Abstract
In vitro stem cell-derived embryo and organ models, termed embryoids and organoids, respectively, provide promising experimental tools to study physiological and pathological processes in mammalian development and organ formation. Most of current embryoid and organoid systems are developed using conventional three-dimensional cultures that lack controls of spatiotemporal extracellular signals. Microfluidics, an established technology for quantitative controls and quantifications of dynamic chemical and physical environments, has recently been utilized for developing next-generation embryoids and organoids in a controllable and reproducible manner. In this review, we summarize recent progress in constructing microfluidics-based embryoids and organoids. Development of these models demonstrates the successful applications of microfluidics in establishing morphogen gradients, accelerating medium transport, exerting mechanical forces, facilitating tissue co-culture studies, and improving assay throughput, thus supporting using microfluidics for building next-generation embryoids and organoids for fundamental and translational research.
Keywords: Microfluidics, embryoid, organoid, mammalian development, stem cell
Introduction
Stem cell-based, in vitro models of mammalian developments and organ formation are becoming indispensable tools for advancing mammalian developmental biology and disease modeling [1–6]. This is particularly true for understanding human development, given our limited access to and bioethical constraints in human embryonic tissues. Till now, there are various models of mammalian embryo and organ developments, termed embryoids and organoids, respectively, that have been reported [2,3,5,7–9]. Embryoids have been developed to recapitulate early embryogenic events, from pre-implantation blastocyst formation, to peri-implantation and peri-gastrulation development, all the way up to early organogenesis [1–6]. For organoids, there are numerous organoids available now to model the development, homeostasis, and pathology of organs associated with the three definitive germ layers [7–9]. Researchers continuously develop improved embryoids and organoids with enhanced maturity, functions, complexity, structural fidelity, and disease or developmental relevance.
Bioengineering technologies have been used successfully in the development of embryoids and organoids [1,2,4–6,10–21]. These technologies include genetic engineering tools [5,6,10–13], functional biomaterials [14–16,19,20], and bioengineering tools [1,2,5,6,10,11,21] that can efficiently modulate spatiotemporal local tissue microenvironment. Genetic engineering tools are utilized to generate signaling and lineage reporter lines, allowing monitoring of intracellular signaling dynamics and cell fate decisions during embryoid and organoid developments [4,10,12,13]. Genetic technologies have also been utilized to direct cells to interact efficiently with specific chemical cues [5,6,10] or local light illuminations [11,12]. Functional biomaterials, such as synthetic hydrogels [14,15,19,20] and natural extracellular matrix (ECM) proteins [16], have also been used for embryoid and organoid developments, either directly in conventional three-dimensional (3D) tissue cultures [14,15] or in bioprinting [17,18] and microfluidics [19,20]. There are other bioengineering tools utilized to control the size and shape of initial cell clusters for embryoid and organoid developments, such as micropatterning [11,21], AggreWell [2,5,6,10], and microwells [1]. For prolonged embryoid and organoid cultures, tissue culture shakers [6,22] and ex utero culture instruments [5,6] have been utilized.
In this review, we focus on discussing promising applications of microfluidics in embryoid and organoid developments. Microfluidic devices can generate gradients of chemical signals, useful for tissue patterning and symmetry breaking. Through precise controls of microfluidic environments, physical signals, such as gas composition, pressure and shear stress, can be modulated for embryoid and organoid developments. Since microfluidic devices contain prescribed chambers and channels, useful for loading and positioning different types of cells, microfluidic devices are useful for controlling and studying cell-cell interactions during embryoid and organoid developments. There are also important efforts in developing automated, high-throughput microfluidic devices for embryoid and organoid developments, promising for translational screening applications.
Microfluidic gradients inducing tissue patterning and symmetry breaking
During development, tissue patterning is achieved through specification and differentiation of embryonic progenitor cells into functional tissue cell types in a well-orchestrated manner. The importance of chemical signals, including morphogens, has been well established in tissue patterning. Morphogen gradients in the extracellular space provide positional information, to which embryonic progenitor cells respond in a dose-dependent manner. Microfluidics offers a convenient platform to create and control graded chemical environments to induce tissue patterning in embryoids and organoids.
Passive diffusion remains the most straightforward way for generating microfluidic gradients. Often time cells are cultured in a microfluidic chamber connected to source and sink reservoirs, which establishes a concentration gradient in the cell chamber following the classic source-sink model of Fickian diffusion. Hydrogels are often added into the cell chamber or between the cell chamber and source and sink reservoirs to prevent advection flows that might cause undesirable effects on cells. Using microfluidic chemical gradients generated using passive diffusion, a broad concentration range of different chemicals have been screened for inducing motor neuron differentiation from mouse embryonic stem cells (mESCs; Figure 1a) [23]. Microfluidic chemical gradients based on passive diffusion have also been integrated with a 2D micropatterned human gastrulation model to achieve in vivo-like axial germ layer patterning, highlighting the importance of combining exogenous bioengineering controls and intrinsic stem cell self-organization to build embryoids and organoids with heightened complexity and in vivo relevance [21].
