Abstract
The envelopment of the Sindbis virus nucleocapsid in the modified cell plasma membrane involves a highly specific interaction between the capsid (C) protein and the endodomain of the E2 glycoprotein. We have previously identified a domain of the Sindbis virus C protein involved in binding to the E2 endodomain (H. Lee and D. T. Brown, Virology 202:390–400, 1994). The C-E2 binding domain resides in a hydrophobic cleft with C Y180 and W247 on opposing sides of the cleft. Structural modeling studies indicate that the E2 domain, which is proposed to bind the C protein (E2 398T, 399P, and 400Y), is located at a sufficient distance from the membrane to occupy the C protein binding cleft (S. Lee, K. E. Owen, H. K. Choi, H. Lee, G. Lu, G. Wengler, D. T. Brown, M. G. Rossmann, and R. J. Kuhn, Structure 4:531–541, 1996). To measure the critical spanning length of the E2 endodomain which positions the TPY domain into the putative C binding cleft, we have constructed a deletion mutant, ΔK391, in which a nonconserved lysine (E2 K391) at the membrane-cytoplasm junction of the E2 tail has been deleted. This mutant was found to produce very low levels of virus from BHK-21 cells due to a defect in an unidentified step in nucleocapsid binding to the E2 endodomain. In contrast, ΔK391 produced wild-type levels of virus from tissue-cultured mosquito cells. We propose that the phenotypic differences displayed by this mutant in the two diverse host cells arise from fundamental differences in the lipid composition of the insect cell membranes which affect the physical and structural properties of membranes and thereby virus assembly. The data suggest that these viruses have evolved properties adapted specifically for assembly in the diverse hosts in which they grow.
Sindbis virus, the prototype of the alphaviruses, has a precise and complex three-dimensional structure. The virion is made up of 240 copies of each of three structural proteins (E1, E2, and capsid [C] protein) in a 1:1:1 stoichiometric arrangement, a membrane bilayer, and a single copy of plus polarity single-stranded RNA. The three virus proteins are organized as a double-shelled icosahedron (32, 33). The envelope glycoproteins E1 and E2 are organized as heterotrimers. Eighty of the E1-E2 heterotrimers are organized in a T=4 icosahedral lattice through E1-E1 protein associations which interconnect all of the heterotrimers (1, 2). The membrane bilayer is derived from the host cell during virus assembly and is situated between the outer T=4 icosahedral protein shell of E1 and E2 glycoproteins and the inner T=4 icosahedral shell composed of C protein (7, 11, 32, 34). The envelope glycoproteins are both type 1 membrane-spanning proteins, and the E2 glycoprotein has a 33-amino-acid endodomain which specifically interacts with the C protein, locking the outer protein shell to the inner protein shell (18, 19, 25). The alphaviruses are not typical of membrane-containing viruses, the majority of which are well described as membrane bilayers with associated virus proteins. The alphaviruses are protein icosahedra with an associated lipid bilayer.
The assembly of the complex, double-shell, membrane-containing structure involves numerous and highly specific protein-protein interactions occurring through two separate pathways (45). The virus structural proteins are synthesized from a subgenomic polycistronic RNA with the potential of producing all of the structural proteins as a single polypeptide. As the nascent protein is produced, the C protein located at the NH-terminal end cuts itself from the developing polyprotein utilizing a proteolytic activity contained in its structure. The freed C protein assembles together with progeny RNA to produce a nucleocapsid which comprises the inner protein shell. The remainder of the polyprotein containing the sequences of glycoproteins E1 and E2 is integrated into the membranes of the cell endoplasmic reticulum (ER). In the ER, the proteins are proteolytically processed to form PE2, the precursor to E2, and E1. The glycoprotein E1 folds from a fully extended form into a more compact form and then binds PE2 to create a heterodimer. The heterodimer forms trimers (three copies each of E1 and PE2), and E1 continues folding through disulfide-bridged intermediates into a compact, energy-rich conformation (6, 30). After formation of the heterotrimer and E1 folding is complete, the glycoprotein trimers are exported from the ER to the plasma membrane. En route, PE2 is converted to E2 by a furin-like protease in the trans-Golgi network and E1 is converted from a stable to a metastable form.
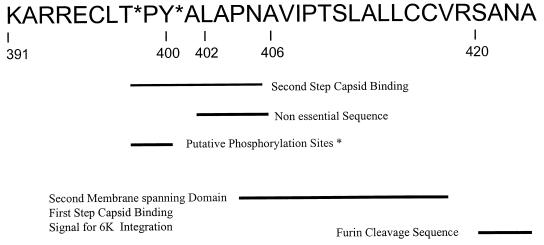
The plasma membrane is the point at which the final events in alphavirus assembly take place. The assembled nucleocapsid binds to the endodomain of the E2 glycoprotein in a two-step interaction which is highly specific (22, 23, 25). The 33-amino-acid E2 endodomain is a multifunctional structure (Fig. 1). The E2 glycoprotein begins its maturation as PE2, which, in the ER, has two membrane-spanning domains (21). The first membrane-spanning domain is composed of amino acids 365 to 390 and is the anchor domain for this glycoprotein (36). The second membrane-spanning domain is predicted to be composed of amino acids 405 to 418 (39) and contains a sequence which is essential for the specific recognition of virus C protein. Binding within this domain is the first of two steps in the binding of nucleocapsids to the E2 endodomain (25). Nucleocapsid binding requires that this domain be removed from the cell membrane and exposed to the cytoplasm. We have demonstrated that this event occurs after the maturation of the spike trimer in the ER and its export from the ER (24). The precise point in the secretory pathway at which the second domain is withdrawn from the cell membrane is not known. The removal of a hydrophobic domain from a membrane bilayer is a very energetically unfavorable event. We have shown that a phosphorylation-dephosphorylation event accompanies this process; however, we have not been able to demonstrate that the event is phosphorylation dependent (23). The sequence of amino acids connecting the two membrane-spanning domains, the cytoplasmic loop, has been implicated as containing the sites for phosphorylation and as encoding the sequence involved in the second tight binding domain for nucleocapsid binding (19).
FIG. 1.
Functional map of the E2 endodomain. See the text for details.
The interaction of nucleocapsid with E2 is virus strain specific, which implies that a specific C protein site exists for E2 binding. Using site-directed mutagenesis, we have identified a domain in the C protein of Sindbis virus which binds to the E2 endodomain (18). The Y180S/E183G mutation in the C protein resulted in the production of virus particles with reduced infectivity and an altered interaction of C protein with the E2 endodomain. Membrane glycoproteins of the C protein mutant had normal functional properties of attachment and membrane fusion, which led us to propose that the defect in this mutant was a failure to uncoat the nucleocapsid as the process of virus membrane-host membrane fusion took place. We postulated that this failure resulted from the altered (weakened) E2-C association (18). In the X-ray crystallographic structure produced by Choi et al. (7), the C protein domain which we identified as critical for E2-C association has a fold which places aromatic ring structures on opposite sides of a hydrophobic cleft. A conserved tyrosine at position 400 in the E2 tail, accessible to binding in the C protein hydrophobic cleft, suggested that an aromatic interaction was possible between E2 Y400, C Y180, and W247 (19). If such an interaction existed, it would suggest that the loss of one of the three aromatic structures participating in the association would significantly weaken the strength of the E2-C association.
