Abstract
We have previously shown that many cell cycle regulatory gene products are markedly affected by infection of primary fibroblasts with human cytomegalovirus (HCMV) (F. M. Jault, J. M. Jault, F. Ruchti, E. A. Fortunato, C. Clark, J. Corbeil, D. D. Richman, and D. H. Spector, J. Virol. 69:6697–6704, 1995). One of these proteins, cyclin E, is a key determinant of cell cycle progression during G1, and its mRNA levels are significantly increased in HCMV-infected fibroblasts (B. S. Salvant, E. A. Fortunato, and D. H. Spector, J. Virol. 72:3729–3741, 1998). To determine the molecular basis of this effect, we have examined the events that occur at the endogenous cyclin E promoter during the course of infection. In vivo dimethyl sulfate footprinting of the cyclin E promoter revealed several regions of protection and hypersensitivity that were unique to infected cells. In accord with this observation, we find that the virus-induced cyclin E transcripts initiate downstream of the start site identified in mock-infected cells, in regions where these newly appearing protected and hypersensitive sites occur. Viral gene expression is required for this induction. However, the viral immediate-early proteins IE1-72 and IE2-86, either alone or in combination, cannot induce expression of the endogenous cyclin E. The virus must progress past the immediate-early phase and express an early gene product(s) for activation of cyclin E expression. Moreover, IE1-72 does not appear to be required, as infection of cells with an HCMV mutant containing a deletion in the IE1-72 gene leads to full upregulation of cyclin E expression. Using electrophoretic mobility shift assays with infected cell extracts and a region of the cyclin E promoter that includes two previously defined E2F sites as the probe, we detected the appearance of an infection-specific banding pattern. One of the infection-specific bands contained the proteins E2F-4, DP-1, and p130, which were maintained in the infected cells as uniquely phosphorylated species. These results suggest that an altered E2F-4–DP-1–p130 complex along with viral early gene expression may play a role in the transcriptional regulation of cyclin E mRNA during HCMV infection.
Human cytomegalovirus (HCMV), a member of the betaherpesvirus family, exerts many different effects on the host cell during infection (15). The virus optimizes the conditions of the cell for its replication by activating the expression of genes that will assist in its DNA synthesis. At the same time, the virus inhibits host cell DNA synthesis by an undefined mechanism that coincides with viral gene expression (5, 11, 25, 35). Since the virus is not competing with the host cell for resources, it is able to maximize its replication. Our interest lies in how HCMV alters the cell cycle at the transcriptional level in order to achieve this effect.
The cell cycle is a dynamic process requiring that all components adhere to strict temporal and spatial requirements. The cyclins are key regulatory factors that are controlled by a variety of mechanisms including phosphorylation, association with their respective cyclin-dependent kinases (cdk's), gene expression, protein stability, subcellular localization, and degradation (2, 14, 21, 51, 54, 55). One of these cyclins, cyclin E, is expressed in the G1 phase of the cell cycle and associates with cyclin-dependent kinase 2 (cdk2). This association along with phosphorylation of the kinase by cyclin-activating kinase activates cdk2. The activated kinase complex is then responsible for several phosphorylation events that are required for progression into the S phase of the cell cycle.
The E2F family of transcription factors also plays a central role in cell cycle control. These transcription factors regulate expression of genes involved in cell cycle progression (cyclin E, cyclin A, cdc2, and cdc25), transcription (E2F-1, b-myb, p107, and c-myc), and DNA synthesis (DNA polymerase α, cdc6, dihydrofolate reductase [DHFR], thymidine kinase, HsOrc1, and H2A) (10, 18, 20, 22–24, 26, 29, 34, 43–45, 47, 48, 52, 56, 58, 63). In the classic model of E2F site regulation, an E2F family member, E2F-1 to -6, forms a heterodimer with a DP family member, DP-1 to -3, and this complex binds to an E2F site on the DNA (consensus TTTCGCGC). The presence of a pocket protein (Rb, p130, or p107) is thought to be repressive, and the sequential phosphorylation of the pocket protein by cyclin D-cdk4/6 and cyclin E-cdk2 results in its release from the complex and activation of transcription by the remaining heterodimeric complex. Subsequent phosphorylation of the heterodimeric complex by cyclin A-cdk2 is believed to lead to the release of the complex from the DNA.
Rb, p107, and even p130, which is most often associated with a repressive G0 complex, have all been detected in E2F-containing complexes in S-phase cells, suggesting that the presence of a pocket protein is not always an indicator of inhibition (2, 33, 40). In addition to their role in transcription, the pocket proteins p107 and p130 can bind to cyclin E or A-cdk2 complexes and inhibit their kinase activity (7, 62). The role of p107 and p130 in the cell cycle is further complicated by the fact that they can simultaneously interact with E2F transcription factors and cyclin-cdk complexes. It is not clear what role the E2F-cyclin-cdk-pocket protein complexes may play in a cycling cell. These types of complexes are commonly detected in electrophoretic mobility shift assays (EMSAs) using extracts from S-phase cells, suggesting that these complexes are not inhibitors of transcription (33).
Our lab and others have shown that HCMV infection affects many factors involved in the regulation of the cell cycle, leading to arrest of the cell prior to mitosis (3–6, 11, 16, 25, 35, 42, 50). These effects include upregulation of cyclin E and its associated kinase activity, downregulation of cyclin A and its associated kinase activity, sustained levels of cyclin B, hyperphosphorylation of Rb, stabilization of p53, altered subcellular localization of cdk2, and sequestration of p53 and replication protein A into viral replication centers.
Recently, we demonstrated that cyclin E upregulation in HCMV-infected cells occurs at the transcriptional level (50). In uninfected cells, E2F factors appear to regulate the transcription of cyclin E in a cell cycle-dependent manner through specific binding sites in the promoter, which are located approximately 100, 600 (two sites), 1,000 (two sites), and 1,100 bp from the translation start site (18, 45). However, it is still not certain which of these sites are most important for regulation of cyclin E transcription, as two independent groups have arrived at different conclusions based on the results of mutagenesis experiments and transient-transfection studies (18, 45). In addition, these two groups detected transcription start sites different from those of one another. Different transformed cell lines were used by the two groups, and this may explain the discrepancies. In a separate study, Bresnahan et al. (3) used cyclin E promoter constructs and transient-transfection assays to examine the regulation of this promoter by HCMV. The results of their transfection studies suggested that the viral immediate-early (IE) protein IE2-86 was the major transactivator of the cyclin E promoter and that the E2F sites were not required.
The goal of the work presented here was to determine the molecular basis of the upregulation of cyclin E transcription by HCMV when the endogenous promoter is in its normal chromatin context. We show that the primary HCMV-induced cyclin E transcripts initiate further downstream than the cyclin E transcripts in mock-infected cells. In accord with these observations, we demonstrate, by in vivo footprinting of the cyclin E promoter, protection as well as hypersensitivity of downstream sites in infected cells. Some of these sites overlap the region that contains the two E2F sites that were shown by Ohtani et al. (45) to be important for cell cycle-mediated regulation of cyclin E transcription. At the same time, we observed that the steady-state levels and patterns of phosphorylation of E2F and Rb family members are specifically altered by infection. We further find that virus-induced complexes, one of which consists of E2F-4, DP-1, and p130, bind to this region of the promoter with kinetics that parallel the induction of the cyclin E transcript. We also show that the induction of cyclin E transcription requires viral gene expression. However, in contrast to what was suggested by the results of Bresnahan et al. (3), we find that IE2-86 alone cannot induce the endogenous cyclin E promoter. Another major viral IE protein, IE1-72, also cannot activate the endogenous cyclin E promoter when expressed alone or in combination with IE2-86. Rather, the results of kinetic experiments suggest that a viral gene product(s) in the early class, expressed after the IE phase, is necessary for the induction of cyclin E expression. Moreover, based on the results of infection with an HCMV mutant virus containing a deletion of the IE1-72 gene, it appears that this gene product is not required for the virus-mediated upregulation of cyclin E.
MATERIALS AND METHODS
Cell culture and virus.
Human foreskin fibroblasts were obtained from the University of California, San Diego, Medical Center and cultured in minimum essential medium with Earle's salts (MEM) (Gibco-BRL) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Summit Biotechnology), 50 μg of gentamicin sulfate (Omega Scientific) per ml, 1.5 μg of amphotericin B (Omega Scientific) per ml, 2 mM l-glutamine (Bio-Whittaker), 200 U of penicillin (Omega Scientific) per ml, and 200 μg of streptomycin (Omega Scientific) per ml. Cells were kept in incubators maintained at 37°C and 7% CO2. The Towne strain of HCMV was obtained from the American Type Culture Collection (VR 977) and propagated as previously described (57). The RC303ΔAcc strain of HCMV contains a deletion in exon 4 of UL122-123 and was a generous gift from Edward Mocarski (41).
Synchronization and infections.
Cells were synchronized in G0 phase by allowing them to grow to confluence as previously described (16). Three days after confluence, the cells were trypsinized, replated at a lower density to allow progression into the cell cycle, and infected at a multiplicity of infection of 5 or mock infected with tissue culture supernatants. At appropriate times postinfection (p.i.), cells were washed with phosphate-buffered saline (PBS), scraped, and processed as described for each type of experiment. Alternatively, cells were synchronized by serum starvation for 48 h in medium with no serum and kept in medium with no serum during the course of the infection. The method of synchronization used for each experiment is given in the text.
Construction of recombinant baculoviruses that express the viral IE proteins IE1-72 and IE2-86 is described elsewhere (R. S. Dwarakanath et al., unpublished data). Human fibroblasts were infected with recombinant baculoviruses at a multiplicity of infection of approximately 200. For the experiments depicted in Fig. 8 and 9A, the cells were also infected with a recombinant baculovirus that contained the UL112-113 gene driving chloramphenicol acetyltransferase expression to allow the assessment of activation by viral IE proteins. Inoculum was diluted in MEM plus 10% FBS and left on the cells for 1 h at room temperature during rocking. The inoculum was then removed, and cells were washed in PBS and refed with MEM plus 10% FBS until the time of harvest. At appropriate times p.i., cells were trypsinized, pelleted, and processed as described for each type of experiment. In instances where cells were infected with both recombinant baculoviruses and UV-inactivated virus, cells were first infected with recombinant baculoviruses and then infected with tissue culture supernatants (mock) or with UV-inactivated virus. UV inactivation of virus was performed as previously described (28).
FIG. 8.
