Abstract
Homeodomain transcription factor A9 (HOXA9) is a member of the HOX cluster family of transcription factors that are crucially involved in embryo implantation, morphogenesis, body axis development, and endothelial cell differentiation. Despite numerous reports on its aberrant expression in a few malignancies, the molecular and functional complexity of HOXA9 across cancers remains obscure. We aimed to analyze the dynamic role of HOXA9 across cancers by identifying, analyzing, and understanding its multiple modes of regulation and functional implications and identifying possible therapeutic avenues. We conducted a comprehensive analysis to determine the role of HOXA9 across cancers. This approach involved the integration of large-scale datasets from public repositories such as the Genomic Data Commons, specifically the Cancer Genome Atlas (GDC-TCGA), across 33 different cancer types. The multiple modes of HOXA9 regulation by genetic and epigenetic factors were determined using online tools, which comprised experimentally validated observations. Furthermore, downstream pathways were identified by predicting the targets of HOXA9 and by performing functional enrichment analysis. We also assessed the clinical significance of HOXA9 in terms of prognosis and stage stratification. This study evaluated the correlation between HOXA9 and tumor-infiltrating molecules and discussed its association with therapeutically approved antineoplastic drugs. HOXA9 was significantly upregulated in 9 tumors and downregulated in 2 cancers. The deregulation of HOXA9 is primarily attributed to epigenetic factors, including promoter DNA methylation and noncoding RNAs (ncRNAs). The HOXA9 transcription factor interacts with PBX/MEIS cofactors and regulates multiple genes involved in cancer-associated EMT, autophagy, the cell cycle, metabolic pathways, Wnt signaling, TGF-β signaling, the AMPK pathway, PI3K/AKT signaling, and NF-κB signaling, thereby establishing control over downstream mechanisms. Differential expression in various clinical stages across cancers was shown to have prognostic significance and to be correlated with tumor-infiltrating immune molecules. The assessment of the correlation of HOXA9 expression with approved antineoplastic drugs revealed that targeting HOXA9 could be the most reliable strategy for preventing cancer progression. HOXA9 is upregulated in the majority of malignancies and drives cancer progression by regulating multiple signaling mechanisms. Hence, HOXA9 could be a reliable diagnostic indicator and a potential therapeutic candidate for solid cancer types.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10238-024-01389-x.
Keywords: HOXA9, TCGA dataset, Cancer progression, Immune infiltration, Therapeutic drugs
Introduction
Homeobox (HOX) genes are an evolutionarily conserved, unique set of genes embedded in four different HOX clusters that mainly function as regulators of morphogenesis and body axis specifications [1, 2]. All 39 HOX genes function as homeodomain transcription factors to regulate diverse biological targets. HOXA9, a member of the HOXA cluster, plays a crucial role in various biological processes, including body axis specification, morphogenesis, endothelial cell proliferation, and embryo implantation [3–6]. Its involvement is not only limited to normal developmental processes but also extends to its aberrant expression in both solid cancers and hematological malignancies [7]. As a HOX protein, it has a unique “DNA binding homeodomain”, which helps in binding to target genes with HOXA9-specific consensus sequences. As a transcription factor, HOXA9 exerts its influence by regulating a diverse set of targets that contribute to cancer-related events, further emphasizing its significance in the context of oncology. Hence, the dysregulation of HOXA9 expression underscores its potential as a key player in the molecular mechanisms associated with cancer progression. To the best of our knowledge, its functional role in cancer progression, its association with immune subtypes, and its clinical importance across cancers have not yet been explored.
As published previously, HOXA9 plays a profound role in regulating cancer-associated biological events when aberrantly expressed in cancer [8]. In most solid cancer types, aberrant expression is primarily attributed to epigenetic factors, while in hematological malignancies, HOXA9 gene fusion is more frequent [7, 9]. Consequently, downstream pathways contribute to the progressive development of tumors. It functions either as a tumor promoter or as a tumor suppressor, depending on the tissue-specific cancer type. Researchers have shown that HOXA9 induces cell proliferation, stemness, angiogenesis, invasion, and metastasis in several cancer types, including colorectal cancer (CRC)[10–12], pancreatic cancer (PC)[13], prostate cancer (PCa)[14], osteosarcoma[15], and glioma (GBM)[16–18], where it also confers resistance to drugs and promotes cancer recurrence. In vitro studies have shown that HOXA9 functions as a tumor suppressor in cutaneous squamous cell carcinoma (CSCC) [19, 20], cervical cancer (CC) [21], breast cancer (BC) [22, 23], non-small cell lung cancer (NSCLC) [24–27], epithelial ovarian cancer (EOC) [28], high-grade noninvasive bladder cancer (HGNOC) [29], uveal melanoma [30] and lung adenocarcinoma (LUAD) [31]. Nevertheless, the molecular mechanism underlying its aberrant expression and its functional implications in all these cancer types have not been elucidated.
In the present study, we systematically analyzed the expression status of HOXA9 in 33 different cancer types using datasets from GDC-TCGA. Multiple modes of HOXA9 regulation have been determined through a comprehensive analysis of genetic and epigenetic data, which revealed that HOXA9 is correlated with gene expression status. To assess the functional role of this gene as a transcription factor, we predicted diverse biological targets, which were subsequently subjected to functional enrichment analysis. In addition to investigating the clinical significance of HOXA9 in prognosis and cancer stages, we identified a potential association between HOXA9 expression and various immune subtypes across cancers, enabling us to correlate HOXA9 expression with individual immune cell types. Hence, this study was conducted to explore the significant role of HOXA9 in multiple cancer types, with a particular focus on its involvement in immunological response, thereby offering new insights into anticancer therapy.
Materials and methods
Data acquisition, processing, and cross-validation
The gene expression data for HOXA9 from 50 different normal tissues were obtained from the consensus dataset in the Human Protein Atlas (HPA) (https://www.proteinatlas.org/) [32]. The differential expression of HOXA9 between solid tissue normal and primary tumor samples in multiple cancer types was visualized as a box plot in the TIMER2.0 database under the cancer exploration ‘Gene DE’ module (http://timer.cistrome.org/) [33]. Additionally, it consists of modules that systematically provide clinical, immunological, and genomic features of a specific gene in the pan-cancer TCGA cohort. The HOXA9 gene expression in 33 individual cancer types was retrieved from RNA-seq data of GDC-TCGA datasets and reposited in the UCSC Xena browser, where the values are represented in log2 (fpkm-uq + 1) (https://xena.ucsc.edu/) [34]. The differential expression in each cancer type was further verified by performing the Wilcoxon test, and the data are represented in box plots using SRplot (https://www.bioinformatics.com.cn/srplot) [35]. A pvalue < 0.05 was considered to indicate statistical significance. Similarly, HOXA9 expression in cancer cell lines was retrieved using the Cancer Cell Line Encyclopedia (CCLE) portal (https://sites.broadinstitute.org/ccle/) [36]. The analyzed data were further cross-validated with samples of Affymetrix U133A (GPL96) and U133Plus2 (GPL570) microarray platforms conjugated in another database, namely GENT2, which collects data from the National Centre for Biotechnology Information-Gene Expression Omnibus (NCBI-GEO) public datasets and processes them using the Bioconductor package with the MAS5 algorithm [37]. Moreover, we evaluated the protein expression of HOXA9 in different tumor types with respect to that in normal tissues from immunohistochemistry (IHC) data in the HPA dataset.
Genetic and epigenetic alteration analysis
Mutations at the HOXA9 locus in TCGA-cancer types were retrieved from the TIMER2.0 database. This database provides the percentage of samples with HOXA9 mutations in TCGA cancer types in the form of a bar plot. Furthermore, single-nucleotide variations (SNVs), including missense mutations, nonsense mutations, and frameshift deletions, represented the percent frequency of mutations in tumor samples. Copy number variations (CNVs), such as homologous-heterologous amplifications and deletions, were determined using the GSCA (Gene Set Cancer Analysis) database (http://bioinfo.life.hust.edu.cn/GSCA/#/) [38]. Using the same database, the correlation between CNVs and the expression of HOXA9 was determined. The epigenetic alterations of HOXA9 were further examined using TCGA datasets. Differential patterns of HOXA9 promoter DNA methylation in tumor samples compared with normal samples and their correlation with gene expression were determined by retrieving and analyzing the methylation data in the same GDC-TCGA datasets from the UCSC Xena browser, which was utilized for gene expression analysis. Furthermore, we retrieved the miRNAs sponging HOXA9 from 3 different databases, namely miRTarBase [39], miRBase [40], and starBase v2.0 [41], which provide experimentally validated miRNA‒mRNA sponges. After retrieving all the miRNAs from the three different databases, overlapping analysis was performed using the Venny 2.1 tool (https://bioinfogp.cnb.csic.es/tools/venny/). The gene-miRNA interaction network was plotted using Cytoscape 3.10.1 (https://cytoscape.org/release_notes_3_10_1.html) [42]. The correlations between these 12 miRNAs and HOXA9 expression in the pan-cancer TCGA dataset were retrieved from starBase v2.0.
