Abstract
Human papillomaviruses (HPVs) infect keratinocytes and induce proliferative lesions. In infected cells, viral gene products alter the activities of cellular proteins, such as Rb and p53, resulting in altered cell cycle response. It is likely that HPV gene products also alter expression of cellular genes. In this study we used microarray analysis to examine the global changes in gene expression induced by high-risk HPV type 31 (HPV31). Among 7,075 known genes and ESTs (expressed sequence tags) tested, we found that 178 were upregulated and 150 were downregulated twofold or more in HPV31 cells compared to normal human keratinocytes. While no specific pattern could be deduced from the list of genes that were upregulated, downregulated genes could be classified to three groups: genes that are involved in the regulation of cell growth, genes that are specifically expressed in keratinocytes, and genes whose expression is increased in response to interferon stimulation. The basal level of expression of several interferon-responsive genes was found to be downregulated in HPV31 cells by both microarray analysis and Northern blot analysis in different HPV31 cell lines. When cells were treated with alpha or gamma interferon, expression of interferon-inducible genes was impaired. At high doses of interferon, the effects were less pronounced. Among the genes repressed by HPV31 was the signal transducer and activator of transcription (Stat-1), which plays a major role in mediating the interferon response. Suppression of Stat-1 expression may contribute to a suppressed response to interferon as well as immune evasion.
Human papillomaviruses (HPVs) are small DNA tumor viruses which infect epithelial tissue and induce hyperproliferative lesions. Infection by high-risk genital HPVs (HPV type 16 [HPV16], HPV18, HPV31, HPV33, and HPV54) is associated with the development of anogenital cancers, while the low-risk HPVs (HPV6 and HPV11) more commonly induce benign genital warts (21, 29, 34, 66). Papillomavirus infection can be divided into several stages. Primary infection occurs at the basal layer of epidermis; following entry, HPV genomes are replicated in S phase as extrachromosomal elements. Genome copy number is maintained at a constant level in these cells, and a low level of transcripts is expressed from the early promoter. As infected keratinocytes divide, they stratify and differentiate. Concurrent with differentiation, HPV genomes are amplified and the late promoter initiates capsid gene expression (8, 13, 23). This results in encapsidation of viral DNA and production of infectious viruses.
Since HPVs encode only 8 to 10 proteins, they must rely extensively on cellular factors to regulate viral transcription and replication. The interaction of viral proteins with the host cell factors is therefore essential for the productive life cycle. Several papillomavirus proteins can directly activate transcription, while others act indirectly by altering the activity of cellular factors. Members of the former group include the E2 protein, which can activate transcription of reporters with multimerized upstream E2 binding sites in transient assays (4, 36). The latter group includes the two oncoproteins, E6 and E7. E6 binds a cellular ubiquitin ligase, which then targets the transactivator p53 for degradation (22, 49, 50, 62). E7 binds the Rb family of proteins, resulting in altered regulation of E2F-inducible genes which control S-phase entry (5, 9, 35, 41). It seems likely that during an HPV infection, alterations in cellular gene expression occur.
Many studies investigating the functions of viral proteins have not been done within the context of the entire viral genome and may not reflect what occurs at physiological concentration as in viral infection. In addition, cervical cancer cell lines that harbor integrated HPV genomes have been used in many studies, and these may not accurately reflect virus-cell interactions during the normal course of infection. Recently, a tissue culture model that simulates the latent stage of HPV infection has been developed. Cloned HPV31 and HPV18 genomes have been used to transfect normal human keratinocytes (NHKs), resulting in cell lines that exhibit characteristics of basal keratinocytes infected with HPV (12, 13, 40). Once induced to differentiate, these cell lines are able to activate late viral functions characteristic of a productive infection (13). This system has been used extensively for studying virus-cell interactions under physiologically relevant conditions.
In recent years, the development of microarrays, or gene chips, has provided a powerful tool to study complicated biological process which results in altered global gene expression (16, 25, 51, 65). Fluorescence-labeled cDNAs derived from two samples are hybridized to a microarray which contains thousands of oligonucleotides or cDNAs. Quantitative measurement of binding allows one to determine the changes in the transcriptional profile of many genes simultaneously. In this study, we used this methodology to examine the effect of high-risk HPV31 on cellular gene expression in keratinocytes. Transcriptional profiles in HPV31-positive cells were compared to those of matched NHKs, using microarray analysis. We found that HPV31 causes major reductions in the expression of genes involved in cell proliferation and interferon response, as well as genes specifically expressed in keratinocytes.
MATERIALS AND METHODS
Cell culture.
NHKs were isolated from human foreskin tissue as described previously (48). Wild-type HPV31 genome was used to immortalize NHKs as described elsewhere (13). The established HPV lines were maintained in serum containing medium (E medium) supplemented with mouse epidermal growth factor (EGF; 5 ng/ml; Collaborative Biomedical Products, Bedford, Mass.) in the presence of murine 3T3 J2 fibroblast feeders (38, 39). Two independently derived HPV lines were used in these studies; the T31 line was used in the microarray analysis, and the LKP31 line (13) was used in Northern and Western blot analysis. NHKs were maintained in E medium in the presence of feeders and EGF for microarray, Northern blot, or Western blot analyses. The growth conditions for NHKs and HPV31-containing cells were identical for all assays. Three NHK lines were used in these studies; TP was used in microarray analysis, and 407 and JP were used in other analyses. All cell lines tested had been frozen and thawed. 3T3 J2 fibroblast feeder cells were maintained in Dulbecco modified Eagle medium supplemented with 10% calf serum (Gibco BRL, Grand Island, N.Y.). Alpha and gamma interferon (IFN-α and -γ) were purchased from BioSource (Camarillo, Calif.).
Microarray analysis.
NHKs or HPV cells were grown in E medium supplemented with mouse EGF in the presence of fibroblast feeders and harvested at 80% confluence. Fibroblast feeders were first removed by versene (phosphate-buffered saline [Gibco BRL], with 0.5 mM EDTA) prior to RNA isolation. Total RNA was isolated by lysing cells in Trizol reagent (Gibco BRL), and poly(A) RNA was affinity purified on an Oligotex column (Qiagen, Valencia, Calif.). Generation of cDNA, fluorescent labeling, and hybridization to the gene chip were performed by Genome Systems (St. Louis, Mo.). Briefly, mRNA was isolated and reverse transcribed with 5′ Cy3- or Cy5-labeled random 9-mer (Operon Technologies, Inc., Alameda, Calif.). The paired reactions were combined and purified with a TE-30 column (Clontech, Palo Alto, Calif.). Fluorescently labeled probe was then applied to the array for hybridization at 60°C for 6.5 h. After hybridization, the glass slides were washed with decreasing ionic strength and scanned with 10-μm resolution to detect Cy3 and Cy5 fluorescence. We examined 7,075 human genes and ESTs (expressed sequence tags) on the array Human UniGEM V (Incyte Pharmaceuticals, Inc., Palo Alto, Calif.) and analyzed the results with the software GEMtool 2.4. A gridding and region detection algorithm was used to determine each element. The area surrounding each element image was used to calculate a local background and was subtracted from the total element signal. Background-subtracted element signals were used to calculate Cy3/Cy5 ratios. The average of the resulting total Cy3 and Cy5 signal gives a ratio that was used to balance or normalize the signals.
