Abstract
Maintaining macrophage (MΦ) heterogeneity is critical to ensure intestinal tissue homeostasis and host defense. The gut microbiota and host factors are thought to synergistically guide intestinal MΦ development, although the exact nature, regulation, and location of such collaboration remain unclear. Here, we report that microbial biochemical energy metabolism promotes colony-stimulating factor 2 (CSF2) production by group 3 innate lymphoid cells (ILC3s) within solitary isolated lymphoid tissues (SILTs) in a cell-extrinsic, NLRP3/P2X7R-dependent fashion in the steady state. Tissue-infiltrating monocytes accumulating around SILTs followed a spatially constrained, distinct developmental trajectory into SILT-associated MΦs (SAMs). CSF2 regulated the mitochondrial membrane potential and reactive oxygen species production of SAMs and contributed to the antimicrobial defense against enteric bacterial infections. Collectively, these findings identify SILTs and CSF2-producing ILC3s as a microanatomic niche for intestinal MΦ development and functional programming fueled by the integration of commensal microbial energy metabolism.
INTRODUCTION
Intestinal macrophages (MΦs) represent a large proportion of the innate immune system in the gut and are critical mediators of host defense and tissue homeostasis. Although their heterogeneity is widely appreciated, the mechanisms regulating MΦ heterogeneity within microanatomic environments of intestinal lamina propria (LP) remain enigmatic (1). Further complicating this matter are the classification strategies for intestinal MΦs. As in other organs, intestinal MΦ subpopulations can be distinguished on the basis of their expression of the markers T cell immunoglobulin and mucin domain containing 4 (Tim-4) and C-C motif chemokine receptor 2 (CCR2) to denote fetal-derived long-lived, self-renewing cells and monocyte-derived ones, respectively (2, 3). Others have demarcated gut MΦs using Tim-4 and CD4 into three subpopulations: Tim-4−CD4− and Tim-4−CD4+ MΦs with rapid monocytic turnover and long-lived Tim-4+CD4+ MΦs with slow monocytic turnover, each subset capable of associating with either neurons, vasculature, and other immune cells within distinct regions to adopt transcriptional profiles and functions tailored to these microenvironments (4, 5) (6-8).
In contrast to other organs, intestinal MΦs integrate signaling by microbial metabolites derived from the commensal microflora into their homeostatic function (7, 9). For example, polysaccharides produced by Helicobacter hepaticus and commensal bacteria-derived short-chain fatty acids (SCFAs) promote tolerogenic MΦs (10-12). Bacteria-metabolized dietary tryptophan controls monocyte differentiation in an aryl hydrocarbon receptor–dependent manner (13). Colonization with the protozoan commensal Tritrichomonas musculis (T. mu) was recently shown to induce monocyte infiltration in the gut by increasing luminal extracellular adenosine 5′-triphosphate (ATP) levels (14). Collectively, these studies raise the question of whether a ubiquitously produced metabolite conserved across microbial kingdoms may serve as a molecular motif for imprinting MΦ heterogeneity.
Microbiota- and host-derived factors collaborate in orchestrating gut MΦ composition and function. Deficiency in the host-derived myeloid growth factor colony-stimulating factor 1 (CSF1) results in a systemic decrease in MΦs, with a less pronounced effect on the intestinal tract, suggesting compensatory growth factors (15-17). Interleukin-34 (IL-34), transforming growth factor–β, or CSF2 has been reported to affect MΦ development in many organs, including the intestinal tract (18-20). In the gut, tissue-resident group 3 innate lymphoid cells (ILC3s) produce large quantities of CSF2 in a microbiota-dependent manner within cryptopatches and isolated lymphoid follicles (ILFs), collectively known as solitary isolated lymphoid tissues (SILTs). CSF2 production in ILC3s was dependent on the IL-1 receptor and MΦ-derived IL-1β (9). Csf2−/− mice displayed a partial reduction in intestinal MΦs, suggesting that MΦ development may, in part, depend on this growth factor (9). CSF2 has recently been shown to license the effector profile of MΦs in the inflamed brain, implicating an impact on MΦ function in addition to development (21). Whether these observations on CSF2 extend to MΦ development and function in the intestine remain unknown.
Here, we report a molecular and spatial framework governing cross-talk between host and microbiota that regulates MΦ heterogeneity. We identified SILTs as a critical niche for the developmental and functional programming of monocyte-derived SILT-associated MΦs (SAMs). Microbe-derived extracellular ATP (ATPex), acting through purinergic receptor P2X 7 (P2X7R)– and NLR family pyrin domain containing 3 (NLRP3)–expressing myeloid cells, drives CSF2 production by ILC3s to induce the developmental and functional transition of monocytes to SAMs along a differentiation pathway distinct from LP-associated MΦs (LAMs). CSF2-dependent SAMs mediated antimicrobial host protection, implicating SILTs as previously underappreciated, broadly scattered myeloid reservoirs across the gut. Collectively, our findings identify a molecular and spatial framework for the anatomically contained differentiation of intestinal MΦs, centered around a microbiotafueled, ILC3-driven, and CSF2-dependent axis that integrates signals indicative of microbial energy into the developmental and functional heterogeneity of MΦs.
RESULTS
CSF2 maintains intestinal monocyte-derived MΦs
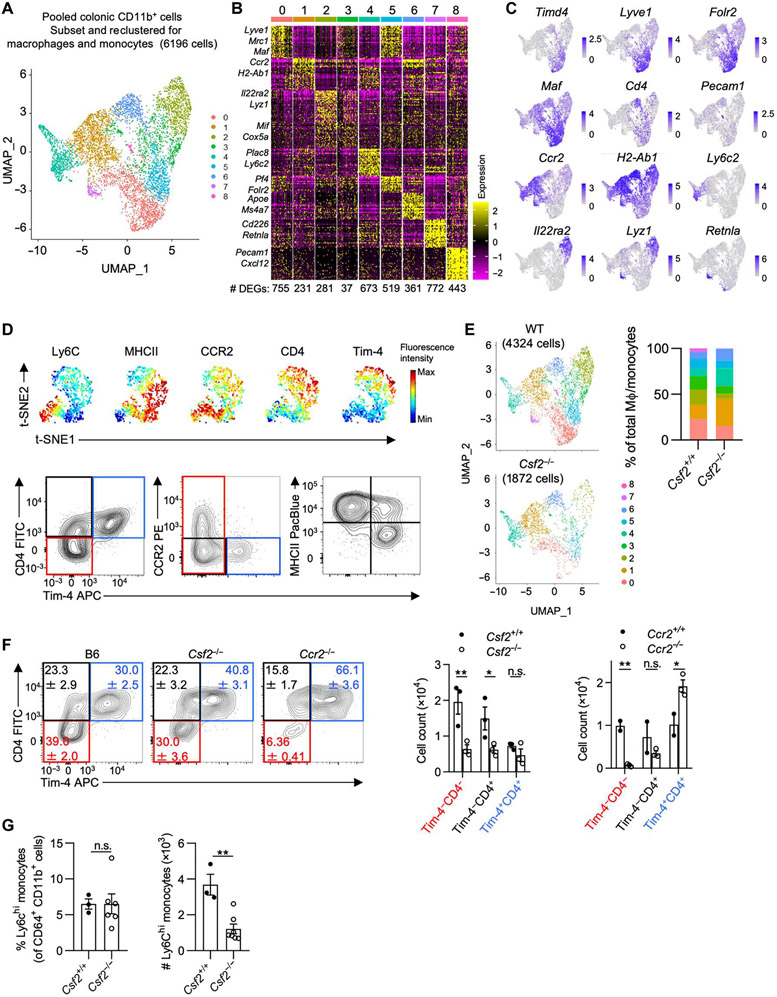
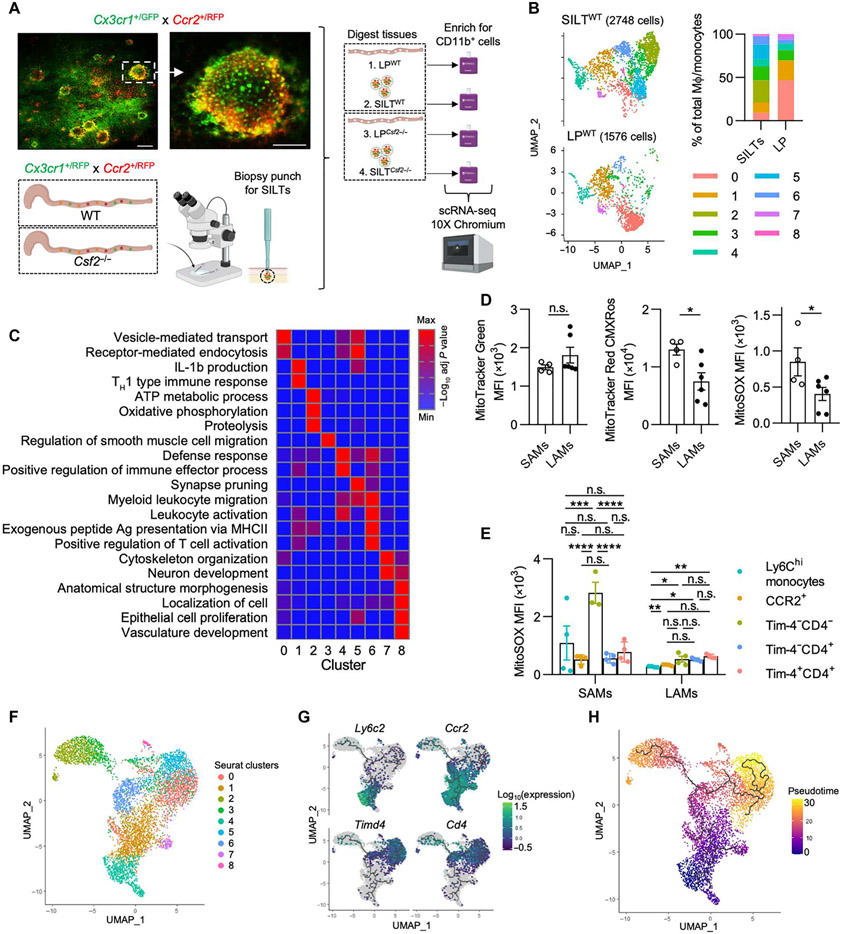
Tissue-resident MΦs in extraintestinal organs group into monocyte- or fetal-derived subpopulations based on cross-organ conserved expression of the following markers: the chemokine receptor CCR2, the phosphatidylserine receptor Tim-4, lymphatic vessel endothelial hyaluronan receptor 1 (LYVE1), and major histocompatibility complex 2 (MHCII) (2). In the gut, MΦs have been classified by Tim-4 and CD4 expression and the “monocyte waterfall” gating strategy, the latter tracking the transition of monocyte into MΦ using the markers Ly6C and MHCII (4, 22). To determine the full scope of gut MΦ heterogeneity, we performed single-cell RNA sequencing (scRNA-seq) on CD11b-enriched cells isolated from the colonic LP of healthy adult mice (8 to 12 weeks old). Subsetting and re-analysis of the MΦ/monocyte clusters, identified based on their expression of Csf1r, Cx3cr1, and C1qa and the absence of dendritic cell (DC) markers, resulted in nine distinct clusters, revealing an anticipated heterogeneity within the colonic MΦ pool (Fig. 1A and fig. S1, A to D). MΦs in clusters 0 and 5 highly expressed Lyve1, Mrc1, Maf, and Timd4 (Fig. 1, B and C) (2, 6, 23). MΦs in clusters 0 and 5 coexpressed Cd4 but virtually no Ccr2. Cluster 4 was marked by high expression of Ly6c2, identifying them as monocytes, whereas cells in clusters 1 and 6 were annotated as monocyte-derived MΦs based on their high Ccr2 expression (Fig. 1, B and C). Lyz1 expression almost exclusively overlapped with Il22ra2 expression in cluster 2, both genes previously reported to be also expressed in DCs located within small intestinal SILTs (24). Cluster 7 MΦs expressed Retnla, indicative of RELMα+ MΦs. Cluster 8 cells showed an unexpected expression of Pecam1, suggesting the existence of a MΦ population possibly involved in the interactions with endothelial or epithelial cells (Fig. 1, B and C). Multiparameter flow cytometry on colonic LP CD64+ CD11b+ cells recapitulated the distinct and partial overlap in the expression of Tim-4, CD4, CCR2, MHCII, and Ly6C across MΦs aligning with the previous classification into Tim-4+CD4+, Tim-4−CD4+, and Tim-4−CD4− MΦs (Fig. 1D). Using flow cytometry, Tim-4−CD4− MΦs were further delineated into Ly6C+, CCR2+, and CCR2− MΦs, suggesting a developmental relationship to monocytes and confirming the suitability of these markers for the analysis of colonic MΦs (Fig. 1D and fig. S1, E and F) (2, 4).
Fig. 1. Developmental requirements and kinetics of colonic LP MΦs.
