Abstract
While TGF-β signaling is essential for microglial function, the cellular source of TGF-β1 ligand and its spatial regulation remains unclear in the adult CNS. Our data supports that microglia but not astrocytes or neurons are the primary producers of TGF-β1 ligands needed for microglial homeostasis. Microglia-Tgfb1 KO leads to the activation of microglia featuring a dyshomeostatic transcriptome that resembles disease-associated, injury-associated, and aged microglia, suggesting microglial self-produced TGF-β1 ligands are important in the adult CNS. Astrocytes in MG-Tgfb1 inducible (i)KO mice show a transcriptome profile that is closely aligned with an LPS-associated astrocyte profile. Additionally, using sparse mosaic single-cell microglia KO of TGF-β1 ligand we established an autocrine mechanism for signaling. Here we show that MG-Tgfb1 iKO mice present cognitive deficits, supporting that precise spatial regulation of TGF-β1 ligand derived from microglia is required for the maintenance of brain homeostasis and normal cognitive function in the adult brain.
Subject terms: Microglia, Neuroimmunology
TGF-β signaling is required for microglial homeostasis, however the source of ligands in the adult brain is unknown. Here, the authors show that microglial homeostasis relies on microglia-derived TGF-β1 ligand via an autocrine mechanism, which is also important for astrocyte homeostasis.
Introduction
Microglia are commonly known as the resident immune cells in the central nervous system (CNS), but their roles expand beyond that of innate immunity. At homeostasis, microglia play a variety of regulatory roles such as surveilling the brain parenchyma for injury or disease, phagocytosis, and synaptic pruning1–4. In addition to their homeostatic role, microglia are vital in inflammatory response initiation and regulation. In the case of injury or inflammation, microglia dynamically alter their function on a spectrum of activation states ranging from the more pro-inflammatory M1-like state to the anti-inflammatory M2-like state5–7. Previous studies have shown that transforming growth factor beta (TGF-β) signaling is required for the development of microglia during the embryonic stage8. Specifically, a cleverly designed “CNS-specific” Tgfb1 knockout (KO) mouse model was developed by overexpressing the Tgfb1 gene in T-cells (via an Il-2 promoter) in a global Tgfb1 KO mouse model, which depletes CNS TGF-β1 constitutively but partially compensates peripheral TGF-β1 levels8. Using this mouse model, it was reported that in the absence of TGF-β1 in the CNS during development, microglia do not establish their signature gene expression, indicating that TGF-β1 is required for normal microglial development8. While this study supported the importance of TGF-β1 in microglial development, whether TGF-β signaling is required in mature microglia to maintain their survival and function in the adult brain is not known. Moreover, serum levels of TGF-β1 in this “CNS” Tgfb1 KO mouse model were undetectable8, resulting in a potential confound due to altered TGF-β1 levels in peripheral tissues and serum that could have indirect effects on microglia maturation.
Regarding whether TGF-β signaling is required for the maintenance of homeostasis in adult microglia, there is some controversy in the literature. Buttgereit et al. 9 reported that Sall1CreERTgfbr2fl/fl mice showed activated morphology in microglia, accompanied by upregulation of CD45/CD11c and certain inflammatory cytokines (Il1b, TNF, and Cxcl10). However, Arnold et al. reported no phenotype in microglia in Cx3cr1CreERTgfbr2fl/fl mice treated with tamoxifen (TAM) at an age of 30 days10. Recently, another study reported an intermediate phenotype (termed “primed”) on evaluating the Cx3cr1CreER-Tgfbr2fl/fl mice11. Whereas they established that TGF-β signaling, via TGF-βR2 in adult microglia, is necessary for maintaining the ramified morphology and certain features of microglial homeostasis11, they further reported that inducible knockout (iKO) of Tgfbr2 in adult microglia only leads to a “primed” state in microglia without effects on many microglia homeostatic signature genes such as P2ry12, Tmem119, Hexb, and Sall111. These studies, while supporting the importance of TGF-β signaling in microglial maturation during developmental stages and maintaining certain features of homeostasis in adulthood, also generate standing questions regarding the requirements and the degree of importance of TGF-β signaling in maintaining microglia homeostatic signature gene expression in the adult CNS. More importantly, the precise cellular source of TGF-β ligands, the spatial/temporal regulation of components of the TGF-β signaling pathway across different cell types in the CNS, and their functional relevance have yet to be identified, leaving some major gaps in our knowledge regarding the regulation of this important signaling pathway in the adult brain.
While it is assumed that many different cell types can be sources of TGF-β ligands in the CNS at homeostasis12–17, the actual production of TGF-β ligands in different cell types in the brain has not been established. Additionally, two previous studies hint at a highly precise spatially localized regulation of the activation of TGF-β ligand through the interaction of leucine-rich repeat protein (LRRC33) and αVβ8 integrin13,18,19. However, whether the TGF-β ligand is regulated by a diffusible paracrine mechanism locally, or whether the ligand is more strictly regulated via an autocrine manner in the CNS is not known. The present study aims to address these major gaps in the field in this study.
Herein, using cell-type-specific conditional or iKO models of the Tgfb1 gene, we demonstrate that microglia- but not neuron- or astrocyte-derived TGF-β1 ligand is required for the maintenance of homeostatic microglia in the early postnatal and adult brain. Furthermore, the loss of microglia-derived TGF-β1 ligands leads to the presence of reactive astrocytes within the brain and causes cognitive deficits in adult mice. Additionally, our study shows that the total TGF-β1 ligand level in the brain is substantially lower compared to that of serum or peripheral tissue, and that the adult brain has accordingly established a precise spatially controlled mechanism to regulate ligand production to maintain homeostasis in individual microglia that is dependent on microglial autocrine TGF-β signaling. Our data also show that TGF-β1 is enriched in microglia, whereas TGF-β2 is instead enriched in astrocytes. Following Tgfb1 gene deletion in microglia, Tgfb3 is upregulated in the Tgfb1 KO microglia. However, at least up to 12 weeks following global microglial Tgfb1 deletion, neither the astrocytic Tgfb2 nor the upregulated Tgfb3 in microglia is able to compensate for the loss of function of Tgfb1 or rescue the dyshomeostatic phenotype in Tgfb1 KO microglia, suggesting distinct expression and functions of the different ligands in the CNS. We also address the questions of microglia-astrocyte crosstalk and the functional relevance of microglial TGF-β signaling in the adult CNS. With the importance of TGF-β signaling becoming more recognized in injury, neurodegeneration, and aging in the CNS, our study provides insights into the mechanisms of how TGF-β signaling can be regulated on a single-cell level via a microglia autocrine mechanism in the adult CNS and expands directions for future studies in understanding how TGF-β1 ligand production and downstream signaling in recipient cells can occur under these conditions.
Results
Abolishing TGF-β1 ligand expression specifically in CNS macrophages
To identify the cell type(s) in the CNS that provide TGF-β1 ligand to microglia and other TGF-β1 responsive cells, we first examined scRNAseq data sets published in previous studies20–24. Highly enriched Tgfb1 mRNA levels in adult mouse microglia are observed in multiple scRNAseq datasets20,21. Further analysis of these data shows that astrocytes, neurons, and oligodendrocytes have minimal Tgfb1 expression. Whereas oligodendrocyte precursor cells (OPCs) and endothelial cells have detectable levels of Tgfb1 expression, these are substantially lower compared to microglia Tgfb1 mRNA levels (Supplementary Fig. 1A, B)20,21. By searching published human scRNAseq datasets, we found that several independent human studies showed similar enrichment of Tgfb1 gene expression in microglia (Supplemental Fig. 1C, D)22–24 while one other independent study showed Tgfb1 enriched expression in microglia only in AD patients but not in control microglia25. To validate microglia Tgfb1 expression in the mouse brain, we undertook a combined RNAscope/IHC analysis to examine the cellular expression pattern of Tgfb1 in the adult mouse brain. Our data show that, indeed, Tgfb1 mRNA is enriched in IBA1-positive microglia but is not detected in neurons (Supplementary Fig. 1E). Tgfb1 mRNA is also detected in a small population of non-IBA1+ cells, which could be endothelial cells or other glial cells. mRNA for the type 1 TGF-β1 receptor (TGF-βR1 or ALK5) is also detected in microglia, supporting that microglia can be both ligand-producing and responding cells for TGF-β1 signaling. To examine whether microglia are a major contributor to TGF-β1 production in the brain, we depleted microglia from the adult mouse brain using the well-established receptor for colony-stimulating factor 1 (CSFR1) antagonist PLX5622 (1200ppm in diet) following our previously published protocol (Supplementary Fig. 1F), which resulted in more than 90% of microglia ablation reported by us and other studies26–28. After successful microglia ablation, we examined the total Tgfb1 mRNA in cortical tissue from the control or microglia-ablated mouse brain. qRT-PCR analysis shows that the PLX5622 treatment leads to a substantial depletion of microglia in the adult brain, indicated by a significant decrease of Iba1 mRNA levels in brain tissue (<5% of WT levels), accompanied by a 70% decrease in total Tgfb1 mRNA levels (Supplementary Fig. 1G). Since microglia compose only 5–10% of total brain cells29,30, microglia ablation leading to a 70% decrease in total Tgfb1 mRNA levels in the brain supports that microglia are a major component for TGF-β1 ligand production.
Next, to establish the direct functional relevance of microglia-produced TGF-β1 ligand to microglia homeostasis, the Cx3cr1CreER line31 was crossed with the Tgfb1fl/fl line to enable TAM-induced TGF-β1 ligand loss in microglia in adulthood. To confirm the efficiency of Tgfb1 gene deletion in microglia in the inducible MG-Tgfb1 KO mice, in our recent study, we sorted microglia using an R26-YFP (yellow fluorescent protein) reporter allele (which labels ~90% of total microglia in the adult mouse brain) at 3 weeks following TAM treatment in Cx3cr1CreERTgfb1wt/wtR26-YFP and Cx3cr1CreERTgfb1fl/flR26-YFP mice32. We demonstrate a significant decrease (99.8%) in Tgfb1 mRNA levels from sorted YFP+ microglia in Cx3cr1CreERTgfb1fl/fl mice in comparison to control microglia (Cx3cr1CreERTgfb1wt/wt)32 (Fig. 1A, right). To examine whether peripheral serum or tissue TGF-β1 levels are also affected in our MG-Tgfb1 iKO mice, we measured the TGF-β1 protein levels in serum and spleen using enzyme-linked immunosorbent assay (ELISA) analysis. In contrast to the previous “CNS-Tgfb1” constitutive KO mouse model in which serum TGF-β1 levels were completely abolished8, our Cx3cr1CreERTgfb1 iKO mice show no difference in serum or spleen TGF-β1 protein levels between Cx3cr1CreER WT and Cx3cr1CreERTgfb1fl/fl mice at 3 weeks after TAM treatment (Supplemental Fig. 2), confirming minimal interference in peripheral TGF-β1 ligand levels. Our ELISA results also show that, compared to the high levels of TGF-β1 ligand in the spleen and serum (control and iKO spleen = 5.7 pg/ug or 5.9 pg/ug protein: control and iKO serum = 1.9 pg/ug or 1.8 pg/ug protein), total brain TGF-β1 levels are dramatically lower (below the detect limit of the ELISA assay), a result that is consistent with a recent independent study33. This suggests that TGF-β ligand production and release in blood/peripheral organs might be distinctly different from that of CNS given the substantial difference in their abundance. Due to the limitation of this analysis method, we were not able to confirm the loss of TGF-β1 ligand at a protein level in our Cx3cr1CreER-Tgfb1 iKO mice directly using an ELISA assay in the light of the low total brain TGF-β protein levels. We next attempted to use fluorescence-activated cell sorting (FACS) analysis on surface TGF-β1 levels which demonstrated similar results. FACS analysis also showed substantially higher cell surface TGF-β1 expression in splenic myeloid cells, as compared to brain myeloid cells (Supplementary Fig. 2). This finding further supports that CNS TGF-β1 levels are substantially lower compared to peripheral tissue, making detection of the TGF-β1 protein in the CNS a challenge using either ELISA or FACS analysis. Our FACS analysis showed the detection of TGF-β1 protein on the surface of CD11b+/CD45+ splenocytes but no difference between control and Cx3cr1CreERTgfb1 iKO mice in this population. This corroborates that our iKO mouse model does not affect TGF-β1 expression in peripheral myeloid cells. However, since CD11b+/CD45+ cells from the brain show very low levels of TGF-β1 antibody binding we could not confidently compare the level of surface TGF-β1 expression on brain myeloid cells between the control and Cx3cr1CreER-Tgfb1fl/fl iKO mice. Nevertheless, qRT-PCR analysis from sorted brain myeloid cells demonstrates that the Cx3cr1CreERTgfb1fl/fl transgenic line can efficiently delete the Tgfb1 gene in these cells32 with minimal interference on systemic serum or spleen TGF-β1 levels. Additionally, RNAseq of the sorted WT and Tgfb1 iKO CNS myeloid cells confirms the loss of the floxed exon 3 from Tgfb1 mRNA in iKO mice while not affecting mRNA counts of exon 4, which is downstream of the 3’ loxP site (Supplementary Fig. 2). qRT-PCR analysis using an exon 3-specific primer/probe set also validated the efficient Tgfb1 gene recombination in sorted microglia in iKO mice (Fig. 1A, right).
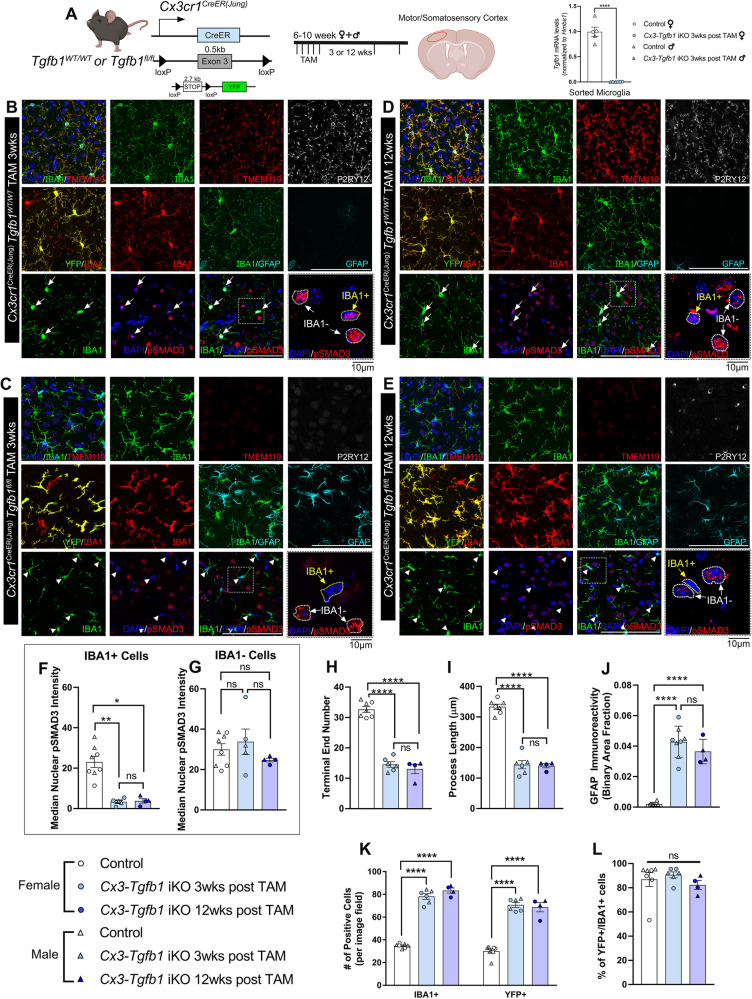
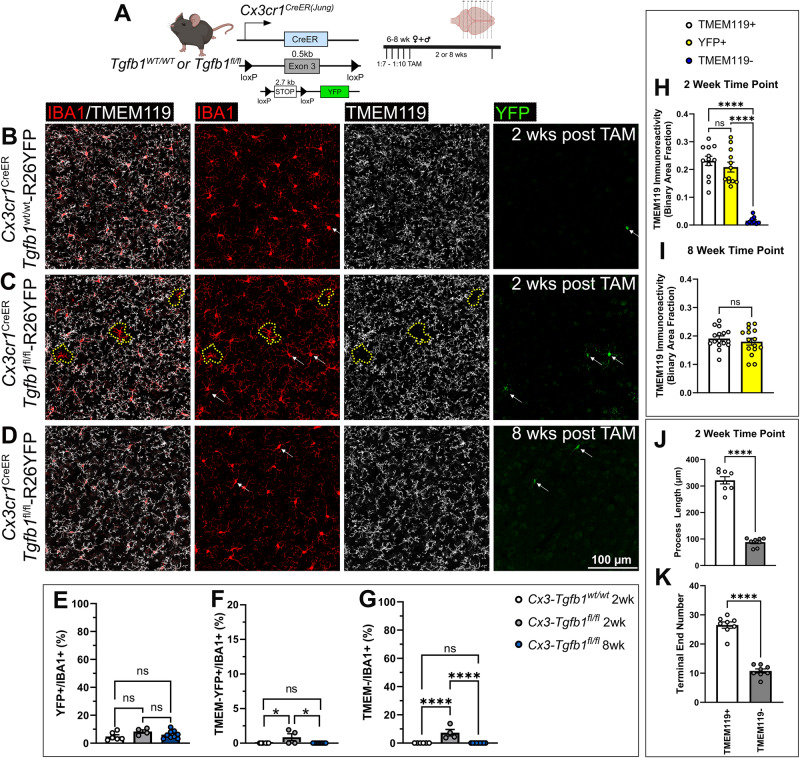
Fig. 1. Microglia-specific Tgfb1 gene deletion in a Cx3cr1CreER(Jung) driver line results in a loss of homeostasis of microglia and an increase in reactive astrocytes in the cortex of the adult mouse brain.
A (left) Mouse model for targeting microglial Tgfb1 and experimental timeline and (right) indicates the gene deletion efficiency in FACS-isolated microglia. B–E Representative images for immunohistochemistry stained for IBA1, TMEM119, P2RY12, YFP, GFAP, and pSMAD3 in B control Cx3cr1CreER(Jung)Tgfb1wt/wt + TAM animals, C Cx3cr1CreER(Jung)Tgfb1fl/fl mice + TAM at 3 weeks after TAM administration, D control Cx3cr1CreER(Jung)Tgfb1wt/wt + TAM animals, and E Cx3cr1CreER(Jung)Tgfb1fl/fl mice + TAM at 12 weeks after TAM administration. F, G Quantification for pSMAD3 fluorescent intensity from F IBA1+ and G IBA1− cells. H, I Microglial morphological quantification of the H terminal end number, and I the summation of process lengths. J GFAP immunoreactivity quantified by binary area fraction. K total IBA1+ or YFP+ cells and L percentage of YFP+ cells among total IBA1+ cells showing no change in % of YFP+ cell even 12 weeks after induction of Tgfb1 KO. A Right (n = 5 for control and n = 6 for KO, p = 0.0004, Welch’s t-test, two-sided); F, G (n = 8 for control, n = 5 for KO 3 week and n = 4 for KO 12 week, **p = 0.006 and *p = 0.029, Kruskal–Wallis test, Dunn’s multiple comparisons test); H, I, K, L (n = 7 for control, n = 6 for KO 3 week and n = 4 for KO 12 week) and J (n = 6 for control, n = 8 for KO 3 week and n = 4 for KO 12 week). Each data point represents the average of a single animal, and the sex of each animal is indicated in the figure legend. Mean ± SE, Scale bar = 100 µm unless otherwise noted. H–L One-way ANOVA, Tukey’s multiple comparisons test. *p < 0.05, **p < 0.01, ****p < 0.0001. A Left, was created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. Source data are provided as a source data file.