A critical role for the interaction of aromatic E2 Y400 with C protein has been supported by experiments in which Y400 has been exchanged for other amino acids. Aromatic substitutions supported virus production, while nonaromatic substitutions resulted in failure to assemble mature virions (13). Molecular modeling of the E2 endodomain into the C cleft containing C Y180 and W247 indicated that E2 Y400 could penetrate to a point at which the postulated aromatic interaction could take place (19). Molecular modeling also implicated a hydrophobic interaction involving E2 L402 as participating in nucleocapsid binding (19).
We have attempted to further assess the role that the aromatic and hydrophobic interactions between residues in E2 and C play in virus maturation by reducing the distance between the point at which the E2 endodomain emerges from the membrane bilayer and E2 Y400. The hypothesis is that shortening of this distance will prevent the specific interaction and prevent virus maturation. To this end, we have produced a deletion in the membrane-proximal region of the E2 endodomain, reducing the distance between E2 Y400 and the membrane by a single amino acid. We have investigated the effects of this deletion on the events leading to the binding of the E2 endodomain to C and the ensuing process of envelopment.
MATERIALS AND METHODS
Na125I, [35S]methionine-cysteine, and [32P]orthophosphate were from New England Nuclear Life Science Products (Boston, Mass.). Restriction enzymes, T4 DNA ligase, and reagents for in vitro transcription (SP6 polymerase, RNasin, etc.) were from New England Biolabs (Beverly, Mass.). Taq DNA polymerase and Moloney murine leukemia virus (MMLV) reverse transcriptase (RT) were from PE Applied Biosystems (Branchburg, N.J.). Sequencing reagents were from United States Biochemical Corporation (Cleveland, Ohio), or for automated sequencing, BigDye terminator chemistry (for plasmids) or dRhodamine terminator chemistry (for PCR products) was used; both were from PE Applied Biosystems. Shrimp alkaline phosphatase was from Amersham-Pharmacia Biotech.
Cell culture, plaque assay, and virus.
Baby hamster kidney (BHK-21) cells were initially provided by Peter Faulkner (Queens University, Kingston, Ontario, Canada). These cells were grown in minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS), 10% tryptose phosphate broth, and 2 mM glutamine as described previously (35). The Aedes albopictus subclones used were all derived from Singh's original larval isolates (43). The U4.4 cells maintained in this laboratory were cloned from cells provided by Sonya Buckley (Yale Arbovirus Research Unit, New Haven, Conn.). This line is maintained in MEM as described above or in Mitsuhashi and Maramorosch (M and M) medium (28). The C7-10 line was provided by Victor Stollar (Rutgers Medical School, New Brunswick, N.J.) and is maintained in MEM as described above and previously (27).
Virus produced from the Toto 1101 cDNA clone of Sindbis virus contains a tyrosine at E2 position 420, S420Y, has been described elsewhere (24), and served as the wild-type template for mutagenesis and as the wild-type cDNA construct for transfection experiments.
Titration of ΔK391 virus grown in U4.4 cells was done on C7-10 cells. For this assay, 1 × 107 to 2 × 107 C7-10 cells/ml are plated onto 25-cm2 flasks in serum-free medium for 1 h prior to infection. Virus dilutions are in phosphate-buffered saline (PBS) with 3% FBS. The remainder of the assay is essentially the same as for BHK cells (35), except that the plaques are allowed to develop for an additional day before staining with 2% neutral red.
Site-directed mutagenesis of Toto S420Y and RT PCR.
Using standard megaprimer site-directed mutagenesis protocols (40) described previously (24) and Taq DNA polymerase, we generated a single mutant with the nonconserved lysine at position 391 in E2 (designated Δ391K) deleted from the S420Y template DNA. The S420Y construct is derived from Toto 1101 full-length Sindbis virus cDNA which contains an SP6 promoter from which full-length infectious transcripts can be generated. The AAA sequence encoding lysine was deleted in the mutagenic primer and amplified using a standard PCR during a second cycle. The resulting product was placed into the wild-type vector using the Bcl (nucleotide 9358) and Spl (nucleotide 10381) sites. After confirmation of the correct sequence throughout the insert, infectious RNA was transcribed in vitro using SP6 polymerase and introduced into cells by electroporation as described below.
Mutant virus sequences were obtained from A. albopictus-grown virus particles. Virus was pelleted, and RNA was extracted and then reverse transcribed and amplified using RT PCR. PCR products were sequenced using Taq automated sequencing. A minimum of 104 PFU of virus was pelleted at 50,000 rpm in an SW 55 Ti rotor for 45 min. The pelleted virus was resuspended in 100 μl containing 10 mM Tris–1 mM EDTA (TE) and solubilized with 100 μl of 2× lysis buffer (100 mM Tris-Cl [pH 7.00], 20 mM EDTA, 1% sodium dodecyl sulfate [SDS]) containing 20 μg of glycogen for 20 min at 37°C. Virus extraction twice with phenol at 60°C is followed by one extraction with phenol-chloroform and one chloroform extraction, and then the virus is precipitated using 20 μg of carrier glycogen. RNA was resuspended in 10 μl of diethyl pyrocarbonate-treated water and transcribed using MMLV RT under the following conditions. PCR buffer contains 10 mM Tris-Cl (pH 8.3), 50 mM KCl, and 5 mM MgCl2 plus 20 U of RNasin, 200 μM each deoxynucleoside triphosphate, 1.0 μM reverse primer, and 50 U of MMLV RT in a final volume of 20 μl. Transcription is at 42°C for 20 min and 99°C for 5 min. After the transcription reaction, the volume of the reaction mixture is increased to 100 μl and the concentrations of the deoxynucleoside triphosphates and MgCl2 are adjusted for the increased volume. Forward primer is added to a final concentration of 2.0 μM, and an additional 1.0 μM reverse primer is added. Taq DNA polymerase is added to a final concentration of 2.5 U/100-μl reaction mixture.
In vitro transcription and RNA transfection.
Full-length mutant and wild-type cDNAs were first linearized using XhoI, treated with proteinase K, phenol extracted, and ethanol precipitated. Templates were then transcribed in vitro as described previously (24, 37). Templates were removed using RNase-free DNase. For BHK cells, electroporation was performed essentially as described by Liljestrom and Garoff (21). For A. albopictus cells, transfections were performed as follows. Cells were pelleted and washed in RNase-free electroporation buffer HBS (20 mM HEPES-HCl [pH 7.0], 137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, and 6 mM dextrose) in diethyl pyrocarbonate-treated water. Washed cells were resuspended in HBS to a concentration of 5 × 107/ml. RNA transcripts in 20 μl were added to 400 μl of washed cells and transferred to a 0.2-cm gap length cuvette. Electroporation conditions were 2.0 kV, 25 μF, and ∞ resistance. Cells were pulsed once. Healthy cells give a time constant of 0.7 s under these conditions. After the pulse, cells were allowed to sit for 10 min before transfer into 10 ml of M and M medium (22). Virus was harvested at 30 h posttransfection.
Metabolic labeling of viral proteins.
Subconfluent 25-cm2 monolayers of transfected BHK-21 cells were metabolically labeled with a [35S]methionine-cysteine protein-labeling mixture at a concentration of 50 μCi/ml. Cultures were labeled at 6 or 16 h after transfection. Monolayers were starved for 30 min in methionine-cysteine-free medium containing 3% FBS and 2 mM glutamine for 30 min prior to addition of the label.
Radioiodination and immunoprecipitation of E2 tail.
