Abstract
Bacterial RecJ exhibits 5′→3′ exonuclease activity that is specific to ssDNA; however, archaeal RecJs show 5′ or 3′ exonuclease activity. The hyperthermophilic archaea Methanocaldococcus jannaschii encodes the 5′-exonuclease MjRecJ1 and the 3′-exonuclease MjRecJ2. In addition to nuclease activity, archaeal RecJ interacts with GINS, a structural subcomplex of the replicative DNA helicase complex. However, MjRecJ1 and MjRecJ2 do not interact with MjGINS. Here, we report the structural basis for the inability of the MjRecJ2 homologous dimer to interact with MjGINS and its efficient 3′ hydrolysis polarity for short dinucleotides. Based on the crystal structure of MjRecJ2, we propose that the interaction surface of the MjRecJ2 dimer overlaps the potential interaction surface for MjGINS and blocks the formation of the MjRecJ2-GINS complex. Exposing the interaction surface of the MjRecJ2 dimer restores its interaction with MjGINS. The cocrystal structures of MjRecJ2 with substrate dideoxynucleotides or product dCMP/CMP show that MjRecJ2 has a short substrate binding patch, which is perpendicular to the longer patch of bacterial RecJ. Our results provide new insights into the function and diversification of archaeal RecJ/Cdc45 proteins.
Keywords: archaeal RecJ, nuclease, GINS, crystal structure, protein complex
The RecJ/cell division cycle 45 (Cdc45) protein occurs broadly across the three domains of life (1). This family of proteins has multiple functions and might be evolved from a common ancestor. Bacterial and archaeal RecJs have seven conserved motifs that are key elements in substrate recognition and the subsequent catalytic reaction, and they participate in several DNA metabolic pathways, including DNA repair and chromosome replication (2, 3, 4). In eukaryotes, Cdc45 interacts with GINS and minichromosome maintenance (MCM) helicase to form the Cdc45-MCM-GINS (CMG) complex, the replicative DNA helicase. Similar to eukaryotic Cdc45, some archaeal RecJs also form a replicative helicase complex via interaction with archaeal GINS and MCM. To elucidate the function of Cdc45/RecJ in different organisms, the structure and function of prokaryotic RecJs and eukaryotic Cdc45 were solved and investigated.
Bacterial RecJs specifically hydrolyze ssDNA from the 5′ end and participate in three DNA repair pathways: homologous recombination, mismatch repair, and base excision repair (1). Bacterial RecJ degrades the DNA strand in coordination with the DNA helicase RecQ or UvrD and generates a long 3′ ssDNA for strand invasion in homologous recombination (2) or a long ssDNA gap for DNA resynthesis by DNA polymerase in mismatch repair (3). The 5′-dRPase activity of RecJ is necessary to promote short patch repair in base excision repair by removing the 5′-deoxyribose phosphate group (4). Generally, most bacterial RecJs feature an N-terminal catalytic core consisting of two domains, Asp-His-His (DHH) and Asp-His-His-associated (DHHA1), and a C-terminal oligonucleotide/oligosaccharide-binding (OB) domain that promotes exonuclease by binding ssDNA. In addition to the common domains, some bacterial RecJs, such as those of Thermus thermophilus and Deinococcus radiodurans, have an additional C-terminal domain (5, 6). The interaction between C-terminal domain IV and the HerA helicase of D. radiodurans RecJ (DraRecJ) can enhance the 5′→3′ nuclease activity by promoting ssDNA substrate binding ability (6). In some bacteria, RecJ is fused with a C-terminal ATPase domain or the RecQ peptide (7).
Many archaeal species, not including those from Crenarchaeota, also encode a RecJ nuclease, which generally degrades ssDNA in the 5′→3′ direction. Compared with bacterial RecJ, archaeal RecJ is a shortened enzyme that includes only the catalytic core and lacks the OB-fold domain. Furthermore, some species from Euryarchaeota, such as Methanocaldococcus jannaschii and Thermoplasma acidophilum, have two RecJs: RecJ1, a 5′→3′ exonuclease, and RecJ2, a 3′→5′ exonuclease. In summary, archaeal RecJs are classified into two groups: 5′ exonuclease and 3′ exonuclease on ssDNA (8, 9, 10, 11, 12). Some of them can also degrade ssRNA (11, 12). The function of 3′ exonuclease on RNA in vivo is not clear. It is suggested that this activity removes 3′-mismatched ribonucleotides from the RNA primers in chromosomal DNA replication or degrades diverse ssRNAs during RNA recycling or maturation (11).
Eukaryotic Cdc45, an essential replication initiation protein, shows structural similarity to the prokaryotic RecJ nuclease subfamily (13, 14, 15). Prokaryotic RecJs and human Cdc45 (hCdc45) all have the domains DHH and DHHA1 (5, 11, 14). Moreover, in comparison with the bacterial RecJ catalytic core (DHH and DHHA1 domains), an additional peptide consisting of approximately 100 amino acid residues is inserted just before the C terminus of the DHH domain of archaeal RecJ and eukaryotic Cdc45 proteins and forms a separate CMG interaction domain (CID) (11). Human Cdc45 also has a fourth domain that is disordered in its crystal structure. However, because Cdc45 has many mutations in the conserved motifs and residues that are essential for RecJ nuclease activity (13, 16), it does not show any nuclease activity and functions instead as a molecular wedge for DNA unwinding (16, 17). In summary, archaeal RecJ and eukaryotic Cdc45 exhibit similar overall topology and functional diversities, indicating their evolution from a common ancestor.
Cdc45 interacts with two subcomplexes, MCM helicase and GINS, to form a multisubunit complex, CMG, the replicative DNA helicase (14, 18). The crystal structure of hCdc45 showed that the CID domain bridges DHH and DHHA1 domains, and it mediates all of the MCM interactions and some of the GINS interactions, crucially in CMG assembly (14). Based on the crystal structure of hCdc45 and the cryo-EM structure of CMG, the following interaction mechanism of hCdc45 with other subunits in the CMG complex has been proposed (14, 19). The CID of hCDC45 interacts with the MCM2 and MCM5 subunits of the MCM helicase, exposing a GINS-interacting element in MCM5, which promotes duplex DNA unwinding (19).
Similar to eukaryotic Cdc45, in addition to nuclease activity, archaeal RecJ also interacts with GINS (11, 20). The nuclease activity of RecJ from Thermococcus kodakarensis was named GAN (GINS-associated nuclease), because it was stimulated by TkoGINS tetramer consisting of two GINS51 and two GINS23 subunits in vitro (20). Similar to Cdc45, archaeal RecJ also forms a multisubunit complex RecJ-MCM-GINS with its subcomplexes GINS and MCM helicase (20) to unwind dsDNA during chromosomal DNA replication (8, 21, 22, 23). The GAN-GINS complex stimulates the helicase activity of MCM, but MCM does not affect the nuclease activity of GAN in vitro (20). The replicative helicase complex without GAN may become unstable and ineffective during replication fork progression. Both GAN and eukaryotic Cdc45 have a CID domain that may be used to activate MCM (14). The C-terminal domain of GINS51 binds to the N-terminal domain of GAN, unlike eukaryotic Cdc45 (22).
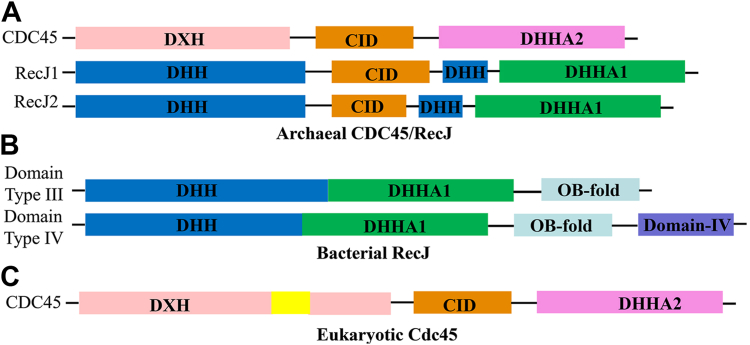
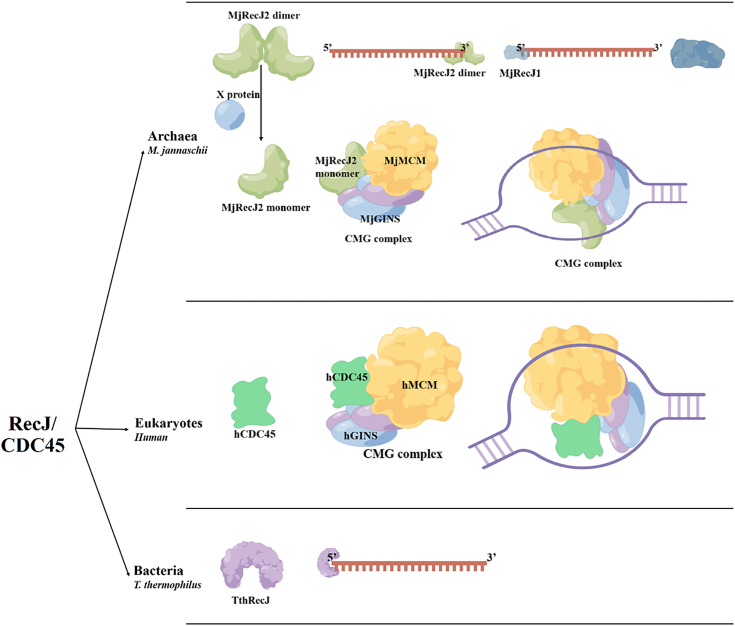
Archaeal and bacterial RecJs or Eukaryotic Cdc45 have clearly different amino acid sequences and domain organization (Fig. 1) (4, 5, 6, 7). Generally, bacterial RecJs hydrolyze ssDNA in the 5′ to 3′ direction (5, 6, 7). The hydrolytic mechanism of bacterial RecJ has been clearly elucidated based on the RecJ structures of T. thermophilus and D. radiodurans (1, 5, 24, 25). Archaeal RecJ nuclease can degrade ssDNA in direction of 5′→3′ or 3′→5′. M. jannaschii have two RecJs, namely MjRecJ1 (5′→3′ exonuclease) and MjRecJ2 (3′→5′ exonuclease). The active sites and GINS interaction surfaces of archaeal 5′-exonuclease RecJ have been identified based on the crystal structures of two archaeal 5′-exonuclease RecJs, PfuRecJ, and TkoGAN (11, 22). Currently, the structure of archaeal RecJ2 has not been demonstrated, which undoubtedly hinders to fully understand the catalytic mechanism of RecJ2. Besides, neither MjRecJ1 nor MjRecJ2 can interact with MjGINS to participate in the formation of CMG complexes (10). To understand the catalytic mechanism of archaeal RecJs in more detail, we solved the crystal structures of an archaeal RecJ with 3′-exonuclease activity, MjRecJ2, and its complex with a substrate dideoxynucleotides or product dCMP/CMP. Interestingly, unlike any previously reported RecJ structure, MjRecJ2 has a dimer topology: one subunit is in an open state, and the other is a closed one. In comparison with reported prokaryotic RecJs, MjRecJ2 has a similar catalytic mechanism and a different substrate binding mode. In other words, bacterial and archaeal RecJs use similar conserved residues for binding metal ions; however, MjRecJ2 has a short substrate binding patch that is perpendicular to that of bacterial RecJ. The results on MjRecJ2 provide the structural basis for its 3′-hydrolysis polarity. Interestingly, the interaction surface of the MjRecJ2 dimer overlaps the potential interaction surface for MjGINS and hinders the formation of the MjRecJ2-GINS complex. The GINS-interacting surface can be exposed by disrupting the dimer, allowing interaction with MjGINS and resulting in an MjRecJ2-GINS complex. Our results provide new insight into the function and evolution of the archaeal RecJ/eukaryotic Cdc45 protein in nucleic acid metabolism.