To further circumvent the challenge of direct detection of low levels of TGF-β1 protein in the CNS, we analyzed the downstream effector of TGF-β1 signaling, i.e., nuclear-localized phosphorylated SMAD3 (pSMAD3) protein levels. Co-immunohistochemistry analysis shows that in control mice (Cx3cr1CreER+/−Tgfb1wt/wt + TAM), pSMAD3 is detected in both IBA1+ microglia cells and IBA1− cells in the brain (Fig. 1A, right and B, E, bottom row). MG-specific deletion of the Tgfb1 gene leads to an explicit and significant decrease of pSMAD3 immunoreactivity exclusively in (Ionized calcium-binding adaptor molecule 1) IBA1+ microglia, without affecting the pSMAD3 immunostaining in IBA1− cells (bottom rows of Fig. 1B–E and quantification in F, G). This specific loss of TGF-β1 signaling (pSMAD3) in microglia from MG-Tgfb1 iKO mice confirms that microglia TGF-β1 signaling depends on microglia produced TGF-β1 ligand that cannot be compensated for by other cells.
Microglial TGF-β1 is required for maintaining adult glia homeostasis
Next, we evaluated whether the loss of microglia-derived TGF-β1 ligand affects microglia morphology and homeostatic status in adult mice (6–10-week-old females and males). At 3 and 12 weeks post TAM administration, substantial morphological changes in microglia in MG-Tgfb1 iKO mice were observed, as compared to control mice (Fig. 1B–E, IBA1 staining). This change in IBA1+ cell morphology in the iKO mice suggests loss of ramification and potential activation of microglia, which prompted us to perform additional detailed morphological analysis and an examination of the homeostatic microglia signature genes. Following microglial Tgfb1 KO, microglia in Cx3cr1CreER(Jung) Tgfb1 iKO mice showed less ramification, as indicated by decreased process terminal end number (Fig. 1H, control = 33, 3 week iKO = 15, 12 week iKO = 13) and total branch length (Fig. 1I, control = 330 µm, 3 week iKO = 149 µm, 12 week iKO = 140 µm) versus control microglia. Moreover, Tgfb1 iKO microglia demonstrated a reduced expression of homeostatic microglia signature genes such as P2ry12 and Tmem119 (Fig. 1B–E, P2RY12, and TMEM119 staining). These results align with an impaired microglial homeostatic status, and are consistent with another independent study using Sall1CreERTgfbr2fl/fl mice that showed similar morphological changes but did not examine the expression of P2ry12 and Tmem1199. Notably, our observed phenotype is more severe than that of two previous studies in which adult microglial TGF-β signaling was abolished via KO of TGF-βR210,11. This could be due to the short distance between the two loxP sites in such Tgfb1 floxed mice, making it highly efficient for the recombination of the Tgfb1 gene32,34,35. These results support the notion that microglial TGF-β1 signaling relies on TGF-β1 ligand produced by microglia, and a loss of microglia-derived TGF-β1 ligand cannot be compensated by TGF-β1 ligand production in other cell types in the adult CNS. Additionally, YFP reporter tracking in the Cx3cr1CreER(Jung) Tgfb1fl/fl-R26YFP and Cx3cr1CreER(Jung) Tgfb1wt/wt-R26YFP mice show that the total number of IBA1+ or YFP+ cells in the brain both increased in the iKO mice (Fig. 1K), while the % of YFP+ among IBA1+ cells in control versus iKO mice at 3- or 12-weeks post TAM remain the same (Fig. 1L). This suggests that the majority of the recombined microglia remain in the CNS and are likely not replaced by YFP- infiltrating monocytes during the weeks following TAM treatment. An increase in reactive astrocytes (indicated by upregulated Glial fibrillary acidic protein (GFAP) expression, Fig. 1B–E, quantification in Fig. 1J) was also observed both at 3- and 12-weeks after TAM treatment in the Cx3cr1CreER(Jung)Tgfb1fl/fl mice, as compared to the control Cx3Cr1CreER(Jung)Tgfb1wt/wt mice. Two independent Cx3cr1CreER mouse drivers were used in this study to confirm the phenotypes. We observed similar phenotypes in the Cx3cr1CreER(Litt)Tgfb1fl/fl mice at 5 and 8 weeks after TAM treatment, but not in the Cx3Cr1CreER(Litt)Tgfb1wt/wt control mice (Supplemental Fig. 3). A similar phenotype for both microglia and astrocytes is observed globally in other brain regions as well (Supplementary Fig. 4 showing hippocampus as another example, note that unlike cortical astrocytes that are mostly GFAP-, many hippocampal astrocytes are already GFAP+ at homeostasis in WT mice). Abolishing TGF-β1 ligand expression in neonatal mice (TAM treatment at P3-P5 in Cx3cr1CreER(Jung)Tgfb1fl/fl iKO) generates a similarly activated microglial phenotype with a reduced ramification morphology as well as significant loss of homeostatic microglia markers throughout the brain (Supplementary Fig. 5, showing representative images in the somatosensory cortical region). This suggests that microglia-derived TGF-β1 ligand is required to maintain homeostasis in the early postnatal mouse brain as well. We observed similar phenotypes in both male and female iKO mice both in neonatal and adult stages (Fig. 1 and Supplementary Fig. 5, no apparent sex differences are noticed, and the sex of each animal is indicated in the figures). Our results are consistent with a recent independent study investigating the origin of TGF-β1 ligands during embryonic development36.
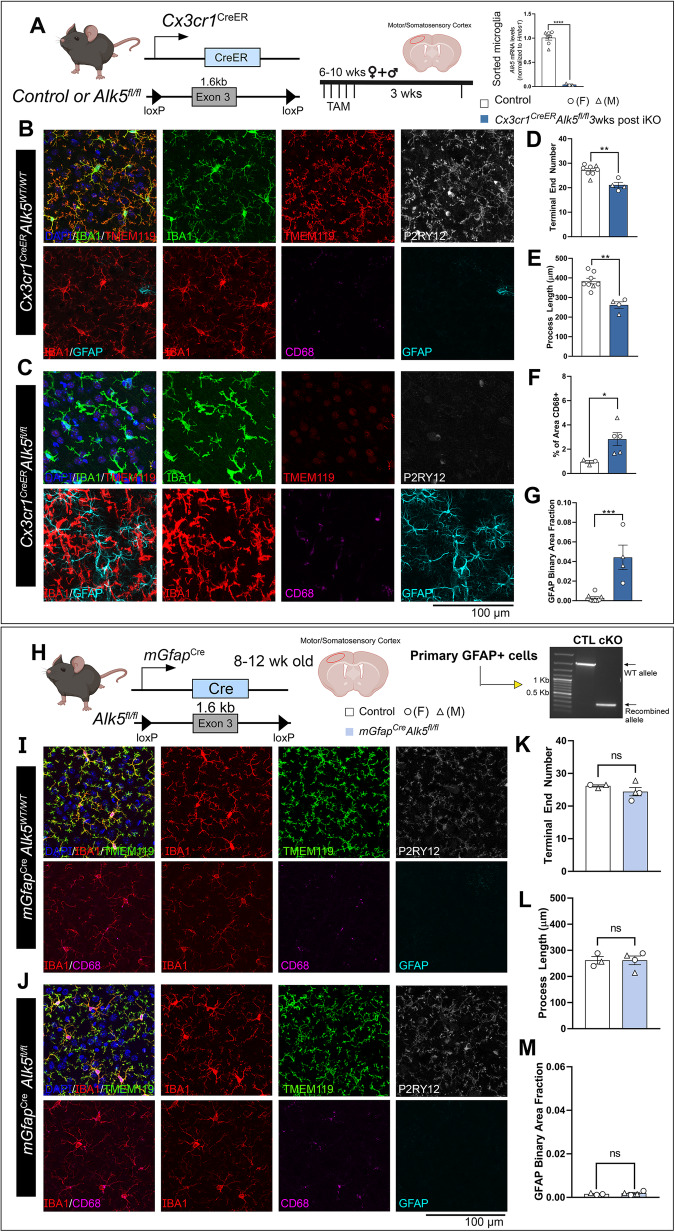
Both microglia and astrocytes are activated in the adult MG-Tgfb1 iKO mice. It is hence unclear whether microglia-derived TGF-β1 ligand loss leads to activation of both cell types directly, due to diminished signaling in each cell type, or whether such loss affected one cell type that indirectly activated the other cell type. To answer this question, we generated microglia-specific or astrocyte-specific TGF-β type 1 receptor (Alk5) iKO mice. Our data show that, loss of TGF-β signaling in microglia (via deletion of Alk5 gene) leads to an activation of microglia, which present with a loss of ramified morphology as well as decreased expression of homeostatic signature genes (P2ry12 and Tmem119) and an increased expression of activated microglia marker (CD68) (Fig. 2A–G). When we deleted TGF-β receptors in microglia, astrocytes were also activated in the MG-Alk5 iKO mice (Fig. 2C, G), which suggests that crosstalk between microglia and astrocytes in the Tgfb ko MG is responsible for the activation of astrocytes. Indeed, when Alk5 is deleted in astrocytes (Fig. 2H, right) via the mGfapCre driver37, neither astrocytes show an increase in GFAP expression phenotype nor microglia show a loss of ramified morphology or decreased homeostatic marker expression in this astrocytic-Alk5 cKO mouse line (Fig. 2H–M).
Fig. 2. ALK5-dependent TGF-β signaling is required for the maintenance of microglial homeostasis but not for astrocytic homeostasis.
A (left) Mouse model for targeting microglial TGF-β type 1 receptor Alk5 and experimental timeline. (Right) indicates the gene deletion efficiency in FACS-isolated microglia (n = 7 control, n = 3 iKO, p < 0.0001). B, C Representative immunohistochemistry images of IBA1, TMEM119, P2RY12, CD68, and GFAP in the cortex of B control animals and C Cx3cr1CreER(Jung)Alk5fl/fl knockouts 3 weeks after TAM administration. Quantification of D microglial process terminal end numbers, E total microglial process length, (D, E n = 8 control, n = 4 iKO, p = 0.001 for (D) and p = 0.0004 for (E)), F % of CD68 immunoreactive positive area (n = 3 control, n = 5 iKO, p = 0.0396), and G GFAP immunoreactive positive area fraction (n = 8 control, n = 4 iKO, p = 0.002, Mann Whitney test, two-sided). H (left) Mouse model for targeting astrocytic Alk5 and experimental timeline. (right) Indicates the gene recombination efficiency in cultured GFAP+ primary cells isolated from control or cKO mice using a PCR primer set that flanks the entire loxP-Alk5-loxP cassette. Cells from three different cKO mice show similar results. I, J Representative immunohistochemistry images of IBA1, TMEM119, P2RY12, CD68, and GFAP in the cortex of I control animals, J mGfapCreTgfb1fl/fl constitutive knockouts at 12-weeks-old. Quantification of K microglial process terminal end numbers, L total microglial process length, and M GFAP immunoreactive positive area fraction (K–M n = 3 control, n = 4 cKO). *p < 0.05, **p < 0.01, ***p < 0.001, ns = not significant. Mean ± SE, All panels are analyzed by two-sided Student’s t-test, except panel (G). (>40 Microglia were quantified for each animal and the average from one mouse was plotted as a single data point in the figure panel and treated as n = 1 for statistical analysis). Scale bar = 100 µm. A, H, Left, were created with BioRender.com and released under a creative commons attribution-noncommercial-noderivs 4.0 International license. Source data are provided as a source data file.
Astrocytic TGF-β1 is not necessary for glial homeostatic maintenance
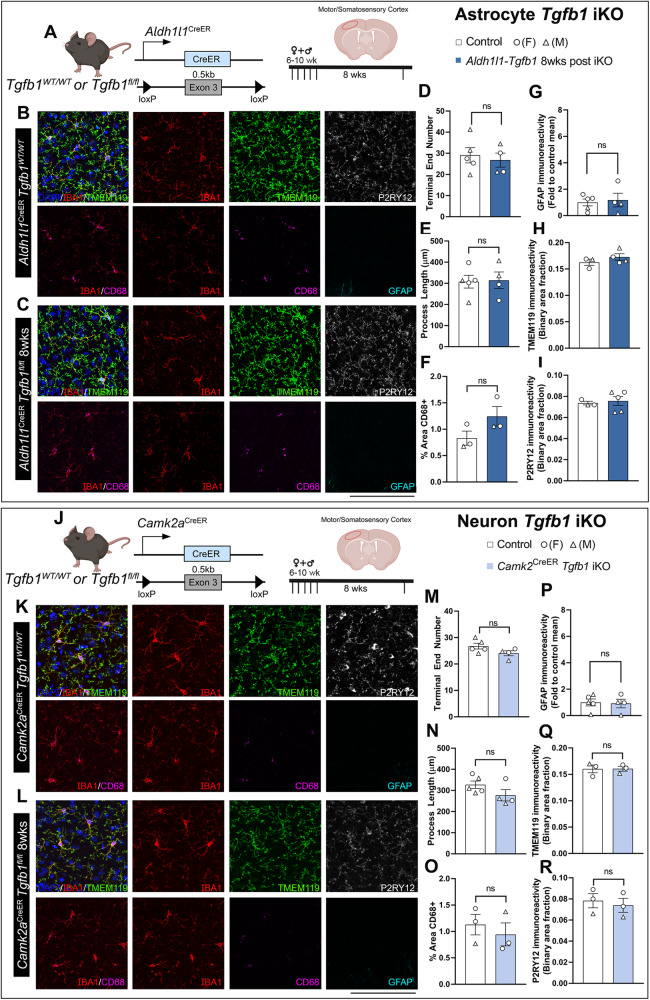
Previous studies suggest that TGF-β ligands produced by astrocytes may play important roles in blood-brain barrier formation, stabilization, and maturation, as well as neuroprotection following injury or disease38–40. We, therefore, next investigated whether astrocyte-specific deletion of the Tgfb1 gene would likewise lead to alterations in microglia morphology and gene expression changes, such as P2ry12, Tmem119, and Cd68. Additionally, we investigated whether loss of the astrocytic Tgfb1 gene could also induce changes in astrocyte reactivity as observed in Cx3cr1CreERTgfb1 or Cx3cr1CreERAlk5 iKO mice. To target adult astrocytes, the well-characterized Aldh1l1CreER mouse strain41 was crossed with Tgfb1fl/fl mice to generate astrocytic Tgfb1 iKO mice (Aldh1l1CreERTgfb1fl/fl). At 8 weeks following TAM administration, microglia morphology and astrocyte state were analyzed. Microglial morphology remained unchanged in Aldh1l1CreERTgfb1fl/fl animals, as compared to control mice (Aldh1l1CreERTgfb1wt/wt mice or Tgfb1fl/fl mice) (Fig. 3A–I for cortex and Supplementary Fig. 6 for hippocampus as an example). Additionally, no changes were observed in homeostatic microglia signature genes (P2ry12 or Tmem119, Fig. 3H, I). Nor did we observe upregulation of CD68 in microglia or an increase in GFAP expression in astrocytes when comparing the Aldh1l1CreERTgfb1fl/fl animals to WT controls at 8 weeks after TAM treatment (Fig. 3 F, G and Supplementary Fig. 6). This result further confirms that astrocytic TGF-β1 production is not required for the maintenance of microglia homeostatic morphology, P2RY12/TMEM119 expression, and suppression of astrocyte GFAP expression.
Fig. 3. Astrocyte-specific or forebrain neuronal specific Tgfb1 gene deletion in the Aldh1l1CreER or Camk2aCreER drivers does not affect the homeostasis of microglia or GFAP expression in astrocytes in adult mouse brain (cortex).
A Astrocyte iKO mouse model and experimental timeline. B, C Representative immunohistochemistry images of cortex from TAM treated (8 weeks post) control B Aldh1l1CreERTgfb1wt/wt and C iKO Aldh1l1CreER Tgfb1fl/fl tissue showing IBA1, TMEM119, P2RY12, CD68, and GFAP immunostaining. Quantification of microglia ramification via D process terminal end numbers, E total process length, and F % of CD68+ immunoreactive area. G Quantification of astrocyte reactivity using GFAP immunoreactive positive area fraction, quantification of H TMEM119, and I P2RY12 immunoreactivity. (D, E, G, n = 5 control, n = 4 iKO) and (F, H, I, n = 3 control and n = 3, 4, and 5 for iKO). J Neuronal iKO mouse model and experimental timeline. K, L Representative images of TAM treated (8 weeks post) control Camk2aCreERTgfb1wt/wt (K) and iKO Camk2CreER Tgfb1fl/fl (L) tissue showing IBA1, TMEM119, P2RY12, CD68, and GFAP immunoreactivity. Quantification of microglia ramification via M process terminal end number, N total process length, and O CD68+ immunoreactive % area. P Quantification of astrocyte reactivity using GFAP+ immunoreactive area fraction, and quantification of Q TMEM119 and R P2RY12 immunoreactivity (M, N, P, n = 5 control and n = 4 iKO) and (O, Q, R, n = 3 for both control and iKO). Mean ± SE (>40 microglia were quantified for each animal and the average from one mouse was plotted as a single data point in the figure panel and treated as n = 1 for statistical analysis). ns = not significant. Two-sided Student’s t-test, scale bar = 100 µm. A, J Created with BioRender.com and released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. Source data are provided as a source data file.
Additionally, we generated constitutive astrocytic Tgfb1 KO mice using the mGfapcre driver line37, which targets a large population of astrocytes constitutively starting from neonatal stages37 to, thereby, ensure that a greater population of astrocytes (95% of cortical astrocytes labeled with Ai14 reporter)42 will have the TGF-β1 ligand KO. Comparing the morphology of mGfapCreTgfb1fl/fl microglia to WT controls, no changes were evident in the ramification of microglia in this independent astrocytic-Tgfb1 cKO mouse line (Supplementary Fig. 7 for cortex and Supplementary Fig. 8 for hippocampus). We next examined the expression of homeostatic microglia signature genes P2ry12 and Tmem119, and did not observe any difference between mGfapcreTgfb1fl/fl mice and control mice (Supplementary Figs. 7 and 8). CD68 expression in microglia and astrocytic GFAP expression also were unchanged. These results, together, support the notion that under normal physiological conditions, adult astrocytes do not produce TGF-β1 ligand necessary for homeostatic maintenance in microglia or quiescence in astrocytes regarding GFAP upregulation. It remains to be determined whether other cellular or functional changes in the astrocytes are altered. Similarly, it is not clear whether, under injury or pathological conditions, astrocytes could upregulate TGF-β1 ligand to modulate glial responses to injury or neurodegeneration.
