Abstract
Mitochondrial aldehyde dehydrogenase (ALDH2) offers proven cardiovascular benefit, although its impact on diabetes remains elusive. This study examined the effects of ALDH2 overexpression and knockout on diabetic cardiomyopathy and the mechanism involved with a focus on mitochondrial integrity. Mice challenged with streptozotocin (STZ, 200 mg/kg, via intraperitoneal injection) exhibited pathological alterations, including reduced respiratory exchange ratio, dampened fractional shortening and ejection fraction, increased left ventricular end-systolic and diastolic diameters, cardiac remodeling, cardiomyocyte contractile anomalies, intracellular Ca2+ defects, myocardial ultrastructural injury, oxidative stress, apoptosis, and mitochondrial damage, which were overtly attenuated or accentuated by ALDH2 overexpression or knockout, respectively. Diabetic patients also exhibited reduced plasma ALDH2 activity, cardiac remodeling, and diastolic dysfunction. In addition, STZ challenge altered expression levels of mitochondrial proteins (PGC-1α and UCP2) and Ca2+ regulatory proteins (SERCA, Na+–Ca2+ exchanger, and phospholamban), dampened autophagy and mitophagy (LC3B ratio, TOM20, Parkin, FUNDC1, and BNIP3), disrupted phosphorylation of Akt, GSK3β, and Foxo3a, and elevated PTEN phosphorylation, most of which were reversed or worsened by ALDH2 overexpression or knockout, respectively. Furthermore, the novel ALDH2 activator torezolid, as well as the classical ALDH2 activator Alda-1, protected against STZ- or high glucose-induced in vivo or in vitro cardiac anomalies, which was nullified by inhibition of Akt, GSK3β, Parkin, or mitochondrial coupling. Our data discerned a vital role for ALDH2 in diabetic cardiomyopathy possibly through regulation of Akt and GSK3β activation, Parkin mitophagy, and mitochondrial function.
Keywords: ALDH2, diabetes, mitophagy, cardiac contraction, GSK3β, mitochondrial function
Introduction
Mitochondrial aldehyde dehydrogenase (ALDH2) possesses a rather important role in the metabolism of acetaldehyde and toxic aldehydes (Zhang and Ren, 2011; Pang et al., 2015; Wu and Ren, 2019; Wu et al., 2022b). More clinical and experimental findings from our group and others have revealed an important role for ALDH2 in cardiovascular diseases, including ischemia injury, sepsis, aging, atherosclerosis, aortic dissection, obesity, arrhythmias, hypertension, and alcoholism (Chen et al., 2008; Doser et al., 2009; Ma et al., 2009; Koda et al., 2010, 2011; Zhang et al., 2017; Wang et al., 2018, 2020; Yang et al., 2018, 2020; Goodnough and Gross, 2020; Jin et al., 2022; Yu et al., 2022; Zhu et al., 2022). More recent evidence has indicated a correlation between ALDH2 polymorphism and increased risk of diabetes mellitus (Guo et al., 2015; Munukutla et al., 2019; Yang et al., 2021), although little knowledge is readily available for the precise role of ALDH2 in cardiovascular anomalies associated with metabolic disorders including diabetes mellitus. As one of the most common metabolic comorbidities worldwide, the prevalence of diabetes is rapidly rising, in particular in East Asian where ALDH2 polymorphism is frequently seen (Xu et al., 2010). This is in line with the findings of compromised ALDH2 expression and/or activity in association with pronounced oxidative stress and myocardial injury in diabetes (Wang et al., 2011). Up-to-date, several scenarios have been put forward for the pathogenesis of diabetic complications including diabetic cardiomyopathy. Of note, oxidative stress, mitochondrial injury, inflammation, and various forms of cell death including apoptosis and ferroptosis may all play vital roles in the etiology of diabetic cardiomyopathy. Given that inactive ALDH2 prompts hyperglycemia and risk of diabetic complications (Matsuoka, 2000), this study was designed to examine the role for ALDH2 in the pathogenesis of diabetic cardiomyopathy and underlying mechanism(s) involved with a particular focus on mitochondrial quality. Both human and experimental models of diabetes were employed. Earlier finding from our laboratory suggested a crucial role of Akt along with its up- or downstream signaling cascades including phosphatase and tensin homologue on chromosome 10 (PTEN, a negative regulator for Akt), glycogen synthase kinase-3β (GSK3β), and forkhead transcriptional factor in ALDH2-initiated cardioprotection against alcoholism and ischemia-reperfusion (Ren, 2007; Doser et al., 2009; Ma et al., 2009, 2010, 2011). More evidence has revealed an important role for mitochondria-selective autophagy—mitophagy in the governance of mitochondrial function in diabetes and other metabolic diseases (Ajoolabady et al., 2022). Therefore, mitophagy was scrutinized along with mitochondrial integrity including mitochondrial membrane potential (MMP), as well as PTEN, Akt, GSK3β, and Foxo3a signaling, which were monitored in wild-type (WT), ALDH2 overexpression, and ALDH2 knockout mice with or without streptozotocin (STZ)-induced diabetes. To understand whether ALDH2 affects myocardial geometry and function in diabetes indirectly through global metabolic changes, global metabolic properties including respiratory exchange ratio (RER), O2 consumption (VO2), CO2 production (VCO2), and total physical activity along with plasma free fatty acid (FFA) profile was monitored in diabetic and non-diabetic mice.
Results
Data processing and weighted correlation network analysis
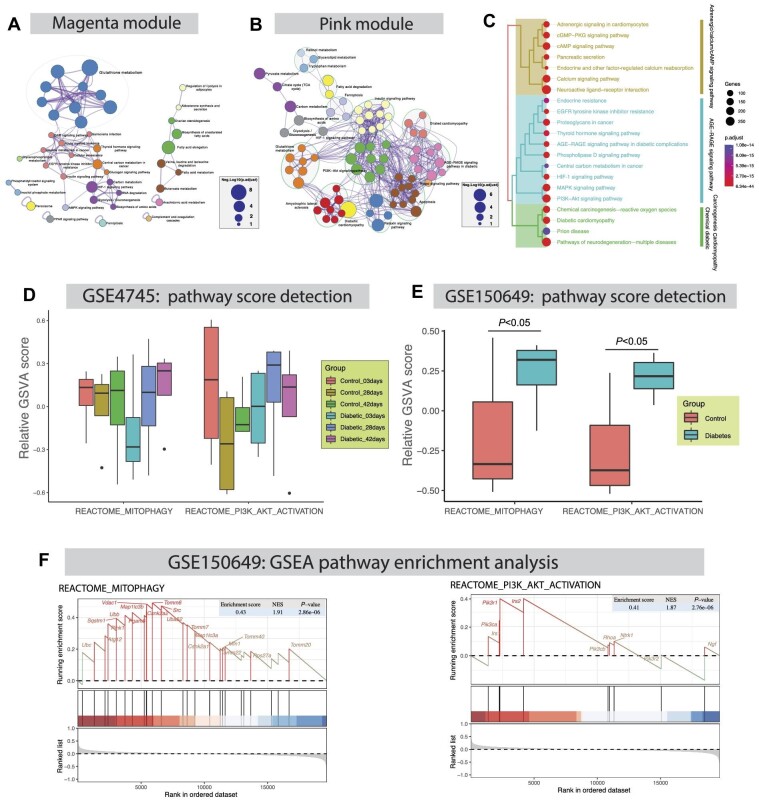
Of note, 24 rat ventricles collected after 3, 28, and 42 days of the STZ (65 mg/kg) or citrate buffer delivery were used to extract total RNA for GSE4745 dataset. Here, rat ventricles at 3 days after STZ injection displayed an initial lesion, the 28-day model exhibited diastolic dysfunction, and 42-day rat ventricles unveiled both diastolic and systolic dysfunction in comparison with control groups. Differential expression analysis was performed across groups of different time points using the ‘Affy’ method. A comparison between 28-day and 3-day rat ventricles revealed 26 differentially expressed genes (DEGs; 6 downregulated and 20 upregulated). In addition, 109 DEGs (46 downregulated and 63 upregulated) were detected between 42-day post STZ injection and control groups. Meanwhile, weighted gene coexpression network analysis (WGCNA) was conducted to discern the correlation between phenotype and gene set with robust multi-array average (RMA)-normalized 7315 coexpressed genes in 24 rat ventricles. In WGCNA, samples were included with multiphenotypes, including diabetic status, STZ injection days (i.e. days after STZ or citrate buffer delivery), body weight, ventricular weight, blood glucose, ventricular weight/body weight ratio, and body weight/control average (Figure 1A). Based on WGCNA, the selected algorithm include the following core parameters: (i) the soft power is 10 to meet the scale-free topology model fitting signed R2 >0.8; (ii) the gene modules are merged with the dynamic tree cutting height at 0.25 (Figure 1B); (iii) MEmagenta and MEpink modules may significantly correlate with diabetic status (Memagenta: correlation = 0.85, P-value = 1e−07; MEpink: correlation = −0.65, P-value = 6e−04) and blood glucose level (Memagenta: correlation = 0.84, P-value = 3e−07; MEpink: correlation = −0.70, P-value = 2e−04) (Figure 1C); and (iv) MEmagenta and MEpink modules have certain correlation variability (Figure 1D).
