Abstract
Despite the expert-level performance of artificial intelligence (AI) models for various medical imaging tasks, real-world performance failures with disparate outputs for various subgroups limit the usefulness of AI in improving patients’ lives. Many definitions of fairness have been proposed, with discussions of various tensions that arise in the choice of an appropriate metric to use to evaluate bias; for example, should one aim for individual or group fairness? One central observation is that AI models apply “shortcut learning” whereby spurious features (such as chest tubes and portable radiographic markers on intensive care unit chest radiography) on medical images are used for prediction instead of identifying true pathology. Moreover, AI has been shown to have a remarkable ability to detect protected attributes of age, sex, and race, while the same models demonstrate bias against historically underserved subgroups of age, sex, and race in disease diagnosis. Therefore, an AI model may take shortcut predictions from these correlations and subsequently generate an outcome that is biased toward certain subgroups even when protected attributes are not explicitly used as inputs into the model. As a result, these subgroups became nonprivileged subgroups. In this review, the authors discuss the various types of bias from shortcut learning that may occur at different phases of AI model development, including data bias, modeling bias, and inference bias. The authors thereafter summarize various tool kits that can be used to evaluate and mitigate bias and note that these have largely been applied to nonmedical domains and require more evaluation for medical AI. The authors then summarize current techniques for mitigating bias from preprocessing (data-centric solutions) and during model development (computational solutions) and postprocessing (recalibration of learning). Ongoing legal changes where the use of a biased model will be penalized highlight the necessity of understanding, detecting, and mitigating biases from shortcut learning and will require diverse research teams looking at the whole AI pipeline.
Keywords: Artificial Intelligence, machine learning
INTRODUCTION
Artificial intelligence (AI) models in medical imaging are able to match expert-level accuracy in multiple diagnostic and prognostic tasks, driven largely by developments in deep learning. For example, AI performance is at par with specialists for the diagnosis of common thoracic pathologies [1] and diabetic retinopathy on fundoscopic images [2] and outperforms single radiologists in detecting abnormalities on screening mammography [3]. Despite this high performance and the clearance of more than 520 algorithms by the FDA [4], the adoption of AI into the clinical workflow is still lagging. Moreover, studies have shown that there is a risk for unintended bias in AI systems affecting individuals unfairly on the basis of race, sex, and other clinical characteristics [5–8]. Although there exists no consensus on a single definition of fairness, there is recognition that bias can arise when AI leverages its ability to recognize patterns in the training data and unintentionally associates certain confounding characteristics with the targeted outcome.
One central observation is that the cause of many prediction failures in AI are not independent phenomena but are instead connected in the sense that AI follows unintended “shortcut” strategies for the targeted task [9,10]. For example, AI can diagnose pneumonia, but it uses portable intensive care unit radiographic markers as surrogates for the task rather than detecting true underlying pathology [11]. Similarly, pneumothorax detection uses shortcuts based on inserted chest tubes [12]. It has been observed that imaging AI models learn spurious age, sex, and race correlations from images when trained for seemingly unrelated tasks [13]. Simultaneously, studies have shown AI imaging models demonstrate bias against historically underserved subgroups of age, sex, and race in disease diagnosis [14]. Therefore, it is concerning that an imaging AI model may take shortcut predictions from these correlations and subsequently generate outcomes that are biased toward certain subgroups (often nonprivileged groups), even when protected attributes are not explicitly used as inputs into the model. In this article, we explore causes of bias that arise from “shortcut learning” and discuss methods of detection and mitigation of these biases.
WHAT TYPES OF AI BIAS ARE OBSERVED IN DIFFERENT STAGES OF AI DEVELOPMENT THAT LEAD TO “SHORTCUT” LEARNING?
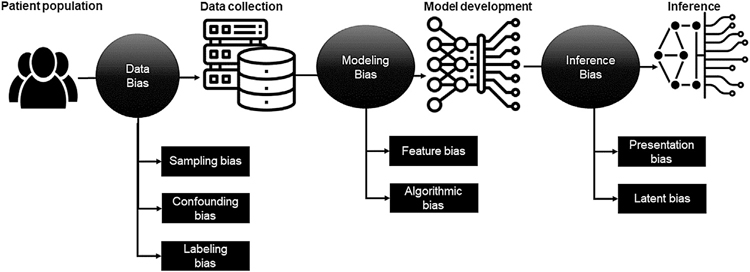
Figure 1 highlights different types of bias that can be introduced at various stages of imaging AI model development and validation, resulting in shortcut learning. These biases tend to be propagated to downstream tasks and ultimately accumulate, leading to biased outcomes [15].
Fig. 1.
Type of bias in different phases of artificial intelligence model development and validation causing “shortcut learning.”
Dataset Bias (Data Collection Bias)
It is critical that training data used in machine learning be representative of the real-world population. Data collection bias can arise from inconsistencies in training data that do not accurately represent a model’s intended use case, resulting in skewed outcomes. Dataset bias can also arise from sampling and labeling biases as well as confounders that are learned by the AI models and used as shortcuts.
Selection or Sampling Bias.