(A) UMAP projection of the combined analysis of WT and Csf2−/− datasets subsetted and reclustered for MΦs and monocytes visualized together. (B) Heatmap depicting the top 30 DEGs for each cluster (log2FC threshold = 0.25, min.pct = 0.25, adjusted P < 0.05), downsampled to 50 cells for visualization. The number of DEGs in each cluster is shown. (C) Feature plots illustrating expression of subset-defining genes. (D) Representative flow cytometry analysis using (top) unbiased t-distributed stochastic neighbor embedding (t-SNE) dimensionality reduction of colonic CD64+ CD11b+ cells showing expression of common MΦ markers or (bottom) Tim-4/CD4,Tim-4/CCR2, and Tim-4/MHCII classification strategies. (E) UMAP projection (left) of MΦs/monocytes in WT versus Csf2−/− colon with relative abundance of each cluster (right). (F) Contour plots (left) and quantification (right) of colonic MΦs in B6, Csf2−/−, and Ccr2−/− mice. (G) Percentage and absolute counts of Ly6Chi monocytes in WT and Csf2−/− mice. Data shown in (A) to (C) and (E) are from the same scRNA-seq experiment obtained from cells pooled from at least three animals per genotype. Two-way analysis of variance (ANOVA) with post hoc Šidák’s multiple comparisons test (F) or unpaired Student’s t test (G) was performed. FITC, fluorescein isothiocyanate; PE, phycoerythrin; APC, allophycocyanin; n.s., not significant.
Previous investigations into Csf2−/− mice did not use this detailed classification of MΦs, prompting us to revisit the requirements for CSF2 on gut MΦ heterogeneity (9). Comparing cluster relative abundance on scRNA-seq datasets generated from wild-type (WT) littermate controls and Csf2−/− mice indicated a reduction of MΦs within clusters 0, 2, 3, 5, 7, and 8 in the absence of Csf2 (Fig. 1E). Analysis of MΦs in the colonic LP using flow cytometry confirmed the reduction in Tim-4−CD4− MΦs and Tim-4−CD4+ MΦs in Csf2−/− mice. The same MΦ populations were similarly reduced in Ccr2−/− mice, implicating the developmental origin of these two MΦ subpopulations as monocyte derived (Fig. 1F and table S1). A decrease in both tissue-infiltrating Ly6Chi monocytes and CCR2+ MΦs suggested a possible developmental blockade along the monocyte-to-MΦ transition in Csf2−/− mice (Fig. 1G and fig. S1G). In contrast to Rag2−/− mice injected with neutralizing anti-CSF2 antibodies, MΦs and monocytes from Csf2−/− and WT mice showed comparable levels of MHCII expression (fig. S1H) (25). In addition to the effects on monocytes and CCR2+ MΦs, Tim-4+ MΦs were partially affected in Csf2−/− mice, suggesting that all MΦ subpopulations variably depend on this growth factor (Fig. 1E).
Monocytes and Tim-4−CD4− MΦs are developmentally related
Tim-4+ MΦs are a long-lived, fetal-derived population with minimal replacement by monocytes. It has been proposed that the expression of Tim-4 may reflect long-term residency within a tissue after differentiation (4, 6, 26-28). To determine the long-term MΦ turnover by infiltrating monocytes, we performed parabiosis of CD45.1 C57BL/6 and CD45.2 Ccr2−/− mice to assess the chimerism of each MΦ subpopulation after 6 or 12 months. As expected, CD45.2 Ccr2−/− parabionts showed CD45.1 frequencies comparable to blood monocytes in both the colonic Tim-4−CD4− and Tim-4−CD4+ MΦ compartments (fig. S1, I and J), consistent with their continual replacement by blood monocytes. Unexpectedly, Tim-4+ MΦs were also replaced by donor CD45.1 monocytes, albeit at a slower rate, suggesting a homeostatic contribution of monocytes to the maintenance of Tim-4+ MΦs (fig. S1, I and J).
To confirm these results, we used a tamoxifen-inducible fate-mapping model. Tamoxifen-containing chow was provided to Ccr2CreERT2 x Rosa26-LSL-tdTomato (Rosa26td) mice during a 1-week pulse phase, followed by a chase period with normal chow for either 1 or 52 week(s). Loss of Tomato labeling was measured in each MΦ subpopulation to assess the replacement of MΦs by newly infiltrated monocytes. Tamoxifen administration labeled ~94% of all blood Ly6Chi monocytes within the 1-week pulse phase, whereas the induced Tomato label was completely lost in monocytes by 52 weeks (fig. S1K). Colonic Ly6Chi monocytes and Tim-4−CD4− MΦs showed labeling efficiency similar to blood monocytes, whereas Tim-4−CD4+ MΦs displayed partial Tomato labeling (64%). Unexpectedly, ~17% of all Tim-4+ MΦs were labeled after the 1-week pulse phase, suggesting either a possible contribution of monocytes to this population during tamoxifen administration or low Ccr2 expression levels and recombinase activity within a subset of Tim-4+ MΦs (fig. S1K). After the first 7 days of the chase period, Tim-4−CD4− MΦs (containing Ly6C+ and CCR2+ cells) displayed replacement by bone marrow–derived (BM) monocytes, as indicated by the loss of Tomato labeling (fig. S1K). Tim-4−CD4+ MΦs and Tim-4+ MΦs did not show signs of replacement, suggesting a slower replacement rate in line with our parabiosis experiments (fig. S1, I to K). After the 52-week chase period, all MΦ subpopulations lost the Tomato label (fig. S1K). These results indicate that all colonic MΦs share a monocytic origin, with Tim-4−CD4− MΦs showing the fastest replacement and strongest reliance on CSF2 (Fig. 1F and fig. S1G).
Diversified microbiotas promote the accumulation of Tim-4−CD4− MΦs
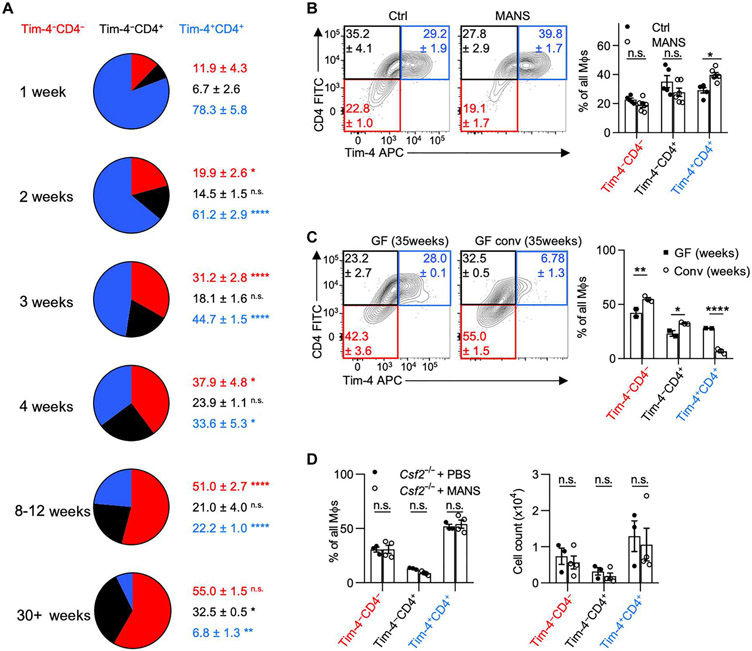
Tissue-resident Tim-4+ MΦs dominate the MΦ pool during embryogenesis and are found in all tissues at early time points of fetal development, whereas Tim-4− MΦs postnatally arise from BM monocytes (2). In the gut, this development requires the microbiota (4, 22). Our fate-mapping and parabiosis data show that colonic MΦ subpopulations display different rates of monocyte replacement, suggesting distinct appearances of the MΦ subpopulations in the neonatal and adult intestine. To delineate the developmental kinetics of MΦs, we assessed the composition of MΦs starting in the neonatal colon, tracking along the first weeks of life until adulthood. In line with previous reports, embryonically derived Tim-4+CD4+ MΦs primarily comprise the colons of newborn mice, followed by increased abundance of Tim-4−CD4− MΦs at 3 to 4 weeks of age. This time corresponds to weaning and the establishment of a diversified microbiota (Fig. 2A) (29). By 8 to 12 weeks after birth, Tim-4−CD4− MΦs comprise the majority of colonic MΦs, suggesting a microbiota-driven adaptation of the MΦ pool. These adaptations in gut MΦs mirror previously reported observations of intestinal CSF2 production, which similarly increased until week 8 in a microbiota-dependent fashion (9).
Fig. 2. The development of colonic Tim-4−CD4− MΦs requires an intact and diversified microbiota.
Colonic MΦ composition in (A) B6 mice across various ages, (B) B6 mice either left untreated or treated with broad-spectrum antibiotics [metronidazole, ampicillin, neomycin, streptomycin (MANS)], and (C) GF mice or GF mice conventionalized with SPF microbiota. (D) Colonic MΦ composition (left) and absolute counts of MΦ populations (right) in Csf2−/− mice either left untreated or treated with MANS for 2 weeks. Multiple unpaired t tests with two-stage Benjamini, Krieger, and Yekutieli false discovery rate test were performed for (A), Q = 5%, reporting q values; each time point compared with previous time point. Two-way ANOVA with post hoc Šidák’s multiple comparisons test was performed for (B) to (D). PBS, phosphate-buffered saline.
Depletion of the microbiota using broad-spectrum antibiotics in adult mice shifted the MΦ pool in favor of Tim-4+CD4+ MΦs, confirming a requirement of the microbiota in regulating gut MΦ composition (Fig. 2B). In contrast, reconstituting germ-free (GF) mice with an adult specific pathogen–free (SPF) microbiota increased Tim-4−CD4− MΦs at the expense of Tim-4+CD4+ MΦs (Fig. 2C). Colonization of adult SPF mice with the protozoan commensal T. mu further increased the abundance of Tim-4−CD4− MΦs (fig. S2A). A comparable shift toward Tim-4−CD4− MΦs was also observed when analyzing “rewilded” mice, i.e., ex-SPF mice that were colonized with the microbiota found in pet store mice (fig S2B). These findings suggest that the increase in Tim-4−CD4− MΦs in the colon may be due to an increase in microbiota-driven monocyte replacement.
To track the rate of monocyte replacement, we labeled all Cx3cr1-expressing cells in tamoxifen-inducible Cx3cr1CreERT2 x Rosa26tdTomato mice and followed the loss of tdTomato labeling in each MΦ population after colonization with T. mu (fig. S2C). Compared with uncolonized littermate controls, T. mu-colonized mice showed reduced percentages of tdTomato+ cells particularly in Tim-4−CD4− MΦs, suggesting an increased replacement of these cells by tdTomato− monocytes (fig. S2D). Monocyte replacement was also elevated in Tim-4−CD4+ MΦs and Tim-4+CD4+ MΦs. Collectively, our data demonstrate that diversifying the gut microbiota promotes MΦ replacement by monocytes and the accumulation of Tim-4−CD4− MΦs. Colonization with T. mu has previously been found to increase the production of CSF2 by intestinal ILC3s, implicating a role for ILC3s in the elevated MΦ replacement rates (30). Csf2−/− mice treated with broad-spectrum antibiotics did not show alterations in Tim-4−CD4− MΦs, suggesting that microbiota-driven changes in the colonic MΦ pool require CSF2 (Fig. 2D).