Figure 1.
WCGNA of GSE4745 dataset constructed in response to the temporal-correlated pathological changes of myocardium in diabetic rattus norvegicus. (A) The heatmap showing the clustering analysis of sample transcript profile and phenotypic traits involving STZ injection days, body weight (BW), ventricular weight (VW), blood glucose (Bglu), ventricular weight/body weight ratio (VW/BW ratio), and body weight/control average (BW/Control). (B) The coexpression gene modules were identified through the hierarchical cluster algorithm and merged by the dynamic tree cut method, in which the cutting height is 0.25. (C) The heatmap showing the Pearson’s correlation analysis among modules and clinical traits. (D) The heatmap showing coexpression correlations among gene modules based on the hierarchical cluster algorithm.
Pathway enrichment analysis
Figure 2A and B show interactive maps of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched with genes in magenta and pink modules, where color code represents the identities of pathways, and node size represents the Benjamini–Hochberg procedure-adjusted P-value (p.adjust) of each gene. These pathways encompass glutathione metabolism (p.adjust = 3.16e−06; enriched genes: Gstm1, Gstp1, Odc1, Gpx4, Gstt2, Mgst1, Mgst3, and Gsta4), metabolism of xenobiotics by cytochrome P450 (p.adjust = 6.31e−05; enriched genes: Gstm1, Gstp1, Cbr1, Gstt2, Mgst1, Mgst3, and Gsta4), fatty acid elongation (p.adjust = 7.94e−05; enriched genes: Acot7, Acot1, Hadha, Hadhb, and Acot2), and HIF-1 signaling pathway (p.adjust = 5.01e−04; enriched genes: Eno1, Eno2, Nppa, Slc2a1, Hk2, Pfkp, and Eif4ebp1), which were primarily enriched in magenta module (Figure 2A), as well as diabetic cardiomyopathy (p.adjust = 1.26e−06; enriched genes: Gapdh, Slc2a4, Smad2, Col1a1, Mtor, Mmp2, Ndufv2, Vdac1, Col3a1, Col1a2, Mpc1, Ndufs8, and Ndufa9), relaxin signaling pathway (p.adjust = 3.89e−04; enriched genes: Kras, Smad2, Col1a1, Mmp2, Col3a1, Col1a2, Vegfb, and Vegfd), pyruvate metabolism (p.adjust = 1.26e−03; enriched genes: Hagh, Mdh1, Pc, Aldh7a1, and Ldhd), carbon metabolism (p.adjust = 1.26e−03; enriched genes: Gapdh, Mdh1, Pgam2, Pc, Eno3, Acads, and Dlst), and PI3K–Akt signaling pathway (p.adjust = 1.58e−03; enriched genes: Kras, Ywhah, Igf1r, Pkn1, Col1a1, Mtor, Col1a2, Vegfb, Lpar1, Vegfd, and Eif4e2), were significantly detected in pink module (Figure 2B). For Gene Ontology (GO) functional enrichment, adrenergic/calcium/cAMP signaling pathway, AGE−RAGE signaling pathway, and chemical diabetic/carcinogenesis/cardiomyopathy were evidently identified, and the subcategories of neuroactive ligand–receptor interaction, cAMP signaling pathway, PI3K–Akt signaling pathway, pathways of neurodegenerative diseases, and diabetic cardiomyopathy were detected (Figure 2C).
Figure 2.
The detection of pathway regulation network, gene set variation score, and gene set enrichment. (A and B) KEGG pathway regulation network was constructed based on the Metascape database involving genes in magenta or pink module. (C) GO functional enrichment map showing that adrenergic/calcium/cAMP signaling pathway, AGE–RAGE signaling pathway, and chemical diabetic/carcinogenesis/cardiomyopathy categories were significantly enriched. (D) GSVA of GSE4745 dataset reveals temporal variation of mitophagy and PI3K–Akt activation pathway scores in response to pathological changes in cardiomyopathy following STZ injection. (E and F) GSVA and GSEA of GSE150649 dataset validate mitophagy and PI3K–Akt activation pathway score variation and enrichment in diabetic hearts of newborn rats.
The gene set variation analysis (GSVA) pathway scores of mitophagy and PI3K–Akt activation exhibited dynamic variation at different time spots in STZ-induced diabetic cardiomyopathy from GSE4745 dataset (Figure 2D) and were overtly suppressed in diabetic hearts from GSE150649 dataset (Figure 2E). In addition, both mitophagy (enrichment score = 0.43, normalized enrichment score (NES) = 1.91, P-value = 2.86e−06) and PI3K–Akt activation (enrichment score = 0.41, NES = 1.87, P-value = 2.76e−06) pathways were also profoundly detected by gene set enrichment analysis (GSEA) of GSE150649 dataset (Figure 2F).
Global metabolic properties of normal and diabetic mice
To evaluate the influence of diabetes and ALDH2 on metabolic indices, calorimetric parameters were assessed in WT, ALDH2 overexpression, and ALDH2−/− mice with or without diabetes using the 6-chamber Comprehensive Laboratory Animal Monitoring System (Oxymax/CLAMS; Columbus Instruments). Our data revealed that diabetes evoked overtly lowered VCO2 and RER (VCO2/VO2) in both light and dark cycles, along with unchanged VO2, heat production, and total physical activity, which were attenuated or accentuated by ALDH2 overexpression or knockout, respectively (Supplementary Figure S1). Notably, little difference was detected for these metabolic parameters in response to either ALDH2 overexpression or knockout itself (Supplementary Figure S1). These data favor that STZ diabetic mice oxidize a lower proportion of carbohydrate compared with non-diabetic mice, representing a higher dependence on fatty acids for energy source over glucose. Thus, ALDH2 expression apparently offers a beneficial effect on energy metabolism in diabetes.
Morphological properties and cell apoptosis of myocardium from normal and diabetic mice
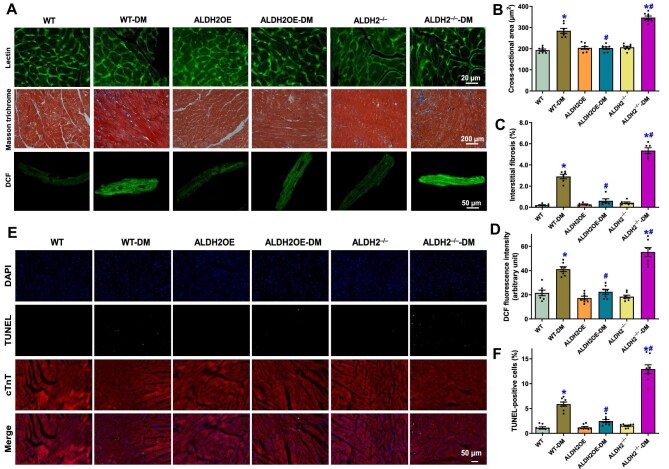
Morphological properties were evaluated using fluorescein isothiocyanate-conjugated wheat germ Lectin staining and Masson trichrome staining in WT, ALDH2 overexpression, and ALDH2−/− mice with or without diabetes. As shown in Figure 3A–C, diabetes enlarged cardiomyocyte cross-sectional area and elevated interstitial fibrosis in mouse hearts, which were obliterated or augmented by overexpression or knockout of ALDH2, respectively. Likewise, STZ-induced diabetes evoked overt oxidative stress evidenced by pronounced dichlorodihydrofluorescein (DCF) fluorescence, which was mitigated or exacerbated by overexpression or knockout of ALDH2, respectively (Figure 3A and D). Assessment of cell apoptosis using the TUNEL assay noted that diabetes significantly elevated apoptosis, which was nullified or intensified by ALDH2 overexpression or knockout, respectively (Figure 3E and F). Neither ALDH2 overexpression nor knockout alone elicited any notable effects on cardiac morphology, oxidative stress, or TUNEL apoptosis (Figure 3). These data support a preservative role for ALDH2 in the maintenance of myocardial morphology and cellular status in diabetes.
Figure 3.