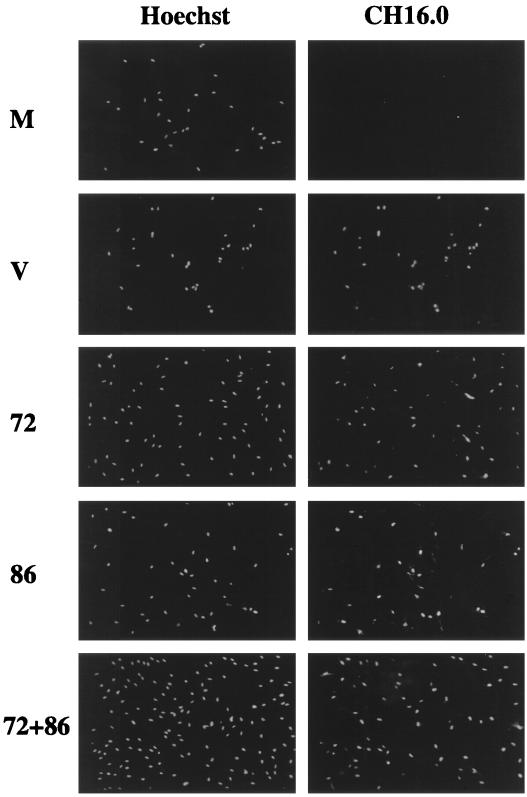
Recombinant baculoviruses express IE1-72 and IE2-86 in human fibroblasts. Cells were confluence synchronized and mock infected (M), infected with HCMV (V), or infected with recombinant baculovirus(es) (72, 86, or 72+86). Coverslips were taken at 24 h p.i., fixed and permeabilized in methanol, and stained with Hoechst stain to identify the total number of cells and CH16.0 to detect cells that expressed IE1-72 or IE2-86.
FIG. 9.
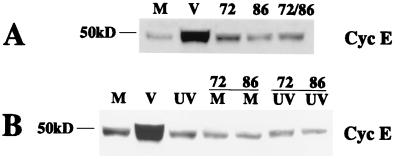
Expression of HCMV IE1-72 and that of IE2-86 are not sufficient to induce endogenous cyclin E. (A) Lysates were prepared at 24 h p.i. from the same experiment that was described in the legend to Fig. 8, and 50 μg of total cell lysate was run on a 10% polyacrylamide gel, transferred to nitrocellulose, and Western blotted for cyclin E. M, mock infected; V, virus infected. (B) Cells were serum starved and were mock infected (M), infected with HCMV (V), infected with UV-irradiated virus (UV), mock infected and infected with recombinant baculovirus (M and 72 or 86), or infected with both UV-irradiated virus and recombinant baculovirus (UV and 72 or 86). Cell lysates were prepared at 24 h p.i., and 50 μg of total cell lysate was run on a 10% polyacrylamide gel, transferred to nitrocellulose, and Western blotted for cyclin E.
Western blot analysis.
Cells were lysed in Laemmli reducing sample buffer (2% sodium dodecyl sulfate [SDS], 10% glycerol, 100 mM dithiothreitol, 60 mM Tris [pH 6.8], 2 μg of aprotinin and leupeptin per ml, 1 mM phenylmethylsulfonyl fluoride [PMSF], 50 mM NaF, 0.5 mM Na3VO4, 4 mM EDTA, 10 mM Na4P2O7, 1 mM benzamidine, and 1 mM NaS2O5). The lysates were then sonicated, boiled for 5 min, and centrifuged for 10 min at 16,000 × g. Samples (100 μg/lane) were run on 10% polyacrylamide gels unless otherwise stated. Proteins were transferred to Immobilon P (Millipore) or Protran (Schleicher & Schuell), and Western blot analysis was performed using the appropriate mouse or rabbit antibody followed by the appropriate horseradish peroxidase-linked secondary antibody (Amersham Life Science Products). Proteins were visualized using Pierce Supersignal or Blaze chemiluminescent detection methods (per manufacturer's instructions).
Cycloheximide block and release experiments.
Confluence-synchronized cells were replated at a lower density in MEM plus 10% FBS and allowed to settle for 1 h prior to the addition of 100 μg of cycloheximide (Sigma) per ml. At 1 h after the addition of cycloheximide, cells were mock or HCMV infected in the presence of 100 μg of cycloheximide per ml. At 3 h p.i., cells were washed three times in MEM plus 10% FBS to release them from the cycloheximide block. At appropriate times post-release from cycloheximide, 20 μg of actinomycin D (Sigma) per ml was added to the culture medium or cells were maintained in 100 μg of cycloheximide per ml for the duration of the experiment. Cells were harvested at 18 h p.i. and processed as described for Western blot analysis.
Immunofluorescence.
Glass coverslips were placed in tissue culture dishes at the time of plating prior to mock, HCMV, or baculovirus infection. At 24 h p.i., coverslips were removed from tissue culture dishes and washed twice in PBS, and cells were simultaneously fixed and permeabilized by placing them in 100% methanol at −20°C for 10 min. Coverslips were washed three times in PBS at this point and before and after all incubations. Coverslips were incubated for 20 min in blocking solution (10% normal goat serum [Jackson Laboratories] in PBS–0.5% bovine serum albumin [BSA]) and then incubated for 10 min in antibody against the IE1-72 and IE2-86 proteins (CH16.0; Goodwin Institute) diluted 1:3,000 in PBS–0.5% BSA. This was followed by a 10-min incubation in fluorescein isothiocyanate-conjugated goat anti-mouse secondary antibody (Jackson Laboratories) diluted 1:500 in PBS–0.5% BSA. Coverslips were then incubated for 10 min with 2 mg of Hoechst (Calbiochem) per ml diluted 1:500 in PBS–0.5% BSA and mounted onto slides using glycerol-paraphenylenediamine (an antiphotobleaching agent) (Sigma). Images were visualized using a Leitz DMRB fluorescent microscope and were photographed using a Leica DMLD slide camera. Images were arranged and labeled using Adobe Photoshop 3.0 and Adobe Illustrator 6.0.2.
Immunoprecipitation.
p130 antibodies (Santa Cruz) or rabbit preimmune serum (collected from a naive New Zealand White female rabbit) was coupled to protein A-Sepharose beads (Pharmacia Biotech) as previously described (30). Cells were lysed in NETN (20 mM Tris [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, 2 μg of aprotinin and leupeptin per ml, 1 mM PMSF, 50 mM NaF, 0.5 mM Na3VO4, 4 mM EDTA, 10 mM Na4P2O7, 1 mM benzamidine, and 1 mM NaS2O5) and subjected to three cycles of freezing and thawing (5 min at −80°C and then 5 min at 37°C). Lysates were centrifuged to pellet debris, and 5 μg of coupled antibody was added to the supernatant and incubated overnight at 4°C during rocking. Coupled antibody was pelleted by centrifugation and washed three times in NETN. The pellet was resuspended in Laemmli reducing sample buffer, boiled, and loaded on a 10% polyacrylamide gel. Gels were transferred and Western blotted as described above, with the exception that protein A- or protein G-horseradish peroxidase was used as the secondary antibody.
Nuclear-cytoplasmic fractionation.
For EMSA, nuclear and cytoplasmic fractionation was performed as follows. Cells were pelleted and resuspended in 1 ml of buffer A (20 mM HEPES [pH 7.9], 10 mM KCl, 1 mM MgCl2, 10% glycerol, 0.1% NP-40, 2 μg of aprotinin and leupeptin per ml, 1 mM PMSF, 50 mM NaF, 0.5 mM Na3VO4, 4 mM EDTA, 10 mM Na4P2O7, 1 mM benzamidine, and 1 mM NaS2O5). This cell lysate was immediately layered onto 1 ml of 37% sucrose and centrifuged for 10 min at 1,320 × g. The top layer (cytoplasmic fraction) was removed and centrifuged for an additional 10 min at 16,000 × g to clear it of membranes. The pellet below the sucrose was lysed in buffer A–300 mM NaCl and subjected to three cycles of freezing and thawing (5 min at −80°C and then 5 min at 37°C). This nuclear fraction was vortexed for 1 min and centrifuged for an additional 10 min at 16,000 × g to clear it of membranes. Protein concentrations were determined using the Bio-Rad protein assay, and lysates were stored at −80°C.
For calf intestinal alkaline phosphatase (CIAP) treatment, 100 μg of nuclear lysates was incubated with 20 U of CIAP (Gibco-BRL) for 30 min at 37°C. Laemmli reducing sample buffer (3×) was added to make the final concentration 1×, and the samples were boiled and loaded on polyacrylamide gels as described above.
EMSA.
Reaction mixtures contained the following: 1× buffer (9), 10% glycerol, 200 ng of sonicated salmon sperm DNA, 5 μg of nuclear extract, 20 fmol of appropriate radiolabeled probe, and in some cases a 30 to 300 molar excess of cold competitor probe or 1 to 2 μg of specific antiserum. Reaction mixtures were incubated for 20 min at room temperature and loaded on a 4% polyacrylamide gel with 5% glycerol in 0.5× Tris-borate-EDTA. Gels were run in 0.5× Tris-borate-EDTA for 2 h at 35 mA per gel, dried down for 1 h at 80°C, and exposed to film.
In vivo DMS footprinting and LMPCR.
For in vivo dimethyl sulfate (DMS) treatment, cells were pelleted and resuspended in 1 ml of medium. Cells were warmed to 37°C, 10 μl of 10% DMS was added, and cells were incubated for 1 min at 37°C. Cells were immediately transferred to 50 ml of ice-cold PBS and pelleted. Cell pellets were washed in PBS and then resuspended in 2 ml of PBS. Genomic DNA was isolated according to the Qiagen Blood and Cell Culture DNA Midi Kit protocol. Isolated DNA was precipitated, resuspended in 10% piperidine, heated to 90°C for 30 min, and then dried in a SpeedVac concentrator overnight. Treated DNA was used in a ligation-mediated PCR (LMPCR) as previously described (1). Primers for cyclin E promoter footprinting were as follows: PE1, 5′ GCG CTC CAG TCC CGG CAG GCG GCG G 3′; PE2, 5′ CGG CGA CGG CAG TGG CGG CGG CGG C 3′; PE3, 5′ CGG CGG CGG CGC CGG GAG TCG GCG G 3′; LMPCR.1, 5′ GCG GCG GCC CCG GCG CTT CGC AGG C3′; and LMPCR.2, 5′ GCC TGC GAA GC 3′. PCR was performed as follows. First, the samples were subjected to 5 min of denaturation at 95°C, 30 min of annealing at 72°C, and 12 min of extension at 76°C. During first-strand synthesis, the reaction mixtures contained 2 μg of the appropriate DNA, 0.2 mM deoxynucleoside triphosphates (dNTPs), 1 U of GC-rich enzyme mix (Roche Biochemicals), 0.5 M GC resolution solution (Roche Biochemicals), 1× reaction buffer (Roche Biochemicals), and 3 pmol of PE1. LMPCR.1 and LMPCR.2 were annealed to make the unidirectional linker that was then ligated to the first-strand synthesis products as previously described (1). For the amplification step, the samples contained 1 M G-C resolution solution (Roche Biochemicals), 1× reaction buffer (Roche Biochemicals), 0.2 mM dNTPs, 2 U of GC-rich enzyme mix (Roche Biochemicals), 10 pmol of PE2, and 10 pmol of LMPCR.1. The samples were denatured for 5 min at 95°C and subjected to 35 cycles of 30 s of denaturation at 95°C, 30 s of annealing at 75°C, and 30 s of extension at 78°C. Finally, the reactions were subjected to a 10-min extension at 72°C. For the end-labeling step, the samples contained 1 M GC resolution solution (Roche Biochemicals), 1× reaction buffer (Roche Biochemicals), 0.2 mM dNTPs, 0.5 U of AmpliTaq DNA polymerase (Perkin-Elmer), and 2 pmol of radiolabeled PE3. The reactions were then denatured at 95°C for 5 min and subjected to nine cycles of 30 s of denaturation at 95°C, 30 s of annealing at 76°C, and 1 min of extension at 79°C. A final 7-min extension was performed at 79°C. For in vitro samples and Maxam-Gilbert reactions, genomic DNA was isolated according to the Qiagen protocol and then treated with the appropriate chemical as previously described (1). The treated samples were subjected to the same LMPCRs as were the in vivo-treated samples. After LMPCR, samples were extracted once with phenol-chloroform-isoamyl alcohol (1) and then precipitated, resuspended in 3 μl of loading buffer, and run on a 6% denaturing polyacrylamide gel.
