SUMMARY
Methionine is an essential branch of the diverse nutrient inputs to dictate mTORC1 activation. In the absence of methionine, SAMTOR binds to GATOR1 and inhibits mTORC1 signaling. However, how mTORC1 is activated upon methionine stimulation remains largely elusive. Here, we report that PRMT1 senses methionine/SAM, by utilizing SAM as a cofactor for an enzymatic activity-based regulation of mTORC1 signaling. Under methionine-sufficient conditions, elevated cytosolic SAM releases SAMTOR from GATOR1, which confers the association of PRMT1 with GATOR1. Subsequently, SAM-loaded PRMT1 methylates NPRL2, the catalytic subunit of GATOR1, thereby suppressing its GAP activity and leading to mTORC1 activation. Notably, genetic or pharmacological inhibition of PRMT1 impedes hepatic methionine sensing by mTORC1 and improves insulin sensitivity in aged mice, establishing the role of PRMT1-mediated methionine sensing at physiological levels. Thus, PRMT1 coordinates with SAMTOR to form the methionine-sensing apparatus of mTORC1 signaling.
In Brief
Jiang et al. reveal that PRMT1 senses methionine levels to activate mTORC1 signaling via inhibition of GATOR1 by taking SAM as a cofactor to facilitate the asymmetric di-methylation of NPRL2, which plays a key role in orchestrating the organismal response to dietary methionine restriction in aged mice.
Graphical Abstract
INTRODUCTION
mTORC1 is a master regulator of cell growth and homeostasis in response to diverse environmental cues, including amino acids.1,2 Dysregulation of mTORC1 signaling has been implicated in a variety of human diseases, including cancer, diabetes, and neurological disorders.3–5 The amino acid signal is sequentially transmitted to mTORC1 through Rag GTPase-mediated lysosomal translocation and Rheb-mediated activation of the mTORC1 complex.6–9 Several protein complexes have been identified to govern the amino acid sensing and subsequent activation of mTORC1 via regulating the activation status of Rag GTPases.10–15 Among these, GATOR1 and FLCN-FNIP have been identified to function as GTPase-activating proteins (GAPs) for RagA/B and RagC/D, respectively, and are frequently mutated in many human diseases.13,16–19 Of note, GATOR1 consists of NPRL2, NPRL3, and DEPDC5, of which NPRL2 catalyzes GATOR1-stimulated GTP hydrolysis via inserting its arginine finger into the nucleotide-binding pocket of RagA/B GTPase.20,21 Furthermore, there are alternative pathways to activate mTORC1 that are independent of Rag proteins.22 For instance, in the context of glutamine-induced mTORC1 activation, Arf1 can serve as a substitute for Rag.23 Additionally, amino acids facilitate the loading of GTP onto Rab1a, subsequently initiating an interaction between Rheb and mTORC1 in the Golgi.24
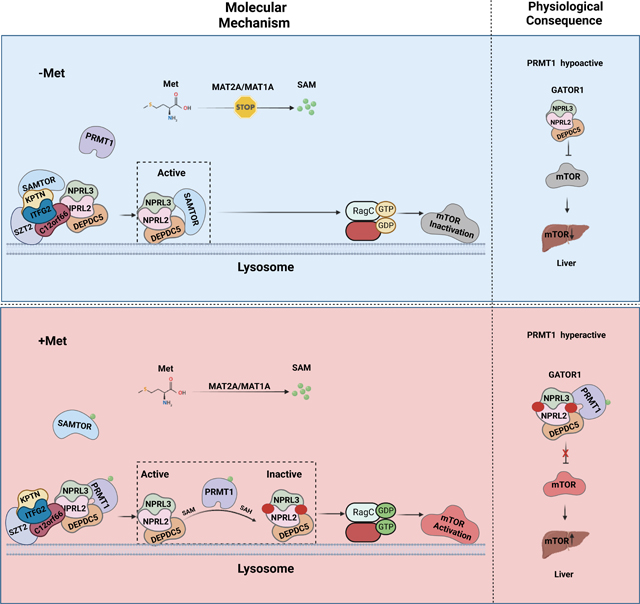
GATOR2 transmits leucine and arginine availability to mTORC1 via directly interacting with the respective sensor proteins, such as Sestrin2, SARB1, and Castor1, and antagonizes the function of GATOR1.25–27 On the other hand, methionine sensing by mTORC1 adopts a distinct mechanism via impinging on GATOR1 instead of GATOR2.28 Under the conditions of methionine scarcity, intracellular SAM is dramatically reduced to a level below its dissociation constant with SAMTOR, and the non-SAM-loaded SAMTOR binds to GATOR1 and inhibits mTORC1 signaling.28 However, SAMTOR’s enzymatic activity is dispensable for its role in inactivating mTORC1 signaling,28 suggesting that additional sensing mechanisms exist in parallel with SAMTOR to suppress the GAP activity of GATOR1 under the methionine-sufficient conditions (Figure. S1A).
Here, we report that PRMT1 is essential for methionine-mediated mTORC1 activation via directly sensing the intracellular levels of SAM. Under methionine-sufficient conditions, elevated cytosolic SAM disassociates SAMTOR from GATOR1, which confers GATOR1 interaction with PRMT1 to methylate NPRL2 and suppress the GAP activity of GATOR1, leading to mTORC1 activation. More importantly, the hepatic Prmt1-Nprl2-mTORC1 axis plays a key role in orchestrating the organismal response to dietary methionine restriction in aged mice, establishing PRMT1 as a physiological SAM sensor of mTORC1 signaling.
RESULTS
PRMT1 dictates methionine-dependent mTORC1 activation
To identify the upstream regulator of GATOR1, we screened for novel GATOR1 interacting proteins using an engineered HEK-293T-NPRL2Flag knock-in cell line, in which a Flag tag was inserted at the N-terminus of endogenous NPRL2 loci in HEK-293T cells using the CRISPR-Cas9 system. Flag-immunoprecipitants prepared from the HEK-293T-NPRL2Flag cells were subjected to mass spectrometric analysis (Figure. S1B), which yielded several arginine methylation-related candidate proteins including PRMT1, PRMT5, and WDR77, with NPRL3 as a known positive control of NPRL2-binding proteins (Figure. S1C). PRMT1 is the major type I arginine methyltransferase that methylates protein substrates to generate monomethyl arginine (MMA) and then asymmetric dimethylarginine (ADMA), whereas PRMT5, WDR77, and CLNS1A form a methylome complex to catalyze the symmetric di-methylation with PRMT5 as the catalytic subunit.29,30 Further GST-pulldown assay demonstrated that only PRMT1, but not PRMT5 or other PRMTs we examined, interacted with NPRL2 in cells (Figure. S1D), therefore excluding PRMT5 and WDR77 as specific NPRL2-interacting proteins, in part because PRMT5 can be enriched by Flag-M2 agarose.31 In further support of the physiological interaction between NPRL2 and PRMT1, we readily detected endogenous PRMT1 in anti-Flag immunoprecipitants from NPRL2Flag knock-in cells, and Flag-tagged endogenous WDR24, a component of the GATOR2 complex,13 coimmunoprecipitated with PRMT1 with a much lower affinity (Figure. S1E).
Given the strong interaction between PRMT1 and the GATOR1 complex, we reasoned that PRMT1 might participate in mTORC1 regulation. Since PRMT1 is essential in human cells,30 we generated a doxycycline-inducible system to acutely suppress PRMT1 expression. Indeed, on day 2 of doxycycline (DOX)-induced suppression of PRMT1 without noticeable cellular stress response or cell proliferation defects, amino acid-induced mTORC1 activation was reduced in PRMT1-deficient cells, confirming that the changes in mTORC1 activity were not likely due to secondary defects in cell proliferation (Figures. S1F–1I). Notably, PRMT1 deficiency specifically affected the regulation of mTORC1 by methionine and, to a lesser extent, leucine and arginine stimulation, but not by any of the other amino acids we tested (Figure. 1A and Figure. S1J). mTORC1 is recruited to the lysosomal surface by Rag GTPase and is a prerequisite step for its activation by Rheb.7,10 Knockdown of PRMT1 impaired the methionine-induced activation of RagA, as indicated by the GTP-form of RagA (Figure. 1A) and lysosomal translocation of mTORC1 (Figures. 1B–1C). To pinpoint where PRMT1 acts in mTORC1 signaling, we performed an epistasis analysis between PRMT1 and the well-established components of mTORC1 signaling in the amino acid sensing pathway. Notably, overexpression of the constitutively active form of Rag GTPases (RagAQ66L/RagCS75N) significantly prevented the impaired mTORC1 activation in PRMT1-deficient cells (Figure. 1D). Furthermore, knockdown of PRMT1 did not perturb mTORC1 signaling in NPRL2 null cells (Figure. 1E and Figure. S1K). Taken together, these results suggest that PRMT1 is crucial for methionine-induced mTORC1 activation and integrates at the upstream of the GATOR1-Rag axis.
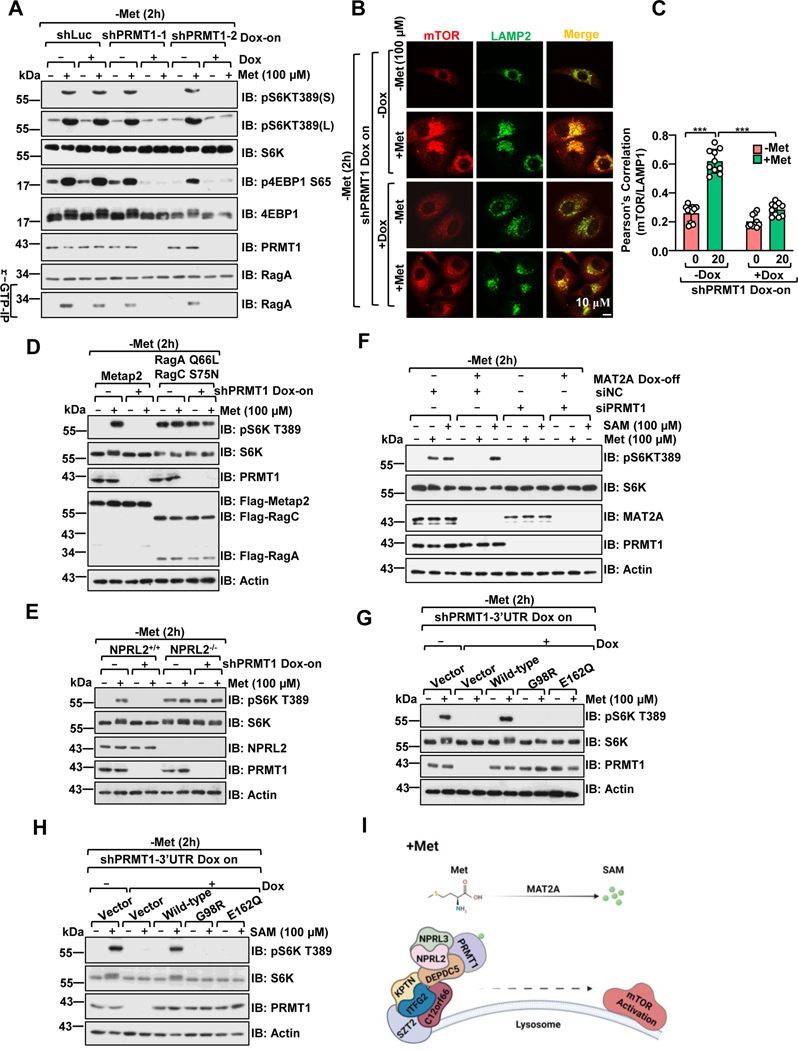
Figure 1. PRMT1 signals methionine availability to regulate mTORC1 activation in a SAM-dependent manner.
A, Control and PRMT1 deficient HEK293 cells were deprived of methionine for 2 hours (h) and restimulated with methionine (100 μM) for 20 min. HEK293 cells were infected with either tet-on-shLuc or tet-on-shPRMT1 lentiviruses and selected with puromycin for 3 days. The stable cell lines were pretreated with or without doxycycline (DOX) for an additional 2 days before methionine starvation and restimulation. Whole-cell lysates (WCL) or r-GTP- immunoprecipitates were analyzed by immunoblotting with the indicated antibodies. pS6K(S): short-term exposure, pS6K(L): long-term exposure. Due to space limitations, we only present the short-term exposure of pS6K in the remaining figures.
B, HEK 293 cells were treated as in A, and the co-localization of mTORC1 and LAMP2 was analyzed via immunostaining. Scale bar, 10 μm.
C, Pearson’s correlation analysis of mTOR and LAMP2 signals in B. 10 cells were analyzed for each condition, ***p<0.001, unpaired, two-tailed Student’s t-test.
D, Control and PRMT1 deficient HEK293 cells were transfected with constructs to express either Metap2 (control) or RagAQ66L+RagCS75N. Cells were treated as described in A, and cell lysates were analyzed by immunoblotting with the indicated antibodies.
E, Wild-type or NPRL2 knockout HEK293 cells were infected with the indicated lentivirus and treated as in A, and cell lysates were analyzed via immunoblotting with the indicated antibodies.
F, PRMT1 is required to signal methionine sufficiency to mTORC1 via MAT2A-mediated SAM production. The MAT2A Dox-off cell line was generated as described in the previous study25 and transfected with siRNA targeting PRMT1, as indicated. Cells were treated with or without DOX for 48 hours, followed by deprivation of methionine for 2 hours and restimulation with methionine (100 μM) for 20 min or SAM (100 μM) for 6 hours. Cell lysates were analyzed via immunoblotting with the indicated antibodies.
G-H, Wild-type PRMT1, but not the SAM-binding-deficient mutants (G98R and E162Q), restored mTORC1 activation upon methionine (G) or SAM (H) stimulation. HEK293 cells expressing tet-on-shPRMT1 targeting the PRMT1–3’UTR were infected with either PRMT1 wild-type, G98R, or E162Q lentiviruses. The stable cell lines were pre-treated with or without DOX for an additional 2 days, deprived of methionine for 2 hours, and restimulated with methionine (100 μM) for 20 min or SAM (100 μM) for 6 hours. Cell lysates were analyzed via immunoblotting with the indicated antibodies.
I, A schematic illustration showing that PRMT1 signals methionine availability to govern mTORC1 activation through the GATOR1 complex.
See also Figure S1.
