Summary
Orofacial clefts (OFCs) are among the most common human congenital birth defects. Previous multiethnic studies have identified dozens of associated loci for both cleft lip with or without cleft palate (CL/P) and cleft palate alone (CP). Although several nearby genes have been highlighted, the “casual” variants are largely unknown. Here, we developed DeepFace, a convolutional neural network model, to assess the functional impact of variants by SNP activity difference (SAD) scores. The DeepFace model is trained with 204 epigenomic assays from crucial human embryonic craniofacial developmental stages of post-conception week (pcw) 4 to pcw 10. The Pearson correlation coefficient between the predicted and actual values for 12 epigenetic features achieved a median range of 0.50–0.83. Specifically, our model revealed that SNPs significantly associated with OFCs tended to exhibit higher SAD scores across various variant categories compared to less related groups, indicating a context-specific impact of OFC-related SNPs. Notably, we identified six SNPs with a significant linear relationship to SAD scores throughout developmental progression, suggesting that these SNPs could play a temporal regulatory role. Furthermore, our cell-type specificity analysis pinpointed the trophoblast cell as having the highest enrichment of risk signals associated with OFCs. Overall, DeepFace can harness distal regulatory signals from extensive epigenomic assays, offering new perspectives for prioritizing OFC variants using contextualized functional genomic features. We expect DeepFace to be instrumental in accessing and predicting the regulatory roles of variants associated with OFCs, and the model can be extended to study other complex diseases or traits.
Keywords: human embryonic craniofacial development, orofacial clefts, convolutional neural network, genome-wide association studies, variant function, epigenomic assay, noncoding variant, SNP activity difference prediction
We developed DeepFace, a convolutional neural network model, to assess the functional impact of variants by SNP activity difference scores. The DeepFace model was trained with 204 epigenomic assays from human embryonic craniofacial developmental stages. It offers insights into the function alteration of craniofacial variants, particularly those that are noncoding.
Introduction
Nonsyndromic orofacial clefts (OFCs) are among some of the most common human birth defects, occurring in 1 in 700 live births worldwide.1 OFCs occur in various forms, including cleft lip alone (CL), cleft palate alone (CP), and a combination of both (CLP), with a spectrum of severity in each case.1,2 Nonsyndromic OFCs arise without accompanying major cognitive or structural abnormalities and exhibit complex etiology. This complexity is due to the interplay of multiple genetic and environmental risk factors contributing to their development.
In recent years, multiple genome-wide association studies (GWASs) have successfully depicted the genetic architecture of OFCs in multiethnic populations.3,4,5,6,7,8,9,10,11,12,13 Although dozens of loci have been identified through GWASs, most genetic discoveries are situated within noncoding and regions of linkage disequilibrium.14 Consequently, delineating the regulatory roles of these associated variants necessitates comprehensive functional genomics data to accurately interpret their biological mechanisms.15
During recent years, large-scale experimental mapping of epigenomic modification assays have been conducted by several large consortia, including the Encyclopedia of DNA Elements (ENCODE)16 and the Roadmap Epigenomics Project,17 which provide insights for annotating the function of noncoding variants by considering their overlap with regulatory elements in related contexts (tissue, cell type, and developmental stage).18,19 Furthermore, convolutional neural network (CNN) models have been recognized as a robust approach for investigating regulatory motifs within the genomic context. They are specifically designed to capture high-level information from long sequences, offering valuable insights into the complex patterns of genomic regulation.20 Currently, many CNN-based frameworks have been implemented to access the function of noncoding variants, such as DeepSEA,21 Basenji,22 ExPecto,23 and our previous work, DeepFun.24,25 These CNN models provide a computational assessment of the regulatory effects resulting from genomic variations by detecting disruptions or creations of regulatory motifs identified through convolutional filters. Consequently, they enable the downstream prediction of chromatin accessibility and regulatory modifications.21,22 However, these current methodologies predominantly focus on proximal sequences adjacent to risk variants, neglecting the potential for cis-regulatory elements to engage in looping interactions extending up to one million base pairs away.26,27 Moreover, the epigenomic regulation of embryonic craniofacial development is highly context specific.28 None of the current methods have trained a craniofacial development model. Therefore, predictions based on the noncontextual model will not reflect the dynamic epigenomic signals during craniofacial development.
To address these challenges, we obtained 204 human craniofacial epigenomic arrays, including datasets across six craniofacial developmental stages and 12 epigenetic indicators for enhancers, promoters, and gene bodies. These chromatin feature annotations could complement the modeling of the epigenomic map in craniofacial development. Moreover, we trained a deep learning model specifically for cleft development, DeepFace, to learn the epigenetic feature association with the long-range DNA sequence feature. Therefore, DeepFace predicts the impact of variants on DNA sequence, enabling us to understand how alterations in the DNA sequence influence epigenetic modifications. Next, we applied the DeepFace model to systematically assess curated CP and cleft lip with or without cleft palate (CL/P) risk variants. Then, we characterized variants with the largest accessibility alteration and the development-specific stage of variants. We anticipated that the CNN model on dense epigenomic maps would be a valuable approach for both gene-regulatory studies and disease studies seeking to elucidate the molecular basis of OFCs.
Material and methods
Primary chromatin feature collection and processing
The 204 chromatin immunoprecipitation of post-translational epigenetic modifications coupled with next-generation sequencing (ChIP-seq) data were collected from human embryonic craniofacial tissues28 and downloaded from the Gene Expression Omnibus (GEO) (accessed on June 8, 2021, GEO: GSE97752).
Briefly, these 204 assays were extracted from 17 individual human embryos during a crucial developmental period. This period encompasses the formation of the human orofacial apparatus, spanning Carnegie stages (CSs) from post-conception weeks (pcw) 4 to pcw 10, including stages CS13, CS14, CS15, CS17, CS20, and F2.29 For each sample, 11 post-translational histone modifications28 were profiled, including the repressive marks (H3K27me3 and H3K9me3), promoter activation marks (H3K4me3 and H3K9ac), transcription regulation marks (H3K36me3, H4K20me1,30,31 and H3K79me232), active regulatory marks (H3K4me1, H3K4me2, H3K27ac, and H2A.Z), and open chromatin signal DNase. Then, we extracted nonoverlapping sequences across the chromosomes, each spanning approximately 131,072 bp (∼131 kb) as the segment length of the input. Sequences with more than 35% unmappable content were discarded, collectively covering approximately 81% of the human genome. Each epigenomic data in bigWig format was further converted and split into these segments, resulting in 14,990 segments for training, 1,805 segments for validation, and 1,798 segments for testing, based on a distribution scheme across various samples in an 8:1:1 ratio.
Curation for OFC-significant variants
We collected a diverse set of orofacial variants from the GWAS Catalog33 (accessed May 16, 2021) using the keyword “oral cleft,” resulting in 306 variants with p value <1 × 10−5 from 33 studies (Table S1). These variants included a total number of 234 unique SNPs. We further obtained two multiethnic craniofacial GWAS datasets for CL/P7,34 and CP13 (available from dbGaP: phs000884.v2.p1). We selected the SNP with at least one SNP with nominal significance p value <1 × 10−5. Our craniofacial-sign (OFC-sign) dataset has 1,787 SNPs in total.
Curation for control variants
From previous studies,35,36,37,38 we observed that trait-related variants tended to manifest their effect in the trait-related tissues. We investigated whether OFC-significant variants exhibit higher absolute SNP activity difference (SAD) scores than variants from unrelated traits using control variant collections equal in size to the OFC-significant collection from two resources: (1) nonsignificant craniofacial development variants and (2) irrelevant trait variant collections. The first variant collection was obtained from the aforementioned two GWAS datasets (CL/P34 and CP only [CPO]13). The OFC-low group was defined as randomly sampled variants with p value >0.5 in both GWAS datasets. The OFC-medium group was defined as randomly sampled variants with p values ranging from 1 × 10−5 to 0.5 in both GWAS datasets. The second irrelevant trait variant was chosen from two GWAS summary statistics datasets: neurodegenerative disease Alzheimer disease39 (AD) and psychiatric disorder schizophrenia40 (SCZ), each with p value <1 × 10−5 significance (AD-sign and SCZ-sign). We further employed random downsampling to ensure that all control datasets contained the same number of SNPs as the OFC-sign collection, thereby enhancing comparability. All the control variant groups were randomly sampled to match the size of the OFC-sign group.