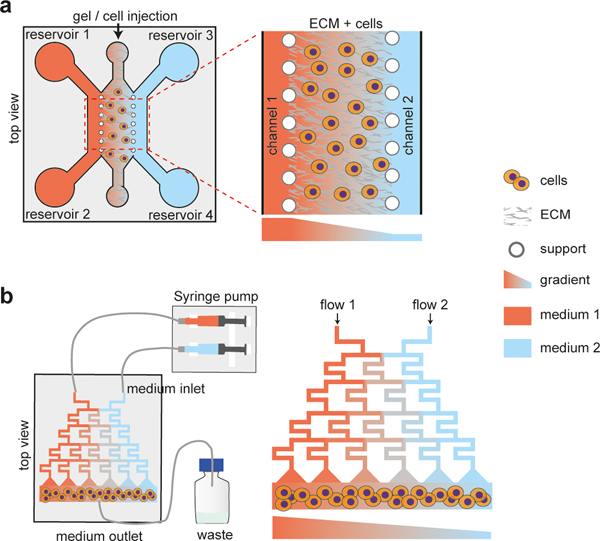
Figure 1.
Morphogen gradient generation by microfluidics.
a. Morphogen gradient generated by diffusion through ECM with cells embedded in ECM [23], with (Left) top view and (Right) zoom-in top view. Culture medium is added into 4 reservoirs. Gel and cells are injected into middle channel. b. Gradient generation by splitting and mixing microfluidic flow [24], with (Left) top view and (Right) zoom-in top view. The flow is driver by syringe pump, and the medium flowing through the microfluidic chip is collected in a waste bottle. Flow 1 and flow 2 in microfluidics systems have different chemical concentrations, which allows for the creation of a concentration gradient within the device.
Microfluidic gradients can also be generated through a series of splitting and mixing of microfluidic flows (Figure 1b). Such microfluidic gradient design has been utilized to establish an exogeneous WNT signal gradient to recapitulate rostral-caudal patterning of the neural tube [24]. Interestingly, an isthmic organizer-like region emerges in the patterned neural tube-like structure at the boundary of putative forebrain and midbrain regions, highlighting the autonomy and modularity during organ development.
Owing to precisely controlled microfluidic environments, embryoids and organoids developed using microfluidics often show improved efficiency and reproducibility. This feature could be best illustrated using the microfluidic post-implantation amniotic sac embryoid (PASE). The PASE was first developed using a conventional 3D culture, in which a small percentage (5–10%) of hPSC clusters would undergo lumenogenesis then symmetry breaking and amniotic patterning, leading to the formation of asymmetric amniotic ectoderm-epiblast pattern that resembles the human amniotic sac [25]. To improve PASE formation efficiency, a microfluidic platform was developed to guide formation of hPSC clusters in prescribed locations before asymmetric morphogen stimulations to drive synchronized PASE formation in a controllable and reproducible manner [3,26].
Microfluidics for controlling material transport and physical environment
Besides chemical signals, other factors, such as nutrients, gases, mechanical forces and geometric topology, also can have an impact on embryoid and organoid development. Controlled flows in microfluidic devices can enhance nutrient and oxygen transport [27], beneficial to tissue growth, survival and maturation [16,27–29]. For example, apoptosis was minimized and proliferation was promoted in microfluidic brain organoid cultures (Figure 2a) [16]. Improved survival and insulin secretion were shown in islet organoids under continuous microfluidic perfusion [27,29]. Microfluidics could also influence embryoid and organoid development by removing secreted factors. For example, in a gut organoid chip with independent controls of fluid flow and mechanical deformation, basal flow in gut organoids was shown to induce villi-like morphogenesis of intestinal epithelium, mainly via removal of WNT antagonists secreted by the tissues themselves [8,30].
Figure 2.
Microfluidics controls material transport and physical environment.
a. Microfluidic flow accelerates material transport for brain organoids [16]. The flow in the microfluidic system is driven by a rocker machine. b. Kidney organoids cultured under microfluidic shear flows [31], which is driven by peristaltic pump. c. Colon tumor organoids embedded in ECM experiencing cyclic pressures through the application of a microfluidic pressure channel [32], with (Upper) overall view and (Lower) zoom-in top view. The pressure in the system is regulated through the pressure channel, where the liquid is subjected to increased pressure using an air compressor and controller. The flow within the medium channel is propelled by a syringe pump. d. Lung tissue experiencing transmural pressure difference established using microfluidics [33]. The pressure difference (Δp) within the microfluidic system is established by the difference in heights (h1 and h2) of the culture medium. e. Microchannel scaffold to guide intestinal epithelial organization and differentiation [34]. In the microfluidic system used for intestinal studies, two independent flows are employed. One flow is responsible for delivering a medium supplemented with nutrients by passive diffusion, while the other flow driven by syringe pump is used for perfusion within the intestinal lumen.
Microfluidics has also been utilized for controlling shear stress and hydrodynamic pressure to promote morphogenesis and maturation during embryoid and organoid developments. It has been shown that kidney organoids exposed to high shear flow exhibited enhanced vascularization and had more mature podocytes and tubular compartments compared with those under static culture (Figure 2b) [31]. Using microfluidics containing a pressure channel, cyclic pressures were applied on colon tumor organoids to mimic peristalsis (Figure 2c) [32]. Applying hydrostatic pressures to mimic transmural pressures on lung explants, transmural pressure was shown to modulate airway branching morphogenesis, airway smooth muscle contraction, and maturation of lung tissues (Figure 2d) [33].
