Abstract
A short sequence, located between the A14L and A15L open reading frames (ORFs) of vaccinia virus, was predicted to encode a hydrophobic protein of 53 amino acids that is conserved in orthopoxviruses, leporipoxviruses, yatapoxiruses, and molluscipoxviruses. We constructed a recombinant vaccinia virus with a 10-codon epitope tag appended to the C terminus of the A14.5L ORF. Synthesis of the tagged protein occurred at late times and was blocked by an inhibitor of DNA replication, consistent with regulation by a predicted late promoter just upstream of the A14.5L ORF. Hydrophobicity of the protein was demonstrated by extraction into the detergent phase of Triton X-114. The protein was associated with purified vaccinia virus particles and with membranes of immature and mature virions that were visualized by electron microscopy of infected cells. Efficient release of the protein from purified virions occurred after treatment with a nonionic detergent and reducing agent. A mutant virus, in which the A14.5L ORF was largely deleted, produced normal-size plaques in several cell lines, and the yields of infectious intra- and extracellular viruses were similar to those of the parent. In contrast, with a mouse model, mutant viruses with the A14.5L ORF largely deleted were attenuated relative to that of the parental virus or a mutant virus with a restored A14.5L gene.
The poxviruses, of which vaccinia virus is the prototype, are large, enveloped, DNA viruses that replicate in the cytoplasm of infected cells (19). Analysis of the genome of vaccinia virus revealed approximately 200 largely nonoverlapping open reading frames (ORFs) of at least 60 amino acids (12). The lower limit for the number of amino acids, however, was arbitrary, and some shorter ORFs may be significant. For example, in the sequenced Copenhagen strain of vaccinia virus, the ORF encoding one of the RNA polymerase subunits was less than 60 amino acids, although it is 63 amino acids long in the WR strain (2). More recently, an analysis of the genome of molluscum contagiosum virus revealed two nonoverlapping ORFs that are conserved in orthopoxviruses and are predicted to encode hydrophobic proteins of less than 60 amino acids (24). In vaccinia virus, one of these putative membrane proteins is 53 amino acids long and is located between ORFs A14L and A15L. This short ORF, named A14.5L, is the subject of the present study.
Poxviruses assemble two types of membranes that form the outer layers of the intracellular mature virion (IMV) and the extracellular enveloped virion (EEV). Thus far, 11 vaccinia virus proteins have been reported to be associated with the IMV membrane (16, 28), and 6 have been associated with the EEV membrane (8, 11, 13, 15, 21–23). Here, we demonstrated that the A14.5L ORF was expressed at late times after vaccinia virus infection and the protein product was incorporated into the IMV membrane. Although the protein was not essential for vaccinia virus replication in tissue culture cells, deletion of the ORF reduced the virulence of vaccinia virus in mice.
MATERIALS AND METHODS
Cells and viruses.
CV-1 (ATCC CCL-70) and BS-C-1 (ATCC CCL-26) cells were grown in Earle's minimal essential medium (Quality Biologicals, Inc.) containing 10% fetal bovine serum in a 5% CO2 atmosphere at 37°C. Vaccinia virus strain WR (ATCC VR-1354) and recombinant vaccinia virus vT7lacOI (1) were propagated in HeLa cells as previously described (9).
Plasmids.
The A14.5L ORF, modified to contain an NdeI restriction endonuclease site at the initiation codon and the influenza virus hemagglutinin (HA) epitope tag sequence YPYDVPDYAS followed by a termination signal and a BamHI site, was generated by PCR with vaccinia virus genomic DNA and oligonucleotide primers TB27 (GGGGGGGGGCATATGATAAGTAATTACGAGCCGTTGC) and TB15 (GGGGGATCCTTAAGATGCATAGTCAGGAACGTCATAAGGATAAATCGCCGAATGAACAAAG). The PCR product was cut with NdeI and BamHI and inserted into the corresponding sites in plasmid pVOTE.2 (30) to produce plasmid pVote-27.
A 518-bp PCR product corresponding to the downstream-flanking region of the A14.5L gene (right flank) was produced from viral genomic DNA with primers TB17 (GGGACTAGTAACGTTAACTCTTTAGCGTTAAAAAATTCCCAAGCGG) and TB18 (GGGGTCGACCGGCTCGTAATTACTTATCATTTACTAGACG), which contain SpeI and SalI sites, respectively, at the 5′ ends. The left end of the right flank was 21 bp upstream of the A14.5L initiation codon and included the stop codon of the A15L gene. This fragment was cloned into SpeI and SalI sites of plasmid pZIPPY, which contains Escherichia coli neomycin resistance (neo) and β-glucuronidase (gus) genes regulated by vaccinia virus promoters and flanked by multiple cloning sites (a gift of T. Shors), producing plasmid pZIPPY-17.
A 586-bp PCR product corresponding to the region upstream of the A14.5L gene (right flank) was produced from viral DNA by using primers TB19 (GGGGAGCTCTTATATGGTTTATATTCCACTTTGTTCATTCGGC) and TB20 (GGGCCGCGGAACTTGGTAT T T T T TGGT TCTGGAGGAGGCG), which contain SstI and SstII sites, at their respective 5′ ends. The right end of this flank region started 57 bp upstream of the A14L initiation codon and included the late promoter sequences of the A14L gene. This DNA was cloned into pZIPPY-17, producing plasmid pZIPPY-1719.