HOXA9 coexpression and construction of a protein–protein interaction network (PPIN)
Furthermore, HOXA9 coexpression analysis was performed using the GeneMANIA [43] and BioGRID [44] databases. Furthermore, the PPIN was generated using the STRING tool (https://string-db.org/) with a confidence score of 0.4 [45]. Common molecules from these three databases were identified, and their degree of correlation with HOXA9 in TCGA cancer types was determined using the ‘Gene Correlation module’ in the TIMER2.0 database.
HOXA9 target prediction and functional enrichment analysis
Computational tools, namely the Gene Transcription Regulation Database (GTRD) (http://gtrd.biouml.org) [46] TFlink (https://tflink.net/) [47], were used to determine the experimentally validated targets of the HOXA9 transcription factor and upstream molecules. The GTRD database identifies the transcription factor-binding sites validated by chromatin immunoprecipitation (ChIP) experiments; these sites were collected from public repositories, namely GEO, ENCODE, the Sequence Read Archive (SRA), and the literature. The TFlink database offers highly accurate and experimentally proven transcription factor-target gene interactions for six organisms, including humans. HOXA9 targets were subjected to functional enrichment analysis using the ShinyGO tool (http://ge-lab.org/go/) [48]. The enrichment fold changes, along with the FDRs and p values of all the pathways, were downloaded, and graphs were generated using SRplot.
The role of HOXA9 in clinical staging and prognosis
The differential expression of HOXA9 in cancer stages (stage I, stage II, stage III, and stage IV) was determined using the same GDC-TCGA datasets from the UCSC Xena browser. One-way ANOVA was performed to determine the statistical significance, and the graphs were generated by using GraphPad Prism 8. Furthermore, the prognostic significance of the HOXA9 gene in TCGA cancer datasets was determined by conducting survival analysis using UALCAN [49] and KMplotter (https://kmplot.com/analysis/) [50]. The overall survival of the patients stratified by stage was also determined.
Correlations of HOXA9 expression with different immune subtypes, immune cell infiltration, and drug-gene interactions
The associations between HOXA9 expression and different immune subtypes of cancer were determined using the TISIDB database (http://cis.hku.hk/TISIDB) [51], which is a well-known repository for information on tumor–immune interactions. Next, we utilized the TIMER 2.0 tool to determine the correlation between HOXA9 expression and major tumor-infiltrating cells, including macrophages, natural killer (NK) cells, CD4+/CD8+ T cells, B cells, and dendritic cells. The raw data were used to construct a heatmap showing the correlation with HOXA9 in TCGA cancer types using SRplot. Accordingly, statistically significant values were represented as asterisks on the heatmap. Furthermore, we explored the GSCA database, which retrieves drug-gene correlation data from the GDSC and CTRP portals. The correlation between the use of therapeutically approved antineoplastic drugs and the expression of HOXA9 has been represented in the form of a bubble plot. The graphs were generated by using SRplot [35] and analyzed by GraphPad Prism 8.
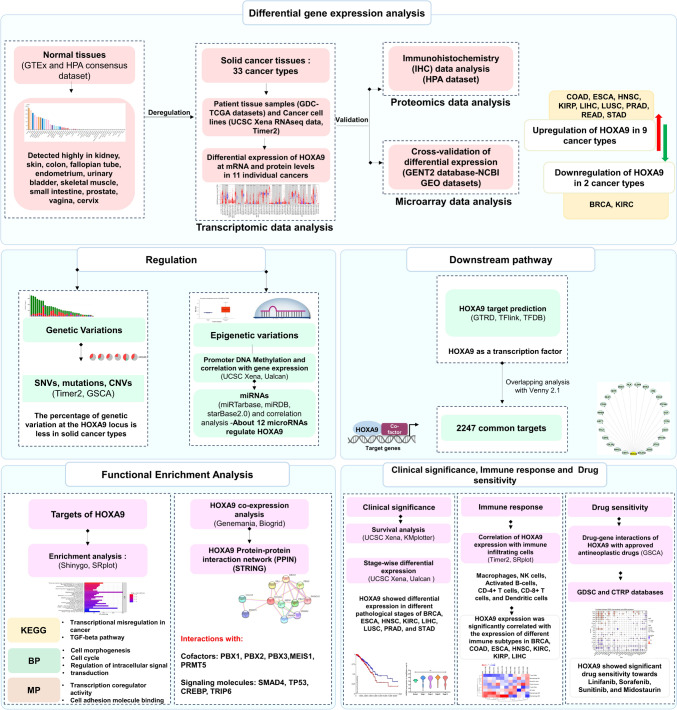
The methodology and results of the analysis are summarized in Fig. 1.
Fig. 1.
Workflow of in silico analysis performed in the present study
Results
HOXA9 is differentially expressed across cancers
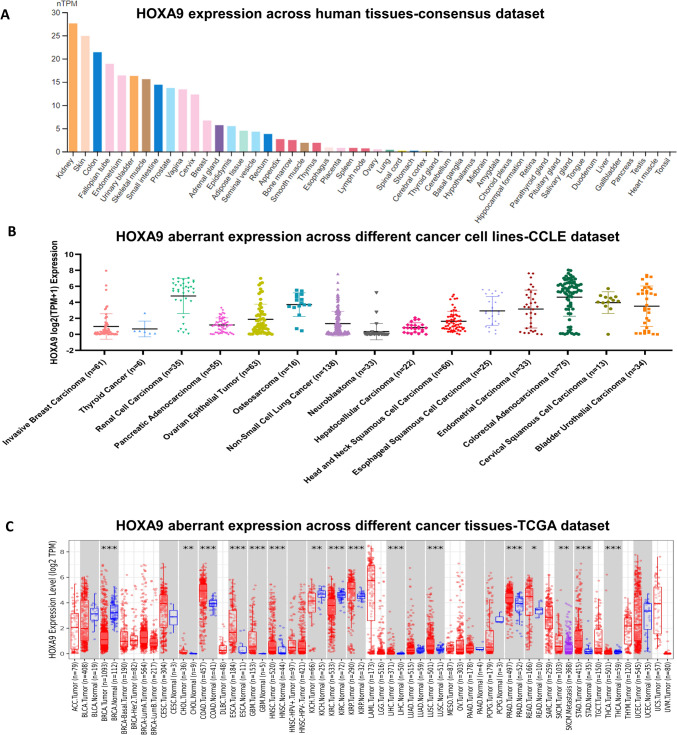
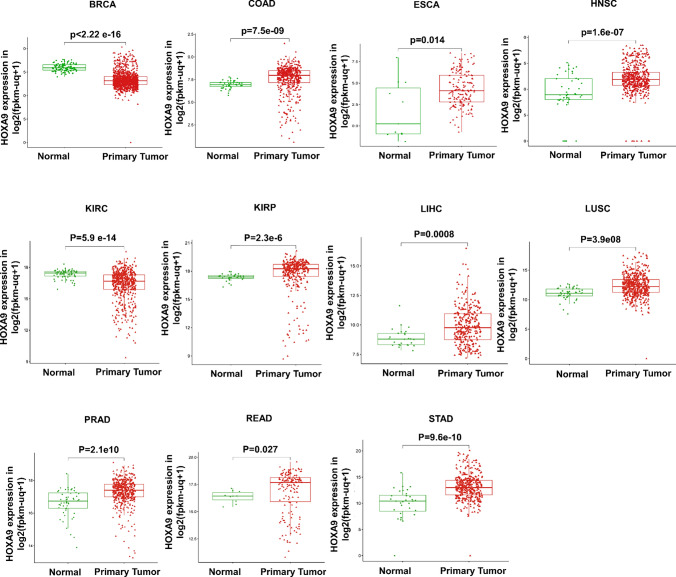
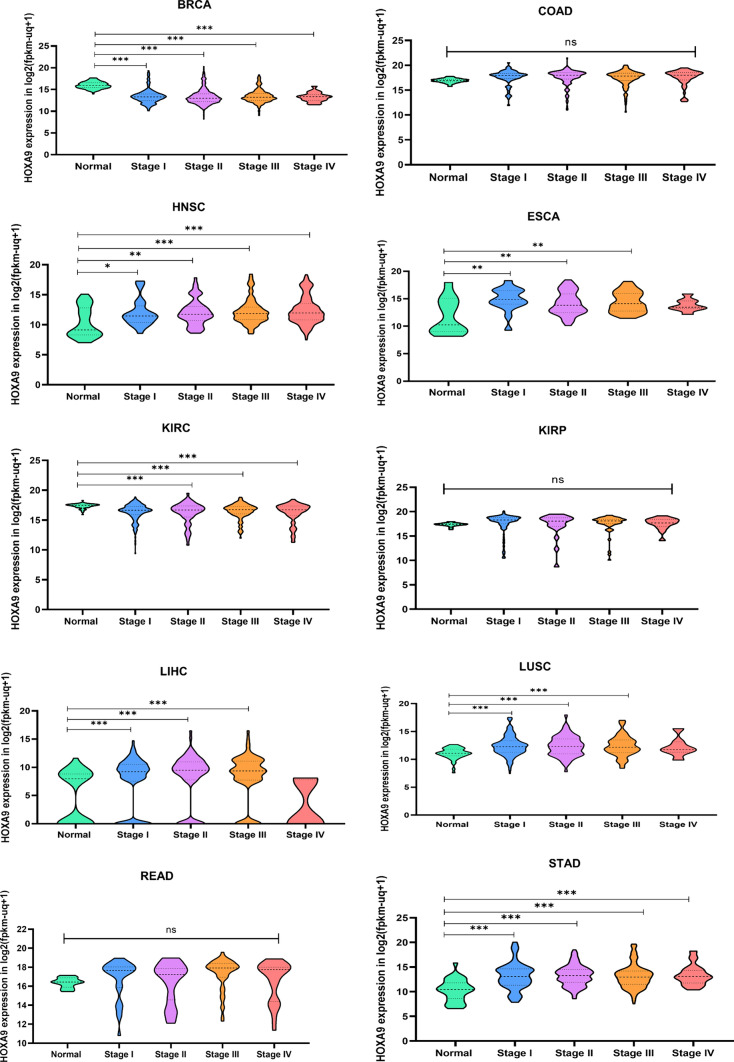
By analyzing gene expression using consensus data from the HPA and GTEx datasets, we discovered that HOXA9 is highly expressed in the kidney and skin under normal physiological conditions (Fig. 2A). Subsequently, we determined the expression of HOXA9 in diverse cell lines associated with each cancer type retrieved from the CCLE portal (Fig. 2B). Initially, we checked the expression status of HOXA9 in TCGA cancer types using the TIMER 2.0 database (Fig. 2C). Notably, HOXA9 was aberrantly expressed in 11 out of 33 GDC-TCGA cancer types. HOXA9 mRNA was highly elevated in the tissues of 9 solid cancers and downregulated in 2 of the cancer types compared to normal tissues. Tumor tissues of colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), head and neck cancer (HNSC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD), rectal adenocarcinoma (READ), and stomach adenocarcinoma (STAD) exhibited increased expression of HOXA9 compared to that of the solid normal samples (p value < 0.05) (Fig. 3). However, HOXA9 expression was significantly downregulated in breast invasive carcinoma (BRCA) and kidney renal clear cell carcinoma (KIRC) (p value < 0.05) (Fig. 3). Furthermore, information on HOXA9 protein expression was available for only a few of the cancer types analyzed using the HPA database and is provided in Supplementary Fig. 1.