Selection or sampling bias occurs because of improper sampling or inclusion of a population in which a certain subgroup is heavily represented while others are not. Often, radiologic images are collected from only a single or a few sites [16–19] and thus lack geographic and racial diversity. The granularity of available images also varies with underlying patterns of systemic racism: Black and Hispanic patients tend to undergo lower quality and nonadvanced imaging for similar presenting symptoms in emergency departments [20]. For example, Black women are less likely to receive advanced technologies such as 3-D tomosynthesis (which has been shown to reduce recall rates) and usually undergo 2-D mammography for breast cancer screening, highlighting a known fact that technological advancements do not always benefit historically vulnerable subpopulations in the early phases of adoption [21]. Other causes of sampling bias include disparities in access, whereby some patients will never be imaged and would hypothetically be included in an ideal dataset [22]. AI models can easily learn these patterns and use them in their predictions [22]. Of more concern is the tendency of AI models to hallucinate and not fail gracefully when they encounter datasets that are out of the distribution of the training dataset [23,24]. Further compounding this is the lack of AI models providing a level of certainty or uncertainty when rendering a prediction. Sampling bias can also result in extreme class imbalance, and hence AI models will learn from the majority case (usually the privileged class) and not the minority classes (usually the nonprivileged classes). These models usually result in good areas under the curve for the majority class but do not generalize to the minority classes. Moreover, strategies to mitigate class imbalance can result in further alteration of the disease distribution of the minority classes. This is important because minority class representation in the overall dataset may be small. However, the burden of disease in the minority classes may be larger as observed in breast cancer screening, in which Black women tend to have more aggressive cancers at diagnosis and at younger ages [25].
Labeling Bias.
Most labels of publicly available datasets are derived using weakly supervised techniques, whereby a subset of labels are generated by a radiologist, which are then used to train a model that is used to label the larger dataset [26]. Although commonly adopted, this strategy can perpetuate hidden signals in the textual reports that are then embedded in the image dataset. Studies have shown that AI models can detect sentiments in text reports, with the ability to even identify a patient’s self-reported race from textual descriptions of clinical notes [27]. Broad classifications of datasets may miss smaller subsets of categories that are embedded in the dataset, causing hidden stratification [28]. This has been documented to result in differences in model performance for pneumothorax for the overall dataset and when analyzed for patients with and without chest tubes [28]. Other causes of reader-based bias include labeling by nonmedical personnel, including workers on Amazon Mechanical Turk, without transparency as to the training and standardization of the labeling process. Delineation or annotation bias occurs because there is significant interreader variability in delineating regions of interest of diseased regions on imaging, which are often used as one of the input channels for deep learning or machine learning. This is amplified when large multisite datasets are leveraged, as several readers are usually involved in the labeling or annotation process, which can introduce their biases into the model [29].
Confounding Bias.
The presence of a confounding attribute can create the illusion of an association between certain variables and the targeted outcome and force the model to learn an incorrect relationship between the studied variable and its outcome, leading to wrong conclusions. For example, Rueckel et al [12,30] demonstrated that AI models trained on open-source chest radiographic data for pneumothorax detection learned the strong association between a confounding attribute, the presence of thoracic tubes, and the diagnosis of pneumothorax. This model makes a systematic error (false negative) when the thoracic tube is not present. An area of ongoing research remains understanding the value of confounders that are helpful in the model prediction and how to harness such features to mitigate disparities. For example, Pierson et al [31] demonstrated that an algorithmic prediction score for the severity of osteoarthritis mitigates known biases in pain and osteoarthritis evaluation, yet the same AI models demonstrate high accuracy in predicting self-reported race of the patients [13]. In such cases, it is unclear as to the contribution of the model’s ability to detect a nonbiologic variable (in this case a confounder) and its contribution to mitigating existing disparities. More work is required to differentiate between spurious confounders such as patient location in the intensive care unit (from radiographic markers) versus significant confounders such as demographics.
Bias in Modeling
During the modeling phase of an AI model, bias arises from systematic errors resulting from erroneous assumptions about the data, which may cause the model to miss a relevant relationship between data inputs (features) and targeted outputs (predictions).
Feature Bias.
Various feature selection methods starting from manual selection on the basis of prior knowledge to automated methods, such as LASSO, minimum redundancy, maximum relevance ensemble [32], and mutual information maximization [33], are used to reduce features (predictors) to the most predictive and robust ones. Such selection techniques can be misleading for targeted tasks and can introduce feature selection bias, which can adversely affect a model’s prediction ability. This occurs because the model overfits the data in the presence of selection bias, causing it to not generalize well. Krawczuk and Łukaszuk [34] demonstrated such feature selection bias using the genomics dataset for well-studied clinical use cases (colon cancer, leukemia, and breast cancer) when the same dataset was used for feature selection and learning. They demonstrated positive feature selection bias in 28 experiments after applying four selection methods (ReliefF; minimum redundancy, maximum relevance; support vector machine recursive feature elimination; and relaxed linear separability), with a difference between validation and test accuracies of between 2.6% to 41.67%. Incorrect assumptions on feature distribution can cause bias. For example, certain feature selection methods assume a continuous and gaussian distribution, but often categorical features or those that are not normally distributed are fed to the feature selection method. This can erroneously remove an important feature or retain a relatively unimportant feature [35,36].
Algorithmic Bias.
“Algorithmic bias” refers to systematic and repeatable errors in an AI model that create unfair outcomes for certain subgroups or individuals as a result of algorithmic design choices during AI model development. Selecting a loss function on the basis of the overall model performance rather than for each subgroup skews performance to the majority group. Design choices that can bias the outcome of an algorithm include the choice of regularization techniques, optimization functions, and use of statistically biased estimators [37]. For example, Ribeiro et al [38] trained a model to discriminate images representing wolves and huskies. Despite showing reasonable accuracy to decide whether the image contained a wolf or not, the model was inferring spurious correlations: the presence or absence of snow in the background.
Bias in Inference or Decision Making
At the final stage of an AI model deployment, bias can be introduced on the basis of how the results of the model are presented to end users.
Presentation Bias.
AI classification models output numeric scores and rankings that are displayed on a user interface for human decision makers. For medical imaging computer vision tasks, it is common to display areas of interest (correlating to areas of high probability) using gradient class activation mapping and saliency maps. Evaluations of these visualization techniques have shown that the utility, repeatability, and reproducibility of these methods are limited [39]. It is more challenging to use these techniques to understand some of the shortcuts that the model is relying on, especially when the evaluator lacks appropriate medical knowledge. Another factor that can introduce bias is related to how and when AI results are presented in the user interfaces. Through visualization, counterexamples, semantics, and uncertainty estimation, we can expect different behaviors from end-user radiologists introducing bias [40].