Antisera.
Antibodies used in Western analysis were E2F-1 (sc-251), E2F-2 (sc-633), E2F-3 (sc-878), E2F-4 (sc-1082), E2F-5 (sc-1083), DP-1 (sc-610), DP-2 (sc-830), p130 (sc-317), p107 (sc-318), cyclin E (sc-198), and CREB binding protein (CBP) (sc-583), all obtained from Santa Cruz Biotechnologies. The Sp-1 antibody was a generous gift from James T. Kadanoga (University of California, San Diego). The G6PD antibody was a generous gift from Rod Nakayama (University of California, Irvine). The UL44 antibody was a generous gift from Lenore Periera (University of California, San Francisco). CH16.0 was obtained from the Goodwin Institute. Antibodies used for supershift analysis were the same as those used for Western analysis with the addition of Rb (14031A [Pharmingen]), E2F-4 (sc-866 [Santa Cruz]), cyclin E (14761C [Pharmingen]), cyclin A (14531C [Pharmingen]), and Sp-1 (sc-420 [Santa Cruz]).
Primer extension analysis.
mRNA was isolated from cells using the Invitrogen FastTrack 2.0 mRNA isolation kit. Primer extension reactions were carried out as described previously (49) using 5 μg of mRNA per reaction and the cyclin E-specific primer E3 (5′ CGG CAG GCG GCG GCG GCG ACG GCA GTG GCG GCG GC 3′). Reaction mixtures were precipitated, resuspended in loading buffer, and run on a 6% denaturing polyacrylamide gel. Sanger sequencing reactions were also performed with E3 using T7 Sequenase version 2.0 (U.S. Biochemical).
RESULTS
HCMV uses a new start site to upregulate cyclin E mRNA.
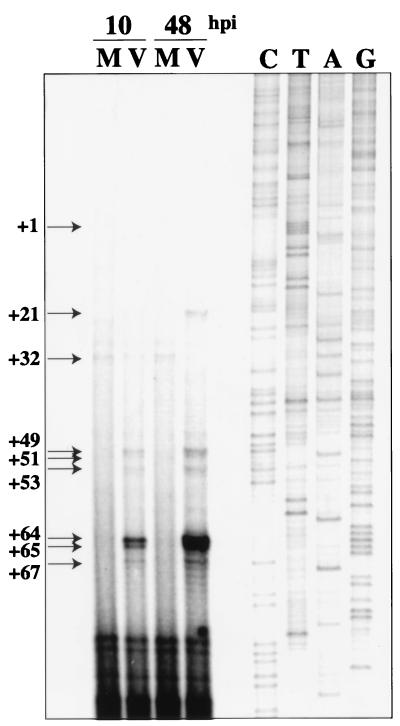
Previously, our lab showed that the effect of HCMV infection on cyclin E expression was at the level of transcription (50). We found that, when G0 synchronized cells were infected with HCMV as they were released from confluence into the G1 phase of the cell cycle, there was a significant induction of cyclin E mRNA by 12 h p.i. that was sustained through 96 h p.i. To map the start sites of transcription from the cyclin E promoter in HCMV-infected cells, synchronized foreskin fibroblasts were again infected upon release into G1. The mRNA was then isolated at 10 and 48 h p.i. and subjected to primer extension analysis. Figure 1 shows that new start sites were present in the infected cells at +21, +49, +51, +53, +64, +65, and +67 relative to the sequence published by Ohtani et al. (45). Mock-infected cells showed only one start site, at +32. Interestingly, most infected cell cyclin E mRNA start sites occurred downstream of the previously identified start site (+1 in Fig. 1) and of the start site that we detected in mock-infected cells (+32 in Fig. 1). As is noted in Fig. 1, we did not detect use of the previously identified start site at +1. Since there are limits to the primer extension assay, we also performed analysis using a primer that was closer to the previously identified start site and again did not detect use of this site in foreskin fibroblasts (data not shown). These results were confirmed by RNase protection analysis (data not shown).
FIG. 1.
Primer extension analysis of cyclin E mRNA at 10 and 48 h p.i. mRNA was isolated using Invitrogen FastTrack 2.0, and primer extension was performed as previously described (49). Arrows indicate locations of transcription start sites relative to the published sequence (45). The site at +1 is marked to indicate the start site that was previously identified by Ohtani et al. (45). We do not detect use of this site in fibroblasts. Sanger sequencing reactions using the same primer as was used in the primer extension were performed in parallel. M, mock-infected cells; V, virus-infected cells.
In vivo footprinting of the cyclin E promoter.
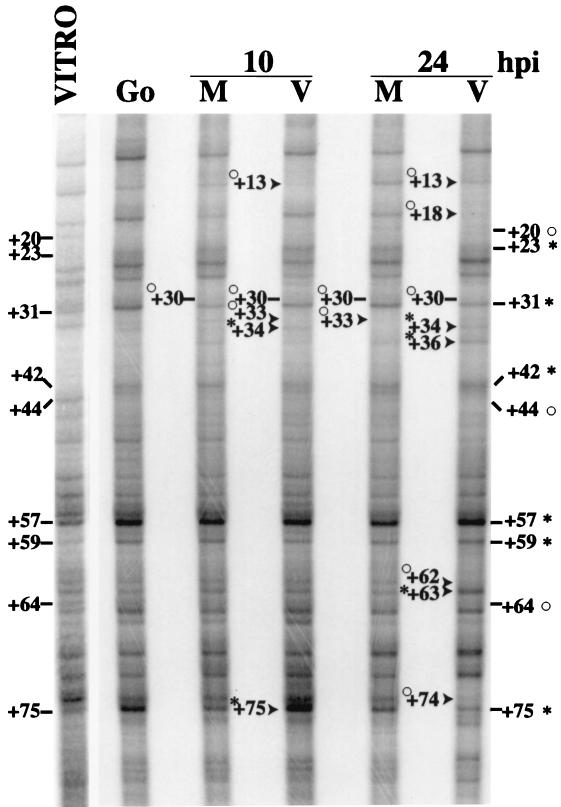
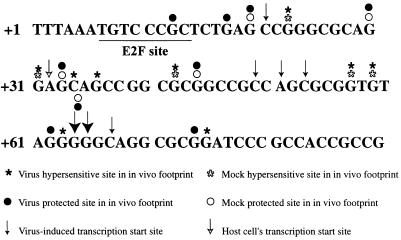
Based on the results from the primer extension analysis, we proceeded to examine the downstream region of the cyclin E promoter at 10 and 24 h p.i. To determine if there was a change in the in vivo site occupancy of the cyclin E promoter during the infection, we used the technique of in vivo footprinting. Cells were confluence synchronized and released into G1 at the time of infection. At 10 and 24 h p.i., cells were harvested, genomic DNA was isolated, and in vivo footprinting was performed as described in Materials and Methods. A large number of changes were observed, and the most notable ones are discussed below. By comparison with the in vitro-treated samples, all in vivo-treated samples showed protection at +20, +44, and +64 and hypersensitivity at +23, +31, +42, +57, and +59 (Fig. 2, part 1). Some level of hypersensitivity was detected in all in vivo samples at +75. However, this hypersensitivity was stronger in G0 and infected cells at 10 h p.i. than in the other in vivo samples. We also found that there was protection at +33 in mock-infected cells, most notably at 24 h p.i., and that this residue was not altered in confluent cells in G0. Changes in this region are consistent with use of the +32 start site that we detected in mock-infected cells.
FIG. 2.
(Part 1) In vivo DMS footprinting analysis of the cyclin E promoter at 10 and 24 h p.i. Cells were treated with 0.1% DMS as described in Materials and Methods. After LMPCR, samples were run out on a 6% denaturing polyacrylamide gel. Hypersensitive sites are indicated by asterisks, and protected sites are identified by open circles. Lines denote changes that were seen in all in vivo samples compared to the in vitro sample. Arrows mark differences between mock (M)- and HCMV (V)-infected samples. In all cases, the corresponding location within the sequence is noted. The lane labeled G0 represents in vivo DMS treatment of confluent uninfected cells. The lane labeled VITRO represents in vitro treatment of DNA from uninfected cells. (Part 2) A summary of the events that occur on the cyclin E promoter. Solid arrows represent virus-induced transcription start sites. The open arrow designates the cellular start site for transcription. Solid asterisks denote hypersensitive sites that were detected in the in vivo footprint of infected cells. Solid circles mark protected sites from the in vivo footprint of infected cells. Open asterisks represent hypersensitivity that was seen in mock-infected cells, and open circles represent protection that was seen in mock-infected cells. The E2F site that is underlined in this panel is a component of the probe that was used in EMSAs.
In infected cells, all of the observed sites of protection and hypersensitivity occurred downstream of the start site that was previously identified (+1 in Fig. 1) (45). At 10 h p.i., protection was seen in infected cells at +13, which is located within a previously defined E2F site (45). This was also apparent at 24 h p.i., relative to the mock-infected sample. Also at 10 h p.i., the hypersensitivity of the +75 site was greatly enhanced in infected cells with respect to both the in vitro sample and the 10-h mock-infected sample. At 10 h p.i., infected cells showed protection at +33 but also began to show the +34 hypersensitivity that was seen in infected cells at 24 h p.i. Three additional sites of protection were seen in infected cells at 24 h p.i. One of these sites occurred at +18, just downstream of the E2F binding site that was also protected at 10 h p.i. The other two sites of protection, at +62 and +74, were further downstream. The protection at +62 was followed by a hypersensitive site at +63 at 24 h p.i. Also at 24 h p.i., there was hypersensitivity at +34 and +36 in infected cells compared to all other in vivo samples as well as the in vitro sample. The downstream changes in protection and hypersensitivity in infected cells occurred in the same regions as the HCMV-induced transcription start sites. The minus strand of cyclin E was also footprinted, and no significant changes were detected, as is often the case in these types of experiments (data not shown).
