Abstract
Background & Aims
Lacticaseibacillus rhamnosus GG (LGG) is the world’s most consumed probiotic but its mechanism of action on intestinal permeability and differentiation along with its interactions with an essential source of signaling metabolites, dietary tryptophan (trp), are unclear.
Methods
Untargeted metabolomic and transcriptomic analyses were performed in LGG monocolonized germ-free mice fed trp-free or -sufficient diets. LGG-derived metabolites were profiled in vitro under anaerobic and aerobic conditions. Multiomic correlations using a newly developed algorithm discovered novel metabolites tightly linked to tight junction and cell differentiation genes whose abundances were regulated by LGG and dietary trp. Barrier-modulation by these metabolites were functionally tested in Caco2 cells, mouse enteroids, and dextran sulfate sodium experimental colitis. The contribution of these metabolites to barrier protection is delineated at specific tight junction proteins and enterocyte-promoting factors with gain and loss of function approaches.
Results
LGG, strictly with dietary trp, promotes the enterocyte program and expression of tight junction genes, particularly Ocln. Functional evaluations of fecal and serum metabolites synergistically stimulated by LGG and trp revealed a novel vitamin B3 metabolism pathway, with methylnicotinamide (MNA) unexpectedly being the most robust barrier-protective metabolite in vitro and in vivo. Reduced serum MNA is significantly associated with increased disease activity in patients with inflammatory bowel disease. Exogenous MNA enhances gut barrier in homeostasis and robustly promotes colonic healing in dextran sulfate sodium colitis. MNA is sufficient to promote intestinal epithelial Ocln and RNF43, a master inhibitor of Wnt. Blocking trp or vitamin B3 absorption abolishes barrier recovery in vivo.
Conclusions
Our study uncovers a novel LGG-regulated dietary trp-dependent production of MNA that protects the gut barrier against colitis.
Keywords: Metabolome, Probiotic, Transcriptome, Tight Junction
Graphical abstract
Summary.
Lactobacillus (Lacticaseibacillus) rhamnosus GG synergizes with dietary tryptophan to increase vitamin B3 metabolism in vivo. Methylnicotinamide, a vitamin B3 degradative product, strongly promotes gut tight junction and barrier function in homeostasis and during experimental colitis.
Unlike tight epithelia, the permeability of the small intestine is primarily defined by the paracellular pathway between epithelial cells lining the lumen.1 This pathway is selectively sealed by tight junctions (TJ) exhibiting barrier properties that can be altered by diet, the gut microbiota and their metabolites, and by host response to these luminal signals. Two distinct routes of small intestinal paracellular flux have been proposed: the “leak” and “pore” pathways mediating the permeability, respectively, of such macromolecules as dextran and of ions and small solutes.2 Both routes are regulated mainly by TJ proteins occludin (Ocln), ZO-1 (Tjp1), and the large family of claudins (Cldns).3, 4, 5 Intestinal expression and intracellular location of Ocln and claudins, both of which constitute intercellular TJ bridges, and of ZO1, which link these bridges to the cytoskeleton, are altered markedly in patients with gut permeability disorders.6,7 Abnormal perturbations in the composition of the gut microbiota, termed gut dysbiosis, can disrupt homeostatic microbiota-host interactions, thereby impacting the expression, integrity, and location of these TJ proteins and, as a consequence, causing inappropriate increases in permeability of the paracellular pathway.8 This leaky gut condition has been associated with inflammatory bowel disease (IBD) and other autoimmune disorders.5,9, 10, 11 Thus, patients with IBD display an increased paracellular permeability with TJ abnormalities, whereas an impaired intestinal barrier precedes and predicts IBD diagnosis by several years.12,13
The harmful association between leaky gut syndrome and IBD is exacerbated by a deficiency in dietary tryptophan (trp, an essential amino acid).11,14 Serum levels of trp are much lower in patients with IBD compared with healthy control subjects,11 reinforcing earlier findings in colitis animal models showing dietary trp levels to be inversely proportional to the magnitude of intestinal inflammation and permeability.15,16 Only 1% of dietary trp is synthesized into serotonin, and most are catabolized in the gut via 2 major pathways: kynurenine and indole. First, in both host and bacteria, the kynurenine pathway synthesizes precursors of vitamin B3 nicotinamide (NAM), and thus, also of NAD+.17 Like trp in IBD serum, NAD+ levels in ileal and colonic tissues of patients with IBD are markedly reduced, indicating a pathologic dysregulation in NAM metabolism.18,19 Second, the gut microbiota, particularly commensal lactobacilli, convert dietary trp into a variety of indoles, which in turn become ligands of the host Aryl Hydrocarbon Receptor found in barrier organ systems, such as the gastrointestinal tract. A common indole, indole-propionic acid, is reduced in the stool of patients with ulcerative colitis (UC).20
Lacticaseibacillus (formerly Lactobacillus) rhamnosus GG (LGG), a widely consumed probiotic strain that is the subject of 210 clinical trials,21 displays promising efficacy in addressing gastrointestinal disorders ranging from diarrhea and necrotizing colitis to autoimmune conditions associated with leaky gut syndrome.22, 23, 24, 25, 26 Despite these findings, conflicting results in trials with patients with irritable bowel syndrome and patients with IBD underscore the complex influence of disease heterogeneity and patient nutrition on LGG efficacy.27,28 Although we recently reported LGG profoundly changing the host's intestinal luminal metabolome while excluding gut colonization of pathobiont bacteria, the identity and health impact of LGG-dependent metabolites remain poorly characterized, emphasizing the need for an understanding of the underlying mechanisms and signaling molecules mediating LGG’s barrier enhancement.29
Here, we obtained comprehensive, untargeted transcriptomic and metabolomic profiles of the ileum and of both feces and serum, respectively, in LGG monoassociated germ-free (GF) mice fed trp-sufficient or trp-free diets. We also characterized LGG-derived metabolites under aerobic and anaerobic culture conditions. Through a multiomic correlation analysis newly developed by us,30 we initially uncovered LGG- and dietary trp-associated metabolites that statistically linked robustly to transcripts belonging to pathways modulating enterocyte TJs and differentiation. We then subsequently showed via in vitro, ex vivo, and in vivo functional experiments that most of the correlated metabolites can actually profoundly benefit the gut barrier. Surprisingly, LGG specifically promotes an intestinal trp-to-NAM metabolic pathway by elevating its key enzymes and transporters. Moreover, we also discovered that methylnicotinamide (MNA), a vitamin B3 metabolite thought to be mainly destined for urinary excretion and thus metabolically inert, actually exhibits strong protection of barrier function in vitro and in vivo. In fact, we also uncovered via in silico analysis that decreased serum MNA is associated with increased disease activity in patients with IBD. Thus, LGG stimulates a dietary trp-dependent vitamin B3 synthesis pathway with MNA being the most barrier-protecting and anticolitogenic metabolite.
Results
LGG and Dietary Tryptophan Synergize to Promote Enterocyte Program
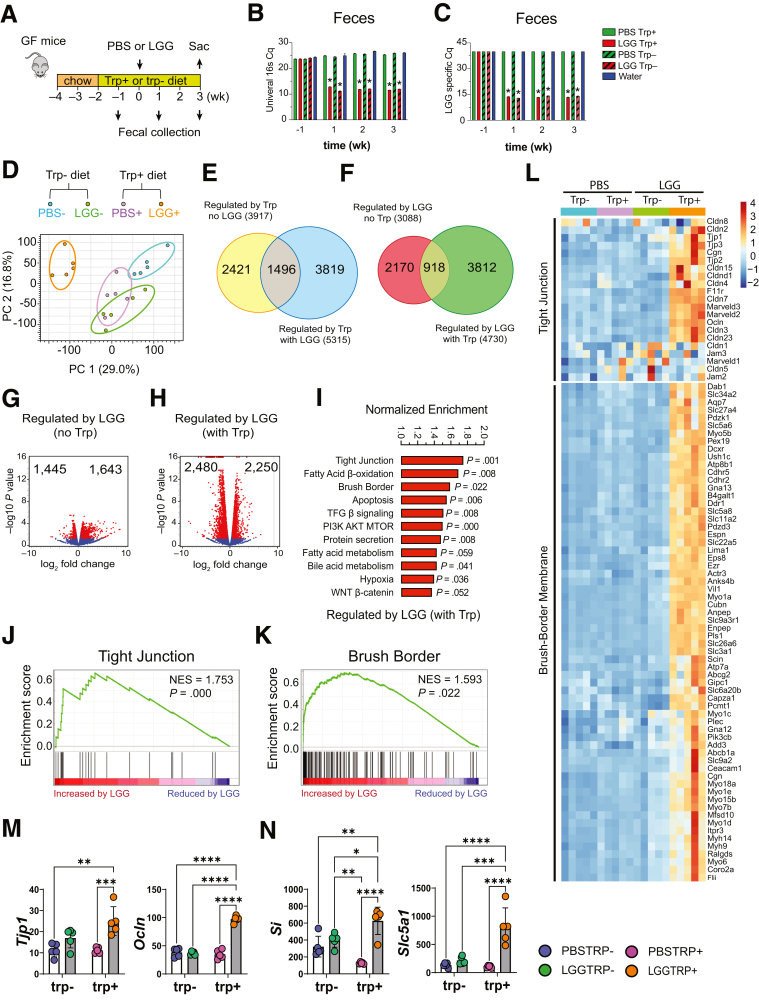
GF mice fed with trp+ and trp- showed a modest weight gain or loss, respectively. Food consumption was slightly greater in trp- mice. Equivalent LGG colonization in trp+ and trp- mice was found by quantitative polymerase chain reaction (qPCR) of fecal DNA using 16S rRNA and LGG-specific primers (Figure 1B and C). Three weeks after inoculation, bulk RNA-sequencing was performed on ilea of GF mice gavaged with phosphate-buffered saline (PBS) and fed trp-free diets (PBS trp-), GF mice gavaged with PBS and fed trp-sufficient diets (PBS trp+), LGG trp-, and LGG trp+ mice (N = 5 each group). Dietary trp alone altered ileal transcriptome in GF mice (Figure 1D). LGG association significantly altered ileal transcriptome under both trp+ and trp- conditions. LGG trp+ transcriptome was noticeably different from the other 3 groups suggesting that LGG and dietary trp drove the largest change in ileal transcriptome.
Figure 1.
LGG and dietary trp promote enterocyte transcriptome. (A) 2 x 2 experimental design: GF (gavaged with PBS) and LGG-monoinoculated mice (LGG) fed a trp-sufficient (trp+) or trp-free (trp-) diet for 3 weeks after inoculation, then sacrificed (Sac). (B) Cq number of RT-PCR analysis using universal 16S primers of fecal samples collected during preinoculation (-1), and at 1-, 2-, and 3-weeks postinoculation. (C) Cq number of RT-PCR using LGG-specific primers at the same time points. (D) Principal component analysis (PCA) of ileal transcriptome from mice fed a trp- diet and gavaged with PBS (PBS-, blue), fed trp+, PBS-gavaged (PBS+, purple), fed trp-, LGG-monocolonized (LGG-, green), and fed trp+, LGG-monocolonized (LGG+, orange). (E) Venn diagram of numbers of ileal transcripts (FC > 1.5, FDR < 0.05) representing trp effect without (PBS+ vs PBS-) and with LGG (LGG+ vs LGG-). (F) Venn diagram of numbers of ileal transcripts (FC > 1.5, FDR < 0.05) representing trp effect without (PBS+ vs PBS-) and with LGG (LGG+ vs LGG-). (G) Volcano plot of genes upregulated and downregulated by LGG without trp, and (H) with trp. (I) Normalized enrichment score following gene set enrichment analysis comparing transcriptomes of trp-sufficient mice with LGG (LGG+) and with trp. Leading edge graphs depicting nonrandom distribution of genes in (J) tight junction and (K) brush border pathways from LGG+ vs PBS+ mice. (L) Heatmap of ileal transcripts from PBS-, PBS+, LGG-, and LGG+ (N = 5 each group), showing only significantly different genes (FDR < 0.05). RPKM of representative transcripts of genes in the (M) tight junction and (N) brush border membrane. White bars, PBS; gray, LGG. Statistical significance among the groups was determined by 2-way analysis of variance, ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001. Ocln, occludin; RPKM, reads per kilobase of transcript, per million mapped reads; Si, sucrase-isomaltase; Slc5a1, Sodium-dependent glucose transporter 1; Tjp1, Tight junction protein 1.
