Abstract
Chemical modifications on mRNA represent a critical layer of gene expression regulation. Research in this area has continued to accelerate over the last decade, as more modifications are being characterized with increasing depth and breadth. mRNA modifications have been demonstrated to influence nearly every step from the early phases of transcript synthesis in the nucleus through to their decay in the cytoplasm, but in many cases, the molecular mechanisms involved in these processes remain mysterious. Here, we highlight recent work that has elucidated the roles of mRNA modifications throughout the mRNA life cycle, describe gaps in our understanding and remaining open questions, and offer some forward-looking perspective on future directions in the field.
Keywords: RNA modifications, RNA metabolism, posttranscriptional regulation, epitranscriptome
1. INTRODUCTION
RNA contains numerous chemical modifications in all organisms. Among the more than 170 chemically distinct RNA modifications known (1), several have been characterized in eukaryotic mRNA. Research on mRNA modifications has expanded exponentially in the past 10 years, fueled by the development of genome-scale methods to map the locations of modified nucleosides such as N6-methyladenosine (m6A), pseudouridine (Ψ), N5-methylcytidine (m5C), N1-methyladenosine (m1A), N4-acetylcytidine (ac4C), dihydrouridine (D), and 2′-O-methyl ribose on any nucleoside (Nm) (Figure 1) (for a review of genome-scale approaches to mapping mRNA modifications, see 2-4). Each of these modifications alters the chemical properties of RNA. The chemical changes caused by modifications affect RNA–RNA (Figure 2) and RNA–protein interactions in various ways that are known, or likely, to affect mRNA metabolism. Here, we review recent literature that connects mRNA modifications to effects on steps in the mRNA life cycle from birth to death, including the interplay between transcription and cotranscriptional RNA modification, capping, splicing, cleavage and polyadenylation, export from the nucleus, translation, and decay (for a review of works before 2018, see 5). The extensive literature on m6A has been reviewed recently (6, 7) and is covered here only in cases where m6A mediates a mechanism of posttranscriptional gene regulation that has not yet been characterized for other mRNA modifications but is likely to occur.
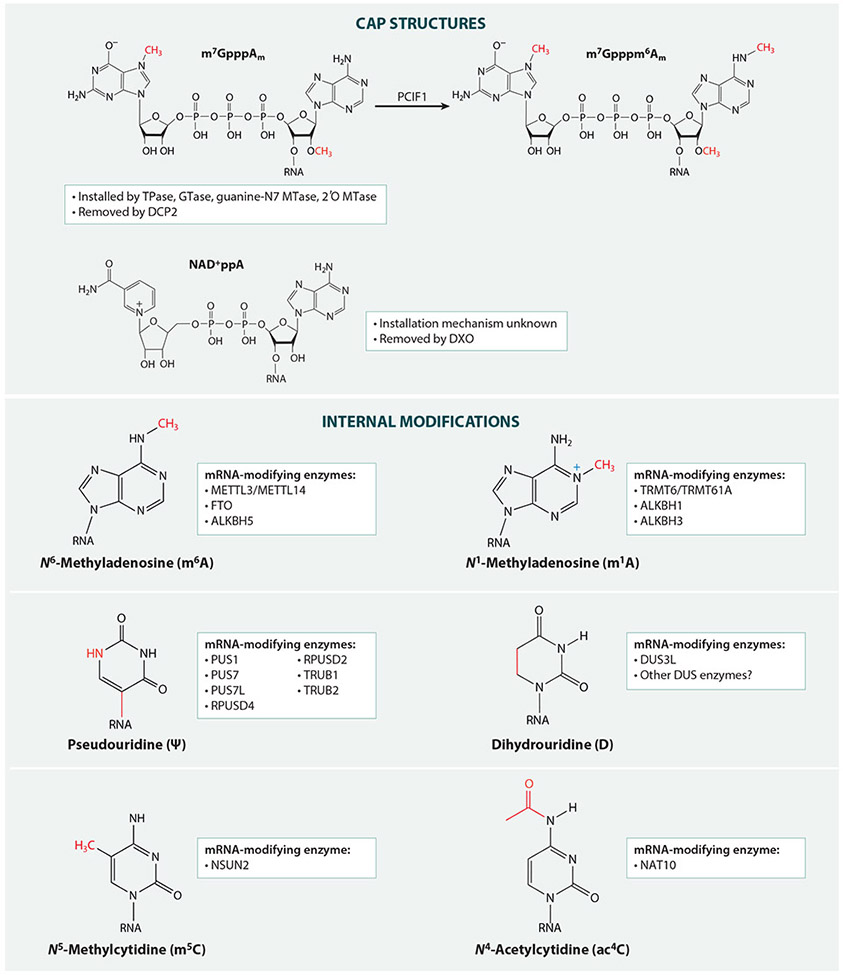
Figure 1.
Chemical structures of the modifications described in this review, as well as known enzymes that install and remove these modifications specifically on mRNA. Abbreviations: Ψ, pseudouridine; ac4C, N4-acetylcytidine; ALKBH, alkB homolog; D, dihydrouridine; DCP2, decapping mRNA 2; DUS, dihydrouridine synthase; DXO, decapping exoribonuclease; FTO, alpha-ketoglutarate dependent dioxygenase; GTase, guanylyl transferase; m1A, N1-methyladenosine; m5C, N5-methylcytidine; m6A, N6-methyladenosine; METTL, methyltransferase like; MTase, methyltransferase; NAD+ppA, NAD+ cap structure with 2′-O-methylated adenosine; NAT10, nuclear acetyltransferase 10; NSUN2, NOP2/Sun RNA methyltransferase 2; PCIF1, phosphorylated CTD interacting factor 1; PUS, pseudouridine synthase; RPUS, RNA pseudouridine synthase; TPase, RNA triphosphatase; TRMT, tRNA methyltransferase; TRUB, TruB pseudouridine synthase family.
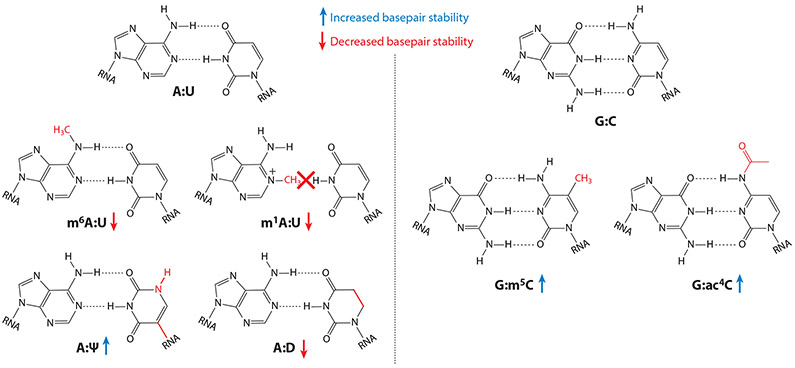
Figure 2.
Chemical structures of A:U and G:C base pairs, showing how the chemical modifications m1A, m6A, Ψ, D, m5C, and ac4C impact base pairing. Arrows indicate increased (blue) or decreased (red) base pair stability as a result of the indicated modification. Abbreviations: Ψ, pseudouridine; ac4C, N4-acetylcytidine; D, dihydrouridine; m1A, N1-methyladenosine; m5C, N5-methylcytidine; m6A, N6-methyladenosine.
2. INTERPLAY BETWEEN TRANSCRIPTION AND RNA MODIFICATION
Key steps in mRNA processing occur cotranscriptionally, including capping, splicing, cleavage, and polyadenylation. Furthermore, proteins that affect later steps in the mRNA life cycle including export from the nucleus, localization in the cytoplasm, translation, and decay are loaded onto nascent pre-mRNAs to assemble functional messenger ribonucleoproteins. It is therefore of interest to know when during mRNA biogenesis RNA modifications are deposited, because this timing constrains the potential effects of RNA modifications on gene expression. Each of the most abundant mRNA modifications is installed by one or more nuclear enzymes, raising the possibility of modification of nascent pre-mRNA at a sufficiently early stage in mRNA biogenesis to affect nuclear processing events and export to the cytoplasm (Figure 3).
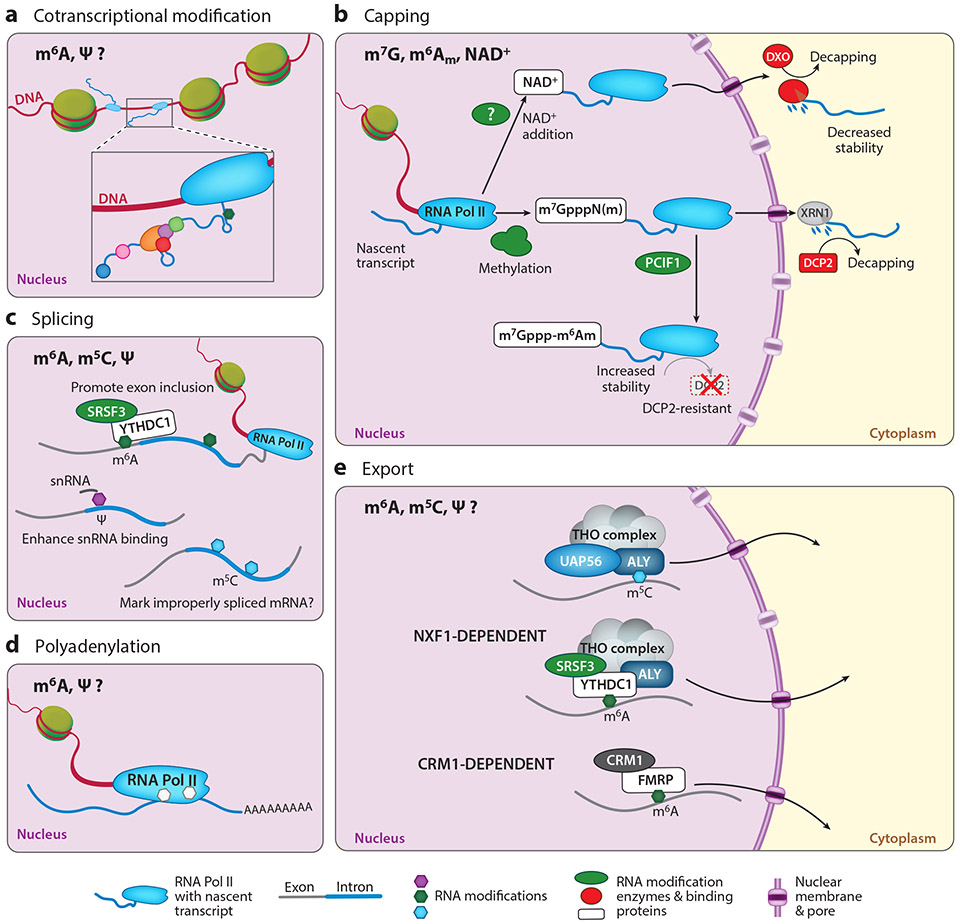
Figure 3.