Figure 2, part 2, summarizes the alterations detected on the cyclin E promoter. In the region of +30 to +36, we saw several different effects. In mock-infected cells, there was protection at +33, and this alteration was correlated with the use of the +32 transcription start site. In infected cells, we saw some +33 protection and +34 hypersensitivity at 10 h p.i., and at this same time point, the use of the +32 start site can be detected in addition to the several downstream start sites. However, by 24 h p.i. in infected cells, there is hypersensitivity at +34 and +36 relative to the in vitro sample, and use of the +32 start site can no longer be detected at 48 h p.i. The changes in the infected cells at +62 and +63 are also consistent with the large increase in cyclin E transcripts that initiate at +64 and +65. Lastly, protection at +13 and +18 in infected cells occurs within a region that contains a previously defined E2F binding site. It should be noted that we examined upstream regions of the cyclin E promoter by in vivo footprinting but did not detect any significant alterations in infected cells (data not shown).
Steady-state levels of the factors that regulate cyclin E transcription.
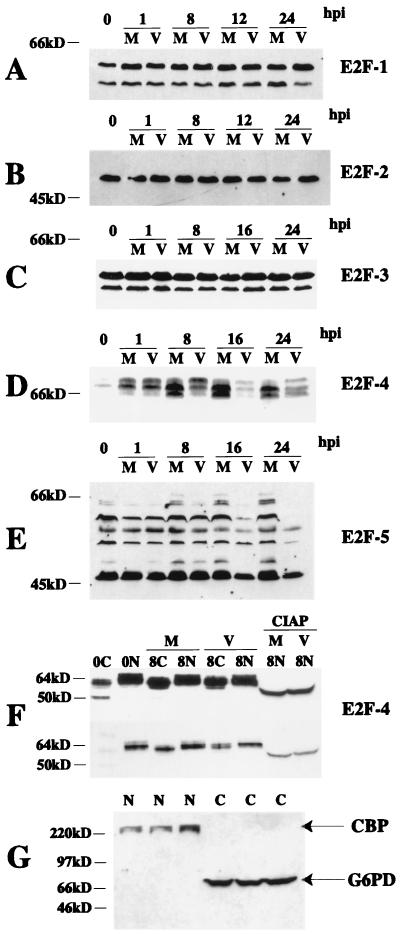
Since we were able to detect in vivo site occupancy of a previously defined E2F site in infected cells, we examined the steady-state levels of the transcription factors that are thought to be responsible for regulation of cyclin E transcription (18, 45). Western blot analysis of members of the E2F family showed no changes in E2F-1, -2, or -3 during a 24-h time course (Fig. 3A to C). As has been observed by others, we detected several differently migrating forms of E2F-5 (59). Figure 3E shows that several of them were preferentially lost in infected cells at 16 h p.i. In contrast, the levels of the different forms of E2F-5 did not vary significantly in mock-infected cells throughout the time course.
FIG. 3.
Western blot analysis of the E2F family of proteins at various times p.i. One hundred micrograms of total cell lysate was run on a 20-cm 10% polyacrylamide gel (0.07% bisacrylamide final concentration) (31). Proteins were transferred to nitrocellulose and analyzed by Western blotting. (A) E2F-1. (B) E2F-2. (C) E2F-3. (D) E2F-4. (E) E2F-5. (F) Nuclear and cytoplasmic lysates were prepared at 0 and 8 h p.i., and 100 μg of nuclear (N), cytoplasmic (C), or nuclear lysates treated with CIAP were run on 10% polyacrylamide gels. Proteins were transferred to nitrocellulose and analyzed by Western blotting for E2F-4. Two different exposures of the same blot are shown. (G) One hundred micrograms of nuclear (N) or cytoplasmic (C) lysates from three independent experiments was run on 10% polyacrylamide gels. Proteins were transferred to nitrocellulose and analyzed by Western blotting for G6PD and CBP. M, mock infected; V, virus infected.
Several species of E2F-4 were detected in both mock- and HCMV-infected cells (Fig. 3D). Nuclear and cytoplasmic fractionation was performed on mock-infected and infected cells to examine the forms of E2F-4 that were present in each fraction. Figure 3G shows a representative example of three independent nuclear-cytoplasmic fractionations that were subjected to Western blot analysis using anti-CBP (nuclear) and anti-G6PD (cytoplasmic) specific antibodies to verify the purity of the fractions. Treatment of nuclear lysates with CIAP revealed that all of these forms represent various phosphorylation states (Fig. 3F). Two exposures of the blot in Fig. 3F are shown so that the mobility of the unphosphorylated form that is detected in the 0-h cytoplasmic fraction, as well as the differences in mobility of the forms of E2F-4 present in mock-infected versus virally infected cells, can be seen more easily. Infected cells showed sustained levels of the two slowest-migrating forms of E2F-4 at 8 h p.i., while mock-infected cells exhibited an increase in the two fastest-migrating forms at this same time point (Fig. 3D). When E2F-4 was examined in nuclear and cytoplasmic fractions at this same time point (Fig. 3F), it was apparent that the most highly phosphorylated form of E2F-4 was located in the nuclear fraction. In infected cells, this highly phosphorylated form was maintained in the nuclear fraction and began to appear in the cytoplasmic fraction. In mock-infected cells, the nuclear E2F-4 was a less-phosphorylated form, and in the cytoplasmic fraction, the highly phosphorylated form was not visible (Fig. 3F). Mock-infected cells maintained high levels of the two fastest-migrating forms of the protein at the 16- and 24-h p.i. time points (Fig. 3D). In infected cells at 16 and 24 h p.i., approximately equivalent amounts of all three forms were seen, but the total concentration of E2F-4 decreased compared to that in mock-infected cells at these time points.
The heterodimeric partners of E2F, the DP proteins, were also examined by Western blotting. The DP-1 in infected cells at 16 and 24 h p.i. consisted of only the slowest-migrating form of the protein, while three differently migrating species of DP-1 were seen in the mock-infected cells throughout the time course (Fig. 4A). In the case of DP-2, the slower-migrating form decreased at 16 and 24 h p.i. in infected cells, while levels of the faster-migrating form of the protein were similar in mock-infected and infected cells during the entire 24-h period (Fig. 4B). For comparison, we show that Sp-1, a transcription factor which can potentially bind to specific sites in the cyclin E promoter, was unaffected by the HCMV infection (Fig. 4C).
FIG. 4.
Western blot analysis of other proteins that might be involved in transcriptional regulation of cyclin E. Lysates were prepared at various times p.i., and 100 μg of total cell lysate was used for each lane. Samples for DP-1 (A) and DP-2 (B) detection were run on a 20-cm 10% polyacrylamide gel (0.07% bisacrylamide final concentration) (31), transferred to nitrocellulose, and analyzed by Western blotting. (C) Samples for Sp-1 detection were run on a 10% minigel (29:1 bis/acrylamide ratio), transferred to Immobilon, and analyzed by Western blotting. Samples for p130 (D) and p107 (E) detection were run on a 7.5% polyacrylamide gel (0.07% bisacrylamide final concentration) (31), transferred to nitrocellulose, and analyzed by Western blotting.
Previously, we reported that, during the HCMV infection, Rb was induced and maintained in its hyperphosphorylated form (25). However, in that earlier study we did not examine the effects of the infection on the steady-state levels of the two Rb-related pocket proteins, p130 and p107. In Fig. 4D, we show that the amount and mobility of the various forms of p130 fluctuated throughout the time course in mock-infected cells, corresponding with phosphorylation and degradation of the protein (12, 17). The p130 in infected cells did not reach the third phosphorylation state and hence did not undergo degradation. The protein was migrating slightly slower than that in uninfected confluent cells, consistent with the form 2 of p130 that others have reported (39). Figure 4E shows that the levels of p107 increased in both mock-infected and infected cells at 16 and 24 h p.i. However, this increase was more striking in mock-infected cells than in infected cells at these same time points.
A virus-induced complex accumulates on the cyclin E promoter.
In order to identify which of the above factors might form a complex with the cyclin E promoter, we performed EMSAs with mock- and HCMV-infected nuclear lysates that were prepared at various times after infection of confluence-synchronized fibroblasts. A region of the cyclin E promoter from nucleotide −21 to +20 was radiolabeled and used as the probe in these experiments. This region of the cyclin E promoter contains the two E2F binding sites located approximately 600 bp upstream from the translation start site that were previously identified by Ohtani and coworkers (45) as being important for cell cycle-dependent regulation of cyclin E transcription. Cells were harvested and fractionated as described in Materials and Methods. Both nuclear and cytoplasmic fractions were subjected to Western blot analysis using anti-CBP (nuclear) and anti-G6PD (cytoplasmic) specific antibodies to verify the purity of the fractions prior to their use in EMSAs (Fig. 3G).
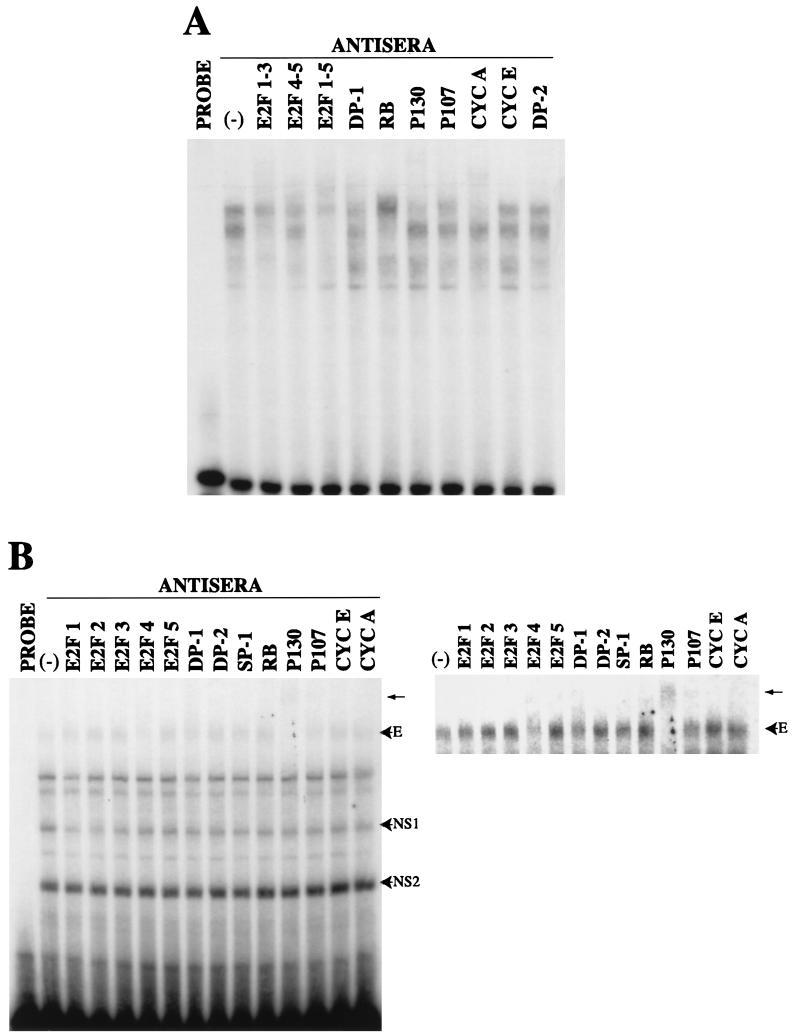
An EMSA performed with nuclear extracts from a time course experiment demonstrated that infected cells contained two complexes that were not observed in mock-infected cells (Fig. 5A). One of the infection-induced complexes was visible as early as 8 h p.i. (complex E), while both complexes (E and L) were visible at 48, 72, and 96 h p.i. While it may appear that complex E is present in mock-infected cells at 8 and 12 h p.i., a separate experiment in which the resolution of the gel was increased verified that these two complexes are migrating differently (data not shown). We believe the faint band shift that is seen in mock-infected cells at 8 and 12 h p.i. represents remnants of the G0 complex, since all cells are not progressing into the cell cycle at exactly the same rate. Several band shifts can be competed with any type of DNA and thus do not represent specific complexes with the cyclin E promoter; these are labeled as nonspecific 1 and 2 (NS1 and NS2).