Dietary trp affected 3917 ileal transcripts in GF mice, and 5315 transcripts in LGG-associated mice (Figure 1E), with 1496 transcripts affected solely by dietary trp. LGG affected 3088 ileal transcripts in trp- mice and 4730 transcripts in trp+ mice (Figure 1F), with 918 transcripts affected solely by LGG. The synergistic effect of LGG and dietary trp was reflected by the number of altered genes and the magnitude of fold changes (Figure 1G and H). These results suggested that LGG and dietary trp robustly changed ileal transcriptome, and the effect was greater than those elicited by dietary trp or LGG alone.
Gene set enrichment analysis revealed that LGG and dietary trp increased transcriptomic programs of enterocyte differentiation and function, including TJ genes, brush border, fatty acid β-oxidation, and apoptosis being impacted the most (Figure 1I-K). Heatmap illustrated a robust elevation of enterocyte genes in LGG trp+ mice, and such elevation strictly depended on dietary trp (Figure 1L). This synergism between LGG and trp promoting the TJ and brush border, respectively, is best exemplified by their genes Ocln and Slc5a1, whose expressions are greatest in LGG trp+ (Figure 1M and N).
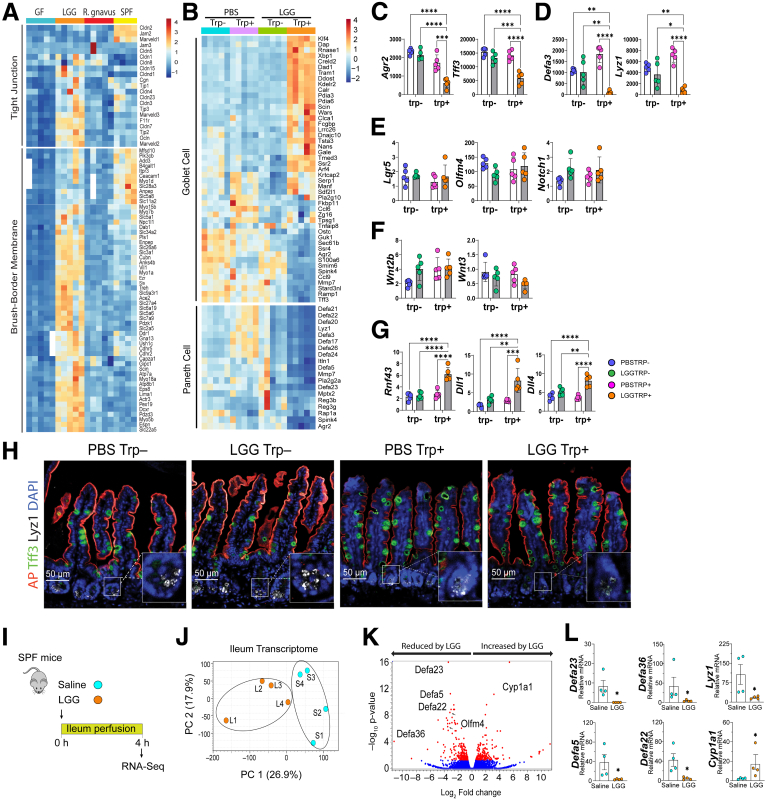
The effects on enterocyte program seemed unique to LGG, because a comparison of ileal transcriptomes of Ruminococcus gnavus–monocolonized and trp-fed GF or specific pathogen-free mice showed that the increased enterocyte genes were predominantly in LGG mice (Figure 2A). The elevated enterocyte program in LGG trp+ seemed to be at the cost of secretory cells (Figure 2B), because a partial goblet cell transcriptome (eg, Tff3, Agr2) (Figure 2C) and almost the entire Paneth cell transcriptome (eg, Lyz1, numerous defensins) (Figure 2D) were reduced in LGG trp+ mice. In contrast, LGG plus dietary trp had no effect on Lgr5 and Olfm4, marker genes of the fast-cycling intestinal stem cells (Figure 2E), or epithelial and stromal Wnt ligand genes (Figure 2F). Importantly, LGG and dietary trp seemed to increase Rnf43, Dll1, and Dll4, regulators of the Wnt and Notch signaling, respectively (Figure 2G). Confocal immunostaining analysis validated a reduced Paneth cell number in LGG trp+ ileum (Figure 2H).
Figure 2.
LGG and dietary trp effects are specific and suppress Paneth cell transcriptome. (A) Representative heatmaps of ileal TJ and brush-border transcripts in GF (blue bar on top of heat map), LGG-monocolonized (orange), Ruminococcus gnavus–monocolonized (red), and specific pathogen free (SPF) (yellow) mice (N = 4–5 each group) suggesting LGG effects are specific. (B) Targeted, representative partial heatmaps of ileal Goblet and Paneth cell transcripts were significantly different (FDR < 0.05) among PBS- (blue), PBS+ (purple), LGG- (green), and LGG+ (orange) mice (N = 5). (C-G) Representative transcripts of goblet cells, Paneth cells, stem cells, Wnt and Notch regulator genes. (H) Immunostaining against Trefoil factor 3 (TFF3, green), alkaline phosphatase (AP, red), Lysozyme 1 (LYZ1, white), DAPI (blue). (I) Conventional mouse intestine was perfused with saline (Ringer) or LGG. (J) Principal component analysis (PCA) showing separation of saline- (blue) and LGG-perfused (orange) ileal transcriptomes (N = 4). (K) Volcano plot of ileal transcriptome perfused with or without LGG. Significantly altered expression is shown in red (|FC| > 1.5, FDR < 0.05). (L) Representative Defa transcripts. Statistical significance among the groups was determined by 1-way analysis of variance, ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001.
LGG colonization in mice consuming trp+ or trp- diets trigger complicated systemic responses to diet-LGG-host interaction. To test if live LGG perfused through the intestinal tract in vivo may acutely elicit a similar intestinal transcriptomic response, we performed intraluminal perfusion using saline or live LGG for 4 hours (Figure 2I). Bulk RNA-Seq on LGG- or saline-perfused ileum showed separated transcriptomic profiles (Figure 2J), with the most reduced transcripts being defensins in LGG-perfused mice (Figure 2K and L). We did not observe changes in enterocyte genes, suggesting that LGG cannot acutely promote enterocyte program within the short time window studied in this perfusion experiment.
LGG Drives Specific Fecal and Serum Metabolome In Vivo
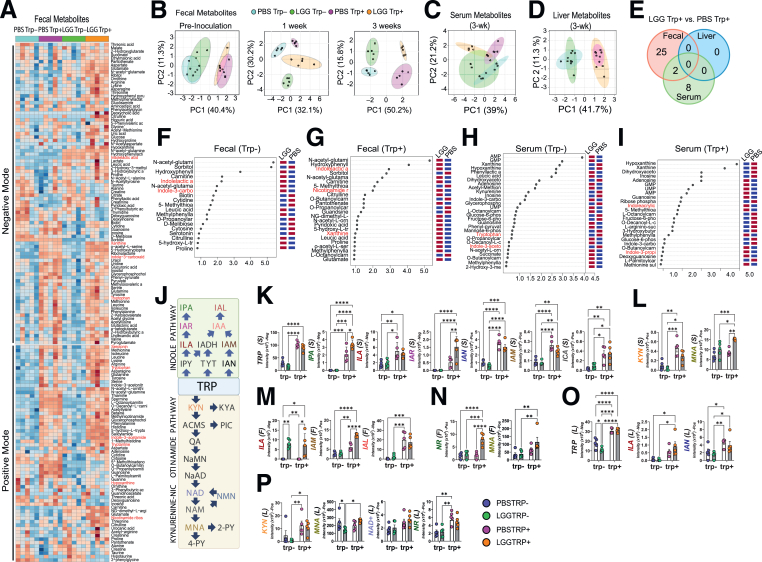
We postulate that LGG may alter the host metabolome, which in turn may regulate intestinal epithelial programming. We performed an untargeted fecal metabolomic analysis using liquid chromatography–mass spectrometry and uncovered approximately 200 polar fecal metabolites that differed in abundance among 4 groups (Figure 3A). In GF mice, dietary trp before colonization markedly separated the host fecal metabolome profile (Figure 3B). Within a week of LGG colonization, fecal metabolite profiles changed (Figure 3B), and the separation persisted after 3 weeks of colonization (Figure 3B). Similar changes were observed for metabolites in the positive mode.
Figure 3.
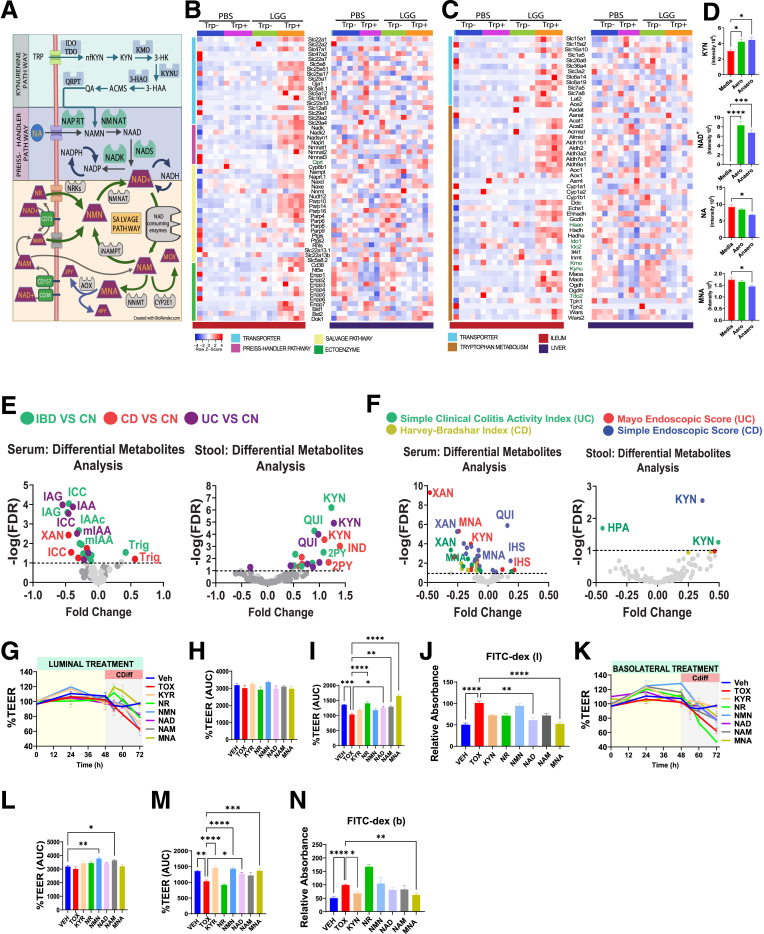
LGG and trp drive indole and kynurenine (KYN)-NAM metabolomic pathways. (A) Heatmap of fecal metabolites analyzed by liquid chromatography–mass spectrometry in negative (top) and positive (bottom) modes. Significant LGG- and trp-induced changes were observed in 121 fecal metabolites (88 negative, 62 positive, 29 in both). Serum had 106 (negative) and 69 (positive) metabolites, whereas the liver had 104 (negative) and 75 (positive). Darker brown indicates upregulation, blue indicates downregulation. (B) Principal component analysis (PCA) of fecal metabolites (negative mode) over time: 1 week before (left), 1 week (middle), and 3 weeks (right) after LGG inoculation (N = 5 each group). PCA of (C) serum and (D) liver metabolites 3 weeks after LGG inoculation. (E) Venn diagram depicting the number of fecal (red), serum (green), and liver (blue) metabolites (negative mode) significantly different between LGG+ vs PBS+ mice (|FC| > 1.5, FDR < 0.05). Twenty-seven fecal metabolites differed between LGG+ and PBS+ mice, 2 also in serum. (F) Variables important in the projection (VIP) scores of fecal metabolites driving intergroup differences between LGG and GF mice deficient in trp (LGG- vs PBS-) and (G) between LGG and GF mice fed trp-sufficient diets (LGG+ vs PBS+). (H) VIP scores of serum metabolites between LGG- and PBS- and between (I) LGG+ vs PBS+. (J) Diagram showing the indole and kynurenine-nicotinamide pathways. Levels of indoles and kynurenine-NAM metabolites in the (K, L) serum, (M, N) fecal, and (O, P) liver. Statistical significance among the groups was determined by 2-way analysis of variance, ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001. IAL, indole-3-carboxyaldehyde; IAM, indole-3-acetamide; IAN, indole-3-acetonitrile; IAR, indole-3-acrylic acid; ILA, indole-3-lactic acid; IPA, indole-3-propionic acid; KYN, kynurenine; NAD+, nicotinamide adenine dinucleotide; NR, nicotinamide riboside.
