buffer. The hybridized blot was washed with 1× SSC buffer containing 0.1% SDS and then with 0.5× SSC buffer containing 0.1% SDS. Each wash was carried out for 15 min at 60°C. Finally, the 32P-labeled DNA fragments were detected by autoradiography.
Preparation of RNA templates.
cRNA and vRNA model templates for RNA synthesis were constructed as described previously (31, 46). Basically, plasmids containing the cDNA for vRNA or cRNA were digested with MboII and then transcribed by T7 RNA polymerase to produce RNA model templates. cDNAs for the synthesis of mutant model templates were generated by PCR. Standard PCR was carried out in a 50-μl reaction mixture containing 25 mM TAPS (pH 9.3); 50 mM KCl; 2 mM MgCl2; 1 mM 2-mercaptoethanol; 20 pmol each of forward and reverse oligonucleotides; 0.25 mM each dATP, dGTP, dCTP, and dTTP; and 2.5 U of Taq polymerase (Takara). PCR products were subjected to electrophoresis on a 2% SeaKem GTG agarose gel, isolated from the gel, and then recovered with SUPREC-01 filter cartridges (Takara). Each purified DNA fragment was digested with PstI and HindIII, cloned into pUC19, and subsequently transformed into E. coli DH5. Following verification of the sequence of each mutant template, large quantities of plasmid DNA were obtained by using the Qiagen plasmid maxikit and used as templates for in vitro transcription by T7 RNA polymerase.
In vitro RNA synthesis.
RNA synthesis in vitro was performed essentially as described previously (11, 46) with a slight modification. In brief, 20 μl of the reaction mixture contained 50 mM HEPES-KOH (pH 7.6); 5 mM magnesium acetate; 100 mM KCl; 2 mM DTT; 0.5 mM each ATP, CTP, and GTP; 50 μM UTP; 5 μCi of [α-32P]UTP (Amersham); 100 U of RNase inhibitor (Takara) per ml; 10 pmol of RNA template (c53, v53, c84, or v84); and 1 mM ApG (Sigma). For mutant templates vM3, cM9, and cM10, primers ApA, ApA, and CpA, respectively, were added to the reaction mixture instead of ApG. About 1 pmol of either the RNP core or the 3P complex, as estimated from the amount of P proteins, was added as the enzyme, and the reaction was carried out at 30°C for 2 h. For capped RNA-primed transcription assays, globin mRNA (Gibco-BRL) was used in place of dinucleotide primers. RNA products were extracted with an equal volume of a 1:1 (vol/vol) mixture of phenol and chloroform-isoamyl alcohol (24:1) and precipitated with ethanol. After centrifugation, samples were dissolved in gel loading buffer, heated at 90°C for 3 min, and subsequently analyzed by electrophoresis on an 8% denaturing polyacrylamide gel in the presence of 8 M urea. Gels were exposed to imaging plates, and the plates were analyzed with a PhosphoImage analyzer (BAS 2000; Fuji, Tokyo, Japan).
RESULTS
Construction of P. pastoris F3P carrying cDNAs for influenza virus RNA polymerase subunits.
The construction of P. pastoris F3P carrying on its chromosome the cDNAs for all three P protein subunits of influenza virus RNA polymerase was performed in two transformation steps. In the first-step transformation, the cDNA for the PB1 subunit protein was inserted into P. pastoris expression plasmid pPic9 to construct pPic9-PB1 (Fig. 1). After treatment with AatI within the HIS gene, the linearized pPic9-PB1 DNA was transfected into P. pastoris KM71 mutS, and His+ transformants were selected. The resulting P. pastoris FPB1 containing the PB1 gene was then transformed with PvuI-treated pPicZ-Amp-HxPB2-PA (the PvuI site is located within the amp gene), which was constructed by inserting the cDNAs for the HxPB2 (Hx tag at the N terminus) and PA proteins, each under the independent control of the alcohol oxidase (AOX1) promoter, into plasmid pPicZ-Amp in tandem (Fig. 1). The selection of P. pastoris F3P containing the HxPB2-PA genes as well as the PB1 gene was carried out in the presence of zeocin. If one copy each of the two plasmids inserted into the P. pastoris chromosome at the expected sites (HIS for the first transformation and amp for the second transformation step), the order of gene integration should be HIS4–Amp–HxPB2-PA–Zeo–Amp–PB1–his, and all three P genes should be located under the independent control of the AOX1 promoter. The gene organization was then checked by Southern blot analysis.
Organization of the PB1, PB2, and PA genes in the P. pastoris F3P chromosome.
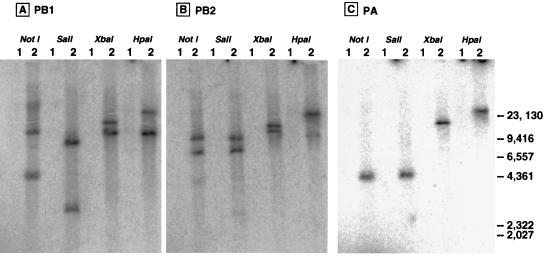
The gene organization and copy numbers of PB1, PB2, and PA cDNAs in the chromosome of P. pastoris F3P were determined by Southern blotting analysis. For this purpose, genomic DNA was isolated from P. pastoris F3P and its parental strain KM71 and digested with NotI, SalI, XbaI, and HpaI (Fig. 1 [bottom] for the restriction enzyme sites). The digested DNA samples were separated by agarose gel electrophoresis, and the gels were subjected to Southern hybridization with 32P-labeled 300-bp-long DNA probes with sequences complementary to the PB1, PB2, and PA genes (see Materials and Methods for probe preparation). The Southern hybridization patterns are shown in Fig. 2. All the probes hybridized to the selected fragments from the F3P genome, whereas none of the DNA fragments from KM71 hybridized with these probes, indicating that the F3P strain carries the sequences for all three P protein genes.
FIG. 2.
Southern blot analyses of F3P genome DNA. Genomic DNA was isolated from both P. pastoris F3P and its parental strain KM71 and digested with NotI, SalI, XbaI, or HpaI at 37°C overnight. The digested DNA samples were separated by electrophoresis on a 0.8% agarose gel and then transferred to Hybond-N+ membranes (Amersham). Southern hybridization was carried out at 60°C overnight with 32P-labeled 300-bp-long DNA probes with sequences complementary to the PB1, PB2, and PA genes. The membranes were washed several times, and the radioactive signals on the membranes were detected with a PhosphorImager analyzer. Lanes 1, DNA from parental strain KM71; lanes 2, DNA from P. pastoris F3P. (A) 32P-labeled PB1 probe; (B) 32P-labeled PB2 probe; (C) 32P-labeled PA probe. The migration positions (in base pairs) of the HindIII-digested λ size markers are shown on the right.