Northern blot analysis.
NHKs and LKP31 cells were cultured to subconfluence in E medium supplemented with EGF. Fibroblast feeders were removed, and total RNA was isolated as described above. Equal amounts of total RNA as determined by UV absorbance were separated by 1% agarose formaldehyde gel electrophoresis and transferred to a nylon membrane (Zeta Probe; Bio-Rad, Hercules, Calif.) in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The membrane was cut into strips and hybridized with cDNA probes labeled with [α32-P]dCTP (Amersham, Buckinghamshire, England) by High Prime (Boehringer Mannheim, Indianapolis, Ind.). Quantitative analysis was done with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Western blot analysis.
NHKs or LKP31 cells were grown to 80% confluence in E medium supplemented with EGF, and the cells were lysed in lysis buffer (10 mM Tris HCl [pH 7.4], 150 mM NaCl, 1% deoxycholate, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate [SDS], 5 mM EDTA, 1% Triton X-100, 1 μg of leupeptin per ml, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM sodium orthovanadate, 20 mM p-nitrophenyl phosphate) on ice for 10 min. Fibroblast feeders were removed as described above before harvesting of keratinocyte lysates. The clear lysate was obtained by centrifugation to remove insoluble cell debris at 12,000 × g for 5 min at 4°C. Equal amounts of protein lysates were separated by SDS–10% polyacrylamide gel electrophoresis (PAGE) and transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Bedford, Mass.). The membrane was then blocked with 5% nonfat dry milk in wash buffer (phosphate-buffered saline, 0.1% Tween 20) for 1 h and incubated with primary antibody (mouse anti-Stat-1α/β; 1 μg/ml; Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) for another hour. The membrane was washed for 1 h with four changes of wash buffer and then incubated with a sheep anti-mouse secondary antibody conjugated with horseradish peroxidase (Amersham). After a second wash as described above, the signal was visualized by enhanced chemiluminescence (Amersham). The levels of Stat-1 protein were determined by densitometric analysis.
RESULTS
Microarray analysis of gene expression in normal and HPV31-transfected keratinocytes.
To identify genes whose expression is altered by HPV31 during conditions similar to infection of basal cells, we examined a matched set of normal and HPV31-transfected human keratinocytes derived from a common donor. Efficient infection of tissue culture cells by HPV is not feasible, and so we examined transfected keratinocytes that stably maintain viral DNA as episomes (13). Human foreskin keratinocytes, derived from a single anonymous donor (designated TP), were transfected with recircularized cloned HPV31 along with a neomycin selectable marker. Following G418 selection, resistant colonies were pooled and expanded as combined culture (designated T31) to ensure that any observed effects were not specific to a single clonal cell line. The episomal state of viral DNA in the T31 pooled culture was confirmed by Southern analysis, and the expression of HPV31 early and late genes was found to be similar to that previously described for HPV31-positive cell lines (data not shown) (13, 48).
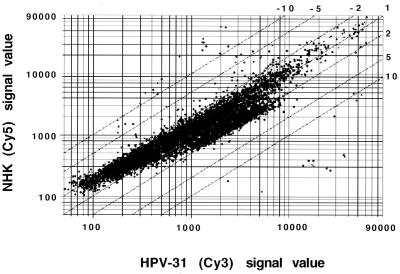
To identify genes whose expression was altered by HPV31 gene products, we used microarray analysis of 7,075 sequence-verified ESTs which included about 4,000 known genes (Genome Systems). Poly(A)-selected RNA was isolated from T31 and TP cells, and fluorescent dye-tagged cDNAs (Cy3 and Cy5, respectively) were generated. These fluorescent cDNAs were simultaneously hybridized to probe sequences on the microarray, and the amount of fluorescence seen with the individual dyes was determined by confocal microscopy. The differential expression of each EST was then calculated from the relative intensity of the Cy3-versus-Cy5 fluorescent signal. The plot of the differential expression of the 7,075 genes is shown in Fig. 1. Overall, most ESTs were not altered in expression by HPV31. Approximately 2.5% (i.e., 178) of ESTs examined were increased in expression between 2- and 3-fold, with no gene activated more than 3.2-fold. In the same analysis, 150 genes were found to be repressed at least twofold by HPV31 gene products. Twenty genes were repressed between 11.9- and 3.2-fold, while 130 genes were repressed 3.2- to 2.0-fold. Genes whose expression was altered more than 2.3-fold are listed in Table 1. The complete list of genes examined and their level of induction or suppression will be available at the web site http://bugs.mimnet.nwu.edu/laimins_lab/chang.
FIG. 1.
Global comparison of gene expression in HPV31 and NHKs. Each dot corresponds to the Cy3 (x axis) and Cy5 (y axis) fluorescent intensity of one single element on the microarray. Twofold, 5-fold, and 10-fold changes in expression are indicated as parallel lines. Dots that represent more than 11.5-fold changes are internal controls for hybridization.
TABLE 1.
