Abstract
The filamentous bacteriophage infects Escherichia coli by interaction with the F pilus and the TolQRA complex. The virus-encoded protein initiating this process is the gene 3 protein (g3p). The g3p molecule can be divided into three different domains separated by two glycine-rich linker regions. Though there has been extensive evaluation of the importance of the diverse domains of g3p, no proper function has so far been assigned to these linker regions. Through the design of mutated variants of g3p that were displayed on the surface of bacteriophage, we were able to elucidate a possible role for the distal glycine-rich linker region. A phage that displayed a g3p comprised of only the N1 domain, the first linker region, and the C-terminal domain was able to infect cells at almost the same frequency as the wild-type phage. This infection was proven to be dependent on the motif between amino acid residues 68 and 86 (i.e., the first glycine-rich linker region of g3p) and on F-pilus expression.
Filamentous bacteriophage is a nonlytic DNA virus that infects Escherichia coli cells harboring an F episome (19, 20, 26). In order for bacteriophage infection to occur, including the translocation of the single-stranded phage DNA over the cell membrane, the expression of two different receptor systems in the gram-negative host cell is critical. The primary receptor, i.e., the F pilus, mediates the adsorption of the phage to the bacterial cell (6, 12). This interaction can take place at a long distance from the cell surface, since the pilus molecule extrudes away from the bacteria. Through the depolymerization of the pilus, the bacteriophage is brought close to the cell surface of the host, where the interaction with the coreceptor takes place (5, 31). The coreceptor, i.e., the TolQRA complex, is localized in the inner membrane and the periplasmic space and is attached to the inside of the outer membrane (9, 16). It was recently shown that it is the C-terminal part of the TolA domain that interacts with the bacteriophage adsorption protein (7, 27). Upon binding to the TolA domain, the viral DNA translocates into the host cell by an as-yet-unknown mechanism.
Expression of the gene 3 viral adsorption protein, g3p, on the surface of the bacteriophage is essential for normal infection to occur (1). The minor coat protein g3p is located together with g6p in three to five copies at one end of the filamentous phage (10). Mature g3p consists of 406 amino acid residues separated in several distinct regions (2). The N-terminal part can be divided into two different domains, N1 and N2, mediating penetration and adsorption during infection, respectively (1, 30). The C-terminal part is responsible for the interaction with g6p, thereby anchoring the g3p in the membrane of the phage coat (1, 14, 21). Furthermore, the g3p has a crucial role in the assembly of the filamentous phage, since in the absence of g3p, the proper termination of the assembly and the following release of the phage will not take place (24).
Two glycine-rich linker regions divide the three domains in g3p. The linkers are comprised of amino acid residues 68 to 86 and 218 to 256 of the mature protein. The first linker consists of repeats of the sequence Glu-Gly-Gly-Gly-Ser, and the second linker consists of repeats of the sequence Gly-Gly-Gly-Ser. There have been reports showing that the first linker can have an effect on the outer membrane, resulting in β-lactamase leakage, impaired F pili, and tolerance to certain colicins (4, 30). Apart from these observations, no proper function has so far been assigned to these regions, which are believed to mainly convey flexibility to the other domains of g3p.
In order to elucidate the functions of the distinct domains in g3p, including the glycine-rich linker regions, differently mutated versions of g3p were designed and displayed on the surface of the bacteriophage. These engineered variants were analyzed for their functional role in the infection process. Our data indicate a new and significant function for the first glycine-rich stretch of g3p.
MATERIALS AND METHODS
Bacterial strains.
The following E. coli strains were used for the cloning, propagation and infection experiments: strain Top 10F′ [F′ lacIq Tn10 (Tetr) mcrAΔ(mrr-hsdRMS-mrcBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG], strain XL1-Blue [F′::Tn10 proA+B+ lacIq Δ(lacZ)M15/recA1 endA1 gyrA96 (Nalr) thi hsdR17 (rK− mk+) supE44 relA1 lac], strain K561 (HfrC λ+ relA1 spoT1 T2r(ompF627 fadL701) lacIq), strain BL21 DE3 (F− ompT[lon] hsdSB (rb− mb−) (an E. coli B strain), and strain K17tolADE3 (F− C600 Strr lac), which was a generous gift from R. E. Webster (29).
Bacteriophage.
The R408 g3p-deleted helper DNA bacteriophage (R408d3) and the R408 wild-type DNA bacteriophage were kindly supplied by J. Rakonjac (23). The R408d3 helper phage was propagated in E. coli K1762, which is strain K561 transformed with the vector constructs pJARA112 and pJARA131. The two vector systems complement the deleted helper phage through the expression of g3p, which is under the control of a phage shock protein promoter (psp). The psp promoter is activated in the presence of bacteriophage g4p, and is therefore silent until the moment of infection. The RNA bacteriophage MS2 and the fd-D13 phage were kind gifts from K. Fridborg and P. Holliger, respectively.
Vector constructs.