Variant annotation
SNPs were annotated with the ANNOVAR function “table_annovar.pl” (v.4/16/2018) with hg19 reference genome and dbsnp150 version annotation.41 The function of the SNPs was annotated and merged into the following categories: exon (variant overlaps a coding), intergenic (variant is in intergenic region), intronic (variant overlaps an intron), ncRNA_exonic (variant overlaps a transcript coding region without coding annotation in the gene definition), ncRNA_intronic (variant overlaps a transcript intron region without coding annotation in the gene definition), upstream/downstream (variant overlaps 1-kb region upstream or downstream of transcription start site), and untranslated region (UTR) 3′/UTR 5′ (variant overlaps a 3′ or 5′ UTR).
Training the CNN model and model performance evaluation
We utilized the CNN framework Basenji22 and our in-house DeepFun24 to train the 204 epigenomic assays. The CNN architecture consists of seven dilated convolution layers with max pooling (in windows of two, four, four, and four) to obtain representations that describe 128-bp bin size, aligning with the 146-bp distance between nucleosome core particles.22 This design allows information sharing across distal regulatory interactions (128 × 27 × 2 = 32,000). We applied seven layers of dilated convolutions to encompass these 128-bp bin representations, transforming every sequence feature (131,000) and the epigenetic signals into a length of 1,024-bp subsequence representations. Our previous work24 has demonstrated that training on the complete features from the ENCODE dataset42 outperforms the training on individual features. Therefore, we trained these sequence features across all 204 chromatin assays. We evaluated the performance on the validation and testing sets based on the Pearson’s correlation coefficient (r) of predicted and real epigenetics features. Each assay’s predicted epigenomic intensity was computed individually. Lastly, we fine-tuned the hyperparameters, learning rate, and batch size and stopped training when there was no reduction in r in the validation set loss over 15 consecutive epochs.
To evaluate the peak binary classification, we followed the Basenji22 model to evaluate the peak binary classification comparison with one well-known method, Model-based Analysis of ChIP-Seq (MACS2).43 We transformed the training and testing datasets to binary peak calls on shorter sequences. Each 131,000 (∼1,024 binned subsequence × 128 bps/bin) sequence was segmented into subsequences of 1,024 bin features, with each subsequence encapsulating a 128-bp binned representative of the functional element. The aim of this deep learning model is to accurately predict the read coverage in 128-bp bins. We identified peaks within the central 256 bps of the subsequences for each dataset by applying a Poisson model to the smoothed, normalized counts. This model was parameterized by the higher value of a global and local null lambda, akin to the MACS2 methodology. We then established a 0.01 false discovery rate (FDR) cutoff to define the ground truth. The area under the precision-recall curve (AUPRC) was used to measure the model performance of prediction. More details about the model can be found in the Basenji model.22
SAD score
The DeepFace model is crafted to forecast the functional impacts of sequence alterations at a single-nucleotide resolution. For each variant, DeepFace considers contextual information within a 1,024-bp subsequence transformed from a 131-kb sequence, predicting the epigenomic activity probability for sequences containing the reference allele or alternative allele. In this context, activity denotes the binding affinity for DNase-seq or histone modifications, respectively. To assess the variant’s impact, we employed the SAD, , where and are from the predicted matrices, to represent the predicted SNP activity for the alternative allele and the reference allele sequence, respectively. An elevated positive SAD score for genetic variants denotes that the alternative allele augments the epigenetic signal in comparison to the reference allele. Conversely, a negative SAD value denotes a diminution of the epigenetic signal. Notwithstanding the collective training of DeepFace models utilizing an extensive dataset, the functional score predicted for each variant is distinct and autonomous.
Motif mapping and visualization
We used the R package “atSNP” to search the potential transcription factor (TF) binding motif of variants in the JASPAR44 and ENCODE42 motif databases. We utilized the “ComputePValues” function within the atSNP toolkit to calculate the p values for all potential motifs. We identified significant motifs as those with Benjamini-Hochberg procedure-adjusted p values <0.05 in either the JASPAR or ENCODE database. Additionally, we employed the “plotMotifMatch” function from the atSNP package to visualize the motif pattern of the significant SNPs.
Cell-type specificity analysis of OFC-sign SNP set
Considering the epigenomic data of DeepFace are limited to the tissue of embryonic craniofacial development, we used two in-house methods, web-based cell-type-specific enrichment analysis of genes (WebCSEA)37 and DeepFun24,25 to contextualize the most relevant cell types of OFC-sign genes. WebCSEA (https://bioinfo.uth.edu/webcsea/) curated a total of 111 single-cell RNA-seq panels of human tissues and 1,355 tissue cell types from 61 different general tissues across 11 human organ systems and used the decoding tissue specificity algorithm35 to measure the enrichment for each cell type.37 We input the most nearby genes of the OFC-sign SNP set and visualized the most enriched cell type with a nominal significance of 1 × 10−3.
The DeepFun web server (https://bioinfo.uth.edu/deepfun/) leverages a CNN architecture trained on approximately 8,000 chromatin feature assays from 225 distinct tissues or cell types from the ENCODE and Roadmap projects. We input all OFC-sign SNPs to the DeepFun web server to assess the SAD scores, which is the normalized version (range from −1 to 1) of SAD used in this study. For every SNP in each cell type, we calculated the mean absolute SAD and then identified the cell types with the highest absolute SAD values across the OFC-sign SNPs. The top count of cell types was defined as the cell type most related to the OFC-sign SNP set.
Results
Narrow peak epigenetic chromatin features had better prediction than broad peak features
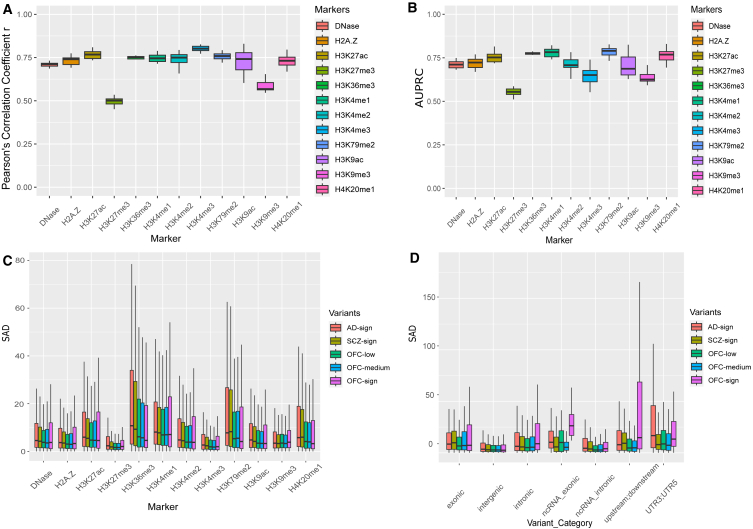
Following the DeepFace design in Figure 1, the trained 204 chromatin feature assays were evaluated for the prediction performance on the r of predicted and real continuous epigenetics features (median ranging from 0.50 to 0.83) and the AUPRC of the binary predicted peak and real peak called by MACS (ranging from 0.54 to 0.81). As shown in Figures 2A and 2B, both continuous and binary epigenomic features shared the same trend over the chromatin features. Specifically, H3K4me3 and H3K79me2 are on the top in Pearson’s r and AUPRC values, respectively. The two broad repressive marks, H3K27me3 and H3K9me3, have the lowest medium performance across the samples over development stages, suggesting that the narrow peak histone modification features tend to have a more accurate prediction than the broad histone modification features.24
Figure 1.