A 509-bp PCR product corresponding to the region downstream of the A14.5L gene (right flank) was produced from viral DNA by using primers TB38 (GGGGGGACTAGTACATTGGTTGTAATATCTGTTATTTTC) and TB67 (GGGGGGG TCGAC T TAAGATGCATAG TCAGGAACG TCATAAGGATAAATCGCCGAATGAACAAAGTGGAATATAAACC), which contain SpeI and SalI sites, respectively, at the 5′ ends. The left end of this region contained the HA epitope sequence followed by the stop codon of the A14.5L gene. This DNA was inserted into the SpeI and SalI regions of the plasmid pZIPPY, producing plasmid pZIPPY-67. The left flank sequence, described above, was cloned into SstI and SstII regions of the plasmid pZIPPY-67, producing plasmid pZIPPY-6719.
Construction of recombinant viruses.
Recombinant viruses were made essentially as described previously, and the genome modifications were confirmed by PCR (10). vT7lacOI-A14.5LHA (code no. v27) was derived by infection of CV-1 cells with vT7lacOI, transfection with pVOTE-27, and selection with mycophenolic acid. vA14.5LHA (code no. v67) and vA14.5LΔneo/gus (code no. v17) were derived by infecting CV-1 cells with vaccinia virus strain WR and transfecting them with pZIPPY-6719 and pZIPPY-1719, respectively, selecting for two rounds of replication in the presence of 2 mg of Geneticin (Gibco Life Technologies) per ml, and picking plaques that stained blue with 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Glu). vA14.5+ (code no. v207) was derived by infecting BS-C-1 cells with vA14.5LΔneo/gus, transfecting them with a 1,440-bp PCR product containing the intact A14.5L gene obtained with primers TB149 (TCGATATAATCATCGTAACTGCTTCTAACGG) and TB152 (GTTACATGAGTGCACCGACGAATCTAG), and isolating plaques that did not stain with X-Glu. vA14.5LΔ (code number vΔ10) was derived by infecting BS-C-1 cells with vA14.5LΔneo/gus and transfecting them with a 1,327-bp PCR product obtained with primers TB149 and TB150 (CGTCTAGTAAATGATAAGTAATTTGGTTTATATTCCACTTTGTTC) and TB152 and TB151 (GAACAAAGTGGAATATAAACCAAATTACTTATCATTTACTAGACG) and then using both products for PCR with primers TB149 and TB152 and isolating plaques that did not stain with X-Glu.
SDS-PAGE and immunoblotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12.5% polyacrylamide) was performed as described previously (17), and proteins were electrophoretically transferred to a nitrocellulose membrane. The blot was incubated in 5% nonfat milk in phosphate-buffered saline (PBS) overnight at 4°C and then for 1 h with monoclonal antibody (MAb) 12CA5 to the HA epitope tag (Covance, Richmond, Calif.) or with an antipeptide serum to the predicted N-terminal amino acids of the mature A17L protein (4) in PBS. The blot was washed three times in PBS before incubation for 1 h with secondary antibody (antirabbit or antimouse) coupled to alkaline phosphatase (Sigma) or with 125I-labelled protein A (Amersham Pharmacia). The blot was washed several times with PBS and developed with the BCIP-NBT (5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium) phosphatase substrate system (Kirkegaard & Perry Laboratories, Inc.) or exposed to Kodak Biomax MS film.
Analysis of virion extracts.
Vaccinia virus virions, purified by sucrose gradient centrifugation (9) from cells infected with vaccinia virus WR or vA14.5LHA, were incubated at 37°C for 30 min in 10 mM Tris (pH 7.4) or in 1% NP-40 in 10 mM Tris (pH 7.4) in the presence or absence of 10 mM dithiothreitol. Soluble and insoluble fractions were separated by centrifugation in a 42.2 Ti rotor at 30,000 × g for 30 min and resuspended to equal volumes in SDS-containing sample buffer. Equivalent amounts of each fraction were loaded on a 12.5% polyacrylamide gel and subjected to electrophoresis. The separated proteins were transferred to a nitrocellulose membrane and analyzed by Western blotting with anti-HA MAb 12CA5. The blot was washed and incubated with sheep anti-mouse antiserum conjugated to horseradish peroxidase (Amersham Life Sciences) followed by Supersignal West Pico chemiluminescent substrate (Pierce, Rockford, Ill.). The signal was visualized by exposing the blot to Biomax ML film (Kodak).
Electron microscopy.
BS-C-1 cells were grown in 60-mm-diameter dishes and infected with either vA14.5LHA or vaccinia virus strain WR at a multiplicity of infection of 10. After 18 h, the cells were fixed and prepared for freezing as previously described (33). Ultrathin sections were cut with a Leica/Reichert Ultracut FSC, collected on formvar-coated grids and stained using standard protocols. The primary antibody used was mHA.11, a mouse monoclonal immunoglobulin G1 (IgG1) to the HA tag (Covance). Grids were subsequently incubated with a secondary antibody, rabbit IgG fraction to whole mouse IgG (Cappel-ICN Pharmaceuticals, Aurora, Ohio), followed by protein A conjugated to 10-nm-diameter colloidal gold particles (Department of Cell Biology, Utrecht University School of Medicine, Utrecht, The Netherlands). Immunostained sections were viewed with a Philips CM100 transmission electron microscope.
Determination of virulence in mice.
The virulence of vaccinia virus was determined essentially as described previously (18, 29, 32). Groups (n = 4) of 5-week-old female BALB/c mice weighing between 15 and 20 g were anesthetized and inoculated intranasally with 103 to 106 PFU of purified vaccinia virus strain WR or mutant viruses in 10 μl of 10 mM Tris-HCl (pH 9.0). Titers of aliquots of the viruses used for infection were redetermined in order to confirm the doses administered. Individual animals were weighed daily, and those that lost greater than 30% of their original body weight were terminated.
RESULTS
Expression of the A14.5L ORF.