The PB1 probe hybridized with two bands of F3P DNA digested with each restriction enzyme (Fig. 2A). Likewise, the PB2 probe hybridized with two bands of F3P DNA digested with all four enzymes (Fig. 2B). With the PA probe, however, a single hybridizing fragment of F3P was detected for each restriction enzyme (Fig. 2C). Based on knowledge of the locations of the cleavage sites on the PB1, PB2, and PA sequences by the four enzymes NotI, SalI, XbaI, and HpaI, and judging from the Southern hybridization patterns, we propose that the F3P chromosome contains two copies of the PB1 gene, two copies of the PB2 gene, and one copy of the PA gene, in the order illustrated in Fig. 1 (bottom panel). Possible mechanisms for the integration are discussed below.
Expression of 3P proteins in P. pastoris F3P.
The expression of 3P proteins in P. pastoris F3P was examined by adding the inducer methanol to activate the alcohol oxidase promoter, which controls the P protein genes. The induced cell lysates were positive for P protein expression by Western blotting against specific anti-P protein antibodies, which were raised in rabbits against each of the three P proteins purified from E. coli. Since these antibodies showed little cross-reaction with each other (45), PB2 and PA, which apparently migrate to the same positions on SDS-PAGE gels, could be detected separately. In order to identify the optimum conditions for maximum expression of the three P proteins, induction was initiated at two different cell densities, 7 × 107 and 1.2 × 108 cells/ml, by transfer of the culture into induction medium containing various concentrations of methanol. At various times after induction, cell lysates were prepared and subjected to Western blotting against each of the anti-P protein antisera.
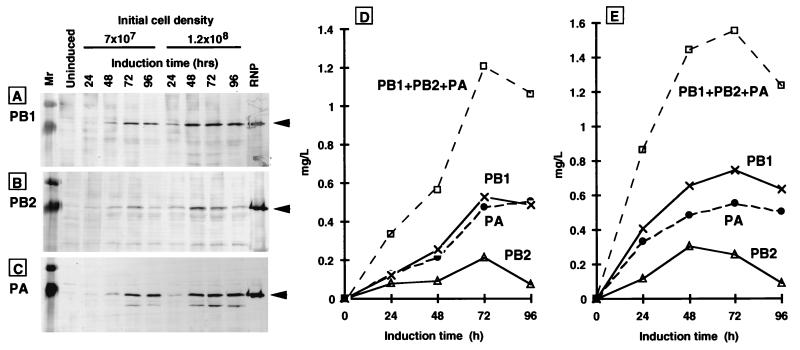
As shown in Fig. 3A, B, and C, the induced cell lysates gave immunostained bands with approximate molecular masses of 90, 86, and 82 kDa with the anti-PB1 (Fig. 3A), anti-PB2 (Fig. 3B), and anti-PA (Fig. 3C) antisera, respectively. The migration positions of these cross-reactive proteins are consistent with the authentic PB1, PB2, and PA proteins (data not shown). Neither the control F3P culture in the absence of methanol nor the parental KM71 in the presence or absence of methanol gave cross-reactive bands with anti-P protein antibodies (data not shown). Thus, we concluded that all the 3P proteins were expressed in P. pastoris F3P. For quantification, the Western blot intensity of each subunit band was measured using different volumes of the cell lysates and converted into the amount of P protein by using a standard curve prepared from a known amount of RNP or purified P protein. Figures 3D and 3E show the amounts of each of the 3P proteins in cell lysates prepared at different induction times. The expression levels were generally higher for the culture with a high initial cell density (Fig. 3E). The time-dependent expression pattern was essentially the same among the three P proteins, but the expression level of PB2 was always lower than that of PB1 and PA. The maximum expression of 3P proteins was obtained when the F3P cell culture was induced at a cell density of 1.2 × 108 cells/ml for 72 h in minimal medium containing 0.5% methanol. Under the best induction conditions thus established, the maximal yield of combined 3P proteins was approximately 1.57 μg/ml of culture (Fig. 3E). This value corresponds to 0.621% of the total cell lysate proteins. Afterwards, accumulation of the three P proteins decreases (Fig. 3D and 3E), but due to the decrease in total protein, the relative content of the combined P proteins increases to 1.475% (initial cell density of 7 × 107 cells/ml) or 0.655% (1.2 × 108 cells/ml) at 96 h after induction.
FIG. 3.
Western blotting analyses of the expression of 3P in P. pastoris. Cells were grown to a concentration of 7 × 107 or 1.2 × 108 cells/ml and then induced for P protein expression by adding 0.5% methanol. Methanol was added every 24 h to give a concentration of 0.5%, and cells were harvested at 24, 48, 72, and 96 h after methanol addition. The control uninduced cells were grown in a medium containing 0.5% glycerol in place of methanol. The cell lysates were analyzed by SDS-PAGE followed by Western blotting with anti-PB1 (A), anti-PB2 (B), and anti-PA (C) antibodies. Lanes: Mr, broad-range molecular weight markers (Bio-Rad); uninduced, a cell lysate at 72 h of culture in glycerol medium; cell lysates at 24, 48, 72, and 96 h after methanol addition; RNP, native RNP isolated from purified influenza virions. The migration positions of the three P subunits are indicated by arrowheads. The yields of each P protein and of the combination of all three P proteins were quantified from the intensities of Western blotting patterns using known amounts of P proteins as references. Symbols: ×, PB1 subunit; ▵, PB2 subunit; ●, PA subunit; □, combined three P proteins. The cell lysates were prepared from two different cell cultures with initial cell densities of 7 × 107/ml (D) and 1.2 × 108/ml (E).
The effect of the methanol concentration on P protein expression was also examined. The results indicated that maximum expression of the three P proteins was obtained at a concentration of methanol between 0.5 and 1.0% (data not shown).
Purification of the 3P protein complex.
To purify the three P proteins from P. pastoris F3P, a 100-ml culture was induced for P protein expression, and the crude cell lysate was subjected to a two-step purification, ammonium sulfate fractionation and Ni2+-NTA-agarose column chromatography. Samples collected from each step of the purification were analyzed by SDS-PAGE and Western blotting. As shown in Fig. 4A, the crude cell lysate contained all three P proteins (lane 2), which were recovered at 60 to 77% saturation of the ammonium sulfate precipitates (lane 3). The ammonium sulfate precipitates were dissolved in Ni2+-NTA affinity binding buffer. The dialysate (Fig. 4A, lane 4) was applied to an Ni2+-NTA-agarose column, and the column was washed in two steps: a first wash with washing buffer containing 5 mM imidazole, and the second wash with elution buffer containing 60 mM imidazole. The initial wash fraction did not contain the 3P proteins (Fig. 4A, lane 6), but the elution fraction with 60 mM imidazole contained the 3P proteins (lane 7). Upon increasing the imidazole concentration in the elution buffer to 200 mM, little P protein was eluted (Fig. 4A, lane 8), indicating that the majority of the 3P protein complex was eluted at 60 mM imidazole. The affinity for the Ni2+-NTA column may not be high enough for the Hx tag sequence used in this study. We then inserted the second wash step with a wash buffer containing 25 mM imidazole prior to elution of the 3P protein complex with elution buffer containing 60 mM imidazole. For initial characterization of the enzymatic activities of the 3P protein complex, the 60 mM imidazole elution fractions were pooled, concentrated, and dialyzed against storage buffer containing 50% glycerol for storage.