Fig. 2.
Gene expression analysis of HOXA9: A Expression of HOXA9 in human tissues retrieved from a consensus dataset in the HPA database. B Expression of HOXA9 across different cancer cell lines retrieved from the CCLE database. C Expression of HOXA9 across different cancer tissues visualized by the TIMER 2.0 database
Fig. 3.
HOXA9 gene expression in the TCGA RNA-Seq dataset retrieved from the UCSC Xena browser: Increased expression of HOXA9 in COAD, ESCA, HNSC, KIRP, LIHC, LUSC, PRAD, READ, and STAD was observed in primary tumors compared to that in normal solid tissue. Downregulation of HOXA9 expression was observed in BRCA and KIRP primary tumor tissues compared to normal solid tissue
The GPL570 dataset showed differential gene expression in tumor tissues of the breast, colon, esophagus, head and neck, kidney, liver, lung, prostate, and stomach. The GPL96 dataset showed differential HOXA9 expression in the breast, colon, esophagus, kidney, lung, and stomach (Supplementary Fig. 2). These findings suggest that HOXA9 may play a role in promoting carcinogenesis in multiple tumor types. Therefore, additional research is necessary for its clinical significance.
Regulation of HOXA9 across cancers
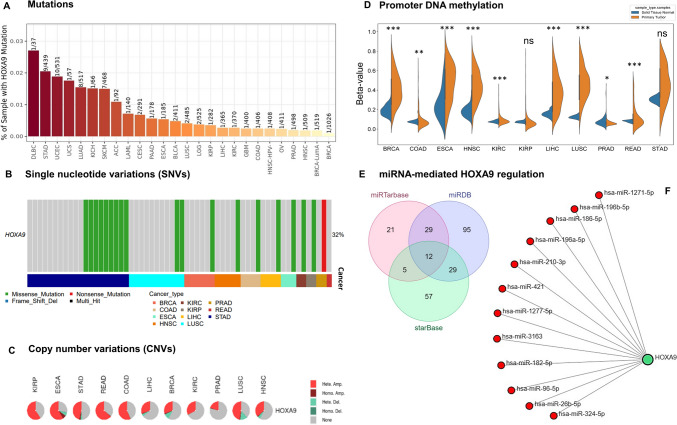
Among the 11 TCGA cancer types, HOXA9 exhibited the highest mutation frequency, with 9% in STAD, 2% in LUSC, and 1% in the remaining tumors. Despite missense mutations frequently occurring SNVs in almost all cancer types, the overall mutation rate across cancers was only 32% (Fig. 4A, B), indicating that other factors might regulate the expression of HOXA9. Subsequently, we checked whether the upregulation of HOXA9 in 9 TCGA cancer types was attributed to CNVs at this locus. Interestingly, the HOXA9 locus exhibited heterozygous amplification in more than 50% of patients across the 6 TCGA cancer types, with READ (64.24%) and ESCA (65.21%) showing the highest percentages (Fig. 4C). When we correlated CNVs with the expression of HOXA9, we found a strong positive correlation for KIRP (Spearman R-coefficient 0.513 and p value 7.4E−18) (Supplementary Table 1).
Fig. 4.
Genetic and epigenetic variation analysis of the HOXA9 gene: A Percentage of mutations at the HOXA9 locus visualized by the Timer2.0 database. B, C Types of SNVs and CNVs observed at the HOXA9 locus in different cancer types visualized by the GSCA database. D Promoter DNA methylation of the HOXA9 promoter was analyzed using TCGA datasets from the UCSC Xena browser and plotted using an SR plot. E Overlapping analysis of miRNAs regulating HOXA9 using 3 different databases and the Venny 2.1 tool. F. Network of 12 miRNAs regulating HOXA9, generated using Cytoscape 3.10.1
Then, we investigated the potential association between the upregulation of HOXA9 expression and epigenetic variation by examining the promoter DNA methylation status across all 11 TCGA cancer types using the same GDC-TCGA datasets employed for gene expression analysis. Among the 11 TCGA cancer types, we found a significant difference in promoter DNA methylation in 9 cancer types (p value < 0.05) (Fig. 4D). Correlation analysis revealed that the reduced expression of HOXA9 in BRCA and KIRC patients resulted from promoter DNA hypermethylation, while the elevated expression in PRAD patients was attributed to promoter DNA hypomethylation (Supplementary Table 2). Our study revealed that the upregulation of HOXA9 in KIRP could mainly be due to CNVs because no differential methylation was detected between normal and primary tumor samples. However, there was no significant correlation between HOXA9 expression and methylation in LUSC, and a weak correlation was observed in HNSC and LIHC (Supplementary Table 2), suggesting the role of other epigenetic factors. Despite being upregulated in HNSC, LIHC, COAD, ESCA, and READ primary tumors, HOXA9 showed hypermethylation compared to that in normal solid tissue. This paradoxical association indicated complex and context-dependent regulation and has recently been reported in a few cancer types [52]. According to the correlation analysis performed in the present study, an inverse correlation was observed between expression and methylation in these cancer types, suggesting a strong need for experimental validation to pinpoint the precise promoter and its methylation status governing the expression in each of the cancer types. Analyzing the deposition pattern of histone marks on promoter regions could also provide valuable insights into their regulation.
We further performed a thorough analysis of three different databases with experimentally validated miRNA‒target gene interactions and identified 12 miRNAs as upstream epigenetic regulators of HOXA9 that play a role in the regulation of its gene expression, namely hsa-miR-1271-5p, hsa-miR-1277-5p, hsa-miR-182-5p, hsa-miR-186-5p, hsa-miR-210-3p, hsa-miR-26b-5p, hsa-miR-3163, hsa-miR-324-5p, hsa-miR-421 and hsa-miR-96-5p. Notably, two HOX cluster-embedded miRNAs, namely hsa-miR-196a-5p and hsa-miR-196b-5p, were also found. The results of the correlation analysis of miRNA gene expression are summarized in Supplementary Table 3. However, Table 1 summarizes the malignancies in which these miRNAs showed an inverse correlation with HOXA9, which might be one of the possible driving factors for HOXA9 regulation. Hence, experimental investigation is essential to understand the underlying driving epigenetic mechanism behind the aberrant expression of HOXA9.
Table 1.