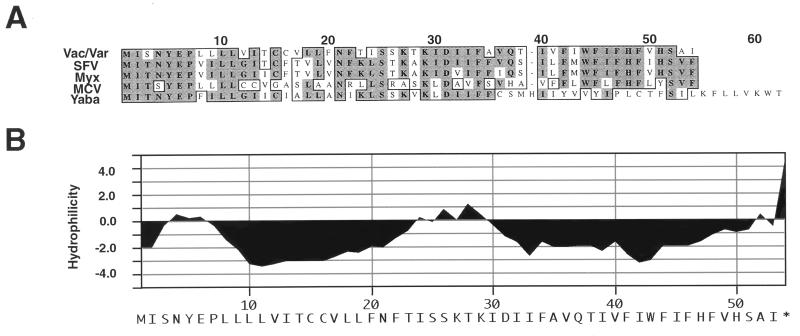
The 53-amino-acid coding sequence, located between the vaccinia virus A14L and A15L ORFs, was designated A14.5L. Analysis of the DNA sequences of two isolates of vaccinia virus (3, 12), two isolates of variola major virus (26), and a variola minor virus isolate (27) indicated the presence of ORFs of the same length and sequence indicating a high degree of conservation among orthopoxviruses. In addition closely related proteins were predicted to be encoded by members of other Chordipoxvirus genera, including two leporipoxviruses (6, 31), a yatapoxvirus (Medline accession no. AB015885), and a molluscipoxvirus (25) (Fig. 1A). As indicated in Fig. 1B, the A14.5L protein was predicted to be largely hydrophobic. Even though such hydrophobic proteins are usually poorly immunogenic, peptides corresponding to the N terminus, C terminus, or the short, central, slightly hydrophilic region were conjugated to keyhole limpet hemocyanin and used to immunize rabbits. Disappointingly, when blots containing lysates of vaccinia virus-infected cells were probed with the antisera, no protein of the expected size was detected. Therefore, we tried another way to identify the putative A14.5L protein.
FIG. 1.
Sequence homology and predicted hydrophobicity of the A14.5L ORF. (A) The sequence of the vaccinia virus (Vac) A14.5L ORF and the identical variola virus (Var) ORF is shown aligned with the orthologous ORFs of Shope fibroma virus (SFV), myxoma virus (Myx), molluscum contagiosum virus (MCV), and Yaba monkey tumor virus (Yaba). The numbers correspond to the codons of the longest ORF. (B) Hydrophilicity plot of the vaccinia virus A14.5L ORF.
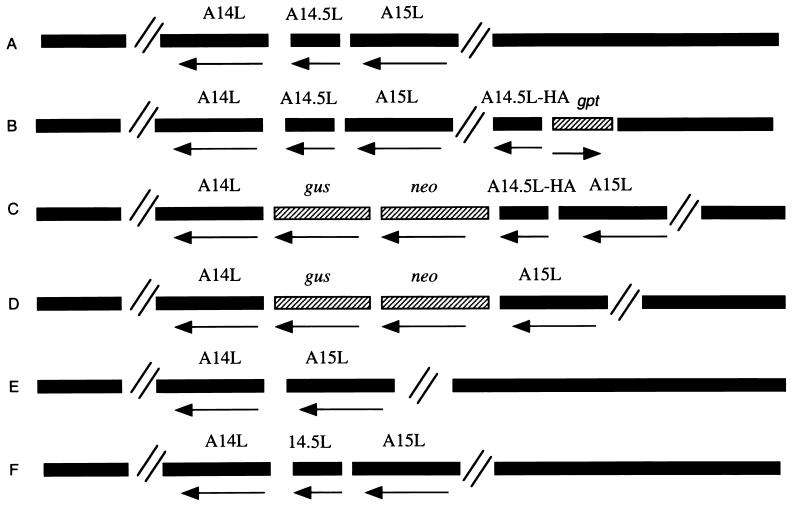
Ten codons representing the influenza virus HA epitope tag were inserted between the C-terminal and stop codons of the A14.5L ORF so that the protein product could be detected with an available MAb. For the initial experiment, an inducible vaccinia virus expression system (30) was chosen to provide a high level of protein synthesis, permit the retention of the unmodified A14.5L ORF in case it was essential, and allow stringent regulation should the HA-tagged version of the A14.5L protein exert a dominant-negative effect on virus replication. The HA-tagged A14.5L ORF was constructed and amplified by PCR and cloned into the pVOTE.2 transfer vector (30). The gene was then inserted by homologous recombination into vaccinia virus vT7lacOI, which contains an inducible copy of the T7 RNA polymerase gene and the E. coli lac repressor under control of a constitutive early/late promoter (1). The recombinant virus vT7lacOI-A14.5LHA was isolated by selection with mycophenolic acid and repeatedly plaque purified. The genomes of the parental vaccinia virus strain WR and vT7lacOI-A14.5LHA are represented in Fig. 2A and B, respectively.
FIG. 2.
Representations of parental and recombinant viral genomes. (A) Parental vaccinia virus strain WR. (B) vT7lacOI-A14.5LHA. (C) vA14.5LHA. (D) vA14.5LΔneo/gus. (E) vA14.5LΔ. (F) vA14.5L+. The vaccinia virus ORFs A14L, A14.5L, and A15L; the antibiotic selection markers neo and gpt; and the reporter gene gus are indicated above the bars. An arrow below the bar indicates the direction of transcription of each ORF. The symbol // signifies additional portions of the vaccinia virus genome. vT7lacOI-A14.5LHA contains a T7 RNA polymerase gene, an E. coli lac repressor gene, and a T7 promoter and lac operator regulating expression of A14.5L-HA, which are not shown. VA14.5L+ is a revertant of vA14.5LΔneo/gus and has the same gene arrangement as the parental vaccinia virus.