Microfluidic devices can also provide precise topologies useful for guiding tissue morphogenesis and differentiation. Laser micromachining was applied to fabricate a microfluidic channel in hydrogels for the development of a gut model suitable for long-term homeostatic culture under an external perfusion pump [34]. Topological features of the microfluidic mini-gut model guided the development of intestinal epithelial tissues, leading to the formation of a tube-shaped structure with crypt- and villus-like domains (Figure 2e). Importantly, intestinal stem cells and Paneth cells were exclusively found in crypt-like regions, whereas enterocytes, enteroendocrine cells and goblet cells were exclusively located in villus-like regions (Figure 2e), mimicking spatial cell organizations in intestinal epithelial tissues.
Microfluidics for controlling tissue-tissue interactions
Tissue-tissue interactions are manifested in every step of mammalian development and organ formation. Microfluidics provides a convenient platform for positioning different tissue cell types at prescribed locations inside a controlled microfluidic environment, imitating in vivo-relevant tissue-tissue interactions. To model invasion of extravillous trophoblasts (EVTs) into maternal uterus during the placentation, a maternal-fetal interface was established by seeding EVTs and endothelium cells into two parallel microfluidic channels separated by ECM or a pillar barrier array [35,36] (Figure 3a). The barrier function of placenta was also imitated by placing trophoblast cells or embryoid bodies in one microfluidic channel to model the embryonic compartment of the fetal-maternal interface and endothelial cells in a separate adjacent channel to model the maternal compartment [37–39]. Similarly, by placing different tissue cell types into opposing microfluidic channels (Figure 3b), intra-organ models were constructed, such as a liver model with hepatocytes interfaced with liver sinusoidal endothelial cells, Kupffer cells and hepatic stellate cells [40], and a pancreas model with pancreatic ductal epithelial cells interfaced with islet cells [41]. Vascular and immune systems have also been incorporated into microfluidic organoid cultures (Figure 3c), such as cerebral [42] and hepatic organoids [43,44].
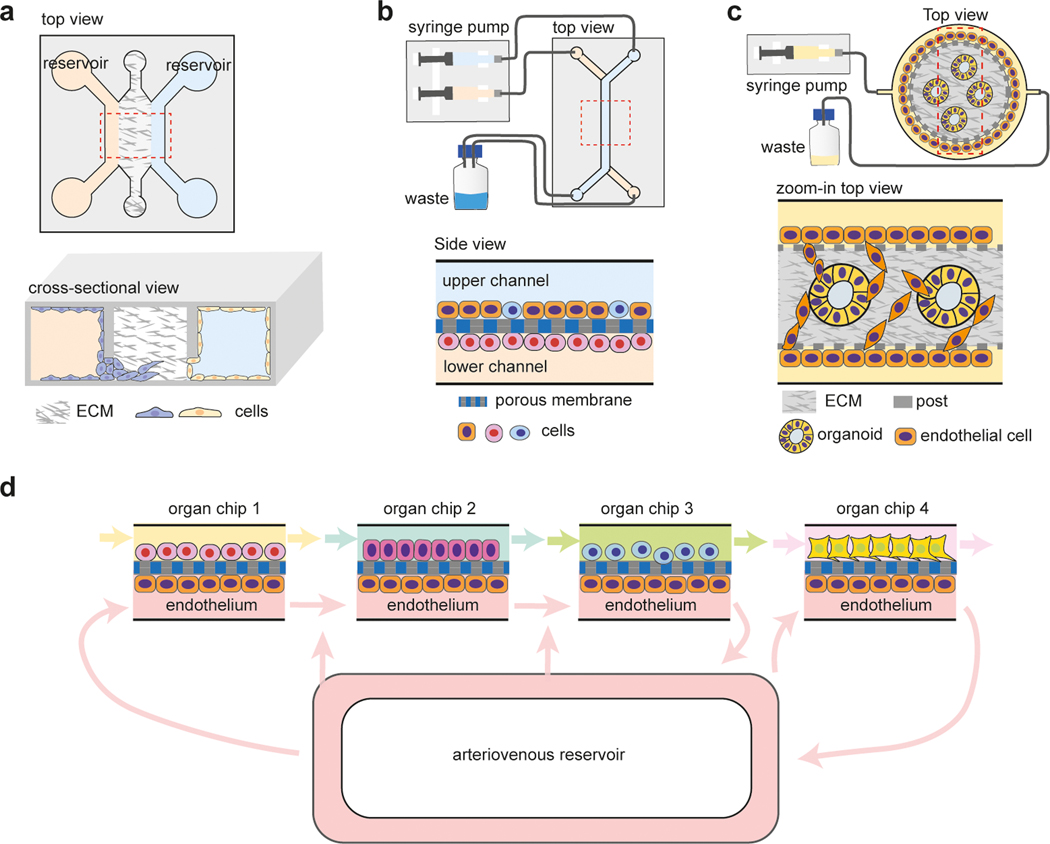
Figure 3.