FIG. 5.
EMSAs using sequences from the cyclin E promoter. (A) Nuclear extracts were prepared at various times p.i. Five micrograms of nuclear extract was combined with the radiolabeled probe CYCE E2F DS (5′ CT AGC GCC GGT TCC GCG CGC AGG GAT TTT AAA TGT CCC GCT CTG AG 3′) as described in Materials and Methods. The letter E designates the early virus- induced complex. The letter L designates the late virus-induced complex. The letter R designates the Rb-containing band shift. The letters NS designate nonspecific complexes. M, mock infected; V, virus infected. (B) Five micrograms of nuclear extract from infected cells at 18 h p.i. was combined with the same radiolabeled probe as in panel A. The indicated amount of competitor probe in molar excess was added to the reaction. The sequences of the competitor probes were as follows: FLAG, 5′ GAT CTA TGG ACT ACA AGG ACG ACG ACG ACA AGG G 3′; CYCE DS MUT, 5′ GCC GGT TCC GAT CGC AGG GAT TTT AAA TGT CAT GCT CTG AG 3′; DHFR E2F, 5′ GGG CGG GGC GGC CAC AAT TTC GCG CCA AAC TTG ACC GCG CG 3′; CYCA CDE/CHR, 5′ CCA TTT CAA TAG TCG CGG GAT ACT TGA ACT GCA AGA ACA GC 3′; and E2F CONS, 5′ GAT CTA TGG ATT TAA GTT TCG CGC CCT TTC TCA TAC TA 3′.
Competition studies using infected cell extracts from 18 h p.i. showed that complex E could be competed by the wild-type −21 to +20 probe but not by a probe that had the E2F sites mutated, indicating that complex E was E2F specific (Fig. 5B). In addition, several other known E2F sites were able to compete for complex E, supporting the notion that it is a bona fide E2F complex. In a separate experiment, using extracts from 96 h p.i., complex L was competed by the E2F mutant probe, indicating that it is not E2F specific (data not shown). Probes corresponding to regions downstream of +20 on the cyclin E promoter did not show any significant complex formation in EMSAs during the first 24 h p.i. (data not shown).
To determine which proteins were present in the above complexes, we used supershift analysis with specific antibodies. For reference and comparison, a radiolabeled probe containing the DHFR dyad E2F site was used in an EMSA with 24-h-p.i. mock-infected nuclear lysates. The banding patterns that were seen with this probe were supershifted by a combination of antibodies to E2F-1, -2, and -3 and E2F-4 and -5 as well as cyclin A, p130, Rb, and DP-1 antibodies (Fig. 6A). With antibodies to individual E2F species, we were able to show that the complexes contained E2F-2, E2F-3, and E2F-4 (data not shown). This experiment was performed to demonstrate that the complexes that are detected on the cyclin E promoter reflect the differences in the E2F sites on that promoter compared with the DHFR promoter rather than some aspect of how the lysates were prepared or the state of the cells. These results using the DHFR promoter as a probe document that the cells do contain these E2F complexes and that they can be supershifted by the antibodies used in the assays. Hence, our inability to detect certain forms of E2F binding to the cyclin E promoter is due to differences in the promoter.
FIG. 6.
Comparison of the E2F complexes that are present in mock- and HCMV-infected cells. (A) The DHFR probe was radiolabeled and combined with 5 μg of nuclear lysate obtained from mock-infected cells at 24 h p.i. The indicated antiserum was added to the reaction to determine if the band shifts could be supershifted or abrogated by the specific antiserum. (B) The CYCE E2F DS probe was radiolabeled and combined with 5 μg of nuclear lysate obtained from HCMV-infected cells at 18 h p.i. The indicated antiserum was added to the reaction to determine if complex E could be supershifted or abrogated by the specific antiserum. The supershift that was produced by the addition of p130 antibody is marked by an arrow. A portion of the gel was cropped and the contrast was adjusted in order to better visualize the effects of the antiserum.
Supershifting studies, using infected cell nuclear extracts from 18 h p.i. and the cyclin E probe, demonstrated that complex E contained p130 (Fig. 6B). The addition of antibodies against E2F-4 or DP-1 resulted in a loss or decrease in the intensity of complex E. A portion of this EMSA is shown with the contrast adjusted so that the supershift or loss in intensity can be more clearly visualized. These data indicate that complex E contains E2F-4, DP-1, and p130. Although Rb could be detected in a complex with the cyclin E probe when both mock-infected and infected cell extracts at later time points were used, this band shift (R) was not altered by HCMV infection (data not shown and Fig. 5A). The pocket protein p130 was also detected in a band shift in confluent uninfected cells, but this complex migrated faster than the one that we detected in infected cells (complex E). This suggests that complex E may contain other as-yet-unidentified factors or that the different phosphorylation states of E2F-4, DP-1, and p130 are affecting the mobility of the complex.
p130 and E2F-4 remain associated in vivo in their specific states of phosphorylation in infected cells.
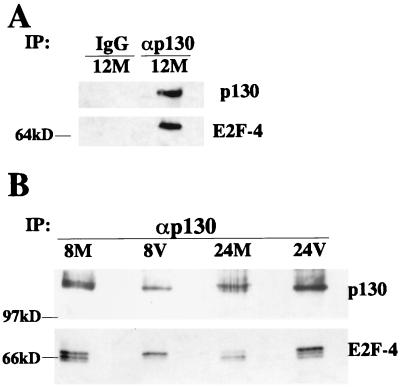
The above experiments demonstrated that E2F-4 and p130 were maintained in specific phosphorylation states in infected cells. Moreover, these two proteins were present in nuclear lysates from infected cells and were shown to associate with the cyclin E promoter in EMSAs. To determine if these proteins were associating in vivo, a coimmunoprecipitation assay was performed. Rabbit preimmune serum or p130 antibody was incubated with total-cell lysates from mock-infected cells from 12 h p.i.; immunoprecipitates were subjected to SDS gel electrophoresis and Western blotted for p130 and E2F-4. Figure 7A shows that the p130-specific antibody, but not rabbit preimmune serum, was able to immunoprecipitate these proteins, demonstrating both their in vivo association and the specificity of the interaction with the p130 antibody. To determine which forms of these proteins were associating in vivo, total-cell lysates were prepared from mock- and HCMV-infected cells at 8 and 24 h p.i. The p130 was immunoprecipitated from the lysates using a p130 antibody, and the immunoprecipitates were subjected to SDS gel electrophoresis. The proteins were then analyzed by Western blotting with antibodies to p130 and E2F-4. Figure 7B demonstrates that there is an in vivo association between E2F-4 and p130 in mock-infected and infected cells at 8 and 24 h p.i. This association was clearly seen in both mock-infected and infected cells at 8 h p.i., with the infected cells showing association with only the highly phosphorylated form of E2F-4. However, at 24 h p.i. in mock-infected cells the association of the two proteins was greatly diminished as the cells were entering into S phase. In contrast, the association was maintained in infected cells, and the majority of the p130-associated E2F-4 was in the highly phosphorylated form.
FIG. 7.
In vivo association between p130 and E2F-4. (A) Mock-infected cell lysates (M) from 12 h p.i. were immunoprecipitated (IP) with p130 antibody or rabbit preimmune serum coupled to protein A-Sepharose. IgG, immunoglobulin G. (B) Mock-infected (M) or infected (V) cell lysates from 8 and 24 h p.i. were immunoprecipitated (IP) with p130 antibody coupled to protein A-Sepharose. Immunoprecipitates were run on a 7.5% polyacrylamide gel. Proteins were transferred to nitrocellulose and Western blotted for p130 and E2F-4.
HCMV IE proteins IE1-72 and/or IE2-86 is not sufficient to induce expression of endogenous cyclin E.
Previously published data implicated the viral IE protein IE2-86 as playing a role in the HCMV-mediated upregulation of cyclin E (3). In that study, it was demonstrated that the viral IE protein IE2-86 but not IE1-72 could upregulate the cyclin E promoter in a transient-transfection assay in an E2F-independent manner (3). Using an in vitro footprinting assay, this group also showed that recombinant IE2-86 could bind to a region of the cyclin E promoter from +35 to +66. Interestingly, that region not only coincided with the region where we detected alterations in our in vivo footprint but flanked the initiation sites of the major HCMV-induced cyclin E transcripts. Because these prior experiments were performed as transient expression assays in which the cyclin E promoter linked to a reporter gene was transfected into U373 cells, an astrocytoma cell line, we felt that it was important to determine if IE2-86 could be playing a role in the upregulation of cyclin E in an in vivo context. We chose to do so by examining the effects of the viral IE proteins IE1-72 and IE2-86 on the endogenous cyclin E promoter in permissive human fibroblasts.
In order to achieve high efficiency of expression of the viral IE proteins in primary fibroblasts, we utilized a recombinant baculovirus system. Recombinant baculoviruses were constructed that expressed the genes of interest under the control of the chicken actin promoter; this promoter is transcriptionally silent in Sf9 cells but active in mammalian cells, resulting in a high level of expression of the gene of interest (53). Construction of recombinant baculoviruses that express IE1-72 or IE2-86 is described elsewhere (R. S. Dwarakanath et al., unpublished data).