Overview of DeepFace workflow
DeepFace is a dilated convolutional neural network (CNN) framework to contextualize the function of common orofacial cleft (OFC) variants trained from 204 human embryonic craniofacial epigenomic arrays (six stages of craniofacial development and 12 histone modification markers for enhancers, promoters, and gene bodies, Wilderman et al.28).
Figure 2.
DeepFace performance
Boxplot indicates the minimum, 25th percentile, median, 75th percentile, and maximum values for each category.
(A) The Pearson’s correlation coefficient (r) of predicted and real epigenetics features among samples across developmental stages by each chromatin feature.
(B) The area under the precision-recall curve (AUPRC) for predicted peak and real peaks called by MACS among the samples across developmental stages by each chromatin feature.
(C) SNP activity difference (SAD) scores of the variants in five different variant categories: AD-sign, SCZ-sign, OFC-low, OFC-medium, and OFC-sign stratified by each chromatin feature.
The function epigenetic features in (A)–(C) are summarized: repressive marks (H3K27me3 and H3K9me3), promoter activation marks (H3K4me3 and H3K9ac), transcription regulation marks (H3K36me3, H4K20me1, and H3K79me2), active regulatory marks (H3K4me1, H3K4me2, H3K27ac, and H2A.Z), and open chromatin signal DNase.
(D) SAD scores of variants from five genetic background categories: AD-sign, SCZ-sign, OFC-low, OFC-medium, and OFC-sign stratified by variant category.
OFC-sign variants show greater enrichment in embryonic craniofacial development than other sets of variants
We implemented our pretrained DeepFace model to predict the SAD scores of curated SNP sets (OFC-sign, OFC-medium, OFC-low, AD-sign, and SCZ-sign, see material and methods). As shown in Figure 2C, while most SNP sets showed minimal variation in absolute SAD scores for histone modifications, notable exceptions were found in transcription regulation markers (H3K36me3, H3K79me2, and H4K20me1). Among them, the AD-sign and SCZ-sign SNP sets exhibited significantly higher median absolute SAD scores. Conversely, Figure 2D revealed that the OFC-sign group consistently presented higher median absolute SAD scores across various variant categories, particularly in intronic (OFC vs. AD pFDR = 0.02; OFC vs. SCZ pFDR = 3.51 × 10−8), ncRNA_exonic (OFC vs. AD pFDR > 0.05; OFC vs. SCZ pFDR = 0.02), and upstream/downstream regions (OFC vs. AD pFDR = 1.01 × 10−5; OFC vs. SCZ pFDR = 6.82 × 10−9) (Figure S1). Meanwhile, OFC-low or OFC-medium tends to be the lowest absolute SAD score in any category. This observation suggested a higher enrichment of functionally variant OFC-sign sets affecting SAD scores when compared to the other groups. Interestingly, the AD-sign set stood out with higher median absolute SAD scores in the UTR3/UTR5 regions. Moreover, the AD-sign set stood out with higher median absolute SAD scores in the upstream; downstream category as well. Figure S2 indicated a higher prevalence of SNPs in the exonic, ncRNA_exonic, UTR3/UTR5, and upstream/downstream categories for both AD and SCZ groups, as these variants may play a more significant role in altering gene function. Coherent with Figure 2C, these genomic regions are typically enriched with transcription regulation signals (H3K36me3, H3K79me2, and H4K20me1). Consequently, the elevated medium absolute SAD scores of transcription regulation markers in AD and SCZ could be attributed to the transcription regulation within the upstream, downstream, and UTR3, and UTR5 regions.
SAD scores offer the promise of interpretation function of known OFC-related variants
SAD scores could link the function to OFC-sign variants (Table S2). For example, rs117496742 (risk-allele A, lead SNP) is an intronic variant located within the YAP1 on chromosome 11q22.1. This variant has garnered genome-wide significance in European populations, as documented in CPO.14 Notably, within the CS20 stage, characterized by active regulatory mark H3K4me1, rs117496742 boasts the highest absolute SAD score (75.44), underscoring its potential regulatory impact. In contrast, during the CS14 stage, the same variant exhibits the lowest SAD score (70.9) within the H3K4me1 profile. Similarly, rs12543318 (risk-allele C, lead SNP) is an intergenic variant proximal to DCAF4L2 and MMP16 on chromosome 8q21. This variant has been identified as nominally significant in multiethnic populations, as reported in CL/P.7 Noteworthy is its behavior within the CS13 stage, marked by active regulatory mark H3K4me2, where it registers the highest absolute SAD score (52.78), indicating its potential regulatory influence. Conversely, in the CS15 stage, this variant displays the lowest SAD score (36.0) within the H3K4me2 profile.
SAD scores of SNPs may reflect temporal regulation roles
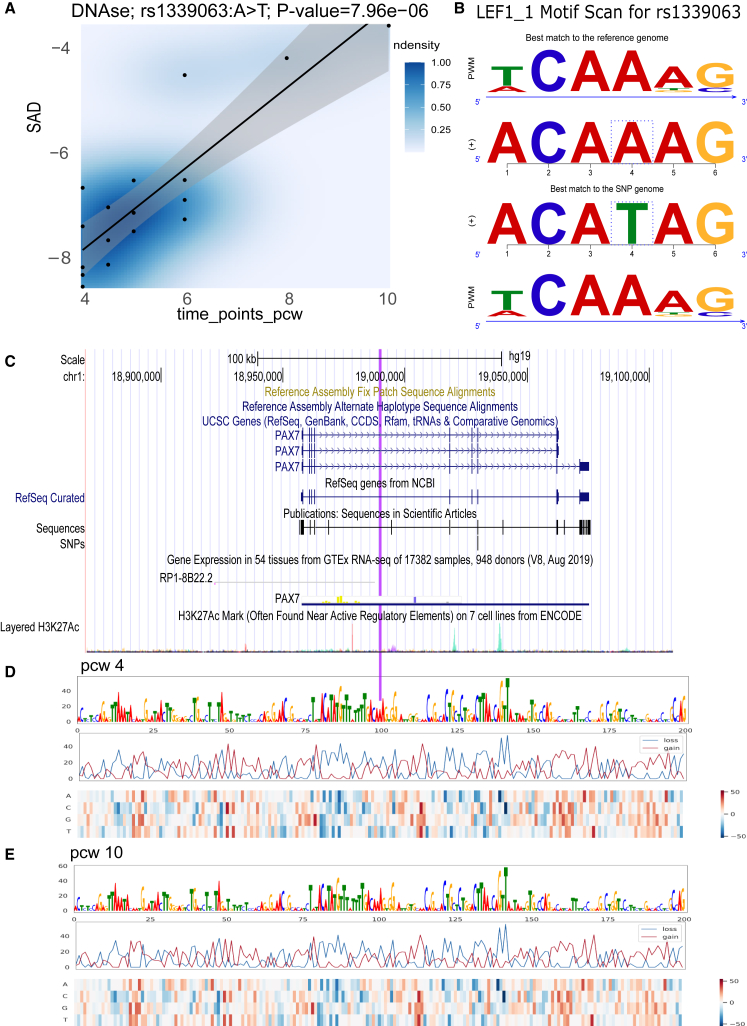
The epigenomic assays from different developmental stages provided us with opportunities to explore the potential temporal epigenetic alteration by variant. We hypothesized whether the SAD scores are associated with the temporally regulatory roles of SNPs. Here, we mainly explored the potential linear regulatory roles of SNPs across the craniofacial development course. To this end, we employed a generalized linear model to assess whether the SAD scores for each SNP exhibited a significant linear relationship with various developmental stages. The linear model coefficient p values were further adjusted by Bonferroni correction of the 1,590 nonzero SAD SNPs from 1,787 OFC-sign SNPs. This procedure revealed six SNPs with significant linear association, suggesting their roles in influencing features throughout the course of craniofacial development. As illustrated in Figure 3, rs1339063 (A>T) is an intronic variant in gene PAX7. The predicted SAD in H3K27me3 showed a significant decrease during the pcw trajectory. The mapped motif is LEF1, a member of the T cell factor/LEF1 family of high-mobility group TFs, which is a downstream mediator of the Wnt/β-catenin signaling pathway.45 The alternative allele (T) decreased the binding affinity of LEF1 on this site. Figures 3D and 3E illustrate that pcw 10 exhibited a lower SAD score when compared to pcw 4, suggesting that the variant had a stronger effect during pcw 4. Both LEF1 and PAX7 were actively expressed in craniofacial tissue from CS13 to CS22, as shown in Cotney lab’s craniofacial Genome Browser46 (Figures S3 and S4).