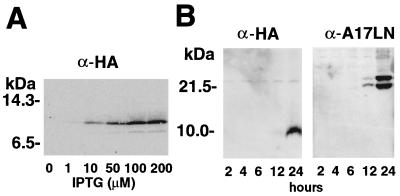
Lysates of cells infected with vT7lacOI-A14.5LHA in the presence of various concentrations of IPTG (isopropyl-β-d-thiogalactopyranoside) were subjected to SDS-PAGE and the proteins were transferred to a membrane and probed with an anti-HA MAb. An IPTG-inducible polypeptide of 10 kDa was detected, somewhat larger than the 7 kDa predicted for the HA-tagged protein (Fig. 3A). Maximal expression occurred at 100 to 200 mM IPTG. The mobility of the A14.5L-HA protein was unchanged by treatment with endo-F–peptide–N-glycosidase F (Oxford Glycosystem, Rosedale, N.Y.) for 3 h at 37°C in 1% NP-40, indicating that the potential N-glycosylation site (NFT, Fig. 1A) was not utilized (data not shown).
FIG. 3.
Expression of the HA-tagged A14.5L ORF. (A) Expression of the epitope-tagged A14.5L protein from an IPTG-inducible bacteriophage T7 promoter. BS-C-1 cells, in a six-well plate, were infected with 10 PFU of vT7lacOI-A14.5LHA per cell. After 1 h at 37°C, fresh medium containing 0 to 200 μM IPTG was added to each monolayer. The cells were harvested at 18 h and lysed in 50 μl of extraction buffer (4), and 20 μl of cleared supernatant was analyzed by SDS-PAGE and Western blotting with MAb 12CA5 to the HA epitope. (B) Expression of the epitope-tagged A14.5L protein from the natural A14.5L promoter. A Western blot of lysates from BS-C-1 cells harvested at 2 to 24 h after infection with vA14.5LHA is shown. BS-C-1 cells were infected with vA14.5LHA, as in panel A, except that no IPTG was added. At intervals from 2 to 24 h, cells from duplicate wells were harvested and analyzed by Western blotting with MAb 12CA5 or polyclonal antibody to the A17L protein. The positions and masses of marker proteins are indicated to the left of each panel.
The results obtained thus far indicated that the A14.5L-HA protein could be synthesized in a relatively stable and nontoxic form from a gene with a T7 promoter in vaccinia virus. The expression of the ORF under endogenous transcriptional signals, however, remained to be demonstrated. We noted a typical late promoter sequence (7) just upstream of the A14.5L ORF. To determine whether the A14.5L ORF can be expressed under its natural promoter, another recombinant vaccinia virus was constructed in which the HA tag was inserted between the C-terminal and stop codons of the endogenous A14.5L ORF so that the gene remained under its own transcriptional and translational regulatory signals. The transfer plasmid used to append the HA tag also contained the neo and gus genes between the A14.5L and A14L ORFs, as indicated in Fig. 2C, to allow selection and detection of the recombinant virus vA14.5LHA. Positive plaques were picked three times in succession, and the presence of the HA tag was demonstrated by PCR. When BS-C-1 cells were infected with vA14.5LHA, the HA-tagged protein was detected at 12 h and accumulated through 24 h, similar to that of the previously described late membrane protein A17L (Fig. 3B). Synthesis of the A14.5L-HA protein was not detected in the presence of a 40-μg/ml concentration of AraC, an inhibitor of DNA replication, consistent with late promoter regulation (data not shown). Because the A14.5L protein was predicted to contain multiple serines and threonines, as well as one tyrosine, we attempted to label it with 32Pi; the results, however, were negative, suggesting that the A14.5L protein is not phosphorylated (data not shown).
Partition of the A14.5L-HA protein in the Triton X-114 detergent phase.
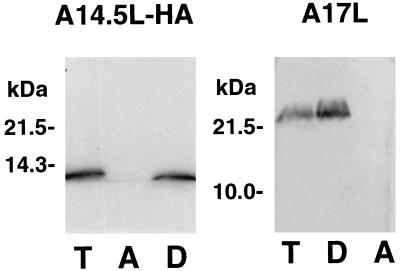
The potential of the A14.5L-HA protein to associate with membranes was evaluated by Triton X-114 partitioning (5) of extracts of cells infected with vA14.5LHA. After temperature-induced phase separation, the location of the HA-tagged protein was determined by SDS-PAGE and immunoblotting. As a control, the blot was also probed with antibody to the previously characterized A17L membrane protein. Both proteins were detected exclusively in the detergent-rich phase, indicating sufficient hydrophobicity for membrane association (Fig. 4).
FIG. 4.
Partition of the A14.5L-HA protein in the Triton X-114 detergent phase. BS-C-1 cells were infected with vA14.5LHA (10 PFU/cell). After 18 h, the cells were washed with PBS and resuspended in 0.2 ml of 10 mM Tris-HCl, 0.15 M NaCl, and 1% (vol/vol) Triton X-114 (pH 7.4). The phase separation was carried out as described previously (14). Equal volumes of total extract (T), the aqueous phase (A), and the detergent phase (D) were analyzed by SDS-PAGE and Western blotting with MAb 12CA5 or polyclonal antibody to the A17L protein. The positions and masses of marker proteins are indicated to the left.
Association of the A14.5L-HA protein with purified virus particles.