These recombinant baculoviruses were used to express the viral IE proteins IE1-72 and IE2-86 in fibroblasts. Cells were confluence synchronized, trypsinized, replated, and allowed to settle for 1 h before mock, HCMV, or baculovirus infection. Infections were performed as described in Materials and Methods. At 24 h p.i., cells were processed for Western blot analysis or immunofluorescence. The level of expression of IE1-72 and IE2-86 was determined by staining coverslips with CH16.0, an antiserum which will recognize both IE proteins; cells were counterstained with Hoechst stain to identify the total number of cells. Figure 8 demonstrates that the majority of the baculovirus-infected cells were expressing IE protein(s) and that there were approximately equivalent amounts of IE protein expression in recombinant baculovirus-infected cells and in HCMV-infected cells. In a separate work (R. S. Dwarakanath et al., unpublished data), we document that the baculovirus-expressed IE2-86 and IE1-72 are fully functional. The IE2-86 expressed from baculovirus is able to efficiently transactivate the viral UL112-113 early gene promoter driving the chloramphenicol acetyltransferase gene that is on a separate baculovirus vector. In addition, the baculovirus expressing IE1-72 is able to complement an HCMV mutant virus containing a nonfunctional IE1-72 gene. (R. S. Dwarakanath et al., unpublished data).
Western blot analysis of the lysates from the above experiment demonstrated that expression of IE1-72 or IE2-86 did not lead to an increase in the levels of cyclin E either alone or in combination (Fig. 9A). Since it is known that viral gene expression is required for the HCMV-mediated upregulation of cyclin E, we wanted to determine if input virus particles coupled with IE1-72 or IE2-86 expression were sufficient to induce cyclin E to the levels that are seen during an infection. Cells were serum starved as described in Materials and Methods and infected with recombinant baculoviruses followed by mock or UV-irradiated virus infection. At 24 h p.i., cells were processed for Western blot analysis. Figure 9B demonstrates that UV-inactivated virus alone or with IE1-72 or IE2-86 expression did not result in an increase in the levels of cyclin E. Hence, the HCMV-mediated upregulation of cyclin E cannot be recapitulated by expression of IE1-72 or IE2-86 in combination with input virus particles.
IE1-72 is not required for the HCMV-mediated upregulation of cyclin E.
While the above experiments demonstrated that expression of IE1-72 or IE2-86 does not lead to an increase in the levels of cyclin E, they did not address whether either of these proteins was required for the HCMV-mediated upregulation of cyclin E. To address this question, the use of a mutant virus was employed. RC303ΔAcc is missing exon 4 of the UL122-123 coding region, which results in a virus that does not express the IE protein IE1-72 (41). Cells were serum starved and mock, HCMV, or RC303ΔAcc infected. They were then harvested for Western blot analysis at 24 h p.i. Figure 10 shows that infection with RC303ΔAcc led to an increase in the levels of cyclin E that was comparable to that seen in cells infected with the wild-type Towne strain. Hence, IE1-72 was not required for the HCMV-mediated upregulation of cyclin E.
FIG. 10.
IE1-72 is not required for the HCMV-mediated upregulation of cyclin E. Cells were serum starved and were mock infected (M), infected with RC303ΔAcc (Δ72), or infected with the Towne strain (V). Cell lysates were prepared at 24 h p.i., and 100 μg of total cell lysate was run on a 10% polyacrylamide gel, transferred to nitrocellulose, and Western blotted for HCMV IE1-72–IE2-86 and cyclin E.
Early gene expression is required for the HCMV-mediated upregulation of cyclin E.
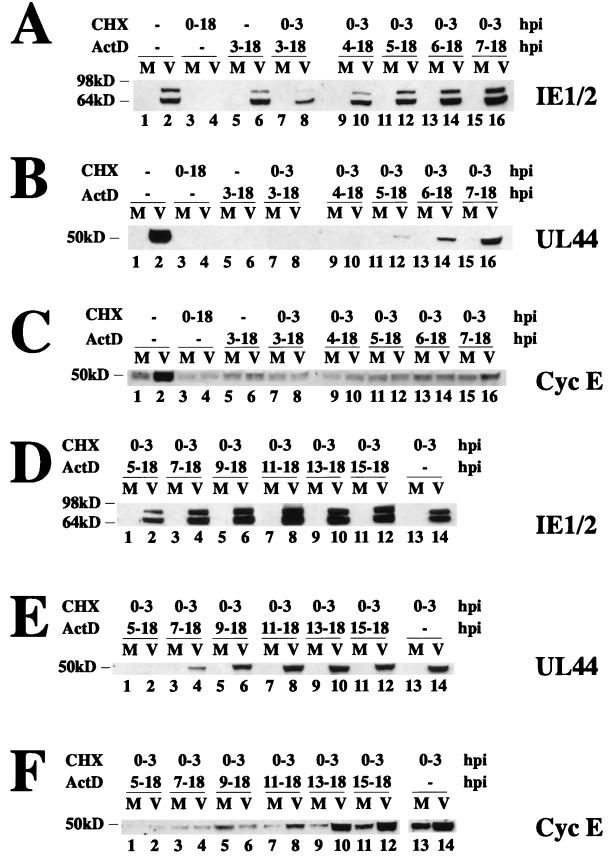
Since our observations suggested that IE1-72 and IE2-86 were not directly responsible for the HCMV-mediated upregulation of cyclin E, we sought to determine which phase of viral gene expression was temporally associated with the upregulation of cyclin E. Cells were confluence synchronized and replated in MEM plus 10% FBS in the presence of cycloheximide in order to halt protein synthesis. They were then mock or HCMV infected in the presence of cycloheximide to allow for accumulation of viral IE transcripts and released from cycloheximide for various times prior to the addition of the transcription inhibitor, actinomycin D. Lysates were prepared from cells at 18 h p.i.; the proteins were subjected to SDS electrophoresis and analyzed by Western blotting for the presence of the HCMV IE1-72 and IE2-86 proteins (Fig. 11A and D), the HCMV 50-kDa UL44 early gene product (Fig. 11B and E), and cyclin E (Fig. 11C and F). Viral IE proteins were detected after release from the cycloheximide block (Fig. 11A, lanes 8, 10, 12, 14, and 16, and 11D, lanes 2, 4, 6, 8, 10, 12, and 14) but not in cells that were maintained in cycloheximide (Fig. 11A, lane 4), demonstrating that protein synthesis was completely inhibited. The activation of early genes by the IE factors was detected by the expression of UL44 at various times post-release from cycloheximide block. The level of this protein increased as the time interval between cycloheximide release and the addition of actinomycin D was lengthened (Fig. 11B, lanes 12, 14, and 16, and 11E, lanes 4, 6, 8, 10, 12, and 14). However, despite the appearance of the IE gene products and the demonstration of their ability to activate UL44 early gene expression, levels of cyclin E were not increased under these conditions (Fig. 11C). Only when cycloheximide was released at 3 h p.i. and actinomycin D addition was delayed until 7 h p.i. or later was there HCMV-induced cyclin E expression (Fig. 11C, lane 16, and 11F, lanes 8, 10, 12, and 14). As expected, HCMV-infected cells that were not treated with either drug showed a marked increase in the levels of cyclin E (Fig. 11C, lane 2). Taken together, these data suggest that viral early gene expression is required for the induction of cyclin E expression.
FIG. 11.
Viral early gene expression is required for the HCMV-mediated upregulation of cyclin E. Cells were confluence synchronized and mock (M)- or HCMV (V)-infected in the presence of cycloheximide (CHX). Cells were released from the cycloheximide block at 3 h p.i., and actinomycin D (ActD) was added at various times postrelease (0 to 15 h p.i.). Lysates were prepared at 18 h p.i., and 50 μg of total cell lysate was run on a 10% polyacrylamide gel, transferred to nitrocellulose, and Western blotted for IE1-72 and IE2-86 (CH16.0) (A and D), UL44 (B and E), or cyclin E (C and F). It should be noted that the apparent small increase in cyclin E in the mock-infected cells in panel F, lane 5, is due to the presence of BSA (due to inefficient washing of the cells prior to lysis) that comigrated at this position and nonspecifically bound some of the cyclin E antibody.
DISCUSSION
Downstream regions of the cyclin E promoter are important for HCMV-mediated upregulation.
We have shown that the HCMV-induced cyclin E transcripts initiate further downstream than the cyclin E mRNA present in mock-infected cells. This change in start site is coincident with the appearance of protection and hypersensitivity in the in vivo footprint of infected cells. In G0 cells, we detected protection at several sites, +20, +44, and +64, and hypersensitivity at +23, +31, +42, +57, +59, and +75. However, with the exception of the +75 hypersensitive site, the same changes were seen in all in vivo samples compared to the in vitro sample. Differences that were detected in the +33 to +36 region correlate with the use of the +32 start site in mock-infected cells. In accord with these results, mock-infected cells showed protection at +33, which was very near the transcription start site that was utilized in these cells. At 24 h p.i. in infected cells, we did not observe protection of the +33 site, but we did observe hypersensitivity of both the +34 and the +36 sites. This result was consistent with this region of the promoter not being utilized in the same manner in infected cells as it was in mock-infected cells.
At 10 h p.i. in HCMV-infected cells, a protected site within the downstream E2F site at +13 was detected. This was also apparent at 24 h p.i., with the addition of a protected site at +18. The +13 and +18 protected sites in infected cells correlate well with the presence of an E2F-4–DP-1–p130 complex bound to that region, as was demonstrated by EMSA. Moreover, the downstream sites of hypersensitivity and protection in infected cells (+62, +63, +74, and +75) occurred in the region where the major virus-induced transcripts initiated.
E2F phosphorylation and complex formation during HCMV infection.
HCMV infection altered the steady-state levels of several factors that are thought to regulate cyclin E gene expression. In HCMV-infected cells, DP-1 levels were maintained in a slower-migrating form. E2F-4 was maintained in a slower-migrating form in infected cells throughout the time course, while in mock-infected cells it shifted to a faster-migrating form at 8 h p.i. It remains to be determined whether the various forms of E2F-4, which arise from differential phosphorylation, differ in their transcriptional activity.
The pocket protein p130 exhibited increased levels of a partially phosphorylated form of the protein in infected cells throughout the time course. The temporal regulation of p130 phosphorylation that was seen in mock-infected cells was typical of cells leaving quiescence and traversing into S phase, at which point the protein was degraded. In a recent study, it was shown that the murine cyclin E promoter was negatively regulated by E2F-4–DP-1–p130 complexes and that entry into the cell cycle resulted in the release of this complex from the DNA (32). Consistent with these data, we detected p130 in gel shift complexes from quiescent, confluent cells. However, we were also able to detect p130, E2F-4, and DP-1 in the infection-induced complex (E) as early as 8 h p.i. In addition, complex E migrated slower than the quiescent p130 complex, indicating that there may be an additional factor or factors in this infection-induced complex. Alternatively, since we have shown via Western blotting that both p130 and E2F-4 are sustained in specific phosphorylation states in infected cells, the change in mobility may be related to the differential phosphorylation of these proteins. In contrast to the persistence of the E2F-4–DP-1–p130 complex on the cyclin E promoter with lysates from infected cells, these proteins in the mock-infected-cell lysates do not form a complex with the cyclin E promoter after release from confluence.