FIG. 4.
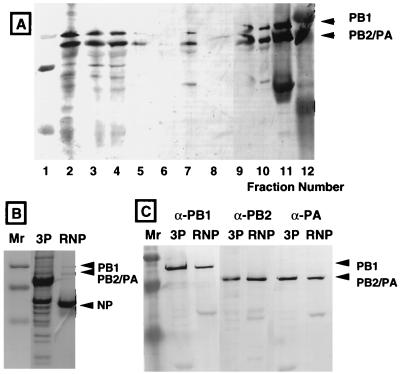
Purification of the 3P complex. (A) The P proteins were precipitated from induced cell lysates with (NH4)2SO4, dissolved in 1× Ni2+-NTA binding buffer, dialyzed against the same buffer, and subjected to Ni2+-NTA-agarose affinity column chromatography. Fractions from each step were analyzed by Western blotting with anti-PB1, anti-PB2, and anti-PA antibodies. Stained bands were quantified by using a standard curve prepared from a known amount of RNP. Lane 1, low-range molecular weight markers (Bio-Rad); lane 2, cell lysate; lane 3, precipitates from (NH4)2SO4 fractionation; lane 4, dialysate in 1× binding buffer; lane 5, flowthrough fraction of the Ni2+ column; lane 6, wash fraction with 1× binding buffer; lane 7, fraction eluted with a buffer containing 60 mM imidazole; lane 8, fraction eluted with a buffer containing 200 mM imidazole; lane 9, precipitates from the second (NH4)2SO4 fractionation; lane 10, the 3P complex in storage buffer; lane 11, RNP; lane 12, broad-range molecular weight markers. See also Table 2 for yields of the three P proteins at each purification step. (B) The partially purified 3P complex was separated, in parallel with RNP, by SDS-PAGE, and the gel was stained with Coomassie brilliant blue. Lane Mr, size markers. NP, influenza virus nucleoprotein. (C) The partially purified 3P complex was separated, in parallel with RNP, by SDS-PAGE, and the gel was immunostained with specific antibodies against PB1, PB2, and PA proteins.
All three P proteins were detected by staining of the partially purified 3P complex with Coomassie brilliant blue (Fig. 4B) (note that PB2 and PA migrate to the same position under the electrophoresis conditions employed, but the presence of both P proteins was confirmed by immunostaining with specific antibodies). After the intensities of the Western blot bands were scanned with a densitometer (Fig. 4C), the concentrations of the three P proteins were estimated from the standard curve obtained by using the RNP core with known amounts of P proteins. The recovery of P proteins at each purification step thus determined is summarized in Table 2. Although the content of PB2 was the lowest among the three P proteins in cell lysates, the stored 3P complex fraction contained three P proteins essentially at the stoichiometric molar ratio, indicating that the influenza virus RNA polymerase formed in P. pastoris is composed of one molecule each of the three P proteins, as in the case of virus-associated RNA polymerase (11). The final yield of 3P proteins was approximately 13.4% of the combined 3P proteins in the cell lysate used (Table 2).
TABLE 2.
Purification of 3P proteins from P. pastoris F3Pa
| Fraction | Amt of 3P protein (μg) | Yield (%) |
|---|---|---|
| Soluble cell lysate | 173.8 | 100 |
| First AS precipitate before dialysis | 129.7 | 74.6 |
| First AS precipitate after dialysis | 130.2 | 74.8 |
| Ni2+-NTA eluate | 43.7 | 25.3 |
| Second AS precipitate after dialysis | 23.3 | 13.4 |
Cell lysate was prepared from an induced P. pastoris culture (100 ml). The 3P proteins were precipitated with (NH4)2SO4 (AS) and subjected to Ni2+-NTA chromatography. The 3P complex in the imidazole eluate fraction was precipitated by adding (NH4)2SO4, dissolved in storage buffer, and stored at −80°C after dialysis against the storage buffer. A fraction from each step of the purification procedure was subjected to SDS-PAGE, and the gel was analyzed by quantitative Western blotting as shown in Fig. 4. The amounts of 3P proteins were determined by quantitative Western blotting with anti-PB1, anti-PB2, and anti-PA antibodies. RNP was used as a reference control for 3P measurements.
RNA synthesis in vitro by the 3P protein complex.
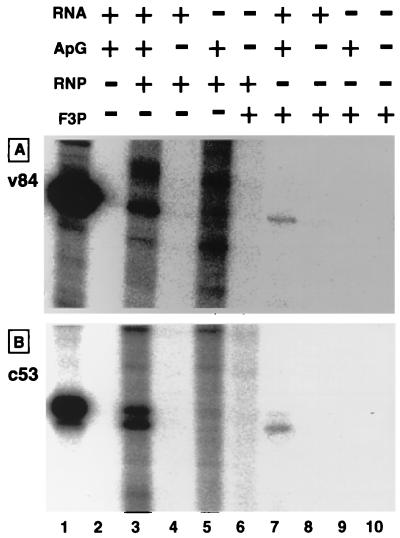
Model RNA templates were developed for detection of in vitro RNA synthesis activity by such template-free RNA polymerases as the solubilized RNA polymerase from RNP (31) and the reconstituted RNA polymerase (20). The RNA synthesis activity of the partially purified 3P protein complex from P. pastoris F3P was examined using a pair of negative-sense model RNAs (v53 and v84) and a pair of positive-sense model RNAs (c53 and c84), each carrying 5′- and 3′-terminal conserved sequences of vRNA or cRNA (note that v and c represent the viral and complementary strand [or negative and positive strand], respectively). The 3P complex exhibited the catalytic activity of RNA synthesis only when a template was added, indicating that the 3P complex is free of any RNA with template activity. Figure 5 shows transcripts directed by the v84 (A) and c53 (B) model templates. The 3P complex catalyzed v84-directed ApG-primed synthesis of RNA (Fig. 5A, lane 7), which migrated on urea-PAGE to the same position as the template v84 (Fig. 5A, lane 1). The synthesis of this template-sized transcript was not detected in the absence of ApG primer (Fig. 5A, lane 8) or v84 template (Fig. 5A, lane 9). RNP added as a control produced several transcripts (Fig. 5A, lane 3), one of which migrated to the same position as the template v84 (Fig. 5A, lane 1). This v84-sized product was not synthesized without addition of the v84 template (Fig. 5A, lane 5). The products formed by RNP in the absence of v84 addition represent those initiated on the endogenous vRNA templates, but no RNP transcripts were detected in the absence of ApG primer (Fig. 5A, lane 4). From the definition of transcriptase (requirements for vRNA template and primers) (see references 7 and 13), the 3P complex and the RNP-associated RNA polymerase might be the transcriptase form.
FIG. 5.
In vitro RNA synthesis by the 3P complex. The 3P complex, prepared as described in the legend to Fig. 4, was subjected to RNA synthesis in vitro using both a negative-sense v84 template (A) and a positive-sense c53 template (B). The reaction mixture contained, in 20 μl, 10 pmol of each template, 1 mM ApG primer, and approximately 1 pmol of the RNP or the 3P complex, as estimated from the contents of 3P proteins. RNA synthesis was carried out at 30°C for 2 h. RNA products were ethanol precipitated and analyzed by electrophoresis on an 8% polyacrylamide sequencing gel. Lane 1, molecular weight marker RNA, v84 (A) and c53 (B).