BHK-21 cells were transfected with mutant or wild-type transcripts as described above. Cycloheximide was added at 75 μg/ml to the cell monolayers for 1 h at 16 h posttransfection. The cells were then removed from the flask in 10 ml of lifting buffer (10 mM HEPES [pH 7.2], 15 mM KCl, 1 mM EDTA, 1 mM EGTA, 0.25 M sucrose). We disrupted the cells by passing the suspension about 20 times through a 27-gauge needle. Nuclei and any remaining intact cells were pelleted by centrifugation at 11,300 × g for 1 min. The supernatant contained membrane vesicles of intracellular organelles and plasma membrane. Half of the membrane vesicles were kept intact on ice, while the other half was dissolved in 1% NP-40. Intact or solubilized membrane vesicles were radioiodinated with Na125I using the Iodo-bead iodination reagent (immobilized N-chlorobenzenesulfonamide; Pierce) in accordance with the manufacturer's instructions. Iodo-beads were resuspended in a 1.5-ml microcentrifuge tube with 600 μl of phosphate-buffered saline without CA2+ and Mg2+ and 400 μCi of Na125I and incubated at room temperature for 5 min, and 500 μl of intact or solubilized membrane vesicles was then added to the reaction mixture. After a 10 min incubation, the beads were removed to terminate iodination. Virus protein was immunoprecipitated using anti-E2 antibody (Ab) as described previously (24), with the following modifications. Absorptions were done in lysis buffer (0.02 M Tris-HCl [pH 7.4], 0.5% NP-40, 0.05 M NaCl) with 2% bovine serum albumin. After absorption, the Ab-beads were washed 4 times with 1 ml of 1 M NaCl in PBS and then once in 2% SDS in water. The washed beads were divided, and half were cut with 5 μg of chymotrypsin (0.6 U) at room temperature for 30 min. After proteinase treatment, phenymethylsulfonyl fluoride was added to 1 mM and the digestion products were absorbed a second time using anti-PE2 tail Ab-beads as detailed previously (24) in lysis buffer with 2% bovine serum albumin. After overnight absorption, the beads were washed as described above, boiled in polyacrylamide gel electrophoresis (PAGE) sample buffer (12.5 mM Tris-HCl [pH 6.8], 10% glycerol 1% SDS, 1% β-mercaptoethanol, 0.01% bromophenol blue), and separated by centrifugation prior to loading onto a 16% Tricine gel as detailed previously (24).
PAGE.
PAGE of radiolabeled proteins was carried out under denaturing conditions in 10.8% polyacrylamide (41) for 4 h at a constant power of 5 W. Tricine gel electrophoresis was done as described above. Fluorography was performed as described previously (2a), and gels were exposed to Kodak XAR-5 film and scanned using a Microtek Scanmaker 5, and the image was printed using Photoshop 5.0 software. Prints were generated on a Codonics 1600 dye sublimation printer.
Low-pH-mediated fusion from within (FFWI).
BHK-21 cells were transfected as described above and plated onto 24-well plates. After 10 h, the transfected cell monolayers were washed twice with PBS containing 2% FBS as described previously (26). The fusion medium used was PBS containing 10 mM HEPES, 10 mM morpholineethanesulfonic acid (MES), 1% sucrose, 1× MEM amino acids, and 1× MEM vitamins. The pH of the medium was adjusted to 5.3 (fusion medium) or 7.2 (control medium). Fusion medium (pH 5.3) or control medium (pH 7.2) was added to the cells at 10 h posttransfection for 1 min. The cells were then placed into MEM and incubated at 37°C for 1 h. Fusion was assessed using light microscopy.
Transmission electron microscopy (TEM).
At 18 h posttransfection, BHK cells were scraped from the flasks and pelleted by low-speed centrifugation. Cell pellets were washed twice in PBS and fixed in 3% glutaraldehyde at 4°C overnight. The cells were then washed three times with 0.1 M cacodylate buffer (pH 7.4), postfixed with 1% osmium tetroxide for 1 h at room temperature, and washed again three times in cacodylate buffer. The cells were stained en bloc for 2 h at room temperature with 0.5% uranyl acetate. After three washes, cell pellets were embedded in 1% agarose and dehydrated through a graded ethanol series. Final embedding was in Spur's resin overnight at 70°C. Ultrathin sections were cut on an LKB Nova microtome and collected on 200-mesh copper grids. Sections were stained with 1% uranyl acetate and Reynolds lead citrate and photographed using a JEOL 100S transmission electron microscope. Negatives were scanned using a Microtek Scanmaker 5, and prints were generated on a Codonics 1600 printer.
RESULTS
Construction of Sindbis virus mutant ΔK391.
Analysis of images of Sindbis virus produced by electron cryomicroscopy (19, 32) has predicted that a critical spanning distance exists between the endodomain of E2 and the hydrophobic C protein cleft thought to be the binding site of the E2 tail. A conserved TPY(A)L (A at 401 is not conserved) motif has been proposed to interact within a hydrophobic pocket of the C protein containing C residues Y180, E133, M132, W247, K135, M137, and F166 (19). The proposed positioning of these two domains places A401 and L402 of E2 in the pocket lying between Y180, W247, and F166 of C. It has also been proposed that an interaction between E2 Y400 and C Y180 and W247 is critical for stabilizing the E2-C association. These models predict that a shift in position of these critical amino acids toward the membrane by as little as the spanning length of one amino acid (4 Å) could disrupt the binding of the E2 tail with C sufficiently to abrogate assembly of the virus. We sought to test this hypothesis by deleting K at E2 position 391 and shifting the position of the TPYAL domain toward the modified membrane. This was done using site-directed PCR mutagenesis employing the megaprimer technique (22, 40) and a cDNA construct described previously which contains a silent mutation, Y at E2 position 420, which serves to assay for the extraction of the distal region of the E2 endodomain from the membrane bilayer prior to envelopment. We have previously reported that the deletion of a five-amino-acid sequence (L402APNA406) downstream from the E2 TPY domain does not interfere with virus production (22); however, deletions upstream of this motif have not been reported.
Mutant E2 ΔK391 is blocked in assembly of Sindbis virus from BHK-21 cells.
To determine the phenotype of the mutant ΔK391, BHK-21 cells were transfected with RNA from ΔK391 or S420Y cDNA and virus was harvested at 8 and 20 h posttransfection as described in Materials and Methods. Virus production was determined by plaque assay on BHK cells as described above, and the results are shown in Table 1. Little infectious ΔK391 was detected from BHK monolayers early in infection (∼102), while less than 1 PFU of infectious virus per cell was detected for the deletion mutant at later times after transfection. By contrast, the cells transfected with E2 S420Y produced wild-type virus levels. The morphology of the plaques produced by ΔK391 was similar to that produced by the parent S420Y.
TABLE 1.
Production of Sindbis virus mutant ΔK391 in RNA-transfected BHK cellsa
| Virus | Virus titer (PFU/ml)
|
|
|---|---|---|
| 8 h | 20 h | |
| S420Y | 1.9 × 107 | 2.1 × 109 |
| Δ391K | <102 | 1.0 × 104 |
Virus titers were determined on BHK-21 cells as described in Materials and Methods.