In a previous report, it was noted that an HCMV-induced E2F-containing complex could associate with the DHFR promoter (61). The DHFR complex was different from the one that we have detected on the cyclin E promoter. It contained components of an S-phase-specific complex, while the infection-induced complex associating with the cyclin E promoter contains components of a G0-phase complex. It seems very likely that HCMV utilizes different methods to control these two E2F-regulated promoters. These data also support the notion that E2F regulation of transcription is not the same on all promoters.
The temporal kinetics of the appearance of an infection-induced complex in EMSAs correlates with the HCMV-induced upregulation of cyclin E mRNA. However, in the past, the presence of an E2F-4–DP-1–p130 complex has been considered to be an indicator of inhibition. This paradox can be explained by the altered phosphorylation states of E2F-4 and p130 that are seen in infected cells. It has been demonstrated that phosphorylation of the pocket domain of p130 abrogates its ability to act as a repressor (8, 27, 37, 39). In an uninfected cell, p130 reaches a third phosphorylation state in S phase and is targeted for degradation (12, 17). The release of pocket protein-mediated repression has always been associated with the hyperphosphorylation of the pocket protein which results in its disassociation from the E2F complex. However, moderate phosphorylation could interfere with the repressive function of a pocket protein while still allowing it to remain in a complex with E2F. More evidence for this model comes from data that show that cyclin E- or cyclin A-associated cdk activity can relieve p130-mediated repression but that only cyclin A can efficiently phosphorylate E2F-DP complexes and reduce their DNA binding affinity (13). Since it has been established that cyclin A-associated kinase activity is inhibited during HCMV infection, the cyclin E activity could relieve the p130-mediated repression without causing release of the complex from the DNA. Whether the infection-induced complex is responsible for the HCMV-mediated upregulation of cyclin E remains to be determined. However, additional evidence suggesting that the E2F-4–p130 complex may be important in HCMV-mediated upregulation of cyclin E is the observation that infection with UV-inactivated virus has no effect on E2F-4 or p130 and does not upregulate cyclin E. Further studies utilizing stable integrated reporter constructs to assess the role of various cis-acting elements within the cyclin E promoter will be necessary to address this question.
The localization of E2F-4 has been shown to be cell cycle regulated; it is localized to both the cytoplasm and the nucleus during G1, but as the cells progress into S phase E2F-4 becomes primarily cytoplasmic (60). It has also been reported previously that the association of the pocket protein p130 with E2F-4 will facilitate its transport into the nucleus (38). The slowest-migrating form of E2F-4 remained in the nucleus in HCMV-infected cells. This suggests that the association of E2F-4 with p130 may play a role in retaining the complex in the nucleus. Interestingly, another member of the herpesvirus family, herpes simplex virus type 1, has recently been shown to induce nuclear localization of a phosphorylated form of E2F-4 (46). In addition, it was reported that this modified E2F-4 was associated with the pocket protein p107. The similarity of these results to what we have observed raises the possibility that herpesviruses may use a common mechanism to alter the transcriptional activity of the E2F and pocket protein families.
Although initiation downstream will not affect the protein product of the cyclin E gene, it does suggest that a new type of transcriptional regulation is occurring in HCMV-infected cells. Since cyclin E is normally regulated by the transcription factor E2F, it is tempting to speculate that the alterations of the E2F family that are seen in infected cells may be playing a role in this new type of transcriptional regulation. Due to the close proximity of the virus-induced transcription start sites to the mock-infected cell start site, the transcription machinery may be utilizing downstream start sites because the AT-rich region at +1 (Fig. 2, part 2) is blocked by the E2F complexes bound to the E2F sites flanking this AT-rich region. If the E2F-4–DP-1–p130 complex is acting in a repressive manner to inhibit the use of the +32 cellular start site, the basal transcription factors would then initiate transcription downstream in a viral factor- or virus-induced cellular factor-dependent manner. However, additional support for a model in which the E2F complex is playing an activating or a neutral rather than a repressive role in HCMV-infected cells is provided by the observation that at 48 h p.i. an additional transcription start site is seen at +21. If the E2F complex were playing a repressive role, then it seems unlikely that a new start site would be located proximal to the complex, especially since this new start site is closer to the E2F complex than is the +32 start site.
Expression of HCMV IE proteins IE1-72 and/or IE2-86 does not result in increased levels of endogenous cyclin E—requirement for early gene expression.
Recently, transient-transfection experiments using a plasmid consisting of the cyclin E promoter and a luciferase reporter suggested that HCMV-mediated upregulation of cyclin E was E2F site independent (3). In that same study, it was reported that cotransfection with a plasmid expressing the HCMV IE protein IE2-86 could transactivate this reporter construct. Because there are many examples where transient-expression assays do not reflect the in vivo situation (19, 28), it is difficult to directly compare the prior work with our studies, where the endogenous cyclin E promoter was analyzed in an in vivo chromatin context. In order to determine if IE2-86 was indeed playing a role in upregulation of cyclin E in vivo, we asked if expression of IE1-72 or IE2-86 could lead to an increase in endogenous cyclin E levels. As our data clearly show, expression of either protein alone, of the two proteins together, or of the protein or proteins combined with infection with UV-inactivated virus does not lead to an increase in endogenous cyclin E levels. It is interesting to note that the region of the promoter that was identified in our in vivo footprinting studies contains the domain that appeared to form a complex with IE2-86 in an in vitro assay using the purified protein (3). However, the binding of IE2-86 to the cyclin E promoter has not yet been demonstrated in vivo. Moreover, since our studies demonstrate that IE2-86 cannot induce expression of endogenous cyclin E, the relevance of the reported in vitro binding is uncertain. The cycloheximide block and release experiment that was depicted in Fig. 11 demonstrated that, although IE gene products were present in amounts that equaled or exceeded those which are seen during HCMV infection, there was no induction of cyclin E. Only after early gene expression was observed were we able to detect an HCMV-induced increase in levels of cyclin E (Fig. 11E and F). These data indicate that IE gene expression is not sufficient for the HCMV-mediated upregulation of cyclin E but rather that some early gene product(s) or downstream effector of that product(s) is required for the increased cyclin E expression in infected cells. It should be noted that the possibility of a cooperative effect of an early gene product with an IE gene product cannot be excluded by the results of this experiment. However, it is unlikely that the viral IE protein IE1-72 plays a role in the HCMV-mediated upregulation of cyclin E, since infection with a mutant virus that was missing the IE1-72 gene product was able to efficiently induce cyclin E.
Taken together, the results of our studies show that, once HCMV enters the cell and begins its gene expression, the following chain of events occur: there is an accumulation of a virus-induced complex on the cyclin E promoter as measured by EMSA, downstream start sites are utilized for cyclin E transcription, there is the appearance of more hypersensitive and protected sites as time progresses, and high levels of cyclin E mRNA continue to accumulate. It is no coincidence that the virus upregulates cyclin E and its associated kinase activity. In fact, it has been reported elsewhere that cdk2 activity is required for viral replication (4). The obvious advantage of upregulating cyclin E expression would be to artificially push the cell into an S-like phase, in which the cell would express DNA replication enzymes and substrates that the virus could use for its replication. Support for this comes from a recent study which has shown that cyclin E expression can push the cell into S phase independently of E2F activity (36).
The work presented here suggests a role for viral early gene products as well as E2F-4–DP-1–p130 complexes in HCMV-induced upregulation of cyclin E mRNA. Future studies focusing on the functional activity of these complexes in infected cells as well as the identity and role of viral early gene products may help to further define the mechanism by which this upregulation occurs.
ACKNOWLEDGMENTS
We thank members of the Spector lab for helpful discussions and Elizabeth Fortunato, Charles Clark, Christopher Morello, David Kim, and Antoanella Bardan for critical reading of the manuscript.
This investigation was supported by NIH grants CA 34729 and CA 73490 and NIH training grant CA 09345.