Microfluidics for tissue co-culture studies.
a. Cells are seeded into two parallel channels separated by an ECM barrier to study cell migration and invasion [35], shown in (Upper) top view and (Lower) side view. b. Cells are seeded at the opposite sides of a porous membrane to study cell-cell interaction by soluble molecules [40,41], shown in (Upper) top overall view and (Lower) zoom-in side view. c. Endothelial cells invading into a center channel containing ECM and organoids for vascularization of organoids [42,43], shown in (Upper) top overall view and (Lower) zoom-in top view. d. Microphysiological systems constructed with multiple organ models on the same chip. Interconnections between organ models are established through arteriovenous reservoir [47,49].
Microphysiological systems containing multiple organ models have been established using microfluidics to study inter-organ communications and model multi-organ processes and systematic diseases [45–49]. Each organ model in the microphysiological system can be maintained in its own optimal condition, and interconnections between organ models are established based on their in vivo relationships [47–49] (Figure 3d).
Microfluidics for scalable translational applications
Microfluidics is intrinsically a scalable technology compatible with translational screens. As a potent high-throughput technology, droplet microfluidics, for example, has been used to generate embryoids and organoids with simplified procedures, great throughput, and low variability. So far, droplet microfluidics has been used for the developments of epiblast spheroids [50], liver organoids [51,52], lung organoids [51,53], kidney organoids [51], islet organoids [19,20], mesenchymal bodies [54], and tumor organoids [51,53,55] (Figure 4a). Some droplet microfluidics-based organoid tools have been utilized for large-scale drug screens [51,55]. In another example, an automated microfluidic culture was developed for pancreatic tumor organoids. This system was applied to test up to 20 regimens and 10 patient samples in parallel, offering a promising platform for individual, combinatorial, and sequential drug screens on pancreatic tumor organoids [56] (Figure 4b).
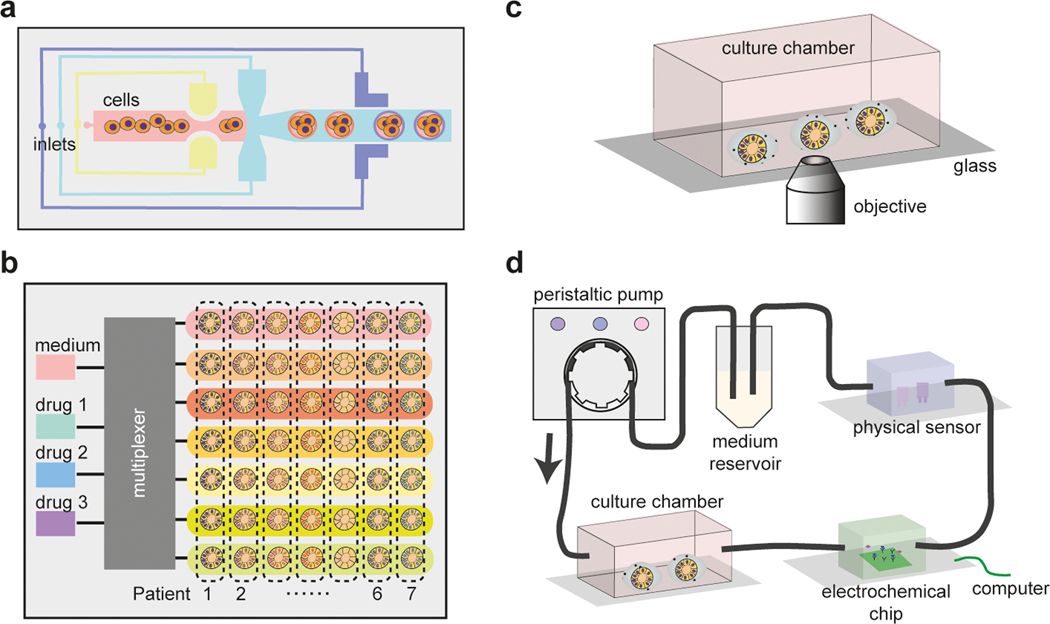
Figure 4.
Microfluidics for scalable productions of embryoids and organoids.
a. Droplet microfluidics for embryoid and organoid generations [19,20]. b. An automated microfluidic system for patient sample screening [56]. The multiplexer is utilized to generate the medium containing various drugs and concentrations. c. Microfluidic chip incorporated with other equipment to be compatible with imaging. d. The microfluidic chip features an integrated sample culture chamber coupled with electrochemical biosensors and physical sensors for real-time monitoring [58]. The flow within the chip is driven by a peristaltic pump.
Live imaging is commonly used for analyzing microfluidic organoid and embryoid cultures, given the controlled positioning and orientations of organoids and embryoids in microfluidic devices [3,21,34,56,57] (Figure 4c). In situ biochemical sensors can also be integrated with microfluidics, to continually monitor relevant culture signals in microfluidic organoid and embryoid cultures [58] (Figure 4d). These sensors include those for monitoring extracellular microenvironment parameters such as pH, oxygen level and temperature. Additionally, electrochemical sensors can be utilized to measure soluble protein biomarkers in microfluidic organoid and embryoid cultures. Thus, integration of biosensing technologies with microfluidic organoid and embryoid cultures offer enhanced capabilities for continuous medium supply, automated sampling and real-time sensing, and precise controls of culture conditions including physiological and mechanical forces, for long-term culture of organoids and embryoids.