Latent Bias.
In latent bias, models may incorrectly label something on the basis of historical data or because of a stereotype that already exists in society [41]. For example, an algorithm to predict treatment outcomes could learn and predict differing outcomes on the basis of patient race, ethnicity, and socioeconomic factors instead of clinically relevant information [41].
HOW TO MEASURE BIAS IN AI MODELS?
Bias arising from shortcut learning can be difficult to assess and requires a combination of domain expertise and technical ability. Today, imaging AI model performance is measured primarily in terms of overall accuracy or ratio between sensitivity and specificity (area under the receiver operating characteristic curve). However, on a test dataset with a <10% positivity rate, a biased model may provide 90% accuracy but only 50% sensitivity. Caution should be given to the selection of metrics to evaluate the performance of AI algorithms, as most of them may not be appropriate and, in turn, may result in a biased estimate of their performance [8,42]. It is important to record disparity rates (eg, true positive, false positive) of the nonprivileged subgroups to comprehend the model performance before deployment. Table 1 summarizes available open-source tool kits for bias detection. These have been used for general AI evaluation from 2015 and their use in health care machine learning is still limited.
Table 1.
Available open-source tool kits for bias detection and mitigation
| Tool | Year | Description | Target | GitHub | License | Paper | Bias Detection | Bias Mitigation |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| AIF360 [79] | 2019 | Open-source Python tool kit | Performance benchmarking | https://github.com/Trusted-AI/AIF360 | Apache version 2.0 | https://ieeexplore.ieee.org/abstract/document/8843908 | Yes | Yes |
| Themis-ML [80] | 2017 | Open-source Python tool kit | Measure fairness for binary classification | https://github.com/cosmicBboy/themis-ml | MIT open-source | https://arxiv.org/abs/1710.06921 | Yes | Yes |
| Fairlearn [81] | 2020 | Open-source Python tool kit | Interactive visualization dashboard and unfairness mitigation | https://github.com/fairlearn/fairlearn | MIT open-source | https://www.microsoft.com/en-us/research/publication/fairlearn-a-toolkit-for-assessing-and-improving-fairness-in-ai/ | Yes | Yes |
| FairML [82] | 2016 | Open-source Python tool kit | Audit cumbersome predictive models | https://github.com/adebayoj/fairml | MIT open-source | https://dspace.mit.edu/handle/1721.1/108212 | Yes | No |
| Aequitas [83] | 2018 | Open-source Python tool kit | Visualization of bias metrics | https://github.com/dssg/aequitas | MIT open-source | https://arxiv.org/abs/1811.05577 | Yes | No |
| VerifyML [84] | 2022 | Open-source Python tool kit | Computing fairness degree | https://github.com/cylynx/verifyml | Apache version 2.0 | https://arxiv.org/abs/2210.08418 | Yes | No |
| Fairkit-Learn [85] | 2022 | Open-source interactive Python tool kit | Understand fairness and support training | https://github.com/INSPIRED-GMU/fairkit-learn | – | https://people.cs.umass.edu/~brun/pubs/pubs/Johnson22.pdf | Yes | Yes |
| Fairness comparison [86] | 2018 | Open-source Python + R | Access to fairnessenhancing classification algorithms | https://github.com/algofairness/fairness-comparison | Apache version 2.0 | https://arxiv.org/abs/1802.04422 | Yes | Yes |
| Fairness Measures [87] | 2017 | Open-source Python | Measure fairness with new metrics | https://github.com/FairnessMeasures/fairness-measures-code | GPL-3.0 | https://link.springer.com/article/10.1007/s10618-017-0506-1 | Yes | No |
| FairTest [88] | 2015 | Open source Python + MongoDB + Python R interface | Discover confounding factor | https://github.com/columbia/fairtest | Apache version 2.0 | https://arxiv.org/abs/1510.02377 | Yes | No |
| Google’s What-If Tool [89] | 2020 | Open source Interactive Python toolbox | Understanding of a black-box classification or regression ML model | https://github.com/pair-code/what-if-tool | Apache version 2.0 | https://pair-code.github.io/what-if-tool/ | Yes | No |
| IBM Watson OpenScale [90] | 2022 | Cloud engine: free basic | Monitor AI models for bias, fairness, and trust | https://github.com/IBM/watson-openscale-samples | Apache version 2.0 | https://www.ibm.com/docs/en/cloud-paks/cp-data/3.5.0?topic=services-watson-openscale | Yes | Yes |
HOW TO MITIGATE BIAS IN AN AI MODEL?
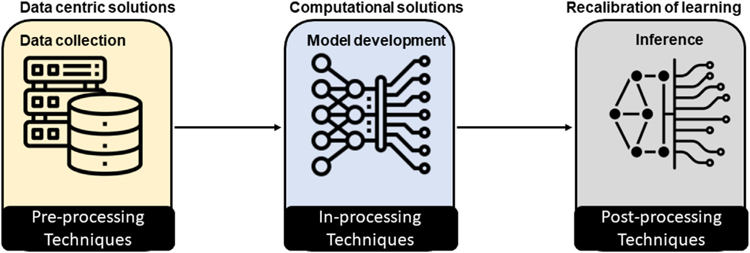
There are several ways to combat bias in AI models, which is traditionally known as debiasing or “fair” AI model development, starting with data-centric approaches to computation methods (Fig. 2). In the following subsection, we group the methods on the basis of their applicability to different phases of the AI model development.
Fig. 2.
Bias mitigation techniques at different phases of artificial intelligence model development.