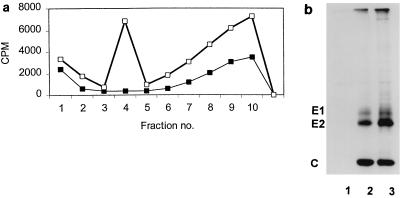
The data presented above show that the E2 deletion mutant ΔK391 is restricted in its ability to produce infectious virus in BHK-21 cells. The possibility that ΔK391 produced noninfectious virus, as did the C protein mutation previously reported, Y180S/E183G, was tested by labeling transfected BHK-21 cells with [35S]methionine-cysteine as described above. Monolayers (4 × 106 cells) transfected with either ΔK391 or S420Y RNA were labeled with 50 μCi of [35S]methionine-cysteine per ml at 5 h posttransfection. Medium and cells were harvested from the transfected cultures after 24 h. The labeled medium was layered onto a 15 to 35% potassium tartrate gradient in PBS-D and centrifuged at 110,000 × g and 4°C for 18 h (22). Gradients were fractionated, and 10 μl of each fraction was counted using scintillation spectrometry. The gradient profiles shown in Fig. 2a demonstrate that virus particles cannot be detected from the mutant-transfected cells, while the wild-type control shows a significant labeled-virus peak sedimenting to virus density (1.19 gm/cm3). These data show that the mutant ΔK391 does not produce large amounts of noninfectious virus and suggest that the deletion at K391 in the E2 tail blocks virus production at a step prior to the assembly of virus particles.
FIG. 2.
Production of virus particles and virus proteins by ΔK391-transfected BHK-21 cells. BHK-21 cells were transfected with mutant ΔK391 or wild-type S420Y as described in Materials and Methods. At 6 h posttransfection, cells were starved in medium lacking cysteine and methionine for 1 h. After the starvation period, [35S]cysteine-methionine was added and the medium and cells were harvested at 20 h posttransfection. (A) Medium from transfected cells was layered onto 15 to 35% potassium tartrate gradients and centrifuged for 18 h at 110,000 × g. Label distribution in the gradient fractions was determined by scintillation counting. Fraction 4 corresponds to virus density (1.19 g/cm3). No virus particles were detected for the ΔK391 mutant (■), while the incorporation of label was at wild-type levels for the S420Y transfection (□). (B) 35S-labeled cells from the experiment described above were harvested with lysis buffer and immunoprecipitated with anti-whole-virus Ab. The precipitates were analyzed by SDS-PAGE as described in Materials and Methods. Lane 1 contains material precipitated from the control (no virus RNA) transfection; no nonspecific binding of cellular proteins is detected. Lanes 2 and 3, respectively, show the Sindbis virus structural proteins precipitated from equal volumes of ΔK391- and S420Y-transfected cells. All three proteins, E1, E2, and C, are produced in roughly equivalent quantities in both the mutant and S420Y.
Protein production and intracellular trafficking of virus structural proteins.
The defect in the production of virus particles described above could result from a failure to synthesize virus structural proteins or to properly transport and process virus proteins. To test these possibilities, cells transfected with mutant or wild-type RNA were labeled with [35S]methionine-cysteine and harvested 24 h posttransfection. The transfected cells were prepared for SDS-PAGE as described in Materials and Methods, and the resulting gel is shown in Fig. 2b. This experiment shows that virus structural proteins produced from both ΔK391 and S420Y RNAs were processed normally by the cells. These data indicate that although no virus particles are produced from the ΔK391 transfection of BHK cells, both virus transcripts (ΔK391 and S420Y) generate the same amount of labeled protein and that processing of PE2 to E2 proceeds normally in ΔK391-transfected cells.
Transport of virus glycoproteins to the plasma membrane was assayed by evaluating the ability of the virus proteins to induce FFWI using the protocol developed by Mann et al. (26). This event has been shown by our laboratory (26) and others (14) to be mediated by envelope proteins which are expressed at the cell surface. The ability of virus glycoproteins to induce cell-cell fusion in host cell plasma membranes mimics their properties in mature virions and suggests that they have matured properly. E1, which mediates Sindbis virus fusion (20, 31), not only must reach the cell surface but must also fold and oligomerize correctly with PE2 prior to being exported from the rough ER (5).
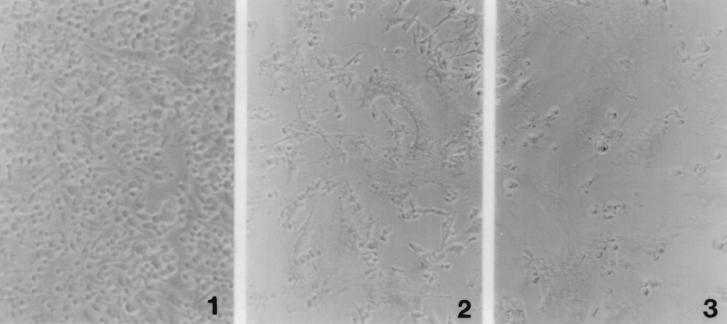
The maturation of ΔK391 proteins was examined in BHK-21 cell monolayers which were transfected with ΔK391 or wild-type RNA and allowed to incubate at 37°C for 10 h. At this time, both cultures, as well as a mock-transfected control culture, were treated with fusion medium at pH 5.3 or control medium at pH 7.2 as detailed in Materials and Methods and returned to pH 7.2 growth medium. Figure 3 shows that the extents of FFWI from ΔK391 and S420Y transfections are roughly equivalent. These data indicate that the defect in virus assembly by the mutant virus is not the result of failure to transport or process the ΔK391 virus glycoproteins. The block is therefore at some stage after virus modification of the plasma membrane.
FIG. 3.
FFWI induced by plasma membrane-localized Sindbis virus glycoproteins. BHK-21 cells transfected with ΔK391, S420Y, or nonviral RNA were processed for fusion at 10 h posttransfection as described in Materials and Methods. Cells were washed with PBS-D and treated for 1 min with pH 5.4 fusion medium at 37°C. Cells were returned to normal medium (pH 7.2) and scored for fusion 1 h later. Panel 1 shows that fusion is not detected in the nonviral RNA-transfected cells. Panels 2 and 3 show the levels of fusion seen in the ΔK391 and S420Y transfections, respectively. The ΔK391- and S420Y-transfected cells demonstrate comparable high levels of cell fusion.
Nucleocapsids do not associate with membranes modified with protein from ΔK391.