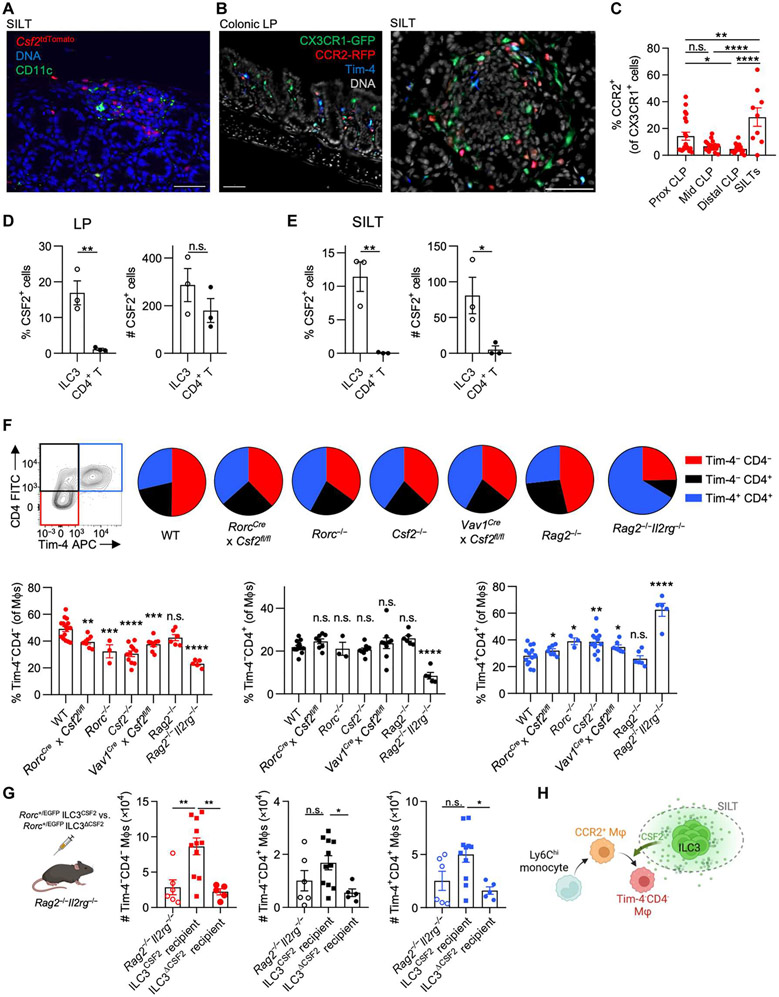
CSF2-producing ILC3s support intestinal CCR2+ Tim-4−CD4− MΦs
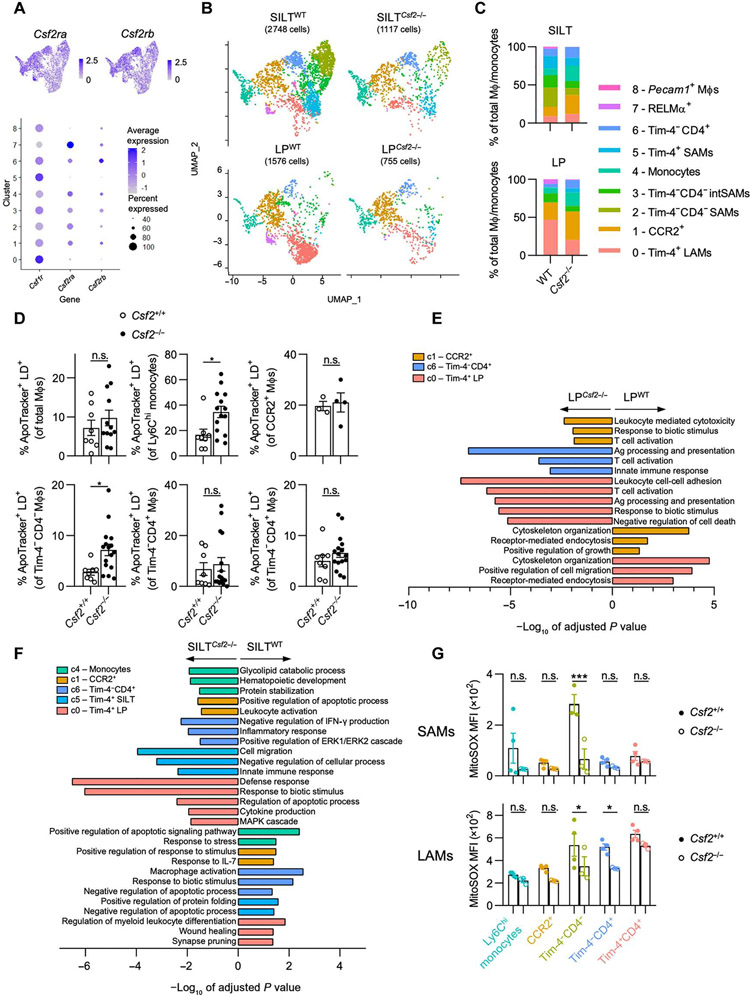
Microenvironmental cues within anatomical niches are critical for imprinting tissue-resident MΦ identity in various organs (31). However, less focus has been placed on niches for monocyte-derived MΦs. In the gut, CSF2-producing ILC3s are abundantly found within postnatally formed SILTs (9). Csf2tdTomato reporter mice revealed an accumulation of tdTomato+ cells within SILTs, prompting us to determine whether CSF2-producing ILC3s constitute supporting cells for monocyte-derived MΦs (Fig. 3A) (9). Quantification of CX3CR1+CCR2+ MΦs in the colonic LP or SILTs of Cx3cr1+/EGFPCcr2+/RFP mice uncovered an accumulation of monocyte-derived CCR2+ MΦs around SILTs compared with the surrounding LP (Fig. 3, B and C). SILTs contain B cells, ILC3s, and T cells, the latter two being possible sources of CSF2 in the LP (9, 32). We compared CSF2 production by T cells and ILC3s within SILTs or the surrounding LP by intracellular cytokine staining. T cells and ILC3s contributed equally to the pool of spontaneous CSF2 within the LP, but ILC3s constituted the predominant source of CSF2 within SILTs (Fig. 3, D and E). To confirm the role of ILC3s as a source for CSF2, we analyzed the colonic MΦ composition across WT, Rag2−/−, RorcCre × Csf2fl/fl, Rorc−/−, Csf2−/−, Vav1Cre × Csf2fl/fl, and Rag2−/−Il2rg−/− mice. Only Rag2−/−Il2rg−/− mice (lacking all lymphocytes), but not Rag2−/− mice (sufficient in ILCs), displayed a reduction in Tim-4−CD4− MΦs, comparable to the decrease observed in Csf2−/− or Rorc−/− mice (lacking ILC3s and Rorc-expressing T cell subsets). Animals lacking Csf2 in Rorc-expressing ILC3s and T cells (RorcCre × Csf2fl/fl) or all hematopoietic cells (Vav1Cre × Csf2fl/fl) recapitulated the altered MΦ composition found in both total body Csf2−/− and Rorc−/− mice but did not reach alterations observed in Rag2−/−Il2rg−/− mice. This indicates that ILC3s, rather than T or B cells, regulate the homeostatic CSF2-dependent MΦ composition in the colon, possibly supplemented by CSF2-producing ILC1s, ILC2s, or natural killer cells in the absence of RAR-related orphan receptor gamma t (RORγt) (Fig. 3F).
Fig. 3. CSF2-producing ILC3s in SILTs induce CCR2+ Tim-4−CD4− MΦs.
(A) Representative immunofluorescence image of a colonic SILT of Csf2tdTomato mouse stained for CD11c and DNA [4′,-diamidino-2-phenylindole (DAPI)]. (B) Representative immunofluorescence images of colonic LP and SILT of Cx3cr1+/GFP Ccr2+/RFP mice stained for Tim-4 and DNA. Scale bars, 50 μm (A and B). (C) Proportion of CCR2-RFP+ of CX3CR1-GFP+ cells based on CellProfiler quantification of images in each colonic LP (CLP) region and SILTs, as indicated. (D and E) Percentage and absolute counts of CSF2+ ILC3s and CD4+ T cells (D) in colonic LP and (C) within SILTs. (F) Colonic MΦ composition as indicated. (G) A total of 104 FACS-sorted small intestinal Rorc+/EGFP ILC3s from either WT or Csf2−/− mice were adoptively transferred into Rag2−/−Il2rg−/− mice, and recipients were analyzed at 6 weeks (left). Quantifications (right) of colonic MΦ populations after transfer, as indicated. (H) Schematic overview of how SILT-enriched, CSF2-producing ILC3s affect MΦ development. One-way ANOVA with post hoc Tukey’s test (C and G), unpaired Student’s t test (D and E), or one-way ANOVA with Dunnett’s test compared with WT (F) was performed.
To further validate the contribution of ILC3s to the regulation of colonic MΦs, we adoptively transferred 104 fluorescence-activated cell sorting (FACS)–purified Rorc+/EGFP ILC3s from WT (ILC3CSF2) or Csf2−/− (ILC3ΔCSF2) mice into Rag2−/−Il2rg−/− recipients. We observed accumulation of Tim-4−CD4− MΦs in the colonic LP after 6 weeks, indicating the dependency of Tim-4−CD4− MΦs on ILC3-derived CSF2 (Fig. 3G and fig. S3, A, and B). Numbers of Tim-4−CD4+ and Tim-4+CD4+ MΦs were also slightly increased in Rag2−/−Il2rg−/− mice after transfer of ILC3CSF2, suggesting their partial dependency on CSF2-producing ILC3s (Fig. 3G). These findings demonstrate that CCR2+ MΦs accumulate around colonic SILTs rich in CSF2-producing ILC3s that support Tim-4−CD4− MΦ development (Fig. 3H).
CSF2 expression by ILC3s has previously been demonstrated to require an intact microbiota and the production of IL-1β (9). We observed a microbiota-dependent increase in monocyte-derived MΦs in the context of microbiotas complexified with nonbacterial commensals and, conversely, a decrease in the absence or strong reduction of gut commensal microbes (Fig. 2, A to G). These observations argue in favor of a conserved mechanism that integrates the diversification of the microbiota into CSF2-dependent regulation of colonic MΦs.
Microbial ATPex regulates MΦ composition and CSF2 production
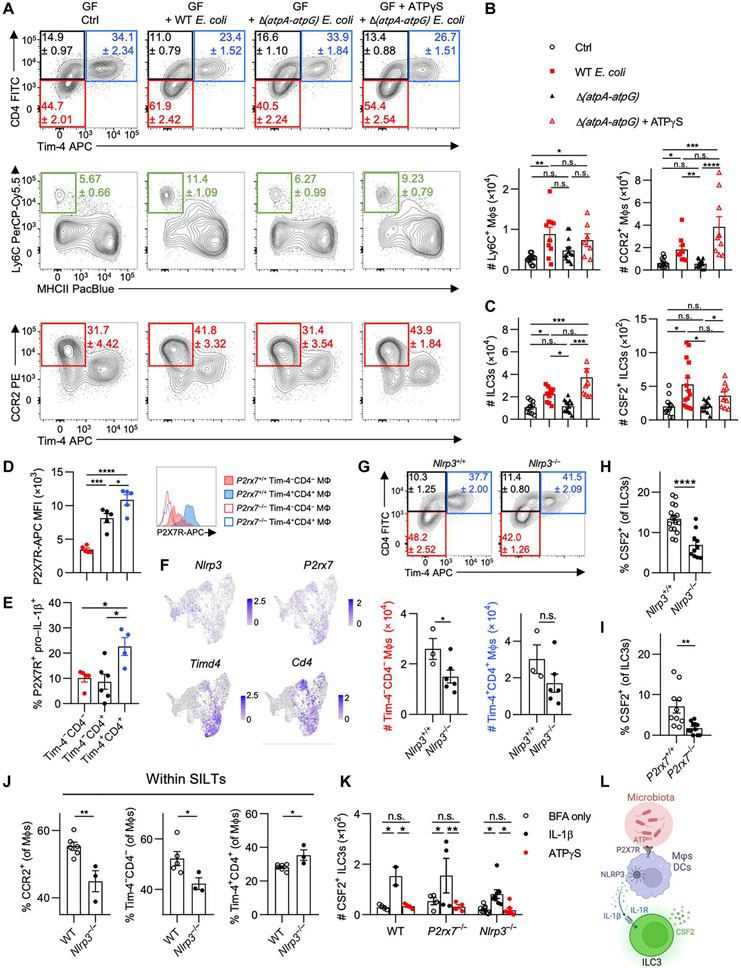
Diversification of the microbiota generates multiple new features within the microbial community, including adaptations to nutrients and synthesis of different metabolites (33). Such changes may not apply to individual microbial species but rather reflect a feature of complex community interactions (34). This raises the possibility for a conserved ubiquitously produced metabolite across all living microbes that indicates microbial vitality but, at the same time, serves as a molecular motif for immune recognition and activation that can indirectly affect MΦ homeostasis. ATP is one such metabolite capable of promoting intestinal immunity (35). We recently demonstrated that colonization with the protozoan commensal T. mu induces immune activation in the colon, including increased monocyte infiltration, regulated by elevated intestinal ATPex levels and inflammasome activation (14). ATPex is a common danger-associated molecular pattern that correlates with the presence of the microbiota and subsequently regulates local adaptive immune cells through P2X7R-dependent recognition by CD11c+ myeloid cells (35, 36). Thus, we asked whether ATPex might serve as a rheostatic indicator of the microbiota, capable of regulating colonic MΦ composition.
In line with previous studies, we first confirmed that ATPex levels in the gut lumen corresponded to microbiota abundance by comparing fecal ATPex in SPF, antibiotics-treated, and germ-free mice (fig. S4A). ATP was shown to be released by multiple bacterial species through an unknown mechanism while undergoing cellular respiration during the growth phase in vitro (37). To specifically investigate the role of bacteria-derived ATP, we used a mutant strain of commensal Escherichia coli MG1655 lacking the operons encoding the ATP synthase subunits A-G [Δ(atpA-atpG)] (38). In contrast to WT E. coli or a mutant lacking nitrate reductase genes [ΔnarG ΔnarZ Δ(napD-napA)], the adenosine triphosphatase (ATPase)–deficient mutant was unable to secrete ATP during growth in vitro, confirming an ATPase-dependent increase in ATPex (fig. S4, B and C). Transformation of the Forster resonance energy transfer–type ATP biosensor ATeam 3.10 into these E. coli strains enabled the quantification of intracellular ATP (ATPint) levels (fig. S4D) (39). Accordingly, less pronounced ATPint levels were observed in the ATPase-deficient E. coli in vitro, confirming its metabolic impairment and release of ATP (fig. S4, E and F).