When RNA synthesis was carried out with the plus-strand c53 template, the 3P complex produced two products (Fig. 5B, lane 7). The minor transcript migrated on urea-PAGE as fast as the c53 template, while the major RNA migrated slightly faster than c53. The synthesis of both RNA products was observed only in the presence of ApG primer (Fig. 5B, compare lanes 7 and 8). The synthesis of small-sized RNA may be due to either internal initiation within the c53 template or termination prior to the 5′ end of c53 RNA. The synthesis of RNAs shorter than the templates is often observed, even with the use of RNP as the enzyme, in in vitro RNA synthesis directed by cRNA templates but not by vRNA templates (46).
Kinetic properties of the 3P RNA polymerase.
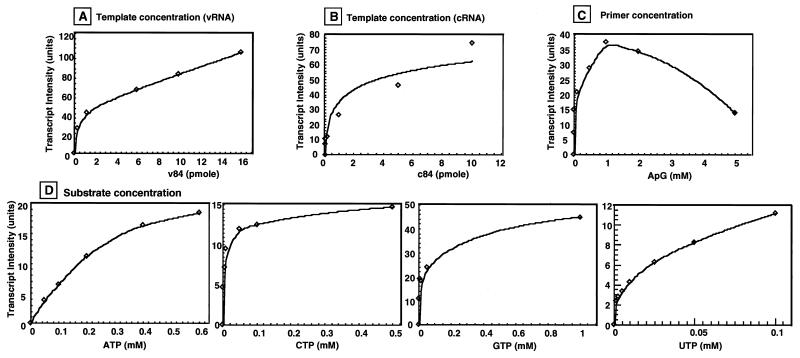
Here we obtained for the first time a large amount of RNA-free functional RNA polymerase with which to measure the kinetic parameters of RNA synthesis. First, we investigated the effects of increasing concentrations of template (v84 and c84), ApG primer, and nucleoside 5′-triphosphate substrates on RNA synthesis (Fig. 6). The minimum amounts of these reaction components required to give the maximum catalytic activity of RNA synthesis were calculated after replotting the data to double-reciprocal graphs. The amounts of v84 and c84 templates that give maximum transcription are apparently different: the Km value for the v84 template was 12.5 nM, while that for c84 was 36.5 nM (Table 3). The Vmax was apparently similar for the two templates, indicating that the RNA polymerase has a greater affinity for the vRNA template than for the cRNA template, but once initiated, the rate of RNA synthesis is the same for both the vRNA and cRNA templates. The high affinity for vRNA is a reaction property expected for the transcriptase.
FIG. 6.
Effects of template, primer, and substrate concentrations on RNA synthesis by the 3P complex. The conditions for the in vitro RNA synthesis reaction were the same as those in Fig. 5 except that the concentration of one of the following reaction components was varied as indicated: (A) v84 template (pmol per 20-μl reaction mixture); (B) c84 template (pmol per 20-μl reaction mixture); (C) ApG primer; and (D) nucleotide substrates (ATP, CTP, GTP, and UTP). The results were replotted into double-reciprocal forms for estimation of the kinetic parameters, as summarized in Table 3.
TABLE 3.
Kinetic parameters for RNA synthesis by F3P proteinsa
| Reaction component | Km (nM) | Vmax (fmol) |
|---|---|---|
| Template | ||
| v84 | 0.0125 | 29.9 |
| c84 | 0.0365 | 36.6 |
| Primer (ApG) | 15 | 20.5 |
| Substrate | ||
| ATP | 340 | 18.8 |
| UTP | 18 | 7.5 |
| GTP | 9.3 | 27.4 |
| CTP | 9.3 | 8.9 |
The effects of varying the concentrations of templates, primers, and substrates on RNA synthesis in vitro by the 3P complex were analyzed. The amount of 3P complex used was approximately 1.2 pmol each of the three P proteins. The standard reaction mixture included ATP, CTP, and GTP, each at 1 mM; [α-32P]UTP, 0.1 mM; ApG primer, 1 mM; and v84 or c84 template, 10 pmol. Vmax represents the amount of 32P-labeled UTP incorporated under the standard reaction conditions.
The Km value for primer ApG was 0.015 mM, which gave maximum catalytic activity of RNA synthesis by the 3P complex (Table 3). This value is close to that observed using the RNP (data not shown). Other dinucleotides show higher Km and lower Vmax values, as in the case of RNP-dependent transcription (8). cRNA template-dependent RNA synthesis also depended on the addition of dinucleotide primers. This observation again supports the prediction that the 3P complex is the transcriptase form. The substrate specificity of the 3P complex was analyzed by measuring v84-directed ApG-dependent RNA synthesis in the presence of increasing concentrations of the substrates ATP, CTP, GTP, and UTP. As summarized in Table 3, the order of Km values was ATP > UTP > GTP = CTP. The Km for ATP is much higher than that for CTP, GTP, and UTP, indicating that the affinity of ATP for the 3P complex is significantly lower than that of the other substrates. This finding may suggest that ATP plays an as yet unidentified role besides being the substrate for RNA polymerization. However, it cannot be excluded yet that the 3P complex preparations used were contaminated with ATP hydrolysis activity.
Sequences on the templates recognized by the RNA polymerase.
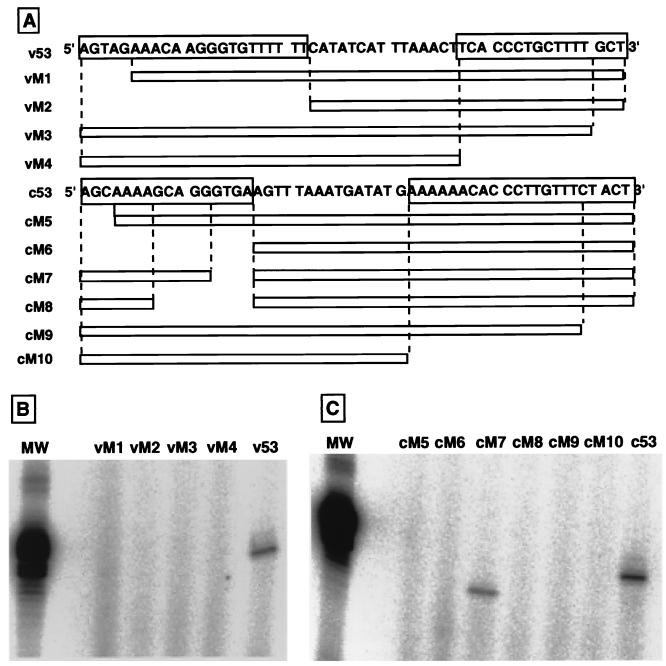
One type of experiment that can be done using RNA-free RNA polymerase is to examine the template recognition specificity of the viral RNA polymerase. As an initial attempt, we constructed various types of deletion mutant model templates, as illustrated in Fig. 7A, and tested whether these mutant templates can be used as templates for RNA synthesis by the 3P complex from P. pastoris F3P. All the mutant v53 templates were virtually inactive in directing RNA synthesis (Fig. 7B, lanes vM1 to vM4). Deletion of the first several nucleotides from either the 3′ or the 5′ terminus made the v53 template inactive, indicating that both 5′- and 3′-terminal conserved sequences are required for the vRNA to function as the template for the influenza virus RNA polymerase. The recognition specificity of the vRNA template by the 3P complex is essentially identical to those of virus-associated or recombinant RNA polymerases (30, 33, 38, 44).