Serum metabolomic profiles were modestly separated with a noticeable overlap between LGG trp- and PBS trp- mice (Figure 3C). In the presence of dietary trp, LGG induced a marked change in serum metabolome, suggesting that LGG impacts the host serum metabolites only in the presence of dietary trp. In contrast, the liver metabolome was only affected by dietary trp, with little impact from LGG (Figure 3D). Cross-comparing the fecal, serum, and liver metabolites suggested that the metabolites modulated by LGG were compartment-specific (Figure 3E), suggesting that LGG exerts a delicate modulation of the host leading to specific fecal and serum metabolomic profiles.
Variables important in the projection analysis revealed the identities of the most significant fecal (Figure 3F and G) or serum (Figure 3H and I) metabolites that were increased or decreased by LGG under trp- (Figure 3F and H) or trp+ (Figure 3G and I) conditions, respectively. Dietary trp can be metabolized in host cells into kynurenine and NAM or converted by gut bacteria into various indoles (Figure 3J). We then specifically examined indole pathway and kynurenine pathway metabolites to identify those showing a synergistic response to LGG and dietary trp. Dietary trp elevated multiple serum indoles and kynurenine, with serum indole-3-acrylic acid showing a synergistic response to LGG and trp (Figure 3K and L). Interestingly, serum MNA, a downstream degradation product of kynurenine-to-NAM pathway (Figure 3J), was also significantly elevated primarily in LGG trp+ mice (Figure 3L). Fecal indole lactic acid was increased by LGG, independent of dietary trp, whereas indole-3-acetamide increased in LGG trp+ mice (Figure 3M). Fecal NAM riboside and MNA were both increased in LGG trp+ mice (Figure 3N). Again, liver indoles and kynurenine were only elevated by dietary trp with limited influence from LGG (Figure 3O and P).
Identification of LGG-Regulated Barrier-Promoting Metabolites
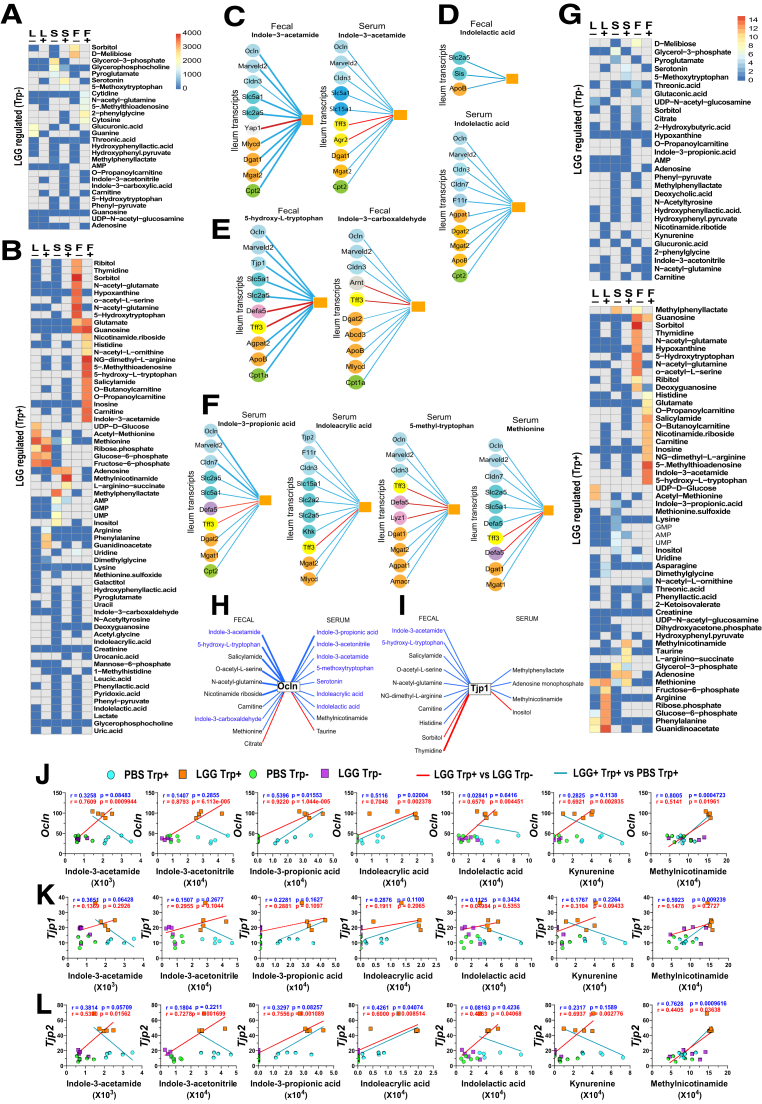
We developed a high-throughput metabolome-transcriptome correlation analysis (METRCA), which identified significant correlations between the 2 omics datasets.30 Comparing LGG trp- and PBS trp- mice, METRCA uncovered 28 LGG-regulated metabolites in fecal, serum, and liver that each correlated with relatively few specific ileal transcripts (Figure 4A). In trp+ mice, METRCA uncovered a marked increase in LGG-regulated metabolites that significantly correlated with numerous ileal transcripts (Figure 4B). Several indoles, among other metabolites, were identified when comparing PBS trp+ and LGG trp+ mice (Figure 4B). Interestingly, both fecal and serum indole-3-acetamide correlated with similar sets of representative TJ, brush border, and lipid metabolism genes (Figure 4C). However, fecal and serum indolelactic acid not only correlated with distinct ileal transcripts (Figure 4D) but also with different numbers of genes. Various trp and non-trp metabolites, although positively correlating with enterocyte gene networks, exhibited negative correlations with goblet or Paneth cell genes (Figure 4C-F).
Figure. 4.
Identification of LGG-regulated barrier-correlating metabolites. (A) Untargeted heatmap determined by the METRCA algorithm to depict the number of transcripts whose expression levels correlated with levels of specific metabolites (negative [-] and positive [+] modes) in the liver (L), serum (S), and feces (F) of LGGtrp- vs PBStrp- mice. (B) Untargeted heatmap calculated by METRCA for LGGtrp+ vs PBStrp+. Correlations between 10 representative transcripts (circles) with an individual fecal metabolite (orange square). Representative transcripts predicted to be modulated by (C) fecal and serum IAM, (D) fecal and serum indolelactic acid, (E) other fecal indoles and nonindoles, and (F) other serum indoles and nonindoles metabolites. Blue line indicates positive correlation; red, negative. A thick line indicates regulation by both LGG in trp-sufficient mice (LGGtrp+ compared with PBStrp+), and by trp in LGG mice (LGGtrp+ compared with LGGtrp-), a thin line by only 1 comparison. Colored circles on genes depict functional groups (light blue, TJ; dark blue, brush-border; gray, proliferation; orange, fatty acid synthesis and metabolism; green, lipid beta-oxidation; yellow, goblet cell; and pink, Paneth cell marker). Targeted heatmap (TJ genes vs metabolites) of number of correlations regulated by LGG (G) without trp (top) and with trp (bottom). (H, I) Correlations between representative TJ transcripts and serum and fecal metabolites whose levels increase primarily with LGG-trp+. Individual correlations between trp-NAM metabolites with (J) Ocln, (K) Tjp1, and (L) Tjp2 genes.
We next conducted a targeted query for metabolites that were correlated with TJ genes. In trp- mice, few LGG-regulated metabolites were correlated and those few were mostly linked with a small number of TJ genes (Figure 4G). In trp+ mice, however, we uncovered a greater number of trp or non-trp metabolites correlating with TJ genes (Figure 4G). Metabolites positively or negatively correlated with Ocln (Figure 4H) or Tjp1 (Figure 4I) were revealed. Several metabolites were found to be correlated with multiple TJ genes, with fecal indole-3-acetamide, serum indole-3-propionic acid, and serum MNA being the most highly ranked positive correlators (Figure 4H-J). These correlations were validated by representative TJ-correlating plots for trp and non-trp derivatives (Figure 4J-L). Because there are sealing claudins that reduce and leaky claudins that enhance paracellular permeability, it is interesting to note that numerous LGG metabolites positively correlated with sealing claudin 3 exhibits either no correlation (eg, fecal NAM riboside, fecal indole-3-acetamide) or negative correlation (eg, fecal salicylamide, fecal sorbitol) with leaky claudin 2 (not shown). Simultaneously, LGG metabolites negatively correlated with claudin 3 (eg, fecal acetyl-glutamine, fecal acetyl-glutamate) demonstrate a positive regulation with claudin 2. Most LGG serum metabolites, including numerous indoles positively correlated with claudin 3, do not correlate with claudin 2.