mRNA modifications can be installed cotranscriptionally and regulate multiple steps in nuclear processing and export. (a) Some modifications are installed cotranscriptionally via direct recruitment of modification enzymes to RNA Pol II as it synthesizes nascent transcripts. (b) mRNA capping involves multiple modifications of the 5′ end of transcripts, which can directly modulate mRNA stability by altering susceptibility to decapping and degradation enzymes. (c) Splicing also often occurs cotranscriptionally and has been shown to be regulated by multiple modifications that can alter mRNA–snRNA interactions, recruit proteins that regulate exon inclusion, or mark improperly spliced transcripts. (d) Less is known about how mRNA modifications influence polyadenylation, but both m6A and Ψ may play important roles. (e) RNA modifications likely also influence the nuclear export of properly processed transcripts through both the NXF1- and CRM1-dependent pathways. Abbreviations: Ψ, pseudouridine; ALY, Aly/REF export factor; CRM1, exportin 1; DCP2, decapping mRNA 2; DXO, decapping exoribonuclease; FMRP, fragile X messenger ribonucleoprotein; m5C, N5-methylcytidine; m6A, N6-methyladenosine; m6Am, N6-2′-O-dimethyladenosine; m7G, N7-methylguanosine; NAD+, nicotinamide-adenine dinucleotide; NXF1, nuclear RNA export factor 1; PCIF1, phosphorylated CTD interacting factor 1; RNA Pol II, RNA polymerase II; snRNA, small nuclear RNA; SRSF3, serine and arginine rich splicing factor 3; THO, THO nuclear export complex; UAP56, DExD-box helicase 39B; XRN1, 5′–3′ exoribonuclease 1; YTHDC1, YTH domain containing protein 1.
2.1. Cotranscriptional pre-mRNA Modification
Cotranscriptional deposition has been demonstrated for the two most abundant modifications found in mature mRNA in human cells, m6A and Ψ. Ke et al. (8) exploited the stable association of nascent RNA attached to elongating RNA polymerase with chromatin to compare the m6A landscape in nascent chromatin-associated pre-mRNA to cytoplasmic mRNA (9). This work concluded that m6A is predominantly installed cotranscriptionally, although it did not exclude the possibility of dynamic changes in mature mRNA methylation under different conditions. A different approach used pulse-chase metabolic labeling with bromouridine to show that m6A is rapidly installed in nascent pre-mRNA in HEK293 cells (10). The m6A methyltransferase complex, METTL3–METTL14, interacts with RNA Pol II (11) and is thus poised to modify nascent pre-mRNA as it emerges from the polymerase. RNA Pol II elongation speed varies across genes (12), and slow elongation through exons may underlie the observed enrichment of m6A in exons (13), despite the abundance of potential m6A sites (RRACH motifs) in introns (8).
Martinez et al. (14) performed biochemical enrichment of pre-mRNA from the chromatin fraction to show that Ψ is installed cotranscriptionally in human cells. Unlike m6A, Ψ is more evenly distributed between exons and introns. Pseudouridine synthase 7 (PUS7) was found to copurify with components of actively transcribed chromatin (15), consistent with its modification of unspliced pre-mRNA. Several additional PUSs were identified as having pre-mRNA targets (14), but the biochemical basis for cotranscriptional recruitment of these enzymes is unknown. It is unclear whether specific recruitment of PUSs to chromatin is necessary to explain the observed distribution of Ψ sites. Notably, all tested PUSs could modify specific intronic target sites in a reconstituted assay containing only minimal RNA and purified PUSs (14). Thus, direct recruitment to elongating RNA polymerase is not strictly required for cotranscriptional RNA modification, although it may increase modification of specific targets, which could contribute to tissue-specific patterns of mRNA modification.
The presence of modified sites within introns provides suggestive evidence that nascent pre-mRNA is a target. Intronic sites were mapped for m5C in mouse embryonic stem cells (mESCs) and brain tissue (16), hm5C in Drosophila S2 cells (17) and mESCs (18), m1A in HEK293T cells (19), and D in budding yeast (20). However, most intronic modified sites were mapped in poly(A)-selected RNA, which may include intron-retained mature mRNA. The observation that short-lived RNAs transcribed from enhancers contain m5C provides further evidence that cotranscriptional deposition of this modification is likely (21). The TRMT6–TRMT61A methyltransferase that installs m1A in mRNA and tRNA is nuclear in human cells and interacts with splicing factors in high-throughput studies (22), but it is not known whether this interaction takes place in the context of nascent pre-mRNA. D was identified in a few introns in poly(A)-selected RNA from Saccharomyces cerevisiae (20), but it was not determined when in the mRNA life cycle these intronic sites were modified. For the intron-modified RPL30 gene, deletion of D synthases resulted in a modest accumulation of unspliced RNA, which could reflect impaired splicing or greater stability of unspliced RNA. Application of appropriate mapping techniques to purified chromatin-associated or metabolically labeled nascent RNA will clarify whether intronic m5C, hm5C, m1A, or D modifications are installed cotranscriptionally and whether they have the potential to affect nuclear pre-mRNA processing.
ac4C is installed in mRNA by the nuclear acetyltransferase NAT10 (23). Mapping of ac4C has been limited to mature poly(A)-selected RNA from total RNA thus far, and ac4C has not been reported in introns, which were depleted from the sequenced sample, as expected for mature mRNA. The basis for recruitment of NAT10 to specific targets, nascent or otherwise, remains to be determined. It is plausible that ac4C could be introduced in pre-mRNA during transcription, as NAT10 also modifies nascent pre-rRNA (24).
Several factors install 2′-O-methyl ribose in noncoding RNA, including the protein-only enzymes FTSJ3, TARBP1/TRMT3, FTSJ1/TRMT7, TRMT13, and TRMT44 and fibrillarin, which is guided to specific targets by base-pairing with C/D-box small nucleolar RNAs (25). The presence of 2′-O-methylated nucleosides in poly(A)-selected mRNA is supported by mass spectrometry analysis of human HeLa cells (26) and mouse liver (27). Initial maps have been reported using single-nucleotide-resolution methods (26), but the enzymes responsible have not been determined for the vast majority of mRNA sites. Both fibrillarin and FTSJ3 modify nascent pre-rRNA, consistent with the possibility for cotranscriptional mRNA methylation on ribose. Overall, it is likely that many mRNA modifications in addition to m6A and Ψ are deposited co-transcriptionally, endowing them with the potential to influence early steps in mRNA biogenesis. However, determining how this deposition is regulated requires detailed further analysis, as different modification enzymes may interact with the transcription machinery and nascent transcripts via different mechanisms.
2.2. Feedback from RNA Modification to Transcription
Transcription and RNA modification are coupled processes when modifying enzymes target nascent pre-mRNA. Functional coupling may occur in the opposite direction via feedback from pre-mRNA modification to the transcriptional machinery. For example, the METTL3–METTL14–WTAP m6A methyltransferase complex localizes to promoter and enhancer regions of actively transcribed DNA in human and Drosophila cells, and depletion of methyltransferase components or nuclear m6A-binding proteins affects transcription (28-30). Notably, tethering METTL3 was sufficient to affect RNA Pol II pausing in Drosophila cells, and this was not observed with catalytically dead METTL3, consistent with a direct role for m6A in RNA (29).
Mechanistically, m6A has been proposed to affect transcriptional regulation by several mechanisms that may generalize to other modifications. Work by Xu et al. (28) uncovered an antagonistic relationship between nascent m6A modification and premature transcription termination by the Integrator complex. They proposed that m6A deposited near the 5′ end of nascent pre-mRNAs promotes productive transcription elongation by recruiting RNA-binding proteins (RBPs) such as hnRNPG and YTHDC1 that compete with binding by Integrator. m1A also binds YTH proteins (31, 32) and so potentially regulates transcription processivity versus premature termination by the same mechanism as m6A. In some cells, m1A was found to be enriched near the 5′ ends of mRNAs. Other pre-mRNA modifications could suppress or enhance premature transcription termination by affecting binding of various RBPs that compete with Integrator.