FIG. 7.
Template activities of deletion mutant model RNAs at either 5′ or 3′ conserved sequences. A set of 5′- and 3′-terminal deletion mutants of both the v53 and c53 model templates were constructed by PCR and then cloned into the pUC19 vector between the PstI and HindIII sites. The sequences of these cDNA deletion mutants were verified by sequencing with DSQ-500L (Shimadzu, Kyoto, Japan). Model RNA templates were generated by transcribing the linearized v53 or c53 deletion mutant cDNA with T7 RNA polymerase. (A) Construction of v53 (vM1 through vM4) and c53 (cM5 through cM10) mutant templates. The sequences of the original v53 and c53 are shown, in which the 5′ and 3′ conserved sequences are boxed. The sequences remaining in the deletion mutant templates are shown by open boxes. (B) Dinucleotide-primed RNA synthesis in vitro was carried out using approximately 10 pmol of v53 and its deletion mutant templates and the 3P complex as the enzyme. (C) Dinucleotide-primed RNA synthesis by the 3P complex was carried out using 10 pmol each of c53 and its deletion mutants. Lane MW, size marker RNA (53 nucleotides in length).
On the other hand, among the deletion mutant cRNAs, template activity was observed for one mutant, cM7, which lacked the internal four nucleotides between nucleotides 13 and 16 from the 5′ terminus (Fig. 7C). However, further extension of the deletion (nucleotides 8 to 16, cM8) completely abolished the template activity. Removal of the first three nucleotides (cM5) or the full conserved sequence (cM6) of the cRNA 5′ terminus resulted in complete loss of template activity. Likewise, deletion of the first five nucleotides (cM9) or the full conserved sequence (cM10) of the cRNA 3′ terminus also resulted in complete loss of transcription activity (Fig. 7C). Thus, we concluded that the essential signal for recognition by the viral RNA polymerase is included in both the 5′- and 3′-terminal conserved sequences. The specificity of cRNA recognition by the 3P complex is also in agreement with that of viral RNA polymerase analyzed in vitro (34, 44) and in vivo (17).
Globin mRNA-directed transcription by the RNA polymerase.
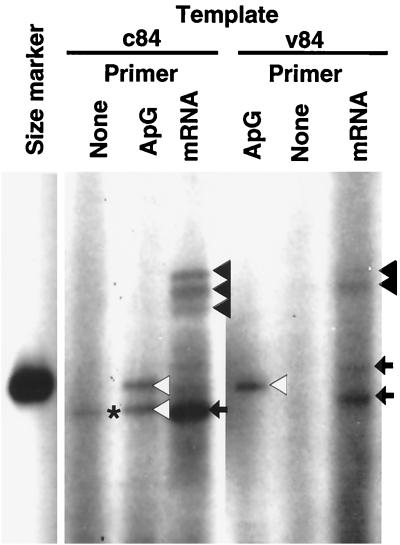
Finally, we examined whether the 3P complex from P. pastoris is able to utilize capped RNA as a primer for transcription. For this purpose, we used globin mRNA, which is known to be a good natural primer (1). In the presence of ApG primer and v84 template, a template-sized transcript was synthesized (Fig. 8, v84 template, ApG lane, open triangle). In the presence of globin mRNA instead of ApG primer, the 3P complex produced transcripts about 10 nucleotides longer than the ApG-primed transcript (Fig. 8, v84 template, mRNA lane, solid triangles). Thus, we concluded that the 3P complex is able to utilize capped RNA as a primer. The detection of capped RNA-primed transcription activity itself indicates the association of capped RNA endonuclease activity with the 3P complex. In addition to this globin mRNA-primed transcript of expected size, smaller transcripts migrating near the v84 size marker were detected (mRNA lanes, arrows). Since no transcript was detected in the absence of primer addition (Fig. 8, v84 template, None lane), it is unlikely that these represent products of unprimed RNA synthesis, but they may be either degradation products of globin mRNA-primed transcripts or internal-initiation transcripts.
FIG. 8.
Capped RNA-primed transcription by the 3P complex. RNA synthesis in vitro by the 3P complex was carried out in a 20-μl reaction mixture containing 15 pmol of either v84 or c84 template and either 20 nmol of ApG (1 mM) or 0.25 μg of globin mRNA as the primer. RNA products were fractionated, together with labeled v84 marker, by 8% PAGE in the presence of 8 M urea. Solid triangles point to globin mRNA-primed transcripts (for mRNA lanes), and open triangles indicate ApG-primed transcripts. The bands marked by arrows might represent degradation and/or internal-initiation products (mRNA lanes). The band produced in the absence of primer addition, marked with a solid star, represents a putative unprimed transcript (for the c84 template).
When c84 was used as a template, we also detected several bands of globin mRNA-primed transcript (Fig. 8, c84 template, mRNA lane, solid triangles), which were as long as the v84-directed globin mRNA-primed transcripts (Fig. 8, v84 template, mRNA lane). In addition, a small RNA was detected (arrow), which migrated faster than the c84 size marker. A similar-size RNA was detected for both the unprimed reaction (Fig. 8, c84 template, None lane, star) and the ApG-primed reaction (Fig. 8, c84 template, ApG, open triangle). In the absence of primer addition, this RNA may represent unprimed initiation product, but in the case of ApG- or globin mRNA-primed reactions, the small transcript may represent internal initiation within the template, internal termination prior to the RNA 5′ end, or degradation of full-sized transcripts.
DISCUSSION
Several different approaches have been employed for the expression of influenza virus RNA polymerase proteins in various organisms. Animal cell lines transformed by recombinant viruses were effectively used for the expression of all three P proteins and for testing each P protein functions in vivo (22, 29), but the expression levels in all these cases were too low for purification and biochemical characterization of the RNA polymerase. In order to increase the expression level, the lytic infection system of recombinant viruses was also established using vaccinia virus (mammalian cell system) (25, 41, 48), simian virus 40 (mammalian cell system) (3), or baculovirus vectors (insect cell system) (20, 39). Recombinant virus infection system have the advantages that (i) all three individual P proteins can be expressed simultaneously or in various combinations, (ii) the three P proteins can be expressed at different ratios and at different times, and (iii) mutations can be introduced into each P protein gene on the recombinant virus vectors. Previously, we established a high-level expression system for individual P proteins using recombinant baculoviruses (20), but the reconstitution efficiency of functional RNA polymerase from isolated P proteins was not high enough for detailed biochemical analyses. However, after coinfection of all three kinds of recombinant baculovirus, each expressing one of the three P proteins, into the same cells, the 3P complex was formed, which showed the activity of model RNA-dependent RNA synthesis (A. Honda, A. Endo, and A. Ishihama, submitted for publication).