Conclusions and future directions
Over the last two decades, a vast array of microfluidic technologies have been developed, with some of them even targeting single-cell and single-molecule analyses [REF]. For more detailed discussions on available microfluidic technologies for bio-related applications, readers are directed to some excellent recent reviews [REF]. Microfluidic tools compatible with mammalian cell cultures are particularly attractive for the development of next-generation embryoid and organoid cultures. Since such efforts are still at exploratory stages in research laboratory settings, polydimethylsiloxane (PDMS)-based microfluidic technologies, such as those based on soft lithography, remain the most versatile and popular ones given the compatibility of PDMS with rapid prototype device fabrication, mammalian cell culture and live imaging. Nonetheless, changes in device material, surface coating, cell number per unit surface area or per unit medium volume may all affect the outcome of otherwise standard embryoid or organoid protocols that have been established using conventional culture tools. Spatial constraints in microfluidics might also present a physical limitation for long-term cultures of embryoids and organoids. Thus, it is important to fully characterize and optimize microfluidic embryoid and organoid development protocols. Future directions in this area include applying microfluidic innovations to obtain embryoid and organoid systems with enhanced maturity, functions, complexity, structural fidelity, and disease or development relevance. Microfluidics can provide a more in vivo-like environment through dynamic spatiotemporal controls of chemical signals, morphogen gradients, material transports, mechanical forces, and tissue topology and orientation. The other direction is to apply microfluidics to improve the efficiency, reproducibility, and scalability of embryoid and organoid cultures, necessary for translational screens. A widely recognized challenge in embryoid and organoid cultures is the intra- and inter-batch variability. Microfluidics can reduce such variability through implementations of precisely controlled spatiotemporal signals to modulate embryoid and organoid developments.
Acknowledgements
We acknowledge the support from the Michigan-Cambridge Research Initiative, the National Institutes of Health (R21 NS113518, R21 HD100931, R21 HD105126, R21 HD105192, R21 HD109635 and R01 GM143297), the National Science Foundation (CMMI 1917304, CBET 1901718 and PFI 2213845), and the 21st Century Jobs Trust Fund received through the Michigan Strategic Fund from the State of Michigan (CASE315037).
Footnotes
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
No original data is used for this review article.
References
- 1.Kagawa H, Javali A, Khoei HH, Sommer TM, Sestini G, Novatchkova M, Reimer YSO, Castel G, Bruneau A, Maenhoudt N, Lammers J, Loubersac S, Freour T, Vankelecom H, David L, Rivron N. Human blastoids model blastocyst development and implantation. Nature 2022, 601:600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu LQ, Wei YL, Duan JL, Schmitz DA, Sakurai M, Wang L, Wang KH, Zhao SH, Hon GC, Wu J. Blastocyst-like structures generated from human pluripotent stem cells. Nature 2021, 591:620–626. [DOI] [PubMed] [Google Scholar]
- 3.Zheng Y, Xue XF, Shao YE, Wang SC, Esfahani SN, Li ZD, Muncie JM, Lakins JN, Weaver VM, Gumucio DL, Fu JP. Controlled modelling of human epiblast and amnion development using stem cells. Nature 2019, 573:421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moris N, Anlas K, van den Brink SC, Alemany A, Schroder J, Ghimire S, Balayo T, van Oudenaarden A, Arias AM. An in vitro model of early anteroposterior organization during human development. Nature 2020, 582:410–415. [DOI] [PubMed] [Google Scholar]
- 5.Amadei G, Handford CE, Qiu CX, De Jonghe J, Greenfeld H, Tran M, Martin BK, Chen DY, Aguilera-Castrejon A, Hanna JH, Elowitz MB, Hollfelder F, Shendure J, Glover DM, Zernicka-Goetz M. Embryo model completes gastrulation to neurulation and organogenesis. Nature 2022, 610:143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarazi S, Aguilera-Castrejon A, Joubran C, Ghanem N, Ashouokhi S, Roncato F, Wildschutz E, Haddad M, Oldak B, Gomez-Cesar E, et al. Post-gastrulation synthetic embryos generated ex utero from mouse naive ESCs. Cell 2022, 185:3290–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anand GM, Megale HC, Murphy SH, Weis T, Lin Z, He Y, Wang X, Liu J, Ramanathan S. Controlling organoid symmetry breaking uncovers an excitable system underlying human axial elongation. Cell 2023, 186:497–512 e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shin W, Kim HJ. 3D in vitro morphogenesis of human intestinal epithelium in a gut-on-a-chip or a hybrid chip with a cell culture insert. Nat Protoc 2022, 17:910–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Revah O, Gore F, Kelley KW, Andersen J, Sakai N, Chen X, Li MY, Birey F, Yang X, Saw NL, et al. Maturation