Figure 3.
DeepFace variant rs1339063: SAD and motif analysis
(A) SAD for rs1339063 (A>T) associated with the post-conception week (pcw) trajectory using DNAse data. The blue area of intensity is positively correlated with the SAD score point density. The black line and gray confidence interval (95%) model the linear relationship between SAD scores and pcw trajectory.
(B) Sequence logo stacks from top to bottom: sequence logo of reference allele matching position weight matrix, reference subsequences, alternative allele subsequences, and sequence logo of SNP allele matching position weight matrix.
(C–E) The best match reference sequence and alternative allele sequence for the motif LEF1 were visualized in (C) UCSC genome browser for the rs1339063 and its surrounding genes. The purple vertical line indicates the exact genomic region of 200 bps for pcw 4 (D) and pcw 10 (E). (D) and (E) show the dynamic gain and loss of SAD scores for all possible substitutions in each of the 200-bp genomic positions around the rs1339063 in pcw 4 and pcw 10, respectively. The alteration between (D) and (E) is relatively small in the figure. These SAD score dynamics were visualized in three rows. Top row: sequence logo weight by the loss of SAD across 200-bp sequence; middle row: the blue and red lines indicating the minimum (loss) and maximum (gain) changes among the possible substitutions from reference allele; and bottom: the quantities in the heatmap, which reflects the change in SAD after substituting the reference allele with the alternative allele.
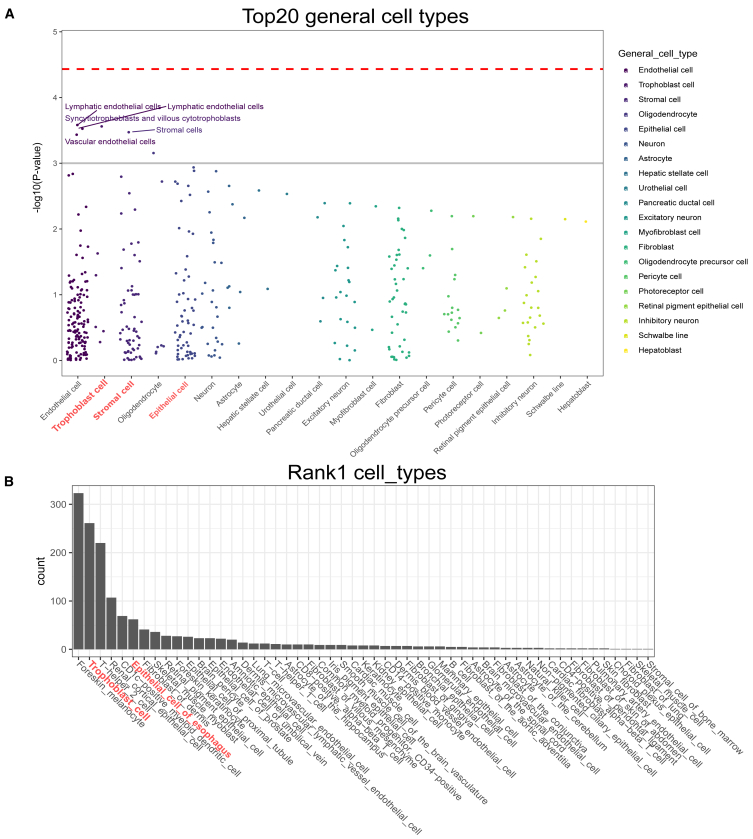
Cell-type specificity analysis of OFC-sign SNP set
The pleiotropic nature of OFC genes underscores the necessity of understanding the specific tissue cell types and contexts where disease-related variants predominantly exert their effects. So far, no human craniofacial context epigenomic data have been available in the current deep learning framework. Therefore, we applied two alternative methods, WebCSEA37 and DeepFun.24,25 The top three enriched cell types identified by WebCSEA were endothelial cells, trophoblast cells, and stromal cells (Figure 4A). The top three ranked primary cell types by DeepFun were foreskin_melanocyte, trophoblast_cell, and T-helper_2_cell (Figure 4B). Both methods identified trophoblast cells among the top three ranked cell types, suggesting that OFC-risk genes manifest their function most during the embryonic developmental stage.47 This cell type is associated with the embryonic stage of craniofacial development,48 aligning with the finding of their similarity to stem cells revealed by Wilderman et al.28 The melanocyte cell originates from the neural crest, which itself emerges from the neural tube. After formation, neural crest cells undergo a process known as the epithelial-to-mesenchymal transition, during which they detach from the uppermost part of the neural tube.49 Recent studies50,51 have revealed that endothelial cells and the vasculature play a pivotal role in guiding tissue morphogenesis and cell differentiation in various cranial structures. Additionally, genes from the vascular endothelial growth factor (VEGF) family have been observed in the mesenchyme surrounding Meckel’s cartilage.52 Furthermore, rare variants in the VEGFA gene have been associated with nonsyndromic CL/P,53 underscoring the significant role of endothelial cells in craniofacial development. In addition, our enrichment analyses identified epithelial cells and stromal cells (mesenchymal cells), both well documented for their involvement in OFC disorders,54 as top related cell types. Specifically, epithelial cells were ranked fifth and fourth in the WebCSEA and DeepFun analyses, respectively. This high ranking suggests that their signals are prominent, as many genes play pleiotropic roles across various cell types. Furthermore, the stromal cell type, a subset of mesenchymal cells crucial for structural support and craniofacial development, was ranked third in the WebCSEA analysis, underscoring their importance in the context of OFCs.
Figure 4.
Cell-type specificity analysis for OFC variants
(A) WebCSEA result: the red dashed line represents the Bonferroni-corrected significance threshold at −log10p value cutoff 3.69 × 10−5. The gray solid line marks the nominal significance level at −log10p value cutoff 1 × 10−3. In every general cell-type category, each dot represents a specific tissue cell type within that category, differentiated by color according to the column it belongs to. We highlighted the top five tissue cell types.
(B) DeepFun results: for each of the SNPs from the OFC-sign SNP set, we calculated the mean absolute SAD and then identified the cell types with the highest absolute SAD values. The count of primary cell types with the highest absolute SAD values is visualized in the bar plot.
Discussion
So far, GWASs from both genotyping and genome sequencing have been extensively performed, leading to many thousands of variants with association signals of the disease or traits under investigation. However, great challenges remain because the roles of most of these variants are not clear, impeding the understanding of molecular mechanisms of disease and further development of disease prevention and therapeutic strategies. Therefore, prioritizing potential causal variants, particularly the thousands of noncoding variants with association signals, is crucial for fully understanding pathogenic mechanism of OFCs. In this work, we aimed to contextualize the function of a comprehensive collection of OFC-related variants during human craniofacial development. To achieve this, we built a deep-learning-based framework, namely DeepFace, by leveraging a spectrum of epigenetics features during the key human embryonic craniofacial development stages. Our DeepFace model pinpointed the high-risk OFC coding and noncoding variants that tended to have the largest predicted SAD scores in several variant categories, including intronic, ncRNA_exonic, and upstream/downstream. Our temporal association analysis further identified six high-risk craniofacial SNPs that exhibited a significant linear relationship between epigenetic impact and the craniofacial developmental process. Overall, DeepFace leveraged the cis-regulatory features to provide a high-resolution prediction on the functional changes caused by OFC-related variants during human craniofacial development. To our knowledge, this is the first deep learning model specifically for craniofacial development by leveraging 204 human functional genomics datasets.