Forebrain excitatory neurons do not produce TGF-β1 for microglia
Next, we investigated whether neurons are an additional source for TGF-β1 ligand production for adult microglia. To this end, we generated a forebrain excitatory neuron-specific Tgfb1 iKO mouse model. To target forebrain neurons, a Camk2aCreER line43 was crossed with the Tgfb1fl/fl line to induce TGF-β1 ligand KO in excitatory neurons43. Camk2aCreER has been reported to recombine in a widespread manner in the cortex, hippocampus, and striatum43. Eight weeks after TAM administration, microglial morphology remained unchanged in the Camk2aCreERTgfb1fl/fl mice compared to WT controls (Fig. 3J–R for cortex and Supplementary Fig. 9 for hippocampus). Additionally, no alterations in TMEM119 or P2RY12 expression in microglia nor an increase in GFAP expression was observed. This supports the notion that microglia do not rely on neuronal TGF-β1 ligands to maintain homeostasis in the adult brain.
Mosaic MG-specific Tgfb1 KO results in patchy activation of glia
The Cx3cr1CreER line has previously been reported to also target border-associated macrophages (BAMs) that reside in the pia and vasculature32,44. Additionally, the two Cx3cr1CreER mouse lines replace the endogenous Cx3cr1 gene with the cre expression cassette, resulting in heterozygosity for the Cx3cr1 gene in both controls (Cx3cr1CreERTgfb1wt/wt) and in iKO (Cx3cr1CreERTgfb1fl/fl) mice31,45. In an effort to further improve the targeting specificity for parenchymal microglia, the Tmem119CreER and P2ry12CreER mouse lines have recently been generated, utilizing homeostatic microglia signature gene promoters to drive CreER cassette expression without affecting the endogenous gene expression of Tmem119 or P2ry1246,47. Our and other recent studies report that the Tmem119CreER and the P2ry12CreER mouse lines show less TAM-independent “leaky” recombination events, but have the drawback of a lower recombination efficiency, which leads to only a subset of microglia being recombined in these two mouse lines (based on R26-YFP reporter gene expression and qRT-PCR results in sorted microglia, Fig. 4A, right)32,44. Two copies of the CreER alleles in the homozygous in the P2ry12CreER(mut/mut) mouse lines47 increased the total recombination efficiency but not to the extent of the Cx3cr1CreER line (Fig. 4A, right)32,45. To examine whether mosaic gene deletion of the TGF-β1 ligand in a subset of parenchyma microglia would generate any phenotype in microglia, we crossed Tmem119CreER and P2ry12CreER (heterozygous cre or homozygous cre) with the Tgfb1fl/fl line to generate different parenchymal-microglia specific Tgfb1 iKO lines with variety degree of mosaic MG-Tgfb1 deletion.
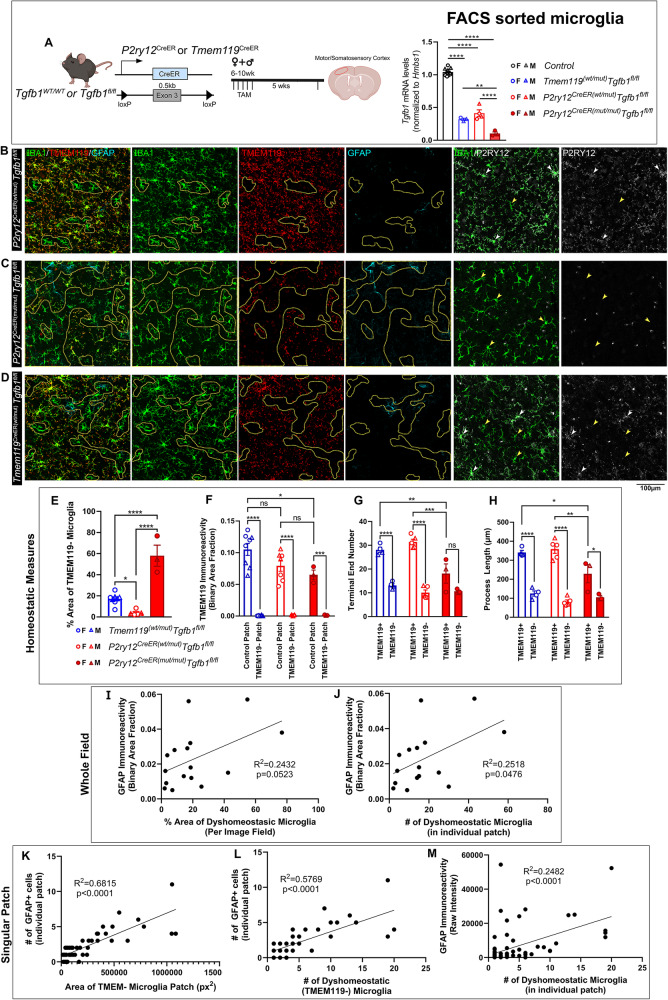
Fig. 4. Mosaic deletion of the Tgfb1 gene in subsets of parenchymal microglia in the P2ry12creERTgfb1fl/fl or the Tmem119CreERTgfb1fl/fl iKO mice leads to distinct patches of dyshomeostatic microglia in the adult mouse brain.
A (left) P2ry12CreER or Tmem119CreER mouse driver to induce Tgfb1 KO in P2RY12+ or TMEM119+ microglia and experimental timeline. A (right) Indicates the gene deletion efficiency in FACS-isolated microglia in all the different mouse lines (n = 10, 3, 4, 3 for each group presented, ****<0.0001 and **p = 0.0038). TAM treated, B P2ry12CreER(wt/mut)Tgfb1fl/fl, C P2ry12CreER(mut/mut)Tgfb1fl/fl, and D Tmem119CreER(wt/mut)Tgfb1fl/fl iKO representative images showing immunohistochemistry for IBA1, TMEM119, P2RY12, and GFAP. Yellow dotted outlines indicate microglia regions with downregulated TMEM119 expression. White arrows depict homeostatic IBA1+ cells that still express P2RY12 expression. Yellow arrowheads show IBA1+ cells that are no longer expressing P2RY12. E Quantification of % area of TMEM− regions across all three lines in the whole image field (n = 8, 5, 3 for each group presented, ****<0.0001 and *p = 0.0312, panel A, right and E are analyzed by one way ANOVA, two-sided, Tukey’s multiple comparisons). F Quantification of TMEM119 expression in P2ry12CreER(wt/mut)Tgfb1fl/fl, P2ry12CreER(mut/mut)Tgfb1fl/fl, and Tmem119CreERTgfb1fl/fl (n = 8, 7, 3 for each group presented, *p = 0.0162, ***p = 0.0008, and ****<0.0001). G, H Quantification of microglia morphology across the three different mouse lines for G terminal number and H process length (n = 4, 5, 3 for each group, *p = 0.0306 and 0.0237, **p = 0.0078 and 0.0016, ***p = 0.0002 and ****p < 0.0001. F–H Analyzed by two-way ANOVA, two-sided, Tukey’s multiple comparison). I, J Correlation of total GFAP immunoreactivity vs % area or a number of dyshomeostatic microglia based on TMEM− the area from representative images across all 3 mouse lines. K–M Correlations of different parameters within an individual dyshomeostatic patch comparing K number of GFAP+ cells vs % area of TMEM− microglia, L number of GFAP+ cells vs number of TMEM− dyshomeostatic microglia, and M GFAP immunoreactivity vs a number of dyshomeostatic TMEM− microglia. Mean ± SE, for correlations, a simple linear regression was used for analysis. Scale bar = 100 µm. A Left, was created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. Source data are provided as a source data file.
To investigate whether partial deletion of microglial Tgfb1 could still produce the disruption of microglial homeostasis we observed in the Cx3Cr1CreERTgfb1 iKO lines, we next evaluated microglial morphology in Tmem119CreER (heterozygous cre) and P2ry12CreER (heterozygous and homozygous cre) Tgfb1 iKO mice. Consistent with the mosaic recombination of the R26-YFP reporter gene and the partial reduction of Tgfb1 mRNA levels in adult microglia in the Tmem119CreER and P2ry12CreER mouse line (Fig. 4A, right), 5 weeks after TAM administration, we observed distinct islands of microglia patches with an activated morphology in both the Tmem119CreERTgfb1 and P2ry12CreERTgfb1 iKO mice (Fig. 4B–D) with a varying degree of % of activation in microglia consistent with the degree of Tgfb1 gene deletion efficiency (Fig. 4A, right, E)32. In addition to the mosaic and patchy morphological changes, we also observed a decrease in morphological ramification and expression of homeostatic microglia signature genes P2ry12 and Tmem119 in these patches of microglia (Fig. 4B, C, quantification Fig. 4F–H). With two copies of the CreER alleles in the P2ry12CreER(mut/mut)Tgfb1fl/fl iKO mice, the phenotype is more severe with >50% of the microglia losing TMEM119 expression (Fig. 4E) and moderate but significant degree of TMEM119 protein downregulation (Fig. 4F) and partial loss of ramification (Fig. 4G, H) even in the adjacent TMEM119+ patches. The morphological changes and loss of P2ry12 and Tmem119 expression were not observed in control Tmem119CreERTgfb1wt/wt or P2ry12CreERTgfb1wt/wt TAM mice (Supplementary Fig. 10). To investigate whether Tgfb1 KO microglia die over time and whether the activated IBA1+ cells are instead infiltrated monocytes, we crossed the R26YFP reporter allele in these mouse lines. Our data show that total IBA1+ cells increased in the P2ry12CreER(mut/mut)Tgfb1fl/fl iKO mice, consistent with a more severe phenotype in this line that affects a higher population of microglia (Supplementary Fig. 10B and 10D). However, the percentage of YFP+ cells among total IBA1+ cells remains the same in WT and iKO mice in all the above three mouse lines (Supplementary Fig. 10A–C, quantification panel E, and panel G). Notably, not all iKO microglia would be expected to be labeled by YFP reporter due to the independence of recombination in distinct floxed alleles, especially given that the floxed Tgfb1 allele is much shorter than the R26-YFP cassette. However, the activated YFP+ microglia patches and the similar % of YFP+ microglia in these microglia-specific CreER lines support the notion that it is, indeed, the parenchymal microglia that become activated and are not replaced by infiltrating YFP-negative peripheral monocytes.
We also observed increased activation of astrocyte patches (visualized by GFAP immunoreactivity) in the cortex of these mosaic microglia-Tgfb1 ligand iKO mouse brains (Fig. 4B–D). On a populational level (analyzed using the entire imaging field), we observed a moderate correlation between the degree of microglial activation and the astrocyte GFAP immunoreactivity (Fig. 4I, R2 = 0.2432, p = 0.0523 and J, R2 = 0.2518, p = 0.0476), consistent with our hypothesis that loss of TGF-β1 ligand from microglia leads to loss of homeostasis in microglia which, in turn, activates astrocytes. To further investigate whether there is a close spatial correlation between local dyshomeostatic microglia and activated astrocytes, we analyzed each individual patch of dyshomeostatic TMEM119- microglia (area of the individual patch and number of dyshomeostatic microglia in each patch) and the local astrocytes activation (GFAP immunoreactivity within the individual patches) or GFAP+ astrocytes in physical contact with the dyshomeostatic microglia territory (measured by the number of GFAP+ astrocytes that are in contact with the individual dyshomeostatic TMEM119− microglia patches). Our data show that there is a positive correlation with all of the parameters analyzed regarding the local astrocyte activation in relationship to local mosaic microglia activation (Fig. 4KR2 = 0.6815, p < 0.0001, Fig. 4LR2 = 0.5769, p < 0.0001, Fig. 4MR2 = 0.2482, p < 0.0001), supporting precise local crosstalk between microglia and astrocytes.
Sparse individual iKO microglia reveal an autocrine signaling mechanism
Notably, in the Tmem119CreERTgfb1fl/fl and P2ry12CreERTgfb1fl/fl heterozygous CreER mice (which had mosaic Tgfb1 gene deletion) the distinct mosaic patches of the few Tgfb1 KO microglia that showed altered morphology and loss of expression of homeostatic microglia signature genes are surrounded by wildtype microglia cells which can produce the TGF-β1 ligand (Fig. 4). This raises the interesting question of whether individual microglia rely on self-produced TGF-β1 ligand that is secreted in an autocrine manner or whether individual microglia could utilize the TGF-β1 ligand from neighboring microglia via a paracrine mechanism.
To further investigate the spatial resolution of TGF-β1 ligand production by individual microglia and investigate whether individual microglia rely on self-produced TGF-β1 ligand using an autocrine mechanism, we designed a mosaic sparse recombination strategy using the Cx3cr1CreER(Jung)Tgfb1fl/fl line with a titrated TAM dilution. To accomplish sparse recombination, we first tested it in the Cx3cr1CreERR26-YFP reporter line. Utilizing this reporter mouse line, we tested the TAM dosage of 1:50 and 1:7–1:10 of the concentration (180 mg/kg) that is utilized in our full dose recombination. Our results (Supplementary Fig. 11) show that for both the 1:50 and 1:7–1:10 TAM dosages, we observed very sparse YFP+ cells in the parenchyma that are also P2RY12+ (suggesting sparse recombination can occur in parenchymal microglia instead of BAMs). This result supports the feasibility of inducing sparse gene deletion in individual microglia surrounded by WT microglia using a titration of TAM dosage. Since the 1:7–1:10 dosage range provided recombination events that were sufficiently sparse, we carried out our subsequent experiments with this range of dosage. A diluted dose of 1:7 (25 mg/kg) of TAM was given over the course of 3 days to Cx3cr1CreER(Jung)Tgfb1fl/flR26-YFP or control mice. This resulted in sparse labeling of microglia in the Cx3cr1CreER(Jung) line, allowing for single-cell analysis of whether microglia depend on self-secreted TGF-β1 ligands to maintain their homeostasis. Remarkably, at 2–3 weeks post TAM administration, we observed isolated sparse individual IBA1+ cells in the parenchyma of the Cx3cr1CreER(Jung)Tgfb1fl/flR26-YFP TAM-treated mice (Fig. 5C, G <10% of total IBA1+ cells) that presented with altered morphology (less ramified, Fig. 5C, J, and K), accompanied by a decrease in TMEM119 expression (Fig. 5C, H blue bar and Supplementary Video 1). These sparse mutated single IBA1+ cells are in the brain parenchyma and do not show typical blood vessel-associated macrophage morphology (Fig. 5C). However, the percentage of YFP+/TMEM119−/IBA1+ cells is very low (Fig. 5F), suggesting that at this low level of TAM dosage, on a single cell level, the recombination of the R26-YFP reporter allele can happen independently from the deletion of targeted floxed genes, a result that is also supported by our recent study using dual reporter alleles in microglia32. In the Cx3cr1CreERTgfb1wt/wtR26-YFP TAM-treated mice, we observed a similar frequency of sparse YFP+ microglia (Fig. 5E) but did not find any individual non-BAM microglia that show this phenotype of activated microglia (marked by loss of TMEM119 expression and altered morphology, Fig. 5B, G), suggesting that the sparse individual “activated” microglia in the Cx3cr1CreERTgfb1fl/fl mice is due to loss of TGF-β1 ligand in the single sparse microglia.
Fig. 5. Sparse-induced knockout of the Tgfb1 gene in individual adult microglia supports an autocrine mechanism of microglial TGF-β ligand production and signaling regulation.
A Mouse model was used to induce Tgfb1 KO in mosaic sparse individual microglia and an experimental timeline depicting a titrated dose of TAM. B–D Representative images showing IBA1, TMEM119, and YFP expression and co-localization in B control tissue at 2 weeks post TAM, C sparse iKO tissue at 2 weeks showing loss of TMEM119 expression in sparse individual microglia, and D absence of TMEM119−/IBA1+ parenchyma microglia in the sparse Tgfb1 iKO brain at 8 weeks post TAM. The yellow dotted outline in C highlights singular microglia showing loss of homeostatic TMEM119 expression. White arrows highlight YFP+ cells showing no loss of homeostatic TMEM119 expression. Note that at this low dosage of TAM, the recombination of individual floxed alleles (R26-YFP reporter or the floxed Tgfb1 gene) occurs independently of each other, therefore YFP+ cells could not track a sparse Tgfb1 KO microglia, consistent with our recent study32. E, F Quantification of percentages of cell populations for E YFP+, F TMEM− YFP+, and G TMEM− cells out of total IBA1+ cells at 2 and 8 weeks post sparse TAM administration (for E–G, n = 6, 4, and 9 for each group presented, *p = 0.0195 and 0.0123 for (F) and ****p < 0.0001, E–G analyzed by one way ANOVA, two-sided, Tukey’s multiple comparison). H, I Quantification of TMEM119 expression at H 2 weeks post (n = 12, 12, and 9 for each group, ****p < 0.0001, one-way ANOVA, two-sided, Tukey’s multiple comparisons) and I 8 weeks post (n = 16 and 15 for each group, not significant) low dose TAM administration from individual TMEM+ cells, TMEM− cells, and YFP+ cells. J–K Detailed morphological analysis of individual microglia in sparse iKO mice at 2 weeks post TAM characterizing J the total process length and K the total terminal end number of individual TMEM119+ or TMEM119− microglia (n = 8 for each group for J and K, ****p < 0.0001, I–K analyzed by Student’s t-test, two-sided). Animals pooled from different cohorts of TAM treatment. Mean ± SE. Scale bar = 100 µm. A Created with BioRender.com and released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. Source data are provided as a source data file.