Methionine is catabolized and recycled into a series of metabolites, including SAM, SAH, and homocysteine, termed the methionine cycle (Figure. S1L).32,33Methionine adenosyltransferase (MAT1A/MAT2A) is a rate-limiting enzyme for the methionine cycle and converts methionine to SAM, 33,34 located in the cytosol, nucleus and mitochondria, providing the local source of SAM. 35–38 Since mTORC1 mainly senses cytosolic and lysosomal amino acid,39 we analyzed the local concentration of SAM in the cytosol. Consistent with the previous studies analyzed by LC/MS,28,40 both the total and cytosolic methionine and SAM levels were reduced upon methionine starvation (Figures. S1M–1N). Interestingly, we found that despite SAH dissociating SAMTOR from GATOR1, SAH could not activate mTORC1 signaling as methionine did (Figure. S1O), consistent with previous studies demonstrating that SAH suppresses autophagy in an mTORC1-independent manner.41,42 These results further reveal that the dissociation of SAMTOR from GATOR1 is necessary, but not sufficient, to activate mTORC1 signaling, indicating that an additional SAM-dependent mechanism is required to suppress the GAP activity of GATOR1. Furthermore, knockdown of PRMT1 blunted the methionine- and SAM-mediated mTORC1 activation (Figure. S1P), suggesting that PRMT1 senses intracellular methionine or SAM levels to modulate mTORC1 activity.
Given that MAT2A is an essential gene for cell survival, 32,34,35 we utilized the reported doxycycline-repressible system to manipulate MAT2A expression.28 Notably, loss of MAT2A strongly impaired methionine-, but not SAM-induced mTORC1 activation (Figure. 1F) without dramatically affecting cell proliferation on day 2 of repression (Figure. S1Q–S1R). These results indicate that methionine-derived SAM, but not methionine itself, is directly sensed by mTOR signaling. Moreover, PRMT1 deficiency attenuated SAM-dependent mTORC1 activation in both MAT2A-intact and -deficient cells (Figure. 1F), suggesting that PRMT1 is likely essential for SAM-dependent mTORC1 activation downstream of MAT2A. The dissociation constants (Kd) for human SAM-SAMTOR and SAM-PRMT1 interactions were approximately 7 μM and 26 μM, respectively (Figure. S1S). This is consistent with previous studies 28,43 and compatible with the oscillation range of intracellular SAM concentrations between methionine deprivation and replenishment conditions in the cytosol (Figure. S1T).28 In comparison with wild-type PRMT1, the SAM binding-deficient mutant forms of PRMT1 (G98R and E162Q) 44 failed to restore the mTORC1 activity in response to methionine or SAM stimulation in PRMT1-depleted cells, as evidenced by the phosphorylation of S6K (Figures. 1G–H) and the recruitment of mTORC1 complex onto lysosome (Figure. S1U). Taken together, PRMT1 activates mTORC1 signaling in a SAM-dependent manner in response to methionine stimulation (Figure. 1I).
PRMT1 orchestrates methionine sensing in concert with SAMTOR
Under conditions of methionine deficiency, non-SAM-loaded SAMTOR binds to GATOR1 and suppresses mTORC1 signaling.28 We next explored whether and how PRMT1 is integrated with SAMTOR-mediated methionine sensing. Methionine replenishment triggered the dissociation of SAMTOR from GATOR1 and reinforced the interaction between PRMT1 and GATOR1 in a time-dependent manner (Figures. 2A–2B). We then varied the medium methionine concentration in methionine-starved cells within the physiological range (10–500 μM) and analyzed the dose-response changes in the association of GATOR1, SAMTOR and PRMT1 with variations in the availability of exogenous methionine. Notably, the mode of interaction was altered at concentrations10–25 μM (Figure. S2A), consistent with the cytosolic levels of SAM, but not methionine (Figure. S2B–2C), indicating that SAM, but not methionine, is directly involved in mediating the interplay between PRMT1, GATOR1 and SAMTOR. In support of this notion, methionine stimulation failed to disrupt the binding between SAMTOR and GATOR1 in MAT2A deficiency cells, under which condition methionine is unable to be converted to SAM, while replenishment of SAM rescued this phenotype by releasing SAMTOR from the GATOR1 complex (Figures. 2C–2D). Concomitantly, the interaction between PRMT1 and GATOR1 was enhanced by methionine in MAT2A intact cells and by SAM in MAT2A deficient cells (Figures. 2C–2D). Thus, the interaction of the GATOR1 complex with SAMTOR and PRMT1 is mutually exclusive in concert with the cytosolic SAM levels.
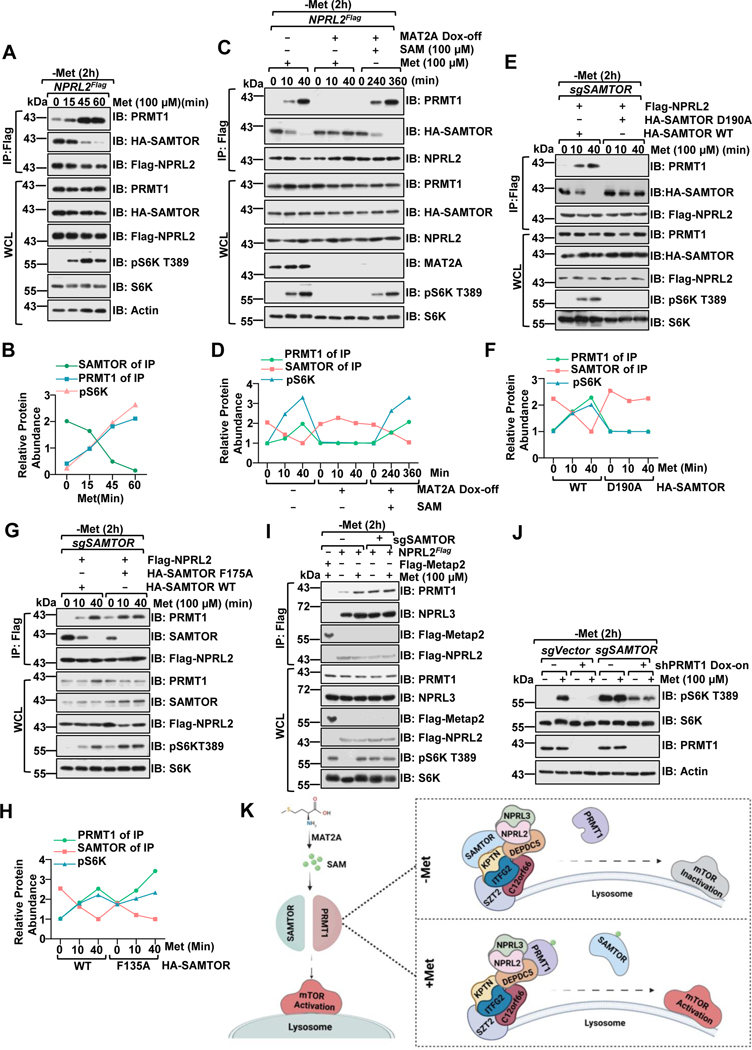
Figure 2. PRMT1 coordinates with SAMTOR to dictate mTORC1 activation.
A-B, NPRL2Flag knock-in HEK293T cells were starved of methionine for 2 hours and restimulated with methionine (100 μM) for the indicated time. Whole-cell lysis (WCL) and anti-FLAG immunoprecipitates (IPs) were analyzed via immunoblotting with the indicated antibodies. Quantified values of the immunoblotting of A were shown as in B. Data are representative one repeat.
C-D, NPRL2Flag knock-in HEK293T MAT2A Dox-off cell lines were generated as described in the previous study25. The stable cell lines were pre-treated with or without doxycycline (DOX) for an additional 2 days, starved of methionine for 2 hours, and restimulated with either methionine (100 μM) or SAM (100 μM) for the indicated time. WCL and anti-FLAG IPs were analyzed via immunoblotting with the indicated antibodies. Quantified values of the immunoblotting of C were shown in D. Data are representative one repeat.
E-H, HEK293 SAMTOR-null cells were transfected with Flag-NPRL2 and wild-type SAMTOR or the indicated D190A (E) or F135A (G) mutants, starved of methionine for 2 hours and restimulated with methionine (100 μM) for the indicated times. The WCL and anti-FLAG IPs were analyzed via immunoblotting with the indicated antibodies. Quantified values of the immunoblotting of E and G were shown as in F, H. Data are representative one repeat.
I, NPRL2Flag knock-in HEK293T cells expressing with or without sgSAMTOR were starved of methionine for 2 hours and restimulated with methionine (100 μM) for 20 min. The WCL and anti-FLAG IPs were analyzed via immunoblotting with the indicated antibodies.
J, SAMTOR and PRMT1 function in parallel to sense methionine availability to mTORC1. HEK293 cells were infected with sgControl, sgSAMTOR, or tet-on-shPRMT1 as indicated and selected with puromycin (1 μg/mL) for 4 days. Cells were pre-treated with or without DOX for 2 days to suppress PRMT1 expression and then challenged as in I, followed by lysis and immunoblotting with the indicated antibodies.
K, A schematic illustration of PRMT1-GATOR1-SAMTOR interaction and mTORC1 regulation in response to methionine signals.
See also Figure S2.
Since methionine promotes the interaction between PRMT1 and GATOR1 in cells in a SAM-dependent manner, we next examined whether SAM binding directly regulates the interaction between PRMT1 and GATOR1 in vitro. Notably, preincubation of PRMT1 with SAM had no obvious effect on its interaction with the GATOR1 complex in vitro (Figures S2D–2E). Moreover, the SAM-binding-deficient PRMT1 mutants (G98R and E162Q) coimmunoprecipitated comparable amounts of endogenous GATOR1 compared to wild-type PRMT1 (Figure. S2F). These results suggest that SAM indirectly promotes the association of PRMT1 with GATOR1 in cells and that PRMT1 adopts a distinct model for sensing methionine/SAM by using SAM as a cofactor for enzymatic activity-based, rather than protein-protein interaction (PPI)-based regulation of GATOR1. In support of this hypothesis, either decreasing the SAM/SAH ratio by inhibiting SAH hydrolase via DZNeP45 (Figure. S2G) or pharmacological inhibition of PRMT1 methyltransferase activity (Figures. S2H–2J) impaired methionine-induced mTORC1 activation in a GATOR1-dependent manner.
Next, we explored how SAM indirectly affects the binding between PRMT1 and GATOR1 in cells. PRMT1 binds to SAM (Kd of around 26 μM) with lower affinity than SAMTOR (Kd of about 7 μM). Thus, we hypothesized that there might be a sequential SAM sensing mode. This process is initially elicited by the dissociation of SAM-loaded SAMTOR (which senses the lower intracellular levels of SAM) from GATOR1, and these two sensors interplay with each other. Indeed, we found that overexpression of SAMTOR disrupted the interaction between PRMT1 and GATOR1 in a dose-dependent manner (Figure. S2K). Consistently, unlike wild-type SAMTOR, the SAM-binding-deficient mutant SAMTOR (D190A), 28 which constitutively interacts with the GATOR1 complex, disrupted the association between PRMT1 and GATOR1 in SAMTOR-null cells (Figures. 2E–2F). On the other hand, the GATOR1 and KICSTOR binding-deficient mutant F175A 46 antagonized the interaction between PRMT1 and GATOR1 to a lesser extent in comparison with the wild-type SAMTOR (Figures. 2G–2H). More importantly, methionine stimulation induced the interaction between PRMT1 and GATOR1 in SAMTOR-intact cells, whereas the interaction between PRMT1 and GATOR1 was enhanced in SAMTOR-null cells and displayed no further responsiveness to methionine variation (Figure. 2I).
However, depletion of PRMT1 did not alter the interaction between SAMTOR and GATOR1 (Figure. S2L). Moreover, PRMT1 binds to GATOR1 via the DEPDC5 subunit (Figure. S2M) and associates with SAMTOR in a GATOR1-dependent manner (Figure. S2N). Given that SAMTOR only interacts with DEPDC5 in the presence of KICSTOR, 28 it is possible that PRMT1 and SAM-SAMTOR do not compete for the same region of GATOR1 for interaction (Figure. S2O), and SAMTOR blunts the interaction between PRMT1 and GATOR1 via an indirect mechanism. Structural insights will be further needed to reveal the exact underlying mechanism.
To further understand how methionine/SAM-induced PRMT1-mediated regulation is involved in mTORC1 signaling, we monitored the localization of PRMT1 in response to methionine stimulation. Notably, methionine increased lysosomal accumulation of both PRMT1 and mTOR and concomitantly elevated the total ADMA modification of proteins in lysosomes (Figure. S2P). Next, we asked whether GATOR1 is required for PRMT1 localization to the lysosomal surface. Interestingly, PRMT1 failed to translocate to lysosomes in NPRL2-deficient cells (Figure. S2Q), whereas PRMT1 constitutively localized to the lysosomal surface in SAMTOR-null cells (Figure. S2R), indicating that SAMTOR impedes the lysosomal translocation of PRMT1 via interfering with its binding to GATOR1. Mutation of the two residues that are essential for SAM binding in PRMT1 did not alter the lysosomal translocation of PRMT1 (Figure. S2S), which is consistent with the observed comparable binding affinity to GATOR1 for these two mutants versus WT-PRMT1 (Figure. S2F). Thus, upon methionine stimulation, PRMT1 translocates to the lysosomal surface and activates mTORC1 in a GATOR1-dependent manner, which is blocked by SAMTOR via impairing its interaction with GATOR1. Finally, via a genetic approach, we explored the relationship between SAMTOR and PRMT1 in methionine sensing. Consistent with the established role of SAMTOR, methionine starvation failed to inactivate mTORC1 in cells lacking SAMTOR. 28 On the other hand, knockdown of PRMT1 reduced mTORC1 activation in SAMTOR-null cells (Figure. 2J). It is likely that PRMT1 functions in parallel with SAMTOR to mediate the methionine sensing by mTORC1, and an additional mechanism possibly contributes to aberrant activation of mTORC1 in SAMTOR-depleted cells. Hence, our results suggest that SAMTOR binds to GATOR1 under methionine-starved conditions and perturbs the interaction of PRMT1 with the GATOR1 complex. Upon methionine stimulation, SAMTOR first dissociates from GATOR1 and confers its subsequent accessibility to SAM-loaded PRMT1, resulting in the activation of mTORC1 signaling (Figure. 2K). Thus, PRMT1 likely coordinates with SAMTOR to form the methionine-sensing apparatus and sequentially connects methionine metabolism with mTORC1 activation (Figure. S2T).