RNA modifications may affect transcription through the formation of RNA-dependent phaseseparated condensates at sites of active transcription. Lee et al. (33) linked m6A-dependent condensate formation at enhancers to transcription activity. They showed that the nuclear m6A-binding protein YTHDC1 formed condensates in vitro that were enlarged in the presence of m6A-modified enhancer RNAs (eRNAs) but not unmodified eRNAs of the same sequence. They hypothesized that these condensates could promote the formation of other enhancer-associated condensates, which have been linked to enhancer activity and gene activation (34). Consistent with this hypothesis, the endogenous transcriptional coactivator BRD4 formed reduced numbers of nuclear foci when YTHDC1 or METTL3 was depleted and showed reduced association with enhancer DNA by chromatin immunoprecipitation sequencing. It will be interesting to test whether m1A, which also binds YTH proteins (31, 32), is present in eRNAs and similarly promotes condensate formation and gene activation.
The effects of m6A on transcription do not always increase mRNA production. m6A deposition was associated with stabilization of chromatin-associated regulatory RNAs and proposed to repress transcription by altering the local chromatin state (35). Because nascent chromatin-associated RNAs are short-lived and require specialized sequencing strategies, the landscape of such nuclear RNA modifications is poorly defined. Feedback from nascent RNA modifications to transcriptional output and regulation is a rapidly emerging topic in the m6A field that should be explored for other cotranscriptional RNA modifications.
3. CAPPING
Processing of the 5′ end of mRNA transcripts is critical for their stability and translation. In eukaryotic mRNA, the majority of transcripts contain a canonical cap containing a terminal N7-methylguanosine (m7G) linked to the next nucleotide via a 5′-to-5′ triphosphate linkage (36). This canonical cap is formed cotranscriptionally via a multistep process that includes an RNA triphosphatase to remove the terminal phosphate, an RNA guanylyltransferase to attach a terminal guanine, and an m7G methyltransferase to install the methyl group (37). This canonical cap is often also 2′-O-methylated at the first and sometimes second adjacent nucleotide by an m7G-specific 2′-O-methyltransferase (38). This canonical cap is thought to both stabilize the mRNA 5′ end by protecting it from exonucleolytic cleavage and enhance binding of translation machinery via direct interactions with the eukaryotic initiation factor (eIF) eIF4E (39). Though much less is known about their functions, additional noncanonical cap structures have been identified, including alternative terminal groups such as the metabolite nicotinamide-adenine dinucleotide (NAD+) (40, 41), and additional modification of cap-adjacent nucleotides, as in the case of N6-methylation of Am at the cap +1 position (42).
3.1. mRNA Capped with NAD+
Though initially identified in prokaryotic RNAs (43, 44), we now know that eukaryotic mRNA can also be capped with NAD+ (40, 41). S. cerevisiae and human NAD+-capped transcripts were identified by taking advantage of NAD+ reactivity with ADP ribosylcyclase, which allowed for biotin incorporation via click chemistry and subsequent capture with streptavidin. Both studies demonstrated extensive overlap between the pools of m7G- and NAD+-capped transcripts, suggesting that there are very few (if any) uniquely NAD+-capped mRNAs. Though difficult to quantify, current estimates suggest that approximately 1–5% of a given transcript has an NAD+ cap, while the rest likely carries a canonical cap structure (41). The mechanism of NAD+ cap addition remains unclear. While it has been demonstrated that both prokaryotic and eukaryotic RNA polymerases can use NAD+ to initiate transcription, the fact that small RNAs derived from intron cleavage can also be NAD+-capped suggests that a posttranscriptional mechanism also exists (40). NAD+ cap removal is better characterized, with the DXO and SpRai1 enzymes identified as the human and Schizosaccharomyces pombe NAD+-decapping enzymes, respectively (40) (Figure 3). In addition to its decapping activity, DXO has 5′-to-3′ exonuclease activity, which means it can generate its own RNA degradation substrate via its NAD+-decapping activity. As a result, removal of these decapping enzymes effectively stabilizes NAD+-capped transcripts. While NAD+-capped transcripts are not efficiently translated via canonical cap-dependent mechanisms, it remains to be seen whether protein can be synthesized via cap-independent mechanisms. Taken together, NAD+-capped transcripts are less stable and inefficiently translated, in direct contrast to the canonical m7G cap, which stabilizes transcripts against decay and increases translation efficiency.
Given that the same transcripts can be capped with NAD+ or m7G, how the distribution of each cap is regulated and how this impacts RNA and protein levels downstream need to be determined. It should be noted that Walters et al. (41) did observe functional enrichment for mitochondrial-encoded transcripts and nuclear-encoded ribosomal protein transcripts and that the levels of NAD+-capped mRNAs were higher in yeast grown in synthetic media compared to yeast grown in rich media. Since both mitochondrial function and ribosome biogenesis are sensitive to cellular metabolic state, it is possible that NAD+-cap addition serves as an indicator of this metabolic state. Since enzyme-mediated decapping can lead directly to 5′-mediated RNA decay, NAD+ could mark transcripts for rapid decay, similar to how uridylation can initiate 3′-mediated decay (45). Taken together, one could envision a regulatory mechanism by which a relative lack of nutrients (e.g., in synthetic media) would trigger increased NAD+-capping and subsequent degradation of transcripts related to mitochondrial and ribosomal function, reducing the energy burden on the cell. This is speculative, however, and significant additional work is needed to reveal the mechanisms of NAD+-cap installation and function.
3.2. Cap-Adjacent N6-2′-O-Dimethyladenosine
In addition to variation at the 5′ terminal position of mRNA transcripts, cap-adjacent nucleotides can also carry additional modifications. The canonical cap is often 2′-O-methylated at the adjacent two nucleotides. When the cap-adjacent nucleotide is an Am, it can be further N6-methylated to form m6Am (38). This additional methylation renders the cap resistant to DCP2-mediated removal, and initial characterization of this modification suggested that it modulates transcript stability (42). Multiple studies later identified PCIF1 as the methyltransferase that N6-methylates cap-adjacent Am nucleotides via recruitment to serine-5-phosphorylated RNA Pol II (46-49). Though the identification of PCIF1 facilitated more detailed study of m6Am function, these studies came to conflicting conclusions as to whether m6Am impacts transcript stability or translation. This may be in part due to confounding variables that need be disentangled: For instance, Boulias et al. (47) showed that while the first transcribed nucleotide of particularly stable transcripts with a half-life >24 h is often m6Am, the N6-methylation may not be the cause of but simply correlated with this phenomenon. Further work is needed to identify the correlative versus causative effects and to determine the effect of cap-adjacent m6Am. But given that it renders transcripts resistant to DCP2-mediated degradation (Figure 3), it is possible that it represents a mechanism to stabilize transcripts beyond the typical mRNA half-life of minutes to a few hours (50).
While these modifications represent just two that diverge from the canonical m7G cap, technical advances are revealing additional noncanonical caps in many different organisms (51). It should also be noted that the terminal cap modification, m7G, has been identified as an internal mRNA modification by mass spectrometry and sequencing studies (52, 53). While enzymatic tricks were used to isolate internal m7G from cap m7G, it remains difficult to accurately measure internal m7G abundance by mass spectrometry. However, these two studies leveraged both specific chemical reactivity and antibodies to map m7G sites and found that the methyltransferase METTL1, in a complex with WDR4, is responsible for internal m7G installation. While the prevalence and location of internal m7G sites on mRNA remain controversial (54), how such internal m7G sites might interact with elongating ribosomes and mRNA-binding proteins remains intriguing.
The chemical diversity of cap structures adds additional layers of complexity to transcriptional and posttranscriptional regulation of gene expression and is likely to have a significant impact on cellular transcriptomes and proteomes. However at present, very little is known about how different caps impact the interactions of mRNA transcripts with the translation machinery and other RBPs. Disentangling the effects of alternative caps on stability and/or translation, as well as distinguishing correlation from causation, is critical to progress in our understanding of these mechanisms. To that end, this is an area that could benefit greatly from biochemical and other in vitro experimental approaches to reveal the molecular players and interactions involved.
4. SPLICING
Intron removal by pre-mRNA splicing is an essential step in eukaryotic gene expression. Splicing is also highly regulated in human cells to produce alternative mRNA isoforms that encode distinct protein variants or control different levels of protein expression (55-57). RNA modifications that are installed cotranscriptionally on nascent pre-mRNA have the potential to directly influence splicing by affecting RNA–RNA and RNA–protein interactions (13). The role of pre-mRNA modifications in alternative splicing is only beginning to be investigated for modifications other than m6A. In Sections 4.1-4.3, we summarize suggestive evidence for modification-sensitive splicing, highlighting important gaps in knowledge and suggesting next steps for the field. For a more extensive treatment of the literature on m6A and splicing, see recent reviews (7, 13).
4.1. Evidence Linking pre-mRNA Modifications to Splicing and Recommended Approaches
Widespread changes in alternative splicing have been observed following genetic depletion of RNA-modifying enzymes including the m6A methyltransferase METTL3 (7); the demethylase FTO (58); and several PUSs, PUS1, PUS7, and RPUSD4 (14). Direct effects of site-specific pre-mRNA modifications on splicing have been shown in a handful of cases for m6A (reviewed in 7, 13) and Ψ (14). Recent work combined nascent RNA labeling with m6A immunoprecipitation and sequencing to relate the kinetics of splicing to the locations and extent of pre-mRNA methylation and proposed that m6A deposition near splice junctions was associated with faster splicing (10). By contrast, enrichment of m6A in introns was associated with slow splicing and alternative splicing. It is not clear whether this correlation reflects a widespread and direct effect of m6A on splicing kinetics, or if an upstream event, such as RNA polymerase elongation speed, independently affects m6A deposition and splicing efficiency.