To determine whether impaired ATP metabolism in commensals affects MΦ heterogeneity in the gut, GF mice were monocolonized with control E. coli, Δ(atpA-atpG) E. coli, or Δ(atpA-atpG) E. coli supplemented with the ATP derivative γS-ATP (ATPγS) via daily rectal enemas. Untreated groups received daily enemas of sterile phosphate-buffered saline (PBS). Colonic MΦ composition was assessed 7 days after engraftment to avoid confounding effects of adaptive immune cells on the colonizing microbes (40, 41). Despite equal colonization, Δ(atpA-atpG) E. coli produced less ATPint at the time of analysis compared with the WT strain (fig. S4, G and H). Both E. coli strains displayed equal and preferred localization in the lumen of the gut with minimal mucosal localization (fig. S4, I and J). Nevertheless, mice colonized with control E. coli showed higher infiltration of Ly6C+ and CCR2+ MΦs in comparison with Δ(atpA-atpG) E. coli–colonized mice (Fig. 4, A and B). Application of rectal ATPγS in GF mice colonized with Δ(atpA-atpG) E. coli recapitulated the increase in Ly6C+ monocytes and CCR2+ MΦs, demonstrating the requirement of bacterial-derived ATPex as a driver of monocyte infiltration and MΦ differentiation (Fig. 4, A and B). No impact was observed on other MΦ subsets, although it is possible that these may manifest after a longer time of colonization, given their slower turnover rates from monocytes (fig. S4K). CSF2 governed the abundance of CCR2+ MΦs and has previously been reported to be produced by ILC3s in a microbiota-dependent manner (9, 42). To determine the impact of ATPex on ILC3s, we also analyzed CSF2 production and ILC3 counts in the colonic LP. Mice gavaged with either WT E. coli or Δ(atpA-atpG) supplemented with ATPγS showed an increase in ILC3 numbers and CSF2+ ILC3s when compared with untreated GF mice or GF mice colonized with Δ(atpA-atpG) E. coli (Fig. 4C). These findings implicate the regulation of CSF2-producing ILC3s by microbe-derived ATPex and led us to investigate how ATPex may activate ILC3s.
Fig. 4. Microbial ATP regulates MΦ composition and drives CSF2 production by ILC3s through P2X7R and NLRP3 inflammasome signaling.
(A to C) GF mice were orally gavaged with WT or Δ(atpA-atpG) Strr E. coli MG1655, with a group of Δ(atpA-atpG) Strr E. coli MG1655 recipients receiving daily intrarectal applications of ATPγS. Mice were analyzed 1 week later. (A) Representative flow cytometry plots show colonic monocyte and MΦ composition. (B) Quantifications of Ly6C+ and CCR2+ MΦs. (C) Quantifications of total ILC3s and CSF2-producing ILC3s in the colon. (D and E) Each WT colonic MΦ population was assessed for (D) median fluorescence intensity (MFI) of P2X7R staining, with histograms (right) showing staining in P2rx7+/+ versus P2rx7−/− mice, or (E) percentage of P2X7R+ pro–IL-1β+ cells. (F) Feature plots illustrating expression of indicated genes in colonic MΦs/monocytes. (G) Contour plots (top) and quantification (bottom) of colonic MΦs in B6 and Nlrp3−/− littermates. (H and I) Percentage of CSF2-producing colonic ILC3s in Nlrp3−/− and P2rx7−/− mice with respective littermate controls. (J) WT and Nlrp3−/− colonic SILTs were isolated and assessed for percentage of each MΦ population. (K) Colonic LP leukocytes were treated with vehicle control, IL-1β, or ATPγS in the presence of brefeldin A (BFA) and quantified for CSF2+ ILC3s. (L) Schematic overview of how microbiota-derived ATPex regulates ILC3-derived CSF2. For (J), each data point represents pooled SILTs from two or three mice. One-way ANOVA with post hoc Tukey’s test (B to E), two-way ANOVA with post hoc Šidák’s multiple comparisons test (K), or unpaired Student’s t test (G to J) was performed.
ATPex activates purinergic receptors on myeloid cells to promote ILC3-derived CSF2
Colonization of the gut by commensal microbes induces the release of IL-1β by MΦs (43). ATPex engages P2X7R, which can activate NLRP3 to result in the processing of pro–IL-1β release of bioactive IL-1β (44). To identify cells that could respond to ATPex stimulation, we analyzed P2X7R expression on colonic lymphoid and myeloid cells and found that MΦs expressed the highest levels of P2X7R (fig. S4, L to N). P2X7R expression on ILC3s was extremely low (fig. S4N). At the subpopulation level, Tim-4+ and Tim-4−CD4+ MΦs expressed the highest levels of P2X7R. We confirmed that P2X7R staining levels were not an artifact of MΦ autofluorescence, because fluorescence was not detected in P2rx7−/− MΦs (Fig. 4D). A higher proportion of Tim-4+ MΦs coexpressed pro–IL-1β protein compared with Tim-4−CD4− and Tim-4−CD4+ MΦs despite lower mRNA Il1b transcripts (Fig. 4E and fig. S4P). We then determined whether colonic MΦs expressed Nlrp3 and found low but detectable expression across all MΦ subpopulations, including fetal-derived Tim-4+ MΦs (Fig. 4F). DCs expressed Nlrp3, P2rx7, and Il1b but showed lower P2X7R protein levels across all subsets when compared with colonic MΦs (fig. S4, M and O). Therefore, MΦs, most likely Tim-4+ MΦs, rather than DCs or monocytes contribute to steady-state P2X7R-mediated ATPex sensing. These findings also complement previous reports showing that MΦ depletion in the steady state alters microbiota-dependent, IL-1β-driven CSF2 production by ILC3s (9, 25).
To determine whether the absence of Nlrp3 affects the colonic composition of MΦs, we analyzed MΦ subsets in Nlrp3−/− mice and their littermate controls and observed a decrease in Tim-4−CD4− MΦ numbers (Fig. 4G). In line with a decrease in CSF2-dependent MΦs in these mice, Nlrp3−/− and P2rx7−/− mice showed a reduction in ILC3-derived CSF2 (Fig. 4, H and I). Monocyte-derived CCR2+ MΦs and Tim-4−CD4− MΦs were decreased within SILTs of Nlrp3−/− mice (Fig. 4J). To determine whether ILC3s could directly sense and respond to ATPex via CSF2 production despite their low P2X7R levels, we stimulated ILC3s from WT, P2rx7−/−, or Nlrp3−/− mice with either ATPγS or IL-1β. Although IL-1β expectedly increased CSF2-producing ILC3s, stimulation with ATPγS failed to activate ILC3s (Fig. 4K). This demonstrates that ILC3-derived CSF2 production requires NLRP3 and P2X7R expression, likely by MΦs or DCs (Fig. 4L). Together, these data indicate that microbe-derived ATPex regulates colonic monocyte-derived MΦs via P2X7R-dependent NLRP3-mediated IL-1β activation in myeloid cells to induce CSF2 production by ILC3s.
scRNA-seq reveals unique SAM populations
The analysis of MΦ populations in Nlrp3−/− mice suggested differential regional effects on MΦs within LP and SILTs. To incorporate spatial information into the actions of CSF2 on colonic MΦs at higher granularity, we performed scRNA-seq analysis of MΦs isolated from either SILTs or LP of WT or Csf2−/− mice. Live Cx3cr1+/EGFPCcr2+/RFP colonic tissues enabled visual demarcation of SILTs and LP using a fluorescent stereomicroscope (Fig. 5A). Biopsy punches containing colonic SILTs or SILT-free LP were isolated and digested before enrichment for CD11b+ cells by magnetic beads (>90% purity) and subsequent scRNA-seq analysis (Fig. 5A). Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction and combined analysis of all four groups (LPWT, SILTWT, LPCsf2−/−, and SILTCsf2−/−) followed by subsetting and re-analysis of the MΦ/monocyte populations, based on parameters previously detailed, resulted in nine distinct clusters as seen before (Figs. 1A and 5B). Cells within clusters 0, 1, and 7 were enriched in the LP, whereas SILTs primarily comprised clusters 2, 3, 5, and 6, indicating preferential localization of some MΦ subpopulations within SILTs (Fig. 5B). Annotation of the nine clusters based on their differentially expressed genes (DEGs) prompted us to label MΦ clusters 0 and 5, which highly expressed Lyve1, Mrc1, Maf, and Timd4, as Tim-4+ LAMs and Tim-4+ SAMs, respectively. We also identified clusters corresponding to CCR2+ MΦs (cluster 1), monocytes (cluster 4), Tim-4−CD4+ MΦs (cluster 6), RELMα+ MΦs (cluster 7), and epithelium/endothelium-associated Pecam1+ MΦs (cluster 8) (Fig. 1B and fig. S5A). Although previous reports identified Tim-4−CD4+ MΦs and investigated their developmental kinetics, the functions of this subpopulation remain unclear (4). Cluster 6 MΦs, corresponding to Tim-4−CD4+ MΦs, displayed a gene expression pattern similar to that reported in inflammatory microglia and border-associated MΦs, including the expression of Apoe, Ms4a7, and Tmem119 (6, 21, 45, 46). Biological pathways enrichment analysis on MΦ cluster 6 indicated that Tim-4−CD4+ MΦs are involved in leukocyte activation, antigen presentation, and T cell activation, suggesting a proinflammatory role for this subpopulation (Fig. 5C).
Fig. 5. scRNA-seq analysis of colonic LP MΦs inside and outside SILTs predicts their different functions based on localization.
(A) Representative live images [scale bars, 200 μm (left) and 100 μm (right)] of a colonic SILT in Cx3cr1+/GFP Ccr2+/RFP mice and experimental scheme for scRNA-seq setup. Data are subsetted and reclustered for MΦs and monocytes as in Fig. 1A. (B) UMAP projection of MΦs/monocytes of SILTWT and LPWT (left) with quantification of the relative cluster abundance for each sample (right). (C) Pathway enrichment analysis (gProfiler, GO biological processes) using DEGs for each cluster (from Fig. 1B). (D and E) Colonic SAMs and LAMs were isolated, stained with mitochondrial dyes, and assessed for staining MFI as indicated. TH1, T helper 1. (D) MFIs of mitochondrial dyes in total SAMs and LAMs. (E) MitoSOX MFI of each MΦ population in each colonic region. (F) UMAP dimensionality reduction using Monocle 3 was performed and visualized with overlaid Seurat annotations from Fig. 1A. (G and H) Trajectory analysis was performed using Monocle 3 as indicated by solid black lines. (G) Changes in expression of subset-defining genes were visualized in conjunction with trajectory analysis. (H) Pseudotime analysis was performed and visualized using Monocle 3. Unpaired Student’s t test (D) or one-way ANOVAwith post hoc Tukey’s test (E) was performed.
Cluster 2 and 3 MΦs were found almost exclusively in SILTS and expressed high levels of Il22ra2 and Lyz1, markers reported for SILT-residing CD11c+MHCII+CD11b−CD103− DCs in the small intestine (24). However, MΦ identity for cluster 2 and 3 cells was confirmed by their expression of MΦ markers (Csf1r, Cx3cr1, C1qa, Adgre1, and Fcgr1) and the absence of DC markers (Flt3, Dpp4, and Zbtb46) (fig. S1, B to D). The absence of Timd4 and Cd4 expression and the low level of Ccr2 expression in cluster 2 and 3 MΦs suggest that these cells correspond to a subset of the Tim-4−CD4− MΦs (Fig. 1B). Cluster 2 and 3 SAMs were enriched in ATP metabolism, oxidative phosphorylation (OXPHOS), and phagocytic pathways, indicative of a population high in energy demand and possibly involved in antimicrobial host defense (Fig. 5C). This prompted us to characterize mitochondria, as a major bioenergetic organelle, in LAMs and SAMs. Comparing mitochondrial mass (MitoTracker Green), mitochondrial membrane potential (MitoTracker Red) as an indicator of energy metabolism, and the production of reactive oxygen species (ROS) particularly superoxide (SOX) (MitoSOX) in LAMs and SAMs, we found equal mitochondrial numbers in MΦs across LP and SILTs but higher mitochondrial membrane potential and ROS production in SAMs (Fig. 5D). Tim-4+ MΦs had higher numbers of mitochondria compared with all other MΦ subpopulations, as previously reported (47), regardless of colonic localization (fig. S5B). The higher MitoSOX staining in SAMs was attributed to a greater staining in Ly6C+ monocytes and especially Tim-4−CD4− SAMs, in line with the pathway analysis of the scRNA-seq data (Fig. 5, C and E).