Influence of ALDH2 overexpression (ALDH2OE) and knockout on diabetes mellitus (DM)-induced changes in myocardial morphology, oxidative stress, and apoptosis. (A) Representative Lectin (top), Masson trichrome (middle), and DCF (bottom) staining micrographs depicting cardiomyocyte cross-sectional area, interstitial fibrosis, and oxidative stress, respectively. (B–D) Quantitative analysis of cardiomyocyte cross-sectional area (B), interstitial fibrosis (C), and DCF fluorescence intensity (D). (E) Representative TUNEL/DAPI/cardiac troponin T (cTnT) staining micrographs depicting cardiomyocyte apoptosis. (F) Quantitative analysis for the percentage of TUNEL-positive cells. Mean ± SEM, n = 6–8 mice or myocardial sections per group; *P < 0.05 vs. WT group, #P < 0.05 vs. WT-DM group.
Biometric and echocardiographic properties of normal and diabetic mice
Diabetes induced subtle but significant loss in body weight along with decreased plasma insulin and elevated blood glucose levels (with exception of body weight in ALDH2 overexpression mice), and neither ALDH2 overexpression nor knockout affected body weight and plasma profiles in WT mice with or without diabetes (Supplementary Figure S2A–C). Myocardial function was further monitored using echocardiography. Diabetes overtly increased left ventricular (LV) end-diastolic diameter (LVEDD), LV end-systolic diameter (LVESD), LV mass (absolute or normalized), and heart weight normalized to body weight, suppressed fractional shortening and ejection fraction, but did not affect LV wall thickness and heart rate (Supplementary Figure S2D–L). ALDH2 overexpression or knockout itself did not affect any echocardiographic indices in the absence of diabetes but effectively alleviated or accentuated, respectively, diabetes-induced anomalies in myocardial geometry and function (Supplementary Figure S2D–L).
Cardiomyocyte contractile and intracellular Ca2+ properties in normal and diabetic mice
Neither diabetes nor ALDH2 manipulation affected resting cell length, while diabetes overtly suppressed peak shortening (PS), maximal velocity of shortening/relengthening (±dL/dt), and time-to-90% relengthening (TR90) without affecting time-to-PS (TPS) in cardiomyocytes from WT mice (Supplementary Figure S3A–F), consistent with our earlier reports (Wold et al., 2006; Ren et al., 2008). ALDH2 overexpression or knockout itself did not exert any effects on cardiomyocyte mechanics in the absence of diabetes but obliterated or accentuated, respectively, diabetes-induced anomalies in cardiomyocyte function (Supplementary Figure S3A–F). Furthermore, Fura-2 fluorescence was utilized to monitor intracellular Ca2+ homeostasis. As shown in Supplementary Figure S3G–I, intracellular Ca2+ rise in response to electrical stimulus evidenced by rise in Fura-2 fluorescence intensity (ΔFFI) and intracellular Ca2+ clearance in cardiomyocytes from diabetic hearts were overtly compromised, which was negated or worsened by ALDH2 overexpression or knockout, respectively. Either diabetes or ALDH2 manipulation had little effect on baseline FFI. ALDH2 manipulation itself did not alter any of these intracellular Ca2+ indices.
Cardiomyocyte mitochondrial ultrastructure and function in normal and diabetic mice
To assess mitochondrial integrity and function, we carried out transmission electron microscopy (TEM) and 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide (JC-1) assays. TEM analysis revealed pronounced cytoarchitectural aberrations including mitochondrial swelling, fragmentation of cristae, distortion of sarcomeres, and myocardial filaments in diabetic hearts (Figure 4A). Quantitative assessment of TEM ultrastructural images depicted enlarged mitochondrial area and mitochondria circularity (short axis-to-long axis ratio), consistent with downregulated mitochondrial proteins PGC-1α and UCP2, decreased aconitase activity (denoting mitochondrial dysfunction), and collapsed MMP measured using JC-1 fluorescence in diabetic hearts (Figure 4B–H). ALDH2 overexpression or deletion did not elicit any notable effect on mitochondrial ultrastructure and function but could rescue or exacerbate, respectively, diabetes-induced mitochondrial anomalies (Figure 4). These findings suggest the involvement of mitochondrial integrity and function in ALDH2-offered protection against diabetic cardiomyopathy.
Figure 4.
Influence of ALDH2 overexpression and knockout on diabetes mellitus-induced changes in mitochondrial ultrastructure, protein expression, and function. (A) Representative TEM ultrastructural images. (B) Mitochondrial area (% total area). (C) Mitochondrial circularity index (major axis length/minor axis length). (D) Representative immunoblot and protein level of PGC-1α (GAPDH as loading control). (E) Representative immunoblot and protein level of UCP2 (α-tubulin as loading control). (F) Aconitase activity. (G) Representative JC-1 staining (aggregate, monomer, and merged) micrographs. (H) Quantitative analysis of JC-1 staining for MMP. Mean ± SEM, n = 5 ultrastructural images (B), 42–54 mitochondria (C), or 6–8 mice or myocardial micrographs (D–F and H) per group; *P < 0.05 vs. WT group, #P < 0.05 vs. WT-DM group.
ALDH2 activity and cardiac function in diabetic patients and protein levels of SERCA2a, Na+–Ca2+ exchanger, and phospholamban in diabetic mouse hearts
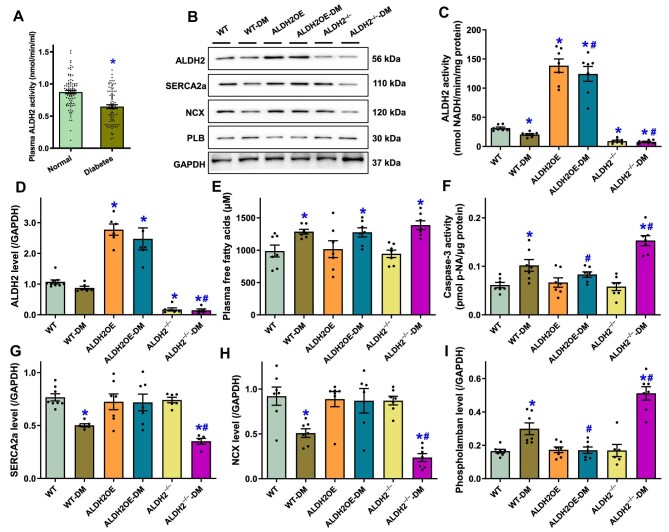
To explore possible interplay between ALDH2 activity and cardiac function in human subjects, a cohort of 273 diabetic patients and 2080 non-diabetic individuals were recruited to our hospital for an assessment of plasma ALDH2 activity and echocardiographic function. Anthropometric information is provided in Supplementary Table S1. In our cohort study, diabetes was confirmed in 11.6% (273/2353) participants and more likely associated with hypertension, coronary artery disease, obesity, and dyslipidemia. More importantly, diabetic patients exhibited significantly higher systolic and diastolic blood pressures and body mass index and also tended to possess higher levels of triglycerides and urine microalbumin as well as lower level of high-density lipoprotein cholesterol. Echocardiographic results indicated the elevated interventricular septal thickness (IVST), left atrial diameter, LV mass index (LVMI), and peak early diastolic (A) velocity, along with the decreased peak early/late diastolic velocity (E/A) ratio and septal peak early diastolic mitral annular (e′) velocity in diabetic patients (denoting diastolic dysfunction) (Supplementary Table S1). In addition, diabetic patients exhibited overtly decreased plasma ALDH2 activity (Figure 5A). Myocardial ALDH2 expression and activity were also dampened in diabetic mouse hearts, which was masked (drastically elevated or decreased) by ALDH2 gene manipulation, while the significantly increased plasma FFA levels by diabetes were unaffected by ALDH2 manipulation (Figure 5B–E). Further assessment of apoptosis by examining caspase-3 activity revealed the increased caspase-3 activity in diabetic hearts, which was alleviated or accentuated by ALDH2 overexpression or knockout, respectively (Figure 5F). Western blot analysis of intracellular Ca2+ regulatory proteins indicated significantly downregulated levels of SERCA2a and Na+–Ca2+ exchanger along with upregulated level of phospholamban in diabetic hearts. ALDH2 overexpression or knockout did not alter the expression of SERCA2a, Na+–Ca2+ exchanger, and phospholamban but nullified or exacerbated, respectively, diabetes-induced abnormalities in these protein levels (Figure 5B and G–I).
Figure 5.
ALDH2 overexpression and knockout on diabetes mellitus-induced changes in myocardial ALDH2, apoptosis, and Ca2+ regulatory proteins as well as plasma FFAs. (A) Plasma ALDH2 activity in normal individuals and patients with diabetes mellitus. (B–I) WT, ALDH2OE, and ALDH2−/− mice were challenged with or without STZ. (B) Representative immunoblots depicting ALDH2, SERCA2a, Na+–Ca2+ exchanger (NCX), and phospholamban (PLB) (GAPDH as loading control). (C) ALDH2 activity in the heart. (D) ALDH2 protein levels in hearts. (E) Plasma FFA levels. (F) Myocardial capsase-3 activity. (G–I) Levels of Ca2+ regulatory proteins SERCA2a, NCX, and PLB in hearts. Mean ± SEM, n = 5 patients (A) or 6–8 mice (C–I) per group; *P < 0.05 vs. normal or WT group, #P < 0.05 vs. WT-DM group.