The extent to which pre-mRNA modifications directly alter the splicing outcome is unclear from most studies. Typical experiments attempt to correlate changes in splicing following genetic manipulation of RNA-modifying enzymes with the locations of modified nucleotides within or adjacent to alternatively spliced regions of transcripts. However, data are lacking for the locations of most modified nucleosides within nascent pre-mRNA introns where many splicing regulatory elements reside.
Nascent pre-mRNA maps of m6A have been determined by two approaches: sequencing of chromatin-associated RNA (8) and sequencing of nascent RNA metabolically labeled with bromouridine (10). The map of Ψ in pre-mRNA from HepG2 cells was limited to highly expressed genes by the high sequence coverage required to distinguish Ψ sites from noisy data: Only ~1% of uridine positions were assessed for the presence of Ψ. Thus, no Ψ data were available for most PUS-sensitive alternatively spliced transcripts. Although antibody-based modification profiling such as for m6A has the potential to capture sites in unspliced pre-mRNA, the relative abundance of mature mRNA leads to underrepresentation of intronic regions in profiles of total cellular RNA. Future work combining enrichment of nascent pre-mRNA with enrichment of modified RNA could provide comprehensive maps of intronic pre-mRNA modification.
A limitation of genetic depletion studies to study the role of RNA modifications in splicing is that indirect effects may predominate. This is particularly the case when the RNA-modifying enzyme has been depleted for a long time (e.g., by genetic knockout or by constitutively expressed short hairpin RNAs). Only 20–30% of alternatively spliced exons that responded to METTL3 or FTO expression were found to contain m6A (8, 58, 59). Some of these splicing changes may be due to intronic m6A sites that were not mapped, but it is likely that many reflect indirect effects downstream of m6A-dependent changes in gene expression. Similarly, RNA sequencing showed splicing alterations in cells depleted of ac4C (NAT10−/− HeLa cells), but acetylated transcripts did not show more splicing changes than similarly expressed unmodified transcripts (23), suggesting a preponderance of indirect effects of NAT10 on splicing.
The RNA-modifying enzymes that install Ψ, m5C, m1A, and D in mRNA also modify tRNAs, and the absence of tRNA modifications can affect tRNA stability and function (60). Effects on the tRNA pool are likely to cause many indirect changes to splicing by affecting the production of splicing factors. Rapid inactivation using chemical inhibitors of catalytic activity, such as recently reported for METTL3 (61), is a promising approach to identify direct effects on splicing. A generalizable approach—as inhibitors are currently unavailable for most RNA-modifying enzymes—is the use of genetically encoded protein degrons for acute enzyme depletion prior to RNA sequencing (62). Wei et al. (63) used this strategy to distinguish splicing changes that are direct effects of METTL3 acting on pre-mRNA targets from indirect effects. They found that indirect effects predominate in steady state: Remarkably few alternative splicing events that were detected in METTL3 knockout mESCs (8) were recapitulated by acute degradation of METTL3 protein.
4.2. Mechanisms of Modification-Sensitive Splicing
Pre-mRNA modifications are likely to affect splicing by stabilizing or destabilizing RNA–RNA and RNA–protein interactions (13). Most modifications affect the stability of RNA base pairing (3) (Figure 2). Splicing requires base pairing between spliceosomal small nuclear RNAs (snRNAs) and intronic sequences, and Ψ modifications of snRNAs in the regions that base pair with pre-mRNA are known to affect splicing (64). It is plausible that Ψ modifications discovered within 5′ splice site and branch site regions (14) could directly affect splice site recognition by U1 and U2 snRNAs, respectively. In addition, pre-mRNA modifications can affect splicing by altering intramolecular RNA structures, as shown for m6A (59).
Clear examples of splicing regulation via modification-sensitive RBPs include regulation of splicing in human cells and in Drosophila by the m6A-binding protein YTHDC1. YTHDC1 was shown to bind methylated exons and promote binding of the splicing factor SRSF3 to promote exon inclusion (65). The Drosophila ortholog of YTHDC1, YT521-B, binds to m6A-methylated introns flanking a regulated sex-specific exon in Sex-lethal and represses inclusion of that exon (66, 67). YTHDC1 may be a general mediator of m6A-sensitive splicing given the broad similarity between splicing changes caused by acute depletion of METTL3 and conditional knockout of Ythdc1 in mESCs (35, 63). Binding of YTH proteins to m1A in addition to m6A (31, 32) raises the possibility that m1A could similarly affect splicing via YTHDC1.
Comparing the locations of pre-mRNA modifications to maps of binding sites for regulatory RBPs is a general strategy to identify candidate regulators of modification-sensitive alternative splicing. For example, m5C sites were identified that overlapped binding sites for splicing factors SRSF3 and SRSF4, with SRSF3 showing the most overlapping sites (16). The Encyclopedia of DNA Elements (ENCODE) project has generated a large data set of binding sites for more than one hundred RBPs in two human cell lines, HepG2 and K562, using enhanced crosslinking immunoprecipitation and sequencing (eCLIP) (68). Comparing these RBP maps to pre-mRNA Ψ sites mapped in HepG2 identified hundreds of overlaps including in introns flanking alternatively spliced regions (14).
4.3. Effects on Splicing or Nuclear Retention and Stability?
Work by Amort et al. (16) on m5C suggests another possible role for intronic RNA modifications: marking improperly spliced RNAs. Substantial fractions of m5C sites identified in nuclear poly(A)-selected RNA map to introns in mouse embryonic stem cells (44%) and mouse brain (70%). In contrast, comparatively few intronic m5C sites (11% and 3.4% of m5C sites mapped to introns in mESCs and brain, respectively) were detected in total cellular poly(A)+ RNA, which is mostly cytoplasmic. This disparity suggests that m5C-modified transcripts with retained introns are held in the nucleus and degraded without export to the cytoplasm, or alternatively that they are rapidly degraded upon entrance to the cytoplasm. It will be interesting to determine the point during or after the splicing process at which these intronic m5C sites are modified and whether the presence of m5C affects their fate of retention in the nucleus and/or rapid decay in the cytoplasm.
Taken together, there are examples suggesting that pre-mRNA modifications can directly impact splicing, motivating further work to expand this to additional modifications. However, studies that rely on genetic knockouts of modification enzymes are prone to indirect effects, due to changes in the kinetics of transcription and the effects of modification loss on other RNA populations such as tRNA, which can impact the production of the splicing machinery itself. Moving forward, acute perturbations to enzyme activity combined with sequencing approaches that specifically target mRNA are essential to distinguishing direct from indirect effects of modifications on pre-mRNA splicing.
5. POLYADENYLATION
Nuclear mRNA biogenesis ends with the cotranscriptional cleavage of nascent transcripts followed by addition of a nontemplated poly(A) tail. Processing of the 3′ end is an essential step in mRNA biogenesis that is extensively regulated to produce mRNA isoforms that differ in their 3′ untranslated regions (UTRs). Alternative 3′ UTRs affect most posttranscriptional steps in eukaryotic gene expression, including mRNA localization, stability, translation, and regulation by microRNAs (69). As noted in Section 2.1, m6A and Ψ have been established as cotranscriptional modifications of pre-mRNA, and indirect evidence makes it likely that additional mRNA modifications are installed in nascent pre-mRNA. The potential for cotranscriptional pre-mRNA pseudouridylation to affect alternative 3′-end processing was suggested by widespread changes in 3′ UTR isoforms following depletion of PUSs that modify nascent pre-mRNA (14). Although this work did not establish a direct role of specific Ψ in affecting cleavage and polyadenylation, Ψ was identified in the binding sites of cleavage and polyadenylation factors, suggesting a likely mechanism. Analysis of steady-state mRNA isoform abundance as a proxy for alternative cleavage and polyadenylation is confounded by the potential for mRNA modifications to affect transcript half-life. This is of particular concern in the case of a modification like m6A, which is known to destabilize transcripts. Overall, the role of cotranscriptional RNA modification in nuclear pre-mRNA processing is understudied.
6. EXPORT
Export of mRNAs from their nuclear transcription sites out to the cytoplasm for translation is intricately regulated and requires that many of the preceding processing steps discussed here have been properly carried out. Methylation of both cytidine (m5C) and adenosine (m6A) have thus far been implicated in the regulation of mRNA export via the NXF1-dependent export pathway (70, 71) (Figure 3). This pathway involves direct mRNA binding by the TREX complex via ALYREF, which functions as an adapter protein. ALYREF itself is a reported m5C-binding protein that facilitates the export of m5C-methylated mRNAs (71). While ALYREF shuttles between the nucleus and cytoplasm, loss of function of the m5C methyltransferase NSUN2 results in retention of more ALYREF in nuclear speckles, which led to the suggestion that loss of m5C-binding sites is sufficient to reduce its shuttling to the cytoplasm. While rescue experiments using both wild type and catalytically inactive NSUN2 support this claim, they do not take into consideration the non-mRNA-targeted effects of NSUN2. Nuclear speckles can accumulate RNA-processing factors under conditions that induce cellular stress. Such conditions could include loss of tRNA methylation that results in aberrant tRNA processing or fragmentation, which is likely to occur upon NSUN2 loss (72). In addition, while ALYREF has a higher apparent binding affinity for m5C-methylated mRNAs, it does bind unmethylated mRNAs, and mapping studies of ALYREF-binding sites in human cells reveal both motifs that contain C and those that lack C (73). Methylation is also not broadly required for the export of all mRNAs, suggesting that it may alter the export efficiencies of only subsets of transcripts. Therefore, there are likely specific conditions or sequence contexts in which ALYREF regulates the export of m5C-containing RNAs that still need to be elucidated.