Considering these notable differences between LAM and SAM populations, we sought to determine the developmental relationship between each MΦ cluster from our scRNA-seq data and thus performed trajectory analysis using Monocle 3 with Seurat-generated clusters overlaid (Fig. 5F). This analysis predicted a common origin for all LAMs and SAMs within the monocyte cluster 4, supporting our fate-mapping and parabiosis studies (Fig. 5G and fig. S1, I to K). Pseudotime analysis revealed a gradual loss of Ly6c2 and Ccr2 expression and gain of Cd4 and Timd4 expression as cells transitioned from monocytes toward differentiated MΦs (Fig. 5G). Tim-4−CD4+ MΦs represent a differentiation branching point, at which cells transition either into Tim-4+ MΦs (clusters 0 and 5) or into SAMs (first to cluster 3 and then cluster 2) (Fig. 5, F to H). Given that Tim-4−CD4+ MΦs were least affected by perturbations in the microbiota (Fig. 2, A to E), their status as a defining branch point for tissue-resident MΦs may be worth additional investigation in the future. In summary, our scRNA-seq analyses reveal a previously undescribed SILT-associated Tim-4−CD4− MΦ population, high in energy metabolism and ROS production, possibly originating from Tim-4−CD4+ MΦs along a distinct differentiation pathway and within a distinct anatomic location.
CSF2 is a spatial determinant of MΦ development and function in the gut
To determine whether LAMs and SAMs are differentially regulated by CSF2, we first identified MΦ clusters that are most likely to be CSF2 responsive based on their Csf2ra and Csf2rb expression (Fig. 6A). All MΦ clusters except for Timd4+Lyve1+ MΦs (clusters 0, 5, and 8) expressed detectable levels of Csf2ra and Csf2rb (Fig. 6A). Comparison of MΦ cluster composition in LP and SILTs revealed a loss in the relative abundance of cluster 2 and 3 MΦs in Csf2−/− compared with WT (Fig. 6, B and C). Tim-4+ LAMs (cluster 0) were reduced in the LPCsf2−/− (Fig. 6, B and C). We hypothesized that CSF2 deficiency renders colonic monocytes and CCR2+ MΦs unable to differentiate and undergo apoptosis based on CSF2’s role as a prosurvival factor for myeloid cells (48). An assessment of apoptosis in colonic MΦs revealed an increase in apoptotic Ly6C+ and Tim-4−CD4− MΦs in Csf2−/− mice, the latter likely corresponding to the loss of SAMs in SILTCsf2−/− (Fig. 6D).
Fig. 6. CSF2 deficiency results in a loss of Tim-4−CD4− SAMs and functional dysregulation of colonic MΦs.
(A) Feature plots (top) and dot plot (bottom) showing expression of Csf1r, Csf2ra, and Csf2rb in each cluster from the merged data. Color denotes expression level, and dot size indicates percentage of cells within the cluster expressing the gene, as indicated. (B) UMAP projection of MΦs/monocytes of each sample. (C) Quantification of the relative abundance of each cluster for each sample. (D) Quantification of apoptotic cells within each colonic MΦ population in sex-matched littermate WT versus Csf2−/− mice by ApoTracker staining, as indicated. (E and F) Pathway enrichment analysis (gProfiler, GO biological processes) using DEGs differentially regulated between WT versus Csf2−/− in (E) LP and (F) SILT for selected clusters, as indicated. Negative values of −log10 of adjusted P value indicate up-regulation in Csf2−/−, and positive values indicate up-regulation in WT. (G) MitoSOX MFI for each SAM population (top) and LAM population (bottom) in sex-matched littermate WT versus Csf2−/− mice. Each SAM data point represents pooled SILTs from two or three mice. Unpaired Student’s t test (D) or two-way ANOVA with post hoc Šidák’s multiple comparisons test (G) was performed. IFN-γ, interferon-γ; MAPK, mitogen-activated protein kinase; ERK1, extracellular signal–regulated kinase 1.
CSF2 promotes the transition of monocyte to MΦs in the inflamed brain, specifically by supporting disease-promoting functions in MΦs (21). These findings are in contrast to our data showing CSF2-driven pathways in steady-state colonic MΦs. Pathway analysis based on DEGs of each cluster in each region of WT versus Csf2−/− mice revealed differences in multiple MΦ clusters, including those devoid of Csf2ra or Csf2rb mRNAs (Fig. 6, A, E, and F). Within the LPCsf2−/−, Tim-4+ LAMs (red) and CCR2+ MΦs (orange) were impaired in cytoskeleton organization, migration, and endocytosis but enriched in proinflammatory processes such as T cell activation, antigen activation, leukocyte-mediated cytotoxicity, and the response to biotic stimuli (Fig. 6E). An even larger number of MΦ clusters were affected in SILTCsf2−/−. Similar to LPCsf2−/− Tim-4+ LAMs, homeostasis of SILTCsf2−/− Tim-4+ SAMs (aqua) was impaired as shown by altered synapse pruning and wound healing functions, as well as up-regulated proinflammatory pathways (Fig. 6F). Tim-4−CD4+ MΦs (dark blue) followed these trends toward altered homeostasis and increased inflammation. Conversely, clusters corresponding to monocytes (green) and CCR2+ MΦs (orange) down-regulated pathways involved in response to stimuli and were enriched in pathways related to cell survival, glycolipid catabolism, and protein stabilization in SILTCsf2−/−, confirming the increased apoptosis in Csf2−/− mice (Fig. 6, D to F). It is possible that CSF2 itself or its effects on MΦ gene expression profiles as described could affect SILT formation and function. This prompted us to quantify SILT numbers and diameters, revealing higher numbers of smaller SILTs in Csf2−/− mice (fig. S6A). However, these alterations had no significant impact on the production of immunoglobulin A (IgA) in the colon during steady state (fig. S6B) (49, 50). Instead, mitochondrial homeostasis of SAMs was impaired in Csf2−/− animals, indicated by lower ROS production in SILT-associated Ly6C+ monocytes and Tim-4−CD4− SAMs, as well as reduced mitochondrial membrane potential in CCR2+ SAMs (Fig. 6G and fig. S6C). These data show that CSF2 controls mitochondrial functions implicated in energy generation and antimicrobial activity particularly within SAMs. As a result, Csf2−/− colons are enriched in MΦs skewed toward proinflammatory processes, whereas down-regulating functions were attributed to antimicrobial host defense and bioenergy metabolism. Collectively, these findings demonstrate that CSF2-rich SILTs constitute microanatomic niches for the homeostatic development and functional programming of MΦs in the colon.
CSF2-dependent Tim-4−CD4− MΦs are required for host defense against enteric infections
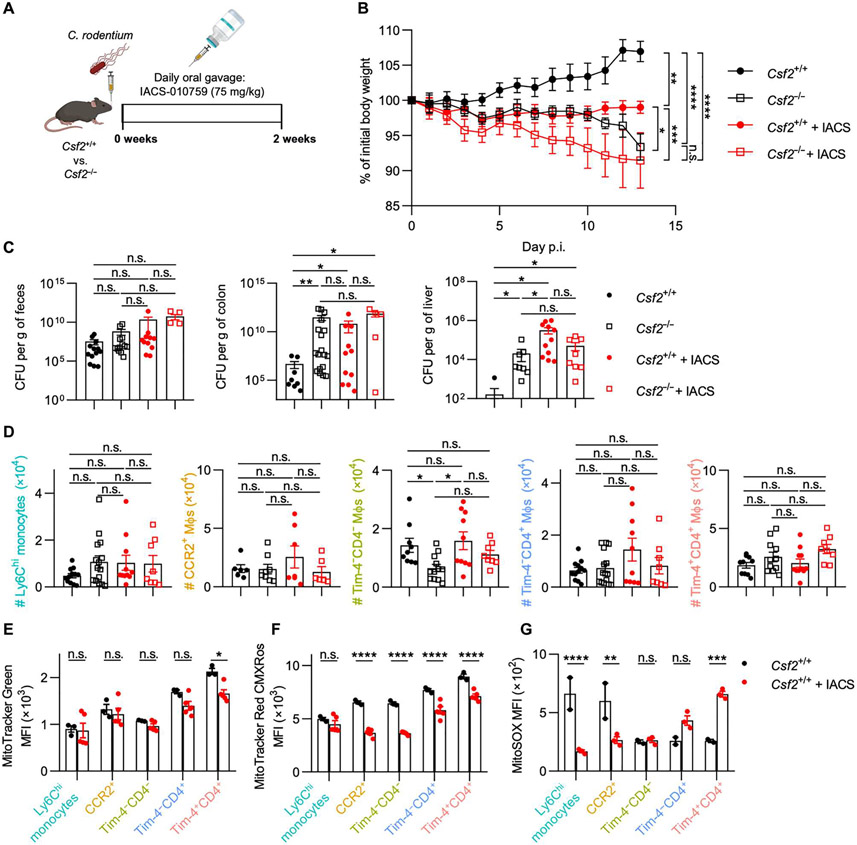
ILC3s are central for the innate immune response against attaching and effacing enteric pathogens such as Citrobacter rodentium, the murine counterpart of human enteropathogenic E. coli (51). CSF2 has been implicated in supporting antimicrobial host defense against C. rodentium through the modulation of CD11c+ myeloid cells (52). In line with this, CD11c-expressing DCs are impaired in their development in Csf2−/− mice as previously reported (fig. S7A) (9). However, in contrast to previous reports using C. rodentium infections, CD11c+MHCII+CD64− DCs did not significantly differ between Csf2−/− and littermate controls after infection, likely because of a CSF2-independent increase in CD103−CD11b+ DCs (fig. S7, B and C). C. rodentium–infected mice had elevated levels of luminal ATPex (fig. S7D). These findings prompted us to revisit whether CSF2-dependent MΦs contribute to the defense against C. rodentium.
Tim-4−CD4− SAMs, the MΦ population most affected in Csf2−/− mice, were characterized by their expression of genes related to OXPHOS, high mitochondrial membrane potential, and elevated ROS production, which were affected in the absence of CSF2. We reasoned that inhibition of OXPHOS in WT mice would partly recapitulate the observed mitochondrial alteration in colonic MΦs of Csf2−/− mice (Figs. 5 and 6). To validate this hypothesis, we compared C. rodentium–infected Csf2−/− and littermate control mice treated daily with IACS-010759 (IACS), an established oral inhibitor of OXPHOS, or vehicle control (Fig. 7A). In line with previous reports, Csf2−/− mice showed greater weight loss and higher pathogen burden and dissemination compared with their WT littermates despite comparable infection efficiency (Fig. 7, B and C) (52). IACS-treated WT mice phenocopied untreated Csf2−/− mice, but disease severity was not exacerbated when OXPHOS was blocked in Csf2−/− mice (Fig. 7, B and C). IACS acted downstream of CSF2 production by ILC3s, because treatment had no effects on the numbers of total or CSF2-producing ILC3s (fig. S7D). More strikingly, OXPHOS inhibition resulted in a decrease in Tim-4−CD4− MΦs, recapitulating the lower Tim-4−CD4− MΦ numbers observed in Csf2−/− mice (Fig. 7D and fig. S7E). In line with the characterization of MΦs in Csf2−/− mice, OXPHOS inhibition had a major impact on the mitochondrial membrane potential across most colonic MΦ subsets and more specific impact on ROS production in Ly6Chi monocytes and CCR2+ MΦs, immediate precursors of Tim-4−CD4− MΦs (Fig. 7, E to G). In summary, these data show that CSF2-dependent host defense against enteric pathogens operates via the regulation mitochondrial energy homeostasis and ROS production imprinted preferentially in SAMs.
Fig. 7. Host defense against C. rodentium requires CSF2-dependent Tim-4−CD4− MΦs and OXPHOS activity.
(A) Experimental set-up of C. rodentium infection and daily oral treatment of OXPHOS inhibitor IACS-010759 (IACS). (B) Body weight was tracked daily during the course of infection. (C) Quantification of C. rodentium in the feces, colon, and liver at end point. (D) Quantification of colonic MΦ populations at end point. (E) Mitochondria counts, (F) mitochondrial membrane potential, and (G) superoxide production of each MΦ population in infected control mice or IACS-treated mice. Mixed-effects analysis with post hoc Šidák’s multiple comparisons test (B), one-way ANOVA with post hoc Tukey’s test (C and D), or two-way ANOVA with post hoc Šidák’s multiple comparisons test (E to G) was performed.
DISCUSSION
Intestinal MΦs are critical for gut homeostasis and host defense. Uncovering the mechanisms regulating their developmental and functional heterogeneity is a critical step in our understanding of these cells. Here, we provide a molecular and cellular framework that governs microbiota- and host-regulated MΦ development in the gut. We demonstrate that microbiota-derived ATPex fuels the production of CSF2 by ILC3s via P2X7R and NLRP3 in myeloid cells and identify SILTs as a distinct niche for CSF2-dependent development and functional programming of SAMs that support antimicrobial defense (fig. S8).
Intestinal SILTs, in contrast to tertiary lymphoid organs (TLOs), form postnatally and mature depending on signals by the gut microbiota (50, 53, 54). Best known to support B cell maturation, our findings indicate that SILTs constitute a developmental hub for monocyte-derived SAMs that support antimicrobial host defense (55). Although ILF-residing Cxcl13-expressing MΦs were shown to support IgA-producing B cells during Salmonella infection, their development is unlikely to be affected by CSF2, because IgA titers were unchanged in Csf2−/− mice (50). We show that monocyte transition into SAMs requires CSF2-dependent regulation of their high mitochondrial membrane potential and ROS production to support host defense against infections. However, their role in regulating adaptive immunity within SILTs remains an open question.