In silico prediction of miRNAs and their inverse correlation with HOXA9 expression in pan-cancer
| SI. no | miRNAs inversely correlate with HOXA9 | Cancer types | Coefficient-R | P value |
|---|---|---|---|---|
| 1 | hsa-miR-1271-5p | Prostate adenocarcinoma | − 0.113 | 1.22E− 02 |
| Rectum adenocarcinoma | − 0.247 | 1.56E− 03 | ||
| 2 | hsa-miR-1277-5p | Kidney renal clear cell carcinoma | − 0.109 | 1.32E− 02 |
| 3 | hsa-miR-182-5p | Breast invasive carcinoma | − 0.105 | 5.25E− 04 |
| 4 | hsa-miR-186-5p | Breast invasive carcinoma | − 0.081 | 7.84E− 03 |
| Kidney renal papillary cell carcinoma | − 0.2 | 6.21E− 04 | ||
| Rectum adenocarcinoma | − 0.16 | 4.31E− 02 | ||
| 5 | hsa-miR-196a-5p | None | – | – |
| 6 | hsa-miR-196b-5p | None | – | – |
| 7 | hsa-miR-210-3p | Colon adenocarcinoma | − 0.094 | 4.55E− 02 |
| Breast invasive carcinoma | − 0.17 | 1.61E− 08 | ||
| 8 | hsa-miR-26b-5p | Kidney renal papillary cell carcinoma | − 0.123 | 3.74E− 02 |
| 9 | hsa-miR-3163 | None | – | – |
| 10 | hsa-miR-324-5p | Breast invasive carcinoma | − 0.147 | 1.19E− 06 |
| 11 | hsa-miR-421 | Prostate adenocarcinoma | − 0.092 | 4.11E− 02 |
| Colon adenocarcinoma | − 0.099 | 3.55E− 02 | ||
| 12 | hsa-miR-96-5p | Breast invasive carcinoma | − 0.142 | 2.75E− 06 |
The HOXA9 transcription factor interacts with cofactors to strengthen DNA binding
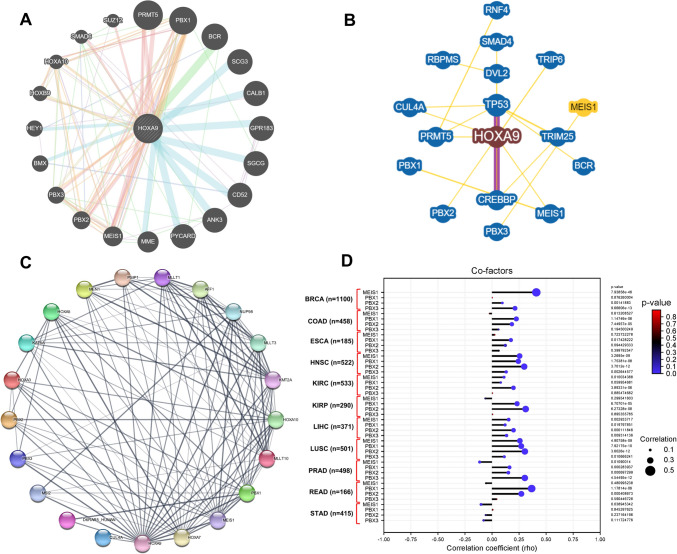
Coexpression analysis of the coexpressed genes using GeneMANIA and BioGRID revealed several interacting factors (Fig. 5A, B). Our overlapping analysis revealed that HOXA9 is primarily coexpressed with five genes: MEIS1, PBX1, PBX2, PBX3, PRMT5, and PBX3. Additionally, the interaction of the HOXA9 protein with the cofactors PBX1, PBX2, PBX3, and MEIS1 was observed and visualized in the PPIN (p value < 1.0e−16) (Fig. 5C). It was also shown to interact with other crucial regulators, namely CUL4A, D6RAR5, HOXA10, HOXA3, HOXA5, HOXA7, MLLT1, KAT6A, MSI2, MLLT10, NUP98, MEN1, PSIP1, MLLT3 and KMT2A. Analysis of the HOXA9 gene revealed a positive correlation with PBX1, PBX2, PBX3, and MEIS1 in all cancer types except for STAD among the 11 studied genes, suggesting enhanced binding of HOXA9 to the promoter region of target genes (Fig. 5D).
Fig. 5.
Coexpression analysis of HOXA9 using the A GeneMANIA, B Biogrid and C STRING databases. D Lollipop plot showing the correlation of HOXA9 with PBX1, PBX2, PBX3 and MEIS1 in different cancer types
The HOXA9 transcription factor (TF) regulates important signaling molecules associated with cancer-related biological events
The ChIP-seq data derived from the TFlink database revealed the list of TFs that regulate HOXA9 expression, and the targets regulated by the HOXA9 TF (Supplementary Table 4). However, our downstream target analysis revealed that HOXA9 functions as a transcription factor for 2247 genes (Supplementary Table 4). Angiogenic markers such as GRB2, MAPK8, FGFR1, PI3KCA, and HIF1A, as well as Wnt/β-catenin pathway markers such as CREBBP, MYC, and NLK, were identified as targets of HOXA9. Other targets were found to be involved in TGF-β signaling, PI3K signaling, NF-κB signaling, and VEGF signaling, all of which are associated with cancer progression. Notably, we identified the following EMT markers as HOXA9 targets: ESR1, BIRC3, COL5A1, CDH2, FHL1, KRT7, INHBA, CD36, DLG1, MITF, DESI1, NLK, CLDN4, MTA3, COL5A2, VIM, OCLN, PTP4A1, EGF, TCF4, CD47, VPS13A, PTK2 and ZEB2.
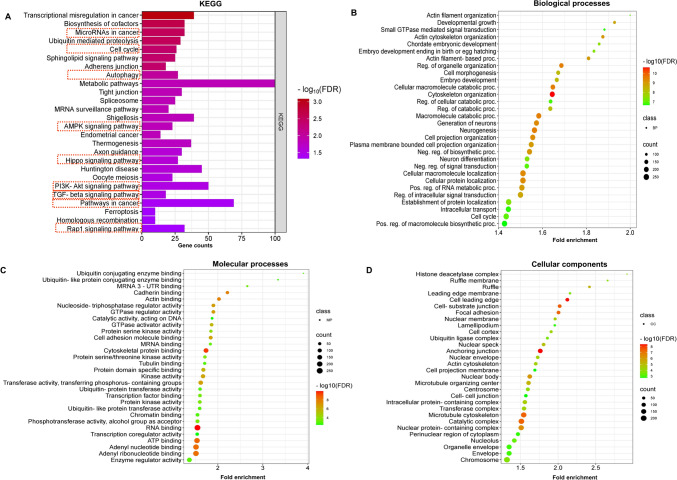
All 2247 targets of HOXA9 were subjected to enrichment analysis to assess their biological significance in cancer progression. KEGG pathway analysis revealed that the targets were enriched in autophagy, the cell cycle, metabolic pathways, the AMPK pathway, Hippo signaling, PI3K/AKT signaling, TGF-β signaling, Rap1 signaling, and transcriptional misregulation in cancer (Fig. 6A). Notably, the pathways represented by these targets were reported to be significantly involved in acquiring cancer hallmarks, indicating the ability of HOXA9 to regulate downstream pathways to promote cancer progression. The top biological process (BP) terms included actin filament organization, cell morphogenesis, cell cycle, negative regulation of signal transduction, and regulation of intracellular transduction (Fig. 6B). In addition, targets with the greatest enrichment in molecular processes (MPs), such as cadherin binding, GTPase regulator activity, protein serine-kinase activity, cell adhesion molecule binding, protein domain-specific binding, transcription factor binding, and transcription coregulator activity, were the most enriched (Fig. 6C). These targets were particularly localized in the cell‒cell junction, nuclear membrane, perinuclear region of the cytoplasm, and microtubule cytoskeleton (Fig. 6D). All the GO terms associated with HOXA9 targets are represented in the form of bubble plots in Fig. 6.
Fig. 6.
Functional enrichment analysis of HOXA9 using the ShinyGO tool. A KEGG pathway analysis. B Biological processes. C Molecular processes. D Cellular components
Clinical significance of HOXA9 across cancers
The prognostic significance of HOXA9 in GDC-TCGA datasets was determined by using the UALCAN database, which analyzes the correlation between the HOXA9 gene and the prognosis of patient samples through follow-up studies. However, there was no significant difference between the expression of HOXA9 and the prognosis of patients with any of the cancer types (Supplementary Fig. 3).
To evaluate the clinical utility of HOXA9 in stage stratification, we analyzed gene expression data across different clinical stages (stages I-IV) in 11 cancer types and revealed significant differential expression in BRCA, HNSC, ESCA, KIRC, LIHC, LUSC, and STAD (p value < 0.05), but not in COAD, KIRP, READ (not reported), or PRAD (Fig. 7). Assessing the expression of HOXA9 in the different clinical stages might aid in preventing cancer progression. Hence, we performed survival analysis for patients stratified by stage I to IV disease. The prognostic significance was observed only for KIRC and KIRP in the 11 cancer types that we studied. Interestingly, over time, we observed better survival in KIRC patients with downregulated HOXA9 expression and KIRP with upregulated HOXA9 expression (p value < 0.05) (Supplementary Fig. 4). The upregulation (downregulation) of genes associated with improved survival outcomes was attributed to activation of the antitumor immune response [53, 54]. There could also be several possible reasons for this contradictory observation beyond the direct immune response, which has recently been reported in solid malignancies [55, 56]. Hence, this observation prompted us to investigate the underlying mechanism of the immune response.