In addition to m5C, m6A has also been implicated in the regulation of mRNA export via binding of YTHDC1 (74) and the TREX complex (70). This likely involves a complex containing both YTHDC1 and SRSF3, and knockdown of either YTHDC1 or SRSF3 increases the abundance of m6A-containing mRNAs in the nucleus (74). While this initial study did not detect an interaction between either YTHDC1 or SRSF3 and components of the NXF1-export pathway, NXF1 did slightly enrich for m6A as measured by immunoprecipitation followed by mass spectrometry of NXF1-bound RNA. Subsequent work, however, suggests that the m6A methyltransferase complex recruits the TREX complex to mRNA (70). Simultaneous knockdown of m6A methyltransferase complex components KIAA1429 and WTAP results in nuclear retention of m6A-methylated transcripts and a reduction of the association of methylated transcripts with the TREX components. In contrast to previous work, this study did detect interactions between YTHDC1, multiple TREX subunits, and NXF1. However, detection of these interactions by immunoprecipitation varied depending on the antibodies used and the configuration of the experiment, suggesting that the antibodies used in the earlier study may have masked the YTHDC1–NXF1 interaction interface. As with m5C, m6A is not strictly required for mRNA export, and it remains to be determined how and why m6A influences the export of some transcripts but not others. Since both the m6A- and m5C-mediated mechanisms converge on the TREX complex via different adaptor proteins (ALYREF and YTHDC1), it remains possible that other modifications also impact export via a similar mechanism.
Though mRNAs are primarily transported via the NXF1-dependent export pathway, some mRNAs can also be shuttled to the cytoplasm via the CRM1-dependent export pathway that is more typically associated with rRNA and snRNA transport. CRM1-dependent mRNA transport may also be regulated by m6A through its interaction with FMRP (75). While the difference in FMRP affinity for m6A-methylated versus unmethylated RNA is small, nuclear retention of m6A-methylated transcripts can be observed upon Fmr1 knockout in mice, and Fmr1 knockout mice phenocopy Mettl14 conditional knockout mice, suggesting a connection between m6A methylation, FMRP, and nuclear export of mRNAs. Work in murine leukemia virus has also demonstrated that both the NXF1 and CRM1 export pathways play important roles in the viral life cycle, and it has been speculated that methylation of viral RNA may play a role in this process as well (76, 77).
While there is tantalizing evidence suggesting that chemical modifications can regulate mRNA export, the reliance on genetic knockout of enzymes that target multiple types of RNA remains a critical issue. Genetic perturbation of enzymes such as NSUN2, which also modify tRNAs, can have wide-ranging impacts on the cellular transcriptome and proteome that can indirectly alter mRNA export. Distinguishing direct interactions between RNA modifications and export machinery from indirect downstream effects is critical for progress in this area. While some tools exist to perturb RNA modifications on specific transcripts (e.g., 78), they vary in efficiency and specificity, so additional technical advances are likely to be required to do this broadly.
7. TRANSLATION
The presence of modified nucleosides within mature mRNA invites the question: What do ribosomes do when they encounter modified mRNA? Single modified sites within coding sequences can affect the accuracy and rate of elongation either positively or negatively. This work has been recently reviewed (79). Here we focus on the effects of mRNA modifications on translation initiation, which is highly regulated and usually the rate-limiting step in protein production in eukaryotes.
Translation initiation requires mRNA recognition by eIFs for recruitment of a 48S preinitiation complex (PIC) consisting of a small ribosomal subunit complexed with initiator tRNA and additional eIFs (39). For most mRNAs, recognition of the m7G cap promotes ribosome recruitment near the 5′ end of the mRNA, and 48S PICs must scan the 5′ UTR to reach the translation initiation site (TIS). Sequences within 5′ UTRs control the rate of initiation by affecting cap recognition, eIF binding, and scanning of the 48S PIC. Each of the abundant mRNA modifications including m6A, Ψ, m5C, m1A, ac4C, and 2′-O-methyl ribose has been identified in the 5′ UTR of eukaryotic mRNAs. Modified nucleosides within 5′ UTRs have the potential to affect initiation directly, by affecting interactions with initiation factors and other RBPs, or indirectly, by altering the stability of RNA structures within the 5′ UTR. The effects of m6A on translation initiation have been recently reviewed (7). We therefore focus on the known and likely effects of other modifications present within 5′ UTRs, emphasizing ac4C as an example for which mechanistic details are emerging.
7.1. Position of N4-Acetylcytidine Within 5′ UTRs Determines Impact on Translation Initiation
Translation assays in vitro and in cells have revealed position-dependent effects of ac4C (Figure 4). Initial mapping of ac4C using an antibody to enrich modified fragments showed a biased distribution of ac4C in HeLa mRNAs with enrichment around translation start codons (23). Subsequent mapping to single-nucleotide resolution identified more than 400 sites in HeLa 5′ UTRs, with estimated occupancy of ac4C of ~10–40% based on the RedaC:T-seq signal at rRNA sites known to be acetylated at 80% and 100% from mass spectrometry experiments (80). A substantial fraction (~20%) of the ac4C modifications mapped to 5′ UTRs occurred within the ribosome footprint of initiating ribosomes at annotated start sites (80). Nucleotides immediately flanking the initiation codon (AUGi) are known as the Kozak sequence and make specific contacts with initiating ribosomes and eIFs and affect the efficiency and fidelity of start codon recognition (39, 81). Including ac4C at −1 and −2 with respect to the AUGi in site-specifically acetylated reporter mRNAs reduced nanoluciferase synthesis by more than 30% in rabbit reticulocyte lysate (RRL) and in transfected HeLa cells. Further experiments showed that endogenous mRNAs with acetylated Kozak sequences, IRF1 and KDM4B, showed increased ribosome density at their initiation codons and increased protein levels in NAT10−/− cells, consistent with a repressive effect of ac4C within the Kozak sequence.
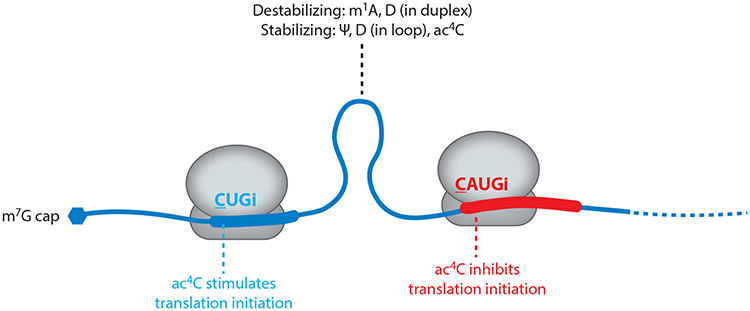
Figure 4.
mRNA modifications can regulate translation via multiple mechanisms. Installation of different chemical groups can stabilize or destabilize structured 5′ untranslated regions, which in turn influences the efficiency with which the ribosome can scan those regions. Modifications at canonical (AUG) or near-cognate (CUG) translation initiation sites can also directly impact codon–anticodon interactions. Abbreviations: Ψ, pseudouridine; ac4C, N4-acetylcytidine; D, dihydrouridine; m1A, N1-methyladenosine; m7G, N7-methylguanosine.
The effect of ac4C on initiation depended on the specific location of the modified nucleoside. Inclusion of ac4C in the 5′ UTR between an upstream TIS and the main TIS decreased protein production, consistent with reduced scanning and increased initiation at a competitive upstream TIS. In contrast, including ac4C in a CUG near-cognate initiation codon increased nanoluciferase synthesis from uncapped CUG by more than 50% in RRL and from capped CUG mRNA in transfected HeLa cells. However, ac4CUG did not significantly increase nanoluciferase synthesis from capped mRNA in RRL for reasons that were not explained. Although acetylated CUG codons were rare, two ac4C sites in upstream TISs with CUG initiation were supported by harringtonine ribosome sequencing of initiating 80S ribosomes (80). It will be interesting to see whether conditions with elevated expression of the mRNA-modifying enzyme NAT10 lead to translational control via increased upstream initiation at acetylated CUG codons within 5′ UTRs.
These reporter studies illustrate the importance of testing site-specific RNA modifications in endogenously modified positions: Moving the ac4C by 1 nucleotide—from the C at +1 in a near-cognate CUG initiation codon to −1 upstream of an AUGi—changed the modification from an activator of initiation to an inhibitor. This discovery was made possible by the use of splint ligation to construct site-specifically modified mRNAs for translation in RRL and in HeLa cells by transfecting mRNA. Splint ligation is significantly more demanding technically than incorporating modified nucleosides throughout an mRNA during in vitro transcription, but the results are more informative because physiological mRNA modification is sparse.
7.2. Enrichment of N5-Methylcytidine Near Translation Start Sites
Enrichment near translation start codons is a striking feature of m5C maps from diverse organisms and cell types, including in mESCs and brain tissue (16), in primary cultured mouse neurons (82), in CD4+ T cells from patients (83), in HeLa cells (84), and in Arabidopsis thaliana (85). However, no one has measured the effect of m5C in the start codon region on translation initiation with a single, endogenous m5C site modified. Delatte et al. (17) showed that full substitution of m5C for C reduced production of radiolabeled firefly luciferase protein in RRL by ~50%, which may be due to negative effects of m5C on elongation (86). Therapeutic mRNA studies have tested m5C-substituted mRNAs and observed increased protein production in some cases, likely due to reduced recognition by innate immune sensors (87). Although m5C likely affects multiple steps, this suggests that m5C is compatible with reasonably efficient translation initiation in mammalian cells. Because m5C is installed in mRNA by enzymes that also modify tRNA, global analysis of the effects of genetically depleting m5C on translation (e.g., by ribosome profiling analysis comparing m5C-modified to unmodified mRNAs) can provide only suggestive evidence that the presence of the modification in mRNA affects translation initiation. A promising approach to identify specific m5C sites that affect translation initiation is to compare modification levels across polysome gradients that separate cellular mRNAs according to the number of translating ribosomes (84). Most sites that showed differential methylation between fractions were less methylated in the well-translated mRNA population, consistent with reduced initiation by mechanism(s) that remain to be explored.