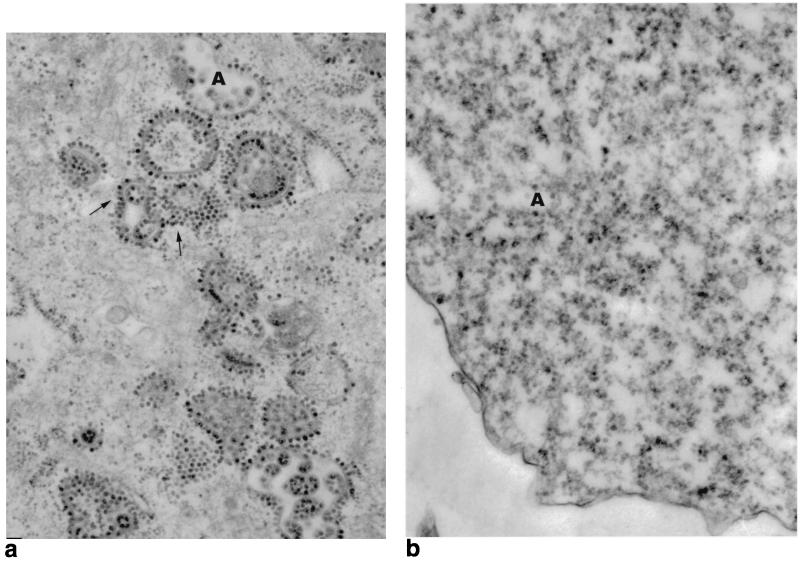
Nucleocapsid structures can be seen by TEM to accumulate along virus-modified plasma membranes at intermediate to late times after infection (4). Toward the later part of the infection, nucleocapsids are seen accumulating on internal vesicles, probably from an intracellular accumulation of modified membrane. Binding of nucleocapsids to modified membranes is a requisite step in the envelopment process. Failure of ΔK391 to bind nucleocapsids would result in the failure to produce virions. Liu and Brown (23) reported that binding of nucleocapsids to virus protein-modified membranes involves at least two steps, extraction and reorientation of the E2 tail, mediated by or concomitant with phosphorylation of the endodomain TPY motif, followed by dephosphorylation. Failure to dephosphorylate wild-type E2 tails in the presence of the phosphatase inhibitor okadaic acid resulted in binding of the nucleocapsids to modified membrane, but envelopment of the virus particles did not proceed. To examine the nucleocapsid binding phenotype of cells infected with the ΔK391 mutant, mutant and wild-type RNAs were used to transfect BHK-21 cells and the cells were prepared for TEM at 18 h posttransfection. Figure 4 shows micrographs of ΔK391- and S420Y-transfected BHK-21 cells. The wild-type RNA transfection shown in Fig. 4A displays extensive alignment of nucleocapsid structures along modified membranes, while in Fig. 4B, the ΔK391 mutant transfection shows nucleocapsids scattered throughout the cell cytoplasm, not associated with membranes. We concluded that membranes modified by the glycoproteins of Sindbis virus ΔK391 are defective in the ability to bind nucleocapsids.
FIG. 4.
TEM of BHK-21 cells transfected with S420Y (a) or ΔK391 (b) RNA. Cell monolayers were prepared for TEM after 20 h of transfection as described in Materials and Methods. (a) Cell transfected with wild-type S420Y RNA. The cytoplasm contains a number of modified membranes with associated nucleocapsids (arrows). The uppercase A denotes a vesicle containing mature S420Y virus particles. (b) Cell transfected with mutant ΔK391 RNA. Nucleocapsids are not seen attached to membranes but are found free in the cytoplasm (A).
It is interesting that when nucleocapsids fail to associate with cell membranes, the cytoplasmic membranes are not seen. This is the case in the experiments presented here (Fig. 3) and in all of the other experiments we have conducted in which nucleocapsid membrane association does not occur. This observation has been made with Sindbis virus temperature-sensitive mutant ts23 (4), with inhibitors which block the reorganization of the E2 tail (23), and with mutations produced in the E2 tail by molecular cloning (22). It appears that C-membrane interaction may stimulate the formation of these membrane structures. The source of these membranes is also unknown. It seems that they do not derive from the ER, as the E2 tail is not exposed or available for binding of C as long as the proteins are in the ER (24).
The E2 tail of the ΔK391 mutant is reoriented into the cytoplasm.
We have previously shown that an E2 double mutant, T398A/Y400N, defective in both potential phosphorylation sites in the E2 endodomain failed to withdraw the distal amino acids of the E2 endodomain from the membrane bilayer (22, 24). The failure to expose this sequence of amino acids to the cell cytoplasm blocked the binding of nucleocapsids and virus maturation. The inability of the ΔK391 mutant to bind nucleocapsids could, therefore, be the result of a failure of this mutant to have the hydrophobic C terminus of the E2 tail extracted from the virus-modified cell membrane. ΔK391 was constructed from Toto 1101 carrying tyrosine at position 420 (S420Y) as described above. Position 420 in the E2 tail is initially oriented toward the lumen of the ER and is proximal to the signalase cleavage site. Prior to virus budding, the E2 tail must be extracted from the membrane to become exposed to the cell cytoplasm for envelopment to occur (24). The introduction of the silent mutation S420Y makes it possible to determine the orientation of this part of the E2 endodomain relative to the membrane bilayer by determining if Y420 is exposed for iodination by a membrane-impermeable iodinating agent.
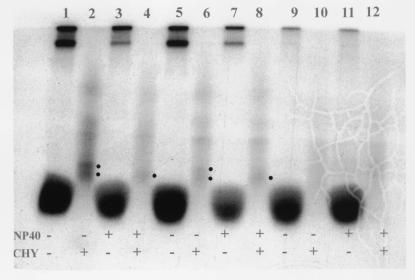
To test the orientation of the E2 tail in cells transfected with the ΔK391 mutant, transfected monolayers were assayed as detailed in Materials and Methods. Cycloheximide was added to the monolayers for 1 h at 16 h posttransfection to inhibit protein synthesis and allow viral proteins to be exported from the rough ER. Transfected BHK cells expressing ΔK391, wild-type virus, or nonvirus control RNA were processed for iodination as described in Materials and Methods (24). Cell monolayers were harvested using 4°C lifting buffer, and membrane vesicles containing the virus membrane glycoproteins were prepared by homogenization of cells through a 27-gauge needle. Iodination of the cell extracts was done in the presence or absence of the detergent NP-40 using Iodo-beads, a membrane-impermeable iodination reagent. The E2 Y420 tail has two tyrosine residues (Y400 and Y420) which can be iodinated once the tail has been extracted from the cell membrane. To determine which positions are iodinated, a chymotryptic digest (cleaving on the C-terminal side of Y) was made of the iodinated E2 tail and a peptide fragment of 2,087 Da was immunoprecipitated and resolved on a Tricine gel. The results of this experiment are shown in Fig. 5 and demonstrate that although the ΔK391 mutant does not bind nucleocapsids, the E2 tail is extracted from the membrane. As shown in lanes 2 and 4 of the wild-type transfection, the fragment containing Y420 is visible without (lane 2) or with (lane 4) NP-40, demonstrating that the tail is extracted and accessible to label without detergent treatment. As is also demonstrated, the mutant ΔK391 displays the same peptide fragments upon treatment with chymotrypsin as does the wild type, shown in lanes 6 (without NP-40) and 8 (with NP-40). Identical processing of transfections with nonviral RNA, lanes 9 to 12, did not generate any of the peptides seen in the virus-transfected lanes.
FIG. 5.
Translocation of the carboxy terminus of the E2 glycoprotein from the plasma membrane occurs in the ΔK391 mutant. BHK-21 cells were transfected with ΔK391, S420Y, or nonviral control RNA, and membrane vesicles were prepared at 20 h posttransfection as described in Materials and Methods. Cell vesicles were iodinated with Na125I in the presence (+) or absence (−) of 0.5% NP-40. Iodinated samples were immunoprecipitated once with anti-whole-virus Ab, digested with chymotrypsin (CHY) (+) or not digested (−), and precipitated a second time with an Ab specific for the 33-amino acid E2 tail. Precipitated E2 proteins and digests were run on a 16.5% Tricine gel. Samples in even-numbered lanes were treated with chymotrypsin, while those in odd-numbered lanes remained undigested. Lanes 1 to 4 represent iodinated E2 from S420Y-transfected cells. As seen in lanes 2 and 4, peptides corresponding to the correct molecular weights are highlighted by the closed circles. Lanes 5 to 8 represent iodinated E2 from the ΔK391 mutant transfections. Lanes 6 and 8 show peptides corresponding to the translocated E2 tail, which are also highlighted by closed circles. Lanes 9 to 12 contain nonviral-RNA controls and demonstrate that no nonspecific binding of cellular proteins was immunoprecipitated by the Abs used. The presence of NP-40 had no effect on iodination of the tail, since these proteins have left the rough ER (lanes 3 and 4, S420Y; lanes 7 and 8, ΔK391). Digestion with chymotrypsin under the conditions employed produced partial digests of the E2 tail, as can be seen by the peptide fragment corresponding to the fully cleaved tail (A401 to Y 420; 2,087 Da) or the partially digested fragment (A401 to A423; 2,268 Da; highlighted by the lower and upper closed circles, respectively). Even-numbered lanes were subjected to a second immune precipitation after chymotrypsin treatment, which eliminated the low-molecular-weight label migrating near the dye front in odd-numbered lanes. The odd-numbered lanes received equal counts of nondigested sample, and the even-numbered lanes received equal counts of digested sample.