and circuit integration of transplanted human cortical organoids. Nature 2022, 610:319–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amadei G, Lau KYC, De Jonghe J, Gantner CW, Sozen B, Chan C, Zhu M, Kyprianou C, Hollfelder F, Zernicka-Goetz M. Inducible Stem-Cell-Derived Embryos Capture Mouse Morphogenetic Events In Vitro. Dev Cell 2021, 56:366–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hellwarth PB, Chang Y, Das A, Liang P-Y, Lian X, Repina NA, Bao X. Optogenetic-mediated cardiovascular differentiation and patterning of human pluripotent stem cells. Advanced Genetics 2021, 2:e202100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miura Y, Li MY, Birey F, Ikeda K, Revah O, Thete MV, Park JY, Puno A, Lee SMH, Porteus MH, Pasca SP. Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat Biotechnol 2020, 38:1421–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simunovic M, Metzger JJ, Etoc F, Yoney A, Ruzo A, Martyn I, Croft G, You DS, Brivanlou AH, Siggia ED. A 3D model of a human epiblast reveals BMP4-driven symmetry breaking. Nat Cell Biol 2019, 21:900–910. [DOI] [PubMed] [Google Scholar]
- 14.Treacy NJ, Clerkin S, Davis JL, Kennedy C, Miller AF, Saiani A, Wychowaniec JK, Brougham DF, Crean J. Growth and differentiation of human induced pluripotent stem cell (hiPSC)-derived kidney organoids using fully synthetic peptide hydrogels. Bioact Mater 2023, 21:142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye SC, Boeter JWB, Mihajlovic M, van Steenbeek FG, van Wolferen ME, Oosterhoff LA, Marsee A, Caiazzo M, van der Laan LJW, Penning LC, Vermonden T, Spee B, Schneeberger K. A Chemically Defined Hydrogel for Human Liver Organoid Culture. Adv Funct Mater 2020, 30:2000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho AN, Jin Y, An Y, Kim J, Choi YS, Lee JS, Kim J, Choi WY, Koo DJ, Yu W, et al. Microfluidic device with brain extracellular matrix promotes structural and functional maturation of human brain organoids. Nat Commun 2021, 12:4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q, Qi GY, Liu XM, Bai JF, Zhao JK, Tang GS, Zhang YS, Chen-Tsai R, Zhang M, Wang DH, Zhang YY, Atala A, He JQ, Sun XZSS. Universal Peptide Hydrogel for Scalable Physiological Formation and Bioprinting of 3D Spheroids from Human Induced Pluripotent Stem Cells. Adv Funct Mater 2021, 31:2104046. [Google Scholar]
- 18.Kapr J, Petersilie L, Distler T, Lauria I, Bendt F, Sauter CM, Boccaccini AR, Rose CR, Fritsche E. Human Induced Pluripotent Stem Cell-Derived Neural Progenitor Cells Produce Distinct Neural 3D In Vitro Models Depending on Alginate/Gellan Gum/Laminin Hydrogel Blend Properties. Adv Healthc Mater 2021, 10:2100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Liu HT, Zhang X, Wang YQ, Zhao MQ, Chen WW, Qin JH. One-Step Generation of Aqueous-Droplet-Filled Hydrogel Fibers as Organoid Carriers Using an All-in-Water Microfluidic System. Acs Appl Mater Inter 2021, 13:3199–3208. [DOI] [PubMed] [Google Scholar]
- 20.Liu HT, Wang YQ, Wang H, Zhao MQ, Tao TT, Zhang X, Qin JH. A Droplet Microfluidic System to Fabricate Hybrid Capsules Enabling Stem Cell Organoid Engineering. Adv Sci 2020, 7:1903739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manfrin A, Tabata Y, Paquet ER, Vuaridel AR, Rivest FR, Naef F, Lutolf MP. Engineered signaling centers for the spatially controlled patterning of human pluripotent stem cells. Nat Methods 2019, 16:640–648. [DOI] [PubMed] [Google Scholar]
- 22.Olmsted ZT, Paluh JL. Co-development of central and peripheral neurons with trunk mesendoderm in human elongating multi-lineage organized gastruloids. Nat Commun 2021, 12:3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demers CJ, Soundararajan P, Chennampally P, Cox GA, Briscoe J, Collins SD, Smith RL. Development-on-chip: in vitro neural tube patterning with a microfluidic device. Development 2016, 143:1884–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rifes P, Isaksson M, Rathore GS, Aldrin-Kirk P, Moller OK, Barzaghi G, Lee JL, Egerod KL, Rausch DM, Parmar M, Pers TH, Laurell T, Kirkeby A. Modeling neural tube development by differentiation of human embryonic stem cells in a microfluidic WNT gradient (vol 15, pg 513, 2020). Nat Biotechnol 2020, 38:1357–1357. [DOI] [PubMed] [Google Scholar]
- 25.Shao Y, Taniguchi K, Townshend RF, Miki T, Gumucio DL, Fu JP. A pluripotent stem cell-based model for post-implantation human amniotic sac development. Nat Commun 2017, 8:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zheng Y, Yan RZ, Sun S, Kobayashi M, Xiang L, Yang R, Goedel A, Kang Y, Xue X, Esfahani SN, et al. Single-cell analysis of embryoids reveals lineage diversification roadmaps of early human development. Cell Stem Cell 2022, 29:1402–1419. Zheng et al. developed a microfluidic post-implantation amniotic sac embryoid (μPASE) with high efficiency and controllability, by using microfluidics to direct formation of hPSC clusters in prescribed locations before asymmetric morphogen stimulations to drive PASE formation.