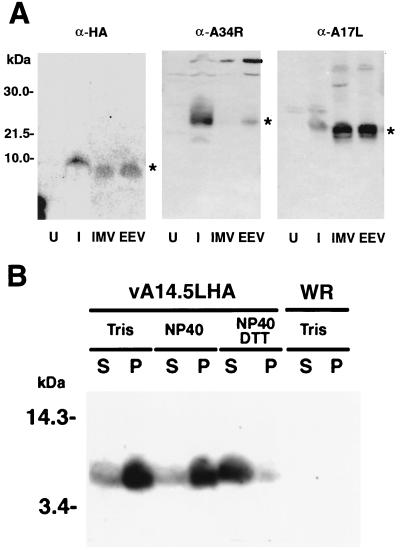
Virus particles, from the lysate and medium of cells infected with vA14.5LHA, were purified by sedimentation through a sucrose cushion followed by CsCl gradient centrifugation. Because of additional membranes, the intracellular enveloped virions (IEV) and EEV have a lower buoyant density than the IMV. Fractions were collected from the CsCl gradient and the fractions corresponding in density to IMV and IEV or EEV were analyzed by Western blotting after SDS-PAGE. Replicate samples were probed with antibodies to the HA tag, the A34R EEV membrane protein, and the A17L IMV membrane protein. The A14.5L-HA protein, like the A17L protein, was in both the IMV and IEV or EEV, whereas the A34R protein was only in the latter (Fig. 5A). Since IMV proteins are also present in EEV, these data suggested that the A14.5L protein is a component of the IMV.
FIG. 5.
Association of the A14L-HA protein with the membrane fraction of virus particles. (A) BS-C-1 cells were mock infected or infected with 10 PFU of vA14.5LHA per cell. After 18 h, the medium was harvested, and the cells were disrupted with a Dounce homogenizer. Virus particles were purified by sedimentation through a sucrose cushion and CsCl centrifugation (23). The virus band was diluted with Tris-HCl (pH 9), collected by centrifugation, and resuspended in Tris buffer. The lysates of mock-infected cells (U) or infected cells (I) were analyzed in parallel with CsCl gradient fractions by SDS-PAGE and Western blotting with MAb 12CA5 (α-HA) or polyclonal antibody to the A34R protein (α-A34R) or A17L protein (α-A17R). Fractions containing IMV and IEV or EEV are indicated. Small differences in the electrophoretic mobilities of proteins from lysates and CsCl gradients could be due to the ionic strengths. The bands corresponding to A14.5L-HA, A34R, and A17L are indicated by asterisks. The positions and masses of marker proteins are indicated to the left. (B) Vaccinia virions, purified by sucrose gradient centrifugation (9) from cells infected with vaccinia virus WR or vA14.5LHA, were incubated with 10 mM Tris (pH 7.4) or 1% NP-40 in 10 mM Tris (pH 7.4) in the presence or absence of 10 mM dithiothreitol. Soluble (S) and insoluble (P) fractions were separated by centrifugation and resuspended to equal volumes in SDS-containing sample buffer. An equivalent amount of each fraction was subjected to electrophoresis, and the separated proteins were transferred to a membrane and analyzed by Western blotting with anti-HA MAb 12CA5.
Membrane proteins can usually be extracted from purified vaccinia virions with either a nonionic detergent or a combination of detergent and reducing agent. The A14.5L protein required dithiothreitol in addition to NP-40 for efficient release into the supernatant fraction (Fig. 5B).
Immunogold electron microscopy.
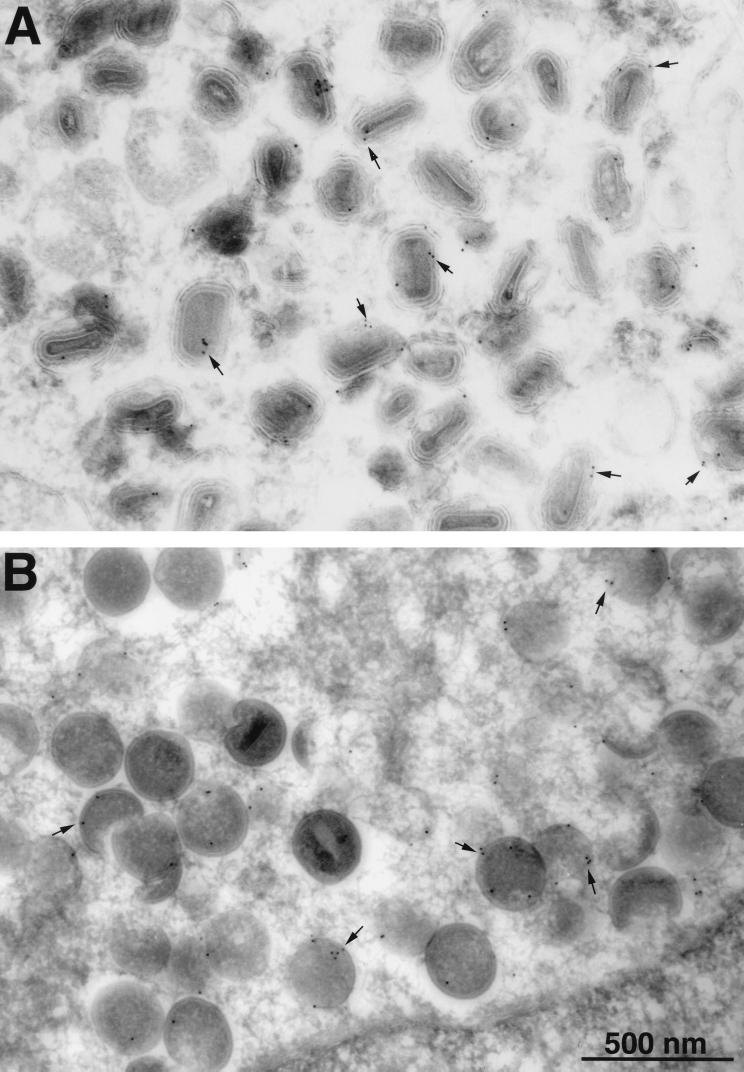
The localization of the A14.5L-HA protein was further investigated by successively incubating ultrathin sections of infected BS-C-1 cells with an anti-HA MAb, a secondary antibody to mouse IgG, and protein A conjugated to colloidal gold. As expected, clusters of immature virions were typically found in virus factory areas near the nucleus, whereas IMV and IEV were in more peripheral parts of the cytoplasm. The gold particles were associated with the membranes of both the mature (Fig. 6A) and immature (Fig. 6B) virions. In control cells infected with vaccinia virus lacking the HA-tagged A14.5L ORF, only occasional, widely dispersed gold particles were detected (data not shown).