Fig. 7.
Expression of HOXA9 in different clinical stages of TCGA cancers: Raw data retrieved from the UCSC Xena RNA-seq dataset were analyzed and plotted using GraphPad Prism 8
HOXA9 expression is correlated with immune cell infiltration across cancers
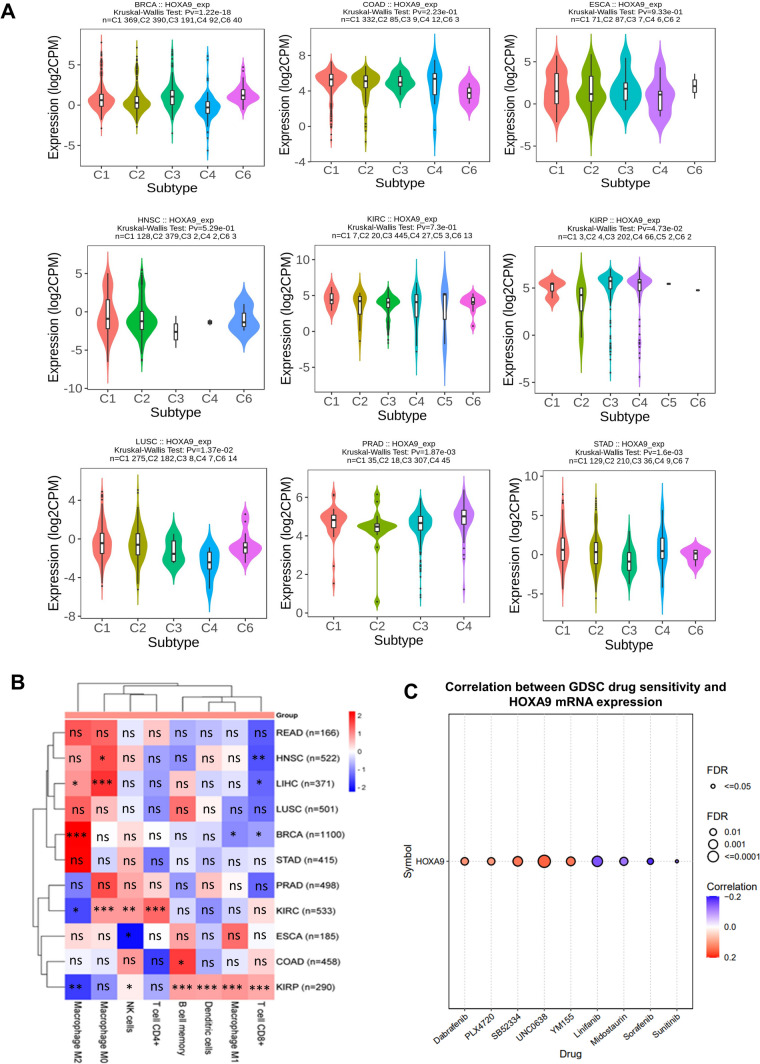
HOXA9 expression in various immune subtypes of each cancer type was assessed using the TISIDB database, categorizing immune subtypes into six groups based on the TISIDB algorithm: wound healing (C1), IFN-gamma dominant (C2), inflammatory (C3), lymphocyte depleted (C4), immunologically quiet (C5), and TGF-β dominant (C6). The results indicated that HOXA9 exhibited differential expression in the immune subtypes BRCA, KIRP, LUSC, PRAD, and STAD (Fig. 8A).
Fig. 8.
HOXA9 and immune interactions: A Expression of HOXA9 in different immune subtypes visualized in the TISIDB database. B Heatmap showing the correlation between HOXA9 expression and immune cells in different cancer types retrieved from the TIMER 2.0 database. C Bubble plot showing the correlation between HOXA9 expression and sensitivity to the antineoplastic drugs from the GSCA database
Building on the findings of HOXA9 expression with respect to immune subtypes, we conducted an extensive analysis to investigate its association with various immune cells, including CD4+ T cells, CD8+ T cells, B cells, dendritic cells, macrophages, and natural killer (NK) cells, in 11 TCGA cancer types. Our analysis using the TIMER2.0 database revealed that the overexpression of HOXA9 was accompanied by the infiltration of immune cells (Fig. 8B). In particular, in KIRP, high expression of HOXA9 was significantly positively correlated with the infiltration of CD8+ T cells, B-memory cells, dendritic cells, NK cells and M1 macrophages, which are all actively involved in promoting antitumor activity. In contrast, there was a negative correlation between HOXA9 expression and protumorigenic M2 macrophages in KIRC. This finding strengthens our previous observation of HOXA9-mediated better survival outcomes in advanced stages of KIRC and KIRP, which might be mainly due to the involvement of HOXA9 in triggering immune response pathways and inducing immune cell activation and infiltration.
HOXA9-drug interactions
According to previous reports, in a few cancer types, HOXA9 is known to induce resistance to therapeutically approved drugs, namely erlotinib, bevacizumab, and temozolomide [57, 58]. However, further studies are needed to determine the effectiveness of other antineoplastic drugs that target HOXA9 in cancer treatment. According to the GDSC, HOXA9 showed a positive correlation with drugs, namely SB52334, dabrafenib, PLX4720, YM155, and UNC0638, indicating that HOXA9 could induce drug resistance in cancer. Notably, we found a negative correlation between HOXA9 expression and linifanib, sorafenib, sunitinib, and midostaurin (Fig. 8C). This indicates that these drugs could increase the chemosensitivity of cells in elevated levels of HOXA9. Based on these observations, researchers should focus on utilizing these approved drugs to target HOXA9 to restrain cancer progression.
Discussion
Based on previous reports that suggested the role of HOXA9 in human cancers, we systematically performed a pan-cancer analysis to determine the differential expression of HOXA9, its mode of regulation, its molecular mechanisms, and its functional implications using valid computational databases. Furthermore, its role as a biomarker for prognosis, stage stratification, correlation with immune cells of the tumor microenvironment, interactions with cofactors, and drug-gene interactions was also determined, aiming to understand the potential role of HOXA9 in cancer progression.
Among the 33 TCGA cancer types analyzed using the RNA-Seq dataset, HOXA9 was found to be highly upregulated in 9 cancer types and downregulated in 2 of the cancer types compared to normal tissues, indicating that HOXA9 acts as an oncogene in the majority of cancer types. Apart from having prognostic significance, it indeed has clinical significance in differentiating the clinical stages of BRCA, HNSC, ESCA, KIRC, LIHC, LUSC, and STAD. These results suggest that HOXA9 might serve as a diagnostic biomarker for these cancers (Table 2).
Table 2.
HOXA9 as a pan-cancer diagnostic indicator
| S.No | Cancer type | Expression of HOXA9 | Genetic alterations | Epigenetic alterations | Clinical significance |
|---|---|---|---|---|---|
| 1 | BRCA | Low↓ | – | Hypermethylation | Differential expression in stages and immune subtypes |
| 2 | COAD | High↑ | – | – | – |
| 3 | ESCA | High↑ | Heterozygous amplification (65.21%) | – | Stages |
| 4 | HNSC | High↑ | – | – | Stages |
| 5 | KIRP | High↑ | Stages and immune subtypes, Immune infiltration | ||
| 6 | KIRC | Low↓ | – | Hypermethylation | Stages |
| 7 | LIHC | High↑ | – | – | Stages |
| 8 | LUSC | High↑ | – | – | Stages and immune subtypes |
| 9 | PRAD | High↑ | – | Hypomethylation | Stages and immune subtypes |
| 10 | READ | High↑ | Heterozygous amplification (64.24%) | – | Stages |
| 11 | STAD | High↑ | – | – | Stages and immune subtypes |
Previous studies have shown that the aberrant expression of HOXA9 in solid cancers primarily results from epigenetic factors. Our data reflect the minimal variation in genetic factors at the HOXA9 locus; specifically, SNVs account for 32% of all cancers. Conversely, we observed a significant incidence of CNVs in READ (64.24%) and ESCA (65.21%). This emphasizes the necessity for focused research to validate and assess the impact of CNVs in these specific cancers. Researchers have demonstrated that HOXA9 promoter methylation is responsible for its aberrant expression in lung adenocarcinoma (LUAD) [31], non-small cell lung cancer (NSCLC) [24, 25], high-grade noninvasive bladder cancer [29], and HNSC [59, 60]. Notably, research on cervical cancer (CC) revealed that the repression of HOXA9 results from increased methylation of its first exon. Restoring its expression could mitigate cancer-associated biological processes [21]. The prognostic significance of HOXA9 methylation has been well-studied in solid cancers [61]. Indeed, in our BRCA and KIRC datasets, the decreased expression of HOXA9 may be linked to hypermethylation at the HOXA9 promoter, while the increased HOXA9 expression in PRAD is a result of promoter DNA hypomethylation. The usual correlation between promoter DNA methylation and gene repression does not always follow the conventional trend. Growing evidence has shown that promoter DNA methylation is also linked to gene activation in different biological contexts, including normal development and metastatic malignancies [52]. CpG promoter DNA methylation either facilitates the binding of enhancers to methylated DNA or blocks the binding of potential repressors to transcription start sites (TSSs), thereby inducing gene transcription [52]. Nevertheless, studies have demonstrated that transcription can also be governed by alternative promoters situated at considerable distances upstream from the TSS [52]. Currently, activation of HOX genes through DNA hypermethylation has been widely regarded as a novel epigenetic mechanism in cancer [62]. In this study, we observed a consistent relationship between promoter DNA hypermethylation and the upregulation of HOXA9 in COAD, ESCA, HNSC, LIHC, and READ. This emphasizes the need for in-depth exploration of the molecular mechanisms involved to elucidate the regulatory pathways governing the transcriptional activation of genes.