Figure 1.
Schematic figures of RecJ/CDC45 nucleases from bacteria, archaea, and eukaryotes.A, the schematic figure of archaeal CDC45 or RecJ. B, the schematic figures of Bacterial RecJ. C, the schematic figures of Eukaryotic CDC45. Cdc45, cell division cycle 45.
Results
MjRecJ1 and MjRecJ2 are DHH phosphodiesterases with opposite digestion directions
Some archaeal species, such as M. jannaschii DSM 2661, contain two RecJ nuclease genes (10). Although the enzyme activities of MjRecJ1 and MjRecJ2 were previously assayed (10), their preference to ssDNA and ssRNA substrates were again confirmed to check the repeatability of results between the two experiments. The results confirmed that both MjRecJs have nuclease activity. M. jannaschii RecJ1 (MjRecJ1) is a 5′ to 3′ nuclease, while RecJ2 (MjRecJ2) is a 3′ to 5′ nuclease (Fig. S1A). In addition, both MjRecJs are specific to the single-strand substrates and nearly do not digest dsDNA (Fig. S1B), suggesting that they are responsible for hydrolyzing single-stranded nucleic acids in vivo. Considering that some nucleases hydrolyze ssDNA and ssRNA at the same time, ssDNA and ssRNA are used as substrates to study their ribose dependence. MjRecJ1 can hydrolyze both ssDNA and ssRNA from the 5′ end, but it strongly prefers to ssDNA (Fig. S1C, left). However, MjRecJ2 favored ssRNA hydrolysis compared with the ssDNA substrate, implying that it has a slight ribose preference (Fig. S1C, right). These results are consistent with our previous enzyme activity assays (10). The two RecJ nucleases of M. jannaschii have classical domains, including the CID domain, DHH domain and DHHA1 domain (Fig. S2A). Although M. jannaschii RecJ1 and RecJ2 have the same domain composition and conserved motifs I-VII, including the DHH motif (motif III) and DHHA1 motif (motif VII), they show lower similarity (approximately 29%) to each other (Fig. S2B).
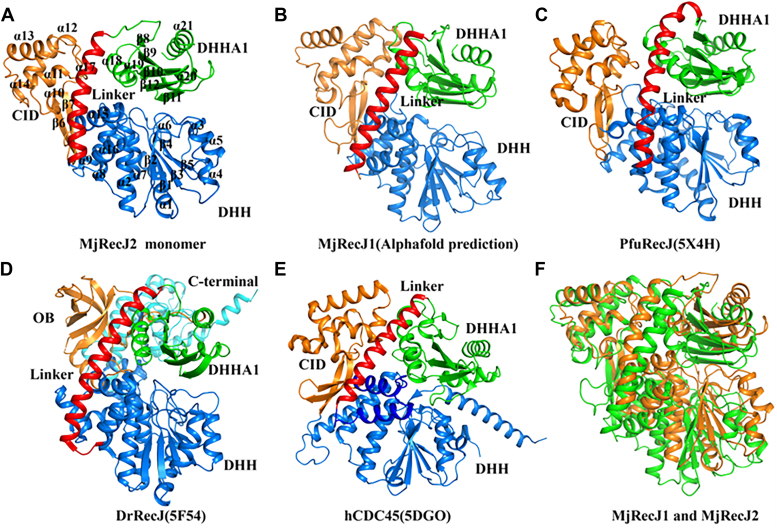
MjRecJ2 monomer adopts a topology similar to that of PfuRecJ and human CDC45
To investigate the functions of MjRecJ2, we determined and analyzed the crystal structure of MjRecJ2, which contained two protomers per asymmetric unit. Detailed structural parameters are shown in Table 1. It is in the dimerized state in the determined structure of MjRecJ2. To understand the structure of MjRecJ2 more clearly, the monomer of MjRecJ2 was extracted from its dimer structure (Fig. 2A). Each monomer is composed of an N-terminal DHH domain (residues 1–187), CID (residues 188–275), DHH domain (residues 276–302), and C-terminal DHHA1 domain (residues 327–429). The DHH domain contains 11 helices (α1-α9, α15, and α16) and 5 β-sheets (β1-β5). The CID domain consists of five helices (α10-α14) and 2 β-sheets (β6 and β7). The DHHA1 domain includes 4 helices (α18-α21) and 5 β sheets (β8-β12). The DHH domain (α16) is connected to the DHHA1 domain (α18) via a long linker (α17, residues 303–326). Although the CID is between the DHH and DHHA1 domains, it is actually inserted into the DHH domain as a separate part, which is very similar to the modeled MjRecJ1 (Fig. 2B) and PfuRecJ (Fig. 2C) (11). Besides, the structural arrangement of MjRecJ2 is similar to that of modeled MjRecJ1 (Fig. 2B), PfuRecJ (Fig. 2C) and human CDC45(Fig. 2E). Archaeal RecJs and eukaryotic Cdc45 lack the OB domain of DraRecJ. The structures of the two MjRecJs were superimposed, and their structural domains are basically the same (Fig. 2F).
Table 1.
Data collection and refinement statistics
| Parameter | MjRecJ2 | MjRecJ2-Mn2+ | MjRecJ2-CMP | MjRecJ2-dCMP | MjRecJ2-dApdA | MjGINS |
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Wavelength (Å) | 0.9792 | 0.9792 | 0.9792 | 0.9792 | 0.9792 | 0.9792 |
| Space group | P 21 21 2 | P 21 21 2 | P 21 21 2 | P 21 21 2 | P 21 21 2 | P 62 2 2 |
| Cell dimensions | ||||||
| a, b, c (Å) | 85.52, 177.82, 66.30 | 83.91, 175.82, 66.19 | 83.83, 175.63, 66.44 | 83.60, 175.17, 66.18 | 83.48, 175.40, 66.26 | 67.41, 67.41, 236.56 |
| β (°)b | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90, 90, 120 |
| Resolution (Å) | 29.64–2.19 (2.25–2.19) | 175.82–2.17 (2.23–2.17) | 175.63–2.26 (2.32–2.26) | 175.17–2.53 (2.60–2.53) | 175.4–2.04 (2.09–2.04) | 33.79–2.17 (2.23–2.17) |
| No. reflections | 52,386 | 52,541 | 46,416 | 33,171 | 62,817 | 17,808 |
| Rmerge (%)a | 8.4 (77.1) | 7.8 (61.1) | 7.4 (58.1) | 8.2 (57.9) | 13.8 (102.4) | 4.8 (154.4) |
| Mean I/σ(I)a | 22.2 (3.4) | 12.6 (2.8) | 12.1 (2.7) | 12 (2.6) | 12.1 (2.3) | 26.7 (2.5) |
| Completeness (%)a | 99.9 (87.9) | 99.7 (100) | 99.8 (100) | 99.8 (99.9) | 100 (99.9) | 99.7 (99.7) |
| Redundancya | 7.4 (7.3) | 4.8 (4.9) | 4.8 (4.9) | 4.8 (4.9) | 7.3 (7.4) | 35.9 (30.5) |
| Refinement | ||||||
| Rwork/Rfree (%)b | 19.86/24.95 | 20.63/24.06 | 20.38/25.09 | 21.18/25.00 | 18.41/23.24 | 22.06/24.85 |
| No. atoms | ||||||
| Protein | 7056 | 6919 | 6911 | 6955 | 7526 | 1650 |
| Water | 229 | 90 | 42 | 67 | 543 | 51 |
| Ligand | - | 2 | 2 | 4 | 9 | |
| R.M.S. Deviation | ||||||
| Bond lengths (Å) | 0.01 | 0.01 | 0.01 | 0.01 | 0.02 | 0.01 |
| Bond angles (°) | 0.94 | 0.9 | 0.92 | 1.1 | 1.73 | 0.83 |
| Ramachandran plot (%) | ||||||
| Favored | 95.09 | 95.44 | 95.68 | 96.85 | 97.56 | 97.25 |
| Allowed | 3.97 | 3.74 | 3.74 | 2.68 | 2.21 | 2.20 |
| Outliers | 0.93 | 0.82 | 0.58 | 0.47 | 0.23 | 0.55 |
The values in parentheses are for the outermost shell.