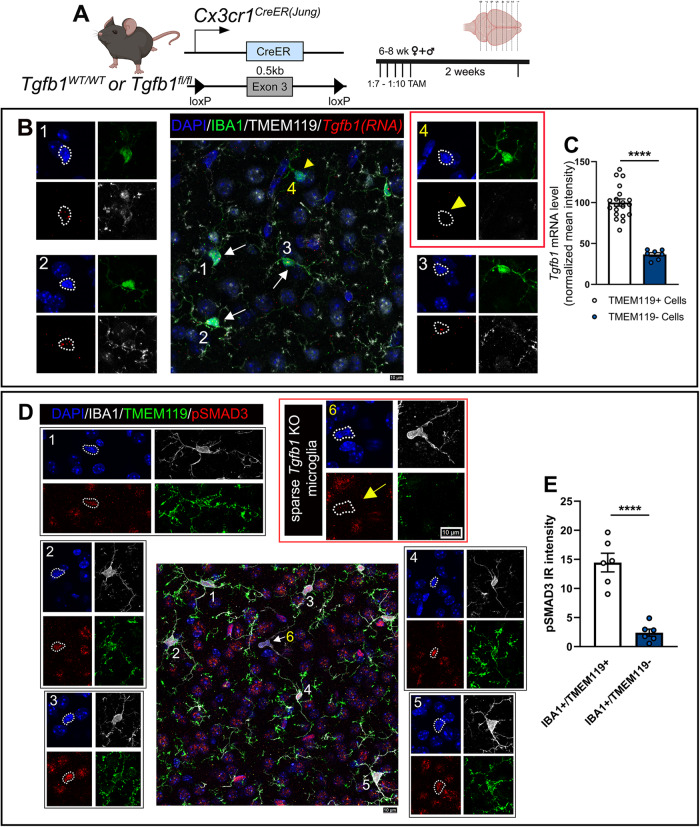
To further confirm this hypothesis, we carried out a combined immunohistochemistry (IHC)/RNAscope using the same Tgfb1 RNAscope probe we used to confirm microglial TGF-β1 expression (Fig. 6A, B). Indeed, we observed significantly decreased Tgfb1 RNAscope probe hybridization in the individual microglia that specifically showed altered morphology accompanied by the loss of TMEM119 expression, while surrounding IBA1+/TMEM119+ microglia showed normal TGF-β1 RNAscope signal (Fig. 6B, C). We further carried out immunostaining for the detection of the downstream signaling effector of TGF-β1 signaling (pSMAD3) in the mosaic sparse MG-Tgfb1 iKO brain and confirmed that loss of pSMAD3 is detected specifically in sparse individual microglia that are TMEM119- and morphologically altered (Fig. 6D, E and Supplementary Fig. 12 for additional image examples). Therefore, our results suggest that microglia show a precise spatial regulation of autocrine TGF-β1 signaling reliant on self-produced TGF-β1 ligands under homeostatic physiological conditions. We next asked whether this loss of homeostasis in individual Tgfb1 KO microglia at 2–3 weeks post TAM is sustained with time or whether, with surrounding wildtype microglia, the individual Tgfb1 KO microglia can regain homeostasis. At 8 weeks post TAM treatment in sparse MG-Tgfb1 iKO mice, the phenotype of sparse TMEM119 negative and morphologically altered microglia is no longer observed (Fig. 5D, F, I). This suggests that these sparse Tgfb1 KO microglia can potentially regain homeostasis (indicated by normal ramified morphology and restoration of TMEM119 expression) at 8 weeks after TAM treatment in a sparse mosaic MG-Tgfb1 gene deletion model. Due to the inability of the R26-YFP reporter allele to simultaneously label the very sparse Tgfb1 KO microglia, we are not able to use the YFP reporter to track the individual Tgfb1 KO microglia longitudinally. However, in the full dosage of TAM-treated Cx3cr1CreER-Tgfb1 iKO mice, we do not see decreased YFP+ cells due to cell death up to 12 weeks after TAM. Additionally, we observed sustained activated phenotype in the majority of the microglia (YFP+) at up to 12 weeks post TAM treatment (Fig. 1E), suggesting that when the majority of the microglia lose TGF-β1 ligand it could be more difficult to recover compared to individual sparsely activated microglia.
Fig. 6. In situ RNA-scope and IHC double labeling confirm loss of Tgfb1 mRNA and downregulation of TGF-β downstream signaling (pSMAD3) in dyshomeostatic individual microglia in the sparse Tgfb1 iKO model.
A Mouse model was used to examine sparse iKO in microglia and the experimental timeline with TAM dosage. B Representative image showing combined immunohistochemistry staining (for IBA1, TMEM119, and DAPI) and Tgfb1 RNA-scope hybridization. (B1–3) Surrounding normal microglia showing TMEM119 expression and Tgfb1 mRNA presence. (B4) A single microglia cell with loss of TMEM119 expression and loss of Tgfb1 mRNA. White arrows were used to mark normal cells in the central panel. Yellow arrowhead is used to mark individual iKO microglia. Note that tissue treatment for RNAscope analysis makes the IHC condition less ideal for morphology evaluation than regular IHC staining, however, IBA1 and TMEM119 expression are still distinguishable for individual WT or iKO microglia. C Quantification of RNAscope signal intensity for Tgfb1 probe in TMEM119+ and TMEM119− cells (n = 20 and 7 for control and TMEM119- group, ****p < 0.0001, Welch’s t-test, two-sided). D Representative image showing co-immunohistochemical staining with DAPI, IBA1, TMEM119, and pSMAD3. (D1–5) Surrounding normal microglia showing TMEM119 expression and pSMAD3 immunostaining. (D6) A single microglia cell with loss of TMEM119 expression and loss of pSMAD3 labeling. The yellow arrow (microglia #6) marks the individual iKO microglia. E Quantification of pSMAD3 immunoreactive intensity in TMEM119+ and TMEM119− cells (n = 6 mice for each group, ****p < 0.0001, Student’s t-test, two-sided). Scale bar = 10 µm. Mean ± SE. For additional representative images see Supplementary Fig. 12. A Created with BioRender.com and released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. Source data are provided as a source data file.
MG-Tgfb1 iKO leads to dyshomeostasis glial transcriptomics
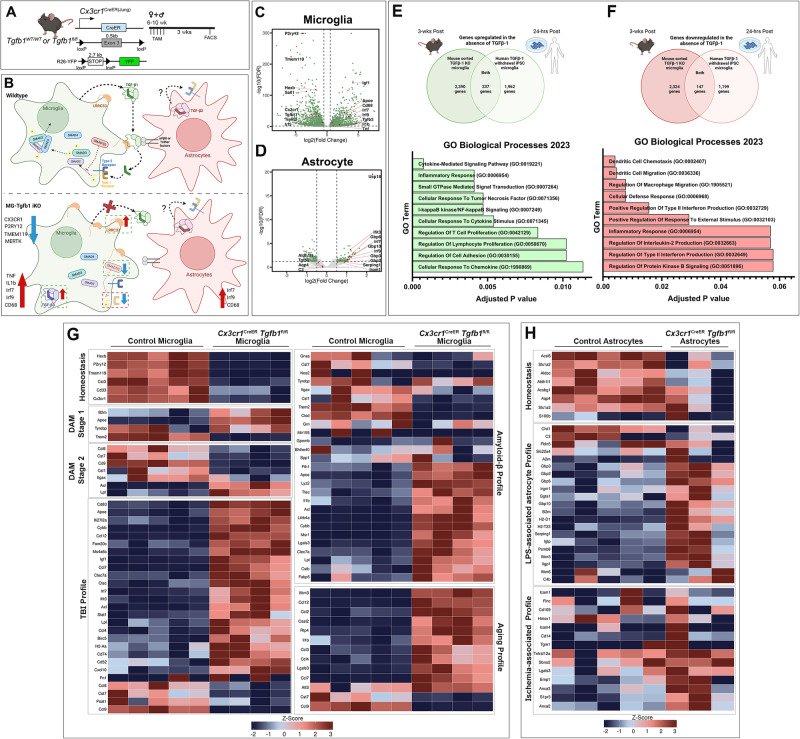
To further characterize the transcriptional changes following the loss of microglial TGF-β1 ligand, microglia, and astrocytes were sorted from the Cx3cr1CreER(Jung)Tgfb1fl/flR26-YFP and Cx3cr1CreER (Jung)Tgfb1wt/wtR26-YFP animals 3 weeks after TAM administration based on YFP expression or ASCA2 immunolabeling (astrocyte staining, Supplementary Figs. 13 and 7)48 and subjected to RNAseq analysis. The purity of samples collected using this sorting method is validated by qRT-PCR for microglia and astrocytic signature genes, respectively (Supplementary Fig. 13). We sorted brain microglia based on recombined YFP reporter expression (which labels about 90% parenchyma microglia in the whole brain) instead of CD11b+/CD45low to avoid the potential caveat that loss of TGF-β signaling in microglia may increase CD45 expression9 and selectively enrich for a subpopulation of microglia in the KO brain. Also, due to the immunostaining data showing the downregulation of homeostatic markers such as P2RY12 or TMEM119 in the KO microglia, we could not use these markers to distinguish microglia vs BAMs. Therefore, we gated CNS myeloid cells based on the YFP signal, which could contain both CNS microglia and BAMs. Principal component analysis (PCA) shows that wildtype microglia samples and Tgfb1 iKO microglia distinctively clustered together (Supplementary Fig. 14). The heatmap shows significantly differentially expressed genes (fold change ≥ |1.5| and adj. p value < 0.05, Supplementary Fig. 14 and Supplementary Data 1 and 2). In contrast to a recent study using Cx3cr1CreERTgfbr2fl/fl receptor inducible KO mice, which reported no changes in many homeostatic microglia signature genes in KO mice11, we observed a large set of differentially expressed genes including downregulation of many microglia homeostatic signature genes (P2ry12, Tmem119, Sall1, etc. Fig. 7C) and upregulation of immune response regulating genes (TNF, Il1b, interferon responsive genes). Using gene set enrichment analysis (GSEA) we observed the upregulation of several pathways related to immune response, immune cell recruitment, and interferon response (Supplemental Fig. 15 and Supplementary Data 3 and 4). We also observed downregulation in platelet aggregation pathway genes (Supplemental Fig. 15). For astrocytes, we observed upregulation of multiple LPS-associated astrocytic genes (Serping1, Ifit3, Gbp3, Fig. 7D, H) and we also observed an increased interferon response (Irf7, Irf9, Fig. 7D). Consistently, GSEA analysis also showed increased interferon activity and decreased metabolic functions (NADH, mitochondria, acetyl CoA, Supplemental Fig. 16 and Supplementary Data 5 and 6) which suggests a transition from metabolic support functions to an activated pro-inflammatory state in astrocytes from the MG-Tgfb1 iKO brain. These data suggest that microglia and astrocytes had disrupted homeostatic functional activity after the loss of microglial TGF-β1.
Fig. 7. Transcriptomic analysis of microglia and astrocyte cells sorted from Cx3cr1CreER(Jung)Tgfb1 iKO mice.
A Mouse model was used to induce Tgfb1 KO and YFP reporter in microglia. B Schematic model showing the summary of transcriptomic changes in microglia or astrocytes pertaining to both inflammatory responses and critical TGF-β signaling component genes, for individual gene list see supplemental information. C, D Volcano plot showing expression log fold changes in microglia or astrocytes comparing iKO vs Control mice. E, F Upregulated and downregulated genes common to this bulk RNA-seq data set and the sequencing results from Abud et al. (human microglia-like cells derived from iPSCs subjected to TGF-β withdrawal for 24 h) and gene ontology (GO) term analysis of overlapping genes from the two data sets. GO analysis was performed using the Enrichr online database. p Values were calculated using Fisher’s exact test, and adjustments for multiple comparisons were made using the Benjamini–Hochberg method. G Microglial differential gene expression observed across various gene sets including, homeostatic microglia genes50–53, stage 1 and 2 disease-associated microglia (DAM) genes51, injury exposed microglial (TBI)53, amyloid beta exposed microglia50,52,53, and aged microglia52,53. H Astrocytic differential gene expression was observed across different gene sets including, homeostatic astrocyte genes, LPS-associated, and ischemia-associated astrocytic genes. Z-scores were calculated and plotted to display differential gene expression54. The astrocyte sample that had an RIN < 8 was excluded from this analysis. A, B, E, and F Created with BioRender.com and released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. Source data are provided as a source data file.
Recently, in a human cellular context, Abud et al. have characterized the transcriptomic profile of human microglial-like cells (iMGLs) derived from human iPSCs that underwent 24 h of TGF-β ligand withdrawal49, which represent human TGF-β signaling targets that are in the acute stage and are potentially direct targets of the signaling pathway. Given that several independent human scRNAseq datasets suggest similar enrichment of TGF-β signaling components in human microglia22–24, we analyzed our dataset and the human TGF-β-deprived iMGLs dataset and identified overlapping genes significantly upregulated or downregulated in the absence of TGF-β signaling in both datasets. We identified 2390 upregulated and 2324 downregulated genes in Cx3cr1CreERTgfbr2fl/fl mice, and there were 1962 upregulated and 1199 downregulated genes reported in the iMGL dataset49. From these genes, we identified 237 upregulated and 147 downregulated genes common to both our dataset and the iMGL dataset (Fig. 7E, F). Gene ontology (GO) analysis was performed for both the upregulated and downregulated gene lists. The upregulated genes were enriched for genes involved in cytokine signaling, inflammatory response, small GTPase and NF-kappa B signaling pathways, response to TNF, and regulation of T cells (Fig. 7E and Supplementary Data 7). The downregulated genes were enriched for genes involved in dendritic cell chemotaxis and migration, macrophage migration, cellular defense response, positive regulation of type II IFN production, and inflammatory response (Fig. 7F and Supplementary Data 7). A number of chemokine receptors are downregulated in the absence of TGF-β signaling (Ccr1, Ccr2, Ccr5, Ccr6, and Cx3cr1) while a number of chemokine ligands are upregulated (Ccl5, Ccl24, Cxcl1, Cxcl9, Cxcl10, and Cxcl16). The presence of chemokine-related genes in both lists explains why there is some overlap in GO terms between the upregulated and downregulated gene lists.
We next wanted to compare the transcriptomic profile of the Cx3cr1CreERTgfb1fl/fl microglia and astrocytes in relation to previously characterized non-homeostatic microglial states. For microglia, we censured multiple previous studies to generate a list of signature genes associated with aging, CNS injury (Traumatic brain injury-TBI), and amyloid-beta pathological conditions50–54. We observed that three weeks after the loss of microglial TGF-β1 ligand, microglia showed downregulation of microglial homeostatic genes (Hexb, P2ry12, Tmem119, Cst3, Cd33, Cx3cr1, Fig. 7G) suggesting dyshomeostasis. We also observed increased expression of aging microglia signature genes (ex. Ifitm3, Ccl12, Il1b Ccl2, Lgals3), and injury-associated TBI signature genes such as Irf7, Igf1, Cxcl10, Ccl12, Axl, Cd63, and Cybb (Fig. 7G). Recently amyloid beta-induced microglial transcriptomic changes have been profiled into DAM 1 and DAM 2 stages, the transition of which depends on TREM2 signaling50–53. Tgfb1 iKO microglia resembles the upregulation of a subset of amyloid beta-associated microglia profile genes, while showing downregulation of other amyloid-beta profile genes. Upon further examination, we noted that the upregulated genes in iKO microglia represent DAM 1 signature genes (B2m, Apoe, Tyrobp, but a decrease in Trem2 levels) while downregulated genes in iKO microglia represent DAM 2 signature genes (Ccl6, Cst7, Cd9, Csf1, Itgax) which correlates well with the downregulation of TREM2 in iKO microglia (Fig. 7G). Additionally, after the loss of microglial TGF-β1, consistent with the observed reactivity in astrocytes by upregulation of GFAP protein, we observed downregulation of some astrocytic homeostasis genes (Aldh1l1, Acsl6, Aldoc), upregulation of in vivo LPS-associated genes (Gbp3, Gbp2, Gbp6, Psmb8, Fig. 7H), and no discernable changes in the ischemia-associated genes in astrocytes from MG-Tgfb1 iKO brains (Fig. 7H).
To further analyze potential ligand-receptor signaling that could mediate the microglia-astrocytes crosstalk and interactions, we next analyzed potential ligands from microglia, potential receptors from astrocytes, and potential target genes in astrocytes-target genes that are differentially expressed in our RNAseq dataset. NicheNet package v1.1.1 in R v4.0.2 was implemented to infer ligand-receptor interactions using 1754 and 100 differentially expressed genes (FC ≥ |1.5| and FDR < 0.5) from microglia and astrocytes, respectively. The curated ligand-receptor interactions from the NicheNet database were used as a reference. We identified seven main ligand targets from microglia (Jam2, Apoe, Trf, Vcam1, Lgals3, Adam17, Adam9) that could be interacting with two astrocyte receptors (Itgb1 and Ldlr) (Supplementary Fig. 17). The downstream gene targets that showed the greatest difference after TGF-β1 ligand loss were Dusp10, Lmna, Cd320, Ldlr, Mfge8, Nek8 and Tm4sf1 in astrocytes. These ligand, receptor, and gene target changes are potential TGF-β dependent pathways that are used in the activation of astrocytes by microglia.
Transcriptomic data from microglia and astrocytes also reveal interesting expression patterns of signaling components of the TGF-β signaling pathways. Consistent with qRT-PCR data from sorted microglia and astrocytes (Supplementary Fig. 13D) and our data showing no observable morphological or immunohistochemical changes in the markers examined following astrocytic-Tgfb1 KO (Fig. 3 and Supplemental Figs. 6–8), RNA-seq data shows Tgfb1, Tgfbr1, Tgfbr2, and Lrrc33 (a protein that is necessary for latent TGF-β1 ligand activation) are all significantly enriched in microglia compared to astrocytes (Supplementary Fig. 18, % mRNA levels in microglia vs astrocytes: Tgfb1 = 500%, Tgfbr1 = 9500%, Tgfbr2 = 2600%, Lrrcc33 = 500%). Instead, astrocytes express Tgfb2, which is absent in microglia, suggesting Tgfb2 might have a potential role in astrocyte function (Supplementary Fig. 18 or Supplementary Data 2). We observed multiple compensatory mechanisms in response to MG-Tgfb1 deletion: (1) an up-regulation of Tgfb3 gene in microglia and no change of Tgfb2 levels in astrocytes, (2) the upregulation of Lrrc33 in microglia, a gene that has been demonstrated to be required in activating the latent TGF-β ligand18, and (3) downregulation of Smad7, which is a negative regulator of the TGF-β signaling pathway. Moreover, we observed a downregulation of Smad3 and Tgfbr1 but not Tgfbr2 mRNA, suggesting a TGF-β signaling-dependent feedforward regulation of Smad3 and Tgfbr1 expression (Fig. 7B and Supplementary Fig. 18). Since our bulk RNAseq analysis used mixed female and male samples, we further validated several key differentially expressed genes in sorted microglia from an additional independent cohort of female and male WT and MG-Tgfb1 iKO mice. qRT-PCR data from sorted microglia confirms that we do not observe a sex difference in all the examined genes (both upregulated and downregulated DEGs) in female or male Tgfb1 iKO mice (Supplementary Fig. 19).