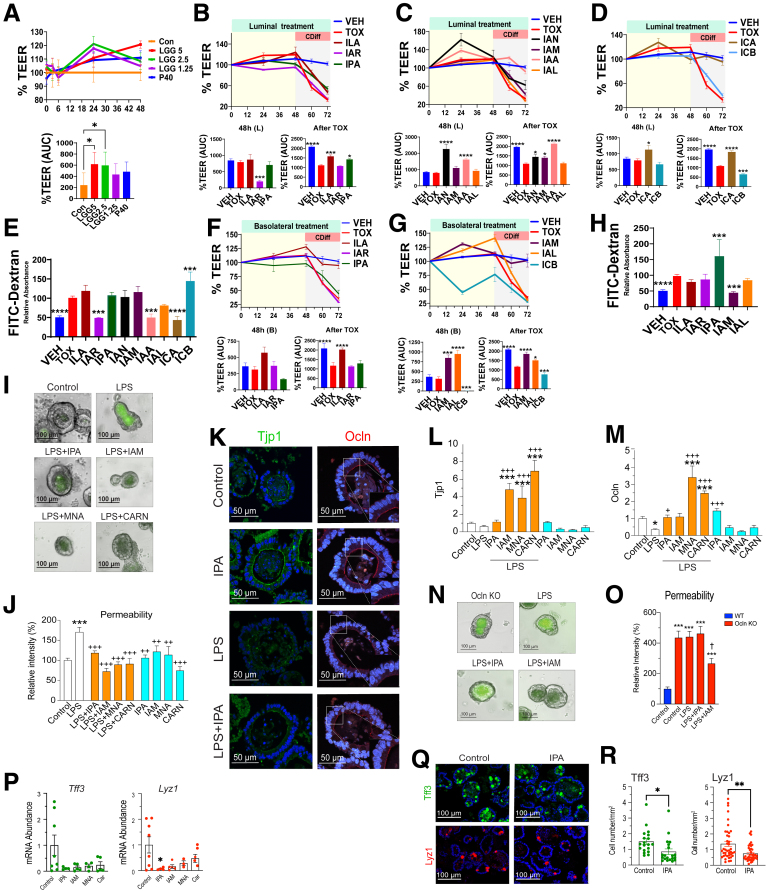
To functionally test if the previously mentioned TJ-correlating fecal and serum metabolites may influence barrier function, we used the polarized Caco-2 monolayers for transepithelial electrical resistance (TEER) and fluorescein isothiocyanate (FITC)-dextran permeability assays. The addition of 2.5%–5% of LGG supernatant on Caco-2 monolayer resulted in a 2-fold increase in TEER (P = .01) compared with the control (Figure 5A). This enhancement exceeded the efficacy of p40, an LGG-derived peptide, suggesting additional barrier-promoting factors in LGG spent medium. We thus used this system to test highly ranked barrier-correlating LGG-regulated metabolites, by applying them to either the luminal or the basolateral sides of the monolayer. Luminal administration of indole-3-acetonitrile, indole-3-acetic acid, and indole-3-carboxylic acid improved TEER by 168%, 91%, and 39%, respectively, after 48 hours of incubation compared with vehicle (Figure 5C and D). Treating cells with a cell free Clostridium difficile supernatant provoked a 46% reduction in TEER, consistent with an increased permeability of the paracellular pathway (Figure 5B). Under Crohn's disease (CD)-treated condition, the luminal application of indoles (indole-3-lactic acid, indole-3-propionic acid, indole-3-acetonitrile, indole-3-acetamide, indole-3-acetic acid, and indole-3-carboxylic acid) restored TEER with efficacies ranging from 29% to 96% (Figure 5B-D). Moreover, basolateral treatment of indole-3-acetamide and indole-3-carboxyaldehyde exhibited a remarkable increase in baseline TEER, with 1.3- and 1.6-fold enhancements, respectively, in comparison with the vehicle. Under CD treatment, the basolateral indole-3-lactic acid, indole-3-acetamide, and indole-3-carboxyaldehyde restored TEER by 57.5%–72.4% (Figure 5F and G). FITC-dextran permeability assays were largely consistent with TEER results (Figure 5E and H).
Figure 5.
LGG-regulated TRP-NAM metabolites promote and protect the gut barrier. (A) Top shows luminal treatment of Caco2 cells with LGG cell-free supernatant at different concentrations (%v/v) or with P40 (50 ng/mL) over 48 hours (N = 6 per group) Control (con), LGG medium or PBS. % TEER was normalized to that of initial. Bottom shows area under the curve (AUC) of TEER from 0–48 hours, normalized to that of Con. (B-D) Top shows luminal treatment of Caco2 with LGG-derived indoles showing % (of initial) TEER. Bottom depicts AUC from 0–48 hours (left) and after Clostridium difficile supernatant treatment (right) with (E) quantified paracellular permeability using FITC-dextran. (F-H) Similar experiments were done with metabolites applied to the basolateral compartment. (I) FITC-dextran permeability in control (media only) or LPS-treated mouse organoids. Selected metabolites were evaluated for their efficacy in protecting the permeability of LPS-treated organoids. (J) Image J-quantified FITC-dextran fluorescence in the lumen normalized to that of control. (K) Representative images showing fluorescent staining of Tjp1 (left) and Ocln (right) of organoids treated with IPA. Expression of (L) Tjp1 (ZO-1) and (M) Ocln in control, LPS-, LPS+metabolite-, and metabolite-treated organoids. (N) Representative images and (O) quantified FITC-dextran fluorescence of organoid permeability of Ocln KO organoids treated with metabolites. (P) Expression of Tff3 (left) and Lyz1 (right) with selected metabolites. (Q) Representative immunofluorescent images of TFF3 (left) and LYZ1 (right) of control organoids and those treated with IPA. (R) Number of TFF3-positive (left) and LYZ1-positive (right) cells per organoid area. Statistical significance among the groups was determined by 1-way analysis of variance, ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001. CARN, carnosine; IAA, indole-3-acetic acid; IAL, Indole-3-carboxyaldehyde; IAM, indole-3-acetamide; IAN, indole-3-acetonitrile; IAR, indole-3-acrylic acid; ICA, indole-3-carboxylic acid; ICB, indole-3-carbinol; ILA, indole-3-lactic acid; IPA, indole-3-propionic acid; VEH, DMSO.
We then evaluated candidate metabolites on enteroids from specific pathogen free mice. Following exposure to lipopolysaccharide (LPS), epithelial permeability increased by 1.75-fold quantified as FITC-dextran intensity within the enteroid. Indole-3-propionic acid, indole-3-acetamide, MNA, and carnosine attenuated LPS induced permeability (Figure 5I and J). Furthermore, we found by immunofluorescent staining and RT-qPCR, indole-3-propionic acid, MNA, indole-3-acetamide, and carnosine significantly increased Tjp1 and Ocln expression compared with the vehicle and LPS group (Figure 5K-M). To test if the barrier-promoting effects of the metabolites may rely on TJ function, we did the experiments with OclnKO enteroids, which exhibited a 320% increase in permeability compared with the wild-type enteroids. LPS did not exacerbate the permeability, nor did the metabolites rescue the permeability in OclnKO enteroids (Figure 5N-O), suggesting that Ocln is required for the barrier restoration by the tested metabolites. In addition, these metabolites also reduced Tff3 and Lyz1 in treated enteroids (Figure 5P-R). Thus, we validated a panel of METRCA-identified, LGG-regulated barrier-enhancing metabolites ex vivo that also affected intestinal cell differentiation.
LGG Stimulates TRP-NAM Pathway and MNA Promotes Gut Barrier
Among the tested metabolites, MNA exhibited a robust barrier-promoting efficacy especially increasing Ocln expression in vitro (Figure 5J and M). MNA is a downstream degradation product of the de novo NAD synthesis (also known as kynurenine) pathway from trp (Figure 6A). Two other NAD synthesis pathways include the Preiss-Handler synthesis from nicotinic acid, and the salvage pathway from NAM mononucleotide and NAM riboside (Figure 6A). NAD catabolism involves at least 6 enzyme families, including sirtuins, CD38 and poly(ADP-ribose) polymerases whose by-product is NAM, which is either recycled back to NAD or methylated into MNA that is, in turn, oxidized into the final excretory products N-methyl-2-pyridone-5-carboxamide (2-PY) and N-methyl-4-pyridone-3-carboxamide (4-PY) (Figure 6A). Targeted transcriptomic analysis revealed NAM metabolism pathway upregulation in the ileum but not in liver of LGG trp+ mice (Figure 6B). We also observed increased trp metabolism gene network in LGG trp+ ileum but not liver (Figure 6C). Specifically, most enzymes and transporters of the trp-NAM pathway were significantly increased (Figure 6B and C). To determine if LGG directly makes trp-NAM metabolites, liquid chromatography–mass spectrometry metabolomics performed on overnight cultured LGG in aerobic or anaerobic conditions, revealed that kynurenine and NAD+ were synthesized (Figure 6D). Nicotinate and MNA decreased in culture, suggesting that the augmented serum MNA in LGG trp+ mice reflect a result of LGG-stimulated trp to NAM metabolism. Despite the impermeable nature of NAD+, the significantly increased ectoenzymes, Cd38, Nt5e (Cd73), Bst1 (Cd157), and various transporters in the LGG trp+ mice (Figure 6A and B) support an enhanced influx of extracellular NAM precursors that were taken into the NAD+-NAM-MNA intracellular metabolic pathways (Figure 6A).
Figure 6.
Serum MNA is reduced in IBD while exogenous MNA robustly promotes barrier function. (A) Graphical illustration depicting the metabolic fate of TRP and NAM metabolites, highlighting enzymes involved in different pathways. (B) Heatmaps illustrating NAM metabolism-related genes associated with metabolite transport (blue), Preiss-Handler pathway (violet), salvage pathway (yellow), and ectoenzyme (green) in the ileum (left) and liver (right). (C) Heatmaps depicting TRP metabolism-related genes associated with TRP transport (blue) and TRP metabolism (brown) in the ileum (left) and liver (right). (D) Liquid chromatography–mass spectrometry analysis of KYN-NAM derived metabolites in the cell-free culture supernatant of LGG (N = 3 per group). (E) Volcano plot displaying results of differential metabolite analysis (DMA) targeting trp-NAM, derived from in silico IBD analysis in serum and stool of patients. Comparisons include IBD vs CN, CD vs CN, and UC vs CN. (F) DMA analysis of trp-NAM metabolites associations with clinical disease activity indices, including Simple Clinical Activity Index and Mayo endoscopic score for UC, and Harvey Bradshaw Index, and Simple Endoscopic Score for CD, with FDR <0.10 threshold indicated by a dashed black line. (G-I) Luminal treatment of metabolites in a time-dependent manner, showing TEER of Caco-2 cells normalized to Veh (N = 5 per group), area under the curve (AUC) of TEER from 0–48 hours of metabolite treatment before Clostridium difficile toxin challenge, and AUC of % TEER 24 hours after C difficile treatment. (J) Quantified paracellular permeability using FITC-dextran levels. (K-N) Basolateral treatment of metabolites in a time-dependent manner, showing AUC of TEER from 0–48 hours of metabolite treatment before C difficile challenge, AUC of % TEER 24 hours after C difficile treatment, and quantified paracellular permeability using FITC-dextran levels. Statistical significance among the groups was determined by 1-way analysis of variance, ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001. 2PY, N1-Methyl-2-pyridone-5-carboxamide; I7AA, 1H-indole-7-acetic acid; HPA, 5-hydroxypicolinic acid; IGA, 3-indoleglyoxylic acid; IHS, 6-hydroxyindole sulfate; IND, indole; KYN, kynurenine; MOX, nicotinamide-N-oxide; NA, nicotinic acid; NAD, NAD+; NAM, nicotinamide; nKYN, n-Acetylkynurenine; NMN, nicotinamide mononucleotide; NR, nicotinamide riboside; QUI, quinolinate; Trig, trigronelline; XAN, xanthurenate.
Within the integrative analysis of 1300 serum metabolites in 484 patients with UC, 464 patients with CD, and 365 controls subject,31 we searched for trp-NAM metabolites in stool and serum samples of patients with IBD, including CD and UC. Significantly lower levels of certain indoles were detected in the serum of patients with IBD compared with healthy control subjects (Figure 6E). Examining the relationship of metabolites with disease indices highlighted the association of low serum MNA and xanthenurate levels with increased disease severity in UC and CD, whereas high fecal kynurenine is linked with the severity of CD (Figure 6F). MNA is the strongest negative correlator with the Mayo endoscopic score for UC (q-value, 5.08E-6) (Figure 6F). In addition, indole acetotylcarnitine consistently displayed significantly lower levels in IBD, whereas indoleacetylglutamine was significantly lower in UC and IBD (Figure 6E). Indole acetic acid derivatives, such as indole-7-acetic acid and methyl indole-3-acetate, exhibited reduced levels in patients with UC and patients with IBD. These findings emphasize that dysregulation in trp-NAM metabolism may play a contributory role in both the onset and progression of IBD.