FIG. 6.
Immunogold labeling of viral membranes with antibody to the HA tag. BSC-1 cells that had been infected with vA14.5L-HA for 24 h were fixed in paraformaldehyde, cryosectioned, and incubated with a mouse MAb to the HA tag, rabbit IgG to mouse IgG, and then with 10-nm-diameter gold particles conjugated to protein A. Electron microscopic images are shown with a 500-nm marker. (A) Field containing large numbers of IEV. (B) Field containing large numbers of immature virons. Arrows point to examples of gold particles.
The A14.5L ORF is nonessential for replication in vitro.
To determine whether the A14.5L ORF is essential for vaccinia virus replication, a recombinant virus, vA14.5LΔneo/gus, was constructed (Fig. 2D). The A14.5L ORF was deleted, except for 23 and 47 bp at either end, and replaced with the neo and gus genes. The recombinant virus was isolated by antibiotic selection and detected by staining with X-Glu. Deletion of the A14.5L ORF was confirmed by PCR analysis (data not shown).
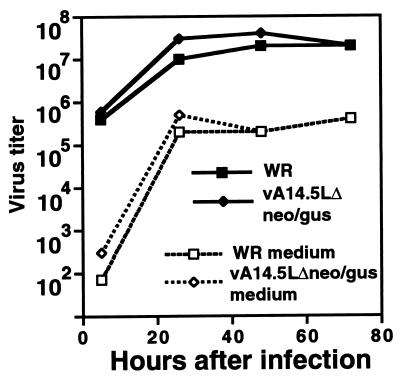
The vA14.5LΔneo/gus virus formed normal-size plaques in a variety of cell lines (monkey BS-C-1 and CV-1, human TK−143B and A549, rabbit kidney 13, and baby hamster kidney 21 cells). Moreover, the yields of intra- and extracellular vA14.5LΔneo/gus and the parental strain WR virus were similar in BS-C-1 cells (Fig. 7). Thus, the A14.5L protein is nonessential for vaccinia virus replication in those tissue culture cells examined.
FIG. 7.
Replication of an A14.5L deletion mutant. B-SC-1 cells in a six-well plate were inoculated with 10 PFU of vaccinia virus strain WR or vA14.5LΔneo/gus per cell for 1 h at 37°C. The cells were then washed twice with medium. At appropriate times, the medium was removed and saved, and the cells were harvested by scraping. The media and cells were centrifuged, and the virus titers in the supernatants of the former (dashed lines) and the pellets of the latter (solid lines) were determined by plaque assay on BS-C-1 cells.
Effect of the A14.5L deletion on the virulence of vaccinia virus for mice.
When administered intranasally, the WR strain of vaccinia virus causes weight loss and death in BALB/c mice. Initial experiments indicated that vA14.5LΔneo/gus was significantly attenuated in this respect. Although this finding suggested that the A14.5L gene was required for virulence, we needed to rule out other explanations. To eliminate the possibility that expression of the neo and gus genes was attenuating, either because of the gene products themselves or polar effects on expression of neighboring genes, the neo and gus genes were deleted from vA14.5LΔneo/gus without restoring the A14.5L ORF. Plaques that did not stain blue were purified, and the loss of the neo and gus genes was confirmed by PCR. This mutant virus, called vA14.5LΔ, retained only 13 bp of the start and 37 bp of the end of the A14.5L ORF (Fig. 2E). Because of the large size of the vaccinia virus genome, we also considered that vA14.5LΔneo/gus might have an unrelated attenuating mutation. To rule this out, vA14.5L+ with a restored A14.5L ORF was constructed from vA14.5LΔneo/gus by recombination (Fig. 2F). Again, plaques that did not stain blue were purified, and an intact A14.5L gene was confirmed by PCR.
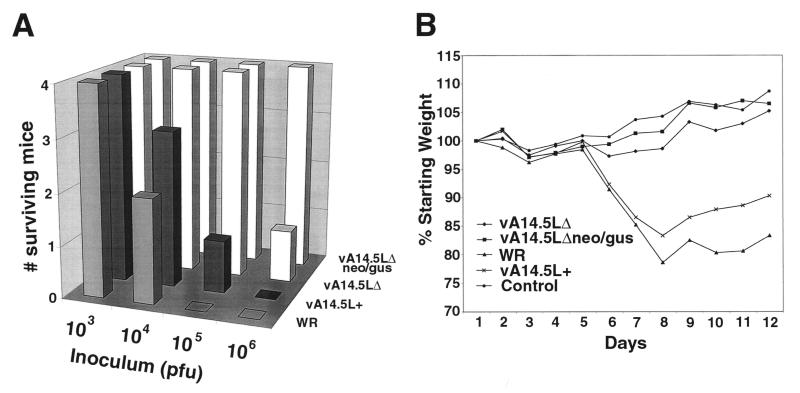
Groups of 5-week-old female BALB/c mice were anesthetized and inoculated intranasally with diluent or with 103 to 106 PFU of vaccinia virus strain WR, vA14.5LΔneo/gus, vA14.5LΔ, or vA14.5L+. Of these viruses, WR and vA14.5L+ contained the A14.5L gene whereas vA14.5LΔneo/gus and vA14.5LΔ did not. The virulence of the different viruses was assessed by loss of weight and death, which occurred between 7 and 10 days after infection. With viruses containing A14.5L, deaths were noted at 104 PFU, and a majority or all of the mice were deceased at 105 PFU and above (Fig. 8A). With viruses lacking A14.5L, deaths were first noted at 106 PFU (Fig. 8A). Correspondingly, more severe weight losses occurred in mice infected with viruses containing A14.5L than with viruses lacking A14.5L. The weight changes for the 103-PFU challenges, which were nonlethal for all groups, are shown in Fig. 8B. The gains in the mean weights of the mice infected with vA14.5LΔneo/gus or vA14.5LΔ virus were similar to those of uninfected controls, whereas the mice infected with intact A14.5L genes suffered appreciable weight losses (Fig. 8B). Although some weight loss occurred when 104 or 105 PFU of the viruses lacking A14.5L was administered, by day 12, the weights had rebounded and approached or exceeded the levels at the start of the experiment (data not shown).