PRMT1 methylates NPRL2 at the R78 residue
Given that PRMT1 physically interacts with GATOR1 and that the ADMA levels of lysosomal proteins are increased upon methionine stimulation, we sought to investigate whether PRMT1 methylates component(s) of the GATOR1 complex in vitro (Figures. S3A–3B).47 Unlike SAMTOR, which exhibits no methyltransferase activity toward GATOR1, PRMT1 promoted the methylation of the purified GATOR1 complex as evidenced by the increased generation of SAH with Histone H4 as a positive control (Figure. 3A and Figure. S3C).48 Next, we analyzed which component(s) of the GATOR1 complex was methylated by PRMT1 with two different ADMA antibodies to exclude the potential artifact of the pan-antibody.49 Notably, only NPRL2, but not NPRL3 or DEPDC5, was readily methylated by PRMT1 (Figure. 3B and Figures. S3D–3E). Consistently, overexpression of PRMT1 promoted the asymmetric di-methylation of NPRL2, but not of NPRL3 or DEPDC5 in cells (Figure. S3F).
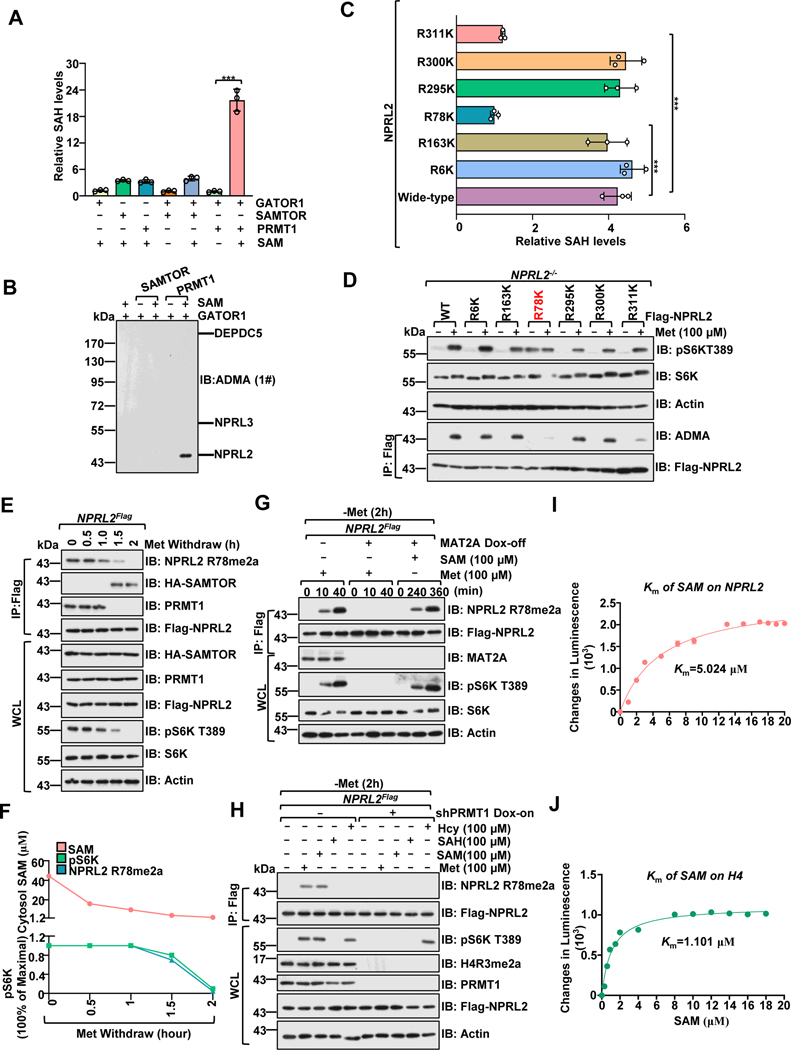
Figure 3. PRMT1 methylates NPRL2 at R78 residue.
A, PRMT1, but not SAMTOR, promotes the methylation of the GATOR1 complex. Purified GATOR1 (250 nM) was subjected to in vitro methylation assays for 1 hour in the presence of SAM (1 μM), PRMT1 (100 ng) and SAMTOR (100 ng), and the generation of SAH was analyzed via the MTase-Glo™ Methyltransferase Assay kit.
B, Immunoblotting with the ADMA levels of GATOR1 complex components from A using a panADMA antibody.
C, GST-NPRL2 (wild-type and mutants) were subjected to in vitro methylation analysis as in A.
D, The methylation of NPRL2 (wild-type and mutant) was analyzed with ADMA antibody in cells.
E, Methionine removal inhibited NPRL2 R78me2a methylation. Cells were subjected to anti-Flag immunoprecipitation and analyzed via immunoblotting with NPRL2 R78me2 antibody.
F, The correlation of cytosolic SAM levels, NPRL2 R78me2, and mTORC1 activity (pS6K) from E are presented in F. Data are representative one repeat.
G, NPRL2Flag knock-in MAT2A Dox-off HEK293T cell lines were generated as described in the previous study25. Cells were pretreated with doxycycline (DOX) for 2 days, starved of methionine for 2 hours, and restimulated with methionine (100 μM) or SAM (100 μM) for the indicated time. The WCL and anti-Flag IPs were analyzed via immunoblotting with the indicated antibodies, including a homemade-specific antibody against NPRL2 R78me2a.
H, NPRL2Flag knock-in HEK293T cells expressing tet-on-shPRMT1 were pretreated with DOX for 2 days to knockdown PRMT1, starved of methionine for 2 hours, and restimulated with methionine (100 μM) for 20 min or SAH (100 μM), SAM (100 μM), and Hcy (100 μM) for 6 hours. The WCL and anti-Flag IPs were analyzed via immunoblotting with the indicated antibodies.
I-J, The Km of SAM for human PRMT1 to mediate the methylation of NPRL2 (I) and H4 (J). Purified NPRL2 proteins (10 ng) were incubated with PRMT1 (20 ng) together with the indicated concentrations of SAM, and the level of SAH generation was analyzed as described in A. n=3 biological repeats.
See also Figure S3.
Next, we sought to identify the arginine residues that are critical for the asymmetric di-methylation event of NPRL2 via mass spectrometry (Figures. S3G–3H).50,51 Our results showed that PRMT1 methylates NPRL2 at six arginine sites, including di-methylation at R6 and R78 and mono-methylation at R163, R295, R300, and R311 (Figures. S3I–3J). The arginine (R) to lysine (K) mutation is commonly used as an unmethylated mimetic because it preserves the positive charge of arginine but cannot be modified by PRMT1.52,53 Of note, replacement of residues R78 or R311 with lysine, but not the other four residues, reduced the asymmetric di-methylation levels of NPRL2 in vitro and in cells (Figures. 3C–3D), indicating that R78 and R311 may be the major methylation sites of NPRL2 catalyzed by PRMT1. Consistent with a previous study,21 mutation of R311 did not affect methionine-induced mTORC1 activation (Figure. 3D). Thus, R78 is likely the major functional methylation site catalyzed by PRMT1.
To further interrogate this modification in cells, we generated and validated an antibody that specifically recognizes NPRL2 R78 asymmetric di-methylation (Figures.S3K–3L). Reducing the SAM/SAH ratio by DZNeP or pharmacological inhibition of PRMT1 inhibited the endogenous R78 asymmetric di-methylation of NPRL2 (Figures.S3M–3N). Consistently, the catalytically dead PRMT1 mutants (G98R and E162Q) failed to methylate NPRL2 in cells without affecting the interaction between GATOR1 and SAMTOR (Figure. S3O). Furthermore, SAMTOR exhibited a dose-dependent inhibition of NPRL2 methylation levels and the interaction between PRMT1 and GAOTR1 (Fig. S3P).
We next investigated whether the methionine availability affects the asymmetric di-methylation at R78 of NPRL2. The removal of methionine inhibited the asymmetric di-methylation of R78 in a time-dependent manner (Figures. 3E and Figures. S3Q–S3R), which correlates with mTORC1 activation and cytosolic SAM levels, but not with methionine levels (Figure. 3F). The addition of methionine restored the asymmetric di-methylation at R78 in methionine-starved cells (Figure. 3G). Inhibition of SAM production via knockdown MAT2A strongly blunted methionine-induced NPRL2 asymmetric di-methylation at R78. Of note, the addition of SAM restored the methylation level of NPRL2 in MAT2A-deficient cells (Figure. 3G). Furthermore, knockdown of PRMT1 suppressed methionine- and SAM-induced R78 asymmetric di-methylation of NPRL2 (Fig. 3H). The Michaelis constant (Km) of SAM for NPRL2 was 5 μM (Figure. 3I) and higher than that of H4 (1 μM) (Figure. 3J). The Km of SAM for NPRL2, but not H4, was compatible with the concentration of SAM (Figure. S3O), which may explain why SAM availability can directly affect the asymmetric di-methylation of NPRL2 instead of H4 in our experimental settings (Figure. 3H). Thus, the SAM-sensitive asymmetric di-methylation of NPRL2 by PRMT1 reveals that PRMT1 has an affinity and catalytic property for SAM that is compatible with the cytosolic SAM concentrations, supporting the conclusion that PRMT1 can serve as a direct sensor of SAM levels for enzymatic activity-based regulation of the mTORC1 pathway.
PRMT1 inhibits the GAP activity of GATOR1
We next sought to investigate how PRMT1 affected GATOR1’s biological functions. Knockdown of PRMT1 did not obviously affect the integrity of NPRL2 in complex with other GATOR1 subunits (Figure.S4A). Given that PRMT1 methylates NPRL2 at R78, which is the arginine finger that carries out GAP activity of GATOR1 (Figure. 4A),21 we reasoned that PRMT1 might directly modulate the GAP activity of GATOR1 via methylating NPRL2 at R78. To test this hypothesis, we purified the GATOR1 complex from HEK-293T cells, performed an in vitro methylation assay, and then measured its GAP activity on Rag A GTPase (Figures. S4B–4D). Direct incubation of GATOR1 with PRMT1 or SAMTOR, without inducing any methylation event, did not affect the GAP activity of GATOR1 (Figure.S4E), indicating that PRMT1 and SAMTOR cannot directly modulate GATOR1 activity via binding to GATOR1. Our results further reveal that PRMT1 methylated NPRL2 and consequently blunted the GAP activity of GATOR1, while the inhibitory effect could be blocked by inhibiting the methyltransferase activity of PRMT1 (Figures. 4B–4D).
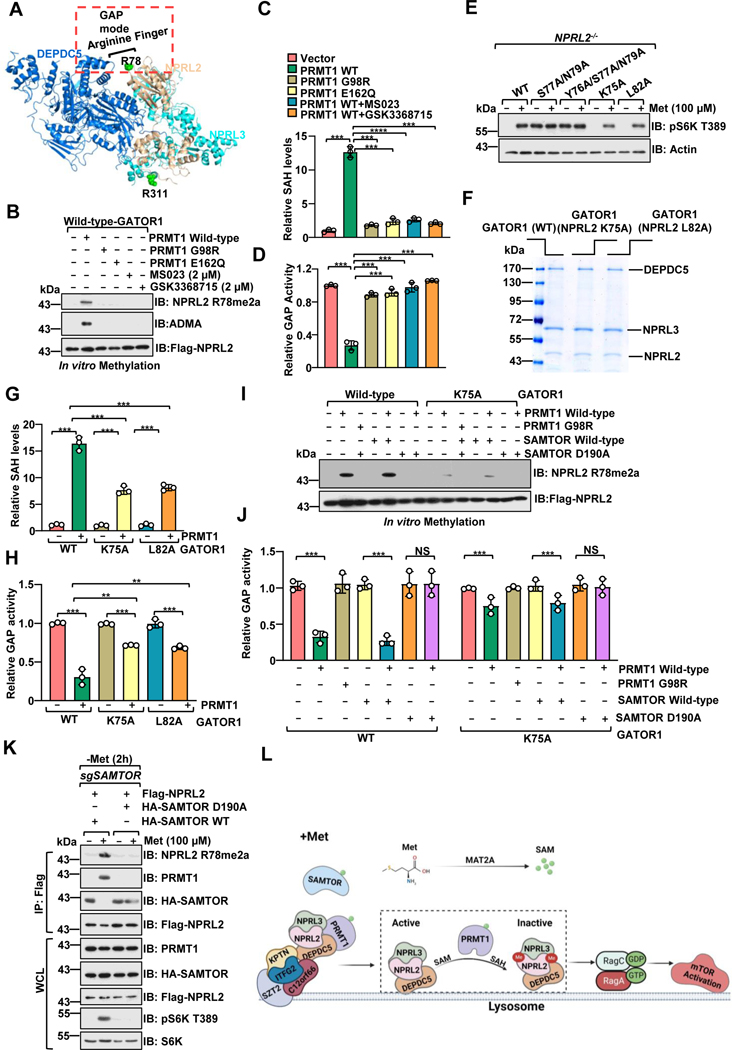
Figure 4. PRMT1 inhibits the GAP activity of GATOR1.
A, Structure of the GATOR1 complex (Protein Data Bank code 6CET) displayed by PyMOL.
B-C, PRMT1 methylates NPRL2 and antagonizes the GAP activity of the GATOR1 complex. Purified GATOR1 complex was subjected to in vitro methylation assays with PRMT1 WT or mutants (G98R, E166Q), with or without PRMT1 inhibitors, and analyzed via the MTase-Glo™ Methyltransferase Assay kit (B) or immunoblotting with the specific antibody for NPRL2 R78me2a (C). n=3 biological repeats, ***p<0.001, unpaired, two-tailed Student’s t-test.
D, PRMT1-mediated asymmetric di-methylation of NPRL2 impairs the GAP activity of the GATOR1 complex. The purified GATOR1 complex was subjected to in vitro methylation assays as in f and was further subjected to GAP activity analysis. n=3 biological repeats, ***p<0.001, unpaired, two-tailed Student’s t-test.
E, The activation of mTORC1 by methionine is decreased in cells expressing the NPRL2 K75A and L82A mutants. NPRL2 null HEK293T cells were infected with either wild-type NPRL2 or the indicated mutants, starved of methionine for 2 hours, restimulated with methionine (100 μM) for 20 min, and analyzed via immunoblotting with the indicated antibodies.
F, Coomassie blue staining of the purified GATOR1 complex containing either wild-type NPRL2 or the indicated mutants.
G, Mutation of the flanking residues near R78 in NPRL2 protein attenuated PRMT1-mediated methylation. The purified GATOR1 complex containing either wild-type NPRL2 or the indicated mutants were subjected to in vitro methylation assays as described in B. n=3 biological repeats, ***p<0.001, unpaired, two-tailed Student’s t-test.
H, Methylation of R78 in NPRL2 protein is essential for PRMT1-mediated inhibition of the GAP activity of GATOR1. The purified GATOR1 complex containing either wild-type NPRL2 or the indicated mutants were subjected to in vitro methylation assays as described in B. n=3 biological repeats, ***p<0.001,**p<0.01, unpaired, two-tailed Student’s t-test.