Preprocessing Techniques
Preprocessing techniques, particularly those categorized as “data-centric” approaches, mitigate bias (eg, sampling bias, confounding bias) in the training data. There are many general preprocessing techniques prescribed for AI, including reweighting [43], disparate impact removal [44], learning fair representation [45], optimized preprocessing [46], and the maximum entropy approach [47]. Apart from these general preprocessing techniques, cross-population training and testing is the most adopted solution for imaging AI, in which datasets from multiple institutions are combined to train a model and validate its performance on a heterogeneous population. For example, Das et al [48] trained a convolution model on a mixture of two chest radiographic datasets (with tuberculosis) and demonstrated that it contributed to a greater prevalence of positive findings. Similarly, Zech et al [11] studied the ability of models to detect pneumonia on chest radiographs by training and testing on data from different hospital systems in the United States and showed that training sets with equal incidence across sites achieved the best performance on the testing set. Larrazabal et al [49] showed that sex-balanced training datasets presented minimal bias toward nonprivileged subgroup. However, the collection of a large, balanced, multi-institutional dataset is always challenging, and it does not guarantee the inclusion of every variation and factor of the targeted population.
In contrast to cross-population training, developing subgroup-specific modeling has been experimented with for medical imaging. For example, Puyol-Antón et al [50] used two preprocessing approaches for bias mitigation in cardiac MRI segmentation: stratified batch sampling and protected group models. In stratified batch sampling, the data are stratified by the protected attribute(s) for each training batch, and samples are selected to ensure that each protected group is equally represented. The protected group models approach trains a different segmentation model for each protected group. Although training an individualized model for each population subgroup is technically demanding (especially when there are multiple subgroups), this approach is necessary when there are biologic differences among various subgroups, rather than trying to achieve good general performance or making a diverse dataset [51–53].
Imaging preprocessing techniques are also applied as preprocessing bias reduction solutions. For example, Rueckel et al [30] demonstrated that including in-image pixel annotations of dehiscent visceral pleura for pneumothorax detection on chest radiography significantly improved algorithm performance and reduced the confounding bias caused by inserted thoracic tubes. To mitigate skin-tone bias while diagnosing diabetic retinopathy, Burlina et al [54] proposed debiasing by altering the retinal appearance through augmentation of the training data via controlled synthetic image generation to include more data from underrepresented subgroups of the population.
In-Processing Techniques
There are multiple in-processing techniques prescribed for AI debiasing for generic image analysis (eg, facial images, natural images), including meta-fair classifier [55], prejudice remover [56], grid search reduction and exponentiated gradient reduction [57], GerryFair classifier [58], adversarial debiasing [59,60], and adding fairness constraints [61–64]. An example of application of an in processing technique is the work of Dinsdale et al [65], who constructed a multi-institutional AI model for detecting age on brain MR images and identified that a model is biased toward the data source and MR scanner subtypes. They were able to improve the classification performance of the model by domain adaptation, whereby they removed confounding factors by creating a feature space that was invariant to the acquisition scanner. After this debiasing, the model’s ability to identify the site of origin decreased from 96% accuracy to 56%, with only a slight decrease in the model task performance.
Correa et al [66] developed a two-step adversarial debiasing approach with partial learning that reduced disparity while preserving the performance of the targeted diagnosis or classification task. They experimented with two independent medical image case studies and showed bias reduction while preserving the targeted performance on both internal and external datasets in radiology and dermatology. Puyol-Antón et al [50] added a meta-fair classifier to the segmentation network, which classified protected attributes along with the cardiac MRI segmentations.
Researchers have also experimented by combining preprocessing and in-processing techniques through federated learning, in which a model is trained in a distributed, collaborative fashion on decentralized data distribution, without having direct access to patient-sensitive data [67]. However, existing federated learning methods focus on minimizing the average aggregated loss functions [68,69], leading to a biased model that performs well for some hospitals while exhibiting undesirable performance for other sites [70]. Recently, Hosseini et al [71] proposed a new federated learning scheme, Prop-FFL, for “fair” AI model training, which uses a novel optimization objective function to decrease performance variations among participating hospitals. There is promise in using texture-agnostic imaging biomarkers that are less sensitive to scanner and site specific variations [72].
Postprocessing Techniques
The most common postprocessing techniques for AI debiasing are equalized odds/calibrated equalized odds [73,74], reject option classification [43], and discrimination-aware ensemble [43]. Marcinkevics et al [75] proposed a debiasing technique of an already trained network for CXR classification on the basis of fine-tuning and pruning to minimize unknown sources of bias and demonstrated that this method reduces the classification disparity. In the task of real age estimation from human facial images, Clapés et al [76] used a simple postprocessing technique for bias correction by shifting apparent age toward the corresponding real age value.
DISCUSSION AND CONCLUSION
Shortcut learning, especially for protected attributes such as demographics (rather than learning true disease characteristics) that are barely, if at all, perceptible to the clinician interpreting medical images, affect performance of the affected subgroups, causing bias. Notably, this occurs even when the model input does not include the protected attribute, as shown in the work of Obermeyer et al [8], in which race is not included as input in the model. The challenge of these proxies is that they are difficult to audit and remove from datasets, as demonstrated by the work of Gichoya et al [13] on AI’s ability to recognize self-reported races without a clear explanation. Recent changes in legislation calling for health care organizations to be penalized for using biased models [77,78] highlight the challenge of evaluating when shortcut learning is the root cause of biased outcome. It is important for the AI community to design AI solutions with bias in mind from the point of idea development, data acquisition and curation, model development and evaluation, and at the point of deployment. To mitigate errors from shortcuts, the AI team must be diverse, combining both domain knowledge and technical expertise to evaluate and then subsequently mitigate bias. We also challenge the community to develop mechanisms through which useful shortcuts that mitigate existing disparities are harnessed to develop more equitable algorithms that work for everyone.