Collectively, our findings propose a mechanistic model explaining how the microbiota, irrespective of strain or kingdom, contributes to the steady-state heterogeneity of intestinal MΦs. In contrast to specific bacteria-derived metabolites such as tryptophan or SCFAs, ATP is abundantly produced across all microbial kingdoms. Here, we demonstrate that ATPex governs the steady-state activation of ILC3s via an NLRP3- and P2X7R-dependent activation of myeloid cells. This is in line with previous reports showing that intestinal MΦs are a major source of steady-state IL-1β and central for the activation of ILC3s (9, 25, 43). ILC3s do not respond to direct stimulation with ATPex but secrete CSF2 in response to IL-1β. Tim-4+ MΦs among other myeloid populations coexpressed the highest levels of P2X7R and pro–IL-1β, making them one possible mediator between microbial ATPex and ILC3 activation. However, Il1b, P2rx7, and Nlrp3 transcripts and P2X7R protein were also detected in DCs and other MΦ subpopulations, opening the possibility for collaboration between these myeloid cells to control ILC3-derived CSF2 via IL-1β. Our findings indicate that ATPex, as a measurement for universal microbial energy metabolism, serves as a rheostat for the control of gut MΦ development in the steady state. However, cell stress or death can contribute to increases in ATPex concentration not originating from the microbiota, which could affect the production of ILC3-derived CSF2. The instability of ATP in the extracellular space and expression of ectonucleotidases by the intestinal epithelium may be a rate-limiting step in the activation of ILC3s and the regulation of MΦ heterogeneity, suggesting a possible feedback loop controlling these interactions.
Although ATPex-mediated activation of the inflammasome drives disease in extraintestinal locations, it constitutes a critical element to tune immune homeostasis in the gut (56). In contrast to its redundant and often proinflammatory role in other organs, CSF2 governs the steady-state transition of monocytes to MΦs in the intestine and controls essential homeostatic functions and defense pathways in gut MΦs (57). We found that ILC3-derived CSF2 promotes survival and differentiation of monocytes and shapes the metabolic and functional program of MΦs within SILTs. Moreover, SAMs followed a distinct developmental trajectory compared with LAMs, diverging from Tim-4−CD4+ MΦs. Deficiency in Csf2 resulted in the loss of SAMs and promoted inflammatory pathways in LAMs even if our gene expression analysis suggests a lack of responsiveness to CSF2. This enrichment of inflammatory pathways in Csf2-deficient MΦs is possibly the result of a misdirected strategy to reinstate antimicrobial host defense and is of translational relevance, considering the presence of mutations in CSF2RB and neutralizing anti-CSF2 autoantibodies in complicated Crohn’s disease (CD) (58, 59). Neutralizing anti-human CSF2 autoantibodies precede the onset of CD by several years and are implicated in elevated subclinical inflammation before diagnosis and disease complications at the time of diagnosis (60). It remains possible that systemic CSF2 deficiency elicits its effects on the intestinal MΦ population indirectly by perturbing critical systemic homeostasis; however, our studies selectively deleting CSF2 in RORγt-dependent ILC3s, highly enriched in the intestine, suggests an important role of CSF2 in maintaining gut-resident MΦs.
In summary, our results suggest that abrogating the steady-state, microbiota-driven, ILC3-CSF2 niche for intestinal MΦ development and function raises the susceptibility to enteric infections that can serve as a trigger for the onset of CD (60). The analysis of MΦs and DCs in C. rodentium–infected Csf2−/− mice refines previous reports and indicates responsiveness of DCs even in the absence of CSF2, whereas MΦs fail to expand and execute their antimicrobial activity because of impaired CSF2-dependent OXPHOS and mitochondrial ROS generation. This is in line with a recent report demonstrating OXPHOS as a distinguishing factor for proinflammatory MΦs, with impairment of OXPHOS leading to cellular stress and reduced MΦ numbers, particularly of alveolar MΦs that are also CSF2 dependent (61, 62). Imprinting of these metabolic functions is predominant in monocytes and SAMs, pointing to a need to develop new tools to refine functional characterization of MΦs within distinct microanatomic locations. Collectively, our data identify an underappreciated role for microbe-derived ATP as a regulator of MΦ development within SILTs guided by ILC3-derived CSF2 and classify SILTs as microanatomic niches for MΦ development to constitute an antimicrobial firewall.
MATERIALS AND METHODS
Study design
We aimed to identify pathways that integrate microbiota- and host-derived signals into the development and function of intestinal MΦs. We used gnotobiotic and transgenic mouse lines to dissect MΦ ontogeny, myeloid growth factor expression, and gene expression of MΦs within spatial niches. We paired mitochondrial characterization and enteric infections to demonstrate that ILC3-derived CSF2, relying on myeloid purinergic signaling triggered by microbiota-derived ATP, supports the differentiation of monocytes to MΦs in the gut. Experiments across this study were conducted at least two to three times on groups of three to five mice of both sexes using age-matched littermate controls. Sequencing data and code are publicly available, and materials used to generate the data are fully disclosed. Figures include representative experiments or pooled data; no outliers were excluded.
Animals
All mice were purchased from the Jackson Laboratory and subsequently bred in house under SPF conditions at the University of Toronto, Division of Comparative Medicine. Unless otherwise stated, all experiments were conducted using 8- to 10-week-old age- and sex-matched littermates of both sexes. Germ-free animals were maintained in the gnotobiotic facility at the University of Toronto, Division of Comparative Medicine. To obtain rewilded mice, pet store mice were purchased from High Oak Ranch Ltd. (Baden, ON) and bred in our mouse facility in a containment room (bioBUBBLE Inc., Fort Collins, CO). C57Bl/6 (B6) pups were cohoused with pet store pups from 3 to 7 weeks of age, separated, and subsequently bred. B6 pregnant dams were gavaged with cecal content from pet store female mice 2 to 3 days before delivery. The pups were used to establish a rewilded colony for experiments. Csf2flox-tdTomato (Csf2fl) were recently described, crossed to Vav1iCre [B6.Cg-Commd10Tg(Vav1-iCre) A2Kio/J; 008610] or RorcCre [B6.FVB-Tg(Rorc-cre)1Litt/J; 022791], and housed at the University of Zurich Laboratory Animal Sciences Center in Zurich, Switzerland (63). All other strains were obtained from the Jackson Laboratory. All experiments were approved by the Faculty of Medicine and Pharmacy Animal Care Committee at the University of Toronto (animal use protocols 20011887 and 20012454 to T.M. and 20012400 to A. Mortha) or the cantonal veterinary office of Zurich (permit number ZH134/2019 to C.S.).
Microbes
All E. coli MG1655 strains were provided by T. Conway (Oklahoma State University). C. rodentium ICC180 was a gift from D. Philpott (University of Toronto). The plasmid encoding the ATP biosensor (pRSET-AT3.10) was a gift from H. Imamura (Kyoto University).
Isolation of intestinal LP leukocytes
Colonic or small intestinal LP cells were isolated as previously described (64). Briefly, intestines were washed in Hanks’ balanced salt solution (HBSS) plus 5 mM EDTA and 10 mM Hepes to strip the epithelium. Tissues were then minced and shaken at 37°C for 20 min in digestion buffer [HBSS with calcium and magnesium supplemented with 10 mM Hepes, 4% fetal bovine serum, penicillin-streptomycin (Sigma-Aldrich), deoxyribonuclease I (0.5 mg/ml; Sigma-Aldrich), and collagenase (0.5 mg/ml; Sigma-Aldrich)]. Supernatants were collected and enriched for leukocytes using a 40/80% Percoll gradient, after which cells were ready for downstream use. To isolate SILTs, intestines were placed under a Zeiss AxioZoom.V16 fluorescent macroscope for live imaging after epithelium stripping. SILTs were isolated using a 1.25-mm biopsy puncher. SILTs and remaining punched out colons were placed separately into R-10+ media until tissue digestion.
C. rodentium infection and pathological assessment
Groups of age- and sex-matched littermates were infected with C. rodentium ICC180 [~2 × 108 colony-forming units (CFUs)] by oral gavage as previously described (65). Mice were weighed daily to monitor disease progression and euthanized at 2 weeks post infection (p.i.) Colons were harvested for LP leukocyte isolation and downstream analysis. CFUs of C. rodentium in the feces, colon, and liver were measured on MacConkey agar plates containing kanamycin (100 μg/ml). To minimize contamination, colons were immediately removed of their luminal content and rinsed in PBS before processing, as previously described (65). The OXPHOS inhibitor IACS-010759 (Chemietek) was dissolved in dimethyl sulfoxide, diluted in 5% methyl cellulose, and administered (7.5 mg/kg) via oral gavage to experimental groups daily, starting 4 hours before C. rodentium infection until the time of analysis.
Intrarectal application of ATPγS
ATPγS tetralithium salt (4080, Bio-Techne) was dissolved in ddH2O and immediately stored at −20°C to avoid degradation. For in vivo application, mice were first anesthetized with isoflurane, and a plastic feeding needle lubricated with sterile jelly gel was used to slowly apply 100 μl of ATPγS (12.5 mg/ml) intrarectally, 4 cm into the colon. Mice were held in an inverted position for at least 30 s to ensure solution remained in the colon before removal of the needle. ATPγS was administered to mice daily from day of E. coli colonization until the day of analysis. Mice in the control group were administered sterile PBS intrarectally.
Luminal ATP measurement
Fecal samples were collected, homogenized in PBS plus 0.01% NaN3 using the Omni Bead Ruptor 24, and centrifuged twice (800g followed by 10,000g) to remove debris and microbes. Supernatants were filtered through a 0.2-μm filter and Amicon Ultra-0.5 centrifugal filter unit (MilliporeSigma) and then analyzed for ATP levels using the ENLITEN ATP Assay System Bioluminescence Detection Kit (Promega) according to the manufacturer’s instructions.
Colonization of germ-free mice with E. coli strains
Groups of age- and sex-matched littermate germ-free mice were orally gavaged with ~103 CFU of E. coli MG1655 WT or E. coli MG1655 ΔnarG ΔnarZ Δ(napD-napA) or ~104 CFU of E. coli MG1655 Strr Δ(atpA-atpG). Differences in starting CFUs accounted for the slower growth rate of E. coli Δ(atpA-atpG) to ensure equal colonization efficiency at the time of analysis. Mice were analyzed 1 week later. Fecal pellets were collected both before gavage and at the time of harvest to confirm colonization.
To determine the localization of the E. coli strains in vivo, SPF mice were treated with ampicillin (1 g/liter) ad libitum for 1 week via drinking water. Mice were then orally gavaged with E. coli MG1655 WT or E. coli MG1655 Strr Δ(atpA-atpG). One week later, fecal content or mucosal scrapings were collected and quantified for ATeam+ bacterial cells.
scRNA sequencing
Sample preparation
Colons (n = 5 per group) from age- and sex-matched Cx3cr1GFP/+ Ccr2RFP/+ WT versus Csf2−/− littermate mice were isolated and stripped of epithelium as detailed above and then placed under a Zeiss AxioZoom.V16 fluorescent macroscope for live imaging. SILTs were identified on the basis of green fluorescent protein (GFP) and red fluorescent protein (RFP) expression and isolated using a 1.25-mm biopsy puncher. SILTs and remaining punched out colons were pooled and placed separately into R-10+ media. Samples were digested and enriched for leukocytes as detailed above. Samples were then enriched for CD11b+ cells using the EasySep Mouse CD11b Positive Selection Kit II (STEMCELL Technologies) as per the manufacturer’s protocol. Purified single-cell suspensions (>90% purity) were resuspended in R-10+ media for 10x Genomics scRNA-seq.
Library preparation, sequencing, preprocessing, and quality control
Single-cell suspensions were prepared and loaded onto the v3 10x Chromium for the generation of sequencing libraries and processing as described by 10x Genomics. CellRanger (10x Genomics) was used to preprocess sequenced cells, align reads, and generate expression matrices. Seurat (v.4.0) was used for all preprocessing, filtering, and downstream analyses (66). Low-quality cells expressing fewer than 200 genes were removed. Doublets and dead cells were excluded on the basis of high number of genes (>6000) and high percentage (>9%) of transcripts mapping to mitochondrial genes, respectively. Cells with high percentage (>20%) of transcripts mapping to dissociation-associated genes, as previously described, were also removed (67).
Normalization, dimensionality reduction, clustering, and cell annotation
To remove technical variation, data were normalized using SCTransform, which uses negative binomial regression to normalize the data, find variable features, and scale the data (68). The variance-stabilizing transformation method in SCTransform was used to select 3000 highly variable features. Mitochondrial gene percentage and number of counts (nCount_RNA) were regressed out. Dimensionality reduction was performed using principal components analysis (PCA), and an elbow plot was used to determine the number of statistically significant PCs for subsequent clustering. FindNeighbors and FindClusters functions were used to perform graph-based clustering. Nonlinear dimensionality reduction and visualization was performed using the UMAP method. Clusters were identified and annotated on the basis of differential gene expression testing using the Wilcoxon rank sum test, with the following parameters in the FindAllMarkers function: min.pct = 0.25, log fold change (logFC) threshold = 0.25, and adjusted P value < 0.05. For heatmaps, each cluster was downsampled to 50 cells for visualization, showing the top 30 DEGs of each cluster.