Cells transfected with the RNA of ΔK391 do produce virus, albeit 5 orders of magnitude less than those transfected with the RNA of the wild type. This suggests that there is limited interaction of C protein with E2, allowing virus assembly. It is possible, therefore, that inefficient exposure of the E2 tail to the cell cytoplasm occurs in the mutant, but all of the E2 tail that is correctly configured is incorporated into virus. Although the amount of label in the E2 peptide containing Y420 is less in the mutant than in the wild type, it is not reduced enough to account for the 5-order-of-magnitude reduction of virus production seen in the mutant-transfected cells. The data suggest that most of the E2 which has reoriented the E2 tail to the cytoplasm is not incorporated into virus. The reduced amount of label may therefore result from a somewhat reduced efficiency of reorganization of the E2 endodomain. These data demonstrate that although nucleocapsids do not attach to the membranes modified with ΔK391 proteins, the specific defect in assembly is due not to an inability of the E2 tail to reorient but to an additional, unidentified step in the envelopment process.
We have previously shown that extraction of the E2 tail correlates with its phosphorylation (23). To determine if the E2 protein of mutant ΔK391 is subjected to phosphorylation, transfected BHK cells were treated with okadaic acid and phosphorylation was determined as described previously (22, 23) and in Materials and Methods. PAGE analysis revealed equal phosphorylation levels in ΔK391 and S420Y transfections (data not shown), indicating that the deletion of E2 K391 does not effect this process.
The ΔK391 mutant produces wild-type levels of virus when grown in A. albopictus U4.4 cells.
Insect and mammalian cells differ dramatically in their physiological, genetic, and biochemical properties. Dramatic differences have been demonstrated both in the way Sindbis virus replicates in these two cell types and in the manner with which the two cell types respond to virus infection (9, 10, 12, 15–17, 27, 35, 38, 42). Because of the striking differences in the virus-cell interactions in these two cell types, we examined the phenotype of the mutant ΔK391 in the insect host cell.
To assess the growth of the mutant ΔK391 in U4.4 cells, we transfected both mutant and wild-type RNAs into mosquito U4.4 cells as described in Materials and Methods. The results are shown in Table 2. Whereas ΔK391 produced very little virus from BHK cells, compared to wild-type virus, yields of this mutant were at wild-type levels when the virus was grown in mosquito cells. Thus, the defect in assembly resulting from the deletion of E2 K391 is a host range mutation.
TABLE 2.
Production of ΔK391 virus in RNA-transfected BHK-21 and U4.4 cellsa
| Virusb | Virus production (PFU/ml) from:
|
|||
|---|---|---|---|---|
| BHK-21 cells
|
U4.4 cells
|
|||
| BHK-21 indicator cells | C7-10 indicator cells | BHK-21 indicator cells | C7-10 indicator cells | |
| ΔK391 | 3.0 × 103 | 8.0 × 104 | 1.0 × 108 | 2.0 × 109 |
| S420Yc | 1.5 × 109 | 1.5 × 109 | 5.0 × 108 | 1.0 × 109 |
| SVHRd | 2.0 × 109 | 1.5 × 109 | ||
Virus was harvested 16 h posttransfection in the case of BHK-21 cells and 28 h posttransfection in the case of U4.4 cells.
RNA transcripts from a cDNA clone of either mutant were used to transfect BHK-21 or U4.4 cells. Virus titers were determined in parallel on BHK-21 or mosquito C7-10 cells.
S420Y contains an S→Y mutation at position 420 in E2 and is the parent of the ΔK391 mutant.
SVHR is the heat-resistant strain of Sindbis virus.
Titers of virus produced from both vertebrate and invertebrate cell types were determined on BHK-21 or C7-10 cells. The virus titer produced by the ΔK391 mutant in C7-10 cells was consistently higher, regardless of the host cell used to produce it. The wild-type SVHR strain of Sindbis virus and the S420Y cDNA clone used as the wild type show equivalent titers on BHK-21 or C7-10 cells. The ΔK391 mutant, however, produces virus particles which display differential titers from the two indicator cell lines used. Titers of mosquito-grown ΔK391 on BHK cells are 5% of that recorded on A. albopictus cells. Despite its host-specific restriction in assembly, ΔK391 can infect BHK-21 cells and the resulting low level of progeny virus forms plaques on BHK-21 cells at about 4% of the level obtained with C7-10 cells. This effect is not the result of a difference in the level of plaque-forming efficiency in these two cell lines, as seen from the titers of the SVHR and S420Y strains in both BHK-21 and C710 cells (Table 2). These results were not due to instability of the virus or an effect of temperature sensitivity (data not shown). The progeny virus recovered from ΔK391 RNA-transfected BHK or U4.4 cells was found to retain the mutant sequence by sequencing of RT PCR products, and the ΔK391 plaques from BHK-21 cells retained the large-plaque phenotype of the S420Y parent (data not shown). Collectively, these data indicate that the mutant ΔK391 is restricted in the ability to produce infectious virions in a host-dependent fashion.
DISCUSSION
Structural details of Sindbis virus have been obtained by electron cryomicroscopy of intact virions and by X-ray crystallography of expressed C protein (7, 19, 32, 33). These studies show Sindbis virus to have a highly ordered structure consisting of nested interconnected T=4 icosahedra with an associated membrane. Our laboratory has investigated the assembly of this structure for over 30 years, and we have particularly focused on the specific structural requirements for assembly. An important event and interaction in Sindbis virus structure and assembly are maturation of the E2 cytoplasmic tail and its binding interaction with the nucleocapsid, respectively. This interaction plays a pivotal role in initiating C envelopment in the virus-modified host cell membrane. The endodomain of Sindbis virus E2 is 33 amino acids in length and contains the sequence KARRECLTPYALAPNAVIPTSLALLCCVRSANA (amino acids 391 to 423, Fig. 1) (36). The carboxy terminus is a multifunctional domain (Fig. 1) and contains the hydrophobic signal sequence for the integration of the adjacent 6,000-molecular-weight (6K) protein into the ER, as well as the signalase cleavage site which separates PE2 from 6K. Algorithms which predict protein sequences as existing as transmembranal domains (39) place amino acids N405 to V418 of PE2 in the ER membrane.
The E2 endodomain sequence TPYALAPNA has been implicated as critical for the binding of C protein by molecular modeling and genetic analysis (13, 22, 25). We have demonstrated that the sequence LAPNA beginning at position L402 is not essential for virus growth in either BHK-21 or U4.4 cells (22), even though L402, P404, and A406 are completely conserved among the alphaviruses. The reason why this deletion is not lethal may be that the hydrophobic properties of this sequence and those of the immediately following V407IPTS sequence are redundant and this sequence may perform the function of LAPNA in C binding. The TPY domain is also completely conserved and contains T398 and Y400, which are both possible substrates for phosphorylation, a modification which occurs concurrently with extraction of the tail domain (23). These residues are implicated as critical for C binding through an aromatic interaction with C Y180 and W247 (19, 44) and E2 Y400.