I-J, SAMTOR blocked PRMT1-mediated inhibitory effect on GATOR1 activity in vitro. The purified GATOR1 complex was subjected to in vitro methylation assays as indicated and was analyzed via the MTase-Glo™ Methyltransferase Assay kit (I) or immunoblotting with the specific antibody for NPRL2 R78me2a (J). n=3 biological repeats, ns, no significant difference, ***p<0.001, **p<0.01, unpaired, two-tailed Student’s t-test.
K, SAMTOR D190A mutant, but not wild-type, blunted PRMT1-mediated NPRL2 asymmetric di-methylation in cells.
L, A schematic model depicting PRMT1-mediated NPRL2 asymmetric di-methylation in methionine sensing of mTORC1 signaling.
See also Figure S4.
We then explored whether we could recapitulate PRMT1’s function using the NPRL2 methylation-deficient mutant. However, the unmethylated mimetic R78K was unresponsive to methionine starvation (Figure. 3D). This finding, along with previous reports,21,54 suggests that replacing R78 with any other amino acids might entirely disrupt the insertion of arginine finger into the nucleotide-binding pocket of RagA/B,21 leading to the catalytic inactivation of GATOR1. Importantly, the amino acids surrounding the methylation site are critical for its modification by PRMT1.55,56 In order to identify mutants that interfere with PRMT1-mediated methylation but retain the intact arginine finger, we mutated the neighboring residues of R78 and identified two evolutionarily conserved residues (K75 and L82) that are required for PRMT1-mediated methylation (Figures. S4F–4G). These two NPRL2 methylation-deficient mutants exhibited reduced mTORC1 activation in comparison with the wild-type reconstituted cells, recapitulating PRMT1 deficiency without affecting its interaction with SAMTOR (Figure. 4E and Figure. S4H). Consistent with a previous result,54 despite the mutants containing the N79A mutation that exhibited reduced R78 asymmetric di-methylation, they failed to sensitize the cells to methionine, suggesting its necessity in mediating arginine finger function. We next recombinantly purified GATOR1 containing NPRL2 (WT versus K75A or L82A mutant) and compared their ability to stimulate GTP hydrolysis. Notably, the methylation deficient NPRL2 mutants (K75A and L82A) did not affect the integrity of the GATOR1 complex (Figure. 4F), but reduced PRMT1-mediated methylation in vitro (Figure. 4G). Consistently, NPRL2 mutants (K75A or L82A) partially escaped from the inhibitory effect of PRMT1 on its GAP activity (Figure. 4H). These data suggest that R78 is a functional methylation site by PRMT1. Mutation of arginine (R) to the bulky hydrophobic phenylalanine (F) is commonly used as a constitutive methylated mimetic.53,57 In support of this notion, GATOR1 that contains the NPRL2 (R78F) mutant failed to stimulate GTP hydrolysis by the Rag GTPases (Figure. S4I). Moreover, genetic inhibition of PRMT1 in the methylated mimetic reconstituted NPRL2-R78F cells was unable to perturb mTORC1 signaling (Figure. S4J). The SAM-binding-deficient mutant SAMTOR (D190A), which constitutively interacts with the GATOR1 complex, blunted the methylation levels of NPRL2 and the inhibitory effect of PRMT1 on GATOR1 (Fig. 4I–K). Taken together, these data indicate that SAM-loaded PRMT1 catalyzes the asymmetric di-methylation of NPRL2 at R78, thereby blocking the GAP activity of GATOR1 and consequently inducing timely mTORC1 activation under methionine-sufficient conditions (Figure. 4L).
PRMT1 has a conserved role in Drosophila
We tested whether PRMT1 modulates mTORC1 signaling in Drosophila. As in mammalian cells, knockdown of dDart1 (ortholog of human PRMT1) 58 impaired methionine-stimulated dTOR signaling in Drosophila S2 cells, and moderately affected leucine- and arginine-induced dTOR signaling (Figures. S4K–S4L). dDart1 also controlled dTOR signaling in a Gator1-dependent manner (Figures. S4M–S4N). Moreover, dSamtor and dDart1 bound to dGator1 in a mutually exclusive and methionine-sensitive manner (Figure. S4O). dDart1, but not dSamtor, promoted the asymmetric di-methylation of dNprl2 (Figure. S4P), the methionine-sensitive methylation of dNprl2 was impaired in dDart1 deficient cells (Figures. S4Q–4R). Furthermore, inhibition of dDart1 in cells expressing the dNprl2 methylated mimetic mutant (R112F, corresponding to R78 of human NPRL2) did not inhibit dTOR signaling (Figures. S4S–4T). Thus, PRMT1 has a conserved role in methionine sensing in Drosophila.
PRMT1 is a physiological methionine/SAM sensor for mTORC1
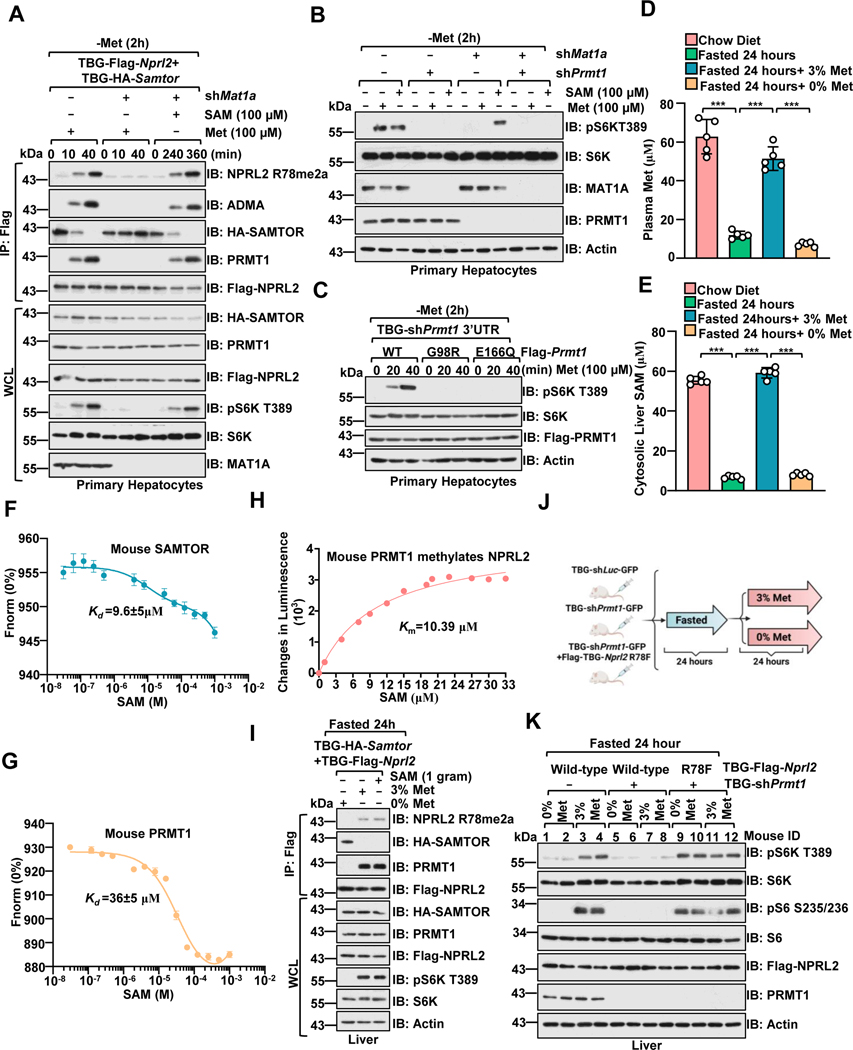
mTORC1 signaling is critical for the homeostatic maintenance of liver metabolism, and the liver is acutely sensitive to changes in nutrients, including glucose and amino acids.59,60 We then tested whether PRMT1 has a methionine/SAM-sensing role in the mouse liver. MAT1a is required for SAM biosynthesis in the adult liver. 32,34 As in HEK293 cells, Samtor perturbed the association between Prmt1 and Gator1 in a SAM-sensitive manner, and deficiency of SAM biosynthesis via inhibiting Mat1a suppressed methionine-induced R78 asymmetric di-methylation of Nprl2 and subsequent mTORC1 activation in primary hepatocytes (Figure. 5A). Knockdown of Prmt1 impaired both methionine- and SAM-dependent mTORC1 activation in Mat1a-intact and -deficient cells (Figure. 5B). The SAM binding-deficient mutants (G98R and E166Q) inhibited methionine-stimulated mTORC1 activation (Figure. 5C). Deficiency of Prmt1 partially suppressed mTORC1 signaling in Samtor-deficient cells (Figure. S5A).
Figure 5. PRMT1 is a physiological methionine/SAM sensor for mTORC1.
A, Primary hepatocytes expressing Flag-Nprl2/HA-Samtor were infected with TBG-shLuc or TBG-shMat1a shRNA. At 72 hours post-infection, cells were deprived of methionine for 2 hour and restimulated with methionine (100 μM) or SAM (100 μM) for the indicated time. The WCL and anti-FLAG IPs were analyzed via immunoblotting with the indicated antibodies.
B, Primary hepatocytes were infected with the indicated shRNA. Cells were then deprived of methionine for 2 hours and restimulated with methionine (100 μM) for 20 mins or SAM (100 μM) for 6 hours. Cell lysates were analyzed by immunoblotting with the indicated antibodies.
C, Primary hepatocytes were infected as indicated. After 72 hours, cells were treated and analyzed as described in A.
D-E, Methionine deprivation significantly reduced SAM levels in the liver. Mice were fasted for 24 hours and refed with 3% or 0% methionine diet for 24 hours. Plasma methionine levels were analyzed via ELISA. (D) Liver tissues were collected for cytosolic SAM level measurement by ELISA (E) (n=6 per group, ***p<0.001, unpaired, two-tailed Student’s t-test).
F-G, The Kd of mouse SAM-SAMTOR and SAM-PRMT1 as determined by the MST assay. n=3 biological repeats were presented.
H, The Km of SAM for mouse PRMT1 to mediate the methylation of NPRL2. Purified NPRL2 proteins (10 ng) were incubated with mouse PRMT1 (20 ng) together with the indicated concentrations of SAM, and the level of SAH generation was analyzed via the MTase-Glo™ Methyltransferase Assay kit. n=3 biological repeats were presented.
I, Dietary methionine regulates Prmt1-Nprl2-Samtor interaction and Prmt1-mediated Nprl2 methylation in the liver. Wild-type male mice with hepatic expression of TBG-Flag-Nprl2 and TBG-HA-Samtor were treated as in D, and the liver lysates were subjected to anti-Flag immunoprecipitation (IP). WCL and IPs were analyzed by immunoblotting with the indicated antibodies.
J-K, A schematic illustration of the experimental setup for studying methionine sensing in vivo. Mice were injected with the respective adenovirus for 21 days to knock down endogenous Prmt1 and overexpress Nprl2 (wild-type or R78F mutant) in the liver as indicated. Mice were then fasted for 24 hours and refed with 3% or 0% methionine diet for 24 hours (J). Liver tissues prepared from the indicated mice were analyzed by immunoblotting with the indicated antibodies (K) (n=2 per group).
See also Figure S5.
We further investigated the methionine sensing of mTORC1 in vivo. Consistent with previous studies analyzed by LC/MS,61,62 we fasted and refed mice with 3% and 0% methionine diets to limit the consequences of fluctuations in other nutrients. Notably, we found that the plasma methionine concentration was dramatically reduced in the 0% methionine-fed mice (Figure. 5D and Figure. S5B), and consequently, the cytosolic concentration of SAM in the liver was reduced in these mice (Figure 5E). Furthermore, our results provide additional support for this finding, with the dissociation constant (Kd) for mouse SAM-Samtor and SAM-Prmt1 estimated to be approximately 10 μM and 36 μM, respectively (Figures. 5F–5G). The Km of SAM for NPRL2 catalyzed by mouse Prmt1 was found to be 10 μM (Figure. 5H), and all these values are in line with the variation range of SAM levels in liver cells. As observed in cultured cells, methionine and SAM regulated the binding of Prmt1 and Samtor to Gator1 and R78 asymmetric di-methylation of Nprl2 in the liver (Figure 5I and Figure S5C). We next determined whether Prmt1 is required for the regulation of mTORC1 by dietary methionine. In fasted mice, mTORC1 was inactivated, as demonstrated by the phosphorylation level of S6, while refeeding led to an increase in phospho-S6 level that was blocked by the PRMT1 inhibitor, GSK3368715, and the mTORC1 inhibitor, rapamycin (Figures. S5D–5G). Hepatic deletion of Nprl2 blunted the inhibitory effects of PRMT1 inhibitor on mTORC1 signaling as indicated by the phosphorylation level of S6 (Figures. S5H–5K). Consistently, deletion of Prmt1 blocked methionine-induced mTORC1 activation in the liver, while co-deletion of Nprl2 or overexpression of the methylated mimetic form of Nprl2 (R78F) strongly blocked the reduction in mTORC1 activity (Figures. 5J–5K and Figures. S5L–5O). Taken together, our results suggest that Prmt1 and Samtor coordinately transmit the methionine availability to the mTORC1 pathway via sensing SAM levels in mouse liver.