In addition to promoter CpG methylation, HOXA9 is regulated by miRNAs in NSCLC [26, 27], acute myeloid leukemia (AML) [63, 64], osteosarcoma [15], epithelial ovarian cancer [28], CRC [12], glioma [16, 65], and uveal melanoma [30]. The sponging of particular miRNAs to the HOXA9 3’UTR not only contributes to increased proliferation, migration, invasion, and metastasis but also facilitates tumor recurrence. Hence, it is worth studying the miRNA-mediated regulation of HOXA9 in different cancer types. In the present study, we identified 12 miRNAs, including two HOX cluster-embedded miRNAs, namely hsa-miR-196a-5p and hsa-miR-196b-5p, that correlate the expression of HOXA9 across cancers. Validating these findings via in vitro studies might help researchers unravel the mode of regulation in several cancer types.
The HOXA9 transcription factor is frequently associated with its cofactors, namely MEIS1 and PBX, which are TALE homeodomain-containing cofactors, thereby enhancing its DNA binding ability [66, 67]. Similarly, we observed an association of HOXA9 with PBX1, PBX2, PBX3, and MEIS1 in all 10 cancer types except for STAD. As a transcription factor, it has an inherent ability to bind to target genes involved in various normal developmental processes [68–72] as well as during cancer progression [17–22, 73–76]. The tumor-promoting property of HOXA9 in EOC cells is mediated through the activation of TGF-β2 [77]. Moreover, it induces an aggressive phenotype in OVC cells via transcriptional activation of its target gene, CDH3 (P-cadherin) [78]. Studies have also determined the functional role of HOXA9 in the regulation of target genes, namely eNOs (endothelial nitric oxide synthase), CDH5 (VE-cadherin), and VEGFR2, to promote angiogenesis [79]. Our analysis revealed the presence of 2247 targets of HOXA9, including angiogenic markers, EMT markers, and molecules of the Wnt/β-catenin, TGF-β, PI3K, NF-κB, and VEGF signaling pathways. Moreover, KEGG pathway analysis revealed the involvement of HOXA9 targets in the cell cycle, autophagy, metabolism, the AMPK pathway, Hippo signaling, PI3K/AKT signaling, TGF-β signaling, Rap1 signaling and transcriptional misregulation in cancer, indicating the potential role of those targets in cancer-associated biological processes.
According to our GO terms associated with HOXA9 targets, we determined that HOXA9 was localized mainly to the nucleus, cytoplasm, and microtubules. These cellular compartments control vital biological processes such as the cell cycle, actin filament organization, cell morphogenesis, and intracellular signal transduction. This regulation involves functions such as cadherin binding, GTPase regulator activity, protein serine-kinase activity, cell adhesion molecule binding, protein domain-specific binding, transcription factor binding, and transcription coregulator activity. Hence, additional studies are needed to assess the differential expression patterns of these targets in various cancers and validate the involved pathways.
Interestingly, the present study revealed that aberrant expression of HOXA9 in KIRC and KIRP patients was associated with improved survival over time. This is due to the HOXA9-mediated activation of the antitumor immune response [53, 54]. Immune cells frequently interact with growing cancer cells in the microenvironment and play an influential role in tumor progression. To understand these dynamics, it is imperative to establish an association between the expression levels of various genes and immune infiltrating molecules. It has been reported that during the progression of prostate cancer (PCa), aberrantly expressed HOXA2, HOXA9, and HOXA10 facilitate the infiltration of dendritic cells, macrophages, and mastocytes [80]. In EOC, HOXA9 facilitates the transcriptional activation of TGF-β2, which in turn triggers the efflux of chemokines such as CXCL12, IL-6, and VEGF-A in peritoneal fibroblasts [73, 81]. In our analysis, we observed variations in the expression of HOXA9 in the immune subtypes BRCA, KIRP, LUSC, PRAD, and STAD. Elevated levels of HOXA9 were significantly correlated with immune cell infiltration in most cancer subtypes, particularly in KIRP. This finding strengthens our previous observation that better survival in KIRP patients could be mainly due to the HOXA9-mediated induction of immune cell infiltration.
In summary, our study clarified the effect of HOXA9 on the tumor immune response across various cancer types. Having elucidated the dynamics of cancer pathways, we identified drugs that target aberrantly expressed HOXA9. Our analysis of drug‒gene interactions revealed that drugs such as linifanib, sorafenib, sunitinib, and midostaurin have the potential to target HOXA9 in cancer, thereby increasing chemosensitivity in cancer cells. Therefore, additional clinical trials are necessary in the future to assess the effectiveness of these drugs in combating aggressive phenotypes. Through our extensive computational analysis, we deduced that HOXA9 serves as a potential biomarker across various cancers, laying the groundwork for further investigation into its role in specific cancer types. Despite conducting a thorough analysis, our study has a few limitations. The specific mechanism through which HOXA9 contributes to oncogenicity in distinct cancer types is still not understood. There is a need to validate these findings through experimental work, both in vitro and in vivo, to elucidate the HOXA9-mediated dynamics of cancer progression. We recently identified the correlation between HOXA9 and its targets, as well as its association with immune cells. More studies are needed to understand the exact pathways involved in cancer progression and immune infiltration. To target HOXA9 therapeutically, clinical trials for the newly proposed drugs must be carried out, considering the specific cancer types.
Conclusion
This is the first comprehensive pan-cancer analysis of HOXA9 to reveal its crucial role in human cancers. Through extensive computational analysis using experimentally validated datasets, we concluded that HOXA9 could serve as a potential diagnostic biomarker for cancer detection and stage stratification. Moreover, our investigation revealed the impact of HOXA9 on various signaling pathways and cancer hallmarks. Understanding the correlation between HOXA9 expression and various immune cells has paved the way for clinicians to consider immunotherapy in various cancer types. Targeting HOXA9 has emerged as a promising strategy with the potential to impede cancer progression, offering new perspectives for future research and clinical interventions in diverse cancer types.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the Wellcome Trust DBT India Alliance, Government of India (Grant No. IA/CPHI/18/1/503927), Joint CSIR-UGC NET Senior Research Fellowship, Government of India (File No. 09/1165(0011)/2020-EMR-I) for financial assistance and the Manipal School of Life Sciences for infrastructure support. This work was supported by the DBT/Wellcome Trust India Alliance Fellowship (Grant number—IA/CPHI/18/1/503927) awarded to Raghu Radhakrishnan.
Abbreviations
- GDC
Genomic Data Commons
- TCGA
The Cancer Genome Atlas
- ncRNA
Noncoding RNA
- CRC
Colorectal cancer
- PC
Pancreatic cancer
- PCa
Prostate cancer
- GBM
Glioblastoma
- CSCC
Cutaneous squamous cell carcinoma
- CC
Cervical cancer
- BC
Breast cancer
- NSCLC
Non-small cell lung cancer
- EOC
Epithelial ovarian cancer
- HGNOC
High-grade noninvasive bladder cancer
- LUAD
Lung adenocarcinoma
- HPA
Human protein atlas
- IHC
Immunohistochemistry
- CCLE
Cancer cell line encyclopedia
- CNV
Copy number variations
- SNV
Single-nucleotide variations
- GSCA
Gene set cancer analysis
- PPIN
Protein‒protein interaction network
- ChIP
Chromatin immunoprecipitation
- COAD
Colon adenocarcinoma
- ESCA
Esophageal carcinoma
- HNSC
Head and neck cancer
- KIRP
Kidney renal papillary cell carcinoma
- LIHC
Liver hepatocellular carcinoma
- LUSC
Lung squamous cell carcinoma
- PRAD
Prostate adenocarcinoma
- READ
Rectal adenocarcinoma
- STAD
Stomach adenocarcinoma
- BRCA
Breast invasive carcinoma
- KIRC
Kidney renal clear cell carcinoma
Author contributions
U.S.S. conceived and designed the study. U.S.S., D.S.B., and R.R. made substantial contributions to the analysis and interpretation of the data and the drafting of the article. S.P.K. and R.R. critically revised the manuscript. All the authors were involved in the final approval of the version to be submitted.