These data suggest that recognition and binding of the nucleocapsid by the E2 tail involve more than one domain in the E2 tail. The region involved in binding specificity mapped genetically to the E2 carboxy terminus and comprises one domain, while current models predict that the TPYAL region is bound in the C protein binding pocket (19), predicting a second C protein binding domain. Collectively, these data begin to outline several overlapping structural and functional domains contained in the E2 tail (Fig. 1). These domains include (i) tail extraction, (ii) phosphorylation-dephosphorylation, (iii) nucleocapsid binding specificity, (iv) a redundant and nonessential sequence, and (v) a signal for 6K integration. Some of these domains appear to function in vastly different contexts in mammalian and insect cells.
The phenotypic difference between the growth of the ΔK391 mutant in a vertebrate host and its growth in an invertebrate host is very significant. We detected very little mutant virus by plaque assay and none by metabolic labeling of BHK-21 cells, while wild-type virus levels were produced by this deletion mutant in mosquito cells. Although little virus is produced in vertebrate cells, all structural proteins are synthesized and trafficked normally. Membrane glycoproteins are processed normally, and E1 is functional for fusion; however, virus envelopment does not take place in the vertebrate cell.
Among the many physiological and biochemical differences between mammalian and insect cells are the differences in cell membranes. Insect and mammalian cell membranes differ in lipid composition, and their physical and structural properties differ because of the differences in composition. The mosquito cell membrane, therefore, presents an environment to the Sindbis virus glycoproteins which differs greatly from that in the mammalian cell. Insect cells do not synthesize cholesterol (8, 29), and it is unclear how much, if any, dietary cholesterol is incorporated into insect membranes. Cholesterol (3, 8) is known to cause cell membranes to thicken, and the degree of thickening is proportional to the amount of cholesterol present (8). The addition of an equimolar amount of cholesterol to phosphatidylcholine, reflecting roughly the plasma membrane composition in vertebrate cells, increases the bilayer thickness from 25 to 31Å (8). The presence of cholesterol also increases membrane viscosity and reduces membrane permeability.
As the membrane bilayer is the major unique host cell component in the envelopment process, it appears that deletion of the amino acid K at E2 position 391 affects the interaction(s) of the E2 tail with the cell membranes of vertebrate and invertebrate hosts in a differential manner. Deletion of E2 K391 at the membrane junction leaves A392 at the membrane interface. It is plausible that, in BHK-21 cells, this aliphatic amino acid is pulled into the membrane, leaving the basic arginine pair (E2 R393-R394) juxtaposed with the membrane. In this scenario, the spanning distance from the membrane to the TPYAL nucleocapsid binding motif is shortened not by one amino acid but effectively by two. This circumstance may render the distance from the membrane to the TPYAL sequence insufficient to reach the critical binding sequences in the hydrophobic pocket in the C protein, preventing the second nucleocapsid binding step. However, this model predicts that the first binding step should still occur. In fact, nucleocapsid binding is not observed in ΔK391-infected BHK-21 cells. This is not a result of failure of the ΔK391 mutant to perform the functions related to the exposure of the first-step binding domain to the cell cytoplasm (see above). It is possible that the misconfiguration of the E2 transmembrane domain prevents some critical configuring of the trimeric structure of the spike structure at the cytoplasmic side of this structure. This alteration may prevent C binding in BHK cells. The observation that a small amount of virus (<1 PFU/cell) is produced by ΔK391-transfected cells may be explained if association with C stabilizes an assembly-competent form of the spike glycoprotein which, in the absence of C binding, is rapidly converted into a defective configuration.
Because of the differences in composition of the insect cell membrane, this hypothesized misconfiguration does not occur and sufficient E2 endodomain is available to allow efficient virus assembly to proceed in this host. These data suggest that exposure of the E2 tail carboxy terminus to the cell cytoplasm is not a sufficient condition for binding of nucleocapsids to virus-modified membranes and that other requirements are placed upon the virus spike glycoprotein complex for the conditions required for nucleocapsid binding to be met. These data imply that membrane composition plays critical roles in establishing these conditions. This hypothesis is being tested in our laboratory.
ACKNOWLEDGMENTS
This research was supported by a grant from The Foundation for Research, Carson City, Nev., and by grant AI42775 from the National Institutes of Health.
REFERENCES
- 1.Anthony R P, Brown D T. Protein-protein interactions in an alphavirus membrane. J Virol. 1991;65:1187–1194. doi: 10.1128/jvi.65.3.1187-1194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony R P, Paredes A M, Brown D T. Disulfide bonds are essential for the stability of the Sindbis virus envelope. Virology. 1992;190:330–336. doi: 10.1016/0042-6822(92)91219-k. [DOI] [PubMed] [Google Scholar]
- 2a.Bonner W M, Laskey R A. A film detection method for tritium-labeled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974;46:83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- 3.Bretscher M. Cholesterol and the Golgi apparatus. Science. 1993;261:1280. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 4.Brown D T, Smith J F. Morphology of BHK-21 cells infected with Sindbis virus temperature-sensitive mutants in complementation groups D and E. J Virol. 1975;15:1262–1266. doi: 10.1128/jvi.15.5.1262-1266.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carleton M, Brown D T. Events in the endoplasmic reticulum abrogate the temperature sensitivity of Sindbis virus mutant ts23. J Virol. 1996;70:952–959. doi: 10.1128/jvi.70.2.952-959.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carleton M, Lee H, Mulvey M, Brown D T. Role of glycoprotein PE2 in formation and maturation of the Sindbis virus spike. J Virol. 1997;71:1558–1566. doi: 10.1128/jvi.71.2.1558-1566.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi H K, Tong L, Minor W, Dumas P, Boege U, Rossmann M G, Wengler G. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature. 1991;354:37–43. doi: 10.1038/354037a0. [DOI] [PubMed] [Google Scholar]
- 8.Clayton R B. The utilization of sterols by insects. J Lipid Res. 1964;5:3–19. [PubMed] [Google Scholar]
- 9.Condreay L D, Adams R H, Edwards J, Brown D T. Effect of actinomycin D and cycloheximide on replication of Sindbis virus in Aedes albopictus (mosquito) cells. J Virol. 1988;62:2629–2635. doi: 10.1128/jvi.62.8.2629-2635.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Condreay L D, Brown D T. Exclusion of superinfecting homologous virus by Sindbis virus-infected Aedes albopictus (mosquito) cells. J Virol. 1986;58:81–86. doi: 10.1128/jvi.58.1.81-86.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coombs K, Brown D T. Organization of the Sindbis virus nucleocapsid as revealed by bifunctional cross-linking agents. J Mol Biol. 1987;195:359–371. doi: 10.1016/0022-2836(87)90657-7. [DOI] [PubMed] [Google Scholar]