Genes whose expression was altered more than 2.3-fold
| GenBank accession no. | Gene name or product | Difference in expression (fold) | |
|---|---|---|---|
| M33882 | Myxovirus (influenza virus) resistance 1 | −11.5 | |
| X67325 | IFN-α-inducible 11.5-kDa protein | −10.1 | |
| X03557 | Interferon-inducible protein 56 | −9.6 | |
| W76205 | ESTs | −6.9 | |
| S73288 | Small proline-rich protein SPRK | −5.4 | |
| M20030 | SprII | −5.1 | |
| W73855 | ESTs | −3.9 | |
| U65590 | Interleukin 1 receptor antagonist | −3.9 | |
| L33404 | Protease, serine, 6 | −3.8 | |
| M34715 | Pregnancy-specific beta-1-glycoprotein | −3.6 | |
| X04470 | Secretory leukocyte protease inhibitor | −3.6 | |
| M87284 | 2′,5′-Oligoadenylate synthetase 2 | −3.5 | |
| AA741307 | ESTs | −3.5 | |
| M97935 | Stat-1α/β | −3.5 | |
| Y00630 | Plasminogen activator inhibitor type II | −3.4 | |
| L33930 | Human CD24 gene | −3.3 | |
| J04164 | Interferon-inducible protein 1-8U | −3.3 | |
| U50931 | Defensin, beta-1 | −3.2 | |
| X59770 | Interleukin 1 receptor, type II | −3.2 | |
| M95787 | Transgelin | −3.2 | |
| X99133 | Gelatinase-associated neutrophil Lipocalin precursor | −3.1 | |
| X74330 | Primase, polypeptide 1 | −3.0 | |
| X56807 | Desmocollin 2 | −2.9 | |
| X82200 | Human Staf50 mRNA | −2.8 | |
| M30818 | Myxovirus (influenza virus) resistance 2 | −2.8 | |
| U92314 | Sulfotransferase family 2B, member 1 | −2.7 | |
| U09364 | Zinc finger protein 136 | −2.6 | |
| X04327 | 2,3-Bisphosphoglycerate mutase | −2.6 | |
| U72882 | Interferon-inducible leucine zipper protein IFP35 | −2.6 | |
| X86809 | Phosphoprotein enriched in astrocytes 15 | −2.6 | |
| Z85996 | CKI (p21) | −2.6 | |
| L06895 | Max dimerization protein (Mad) | −2.5 | |
| M22612 | Trypsin1 | −2.5 | |
| U79725 | Human A33 antigen precursor | −2.5 | |
| AJ000480 | C8FW phosphoprotein | −2.5 | |
| M77830 | Desmoplakin I and II | −2.5 | |
| X89960 | Mitochondrial capsule selenoprotein | −2.5 | |
| X57348 | Stratifin | −2.5 | |
| M23263 | Androgen receptor | −2.4 | |
| AA100757 | ESTs | −2.4 | |
| AF013970 | Myeloid translocation gene-related protein 1 | −2.4 | |
| AB001928 | Cathepsin L2 | −2.4 | |
| X13916 | Low-density lipoprotein-related protein 1 | −2.4 | |
| L34155 | Laminin, alpha 3 | −2.4 | |
| V01512 | v-fos FBJ murine osteosarcoma virus oncogene homolog | −2.4 | |
| X97198 | Receptor phosphatase PCP-2 | −2.4 | |
| X77533 | Activin A receptor, type IIB | −2.3 | |
| D28915 | Hepatitis C-associated microtubular aggregate protein p44 | −2.3 | |
| AF034840 | CD39-like 3 | −2.3 | |
| U49260 | Mevalonate decarboxylase | −2.3 | |
| U62801 | Protease, serine 9 | −2.3 | |
| AF043325 | N-Myristoyltransferase 2 | −2.3 | |
| M55542 | Interferon-inducible guanylate binding protein 1 | −2.3 | |
| X04741 | Ubiquitin carboxyl-terminal esterase L1 | +3.2 | |
| V00571 | Corticotropin-releasing hormone | +3.1 | |
| T91294 | ESTs | +3.1 | |
| X16832 | Cathepsin H | +2.9 | |
| M69199 | Human GOS2 gene | +2.9 | |
| M73548 | Adenomatosis polyposis coli | +2.9 | |
| AA085711 | ESTs | +2.8 | |
| AA477828 | ESTs | +2.8 | |
| AA705034 | ESTs, weakly similar to reverse transcriptase | +2.7 | |
| R55750 | ESTs | +2.7 | |
| AA018443 | ESTs | +2.7 | |
| W67951 | ESTs | +2.7 | |
| AA731863 | ESTs | +2.7 | |
| AA134111 | ESTs | +2.6 | |
| AA705184 | ESTs | +2.6 | |
| AA524538 | Succinate-CoA ligase, ADP-forming, beta subunit | +2.6 | |
| AA633231 | ESTs | +2.6 | |
| AA040834 | ESTs, weakly similar to collagens | +2.6 | |
| H18233 | ESTs | +2.5 | |
| AA150502 | ESTs | +2.5 | |
| AA575973 | ESTs | +2.5 | |
| AI033548 | ESTs | +2.5 | |
| W15253 | ESTs | +2.5 | |
| N57571 | ESTs | +2.5 | |
| R55697 | ESTs | +2.5 | |
| D83884 | ESTs | +2.5 | |
| N22132 | ESTs | +2.5 | |
| AA425325 | ESTs | +2.5 | |
| AA805921 | ESTs | +2.5 | |
| AA527448 | ESTs | +2.5 | |
| AA134926 | ESTs | +2.4 | |
| AA683531 | ESTs | +2.4 | |
| AA648117 | ESTs | +2.4 | |
| AA733074 | ESTs, weakly similar to C15H9.5 (Caenorhabditis elegans) | +2.4 | |
| AA280262 | ESTs | +2.4 | |
| N52534 | ESTs | +2.4 | |
| U39905 | Solute carrier family 18 | +2.4 | |
| AA056410 | ESTs, weakly similar to immunoglobulin E receptor beta subunit | +2.4 | |
| AA190841 | ESTs | +2.4 | |
| AA846757 | ESTs | +2.4 | |
| H98977 | ESTs | +2.4 | |
| N49233 | ESTs | +2.4 | |
| AA557324 | ESTs, highly similar to cytochrome P450 IVA2 | +2.4 | |
| N51427 | ESTs | +2.4 | |
| AI025984 | ESTs | +2.4 | |
| R68857 | ESTs | +2.4 | |
| J04456 | Galectin1 | +2.4 | |
| N34956 | Frizzled homolog 7 | +2.4 | |
| U04343 | T-lymphocyte activation antigen CD86 precursor | +2.4 | |
| AA287975 | ESTs | +2.3 | |
| Y15228 | Leukemia-associated gene 2 | +2.3 | |
| AA6591191 | ESTs | +2.3 | |
| N51263 | ESTs | +2.3 | |
| U04636 | Prostaglandin-endoperoxide synthase 2 | −2.3 | |
| X90840 | Axonal transport of synaptic vesicles | −2.3 | |
| V00497 | Hemoglobin, beta | −2.3 | |
| AI004422 | ESTs | −2.3 | |
| M21054 | Phospholipase A2, group IB | −2.3 | |
| AA521454 | ESTs | −2.3 | |
| N24715 | ESTs | −2.3 | |
| X04707 | Thyroid hormone receptor beta | −2.3 | |
| AA780043 | ESTs | −2.3 | |
| U67615 | Chediak-Higashi syndrome 1 | −2.3 | |
| M14660 | Interferon-inducible protein 54 | −2.3 | |
| AA255824 | ESTs | −2.3 | |
| AA905409 | ESTs | −2.3 | |
| AF002668 | Membrane fatty acid desaturase | −2.3 | |
| U05291 | Fibromodulin | +2.3 | |
| W46356 | ESTs | +2.3 | |
| N24732 | GA-binding protein transcription factor, alpha subunit | +2.3 | |
| U70310 | X-ray repair complementing defective repair in Chinese hamster cells 9 | +2.3 | |
| AA417890 | ESTs | +2.3 | |
| AA759306 | ESTs | +2.3 | |
| Aa657981 | ESTs | +2.3 | |
| AA046743 | ESTs, weakly similar to rho type GTPase-activating protein rhoGAPX-1 | +2.3 | |
| AA855042 | ESTs | +2.3 | |
| M34057 | Latent transforming growth factor beta Binding protein 1 | +2.3 | |
| M13955 | Keratin, type II | +2.3 | |
| D83664 | CAAF-1 | +2.3 | |
| M13699 | Ceruloplasmin | +2.3 | |
| N70701 | ESTs | +2.3 | |
| J03810 | Solute carrier family 2 | +2.3 | |
| AA126720 | ESTs | +2.3 | |
| N67756 | ESTs | +2.3 | |
| AA631112 | ESTs | +2.3 | |
| N31946 | ESTs | +2.3 | |
| AA054397 | ESTs | +2.3 | |
| AA281427 | ESTs | +2.3 | |
| AI034154 | ESTs | +2.3 | |
| AA115328 | ESTs | +2.3 | |
| N37009 | ESTs | +2.3 | |
| AA053711 | ESTs | +2.3 | |
| AA176450 | ESTs | +2.3 | |
| N53491 | ESTs | +2.3 | |
| AA401234 | ESTs | +2.3 | |
| AA536176 | ESTs | +2.3 | |
| N25950 | ESTs | +2.3 | |
| AA884260 | ESTs | +2.3 | |
| R85437 | ESTs | +2.3 | |
| AA410580 | ESTs | +2.3 |
The majority of genes whose expression was increased by more than twofold by HPV31 are ESTs of unknown identity (Table 1). Several known genes such as those encoding ubiquitin carboxyl-terminal esterase, cathepsin H, adenomatosis polyposis coli, and corticotropin-releasing hormone were increased approximately threefold in expression. Overall, no obvious patterns of activation of specific gene families by HPV31 could be identified. Interestingly, no E2F-regulated gene examined was found to be activated more than twofold. For example, cyclin E was found to be altered slightly (1.1- to 1.3-fold) in HPV 31 cells.