Western blot analysis for autophagy, mitophagy, Akt, GSK3β, PTEN, and Foxo3a signaling
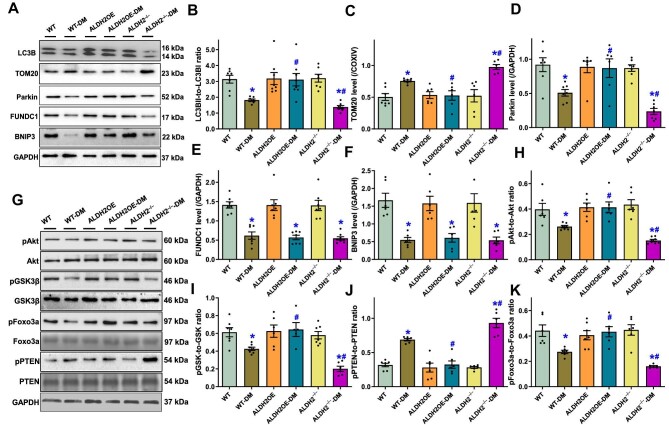
Given that GSVA pathway score identified PI3K–Akt signaling and mitophagy in STZ-induced diabetic hearts, we next examined the protein levels of Akt and its up- or downstream signaling molecules PTEN (a negative regulator of Akt signaling), GSK3β, and Foxo3a, as well as mitophagy (heavily regulated by Akt, GSK3β, and Foxo3a) signaling molecules, in mice with ALDH2 manipulation. Western blot analysis noted that diabetes significantly dampened autophagy (LC3BII-to-LC3BI ratio) and mitophagy (TOM20, Parkin, FUNDC1, and BNIP3), while ALDH2 overexpression or knockout itself did not affect these autophagy and mitophagy protein marker levels but abolished or worsened, respectively, diabetes-induced changes in LC3B ratio, TOM20, and Parkin (without affecting FUNDC1 and BNIP3) (Figure 6A–F). Moreover, diabetes overtly suppressed phosphorylation of Akt, GSK3β, and Foxo3a but increased phosphorylation of PTEN, which was mitigated or intensified by ALDH2 overexpression or knockout, respectively (Figure 6G–K). ALDH2 manipulation itself failed to affect phosphorylation of these signaling molecules in the absence of diabetes and neither diabetes nor ALDH2 manipulation alone affected total protein expression of Akt, GSK3β, PTEN, and Foxo3a (Figure 6G–K).
Figure 6.
Influence of ALDH2 overexpression and knockout on diabetes mellitus-induced changes in autophagy/mitophagy and related regulatory post-insulin receptor signaling. (A) Representative immunoblots depicting autophagy/mitophagy protein markers (GAPDH as loading control). (B) LC3BII-to-LC3BI ratio. (C–F) Protein levels of TOM20 (C), Parkin (D), FUNDC1 (E), and BNIP3 (F). (G) Representative immunoblots depicting total and phosphorylated Akt, GSK3β, PTEN, and Foxo3a (GAPDH as loading control). (H) pAkt-to-Akt ratio. (I) pGSK3β-to-GSK3β ratio. (J) pPTEN-to-PTEN ratio. (K) pFoxo3a-to-Foxo3a ratio. Mean ± SEM, n = 6–8 mice per group; *P < 0.05 vs. WT group, #P < 0.05 vs. WT-DM group.
Screening of potential ALDH2 activator by molecular docking and effects of torezolid on STZ- or high glucose-induced myocardial contractile dysfunction
Initial screening of the FDA-approved database yielded 49 candidate ALDH2 agonists, according to the LibDockScore and PyRx binding energy scores (Supplementary Table S2). The binding ability and interaction mode of the top-ranked molecule torezolid with ALDH2 were further verified using autodock molecular docking. As shown in Figure 7A, small molecules formed traditional hydrogen bonds with GLU268, GLN349, and ASP346 and π–π conjugate interactions with TRP168. In addition, small molecules also interacted with several amino acids at the active site of protein through van der Waals force and hydrophobic action. The docking and binding energy was –7.53, indicating strong binding capacity between torezolid and ALDH2. To examine potential effects of torezolid on diabetic cardiomyopathy, cardiomyocytes from WT or Parkin knockout (Wu et al., 2022a) mice were exposed to normal glucose (5.5 mM) or high glucose (25.5 mM) in the absence or presence of torezolid (5 μM) (Sharon et al., 2019) for 24 h. Our results revealed that torezolid overtly elevated ALDH2 activity in both normal and high glucose-incubated murine cardiomyocytes, which was suppressed by the ALDH2 inhibitor Daidzin (Figure 7B). Torezolid treatment in vivo effectively alleviated diabetes-evoked pathological changes in LVEDD, LVESD, fractional shortening, ejection fraction, and LV mass (data not shown), without affecting LV wall thickness and heart rate, while torezolid failed to elicit any effects by itself (Figure 7C–H). Incubation of cardiomyocytes with high glucose mimicked diabetes-induced cardiac anomalies manifested as compromised PS and ±dL/dt, prolonged TR90, and unchanged TPS, which were mitigated by torezolid treatment in vitro, though torezolid did not exert any effect on cardiomyocyte mechanics under normal glucose incubation (Figure 7I–M). Interestingly, torezolid-offered beneficial effect against high glucose was vanished by Parkin ablation, while Parkin deletion did not alter torezolid response in normal glucose environment (Figure 7I–M).
Figure 7.
Influence of the newly screened ALDH2 agonist torezolid on STZ- or high glucose (HG)-induced myocardial contractile dysfunction. (A) Molecular docking simulation of the binding between torezolid and ALDH2. (B) ALDH2 activity in cardiomyocytes from WT mice exposed to normal glucose (NG, 5.5 mM) or HG (25.5 mM) in the absence or presence of torezolid (5 μM) or the ALDH2 inhibitor Daidzin (120 μM). (C–H) Echocardiographic properties in control or STZ diabetic mice treated with or without torezolid (10 mg/kg, i.p., twice daily for 4 weeks, initiated concurrently with STZ injection). (C) LVEDD. (D) LVESD. (E) Fractional shortening. (F) Ejection fraction. (G) LV wall thickness. (H) Heart rate. (I–M) In vitro mechanical properties in cardiomyocytes from WT or Parkin knockout (PRKKO) mice exposed to NG or HG in the presence or absence of torezolid for 24 h. (I) PS (normalized to cell length). (J) +dL/dt. (K) –dL/dt. (L) TPS. (M) TR90. Mean ± SEM, n = 5 isolations (B), 6 mice (C–H), or 60 cells (I–M) per group; *P < 0.05 vs. NG or control group, #P < 0.05 vs. DM or HG group.
Influence of ALDH2 overexpression, Alda-1, mitochondrial uncoupling, and Akt/GSK3β manipulation on high glucose-induced cardiomyocyte contractile and mitochondrial dysfunctions
To evaluate the impact of ALDH2 on mitochondrial function, H9C2 cells were transfected with or without ALDH2 and then incubated with normal glucose (5.5 mM) or high glucose (25.5 mM) for 24 h. Assessment of MMP using tetramethylrhodamine ethyl ester (TMRE) revealed that ALDH2 transfection rescued high glucose-induced collapse in MMP (Figure 8A and B). To discern a causal role of Akt–GSK3β in ALDH2-induced mechanical responses in diabetes, cardiomyocytes from WT mice were exposed to normal glucose (5.5 mM) or high glucose (25.5 mM) for 24 h in the absence or presence of the classical ALDH2 activator Alda-1 (20 μM), the Akt inhibitor AKTi (1 μM), the GSK3β activator LY294002 (10 μM), or the mitochondrial uncoupler FCCP (1 μM) (Matsuoka, 2000; Morino et al., 2005; Koda et al., 2010; Zhang et al., 2013) prior to assessment of cardiomyocyte mechanical properties. Figure 8C–H depict that high glucose overtly suppressed cardiomyocyte contractile function (decreased PS, compromised ±dL/dt, and prolonged TR90), which was abolished by Alda-1, while AKTi, LY294002, and FCCP all abolished Alda-1-offered beneficial effects without eliciting any notable effects by themselves. These findings support an obligatory role for Akt, GSK3β, and mitochondrial integrity in Alda-1-elicted cardioprotection.
Figure 8.