Hepatic Prmt1-Nprl2-mTORC1 axis dictates insulin sensitivity to dietary methionine restriction in aged mice
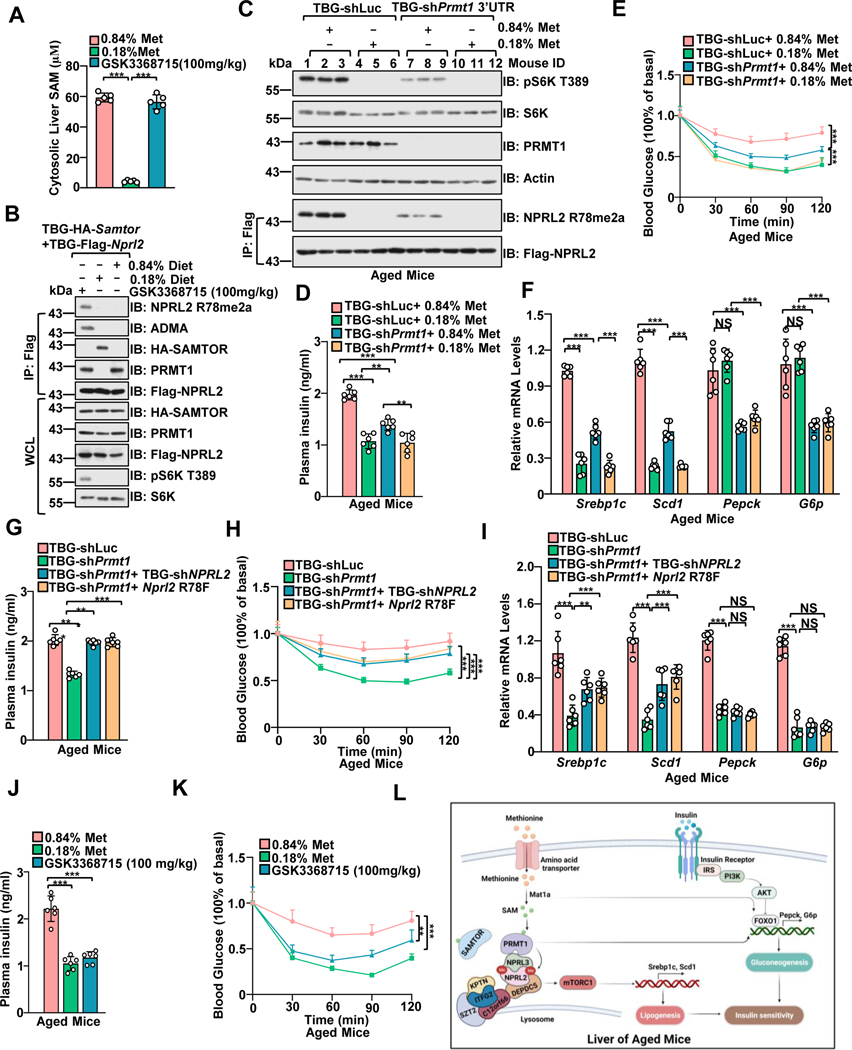
Complete dietary removal of methionine causes rapid weight loss and deterioration of animal health,63 while methionine restriction has a profound effect on physiological responses, including improving insulin sensitivity and extending the lifespan of both Drosophila and rodents.64–66 Using a methionine-restricted diet (MRD), we further interrogated the methionine sensing by mTORC1 in the longer-term physiological setting. Consistent with a previous study,40 MRD reduced the cytosolic levels of SAM in the liver (Figure. 6A and Figure. S6A), which is below the dissociation constant (Kd) of mouse Prmt1 and Samtor to SAM and the Km of SAM on NPRL2 catalyzed by mouse Prmt1. Moreover, methionine restriction promoted the interaction of Samtor with Gator1 and suppressed Prmt1 mediated R78 asymmetric di-methylation of Nprl2 in the liver (Figure. 6B), indicating that the changes in SAM levels observed in mouse liver are sufficient to induce the alterations in Nprl2 methylation and mTORC1 activity.
Figure 6. Hepatic Prmt1-Nprl2-mTORC1 dictates insulin sensitivity to dietary methionine restriction in aged mice.
A, Mice with a hepatic expression of TBG-Flag-Nprl2/TBG-HA-Samtor were fed with the indicated methionine diet (0.84% or 0.18%) or orally administrated with PRMT1 inhibitor (GSK3368715) for 50 days The liver lysates were prepared for SAM level measurement by ELISA (n=6 per group, ***p<0.001, unpaired, two-tailed Student’s t-test).
B, The WCL and IPs of the liver lysates from A were analyzed by immunoblotting with the indicated antibodies.
C-F, Aged mice (13-month-old) were injected with shLuc or shPrmt1 adenoviruses to knock down endogenous Prmt1. The mice were fed 0.84% or 0.18% methionine diets for 50 days. Liver lysates were subjected to immunoblotting with the indicated antibodies (C) (n=3 per group). The plasma insulin levels were determined by ELISA (n=6 per group) (D). The Intraperitoneal Insulin Tolerance Test (ITT) assay was performed with the indicated mice (n=6 per group) (E). The expression levels of the indicated genes were measured by qPCR (n=6 per group) (F). **p<0.01, ***p<0.001.
G-I, Mice were infected with the indicated adenoviruses for 21 days. Plasma insulin levels were determined by ELISA (n=6 per group) (G). The ITT was performed on the indicated mice (n=6 per group) (H). The expression levels of the indicated genes were measured by qPCR (n=6 mice) (I). J-K, Mice were treated as in C. Plasma insulin levels were measured by ELISA (n=6 mice) (J). The ITT was performed on the indicated mice (n=6 per group) (K). ns, no significant difference, **p<0.01, ***p<0.001.
L, A schematic model depicting PRMT1-mediated NPRL2 methylation in methionine sensing of mTORC1 signaling to regulate insulin sensitivity in aged mice.
See also Figure S6.
We next examined the physiological importance of methionine sensing by mTORC1. Insulin resistance is a hallmark of aging67 (Figures. S6B–6C) and mTORC1 hyperactivation.68 Dietary methionine restriction reprograms lipid metabolism via inhibiting the expression of lipogenesis genes (Scd1 and Srebp1c),69,70 which are downstream targets of mTORC1 and critical for hepatic insulin sensitivity.70–73 Hepatic Mat1a deficiency inhibits hepatic de novo lipogenesis and improves insulin resistance in obese mice.74 We further investigated whether Samtor- and Prmt1-mediated SAM sensing by mTORC1 plays a role in the methionine-sensitive response in the livers of aged mice. The MRD-induced decrease in mTORC1 activity, lipogenesis gene levels (Scd1 and Srebp1c), plasma insulin levels, and the degree of insulin resistance were partially prevented when MRD-fed mice were deficient in Samtor in the liver (Figures. S6D–S6H). It is possible that other tissues, such as adipocytes and skeletal muscle, mediate insulin resistance and play a role in the overall response to methionine restriction. Moreover, inhibition of Prmt1 suppressed R78 asymmetric di-methylation of Nprl2 and mTORC1 signaling (Figure. 6C). Hepatic suppression of Prmt1 inhibited plasma insulin levels, ameliorated insulin resistance, and partially blunted the decrease in lipogenic gene levels (Scd1 and Srebp1c), plasma insulin levels, and insulin resistance grade of mice fed with MRD (Figures. 6D–6F and Figure. S6I). Prmt1 possibly regulates gluconeogenesis gene expression via modulating Foxo1 (Figure. 6F).75,76 In cultured cells, point mutations G98R and E166Q in Prmt1 impaired methionine-sensitive regulation of mTORC1 signaling via abolishing the affinity of Prmt1 to SAM. Similarly, hepatic overexpression of Prmt1 G98R and Prmt1 E166Q conferred the same phenotype as hepatic deficiency of Prmt1 (Figures. S6J–6M). As in hepatic Samtor-deficient mice, the MRD-induced decrease in mTORC1 activity, lipogenesis gene levels (Scd1 and Srebp1c), plasma insulin levels, and insulin resistance extent were also prevented when MRD-fed mice expressing methylated mimetic mutant Nprl2 (R78F) (Figures. S6N–6R). Co-deletion of Nprl2 or overexpressing R78F Nprl2 partially restored the effect caused by hepatic deficiency of Prmt1 in aged mice (Figures. 6G–6I, and Figure. S6S), indicating that Prmt1 regulates the methionine-sensitive response of mTORC1 signaling depending on its modification of Nprl2. Moreover, pharmacological inhibition of PRMT1 suppressed hepatic mTORC1 signaling, expression levels of lipogenesis genes, plasma insulin levels, improved insulin sensitivity, and phenocopying the treatment of MRD, but without dramatically affecting body weight (Figures. 6J–6K and Figures. S6T–6V). Hence, we conclude that Samtor and Prmt1 mediated regulation of mTORC1 is required to maintain the physiological response to MRD in aged mice (Figure. 6L).
DISCUSSION
In conclusion, our study reveals a novel function of PRMT1 in the methionine sensing by mTORC1 signaling via utilizing SAM as a cofactor to methylate NPRL2 and antagonize the GAP activity of GATOR1, establishing PRMT1 as a conserved physiological methionine/SAM sensor of mTORC1 signaling to dictate hepatic insulin sensitivity. Unlike previously reported amino acid sensors, including SAMTOR, Sestrin2, Castor1, and SAR1B,25–27 that use a passive mode to regulate mTORC1 signaling, PRMT1 regulates mTORC1 signaling in an enzyme-dependent manner. PRMT1 coordinates with SAMTOR to form a methionine-sensing machine and sequentially transmits the methionine availability to mTORC1, ensuring this process is tightly regulated.
Notably, PRMT1 is also reported to affect the mTORC1 signaling pathway by methylating the GATOR2 component WDR24 on R329.77 However, the R239 residue is conserved only in humans and mice, but not in other species (Figure. S6W). Moreover, we found that the inhibition of WDR24 methylation via mutating R329 to lysine did not affect the methionine sensing of mTORC1 in our experimental setting (Figure. S6W). These results indicate that NPRL2, but not WDR24, is likely the specific substrate of PRMT1 to dictate the methionine sensing of mTORC1. In support of this notion, amino acid-sensing pathways are conserved in many eukaryotic organisms,78 and our proposed methionine-sensing pathway is found to be conserved in Drosophila, mice, and humans (Figures. S4). Hence, it is possible that the PRMT1-NPRL2 axis evolutionarily preceded the emergence of PRMT1-WDR24, and that these two proposed mechanisms might coordinately regulate amino acid sensing of mTORC1 in mammals. Given that leucine and arginine sensing impinge on GATOR2, which functions upstream of GATOR1, PRMT1 loss may affect leucine and methionine sensing via modulating the GAP activity of GATOR1. This further indicates the potential crosstalk of different amino acid sensing machineries, which warrants further in-depth investigation (Figure. S6X).
Hepatic SAM levels and mTORC1 activity are increased during aging in Drosophila and rodents. 59,79,80 However, prolonged treatment with rapamycin leads to insulin resistance due to the off-target inhibition of mTORC2, 81 and a new way of selectively inhibiting mTORC1 signaling from intervening in aging and lifespan is urgently needed. Methionine restriction diet improves health and extends lifespan in many organisms.65,66,82 Our findings affirm the crucial role of hepatic SAMTOR-PRMT1-NPRL2-mTORC1 in the response to methionine restriction in aged mice, providing a novel therapeutic strategy to inhibit hepatic lipogenesis and enhance insulin sensitivity in aged mice through the selective mTORC1 inhibition by PRMT1 inhibitors. Many tissues, including adipocytes, liver, and skeletal muscle, exhibit resistance to insulin. We specifically analyzed MRD-mediated responses in the livers of aged mice, and it is possible that our observed mechanism also plays a role in mediating the methionine-sensitive organismal response in other organs. Furthermore, it will be interesting to investigate whether PRMT1 inhibitors could ameliorate other metabolic syndromes, such as obesity, with over-activation of mTORC1 signaling in future studies.
Limitations of the study
There exist several constraints in our study. First, as SAM functions both as a substrate for PRMT1 and as a molecule sensed by SAMTOR, the exact molecular mechanism underlying how PRMT1 collaborates with SAMTOR to monitor methionine levels remains elusive and needs to be further investigated in a separate biochemistry-focused study. Secondly, to determine the exact composition of the PRMT1/SAMTOR/GATOR1 sensing mechanism, it is necessary to obtain structural insights to uncover the fundamental molecular mechanisms involved in this dynamic process. Thirdly, the decrease in mTORC1 activation upon PRMT1 knockdown in SAMTOR-null cells implies the presence of an additional mechanism that could also play a role in conferring aberrant mTORC1 activation in the absence of SAMTOR. Hence, it warrants further investigation to determine if there is an additional pathway besides PRMT1 in suppressing GATOR1 and if this pathway also orchestrates with SAMTOR to confer timely sensing of methionine. Fourth, since asymmetric di-methylation is a reversible process,30 it will be interesting to investigate which enzyme is required to mediate the de-methylation of NPRL2, thereby creating the dynamic and reversible process to allow cells to better sense the changes in methionine levels to subsequently relay to the timely control of mTORC1 signaling.
STAR ★ METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Wenyi Wei (wwei2@bidmc.harvard.edu).
Materials availability
All unique reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
Unprocessed western blotting data in the manuscript can be found in Data S1–Source Data.
Excel datasheets including values underlying each graph are provided in Data S2–Source Data.
Figures S1–S6 are available as supplemental information.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
HEK293, HEK293T, HeLa, MDA-MB-231and MCF7 cells were maintained in DMEM containing 100 units/ml penicillin and 100 μg/ml streptomycin supplemented with 10% FBS. HCC1500 cells were a gift from Dr. Piotr Sicinski (DFCI, Boston, MA) and maintained in RPM1 1640 containing 100 units/ml penicillin and 100 μg/ml streptomycin supplemented with 10% FBS. NPRL2−/− HEK293T cells and NPRL2Flag HEK293T cells were a gift from the lab of Dr. David M. Sabatini. The MAT2A dox-off cell line was generated and used as previously described25.
Primary hepatocytes were isolated as previously described71 and cultured in the Medium 199 (Life Technologies) supplemented with 5% (vol/vol) FBS, 100 units ml−1 penicillin and 100 μg ml−1 streptomycin sulfate. Cells were then infected with the indicated adenovirus and subjected to further analysis 72 hours after infection.
S2 cells were cultured in Drosophila Schneider’s Medium (Invitrogen) with 10% fetal bovine serum, 100 U/ml of penicillin, and 100 μg/ml of Streptomycin at 25°C and 5% CO2. Transfection was carried out using Lipo3000 according to the manufacturer’s instructions. A ubiquitin-Gal4 construct was cotransfected with pUAST expression vectors for overexpression experiments. siRNA was transfected into cells by RNAiMAX transfection reagent (Thermo,13778150) to knock down the indicated genes, and cells were assayed 48 hours after siRNA delivery.
Animals
The 8-week-old male C57BL/6 or BALB/c nude female mice were purchased from Shanghai Laboratory Animal Center (Shanghai, China). All mice were housed in the animal facility at Tongji University at the temperature maintained at 21–23 °C under a 12 h:12 h light: dark cycle. All mice had ad libitum access to a standard chow diet (Research Diets, D10001). All animal care and use procedures followed the guidelines of the Animal Care and Use Committee of Tongji University. The Nprl2 (pAAV-TBG-sfGFP-3xFLAG-miR30 shRNA (Nprl2)-WPRE, targeting sequence: GAGTATGATGTGCCCGTCTTT), Prmt1(pAAV-TBG-sfGFP-3xFLAG-miR30 shRNA (Prmt1)-WPRE, targeting sequence: CGCAACTCCATGTTTCACAAT) RNAi adeno-associated viruses were generated by Obie Technology (Shanghai, China). The Nprl2 RNAi, Prmt1 RNAi, or control adeno-associated virus was delivered by tail-vein injection to mice. At 21 days after viral infection, mice were subjected to the indicated analysis. For the fasting and refeeding experiments, mice were fasted for 24 hours. The refed group was fed the standard chow diet for 24 hours, or a 3% methionine or 0% methionine-containing diet (Trophic Animal Feed High-Tech Co., Ltd, China). Rapamycin (10 mg/kg) was administered to the mice via intraperitoneal injections 4 hours before refeeding for 6 hours. PRMT1 inhibitor (GSK3368715) was administered via oral gavage to mice for 2 consecutive days at a dose of 100 mg/kg.