As summarized in Table 1, two SNPs, rs1339063 and rs56675509, were in the intronic region of gene PAX7, which encodes paired box 7. PAX7 belongs to the paired box gene family and plays a role in neural crest development, contributing to various tissues, including craniofacial bones and cartilage.57,58 Several SNPs have been reported to increase the risk of nonsyndromic CL/P.59,60 Our finding thus supported the utilities of the DeepFace model; however, further experiment validation of the regulatory roles of these two SNPs will be warranted. We further discuss their roles below.
Table 1.
Summary of six SNPs with significant linear association
| SNP ID | Chr | Posa | Minor allele | Gene | Motif | Trait | Reference (PubMed) |
|---|---|---|---|---|---|---|---|
| rs56675509 | 1 | 18971634 | C | PAX7 | ZBTB14 | CL/P_all_pop | Mukhopadhyay et al.34 |
| rs1339063 | 1 | 18989575 | T | PAX7 | LEF1 | CL/P_all_pop | Mukhopadhyay et al. 34 |
| rs2302304 | 19 | 3733651 | A | TJP3 | Nkx2-5 | cleft lip with or without cleft palate x maternal periconceptional vitamin use interaction (parent of origin effect) | Haaland et al.55 |
| rs6495117 | 15 | 74899500 | A | CLK3 | EGR1 | nonsyndromic cleft lip with cleft palate | Yu et al.56 |
| rs11787407 | 8 | 129985440 | G | LINC00976/CCDC26 | JUN/FOS | csa_CL/P,eur_CL/P, CL/P_all_pop | Mukhopadhyay et al.34 |
| rs12075674 | 1 | 209995470 | A | IRF6 | AFP | csa_CL/P, CL/P_all_pop | Mukhopadhyay et al.34 |
csa_CL/P, cleft lip with or without palate in Central/South Asian ancestry; eur_CL/P, cleft lip with or without palate in European ancestry; CL/P_all_pop, cleft lip with or without palate in all populations (European and Central/South Asian).
hg19.
The two representative significant motifs on rs1339063 and rs56675509 are LEF1 and ZBTB14, respectively (Figures S4 and S5). Gene LEF1 is expressed in the neural crest61 and plays a role in patterning the mesoderm and ectoderm in Xenopus.62 In mice, Lef1 plays an important role in epithelial-mesenchymal transition during palatal fusion.63 ZBTB14 belongs to the zinc-finger and BTB/POZ (broad-complex, tramtrack, and bric-a-brac/poxvirus and zinc-finger) domain-containing protein family, which regulates organ morphogenesis and development.64,65 In Xenopus, Zbtb14 plays a crucial role in the formation of dorsal-ventral and anterior-posterior axes by regulating BMP and Wnt signaling pathways, both of which are crucial to midfacial development.66,67
SNP rs2302304 (Figure S6) is an intronic variant in gene TJP3 encoding tight junction protein 3, which is a member of the family of membrane-associated guanylate kinase-like proteins that are associated with intracellular junctions.68 Silencing tjp3/zo-3 using morpholinos leads to edema, loss of blood circulation, and tail fin malformations in zebrafish embryos.69 The TF binding motif of this variant is NK2 homeobox 5 (Nkx2-5), which has been reported to play an important role in craniofacial development in zebrafish through regulating the endothelin.70 Funato et al.71 also found that NKX2-5 is involved in molecular function and biological pathways of CPO, incomplete CP, and submucous CP.
SNP rs6495117 (Figure S7) is an intronic variant in gene CLK3 encoding CDC-like kinase 3, which is a member of the cdc2-like kinases with four isoforms.72 In Xenopus, Clk3 knockdown leads to severe developmental defects such as reduced head and eye size and a shortened anterior-posterior axis.73 The TF binding motif of this variant is early growth response 1 (EGR1), which is an EGR gene that regulates the skeleton’s normal development.74,75 In our previous work,76 using a developmental-stage-specific network approach integrating TFs and microRNAs, our results showed that Egr1 was a crucial regulator in mice embryogenesis from embryonic day (E) 11.5 to 13.5.
SNP rs11787407 (Figure S8) is an intergenic variant nearby gene LINC00976/CCDC26, a long noncoding RNA that is related to cancers,77 though its functions remain to be elucidated. It is suggested that rs987525, located near CCDC26, increases the risk of nonsyndromic CL/P.78,79,80 The motif of the variant is FOS/JUN, which is a transcriptional regulator consisting of members of the Fos and Jun families.81 Fos disruption causes craniofacial anomalies in zebrafish.82 The recent single-cell RNA-seq and single-cell multiome studies in mice also showed that Fos and Jun were involved in secondary palate development54 and all-trans retinoic-acid-induced CP.83
SNP rs12075674 (Figure S9) is an intronic variant in gene IRF6 encoding interferon regulatory factor 6, which is one of nine TFs that share a highly conserved helix-turn-helix DNA-binding domain.84 IRF6-related disorders, which are caused by both common and rare variants, have a wide variety of symptoms, including nonsyndromic CL/P and CPO and Van der Woude syndrome (MIM: 119300) at the mild end to the more severe popliteal pterygium syndrome (MIM: 119500).85,86 The alpha-fetoprotein enhancer binding protein (AFP-1) motif currently lacks direct evidence linking it to craniofacial development.
In summary, our DeepFace framework provided a quantitative measurement of craniofacial-related SNPs during craniofacial development stages. We acknowledged that these six SNPs only represent a monotonic trend of regulatory role. Although specifically trained and applied to craniofacial development, DeepFace is limited by several factors, including a limited number of functional genomics datasets, low prediction performance on broad repressive marks (H3K27me3 and H3K9me3), and a lack of extensive comparison with many other tissues or development stages. Although significant SNPs with temporal effects were observed, their impacts were relatively small. It is expected that many more variants with significant correlations between SAD score trends and developmental stages can be identified. There are additional facets of SNP characteristics that warrant further exploration. This includes those SNPs with the strongest impact, those with specific influences at specific development stages or cell types, and those related to particular chromatin features, all of which could be investigated in future studies. Therefore, we provide the OFC-sign SAD matrix (Table S2) to the research community, which is composed by SAD scores for 204 epigenomic features by 1,787 OFC-sign SNPs. Finally, as sequencing technologies are evolving quickly, we expect that many more genomics datasets will be generated, especially those by assay for transposase-accessible chromatin with sequencing (ATAC-seq) and single-cell multiome,54 in craniofacial development. Such data will allow us to refine the DeepFace model for both accuracy and precision toward the development stages and tissue and cell types.
Conclusion
We trained a deep-learning-based model to in silico evaluate the SNP alleles on epigenomic alteration across human craniofacial development during embryonic stages from pcw 4 to pcw 10. Our deep learning model, DeepFace, identified that the OFC-related significant SNP set tended to have stronger SAD scores in several variant categories than other groups, suggesting that these high-risk variants manifest their functional impact during these development stages. We pinpointed six SNPs with a significant linear relationship with SAD scores across developmental progression. Those SNPs may have critical roles in OFCs, and further investigation is warranted. Our study demonstrates that DeepFace has great potential to harness the long-range regulatory element signals from comprehensive epigenomic assays and thus to prioritize, interpret, and decode the dynamic influence of variants related to OFCs and other traits.
Data and code availability
All datasets analyzed in this study are publicly available. The 204 ChIP of post-translational histone modifications from human embryonic craniofacial tissues were obtained from GEO: GSE97752. The OFC-related variants were obtained from the GWAS Catalog (https://www.ebi.ac.uk/gwas/). Multiethnic craniofacial raw data for CL/P and CP are available from dbGaP: phs000884.v2.p1. Other data can be accessed from public resources described in the material and methods. The source code for the pretrained DeepFace model and SAD scores are available at the following GitHub repository: https://github.com/bsml320/DeepFace/.