- 12.Erwin C, Brown D T. Requirement of cell nucleus for Sindbis virus replication in cultured Aedes albopictus cells. J Virol. 1983;45:792–799. doi: 10.1128/jvi.45.2.792-799.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaedigk-Nitschko K, Schlesinger M J. Site-directed mutations in Sindbis virus E2 glycoprotein's cytoplasmic domain and the 6K protein lead to similar defects in virus assembly and budding. Virology. 1991;183:206–214. doi: 10.1016/0042-6822(91)90133-v. [DOI] [PubMed] [Google Scholar]
- 14.Gallaher W R, Levitan D B, Kerwin K S, Blough H A. Molecular and biological parameters of membrane fusion. London, England: Academic Press, Inc.; 1980. [Google Scholar]
- 15.Gliedman J B, Smith J F, Brown D T. Morphogenesis of Sindbis virus in cultured Aedes albopictus cells. J Virol. 1975;16:913–926. doi: 10.1128/jvi.16.4.913-926.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karpf A R, Blake J M, Brown D T. Characterization of the infection of Aedes albopictus cell clones by Sindbis virus. Virus Res. 1997;50:1–13. doi: 10.1016/s0168-1702(97)01461-5. [DOI] [PubMed] [Google Scholar]
- 17.Karpf A R, Brown D T. Comparison of Sindbis virus-induced pathology in mosquito and vertebrate cell cultures. Virology. 1998;240:193–201. doi: 10.1006/viro.1997.8914. [DOI] [PubMed] [Google Scholar]
- 18.Lee H, Brown D T. Mutations in an exposed domain of Sindbis virus capsid protein result in the production of noninfectious virions and morphological variants. Virology. 1994;202:390–400. doi: 10.1006/viro.1994.1355. [DOI] [PubMed] [Google Scholar]
- 19.Lee S, Owen K E, Choi H K, Lee H, Lu G, Wengler G, Brown D T, Rossmann M G, Kuhn R J. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure. 1996;4:531–541. doi: 10.1016/s0969-2126(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 20.Levy-Mintz P, Kielian M. Mutagenesis of the putative fusion domain of the Semliki Forest virus spike protein. J Virol. 1991;65:4292–4300. doi: 10.1128/jvi.65.8.4292-4300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lilijestrom P, Garoff H. Internally located cleavable signal sequences direct the formation of Semliki Forest virus membrane proteins from a polyprotein precursor. J Virol. 1991;65:147–154. doi: 10.1128/jvi.65.1.147-154.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L N, Lee H, Hernandez R, Brown D T. Mutations in the endo domain of Sindbis virus glycoprotein E2 block phosphorylation, reorientation of the endo domain, and nucleocapsid binding. Virology. 1996;222:236–246. doi: 10.1006/viro.1996.0414. [DOI] [PubMed] [Google Scholar]
- 23.Liu N, Brown D T. Phosphorylation and dephosphorylation events play critical roles in Sindbis virus maturation. Virology. 1993;196:703–711. doi: 10.1006/viro.1993.1527. [DOI] [PubMed] [Google Scholar]
- 24.Liu N, Brown D T. Transient translocation of the cytoplasmic (endo) domain of a type I membrane glycoprotein into cellular membranes. J Cell Biol. 1993;120:877–883. doi: 10.1083/jcb.120.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez S, Yao J-S, Kuhn R J, Strauss E G, Strauss J H. Nucleocapsid-glycoprotein interactions required for assembly of alphaviruses. J Virol. 1994;68:1316–1323. doi: 10.1128/jvi.68.3.1316-1323.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mann E, Edwards J, Brown D T. Polycaryocyte formation mediated by Sindbis virus glycoproteins. J Virol. 1983;45:1083–1089. doi: 10.1128/jvi.45.3.1083-1089.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller M L, Brown D T. Morphogenesis of Sindbis virus in three subclones of Aedes albopictus (mosquito) cells. J Virol. 1992;66:4180–4190. doi: 10.1128/jvi.66.7.4180-4190.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitsuhashi J, Maramorosch K. Leafhopper tissue culture: embryonic, nymphal and imaginal tissues from aseptic insects. Contrib Boyce Thompson Inst. 1964;22:435. [Google Scholar]
- 29.Mitsuhashi J, Nakasone S, Horie Y. Sterol-free eukaryotic cells from continuous cell lines of insects. Cell Biol Int Rep. 1983;7:1057–1062. doi: 10.1016/0309-1651(83)90011-5. [DOI] [PubMed] [Google Scholar]
- 30.Mulvey M, Brown D T. Assembly of the Sindbis virus spike protein complex. Virology. 1996;219:125–132. doi: 10.1006/viro.1996.0229. [DOI] [PubMed] [Google Scholar]
- 31.Omar A, Koblet H. Semliki Forest virus particles containing only E1 envelope glycoprotein are infectious and can induce cell-cell fusion. Virology. 1988;166:17–23. doi: 10.1016/0042-6822(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 32.Paredes A M, Brown D T, Rothnagel R, Chiu W, Schoepp R J, Johnston R E, Prasad B V. Three-dimensional structure of a membrane-containing virus. Proc Natl Acad Sci USA. 1993;90:9095–9099. doi: 10.1073/pnas.90.19.9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paredes A M, Heidner H, Thuman-Commike P, Prasad B V V, Johnston R E, Chiu W. Structural localization of the E3 glycoprotein in attenuated Sindbis virus mutants. J Virol. 1998;72:1534–1541. doi: 10.1128/jvi.72.2.1534-1541.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paredes A M, Simon M N, Brown D T. The mass of the Sindbis virus nucleocapsid suggests it has T = 4 icosahedral symmetry. Virology. 1992;187:329–332. doi: 10.1016/0042-6822(92)90322-g. [DOI] [PubMed] [Google Scholar]
- 35.Renz D, Brown D T. Characteristics of Sindbis virus temperature-sensitive mutants in cultured BHK-21 and Aedes albopictus (mosquito) cells. J Virol. 1976;19:775–781. doi: 10.1128/jvi.19.3.775-781.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice C M, Bell J R, Hunkapiller M W, Strauss E G, Strauss J H. Isolation and characterization of the hydrophobic COOH-terminal domains of the Sindbis virion glycoproteins. J Mol Biol. 1982;154:355–378. doi: 10.1016/0022-2836(82)90069-9. [DOI] [PubMed] [Google Scholar]
- 37.Rice C M, Levis R, Strauss J H, Huang H V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987;61:3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riedel B, Brown D T. Novel antiviral activity found in the media of Sindbis virus-persistently infected mosquito (Aedes albopictus) cell cultures. J Virol. 1979;29:51–60. doi: 10.1128/jvi.29.1.51-60.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rost B, Casadio R, Fariselli P, Sander C. Transmembrane helices predicted at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarkar G, Sommer S S. The “megaprimer” method of site directed mutagenesis. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 41.Scheefers H, Scheefers-Borchel U, Edwards J, Brown D T. Distribution of virus structural proteins and protein-protein interactions in plasma membrane of baby hamster kidney cells infected with Sindbis or vesicular stomatitis virus. Proc Natl Acad Sci USA. 1980;77:7277–81. doi: 10.1073/pnas.77.12.7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheefers-Borchel U, Scheefers H, Edwards J, Brown D T. Sindbis virus maturation in cultured mosquito cells is sensitive to actinomycin D. Virology. 1981;110:292–301. doi: 10.1016/0042-6822(81)90061-1. [DOI] [PubMed] [Google Scholar]
- 43.Singh K R, Pavri K M. Experimental studies with chikungunya virus in Aedes aegypti and Aedes albopictus. Acta Virol. 1967;11:517–526. [PubMed] [Google Scholar]
- 44.Skoging U, Vihinen M, Nilsson L, Liljestrom P. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure. 1996;4:519–529. doi: 10.1016/s0969-2126(96)00058-5. [DOI] [PubMed] [Google Scholar]
- 45.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]