Expression of interferon-inducible genes is repressed by HPV31 gene products.
In contrast to the upregulated genes, those genes whose expression was repressed by HPV proteins could be grouped into three categories: regulators of cell growth, keratinocyte-specific genes, and factors mediating the interferon response. The p21 gene, whose expression is regulated by p53, was found to be reduced 2.6-fold, consistent with the increased turnover of p53 induced by high-risk E6 (10, 47, 50). Two other genes, encoding Mad and transgelin, were also downregulated in HPV cells. Mad is a negative regulator of Myc and Max activities (2, 24), while transgelin is highly expressed in senescent cells (56). Several genes encoding keratinocyte-specific proteins such as SPRK, human small proline-rich protein II (SprII), defensin, desmocollin 2, desmoplakin, and stratifin, were also downregulated by HPV gene products (Table 1) (26, 30, 37, 42, 58, 64).
The most pronounced reductions in gene expression were observed for interferon-responsive genes. At least 10 of the 67 genes downregulated more than 2.3-fold were interferon inducible and included those with the most significant decreases. The interferon-responsive genes whose basal levels of expression were repressed by HPV31 included those encoding myxovirus (influenza virus) resistance 1, IFN-α-inducible 11.5-kDa protein, interferon-inducible protein 56, 2′,5′-oligoadenylate synthetase 2, Stat-1, interferon-inducible protein 1-8U, Staf50, myxovirus resistance 2, interferon-inducible leucine zipper protein IFP35, interferon-inducible guanylate binding protein 1, and interferon-inducible protein 54 (1, 18, 19, 33, 44, 45, 52, 57, 60, 61).
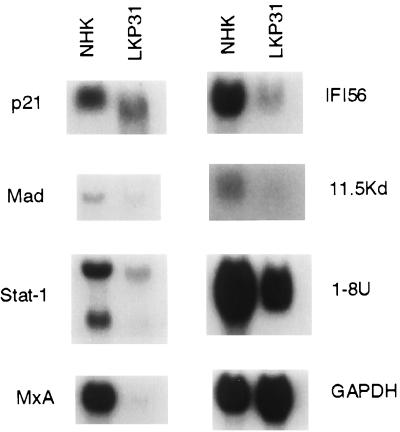
We next wanted to confirm that genes whose expression was reduced by HPV31 by microarray analysis were also reduced when examined by Northern analysis. In addition, it was important to ensure that the changes observed were not specific to the matched set of keratinocytes examined by microarray. Equal amounts of total RNAs from another HPV31-positive cell line (LKP31) as well as RNA from a second NHK donor (designated 406) were isolated and examined by Northern analysis. The results for Northern analysis of seven of the repressed genes are shown in Fig. 2, and the levels of reduction were consistent with those observed in the microarray analysis. Subsequently, we examined the expression levels of MxA and Stat-1 in three other transfected keratinocyte lines containing episomal HPV31 DNA, as well as two more NHK lines, and obtained similar results (data not shown). In our studies, the repression of interferon-inducible genes was observed in five different HPV31 lines and five different NHK lines with passage number ranging from 4 to 13 (HPV31 cells) and from 2 to 5 (NHKs).
FIG. 2.
Level of expression in genes suppressed in HPV31 cells demonstrated by microarray analysis is confirmed by Northern blot analysis. Total RNAs (20 μg) from NHKs or LKP31 cells were separated by formaldehyde agarose electrophoresis, transferred to a membrane, and hybridized with various cDNA probes. The slightly faster mobility of p21 and Stat-1 signals in LKP31 cells is likely due to electrophoretic artifact since it was not observed in separate experiments.
Repression of interferon-inducible genes by HPV31 results in an altered interferon response.
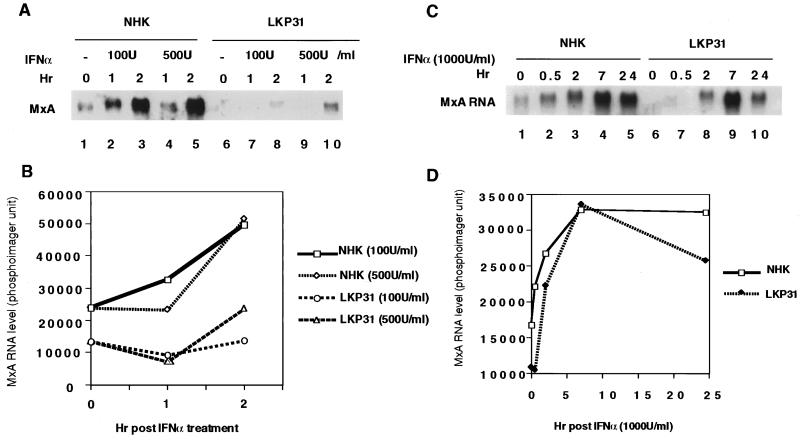
One of the major changes in gene expression identified by our microarray analysis was the repression by HPV31 of a large number of interferon-inducible genes compared to NHKs. This repression was observed in the absence of any interferon treatment. We wanted to determine if this basal repression would result in an impaired response of HPV31-positive cells to interferons. This was first examined by treating LKP31 cells and NHKs with IFN-α and determining the expression of a representative interferon-inducible gene, MxA, which encodes the interferon-responsive myxovirus resistance gene 1. MxA is primarily induced by IFN-α (1), and its basal level of expression was lower in LKP31 cells than in NHKs (Fig. 3A, lanes 1 and 6). Subconfluent cultures of LKP31 cells and NHKs were treated with IFN-α at one of two different concentrations (100 or 500 U/ml), and MxA expression was analyzed by Northern analysis. As seen in Fig. 3A, MxA RNA was induced upon interferon treatment in NHKs as well as in LKP31 cells, but the level of induction was reduced in LKP31 cells, especially with the lower dose of IFN-α tested (100 U/ml [Fig. 3B]). Upon treatment with higher levels (1,000 U/ml) of IFN-α, the induction of MxA RNA in LKP31 cells was also reduced, especially at earlier time points (Fig. 3C, lanes 1, 2, 6, and 7). However, after the initial delay, the amounts of MxA RNA in LKP31 cells and NHK reached comparable levels at 7 h posttreatment (Fig. 3D).