7.3. RNA Modifications May Affect Translation by Changing the Stability of 5′ UTR Structures
Modified nucleosides affect the stability of intramolecular RNA folding (Figure 2) (see Section 4.2), which is likely to affect translation initiation. Stable stems within 5′ UTRs impede cap binding, cap-dependent ribosome loading, and scanning to varying degrees depending on their location (Figure 4) (39, 88). Duplex destabilizing modifications such as m1A, which blocks Watson–Crick base pairing, could enhance cap accessibility when present near the mRNA 5′ end. By contrast, Ψ stabilizes RNA duplexes by 1–2 kcal/mol (89). The overall effect of a modification on RNA folding depends on the specific location. For example, D disfavors base pairing compared to U but promotes hairpin formation when present in the loop (90, 91).
The relationship between 5′ UTR structure and site-specific RNA modification has been investigated in reporter assays for ac4C, which stabilizes RNA duplexes and increases their melting temperature (92). Arango et al. (80) hypothesized that ac4C stabilizes an RNA structure that impedes 48S scanning and thereby favors initiation at upstream TISs in weak contexts; consistent with this hypothesis, the inhibition of downstream (main TIS) initiation and nanoluciferase synthesis was observed for a structured 5′ UTR with ac4C but not an unstructured 5′ UTR with ac4C. Globally, ac4C-modified 5′ UTRs (ac4C+) have more stable predicted folds compared to ac4C− 5′ UTRs.
Aided by methodological advances, recent work has begun to reveal what ribosomes do when they encounter modified mRNA. Particularly in the case of translation initiation, the specific position of a given modification can be critical. Further development of approaches to introduce modifications at specific sites is essential to testing the functions of other modifications via in vitro translation experiments. Combining this with cellular experiments comparing modification occupancies across polysome gradients could reveal the involvement of additional modifications in translation initiation.
8. STABILITY AND DECAY
mRNA modifications can alter transcript structure and half-life via multiple routes that are not mutually exclusive and can act in concert with one another. The addition of chemical moieties alters RNA chemistry, influencing backbone hydrolysis, base-pairing strength and specificity, and structure (Figure 2) (93). For instance, 2′-O-methylation stabilizes the RNA backbone by slowing hydrolysis via 2′ attack on the phosphodiester linkage. In the context of the cell, additional protein factors come into play that influence how chemical modifications alter RNA stability: Modifications can alter susceptibility to nucleases and which binding proteins interact with the RNA in question. m6A is an illustrative example of this: m6A in the 3′ UTRs of transcripts can recruit binding proteins such as YTHDF2, which in turn can recruit decay machinery to those transcripts (94, 95). This has been reviewed extensively elsewhere (6, 7), so we highlight a few other examples from the recent literature that illustrate these mechanisms.
8.1. Stabilization by N5-Methylcytidine
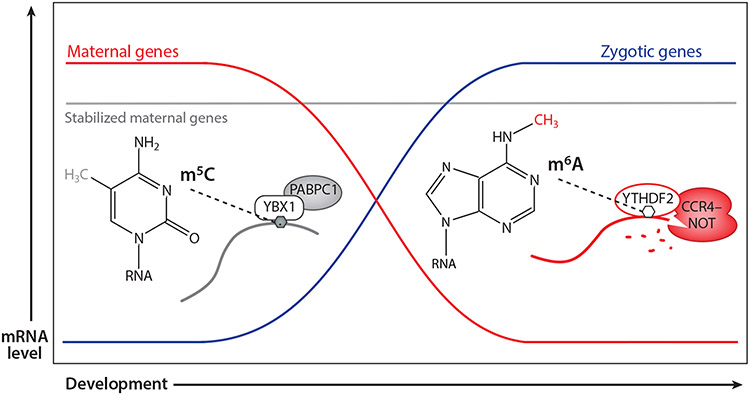
Early bisulfite-based sequencing studies of m5C yielded highly divergent information with respect to its prevalence and distribution (16, 96-98). Methods have since improved and allowed for more mechanistic dissection of this modification (99), and recent studies of m5C function now suggest that it can stabilize mRNA. The maternal-to-zygotic transition (MZT) is a critical developmental stage during which expression of maternal mRNAs broadly declines and zygotic genes must be properly activated (100). In zebrafish, m5C is installed by the NSUN2 methyltransferase and stabilizes a subset of maternal mRNAs related to mRNA metabolism and cell cycle regulation (101). This effect is due at least in part to the binding of YBX1 to m5C-containing mRNAs, which in turn recruits PABPC1 and stabilizes these transcripts. Loss of YBX1 in zebrafish results in developmental arrest at 6 h postfertilization and subsequent lethality, suggesting that m5C-mediated maternal mRNA stabilization is critical for proper development. However, YBX1 does have other documented critical roles in development that may be unrelated to m5C (e.g., 102). Stabilization of mRNAs by m5C has also been observed in urothelial carcinoma of the bladder (UCB). In a recent study of a UCB patient cohort, significantly more m5C sites were found in tumor samples relative to controls (103). This higher prevalence of m5C correlated with the increased stability of several oncogenes known to drive metastasis and invasiveness. While to date this has been shown only in UCB, the same principle could extend to other cancers as well.
This m5C-mediated stabilization of mRNA transcripts may provide an interesting countermechanism to m6A-mediated mRNA decay (Figure 5). Thus far, m5C and m6A have been characterized in equivalent (or at least similar) biological systems but in separate studies. For instance, m6A destabilizes maternal mRNA transcripts during the MZT in zebrafish (104). During the MZT, m6A could facilitate the decay of maternal transcripts while m5C ensures that a subset of transcripts remain sufficiently stable through the appropriate developmental stages. Similarly, m6A has also been shown to destabilize oncogenes in multiple cancers (105, 106), so this interplay between stabilizing and destabilizing modifications could be a more generally applicable mechanism in other biological contexts.
Figure 5.
m6A and m5C can act as opposing regulatory marks during biological processes, such as the maternal-to-zygotic transition. While m6A destabilizes maternal mRNAs via recruitment of mRNA decay machinery, m5C protects a subset of maternal transcripts from premature degradation by recruiting YBX1 and PABPC1. Abbreviations: m5C, N5-methylcytidine; m6A, N6-methyladenosine.
8.2. Context-Dependent Effects of Pseudouridine on mRNA Stability?
As described in Sections 4.1, 4.2, and 7.3, recent work has shown that Ψ can affect mRNA splicing and translation. While its potential roles in mRNA stability are still less clear, there remain some intriguing lines of evidence indicating that it may also stabilize mRNA transcripts. Of the initial sequencing studies that mapped Ψ sites in yeast and human transcriptomes (107-109), one study did find that yeast transcripts modified by the Pus7p enzyme had reduced mRNA expression levels in a Pus7p mutant strain (108), suggesting that the presence of Ψ may stabilize at least a subset of transcripts. It has since been demonstrated that human PUS7 regulates alternative splicing [as do other PUS enzymes (14)], so it is possible that changes in the observed equilibrium levels of some Pus7p targets were an indirect result of changes in splicing. Notably, similar Ψ mapping studies in the parasite Toxoplasma gondii also revealed a modest but statistically significant change in mRNA stability in strains with the mutated PUS enzyme TgPUS1 (110). However in this case, the effect was in the opposite direction: The presence of Ψ resulted in reduced mRNA stability, and moreover, the effect was not specific to the location of the Ψ site within the transcript (5′ UTR, coding sequence, or 3′ UTR). Thus, while there are some intriguing possible roles for Ψ in the regulation of mRNA stability, more detailed study is required to disentangle the observed effects from its other well-documented functions in mRNA splicing and translation.
While the mechanisms of m6A-mediated mRNA decay have now been demonstrated in many contexts, much work remains to reveal the roles of additional modifications in regulating mRNA decay and/or stability. As we have highlighted in other sections, correlation must be distinguished from causality, particularly considering that the stability of a transcript can be influenced by how it was spliced and otherwise processed in the first place. For instance, loss of a modification that influences intron retention would likely also impact the stability of the transcript, without directly interfacing with the decay machinery. A broader investigation of direct interactions between modifications and the decay machinery might yield interesting leads in this regard. The ability to monitor multiple modifications in a single experiment would also be tremendously valuable for directly investigating possible mechanisms of coordination or antagonism within the same biological system.
9. CONCLUSIONS AND PERSPECTIVES
Emerging maps of modified nucleosides suggest their potential to affect every step of mRNA bio-genesis, function, and decay. Profiling nascent pre-mRNA has identified m6A and Ψ modifications that are installed cotranscriptionally and are poised to influence nuclear RNA processing. Other mRNA modifications are installed by nuclear enzymes, and applying sequencing-based modification profiling to previously uncharted classes of RNA such as pre-mRNA is likely to reveal further examples of modification-sensitive alternative splicing and 3′-end processing. The complete landscape of alternative mRNA 5′ caps is currently unknown and may be complex. New methods and tools are becoming increasingly important for revealing molecular mechanisms, particularly when disentangling direct effects of modifications from indirect and downstream consequences of altering modifications or their regulatory enzymes. In this regard, the development of chemical inhibitors for acute inhibition of additional RNA-modifying enzymes, as recently done for the METTL3 m6A methyltransferase, is a promising direction for the field. Detailed mechanistic explanations of regulation by site-specific RNA modifications are few, and more examples are needed to establish paradigms. It is becoming increasingly clear that context matters, as the same modification in different parts of a transcript can have very different functional outcomes. In a similar vein, cellular context also matters. As we dive deeper into mechanisms, more examples of systems where multiple modifications coordinate to regulate cellular processes—such as the opposing effects of m6A and m5C on mRNA stability—are likely to emerge.