We then assessed by TEER and dextran permeability all kynurenine-NAM-related metabolites for their effects on the gut barrier. Luminal administration of individual metabolites for 48 hours did not induce significant changes in baseline TEER (Figure 6G and H). However, in the presence of a barrier disruptor, C difficile spent medium, NAM riboside, NAD+, NAM, and MNA ameliorated the disrupted TEER by 25%–60% (Figure 6I). Notably, the marked increase in dextran permeability caused by C difficile was effectively mitigated by luminal kynurenine, NAM riboside, NAD+, NAM, and MNA by 30%–50% (Figure 6J). Before C difficile, basolateral application of NAM mononucleotide and NAM increased baseline TEER modestly after 48 hours (Figure 6K and L). In the presence of C difficile, basolateral kynurenine, NAM mononucleotide, NAM, NAD+, and MNA rescued the TEER by 18%–42% (Figure 6K and M), whereas basolateral kynurenine and MNA prevented marked increases in dextran permeability (Figure 6N). These findings highlight the contribution of trp-NAM metabolic products to enhancing and protecting the gut barrier, with MNA unexpectedly being the most robust candidate.
MNA Protects Barrier Function and Promotes Colonic Healing During Colitis
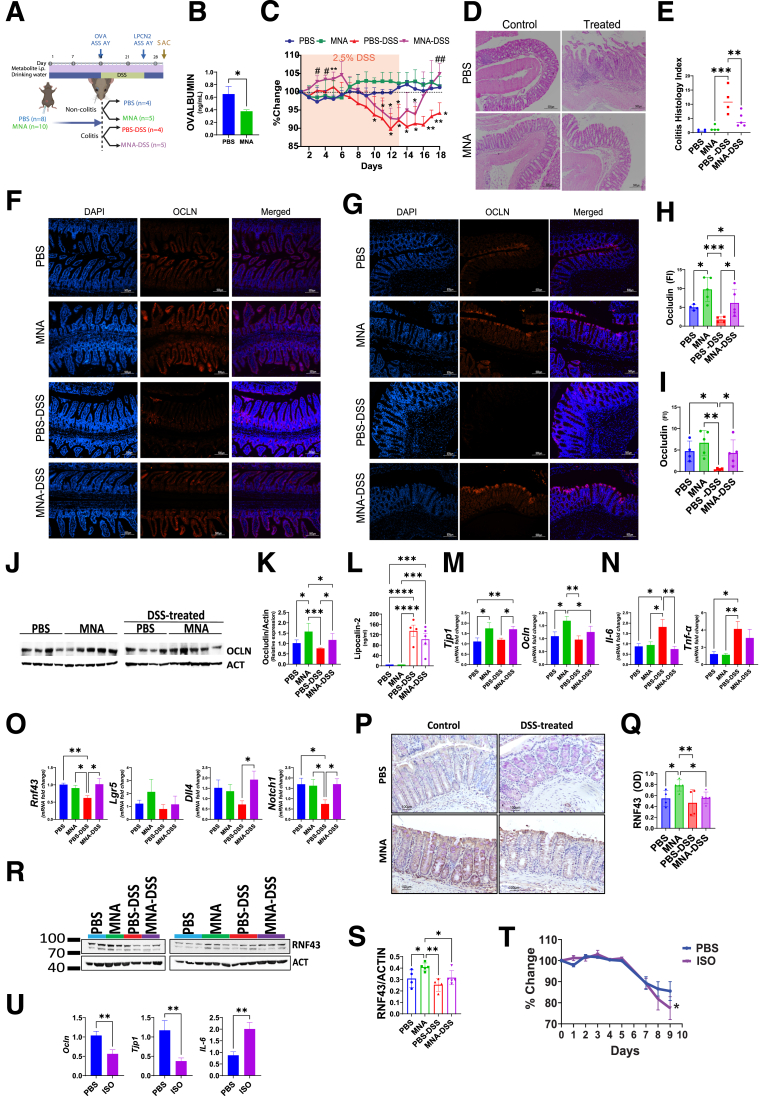
With the compelling in vitro observation of MNA efficacy in barrier enhancement and protection, we next investigated MNA’s effect in vivo. Exogenous MNA, when administered intraperitoneally to specific pathogen-free mice every other day for 2 weeks, significantly increased by 2-fold barrier function as assessed by ovalbumin flux from lumen to plasma, an assay that estimates permeability of the gut paracellular pathway (Figure 7A and B). We then tested MNA effects in 4 groups of mice that were treated with PBS, MNA, PBS– dextran sodium sulfate (DSS), and MNA-DSS (Figure 7C). Although no significant changes in body weight were observed in the noncolitis groups, that of the DSS-treated group declined markedly after 8 days. Remarkably, during the recovery phase, the MNA-treated but not the PBS-treated DSS mice exhibited a robust and rapid recovery, reaching 100% of initial body weight within 4 days, suggesting a remarkable efficacy of MNA in promoting colonic healing. This finding is supported by histologic analysis indicating a substantial amelioration of colonic inflammation and tissue damage in MNA-treated DSS mice (Figure 7D and E).
Figure 7.
MNA mitigates DSS colitis and promotes colonic healing in mice. (A) Experimental design for assessing MNA efficacy against 2.5% DSS-induced colitis. (B) Serum ovalbumin levels (ng/mL) following 2 weeks of MNA (N = 10) or PBS (N = 8) administration (intraperitoneal, every other day). (C) Body weight changes (% of initial) of mice during DSS induction and recovery periods. Data are mean ± standard error. Significant differences are denoted as ∗P < .05, ∗∗P < .01 vs PBS group; and #P < .05, ##P < .01 for PBS-DSS vs MNA-DSS groups. (D) Representative images of colon sections stained with hematoxylin and eosin. (E) Blinded assessment of Mouse Colitis Histology Index (MCHI) score following the recovery period from DSS treatment. Representative images of immunofluorescent staining in (F) ileum and (G) colon, with quantitative analysis of Ocln expression in the (H) ileum and (I) colon. (J) Western blot analysis of Ocln and β-actin (Act) in the ileum, and (K) corresponding quantification. (L) Fecal lipocalin-2 (lpcn2) levels in different groups after DSS treatment. (M-O) Quantitative reverse transcription polymerase chain reaction analysis of gene expression (Tjp1, Ocln, Il-6, Tnf-α, Rnf43, Lgr5, Dll4, Notch1) in the colon at sacrifice. (P) Representative IHC images, and (Q) quantitative analysis of Rnf43 in the colon. (R) Western blot analysis of colon RNF43, and (S) corresponding quantification. (T) Evaluation of trp-NAM deficiency via ISO treatment, including changes in body weight (N = 4 for both PBS =and ISO treatment groups). (U) Expression of Ocln, Tjp1, and Il-6 in the colon at sacrifice. Statistical significance among the groups was determined by 1-way analysis of variance, ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001.
To investigate the protective mechanisms underlying MNA effects, we determined Ocln expression. At steady state, MNA treatment alone led to a 2-fold increase in ileal and colon Ocln levels (Figure 7F-I). DSS treatment markedly reduced Ocln levels, and MNA but not PBS injection rescued significantly Ocln expression (Figure 7D-G). Notably, MNA administration not only rescued colon Ocln levels from the impact of DSS but also maintained them at levels comparable with those of the PBS group (Figure 7F-I). These MNA-mediated effects on Ocln were validated by Western blot analysis (Figure 7J and K).
Moreover, MNA treatment reduced fecal lipocalin levels compared with those of PBS-treated DSS mice, further supporting observations that MNA affords mucosal protection against inflammation (Figure 7L). MNA alone increased Tjp1 and Ocln by >50% at steady state before DSS, whereas MNA treatment after DSS administration prevented DSS-induced reductions in Ocln levels (Figure 7M). Finally, interleukin-6 and tumor necrosis factor-α levels that were low before DSS administration increased markedly in PBS-injected mice, an increase prevented or mitigated by MNA (P < .05) (Figure 7N).
To explore the mechanism of MNA-increased colonic healing, we examined intestinal epithelial differentiation pathway regulators and observed a DSS-induced 40% decrease in intestinal Rnf43 transcripts. This Rnf43 reduction was completely rescued by MNA. Dll4 and Notch1 displayed the same trend as Rnf43 of being promoted by MNA in DSS mice (Figure 7O). In contrast, Lgr5 expression seemed independent of treatment. We then validated by immunohistochemistry (Figure 7P and Q) and Western blot (Figure 7R and S) that MNA significantly increased intestinal Rnf43 in homeostasis and during the DSS colitis recovery phase. These results collectively suggest that MNA may promote epithelial recovery by promoting enterocyte differentiation via regulating key epithelial regenerating pathways involving RNF43 and Notch.
Next, we sought to investigate to what extent the trp-NAM pathway confers protection. We treated mice with isoniazid (ISO), a known antituberculosis drug that interrupts trp absorption and NAM production.32 For 4 days before DSS-induced colitis, we administered ISO once a day (1mg/mouse). ISO treatment led to a significant decrease in body weight in ISO-DSS mice compared with PBS-DSS mice (Figure 7T). ISO exacerbated the colitis by further decreasing colonic Ocln and Tjp1 while increasing colonic IL-6 (Figure 7U). These findings collectively suggest that blocking trp-NAM absorption and metabolism exacerbated colitis.
Discussion
Using our recently developed innovative METRCA algorithm, we uncovered, then validated, specific serum and intestinal metabolites that promote or impair barrier function. Many of the metabolites we discovered correlate positively not only with barrier-promoting Ocln and Tjp1, but interestingly also with sealing claudins but not with leaky claudins. Although expression levels of these TJ components do not equate to barrier function, these results nevertheless demonstrate for the first time that LGG synergizes with a key dietary component to regulate gut epithelial differentiation and permeability by altering the abundances of barrier-modulating metabolites.
We also demonstrate here that LGG drives a dietary trp-dependent metabolic pathway involving NAM that effectively protects the gut barrier against colitogenic insult. Inflamed gut and colitis have been associated with a disrupted trp-NAM metabolism.33,34 We found that serum and fecal MNA, a by-product of NAM metabolism, are elevated in LGG trp+ mice. Our in silico analysis of a published IBD metabolomic dataset31 revealed that serum MNA is strongly and inversely correlated with disease activities in patients with IBD. Additionally, low MNA levels were reported in the urine of IBD cohorts35 further substantiating a dysfunctional NAM metabolism during intestinal inflammation. The positive correlation of serum MNA with 10 TJ genes and its robust promotion of TEER and barrier protection ex vivo and in vivo, collectively underscore its strong barrier-promoting effects. Some strains of lactobacilli make NAM-metabolites,36 and we found that LGG makes kynurenine and NAD+ in vitro, which may fuel intestinal NAM metabolism in the host cells. Indeed, most enzymes and transporters involved in de novo NAM synthesis, recycling, and scavenger pathways are elevated in mouse ileum in response to LGG and dietary trp. Administering exogenous MNA alleviates colitis, whereas administering inhibitors of trp-NAM absorption exacerbates colitis. Thus, we document for the first time that LGG promotes trp-NAM metabolism and that MNA is a barrier protector. Our finding may have implications for precision probiotic intervention based on patient serum MNA abundances.