FIG. 3.
Induction of MxA RNA by IFN-α is impaired in HPV31 cells. (A) Northern blot analysis of 8 μg of total RNA from NHK or LKP31 cells treated with IFN-α (100 or 500 U/ml) with MxA cDNA as a probe. MxA RNA signals in panel A were quantified by a PhosphorImager, and the results are shown graphically in panel B. The induction of MxA RNA by IFN-α reaches comparable levels in LKP31 cells and NHKs at a higher dose (1,000 U/ml) and longer time exposure. (C) Northern blot analysis of 10 μg of total RNA from NHKs and LKP31 cells treated with IFN-α with MxA cDNA as a probe. MxA RNA signals in panel C were quantified by a PhosphorImager, and the results are shown graphically in panel D.
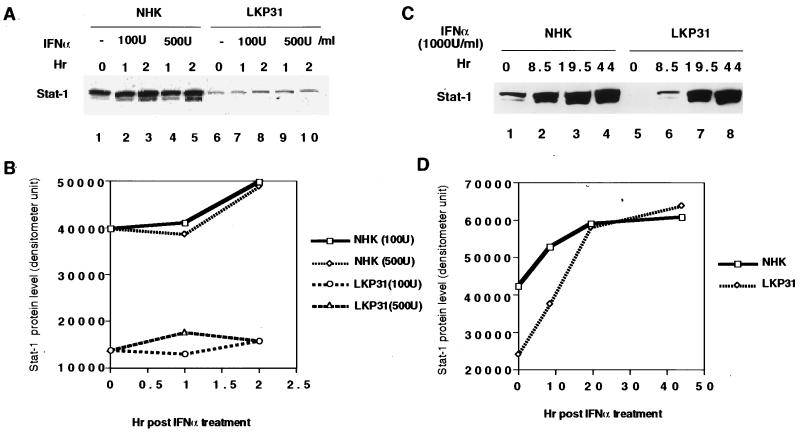
Among the interferon-inducible genes whose expression was downregulated by HPV31 was Stat-1. Stat-1 is a major regulator of interferon-stimulated transactivation, and the downregulation of other interferon-inducible genes could be the result of reduced Stat-1 expression (7, 20). In addition, the expression of Stat-1 itself is activated by interferon treatment. We therefore examined Stat-1 protein levels in response to interferon treatment. As seen in Fig. 4A, the levels of Stat-1 protein were significantly reduced in untreated LKP31 cells (lanes 1 and 6). Following treatment with IFN-α (100 and 500 U/ml), there was an induction in Stat-1 protein but levels remained reduced in LKP31 cells and did not reach levels seen in NHKs (Fig. 4B). Upon treatment with higher doses of IFN-α (1,000 U/ml), the response was still reduced until 20 h posttreatment, when the levels of Stat-1 reached those seen in NHKs (Fig. 4C, lanes 3, 4, 7, and 8; Fig. 4D). These results suggest that the response to IFN-α was reduced in HPV31-positive cells. At higher doses and at longer times of exposure to IFN-α, the effects were less pronounced.
FIG. 4.
The induction of Stat-1 protein by IFN-α is impaired in HPV31 cells. (A) Western blot analysis of equal amounts of protein lysates from NHKs and LKP31 cells treated with IFN-α (100 or 500 U/ml) to detect Stat-1 protein level. Stat-1 protein migrates in SDS-PAGE as a doublet with molecular weights of 91,000 and 84,000. Quantitative estimation of the results in panel A was done with a densitometer and shown graphically in panel B. The induction of Stat-1 protein by IFN-α reaches comparable levels in LKP31 cells and NHKs with a higher dose (1,000 U/ml) and longer time. (C) Western blot analysis of equal amount of protein lysates from NHKs or LKP31 cells treated with IFN-α to detect Stat-1 protein. Quantitative estimation of the results in panel C was done with a densitometer and shown graphically in panel D.
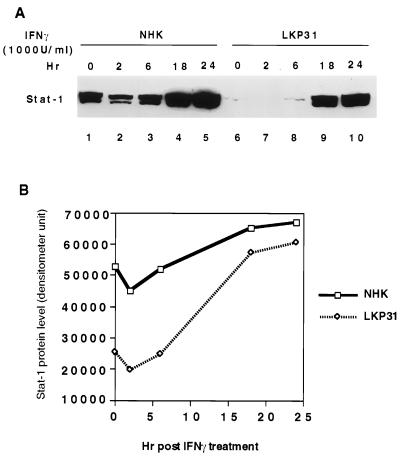
Stat-1 plays a major role in both IFN-α/β and IFN-γ pathways. Upon exposure to IFN-α, Stat-1 is phosphorylated by Tyk2 and Jak1 in the cytoplasm and forms heterodimer with phosphorylated Stat-2. The heterodimer translocates to the nucleus and forms the ISGF-3 (interferon-stimulated gene factor 3) complex with p48, leading to transactivation of IFN-α/β-responsive genes. When cells are treated with IFN-γ, Stat-1 is phosphorylated by Jak1 and Jak2, forms a homodimer, and translocates to the nucleus to activate gene expression. Since the basal level of Stat-1 is downregulated in HPV31 cells, we next examined if the IFN-γ response was also altered by HPV31. For this, we examined the induction of Stat-1 protein by IFN-γ. As shown in Fig. 5, the levels of Stat-1 protein were low in LKP31 cells, and while they increased upon IFN-γ treatment, they remained significantly below the levels seen in NHKs at earlier time points (Fig. 5A, lanes 1 to 3 and 6 to 8). However, at 18 and 24 h after IFN-γ treatment, the amount of Stat-1 protein reached a level comparable to that seen in NHKs (Fig. 5A, lanes 4, 5, 9, and 10; Fig. 5B). We conclude that in response to IFN-α and -γ stimulation, HPV31 cells exhibit a lag in activation compared to NHKs. Eventually this lag in response could be restored upon a longer exposure to interferon or at a higher dose of interferon. The interferon response is complex, involving multiple factors. The initial delay observed in HPV31 cells maybe explained, in part, by a reduced basal level of Stat-1.
FIG. 5.
The induction of Stat-1 protein by IFN-γ is impaired in HPV31 cells. (A) Western blot analysis of equal amounts of protein lysates from NHKs and LKP31 cells treated with IFN-γ (1,000 U/ml) to detect Stat-1 protein level. The image in panel A was quantified by a densitometer, and the results are shown graphically in panel B.