To achieve acute liver-specific deletion Samtor, Mat1a, Prmt1 or overexpression of Prmt1(wild-type, G98R, E166Q), HA-Samtor, Flag-Nprl2 (wild-type and R78F), AAV expressing shRNA or indicated genes driven by the liver-specific TBG promoter (AAV8-TBG-shRNA/overexpression gene, Obio Technology) was administered to 13 months aged mice by IP injection (2 × 1011 vg/mouse), 2.5 weeks prior to initiation of the dietary treatment. The indicated mice were fed with 0.84% methionine or 0.18 % methionine-containing diet (Trophic Animal Feed High-Tech Co., Ltd, China), accordingly. Fifty days later, mice were subjected to the indicated analyses.
METHOD DETAILS
Immunoblotting and immunoprecipitations
Cells or tissues were collected and lysed in lysis buffer (50 mM Tris pH 7.5, 120 mM NaCl, 10% glycerol, 0.5% NP-40, 0.1% SDS) supplemented with protease inhibitors (Complete Mini, Roche) and phosphatase inhibitors (phosphatase inhibitor cocktail set I and II, Calbiochem). Cell lysates were cleared by centrifugation in a microcentrifuge (12,000 rpm for 15 mins at 4°C), and protein concentrations of lysates were determined using the BCA method (Bio-Rad). Cell lysates were prepared by adding 4X loading buffer (0.2 M Tris-HCI, 0.4 M DTT, 8% SDS, 6 mM Bromophenol blue, 4.3 M Glycerol) and boiling at 98°C for 10 min. The equal amounts of total proteins were resolved by 8–15% SDS-PAGE and transferred to PVDF membranes. Immunoblots were blocked with 5% non-fat milk in Tris-buffered saline (TBS) containing 0.1% and then probed with primary antibodies overnight at 4 °C and secondary antibodies (1:5000). Immunoreactivity was developed with enzymatic detection via Pierce ECL Plus western blotting substrate (Boston Bioproducts Ins. Cat#WB-100L).
For immunoprecipitation, cells were lysed in EBC buffer (50 mM Tris pH 7.5, 120 mM NaCl, 0.5% NP-40) supplemented with protease inhibitor (Complete Mini, Roche), anti-Flag M2 affinity Gel (A2220, Sigma) or anti-HA (A2095) was washed three times and then resuspended to lysis buffer, 30 μL of the mixed slurry were added to the cleared lysates and incubated at 4°C in a shaker for 120 minutes. Immunoprecipitants were washed four times, once with EBC buffer and twice with EBC buffer with 500 mM NaCl. Immunoprecipitated proteins were denatured by adding 60 μL of 2× SDS loading buffer, boiled for 5 minutes, and analyzed by immunoblotting.
Cell transfection, virus infection, and stable cell line generation
For transient transfection, the encoding plasmids were transfected into HEK293T cells with polyethylenimine (PEI) (Polysciences). To generate the lentivirus, HEK293T cells were transfected with the sgRNA, shRNA, or pLenti-encoding plasmids along with psPAX2 and PMD2G. Sixteen hours post-transfection, the medium was changed to DMEM with 10% FBS. Thirty-six hours later, the supernatants were collected and passed through a 0.45 μm filter. The targeted cells were infected with the indicated virus-containing medium. Forty-eight hours later, cells were selected with puromycin (1 μg/mL), and surviving cells were expanded and subjected to further analysis. Guide RNAs targeting the indicated genes were cloned into lentiCripsr-V2-Puro (Addgene 52961). Tet-On inducible shRNAs were cloned into the pLKO-Tet-on vector. The shRNAs were purchased from Sigma-Aldrich. The target sequences are listed in Table S1.
Lysosome purification
Lysosome purification was performed as described previously 12. Briefly, HeLa cells infected with Flag-RFP-LAMP1 virus were rinsed twice in ice-cold PBS and then lysed in the lysosome fractionation buffer (90 mM K-Gluconate, 50 mM KCl, 1 mM EGTA, 5 mM MgCl2, 50 mM Sucrose, 20mM HEPES, pH 7.4, supplemented with 2.5 mM ATP, 5 mM Glucose and protease inhibitors), cells were mechanically broken with a 23G needle attached to a 1ml syringe for 6–8 times and then centrifuged at 2000 g for 10 mins. The supernatant was then subjected to anti-Flag immunoprecipitation at 4 °C for 3 hours. The captured lysosomes were lysed directly with 2×SDS loading buffer and subjected to immunoblotting.
Immunostaining
HEK293T and HeLa cells were cultured on gelatin-coated coverslips in 12-well plates (100,000 cells/well). After methionine treatments, cells were washed twice with PBS, fixed with 4% paraformaldehyde, washed twice with PBS, and permeabilized with 0.1% Triton X-100 in PBS. Cells were washed and blocked in PBS containing 1% BSA for 1 hour at room temperature. The coverslips were incubated overnight with primary antibodies for LAMP1 (1:50) and mTOR (1:100) at 4°C. Cells were then rinsed with PBS 4 times and incubated with secondary antibodies in PBS (1:1000 dilution) for 2h at room temperature. After washing, the coverslips were mounted on slides using a mounting buffer containing DAPI (Invitrogen). Images were acquired on a Zeiss LSM 510 Meta confocal system.
GATOR1 and RagA-RagC complex purification
For the purification of the GATOR1 complex, the GATOR1 complex (pRK5-HA-NPRL3, pRK5HA-DEPDC5, pRK5-Flag-NPRL2) and RagA-RagC complex (Flag-RagCS75N, GST-RagA) were co-transfected into HEK293T cells. Twenty-four hours post-transfection, cells were lysed in EBC buffer, the supernatant was further spun at 100,000g for 1h and filtered with 0.45-μm membrane, incubated with 100μl anti-Flag M2 beads for 4 hours with rotation at 4°C. The beads were washed three times with EBC buffer with 500 mM NaCl. The bound proteins were eluted with 0.5 mg/mL Flag peptide (Sigma, F3290) for 30 min with rotation. The eluted proteins were concentrated to 100 μl with a centrifugal filter with a 30 kDa cut-off (Millipore, MPUFC503024), of which 10 μl was used for SDS-PAGE and Coomassie Blue staining.
The purification of SAMTOR and PRMT1 proteins
Human and mouse SAMTOR and PRMT1 genes were synthesized (Tsingke, Shanghai) and subcloned into a modified pMLink vector with protein A tags in the N-terminus followed by a SUMO tag and a 3C cleavage site. To purify SAMTOR and PRMT1 proteins, human Expi 293F cell expressing system was used. Briefly, human Expi 293F cells were cultured in serum-free medium (Sino Bio., Beijing) until the cell density reached 1.2 × 106 cells/mL. The pMLink plasmids containing the desired protein gene were transfected into cells using PEI at a 1:4 ratio and, in total 4 mg plasmids per liter. Transfected cells were cultured for another 72 hours before harvesting.
Transfected cells were centrifuged at 3000 rpm and resuspended in lysis buffer containing 30 mM HEPES pH 8.0, 300 mM NaCl, 0.25% CHAPS, 0.2 mM EDTA, 2 mM DTT, 10% glycerol (v/v), 1 mM PMSF, 1 μg/mL Aprotinin, 1 μg/mL Pepstatin, and 1 μg/mL Leupeptin. After incubation on an overhead rotor at 4 °C for 30 min, the cell debris was removed by centrifugation at 15,000 rpm for 30 min. The supernatant was incubated with an IgG affinity gel (Smart-Lifesciences, Changzhou, China) for 2 hours at 4 °C. The resin was washed three times using lysis buffer (containing 0.1% CHAPS). The desired proteins were eluted by on-column overnight digestion using Ulp1 protease. To improve the purity, the proteins were further purified using gel filtration. Finally, all four proteins were concentrated and stored in a buffer containing 20 mM HEPES pH 7.4, 300 mM NaCl.
Microscale thermophoresis (MST) assay
Recombinant proteins dialyzed against MST Buffer (25 mM HEPES pH 7.4, 50 mM NaCl, 2.5 mM MgCl2, 0.025% NP-40) were labeled with MST fluorescence dye (NanoTemper MO-L011) according to manufacturer’s instructions. The binding of labeled proteins to leucine, arginine, or isoleucine was measured on a Monolith NT.115 machine (NanoTemper) with the software MO.Control. The protein concentration was set to 100 nM. SAM gradient was set as: 1 mM, 1/2 mM, 1/4 mM, 1/6 mM, 1/8 mM, 1/10 mM, 1/12 mM, 1/14 mM, 1/16 mM, 1/18 mM, 1/20 mM, 1/22 mM, 1/24 mM, 1/26 mM, 1/28mM, and 1/30 mM. Data from three independent biological replicates were analyzed by GraphPad Prism 8.0.
In vitro methylation assay
For the in vitro methylation of NPRL2, a 50 μL mixture in PBS buffer consisting of 250 nM GATOR1 complex and 40 μM SAM (final concentration) was initiated by adding 1 μg of recombinant PRMT1 (31411, Active Motif) and incubating at 30 ºC for 1–1.5 hour. The reaction was terminated by adding 10 μL 4×SDS loading buffer. The reaction mixture was analyzed by 10% SDS-PAGE, and the methylation levels were immunoblotted with ADMA antibody. The generation of SAH and Km of SAM were analyzed using the MTase-Glo™ Methyltransferase Assay (V7601) kit according to the manufacturer’s instructions.
Stimulated GTP hydrolysis assay.
The GAP activity of GATOR1 was determined via the GTPase-GloTm assay (V7681, Promega) according to the manufacturer’s instructions. Briefly, a 2× GTP solution containing 10 μM GTP and 1mM DTT in GTPase/GAP Buffer was prepared, and 50 nM RagA/RagC GTPase and 1 μM GATOR1 complex were added to a final volume of 25 μL. The reaction mixture was then incubated at room temperature (25°C) for 120 minutes. Then, 25 μL of reconstituted GTPase-Glo™ Reagent was added to terminate the GTPase reaction. The reaction mixture was incubated with shaking for 30 minutes at room temperature (25°C). Finally, 50 μl of Detection Reagent was added to the reaction mixture and incubated for 10 minutes at room temperature (25°C), and luminescence intensities were measured.
Methionine, SAM, and insulin measurement
Cytosolic or plasma methionine concentrations were measured using the methionine assay kit (Sigma, MAK-347), and SAM levels were determined via the SAM ELISA Assay Kit (Eagle Bioscience, SAM31-K01). The plasma insulin levels were analyzed using the Insulin Mouse ELISA Kit (Thermo Fisher, 0534063022). All the measurements were performed according to the manufacturer’s instructions.
Intraperitoneal Insulin Tolerance Test (ITT) analysis
For the ITT analysis, mice were fasted for 6 hours and received an intraperitoneal injection of insulin (0.75 U kg−1 body weight). Glucose levels (obtained from tail blood by cutting < 5 mm from the tail tip) were measured at 0, 15, 30, 60, 90, and 120 mins after injection using the OneTouch Ultra blood glucose monitoring system (Johnson).
Cell growth and viability assay
The indicated MDA-MB231 cells were plated on 96-well plates (2000 per well). Cells were incubated for 10 min with CellTiter-Glo (Promega, G7573) at the indicated times, and luminescence was measured using a 96-well plate reader (GloMax-96 microplate luminometer; Promega). The luminescence values were normalized to those of the control and are shown as relative viability (%).
Mass spectrometry analysis
To identify the binding partner of NPRL2, five 15-cm dishes NPRL2Flag knock-in HEK293T cells were lysed in EBC Lysis Buffer, cell lysates were clarified by centrifugation. Clarified lysate from each group was incubated with 300 μl anti-Flag resin (Sigma) by rotating at 4 °C for 2 hr. To identify the arginine methylation site of NPRL2, GST-NPRL2 were transfected with GFP-PRMT1 into ten 10-cm HEK293T cells. 48 hours post-transfection, cells were lysed and subjected to GST-pull down with GST Agarose Beads (Sigma) by rotating at 4 °C for 3 hr. Proteins immobilized on the resins were denatured and dissolved in Laemmli Sample Buffer by heating. Proteins were separated by SDS-PAGE, Coomassie blue-stained, and identified by mass spectrometry. Briefly, the NPRL2 band was treated with 10 mM DTT for 30 minutes to reduce it, followed by alkylation using 55 mM iodoacetamide for 45 minutes. The treated sample was then digested using trypsin/LysC enzymes. The resulting peptides were extracted from the gel and subjected to microcapillary reversed-phase liquid chromatography-tandem mass spectrometry (LC-MS/MS) using a high-resolution QExactive HF Orbitrap in positive ion DDA mode (Top 8). Fragmentation was achieved using higher energy collisional dissociation (HCD) via a Proxeon EASY-nLc1200 UPLC nano-HPLC system. MS/MS data were matched against the UniProt Human protein database (version 2021_0616) using Mascot 2.7, and data analysis was performed using Scaffold Q+S 5.0 software to identify peptides and modified peptides. Peptides meeting a 1% false discovery rate (FDR) threshold were accepted for further analysis. To exclude the potential artifact as we have detected in PRMT5’s interaction with GATOR1, we highly recommend validating the mass spectrometry data via different experimental approach in the following up study.