Rwork=∑hkl||Fobs|−|Fcalc||/∑hkl|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively. Rfree, calculated the same as Rwork, but from a test set containing 5% of data excluded from the refinement calculation. Rmerge=∑hkl∑i|Ii(hkl) – <I(hkl)>|/∑hkl∑i|Ii(hkl), where <I(hkl)> is the mean intensity of a set of equivalent reflections.
Figure 2.
Topology of MjRecJ2 and its similarity to RecJ/Cdc45. The overall topology of (A) MjRecJ2 monomer, (B) MjRecJ1, whose structure was folded using the AlphaFold program, The AlphaFold2 software was acquired from its repository on GitHub (https://github.com/deepmind/alphafold) and subsequently installed following the provided guidelines. Utilizing the prescribed algorithms, protein structure prediction was executed. Multiple sequence alignment (MSA) was conducted through HHblits, with the E-value parameter established at 1E-10. Sequences characterized by coverage levels below 70% and Gap values surpassing 75% in the MSA were subjected to exclusion. C, PfuRecJ (PDB 5X4H), (D) DraRecJ (5F54), and (E) hCdc45(5DGO). Various domains are displayed as follows: the DHH domain is shown in marine, the CID or OB domain in orange, the linker in red, the C-terminal domain in cyan and the DHHA1 domain in green. F, structure comparison of MjRecJ1 and MjRecJ2. MjRecJ1 is shown in green, and MjRecJ2 is shown in orange. Cdc45, cell division cycle 45; CID, CMG interaction domain; DHH, Asp-His-His; DHHA1, Asp-His-His-associated; OB, oligosaccharide-binding; PDB, Protein Data Bank.
To characterize the unique difference of MjRecJ2 from other archaeal RecJs, multiple sequence alignment of archaeal RecJs was performed. The results show that all archaeal RecJs are relatively conserved in the DHH domain (including motifs I-IV) and DHHA1 domain (containing motifs VI and VII); however, the sequence difference in motif V and the CID domain is obvious (Fig. S3), which also indicates that various archaeal RecJs differ in GINS interaction and hydrolysis direction.
To more clearly explain the evolutionary relationship between MjRecJ2 and prokaryotic RecJ and eukaryotic Cdc45, MjRecJ2 and several typical RecJ/Cdc45 proteins are structurally superimposed. The results show that the DHHA1 domains of MjRecJ2 and TkoRecJ are quite different (Fig. S4A), and the structural arrangement of MjRecJ2 is more similar to that of PfuRecJ (Fig. S4B). However, MjRecJ2 cannot interact with MjGINS, which may be related to the dimer form of MjRecJ2. In comparison with bacterial RecJs, most archaeal RecJs, including MjRecJ2, lost the OB domains of bacterial RecJs (Fig. S4, C and D). Although the secondary structure of MjRecJ2 is similar to that of hCDC45 and EhiCDC45, there are slight differences in the overall structural orientation (Fig. S4, E and F).
MjRecJ2 dimer hinders its interaction with MjGINS
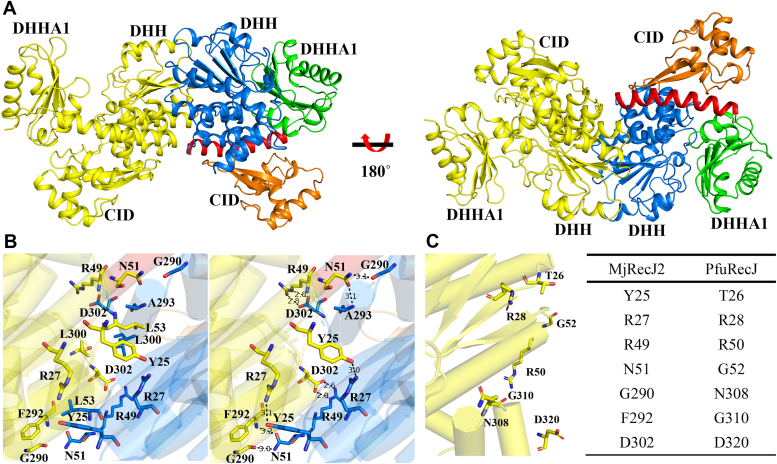
Since MjRecJ2 is a dimer (Fig. 3A), the amino acids that are potentially involved in dimer formation were analyzed. Four groups of hydrogen bond interactions, including Tyr25 and Arg27, Arg49 and Asp302, Asn51 and G290, and Asn51 and Ala293, are the main components for forming dimers (Fig. 3B). In addition, Arg49 and Asp302 and Asp302 and Lys45 can also form salt bridges between the two MjRecJ2 domains (Fig. 3B).
Figure 3.
The potential interaction model of MjRecJ2 and MjGINS.A, MjRecJ2 dimer conformation (rotated 180 degrees). One subunit is shown in yellow, and the other subunit is marked in the same color as in Figure 1A. B, MjRecJ2 dimer interaction interface analysis (left). Hydrogen bonding interactions at the MjRecJ2 dimer interface (right). The subunit is marked in the same color as in Figure 3A. Key interactional residues are shown in stick representation. C, potential PfuRecJ dimer interaction residues (left) and comparison of the potential dimer interaction residues of PfuRecJ and MjRecJ2 (right).
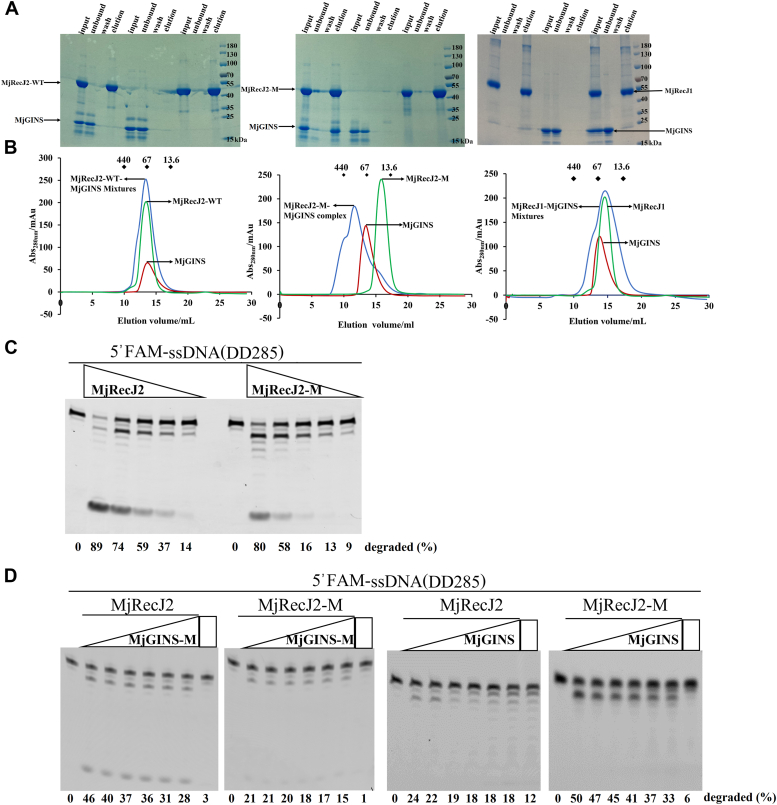
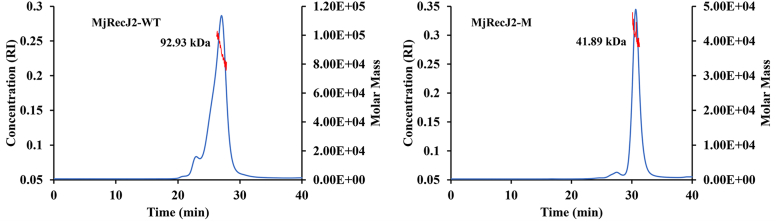
TkoRecJ nuclease (GAN) and TkoGINS form a complex through the DHH domain of RecJ and the C-terminal β-sheet domain of the GINS51 subunit (22). Pyrococcus furiosus RecJ and Thermoplasma acidophilum RecJ2 also form a complex with their GINS51 (11, 12). Interestingly, our previous gel filtration and pull-down experiments showed that neither MjRecJ1 nor MjRecJ2 formed a complex with MjGINS51 (10). Since both TkoGAN and PfuRecJ, which can interact with GINS, are monomeric forms, we speculated that the dimer interaction surface hides some key residues for interaction between MjRecJ2 and MjGINS. To further analyze the reason for the inability of the MjRecJ2 dimer to interact with MjGINS, we first compared the interactional residues of the MjRecJ2 dimer with those of PfuRecJ (Fig. 3C, left panel). Compared with MjRecJ2, the residues Thr26, Gly52, Asn308, and Gly310 in PfuRecJ are naturally mutated, preventing PfuRecJ from existing in the dimer form and interacting with GINS (Fig. 3C). Given that dimeric structure is a key factor in disrupting the potentially strong interaction between MjRecJ2 and MjGINS, we made a monomeric MjRecj2-M containing multiple site mutations, including Y25A, N51A, L53A, L300A, and D302A, based on dimeric interaction analysis, and purified the mutant protein. From the gel filtration results, the green curve on the left side of Figure 4B shows that the size of MjRecJ2 is 85 kDa, and the green curve in the middle of Figure 4B shows that the size of MjRecJ2-M is 42 kDa. These results indicate that MjRecJ-M is in monomeric form, whereas WT MjRecJ2 is in dimeric form. To further analyze the molecular weight of the WT and mutant MjRecJ2, we conducted size exclusion chromatography with multiangle static light scattering experiments. The results showed that WT MjRecJ2 was eluted as a single peak with an apparent molecular weight of 92.93 kDa, whereas MjRecJ2-M was 41.89 kDa, suggesting that WT MjRecJ2 is a dimer and MjRecJ2-M is a monomer in solution (Fig. 5). Copurification (Fig. 4A) and gel-filtration (Fig. 4B) experiments on MjRecj2-M and MjGINS confirmed that when the dimer is disrupted, the MjRecJ2 monomer can interact with MjGINS, implying that the dimeric form of MjRecJ2 is the main reason for its inability to form the MjRecJ2-GINS complex.