MG-Tgfb1 iKO leads to cognitive, but not general motor deficits
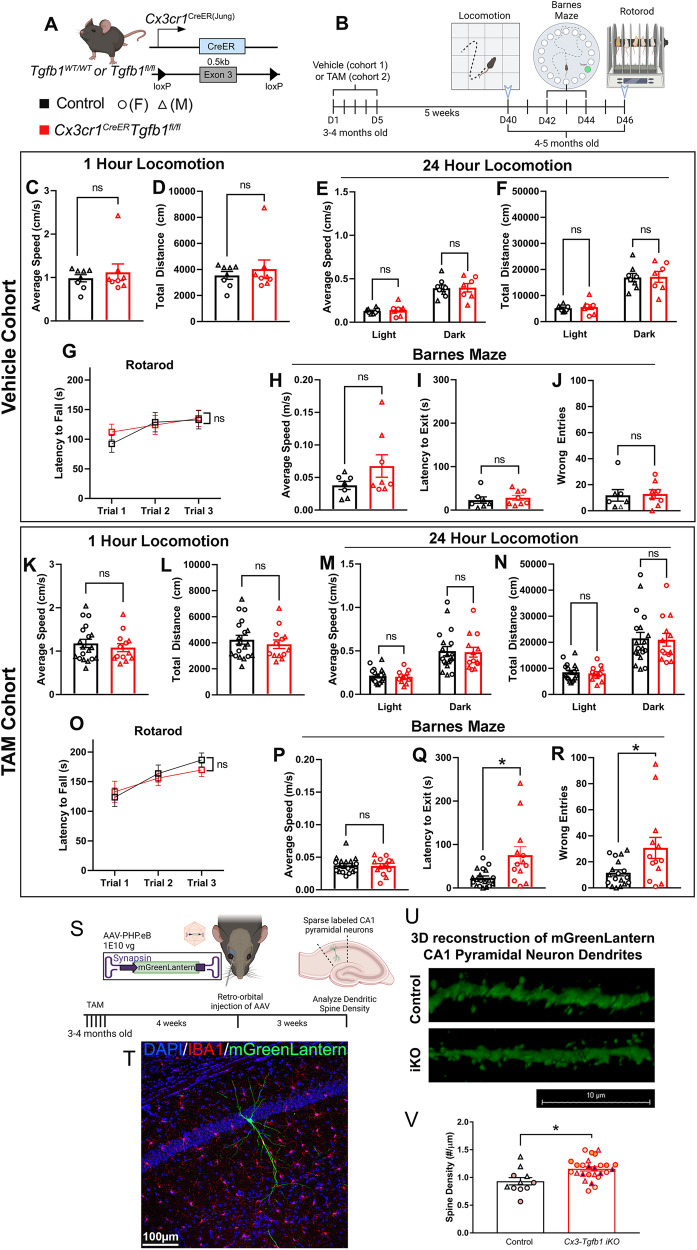
We next investigated whether the DAM-associated and aging-associated microglia profile in the MG-Tgfb1 iKO mice and the presence of reactive astrocytes in these mice could affect neurological function in young adult mice. Full dosage TAM was used in this experiment to achieve maximum changes in microglia and astrocytes in the adult brain (both female and male are used in the behavioral tests). A behavioral battery was used to examine general locomotion, motor coordination/learning, and cognitive function involving learning and memory. We first assessed voluntary movement in control and MG-Tgfb1 iKO mice at 5 weeks after TAM injection using an automated open field locomotion tracking system and monitored mice for 23 h with free access to food and water (Omnitech Electronics INC, Columbus, OH). We did not observe any change in general locomotion in the Cx3cr1CreERTgfb1fl/fl + TAM animals compared to Cx3cr1CreERTgfb1wt/wt + TAM controls during the exploratory phase (1 h after naïve exposure to the chamber), or during the light or the dark cycle (Fig. 8K–N). Next, we carried out an acceleration rotarod test to evaluate motor coordination and motor learning. We specifically used a three-trial acceleration paradigm that starts at 1 rpm and increases to 35 rpm over the course of 5 min to evaluate their starting motor coordination, and how their performance improves over each trial. The Cx3cr1CreERTgfb1fl/fl + TAM animals did not show a difference in performance compared to the control Cx3cr1CreERTgfb1wt/wt + TAM group in the rotarod test (Fig. 8O), suggesting that motor coordination and motor learning are not affected in the MG-Tgfb1 iKO mice at this time point after gene deletion. However, when we evaluated the cognitive function (spatial learning/memory) in control and MG-Tgfb1 iKO mice using a 2-day Barnes Maze learning paradigm, Cx3cr1CreERTgfb1fl/fl TAM group showed an increase in latency to reach the escape hole and higher error trial numbers to locate the hole compared to the control mice (Fig. 8P–R), suggesting impaired spatial learning in the Cx3cr1CreERTgfb1fl/fl iKO mice. Importantly, Cx3cr1CreERTgfb1fl/fl mice that received vehicle treatment do not show any difference compared to control mice in any of the above behavioral tests (Fig. 8C–J), demonstrating that the behavioral deficits in cognitive function measured by Barnes Maze in the Cx3cr1CreERTgfb1fl/fl + TAM mice are specifically caused by TAM-induced deletion of the microglial-Tgfb1 gene in these mice. These data support that microglia-derived TGF-β1 ligand is required in maintaining microglia homeostasis, astrocyte quiescence, and normal cognitive function in the adult brain. We did not observe significant differences between female and male mice in either genotype.
Fig. 8. Behavioral assessment shows normal general motor function and motor learning but defective spatial learning and memory in young adult Cx3cr1CreERTgfb1fl/fl iKO mice and increased dendritic spine density in hippocampal CA1 neurons.
A Mouse model was used to induce Tgfb1 KO in microglia. B Experimental timeline, showing the order of behavioral measurements. C–J Behavioral measurements in vehicle-treated Cx3cr1CreERTgfb1wt/wt or Cx3cr1CreERTgfb1fl/fl mice showing open field test (OFT) of the first hour in locomotion chamber C average speed and D total distance traveled. E, F Average speed and total distance during the light and dark cycles in a 23-h period. G Accelerated rotarod learning test. H–J Barnes maze test showing H average speed during testing, I latency to locating the target hole, and J number of error trails before locating the target hole (n = 8 for each group, Student’s t-test, two-sided). K–R Behavioral measurements from TAM-treated control and iKO mice showing open field test (OFT) of the first hour in locomotion chamber K average speed and L total distance traveled. M, N Average speed and total distance during the light and dark cycles in a 23-h period. O Accelerated rotarod learning test. P–R Barnes maze test showing P average speed during testing, Q latency to locating the target hole, and R number of error trails before locating the target hole. (control n = 19, iKO n = 13) (ns = not significant, *p = 0.0210 for panel Q, and *p = 0.0398 for panel R, Unpaired t-test with Welch’s correction, two-sided). S Experimental design and timeline for systemic AAV for neuronal labeling in MG-Tgfb1 iKO mice. T Representative image of a sparse CA1 hippocampal neuron labeled with the AAV-PHP.eB syanpsin-mGreenLantern virus. U 3D reconstruction of mGreenLantern CA1 pyramidal basal neuron dendrites from control and iKO mice. V Quantification of spine density (n = 11 control and n = 26 individual dendritic segments pooled from n = 3 control mice and n = 4 iKO mice (dendritic segments from the same animal is color coated, circle indicates female, and triangle indicates male. Statistical analysis was carried out using the average from individual mice as a single n, *p = 0.0316, Student’s t-test, two-sided). Mean ± SE. Scale bars: 10 µm or 100 µm as indicated. A, B, and S Created with BioRender.com and released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. Source data are provided as a source data file.
Loss of TGF-β signaling during embryonic development is known to induce neuronal apoptosis55, demyelination, and altered oligodendrocyte lineage as well as loss of somatostatin (SST)+ or Parvalbumin (PV)+ interneurons in neonatal mice10. We next investigated whether a similar phenotype is also observed in adult MG-Tgfb1 iKO mice (TAM administration was given to young adult mice, aged 6–10-weeks-old). Consistent with the lack of major motor deficits in the adult MG-Tgfb1 iKO mice, we did not observe any significant differences in the number of Oligodendrocyte transcription factor 2 (OLIGO2+), neuron-glial antigen 2 (NG2+) or CC1+ cells and myelin basic proteins (MBP) levels in the cortical area (Supplementary Fig. 20). Similarly, there are no differences in the number or distribution of SST+ or PV+ interneurons (Supplementary Fig. 20) in the cortical layers of MG-Tgfb1 iKO mice at 12 weeks post-TAM (a time point when microglia and astrocytes activation persist as shown in Fig. 1). Furthermore, the total NeuN+ neuronal population in all cortical layers at the cortical somatosensory region does not differ between control of MG-Tgfb1 iKO mice at 12 weeks post TAM (Supplementary Fig. 20). These data suggest that, unlike the constitutive or global loss of Tgfb1 ligand or receptors during the developmental stage8,10, adult microglial Tgfb1 gene ablation has less effect on the adult oligodendrocyte lineages, the overall neuronal survival or the cortical interneuron population. This data also suggests that the observed cognitive deficits might be due to more subtle structural or functional changes in neurons in the iKO mice. Given the downregulation of multiple genes that are implicated in neuronal-microglia communications (P2RY12, CX3CR1)56,57 and receptors that are required for phagocytosis function in microglia (P2RY12, MER proto-oncogene tyrosine kinase-MerTK and triggering receptor expressed on myeloid cells 2-TREM258,59), we next hypothesized that synaptic pruning might be decreased in the MG-Tgfb1 ligand knockout mice. To facilitate the measurement of dendritic spine density, neurons were sparsely labeled with the bright, monomeric fluorescent protein mGreenLantern (a bright monomeric fluorescent protein)60 using a retro-orbitally delivered systemic Adeno-associated viruses (AAV) vector61 (AAV-PHP.eB-hSyn-mGreenLantern-WPRE-pA, 1E10 vg/mouse) in WT or iKO mice (Fig. 8S). Given the known role of the hippocampus in learning and memory and the observed cognitive deficits in the iKO mice, we analyzed the dendritic spine density of basal dendrites of the CA1 pyramidal neurons that are mGreenLatern positive (Fig. 8T). Our results show an increased spine density in the CA1 neurons of the MG-Tgfb1 iKO mice (Fig. 8U, V), consistent with the observation of the downregulation of TREM2 and MerTK receptors in Tgfb1 KO microglia.
MG-Tgfb1 iKO repopulate dyshomeostasis microglia after MG ablation
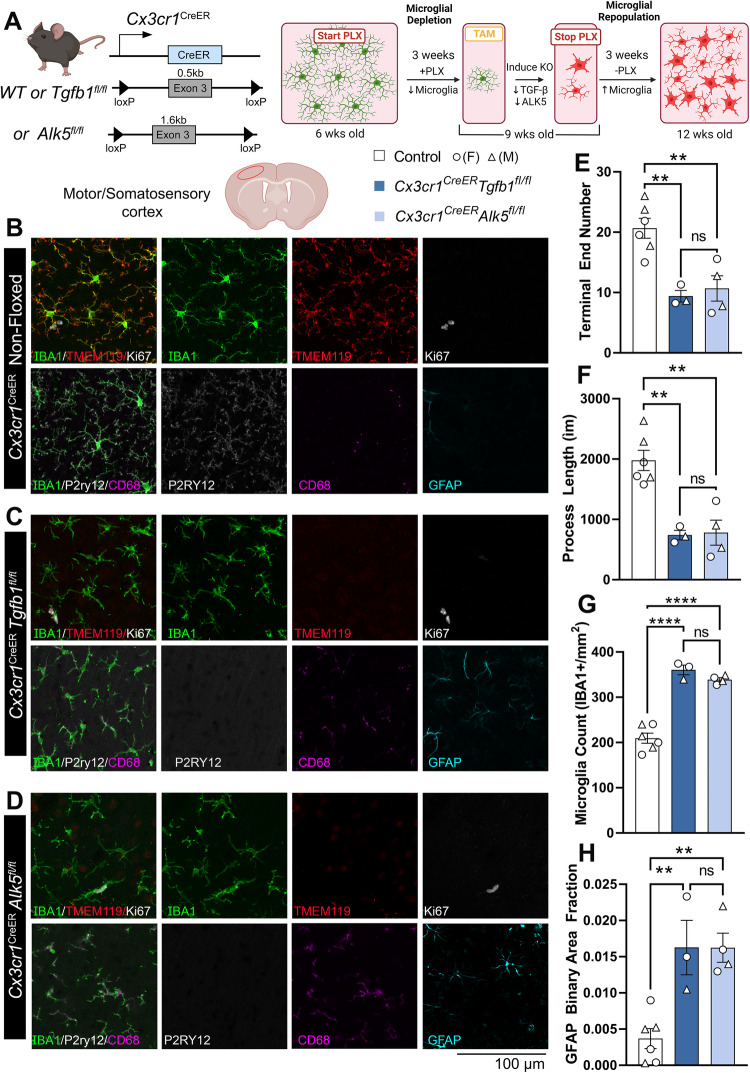
Lastly, ablating and repopulation of microglia in dysregulated CNS have recently been suggested as a strategy to “reset” the inflammatory environment of the CNS28,62. However, since multiple studies have suggested an overall decreased TGF-β signaling in aging or disease brain63–65, whether the compromised TGF-β signaling in microglia would affect the repopulated microglia under these contexts is not known. Next, we tested whether MG-Tgfb1 or MG-Alk5 deletion affects microglia repopulation in the adult brain following pharmacological microglia ablation (via the CSF1R inhibitor, PLX5622). Our data supports that while abolishment of TGF-β signaling does not prevent repopulation of microglia in the CNS after PLX5622 treatment, a homeostatic state is not reached in the repopulated microglia when TGF-β signaling is silenced in microglia via either microglia-specific ligand knockout (MG-Tgfb1 iKO) or receptor knockout (MG-Alk5 iKO). In the absence of microglia-derived TGF-β1 ligand or loss of ALK5 receptors, microglia overpopulate the brain (with an excess number compared to WT mice, Fig. 9A–D, H) and the repopulated microglia show activated morphology (Fig. 9E, F) and lack the homeostatic microglia signature gene expression (Fig. 9B–D). Importantly, the repopulation of dyshomeostatic Tgfb1 or Alk5 knockout microglia also leads to the activation of astrocytes (indicated by GFAP upregulation in astrocytes, Fig. 9G). This indicates that repopulation/resetting of the microglia population in a disease context where TGF-β signaling is diminished might lead to repopulation of non-homeostatic microglia, and may not be an ideal strategy.
Fig. 9. TGF-β signaling via microglia-derived TGF-β1 ligand or ALK5-dependent signaling is required for the repopulation of homeostatic microglia after PLX5622 ablation.
A Mouse model for Tgfb1 and Alk5 iKO in microglia and experimental design. B, C Representative images of B control, C Cx3cr1CreERTgfb1fl/fl, and D Cx3cr1CreERAlk5fl/fl mice showing immunostaining of IBA1, TMEM119, Ki67, P2RY12, CD68, and GFAP. Quantification of microglial morphology by E terminal end number (**p = 0.0044 and p = 0.0053) and F and process length (**p = 0.0022 and p = 0.0014). G Total microglia count (****p < 0.0001). H Quantification of astrocyte reactivity using GFAP immunoreactivity (**p = 0.0057 and p = 0.0032). E–H, n = 6, 3, 4 for each group, one-way ANOVA, two-sided, Tukey’s multiple comparisons). Mean ± SE, each data point represents the average of a single animal. Scale bar = 100 µm. A Created with BioRender.com and released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. Source data are provided as a source data file.
Discussion
Furthering our understanding of how TGF-β1 signaling is precisely regulated in the brain can provide important insight into microglia function during steady state and disease conditions. Our study addresses several gaps in our knowledge about CNS TGF-β ligand production and regulation and sheds light on how alteration of a single cytokine gene (Tgfb1) in microglia could causally contribute to cognitive deficits in young adult mice in the absence of brain injury or other disease-causing stressors.
Currently, the prevailing understanding of the source of TGF-β1 ligands in the CNS has been speculated to be coming from multiple cell types and that TGF-β ligands can be widely shared among different cell types13–16,39,40,47,55,66–68. Several reviews have proposed the sharing of TGF-β1 amongst all the cell types, despite not yet having a well-rounded experimental understanding of TGF-β1 ligand production and distribution12–17. Our data supports that microglia-produced TGF-β1 ligand is required for maintaining microglial homeostasis and subsequent astrocyte quiescence in the adult CNS as early as neonatal stages. We used multiple myeloid- or microglia-CreER drivers to rigorously investigate this phenotype. Two independent Cx3cr1CreERTgfb1 iKO mouse lines both lead to a global loss of microglia homeostasis revealed by morphological changes and downregulation of homeostatic gene expression such as Tmem119 and P2ry12 without affecting serum or spleen levels of TGF-β ligand (demonstrated by both ELISA and FACS). We also show that astrocytic (via either inducible Aldh1l1CreER line or constitutive postnatal deletion via the mGfapCre driver) or neuronal (via inducible Camk2aCreER line) deletion of the Tgfb1 gene does not affect microglia morphology or expression of signature homeostatic microglia genes such as TMEM119 or P2RY12. Recent studies by us and others show that the Cx3cr1CreER mouse lines31,45 recombine a portion of splenocyte macrophages even after the waiting period of >3 weeks, therefore, there is a possibility that the changes in microglia phenotype observed in the Cx3cr1CreERTgfb1 iKO mice could be due to parenchyma microglia population depletion and peripheral macrophage replacement in the brain. Alternatively, the activation of Tgfb1 KO BAMs (which are also targeted by the Cx3cr1CreER lines) could subsequently activate the rest of the parenchyma microglia. However, results from the P2ry12CreER(wt/mut)Tgfb1fl/fl mice and Tmem119CreER(wt/mut)Tgfb1fl/flR26-YFP mice also showed morphological and homeostatic microglial marker expression changes in the mosaic patches of YFP+ cells (indicating they were P2ry12+ or Tmem119+ parenchymal microglia at the time of TAM administration). Additionally, if peripheral monocytes (YFP−) are infiltrated into the brain, we expect to observe a decreased percentage of YFP + /IBA1+ cells in the brain. Instead, our data shows that total YFP+ and total IBA1+ cells in the brain both increased in the Cx3cr1CreERTgfb1 and homozygous P2ry12CreER(mut/mut)Tgfb1 iKO mice while the percentage of YFP+/IBA1+ in total IBA1+ cells remained the same compared to control mice. This suggests that loss of TGF-β1 ligand in microglia does not lead to cell death of microglia or the replacement of microglia by peripheral monocytes but instead suggests increased proliferation of microglia in iKO mice, consistent with a previous study with Tgfbr2 knockout9 and supported by Ki67 gene upregulation in our iKO microglia (RNAseq). In summary, these data support that the parenchymal resident microglia are altering their phenotype in response to the deletion of the Tgfb1 gene, rather than being replaced or indirectly altered by peripheral monocytes or macrophages.