FIG. 8.
Virulence of parental and mutant vaccinia viruses in mice. Groups of four mice were inoculated intranasally with 103 to 106 PFU of purified vaccinia virus strain WR, vA14.5LΔneo/gus, vA14.5LΔ, and vA14.5L+. (A) Death of animals. (B) Percentage of original weight of mice in each group infected with 103 PFU.
DISCUSSION
Comparative genomics is useful for identifying short ORFs that can be overlooked when analyzing a large poxvirus genome. Thus, the potential significance of the A14.5L ORF of vaccinia virus and other orthopoxviruses was first appreciated when a homologous ORF with typical late promoter sequences was detected in a distantly related poxvirus (25). Further analysis indicated that orthologous genes are present in all vertebrate poxviruses examined, including orthopoxviruses, leporipoxviruses, yatapxoviruses, and molluscipox viruses. Here, we have shown that the 53-amino-acid ORF was expressed under its natural promoter as a viral membrane protein and that deletion of the ORF attenuated the virulence of vaccinia virus for mice.
Our inability to generate an antibody to the extremely hydrophobic A14.5L protein made it necessary to rely on a short epitope tag which was added between the C-terminal and stop codons of the A14.5L ORF. Since neither transcriptional nor translational signals were altered by this addition and the ORF remained in its original location within the genome, we are confident that the late expression of the epitope-tagged protein accurately reflected the temporal expression of the unmodified one. Moreover, the tagged protein was specifically incorporated into virus particles.
The hydrophobic nature of the A14.5L protein was predicted from its amino acid composition and confirmed by Triton X-114 phase partitioning. When infected cells were cryosectioned, immunostained, and examined by electron microscopy, the protein A-conjugated gold particles were found in association with immature and mature virus particles, usually on or adjacent to the membrane envelope. Although there is an N-terminal hydrophobic segment, it was not predicted to be a signal peptide (20), and further studies are needed to determine the topology of the A14.5L protein.
The conservation of the A14.5L ORF among distantly related poxviruses, its expression during vaccinia virus infection, and the association of the protein with the membranes of virus particles suggested that it has a significant role and is not a useless remnant of some longer ancestral ORF. Nevertheless, deletion of the A14.5L ORF had no discernible effect on plaque size or virus yield in tissue culture cells. This result was not too surprising, because poxviruses have many genes that are not essential in tissue culture. In some cases, however, the gene deletions decrease virulence in animal models. Our initial studies indicated that the deletion mutant vA14.5LΔneo/gus was attenuated, compared to the parental vaccinia virus strain WR, when administered intranasally to mice. To be sure that the effect was due to the deletion of the A14.5L gene, two additional recombinant viruses were made: one had the neo and gus genes removed from the site of the deletion, and the other had the A14.5L ORF fully restored. The viruses WR and vA14.5L+ with the A14.5L gene present caused significantly more weight loss and death than the viruses vA14.5LΔneo/gus and vA14.5LΔ with the A14.5L gene deleted. Whether the A14.5L membrane protein enhances virus replication and dissemination in vivo or protects the virus against innate or acquired immune responses remains to be determined.
ACKNOWLEDGMENTS
We thank Norman Cooper for providing tissue culture cells, Jerry Sisler for DNA sequencing, Andrea Weisberg for electron microscopy, and Rafael Molina for help with the animal experiments.