TAKE-HOME POINTS.
Models use shortcuts as confounders in their predictions.
These shortcuts are not always obvious, and are often hidden to the human eye which makes their evaluation difficult.
Various bias mitigation strategies including preprocessing, post processing and algorithmic approaches can be applied to remove bias arising from shortcuts.
Acknowledgments
Dr Gichoya declares support from US National Science Foundation (grant number 1928481) from the Division of Electrical, Communication & Cyber Systems, RSNA Health Disparities grant (#EIHD2204), NIH (NIBIB) MIDRC grant under contracts 75N92020C00008 and 75N92020C00021, AIM-AHEAD pilot project award, DeepLook award for validating radiomics breast cancer model, Clarity consortium award and GE Edison; Honoraria by NBER (National Bureau of Economic Research) for authorship in their 2022 conference collection; and leadership roles as SIIM board of director, Member of the HL7 Board, and ACR Advisory Committee. Dr Purkayastha declares leadership roles as DSMB member for Hummer NIMH R61 (#1R61MH119291–01). Dr Shiradkar declares support from NCIR01CA208236, DoD CA210856, Pilot funding from the Winship Cancer Center (ACS-IRG, Winship Invest$). All other authors state that they have no conflict of interest related to the material discussed in this article. The authors are non-partner/non-partnership track/employees.
REFERENCES
- 1.Rajpurkar P, Irvin J, Zhu K, et al. CheXNet: radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv. Published November 14, 2017. Available at: http://arxiv.org/abs/1711.05225. Accessed July 30, 2023. [Google Scholar]
- 2.Ting DSW, Cheung CYL, Lim G, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA 2017;318:2211–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker AS, Marcon M, Ghafoor S, Wurnig MC, Frauenfelder T, Boss A. Deep learning in mammography: diagnostic accuracy of a multipurpose image analysis software in the detection of breast cancer. Invest Radiol 2017;52:434–40. [DOI] [PubMed] [Google Scholar]
- 4.US Food and Drug Administration, Center for Devices and Radiological Health. Artificial intelligence and machine learning (AI/ML)-enabled medical devices. Available at: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-aiml-enabled-medical-devices. Accessed March 23, 2023.
- 5.Parikh RB, Teeple S, Navathe AS. Addressing bias in artificial intelligence in health care. JAMA 2019;322:2377–8. [DOI] [PubMed] [Google Scholar]
- 6.Seyyed-Kalantari L, Zhang H, McDermott MBA, Chen IY, Ghassemi M. Underdiagnosis bias of artificial intelligence algorithms applied to chest radiographs in under-served patient populations. Nat Med 2021;27:2176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whittaker M, Alper M, Bennett CL, et al. Disability, bias, and AI. AI Now Institute Available at: https://ainowinstitute.org/publication/disabilitybiasai-2019. Accessed July 30, 2023. [Google Scholar]
- 8.Obermeyer Z, Powers B, Vogeli C, Mullainathan S. Dissecting racial bias in an algorithm used to manage the health of populations. Science 2019;366:447–53. [DOI] [PubMed] [Google Scholar]
- 9.Geirhos R, Jacobsen JH, Michaelis C, et al. Shortcut learning in deep neural networks. Mat Machine Intel 2020;2:665–73. [Google Scholar]
- 10.DeGrave AJ, Janizek JD, Lee SI. AI for radiographic COVID-19 detection selects shortcuts over signal. Mat Machine Intel 2021;3:610–9. [Google Scholar]
- 11.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med 2018;15:e1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rueckel J, Trappmann L, Schachtner B, et al. Impact of confounding thoracic tubes and pleural dehiscence extent on artificial intelligence pneumothorax detection in chest radiographs. Invest Radiol 2020;55:792–8. [DOI] [PubMed] [Google Scholar]
- 13.Gichoya JW, Banerjee I, Bhimireddy AR, et al. AI recognition of patient race in medical imaging: a modelling study. Lancet Digit Health 2022;4:e406–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seyyed-Kalantari L, Liu G, McDermott M, Chen IY, Ghassemi M. CheXclusion: fairness gaps in deep chest x-ray classifiers. Pac Symp Biocomput 2021;26:232–43. [PubMed] [Google Scholar]
- 15.Hao K This is how AI bias really happens—and why it’s so hard to fix. MIT Technology Review. Published February 4, 2019. Available at: https://www.technologyreview.com/2019/02/04/137602/this-is-howai-bias-really-happensand-why-its-so-hard-to-fix/. Accessed March 23, 2023.