Additionally, by using low TAM dosage to achieve sparsely mosaic Tgfb1 KO in very few individual microglia, our data supports that not only do microglia produce their own ligand to regulate their quiescent state during adulthood, but they likely do so in an autocrine manner since Tgfb1 gene deletion in sparsely distributed individual microglia leads to downregulation in TGF-β signaling (pSMAD3) and phenotypic changes in individual cells despite their surrounding WT microglia population. This data suggests that microglia regulate TGF-β signaling and related downstream pathways in a spatially precise manner which is consistent with the very low concentration of TGF-β1 ligand in brain tissue compared to spleen and serum levels. This mechanism is of particular importance in relation to disease or injury response since the glial activation cascade could be reliant on local fluctuating TGF-β1 levels. This model contrasts with the current prevailing model of shared TGF-β ligand production and signaling among many different cell types in the CNS17. We believe that our model helps to clarify one of the reasons for current controversy regarding the levels and role of TGF-β signaling in disease, aging, and Alzheimer’s disease (AD), which could be at least partially a result of the focus solely on serum or cerebrospinal fluid (CSF) TGF-β levels, which according to our results might not be biologically relevant to local microglia status. While there is some discrepancy in whether human Tgfb1 gene expression also enriches in microglia in the human CNS (with several published datasets supporting enriched Tgfb1 expression in microglia, one study showing differences in control vs AD subjects in microglial-Tgfb1 expression pattern22–25), our mouse MG-Tgfb1 adult iKO transcriptomic analysis show many overlapping upregulated or downregulated DEGs (differentially expressed genes) with a human cultured microglial-like cell transcriptomic dataset after TGF-β withdrawal, supporting the potential relevance of our study in human cells. Importantly, TGF-β is synthesized as a latent form (L-TGF-β) whose activation requires the release of the mature c-terminal domain from non-covalently bound latency-associated peptide (LAP)18,69,70. One recent study also suggested a possible highly localized mechanism responsible for the release of the mature TGF-β1 ligand from the non-covalently bound prodomain (LAP) based on the coordinated molecular action of microglia-expressed LRRC33 (functioning as a LAP binding protein) and αVβ8 integrin18, possibly expressed on other cell types such as astrocytes. Our model agrees with this recent study13,69 that TGF-β ligand release is controlled at the single cell level by multiple precise mechanisms. The Qin et al. paper demonstrated that the activation of active TGF-β ligand from its latent prodomain-bound form is controlled by LRRC33 protein expression on the microglia membrane13. Our study now shows that not only is the unlocking of TGF-beta active ligand controlled by LRRC33 with single-cell precision (shown by Qin et al.), but also even at the production level, TGF-beta ligand produced by single microglia is required for the homeostasis of the same cell. This spatially precise model and mechanism demonstrates another distinct layer of the mechanism of this type of autocrine signaling and is an important addition to the mechanistic repertoire that cells use to refine extracellular signaling.
While this microglial autocrine mechanism appears to be the primary mechanism for TGF-β1 signaling in microglia during homeostasis, we observed that sparse individual KO microglia surrounded by WT microglia could potentially recover to homeostatic state (measured by morphology and expression of TMEM119 and P2RY12) at 8 weeks after the loss of native TGF-β1 ligand production. Note that on the populational level (with full TAM dosage and when most microglia are Tgfb1 KO) even at 12 weeks, microglia in the Cx3cr1CreERTgfb1 iKO mice still show morphological changes and decrease of TMEM119 and P2RY12 expression. This suggests that the milieu environment of surrounding WT microglia is able to “reset” the sparse individual Tgfb1 KO microglia in the sparsely mosaic KO mice but not so efficiently when the majority of the microglia are KO cells that are activated. The mechanism for this recovery and to what extent the remaining “normal” microglia can help “reset” the mosaic-activated microglia is not clear and warrants further investigation in future studies. Our RNAseq data from the full dose TAM treated MG-Tgfb1 iKO mice show that not Tgfb2 but Tgfb3 levels are upregulated in the MG-Tgfb1 KO microglia (Supplementary Data 4), raising the interesting question of whether upregulated microglial TGF-β3 levels are able to compensate for the loss of TGF-β1 ligand in microglia. Previous studies have shown that Tgfb1− and Tgfb3-specific single KO mice have different phenotypes and that the swapping of the code sequence between Tgfb1 and Tgfb3 genes only leads to partial rescue of the phenotypes, suggesting non-overlapping functions of the two ligands, which is also supported by unique biophysical properties between the two ligands in recent studies71. Our results also suggest that upregulation of the TGF-β3 detected at 3 weeks post TAM in Tgfb1-KO microglia was not able to rescue the phenotype in microglia at up to 12 weeks post TAM in full dosage recombined mice. A recent study presented Cryogenic electron microscopy (Cryo-EM) structures which show that LRRC33 only presents L-TGF-β1 but not the -β2 or -β3 isoforms due to the differences of key residues on the growth factor domains18. This molecular selectivity offered by microglia-expressed LRRC33 could possibly explain why upregulated TGF-β3 expression in the TGF-β1 KO microglia could not compensate for the loss of TGF-β1 ligand and rescue the phenotype in microglia in full TAM dosage MG-Tgfb1 iKO mice. RNAseq data from sorted microglia and astrocytes from WT or MG-Tgfb1 iKO brains also reveal interesting cell type-specific transcriptomic regulation of the TGF-β signaling components in different cells during homeostasis or in response to the disturbance of TGF-β signaling. Specifically, we found that TGF-β1 is mainly enriched in microglia while TGF-β2 is enriched in astrocytes. Correspondingly, microglia express LRRC33 which preferentially presents TGF-β1 instead of TGF-β2 or -β3 for ligand activation. These patterns might explain why the deletion of microglial TGF-β1 but not astrocytic TGF-β1 leads to the observed phenotypes in both microglia and astrocytes. Loss of microglial TGF-β1 ligand also leads to downregulation of Smad3 and Tgfbr1 but not Tgfbr2 in microglia. This suggests a feedforward regulation of TGF-β signaling on the expression of SMAD3 and TGF-βR1 facilitating further TGF-β signaling, which agrees with a recent study utilizing Smad4 knockout mice9. Conversely, Tgfb1 KO microglia also upregulate Tgfb3 and Nrros (LRRC33) while downregulating the inhibitory Smad7, reflecting an attempt to compensate for the loss of TGF-β signaling in KO microglia. These gene expression changes reflect a highly dynamic regulation of this signaling pathway and support the precise spatially regulated autocrine mechanism in microglia. Consistently, none of these changes in the TGF-β signaling components are observed in astrocytes, indicating that the transcriptomic changes observed in astrocytes in the MG-Tgfb1 iKO mice are likely not due to direct loss of TGF-β signaling in astrocytes. While Alk5 deletion in astrocytes does not lead to GFAP upregulation in astrocytes or microglia morphological changes, it is not clear whether other transcriptomic or functional changes occur in the Alk5 deleted astrocytes. Additionally, whether genetic deletion of Tgfb2 in astrocytes would lead to activation of astrocytes and transcriptomic changes also warrants further investigation in future studies.
In the absence of the endogenous microglia-derived TGF-β1 ligand, microglia showed reduced ramification, decreased parenchymal homeostatic microglia signature gene expression, increased pro-inflammatory cytokine expression, and upregulation of interferon response genes. The expression profile of the Tgfb1 iKO microglia aligned with disease-associated microglia (DAMs)51,53, which have been described using both injury models (TBI)53 and disease states (amyloid beta pathology)50–53. Additionally, these transcriptomic profile changes also corresponded with observed gene expression changes in aging microglia52,53, suggesting that TGF-β1 signaling in microglia can provide vital insights into injury, neurodegenerative disease, and aging. The microglial Tgfb1 iKO phenotype observed in our study is consistent with that described after constitutive Tgfbr2 deletion in myeloid cells by the Cx3cr1Cre promoter during development10 and a Sall1CreER driver in adult9, which are much more severe than the phenotype observed when Tgfbr2 was deleted at P3010 or in a separate study at 2 months of age using the adult Cx3cr1CreERTgfbr2 inducible mice26. Factors such as the dosage and route of TAM treatment, efficiency of recombination of the floxed genes, and whether cre-mediated recombination leads to the complete absence of the target protein, or a truncated protein can all contribute to the severity of the phenotypes. One specific potential caveat regarding the adult Cx3cr1CreERTgfbr2fl/fl iKO study11 is that KO microglia were sorted from Cx3cr1CreER(+/wt) mice which are heterozygous for the Cx3cr1 gene while control microglia are sorted from Cx3cr1CreER(wt/wt) mice which has both alleles of the Cx3cr1 gene. Heterozygosity of Cx3cr1 has previously been reported to cause changes in gene expression or function in microglia72–76 and therefore might introduce additional confounds to the data interpretation. Additionally, both previous studies10,11 used the same Tgfbr2 floxed mouse line77 which has shown deletion of exons 2/3 does not alter the reading frame of the remaining exons leading to a truncated protein with normal serine/threonine kinase activity. The Sall1CreERTgfbr2fl/fl study9 used a different floxed Tgfbr2 mouse line78 which could potentially explain the differences in the three Tgfbr2 KO studies. In our study, we utilized a Tgfb1 floxed mouse model (065809-JAX with 0.5 kb of floxed region) which leads to a frameshift and results in the complete absence of the active TGF-β1 ligand. This might explain the much more robust phenotypes in our adult iKO mice.
We also observed transcriptomic changes in astrocytes in the MG-Tgfb1 ligand mice, featuring GFAP protein upregulation and increased interferon response genes. While GFAP is a pan-reactive marker, RNAseq data also shows upregulation of multiple previously reported disease-associated potentially detrimental astrocyte markers. Deletion of the Tgfb1 gene in astrocytes via the constitutive mGfapCre driver line or the inducible Aldh1l1CreER line did not induce morphological changes in microglia, nor upregulation GFAP expression in astrocytes, suggesting the reactivity of astrocytes in the MG-Tgfb1 iKO mice is likely secondary to microglial profile change instead of direct loss of TGF-β signaling in astrocytes. This is consistent with the absence of the Tgfb1 gene in astrocytes which instead express Tgfb2. Loss of TGF-β signaling in microglia leads to the upregulation of multiple pro-inflammatory cytokines which could in turn mediate the crosstalk between KO microglia and neighboring astrocytes. Our results in the Alk5 receptor knockout specifically in microglia or astrocytes support a direct role of TGF-β signaling in microglia and subsequent changes in astrocytes due to crosstalk between the knockout microglia and surrounding astrocytes. Indeed, a detailed analysis of the spatial relationship of the activated microglia patches to the GFAP+ astrocyte distribution supports a close spatial correlation of the size and number of dyshomeostatic microglia to local patches of GFAP+ reactive astrocytes that could be mediated through diffusion of local cytokines. One such potential crosstalk can be through TNF signaling since it has been shown that TNF can promote disease-associated astrocyte activation and the TGF-β1 ligand knockout microglia shows an increase in TNFα in KO microglia79. Additional candidates for this crosstalk are also predicted using the ligand-receptor-target gene analysis and suggest that Integrin Subunit Beta 1 (Itgb1) and low-density lipoprotein receptor (Ldlr) are potential candidates for mediating microglial-dependent astrocyte reactivity that warrant further investigation in future studies.
Recently, enforced repopulation of adult microglia in diseased brains has been explored as a potential therapeutic strategy to “reset” homeostasis during neuroinflammation of the CNS28,62. TGF-β signaling-related genes are regulated during this process80. However, whether TGF-β signaling itself plays a role in adult microglia repopulation is not known. Our data show that after PLX5622 ablation of microglia, when TGF-β signaling is silenced in microglia with either the TGF-β1 ligand or the type 1 receptor Alk5 knockout, the microglia population repopulates with excess numbers of microglia and dyshomeostatic microglia with activated morphology and decreased expression of microglia homeostatic signature markers and upregulation of CD68, accompanied by activation of astrocytes with GFAP expression. In pathological and disease contexts where TGF-β signaling has been reported to be diminished63–65, the ablation and “resetting” strategy on the microglia population might not be an optimal strategy due to the repopulation of the dyshomeostatic microglia population.
Lastly, behavioral analysis in the Cx3cr1CreER(Jung)Tgfb1 young adult mice shows that at 5 weeks following TAM treatment, there are significant deficits in the spatial learning and memory of MG-Tgfb1 iKO mice without affecting the general locomotion function or motor learning at the time of assessment. This moderate cognitive phenotype without severe motor deficits in our young adult MG-Tgfb1 iKO mice is an interesting contrast to the much more severe motor deficits and early life lethality in the constitutive gene knockout of Lrrc33−/− mice81 and Itgb8−/− mice82,83, and embryonic microglial deletion of Tgfbr2 via the Cx3cr1Cre driver10. This difference suggests that loss of TGF-β signaling in microglia is more detrimental and causes more severe functional consequences during development vs in the adult CNS, a result that is also consistent with the difference in other cellular phenotypes (loss of myelination and cortical inhibitory interneuron) between embryonic vs adult KO mice. We did not observe any sex differences in our iKO mice using immunohistochemical, transcriptomic, or behavioral analysis. Whether the abolishment of TGF-β signaling in microglia via ligand or receptor gene deletions could cause a more severe cellular or behavioral phenotype in aged or injury/diseased context in different sexes is also an interesting topic that can be further investigated in future studies. During the preparation of this paper, a recent study also reported a deficit in learning using a Morris Water Maze test in Crybb1CreSmad4 cKO mice84. The Crybb1Cre driver targets embryonic macrophages (with some off-target recombination in OPCs and neurons) and therefore the observed learning deficits could result from a deficit of neurons and projections during development84. However, our data show that adult microglia rely on self-derived TGF-β1 ligands to maintain homeostasis and TAM-induced deletion of the TGF-β1 ligand in adult microglia leads to learning deficits in adult mice, suggesting an ongoing reliance on microglia-derived TGF-β1 ligand and TGF-β1 signaling in microglia to maintain normal cognitive function in adulthood. Given the downregulation of multiple receptors such as P2RY12, TREM2, and MERTK which are involved in neuronal activity sensing and phagocytosis-mediated synaptic pruning, we examined the dendritic spine density in the CA1 pyramidal neurons. We chose to analyze CA1 pyramidal neurons because of their well-known roles in spatial memory and learning85–88 which is relevant to the learning deficits we observed in the Barnes Maze. The AAV-PHP.eB Serotype and a low dose titer via systemic delivery were chosen to sparsely label neurons with minimal disturbance on microglia morphology or function which could be a confound using intracerebral injection of virus. Using this method, we observed increased dendritic spines in the CA1 pyramidal neurons of the MG-Tgfb1 iKO mice, consistent with the downregulation of TREM2 and MERTK expression in KO microglia. Our results are further supported by a previous study showing increased dendritic spine density and functional deficits in TREM2 ko mice89. This data establishes a link regarding TGF-β signaling in microglia with TREM2 expression and neuronal function. However, it is possible that peripheral tissue macrophages could also rely on autocrine TGF-β1 ligand for their homeostasis, for example, gut macrophages90, which could also potentially have some impact on the behavioral phenotypes. In summary, our result may have important implications for the role of microglial-TGF-β1 signaling in cognitive deficits observed during aging, neurodegenerative diseases, or after CNS injury. While constant basal TGF-β1 signaling is necessary for microglial homeostasis, TGF-β1 levels change with aging, injury, and disease12,13,15,40. Our results support that microglia dysregulation caused by the loss of TGF-β1 ligand results in transcriptomic features resembling DAMs and aged microglia. This could play a causal role in driving the cognitive deficits observed in disease conditions and targeting TGF-β signaling might be a potential therapeutic strategy to mitigate these deficits.
Methods
Experimental model and subject details
Animals
The University of Cincinnati (UC) Animal Care and Use Program (ACUP) encompasses Laboratory Animal Medical Services (LAMS, animal facilities) and the Institutional Animal Care and Use Committee (IACUC) office. All animal protocols were approved by the IACUC (animal protocol number: 21-03-02-01). Mice were housed in the animal facility of the University of Cincinnati on a 14-h light/10-h dark diurnal cycle. Food and water were provided ad libitum. The Cre-loxP recombination system was utilized to achieve cell-type specific constitutive or inducible knockout of the Tgfb1 or Alk5 gene. Cx3cr1CreER(Jung) (JAX: 02094045), Cx3cr1CreER(Littman) (JAX: 02116031), P2ry12CreER (JAX: 03472747), Tmem119CreER (JAX: 03182046), MgfapCre (JAX: 02409891), Aldh1l1CreER (JAX: 02965541), and Camk2CreER (JAX: 01236243) transgenic mouse lines in which the expression of Cre recombinase are under the control of the Cx3cr1 (myeloid cell), P2ry12 (Microglia), Tmem119 (Microglia and peri-vesicular fibroblast), mouse Gfap (astrocytes and adult neural stem cells), Aldh1l1 (astrocytes), and Camk2 (forebrain excitatory neurons) promoters, respectively were purchased from the Jackson Laboratory. These animals were crossed with floxed Tgfb1 mouse line (JAX: 065809) or with Alk5fl/fl mice (JAX:028701). Among all the mouse cre driver lines used in this study, the MgfapCre mouse line is the only line that is non-inducible and has previously been shown to induce loxP-specific gene recombination at perinatal stages in mice which targets a large percentage of astrocytes (>90%) and a small percentage of cortical neurons (<1.3%) and some oligodendrocytes (<6% of total reporter positive cells)42. All the other Cre driver lines have the CreERT292. For all experiments, mice were euthanized by administration of avertin followed by transcardial perfusion with either PB then 4% PFA (for IHC) or 1xHBSS (for flow cytometry, FACS, RNAseq, and qRT-PCR), followed by removal of essential organs (brain). Avertin 40× stock was prepared by mixing 10 g of 2,2,2-tribromoethanol (Sigma T48402) in 10 ml 2-methyl-2-butanol (Sigma 240486), then diluting 1/40 in sterile saline.
TAM administration
TAM injections were administered based on mouse body weight (BW) as described previously93. One hundred eighty mg/kg of BW was administered via oral gavage for five consecutive days for full dose induction. Young adult mice were treated at 6–10 weeks of age with TAM (180 mg/kg). TAM solution was formulated from 100 μl EtOH and 900 μl of sunflower seed oil diluted with 30 mg of TAM powder. For sparse recombination, mice received 3 days of TAM at th dosage of 1:10 (18 mg/kg BW, we noticed that 1:7 dilution also gave sparse labeled individual cells and similar results to 1:10. Each lab should test the titration of dilution in their own lab). Sparse Tgfb1 KOs were generated using a 1:7–1:10 dilution of TAM in the vehicle (EtOH and sunflower seed oil) to achieve the desired dosage. Note that mice that receive the vehicle, diluted dosage (1:7–1:10), or full dosage of TAM should be housed separately to prevent TAM cross-contamination between different groups. For neonatal TAM treatment, pups are treated on P3–P5 daily by directly injecting TAM solution into the stomach (50 µg/pup/day) and harvested on day P18.
Microglia ablation via PLX5622 administration
Mice were treated with either the PLX5622 diet (AIN-76A rodent diet With 1200 PPM PLX5622, formulated by research diets with PLX5622 provided by Plexxikon) or the control diet (AIN-76A rodent diet, research diets, NJ). Animals had ad libitum access to the diet and water for the entirety of the study. For measuring Tgfb1 mRNA levels after microglia ablation, C57bl6/J wildtype mice were treated with control or PLX5622 diet for 7 days. Brain tissue was harvested and processed for qRT-PCR as described previously94. For the ablation and repopulation experiment as illustrated in the experimental timeline, mice were given ad libitum access to the PLX5622 diet for 21 days and treated with TAM at the end of the PLX 5622 treatment. Mice were then returned to a standard chow diet for the remainder of the experiment to allow for repopulation.