Figure 4.
Interaction of MjGINS with the MjRecJ2 monomeric mutant and its effect on the mutant’s nuclease activity.A, pull-down of MjGINS by each MjRecJ. His-Tag MjRecJ1, MjRecJ2 or MjRecJ2-M was mixed with non-His-tagged MjGINS (input) and loaded onto the Ni-NTA resin. The proteins that flowed out (unbound), the proteins that washed out (wash), and the proteins eluted by buffer containing 300 mM imidazole from resins (elution) were verified by 15% SDS‒PAGE. B, gel-filtration of each MjRecJ and MjGINS. The formation of mixtures between MjGINS and MjRecJ2-M was indicated by an earlier elution peak of proteins. MjRecJ1 and MjRecJ2 did not generate the earlier elution peak. C, nuclease activity comparison of dimer-disrupted MjRecJ2-M and MjRecJ2. Different concentrations (0, 300, 200, 150, 50, and 25 nM) of either MjRecJ2 or MjRecJ2-M were incubated with 200 nM 5′FAM-labeled ssDNA substrates at 55 °C for 20 min in the specified reaction buffer. The extent of substrate degradation was measured and is presented in the bottom of the panel. D, effects of MjGINS on the nuclease enzyme activity of MjRecJ2 and MjRecJ2-M. The increased amount (0, 5, 10, 15, 30, 50, 100, and 200 nM) of MjGINS-M or MjGINS were incubated with MjRecJ2 or MjRecJ2-M, and the enzyme activity was measured in the same conditions as Figure 3C. The extent of substrate degradation was measured and listed at the bottom of the panel. FAM, carboxyfluorescein.
Figure 5.
The determination of protein molecular weight using size exclusion chromatography coupled with multiangle light scattering (SEC-MALS). The molar masses calculated from the SEC-MALS analysis are represented by the red line. The graph also shows the Refractive Index (RI) signal, which correlates with the concentration of the protein in the eluate. By analyzing the intensity of scattered light at multiple angles, SEC-MALS provides an accurate measurement of the molecular weight, independent of the protein's shape and size in solution.
To investigate the effect of dimer disruption on its enzymatic activity, the nuclease activity of MjRecJ2-M was measured. Our results showed that the activity of the MjRecJ2-M mutant was reduced by 60 to 70% (Fig. 4C). Considering that MjRecJ2-M interacts with MjGINS, we assayed the effect of MjGINS on the exonuclease activity of the MjRecJ-M mutant. This result did not conform to our expectation that the activity of monomeric mutant MjRecJ2-M is promoted by MjGINS, despite their established interaction (Fig. 4, B and D).
MjGINS exhibits a tetramer state
Furthermore, we obtained crystals of MjGINS to further investigate the reason why MjRecJ2 cannot interact with MjGINS. Unfortunately, we only obtained the structure of part of MjGINS (domain A, residues 21–115) due to protein degradation during crystallization. The crystal structure shows that MjGINS domain A also exists as a tetramer (Fig. S5A), which is similar to that of intact T. kodakarensis GINS (TkoGINS) (26). Unfortunately, domain A is not the domain interacting with archaeal RecJs. Consequently, we predicted the structure of full-length MjGINS using AlphaFold software (Fig. S5B). The predicted full-length structure shows a long loop between structural domain A and structural domain B of MjGINS, which is the candidate region to degrade during its crystallization. To compare the differences between TkoGINS and our determined MjGINS, we extracted the A domain tetramer of TkoGINS containing 2 A structural domains of GINS51 and 2 A structural domains of GINS23 (Fig. S5C). Furthermore, we superimposed the A domain of monomeric MjGINS with TkoGINS51 (top panel) and TkoGINS23 (bottom panel). The superimposed results show that the A domain of monomeric MjGINS is very similar to the A domain of TkoGINS51 and TkoGINS23 (Fig. S5D).
The interaction interface of the tetramer is shown in Fig. S6, A and B. On the surface of the MjGINS dimer between subunits a and b, hydrogen bonds between Y26 and D48, K30 and D48, K30 and D52, R68 and D47, Y72 and D48, and Y72 and D52 and salt bridges between K30 and D48, K30 and D52, and R68 and D47 are the main interaction strengths. On the surface of the MjGINS dimer between subunits a and c, hydrogen bonds between R77 and A104, R77 and I105, and K58 and E111 and hydrophobic interactions between K74 and E106, K78 and Y86, and I101 and I105 are the main interaction strengths. We designed a dimeric mutant MjGINS-M by mutating the conserved interactional residues between subunits a and c, including K58A, K74A, K78A, I101A, and I105A. Gel-filtration experiments confirmed that these mutations disrupt the tetrameric form of MjGINS (Fig. S6C left, 73.36 kDa), resulting instead in a dimer (Fig. S6C right, 36.02 kDa). Dimeric MjGINS-M was mixed with dimeric MjRecJ2 and monomeric mutant MjRecJ2-M, and the potential interactions were detected by gel filtration chromatography assays. The results showed that dimeric MjGINS-M, similar to tetrameric WT MjGINS, interacts with monomeric MjRecJ-M (Fig. S6D right), not dimeric MjRecJ2 (Fig. S6D left), suggesting that dimeric MjRecJ2, not tetrameric MjGINS, inhibits the interaction of MjRecJ2 with MjGINS. Furthermore, we conducted a pull-down experiment to verify the interaction between MjGINS-M and either WT MjRecJ2 or MjRecJ2-M. The results confirmed that the MjRecJ2-M monomer can interact with MjGINS-M when the dimer is disrupted (Fig. S7A). However, the WT MjRecJ2 dimer does not interact with MjGINS-M (Fig. S7B), suggesting that the dimeric form of MjRecJ2, not the tetrameric MjGINS, hinders the interaction between MjGINS and MjRecJ2.
Divalent metal ion-binding sites of MjRecJ2
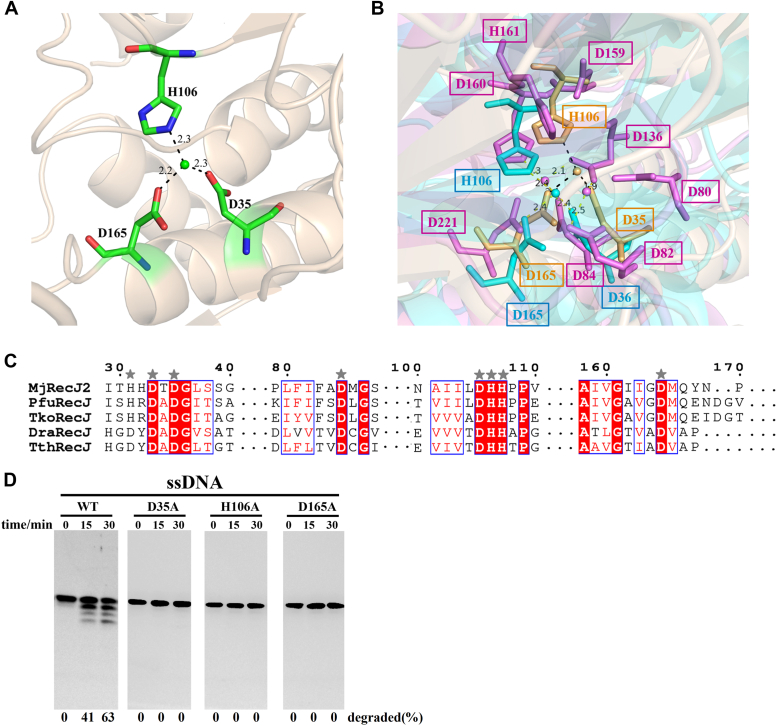
We determined the crystal structure of the MjRecJ2-Mn2+ complex through soaking divalent ion experiments. The structure of the MjRecJ-Mn2+ complex exists in dimer form similar to the apo structure. The three residues binding Mn2+ are Asp35, His106, and Asp165 (Fig. 6A), which are almost identical to the metal ion-binding sites of PfuRecJ (Asp35, His106, and Asp165). Although more residues are responsible for coordinating the catalytic metal ion in bacterial RecJ (DraRecJ), the same conserved amino acids as those of MjRecJ2 are found in its metal ion-binding site (Asp82, His161, Asp221, Fig. 6B). Multiple sequence alignment of secondary structures involved in binding metal ions also showed that these residues are well conserved in both archaea and bacteria (Fig. 6C). When the conserved residues binding divalent metal ions were mutated to alanine, all mutants of MjRecJ2 showed significantly reduced exonuclease activity on ssDNA substrates (Fig. 6D).