- 16.Kaushal A, Altman R, Langlotz C. Geographic distribution of US cohorts used to train deep learning algorithms. JAMA 2020;324:1212–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee RS, Gimenez F, Hoogi A, Miyake KK, Gorovoy M, Rubin DL. A curated mammography data set for use in computer-aided detection and diagnosis research. Sci Data 2017;4:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halling-Brown MD, Warren LM, Ward D, et al. OPTIMAM mammography image database: a large-scale resource of mammography images and clinical data. Radiol Artif Intell 2021;3:e200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaffter T, Buist DSM, Lee CI, et al. Evaluation of combined artificial intelligence and radiologist assessment to interpret screening mammograms. JAMA Netw Open 2020;3:e200265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shan A, Baumann G, Gholamrezanezhad A. Patient race/ethnicity and diagnostic imaging utilization in the emergency department: a systematic review. J Am Coll Radiol 2021;18:795–808. [DOI] [PubMed] [Google Scholar]
- 21.Christensen EW, Waid M, Scott J, Patel BK, Bello JA, Rula EY. Relationship between race and access to newer mammographic technology in women with medicare insurance. Radiology 2023;306:e221153. [DOI] [PubMed] [Google Scholar]
- 22.Celi LA, Cellini J, Charpignon ML, Dee EC. Sources of bias in artificial intelligence that perpetuate healthcare disparities—a global review. PLoS Digital. Available at: https://journals.plos.org/digitalhealth/article?id=10.1371/journal.pdig.0000022&utm. Accessed July 30, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramesh V, Chi NA, Rajpurkar P. Improving radiology report generation systems by removing hallucinated references to non-existent priors. In: Parziale A, Agrawal M, Joshi S, et al. , eds. Proceedings of the 2nd Machine Learning for Health Symposium. Proc Machine Learning Res; 2022;193:456–73. [Google Scholar]
- 24.Bhadra S, Kelkar VA, Brooks FJ, Anastasio MA. On hallucinations in tomographic image reconstruction. IEEE Trans Med Imaging 2021;40:3249–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendrick RE, Monticciolo DL, Biggs KW, Malak SF. Age distributions of breast cancer diagnosis and mortality by race and ethnicity in US women. Cancer 2021;127:4384–92. [DOI] [PubMed] [Google Scholar]
- 26.Irvin J, Rajpurkar P, Ko M, et al. CheXpert: a large chest radiograph dataset with uncertainty labels and expert comparison. AAAI 2019;33:590–7. [Google Scholar]
- 27.Adam H, Yang MY, Cato K, et al. Write it like you see it: detectable differences in clinical notes by race lead to differential model recommendations. arXiv. Published May 8, 2022. Available at: http://arxiv.org/abs/2205.03931. Accessed July 30, 2023. [Google Scholar]
- 28.Oakden-Rayner L Exploring large-scale public medical image datasets. Acad Radiol 2020;27:106–12. [DOI] [PubMed] [Google Scholar]
- 29.Roge A, Hiremath A, Sobota M, et al. Evaluating the sensitivity of deep learning to inter-reader variations in lesion delineations on biparametric MRI in identifying clinically significant prostate cancer. In: Medical imaging 2022: computer-aided diagnosis. Bellingham, WA: SPIE; 2022:264–73. [Google Scholar]
- 30.Rueckel J, Huemmer C, Fieselmann A, et al. Pneumothorax detection in chest radiographs: optimizing artificial intelligence system for accuracy and confounding bias reduction using in-image annotations in algorithm training. Eur Radiol 2021;31:7888–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierson E, Cutler DM, Leskovec J, Mullainathan S, Obermeyer Z. An algorithmic approach to reducing unexplained pain disparities in underserved populations. Nat Med 2021;27:136–40. [DOI] [PubMed] [Google Scholar]
- 32.De Jay N, Papillon-Cavanagh S, Olsen C, El-Hachem N, Bontempi G, Haibe-Kains B. mRMRe: an R package for parallelized mRMR ensemble feature selection. Bioinformatics 2013;29:2365–8. [DOI] [PubMed] [Google Scholar]
- 33.Bachman P, Hjelm RD, Buchwalter W. Learning representations by maximizing mutual information across views. In: Wallach H, Larochelle H, Beygelzimer A, d’Alché-Buc F, Fox E, Garnett R, eds. Advances in neural information processing systems, vol 32. Available at: https://proceedings.neurips.cc/paper_files/paper/2019/file/ddf354219aac374f1d40b7e760ee5bb7-Paper.pdf. Accessed July 30, 2023. [Google Scholar]
- 34.Krawczuk J, Łukaszuk T. The feature selection bias problem in relation to high-dimensional gene data. Artif Intell Med 2016;66:63–71. [DOI] [PubMed] [Google Scholar]
- 35.Yu H, Wang Y, Zeng D. A general framework of nonparametric feature selection in high-dimensional data. Biometrics 2023;79:951–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Climente-González H, Azencott CA, Kaski S, Yamada M. Block HSIC Lasso: model-free biomarker detection for ultra-high dimensional data. Bioinformatics 2019;35:i427–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danks D, London AJ. Algorithmic bias in autonomous systems. In: Proceedings of the Twenty-Sixth International Joint Conference on Artificial Intelligence. International Joint Conferences on Artificial Intelligence Organization; 2017. [Google Scholar]
- 38.Ribeiro MT, Singh S, Guestrin C. “Why should I trust you?” Explaining the predictions of any classifier. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining: KDD ‘16. New York: Association for Computing Machinery; 2016:1135–44. [Google Scholar]