Attempts to express cDNAs for the PB1, PB2, and PA proteins cloned in various conventional E. coli expression vectors were all unsuccessful, but we have succeeded in expressing the P proteins in E. coli by changing the mRNA nucleotide sequences at 5′-terminal proximal regions so as to match the codon usage pattern to the E. coli type (Y. Asano and A. Ishihama, unpublished). Although the expression of all three P proteins increased to detectable levels, the highly expressed P proteins again formed inclusion bodies (10). So far we have failed to reconstitute the functional RNA polymerase using insoluble P proteins purified from E. coli.
In this study, we used the P. pastoris expression system for heterologous proteins for simultaneous expression of the influenza virus P proteins. Previously, expression of functional viral RNA polymerase in the budding yeast S. cerevisiae has been observed for the plant RNA virus brome mosaic virus (BMV) (14). The results herein described were quite remarkable, because three P proteins were expressed at significant levels in P. pastoris and furthermore the expressed P proteins were assembled into functional RNA polymerase. The presence and organization of all the genes for the three P proteins in the genomic DNA of P. pastoris F3P were confirmed by Southern blot analysis (Fig. 2). The gene copy numbers of PB1 and PB2 were twice that of PA (Fig. 1, bottom). A possible mechanism for the generation of such a construct includes four integration events, as shown in Fig. 1: two crossover events at the AatI site within the His4 gene of the plasmid carrying the PB1 clone, and two crossovers at the PvuI site within the amp gene of the plasmid carrying both the PB2 and PA genes. Since P. pastoris F3P contains only a single copy of the PA gene, one PA copy might be deleted after integration of two complete sets of the PB2 and PA genes. However, it is also possible, albeit not as likely, that only the PB2 gene was integrated into the chromosome at one of the two-step integration reactions in the second-step transformation.
The Western blot analysis clearly showed that all three P proteins, PB1, PB2, and PA, were expressed in P. pastoris F3P, and the expressed 3P proteins remained soluble in the cell lysate (Fig. 3). The expression yield (1.57 μg/ml) of 3P was not as high as those of several heterologous proteins, such as human tumor necrosis factor (42), the antigen pertactin (p69) of Bordetella pertussis (35), and glycosylated invertase (47), which have all been expressed in P. pastoris up to several grams per liter of culture. However, the expression levels of P proteins are as high as those of two membrane proteins of influenza virus, neuraminidase (NA) and hemagglutinin (HA), which ranged from 2.5 to 3 μg/ml (24) and 0.375 to 0.675 μg/ml (37), respectively. In the cases of NA and HA expression, the secreted products from P. pastoris were shown to be sufficient to serve as recombinant vaccines that elicit partial or fully protective antibodies in mice. Since the expression levels in yeast cells of proteins bearing a complete nuclear localization signal (NLS) are less than those of the corresponding proteins with deletions at the NLS (26, 36, 40), it appears that the NLS downregulates gene expression, presumably at the step of translation. Since all three P proteins carry the NLS sequence (13), their expression levels could be elevated by introducing mutations in the NLS sequences.
The three P proteins expressed in the P. pastoris F3P strain formed a complex(es) which binds to the Ni2+-NTA-agarose column via the Hx tag added at the N terminus of the PB2 protein, but the affinity of the 3P complex for the Ni2+ resin was not so great as to be used for a single-step purification by elution with a high concentration of imidazole. The affinity for Ni2+-NTA-agarose may be weaker for the Hx-tag sequence used (27) than for the widely used hexahistidine sequence. The subunit-subunit contact network within the influenza virus RNA polymerase is formed through two major contacts, the PB2 N-terminal domain to the PB1 C-terminal domain and the PB1 N-terminal domain to the PA C-terminal domain (45). Since the Hx tag was added at the N terminus of the PB2 protein, the PB2-PB1 contact may interfere with free access of the Hx tag to the Ni2+-NTA-agarose. PB2 carries the NLS (32) and two capped RNA-binding sites (10), which may interact with such cellular components as the nuclear transport machinery and the RNA cap-binding proteins, thereby preventing the Hx tag from binding tightly to the Ni2+ resin.
The catalytic activities of RNA synthesis in vitro were analyzed using the partially purified 3P complex. The 3P complex exhibited RNA synthesis activity only when exogenous model RNAs were added as templates (Fig. 5). The v84 model RNA-directed and ApG-primed activity of template-sized RNA by the 3P complex was about 20% of the RNP activity, as normalized to the content of P proteins (Fig. 4 and 5). The 3P complex has the same enzymatic characteristics as the purified or reconstituted influenza virus RNA: (i) the 3P complex is unable to synthesize RNA in the absence of primer (Fig. 5) (11, 20, 31, 43); (ii) the v-sense RNA is a better template than the c-sense RNA polymerase (Fig. 6 and Table 3) (11, 20, 28, 46); (iii) both 3′ and 5′ conserved sequences are required for transcription initiation (Fig. 7) (5, 31, 33, 38); and (iv) capped RNA such as globin mRNA can be used as primers for model template-dependent RNA synthesis (Fig. 8) (21). These reaction properties are characteristic of the transcriptase. Since the 5′-terminal triphosphate remains associated with the replication products (9), the replicase form of RNA polymerase must be able to initiate RNA synthesis de novo without primers. In addition, the replicase should use both vRNA and cRNA equally as templates. Thus, we concluded that the majority of the 3P complex formed in P. pastoris F3P is the transcriptase form of the viral RNA polymerase. The functional integrity in vivo of the 3P complex in P. pastoris cells is being tested.
The Km for ATP in RNA synthesis by the 3P complex was much higher than the Km values for other ribonucleoside triphosphates. Klumpp et al. (19) showed that the Km for ATP is 10-fold higher than the Km for other ribonucleotide triphosphates in transcription initiation. The high ATP concentration may be required for the functional conversion of RNA polymerase during the transition from transcription initiation to elongation (19). However, it has not yet been excluded that the requirement for a high ATP concentration is due to contamination with cellular ATPase in the 3P complex fraction.
ACKNOWLEDGMENTS
We thank S. Yuasa and Y. Nagami (Mitsubishi Research Institute) for discussions and S. Ueda and A. Iwata (Nippon Institute for Biological Science) for preparation of anti-P protein antibodies.
This work was supported by Grants-in-Aid from the Ministry of Education, Science and Culture of Japan and by Core Research for Evolutional Science and Technology (CREST) of the Japan Science and Technology Corporation. J.-S.H. is a recipient of a Japan Society for Promotion of Science postdoctoral fellowship.