Figure 6.
Mechanism of phosphodiester bond hydrolysis catalyzed by MjRecJ2.A, structure and conserved residues of MjRecJ2 for binding Mn2+. B, comparison of the ion-binding center of MjRecJ2 (orange) with those of archaeal PfuRecJ (5X4I, blue) and bacterial DraRecJ (2XZP, violet). C, partial sequence alignment of MjRecJ2 with TkoRecJ, PfuRecJ, DraRecJ, and TthRecJ focused on the conserved residues responsible for ion binding. D, mutants of key conserved residues for binding divalent metal ions in MjRecJ2 lose their exonuclease activities. Different mutants of MjRecJ2 (100 nM) were incubated with 200 nM 5′FAM-labeled ssDNA substrates at 55 °C for the indicated time (0, 15, and 30 min) in the specified reaction buffer. The extent of substrate degradation was measured and listed at the bottom of each panel. FAM, carboxyfluorescein.
Structure of MjRecJ2 binding a substrate of dApdA
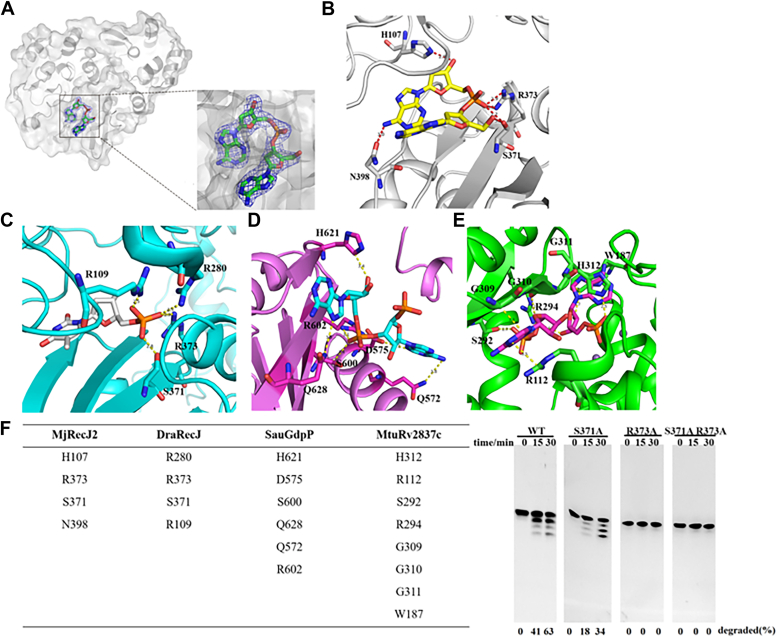
To investigate the catalytic mechanism of MjRecJ2 in degrading oligodeoxynucleotides, we determined the crystal structure of the MjRecJ2-dApdA complex, which is also a dimer structure. MjRecJ2 is preferable to shorter ssDNAs, so the deoxydinucleotide dApdA was used for cocrystallization. In the substrate binding pocket of MjRecJ2 (located between the DHH and DHHA1 domains), a clear view of the electron density of dApdA is observed (Fig. 7A). The two deoxyadenines exist in a near parallel state and are stabilized by interacting with MjRecJ2 (Fig. 7B). In the MjRecJ2-dApdA complex, His107 from the DHH domain forms a hydrogen bond with the hydroxyl group of dApdA. The middle phosphodiester bond of dApdA is firmly anchored by residues Ser371 and Arg373 from the DHHA1 domain. In addition, Asn398 from the DHHA1 domain is attached to the adenine base of dApdA via a hydrogen bond. These four residues form the basis of MjRecJ2 recognition and binding of ssDNA (Fig. 7B).
Figure 7.
Different substrate binding patches between RecJ and NrnA. Comparison of substrate binding patches of MjRecJ2 (A and B), bacterial DraRecJ (C, PDB ID 5XSP), bacterial nanoRNases GdpP from Staphylococcus aureus (D, PDB ID 5XSP) and Rv2837c from Mycobacterium tuberculosis (E, PDB ID 5JJU). F, summarized residues in the binding channel of MjRecJ2, DraRecJ, SauGdpP, and MtuRv2837c (left) and the exonuclease activity of MjRecJ2 and its mutation on the binding phosphodiester bond (right). Variants of MjRecJ2 (100 nM) were incubated with 200 nM 5′FAM-labeled ssDNA substrates at 55 °C for the specified time (0, 15, and 30 min) in reaction buffer. The extent of substrate degradation was measured and listed at the bottom of each panel. FAM, carboxyfluorescein; PDB, Protein Data Bank.
To better understand the difference in the catalytic mechanism between MjRecJ2 and bacterial RecJ, we analyzed the substrate-bound structure of bacterial DraRecJ (Fig. 7C) and two bacterial nanoRNases, which hydrolyze dinucleotides, GdpP from Staphylococcus aureus (Fig. 7D) and Rv2837c from Mycobacterium tuberculosis (Fig. 7E). In the structure of the DraRecJ-dTMP complex, Ser371 and Arg373 are responsible for binding dTMP in a way identical to that of MjRecJ2. Residues Arg109 and Arg280 of DraRecJ interact with the phosphate group of TMP in a way that differs from the interaction of MjRecJ2 with dApdA. In the structure of SauGdpP-pApA, residues Gln572, Asp575, Gln628, and His621 are involved in the interaction with pApA. Although His621 of SauGdpP forms a hydrogen bond with the hydroxyl group of pApA in a similar way to His107 in MjRecJ2, other residues interact with pApA in a way different from that of MjRecJ2. Gln572 forms a hydrogen bond with the first purine base of the substrate, and Asp575 forms a hydrogen bond with a bound water molecule. Gln628 forms a hydrogen bond with the other adenine base of the substrate. Ser600 and Arg602 form a hydrogen bond with the oxygen atom of the phosphate group. MtuRv2837c, the other NrnA, also interacts with pApA. His312 of MtuRv2837c forms a hydrogen bond with the internal phosphate group of pApA. Arg112 interacts with the adenine base of pApA. Ser292 and Arg294 form hydrogen bonds with the terminal phosphate group of pApA. His312 also interacts with the adenine base of pApA.
Similar residues in the binding channels of MjRecJ2, DraRecJ, SauGdpP, and MtuRv2837c were compared (Fig. 7F, left). It appears that the amino acids interacting with substrates in MjRecJ2 are relatively conserved in DraRecJ and less conserved in SauGdpP and MtuRv2837c. Based on the structural analysis of the MjRecJ2-dApdA complex, we performed single and double mutation of amino acids Ser371 and Arg373, which are involved in phosphodiester bond interactions. The R373A mutant was completely inactivated, suggesting that it plays a key role in the binding and catalysis of the substrate (Fig. 7F, right).
Structure of MjRecJ2 in complex with CMP and dCMP
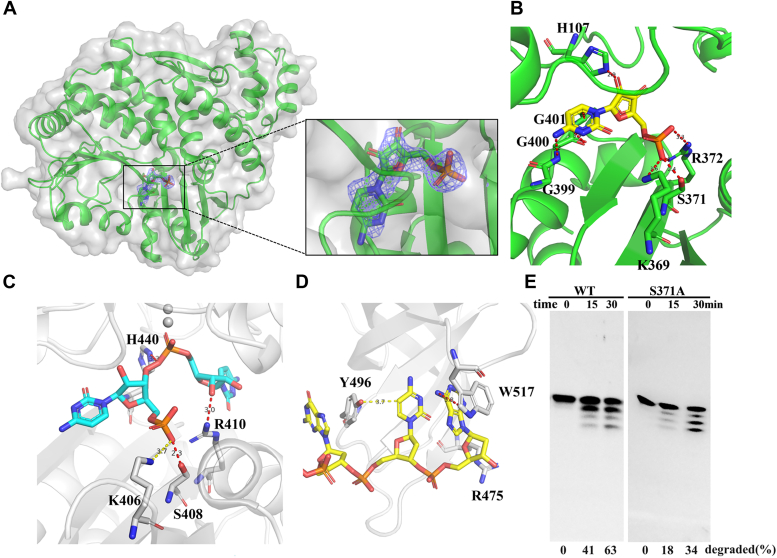
To elucidate the effect of the 5′-phosphate group on MjRecJ2, we determined the structures of two complexes, MjRecJ2-CMP and MjRecJ2-dCMP. In the substrate binding pocket of MjRecJ2, the electron density map of CMP is shown (Fig. 8A). CMP can interact with His107, Lys369, Ser371, Arg372, G399, G400, and G401 to form hydrogen bonds (Fig. 8B). Of these residues, Lys369, Ser371, and Arg372 form hydrogen bonds with the phosphate group of CMP, His107 forms hydrogen bonds with the ribose of CMP, and G399, G400, and G401 form interactions with the pyrimidine bases of CMP (Fig. 8B). The structure of the PfuRecJ and CMP complex shows that Lys406 and Ser408, conserved in MjRecJ2, interact with a phosphate group, His440 interacts with a ribose, and Arg410 interacts with the ribose of another CMP (Fig. 8C). In the DraRecJ-ssDNA complex structure, Arg475, Tyr496, and Trp517 interact with ssDNA, with Tyr496 interacting with the guanine base of ssDNA, but none of these three residues are conserved in MjRecJ2 (Fig. 8D). The activity of the Ser371A mutation decreased only slightly (Fig. 8E), indicating that the binding of the 5′-phosphate group by Ser371 has a minor effect on the degradation of short ssDNA.