- 39.Arun N, Gaw N, Singh P, et al. Assessing the trustworthiness of saliency maps for localizing abnormalities in medical imaging. Radiol Artif Intell 2021;3:e200267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reyes M, Meier R, Pereira S, et al. On the interpretability of artificial intelligence in radiology: challenges and opportunities. Radiol Artif Intell 2020;2:e190043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeCamp M, Lindvall C. Latent bias and the implementation of artificial intelligence in medicine. J Am Med Inform Assoc 2020;27:2020–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Obuchowski NA, Bullen JA. Statistical considerations for testing an AI algorithm used for prescreening lung CT images. Contemp Clin Trials Commun 2019;16:100434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamiran F, Calders T. Data preprocessing techniques for classification without discrimination. Knowl Inf Syst 2012;33:1–33. [Google Scholar]
- 44.Feldman M, Friedler SA, Moeller J, Scheidegger C, Venkatasubramanian S. Certifying and removing disparate impact. In: Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining: KDD ‘15. New York: Association for Computing Machinery; 2015:259–68. [Google Scholar]
- 45.Zemel R, Wu Y, Swersky K, Pitassi T, Dwork C. Learning fair representations. In: Dasgupta S, McAllester D, eds. Proceedings of the 30th International Conference on Machine Learning, Vol 28. Proceedings of Machine Learning Research; 2013:325–33. [Google Scholar]
- 46.Calmon F, Wei D, Vinzamuri B, Natesan Ramamurthy K, Varshney KR. Optimized pre-processing for discrimination prevention. In: Guyon I, Luxburg UV, Bengio S, et al. eds. Advances in neural information processing systems, vol 30. Available at: https://proceedings.neurips.cc/paper_files/paper/2017/file/9a49a25d845a483fae4be7e341368e36-Paper.pdf. Accessed July 30, 2023. [Google Scholar]
- 47.Celis LE, Keswani V, Vishnoi N. Data preprocessing to mitigate bias: a maximum entropy based approach. In: Iii HD, Singh A, eds. Proceedings of the 37th International Conference on Machine Learning, Vol 119. Proceedings of Machine Learning Research; 2020:1349–59. [Google Scholar]
- 48.Das D, Santosh KC, Pal U. Cross-population train/test deep learning model: abnormality screening in chest x-rays. In: 2020 IEEE 33rd International Symposium on Computer-Based Medical Systems. (CBMS; ); 2020:514–9. [Google Scholar]
- 49.Larrazabal AJ, Nieto N, Peterson V, Milone DH, Ferrante E. Gender imbalance in medical imaging datasets produces biased classifiers for computer-aided diagnosis. Proc Natl Acad Sci 2020;117:12592–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puyol-Antón E, Ruijsink B, Piechnik SK, et al. Fairness in cardiac MR image analysis: an investigation of bias due to data imbalance in deep learning based segmentation. In: Medical Image Computing and Computer Assisted Intervention—MICCAI 2021. Cham, Switzerland: Springer International; 2021:413–23. [Google Scholar]
- 51.Morris JC, Schindler SE, McCue LM, et al. Assessment of racial disparities in biomarkers for Alzheimer disease. JAMA Neurol 2019;76:264–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu G, Allen B, Lopez O, et al. Racial differences in gray matter integrity by diffusion tensor in black and white octogenarians. Curr Alzheimer Res 2015;12:648–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Handa VL, Lockhart ME, Fielding JR, et al. Racial differences in pelvic anatomy by magnetic resonance imaging. Obstet Gynecol 2008;111:914–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burlina P, Joshi N, Paul W, Pacheco KD, Bressler NM. Addressing artificial intelligence bias in retinal diagnostics. Transl Vis Sci Technol 2021;10:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Celis LE, Huang L, Keswani V, Vishnoi NK. Classification with fairness constraints: a meta-algorithm with provable guarantees. In: Proceedings of the Conference on Fairness, Accountability, and Transparency: FAT* ‘19. New York: Association for Computing Machinery; 2019:319–28. [Google Scholar]
- 56.Kamishima T, Akaho S, Asoh H, Sakuma J. Fairness-aware classifier with prejudice remover regularizer. In: Machine learning and knowledge discovery in databases. Berlin: Springer; 2012:35–50. [Google Scholar]
- 57.Agarwal A, Beygelzimer A, Dudík M, Langford J, Wallach H. A reductions approach to fair classification. arXiv. Published March 6, 2018. Available at: http://proceedings.mlr.press/v80/agarwal18a/agarwal18a.pdf. Accessed March 25, 2023. [Google Scholar]
- 58.Kearns M, Neel S, Roth A, Wu ZS. Preventing fairness gerrymandering: auditing and learning for subgroup fairness. In: Dy J, Krause A, eds. Proceedings of the 35th International Conference on Machine Learning, Vol 80. Proceedings of Machine Learning Research; 2018:2564–72. [Google Scholar]
- 59.Reimers C, Bodesheim P, Runge J, Denzler J. Towards learning an unbiased classifier from biased data via conditional adversarial debiasing. arXiv. Published March 10, 2021. Available at: http://arxiv.org/abs/2103.06179. Accessed July 30, 2023. [Google Scholar]
- 60.Zhang BH, Lemoine B, Mitchell M. Mitigating unwanted biases with adversarial learning. In: Proceedings of the 2018 AAAI/ACM Conference on AI, Ethics, and Society: AIES ‘18. New York: Association for Computing Machinery; 2018:335–40. [Google Scholar]
- 61.Zafar MB, Valera I, Rodriguez MG, Gummadi KP. Fairness constraints: mechanisms for fair classification. arXiv. Published July 19, 2015. Available at: http://proceedings.mlr.press/v54/zafar17a/zafar17a.pdf. Accessed March 25, 2023. [Google Scholar]
- 62.Zafar MB, Valera I, Gomez-Rodriguez M, Gummadi KP. Fairness constraints: a flexible approach for fair classification. J Mach Learn Res 2019;20:1–42. [Google Scholar]
- 63.Donini M, Oneto L, Ben-David S, Shawe-Taylor J, Pontil M. Empirical risk minimization under fairness constraints. arXiv. Published February 23, 2018. Available at: http://arxiv.org/abs/1802.08626. Accessed July 30, 2023. [Google Scholar]
- 64.Berk R, Heidari H, Jabbari S, et al. A convex framework for fair regression. arXiv. Published June 7, 2017. Available at: http://arxiv.org/abs/1706.02409. Accessed July 30, 2023. [Google Scholar]