Web resources
dbGaP, https://www.ncbi.nlm.nih.gov/gap/
DeepFace, https://github.com/bsml320/DeepFace/
DeepFun, https://bioinfo.uth.edu/deepfun/
GWAS Catalog, https://www.ebi.ac.uk/gwas/
Online Mendelian Inheritance in Man (OMIM), https://omim.org/
WebCSEA, https://bioinfo.uth.edu/webcsea/
Acknowledgments
We are thankful for the National Institutes of Health (NIH) grant R01DE030122 for supporting this project. Z.Z. was partially supported by R01DE029818, R01LM012806, U01AG079847, and Chair Professorship for Precision Health funds. T.I. and F.Y. are CPRIT Postdoctoral Fellow and Predoctoral Fellow in the Biomedical Informatics, Genomics, and Translational Cancer Research Training Program (BIG-TCR) funded by the Cancer Prevention and Research Institute of Texas (CPRIT RP210045), respectively. M.L.M. was partially supported by NIH grants R01DE016148, R37DE008559, R01DE032319, R01DE031261, R01DE031855, R01DE032122, and X01HG007845. We are thankful for the technical support from the Cancer Genomics Core, funded by the Cancer Prevention and Research Institute of Texas (CPRIT RP180734). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. We are thankful for the technical support from Luyao Chen from the Center for Secure Artificial Intelligence for Healthcare, McWilliams School of Biomedical Informatics, the University of Texas Health Science Center at Houston. We thank all members of the Bioinformatics and Systems Medicine Laboratory for discussions.
Author contributions
Z.Z., P.J., Y.D., and G.P. contributed to the conception and design of the study; Y.D., T.I., G.P., N.M., M.L.M., F.Y., and Y.C. collected the data and performed the analysis; Y.D., T.I., G.P., F.Y., Y.C., X.J., and L.M.S. interpreted the results; and Y.D., T.I., G.P., and Z.Z. wrote the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2024.100312.
Supplemental information
References
- 1.Leslie E.J., Marazita M.L. Genetics of cleft lip and cleft palate. Am. J. Med. Genet. C Semin. Med. Genet. 2013;163C:246–258. doi: 10.1002/ajmg.c.31381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dixon M.J., Marazita M.L., Beaty T.H., Murray J.C. Cleft lip and palate: understanding genetic and environmental influences. Nat. Rev. Genet. 2011;12:167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaty T.H., Murray J.C., Marazita M.L., Munger R.G., Ruczinski I., Hetmanski J.B., Liang K.Y., Wu T., Murray T., Fallin M.D., et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat. Genet. 2010;42:525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnbaum S., Ludwig K.U., Reutter H., Herms S., Steffens M., Rubini M., Baluardo C., Ferrian M., Almeida de Assis N., Alblas M.A., et al. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat. Genet. 2009;41:473–477. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- 5.Camargo M., Rivera D., Moreno L., Lidral A.C., Harper U., Jones M., Solomon B.D., Roessler E., Vélez J.I., Martinez A.F., et al. GWAS reveals new recessive loci associated with non-syndromic facial clefting. Eur. J. Med. Genet. 2012;55:510–514. doi: 10.1016/j.ejmg.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grant S.F.A., Wang K., Zhang H., Glaberson W., Annaiah K., Kim C.E., Bradfield J.P., Glessner J.T., Thomas K.A., Garris M., et al. A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J. Pediatr. 2009;155:909–913. doi: 10.1016/j.jpeds.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 7.Leslie E.J., Carlson J.C., Shaffer J.R., Feingold E., Wehby G., Laurie C.A., Jain D., Laurie C.C., Doheny K.F., McHenry T., et al. A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q13. Hum. Mol. Genet. 2016;25:2862–2872. doi: 10.1093/hmg/ddw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mangold E., Ludwig K.U., Birnbaum S., Baluardo C., Ferrian M., Herms S., Reutter H., de Assis N.A., Chawa T.A., Mattheisen M., et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat. Genet. 2010;42:24–26. doi: 10.1038/ng.506. [DOI] [PubMed] [Google Scholar]
- 9.Sun Y., Huang Y., Yin A., Pan Y., Wang Y., Wang C., Du Y., Wang M., Lan F., Hu Z., et al. Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat. Commun. 2015;6:6414. doi: 10.1038/ncomms7414. [DOI] [PubMed] [Google Scholar]
- 10.Wolf Z.T., Brand H.A., Shaffer J.R., Leslie E.J., Arzi B., Willet C.E., Cox T.C., McHenry T., Narayan N., Feingold E., et al. Genome-wide association studies in dogs and humans identify ADAMTS20 as a risk variant for cleft lip and palate. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ludwig K.U., Mangold E., Herms S., Nowak S., Reutter H., Paul A., Becker J., Herberz R., AlChawa T., Nasser E., et al. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat. Genet. 2012;44:968–971. doi: 10.1038/ng.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beaty T.H., Ruczinski I., Murray J.C., Marazita M.L., Munger R.G., Hetmanski J.B., Murray T., Redett R.J., Fallin M.D., Liang K.Y., et al. Evidence for gene-environment interaction in a genome wide study of nonsyndromic cleft palate. Genet. Epidemiol. 2011;35:469–478. doi: 10.1002/gepi.20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leslie E.J., Liu H., Carlson J.C., Shaffer J.R., Feingold E., Wehby G., Laurie C.A., Jain D., Laurie C.C., Doheny K.F., et al. A genome-wide association study of nonsyndromic cleft palate identifies an etiologic missense variant in GRHL3. Am. J. Hum. Genet. 2016;98:744–754. doi: 10.1016/j.ajhg.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu H., Yan F., Hu R., Suzuki A., Iwaya C., Jia P., Iwata J., Zhao Z. CleftGeneDB: a resource for annotating genes associated with cleft lip and cleft palate. Sci. Bull. 2021;66:2340–2342. doi: 10.1016/j.scib.2021.07.008. [DOI] [PubMed] [Google Scholar]
- 15.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J., et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R., et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang C., Thompson P., Wang Y., Yu Y., Zhang J., Kong D., Colen R.R., Knickmeyer R.C., Zhu H., Alzheimer’s Disease Neuroimaging Initiative FGWAS: Functional genome wide association analysis. Neuroimage. 2017;159:107–121. doi: 10.1016/j.neuroimage.2017.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu R., Pei G., Jia P., Zhao Z. Decoding regulatory structures and features from epigenomics profiles: A Roadmap-ENCODE Variational Auto-Encoder (RE-VAE) model. Methods. 2021;189:44–53. doi: 10.1016/j.ymeth.2019.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alipanahi B., Delong A., Weirauch M.T., Frey B.J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 2015;33:831–838. doi: 10.1038/nbt.3300. [DOI] [PubMed] [Google Scholar]
- 21.Zhou J., Troyanskaya O.G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods. 2015;12:931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelley D.R., Reshef Y.A., Bileschi M., Belanger D., McLean C.Y., Snoek J. Sequential regulatory activity prediction across chromosomes with convolutional neural networks. Genome Res. 2018;28:739–750. doi: 10.1101/gr.227819.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou J., Theesfeld C.L., Yao K., Chen K.M., Wong A.K., Troyanskaya O.G. Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk. Nat. Genet. 2018;50:1171–1179. doi: 10.1038/s41588-018-0160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pei G., Hu R., Dai Y., Manuel A.M., Zhao Z., Jia P. Predicting regulatory variants using a dense epigenomic mapped CNN model elucidated the molecular basis of trait-tissue associations. Nucleic Acids Res. 2021;49:53–66. doi: 10.1093/nar/gkaa1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pei G., Hu R., Jia P., Zhao Z. DeepFun: a deep learning sequence-based model to decipher non-coding variant effect in a tissue- and cell type-specific manner. Nucleic Acids Res. 2021;49:W131–W139. doi: 10.1093/nar/gkab429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gasperini M., Tome J.M., Shendure J. Towards a comprehensive catalogue of validated and target-linked human enhancers. Nat. Rev. Genet. 2020;21:292–310. doi: 10.1038/s41576-019-0209-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xi W., Beer M.A. Loop competition and extrusion model predicts CTCF interaction specificity. Nat. Commun. 2021;12:1046. doi: 10.1038/s41467-021-21368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilderman A., VanOudenhove J., Kron J., Noonan J.P., Cotney J. High-resolution epigenomic atlas of human embryonic craniofacial development. Cell Rep. 2018;23:1581–1597. doi: 10.1016/j.celrep.2018.03.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoenwolf G.C., Bleyl S.B., Brauer P.R., Francis-West P.H. Elsevier - Health Sciences Division; 2021. Larsen’s Human Embryology. [Google Scholar]