Figure 8.
Comparison of the binding mode of the 5′-terminal phosphate group by MjRecJ2, PfuRecJ and DraRecJ.A, electron density maps of CMP. 2Fo-Fc(blue) electron density maps were contoured at 1 σ. B, the binding interaction of MjRecJ2 and the single nucleotide product CMP. Details of MjRecJ2 and CMP interaction are shown. The CMP is in stick representation (yellow), Amino acids of MjRecJ2 interacting with CMP are shown in green stick. C, CMP binding mode of PfuRecJ (PfuRecJ-D83A_Zn2+_CMP, PDB: 5X4J). Details of PfuRecJ and CMP interaction are shown. The CMP is in stick representation (cyan), Amino acids of PfuRecJ interacting with CMP are shown in gray stick. D, interaction of DraRecJ with 5′p-ssDNA viewed on the 5′ phosphate group (PDB: 5F55). Details of DraRecJ and CMP interaction are shown. The CMP is in stick representation (yellow), Amino acids of DraRecJ interacting with CMP are shown in gray stick. E, WT and S371A of MjRecJ2 (100 nM) were incubated with 200 nM 5′ FAM-labeled ssDNA substrates at 55 °C for the indicated time (0, 15, and 30 min) in the specified reaction buffer. The extent of substrate degradation was measured and listed at the bottom of each panel. Figure 8E is a partial reuse of Figure 7F, where we have placed the results of WT and the S371A mutation in order to confirm the role of Ser371 in binding the phosphodiester bond of dApdA. FAM, carboxyfluorescein; PDB, Protein Data Bank.
We also determined the complex structure of MjRecJ2 and dCMP. In the substrate binding pocket of MjRecJ2, the electron density map of dCMP is shown (Fig. S8A). Similar to CMP, residues Ser371 and Arg373 form hydrogen bonds with the phosphate group of dCMP (Fig. S8B). However, Asn398 forms a hydrogen bond with the cytosine base, which is different from the interaction shown in the structure of the MjRecJ2 and CMP complex (Figs. 8B and S8B).
Discussion
Bacterial RecJ nuclease specifically degrades ssDNA from the 5′-terminus during various DNA repair pathways. A homolog of archaeal RecJ, CDC45, exists in eukaryotes and unwinds chromosomal DNA in DNA replication by forming a complex with GINS and MCM. The functions and evolution of archaeal RecJ/Cdc45 are diverse and complex. Interestingly, some archaea encode not only 5′-3′ RecJ nucleases but also 3′-5′ RecJ nucleases, such as M. jannaschii. In addition, some archaeal species encode a homolog of the Cdc45 protein (11, 12, 20, 22). Some archaeal RecJs are also able to form a complex with GINS and MCM to participate in DNA replication (12, 20). However, in some archaea, such as M. jannaschii, their RecJs do not interact with GINS, and there is no Cdc45 protein. To further investigate the biological functions of archaeal RecJ in DNA repair and replication, we focused on RecJ2 from M. jannaschii and determined the crystal structures of MjRecJ2(apo), MjRecJ2-Mn2+, MjRecJ2-dApdA, MjRecJ2-CMP, and MjRecJ2-dCMP. Based on these structures and biochemical analyses, we elucidated the mechanism by which MjRecJ2 degrades single-stranded nucleic acids and interacts with MjGINS.
The activity assay showed that MjRecJ2 is a 3′-5′ nuclease (Fig. S1A) that cleaves ssDNA but not dsDNA (Fig. S1B). The preference of MjRecJ2 for ssRNA is not shared by MjRecJ1 (Fig. S1C), suggesting that they have different functions in M. jannaschii. The structure of MjRecJ2 has an additional sequence between motifs IV and V of the DHH domain (Fig. S2), which forms a separate structural domain, CID. Human CDC45 exhibits a structural fold more similar to that of MjRecJ2 (Fig. 2), indicating that they are derived from the same ancestor. CID is a separate domain in archaeal RecJ and eukaryotic CDC45 and is involved in subunit interactions for the formation of the DNA helicase complex CMG. The OB-fold domain of bacterial RecJ has a similar location to the CID domain in archaeal RecJ and eukaryotic Cdc45 (Figs. 2 and S4). Although they are in the same position, they have different functions, particularly their effect on nuclease activity. The bacterial OB-fold domain contributes greatly to enzyme activity by enhancing the ability of RecJ to bind single-stranded DNA. In addition, archaeal RecJs differ significantly in the motif V and CID structural domains (Fig. S3), implying that the CID domain is a major element in the functional diversification of archaeal RecJs. These functional diversities include ribose preference, hydrolysis polarity, and interaction with GINS.
In T. kodakarensis, the gene deletion of gan or fen1 does not show a significant effect on survival and growth at normal temperature, suggesting that the archaeal RecJ may not be essential for the CMG replication helicase (27). However, deletion of gan resulted in the death of T. kodakaraensis cells at high temperatures, probably because the MCM itself could not satisfy the rapid replication of DNA at high temperatures. This result also implies that archaeal RecJ can work synergistically with Fen1 nuclease to remove RNA primers during Okazaki fragment maturation. Similarly, MjRecJ1 and MjRecJ2 have been reported to complement the function of the deleted RecJ gene during DNA recombination repair in Escherichia coli. In these ways, the archaeal RecJs mainly function in DNA repair. Unlike the classical archaeal RecJ nucleases, the RecJdbh protein from Sulfolobus acidocaldarius, also known as archaeal Cdc45, lacks nuclease activity. In S. acidocaldarius, Cdc45 and two subcomplexes of GINS and MCM form the complex CMG, which may function as a replicative DNA helicase. It is inferred that in Archaea, except Crenarchaea, RecJ is not essential for DNA replication but mainly plays an important role in DNA repair, such as recombinational repair and removal of RNA primers. The function of CMG in Crenarchaea must be substantiated by further studies.
Although MjRecJ2 has a CID structural domain, it is unable to interact with GINS to form CMG complexes. MjRecJ2 has a topological structure similar to those of PfuRecJ and TkoRecJ, and the only difference between them is that MjRecJ2 exists as a dimer (Fig. 3A). This difference inspired us to examine the function of the MjRecJ2 dimer. Our speculation is that the dimeric interface of MjRecJ2 and the potential interaction interface of MjGINS overlap with each other, preventing the formation of a complex between MjGINS and MjRecJ2 (Fig. 3B). Subsequently, we disrupted the MjRecJ2 dimer by site-directed mutagenesis of interacting residues, and results of pull-down and gel filtration experiments of MjRecJ2-M and MjGINS supported our speculation (Fig. 4, A and B). Although MjRecJ2-M interacts with MjGINS, the enzymatic activity of monomeric MjRecJ2 is not stimulated by MjGINS, which is different from those of other archaeal RecJs interacting with GINS. This result indicates that MjGINS has little effect on the enzymatic activity of MjRecJ2 and implies that MjRecJ2 exerts its 3′-5′ exonuclease activity in a dimeric form. When M. jannaschii requires a replicative DNA helicase to unwind chromosomal DNA, an unidentified protein X changes dimeric MjRecJ2 into a monomer, and monomeric MjRecJ2 interacts with GINS and MCM to exert helicase activity (Fig. 9). In addition, the MjRecJ2 dimer interaction residues are not conserved in PfuRecJ, explaining how PfuRecJ can interact with PfuGINS as a monomer (Fig. 3B).
Figure 9.
The possible rolesof MjRecJ and its evolutionary derivatives in DNA replication and repair.
The crystal structures and catalytic mechanisms of three 5′-3′ RecJ exonucleases, archaeal PfuRecJ, and bacterial TthRecJ and DraRecJ, have been previously characterized. Here, we determined the crystal structure and preliminarily elucidated the catalytic mechanism of RecJ2 with 3′-5′ exonuclease activity. These crystal structures are from one apo MjRecJ2 structure and four complex structures, MjRecJ2-Mn2+, MjRecJ2-dApdA, MjRecJ2-CMP, and MjRecJ2-dCMP. In the structure of MjRecJ2-Mn2+, Asp35, His106, and Asp165 coordinate with Mn2+ (Fig. 6A), and the metal ion binding residues are also highly conserved in archaea and bacteria (Fig. 6, B and C). We identified the amino acids interacting with dApdA by analyzing the complex structure of MjRecJ2-dApdA. Among them, Ser371 and Arg373 can bind to the phosphate group of dApdA and correspond to the residues binding the phosphate group in the structure of DraRecJ-dTMP (Fig. 7). Asn398 stabilizes 5′-dApdA through the formation of hydrogen bonds with the adenine base of dApdA. In addition, the complex structures of MjRecJ2-(d)CMP also help to elucidate the role of the 5′-phosphate group in the hydrolysis reaction of the phosphodiester bond. The interaction of MjRecJ2 and (d)CMP is weaker, which is useful for faster release of the product (d)CMP.