- 65.Dinsdale NK, Jenkinson M, Namburete AIL. Deep learning-based unlearning of dataset bias for MRI harmonisation and confound removal. Neuroimage 2021;228:117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Correa R, Jeong JJ, Patel B, Trivedi H, Gichoya JW, Banerjee I. Twostep adversarial debiasing with partial learning—medical image case-studies. arXiv. Published November 16, 2021. Available at: http://arxiv.org/abs/2111.08711. Accessed July 30, 2023. [Google Scholar]
- 67.Chang K, Balachandar N, Lam C, et al. Distributed deep learning networks among institutions for medical imaging. J Am Med Inform Assoc 2018;25:945–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brendan McMahan H, Moore E, Ramage D, Hampson S, Agüera y Arcas B. Communication-efficient learning of deep networks from decentralized data. arXiv. Published February 17, 2016. Available at: http://arxiv.org/abs/1602.05629. Accessed July 30, 2023. [Google Scholar]
- 69.Li T, Sahu AK, Talwalkar A, Smith V. Federated learning: challenges, methods, and future directions. IEEE Signal Process Mag 2020;37:50–60. [Google Scholar]
- 70.Peng L, Luo G, Walker A, et al. Evaluation of federated learning variations for COVID-19 diagnosis using chest radiographs from 42 US and European hospitals. J Am Med Inform Assoc 2022;30:54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hosseini SM, Sikaroudi M, Babaie M, Tizhoosh HR. Proportionally fair hospital collaborations in federated learning of histopathology images. IEEE Trans Med Imaging 2023;42(7). [DOI] [PubMed] [Google Scholar]
- 72.Shiradkar R, Ghose S, Mahran A, et al. Prostate surface distension and tumor texture descriptors from pre-treatment MRI are associated with biochemical recurrence following radical prostatectomy: preliminary findings. Front Oncol 2022;12:841801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pleiss G, Raghavan M, Wu F, Kleinberg J, Weinberger KQ. On fairness and calibration. In: Guyon I, Luxburg UV, Bengio S, et al. , eds. Advances in neural information processing systems, vol 30. Available at: https://proceedings.neurips.cc/paper_files/paper/2017/file/b8b9c74ac526fffbeb2d39ab038d1cd7-Paper.pdf. Accessed July 30, 2023. [Google Scholar]
- 74.Hardt M, Price E, Srebro N. Equality of opportunity in supervised learning. In: Proceedings of the 30th International Conference on Neural Information Processing Systems: NIPS’16. Red Hook, NY: Curran Associates; 2016:3323–31. [Google Scholar]
- 75.Marcinkevics R, Ozkan E, Vogt JE. Debiasing deep chest x-ray classifiers using intra- and post-processing methods. arXiv. Published July 26, 2022. Available at: http://arxiv.org/abs/2208.00781. Accessed July 30, 2023. [Google Scholar]
- 76.Clapés A, Anbarjafari G, Bilici O, Temirova D, Avots E, Escalera S. From apparent to real age: gender, age, ethnic, makeup, and expression bias analysis in real age estimation. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops. (CVPRW; ); 2018:2436–243609. [Google Scholar]
- 77.Shachar C, Gerke S. Prevention of bias and discrimination in clinical practice algorithms. JAMA 2023;329:283–4. [DOI] [PubMed] [Google Scholar]
- 78.Goodman KE, Morgan DJ, Hoffmann DE. Clinical algorithms, antidiscrimination laws, and medical device regulation. JAMA 2023;329:285–6. [DOI] [PubMed] [Google Scholar]
- 79.Bellamy RKE, Dey K, Hind M, et al. AI Fairness 360: an extensible toolkit for detecting, understanding, and mitigating unwanted algorithmic bias. arXiv. Published October 3, 2018. Available at: http://arxiv.org/abs/1810.01943. Accessed July 30, 2023. [Google Scholar]
- 80.Bantilan N Themis-ml: a fairness-aware machine learning interface for end-to-end discrimination discovery and mitigation. J Technol Hum Serv 2018;36:15–30. [Google Scholar]
- 81.Bird S, Dudík M, Edgar R, et al. Fairlearn: a toolkit for assessing and improving fairness in AI. Published May 18, 2020. Available at: https://www.microsoft.com/en-us/research/publication/2020/05/Fairlearn_WhitePaper-2020-09-22.pdf. Accessed March 25, 2023.
- 82.Adebayo JA. FairML: toolbox for diagnosing bias in predictive modeling. Available at: https://dspace.mit.edu/handle/1721.1/108212?show=full. Accessed March 25, 2023.
- 83.Saleiro P, Kuester B, Hinkson L, et al. Aequitas: a bias and fairness audit toolkit. arXiv. Published November 14, 2018. Available at: http://arxiv.org/abs/1811.05577. Accessed July 30, 2023. [Google Scholar]
- 84.Xu G, Han X, Deng G, et al. VerifyML: obliviously checking model fairness resilient to malicious model holder. arXiv. Published October 16, 2022. Available at: http://arxiv.org/abs/2210.08418. Accessed July 30, 2023. [Google Scholar]
- 85.Johnson B, Brun Y. Fairkit-learn: a fairness evaluation and comparison toolkit. In: 2022 IEEE/ACM 44th International Conference on Software Engineering: Companion Proceedings. (ICSE-Companion; ); 2022:70–4. [Google Scholar]
- 86.Friedler SA, Scheidegger C, Venkatasubramanian S, Choudhary S, Hamilton EP, Roth D. A comparative study of fairness-enhancing interventions in machine learning. arXiv. Published February 13, 2018. Available at: http://arxiv.org/abs/1802.04422. Accessed July 30, 2023. [Google Scholar]
- 87.Žliobaitė I Measuring discrimination in algorithmic decision making. Data Min Knowl Discov 2017;31:1060–89. [Google Scholar]
- 88.Tramèr F, Atlidakis V, Geambasu R, et al. FairTest: discovering unwarranted associations in data-driven applications. arXiv. Published October 8, 2015. Available at: http://arxiv.org/abs/1510.02377. Accessed July 30, 2023. [Google Scholar]
- 89.What-If Tool. Available at: https://pair-code.github.io/what-if-tool/. Accessed March 25, 2023. [Google Scholar]
- 90.IBM Documentation. Published October 26, 2022. https://www.ibm.com/docs/en/cloud-paks/cp-data/3.5.0?topic=services-watson-openscale. Accessed March 25, 2023.