- 30.Wang Z., Zang C., Rosenfeld J.A., Schones D.E., Barski A., Cuddapah S., Cui K., Roh T.-Y., Peng W., Zhang M.Q., et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beck D.B., Oda H., Shen S.S., Reinberg D. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 2012;26:325–337. doi: 10.1101/gad.177444.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ljungman M., Parks L., Hulbatte R., Bedi K. The role of H3K79 methylation in transcription and the DNA damage response. Mutat. Res. Rev. Mutat. Res. 2019;780:48–54. doi: 10.1016/j.mrrev.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Sollis E., Mosaku A., Abid A., Buniello A., Cerezo M., Gil L., Groza T., Güneş O., Hall P., Hayhurst J., et al. The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res. 2023;51:D977–D985. doi: 10.1093/nar/gkac1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukhopadhyay N., Feingold E., Moreno-Uribe L., Wehby G., Valencia-Ramirez L.C., Muñeton C.P.R., Padilla C., Deleyiannis F., Christensen K., Poletta F.A., et al. Genome-wide association study of non-syndromic orofacial clefts in a multiethnic sample of families and controls identifies novel regions. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.621482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pei G., Dai Y., Zhao Z., Jia P. deTS: tissue-specific enrichment analysis to decode tissue specificity. Bioinformatics. 2019;35:3842–3845. doi: 10.1093/bioinformatics/btz138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai Y., Hu R., Manuel A.M., Liu A., Jia P., Zhao Z. CSEA-DB: an omnibus for human complex trait and cell type associations. Nucleic Acids Res. 2021;49:D862–D870. doi: 10.1093/nar/gkaa1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai Y., Hu R., Liu A., Cho K.S., Manuel A.M., Li X., Dong X., Jia P., Zhao Z. WebCSEA: web-based cell-type-specific enrichment analysis of genes. Nucleic Acids Res. 2022;50:W782–W790. doi: 10.1093/nar/gkac392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia P., Dai Y., Hu R., Pei G., Manuel A.M., Zhao Z. TSEA-DB: a trait–tissue association map for human complex traits and diseases. Nucleic Acids Res. 2019;48:D1022–D1030. doi: 10.1093/nar/gkz957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kunkle B.W., Grenier-Boley B., Sims R., Bis J.C., Damotte V., Naj A.C., Boland A., Vronskaya M., van der Lee S.J., Amlie-Wolf A., et al. Author Correction: Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019;51:1423–1424. doi: 10.1038/s41588-019-0495-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trubetskoy V., Pardiñas A.F., Qi T., Panagiotaropoulou G., Awasthi S., Bigdeli T.B., Bryois J., Chen C.-Y., Dennison C.A., Hall L.S., et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502–508. doi: 10.1038/s41586-022-04434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kheradpour P., Kellis M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014;42:2976–2987. doi: 10.1093/nar/gkt1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castro-Mondragon J.A., Riudavets-Puig R., Rauluseviciute I., Lemma R.B., Turchi L., Blanc-Mathieu R., Lucas J., Boddie P., Khan A., Manosalva Pérez N., et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022;50:D165–D173. doi: 10.1093/nar/gkab1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santiago L., Daniels G., Wang D., Deng F.-M., Lee P. Wnt signaling pathway protein LEF1 in cancer, as a biomarker for prognosis and a target for treatment. Am. J. Cancer Res. 2017;7:1389–1406. [PMC free article] [PubMed] [Google Scholar]
- 46.Yankee T.N., Oh S., Winchester E.W., Wilderman A., Robinson K., Gordon T., Rosenfeld J.A., VanOudenhove J., Scott D.A., Leslie E.J., et al. Integrative analysis of transcriptome dynamics during human craniofacial development identifies candidate disease genes. Nat. Commun. 2023;14:4623. doi: 10.1038/s41467-023-40363-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roberts R.M., Loh K.M., Amita M., Bernardo A.S., Adachi K., Alexenko A.P., Schust D.J., Schulz L.C., Telugu B.P.V.L., Ezashi T., et al. Differentiation of trophoblast cells from human embryonic stem cells: to be or not to be? J. Reprod. Fertil. 2014;147:D1–D12. doi: 10.1530/REP-14-0080. [DOI] [PubMed] [Google Scholar]
- 48.Roth D.M., Bayona F., Baddam P., Graf D. Craniofacial development: Neural crest in molecular embryology. Head Neck Pathol. 2021;15:1–15. doi: 10.1007/s12105-021-01301-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mort R.L., Jackson I.J., Patton E.E. The melanocyte lineage in development and disease. Development. 2015;142:620–632. doi: 10.1242/dev.106567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asrar H., Tucker A.S. Endothelial cells during craniofacial development: Populating and patterning the head. Front. Bioeng. Biotechnol. 2022;10 doi: 10.3389/fbioe.2022.962040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lewis A.E., Hwa J., Wang R., Soriano P., Bush J.O. Neural crest defects in ephrin-B2 mutant mice are non-autonomous and originate from defects in the vasculature. Dev. Biol. 2015;406:186–195. doi: 10.1016/j.ydbio.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiszniak S., Mackenzie F.E., Anderson P., Kabbara S., Ruhrberg C., Schwarz Q. Neural crest cell-derived VEGF promotes embryonic jaw extension. Proc. Natl. Acad. Sci. USA. 2015;112:6086–6091. doi: 10.1073/pnas.1419368112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun B., Liu Y., Huang W., Zhang Q., Lin J., Li W., Zhang J., Chen F. Functional identification of a rare vascular endothelial growth factor a (VEGFA) variant associating with the nonsyndromic cleft lip with/without cleft palate. Bioengineered. 2021;12:1471–1483. doi: 10.1080/21655979.2021.1912547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan F., Suzuki A., Iwaya C., Pei G., Chen X., Yoshioka H., Yu M., Simon L.M., Iwata J., Zhao Z. Single-cell multiomics decodes regulatory programs for mouse secondary palate development. Nat. Commun. 2024;15:1–17. doi: 10.1038/s41467-024-45199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haaland Ø.A., Romanowska J., Gjerdevik M., Lie R.T., Gjessing H.K., Jugessur A. A genome-wide scan of cleft lip triads identifies parent-of-origin interaction effects between ANK3 and maternal smoking, and between ARHGEF10 and alcohol consumption. F1000Res. 2019;8:960. doi: 10.12688/f1000research.19571.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu Y., Zuo X., He M., Gao J., Fu Y., Qin C., Meng L., Wang W., Song Y., Cheng Y., et al. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat. Commun. 2017;8:14364. doi: 10.1038/ncomms14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chi N., Epstein J.A. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 2002;18:41–47. doi: 10.1016/s0168-9525(01)02594-x. [DOI] [PubMed] [Google Scholar]