Bacterial RecJs, such as TthRecJ, act mainly as 5′-3′ exonucleases, while eukaryotic CDC45 mainly participates in DNA replication by interacting with GINS and MCM to form a replicative helicase (CMG complex). Based on these results, we propose the possible functions of MjRecJ in DNA replication and repair (Fig. 9). First, MjRecJ2 functions as an exonuclease in its normal dimeric state to exert 3′-5′ exonuclease activity during DNA repair. MjRecJ1 also participates in the DNA repair process as an ssDNA-specific 5′-3′ exonuclease in a similar mode to bacterial RecJ nuclease. Second, MjRecJ2 participates in DNA unwinding as a subunit of replicative DNA helicase in DNA replication in a similar mode to the eukaryotic Cdc45 protein. Of course, since the MjRecJ dimer does not interact with GINS, it is plausible that there should be an unidentified protein X in M. jannaschii that converts the MjRecJ2 dimer into a monomer for the formation of the CMG complex. Another possibility is that the posttranslational modification may control the proposed monomer/dimer transition of MjRecJ2. All these possibilities will add some complexity to our proposed model, and will be further investigated. Overall, our results expand the diversity of functions and structures of RecJ nuclease family proteins and deepen the understanding of the evolutionary relationship between archaeal RecJ and eukaryotic Cdc45 protein, which is of great importance to DNA unwinding during DNA replication.
Experimental procedures
Materials
Nickel–nitrilotriacetic acid resin was purchased from Bio-Rad. The crystallization kits were purchased from Hampton Research and Molecular Dimensions Company. Oligodeoxyribonucleotides and oligoribonucleotides were synthesized by Sangon Biotech. The expression vectors pDEST17 and pET28-SUMO were used throughout this study. The E. coli strain DH5α was used in gene cloning, and the Rosetta 2(DE3)pLysS strain was used to express recombinant protein. All other chemicals and reagents were of analytical grade.
Preparation of M. jannaschii proteins
The genes encoding MjRecJ1, MjRecJ2 and GINS were amplified from M. jannaschii genomic DNA by PCR using their respective primers (Table S1) and then inserted into pDEST17 and pET28-sumo. Amino acid substitutions were introduced into RecJ2 with the QuikChange Site-Directed Mutagenesis Kit using PrimeSTAR DNA polymerase and the appropriate primers (Table S1). The genes encoding monomeric MjRecJ2-M containing the mutations of Y25A, N51A, L53A, L300A, and D302A and dimeric MjGINS-M containing the mutations of K58A, K74A, K78A, I101A, and I105A were synthesized by Sangon Biotech. Nucleotide sequences were confirmed by DNA sequencing. Recombinant plasmids were introduced into the Rosetta 2(DE3)pLysS strain of E. coli to express recombinant proteins. The expression of recombinant proteins was induced by 0.5 mM isopropylthio-β-galactoside at 20 °C for 16 h. The recombinant proteins were purified via immobilized Ni2+ affinity chromatography. The 6×His tag and SUMO domain were removed from MjGINS by treatment with UBL-specific protease 1 overnight at 4 °C. After verifying the purity of the eluate using 15% SDS‒PAGE, samples were dialyzed against a storage buffer (20 mM Tris–HCl, pH 8.0; 0.1 M NaCl, and 50% glycerol) and stored in small aliquots at −20 °C.
Purification and crystallization of MjRecJ2
To prepare MjRecJ2 for crystallization, it was overexpressed in the form of the fusion protein SUMO-MjRecJ2. After Ni-NTA affinity purification according to the above protocol, the SUMO tag was cut by ULPI protease, and then the His-tag-free MjRecJ was recovered by flowing through a Ni-NTA affinity column. The primarily purified MjRecJ2 was fully dialyzed against buffer A (20 mM Tris–HCl pH 8.0, 50 mM NaCl, 5 mM β-mercaptoethanol, and 5% glycerol) and loaded onto an anion exchange HiTrap SP-Sepharose HP column (GE HealthCare) preequilibrated with buffer A. MjRecJ2 was eluted with a linear gradient from 20 to 1000 mM NaCl. Fractions containing MjRecJ2 were pooled and concentrated using a 10-kDa Amicon Ultra15 centrifugal filter (Millipore) and purified further using a 120 ml Hiload Superdex 200 column (GE HealthCare) with a buffer of 20 mM Hepes (pH 7.0), 100 mM NaCl, 1 mM DTT, 0.1 mM EDTA, and 2% glycerol. The fractions containing MjRecJ2 were pooled and concentrated to 15 mg/ml for crystallization. Selenomethionine-labeled (SeMet) MjRecJ2 was expressed using the methionine auxotrophic E. coli strain B834 (DE3) in a defined medium and purified similarly to the native protein.
The crystals of MjRecJ2 were grown at 18 °C using the hanging drop vapor-diffusion method by mixing equal volumes of protein and reservoir solution. The reservoir solution contained 15% w/v PEG 400 and 50 mM Hepes, pH 7.6. After 1 week, crystals were harvested, then mounted, and flash-frozen in liquid nitrogen for diffraction tests and data collection. Divalent metal ions Mn2+ and Mg2+ were introduced by soaking the crystals in a metal ion solution for 12 h. Cocrystallization of MjRecJ2 with dinucleotides and a single (deoxy)nucleotide was carried out at 4 °C by soaking the MjRecJ2 crystals in the corresponding solution.
Structure determination and refinement
The crystals were stabilized and cryoprotected by the addition of a reservoir solution containing 20% glycerol and then flash cooled in liquid nitrogen. All X-ray diffraction data sets were collected at 100 K at BL17U1 of the Shanghai Synchrotron Radiation Facility. Indexing, integration, scaling, and merging of the diffraction data were performed by using the HKL2000 program suite (28).
The structure of SeMet-labeled MjRecJ2 was determined using single-wavelength anomalous dispersion. The initial structure was solved by the autoSHARP pipeline (29). Then, maximum likelihood-based refinement of the atomic positions and temperature factors was performed with Phenix (https://phenix-online.org/) (30, 31). The atomic model was fit with the program Coot (32, 33). The stereochemical quality of the final model was assessed with MolProbity (34). The data collection statistics and the refinement statistics of the MjRecJ structures are shown in Table 1. Figures were prepared with PyMOL (https://pymol.org/) (35).
Nuclease activity analysis
The exonuclease activities of MjRecJ1 and MjRecJ2 and their mutants were characterized using the synthesized oligoribonucleotides and oligodeoxyribonucleotides (Table S1, 200 nM) in reaction buffer of 20 mM Tris–HCl (pH 8.0), 50 mM NaCl, 1 mM DTT, 2.0 mM MnCl2, and 100 ng/μl bovine serum albumin (BSA) (MjRecJ1) and of 20 mM Tris–HCl (pH 8.5), 50 mM NaCl, 1 mM DTT, 1.0 mM MnCl2, and 100 ng/μl BSA (MjRecJ2 and its mutants) based on the previous results (10). After incubation for a specified time at 50 °C, an equal volume of stopping buffer (90% formamide, 100 mM EDTA, and 0.2% SDS) was added to the reaction. Subsequently, the reactions were subjected to 15% 8 M urea-denatured PAGE. After electrophoresis, images of the gels were scanned and quantitated using an FL9500 Fluorescent Scanner (GE HealthCare).
Characterization of the interaction between MjRecJ2 and MjGINS by copurification
Copurification was used to qualitatively identify the interactions of MjRecJ2 and MjGINS. During copurification, His-tagged WT or mutant MjRecJ2 was mixed with His-tag-free GINS at a molecular ratio of 1:2. The mixtures were incubated at 37 °C for 30 min to allow potential complex formation. Then, the incubated mixtures were subjected to immobilized Ni2+ affinity chromatography to copurify the possible MjGINS subunit.
Gel-filtration chromatography
MjRecJ2, MjGINS, and their mixtures (in a molecular ratio of 1:2) were run in a gel-filtration experiment to identify the possible complex in a buffer of 20 mM Tris–HCl pH 8.0, 1 mM DTT, and 150 mM NaCl, judged by their respective elution times. To determine its possible interaction with MjGINS, MjRecJ2 and its mutants were incubated with MjGINS for 2 h, and then MjRecJ2, its mutants, MjGINS, and the incubated mixtures were applied to the column for gel filtration. Gel-filtration chromatography was performed using a Superdex 200 Increase 10/300 GL column (GE HealthCare) in 20 mM Tris pH 8.0 and 100 mM NaCl for the analysis of the interaction of MjRecJ2 and its mutants with MjGINS. The elution profiles of the samples were monitored by UV absorption at 260 and 280 nm. In a specific application range, the elution volume of a sample corresponds to its molecular weight according to the equation: Log10(MW) = a∗(Velut-Vo)+b. Here, MW represents the molecular weight of protein, Velut is the elution volume, Vo is the dead volume, and a and b are coefficients. By utilizing standard proteins such as ferritin (440 kDa), BSA (67 kDa), and cytochrome C (13.6 kDa), a linear plot of lgMW-Velut was generated. The elution volume of each standard protein is denoted by a diamond shape on the plot for clarity.
Size exclusion chromatography with multiangle light scattering detection
The molecular weight measurements for MjRecJ2-WT and MjRecJ2-M dissolved in the solution were conducted using a multiangle light scattering detector (Wyatt Dawn Heleos-II), coupled with a size exclusion chromatography system featuring a Superdex 200 Increase 10/300 GL column. For this analysis, protein samples of approximately 2.5 mg/ml concentration, with a volume of 100 μl, were injected into a buffered solution that consisted of 25 mM Tris–HCl (pH of 8.0), 150 mM NaCl, and a 5% concentration of glycerin. The resulting data were then processed and analyzed using the ASTRA software (https://www.wyatt.com/products/software/astra.html) developed by Wyatt Technology, which provided the molecular weight estimates for the proteins (36, 37).
Data availability
Structure factors and coordinates have been deposited in the Protein Data Bank under accession codes7YIL, 7YIK, 7YOQ, 7YOR, 7YKV, and 7YOS for MjGINS-domain A, MjRecJ2(apo), MjRecJ2-CMP, MjRecJ2-dCMP, MjRecJ2-dApdA, and MjRecJ2-Mn2+ structures, respectively.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant Nos. 32170097 and U1832161), the Oceanic Interdisciplinary Program of Shanghai Jiao Tong University (Grant No SL2022MS018), and the National Key R&D Program of China (Grant Nos. 2018YFC0310704 and 2018YFC0309806).