DISCUSSION
In this report, we used microarray analysis to examine the effect of HPV31 genome on the transcription of cellular genes. In cells that stably maintain HPV31 episomes, we observed the expression of 178 ESTs to be upregulated and that of 150 to be downregulated at least twofold among the 7,075 verified sequences screened. These cells are believed to mimic infected basal keratinocytes in vivo, as they can induce late functions upon differentiation. In our analysis, no gene was found to be activated by HPV31 gene products more than 3.2-fold, and among these no easily discernible families could be identified. Since the high-risk E7 proteins bind Rb, it was expected that E2F-responsive genes would be substantially increased in expression in HPV31-positive cells. However, none were found to be activated more than twofold. This could be due to the asynchronous nature of the cells examined or other, more complex reasons.
Among the 150 genes that were repressed by HPV31, three groups were identified. The first group included genes involved in regulation of cell growth. The E6 protein of HPV31, like that of HPV16 and -18, increases the turnover rate of p53, resulting in decreased steady-state levels (55). A major transcriptional target of p53 is the p21waf1 gene, which is a negative regulator of cyclin-dependent kinases (15). Consistent with the reduction in p53 levels, the levels of p21 transcripts were found to be reduced by both microarray and Northern analysis. In addition, Mad, the cellular regulator of Myc activity, was found to be reduced in expression by HPV31. Mad proteins form a heterodimeric complex with Max and antagonize the positive effect of Myc-Max heterodimers (2). Myc has been implicated as an activator of telomerase, and in cells expressing HPV E6 and E7, high levels of telomerase activity have been observed (27, 28, 59, 63). In our studies, no consistent transcriptional activation of Myc was detected, and downregulation of Mad expression may provide an alternative mechanism by which Myc function can be increased in infections by high-risk HPV types. A third gene found to be repressed by HPV31 is the transgelin gene. Transgelin is able to cross-link actin and remodel cytoskeleton (43). The expression of transgelin is often increased in senescing cells and is overexpressed in prematurely aging patients with Werner syndrome (53, 56). Downregulation of transgelin may contribute to loss of contact inhibition and enhanced motility characteristic of HPV-transformed cells (43).
The second class of genes found to be repressed by HPV31 were those which are expressed specifically in keratinocytes. These included genes encoding SprII, a small proline-rich protein found in UV-irradiated keratinocytes (26), and defensin, which is highly expressed in airway epithelia and has been suggested to possess antimicrobial activity (37, 64). Another repressed gene encodes stratafin, a member of family of acidic proteins that are enriched in stratifying epithelia (30). The most interesting of the keratinocyte-specific genes found to be repressed were the desmocollin and desmoplakin genes. It is possible that decreased expression of these factors could lead to less stringent cell-cell junctions that may be advantageous to productive viral life cycle.
The most intriguing set of genes found to be downregulated by HPV31 gene products were the interferon-responsive genes. Among these, Stat-1 is a primary regulator of the interferon response pathway. Stat-1 belongs to a family of proteins which normally remain latent in cytoplasm. Stat-1 is phosphorylated and activated upon interferon stimulation and turns on genes under the control of the ISRE element (7, 20) and the GAS element (7). The Stat-2 gene, also examined on the microarray, showed only marginal reduction in HPV31 cells (−1.1-fold [data not shown]). The low basal level of Stat-1 in HPV31 cells could be responsible for the low basal level of various interferon-inducible genes and contribute to the impaired response to interferon stimulation.
Additional mechanisms for altering the cellular response to interferon by HPV gene products have been previously reported. The HPV16 E6 protein has been shown to bind to and inhibit the function of the IRF3 protein (46). The IRF3 protein is an activator of interferon synthesis, and disruption of this activity interferes with the interferon response. In addition, the HPV16 E7 protein has been shown to directly bind to the p48 subunit of the ISGF-3 complex and prevent its nuclear translocation (3). Many viruses target and debilitate the interferon and immune responses during infection. Examples include adenoviruses, herpes simplex viruses (HSV), and hepatitis C viruses (6, 14, 54), all of which inhibit the activity of the double-stranded RNA-dependent kinase, PKR. In addition, adenovirus E1A binds to p48 and inhibits its nuclear translocation (31, 32). With respect to immune surveillance, HSV can downregulate antigen presentation by interfering with peptide translocation via the TAP proteins (17). Thus, it is not surprising that HPV uses multiple mechanisms to interfere with repression of the interferon response. It is interesting that among the 7,075 ESTs examined in this study, the interferon-inducible genes were identified as major targets of HPV action. This underscores the primary importance of suppressing the interferon response during papillomavirus infection.
The repression of the interferon response by HPV31 may contribute to evasion of HPV-infected cells from immune surveillance by the host. Interferon also stimulates expression of class I and II major histocompatibility antigens as well as LMP2 and -7, which are part of the 20S proteasome degradation machinery used to display of antigens on the cell surface. In our HPV-positive cells, we did not detect increased expression of class I or class II genes, suggesting that viral products may impair the activation in response to viral infection (data not shown). It is therefore possible that repression of Stat-1 expression may contribute to the evasion of the immune response during HPV infections. In our studies, the basal levels of Stat-1 were found to be reduced consistent with a repression of transcription initiation. However, it is equally plausible that this reduction is due to increased turnover of Stat-1 mRNA. It will be important to determine which of the HPV gene products are responsible for the repression of Stat-1, the mechanism of action, and whether this property extends to low-risk HPV types.
Previous studies using microarray analysis of a number of ESTs similar to that used in our study demonstrated that infection by human cytomegalovirus (HCMV) altered expression of 248 genes more than 4-fold, with many activated 6- to 15-fold (65). Interestingly, the expression of most interferon-responsive genes was increased upon HCMV infection. Acute viral infection is known to trigger an interferon response, and it is not surprising that cell lines that stably maintain the HPV genome showed an altered response manifested in this case by suppressed basal levels of interferon-inducible genes. This suppression may contribute to evasion of immune surveillance. In our study, no gene was found to be activated more than 3.2-fold, while 22 genes were repressed between 11- and 3-fold. The less severe effects seen in our studies may reflect differences between acute infection by HCMV and latent infection that is modeled by our stable HPV cell lines. Alternatively, it may be the result of more fundamental differences in the biology of these different viruses. Microarray analysis of genes activated by the platelet-derived growth factor receptor following ligand binding revealed no gene to be activated more than twofold (11). It is possible that the changes in expression of less than twofold by HPV31 can lead to profound changes in the cellular environment; however, we did not investigate these genes in detail in this study. This study has identified possible pathways by which HPV modifies the cellular environment to facilitate viral replication and evade detection by the immune system.
ACKNOWLEDGMENTS
We thank G. Sen for valuable advice and reagents, and we thank K. Rundell, R. Longnecker, F. Stubenrauch, and W. Hubert for comments on the manuscript. We also thank R. N. Eisenman and C. M. Horvath for Mad and Stat-1 cDNAs and C. Biaggi, W. Hubert, and A. Merchant for the HPV31 immortalized line T31 and for NHK isolates 407 and TP.