- 58.Murdoch B., DelConte C., García-Castro M.I. Pax7 lineage contributions to the mammalian neural crest. PLoS One. 2012;7 doi: 10.1371/journal.pone.0041089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaczkowska A., Biedziak B., Budner M., Zadurska M., Lasota A., Hozyasz K.K., Dąbrowska J., Wójcicki P., Szponar-Żurowska A., Żukowski K., et al. PAX7 nucleotide variants and the risk of non-syndromic orofacial clefts in the Polish population. Oral Dis. 2019;25:1608–1618. doi: 10.1111/odi.13139. [DOI] [PubMed] [Google Scholar]
- 60.Khan M.I., Cs P., Srinath N. Role of PAX7 gene rs766325 and rs4920520 polymorphisms in the etiology of non-syndromic cleft lip and palate: A genetic study. Glob. Med. Genet. 2022;9:208–211. doi: 10.1055/s-0042-1748531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Genderen C., Okamura R.M., Fariñas I., Quo R.G., Parslow T.G., Bruhn L., Grosschedl R. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- 62.Roël G., Gent Y.Y.J., Peterson-Maduro J., Verbeek F.J., Destrée O. Lef1 plays a role in patterning the mesoderm and ectoderm in Xenopus tropicalis. Int. J. Dev. Biol. 2009;53:81–89. doi: 10.1387/ijdb.072395gr. [DOI] [PubMed] [Google Scholar]
- 63.Shu X., Shu S., Cheng H. Genome-wide mRNA-seq profiling reveals that LEF1 and SMAD3 regulate epithelial-mesenchymal transition through the Hippo signaling pathway during palatal fusion. Genet. Test. Mol. Biomarkers. 2019;23:197–203. doi: 10.1089/gtmb.2018.0221. [DOI] [PubMed] [Google Scholar]
- 64.Lee S.-U., Maeda T. POK/ZBTB proteins: an emerging family of proteins that regulate lymphoid development and function. Immunol. Rev. 2012;247:107–119. doi: 10.1111/j.1600-065X.2012.01116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Siggs O.M., Beutler B. The BTB-ZF transcription factors. Cell Cycle. 2012;11:3358–3369. doi: 10.4161/cc.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takebayashi-Suzuki K., Konishi H., Miyamoto T., Nagata T., Uchida M., Suzuki A. Coordinated regulation of the dorsal-ventral and anterior-posterior patterning of Xenopus embryos by the BTB/POZ zinc finger protein Zbtb14. Dev. Growth Differ. 2018;60:158–173. doi: 10.1111/dgd.12431. [DOI] [PubMed] [Google Scholar]
- 67.Suzuki A., Sangani D.R., Ansari A., Iwata J. Molecular mechanisms of midfacial developmental defects. Dev. Dynam. 2016;245:276–293. doi: 10.1002/dvdy.24368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Itoh M., Furuse M., Morita K., Kubota K., Saitou M., Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J. Cell Biol. 1999;147:1351–1363. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kiener T.K., Selptsova-Friedrich I., Hunziker W. Tjp3/zo-3 is critical for epidermal barrier function in zebrafish embryos. Dev. Biol. 2008;316:36–49. doi: 10.1016/j.ydbio.2007.12.047. [DOI] [PubMed] [Google Scholar]
- 70.Iklé J.M., Tavares A.L.P., King M., Ding H., Colombo S., Firulli B.A., Firulli A.B., Targoff K.L., Yelon D., Clouthier D.E. Nkx2.5 regulates endothelin converting enzyme-1 during pharyngeal arch patterning. Genesis. 2017;55 doi: 10.1002/dvg.23021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Funato N., Nakamura M. Identification of shared and unique gene families associated with oral clefts. Int. J. Oral Sci. 2017;9:104–109. doi: 10.1038/ijos.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jain P., Karthikeyan C., Moorthy N.S.H.N., Waiker D.K., Jain A.K., Trivedi P. Human CDC2-like kinase 1 (CLK1): a novel target for Alzheimer’s disease. Curr. Drug Targets. 2014;15:539–550. doi: 10.2174/1389450115666140226112321. [DOI] [PubMed] [Google Scholar]
- 73.Virgirinia R.P., Nakamura M., Takebayashi-Suzuki K., Fatchiyah F., Suzuki A. The dual-specificity protein kinase Clk3 is essential for Xenopus neural development. Biochem. Biophys. Res. Commun. 2021;567:99–105. doi: 10.1016/j.bbrc.2021.06.005. [DOI] [PubMed] [Google Scholar]
- 74.Sukhatme V.P., Cao X.M., Chang L.C., Tsai-Morris C.H., Stamenkovich D., Ferreira P.C., Cohen D.R., Edwards S.A., Shows T.B., Curran T. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. 1988;53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- 75.Mcmahon A.P., Champion J.E., Mcmahon J.A., Sukhatme V.P. Developmental expression of the putative transcription factor Egr-1 suggests that Egr-1 and c-fos are coregulated in some tissues. Development. 1990;108:281–287. doi: 10.1242/dev.108.2.281. [DOI] [PubMed] [Google Scholar]
- 76.Yan F., Jia P., Yoshioka H., Suzuki A., Iwata J., Zhao Z. A developmental stage-specific network approach for studying dynamic co-regulation of transcription factors and microRNAs during craniofacial development. Development. 2020;147 doi: 10.1242/dev.192948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hirano T., Tsuruda T., Tanaka Y., Harada H., Yamazaki T., Ishida A. Long noncoding RNA CCDC26 as a modulator of transcriptional switching between fetal and embryonic globins. Biochim. Biophys. Acta Mol. Cell Res. 2021;1868 doi: 10.1016/j.bbamcr.2020.118931. [DOI] [PubMed] [Google Scholar]
- 78.Yildirim M., Seymen F., Deeley K., Cooper M.E., Vieira A.R. Defining predictors of cleft lip and palate risk. J. Dent. Res. 2012;91:556–561. doi: 10.1177/0022034512444928. [DOI] [PubMed] [Google Scholar]
- 79.Mostowska A., Hozyasz K.K., Wojcicki P., Biedziak B., Paradowska P., Jagodzinski P.P. Association between genetic variants of reported candidate genes or regions and risk of cleft lip with or without cleft palate in the polish population. Birth Defects Res. A Clin. Mol. Teratol. 2010;88:538–545. doi: 10.1002/bdra.20687. [DOI] [PubMed] [Google Scholar]
- 80.Boehringer S., van der Lijn F., Liu F., Günther M., Sinigerova S., Nowak S., Ludwig K.U., Herberz R., Klein S., Hofman A., et al. Genetic determination of human facial morphology: links between cleft-lips and normal variation. Eur. J. Hum. Genet. 2011;19:1192–1197. doi: 10.1038/ejhg.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zenz R., Eferl R., Scheinecker C., Redlich K., Smolen J., Schonthaler H.B., Kenner L., Tschachler E., Wagner E.F. Activator protein 1 (Fos/Jun) functions in inflammatory bone and skin disease. Arthritis Res. Ther. 2008;10:201. doi: 10.1186/ar2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maili L., Tandon B., Yuan Q., Menezes S., Chiu F., Hashmi S.S., Letra A., Eisenhoffer G.T., Hecht J.T. Disruption of fos causes craniofacial anomalies in developing zebrafish. Front. Cell Dev. Biol. 2023;11 doi: 10.3389/fcell.2023.1141893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang B., Xu M., Zhao J., Yin N., Wang Y., Song T. Single-cell transcriptomics reveals activation of macrophages in all-trans retinoic acid (atRA)-induced cleft palate. J. Craniofac. Surg. 2023;35:177–184. doi: 10.1097/SCS.0000000000009782. [DOI] [PubMed] [Google Scholar]
- 84.Kondo S., Schutte B.C., Richardson R.J., Bjork B.C., Knight A.S., Watanabe Y., Howard E., de Lima R.L.L.F., Daack-Hirsch S., Sander A., et al. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat. Genet. 2002;32:285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schutte B.C., Saal H.M., Goudy S., Leslie E.J. University of Washington, Seattle; 2021. IRF6-Related Disorders. [Google Scholar]
- 86.Zucchero T.M., Cooper M.E., Maher B.S., Daack-Hirsch S., Nepomuceno B., Ribeiro L., Caprau D., Christensen K., Suzuki Y., Machida J., et al. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N. Engl. J. Med. 2004;351:769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets analyzed in this study are publicly available. The 204 ChIP of post-translational histone modifications from human embryonic craniofacial tissues were obtained from GEO: GSE97752. The OFC-related variants were obtained from the GWAS Catalog (https://www.ebi.ac.uk/gwas/). Multiethnic craniofacial raw data for CL/P and CP are available from dbGaP: phs000884.v2.p1. Other data can be accessed from public resources described in the material and methods. The source code for the pretrained DeepFace model and SAD scores are available at the following GitHub repository: https://github.com/bsml320/DeepFace/.