MΦ and monocyte clusters were identified on the basis of expression of MΦ markers (C1qa, Csf1r, Cx3cr1, and Adgre1) and the absence of DC markers (Flt3, Dpp4, and Zbtb46). These clusters were further subsetted (using the “subset” function), and normalization, dimensionality reduction, and clustering were reperformed as described above to obtain specific MΦ and monocyte clusters. Clusters were identified, annotated, and visualized as described above.
Differential gene expression
To compare the gene expression of MΦs between WT versus Csf2−/− [knockout (KO)] LP and TLO, the “subset” function was used to separate each cluster from each dataset. The FindMarkers function (min.pct = 0.25, logFC threshold = 0.25, and adjusted P value < 0.05) was used to compute differentially up-regulated and down-regulated genes for each cluster in KO relative to WT of each region (e.g., KO LP relative to WT LP and KO TLO relative to WT TLO). Resulting genes were used for subsequent pathway enrichment analysis, as indicated.
Pathway enrichment analysis
gProfiler functional profiling (https://biit.cs.ut.ee/gprofiler/gost) was used to measure over-representation of target gene list against the annotated gene database of Gene Ontology (GO; www.geneontology.org). Enriched biological processes of GO (BP, 2019) and enriched Kyoto Encyclopedia of Genes and Genomes pathways were identified and ordered on the basis of enrichment scores (−log10 of the adjusted P value).
Single-cell trajectory analysis
The R package Monocle 3 was used to assess cell trajectories (69-71). Data previously analyzed with Seurat (v.4.0), as described above, were imported into Monocle 3 for reclustering. Briefly, highly variable genes imported from the Seurat analysis were used for PCA dimensionality reduction, followed by UMAP nonlinear dimensionality reduction and subsequent clustering using Leidan community detection (https://arxiv.org/abs/1802.03426). The number of Monocle clusters was similar to Seurat clusters. This method also generates “partitions” representing groups corresponding to separate trajectories. Cell trajectory was assessed using the “learn_graph” function, which uses the DDRTree method to learn tree-like trajectories and further reduce dimensionality. Data were visualized with UMAP embeddings and trajectories derived within Monocle and overlaid with Seurat clusters.
Quantification and statistical analysis
Statistical analysis of nonsequencing data was performed with the GraphPad Prism software (GraphPad), with statistical tests detailed in the figure legends. All data are shown as means ± SEM. Significant differences were defined at P < 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; n.s., not significant. Unless otherwise stated, data shown in each figure are representative of at least three independent experiments with at least three sex- and age-matched mice per group per experiment.
Supplementary Material
Acknowledgments:
We thank all members of #theonlylabever for the support and discussion. We acknowledge the support by the University of Toronto, Temerty Faculty of Medicine Flow Cytometry Core facility, the 10x Genomics staff at the Princess Margaret Genomics Centre, and the Division for Comparative Medicine. We wish to thank J.-C. Zúñiga-Pflücker and A. Gehring for the critical reading of the manuscript.
Funding:
This study was supported by an Ontario Trillium Scholarship and Vanier Canada Graduate Scholarship–NSERC (P.C.). K.B. is supported by a Canadian Institutes of Health Research (CIHR) Banting Postdoctoral Fellowship Program and the Canadian Allergy, Asthma and Immunology Foundation (CAAIF) and Takeda Canada. L.N. is funded by an Ontario Graduate Scholarship and a NSERC-PGS award. S.L.T. is a recipient of the Dr. Edward Ketchum Graduate Student Scholarship and the Canada Graduate Scholarships–Master’s (CGS M) award. H.H. is supported by the Vanier Canada Graduate Scholarship–CIHR. C.S. is supported by the Swiss National Science Foundation (Eccellenza grant 194216), the Peter Hans Hofschneider Professorship for Molecular Medicine, and the SwissLife Jubiläumsstiftung. C.S. and J.G. are supported by the Research Fund of the Swiss Lung Association, Bern. J.G. is an awardee of the UZH Candoc. T.M. is supported by a Canada Research Chair in NKT cell Immunobiology, a CIHR grant (PJT-175055), and a Canada Foundation for Innovation Physical Infrastructure Grant (29186). A. Mortha is supported by the Canadian Foundation for Innovation John R. Evans Leaders Fund, the CIHR (PJT-388337, 6210100847, and 6210100850), and a NSERC-Discovery Grant (RGPIN-2019-04521). A. Mortha is the Tier 2 Canadian Research Chair in Mucosal Immunology and supported by the Tier 2 CRC-CIHR program (CRC-2021-00511).
Footnotes
Data and materials availability: Sequencing data have been deposited on NCBI GEO under accession no. GSE231371. Information required to reanalyze the data reported are available via https://doi.org/10.5281/zenodo.7883947. All other data needed to support the conclusions of the paper are present in the paper or the Supplementary Materials.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Chiaranunt P, Tai SL, Ngai L, Mortha A, Beyond immunity: Underappreciated functions of intestinal macrophages. Front. Immunol 12, 749708 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dick SA, Wong A, Hamidzada H, Nejat S, Nechanitzky R, Vohra S, Mueller B, Zaman R, Kantores C, Aronoff L, Momen A, Nechanitzky D, Li WY, Ramachandran P, Crome SQ, Becher B, Cybulsky MI, Billia F, Keshavjee S, Mital S, Robbins CS, Mak TW, Epelman S, Three tissue resident macrophage subsets coexist across organs with conserved origins and life cycles. Sci. Immunol 7, eabf7777 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Kang B, Alvarado LJ, Kim T, Lehmann ML, Cho H, He J, Li P, Kim BH, Larochelle A, Kelsall BL, Commensal microbiota drive the functional diversification of colon macrophages. Mucosal Immunol. 13, 216–229 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw TN, Houston SA, Wemyss K, Bridgeman HM, Barbera TA, Zangerle-Murray T, Strangward P, Ridley AJL, Wang P, Tamoutounour S, Allen JE, Konkel JE, Grainger JR, Tissue-resident macrophages in the intestine are long lived and defined by Tim-4 and CD4 expression. J. Exp. Med 215, 1507–1518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Z, Gu Y, Chakarov S, Bleriot C, Kwok I, Chen X, Shin A, Huang W, Dress RJ, Dutertre CA, Schlitzer A, Chen J, Ng LG, Wang H, Liu Z, Su B, Ginhoux F, Fate mapping via Ms4a3-expression history traces monocyte-derived cells. Cell 178, 1509–1525.e19 (2019). [DOI] [PubMed] [Google Scholar]
- 6.De Schepper S, Verheijden S, Aguilera-Lizarraga J, Viola MF, Boesmans W, Stakenborg N, Voytyuk I, Schmidt I, Boeckx B, de Casterlé ID, Baekelandt V, Dominguez EG, Mack M, Depoortere I, De Strooper B, Sprangers B, Himmelreich U, Soenen S, Guilliams M, Berghe PV, Jones E, Lambrechts D, Boeckxstaens G, Self-maintaining gut macrophages are essential for intestinal homeostasis. Cell 175, 400–415.e13 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Muller PA, Koscsó B, Rajani GM, Stevanovic K, Berres ML, Hashimoto D, Mortha A, Leboeuf M, Li XM, Mucida D, Stanley ER, Dahan S, Margolis KG, Gershon MD, Merad M, Bogunovic M, Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell 158, 300–313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matheis F, Muller PA, Graves CL, Gabanyi I, Kerner ZJ, Costa-Borges D, Ahrends T, Rosenstiel P, Mucida D, Adrenergic signaling in muscularis macrophages limits infection-induced neuronal loss. Cell 180, 64–78.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, Merad M, Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science 343, 1249288 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danne C, Ryzhakov G, Martínez-López M, Ilott NE, Franchini F, Cuskin F, Lowe EC, Bullers SJ, Arthur JSC, Powrie F, A large polysaccharide produced by helicobacter hepaticus induces an anti-inflammatory gene signature in macrophages. Cell Host Microbe 22, 733–745.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang PV, Hao L, Offermanns S, Medzhitov R, The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. U.S.A 111, 2247–2252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulthess J, Pandey S, Capitani M, Rue-Albrecht KC, Arnold I, Franchini F, Chomka A, Ilott NE, Johnston DGW, Pires E, McCullagh J, Sansom SN, Arancibia-Cárcamo CV, Uhlig HH, Powrie F, The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 50, 432–445.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goudot C, Coillard A, Villani AC, Gueguen P, Cros A, Sarkizova S, Tang-Huau TL, Bohec M, Baulande S, Hacohen N, Amigorena S, Segura E, Aryl hydrocarbon receptor controls monocyte differentiation into dendritic cells versus macrophages. Immunity 47, 582–596.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Chiaranunt P, Burrows K, Ngai L, Cao EY, Liang H, Tai SL, Streutker CJ, Girardin SE, Mortha A, NLRP1B and NLRP3 control the host response following colonization with the commensal protist tritrichomonas musculis. J. Immunol 208, 1782–1789 (2022). [DOI] [PubMed] [Google Scholar]
- 15.Witmer-Pack MD, Hughes DA, Schuler G, Lawson L, McWilliam A, Inaba K, Steinman RM, Gordon S, Identification of macrophages and dendritic cells in the osteopetrotic (op/op) mouse. J. Cell Sci 104, 1021–1029 (1993). [DOI] [PubMed] [Google Scholar]
- 16.Sehgal A, Donaldson DS, Pridans C, Sauter KA, Hume DA, Mabbott NA, The role of CSF1R-dependent macrophages in control of the intestinal stem-cell niche. Nat. Commun 9, 1272 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai XM, Ryan GR, Hapel AJ, Dominguez MG, Russell RG, Kapp S, Sylvestre V, Stanley ER, Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood 99, 111–120 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Guilliams M, de Kleer I, Henri S, Post S, Vanhoutte L, de Prijck S, Deswarte K, Malissen B, Hammad H, Lambrecht BN, Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med 210, 1977–1992 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greter M, Lelios I, Pelczar P, Hoeffel G, Price J, Leboeuf M, Kündig TM, Frei K, Ginhoux F, Merad M, Becher B, Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity 37, 1050–1060 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schridde A, Bain CC, Mayer JU, Montgomery J, Pollet E, Denecke B, Milling SWF, Jenkins SJ, Dalod M, Henri S, Malissen B, Pabst O, Mcl Mowat A, Tissue-specific differentiation of colonic macrophages requires TGFβ receptor-mediated signaling. Mucosal Immunol. 10, 1387–1399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amorim A, de Feo D, Friebel E, Ingelfinger F, Anderfuhren CD, Krishnarajah S, Andreadou M, Welsh CA, Liu Z, Ginhoux F, Greter M, Becher B, IFNγ and GM-CSF control complementary differentiation programs in the monocyte-to-phagocyte transition during neuroinflammation. Nat. Immunol 23, 217–228 (2022). [DOI] [PubMed] [Google Scholar]