Funding
Open access funding provided by Manipal Academy of Higher Education, Manipal. This study was funded by Wellcome Trust DBT India Alliance, Government of India (Grant No. IA/CPHI/18/1/503927).
Availability of data and materials
All data retrieved and analyzed in this study are included in the manuscript, and additional information has been provided as supplementary files.
Code availability
NA.
Declarations
Conflict of interest
The authors declare no conflicts of interest.
Consent for publication
All the authors have read and approved the final manuscript.
Ethical approval
NA.
Consent for participation
NA.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Holland PW, Garcia-Fernàndez J. Hox genes and chordate evolution. Dev Biol. 1996;173:382–95. [DOI] [PubMed] [Google Scholar]
- 2.Cillo C, Cantile M, Faiella A, Boncinelli E. Homeobox genes in normal and malignant cells. J Cell Physiol. 2001;188:161–9. [DOI] [PubMed] [Google Scholar]
- 3.Wellik DM. Hox patterning of the vertebrate axial skeleton. Dev Dyn Off Publ Am Assoc Anat. 2007;236:2454–63. [DOI] [PubMed] [Google Scholar]
- 4.Xu B, Geerts D, Bu Z, Ai J, Jin L, Li Y, et al. Regulation of endometrial receptivity by the highly expressed HOXA9, HOXA11 and HOXD10 HOX-class homeobox genes. Hum Reprod. 2014;29:781–90. [DOI] [PubMed] [Google Scholar]
- 5.Yan J, Chen Y-X, Desmond A, Silva A, Yang Y, Wang H, et al. Cdx4 and menin co-regulate Hoxa9 expression in hematopoietic cells. PLoS ONE. 2006;1:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel CV, Sharangpani R, Bandyopadhyay S, DiCorleto PE. Endothelial cells express a novel, tumor necrosis factor-alpha-regulated variant of HOXA9. J Biol Chem. 1999;274:1415–22. [DOI] [PubMed] [Google Scholar]
- 7.Tang L, Peng L, Tan C, Liu H, Chen P, Wang H. Role of HOXA9 in solid tumors: mechanistic insights and therapeutic potential. Cancer Cell Int. 2022;22:349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shenoy US, Adiga D, Alhedyan F, Kabekkodu SP, Radhakrishnan R. HOXA9 transcription factor is a double-edged sword: from development to cancer progression. Cancer Metastasis Rev. 2023;43:709–28. [DOI] [PMC free article] [PubMed]
- 9.Aryal S, Zhang Y, Wren S, Li C, Lu R. Molecular regulators of HOXA9 in acute myeloid leukemia. FEBS J. 2023;290:321–39. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe Y, Saito M, Saito K, Matsumoto Y, Kanke Y, Onozawa H, et al. Upregulated HOXA9 expression is associated with lymph node metastasis in colorectal cancer. Oncol Lett. 2018;15:2756–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osmond B, Facey COB, Zhang C, Boman BM. HOXA9 Overexpression contributes to stem cell overpopulation that drives development and growth of colorectal cancer. Int J Mol Sci. 2022;23:6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Bu J, Liu X, Wang W, Mai W, Lv B, et al. miR-133b suppresses metastasis by targeting HOXA9 in human colorectal cancer. Oncotarget. 2017;8:63935–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Z, Chen C, Zhou Q, Wang Y, Zhao Y, Zhao X, et al. LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. 2017;410:68–81. [DOI] [PubMed] [Google Scholar]
- 14.Malek R, Gajula RP, Williams RD, Nghiem B, Simons BW, Nugent K, et al. TWIST1-WDR5-Hottip regulates Hoxa9 chromatin to facilitate prostate cancer metastasis. Cancer Res. 2017;77:3181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z-F, Li G-R, Cao C-N, Xu Q, Wang G-D, Jiang X-F. MicroRNA-1294 targets HOXA9 and has a tumor suppressive role in osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22:8582–8. [DOI] [PubMed] [Google Scholar]
- 16.Qin K, Tian G, Chen G, Zhou D, Tang K. miR-647 inhibits glioma cell proliferation, colony formation and invasion by regulating HOXA9. J Gene Med. 2020;22:e3153. [DOI] [PubMed] [Google Scholar]
- 17.Costa BM, Smith JS, Chen Y, Chen J, Phillips HS, Aldape KD, et al. Reversing HOXA9 oncogene activation by PI3K inhibition: epigenetic mechanism and prognostic significance in human glioblastoma. Cancer Res. 2010;70:453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonçalves CS, Xavier-Magalhães A, Martins EP, Pinto AA, Pires MM, Pinheiro C, et al. A novel molecular link between HOXA9 and WNT6 in glioblastoma identifies a subgroup of patients with particular poor prognosis. Mol Oncol. 2020;14:1224–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han S, Li X, Liang X, Zhou L. HOXA9 transcriptionally promotes apoptosis and represses autophagy by targeting NF-κB in cutaneous squamous cell carcinoma. Cells. 2019;8:1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou L, Wang Y, Zhou M, Zhang Y, Wang P, Li X, et al. HOXA9 inhibits HIF-1α-mediated glycolysis through interacting with CRIP2 to repress cutaneous squamous cell carcinoma development. Nat Commun. 2018;9:1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alvarado-Ruiz L, Martinez-Silva MG, Torres-Reyes LA, Pina-Sanchez P, Ortiz-Lazareno P, Bravo-Cuellar A, et al. HOXA9 is underexpressed in cervical cancer cells and its restoration decreases proliferation, migration and expression of epithelial-to-mesenchymal transition genes. Asian Pac J Cancer Prev. 2016;17:1037–47. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert PM, Mouw JK, Unger MA, Lakins JN, Gbegnon MK, Clemmer VB, et al. HOXA9 regulates BRCA1 expression to modulate human breast tumor phenotype. J Clin Invest. 2010;120:1535–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun M, Song C-X, Huang H, Frankenberger CA, Sankarasharma D, Gomes S, et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc Natl Acad Sci U S A. 2013;110:9920–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang J-A, Bin LB, Kim Y, Hong S-H, Kim Y-H, Han J, et al. HOXA9 inhibits migration of lung cancer cells and its hypermethylation is associated with recurrence in non-small cell lung cancer. Mol Carcinog. 2015;54(Suppl 1):E72–80. [DOI] [PubMed] [Google Scholar]
- 25.Ooki A, Maleki Z, Tsay J-CJ, Goparaju C, Brait M, Turaga N, et al. A panel of novel detection and prognostic methylated DNA markers in primary non-small cell lung cancer and serum DNA. Clin Cancer Res an Off J Am Assoc Cancer Res. 2017;23:7141–52. [DOI] [PubMed] [Google Scholar]
- 26.Yu S-L, Lee DC, Sohn HA, Lee SY, Jeon HS, Lee JH, et al. Homeobox A9 directly targeted by miR-196b regulates aggressiveness through nuclear factor-kappa B activity in non-small cell lung cancer cells. Mol Carcinog. 2016;55:1915–26. [DOI] [PubMed] [Google Scholar]
- 27.Gao L, Yan S-B, Yang J, Kong J-L, Shi K, Ma F-C, et al. MiR-182-5p and its target HOXA9 in non-small cell lung cancer: a clinical and in-silico exploration with the combination of RT-qPCR, miRNA-seq and miRNA-chip. BMC Med Genom. 2020;13:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chong GO, Jeon H-S, Han HS, Son JW, Lee YH, Hong DG, et al. Overexpression of microRNA-196b accelerates invasiveness of cancer cells in recurrent epithelial ovarian cancer through regulation of homeobox A9. Cancer Genom Proteom. 2017;14:137–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitchen MO, Bryan RT, Haworth KE, Emes RD, Luscombe C, Gommersall L, et al. Methylation of HOXA9 and ISL1 predicts patient outcome in high-grade non-invasive bladder cancer. PLoS ONE. 2015;10:e0137003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia Z, Yang C, Yang X, Wu S, Feng Z, Qu L, et al. miR-652 Promotes proliferation and migration of uveal melanoma cells by targeting HOXA9. Med Sci Monit Int Med J Exp Clin Res. 2019;25:8722–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lissa D, Ishigame T, Noro R, Tucker MJ, Bliskovsky V, Shema S, et al. HOXA9 methylation and blood vessel invasion in FFPE tissues for prognostic stratification of stage I lung adenocarcinoma patients. Lung Cancer. 2018;122:151–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thul PJ, Lindskog C. The human protein atlas: a spatial map of the human proteome. Protein Sci. 2018;27:233–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol United States. 2020;38:675–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang D, Chen M, Huang X, Zhang G, Zeng L, Zhang G, et al. SRplot: A free online platform for data visualization and graphing. PLoS ONE. 2023;18:e0294236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nusinow DP, Szpyt J, Ghandi M, Rose CM, McDonald ER 3rd, Kalocsay M, et al. Quantitative proteomics of the cancer cell line encyclopedia. Cell. 2020;180:387-402.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park S-J, Yoon B-H, Kim S-K, Kim S-Y. GENT2: an updated gene expression database for normal and tumor tissues. BMC Med Genomics. 2019;12:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C-J, Hu F-F, Xie G-Y, Miao Y-R, Li X-W, Zeng Y, et al. GSCA: an integrated platform for gene set cancer analysis at genomic, pharmacogenomic and immunogenomic levels. Brief Bioinform. 2023;24:bbac5558. [DOI] [PubMed] [Google Scholar]