The essential nature of vitamin B3 and its derivatives as building blocks of NAD for energy is well known, but its role as nonoxidative regulators of physiologic processes is just being appreciated.37,38 This step of NAM-to-MNA methylation is catalyzed by NAM N-methyltransferase as a degradation pathway and is considered irreversible.39 Therefore, MNA has long been viewed as an inert and transient intermediate for eventual disposal via renal excretion. However, MNA is a relatively common metabolite enriched in Judas ear fungus and green tea already consumed by humans, and its potential anti-inflammatory40,41 and vasoprotective activities42 have been proposed by recent studies. The potential transporters for MNA (eg, Mate1, Mate2, OCT1-3) have only been speculated and more studies are needed; however, active uptake of exogenous MNA was shown in canine kidney43,44 and rat liver cells,45 consistent with our observed effects of exogenous MNA in vivo and in vitro. We present compelling data to support MNA’s positive contribution to barrier-strengthening and colonic healing. We found that injecting MNA to wild-type mice is sufficient to elevate RNF43, an E3 ubiquitin ligase that is downstream of AHR signaling and suppresses Wnt signaling by removing Frizzled receptors from cell surfaces.46,47 MNA also rescued DSS-induced reduction in Dll4 and Notch1 in vivo. Interestingly, LGG and dietary trp synergistically promoted RNF43 and Dll ligands, reflecting a Wnt-Off and Notch-On scenario favorable for enterocyte differentiation.48 The Wnt-Off condition is also consistent with a suppression of all Paneth cell transcriptome. Indeed, a robustly promoted enterocyte program and a suppressed Paneth cell program were only found in LGG trp+ mice, suggesting a cellular differentiation program uniquely regulated by LGG in the presence of dietary trp. Our finding of MNA-regulated RNF43 may reflect a previously reported link of the canonical Wnt pathway with NAD+ salvage pathway.49 Our finding of MNA-mediated stabilization of Ocln and RNF43 during DSS colitis indicates a promoted stem cell to enterocyte differentiation and maturation, ultimately strengthening the gut barrier and health.
"Leaky gut” allows the entry of harmful substances from the intestinal lumen into circulation, thereby triggering a cascade of metabolic alterations50 and inflammatory51 response in the host. Because impairment of gut barrier stands as an evolutionarily conserved hallmark of aging,52 the light shed by this study on LGG presents a promising avenue for delaying the age-related decline in gut health. Interestingly, vitamin B3 derivatives, such as NAM riboside, are considered as beneficial supplements for healthy aging and longevity.53 In this context, our use of GF mice provided a controlled environment to dissect a simplified interplay among LGG, dietary trp, and the host. Our findings of trp-NAM were validated in conventional mice that harbor intact gut microbiota, and highlight the reliance of certain LGG's beneficial effects on key dietary components, such as trp. Our metabolome-based observations may apply to other lactobacilli species. Lacticaseibacillus plantarum improves barrier function by enhancing TJ proteins,54 whereas Lacticaseibacillus acidophilus increases mucin production.55 Lacticaseibacillus casei influences antimicrobial peptide production, bolstering host defense,56 and Lacticaseibacillus reuteri regulates cytokine release to influence intestinal epithelium.57 Our study suggests that the salutary effects of probiotics may be enhanced by trp supplementation, and the inconsistent findings may be attributed to dietary differences in previous studies. Furthermore, LGG-derived indoles demonstrate improvements in disrupted barriers in Caco-2 and enteroid models. Noteworthy indoles, such as indole-3-propionic acid,58 indole-3-acetic acid,59 indole-3-lactic acid,60 indole-3-pyruvate,61 and indole-3-aldehyde61 have been shown in various investigations to promote gut homeostasis by activating AHR61 and PXR receptors.62,63 These receptors, in turn, may regulate the integrity of epithelial and immune cells, highlighting the broader implications of probiotic interventions. Future work will delineate how MNA stimulates TJ gene expression and barrier function. We conclude that LGG stimulates dietary trp-dependent production of barrier-protecting MNA.
Materials and Methods
Animal procedures and protocols were in accordance with the Institutional Animal Care and Use Committee, Rutgers New Jersey Medical School.
Experimental Designs
Gnotobiotic study
GF mice (C57BL/6, Charles River Laboratories) were fed either trp-free or trp-sufficient diets.30 The trp-sufficient diet, containing 0.2% trp, had otherwise exactly the same composition as the trp-free. Mice were acclimatized for 2 weeks then divided into 4 groups, 2 of which were fed trp-sufficient, and the other 2 fed trp-free diets for another 2 weeks (Figure 1A). At Week 0, mice in trp-free and trp-sufficient conditions were divided into cohorts that were gavaged with either LGG (108 CFU/mL in 200 μL PBS) or sterile PBS. Mice were continuously fed with trp-free and trp-sufficient diets until sacrifice 21 days after inoculation. Fecal pellets were collected before and after inoculation. Feces, intestine, serum, and liver were collected at sacrifice.
LGG administration and GF monitoring
LGG was cultured (37°C) in MRS broth and centrifuged (1200 × g, 10 minutes) to pellet before gavaging to mice. Body weight and feeding rate were monitored. GF status before inoculation was validated by a commercial vendor (IDEXX) and constantly monitored in-house by fecal DNA analysis of bacterial 16S rRNA. After inoculation, qPCR for fecal DNA with 16S rRNA and LGG-specific primers were used to confirm colonization. Same engraftment efficiency of LGG was found in mice fed with trp+ or trp- diets. LGG qPCR primers are: forward, 5’-CGCCCTTAACAGCAGTCTTC-3’ and reverse, 5’-GCCCTCCGTATGCTTAAACC-3’. The 16S rRNA qPCR primers are forward 5'-AGAGTTTGATCCTGGCTCAG-3’, and reverse, 5'- CTTGTGCGGGCCCCCGTCAATTC-3'.
Intestinal perfusion
Procedures followed previous work.64 Mice were perfused with live LGG (3.3X108 CFU/mL) + Inulin (3.3 mg/mL), or with Krebs-Ringer Buffer + inulin. After 4 hours of perfusion, the intestines were collected for RNA sequencing.
DSS-induced colitis and metabolite treatment
C57BL/6 mice were intraperitoneally injected with PBS or 30 mg/kg MNA every other day for 2 weeks then gavaged with 100 μg/kg chicken ovalbumin (Figure 7A). About 24 hours postgavage, plasma ovalbumin levels were quantified using an enzyme-linked immunosorbent assay kit (abx555335, Abbexa, UK) to determine gut permeability in homeostasis. For DSS experiments, the mice were grouped into PBS, MNA, PBS+DSS, and MNA+DSS cohorts and subjected to 2.5% DSS treatment for 11 days, followed by a 5-day recovery period. MNA was injected every other day after initiation of DSS treatment. After sacrifice, tissues were collected for immunohistochemistry and immunofluorescence staining, qPCR, and Western blot analysis, whereas fecal samples from Day 11 were analyzed for lipocalin 2 levels using enzyme-linked immunosorbent assay (ab199083).
Caco-2 transepithelial electrical resistance and paracellular flux
Cells (20,000 per well) were suspended in Dulbecco's modified eagle medium media (20% fetal bovine serum, 1% Primocin) in 96-well transplates (MilliporeSigma PSHT004R5) and polarized for 21 days. TEER was measured with an EVOM system using an STX100 electrode (World Precision Instruments). Indole and NAM-derived metabolites were added apically or basolaterally (Table 1). To induce barrier disruption and assess efficacy of metabolites to repair disruption, Caco-2 monolayers were treated with 0.15% vol/vol C difficile cell-free supernatant. Paracellular flux was assessed by introducing FITC-dextran (5 mg/mL) to apical chambers, and FITC fluorescence in 10 μL samples from the basolateral chamber was measured in a black 96-well plate (Costar 3915) using a Glomax (Promega, WI) plate reader (excitation: 490 nm, emission: 520 nm).
Table 1.
Metabolite Dosing Information
| Metabolites | Dose | Application |
|---|---|---|
| Kynurenine | 10 mM | Caco-2 luminal/basolateral treatment |
| Nicotinamide riboside | 10 mM | |
| Nicotinamide mononucleotide | 10 mM | |
| NAD+ | 10 mM | |
| Nicotinamide | 10 mM | |
| Methylnicotinamide | 10 mM | |
| Indole-3-acetamide | 500 μM | Caco2 basolateral treatment |
| Indole-3-carboxyaldehyde | 500 μM | |
| Indole-3-lactic acid | 500 μM | |
| Indole-3-propionic acid | 500 μM | |
| Indole-3-acrylic acid | 250 μM | |
| Indole-3-carbinol | 250 μM | |
| Indole-3-acetamide | 250 μM | Caco2 luminal treatment |
| Indole-3-acetic acid | 250 μM | |
| Indole-3-acetonitrile | 250 μM | |
| Indole-3-carboxylic acid | 250 μM | |
| Indole-3-carboxyaldehyde | 100 μM | |
| Indole-3-lactic acid | 50 μM | |
| Indole-3-propionic acid | 100 μM | |
| Indole-3-acrylic acid | 250 μM | |
| Indole-3-carbinol | 100 μM | |
| Indole-3-propionic acid | 0.1 mM | Enteroids |
| Indole-3- acetamide | 200 μM | |
| Methylnicotinamide | 87 nM | |
| Carnosine | 20 mM |
Intestinal organoid treatment with LPS and metabolites
Enteroids were isolated as previously described.48 To grow wild-type and Ocln KO enteroids, Oclnfl/fl and Oclnfl/fl-Vil/CREERT2 mice were injected (intraperitoneally) with tamoxifen and sacrificed 7 days after the injection to establish organoids. Day 0 was when enteroids were seeded. On Day 3, media were replaced with LPS (1 μg/mL) with or without metabolites (indole-3-propionic acid [0.1 mM],65 indole-3-acetamide [200 μM], MNA [12 ng/mL], carnosine [20 mM]), incubated for 48 hours, and analyzed.
Bulk RNA sequencing
Before analyses, the high quality of extracted ileal RNA from all samples was confirmed (RIN, 8.6 ± 0.56). Total RNA was subjected to poly(A) selection using oligo(dT)25 magnetic beads (New England Biolabs). Libraries were constructed using Next (New England Biolabs) Ultra II RNA library preparation with sample purification beads and Next Multiplex Oligos. Poly(A) selection and library quality were assessed using TapeStation 2200 (Agilent) and libraries quantified (Qubit 4.0 fluorometer, ThermoFisher). The prepared libraries were sequenced (Illumina NextSeq 500) then analyzed by CLC Genomics Workbench 20.0.4.
Metabolite extraction and liquid chromatography–mass spectrometry analysis
Feces and liver metabolites were extracted using the reported procedures.29 Serum metabolites were extracted in methanol (ratio of serum:methanol, 1:4) at –20°C for 20 minutes, and centrifuged at 17,000 × g for 2 minutes. Supernatants were stored on ice. The pellet was further extracted in acetonitrile:methanol:water (400:400:200), for 10 minutes on ice, centrifuged at 17,000 × g for 10 minutes, and the supernatant combined with the first extraction. Liquid chromatography–mass spectrometry analysis was performed as previously described.29 The metabolite data were analyzed in MAVEN (Metabolic Analysis and Visualization Engine). Compound identification used the accurate mass and the retention time learned from an in-house chemical library. MetaboAnalyst 5.0 was then used for further statistical and functional interpretation of metabolomics data. Finally, principal component analysis and variables important in the projection (cutoff 1.229) analyses were used to identify those metabolites substantially driving differences between 2 treatments.