RNA isolation and real-time RT-PCR analyses
Total RNA was extracted from homogenized liver or cells using Trizol Reagent (Invitrogen), following the manufacturer’s instructions. Total RNA was digested by RNase-free DNase I (Promega). Quantitative PCR (qPCR) was performed using the Hieff qPCR SYBR Green Master Mix (Takara) and analyzed on a Bio-Rad CFX96 apparatus (Bio-Rad). Primer sequences are listed in Supplementary Table S2.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical data are presented as mean ±SD. All experiments were independently repeated at least twice, and similar results were obtained. All quantification analyses were performed using GraphPad Prism 8. (GraphPad Software). All parameters were tested using unpaired two-tailed Student’s t-test or two-way ANOVA as described in the figure legends. P<0.05 were considered statistically significant. Schematic models were created using Biorender (http://biorender.com/)
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-mTOR | Cell Signaling Technology | Cat#: 2983 |
| Rabbit anti-ADMA | Cell Signaling Technology | Cat#: 13522 |
| Rabbit anti-RagA | Cell Signaling Technology | Cat#: 4357 |
| Rabbit anti-pS6K T389 | Cell Signaling Technology | Cat#: 9234 |
| Rabbit anti-S6K | Cell Signaling Technology | Cat#: 2708 |
| Rabbit anti-S6 | Cell Signaling Technology | Cat#: 2317 |
| Rabbit anti-pS6 S235/236 | Cell Signaling Technology | Cat#: 4858 |
| Rabbit anti-p4EBP1 S65 | Cell Signaling Technology | Cat#: 9451 |
| Rabbit anti-4EBP1 | Cell Signaling Technology | Cat#: 9644 |
| Rabbit anti-NPRL2 | Cell Signaling Technology | Cat#: 37344 |
| Rabbit anti-Flag | Cell Signaling Technology | Cat#: 14793 |
| Rabbit anti-HA | Cell Signaling Technology | Cat#: 3724 |
| Rabbit anti-pCAD | Cell Signaling Technology | Cat#: S1859 |
| Rabbit anti-pdS6K | Cell Signaling Technology | Cat#: 9029 |
| Rabbit anti-PRMT1 | Proteintech | Cat#: 11279 |
| Rabbit anti-WDR24 | Proteintech | Cat#: 20778 |
| Rabbit anti-MTHFD2 | Proteintech | Cat#: 12270 |
| Rabbit anti-MAT2A | Proteintech | Cat#: 55309 |
| Mouse anti-β-Actin | Proteintech | Cat#: 20778 |
| Rabbit anti-NPRL3 | Abcam | Cat#: ab121346 |
| Rabbit anti-MAT1A | Abcam | Cat#: ab129176 |
| Rabbit anti-ADMA (#2) | Dr. Mark T. Bedford UT MD Anderson Cancer Center | N/A |
| Rabbit anti-NPRL2 R78me2a | This paper | |
| HRP-conjugated anti-mouse secondary antibody | Sigma-Aldrich | Cat#: A-4416 |
| HRP-conjugated anti-rabbit secondary antibody | Sigma-Aldrich | Cat#: A-4914 |
| Anti-mouse Alexa Fluor 488 | Life Technologies/Molecular Probes | Cat#: A11001 |
| Anti-Rabbit Alexa Fluor 594 | Life Technologies/Molecular Probes | Cat#: A32740 |
| Bacterial and Virus Strains | ||
| XL10 Gold Escherichia coli | Agilent | Cat #200314 |
| BL21(DE3) Escherichia coli | Dr. William G. Kaelin, Jr., Dana-Farber Cancer Institute | N/A |
| E.coli: One Shot Stbl3 Chemically competent cells | Thermo Fisher Scientific | Cat#C737303 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Anti-FLAG M2 Affinity Gel | Sigma-Aldrich | Cat#: A-2220 |
| Anti-HA M2 Affinity Gel | Sigma-Aldrich | Cat#: A-2095 |
| Full Amino-acids-Deficient Medium | US Biological Life Sciences | Cat#: R8999-04A |
| Methionine-Deficient Medium | US Biological Life Sciences | Cat#: R8999-06 |
| Leucine-Deficient Medium | US Biological Life Sciences | Cat#: R8998-02 |
| Arginine-Deficient Medium | US Biological Life Sciences | Cat#: R8998-01 |
| Dialyzed FBS | Gibco | Cat#: 26400044 |
| MS023 | Selleck Chemicals | Cat#: S8112 |
| Rapamycin | Selleck Chemicals | Cat#: S1039 |
| GSK3368715 | Selleck Chemicals | Cat#: S8858 |
| Doxycycline Hyclate | Selleck Chemicals | Cat#: WC2031 |
| L-Methionine hydrochloride solution | Sigma | Cat#: 50272 |
| L-Homocysteine | Sigma | Cat#: 69453 |
| SAH | Sigma | A9384 |
| SAM | Cayman Chemical | 13956 |
| Experimental Models: Cell Lines | ||
| HEK293 | Dr. William G. Kaelin, Jr., Dana-Farber Cancer Institute | N/A |
| HEK293T | Dr. William G. Kaelin, Jr., Dana-Farber Cancer Institute | N/A |
| NPRL2Flag HEK293T cells | Dr. David M. Sabatini Whitehead Institute | N/A |
| NPRL2−/− HEK293T cells | Dr. David M. Sabatini Whitehead Institute | N/A |
| MDA-MB-231 cells | Dr. Piotr Sicinski Dana-Farber Cancer Institute | N/A |
| MCF7 cells | Dr. Piotr Sicinski Dana-Farber Cancer Institute | N/A |
| HCC1500 cells | Dr. Piotr Sicinski Dana-Farber Cancer Institute | N/A |
| Experimental Models: Organisms/Strains | ||
| C57BL/6 | Shanghai Laboratory Animal Center | N/A |
| Recombinant DNA | ||
| Tet-on-shPRMT1 | This paper | N/A |
| HA-NPRL2-pRK5 | Bar-Peled et al. 2013 | Addgene 100513 |
| Flag-NPRL2-pRK5 | Bar-Peled et al. 2013 | Addgene 46333 |
| HA-NPRL3-pRK5 | Bar-Peled et al. 2013 | Addgene 46330 |
| HA-DEPDC5-pRK5 | Bar-Peled et al. 2013 | Addgene 46327 |
| HA-NPRL2-pRK5 | Bar-Peled et al. 2013 | Addgene 99709 |
| GFP-PRMT9 | Hadjikyriacou et al. 2015 | Addgene 79675 |
| GFP-PRMT1 | Dr. Yanzhong Yang City of Hope | N/A |
| GFP-PRMT2 | Dr. Yanzhong Yang City of Hope | N/A |
| GFP-PRMT3 | Dr. Yanzhong Yang City of Hope | N/A |
| GFP-PRMT4 | Dr. Yanzhong Yang City of Hope | N/A |
| GFP-PRMT5 | Dr. Yanzhong Yang City of Hope | N/A |
| pLVX-CMV-HA-PRMT1 | This paper | N/A |
| pLVX-CMV-HA-PRMT1-G98R | This paper | N/A |
| pLVX-CMV-HA-PRMT1-E162Q | This paper | N/A |
| pLVX-CMV-HA-NPRL2 | This paper | N/A |
| HA-dSamtor | Obio Technology | N/A |
| Flag-dNprl2 | Obio Technology | N/A |
| Flag-dNprl2-R78F | Obio Technology | N/A |
| Myc-dDart1 | Obio Technology | N/A |
| TBG-Flag-Nprl2 | Obio Technology | N/A |
| TBG-Flag-Nprl2-R78F | Obio Technology | N/A |
| TBG-HA-Samtor | Obio Technology | N/A |
| TBG-Flag-Nprl2 | Obio Technology | N/A |
| TBG-Flag-Prmt1 | Obio Technology | N/A |
| TBG-Flag-Prmt1-G98R | Obio Technology | N/A |
| TBG-Flag-Prmt1-E162Q | Obio Technology | N/A |
| TBG-shMat1a | Obio Technology | N/A |
| TBG-shSamtor | Obio Technology | N/A |
| TBG-shNprl2 | Obio Technology | N/A |
| TBG-shPrmt1-3’UTR | Obio Technology | N/A |
| Oligonucleotides | ||
| PRMT1-CDS shRNA targeting sequence | GTGTTCCAGTATCTCTGATTA | N/A |
| PRMT1-3’UTR shRNA targeting sequence | TGAGCGTTCCTAGGCGGTTTC | N/A |
| PRMT1 sgRNA targeting sequence #1 | AAAGCCAACAAGTTAGACCA | N/A |
| PRMT1 sgRNA targeting sequence #2 | GATGGCCGTCACATACAGCG | N/A |
| PRMT1 sgRNA targeting sequence #13 | GAGGCTCATCCCATTAGCCA | N/A |
| SAMTOR sgRNA targeting sequence | GAAATACTGCTCGTGCGCAG | N/A |
| NPRL2 sgRNA targeting sequence | GATGCGGCAGCCGCTGCCCA | N/A |
| MAT2A sgRNA targeting sequence | TTAAAGGAGGTCTGTGCCGG | N/A |
| Mat1a (mouse) shRNA targeting sequence | GCAGGATAATGGTGCAGTCAT | N/A |
| Samtor (mouse) shRNA targeting sequence | GATGTTGGCAGCTGCTTTAAT | N/A |
| sidDart1 sequence | GCAGCGAGGAUACAUACAATT | N/A |
| sidNprl2 sequence | TGGGTTTGTTGTAGTTTGTAA | N/A |
| sidSamtor sequence | TGGAATCCTACAGAGCCGAGGG | N/A |
| F-alpha-dtubulin | 5’-CAACCAGATGGTCAAGTGCG-3’ | N/A |
| R-alpha-dtubulin | 5’-ACGTCCTTGGGCACAACATC-3’ | N/A |
| F-dSamtor | 5’-GACCAACGATGGGAAGGTGG-3’ | N/A |
| R-dSamtor | 5’-GCTCTGTAGGATTCCAGGAGT-3’ | N/A |
| F-dDart1 | 5’-TGAGGGAGTGGACATTATTATTTCC-3’ | N/A |
| R-dDart1 | 5’-TATGTGTTCGACTCCTGAATTTGAG-3’ | N/A |
| F-dNprl2 | 5’-GGCTGCAAGATAAGCTGCCA-3’ | N/A |
| R-dNprl2 | 5’-GCGTTGAAGATGCTGCTTGG-3’ | N/A |
| F-mouse Srebp1c | 5’-GGAGCCATGGATTGCACATT-3’ | N/A |
| R-mouse Srebp1c | 5’-GGCCCGGGAAGTCACTGT-3’ | N/A |
| F-mouse Scd1 | 5’-GCAAGCTCTACACCTGCCTCTT-3’ | N/A |
| R-mouse Scd1 | 5’-CGTGCCTTGTAAGTTCTGTGGC-3’ | N/A |
| F-mouse Pepck | 5’-CGATGACATCGCCTGGATGA-3’ | N/A |
| R-mouse Pepck | 5’-TCTTGCCCTTGTGTTCTGCA-3’ | N/A |
| F-mouse G6p | 5’-GAAGGCCAAGAGATGGTGTGA-3’ | N/A |
| R-mouse G6p | 5’-TGCAGCTCTTGCGGTACATG-3’ | N/A |
Highlights.
PRMT1 senses methionine/SAM levels to mTORC1
PRMT1 orchestrates methionine sensing in concert with SAMTOR
PRMT1 methylates NPRL2 and inhibits GATOR1 activity
PRMT1-NPRL2-mTORC1 axis confer organismal response to dietary methionine restriction
ACKNOWLEDGMENTS
We thank all the members from Dr. Wei and Dr. Xiao’s lab for the critical reading and helpful suggestions. This work was supported by NIH grants (R01CA177910 and R35CA253027 to WW. Shanghai Orthopedic Clinical Medical Research Center Special Program (21MMC1930100) and National Natural Science Foundation of China (31976006, 82173028 and 82141105) to X.J.
Footnotes
DECLARATION OF INTERESTS
W.W. is a co-founder and consultant for the ReKindle Therapeutics. Other authors declare no competing financial interests.