- 39.Huang H-Y, Lin Y-C-D, Li J, Huang K-Y, Shrestha S, Hong H-C, miRTarBase, et al. updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020;2020(48):D148–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J-H, Liu S, Zhou H, Qu L-H, Yang J-H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oughtred R, Rust J, Chang C, Breitkreutz B-J, Stark C, Willems A, et al. The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021;30:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yevshin I, Sharipov R, Valeev T, Kel A, Kolpakov F. GTRD: a database of transcription factor binding sites identified by ChIP-seq experiments. Nucleic Acids Res. 2017;45:D61–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liska O, Bohár B, Hidas A, Korcsmáros T, Papp B, Fazekas D, et al. TFLink: an integrated gateway to access transcription factor-target gene interactions for multiple species. Database (Oxford). 2022;2022:baac083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia (United States). 2017;19:649–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Győrffy B. Transcriptome-level discovery of survival-associated biomarkers and therapy targets in non-small-cell lung cancer. Br J Pharmacol. 2024;181:362–74. [DOI] [PubMed] [Google Scholar]
- 51.Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35:4200–2. [DOI] [PubMed] [Google Scholar]
- 52.Smith J, Sen S, Weeks RJ, Eccles MR, Chatterjee A. Promoter DNA hypermethylation and paradoxical gene activation. Trends Cancer. 2020;6:392–406. [DOI] [PubMed] [Google Scholar]
- 53.Ning S, Li H, Qiao K, Wang Q, Shen M, Kang Y, et al. Identification of long-term survival-associated gene in breast cancer. Aging (Albany NY). 2020;12:20332–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qian Y, Daza J, Itzel T, Betge J, Zhan T, Marmé F, et al. Prognostic cancer gene expression signatures: current status and challenges. Cells. 2021;10:648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deng J-L, Xu Y-H, Wang G. Identification of potential crucial genes and key pathways in breast cancer using bioinformatic analysis. Front Genet. 2019;10:695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y-C, Hsieh H-H, Chang H-C, Wang H-C, Lin W-J, Lin J-J. CDC25B induces cellular senescence and correlates with tumor suppression in a p53-dependent manner. J Biol Chem. 2021;296:100564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andersen LB, Mahler MSK, Andersen RF, Jensen LH, Raunkilde L. The clinical impact of methylated homeobox A9 ctDNA in patients with non-resectable biliary tract cancer treated with erlotinib and bevacizumab. Cancers (Basel). 2022;14:4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ju T, Jin H, Ying R, Xie Q, Zhou C, Gao D. Overexpression of NAC1 confers drug resistance via HOXA9 in colorectal carcinoma cells. Mol Med Rep. 2017;16:3194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou C, Li J, Li Q, Liu H, Ye D, Wu Z, et al. The clinical significance of HOXA9 promoter hypermethylation in head and neck squamous cell carcinoma. J Clin Lab Anal. 2019;33:e22873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guerrero-Preston R, Soudry E, Acero J, Orera M, Moreno-López L, Macía-Colón G, et al. NID2 and HOXA9 promoter hypermethylation as biomarkers for prevention and early detection in oral cavity squamous cell carcinoma tissues and saliva. Cancer Prev Res (Phila). 2011;4:1061–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cai H, Ke Z-B, Dong R-N, Chen H, Lin F, Zheng W-C, et al. The prognostic value of homeobox A9 (HOXA9) methylation in solid tumors: a systematic review and meta-analysis. Transl Cancer Res. 2021;10:4347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su J, Huang Y-H, Cui X, Wang X, Zhang X, Lei Y, et al. Homeobox oncogene activation by pan-cancer DNA hypermethylation. Genome Biol. 2018;19:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ye S, Xiong F, He X, Yuan Y, Li D, Ye D, et al. DNA hypermethylation-induced miR-182 silence targets BCL2 and HOXA9 to facilitate the self-renewal of leukemia stem cell, accelerate acute myeloid leukemia progression, and determine the sensitivity of BCL2 inhibitor venetoclax. Theranostics. 2023;13:77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rawat VPS, Götze M, Rasalkar A, Vegi NM, Ihme S, Thoene S, et al. The microRNA miR-196b acts as a tumor suppressor in Cdx2-driven acute myeloid leukemia. Haematologica Italy. 2020;105:e285–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng D-H, Wang X, Lu L-N, Chen D-L, Chen J-M, Lin F-M, et al. MiR-638 serves as a tumor suppressor by targeting HOXA9 in glioma. Eur Rev Med Pharmacol Sci. 2018;22:7798–806. [DOI] [PubMed] [Google Scholar]
- 66.Lawrence HJ, Rozenfeld S, Cruz C, Matsukuma K, Kwong A, Kömüves L, et al. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia. 1999;13:1993–9. [DOI] [PubMed] [Google Scholar]
- 67.Collins CT, Hess JL. Deregulation of the HOXA9/MEIS1 axis in acute leukemia. Curr Opin Hematol. 2016;23:354–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steger J, Füller E, Garcia-Cuellar M-P, Hetzner K, Slany RK. Insulin-like growth factor 1 is a direct HOXA9 target important for hematopoietic transformation. Leukemia. 2015;29:901–8. [DOI] [PubMed] [Google Scholar]
- 69.Bandyopadhyay S, Ashraf MZ, Daher P, Howe PH, DiCorleto PE. HOXA9 participates in the transcriptional activation of E-selectin in endothelial cells. Mol Cell Biol. 2007;27:4207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gwin K, Frank E, Bossou A, Medina KL. Hoxa9 regulates Flt3 in lymphohematopoietic progenitors. J Immunol. 2010;185:6572–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bei L, Lu Y, Eklund EA. HOXA9 activates transcription of the gene encoding gp91Phox during myeloid differentiation. J Biol Chem. 2005;280:12359–70. [DOI] [PubMed] [Google Scholar]
- 72.Bruhl T, Urbich C, Aicher D, Acker-Palmer A, Zeiher AM, Dimmeler S. Homeobox A9 transcriptionally regulates the EphB4 receptor to modulate endothelial cell migration and tube formation. Circ Res. 2004;94:743–51. [DOI] [PubMed] [Google Scholar]
- 73.Ko SY, Naora H. Adaptation of ovarian cancer cells to the peritoneal environment: Multiple mechanisms of the developmental patterning gene HOXA9. Cancer Cell Microenviron. 2014;1:e379. [PMC free article] [PubMed] [Google Scholar]
- 74.Usui A, Ko SY, Barengo N, Naora H. P-cadherin promotes ovarian cancer dissemination through tumor cell aggregation and tumor-peritoneum interactions. Mol Cancer Res. 2014;12:504–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alvarado-Ruíz L, Aguilar-Lemarroy A, Alejandro B, Jave SL. Determination of biological processes regulated by HOXA9 in cells derived from Cervical, Uterine and its influence on the immune response. Front Immunol. 2015;6.
- 76.Xavier-Magalhães A, Gonçalves CS, Fogli A, Lourenço T, Pojo M, Pereira B, et al. The long non-coding RNA HOTAIR is transcriptionally activated by HOXA9 and is an independent prognostic marker in patients with malignant glioma. Oncotarget. 2018;9:15740–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ko SY, Ladanyi A, Lengyel E, Naora H. Expression of the homeobox gene HOXA9 in ovarian cancer induces peritoneal macrophages to acquire an M2 tumor-promoting phenotype. Am J Pathol. 2014;184:271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ko SY, Naora H. HOXA9 promotes homotypic and heterotypic cell interactions that facilitate ovarian cancer dissemination via its induction of P-cadherin. Mol Cancer. 2014;13:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rössig L, Urbich C, Brühl T, Dernbach E, Heeschen C, Chavakis E, et al. Histone deacetylase activity is essential for the expression of HoxA9 and for endothelial commitment of progenitor cells. J Exp Med. 2005;201:1825–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Song Y-P, Xian P, Luo H, Dai J-Y, Bai Y, Li Y, et al. Comprehensive Landscape of HOXA2, HOXA9, and HOXA10 as Potential Biomarkers for Predicting Progression and Prognosis in Prostate Cancer. J Immunol Res. 2022;2022:5740971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ko SY, Barengo N, Ladanyi A, Lee J-S, Marini F, Lengyel E, et al. HOXA9 promotes ovarian cancer growth by stimulating cancer-associated fibroblasts. J Clin Invest. 2012;122:3603–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data retrieved and analyzed in this study are included in the manuscript, and additional information has been provided as supplementary files.
NA.