- 22.Bain CC, Bravo-Blas A, Scott CL, Gomez Perdiguero E, Geissmann F, Henri S, Malissen B, Osborne LC, Artis D, Mowat AMI, Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat. Immunol 15, 929–937 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moura Silva H, Kitoko JZ, Queiroz CP, Kroehling L, Matheis F, Yang KL, Reis BS, Ren-Fielding C, Littman DR, Bozza MT, Mucida D, Lafaille JJ, c-MAF-dependent perivascular macrophages regulate diet-induced metabolic syndrome. Sci. Immunol 6, eabg7506 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Guendel F, Kofoed-Branzk M, Gronke K, Tizian C, Witkowski M, Cheng HW, Heinz GA, Heinrich F, Durek P, Norris PS, Ware CF, Ruedl C, Herold S, Pfeffer K, Hehlgans T, Waisman A, Becher B, Giannou AD, Brachs S, Ebert K, Tanriver Y, Ludewig B, Mashreghi MF, Kruglov AA, Diefenbach A, Group 3 innate lymphoid cells program a distinct subset of IL-22BP-producing dendritic cells demarcating solitary intestinal lymphoid tissues. Immunity 53, 1015–1032.e8 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Castro-Dopico T, Fleming A, Dennison TW, Ferdinand JR, Harcourt K, Stewart BJ, Cader Z, Tuong ZK, Jing C, Lok LSC, Mathews RJ, Portet A, Kaser A, Clare S, Clatworthy MR, GM-CSF calibrates macrophage defense and wound healing programs during intestinal infection and inflammation. Cell Rep. 32, 107857 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott CL, Zheng F, de Baetselier P, Martens L, Saeys Y, de Prijck S, Lippens S, Abels C, Schoonooghe S, Raes G, Devoogdt N, Lambrecht BN, Beschin A, Guilliams M, Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun 7, 10321 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Theurl I, Hilgendorf I, Nairz M, Tymoszuk P, Haschka D, Asshoff M, He S, Gerhardt LMS, Holderried TAW, Seifert M, Sopper S, Fenn AM, Anzai A, Rattik S, McAlpine C, Theurl M, Wieghofer P, Iwamoto Y, Weber GF, Harder NK, Chousterman BG, Arvedson TL, McKee M, Wang F, Lutz OMD, Rezoagli E, Babitt JL, Berra L, Prinz M, Nahrendorf M, Weiss G, Weissleder R, Lin HY, Swirski FK, On-demand erythrocyte disposal and iron recycling requires transient macrophages in the liver. Nat. Med 22, 945–951 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bleriot C, Chakarov S, Ginhoux F, Determinants of resident tissue macrophage identity and function. Immunity 52, 957–970 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Knoop KA, Gustafsson JK, McDonald KG, Kulkarni DH, Coughlin PE, McCrate S, Kim D, Hsieh CS, Hogan SP, Elson CO, Tarr PI, Newberry RD, Microbial antigen encounter during a preweaning interval is critical for tolerance to gut bacteria. Sci. Immunol 2, eaao1314 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chudnovskiy A, Mortha A, Kana A, Kennard A, Ramirez JD, Rahman A, Remark R, Mogno I, Ng R, Gnjatic S, Amir EAD, Solovyov A, Greenbaum B, Clemente J, Faith J, Belkaid Y, Grigg ME, Merad M, Host-protozoan interactions protect from mucosal infections through activation of the inflammasome. Cell 167, 444–456.e14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guilliams M, Thierry GR, Bonnardel J, Bajenoff M, Establishment and maintenance of the macrophage niche. Immunity 52, 434–451 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Hamada H, Hiroi T, Nishiyama Y, Takahashi H, Masunaga Y, Hachimura S, Kaminogawa S, Takahashi-Iwanaga H, Iwanaga T, Kiyono H, Yamamoto H, Ishikawa H, Identification of multiple isolated lymphoid follicles on the antimesenteric wall of the mouse small intestine. J. Immunol 168, 57–64 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Blaut M, Clavel T, Metabolic diversity of the intestinal microbiota: Implications for health and disease. J. Nutr 137, 751S–755S (2007). [DOI] [PubMed] [Google Scholar]
- 34.Patnode ML, Guruge JL, Castillo JJ, Couture GA, Lombard V, Terrapon N, Henrissat B, Lebrilla CB, Gordon JI, Strain-level functional variation in the human gut microbiota based on bacterial binding to artificial food particles. Cell Host Microbe 29, 664–673.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, Takeda K, ATP drives lamina propria TH17 cell differentiation. Nature 455, 808–812 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Perruzza L, Gargari G, Proietti M, Fosso B, D’Erchia AM, Faliti CE, Rezzonico-Jost T, Scribano D, Mauri L, Colombo D, Pellegrini G, Moregola A, Mooser C, Pesole G, Nicoletti M, Norata GD, Geuking MB, McCoy KD, Guglielmetti S, Grassi F, T follicular helper cells promote a beneficial gut ecosystem for host metabolic homeostasis by sensing microbiota-derived extracellular ATP. Cell Rep. 18, 2566–2575 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mempin R, Tran H, Chen C, Gong H, Kim Ho K, Lu S, Release of extracellular ATP by bacteria during growth. BMC Microbiol. 13, 301 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones SA, Chowdhury FZ, Fabich AJ, Anderson A, Schreiner DM, House AL, Autieri SM, Leatham MP, Lins JJ, Jorgensen M, Cohen PS, Conway T, Respiration of Escherichia coli in the mouse intestine. Infect. Immun 75, 4891–4899 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imamura H, Huynh Nhat KP, Togawa H, Saito K, Iino R, Kato-Yamada Y, Nagai T, Noji H, Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl. Acad. Sci. U.S.A 106, 15651–15656 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hapfelmeier S, Lawson MAE, Slack E, Kirundi JK, Stoel M, Heikenwalder M, Cahenzli J, Velykoredko Y, Balmer ML, Endt K, Geuking MB, Curtiss R III, McCoy KD, Macpherson AJ, Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macpherson AJ, Uhr T, Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 303, 1662–1665 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Satoh-Takayama N, Vosshenrich CAJ, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention JJ, Thiam K, Cerf-Bensussan N, Mandelboim O, Eberl G, di Santo JP, Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity 29, 958–970 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Shaw MH, Kamada N, Kim YG, Nunez G, Microbiota-induced IL-1β, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J. Exp. Med 209, 251–258 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perregaux D, Gabel CA, Interleukin-1 beta maturation and release in response to ATP and nigericin. Evidence that potassium depletion mediated by these agents is a necessary and common feature of their activity. J. Biol. Chem 269, 15195–15203 (1994). [PubMed] [Google Scholar]
- 45.Satoh J, Kino Y, Asahina N, Takitani M, Miyoshi J, Ishida T, Saito Y, TMEM119 marks a subset of microglia in the human brain. Neuropathology 36, 39–49 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Sankowski R, Böttcher C, Masuda T, Geirsdottir L, Sagar, Sindram E, Seredenina T, Muhs A, Scheiwe C, Shah MJ, Heiland DH, Schnell O, Grün D, Priller J, Prinz M, Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nat. Neurosci 22, 2098–2110 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Scott NA, Lawson MAE, Hodgetts RJ, le Gall G, Hall LJ, Mann ER, Macrophage metabolism in the intestine is compartment specific and regulated by the microbiota. Immunology 166, 138–152 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wan CK, Oh J, Li P, West EE, Wong EA, Andraski AB, Spolski R, Yu ZX, He J, Kelsall BL, Leonard WJ, The cytokines IL-21 and GM-CSF have opposing regulatory roles in the apoptosis of conventional dendritic cells. Immunity 38, 514–527 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Velaga S, Herbrand H, Friedrichsen M, Jiong T, Dorsch M, Hoffmann MW, Förster R, Pabst O, Chemokine receptor CXCR5 supports solitary intestinal lymphoid tissue formation, B cell homing, and induction of intestinal IgA responses. J. Immunol 182, 2610–2619 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Koscsó B, Kurapati S, Rodrigues RR, Nedjic J, Gowda K, Shin C, Soni C, Ashraf AZ, Purushothaman I, Palisoc M, Xu S, Sun H, Chodisetti SB, Lin E, Mack M, Kawasawa YI, He P, Rahman ZSM, Aifantis I, Shulzhenko N, Morgun A, Bogunovic M, Gut-resident CX3CR1hi macrophages induce tertiary lymphoid structures and IgA response in situ. Sci. Immunol 5, eaax0062 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song C, Lee JS, Gilfillan S, Robinette ML, Newberry RD, Stappenbeck TS, Mack M, Cella M, Colonna M, Unique and redundant functions of NKp46+ ILC3s in models of intestinal inflammation. J. Exp. Med 212, 1869–1882 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hirata Y, Egea L, Dann SM, Eckmann L, Kagnoff MF, GM-CSF-facilitated dendritic cell recruitment and survival govern the intestinal mucosal response to a mouse enteric bacterial pathogen. Cell Host Microbe 7, 151–163 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eberl G, Lochner M, The development of intestinal lymphoid tissues at the interface of self and microbiota. Mucosal Immunol. 2, 478–485 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Overacre-Delgoffe AE, Bumgarner HJ, Cillo AR, Burr AHP, Tometich JT, Bhattacharjee A, Bruno TC, Vignali DAA, Hand TW, Microbiota-specific T follicular helper cells drive tertiary lymphoid structures and anti-tumor immunity against colorectal cancer. Immunity 54, 2812–2824.e4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsuji M, Suzuki K, Kitamura H, Maruya M, Kinoshita K, Ivanov II, Itoh K, Littman DR, Fagarasan S, Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity 29, 261–271 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Aganna E, Martinon F, Hawkins PN, Ross JB, Swan DC, Booth DR, Lachmann HJ, Bybee A, Gaudet R, Woo P, Feighery C, Cotter FE, Thome M, Hitman GA, Tschopp J, McDermott M, Association of mutations in the NALP3/CIAS1/PYPAF1 gene with a broad phenotype including recurrent fever, cold sensitivity, sensorineural deafness, and AA amyloidosis. Arthritis Rheum. 46, 2445–2452 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Achuthan AA, Lee KMC, Hamilton JA, Targeting GM-CSF in inflammatory and autoimmune disorders. Semin. Immunol 54, 101523 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Han X, Uchida K, Jurickova I, Koch D, Willson T, Samson C, Bonkowski E, Trauernicht A, Kim MO, Tomer G, Dubinsky M, Plevy S, Kugathsan S, Trapnell BC, Denson LA, Granulocyte-macrophage colony-stimulating factor autoantibodies in murine ileitis and progressive ileal Crohn’s disease. Gastroenterology 136, 1261–1271.e3 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chuang LS, Villaverde N, Hui KY, Mortha A, Rahman A, Levine AP, Haritunians T, Evelyn Ng SM, Zhang W, Hsu NY, Facey JA, Luong T, Fernandez-Hernandez H, Li D, Rivas M, Schiff ER, Gusev A, Schumm LP, Bowen BM, Sharma Y, Ning K, Remark R, Gnjatic S, Legnani P, George J, Sands BE, Stempak JM, Datta LW, Lipka S, Katz S, Cheifetz AS, Barzilai N, Pontikos N, Abraham C, Dubinsky MJ, Targan S, Taylor K, Rotter JI, Scherl EJ, Desnick RJ, Abreu MT, Zhao H, Atzmon G, Pe’er I, Kugathasan S, Hakonarson H, McCauley JL, Lencz T, Darvasi A, Plagnol V, Silverberg MS, Muise AM, Brant SR, Daly MJ, Segal AW, Duerr RH, Merad M, McGovern DPB, Peter I, Cho JH, A frameshift in CSF2RB predominant among Ashkenazi jews increases risk for Crohn’s disease and reduces monocyte signaling via GM-CSF. Gastroenterology 151, 710–723.e2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mortha A, Remark R, Del Valle DM, Chuang L-S, Chai Z, Alves I, Azevedo C, Gaifem J, Martin J, Petralia F, Tuballes K, Barcessat V, Tai SL, Huang H-H, Laface I, Jerez YA, Boschetti G, Villaverde N, Wang MD, Korie UM, Murray J, Choung R-S, Sato T, Laird RM, Plevy S, Rahman A, Torres J, Porter C, Riddle MS, Kenigsberg E, Pinho SS, Cho JH, Merad M, Colombel J-F, Gnjatic S, Neutralizing anti-granulocyte macrophage-colony stimulating factor autoantibodies recognize post-translational glycosylations on granulocyte macrophage-colony stimulating factor years before diagnosis and predict complicated Crohn’s disease. Gastroenterology 163, 659–670 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dranoff G, Crawford AD, Sadelain M, Ream B, Rashid A, Bronson RT, Dickersin GR, Bachurski CJ, Mark EL, Whitsett JA, Mulligan RC, Involvement of granulocyte-macrophage colony-stimulating factor in pulmonary homeostasis. Science 264, 713–716 (1994). [DOI] [PubMed] [Google Scholar]
- 62.Wculek SK, Heras-Murillo I, Mastrangelo A, Mañanes D, Galán M, Miguel V, Curtabbi A, Barbas C, Chandel NS, Enríquez JA, Lamas S, Sancho D, Oxidative phosphorylation selectively orchestrates tissue macrophage homeostasis. Immunity 56, 516–530.e9 (2023). [DOI] [PubMed] [Google Scholar]
- 63.Gschwend J, Sherman SPM, Ridder F, Feng X, Liang HE, Locksley RM, Becher B, Schneider C, Alveolar macrophages rely on GM-CSF from alveolar epithelial type 2 cells before and after birth. J. Exp. Med 218, e20210745 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chiaranunt P, Burrows K, Ngai L, Mortha A, Isolation of mononuclear phagocytes from the mouse gut. Methods Enzymol. 632, 67–90 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Bouladoux N, Harrison OJ, Belkaid Y, The mouse model of infection with Citrobacter rodentium. Curr. Protoc. Immunol 119, 19.1511–19.15.25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hao Y, Hao S, Andersen-Nissen E, Mauck WM III, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O’Flanagan CH, Campbell KR, Zhang AW, Kabeer F, Lim JLP, Biele J, Eirew P, Lai D, Pherson AM, Kong E, Bates C, Borkowski K, Wiens M, Hewitson B, Hopkins J, Pham J, Ceglia N, Moore R, Mungall AJ, Alpine JNM; CRUK IMAXT Grand Challenge Team, Shah SP, Aparicio S, Dissociation of solid tumor tissues with cold active protease for single-cell RNA-seq minimizes conserved collagenase-associated stress responses. Genome Biol. 20, 210 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hafemeister C, Satija R, Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL, The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C, Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, Trapnell C, Shendure J, The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.