This work was supported by grants from the National Cancer Institute (CA59655) to L.A.L. and the Illinois Department of Public Health (96190152) to Y.E.C.
REFERENCES
- 1.Aebi M, Fah J, Hurt N, Samuel C E, Thomis D, Bazzigher L, Pavlovic J, Haller O, Staeheli P. cDNA structures and regulation of two interferon-induced human Mx proteins. Mol Cell Biol. 1989;9:5062–5072. doi: 10.1128/mcb.9.11.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayer D E, Kretzner L, Eisenman R N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 3.Barnard P, McMillan N A J. The human papillomavirus E7 oncoprotein abrogates signaling mediated by interferon-α. Virology. 1999;259:305–313. doi: 10.1006/viro.1999.9771. [DOI] [PubMed] [Google Scholar]
- 4.Bouvard V, Storey A, Pim D, Banks L. Characterization of the human papillomavirus E2 protein: evidence of trans-activation and trans-repression in cervical keratinocytes. EMBO J. 1994;13:5451–5459. doi: 10.1002/j.1460-2075.1994.tb06880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng S, Schmidt G D, Murant T, Broker T R, Chow L T. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 1995;9:2335–2349. doi: 10.1101/gad.9.19.2335. [DOI] [PubMed] [Google Scholar]
- 6.Chou J, Chen J J, Gross M, Roizman B. Association of a M(r) 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2 alpha and premature shutoff of protein synthesis after infection with gamma 1 34.5- mutants of herpes simplex virus 1. Proc Natl Acad Sci USA. 1995;92:10516–10520. doi: 10.1073/pnas.92.23.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 8.Dollard S C, Wilson J L, Demeter L M, Bonnez W, Reichman R C, Broker T R, Chow L T. Production of human papillomavirus and modulation of the infectious program in epithelial raft cultures. Genes Dev. 1992;6:1131–1142. doi: 10.1101/gad.6.7.1131. [DOI] [PubMed] [Google Scholar]
- 9.Dyson N, Howley P M, Munger K, Harlow E. The human papillomavirus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 10.El-Deiry W, Tokino T, Velculescu V, Levy D, Parson R, Trent J, Lin D, Mercer W, Kinzler K, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 11.Fambrough D, McClure K, Kazlauskas A, Lander E S. Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell. 1999;97:727–741. doi: 10.1016/s0092-8674(00)80785-0. [DOI] [PubMed] [Google Scholar]
- 12.Frattini M G, Lim H B, Doorbar J, Laimins L A. Induction of human papillomavirus type 18 late gene expression and genomic amplification in organotypic cultures from transfected DNA templates. J Virol. 1997;71:7068–7072. doi: 10.1128/jvi.71.9.7068-7072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frattini M G, Lim H B, Laimins L A. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc Natl Acad Sci USA. 1996;93:3062–3067. doi: 10.1073/pnas.93.7.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghadge G D, Malhotra P, Furtado M R, Dhar R, Thimmapaya B. In vitro analysis of virus-associated RNA I (VAI RNA): inhibition of the double-stranded RNA-activated protein kinase PKR by VAI RNA mutants correlates with the in vivo phenotype and the structural integrity of the central domain. J Virol. 1994;68:4137–4151. doi: 10.1128/jvi.68.7.4137-4151.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harper J, Adami G, Wei N, Keyomarsi K, Elledge S. The p21 cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 16.Heller R A, Schena M, Chai A, Shalon D, Bedilion T, Gilmore J, Woolley D E, Davis R W. Discovery and analysis of inflammatory disease-related genes using cDNA microarrays. Proc Natl Acad Sci USA. 1997;94:2150–2155. doi: 10.1073/pnas.94.6.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. Herpes simplex virus turns off the TAP to evade immunity. Nature. 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 18.Horisberger M A. Interferon-induced human protein MxA is a GTPase which binds transiently to cellular proteins. J Virol. 1992;66:4705–4709. doi: 10.1128/jvi.66.8.4705-4709.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horisberger M A, McMaster G K, Zeller H, Wathelet M G, Dellis J, Content J. Cloning and sequence analyses of cDNAs for interferon- and virus-induced human Mx proteins reveal that they contain putative guanine nucleotide sites: functional study of the corresponding gene promoter. J Virol. 1990;64:1171–1181. doi: 10.1128/jvi.64.3.1171-1181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horvath C M, Darnell J E., Jr The state of the STATs: recent developments in the study of signal transduction to the nucleus. Curr Opin Cell Biol. 1997;9:233–239. doi: 10.1016/s0955-0674(97)80067-1. [DOI] [PubMed] [Google Scholar]
- 21.Howley P M. Papillomavirinae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publisher; 1996. pp. 947–978. [Google Scholar]
- 22.Huibregtse J M, Scheffner M, Howley P M. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hummel M, Hudson J B, Laimins L A. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J Virol. 1992;66:6070–6080. doi: 10.1128/jvi.66.10.6070-6080.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurlin P J, Foley K P, Ayer D E, Eisenman R N, Hanahan D, Arbeit J M. Regulation of Myc and Mad during epidermal differentiation and HPV-associated tumorigenesis. Oncogene. 1995;11:2487–2501. [PubMed] [Google Scholar]
- 25.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C F, Trent J M, Staudt L M, Hudson J J, Boguski M S, Lashkari D, Shalon D, Botstein D, Brown P O. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 26.Kartasova T, van de Putte P. Isolation, characterization, and UV-stimulated expression of two families of genes encoding polypeptides of related structure in human epidermal keratinocytes. Mol Cell Biol. 1988;8:2195–2203. doi: 10.1128/mcb.8.5.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiyono T, Foster S A, Koop J I, K. M J, Galloway D A, Klingelhutz A J. Both Rb/p16ink4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 28.Klingelhutz A J, Foster S A, McDougall J K. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature. 1996;380:79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- 29.Laimins L A. The biology of human papillomaviruses: from warts to cancer. Infect Agents Dis. 1993;2:74–86. [PubMed] [Google Scholar]
- 30.Leffers H, Madsen P, Rasmussen H H, Honore B, Andersen A H, Walbum E, Vandekerckhove J, Celis J E. Molecular cloning and expression of the transformation sensitive epithelial marker stratifin. J Mol Biol. 1993;231:982–998. doi: 10.1006/jmbi.1993.1346. [DOI] [PubMed] [Google Scholar]
- 31.Leonard G T, Sen G. Restoration of interferon responses of adenovirus E1A-expressing HT1080 cell lines by overexpression of p48 protein. J Virol. 1997;71:5095–5101. doi: 10.1128/jvi.71.7.5095-5101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leonard G T, Sen G C. Effects of adenovirus E1A protein on interferon-signaling. Virology. 1996;224:25–33. doi: 10.1006/viro.1996.0503. [DOI] [PubMed] [Google Scholar]