Influence of ALDH2 overexpression or the ALDH2 agonist Alda-1 on HG-induced MMP collapse and cardiomyocyte contractile dysfunction in the absence or presence of various pharmacological inhibitors. (A) Representative flow cytometric histograms of TMRE (1 μl/ml)-loaded H9C2 cells with or without ALDH2 overexpression exposed to NG (5.5 mM) or HG (25.5 mM) for 24 h. (B) Quantitative analysis of TMRE mean intensity for MMP in H9C2 cells. (C–H) Cardiomyocytes from WT mice were exposed to NG or HG for 24 h in the absence or presence of Alda-1 (20 μM), the Akt inhibitor AKTi (1 μM), the GSK3β activator LY294002 (10 μM), or the mitochondrial uncoupler FCCP (1 μM) prior to assessment of mechanical properties. (C) Resting cell length. (D) PS (normalized to cell length). (E) +dL/dt. (F) –dL/dt. (G) TPS. (H) TR90. (I) Scheme depicting the proposed role of ALDH2 in diabetic cardiomyopathy. Diabetes mellitus or hyperglycemia downregulates ALDH2, which promotes activation of PTEN and deactivation of Akt and GSK3β, leading to mitochondrial injury. Meanwhile, diabetes-evoked downregulation of ALDH2 suppresses Foxo3a activation and Parkin-mediated mitophagy, also favoring mitochondrial injury. Torezolid and Alda-1 activate ALDH2 to mitigate hyperglycemia-evoked cardiomyocyte anomalies, while AKTi and LY294002 nullify Akt–GSK3β signaling to disengage ALDH2 activator-offered cardioprotection. Mean ± SEM, n = 6 isolations (B) or 42 cells (C–H) per group, *P < 0.05 vs. NG group, #P < 0.05 vs. HG group, †P < 0.05 vs. HG-Alda-1 group.
Discussion
Salient findings from our present study noted a tight association between lowered plasma ALDH2 activity and compromised myocardial function in patients with diabetes mellitus. This was supported by experimental evidence for a beneficial role of ALDH2 in preservation against diabetes-induced myocardial remodeling and contractile anomalies. ALDH2 overexpression attenuated, while ALDH2 ablation accentuated, respectively, diabetes-induced myocardial anomalies likely through the regulation of mitophagy, mitochondrial integrity and function, and the Akt post-insulin receptor signaling involving PTEN, GSK3β, and Foxo3a. Using virtual screening, we identified a novel ALDH2 activator torezolid, which induces ALDH2 activation along with the protection against STZ- or high glucose-induced cardiomyocyte anomalies in a Parkin-mediated mitophagy-dependent manner, while the classical ALDH2 activator Alda-1 offers protection against glucose toxicity depending on Akt and GSK-3β as well as mitochondrial coupling. Our data also suggested that ALDH2 activates Akt in the face of diabetes through suppressing diabetes-induced PTEN phosphorylation. Analysis of global metabolism suggested that ALDH2 may protect against diabetes mellitus-induced changes in RER but not those in plasma glucose, insulin, and FFA levels or total physical activity. These findings favor a cardioprotective property of ALDH2, which may be either directly cardioprotective or secondary to global metabolic benefit, and thus reveal the therapeutic potential of ALDH2 in myocardial remodeling and contractile anomalies in diabetes.
Cardiac remodeling and compromised contractile function are considered main hallmarks of diabetic cardiomyopathy (Wold et al., 2006; Ren et al., 2008). Here, we noted compromised echocardiographic indices in diabetic patients and diabetic murine hearts. Intriguingly, these anomalies were closely associated with declined plasma ALDH2 activity and myocardial ALDH2 levels and were alleviated or accentuated by ALDH2 overexpression or deletion, respectively. A number of scenarios were postulated for diabetes-elicited cardiomyopathy (Wold et al., 2006; Ren et al., 2008). In this study, we noted overtly elevated oxidative stress and cell apoptosis as well as compromised intracellular Ca2+ handling in diabetic hearts. In addition, mitochondrial injury and compromised mitophagy were evident in diabetic hearts, in line with mitochondrial dysfunction and mitophagy defect in diabetic cardiomyopathy (Ren et al., 2010). These diabetes-induced changes could be restored or worsened by ALDH2 overexpression or knockout, further consolidating that ALDH2 offers protection against diabetic cardiomyopathy. Moreover, ALDH2 manipulation appears to influence diabetes-induced drop in VCO2 and hence RER (VCO2/VO2) without affecting physical activity and plasma levels of glucose, insulin, and FFAs, suggesting that ALDH2 favors energy utilization toward glucose oxidation over fatty acid oxidation, although further study is warranted. The apparent therapeutic potential of ALDH2 in diabetes is consistent with the decreased level and/or enzymatic activity of ALDH2 in diabetic patients and mouse models (Wang et al., 2011) as well as the notion that ALDH2 mutation prompts hyperglycemia and diabetes risk (Dakeishi et al., 2008). Interestingly, various toxic aldehydes appear to serve as the main substrate for ALDH2 to ignite oxidative stress and pathological changes. For example, sublethal levels of aldehydes may compile with moderately decreased ALDH2 to provoke oxidative damage and gene alterations under pathological stress (Sano, 2010; Zhang and Ren, 2011). Furthermore, the beneficial effects of ALDH2 activators torezolid and Alda-1 on STZ-induced diabetes or high glucose toxicity were obliterated by Parkin ablation or mitochondrial uncoupler FCCP, denoting a permissive role of Parkin-mediated mitophagy and mitochondrial integrity in ALDH2-elicited cardioprotection against hyperglycemia (Figure 8I). This is also in line with the previously reported role of mitophagy regulation in ALDH2-offered cardioprotection (Wang et al., 2020; Abudureyimu et al., 2022). Given the apparent toxicity of the classical ALDH2 activator Alda-1 limiting its in vivo usage, identification of torezolid as a novel ALDH2 activator may pave the way for developing ALDH2-associated drugs for cardiovascular and metabolic diseases.
In this study, we noted compromised mitophagy involving Parkin, FUNDC1, BNIP3, and TOM20 in the onset of mitochondrial injury and cardiac dysfunction in diabetes, which is consistent with earlier findings in diabetic hearts (Dewanjee et al., 2021). However, only Parkin-mediated mitophagy (and TOM20 as a global mitophagy marker) was responsive to ALDH2 manipulation under diabetic challenge. Parkin is an E3 ligase that binds to and ubiquitinates mitochondrial substrates and proteins to govern mitochondrial quality (Ajoolabady et al., 2022). Recent findings from our group suggested that Parkin deficiency worsened diet-induced obesity cardiomyopathy via evoking VDAC1-mediated mitochondrial Ca2+ overload (Wu et al., 2022a).
Akt, GSK3β, and Foxo3a function as essential signaling molecules not only for mitochondrial homeostasis but also for mitophagy in various pathological settings (Dakeishi et al., 2008; Peng and Yin, 2009; Miura and Tanno, 2010). Meanwhile, these post-insulin receptor signaling molecules may serve as effectors for mitochondrial injury, as the latter may compromise insulin signaling at post-receptor levels (Morino et al., 2005). Our data revealed that experimental diabetes worsened phosphorylation of Akt, GSK3β, and Foxo3a, which was mitigated or exacerbated by ALDH2 overexpression or knockout, respectively, and dampened phosphorylation of Foxo3a and GSK3β coincided with mitochondrial and Parkin mitophagy changes in diabetes, such as dropped MMP, PGC-1α, UCP2, and aconitase, reminiscent of our earlier reports (Turdi et al., 2007; Ren et al., 2008). GSK-3β is a negative regulator of mitochondrial function and can be inactivated by phosphorylation at Ser9 by oxidative stress (Relling et al., 2006; Zhang et al., 2011). In this study, the decreased GSK3β phosphorylation in diabetes, which would disinhibit GSK3β-evoked cardiac remodeling, was abrogated or aggravated by ALDH2 overexpression or ablation, respectively, accordant with the finding that the GSK-3β activator LY294002 and the Akt inhibitor AKTi nullified Alda-1-offered protection against glucose toxicity. These data suggested a role for Akt–GSK3β and mitochondrial integrity (possibly through Parkin-mediated mitophagy, Figure 8I) in the pathogenesis of diabetic cardiomyopathy (also supported by the results from GSVA analysis) and ALDH2-offered cardioprotection. ALDH2 also suppressed diabetes-evoked phosphorylation of PTEN, suggesting a role for PTEN in ALDH2-rescued Akt activation in diabetic hearts. Although how ALDH2 turns on mitophagy remains unclear, the transcription factor Foxo3a is speculated to serve as a springboard between ALDH2 and Parkin-mediated mitophagy in diabetic hearts. Previous findings from our group and others indicated that Foxo3a plays a role in the governance of Parkin-mediated mitophagy in diabetes (Yu et al., 2017; Huang et al., 2022) and ALDH2 regulates Foxo3a signaling through AMPK phosphorylation (Guo et al., 2015). Further study is warranted to decipher the Foxo3a–Parkin mitophagy regulation in a seemingly Akt–GSK3β-independent manner (the latter would suppress autophagy/mitophagy).