Author contributions
W.-W. W., G.-S. Y., H. Z., Y.-X. Z., and F. Y. investigation; W.-W. W., G.-S. Y., H. Z., and Y.-X. Z., formal analysis; W.-W. W., G.-S. Y., H. Z., Q.-S. W., F. Y., and X.-P. L. conceptualization; W.-W. W., G.-S. Y., and H. Z., methodology; W.-W. W., G.-S. Y., Q.-S. W., and X.-P. L. writing–original draft; G.-S. Y., H. Z., J.-H. H., and X.-P. L. writing–review and editing; Q.-S. W., J.-H. H., F. Y., X. X., and X.-P. L. resources; Y.-X. Z. and F. Y. software; F. Y., X. X., and X.-P. L. supervision; F. Y. validation; X. X. and X.-P. L. project administration; X.-P. L. funding acquisition.
Reviewed by members of the JBC Editorial Board. Edited by Patrick Sung
Footnotes
Present address for Gang-Shun Yi: Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Roosevelt Drive, Oxford OX3 7BN, UK.
Supporting information
References
- 1.Wakamatsu T., Kim K., Uemura Y., Nakagawa N., Kuramitsu S., Masui R. Role of RecJ-like protein with 5′-3′ exonuclease activity in oligo(deoxy)nucleotide degradation. J. Biol. Chem. 2011;286:2807–2816. doi: 10.1074/jbc.M110.161596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thoms B., Borchers I., Wackernagel W. Effects of single-strand DNases ExoI, RecJ, ExoVII, and SbcCD on homologous recombination of recBCD+ strains of Escherichia coli and roles of SbcB15 and XonA2 exoI mutant enzymes. J. Bacteriol. 2008;190:179–192. doi: 10.1128/JB.01052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burdett V., Baitinger C., Viswanathan M., Lovett S.T., Modrich P. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6765–6770. doi: 10.1073/pnas.121183298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dianov G., Sedgwick B., Daly G., Olsson M., Lovett S., Lindahl T. Release of 5′-terminal deoxyribose-phosphate residues from incised abasic sites in DNA by the Escherichia coli RecJ protein. Nucleic Acids Res. 1994;22:993–998. doi: 10.1093/nar/22.6.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wakamatsu T., Kitamura Y., Kotera Y., Nakagawa N., Kuramitsu S., Masui R. Structure of RecJ exonuclease defines its specificity for single-stranded DNA. J. Biol. Chem. 2010;285:9762–9769. doi: 10.1074/jbc.M109.096487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng K., Zhao Y., Chen X., Li T., Wang L., Xu H., et al. A novel C-terminal domain of RecJ is critical for interaction with HerA in Deinococcus radiodurans. Front. Microbiol. 2015;6:1302. doi: 10.3389/fmicb.2015.01302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morimatsu K., Kowalczykowski S.C. RecQ helicase and RecJ nuclease provide complementary functions to resect DNA for homologous recombination. Proc. Natl. Acad. Sci. U. S. A. 2014;111:5133–5142. doi: 10.1073/pnas.1420009111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z., Pan M., Santangelo T.J., Chemnitz W., Yuan W., Edwards J.L., et al. A novel DNA nuclease is stimulated by association with the GINS complex. Nucleic Acids Res. 2011;39:6114–6123. doi: 10.1093/nar/gkr181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Y., Gristwood T., Hodgson B., Trinidad J.C., Albers S.V., Bell S.D. Archaeal orthologs of Cdc45 and GINS form a stable complex that stimulates the helicase activity of MCM. Proc. Natl. Acad. Sci. U. S. A. 2016;113:13390–13395. doi: 10.1073/pnas.1613825113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi G.S., Yang S., Wang W.W., Chen J.N., Wei D., Cao W., et al. Two Archaeal RecJ Nucleases from Methanocaldococcus jannaschii show reverse hydrolysis polarity: implication to their unique function in Archaea. Genes (Basel) 2017;8:211. doi: 10.3390/genes8090211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M.-J., Yi G.-S., Yu F., Zhou H., Chen J.-N., Xu C.-Y., et al. The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45. Nucleic Acids Res. 2017;45:12551–12564. doi: 10.1093/nar/gkx887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogino H., Ishino S., Kohda D., Ishino Y. The RecJ2 protein in the thermophilic archaeon Thermoplasma acidophilum is a 3′-5′ exonuclease that associates with a DNA replication complex. J. Biol. Chem. 2017;292:7921–7931. doi: 10.1074/jbc.M116.767921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez-Pulido L., Ponting C.P. Cdc45: the missing RecJ ortholog in eukaryotes? Bioinformatics. 2011;27:1885–1888. doi: 10.1093/bioinformatics/btr332. [DOI] [PubMed] [Google Scholar]
- 14.Simon A.C., Sannino V., Costanzo V., Pellegrini L. Structure of human Cdc45 and implications for CMG helicase function. Nat. Commun. 2016;7 doi: 10.1038/ncomms11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurniawan F., Shi K., Kurahashi K., Bielinsky A.-K., Aihara H. Crystal structure of Entamoeba histolytica Cdc45 suggests a conformational switch that may regulate DNA replication. iScience. 2018;3:102–109. doi: 10.1016/j.isci.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krastanova I., Sannino V., Amenitsch H., Gileadi O., Pisani F.M., Onesti S. Structural and functional insights into the DNA replication factor Cdc45 reveal an evolutionary relationship to the DHH family of phosphoesterases. J. Biol. Chem. 2012;287:4121–4128. doi: 10.1074/jbc.M111.285395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szambowska A., Tessmer I., Kursula P., Usskilat C., Prus P., Pospiech H., et al. DNA binding properties of human Cdc45 suggest a function as molecular wedge for DNA unwinding. Nucleic Acids Res. 2014;42:2308–2319. doi: 10.1093/nar/gkt1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gambus A., Jones R.C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R.D., et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 19.Abid Ali F., Renault L., Gannon J., Gahlon H.L., Kotecha A., Zhou J.C., et al. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat. Commun. 2016;7 doi: 10.1038/ncomms10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagata M., Ishino S., Yamagami T., Ogino H., Simons J.-R., Kanai T., et al. The Cdc45/RecJ-like protein forms a complex with GINS and MCM, and is important for DNA replication in Thermococcus kodakarensis. Nucleic Acids Res. 2017;45:10693–10705. doi: 10.1093/nar/gkx740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marinsek N., Barry E.R., Makarova K.S., Dionne I., Bell S.D. GINS, a central nexus in the archaeal DNA replication fork. EMBO J. 2006;7:539–545. doi: 10.1038/sj.embor.7400649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oyama T., Ishino S., Shirai T., Yamagami T., Nagata M., Ogino H., et al. Atomic structure of an archaeal GAN suggests its dual roles as an exonuclease in DNA repair and a CMG component in DNA replication. Nucleic Acids Res. 2016;44:9505–9517. doi: 10.1093/nar/gkw789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makarova K.S., Koonin E.V., Kelman Z. The CMG (CDC45/RecJ, MCM, GINS) complex is a conserved component of the DNA replication system in all archaea and eukaryotes. Biol. Direct. 2012;7:1–10. doi: 10.1186/1745-6150-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng K., Xu H., Chen X., Wang L., Tian B., Zhao Y., et al. Structural basis for DNA 5′-end resection by RecJ. Elife. 2016;5 doi: 10.7554/eLife.14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng K., Xu Y., Chen X., Lu H., He Y., Wang L., et al. Participation of RecJ in the base excision repair pathway of Deinococcus radiodurans. Nucleic Acids Res. 2020;48:9859–9871. doi: 10.1093/nar/gkaa714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oyama T., Ishino S., Fujino S., Ogino H., Shirai T., Mayanagi K., et al. Architectures of archaeal GINS complexes, essential DNA replication initiation factors. BMC Biol. 2011;9:1–12. doi: 10.1186/1741-7007-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burkhart B.W., Cubonova L., Heider M.R., Kelman Z., Reeve J.N., Santangelo T.J. The GAN exonuclease or the flap endonuclease Fen1 and RNase HII are necessary for viability of Thermococcus kodakarensis. J. Bacteriol. 2017;199 doi: 10.1128/JB.00141-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:276307–276326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Bricogne G., Vonrhein C., Flensburg C., Schiltz M. PaciorekGeneration, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 2023;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 30.Adams P.D., Grosse-Kunstleve R.W., Li W.H., Ioerger T.R., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 31.Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D. Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeLano W.L., The PyMOL Molecular Graphics System, Schroedinger, LLC; New York, NY
- 36.Wyatt P.J. Light scattering and the absolute characterization of macromolecules. Anal. Chim. Acta. 1993;272:1–40. [Google Scholar]
- 37.Andersson M., Wittgren B., Wahlund K.-G. Accuracy in Multiangle light scattering measurements for molar Mass and Radius Estimations. Model calculations and experiments. Anal. Chem. 2003;75:4279–4291. doi: 10.1021/ac030128+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structure factors and coordinates have been deposited in the Protein Data Bank under accession codes7YIL, 7YIK, 7YOQ, 7YOR, 7YKV, and 7YOS for MjGINS-domain A, MjRecJ2(apo), MjRecJ2-CMP, MjRecJ2-dCMP, MjRecJ2-dApdA, and MjRecJ2-Mn2+ structures, respectively.