- 33.Levy D, Larner A, Chaudhuri A, Babiss L, Darnell J E., Jr Interferon-stimulated transcription: isolation of an inducible gene and identification of its regulatory region. Proc Natl Acad Sci USA. 1986;83:8929–8933. doi: 10.1073/pnas.83.23.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowy D R, Kirnbauer R, Schiller J T. Genital human papillomavirus infection. Proc Natl Acad Sci USA. 1994;91:2436–2440. doi: 10.1073/pnas.91.7.2436. . (Review.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin L G, Demers G W, Galloway D A. Disruption of the G1/S transition in human papillomavirus type 16 E7-expressing human cells is associated with altered regulation of cyclin E. J Virol. 1998;72:975–985. doi: 10.1128/jvi.72.2.975-985.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McBride A A, Romanczuk H, Howley P M. The papillomavirus E2 regulatory proteins. J Biol Chem. 1991;266:18411–18414. [PubMed] [Google Scholar]
- 37.McCray P B J, Bentley L. Human airway epithelia express a β-defensin. Am J Respir Cell Mol Biol. 1997;16:343–349. doi: 10.1165/ajrcmb.16.3.9070620. [DOI] [PubMed] [Google Scholar]
- 38.Meyers C, Frattini M G, Hudson J B, Laimins L A. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science. 1992;257:971–973. doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- 39.Meyers C, Laimins L A. In vitro systems for the study and propagation of human papillomaviruses. Curr Top Microbiol Immol. 1994;186:199–215. doi: 10.1007/978-3-642-78487-3_11. . (Review.) [DOI] [PubMed] [Google Scholar]
- 40.Meyers C, Mayer T J, Ozbun M A. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J Virol. 1997;71:7381–7386. doi: 10.1128/jvi.71.10.7381-7386.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munger K, Werness B A, Dyson N, Phelps W C, Harlow E, Howley P M. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parker A E, Wheeler G N, Arnemann J, Pidsley S C, Ataliotis P, Thomas C L, Rees D A, Magee A I, Buxton R S. Desmosomal glycoproteins II and III. J Biol Chem. 1991;266:10438–10445. [PubMed] [Google Scholar]
- 43.Prinjha R K, Shapland C E, Hsuan J J, Totty N F, Mason I J, Lawson D. Cloning and sequencing of cDNAs encoding the actin cross-linking protein transgelin defines a new family of actin-associated proteins. Cell Motil Cytoskel. 1994;28:243–255. doi: 10.1002/cm.970280307. [DOI] [PubMed] [Google Scholar]
- 44.Rasmussen U B, Wolf C, Mattei M-G, Chenard M-P, Bellocq J-P, Chambon P, Rio M-C, Basset P. Identification of a new interferon-α-inducible gene (p27) on human chromosome 14q32 and its expression in breast carcinoma. Cancer Res. 1993;53:4096–4101. [PubMed] [Google Scholar]
- 45.Robinson P A, Marley J J, High A S, Hume W J. Differential expression of protease inhibitor and small proline-rich protein genes between normal human oral tissue and odontogenic keratinocytes. Arch Oral Biol. 1994;39:251–259. doi: 10.1016/0003-9969(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 46.Ronco L V, Karpova A Y, Vidal M, Howley P M. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruesch M N, Laimins L A. Human papillomavirus oncoproteins alter differentiation-dependent cell cycle exit on suspension in semisolid medium. Virology. 1998;250:19–29. doi: 10.1006/viro.1998.9359. [DOI] [PubMed] [Google Scholar]
- 48.Ruesch M N, Stubenrauch F, Laimins L A. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J Virol. 1998;72:5016–5024. doi: 10.1128/jvi.72.6.5016-5024.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 50.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 51.Schena M, Shalon D, Heller R, Chai A, Brown P O, Davis R W. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci USA. 1996;93:10614–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schindler C, Fu X-Y, Improta T, Aebersold R, Darnell J E., Jr Proteins of transcription factor ISGF-3: one gene encodes the 91- and 84-kDa ISGF-3 proteins that are activated by interferon α. Proc Natl Acad Sci USA. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shapland C, Hsuan J J, Totty N F, Lawson D. Purification and properties of transgelin: a transformation and shape change sensitive actin-gelling protein. J Cell Biol. 1993;121:1065–1073. doi: 10.1083/jcb.121.5.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor D R, Shi S T, Romano P R, Barber G N, Lai M M C. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285:107–110. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- 55.Thomas J T, Laimins L A. Human papillomavirus oncoproteins E6 and E7 independently abrogate the mitotic spindle checkpoint. J Virol. 1998;72:1131–1137. doi: 10.1128/jvi.72.2.1131-1137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thweatt R, Lumpkin C K J, Goldstein S. A novel gene encoding a smooth muscle protein is overexpressed in senescent human fibroblasts. Biochem Biophys Res Commun. 1992;187:1–7. doi: 10.1016/s0006-291x(05)81449-4. [DOI] [PubMed] [Google Scholar]
- 57.Tissot C, Mechti N. Molecular cloning of a new interferon-induced factor that represses human immunodeficiency virus type 1 long terminal repeat expression. J Biol Chem. 1995;270:14891–14898. doi: 10.1074/jbc.270.25.14891. [DOI] [PubMed] [Google Scholar]
- 58.Virata M L A, Wagner R M, Parry D A D, Green K J. Molecular structure of the human desmoplakin I and II amino terminus. Proc Natl Acad Sci USA. 1992;89:544–548. doi: 10.1073/pnas.89.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J, Xie L Y, Allan S, Beach D, Hannon G J. Myc activates telomerase. Genes Dev. 1998;12:1769–1774. doi: 10.1101/gad.12.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang X, Johansen L M, Tae H-J, Taparowsky E J. IFP 35 forms complexes with B-ATF, a member of the AP1 family of transcription factors. Biochem Biophys Res Commun. 1996;229:316–322. doi: 10.1006/bbrc.1996.1799. [DOI] [PubMed] [Google Scholar]
- 61.Wathelet M G, Clauss I M, Content J, Hues G A. The IFI-56K and IFI-54K interferon-inducible human genes belong to the same gene family. FEBS Lett. 1988;231:164–171. doi: 10.1016/0014-5793(88)80724-5. [DOI] [PubMed] [Google Scholar]
- 62.Werness B A, Levine A J, Howley P M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 63.Wu K-J, Grandori C, Amacker M, Simon-Vermont N, Polack A, Lingner J, Dalla-Favera R. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21:220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 64.Yang D, Chertov O, Bykovskaia S N, Chen Q, Buffo M J, Shogan J, Anderson M, Schroder J M, Wang J M, Howard O M Z, Oppenheim J J. β-Defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science. 1999;286:525–528. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]
- 65.Zhu H, Cong J-P, Mamtora G, Gingeras T, Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:14470–14475. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.zur Hausen H, de Villiers E M. Human papillomaviruses. Annu Rev Microbiol. 1994;48:427–447. doi: 10.1146/annurev.mi.48.100194.002235. [DOI] [PubMed] [Google Scholar]