In summary, our study demonstrated a role of ALDH2 in the protection against diabetic cardiomyopathy, possibly through Akt–GSK3β-mediated preservation of mitochondrial integrity and Parkin-dependent mitophagy. Considering the compromised plasma ALDH2 activity and cardiac echocardiographic properties in patients with diabetes, our findings denote a unique role for ALDH2 in the prevalence of diabetic cardiomyopathy as well as therapeutics of diabetic complications. Nonetheless, many questions still remain unanswered, including the control of ‘on-and-off’ switch in diabetic complications in individuals with ALDH2 polymorphism. Moreover, a close scrutiny should be geared toward whether epigenetic modification is involved in ALDH2-evoked metabolic response in high-risk human populations.
Materials and methods
Experimental animals, experimental diabetes, and ALDH2 activity
All animal procedures were approved by the Institutional Animal Care and Use Committee at Zhongshan Hospital Fudan University (Shanghai, China). In brief, global ALDH2 transgenic and ALDH2 knockout mice were generated as described before (Doser et al., 2009; Ma et al., 2010, 2011; Zhang et al., 2013). To establish experimental diabetes, 5-month-old adult C57BL/6 (used as WT), global ALDH2 transgenic, and global ALDH2 knockout mice (male and female at 1:1 ratio) were injected with STZ (200 mg/kg, i.p.) and kept in a temperature-controlled room under a 12-h/12-h light/dark cycle with free access to water and food (Supplementary Figure S4A). Global transgenic and knockout mice were employed to evaluate the whole-body metabolic influence on cardiac responses. Four weeks later, blood glucose levels were examined and mice with fasting blood glucose levels >13 mM were deemed diabetic (Zhang et al., 2012). ALDH2 activity was assessed in 33 mM sodium pyrophosphate containing 0.8 mM NAD+, 15 μM propionaldehyde, and 0.1 ml protein extract. ALDH2 substrate propionaldehyde was oxidized in propionic acid, while NAD+ was reduced to NADH, determined by spectrophotometric absorbance at 340 nm to reflect ALDH2 activity (Wang et al., 2018). For in vivo torezolid treatment, WT control or STZ diabetic mice were administrated with torezolid (10 mg/kg, i.p., twice daily for 4 weeks; S5278, Selleckchem) (Sharon et al., 2019). Torezolid was dissolved in DMSO to generate a stock solution at a concentration of 22.5 mg/ml and then diluted in 0.9% saline to a final concentration of 1 mg/ml for injection (Supplementary Figure S4B).
Differential expression analysis
Two datasets from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), GSE4745 of gene expression profiling in response to temporal-correlated pathological changes of myocardium in diabetic rattus norvegicus (Gerber et al., 2006) and GSE150649 of gene expression profiling in newborn rat hearts in response to control and maternal diabetes (Preston et al., 2020), were employed. In this script, differential expression analysis was performed based on ‘Affy’ package of R software (Pohl et al., 2009) (version 4.1.3), which was curated by data processing principles including expression data background-corrected, log2-transformed data normalization in order to perform a linear model analysis (Pohl et al., 2009). Data normalization, missing value procession, and powerful differential expression analyses were performed by function of RMA, K-nearest neighbor (KNN), and linear models for microarray data (LIMMA) algorithms, respectively. DEGs were identified by adjusted P-value (corrected by the Benjamini–Hochberg method) <0.05 and absolute log2(fold change) (log2FC) >1.0 (Gautier et al., 2004). Herein, the genes were deemed with significantly increased (log2FC > 1.5) or decreased (log2FC < −1.5) expression.
WGCNA
WGCNA was applied to analyze diabetic cardiomyopathy-related modules and genes in coexpression modules of GSE4745. Total genes were selected to construct the interactive network. The ‘hclust’ algorithm was performed to select appropriate samples. Transcriptional expression profiles were included to construct a weighted network based on the soft-threshold method. The analysis parameters were set as follows: (i) the gene adjacency of topological overlap matrix (TOM) and corresponding dissimilarity of coexpression (1 − TOM) were detected; (ii) suitable soft-threshold power was used to detect the gene coexpression adjacency; (iii) the dynamic tree cut method was applied to detect the gene modules with the cut-off height at 0.25; (iv) genes with gene significance for weight (|GS|) >0.40 and module membership (|MM|) >0.70 were considered significant regulators (Langfelder and Horvath, 2008).
Pathway regulation network detection
Genes in hub modules were selected to construct pathway interactive networks based on the Metascape database (https://metascape.org/gp/index.html#/main/step1) (Zhou et al., 2019). The parameters were set as follows: pathway enrichment score >1.5, a minimal number of overlap at 3, and P-value > 0.05. Following enrichment, the gene annotation, functional enrichment, and significant pathway detection results were downloaded to construct an interactive network using the Cytoscape software (Hänzelmann et al., 2013; Zhou et al., 2019). In pathway comparison, the GSVA score was detected using the ssGSEA method based on the transcriptome profile (Hänzelmann et al., 2013). GSEA was applied to analyze the pathway variance in pathological changes of diabetic myocardium (Powers et al., 2018).
Human subjects and echocardiographic assessment
The study received approval from the Institutional Human Research Committee of Zhongshan Hospital Fudan University and was in compliant with ethical guidelines set in the Declaration of Helsinki. A total of 2353 otherwise healthy individuals ≥60 years of age were recruited into Shanghai Heart Health Study (SHHS), and a baseline survey was conducted including general information using self-administered questionnaire. Serum and urine profiles were constructed. Transthoracic echocardiography was performed using an Acuson 218 sector scanner equipped with a 3.5-MHz transducer. Two-dimensional and Doppler images were obtained in the parasternal long and short axes from the apical 4- and 2-chamber long-axis views. M-mode echocardiograms of LV were recorded from the parasternal long-axis view in at least three cardiac cycles. LVEDD, LVESD, IVST, and posterior wall thickness (PWT) were measured using M-mode or 2-D echocardiography from the parasternal long-axis view. LV mass was calculated as 0.80 × (1.04 × (IVST + LVEDD + PWT)3 − (LVEDD)3) + 0.6, which was further divided by body surface area (BSA) for LVMI. LV ejection fraction was calculated from LV end-systolic and end-diastolic volumes measured from the apical 4- and 2-chamber views, using the modified biplane Simpson's method. Left atrial volume was measured using the same method and indexed to BSA as left atrial volume index. Doppler echocardiographic recordings were performed using pulsed wave Doppler with the sample volume at the tips of the mitral valve in the apical 4-chamber view. Both E and A velocities and the E/A ratio were measured as indicators of LV end-diastolic pressure. Assessment of septal e′ velocity was performed by pulsed wave Doppler imaging of the lateral wall in the apical 4-chamber view (Wang et al., 2018).
Blood and plasma profiles
Plasma insulin and FFAs were measured 3 h following removal of diet from animals, using insulin and FFA assay kits (Cayman Chemical). Blood glucose levels were measured using an Accu-Chek glucose analyzer (Hoffmann-La Roche Inc.) (Zhang et al., 2017).
Metabolic measurement
Indirect calorimetry and total physical activity were measured using the Oxymax/CLAMS. VO2, VCO2, RER, and physical activity were measured every 10 min. The result recorded in the first or last half hour were not used (Zhang et al., 2012).
Virtual screening of ALDH2 agonists
The crystal structure of ALDH2 (PDB ID: 3INJ) was obtained from the PDB database and further processed using Discovery Studio V4.5, such as hydrogenation, removal of heteroatoms and water molecules, and repair of incomplete amino acid. Then, an FDA-approved compound library from Targetmol, containing a total of 1436 substances, was used for virtual screening. The ligands were processed, and active sites of protein were obtained through the Prepare ligands and Define site modules of Discovery Studio, respectively. The LibDock module was used for docking, and the results were sorted according to LibDockScore. PyRx V0.8 software was also employed for virtual screening. The Open Babel module of PyRx was applied to optimize the ligand conformations, and active site of protein was acquired through the AutoDock Wizard module. Finally, the docking was proceeded using the Vina Wizard module, and results were sorted according to binding energy scores. The candidate agonists were finally picked according to the rank of LibDockScore and PyRx binding energy scores.
Molecular docking of ALDH2
The structure of Aldh2 protein (PDB ID: 1O02) was obtained from the PDB database. The water molecules and heteroatoms were removed. The sdf file of Torezolid molecule (CAS Registry Number: 856866-72-3) required for the docking was obtained from the Pubchem database and converted into mol2 structure. After hydrogenation and charge pretreatment of small molecules and proteins, AutoDockTool-1.5.6 was applied for molecular docking. Semi-flexible docking and Lamarck Genetic Algorithm were used, and the number of dockings was set to 50. Gridbox coordinates and size were set based on the information of protein active pockets, generated from POCASA1.1 website (http://altair.sci.hokudai.ac.jp/g6/service/pocasa/). Other parameters were set to the default values. Finally, results of molecular docking were visualized through Discovery Studio V4.5.