REFERENCES
- 1.Alexander W A, Moss B, Fuerst T R. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amegadzie B Y, Ahn B-Y, Moss B. Characterization of a 7-kilodalton subunit of vaccinia virus DNA-dependent RNA polymerase with structural similarities to the smallest subunit of eukaryotic RNA polymerase II. J Virol. 1992;66:3003–3010. doi: 10.1128/jvi.66.5.3003-3010.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antoine G, Scheiflinger F, Dorner F, Falkner F G. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology. 1998;244:365–396. doi: 10.1006/viro.1998.9123. [DOI] [PubMed] [Google Scholar]
- 4.Betakova T, Wolffe E J, Moss B. Regulation of vaccinia virus morphogenesis: phosphorylation of the A14L and A17L membrane proteins and C-terminal truncation of the A17L protein are dependent on the F10L protein kinase. J Virol. 1999;73:3534–3543. doi: 10.1128/jvi.73.5.3534-3543.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1990;256:1604–1607. [PubMed] [Google Scholar]
- 6.Cameron C, Hota-Mitchell S, Chen L, Barrett J, Cao J X, Macaulay C, Willer D, Evans D, McFadden G. The complete DNA sequence of myxoma virus. Virology. 1999;264:298–318. doi: 10.1006/viro.1999.0001. [DOI] [PubMed] [Google Scholar]
- 7.Davison A J, Moss B. The structure of vaccinia virus late promoters. J Mol Biol. 1989;210:771–784. doi: 10.1016/0022-2836(89)90108-3. [DOI] [PubMed] [Google Scholar]
- 8.Duncan S A, Smith G L. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J Virol. 1992;66:1610–1621. doi: 10.1128/jvi.66.3.1610-1621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Earl P L, Cooper N, Wyatt S, Moss B, Carroll M W. Preparation of cell cultures and vaccinia virus stocks. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley and Sons; 1998. pp. 16.16–16.16.3. [Google Scholar]
- 10.Earl P L, Moss B, Wyatt L S, Carroll M W. Generation of recombinant vaccinia viruses. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates & Wiley Interscience; 1998. pp. 16.17.1–16.17.19. [Google Scholar]
- 11.Engelstad M, Howard S T, Smith G L. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology. 1992;188:801–810. doi: 10.1016/0042-6822(92)90535-w. [DOI] [PubMed] [Google Scholar]
- 12.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 13.Hirt P, Hiller G, Wittek R. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J Virol. 1986;58:757–764. doi: 10.1128/jvi.58.3.757-764.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hooper N M. Identification of a glycosylphosphatidyl anchor on membrane proteins. In: Hooper N M, Turner A J, editors. Lipid modification of proteins. New York, N.Y: Oxford University Press; 1992. pp. 91–92. [Google Scholar]
- 15.Isaacs S N, Wolffe E J, Payne L G, Moss B. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J Virol. 1992;66:7217–7224. doi: 10.1128/jvi.66.12.7217-7224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen O N, Houthaeve T, Shevchenko A, Cudmore S, Ashford T, Mann M, Griffiths G, Locker J K. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J Virol. 1996;70:7485–7497. doi: 10.1128/jvi.70.11.7485-7497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Moore J B, Smith G L. Steroid hormone synthesis by a vaccinia enzyme—a new type of virus virulence factor. EMBO J. 1992;11:1973–1980. doi: 10.1002/j.1460-2075.1992.tb05251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2637–2671. [Google Scholar]
- 20.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 21.Parkinson J E, Smith G L. Vaccinia virus gene A36R encodes a Mr 43–50 K protein on the surface of extracellular enveloped virus. Virology. 1994;204:376–390. doi: 10.1006/viro.1994.1542. [DOI] [PubMed] [Google Scholar]
- 22.Payne L G, Norrby E. Presence of hemagglutinin in the envelope of extracellular vaccinia virus particles. J Gen Virol. 1976;32:63–72. doi: 10.1099/0022-1317-32-1-63. [DOI] [PubMed] [Google Scholar]
- 23.Roper R L, Payne L G, Moss B. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J Virol. 1996;70:3753–3762. doi: 10.1128/jvi.70.6.3753-3762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Senkevich T G, Bugert J J, Sisler J R, Koonin E V, Darai G, Moss B. Genome sequence of a human tumorigenic poxvirus: prediction of specific host response-evasion genes. Science. 1996;273:813–816. doi: 10.1126/science.273.5276.813. [DOI] [PubMed] [Google Scholar]
- 25.Senkevich T G, Koonin E V, Bugert J J, Darai G, Moss B. The genome of molluscum contagiosum virus: analysis and comparison with other poxviruses. Virology. 1997;233:19–42. doi: 10.1006/viro.1997.8607. [DOI] [PubMed] [Google Scholar]
- 26.Shchelkunov S N, Massung R F, Esposito J J. Comparison of the genome DNA sequences of Bangladesh-1975 and India-1967 variola viruses. Virus Res. 1995;36:107–118. doi: 10.1016/0168-1702(94)00113-q. [DOI] [PubMed] [Google Scholar]
- 27.Shchelkunov S N, Totmenin A V, Loparev V N, Safronov P F, Gutorov V V, Chizhikov V E, Knight J C, Parsons J M, Massung R F, Esposito J J. Alastrim smallpox variola minor virus genome DNA sequences. Virology. 2000;266:361–386. doi: 10.1006/viro.1999.0086. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi T, Oie M, Ichihashi Y. N-terminal amino acid sequences of vaccinia virus structural proteins. Virology. 1994;202:844–852. doi: 10.1006/viro.1994.1406. [DOI] [PubMed] [Google Scholar]
- 29.Turner G S. Respiratory infection of mice with vaccinia virus. J Gen Virol. 1967;1:399–402. doi: 10.1099/0022-1317-1-3-399. [DOI] [PubMed] [Google Scholar]
- 30.Ward G A, Stover C K, Moss B, Fuerst T R. Stringent chemical and thermal regulation of recombinant gene expression by vaccinia virus vectors in mammalian cells. Proc Natl Acad Sci USA. 1995;92:6773–6777. doi: 10.1073/pnas.92.15.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willer D O, McFadden G, Evans D H. The complete genome sequence of Shope (rabbit) fibroma virus. Virology. 1999;264:319–343. doi: 10.1006/viro.1999.0002. [DOI] [PubMed] [Google Scholar]
- 32.Williamson J D, Reith R W, Jeffrey L J, Arrand J R, Mackett M. Biological characterization of recombinant vaccinia viruses in mice infected by the respiratory route. J Gen Virol. 1990;71:2761–2767. doi: 10.1099/0022-1317-71-11-2761. [DOI] [PubMed] [Google Scholar]
- 33.Wolffe E J, Moore D M, Peters P J, Moss B. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J Virol. 1996;70:2797–2808. doi: 10.1128/jvi.70.5.2797-2808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]