Sparse viral labeling of neurons and dendritic spine density analysis
To label neurons, we used Gibson assembly to clone an AAV expression vector utilizing a human synapsin promoter (hSyn) for neuron-specific expression of the fluorescent protein mGreenLantern (pAAV-hSyn-mGreenLantern-WPRE-bGH-polyA). This construct was produced using the AAV-PHP.eB capsid by the Viral Vector Facility at Cincinnati Children’s Hospital Medical Center. To avoid injury-related microglial reactivity, 1E10 vg of the viral construct was administered retro-orbitally in 90 µl of sterile saline. Four weeks after TAM administration, mice were anesthetized with isoflurane during the unilateral retro-orbital injection. Three weeks were allotted for viral transduction. Sparse labeled mGreenLantern positive CA1 pyramidal neurons are imaged at basal dendrites. For quantification of dendritic spines, images were captured on a Leica Stellaris eight confocal microscope with a 63× objective and Z-series (Z-step of 0.2 um) with 2.5× digital zoom and 2048 × 2048 pixels. To assess spine density, basal dendrites from multiple CA1 pyramidal neurons are analyzed for each animal, and the spine number/length of each dendritic segment is averaged for each animal and used as n = 1 for statistical analysis.
qRT-PCR
RNA was isolated from the cortex using the RNAqueous-Micro Total RNA isolation kit (AM1931, ThermoFisher Scientific). cDNA was then generated using superscript III reverse transcriptase (18080044, ThermoFisher Scientific) or iScript cDNA synthesis kit (1708890, BioRad). The cDNA was then used for qRT-PCR using probes for Hmbs1 (hydroxymethylbilane synthase), Hprt1 (hypoxanthine phosphoribosyltransferase 1), Iba1, Tgfb1, Alk5, Tgfbr2, Sall1, Glast, Glt1, and Atp1b2. cDNA levels were quantified using a Roche Light Cycler II 480. Quantification of qRT-PCR values was normalized using the housekeeping gene Hmbs1 CT value, which did not change between groups after manipulation to account for potential variability in cDNA preparations.
ELISA
For ELISA analysis, tissue was collected after perfusion with phosphate buffer solution and flash frozen in cold isopropyl alcohol. Mouse serum is collected by clotted blood without any anticoagulant for 30 min followed by centrifugation at 1500 g for 10 min at 4 °C. Serum is collected from the supernatant and frozen at −80°C. The tissue was sectioned with a cryostat to punch 2 mm punches of tissue. Tissue was placed in RIPA buffer then homogenized using sonication at 30% amplitude, for 3-s pulses with 2-s pauses. The BCA method was used to determine the total protein concentration in the samples and Quantikine ELISA Human TGF-β1 kit (R&D Systems, Minneapolis, MN) was used to analyze TGF-β1 ligand levels following the instruction from the manufacturer.
Tissue collection for flow cytometry or FACS
Mice were transcardially perfused with cold 1× HBSS for 2–3 min. The brains and spleens were extracted and mechanically dissociated with a scalpel before using the papain dissociation kit (9001-73-4, Worthington Biochemical Corporation). For spleens, following dissociation, red blood cells were lysed using ammonium chloride. For the brains, once dissociated, cells were suspended in a 37% percoll solution and spun at 800 g for 20 min to remove excess myelin and debris. The cells were collected, washed, and resuspended in FACS buffer containing PBS with 1% (v/v) fetal bovine serum and 0.1% (w/v) NaN3 (Sigma), and counted. The number of cell subpopulations in the CNS was determined by multiplying the percentage of lineage marker-positive cells by the total number of mononuclear cells isolated from the brain. Transcriptional and translational inhibitors actinomycin, anisomycin, and typtolide were used to prevent activation of microglia during the preparation of tissues as was previously described by Marsh et al. 95. Inhibitors were added to the dissection solution, and the papain enzyme cocktail from the Worthington kit.
Flow cytometry analysis of TGF-β1 expression
To carry out flow cytometry analysis, the Fc receptors were initially blocked using anti-mouse CD16/32 (0.25 μg; ThermoFisher) for 15 min at 4 °C. Cells were then washed with FACS buffer and stained for the surface marker for 30 min at 4 °C using the specified antibodies. These antibodies included: CD45 (clone 30-F11), CD11b (clone M1/70), and TGF-β1 (clone TW7-16B4) (all from Biolegend). Cells were then washed with PBS and viability staining was performed using the LIVE/DEAD fixable dead cell stain kit (Invitrogen). Following viability staining, cells were washed with PBS and resuspended in FACS buffer for flow cytometry analysis. Cells were acquired on a BD Canto II and analyzed using FlowJo X software (vX10). As controls, fluorescence minus one (FMOs) was used to place the gates for analysis. For flow cytometry analysis, cells were first gated according to FSC-SSC, then restricted to single cells and live cells. Myeloid cells were identified as CD45+ CD11b+.
FACS of microglia and astrocytes for qRT-PCR and RNA-seq
Gating was determined using the yellow fluorescent protein expressed by Cx3cr1CreER-R26-YFP for CNS myeloid cell collection, and ASCA-2 APC conjugated antibody (130-117-535, Miltenyi Biotec) for astrocytes. Any double-positive cells were excluded from the gating to improve the purity of the samples.
Bulk RNA-sequencing
Non-directional RNA-seq was performed by the genomics, epigenomics, and sequencing core at the University of Cincinnati. To summarize, the quality of total RNA was QC analyzed by Bioanalyzer (Agilent, Santa Clara, CA). About 100 pg total RNA was used as input for cDNA amplification using NEBNext single cell/low input RNA Library Prep Kit (NEB) under PCR cycle number 15. After Bioanalyzer QC, 20 ng cDNA was used for library construction under PCR cycle number 6. After library QC and quantification via Qubit quantification (ThermoFisher, Waltham, MA), individually indexed libraries were proportionally pooled and sequenced using NextSeq 2000 Sequencer (Illumina, San Diego, CA) under the sequencing setting of PE 2 × 61 bp to generate about 60 M reads. Once the sequencing was completed, fastq files were generated via Illumina BaseSpace Sequence Hub.
RNA-sequencing analysis
RNA-seq reads with adapter sequences or bad-quality segments were trimmed using Trim Galore! v0.4.296 and cutadapt v1.9.197.The trimmed reads were aligned to the reference mouse genome version mm10 with STAR v2.6.1e98. Duplicated aligned reads were removed using Sambamba v0.6.899.Gene-level expression was assessed by counting features for each gene, as defined in the NCBI’s RefSeq database100. Read counting was done using featureCounts v1.6.2 from the Rsubread package101. Raw counts were normalized as transcripts per million (TPM). Differential gene expressions between groups of samples were assessed with R package DESeq2 v1.26.0102. Gene list and log2 fold changes are used for GSEA103,104 analysis using the GO pathway dataset. Plots were generated using the ggplot2105 package and base graphics in R. The PCA analysis comparing astrocytes from control samples and astrocytes from MG-Tgfb1 iKO mice shows one astrocyte sample from iKO mice diverge from other iKO samples and this sample had a lower RNA integrity number (below 8 while all other samples have RIN of >8), suggesting partial RNA degradation. We included this sample in the PCA plot, general DEG heatmap, and volcano plot, however, this sample was excluded for characterizing the astrocytic activation profile. NicheNet package v1.1.1 in R v4.0.2 was implemented to infer ligand-receptor interactions using 1754 and 100 differentially expressed genes (FC ≥ |1.5| and FDR < 0.5) from microglia and astrocytes, respectively. The curated ligand-receptor interactions from the NicheNet database were used as a reference.
RNAseq dataset comparisons method
To generate a list of upregulated and downregulated genes, genes from the DEG analysis were first filtered by adjusted p value (<0.1 for the mouse-sorted microglia dataset and <0.05 for the iMGL dataset). From these genes, upregulated and downregulated genes were sorted based on log2-fold change. To identify genes common to both the mouse-sorted microglia dataset and the human iMGL dataset, human gene names were first converted to orthologous mouse gene names using Ensembl’s Biomart. After the conversion of the human gene names to mouse orthologs, filtering for genes either upregulated in both datasets or downregulated in both datasets revealed 237 upregulated and 147 downregulated genes. GO analysis was conducted on the upregulated and downregulated gene lists using Enrichr.
Immunohistochemistry
Mice were perfused with 4% PFA and drop-fixed overnight before being transferred to 20% sucrose, followed by 30% sucrose once the tissue had sunk. Then tissue was sectioned on a cryostat in 30 μm thickness and subjected to IHC as previously described106. Antibodies for GFP (1:1000, Invitrogen or 1:500, Aves), IBA1 (1:500, Abcam or 1:1000, Wako), P2RY12 (1:500, Biolegend), TMEM119 (1:2000, GeneTex), GFAP (1:1000, Sigma), NEUN (1:1000, Biolegend), CD68 (1:4000, BioRad), MBP (1:250, BioRad), CC1 (1:250, Millipore Sigma), Olig2 (1:500, RND Systems), NG2 (1:250, Millipore), Parvalbumin (1:1000, Swant), Somatostatin (1:250, Santa Cruz), Ki67 (1:300, Invitrogen) and pSMAD3 (1:150, Abcam) were used. Tissue was blocked for 1 h at room temperature (RT) in 4%BSA/0.3% Triton-X100, then incubated overnight at 4 °C in primary antibody. Tissue was then incubated for 2–3 h in appropriate secondary antibodies conjugated with Alexa fluorescence 488, 555, 647, or 790.
RNA-scope
To fluorescently label RNA, RNA-scope was employed using the ACD RNA-scope Multiplex v.2 kit. Samples for RNA-seq were perfused with cold PB and the brain was dissected out and drop fixed in 4% PFA for 7 h before transferring to 20% and 30% sucrose. Brains were sectioned on a Leica Cryostat at the thickness of 16um and directly mounted onto superfrost plus glass slides. RNAscope hybridization steps were carried out following the instructions from the manufacturer. A Tgfb1 probe was used (443571, ACD Biosciences) to label ligand RNA and an ALK5 probe was used (406201, ACD Biosciences) to label the TGF-β1 type I receptor. Immunohistochemistry was carried out after the RNAscope hybridization as described above to identify different cell types or expressions of different homeostatic microglia markers. Confocal images were obtained for IHC and RNAscope/IHC samples at the University of Cincinnati Imaging Core utilizing a Confocal microscope. 3D reconstruction of the Z-stacks was performed in LAS X software. Imaging was also analyzed using the Neurolucida image analysis program (MBF Bioscience) for detailed morphological analysis.
Image analysis
NIS elements were used for microglial morphology quantification. For cortical regions, we used bregma coordinates: AP ~ +0.5 mm. For hippocampal regions, we used bregma coordinates: AP ~−2mm. A trace function was utilized for measuring the microglia process length (μm) and a counting function was utilized to quantify the number of terminal ends for each microglial process. Reactive astrocytes were quantified by measuring the area of GFAP immunoreactive astrocytes in the field via thresholding against the background. CD68 quantification was accomplished through the image processing package of ImageJ, Fiji. Z-stacks from confocal imaging were merged into a max projection on the Leica Application Suite X (LASX), and then exported to Fiji. The threshold function was utilized on the fluorescent staining of CD68 against the background, and averages were taken of the mean area. On average 4–6 images from 3–4 brain sections at similar brain regions were analyzed per mouse and the value from multiple images was averaged for each mouse which is used as a single data point in statistical analysis. Neurolucida (MBF Bioscience) was utilized for 3D microglial reconstructions. Microglia cell bodies were constructed by tracing the outer perimeter with the cell body trace function each time a primary microglia process branched out. Then each process was traced with the process tracer function. For pSMAD3 quantification, ImageJ was used to identify IBA1+ and IBA1- nuclei, the nuclei were then traced, and the median fluorescent intensity was measured. Multiple microglia (7–10 microglia) were analyzed from randomly sampled multiple images in individual mice and the average from multiple cells was used as a single data point in statistical analysis.
Behavioral assays
Locomotion
To measure general locomotion function, an automated 42 × 42 × 31 cm Plexiglas open box for 23 h with ad libitum food and water (on the lid) and fresh bedding on the chamber of the floor (Omnitech Electronics INC, Columbus, OH) with laser sensors were used to monitor animal behavior. Animals were housed in a dual chamber, allowing for scent exchange but not physical interaction. Lighting was set to mimic that of the animals' normal housing rooms, on a 14/10 h light/dark cycle. Data was automatically scored using Fusion (Accuscan, Columbus, OH, USA).
Rotarod
Using the Roto-rod Series 8 apparatus (IITC Life Science Inc., Woodland Hills, CA) mice were given an acceleration paradigm. The rod rotation started at 1 RPM and reached 30 RPM by the end of the 5-min session. The test concluded when the animal fell from the rod but was not returned to the home cage until all animals finished the test. Animals performed three trials and were allotted at least 15 min in their home cage before starting their next trial. Seventy percent ethanol was used to clean the apparatus in between animals.
Barnes maze
Barnes maze apparatus (Stoelting Company, Wood Dale, IL) was designed as a gray circular platform 91 cm in diameter, and 90 cm in height with 20, 5 cm diameter holes equally distributed around the edge of the platform. Of the 20 holes, 19 had 2 cm deep gray trays beneath, with one of the holes having a 5 cm deep escape box. A short challenging Barnes maze learning paradigm was used to assess spatial learning and memory. Animals were given two training sessions, 4 h apart, with each training session ending when the animal located and fully entered the escape box. LED lights, a heat lamp, and a fan were used to motivate escape behavior. The following day (24 h later) a test session occurred, with the test ending when the animal fully entered the escape box. In-between animals the maze was sanitized using 70% ethanol. Data was automatically scored using AnyMaze (Stoelting Company, Wood Dale, IL).
Statistical analysis
All studies were analyzed using SigmaPlot. Results are expressed by mean ± SEM of the indicated number of experiments. Statistical analysis was performed using the Student’s t-test, and one- or two-way analysis of variance (ANOVA), as appropriate, with Tukey post hoc tests. A p value equal to or less than 0.05 was considered significant.). Graphs were made in GraphPad Prism and some portions of figures were generated with Biorender.com.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgments
Y.L. is supported by NIH grants (R01NS125074, R01AG083164, and R21NS127177). A.B. is supported by NIH 1F31NS125930. I.I. is supported by NIH R35GM146890. K.W. is supported by NIH F31 NS129204. J.E.R. is supported by a Cincinnati Children’s Research Foundation Trustee Award and a Simons Foundation Autism Research Initiative (SFARI) Bridge to Independence Award (663007). We thank Chet Closson and the University of Cincinnati live imaging core (supported by NIHS10OD030402) for technical support. We thank Brendan Chestnut for assistance with vector cloning, as well as Thouwa Samake and the Vector Production Facility at Cincinnati Children’s Hospital Medical Center for AAV production. We also thank Dr. Xiang Zhang and the Genomics, Epigenomics, and Sequencing Core at the University of Cincinnati for RNAseq analysis support. All figures were created using Biorender software.
Author contributions
Y.L. conceptualized the study. Y.L., A.B., and E.W. designed the experiments. L.M. maintained all the mouse colonies and genotyped all mice in this study. A.B. and K.W. performed TAM injection and the immunohistochemistry staining with help from M.W., M.K.S., E.W., A.T., and J.D.P. A.B. performed all the behavioral analyses. A.B., M.W., and M.K.S. carried out microglia morphological analysis and astrocyte GFAP quantification. E.W. carried out the ELISA, microglia ablation experiment, and qRT-PCR analysis of the Tgfb1 gene. A.B. and E.W. performed cell sorting of microglia and astrocytes with assistance from the Flow Cytometry core at CCHMC and prepared all RNA samples for RNAseq analysis. R.M.S.G. and J.E.R. designed and packaged the AAV-PHP.eB synapsin mGreenLantern virus at the CCHMC viral vector core. A.P. and K.M.R. assisted in the bioinformatics analysis of the RNAseq data in this study. A.A. and II carried out the flow cytometry analysis of Tgfb1 expression in brain microglia and spleen myeloid cells. NG provided suggestions and consultants on experimental designs. A.B., E.W., N.H.G., and Y.L. drafted and revised the paper. All authors read, edited, and approved the final version of the manuscript.
Peer review
Peer review information
Nature Communications thanks Marianela Traetta, Marie-Eve Tremblay, Björn Spittau, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The RNA-seq data generated in this study have been deposited in the GEO database under accession code GEO: GSE236032 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL30172). Other RNA-seq data presented, but not generated, in this study include: (1) GEO: GSE89189 (generation of human microglia-like cells to study neurological disease Fig. 7), (2) https://singlecell.broadinstitute.org/single_cell/study/SCP1879/synucleinopathy-associated-astrocytes?genes=Tgfb1&tab=distribution#study-visualize (mouse single-cell RNA-seq database S Fig. 1), (3) https://brainrnaseq.org/ (mouse and human RNAseq database S Fig. 1), and (4) https://celltypes.brain-map.org/rnaseq/human_ctx_smart%20seq?selectedVisualization=Scatter+Plot&colorByFeature=Gene+Expression&colorByFeatureValue=GAD1 (human single cell RNAseq database S Fig. 1). Microscopy data and behavioral test data reported in this paper will be shared by the lead contact upon request. Source data are provided with this paper. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-49596-0.