REFERENCES
- 1.Ausubel F M, et al. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1987. [Google Scholar]
- 2.Beijersbergen R L, Bernards R. Cell cycle regulation by the retinoblastoma family of growth inhibitory proteins. Biochim Biophys Acta. 1996;1287:103–120. doi: 10.1016/0304-419x(96)00002-9. [DOI] [PubMed] [Google Scholar]
- 3.Bresnahan W A, Albrecht T, Thompson E A. The cyclin E promoter is activated by human cytomegalovirus 86-kDa immediate early protein. J Biol Chem. 1998;273:22075–22082. doi: 10.1074/jbc.273.34.22075. [DOI] [PubMed] [Google Scholar]
- 4.Bresnahan W A, Boldogh I, Chi P, Thompson E A, Albrecht T. Inhibition of cellular Cdk2 activity blocks human cytomegalovirus replication. Virology. 1997;231:239–247. doi: 10.1006/viro.1997.8489. [DOI] [PubMed] [Google Scholar]
- 5.Bresnahan W A, Boldogh I, Thompson E A, Albrecht T. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology. 1996;224:150–160. doi: 10.1006/viro.1996.0516. [DOI] [PubMed] [Google Scholar]
- 6.Bresnahan W A, Thompson E A, Albrecht T. Human cytomegalovirus infection results in altered Cdk2 subcellular localization. J Gen Virol. 1997;78:1993–1997. doi: 10.1099/0022-1317-78-8-1993. [DOI] [PubMed] [Google Scholar]
- 7.Castaño E, Kleyner Y, Dynlacht B D. Dual cyclin-binding domains are required for p107 to function as a kinase inhibitor. Mol Cell Biol. 1998;18:5380–5391. doi: 10.1128/mcb.18.9.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow K N, Dean D C. Domains A and B in the Rb pocket interact to form a transcriptional repressor motif. Mol Cell Biol. 1996;16:4862–4868. doi: 10.1128/mcb.16.9.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cobrinik D, Lee M H, Hannon G, Mulligan G, Bronson R T, Dyson N, Harlow E, Beach D, Weinberg R A, Jacks T. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 1996;10:1633–1644. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]
- 10.Dalton S. Cell cycle regulation of the human cdc2 gene. EMBO J. 1992;11:1797–1804. doi: 10.1002/j.1460-2075.1992.tb05231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dittmer D, Mocarski E S. Human cytomegalovirus infection inhibits G1/S transition. J Virol. 1997;71:1629–1634. doi: 10.1128/jvi.71.2.1629-1634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong F, Cress W D, Jr, Agrawal D, Pledger W J. The role of cyclin D3-dependent kinase in the phosphorylation of p130 in mouse BALB/c 3T3 fibroblasts. J Biol Chem. 1998;273:6190–6195. doi: 10.1074/jbc.273.11.6190. [DOI] [PubMed] [Google Scholar]
- 13.Dynlacht B D, Flores O, Lees J A, Harlow E. Differential regulation of E2F transactivation by cyclin/cdk2 complexes. Genes Dev. 1994;8:1772–1786. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- 14.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 15.Fields B N, Knipe D M, Howley P M. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2447–2492. [Google Scholar]
- 16.Fortunato E A, Spector D H. p53 and RPA are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus. J Virol. 1998;72:2033–2039. doi: 10.1128/jvi.72.3.2033-2039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garriga J, Limón A, Mayol X, Rane S G, Albrecht J H, Reddy E P, Andrés V, Graña X. Differential regulation of the retinoblastoma family of proteins during cell proliferation and differentiation. Biochem J. 1998;333:645–654. doi: 10.1042/bj3330645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geng Y, Eaton E N, Picón M, Roberts J M, Lundberg A S, Gifford A, Sardet C, Weinberg R A. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene. 1996;12:1173–1180. [PubMed] [Google Scholar]
- 19.Ghazal P, Nelson J A. Interactions between human cytomegalovirus immediate-early proteins and the long terminal repeat of human immunodeficiency virus. Rev Med Virol. 1993;3:47–55. [Google Scholar]
- 20.Hateboer G, Wobst A, Petersen B O, Le Cam L, Vigo E, Sardet C, Helin K. Cell cycle-regulated expression of mammalian CDC6 is dependent on E2F. Mol Cell Biol. 1998;18:6679–6697. doi: 10.1128/mcb.18.11.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helin K. Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev. 1998;8:28–35. doi: 10.1016/s0959-437x(98)80058-0. [DOI] [PubMed] [Google Scholar]
- 22.Henglein B, Chenivesse X, Wang J, Eick D, Bréchot C. Structure and cell cycle-regulated transcription of the human cyclin A gene. Proc Natl Acad Sci USA. 1994;91:5490–5494. doi: 10.1073/pnas.91.12.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiebert S W, Lipp M, Nevins J R. E1A-dependent trans-activation of the human MYC promoter is mediated by the E2F factor. Proc Natl Acad Sci USA. 1989;86:3594–3598. doi: 10.1073/pnas.86.10.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsiao K M, McMahon S L, Farnham P J. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes Dev. 1994;8:1526–1537. doi: 10.1101/gad.8.13.1526. [DOI] [PubMed] [Google Scholar]
- 25.Jault F M, Jault J M, Ruchti F, Fortunato E A, Clark C, Corbeil J, Richman D D, Spector D H. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J Virol. 1995;69:6697–6704. doi: 10.1128/jvi.69.11.6697-6704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson D G, Ohtani K, Nevins J R. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 27.Knudsen E S, Wang J Y. Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol Cell Biol. 1997;17:5771–5783. doi: 10.1128/mcb.17.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koval V, Jault F M, Pal P G, Moreno T N, Aiken C, Trono D, Spector S A, Spector D H. Differential effects of human cytomegalovirus on integrated and unintegrated human immunodeficiency virus sequences. J Virol. 1995;69:1645–1651. doi: 10.1128/jvi.69.3.1645-1651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam E W, Bennett J D, Watson R J. Cell-cycle regulation of human B-myb transcription. Gene. 1995;160:277–281. doi: 10.1016/0378-1119(95)00184-8. [DOI] [PubMed] [Google Scholar]
- 30.Lane D, Harlow E. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 31.Lavoie J N, L'Allemain G, Brunet A, Müller R, Pouysségur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J Biol Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- 32.Le Cam L, Polanowska J, Fabbrizio E, Olivier M, Philips A, Ng Eaton E, Classon M, Geng Y, Sardet C. Timing of cyclin E gene expression depends on the regulated association of a bipartite repressor element with a novel E2F complex. EMBO J. 1999;18:1878–1890. doi: 10.1093/emboj/18.7.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lees E, Faha B, Dulic V, Reed S I, Harlow E. Cyclin E/cdk2 and cyclin A/cdk2 kinases associate with p107 and E2F in a temporally distinct manner. Genes Dev. 1992;6:1874–1885. doi: 10.1101/gad.6.10.1874. [DOI] [PubMed] [Google Scholar]
- 34.Liu N, Lucibello F C, Zwicker J, Engeland K, Müller R. Cell cycle-regulated repression of B-myb transcription: cooperation of an E2F site with a contiguous corepressor element. Nucleic Acids Res. 1996;24:2905–2910. doi: 10.1093/nar/24.15.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu M, Shenk T. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J Virol. 1996;70:8850–8857. doi: 10.1128/jvi.70.12.8850-8857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukas J, Herzinger T, Hansen K, Moroni M C, Resnitzky D, Helin K, Reed S I, Bartek J. Cyclin E-induced S phase without activation of the pRb/E2F pathway. Genes Dev. 1997;11:1479–1492. doi: 10.1101/gad.11.11.1479. [DOI] [PubMed] [Google Scholar]
- 37.Lundberg A S, Weinberg R A. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magae J, Wu C L, Illenye S, Harlow E, Heintz N H. Nuclear localization of DP and E2F transcription factors by heterodimeric partners and retinoblastoma protein family members. J Cell Sci. 1996;109:1717–1726. doi: 10.1242/jcs.109.7.1717. [DOI] [PubMed] [Google Scholar]
- 39.Mayol X, Garriga J, Graña X. G1 cyclin/CDK-independent phosphorylation and accumulation of p130 during the transition from G1 to G0 lead to its association with E2F-4. Oncogene. 1996;13:237–246. [PubMed] [Google Scholar]
- 40.Moberg K, Starz M A, Lees J A. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mocarski E S, Kemble G W, Lyle J M, Greaves R F. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc Natl Acad Sci USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muganda P, Mendoza O, Hernandez J, Qian Q. Human cytomegalovirus elevates levels of the cellular protein p53 in infected fibroblasts. J Virol. 1994;68:8028–8034. doi: 10.1128/jvi.68.12.8028-8034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neuman E, Flemington E K, Sellers W R, Kaelin W G., Jr Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol Cell Biol. 1994;14:6607–6615. doi: 10.1128/mcb.14.10.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohtani K, DeGregori J, Leone G, Herendeen D R, Kelly T J, Nevins J R. Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol Cell Biol. 1996;16:6977–6984. doi: 10.1128/mcb.16.12.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohtani K, DeGregori J, Nevins J R. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci USA. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olgiate J, Ehmann G L, Vidyarthi S, Hilton M J, Bachenheimer S L. Herpes simplex virus induces intracellular redistribution of E2F4 and accumulation of E2F pocket protein complexes. Virology. 1999;258:257–270. doi: 10.1006/viro.1999.9755. [DOI] [PubMed] [Google Scholar]
- 47.Oswald F, Dobner T, Lipp M. The E2F transcription factor activates a replication-dependent human H2A gene in early S phase of the cell cycle. Mol Cell Biol. 1996;16:1889–1895. doi: 10.1128/mcb.16.5.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearson B E, Nasheuer H P, Wang T S. Human DNA polymerase alpha gene: sequences controlling expression in cycling and serum-stimulated cells. Mol Cell Biol. 1991;11:2081–2095. doi: 10.1128/mcb.11.4.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodems S M, Clark C L, Spector D H. Separate DNA elements containing ATF/CREB and IE86 binding sites differentially regulate the human cytomegalovirus UL112-113 promoter at early and late times in the infection. J Virol. 1998;72:2697–2707. doi: 10.1128/jvi.72.4.2697-2707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salvant B S, Fortunato E A, Spector D H. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J Virol. 1998;72:3729–3741. doi: 10.1128/jvi.72.5.3729-3741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sánchez I, Dynlacht B D. Transcriptional control of the cell cycle. Curr Opin Cell Biol. 1996;8:318–324. doi: 10.1016/s0955-0674(96)80004-4. [DOI] [PubMed] [Google Scholar]
- 52.Schulze A, Zerfass K, Spitkovsky D, Middendorp S, Bergès J, Helin K, Jansen-Dürr P, Henglein B. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc Natl Acad Sci USA. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shoji I, Aizaki H, Tani H, Ishii K, Chiba T, Saito I, Miyamura T, Matsuura Y. Efficient gene transfer into various mammalian cells, including non-hepatic cells, by baculovirus vectors. J Gen Virol. 1997;78:2657–2664. doi: 10.1099/0022-1317-78-10-2657. [DOI] [PubMed] [Google Scholar]
- 54.Sidle A, Palaty C, Dirks P, Wiggan O, Kiess M, Gill R M, Wong A K, Hamel P A. Activity of the retinoblastoma family proteins, pRB, p107, and p130, during cellular proliferation and differentiation. Crit Rev Biochem Mol Biol. 1996;31:237–271. doi: 10.3109/10409239609106585. [DOI] [PubMed] [Google Scholar]
- 55.Slansky J E, Farnham P J. Introduction to the E2F family: protein structure and gene regulation. Curr Top Microbiol Immunol. 1996;208:1–30. doi: 10.1007/978-3-642-79910-5_1. [DOI] [PubMed] [Google Scholar]
- 56.Slansky J E, Farnham P J. Transcriptional regulation of the dihydrofolate reductase gene. Bioessays. 1996;18:55–62. doi: 10.1002/bies.950180111. [DOI] [PubMed] [Google Scholar]
- 57.Tamashiro J C, Hock L J, Spector D H. Construction of a cloned library of the EcoRI fragments from the human cytomegalovirus genome (strain AD169) J Virol. 1982;42:547–557. doi: 10.1128/jvi.42.2.547-557.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tommasi S, Pfeifer G P. Constitutive protection of E2F recognition sequences in the human thymidine kinase promoter during cell cycle progression. J Biol Chem. 1997;272:30483–30490. doi: 10.1074/jbc.272.48.30483. [DOI] [PubMed] [Google Scholar]
- 59.Vaishnav Y N, Vaishnav M Y, Pant V. The molecular and functional characterization of E2F-5 transcription factor. Biochem Biophys Res Commun. 1998;242:586–592. doi: 10.1006/bbrc.1997.8010. [DOI] [PubMed] [Google Scholar]
- 60.Verona R, Moberg K, Estes S, Starz M, Vernon J P, Lees J A. E2F activity is regulated by cell cycle-dependent changes in subcellular localization. Mol Cell Biol. 1997;17:7268–7282. doi: 10.1128/mcb.17.12.7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wade M, Kowalik T F, Mudryj M, Huang E S, Azizkhan J C. E2F mediates dihydrofolate reductase promoter activation and multiprotein complex formation in human cytomegalovirus infection. Mol Cell Biol. 1992;12:4364–4374. doi: 10.1128/mcb.12.10.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woo M S, Sánchez I, Dynlacht B D. p130 and p107 use a conserved domain to inhibit cellular cyclin-dependent kinase activity. Mol Cell Biol. 1997;17:3566–3579. doi: 10.1128/mcb.17.7.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu L, Xie E, Chang L S. Differential roles of two tandem E2F sites in repression of the human p107 promoter by retinoblastoma and p107 proteins. Mol Cell Biol. 1995;15:3552–3562. doi: 10.1128/mcb.15.7.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]