RT-qPCR analysis
Primers used include: β-actin (Actb), forward 5’-TTG TTA CCA ACT GGG ACG ACA TGG-3’, reverse 5’- CTG GGG TGT TGA AGG TCT CAA ACA-3’; ZO-1 (Tjp1), forward 5’-GGG AGG GTC AAA TGA AGA CA-3’, reverse 5’-GGC ATT CCT GCT GGT TAC AT-3’; Ocln, forward 5’-ATT CCA TCA GTT TCC TAT CT-3’, reverse 5’-ACC AGG ACC TTT CTT GAC-3’; Lyz1, forward 5’-ATG GCT ACC GTG CT TCA AG-3’, reverse 5’- CGG TCT CCA CGG TTG TAG TT-3’; Tff3, forward 5’- TAA TGC TGT TGG TGF TCC TG-3’, reverse 5’- CAG CCA CGG TTG TTA CAC TG-3’, Dll4, forward 5’-AGC TGG AAG TGG ACT GTG GT-3’, reverse 5’-TAG AGT CCC TGG GAG AGC AA-3’; Rnf43, forward 5’-CCG GGT CAT TTC GTG CCT C-3’, reverse 5’-CCT GGT TCC TGG TAA GAT GGA-3’; Lgr5, forward 5’- TGA GCG GGA CCT TGA AGA TT-3’, reverse 5’ AGG TGC TCA CAG GGC TTG AA-3; Notch1, forward 5’- ATG TCG ATG TTC GAG GAC CAG-3’, reverse 5’ TCA CTG TTG CCT GTC TCA AG-3’; IL-6 forward 5’- ACC ACG AGG ATC AGT ACG AA-3’, reverse 5’- TGT TCFG CAT AAG GGC TCT GT-3’;Tnf- α forward 5’- GAT CTC AAA GAC AAC CAA CAT GTG-3’, reverse 5’- CTC CAG CTG GAA GAC TCC AAB CAG-3’. All samples were normalized to β actin.
Permeability and immunohistochemistry
Permeability and immunohistochemistry of enteroids were described previously.48 For colon and ileal tissues, the sections (5 μm) underwent antigen retrieval in 0.1 M citrate acid buffer (pH 6) at subboiling temperature for 20 minutes. Following blocking with PBS containing 0.1% Triton X-100, 2% normal serum, and 2% bovine serum albumin for 1 hour at room temperature, slides were incubated with primary antibodies overnight at 4°C. After 3 PBS washes, slides were incubated with fluorescent dye-conjugated secondary antibodies for 1 hour at room temperature, followed by DAPI nuclear counterstaining for 10 minutes. Mounted (Prolong Gold Antifade Mountant) slides were imaged at 10X magnification with 3 representative sections per mouse using Nikon TE2000. Quantitative analysis was performed using Image J. Primary antibodies were diluted in blocking buffer. Primary antibodies were rabbit monoclonal OCLN, mouse monoclonal ZO-1 and TFF3 (AB216327, 1A12 and 14475882, respectively, ThermoFisher) and rabbit polyclonal LYZ1 (PU024-5UP, Biogenex).
METRCA
Correlation analysis between metabolome and transcriptome used our newly developed R script.30 Transcriptome data were filtered and only differentially expressed genes (fold change >1.5, FDR-adjusted P < .05) were considered. Pairwise Pearson correlation coefficients were calculated between genes and metabolites, and their statistical significance was obtained via the cor.test function in R. Metabolite-gene pairs with FDR-adjusted P < .05 were significantly associated. Heatmap was generated by pheatmap package in R. Genes were clustered by hierarchical clustering.
Statistical analysis
All results are expressed as mean ± standard error of the mean. Significance was determined by 1-way or 2-way analysis of variance, Student t test, and Tukey post hoc test. Animal numbers were described in each figure panel with individual data points plotted in the graphs.
Acknowledgments
The authors are grateful to Jayanth Ambat, Abby Jones, Paula Lopez, Niyati Boghani, Nisarg Baboo, Aditi Mandaliya, Temitope Ali, Denise Zieba, and Eric Chiles for help in experiments; to Dr Preeti Raju (Harvard Medical School) for help with Ocln-KO colony; to Fang Yan, PhD (Vanderbilt University) for providing p40 as a gift; and the staff of Comparative Medicine Resources, Rutgers University for help in establishing our germ-free colonies. Panan Suntornsaratoon and Jayson M. Antonio contributed equally to this work. Figures created with Biorender.com.
CRediT Authorship Contributions
Panan Suntornsaratoon, PhD (Data curation: Lead; Formal analysis: Lead; Investigation: Equal; Supervision: Supporting; Validation: Equal; Writing – original draft: Equal; Writing – review & editing: Supporting)
Jayson M. Antonio, PhD (Conceptualization: Equal; Data curation: Lead; Formal analysis: Equal; Investigation: Lead; Methodology: Equal; Validation: Equal; Writing – original draft: Equal; Writing – review & editing: Equal)
Juan Flores, PhD (Data curation: Equal; Formal analysis: Equal; Investigation: Equal; Validation: Equal)
Ravij Upadhyay, MS (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Validation: Supporting)
John Veltri, DO (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting)
Sheila Bandyopadhyay, PhD (Funding acquisition: Supporting; Investigation: Supporting; Methodology: Supporting)
Rhema Dadala, BS (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting)
Michael Kim, MS (Investigation: Supporting; Methodology: Supporting; Validation: Supporting)
Yue Liu (Investigation: Supporting; Methodology: Supporting; Validation: Supporting)
Ishwarya Balasubramanian, PhD (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting)
Jerrold R. Turner, PhD (Resources: Equal; Validation: Supporting; Writing – review & editing: Supporting)
Xiaoyang Su, PhD (Data curation: Lead; Formal analysis: Supporting; Methodology: Equal; Validation: Supporting; Writing – review & editing: Supporting)
Wei Vivian Li, PhD (Formal analysis: Lead; Methodology: Lead; Validation: Supporting; Writing – review & editing: Supporting)
Nan Gao, PhD (Conceptualization: Lead; Formal analysis: Equal; Investigation: Equal; Project administration: Lead; Supervision: Equal; Writing – original draft: Equal; Writing – review & editing: Equal)
Ronaldo P. Ferraris, PhD (Formal analysis: Lead; Funding acquisition: Lead; Investigation: Equal; Project administration: Equal; Supervision: Lead; Writing – original draft: Equal; Writing – review & editing: Equal)
Footnotes
Conflicts of interest The authors disclose no conflicts.
FundingNational Institutes of Health grants R01-AT010243 (NG, RPF), R01DK102934 and R01DK119198 (NG), R35GM142702 (WVL), F31 DK121428 (SB), and R01 DK061931 (JRT). NSF Grant No. IOS 1754783 (RPF), NSF Grant 1952823 (NG). Services, results, and products in support of this research were generated by the Rutgers Cancer Institute of New Jersey Metabolomics Shared Resource, supported, in part, with funding from NCI-CCSG P30CA072720-5923.
Contributor Information
Nan Gao, Email: ngao@newark.rutgers.edu.
Ronaldo P. Ferraris, Email: ferraris@njms.rutgers.edu.
References
- 1.Gunzel D., Yu A.S. Claudins and the modulation of tight junction permeability. Physiol Rev. 2013;93:525–569. doi: 10.1152/physrev.00019.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang G.H., Weber C.R. Molecular aspects of tight junction barrier function. Curr Opin Pharmacol. 2014;19:84–89. doi: 10.1016/j.coph.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zihni C., Mills C., Matter K., et al. Tight junctions: from simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol. 2016;17:564–580. doi: 10.1038/nrm.2016.80. [DOI] [PubMed] [Google Scholar]
- 4.Tsukita S., Tanaka H., Tamura A. The claudins: from tight junctions to biological systems. Trends Biochem Sci. 2019;44:141–152. doi: 10.1016/j.tibs.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Horowitz A., Chanez-Paredes S.D., Haest X., et al. Paracellular permeability and tight junction regulation in gut health and disease. Nat Rev Gastroenterol Hepatol. 2023;20:417–432. doi: 10.1038/s41575-023-00766-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciccocioppo R., Finamore A., Ara C., et al. Altered expression, localization, and phosphorylation of epithelial junctional proteins in celiac disease. Am J Clin Pathol. 2006;125:502–511. doi: 10.1309/DTYR-A91G-8R0K-TM8M. [DOI] [PubMed] [Google Scholar]
- 7.Zeissig S., Bürgel N., Günzel D., et al. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn's disease. Gut. 2007;56:61–72. doi: 10.1136/gut.2006.094375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu L.C. Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: exploring a common ground hypothesis. J Biomed Sci. 2018;25:79. doi: 10.1186/s12929-018-0483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehandru S., Colombel J.F. The intestinal barrier, an arbitrator turned provocateur in IBD. Nat Rev Gastroenterol Hepatol. 2021;18:83–84. doi: 10.1038/s41575-020-00399-w. [DOI] [PubMed] [Google Scholar]
- 10.Sayoc-Becerra A., Krishnan M., Fan S., et al. The JAK-inhibitor tofacitinib rescues human intestinal epithelial cells and colonoids from cytokine-induced barrier dysfunction. Inflamm Bowel Dis. 2020;26:407–422. doi: 10.1093/ibd/izz266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikolaus S., Schulte B., Al-Massad N., et al. Increased tryptophan metabolism is associated with activity of inflammatory bowel diseases. Gastroenterology. 2017;153:1504–1516.e2. doi: 10.1053/j.gastro.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 12.Turpin W., Lee S.H., Raygoza Garay J.A., et al. Increased intestinal permeability is associated with later development of Crohn's Disease. Gastroenterology. 2020;159:2092–2100.e5. doi: 10.1053/j.gastro.2020.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Yoo J.H., Donowitz M. Intestinal enteroids/organoids: a novel platform for drug discovery in inflammatory bowel diseases. World J Gastroenterol. 2019;25:4125–4147. doi: 10.3748/wjg.v25.i30.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Owczarek D., Rodacki T., Domagala-Rodacka R., et al. Diet and nutritional factors in inflammatory bowel diseases. World J Gastroenterol. 2016;22:895–905. doi: 10.3748/wjg.v22.i3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hashimoto T., Perlot T., Rehman A., et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim C.J., Kovacs-Nolan J.A., Yang C., et al. l-Tryptophan exhibits therapeutic function in a porcine model of dextran sodium sulfate (DSS)-induced colitis. J Nutr Biochem. 2010;21:468–475. doi: 10.1016/j.jnutbio.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 17.Castro-Portuguez R., Sutphin G.L. Kynurenine pathway, NAD(+) synthesis, and mitochondrial function: targeting tryptophan metabolism to promote longevity and healthspan. Exp Gerontol. 2020;132 doi: 10.1016/j.exger.2020.110841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi J., Toyoshima A., Honda Y., et al. Pharmacokinetic study on acetaminophen: interaction with a Chinese medicine. J Med Dent Sci. 1997;44:31–35. [PubMed] [Google Scholar]
- 19.Roager H.M., Licht T.R. Microbial tryptophan catabolites in health and disease. Nat Commun. 2018;9:3294. doi: 10.1038/s41467-018-05470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vich Vila A., Hu S., Andreu-Sanchez S., et al. Faecal metabolome and its determinants in inflammatory bowel disease. Gut. 2023;72:1472–1485. doi: 10.1136/gutjnl-2022-328048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.ClinicalTrials.gov. ClinicalTrials.gov. National Library of Medicine; 2022. [Google Scholar]
- 22.Capurso L. Thirty years of Lactobacillus rhamnosus GG: a review. J Clin Gastroenterol. 2019;53(Suppl 1):S1–S41. doi: 10.1097/MCG.0000000000001170. [DOI] [PubMed] [Google Scholar]