REFERENCES
- 1.Bouloy M, Plotch S J, Krug R. Globin mRNA are primers for the transcription of influenza viral RNA in vitro. Proc Natl Acad Sci USA. 1978;75:4886–4890. doi: 10.1073/pnas.75.10.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cregg J M, Vedvick T S, Raschke W C. Recent advances in the expression of foreign genes in Pichia pastoris. Bio/Technology. 1993;11:905–910. doi: 10.1038/nbt0893-905. [DOI] [PubMed] [Google Scholar]
- 3.de la Luna S, Martin J, Portela A, Ortin J. Influenza virus naked RNA can be expressed upon transfection into cells co-expressing the three subunits of the polymerase and the nucleoprotein from simian virus 40 recombinant virus. J Gen Virol. 1993;74:535–539. doi: 10.1099/0022-1317-74-3-535. [DOI] [PubMed] [Google Scholar]
- 4.Faber K N, Harder W, Ab G, Veenhuis M. Review: methylotrophic yeasts as factories for the production of foreign proteins. Yeast. 1995;11:1331–1344. doi: 10.1002/yea.320111402. [DOI] [PubMed] [Google Scholar]
- 5.Fodor E, Seong B L, Brownlee G G. Photochemical cross-linking of influenza A polymerase to its virion RNA promoter defines a polymerase binding site at residues 9 to 12 of the promoter. J Gen Virol. 1993;74:1327–1333. doi: 10.1099/0022-1317-74-7-1327. [DOI] [PubMed] [Google Scholar]
- 6.Gellissen G, Hollenberg C P. Application of yeasts in gene expression studies: a comparison of Saccharomyces cerevisiae, Hansenula polymorpha and Kluyveromyces lactis. Gene. 1997;190:87–97. doi: 10.1016/s0378-1119(97)00020-6. [DOI] [PubMed] [Google Scholar]
- 7.Honda A, Ishihama A. The molecular anatomy of influenza virus RNA polymerase. Biol Chem. 1997;378:483–488. [PubMed] [Google Scholar]
- 8.Honda A, Mizumoto K, Ishihama A. RNA polymerase of influenza virus: dinucleotide-primed initiation of transcription at specific positions on viral RNA. J Biol Chem. 1986;261:5987–5991. [PubMed] [Google Scholar]
- 9.Honda A, Mizumoto K, Ishihama A. Identification of the 5′ terminal structure of influenza virus genome RNA by a newly developed enzymatic method. Virus Res. 1998;55:199–206. doi: 10.1016/s0168-1702(98)00048-3. [DOI] [PubMed] [Google Scholar]
- 10.Honda A, Mizumoto K, Ishihama A. Two separate sequences of PB2 subunit constitute the RNA cap-binding site of influenza virus RNA polymerase. Genes Cells. 1999;4:475–485. doi: 10.1046/j.1365-2443.1999.00275.x. [DOI] [PubMed] [Google Scholar]
- 11.Honda A, Mukaigawa J, Yokoiyama A, Kato A, Ueda S, Nagata K, Krystal M, Nayak D P, Ishihama A. Purification and molecular structure of RNA polymerase from influenza virus A/PR8. J Biochem (Tokyo) 1990;107:624–628. doi: 10.1093/oxfordjournals.jbchem.a123097. [DOI] [PubMed] [Google Scholar]
- 12.Honda A, Ueda K, Nagata K, Ishihama A. RNA polymerase of influenza virus: role of NP in RNA chain elongation. J Biochem (Tokyo) 1988;104:1021–1026. doi: 10.1093/oxfordjournals.jbchem.a122569. [DOI] [PubMed] [Google Scholar]
- 13.Ishihama A. A multi-functional enzyme with RNA polymerase and RNase activities: molecular anatomy of influenza virus RNA polymerase. Biochimie. 1996;78:1097–1102. doi: 10.1016/s0300-9084(97)86735-1. [DOI] [PubMed] [Google Scholar]
- 14.Janda M, Ahlquist P. RNA-dependent replication, transcription and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell. 1993;72:961–970. doi: 10.1016/0092-8674(93)90584-d. [DOI] [PubMed] [Google Scholar]
- 15.Kato A, Mizumoto K, Ishihama A. Purification and enzymatic properties of an RNA polymerase-RNA complex from influenza virus. Virus Res. 1985;3:115–127. doi: 10.1016/0168-1702(85)90002-4. [DOI] [PubMed] [Google Scholar]
- 16.Kawakami K, Ishihama A. RNA polymerase of influenza virus. III. Isolation of RNA polymerase-RNA complexes from influenza virus PR8. J Biochem (Tokyo) 1983;93:989–996. doi: 10.1093/oxfordjournals.jbchem.a134254. [DOI] [PubMed] [Google Scholar]
- 17.Kimura N, Fukusha A, Oda K, Nakada S. An in vivo study of the replication origin in the influenza virus complementary RNA. J Biochem (Tokyo) 1993;113:88–92. doi: 10.1093/oxfordjournals.jbchem.a124009. [DOI] [PubMed] [Google Scholar]
- 18.Kimura N, Nishida M, Nagata K, Ishihama A, Oda K, Nakada S. Expression of a recombinant influenza viral RNA in cells permanently expressing the RNA polymerase and NP genes of influenza virus. J Gen Virol. 1992;73:1321–1328. doi: 10.1099/0022-1317-73-6-1321. [DOI] [PubMed] [Google Scholar]
- 19.Klumpp K, Ford M J, Ruigrok R W. Variation in ATP requirement during influenza virus transcription. J Gen Virol. 1998;79:1033–1045. doi: 10.1099/0022-1317-79-5-1033. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi M, Tuchiya K, Nagata K, Ishihama A. Reconstitution of influenza virus RNA polymerase from three subunits expressed using recombinant baculovirus system. Virus Res. 1992;22:235–245. doi: 10.1016/0168-1702(92)90055-e. [DOI] [PubMed] [Google Scholar]
- 21.Krug R M, Alonso-Caplen F V, Julkenun I, Katze M G. Expression and replication of the influenza virus genome. In: Krug R M, editor. The influenza viruses. New York, N.Y: Plenum Press; 1989. pp. 89–101. [Google Scholar]
- 22.Krystal M, Li R, Lyles D, Pavlakis G, Palese P. Expression of the three virus polymerase proteins in a single cell allows growth complementation of viral mutants. Proc Natl Acad Sci USA. 1986;83:2709–2713. doi: 10.1073/pnas.83.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laroche Y, Storme V, De Meutter J, Messens J, Lauwereys M. High-level secretion and very efficient isotopic labeling of tick anticoagulant peptide (TAP) expressed in the methylotrophic yeast, Pichia pastoris. Bio/Technology. 1994;12:1119–1124. doi: 10.1038/nbt1194-1119. [DOI] [PubMed] [Google Scholar]
- 24.Martinet W, Saelens X, Deroo T, Neirynck S, Contreras R, Min Jou W, Fiers W. Protection of mice against a lethal influenza challenge by immunization with yeast-derived recombinant influenza neuraminidase. Eur J Biochem. 1997;247:332–338. doi: 10.1111/j.1432-1033.1997.00332.x. [DOI] [PubMed] [Google Scholar]