SUPPLEMENTAL INFORMATION
Supplemental information includes 6 supplemental figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kim J, and Guan KL (2019). mTOR as a central hub of nutrient signalling and cell growth. Nat Cell Biol 21, 63–71. 10.1038/s41556-018-0205-1. [DOI] [PubMed] [Google Scholar]
- 2.Liu GY, and Sabatini DM (2020). mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol 21, 183–203. 10.1038/s41580-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma XM, and Blenis J. (2009). Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10, 307–318. 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 4.Saxton RA, and Sabatini DM (2017). mTOR Signaling in Growth, Metabolism, and Disease. Cell 169, 361–371. 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 5.Mossmann D, Park S, and Hall MN (2018). mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat Rev Cancer 18, 744–757. 10.1038/s41568-018-0074-8. [DOI] [PubMed] [Google Scholar]
- 6.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, and Guan KL (2008). Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 10, 935–945. 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, and Sabatini DM (2008). The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501. 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inoki K, Li Y, Xu T, and Guan KL (2003). Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 17, 1829–1834. 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, and Manning BD (2014). Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156, 771–785. 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, and Sabatini DM (2010). Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303. 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfson RL, Chantranupong L, Wyant GA, Gu X, Orozco JM, Shen K, Condon KJ, Petri S, Kedir J, Scaria SM, et al. (2017). KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature 543, 438–442. 10.1038/nature21423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng M, Yin N, and Li MO (2017). SZT2 dictates GATOR control of mTORC1 signalling. Nature 543, 433–437. 10.1038/nature21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, and Sabatini DM (2013). A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106. 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, et al. (2015). Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 347, 188–194. 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rebsamen M, Pochini L, Stasyk T, de Araujo ME, Galluccio M, Kandasamy RK, Snijder B, Fauster A, Rudashevskaya EL, Bruckner M, et al. (2015). SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 519, 477–481. 10.1038/nature14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, and Sabatini DM (2013). The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell 52, 495–505. 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Napolitano G, Di Malta C, Esposito A, de Araujo MEG, Pece S, Bertalot G, Matarese M, Benedetti V, Zampelli A, Stasyk T, et al. (2020). A substrate-specific mTORC1 pathway underlies Birt-Hogg-Dube syndrome. Nature 585, 597–602. 10.1038/s41586-020-2444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baldassari S, Picard F, Verbeek NE, van Kempen M, Brilstra EH, Lesca G, Conti V, Guerrini R, Bisulli F, Licchetta L, et al. (2019). The landscape of epilepsy-related GATOR1 variants. Genet Med 21, 398–408. 10.1038/s41436-018-0060-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nickerson ML, Warren MB, Toro JR, Matrosova V, Glenn G, Turner ML, Duray P, Merino M, Choyke P, Pavlovich CP, et al. (2002). Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer Cell 2, 157–164. 10.1016/s1535-6108(02)00104-6. [DOI] [PubMed] [Google Scholar]
- 20.Shen K, Huang RK, Brignole EJ, Condon KJ, Valenstein ML, Chantranupong L, Bomaliyamu A, Choe A, Hong C, Yu Z, and Sabatini DM (2018). Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes. Nature 556, 64–69. 10.1038/nature26158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen K, Valenstein ML, Gu X, and Sabatini DM (2019). Arg-78 of Nprl2 catalyzes GATOR1-stimulated GTP hydrolysis by the Rag GTPases. J Biol Chem 294, 2970–2975. 10.1074/jbc.AC119.007382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimobayashi M, and Hall MN (2016). Multiple amino acid sensing inputs to mTORC1. Cell Res 26, 7–20. 10.1038/cr.2015.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jewell JL, Kim YC, Russell RC, Yu FX, Park HW, Plouffe SW, Tagliabracci VS, and Guan KL (2015). Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science 347, 194–198. 10.1126/science.1259472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas JD, Zhang YJ, Wei YH, Cho JH, Morris LE, Wang HY, and Zheng XF (2014). Rab1A is an mTORC1 activator and a colorectal oncogene. Cancer Cell 26, 754–769. 10.1016/j.ccell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolfson RL, Chantranupong L, Saxton RA, Shen K, Scaria SM, Cantor JR, and Sabatini DM (2016). Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351, 43–48. 10.1126/science.aab2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chantranupong L, Scaria SM, Saxton RA, Gygi MP, Shen K, Wyant GA, Wang T, Harper JW, Gygi SP, and Sabatini DM (2016). The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell 165, 153–164. 10.1016/j.cell.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Ou Y, Luo R, Wang J, Wang D, Guan J, Li Y, Xia P, Chen PR, and Liu Y. (2021). SAR1B senses leucine levels to regulate mTORC1 signalling. Nature 596, 281–284. 10.1038/s41586-021-03768-w. [DOI] [PubMed] [Google Scholar]
- 28.Gu X, Orozco JM, Saxton RA, Condon KJ, Liu GY, Krawczyk PA, Scaria SM, Harper JW, Gygi SP, and Sabatini DM (2017). SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 358, 813–818. 10.1126/science.aao3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henry MF, and Silver PA (1996). A novel methyltransferase (Hmt1p) modifies poly(A)+-RNA-binding proteins. Mol Cell Biol 16, 3668–3678. 10.1128/MCB.16.7.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, and Bedford MT (2013). Protein arginine methyltransferases and cancer. Nat Rev Cancer 13, 37–50. 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 31.Nishioka K, and Reinberg D. (2003). Methods and tips for the purification of human histone methyltransferases. Methods 31, 49–58. 10.1016/s1046-2023(03)00087-2. [DOI] [PubMed] [Google Scholar]
- 32.Lu SC, and Mato JM (2012). S-adenosylmethionine in liver health, injury, and cancer. Physiol Rev 92, 1515–1542. 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanderson SM, Gao X, Dai Z, and Locasale JW (2019). Methionine metabolism in health and cancer: a nexus of diet and precision medicine. Nat Rev Cancer 19, 625–637. 10.1038/s41568-019-0187-8. [DOI] [PubMed] [Google Scholar]
- 34.Lu SC, Alvarez L, Huang ZZ, Chen L, An W, Corrales FJ, Avila MA, Kanel G, and Mato JM (2001). Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci U S A 98, 5560–5565. 10.1073/pnas.091016398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramani K, and Lu SC (2017). Methionine adenosyltransferases in liver health and diseases. Liver Res 1, 103–111. 10.1016/j.livres.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramani K, Mato JM, and Lu SC (2011). Role of methionine adenosyltransferase genes in hepatocarcinogenesis. Cancers (Basel) 3, 1480–1497. 10.3390/cancers3021480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reytor E, Perez-Miguelsanz J, Alvarez L, Perez-Sala D, and Pajares MA (2009). Conformational signals in the C-terminal domain of methionine adenosyltransferase I/III determine its nucleocytoplasmic distribution. FASEB J 23, 3347–3360. 10.1096/fj.09-130187. [DOI] [PubMed] [Google Scholar]
- 38.Barbier-Torres L, Murray B, Yang JW, Wang J, Matsuda M, Robinson A, Binek A, Fan W, Fernandez-Ramos D, Lopitz-Otsoa F, et al. (2022). Depletion of mitochondrial methionine adenosyltransferase alpha1 triggers mitochondrial dysfunction in alcohol-associated liver disease. Nat Commun 13, 557. 10.1038/s41467-022-28201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wyant GA, Abu-Remaileh M, Wolfson RL, Chen WW, Freinkman E, Danai LV, Vander Heiden MG, and Sabatini DM (2017). mTORC1 Activator SLC38A9 Is Required to Efflux Essential Amino Acids from Lysosomes and Use Protein as a Nutrient. Cell 171, 642–654 e612. 10.1016/j.cell.2017.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mentch SJ, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, Gomez Padilla P, Ables G, Bamman MM, Thalacker-Mercer AE, et al. (2015). Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab 22, 861–873. 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang W, Li N, Zhang Y, Wang X, Yin M, and Lei QY (2022). AHCYL1 senses SAH to inhibit autophagy through interaction with PIK3C3 in an MTORC1-independent manner. Autophagy 18, 309–319. 10.1080/15548627.2021.1924038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khayati K, Antikainen H, Bonder EM, Weber GF, Kruger WD, Jakubowski H, and Dobrowolski R. (2017). The amino acid metabolite homocysteine activates mTORC1 to inhibit autophagy and form abnormal proteins in human neurons and mice. FASEB J 31, 598–609. 10.1096/fj.201600915R. [DOI] [PubMed] [Google Scholar]
- 43.Hu H, Luo C, and Zheng YG (2016). Transient Kinetics Define a Complete Kinetic Model for Protein Arginine Methyltransferase 1. J Biol Chem 291, 26722–26738. 10.1074/jbc.M116.757625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, and Cheng X. (2003). Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure 11, 509–520. 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazid MA, Ward C, Luo Z, Liu C, Li Y, Lai Y, Wu L, Li J, Jia W, Jiang Y, et al. (2022). Rolling back human pluripotent stem cells to an eight-cell embryo-like stage. Nature 605, 315–324. 10.1038/s41586-022-04625-0. [DOI] [PubMed] [Google Scholar]
- 46.Tang X, Zhang Y, Wang G, Zhang C, Wang F, Shi J, Zhang T, and Ding J. (2022). Molecular mechanism of S-adenosylmethionine sensing by SAMTOR in mTORC1 signaling. Sci Adv 8, eabn3868. 10.1126/sciadv.abn3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsiao K, Zegzouti H, and Goueli SA (2016). Methyltransferase-Glo: a universal, bioluminescent and homogenous assay for monitoring all classes of methyltransferases. Epigenomics 8, 321–339. 10.2217/epi.15.113. [DOI] [PubMed] [Google Scholar]
- 48.Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, and Zhang Y. (2001). Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293, 853–857. 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Person MD, and Bedford MT (2022). Pan-methylarginine antibody generation using PEG linked GAR motifs as antigens. Methods 200, 80–86. 10.1016/j.ymeth.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bedford MT, and Clarke SG (2009). Protein arginine methylation in mammals: who, what, and why. Mol Cell 33, 1–13. 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bedford MT, and Richard S. (2005). Arginine methylation an emerging regulator of protein function. Mol Cell 18, 263–272. 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 52.McBride AE, Cook JT, Stemmler EA, Rutledge KL, McGrath KA, and Rubens JA (2005). Arginine methylation of yeast mRNA-binding protein Npl3 directly affects its function, nuclear export, and intranuclear protein interactions. J Biol Chem 280, 30888–30898. 10.1074/jbc.M505831200. [DOI] [PubMed] [Google Scholar]
- 53.Wang YP, Zhou W, Wang J, Huang X, Zuo Y, Wang TS, Gao X, Xu YY, Zou SW, Liu YB, et al. (2016). Arginine Methylation of MDH1 by CARM1 Inhibits Glutamine Metabolism and Suppresses Pancreatic Cancer. Mol Cell 64, 673–687. 10.1016/j.molcel.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 54.Egri SB, Ouch C, Chou HT, Yu Z, Song K, Xu C, and Shen K. (2022). Cryo-EM structures of the human GATOR1-Rag-Ragulator complex reveal a spatial-constraint regulated GAP mechanism. Mol Cell 82, 1836–1849 e1835. 10.1016/j.molcel.2022.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gayatri S, Cowles MW, Vemulapalli V, Cheng D, Sun ZW, and Bedford MT (2016). Using oriented peptide array libraries to evaluate methylarginine-specific antibodies and arginine methyltransferase substrate motifs. Sci Rep 6, 28718. 10.1038/srep28718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wooderchak WL, Zang T, Zhou ZS, Acuna M, Tahara SM, and Hevel JM (2008). Substrate profiling of PRMT1 reveals amino acid sequences that extend beyond the “RGG” paradigm. Biochemistry 47, 9456–9466. 10.1021/bi800984s. [DOI] [PubMed] [Google Scholar]
- 57.Campbell M, Chang PC, Huerta S, Izumiya C, Davis R, Tepper CG, Kim KY, Shevchenko B, Wang DH, Jung JU, et al. (2012). Protein arginine methyltransferase 1-directed methylation of Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen. J Biol Chem 287, 5806–5818. 10.1074/jbc.M111.289496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boulanger MC, Miranda TB, Clarke S, Di Fruscio M, Suter B, Lasko P, and Richard S. (2004). Characterization of the Drosophila protein arginine methyltransferases DART1 and DART4. Biochem J 379, 283–289. 10.1042/BJ20031176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sengupta S, Peterson TR, Laplante M, Oh S, and Sabatini DM (2010). mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 468, 1100–1104. 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- 60.Cornu M, Oppliger W, Albert V, Robitaille AM, Trapani F, Quagliata L, Fuhrer T, Sauer U, Terracciano L, and Hall MN (2014). Hepatic mTORC1 controls locomotor activity, body temperature, and lipid metabolism through FGF21. Proc Natl Acad Sci U S A 111, 11592–11599. 10.1073/pnas.1412047111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cangelosi AL, Puszynska AM, Roberts JM, Armani A, Nguyen TP, Spinelli JB, Kunchok T, Wang B, Chan SH, Lewis CA, et al. (2022). Zonated leucine sensing by Sestrin-mTORC1 in the liver controls the response to dietary leucine. Science 377, 47–56. 10.1126/science.abi9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finkelstein JD, and Martin JJ (1986). Methionine metabolism in mammals. Adaptation to methionine excess. J Biol Chem 261, 1582–1587. [PubMed] [Google Scholar]
- 63.Anthony TG, Morrison CD, and Gettys TW (2013). Remodeling of lipid metabolism by dietary restriction of essential amino acids. Diabetes 62, 2635–2644. 10.2337/db12-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orentreich N, Matias JR, DeFelice A, and Zimmerman JA (1993). Low methionine ingestion by rats extends life span. J Nutr 123, 269–274. 10.1093/jn/123.2.269. [DOI] [PubMed] [Google Scholar]
- 65.Miller RA, Buehner G, Chang Y, Harper JM, Sigler R, and Smith-Wheelock M. (2005). Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell 4, 119–125. 10.1111/j.1474-9726.2005.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee BC, Kaya A, Ma S, Kim G, Gerashchenko MV, Yim SH, Hu Z, Harshman LG, and Gladyshev VN (2014). Methionine restriction extends lifespan of Drosophila melanogaster under conditions of low amino-acid status. Nat Commun 5, 3592. 10.1038/ncomms4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Selman C, and Withers DJ (2011). Mammalian models of extended healthy lifespan. Philos Trans R Soc Lond B Biol Sci 366, 99–107. 10.1098/rstb.2010.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saxton RA, and Sabatini DM (2017). mTOR Signaling in Growth, Metabolism, and Disease. Cell 168, 960–976. 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hasek BE, Boudreau A, Shin J, Feng D, Hulver M, Van NT, Laque A, Stewart LK, Stone KP, Wanders D, et al. (2013). Remodeling the integration of lipid metabolism between liver and adipose tissue by dietary methionine restriction in rats. Diabetes 62, 3362–3372. 10.2337/db13-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li S, Brown MS, and Goldstein JL (2010). Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc Natl Acad Sci U S A 107, 3441–3446. 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu TF, Tang JJ, Li PS, Shen Y, Li JG, Miao HH, Li BL, and Song BL (2012). Ablation of gp78 in liver improves hyperlipidemia and insulin resistance by inhibiting SREBP to decrease lipid biosynthesis. Cell Metab 16, 213–225. 10.1016/j.cmet.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 72.Ntambi JM, Miyazaki M, Stoehr JP, Lan H, Kendziorski CM, Yandell BS, Song Y, Cohen P, Friedman JM, and Attie AD (2002). Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc Natl Acad Sci U S A 99, 11482–11486. 10.1073/pnas.132384699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shimomura I, Hammer RE, Richardson JA, Ikemoto S, Bashmakov Y, Goldstein JL, and Brown MS (1998). Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev 12, 3182–3194. 10.1101/gad.12.20.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saenz de Urturi D, Buque X, Porteiro B, Folgueira C, Mora A, Delgado TC, Prieto-Fernandez E, Olaizola P, Gomez-Santos B, Apodaka-Biguri M, et al. (2022). Methionine adenosyltransferase 1a antisense oligonucleotides activate the liver-brown adipose tissue axis preventing obesity and associated hepatosteatosis. Nat Commun 13, 1096. 10.1038/s41467-022-28749-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamagata K, Daitoku H, Takahashi Y, Namiki K, Hisatake K, Kako K, Mukai H, Kasuya Y, and Fukamizu A. (2008). Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell 32, 221–231. 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 76.Choi D, Oh KJ, Han HS, Yoon YS, Jung CY, Kim ST, and Koo SH (2012). Protein arginine methyltransferase 1 regulates hepatic glucose production in a FoxO1-dependent manner. Hepatology 56, 1546–1556. 10.1002/hep.25809. [DOI] [PubMed] [Google Scholar]
- 77.Yin S, Liu L, Ball LE, Wang Y, Bedford MT, Duncan SA, Wang H, and Gan W. (2023). CDK5-PRMT1-WDR24 signaling cascade promotes mTORC1 signaling and tumor growth. Cell Rep 42, 112316. 10.1016/j.celrep.2023.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wolfson RL, and Sabatini DM (2017). The Dawn of the Age of Amino Acid Sensors for the mTORC1 Pathway. Cell Metab 26, 301–309. 10.1016/j.cmet.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Uthus EO, and Brown-Borg HM (2003). Altered methionine metabolism in long living Ames dwarf mice. Exp Gerontol 38, 491–498. 10.1016/s0531-5565(03)00008-1. [DOI] [PubMed] [Google Scholar]
- 80.Obata F, and Miura M. (2015). Enhancing S-adenosyl-methionine catabolism extends Drosophila lifespan. Nat Commun 6, 8332. 10.1038/ncomms9332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, et al. (2012). Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 335, 1638–1643. 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lees EK, Krol E, Grant L, Shearer K, Wyse C, Moncur E, Bykowska AS, Mody N, Gettys TW, and Delibegovic M. (2014). Methionine restriction restores a younger metabolic phenotype in adult mice with alterations in fibroblast growth factor 21. Aging Cell 13, 817–827. 10.1111/acel.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Unprocessed western blotting data in the manuscript can be found in Data S1–Source Data.
Excel datasheets including values underlying each graph are provided in Data S2–Source Data.
Figures S1–S6 are available as supplemental information.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.