References
- 1.Paolicelli RC, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 2.Carroll JA, Race B, Williams K, Striebel JF, Chesebro B. Innate immune responses after stimulation with toll-like receptor agonists in ex vivo microglial cultures and an in vivo model using mice with reduced microglia. J. Neuroinflammation. 2021;18:194. doi: 10.1186/s12974-021-02240-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sierra A, et al. Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell. 2010;7:483–495. doi: 10.1016/j.stem.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 5.Hu X, et al. Microglia/macrophage polarization dynamics reveal novel mechanism of injury expansion after focal cerebral ischemia. Stroke. 2012;43:3063–3070. doi: 10.1161/STROKEAHA.112.659656. [DOI] [PubMed] [Google Scholar]
- 6.Han, Y. et al. TNF-a mediates SDF-1a-induced NF-k B activation and cytotoxic effects in primary astrocytes. J. Clin. Invest108, 425–435 (2001). [DOI] [PMC free article] [PubMed]
- 7.Lian H, Litvinchuk XA, Chiang AC, Aithmitti N, Jankowsky JL. Astrocyte-microglia cross talk through complement activation modulates amyloid pathology in mouse models of Alzheimer’ s. Disease. 2016;36:577–589. doi: 10.1523/JNEUROSCI.2117-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butovsky O, et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buttgereit A, et al. Sall1 is a transcriptional regulator defining microglia identity and function. Nat. Immunol. 2016;17:1397–1408. doi: 10.1038/ni.3585. [DOI] [PubMed] [Google Scholar]
- 10.Arnold TD, et al. Impaired αVβ8 and TGFβ signaling lead to microglial dysmaturation and neuromotor dysfunction. J. Exp. Med. 2019;216:900–915. doi: 10.1084/jem.20181290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zöller T, et al. Silencing of TGFβ signalling in microglia results in impaired homeostasis. Nat. Commun. 2018;9:1–13. doi: 10.1038/s41467-018-06224-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattson MP, et al. Cellular signaling roles of TGFβ, TNFα and βAPP in brain injury responses and Alzheimer’s disease. Brain Res. Rev. 1997;23:47–61. doi: 10.1016/S0165-0173(96)00014-8. [DOI] [PubMed] [Google Scholar]
- 13.Qin Y, et al. A milieu molecule for TGF-B required for microglia function in the nervous system. Cell. 2018;174:156–157. doi: 10.1016/j.cell.2018.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyers EA, Kessler JA. TGF-β family signaling in neural and neuronal differentiation, development, and function. Cold Spring Harb. Perspect. Biol. 2017;9:a022244. doi: 10.1101/cshperspect.a022244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kashima R, Hata A. The role of TGF-β superfamily signaling in neurological disorders. Acta Biochim. Biophys. Sin. 2018;50:106–120. doi: 10.1093/abbs/gmx124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vivien D, Ali C. Transforming growth factor-β signalling in brain disorders. Cytokine Growth Factor Rev. 2006;17:121–128. doi: 10.1016/j.cytogfr.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 17.Spittau B, Dokalis N, Prinz M. The role of TGFβ signaling in microglia maturation and activation. Trends Immunol. 2020;41:836–848. doi: 10.1016/j.it.2020.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Duan Z, et al. Specificity of TGF-β1 signal designated by LRRC33 and integrin αVβ8. Nat. Commun. 2022;13:4988. doi: 10.1038/s41467-022-32655-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarty JH. αvβ8 Integrin adhesion and signaling pathways in development, physiology and disease. J. Cell Sci. 2020;133:jcs239434. doi: 10.1242/jcs.239434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broad Institute. Study: synucleinopathy-associated astrocytes. Single Cell Portalhttps://singlecell.broadinstitute.org/single_cell/study/SCP1879/synucleinopathy-associated-astrocytes (2018).
- 22.Tasic B, et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature. 2018;563:72–78. doi: 10.1038/s41586-018-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Transcriptomics explorer::allen brain atlas: cell types. https://celltypes.brain-map.org/rnaseq/human_ctx_smart-seq?selectedVisualization=Heatmap&colorByFeature=Cell+Type&colorByFeatureValue=GAD1 (2024).
- 24.Zhang Y, et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang AC, et al. A human brain vascular atlas reveals diverse mediators of Alzheimer’s risk. Nature. 2022;603:885–892. doi: 10.1038/s41586-021-04369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bedolla A, et al. Diphtheria toxin induced but not CSF1R inhibitor mediated microglia ablation model leads to the loss of CSF/ventricular spaces in vivo that is independent of cytokine upregulation. J. Neuroinflammation. 2022;19:3. doi: 10.1186/s12974-021-02367-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spiteri AG, et al. PLX5622 reduces disease severity in lethal CNS infection by off-target inhibition of peripheral inflammatory monocyte production. Front. Immunol. 2022;13:851556. doi: 10.3389/fimmu.2022.851556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spangenberg E, et al. Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer’s disease model. Nat. Commun. 2019;10:3758. doi: 10.1038/s41467-019-11674-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dos Santos SE, et al. Similar microglial cell densities across brain structures and mammalian species: implications for brain tissue function. J. Neurosci. 2020;40:4622–4643. doi: 10.1523/JNEUROSCI.2339-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawson LJ, Perry VH, Dri P, Gordon S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience. 1990;39:151–170. doi: 10.1016/0306-4522(90)90229-W. [DOI] [PubMed] [Google Scholar]
- 31.Parkhurst CN, et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 2013;155:1596–1609. doi: 10.1016/j.cell.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bedolla AM, et al. A comparative evaluation of the strengths and potential caveats of the microglial inducible CreER mouse models. Cell Rep. 2024;43:113660. doi: 10.1016/j.celrep.2023.113660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamaguchi M, et al. Circulating transforming growth factor-β1 facilitates remyelination in the adult central nervous system. Elife. 2019;8:e41869. doi: 10.7554/eLife.41869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stifter SA, Greter M. STOP floxing around: Specificity and leakiness of inducible Cre/loxP systems. Eur. J. Immunol. 2020;50:338–341. doi: 10.1002/eji.202048546. [DOI] [PubMed] [Google Scholar]
- 35.Zheng B, Sage M, Sheppeard EA, Jurecic V, Bradley A. Engineering mouse chromosomes with Cre-loxP: range, efficiency, and somatic applications. Mol. Cell Biol. 2000;20:648–655. doi: 10.1128/MCB.20.2.648-655.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKinsey, G. L. et al. Radial glia promote microglial development through integrin αVβ8 -TGFβ1 signaling. Preprint at bioRxiv10.1101/2023.07.13.548459 (2023).
- 37.Garcia ADR, Doan NB, Imura T, Bush TG, Sofroniew MV. GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain. Nat. Neurosci. 2004;7:1233–1241. doi: 10.1038/nn1340. [DOI] [PubMed] [Google Scholar]
- 38.Diniz LP, Matias I, Siqueira M, Stipursky J, Gomes FCA. Astrocytes and the TGF-β1 pathway in the healthy and diseased brain: a double-edged sword. Mol. Neurobiol. 2019;56:4653–4679. doi: 10.1007/s12035-018-1396-y. [DOI] [PubMed] [Google Scholar]
- 39.Cekanaviciute E, et al. Astrocytic TGF-β signaling limits inflammation and reduces neuronal damage during central nervous system toxoplasma infection. J. Immunol. 2014;193:139–149. doi: 10.4049/jimmunol.1303284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doyle KP, Cekanaviciute E, Mamer LE, Buckwalter MS. TGFβ signaling in the brain increases with aging and signals to astrocytes and innate immune cells in the weeks after stroke. J. Neuroinflammation. 2010;7:1–13. doi: 10.1186/1742-2094-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srinivasan R, et al. New transgenic mouse lines for selectively targeting astrocytes and studying calcium signals in astrocyte processes in situ and in vivo. Neuron. 2016;92:1181–1195. doi: 10.1016/j.neuron.2016.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hill SA, et al. Sonic hedgehog signaling in astrocytes mediates cell type-specific synaptic organization. Elife. 2019;8:e45545. doi: 10.7554/eLife.45545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faust, T. E. et al. A comparative analysis of microglial inducible Cre lines. Cell Rep.42, 113031 (2023). [DOI] [PMC free article] [PubMed]
- 45.Yona S, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaiser, T. & Feng, G. Tmem119-EGFP and Tmem119-CreERT2 transgenic mice for labeling and manipulating microglia. eNeuro6, ENEURO.0448-18.2019 (2019). [DOI] [PMC free article] [PubMed]
- 47.McKinsey GL, et al. A new genetic strategy for targeting microglia in development and disease. eLife. 2020;9:1–34. doi: 10.7554/eLife.54590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Batiuk MY, et al. An immunoaffinity-based method for isolating ultrapure adult astrocytes based on ATP1B2 targeting by the ACSA-2 antibody. J. Biol. Chem. 2017;292:8874–8891. doi: 10.1074/jbc.M116.765313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abud EM, et al. iPSC-derived human microglia-like cells to study neurological diseases. Neuron. 2017;94:278–293.e9. doi: 10.1016/j.neuron.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Butovsky O, Weiner HL. Microglial signatures and their role in health and disease. Nat. Rev. Neurosci. 2018;19:622–635. doi: 10.1038/s41583-018-0057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deczkowska A, et al. Disease-associated microglia: a universal immune sensor of neurodegeneration. Cell. 2018;173:1073–1081. doi: 10.1016/j.cell.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 52.Boche D, Gordon MN. Diversity of transcriptomic microglial phenotypes in aging and Alzheimer’s disease. Alzheimer’s Dement. 2022;18:360–376. doi: 10.1002/alz.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pettas S, et al. Profiling microglia through single-cell RNA sequencing over the course of development, aging, and disease. Cells. 2022;11:2383. doi: 10.3390/cells11152383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carroll JA, Race B, Williams K, Striebel J, Chesebro B. RNA-seq and network analysis reveal unique glial gene expression signatures during prion infection. Mol. Brain. 2020;13:71. doi: 10.1186/s13041-020-00610-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brionne, T. C., Tesseur, I., Masliah, E. & Wyss-coray, T. Loss of TGF-β 1 leads to increased neuronal cell death and microgliosis in mouse brain. Neuron40, 1133–1145 (2003). [DOI] [PubMed]
- 56.Yu T, et al. P2Y12 regulates microglia activation and excitatory synaptic transmission in spinal lamina II neurons during neuropathic pain in rodents. Cell Death Dis. 2019;10:165. doi: 10.1038/s41419-019-1425-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Limatola C, et al. Chemokine CX3CL1 protects rat hippocampal neurons against glutamate-mediated excitotoxicity. J. Neuroimmunol. 2005;166:19–28. doi: 10.1016/j.jneuroim.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 58.Diaz-Aparicio I, et al. Microglia actively remodel adult hippocampal neurogenesis through the phagocytosis secretome. J. Neurosci. 2020;40:1453–1482. doi: 10.1523/JNEUROSCI.0993-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takahashi K, Rochford CDP, Neumann H. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J. Exp. Med. 2005;201:647–657. doi: 10.1084/jem.20041611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Campbell BC, et al. mGreenLantern: a bright monomeric fluorescent protein with rapid expression and cell filling properties for neuronal imaging. Proc. Natl. Acad. Sci. USA. 2020;117:30710–30721. doi: 10.1073/pnas.2000942117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chan KY, et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 2017;20:1172–1179. doi: 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Han J, Zhu K, Zhang X-M, Harris RA. Enforced microglial depletion and repopulation as a promising strategy for the treatment of neurological disorders. Glia. 2019;67:217–231. doi: 10.1002/glia.23529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tesseur I, et al. Deficiency in neuronal TGF-β signaling promotes neurodegeneration and Alzheimer’s pathology. J. Clin. Invest. 2006;116:3060–3069. doi: 10.1172/JCI27341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tesseur I, et al. Deficiency in neuronal TGF-β signaling leads to nigrostriatal degeneration and activation of TGF-β signaling protects against MPTP neurotoxicity in mice. J. Neurosci. 2017;37:4584–4592. doi: 10.1523/JNEUROSCI.2952-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Battaglia G, et al. Early defect of transforming growth factor β1 formation in Huntington’s disease. J. Cell. Mol. Med. 2011;15:555–571. doi: 10.1111/j.1582-4934.2010.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buckwalter MS, et al. Chronically increased transforming growth factor-β1 strongly inhibits hippocampal neurogenesis in aged mice. Am. J. Pathol. 2006;169:154–164. doi: 10.2353/ajpath.2006.051272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harvey BK, Hoffer BJ, Wang Y. Stroke and TGF-β proteins: Glial cell line-derived neurotrophic factor and bone morphogenetic protein. Pharmacol. Ther. 2005;105:113–125. doi: 10.1016/j.pharmthera.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 68.Luo J. TGF-β as a key modulator of astrocyte reactivity: disease relevance and therapeutic implications. Biomedicines. 2022;10:1206. doi: 10.3390/biomedicines10051206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peterson AJ, O’Connor MB. Lean on me: cell–cell interactions release TGF-β for local consumption only. Cell. 2018;174:18–20. doi: 10.1016/j.cell.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 70.Massague J. TGF-β signal transduction. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 71.Huang T, Schor SL, Hinck AP. Biological activity differences between TGF-β1 and TGF-β3 correlate with differences in the rigidity and arrangement of their component monomers. Biochemistry. 2014;53:5737–5749. doi: 10.1021/bi500647d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee S, et al. CX3CR1 deficiency alters microglial activation and reduces beta-amyloid deposition in two Alzheimer’s disease mouse models. Am. J. Pathol. 2010;177:2549–2562. doi: 10.2353/ajpath.2010.100265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gyoneva S, et al. Cx3cr1-deficient microglia exhibit a premature aging transcriptome. Life Sci. Alliance. 2019;2:e201900453. doi: 10.26508/lsa.201900453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hickman SE, Allison EK, Coleman U, Kingery-Gallagher ND, El Khoury J. Heterozygous CX3CR1 deficiency in microglia restores neuronal β-amyloid clearance pathways and slows progression of Alzheimer’s like-disease in PS1-APP mice. Front. Immunol. 2019;10:2780. doi: 10.3389/fimmu.2019.02780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rogers JT, et al. CX3CR1 deficiency leads to impairment of hippocampal cognitive function and synaptic plasticity. J. Neurosci. 2011;31:16241–16250. doi: 10.1523/JNEUROSCI.3667-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sellner S, et al. Microglial CX3CR1 promotes adult neurogenesis by inhibiting Sirt 1/p65 signaling independent of CX3CL1. Acta Neuropathol. Commun. 2016;4:102. doi: 10.1186/s40478-016-0374-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chytil A, Magnuson MA, Wright CVE, Moses HL. Conditional inactivation of the TGF-β type II receptor using Cre:Lox. Genesis. 2002;32:73–75. doi: 10.1002/gene.10046. [DOI] [PubMed] [Google Scholar]
- 78.Levéen P, et al. Induced disruption of the transforming growth factor beta type II receptor gene in mice causes a lethal inflammatory disorder that is transplantable. Blood. 2002;100:560–568. doi: 10.1182/blood.V100.2.560. [DOI] [PubMed] [Google Scholar]
- 79.Liddelow SA, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhan L, et al. Proximal recolonization by self-renewing microglia re-establishes microglial homeostasis in the adult mouse brain. PLoS Biol. 2019;17:e3000134. doi: 10.1371/journal.pbio.3000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wong K, et al. Mice deficient in NRROS show abnormal microglial development and neurological disorders. Nat. Immunol. 2017;18:633–641. doi: 10.1038/ni.3743. [DOI] [PubMed] [Google Scholar]
- 82.Aluwihare P, et al. Mice that lack activity of alphavbeta6- and alphavbeta8-integrins reproduce the abnormalities of Tgfb1- and Tgfb3-null mice. J. Cell Sci. 2009;122:227–232. doi: 10.1242/jcs.035246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu J, et al. beta8 Integrins are required for vascular morphogenesis in mouse embryos. Development. 2002;129:2891–2903. doi: 10.1242/dev.129.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brioschi S, et al. A Cre-deleter specific for embryo-derived brain macrophages reveals distinct features of microglia and border macrophages. Immunity. 2023;56:1027–1045.e8. doi: 10.1016/j.immuni.2023.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bendel O, et al. Reappearance of hippocampal CA1 neurons after ischemia is associated with recovery of learning and memory. J. Cereb. Blood Flow. Metab. 2005;25:1586–1595. doi: 10.1038/sj.jcbfm.9600153. [DOI] [PubMed] [Google Scholar]
- 86.Cinalli DA, et al. DREADD‐inactivation of dorsal CA1 pyramidal neurons in mice impairs retrieval of object and spatial memories. Hippocampus. 2023;33:6–17. doi: 10.1002/hipo.23484. [DOI] [PubMed] [Google Scholar]
- 87.Gu, L., Ren, M., Lin, L. & Xu, J. Calbindin-expressing CA1 pyramidal neurons encode spatial information more efficiently. eNeuro10, ENEURO.0411-22.2023 (2023). [DOI] [PMC free article] [PubMed]
- 88.Volpe BT, Davis HP, Towle A, Dunlap WP. Loss of hippocampal CA1 pyramidal neurons correlates with memory impairment in rats with ischemic or neurotoxin lesions. Behav. Neurosci. 1992;106:457–464. doi: 10.1037/0735-7044.106.3.457. [DOI] [PubMed] [Google Scholar]
- 89.Filipello F, et al. The microglial innate immune receptor TREM2 Is required for synapse elimination and normal brain connectivity. Immunity. 2018;48:979–991.e8. doi: 10.1016/j.immuni.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 90.Schridde A, et al. Tissue-specific differentiation of colonic macrophages requires TGF-β receptor-mediated signaling. Mucosal Immunol. 2017;10:1387–1399. doi: 10.1038/mi.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gregorian C, et al. Pten deletion in adult neural stem/progenitor cells enhances constitutive neurogenesis. J. Neurosci. 2009;29:1874–1886. doi: 10.1523/JNEUROSCI.3095-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem. Biophys. Res. Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 93.Jin Y, Barnett A, Zhang Y, Yu X, Luo Y. Poststroke sonic hedgehog agonist treatment improves functional recovery by enhancing neurogenesis and angiogenesis. Stroke. 2017;48:1636–1645. doi: 10.1161/STROKEAHA.117.016650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Luo F, et al. Inhibition of CSPG receptor PTPσ promotes migration of newly born neuroblasts, axonal sprouting, and recovery from stroke. Cell Rep. 2022;40:111137. doi: 10.1016/j.celrep.2022.111137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marsh, S. E. et al. Dissection of artifactual and confounding glial signatures by single-cell sequencing of mouse and human brain. Nat. Neurosci.25, 306–316 (2022). [DOI] [PubMed]
- 96.Krueger, F. Trim Galore: a Wrapper Tool Around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ Files, with Some Extra Functionality for MspI-Digested RRBS-Type (Reduced Representation Bisufite-Seq) Libraries (accessed 28 April 2016); https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/.
- 97.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 98.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032–2034. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.O’Leary NA, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47:e47–e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mootha VK, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 105.Wickham, H. (ed) Ggplot2: Elegant Graphics for Data Analysis. (Springer International Publishing, 2016). 10.1007/978-3-319-24277-4.
- 106.Turcato FC, Wegman E, Lu T, Ferguson N, Luo Y. Dopaminergic neurons are not a major Sonic hedgehog ligand source for striatal cholinergic or PV interneurons. iScience. 2022;25:105278. doi: 10.1016/j.isci.2022.105278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The RNA-seq data generated in this study have been deposited in the GEO database under accession code GEO: GSE236032 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL30172). Other RNA-seq data presented, but not generated, in this study include: (1) GEO: GSE89189 (generation of human microglia-like cells to study neurological disease Fig. 7), (2) https://singlecell.broadinstitute.org/single_cell/study/SCP1879/synucleinopathy-associated-astrocytes?genes=Tgfb1&tab=distribution#study-visualize (mouse single-cell RNA-seq database S Fig. 1), (3) https://brainrnaseq.org/ (mouse and human RNAseq database S Fig. 1), and (4) https://celltypes.brain-map.org/rnaseq/human_ctx_smart%20seq?selectedVisualization=Scatter+Plot&colorByFeature=Gene+Expression&colorByFeatureValue=GAD1 (human single cell RNAseq database S Fig. 1). Microscopy data and behavioral test data reported in this paper will be shared by the lead contact upon request. Source data are provided with this paper. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. Source data are provided with this paper.