- 27.Patel SN, Ishahak M, Chaimov D, Velraj A, LaShoto D, Hagan DW, Buchwald P, Phelps EA, Agarwal A, Stabler CL. Organoid microphysiological system preserves pancreatic islet function within 3D matrix. Sci Adv 2021, 7:eaba5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ao Z, Cai H, Havert DJ, Wu Z, Gong Z, Beggs JM, Mackie K, Guo F. One-Stop Microfluidic Assembly of Human Brain Organoids To Model Prenatal Cannabis Exposure. Analytical Chemistry 2020, 92:4630–4638. [DOI] [PubMed] [Google Scholar]
- 29.Jun Y, Lee J, Choi S, Yang JH, Sander M, Chung S, Lee SH. In vivo-mimicking microfluidic perfusion culture of pancreatic islet spheroids. Sci Adv 2019, 5:aax4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin W, Hinojosa CD, Ingber DE, Kim HJ. Human Intestinal Morphogenesis Controlled by Transepithelial Morphogen Gradient and Flow-Dependent Physical Cues in a Microengineered Gut-on-a-Chip. Iscience 2019, 15:391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Homan KA, Gupta N, Kroll KT, Kolesky DB, Skylar-Scott M, Miyoshi T, Mau D, Valerius MT, Ferrante T, Bonventre JV, Lewis JA, Morizane R. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat Methods 2019, 16:255–262. Homan et al. showed that kidney organoids exposed to microfluidic high shear flows exhibited enhanced vascularization and had more mature podocytes and tubular compartments compared with those under static culture.
- 32.Fang G, Lu H, Al-Nakashli R, Chapman R, Zhang Y, Ju LA, Lin G, Stenzel MH, Jin D. Enabling peristalsis of human colon tumor organoids on microfluidic chips. Biofabrication 2021, 14:015006. [DOI] [PubMed] [Google Scholar]
- 33.Nelson CM, Gleghorn JP, Pang M-F, Jaslove JM, Goodwin K, Varner VD, Miller E, Radisky DC, Stone HA. Microfluidic chest cavities reveal that transmural pressure controls the rate of lung development. Development 2017, 144:4328–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nikolaev M, Mitrofanova O, Broguiere N, Geraldo S, Dutta D, Tabata Y, Elci B, Brandenberg N, Kolotuev I, Gjorevski N, Clevers H, Lutolf MP. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 2020, 585:574–578. Nikolaev et al. established a mini-gut model suitable with long-term homeostatic culture by using a microchannel scaffold to guide intestinal epithelial growth and external pumps for perfusion.
- 35. Park JY, Mani S, Clair G, Olson HM, Paurus VL, Ansong CK, Blundell C, Young R, Kanter J, Gordon S, Yi AY, Mainigi M, Huh DD. A microphysiological model of human trophoblast invasion during implantation. Nat Commun 2022, 13:1252. Park et al. reported a microfluidic maternal-fetal interface to model invasion of extravillous trophoblasts into the uterus by seeding extravillous trophoblast cells and endothelium cells into two adjacent microfluidic channels separated by an ECM barrier.
- 36.Pu Y, Gingrich J, Veiga-Lopez A. A 3-dimensional microfluidic platform for modeling human extravillous trophoblast invasion and toxicological screening. Lab Chip 2021, 21:546–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blundell C, Tess ER, Schanzer ASR, Coutifaris C, Su EJ, Parry S, Huh D. A microphysiological model of the human placental barrier. Lab Chip 2016, 16:3065–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boos JA, Misun PM, Brunoldi G, Furer LA, Aengenheister L, Modena M, Rousset N, Buerki-Thurnherr T, Hierlemann A. Microfluidic Co-Culture Platform to Recapitulate the Maternal-Placental-Embryonic Axis. Adv Biol 2021, 5:2100609. [DOI] [PubMed] [Google Scholar]
- 39.Mosavati B, Oleinikov A, Du E. 3D microfluidics-assisted modeling of glucose transport in placental malaria. Sci Rep-Uk 2022, 12:15278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jang KJ, Otieno MA, Ronxhi J, Lim HK, Ewart L, Kodella KR, Petropolis DB, Kulkarni G, Rubins JE, Conegliano D, et al. Reproducing human and cross-species drug toxicities using a Liver-Chip. Sci Transl Med 2019, 11:eaax5516. [DOI] [PubMed] [Google Scholar]
- 41.Mun KS, Arora K, Huang YJ, Yang FMY, Yarlagadda S, Ramananda Y, Abu-El-Haija M, Palermo JJ, Appakalai BN, Nathan JD, Naren AP. Patient-derived pancreas-on-a-chip to model cystic fibrosis-related disorders. Nat Commun 2019, 10:3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salmon I, Grebenyuk S, Fattah ARA, Rustandi G, Pilkington T, Verfaillie C, Ranga A. Engineering neurovascular organoids with 3D printed microfluidic chips. Lab Chip 2022, 22:1615–1629. [DOI] [PubMed] [Google Scholar]