SUMMARY POINTS.
Many chemical modifications are likely added cotranscriptionally and can impact all phases of the mRNA life cycle.
While many RNA-modification enzymes have been identified, studying their roles via genetic knockout is complicated by the indirect effects of total knockout on cellular transcriptomes and proteomes.
Detailed study of specific modifications and transcripts is revealing nuances of RNA-modification-mediated regulation that would otherwise be missed.
Continued development of tools to install or remove modifications in a targeted manner is critical to advancing our understanding of the molecular mechanisms of chemical modifications.
ACKNOWLEDGMENTS
The authors would like to thank the members of the Gilbert and Nachtergaele Labs for helpful discussions. This work was supported by National Institutes of Health (NIH) grants R01GM101316, R21CA246118, and R21ES031525 to W.V.G. S.N. was supported by a Child Health Research Award from the Charles H. Hood Foundation, a Distinguished Scientist Award from the Sontag Foundation, and NIH grant R01HG011868.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Boccaletto P, Stefaniak F, Ray A, Cappannini A, Mukherjee S, et al. 2022. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 50(D1):D231–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert WV, Bell TA, Schaening C. 2016. Messenger RNA modifications: form, distribution, and function. Science 352(6292):1408–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harcourt EM, Kietrys AM, Kool ET. 2017. Chemical and structural effects of base modifications in messenger RNA. Nature 541:339–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Owens MC, Zhang C, Liu KF. 2021. Recent technical advances in the study of nucleic acid modifications. Mol. Cell 81(20):4116–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nachtergaele S, He C. 2018. Chemical modifications in the life of an mRNA transcript. Annu. Rev. Genet 52:349–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He PC, He C. 2021. m6A RNA methylation: from mechanisms to therapeutic potential. EMBO J. 40(3):e105977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murakami S, Jaffrey SR. 2022. Hidden codes in mRNA: control of gene expression by m6A. Mol. Cell 82(12):2236–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, et al. 2017. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31(10):990–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, et al. 2012. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 150(2):279–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Louloupi A, Ntini E, Conrad T, Ørom UAV. 2018. Transient N-6-methyladenosine transcriptome sequencing reveals a regulatory role of m6A in splicing efficiency. Cell Rep. 23(12):3429–37 [DOI] [PubMed] [Google Scholar]
- 11.Huang H, Weng H, Zhou K, Wu T, Zhao BS, et al. 2019. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 567(7748):414–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jonkers I, Lis JT. 2015. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell. Biol 16(3):167–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez NM, Gilbert WV. 2018. Pre-mRNA modifications and their role in nuclear processing. Quant. Biol 6(3):210–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez NM, Su A, Burns MC, Nussbacher JK, Schaening C, et al. 2022. Pseudouridine synthases modify human pre-mRNA co-transcriptionally and affect pre-mRNA processing. Mol. Cell 82(3):645–59.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji X, Dadon DB, Abraham BJ, Lee TI, Jaenisch R, et al. 2015. Chromatin proteomic profiling reveals novel proteins associated with histone-marked genomic regions. PNAS 112(12):3841–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amort T, Rieder D, Wille A, Khokhlova-Cubberley D, Riml C, et al. 2017. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 18(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, et al. 2016. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 351(6270):282–85 [DOI] [PubMed] [Google Scholar]
- 18.Lan J, Rajan N, Bizet M, Penning A, Singh NK, et al. 2020. Functional role of Tet-mediated RNA hydroxymethylcytosine in mouse ES cells and during differentiation. Nat. Commun 11(1):4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou H, Rauch S, Dai Q, Cui X, Zhang Z, et al. 2019. Evolution of a reverse transcriptase to map N1-methyladenosine in human messenger RNA. Nat. Methods 16(12):1281–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Draycott AS, Schaening-Burgos C, Rojas-Duran MF, Wilson L, Scharfen L, et al. 2022. Transcriptome-wide mapping reveals a diverse dihydrouridine landscape including mRNA. PLOS Biol. 20(5):e3001622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguilo F, Li S, Balasubramaniyan N, Sancho A, Benko S, et al. 2016. Deposition of 5-methylcytosine on enhancer RNAs enables the coactivator function of PGC-1a. Cell Rep. 14(3):479–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huttlin EL, Bruckner RJ, Navarrete-Perea J, Cannon JR, Baltier K, et al. 2021. Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell 184(11):3022–40.e28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, et al. 2018. Acetylation of cytidine in mRNA promotes translation efficiency. Cell 175(7):1872–86.e24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sleiman S, Dragon F. 2019. Recent advances on the structure and function of RNA acetyltransferase Kre33/NAT10. Cells 8(9):1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ayadi L, Galvanin A, Pichot F, Marchand V, Motorin Y. 2019. RNA ribose methylation (2′-O-methylation): occurrence, biosynthesis and biological functions. Biochim. Biophys. Acta Gene Regul. Mech 1862(3):253–69 [DOI] [PubMed] [Google Scholar]
- 26.Dai Q, Moshitch-Moshkovitz S, Han D, Kol N, Amariglio N, et al. 2017. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat. Methods 14(7):695–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elliott BA, Ho HT, Ranganathan SV, Vangaveti S, Ilkayeva O, et al. 2019. Modification of messenger RNA by 2′-O-methylation regulates gene expression in vivo. Nat. Commun 10(1):3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu W, He C, Kaye EG, Li J, Mu M, et al. 2022. Dynamic control of chromatin-associated m6A methylation regulates nascent RNA synthesis. Mol. Cell 82(6):1156–68.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akhtar J, Renaud Y, Albrecht S, Ghavi-Helm Y, Roignant JY, et al. 2021. m6A RNA methylation regulates promoter-proximal pausing of RNA polymerase II. Mol. Cell 81(16):3356–67.e6 [DOI] [PubMed] [Google Scholar]
- 30.Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millán-Zambrano G, et al. 2017. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 552(7683):126–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dai X, Wang T, Gonzalez G, Wang Y. 2018. Identification of YTH domain-containing proteins as the readers for N1-methyladenosine in RNA. Anal. Chem 90(11):6380–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seo KW, Kleiner RE. 2020. YTHDF2 recognition of N1-methyladenosine (m1A)-modified RNA is associated with transcript destabilization. ACS Chem. Biol 15(1):132–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JH, Wang R, Xiong F, Krakowiak J, Liao Z, et al. 2021. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol. Cell 81(16):3368–85.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. 2017. A phase separation model for transcriptional control. Cell 169(1):13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, Dou X, Chen C, Chen C, Liu C, et al. 2020. N6-Methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367(6477):580–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pillutla RC, Yue Z, Maldonado E, Shatkin AJ. 1998. Recombinant human mRNA cap methyltransferase binds capping enzyme/RNA polymerase IIo complexes. J. Biol. Chem 273(34):21443–46 [DOI] [PubMed] [Google Scholar]
- 37.Tsukamoto T, Shibagaki Y, Niikura Y, Mizumoto K. 1998. Cloning and characterization of three human cDNAs encoding mRNA (guanine-7-)-methyltransferase, an mRNA cap methylase. Biochem. Biophys. Res. Commun 251(1):27–34 [DOI] [PubMed] [Google Scholar]
- 38.Langberg SR, Moss B. 1981. Post-transcriptional modifications of mRNA. Purification and characterization of cap I and cap II RNA (nucleoside-2′-)-methyltransferases from HeLa cells. J. Biol. Chem 256(19):10054–60 [PubMed] [Google Scholar]
- 39.Hinnebusch AG, Ivanov IP, Sonenberg N. 2016. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 352(6292):1413–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiao X, Doamekpor SK, Bird JG, Nickels BE, Tong L, et al. 2017. 5′ end nicotinamide adenine dinucleotide cap in human cells promotes RNA decay through DXO-mediated deNADding. Cell 168(6):1015–27.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walters RW, Matheny T, Mizoue LS, Rao BS, Muhlrad D, Parker R. 2017. Identification of NAD+ capped mRNAs in Saccharomyces cerevisiae. PNAS 114(3):480–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, et al. 2017. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541:371–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen YG, Kowtoniuk WE, Agarwal I, Shen Y, Liu DR. 2009. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat. Chem. Biol 5(12):879–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cahová H, Winz M-L, Höfer K, Nübel G, Jäschke A. 2015. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature 519(7543):374–77 [DOI] [PubMed] [Google Scholar]
- 45.Lim J, Ha M, Chang H, Kwon SC, Simanshu DK, et al. 2014. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 159(6):1365–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akichika S, Hirano S, Shichino Y, Suzuki T, Nishimasu H, et al. 2019. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363(6423):eaav0080. [DOI] [PubMed] [Google Scholar]
- 47.Boulias K, Toczydlowska-Socha D, Hawley BR, Liberman N, Takashima K, et al. 2019. Identification of the m6Am methyltransferase PCIF1 reveals the location and functions of m6Am in the transcriptome. Mol. Cell 75(3):631–43.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sendinc E, Valle-Garcia D, Dhall A, Chen H, Henriques T, et al. 2019. PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. Mol. Cell 75(3):620–30.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pandey RR, Delfino E, Homolka D, Roithova A, Chen KM, et al. 2020. The mammalian cap-specific m6Am RNA methyltransferase PCIF1 regulates transcript levels in mouse tissues. Cell Rep. 32(7):108038. [DOI] [PubMed] [Google Scholar]