Data analysis
Data are presented as mean ± standard error of the mean (SEM). All in vivo mouse data represent average of both sexes (with little sex difference noted). Statistical comparison was performed using one-way analysis of variance (ANOVA) (or two-way ANOVA for RER and physical activity studies) followed by the Tukey post hoc test. Significance was set as P < 0.05.
Supplementary Material
Contributor Information
Yingmei Zhang, Department of Cardiology, Shanghai Institute of Cardiovascular Diseases, Zhongshan Hospital Fudan University, Shanghai 710032, China; National Clinical Research Center for Interventional Medicine, Shanghai 200032, China.
Rongjun Zou, Department of Cardiovascular Surgery, Guangdong Provincial Hospital of Chinese Medicine, the Second Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou 510120, China; The Second Clinical College of Guangzhou University of Chinese Medicine, Guangzhou 510405, China.
Miyesaier Abudureyimu, National Clinical Research Center for Interventional Medicine, Shanghai 200032, China; Cardiovascular Department, Shanghai Xuhui Central Hospital, Fudan University, Shanghai 200031, China.
Qiong Liu, Key Laboratory of Resource Biology and Biotechnology in Western China, Ministry of Education, School of Life Sciences and Medicine, Northwest University, Xi'an 710069, China; Xi'an Key Laboratory of Cardiovascular and Cerebrovascular Diseases, Xi'an No.3 Hospital, The Affiliated Hospital of Northwest University, School of Life Sciences and Medicine, Northwest University, Xi'an 710069, China.
Jipeng Ma, Department of Cardiovascular Surgery, Xijing Hospital, Air Force Medical University, Xi'an 710032, China.
Haixia Xu, Department of Cardiology, Shanghai Institute of Cardiovascular Diseases, Zhongshan Hospital Fudan University, Shanghai 710032, China; National Clinical Research Center for Interventional Medicine, Shanghai 200032, China; Department of Cardiology, Affiliated Hospital of Nantong University, Nantong 226001, China.
Wei Yu, School of Pharmacy, Xianning Medical College, Hubei University of Science and Technology, Xianning 437100, China.
Jian Yang, Department of Cardiovascular Surgery, Xijing Hospital, Air Force Medical University, Xi'an 710032, China.
Jianguo Jia, Department of Cardiology, Shanghai Institute of Cardiovascular Diseases, Zhongshan Hospital Fudan University, Shanghai 710032, China; National Clinical Research Center for Interventional Medicine, Shanghai 200032, China.
Sanli Qian, Department of Cardiology, Shanghai Institute of Cardiovascular Diseases, Zhongshan Hospital Fudan University, Shanghai 710032, China; National Clinical Research Center for Interventional Medicine, Shanghai 200032, China.
Haichang Wang, Xi'an International Medical Center Hospital Affiliated to Northwest University, Xi'an 710077, China.
Yang Yang, Key Laboratory of Resource Biology and Biotechnology in Western China, Ministry of Education, School of Life Sciences and Medicine, Northwest University, Xi'an 710069, China; Xi'an Key Laboratory of Cardiovascular and Cerebrovascular Diseases, Xi'an No.3 Hospital, The Affiliated Hospital of Northwest University, School of Life Sciences and Medicine, Northwest University, Xi'an 710069, China.
Xin Wang, Division of Cardiovascular Sciences, Faculty of Biology, Medicine and Health, The University of Manchester, Manchester M13 9GB, UK.
Xiaoping Fan, Department of Cardiovascular Surgery, Guangdong Provincial Hospital of Chinese Medicine, the Second Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou 510120, China; The Second Clinical College of Guangzhou University of Chinese Medicine, Guangzhou 510405, China.
Jun Ren, Department of Cardiology, Shanghai Institute of Cardiovascular Diseases, Zhongshan Hospital Fudan University, Shanghai 710032, China; National Clinical Research Center for Interventional Medicine, Shanghai 200032, China.
Funding
This work was supported in part by the State Key Laboratory of Dampness Syndrome of Traditional Chinese Medicine jointly established by Guangdong Province and the Ministry (SZ2022KF10), Scientific Research Initiation Project of Guangdong Provincial Hospital of Traditional Chinese Medicine (2021KT1709), Guangzhou School (College) Joint Sponsorship Project for Fundamental and Applied Research (202201020605), and Program of Shanghai Academic/Technology Research Leader (20XD1420900).
Conflict of interest: none declared.
Author contributions: Y.Z., R.Z., M.A., Q.L., J.M., H.X., W.Y., J.Y., J.J., S.Q.: data collection and analysis; H.W., X.W., and Y.Y.: helpful advice and comments on manuscript; Y.Z., X.F., J.R.: study design, research funding, and manuscript writing. All authors have read and approved the final manuscript.
References
- Abudureyimu M., Zhao L., Luo X. et al. (2022). Influences of ALDH2 on cardiomyocyte apoptosis in heart failure rats through regulating PINK1–Parkin signaling pathway-mediated mitophagy. Cell. Mol. Biol. 68, 94–102. [DOI] [PubMed] [Google Scholar]
- Ajoolabady A., Chiong M., Lavandero S. et al. (2022). Mitophagy in cardiovascular diseases: molecular mechanisms, pathogenesis, and treatment. Trends Mol. Med. 28, 836–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.H., Budas G.R., Churchill E.N. et al. (2008). Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science 321, 1493–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakeishi M., Murata K., Sasaki M. et al. (2008). Association of alcohol dehydrogenase 2 and aldehyde dehydrogenase 2 genotypes with fasting plasma glucose levels in Japanese male and female workers. Alcohol Alcohol. 43, 143–147. [DOI] [PubMed] [Google Scholar]
- Dewanjee S., Vallamkondu J., Kalra R.S. et al. (2021). Autophagy in the diabetic heart: a potential pharmacotherapeutic target in diabetic cardiomyopathy. Ageing Res. Rev. 68, 101338. [DOI] [PubMed] [Google Scholar]
- Doser T.A., Turdi S., Thomas D.P. et al. (2009). Transgenic overexpression of aldehyde dehydrogenase-2 rescues chronic alcohol intake-induced myocardial hypertrophy and contractile dysfunction. Circulation 119, 1941–1949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gautier L., Cope L., Bolstad B.M. et al. (2004). affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315. [DOI] [PubMed] [Google Scholar]
- Gerber L.K., Aronow B.J., Matlib M.A. (2006). Activation of a novel long-chain free fatty acid generation and export system in mitochondria of diabetic rat hearts. Am. J. Physiol. Cell Physiol. 291, C1198–C1207. [DOI] [PubMed] [Google Scholar]
- Goodnough C.L., Gross E.R. (2020). Precision medicine considerations for the management of heart disease and stroke in East Asians. Cardiol. Plus 5, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Yu W., Sun D. et al. (2015). A novel protective mechanism for mitochondrial aldehyde dehydrogenase (ALDH2) in type i diabetes-induced cardiac dysfunction: role of AMPK-regulated autophagy. Biochim. Biophys. Acta 1852, 319–331. [DOI] [PubMed] [Google Scholar]
- Hänzelmann S., Castelo R., Guinney J. (2013). GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 14, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Yao T., Chen J. et al. (2022). Effect of Sirt3 on retinal pigment epithelial cells in high glucose through Foxo3a/PINK1–Parkin pathway mediated mitophagy. Exp. Eye Res. 218, 109015. [DOI] [PubMed] [Google Scholar]
- Jin J., Chen J., Wang Y. (2022). Aldehyde dehydrogenase 2 and arrhythmogenesis. Heart Rhythm 19, 1541–1547. [DOI] [PubMed] [Google Scholar]
- Koda K., Salazar-Rodriguez M., Corti F. et al. (2010). Aldehyde dehydrogenase activation prevents reperfusion arrhythmias by inhibiting local renin release from cardiac mast cells. Circulation 122, 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Guo R., Yu L. et al. (2011). Aldehyde dehydrogenase 2 (ALDH2) rescues myocardial ischaemia/reperfusion injury: role of autophagy paradox and toxic aldehyde. Eur. Heart J. 32, 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Li J., Gao F. et al. (2009). Aldehyde dehydrogenase 2 ameliorates acute cardiac toxicity of ethanol: role of protein phosphatase and forkhead transcription factor. J. Am. Coll. Cardiol. 54, 2187–2196. [DOI] [PubMed] [Google Scholar]