- 43.Bonanini F, Kurek D, Previdi S, Nicolas A, Hendriks D, de Ruiter S, Meyer M, Cabrer MC, Dinkelberg R, Garcia SB, et al. In vitro grafting of hepatic spheroids and organoids on a microfluidic vascular bed. Angiogenesis 2022, 25:455–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Natarajan V, Simoneau CR, Erickson AL, Meyers NL, Baron JL, Cooper S, McDevitt TC, Ott M. Modelling T-cell immunity against hepatitis C virus with liver organoids in a microfluidic coculture system. Open Biol 2022, 12:210320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tao TT, Deng PW, Wang YQ, Zhang X, Guo YQ, Chen WW, Qin JH. Microengineered Multi-Organoid System from hiPSCs to Recapitulate Human Liver-Islet Axis in Normal and Type 2 Diabetes. Adv Sci 2022, 9:2103495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trapecar M, Communal C, Velazquez J, Maass CA, Huang YJ, Schneider K, Wright CW, Butty V, Eng G, Yilmaz O, Trumper D, Griffith LG. Gut-Liver Physiomimetics Reveal Paradoxical Modulation of IBD-Related Inflammation by Short-Chain Fatty Acids. Cell Syst 2020, 10:223–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novak R, Ingram M, Marquez S, Das D, Delahanty A, Herland A, Maoz B, Jeanty SSF, Somayaji MR, Burt M, et al. Robotic fluidic coupling and interrogation of multiple vascularized organ chips. Nat Biomed Eng 2020, 4:407–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herland A, Maoz B, Das D, Somayaji MR, Prantil-Baun R, Novak R, Cronce M, Huffstater T, Jeanty SSF, Ingram M, et al. Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nat Biomed Eng 2020, 4:421–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ronaldson-Bouchard K, Teles D, Yeager K, Tavakol DN, Zhao YM, Chramiec A, Tagore S, Summers M, Stylianos S, Tamargo M, et al. A multi-organ chip with matured tissue niches linked by vascular flow. Nat Biomed Eng 2022, 6:351–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schindler M, Siriwardena D, Kohler TN, Ellermann AL, Slatery E, Munger C, Hollfelder F, Boroviak TE. Agarose microgel culture delineates lumenogenesis in naive and primed human pluripotent stem cells. Stem Cell Reports 2021, 16:1347–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang SW, Zhao HR, Zhang WJ, Wang JQ, Liu YH, Cao YX, Zheng HH, Hu ZW, Wang SB, Zhu Y, Wang W, Cui SZ, Lobie PE, Huang LQ, Ma SH. An Automated Organoid Platform with Inter-organoid Homogeneity and Inter-patient Heterogeneity. Cell Rep Med 2020, 1:100161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang YQ, Liu HT, Zhang M, Wang H, Chen WW, Qin JH. One-step synthesis of composite hydrogel capsules to support liver organoid generation from hiPSCs. Biomater Sci-Uk 2020, 8:5476–5488. [DOI] [PubMed] [Google Scholar]
- 53.Zhang WJ, Li DH, Jiang SW, Galan EA, Zhang ZY, Huang LQ, Ma SH. Microfluidic droplets as structural templates for Matrigel to enable 1-week large organoid modeling. Chem Eng Sci 2021, 238:116632. [Google Scholar]
- 54.Sart S, Tomasi RFX, Barizien A, Amselem G, Cumano A, Baroud CN. Mapping the structure and biological functions within mesenchymal bodies using microfluidics. Sci Adv 2020, 6:eaaw7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fang GC, Lu HX, de la Fuente LR, Law AMK, Lin GG, Jin DY, Gallego-Ortega D. Mammary Tumor Organoid Culture in Non-Adhesive Alginate for Luminal Mechanics and High-Throughput Drug Screening. Adv Sci 2021, 8:2102418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schuster B, Junkin M, Kashaf SS, Romero-Calvo I, Kirby K, Matthews J, Weber CR, Rzhetsky A, White KP, Tay S. Automated microfluidic platform for dynamic and combinatorial drug screening of tumor organoids. Nat Commun 2020, 11:5271. Schuster et al. developed an automated microfluidic system to perform individual, combinatorial, and sequential drug screens on pancreatic tumor organoids.
- 57.Khan I, Prabhakar A, Delepine C, Tsang H, Pham V, Sur M. A low-cost 3D printed microfluidic bioreactor and imaging chamber for live-organoid imaging. Biomicrofluidics 2021, 15:024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Shaegh SAM, Massa S, Riahi R, Chae S, Hu N, et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. P Natl Acad Sci USA 2017, 114:E2293–E2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berlanda SF, Breitfeld M, Dietsche CL, Dittrich PS. Recent Advances in Microfluidic Technology for Bioanalysis and Diagnostics. Analytical Chemistry 2021, 93:311–331. [DOI] [PubMed] [Google Scholar]
- 60.Jovic D, Liang X, Zeng H, Lin L, Xu FP, Luo YL. Single-cell RNA sequencing technologies and applications: A brief overview. Clin Transl Med 2022, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mi F, Hu CM, Wang Y, Wang L, Peng F, Geng PF, Guan M. Recent advancements in microfluidic chip biosensor detection of foodborne pathogenic bacteria: a review. Anal Bioanal Chem 2022, 414:2883–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No original data is used for this review article.