- 25.Mena I, de la Luna S, Albo C, Martin J, Nieto A, Ortin J, Portela A. Synthesis of biologically active influenza virus core proteins using a vaccinia virus-T7 RNA polymerase expression system. J Gen Virol. 1994;75:2109–2114. doi: 10.1099/0022-1317-75-8-2109. [DOI] [PubMed] [Google Scholar]
- 26.Moreland R B, Nam H G, Herefford L M, Fried H M. Identification of a nuclear localization signal of a yeast ribosomal protein. Proc Natl Acad Sci USA. 1985;82:6561–6565. doi: 10.1073/pnas.82.19.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mori A, Yamada K, Kimura J, Koide T, Yuasa S, Yamada E, Miyamura T. Enzymatic characterization of purified NS3 serine proteinase of hepatitis C virus expressed in Escherichia coli. FEBS Lett. 1996;378:37–42. doi: 10.1016/0014-5793(95)01423-3. [DOI] [PubMed] [Google Scholar]
- 28.Nagata K, Takeuchi K, Ishihama A. In vitro synthesis of influenza viral RNA: biochemical complementation assay of factors required for influenza virus replication. J Biochem (Tokyo) 1989;106:205–208. doi: 10.1093/oxfordjournals.jbchem.a122833. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura Y, Oda K, Nakada S. Growth complementation of influenza virus temperature-sensitive mutants in mouse cells which express the RNA polymerase and nucleoprotein genes. J Biochem (Tokyo) 1991;110:395–401. doi: 10.1093/oxfordjournals.jbchem.a123592. [DOI] [PubMed] [Google Scholar]
- 30.Neumann G, Hobom G. Mutational analysis of influenza virus promoter elements in vivo. J Gen Virol. 1995;76:1709–1717. doi: 10.1099/0022-1317-76-7-1709. [DOI] [PubMed] [Google Scholar]
- 31.Parvin J D, Palese P, Honda A, Ishihama A, Krystal M. Promoter analysis of influenza virus RNA polymerase. J Virol. 1989;63:5142–5152. doi: 10.1128/jvi.63.12.5142-5152.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perales B, de la Luna S, Palacios I, Ortín J. Mutational analysis identifies functional domains in the influenza A virus PB2 polymerase subunit. J Virol. 1996;70:1678–1686. doi: 10.1128/jvi.70.3.1678-1686.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piccone M E, Fernandez-Sesma A, Palese P. Mutational analysis of the influenza virus vRNA promoter. Virus Res. 1993;28:99–112. doi: 10.1016/0168-1702(93)90129-b. [DOI] [PubMed] [Google Scholar]
- 34.Pritlove D C, Fodor E, Seong B L, Brownlee G G. In vitro transcription and polymerase binding studies of the termini of influenza A virus cRNA: evidence for a cRNA panhandle. J Gen Virol. 1995;76:2205–2213. doi: 10.1099/0022-1317-76-9-2205. [DOI] [PubMed] [Google Scholar]
- 35.Romanos M A, Clare J J, Beesley K M, Rayment F B, Ballantine S P, Makoff A J, Dougan G, Fairweather N F, Charles I G. Recombinant Bordetella pertussis pertactin (P69) from the yeast Pichia pastoris: high-level production and immunological properties. Vaccine. 1991;9:901–906. doi: 10.1016/0264-410x(91)90011-t. [DOI] [PubMed] [Google Scholar]
- 36.Rong L, Klein H L. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J Biol Chem. 1993;268:1252–1259. [PubMed] [Google Scholar]
- 37.Saelens X, Vanlandschoot P, Martinet W, Maras M, Neirynck S, Contreras R, Fiers W, Min Jou W. Protection of mice against a lethal influenza virus challenge after immunization with yeast-derived secreted influenza virus hemagglutinin. Eur J Biochem. 1999;260:166–175. doi: 10.1046/j.1432-1327.1999.00150.x. [DOI] [PubMed] [Google Scholar]
- 38.Seong B L, Brownlee G G. Nucleotides 9 to 11 of the influenza A virion RNA promoter are crucial for activity in vitro. J Gen Virol. 1992;73:3115–3124. doi: 10.1099/0022-1317-73-12-3115. [DOI] [PubMed] [Google Scholar]
- 39.Shi L, Galarza J M, Summers D F. Recombinant-baculovirus-expressed PB2 subunit of the influenza A virus RNA polymerase binds cap groups as an isolated subunit. Virus Res. 1996;42:1–9. doi: 10.1016/0168-1702(96)01289-0. [DOI] [PubMed] [Google Scholar]
- 40.Silver P A, Chiang C, Sadler I. Mutations that alter both localization and production of a yeast nuclear protein. Genes Dev. 1988;2:707–717. doi: 10.1101/gad.2.6.707. [DOI] [PubMed] [Google Scholar]
- 41.Smith G L, Levin J Z, Palese P, Moss B. Synthesis and cellular location of the ten influenza polypeptides individually expressed by recombinant vaccinia viruses. Virology. 1987;160:336–345. doi: 10.1016/0042-6822(87)90004-3. [DOI] [PubMed] [Google Scholar]
- 42.Sreekrishna K, Nelles L, Potenz R, Cruze J, Mazzaferro P, Fish W, Fuke M, Holden K, Phelps D, Wood P, Parker K. High-level expression, purification and characterization of recombinant human tumor necrosis factor synthesized in the methylotrophic yeast Pichia pastoris. Biochemistry. 1989;28:4117–4125. doi: 10.1021/bi00435a074. [DOI] [PubMed] [Google Scholar]
- 43.Szewczyk B, Laver W G, Summers D F. Purification, thioredoxin renaturation, and reconstituted activity of the three subunits of the influenza A virus RNA polymerase. Proc Natl Acad Sci USA. 1988;85:7907–7911. doi: 10.1073/pnas.85.21.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tiley L S, Hagen M, Matthews J T, Krystal M. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J Virol. 1994;68:5108–5116. doi: 10.1128/jvi.68.8.5108-5116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toyoda T, Adyshev D M, Kobayashi M, Iwata A, Ishihama A. Molecular assembly of influenza virus RNA polymerase: determination of the subunit-subunit contact sites. J Gen Virol. 1996;77:2149–2157. doi: 10.1099/0022-1317-77-9-2149. [DOI] [PubMed] [Google Scholar]
- 46.Toyoda T, Kobayashi M, Ishihama A. Replication in vitro of the influenza virus genome: selective dissociation of RNA replicase from virus-infected cell ribonucleoprotein complexes. Arch Virol. 1994;136:269–286. doi: 10.1007/BF01321057. [DOI] [PubMed] [Google Scholar]
- 47.Tschopp J F, Sverlow G, Kosson R, Craig W, Grinna L. High level secretion of glycosylated invertase in the methylotrophic yeast Pichia pastoris. Bio/Technology. 1987;5:1305–1308. [Google Scholar]
- 48.Zhang H, Air G M. Expression of functional influenza virus A polymerase proteins and template from cloned cDNAs in recombinant vaccinia virus infected cells. Biochem Biophys Res Commun. 1994;200:95–101. doi: 10.1006/bbrc.1994.1419. [DOI] [PubMed] [Google Scholar]