- Ma H., Yu L., Byra E.A. et al. (2010). Aldehyde dehydrogenase 2 knockout accentuates ethanol-induced cardiac depression: role of protein phosphatases. J. Mol. Cell. Cardiol. 49, 322–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K. (2000). Genetic and environmental interaction in Japanese type 2 diabetics. Diabetes Res. Clin. Pract. 50 Suppl 2, S17–S22. [DOI] [PubMed] [Google Scholar]
- Miura T., Tanno M. (2010). Mitochondria and GSK-3β in cardioprotection against ischemia/reperfusion injury. Cardiovasc. Drugs Ther. 24, 255–263. [DOI] [PubMed] [Google Scholar]
- Morino K., Petersen K.F., Dufour S. et al. (2005). Reduced mitochondrial density and increased IRS-1 serine phosphorylation in muscle of insulin-resistant offspring of type 2 diabetic parents. J. Clin. Invest. 115, 3587–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munukutla S., Pan G., Palaniyandi S.S. (2019). Aldehyde dehydrogenase (ALDH) 2 in diabetic heart diseases. Adv. Exp. Med. Biol. 1193, 155–174. [DOI] [PubMed] [Google Scholar]
- Pang J.J., Barton L.A., Chen Y.G. et al. (2015). Mitochondrial aldehyde dehydrogenase in myocardial ischemia-reperfusion injury: from bench to bedside. Sheng Li Xue Bao 67, 535–544. [PubMed] [Google Scholar]
- Peng G.S., Yin S.J. (2009). Effect of the allelic variants of aldehyde dehydrogenase ALDH2*2 and alcohol dehydrogenase ADH1B*2 on blood acetaldehyde concentrations. Hum. Genomics 3, 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl A.A., Sugnet C.W., Clark T.A. et al. (2009). Affy exon tissues: exon levels in normal tissues in human, mouse and rat. Bioinformatics 25, 2442–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers R.K., Goodspeed A., Pielke-Lombardo H. et al. (2018). GSEA-InContext: identifying novel and common patterns in expression experiments. Bioinformatics 34, i555–i564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston C.C., Larsen T.D., Eclov J.A. et al. (2020). Maternal high fat diet and diabetes disrupts transcriptomic pathways that regulate cardiac metabolism and cell fate in newborn rat hearts. Front. Endocrinol. 11, 570846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relling D.P., Esberg L.B., Fang C.X. et al. (2006). High-fat diet-induced juvenile obesity leads to cardiomyocyte dysfunction and upregulation of Foxo3a transcription factor independent of lipotoxicity and apoptosis. J. Hypertens. 24, 549–561. [DOI] [PubMed] [Google Scholar]
- Ren J. (2007). Acetaldehyde and alcoholic cardiomyopathy: lessons from the ADH and ALDH2 transgenic models. Novartis Found. Symp. 285, 69–76; discussion 76–79, 198–199. [DOI] [PubMed] [Google Scholar]
- Ren J., Duan J., Thomas D.P. et al. (2008). IGF-I alleviates diabetes-induced RhoA activation, eNOS uncoupling, and myocardial dysfunction. Am. J. Physiol. Regul. Integr. Comp. Physiol. 294, R793–R802. [DOI] [PubMed] [Google Scholar]
- Ren J., Pulakat L., Whaley-Connell A. et al. (2010). Mitochondrial biogenesis in the metabolic syndrome and cardiovascular disease. J. Mol. Med. 88, 993–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano M. (2010). Cardioprotection by hormetic responses to aldehyde. Circ. J. 74, 1787–1793. [DOI] [PubMed] [Google Scholar]
- Sharon D., Cathelin S., Mirali S. et al. (2019). Inhibition of mitochondrial translation overcomes venetoclax resistance in AML through activation of the integrated stress response. Sci. Transl. Med. 11, eaax2863. [DOI] [PubMed] [Google Scholar]
- Turdi S., Li Q., Lopez F.L. et al. (2007). Catalase alleviates cardiomyocyte dysfunction in diabetes: role of Akt, Forkhead transcriptional factor and silent information regulator 2. Life Sci. 81, 895–905. [DOI] [PubMed] [Google Scholar]
- Wang J., Wang H., Hao P. et al. (2011). Inhibition of aldehyde dehydrogenase 2 by oxidative stress is associated with cardiac dysfunction in diabetic rats. Mol. Med. 17, 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wang C., Turdi S. et al. (2018). ALDH2 protects against high fat diet-induced obesity cardiomyopathy and defective autophagy: role of CaM kinase II, histone H3K9 methyltransferase SUV39H, Sirt1, and PGC-1α deacetylation. Int. J. Obes. 42, 1073–1087. [DOI] [PubMed] [Google Scholar]
- Wang S., Wang L., Qin X. et al. (2020). ALDH2 contributes to melatonin-induced protection against APP/PS1 mutation-prompted cardiac anomalies through cGAS–STING–TBK1-mediated regulation of mitophagy. Signal Transduct. Target. Ther. 5, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold L.E., Ceylan-Isik A.F., Fang C.X. et al. (2006). Metallothionein alleviates cardiac dysfunction in streptozotocin-induced diabetes: role of Ca2+ cycling proteins, NADPH oxidase, poly(ADP-Ribose) polymerase and myosin heavy chain isozyme. Free Radic. Biol. Med. 40, 1419–1429. [DOI] [PubMed] [Google Scholar]
- Wu N.N., Bi Y., Ajoolabady A. et al. (2022a). Parkin insufficiency accentuates high-fat diet-induced cardiac remodeling and contractile dysfunction through VDAC1-mediated mitochondrial Ca2+ overload. JACC Basic Transl. Sci. 7, 779–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N.N., Ren J. (2019). Aldehyde dehydrogenase 2 (ALDH2) and aging: is there a sensible link? Adv. Exp. Med. Biol. 1193, 237–253. [DOI] [PubMed] [Google Scholar]
- Wu Y.C., Yao Y., Tao L.S. et al. (2022b). The role of acetaldehyde dehydrogenase 2 in the pathogenesis of liver diseases. Cell. Signal. 102, 110550. [DOI] [PubMed] [Google Scholar]
- Xu F., Chen Y., Lv R. et al. (2010). ALDH2 genetic polymorphism and the risk of type II diabetes mellitus in CAD patients. Hypertens. Res. 33, 49–55. [DOI] [PubMed] [Google Scholar]
- Yang K., Ren J., Li X. et al. (2020). Prevention of aortic dissection and aneurysm via an ALDH2-mediated switch in vascular smooth muscle cell phenotype. Eur. Heart J. 41, 2442–2453. [DOI] [PubMed] [Google Scholar]
- Yang M.Y., Wang Y.B., Han B. et al. (2018). Activation of aldehyde dehydrogenase 2 slows down the progression of atherosclerosis via attenuation of ER stress and apoptosis in smooth muscle cells. Acta Pharmacol. Sin. 39, 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.S., Chen Y.H., Hu J.T. et al. (2021). Aldehyde dehydrogenase mutation exacerbated high-fat-diet-induced nonalcoholic fatty liver disease with gut microbiota remodeling in male mice. Biology 10, 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Gao B., Li N. et al. (2017). Sirt3 deficiency exacerbates diabetic cardiac dysfunction: role of Foxo3A–Parkin-mediated mitophagy. Biochim. Biophys. Acta Mol. Basis Dis. 1863, 1973–1983. [DOI] [PubMed] [Google Scholar]
- Yu X., Zeng X., Xiao F. et al. (2022). E-cigarette aerosol exacerbates cardiovascular oxidative stress in mice with an inactive aldehyde dehydrogenase 2 enzyme. Redox. Biol. 54, 102369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Zhang Y., La Cour K.H. et al. (2013). Mitochondrial aldehyde dehydrogenase obliterates endoplasmic reticulum stress-induced cardiac contractile dysfunction via correction of autophagy. Biochim. Biophys. Acta 1832, 574–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Babcock S.A., Hu N. et al. (2012). Mitochondrial aldehyde dehydrogenase (ALDH2) protects against streptozotocin-induced diabetic cardiomyopathy: role of GSK3β and mitochondrial function. BMC Med. 10, 40. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang Y., Ren J. (2011). ALDH2 in alcoholic heart diseases: molecular mechanism and clinical implications. Pharmacol. Ther. 132, 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang C., Zhou J. et al. (2017). Complex inhibition of autophagy by mitochondrial aldehyde dehydrogenase shortens lifespan and exacerbates cardiac aging. Biochim. Biophys. Acta Mol. Basis Dis. 1863, 1919–1932. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xia Z., La Cour K.H. et al. (2011). Activation of Akt rescues endoplasmic reticulum stress-impaired murine cardiac contractile function via glycogen synthase kinase-3β-mediated suppression of mitochondrial permeation pore opening. Antioxid. Redox Signal. 15, 2407–2424. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhou Y., Zhou B., Pache L. et al. (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z.Y., Liu Y.D., Gong Y. et al. (2022). Mitochondrial aldehyde dehydrogenase (ALDH2) rescues cardiac contractile dysfunction in an APP/PS1 murine model of Alzheimer's disease via inhibition of ACSL4-dependent ferroptosis. Acta Pharmacol. Sin. 43, 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.