- 50.Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MSH. 2009. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 16(1):45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Chew BLA, Lai Y, Dong H, Xu L, et al. 2019. Quantifying the RNA cap epitranscriptome reveals novel caps in cellular and viral RNA. Nucleic Acids Res. 47(20):e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pandolfini L, Barbieri I, Bannister AJ, Hendrick A, Andrews B, et al. 2019. METTL1 promotes let-7 microRNA processing via m7G methylation. Mol. Cell 74(6):1278–90.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang LS, Liu C, Ma H, Dai Q, Sun HL, et al. 2019. Transcriptome-wide mapping of internal N7-methylguanosine methylome in mammalian mRNA. Mol. Cell 74(6):1304–16.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Enroth C, Poulsen LD, Iversen S, Kirpekar F, Albrechtsen A, Vinther J. 2019. Detection of internal N7-methylguanosine (m7G) RNA modifications by mutational profiling sequencing. Nucleic Acids Res. 47(20):e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu XD, Ares M. 2014. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet 15(10):689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nilsen TW, Graveley BR. 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463(7280):457–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wright CJ, Smith CWJ, Jiggins CD. 2022. Alternative splicing as a source of phenotypic diversity. Nat. Rev. Genet 23:697–710 [DOI] [PubMed] [Google Scholar]
- 58.Zhao X, Yang Y, Sun B-F, Shi Y, Yang X, et al. 2014. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24(12):1403–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. 2015. N6-Methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 518(7540):560–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phizicky EM, Hopper AK. 2010. tRNA biology charges to the front. Genes Dev. 24(17):1832–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, et al. 2021. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593(7860):597–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kanemaki MT. 2022. Ligand-induced degrons for studying nuclear functions. Curr. Opin. Cell Biol 74:29–36 [DOI] [PubMed] [Google Scholar]
- 63.Wei G, Almeida M, Pintacuda G, Coker H, Bowness JS, et al. 2021. Acute depletion of METTL3 implicates N6-methyladenosine in alternative intron/exon inclusion in the nascent transcriptome. Genome Res. 31(8):1395–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Borchardt EK, Martinez NM, Gilbert WV. 2020. Regulation and function of RNA pseudouridylation in human cells. Annu. Rev. Genet 54:309–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao W, Adhikari S, Dahal U, Chen Y-S, Hao Y-J, et al. 2016. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61(4):507–19 [DOI] [PubMed] [Google Scholar]
- 66.Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, et al. 2016. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540(7632):301–4 [DOI] [PubMed] [Google Scholar]
- 67.Kan L, Grozhik AV, Vedanayagam J, Patil DP, Pang N, et al. 2017. The m6A pathway facilitates sex determination in Drosophila. Nat. Commun 8:15737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Nostrand EL, Freese P, Pratt GA, Wang X, Wei X, et al. 2020. A large-scale binding and functional map of human RNA-binding proteins. Nature 583(7818):711–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mitschka S, Mayr C. 2022. Context-specific regulation and function of mRNA alternative polyadenylation. Nat. Rev. Mol. Cell. Biol 23:779–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lesbirel S, Viphakone N, Parker M, Parker J, Heath C, et al. 2018. The m6A-methylase complex recruits TREX and regulates mRNA export. Sci. Rep 8(1):13827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang X, Yang Y, Sun B-F, Chen Y-S, Xu J-W, et al. 2017. 5-Methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 27(5):606–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, et al. 2014. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 33(18):2020–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi M, Zhang H, Wu X, He Z, Wang L, et al. 2017. ALYREF mainly binds to the 5′ and the 3′ regions of the mRNA in vivo. Nucleic Acids Res. 45(16):9640–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, et al. 2017. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife 6:e31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edens BM, Vissers C, Su J, Arumugam S, Xu Z, et al. 2019. FMRP modulates neural differentiation through m6A-dependent mRNA nuclear export. Cell Rep. 28(4):845–54.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Courtney DG, Chalem A, Bogerd HP, Law BA, Kennedy EM, et al. 2019. Extensive epitranscriptomic methylation of A and C residues on murine leukemia virus transcripts enhances viral gene expression. mBio 10(3):e01209–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mougel M, Akkawi C, Chamontin C, Feuillard J, Pessel-Vivares L, et al. 2020. NXF1 and CRM1 nuclear export pathways orchestrate nuclear export, translation and packaging of murine leukaemia retrovirus unspliced RNA. RNA Biol. 17(4):528–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rauch S, He E, Srienc M, Zhou H, Zhang Z, Dickinson BC. 2019. Programmable RNA-guided RNA effector proteins built from human parts. Cell 178(1):122–34.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Franco MK, Koutmou KS. 2022. Chemical modifications to mRNA nucleobases impact translation elongation and termination. Biophys. Chem 285:106780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arango D, Sturgill D, Yang R, Kanai T, Bauer P, et al. 2022. Direct epitranscriptomic regulation of mammalian translation initiation through N4-acetylcytidine. Mol. Cell 82(15):2797–814.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hinnebusch AG. 2017. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem. Sci 42(8):589–611 [DOI] [PubMed] [Google Scholar]
- 82.Jian H, Zhang C, Qi ZY, Li X, Lou Y, et al. 2021. Alteration of mRNA 5-methylcytosine modification in neurons after OGD/R and potential roles in cell stress response and apoptosis. Front. Genet 12:633681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guo G, Wang H, Shi X, Ye L, Yan K, et al. 2020. Disease activity-associated alteration of mRNA m5 C methylation in CD4+ T cells of systemic lupus erythematosus. Front. Cell Dev. Biol 8:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schumann U, Zhang HN, Sibbritt T, Pan A, Horvath A, et al. 2020. Multiple links between 5-methylcytosine content of mRNA and translation. BMC Biol. 18(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang L, Perrera V, Saplaoura E, Apelt F, Bahin M, et al. 2019. m5C methylation guides systemic transport of messenger RNA over graft junctions in plants. Curr. Biol 29(15):2465–76.e5 [DOI] [PubMed] [Google Scholar]
- 86.Jones JD, Monroe J, Koutmou KS. 2020. A molecular-level perspective on the frequency, distribution, and consequences of messenger RNA modifications. WIRES RNA 11(4):e1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang X, Liu A. 2022. The pivotal role of chemical modifications in mRNA therapeutics. Front. Cell Dev. Biol 10:1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Niederer RO, Rojas-Duran MF, Zinshteyn B, Gilbert WV. 2022. Direct analysis of ribosome targeting illuminates thousand-fold regulation of translation initiation. Cell Syst. 13(3):256–64.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kierzek E, Malgowska M, Lisowiec J, Turner DH, Gdaniec Z, Kierzek R. 2014. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 42(5):3492–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dalluge JJ, Hashizume T, Sopchik AE, McCloskey JA, Davis DR. 1996. Conformational flexibility in RNA: the role of dihydrouridine. Nucleic Acids Res. 24(6):1073–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dyubankova N, Sochacka E, Kraszewska K, Nawrot B, Herdewijn P, Lescrinier E. 2015. Contribution of dihydrouridine in folding of the D-arm in tRNA. Org. Biomol. Chem 13(17):4960–66 [DOI] [PubMed] [Google Scholar]
- 92.Bartee D, Nance KD, Meier JL. 2022. Site-specific synthesis of N4-acetylcytidine in RNA reveals physiological duplex stabilization. J. Am. Chem. Soc 144(8):3487–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Helm M. 2006. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 34(2):721–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, et al. 2014. N6-Methyladenosine-dependent regulation of messenger RNA stability. Nature 505(7481):117–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Du H, Zhao Y, He J, Zhang Y, Xi H, et al. 2016. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun 7:12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hussain S, Aleksic J, Blanco S, Dietmann S, Frye M. 2013. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol. 14(11):215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Khoddami V, Cairns BR. 2013. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol 31(5):458–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, et al. 2012. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 40(11):5023–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang T, Chen W, Liu J, Gu N, Zhang R. 2019. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat. Struct. Mol. Biol 26(5):380–88 [DOI] [PubMed] [Google Scholar]
- 100.Yartseva V, Giraldez AJ. 2015. The maternal-to-zygotic transition during vertebrate development. Curr. Top. Dev. Biol 113:191–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang Y, Wang L, Han X, Yang W-L, Zhang M, et al. 2019. RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol. Cell 75(6):1188–202.e11 [DOI] [PubMed] [Google Scholar]
- 102.Evans MK, Matsui Y, Xu B, Willis C, Loome J, et al. 2020. Ybx1 fine-tunes PRC2 activities to control embryonic brain development. Nat. Commun 11(1):4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen X, Li A, Sun BF, Yang Y, Han YN, et al. 2019. 5-Methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol 21(8):978–90 [DOI] [PubMed] [Google Scholar]
- 104.Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, et al. 2017. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542(7642):475–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, et al. 2018. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell 172(1–2):90–105.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu J, Eckert MA, Harada BT, Liu S-M, Lu Z, et al. 2018. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol 20(9):1074–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. 2014. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515(7525):143–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, et al. 2014. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159(1):148–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lovejoy AF, Riordan DP, Brown PO. 2014. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLOS ONE 9(10):e110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nakamoto MA, Lovejoy AF, Cygan AM, Boothroyd JC. 2017. mRNA pseudouridylation affects RNA metabolism in the parasite Toxoplasma gondii. RNA 23(12):1834–49 [DOI] [PMC free article] [PubMed] [Google Scholar]