- 23.Li Y.T., Xu H., Ye J.Z., et al. Efficacy of Lactobacillus rhamnosus GG in treatment of acute pediatric diarrhea: a systematic review with meta-analysis. World J Gastroenterol. 2019;25:4999–5016. doi: 10.3748/wjg.v25.i33.4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mantegazza C., Molinari P., D'Auria E., et al. Probiotics and antibiotic-associated diarrhea in children: a review and new evidence on Lactobacillus rhamnosus GG during and after antibiotic treatment. Pharmacol Res. 2018;128:63–72. doi: 10.1016/j.phrs.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Szajewska H., Kołodziej M. Systematic review with meta-analysis: Lactobacillus rhamnosus GG in the prevention of antibiotic-associated diarrhoea in children and adults. Aliment Pharmacol Ther. 2015;42:1149–1157. doi: 10.1111/apt.13404. [DOI] [PubMed] [Google Scholar]
- 26.McFarland L.V., Evans C.T., Goldstein E.J.C. Strain-specificity and disease-specificity of probiotic efficacy: a systematic review and meta-analysis. Front Med (Lausanne) 2018;5:124. doi: 10.3389/fmed.2018.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sentongo T.A., Cohran V., Korff S., et al. Intestinal permeability and effects of Lactobacillus rhamnosus therapy in children with short bowel syndrome. J Pediatr Gastroenterol Nutr. 2008;46:41–47. doi: 10.1097/01.mpg.0000304452.92175.f5. [DOI] [PubMed] [Google Scholar]
- 28.Francavilla R., Miniello V., Magista A.M., et al. A randomized controlled trial of Lactobacillus GG in children with functional abdominal pain. Pediatrics. 2010;126:e1445–e1452. doi: 10.1542/peds.2010-0467. [DOI] [PubMed] [Google Scholar]
- 29.Kim J., Balasubramanian I., Bandyopadhyay S., et al. Lactobacillus rhamnosus GG modifies the metabolome of pathobionts in gnotobiotic mice. BMC Microbiol. 2021;21:165. doi: 10.1186/s12866-021-02178-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suntornsaratoon P., Ferraris R.P., Ambat J., et al. Metabolomic and transcriptomic correlative analyses in germ-free mice link Lacticaseibacillus rhamnosus GG-associated metabolites to host intestinal fatty acid metabolism and beta-oxidation. Lab Invest. 2024;104 doi: 10.1016/j.labinv.2024.100330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di'Narzo A.F., Houten S.M., Kosoy R., et al. Integrative analysis of the inflammatory bowel disease serum metabolome improves our understanding of genetic etiology and points to novel putative therapeutic targets. Gastroenterology. 2022;162:828–843. e11. doi: 10.1053/j.gastro.2021.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nabity S.A., Mponda K., Gutreuter S., et al. Isoniazid-associated pellagra during mass scale-up of tuberculosis preventive therapy: a case-control study. Lancet Glob Health. 2022;10:e705–e714. doi: 10.1016/S2214-109X(22)00096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michaudel C., Danne C., Agus A., et al. Rewiring the altered tryptophan metabolism as a novel therapeutic strategy in inflammatory bowel diseases. Gut. 2023;72:1296–1307. doi: 10.1136/gutjnl-2022-327337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen C., Yan W., Tao M., et al. NAD(+) metabolism and immune regulation: new approaches to inflammatory bowel disease therapies. Antioxidants (Basel) 2023;12:1230. doi: 10.3390/antiox12061230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aldars-Garcia L., Gil-Redondo R., Embade N., et al. Serum and urine metabolomic profiling of newly diagnosed treatment-naive inflammatory bowel disease patients. Inflamm Bowel Dis. 2024;30:167–182. doi: 10.1093/ibd/izad154. [DOI] [PubMed] [Google Scholar]
- 36.Sugiyama K., Iijima K., Yoshino M., et al. Nicotinamide mononucleotide production by fructophilic lactic acid bacteria. Sci Rep. 2021;11:7662. doi: 10.1038/s41598-021-87361-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chini C.C.S., Zeidler J.D., Kashyap S., et al. Evolving concepts in NAD(+) metabolism. Cell Metab. 2021;33:1076–1087. doi: 10.1016/j.cmet.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Covarrubias A.J., Perrone R., Grozio A., et al. NAD(+) metabolism and its roles in cellular processes during ageing. Nat Rev Mol Cell Biol. 2021;22:119–141. doi: 10.1038/s41580-020-00313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pissios P. Nicotinamide N-methyltransferase: more than a vitamin B3 clearance enzyme. Trends Endocrinol Metab. 2017;28:340–353. doi: 10.1016/j.tem.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bryniarski K., Biedron R., Jakubowski A., et al. Anti-inflammatory effect of 1-methylnicotinamide in contact hypersensitivity to oxazolone in mice; involvement of prostacyclin. Eur J Pharmacol. 2008;578:332–338. doi: 10.1016/j.ejphar.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 41.Kannt A., Rajagopal S., Kadnur S.V., et al. A small molecule inhibitor of nicotinamide N-methyltransferase for the treatment of metabolic disorders. Scientific Reports. 2018;8:3660. doi: 10.1038/s41598-018-22081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Domagala T.B., Szeffler A., Dobrucki L.W., et al. Nitric oxide production and endothelium-dependent vasorelaxation ameliorated by N1-methylnicotinamide in human blood vessels. Hypertension. 2012;59:825–832. doi: 10.1161/HYPERTENSIONAHA.111.183210. [DOI] [PubMed] [Google Scholar]
- 43.Ross C.R., Pessah N.I., Farah A. Studies of uptake and runout of p-aminohippurate and N-methylnicotinamide in dog renal slices. J Pharmacol Exp Ther. 1968;160:381–386. [PubMed] [Google Scholar]
- 44.Farah A., Frazer M., Porter E. Studies on the uptake of N'-methylnicotinamide by renal slices of the dog. J Pharmacol Exp Ther. 1959;126:202–211. [PubMed] [Google Scholar]
- 45.Moseley R.H., Morrissette J., Johnson T.R. Transport of N1-methylnicotinamide by organic cation-proton exchange in rat liver membrane vesicles. Am J Physiol. 1990;259:G973–G982. doi: 10.1152/ajpgi.1990.259.6.G973. [DOI] [PubMed] [Google Scholar]
- 46.Metidji A., Omenetti S., Crotta S., et al. The environmental sensor AHR protects from inflammatory damage by maintaining intestinal stem cell homeostasis and barrier integrity. Immunity. 2018;49:353–362.e5. doi: 10.1016/j.immuni.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wisniewski P.J., Nagarkatti M., Nagarkatti P.S. Regulation of intestinal stem cell stemness by the aryl hydrocarbon receptor and its ligands. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.638725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearce S.C., Al-Jawadi A., Kishida K., et al. Marked differences in tight junction composition and macromolecular permeability among different intestinal cell types. BMC Biol 2018;16:19. epithelial barrier and mucus barrier. J Agric Food Chem. 2021;69:1487–1495. doi: 10.1186/s12915-018-0481-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bineau E., Rambla J.L., Duboscq R., et al. Inheritance of secondary metabolites and gene expression related to tomato fruit quality. Int J Mol Sci. 2022;23:6163. doi: 10.3390/ijms23116163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stewart A.S., Pratt-Phillips S., Gonzalez L.M. Alterations in intestinal permeability: the role of the "leaky gut" in health and disease. J Equine Vet Sci. 2017;52:10–22. doi: 10.1016/j.jevs.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L., Yan J., Zhang C., et al. Improving intestinal inflammaging to delay aging? A new perspective. Mech Ageing Dev. 2023;214 doi: 10.1016/j.mad.2023.111841. [DOI] [PubMed] [Google Scholar]
- 52.Salazar A.M., Aparicio R., Clark R.I., et al. Intestinal barrier dysfunction: an evolutionarily conserved hallmark of aging. Dis Model Mech. 2023;16 doi: 10.1242/dmm.049969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bita A., Scorei I.R., Ciocilteu M.V., et al. Nicotinamide riboside, a promising vitamin b(3) derivative for healthy aging and longevity: current research and perspectives. Molecules. 2023;28:6078. doi: 10.3390/molecules28166078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karczewski J., Troost F.J., Konings I., et al. Regulation of human epithelial tight junction proteins by Lactobacillus plantarum in vivo and protective effects on the epithelial barrier. Am J Physiol Gastrointest Liver Physiol. 2010;298:G851–G859. doi: 10.1152/ajpgi.00327.2009. [DOI] [PubMed] [Google Scholar]
- 55.Bergstrom A., Kristensen M.B., Bahl M.I., et al. Nature of bacterial colonization influences transcription of mucin genes in mice during the first week of life. BMC Res Notes. 2012;5:402. doi: 10.1186/1756-0500-5-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bai Y., Huang Y., Li Y., et al. The murine Reg3a stimulated by Lactobacillus casei promotes intestinal cell proliferation and inhibits the multiplication of porcine diarrhea causative agent in vitro. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.675263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao J., Cao S., Xiao H., et al. Lactobacillus reuteri 1 enhances intestinal epithelial barrier function and alleviates the inflammatory response induced by enterotoxigenic Escherichia coli K88 via suppressing the MLCK signaling pathway in IPEC-J2 cells. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.897395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alexeev E.E., Lanis J.M., Kao D.J., et al. Microbiota-derived indole metabolites promote human and murine intestinal homeostasis through regulation of interleukin-10 receptor. Am J Pathol. 2018;188:1183–1194. doi: 10.1016/j.ajpath.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang X., Shi L., Wang N., et al. Gut bacterial indole-3-acetic acid induced immune promotion mediates preventive effects of fu brick tea polyphenols on experimental colitis. J Agric Food Chem. 2023;71:1201–1213. doi: 10.1021/acs.jafc.2c06517. [DOI] [PubMed] [Google Scholar]
- 60.Cui Q., Zhang Z., Tian X., et al. Bifidobacterium bifidum ameliorates DSS-induced colitis in mice by regulating AHR/NRF2/NLRP3 inflammasome pathways through indole-3-lactic acid production. J Agric Food Chem. 2023;71:1970–1981. doi: 10.1021/acs.jafc.2c06894. [DOI] [PubMed] [Google Scholar]
- 61.Scott S.A., Fu J., Chang P.V. Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc Natl Acad Sci U S A. 2020;117:19376–19387. doi: 10.1073/pnas.2000047117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Venkatesh M., Mukherjee S., Wang H., et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Illes P., Krasulova K., Vyhlidalova B., et al. Indole microbial intestinal metabolites expand the repertoire of ligands and agonists of the human pregnane X receptor. Toxicol Lett. 2020;334:87–93. doi: 10.1016/j.toxlet.2020.09.015. [DOI] [PubMed] [Google Scholar]
- 64.Jiang L., Ferraris R.P. Developmental reprogramming of rat GLUT5 requires de novo mRNA and protein synthesis. Am J Physiol Gastrointest Liver Physiol. 2001;280:G113–G120. doi: 10.1152/ajpgi.2001.280.1.G113. [DOI] [PubMed] [Google Scholar]
- 65.Li J., Zhang L., Wu T., et al. Indole-3-propionic acid improved the intestinal barrier by enhancing epithelial barrier and mucus barrier. J Agric Food Chem. 2021;69:1487–1495. doi: 10.1021/acs.jafc.0c05205. [DOI] [PubMed] [Google Scholar]