The engineered g3p mutants were constructed by using the Chlr vector pAM18 (pACYC184 derived) and the g3p gene sequence from bacteriophage R408 (18, 28). The expression from the pAM18 vector is under the control of a lac promoter. The different g3p gene sequences were amplified by using specific oligonucleotides containing appropriate restriction sites. The N1-linker 1 (N1L1) construct is comprised of residues 1 to 86 of g3p, where residues 1 to 67 correspond to the N1 domain and residues 68 to 86 correspond to the first glycine-rich linker region (L1). In order to anchor the fragment in the phage coat, N1L1 was fused to the C-terminal domain (CT) corresponding to residues 257 to 406 of g3p. The whole fragment was amplified by using the sequences 5′ CCCGAGCTCGTGAAAAAATTATTATTCGCAATTCTT and 3′ CCCAAGCTTTTAAGACTCCTTATTACGCAGTATGTTAGC as external primers and sequences 5′ GGTTCCGGTGATTTTGATTATGAAAAGATGGCAAAC and 3′ ATAATCAAAATCACCGCCACCCTCAGAACCGCCACCCTCAGAACCGCCACCC TCAGAGCCACCACCC TCAT T T TCAGGGAT as internal primers. The amplified fragment was cloned into the pAM18 vector by using restriction sites SacI and HindIII. The N2-linker 2 (N2L2) construct is comprised of residues 87 to 406 of g3p. Included in this fragment is the N2 domain (residues 87 to 217), the second glycine-rich linker region (L2; residues 218 to 256), and the CT domain (residues 257 to 406). The fragment was amplified by using the sequences 5′ CCCGAGCTCGTGAAAAAATTATTATTCGCAAT TCCT T TAGT TGTTCCT T TCTAT TCTCACTCCACTAAACCTCCTGAGTACGGTGAT and 3′ CCCAAGCTTTTAAGACTCCTTATTACGCAGTATGTTAGC as primers. The amplified fragment was cloned into the pAM18 vector by using restriction sites SacI and HindIII. All the g3p mutant constructs contained the g3p leader sequence in order to direct the protein to the periplasmic space.
The PCR amplification was performed for 25 cycles for 30 s of denaturation at 94°C, 30 s of annealing at 55°C, and 2 min of extension at 72°C and a final extension for 7 min. The same PCR program was used for all the amplifications.
After cloning into the pAM18 vector, the ligations were transformed into chemically competent Top 10F′ cells. All constructs were verified by DNA sequencing by using the BigDye Terminator Cycle Sequencing kit (Perkin-Elmer, Elmerville, Calif.).
Protein constructs.
Four different protein constructs, consisting of the N1 (residues 1 to 67), the N1L1 (residues 1 to 86), the N2 (residues 87 to 217), and the N1L1N2 (residues 1 to 217) domains, respectively, were prepared for inhibition analysis. All of the constructs were preceded by the g3p signal peptide and were followed by two alanine and six histidine residues. The constructs were cloned into the pUC19 expression vector by using appropriate restriction sites and were verified by DNA sequencing with the BigDye Terminator Cycle Sequencing kit (Perkin Elmer).
The protein constructs were transformed into chemically competent Top 10F′ cells then expressed and purified from the E. coli periplasmic space (QIAGEN, Ltd., Crawley, United Kingdom). Isolated and concentrated proteins were adsorbed onto Ni nitriloacetic acid agarose (QIAGEN) and were eluted in 50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, and 250 mM imidazole. In order to determine the purity of the samples, the preparations were separated on a 12.5% polyacrylamide gel. This was performed in the presence of sodium dodecyl sulphate (SDS) (1%) under nonreducing conditions (15). The samples were subsequently blotted onto a nitrocellulose filter membrane by using a semidry transfer cell (Bio-Rad Laboratories, Hercules, Calif.). The blot was performed at 15 V for 30 min with a limiting current of 0.25 A. An anti-His6 antibody (QIAGEN) was used as the primary antibody. This antibody was detected by using peroxidase-conjugated rabbit-anti-mouse antibody (RAM-P260; Dako A/S, Glostrup, Denmark) followed by chemiluminescence detection with the ECL+ system (Amersham Pharmacia Biotech, Little Chalfont, England). The preparations were more than 90% pure. The concentrations of the samples were determined by using spectrophotometry at 280 nm.
Phage propagation.
Phage stocks were prepared from engineered g3p mutants that were cloned in the pAM18 vector and transformed into Top 10F′ bacterial cells. Fresh overnight cultures from single colonies were diluted and vigorously shaken at 37°C until an optical density at 660 nm (OD660) of 0.4 was reached. The cultures were maintained in Terrific broth (TB) medium (10 g of Bacto tryptone, 1 g of yeast extract, 4 g of NaCl, and 1 g of glucose per liter) supplemented with 15 μg of chloramphenicol per ml, 10 μg of tetracycline per ml, and 1% glucose. At the time of logarithmic phase, the cells were infected with R408d3 helper phage at an input ratio of 10 to 100 phages per cell and 1.0 mM of isopropyl-β-d-thiogalactopyranoside (IPTG) was added. The phage infection was carried out for 20 min at 37°C without shaking. Unbound input phage was removed by centrifugation for 10 min at 2,000 × g. The pellet was resuspended in fresh TB medium containing tetracycline, chloramphenicol, and 1.0 mM IPTG. The R408d3 helper phage was complemented by using the different g3p mutants for 6 h at 30°C in a shaker incubator. After propagation, the cells were removed by centrifugation at 2,000 × g for 10 min at room temperature. The phage-containing supernatants were filtered through a 0.2-μm-pore-size Dynagard filter (Microgen Inc., Laguna Hills, Calif.). The phage were precipitated overnight at 4°C with 5% polyethylene glycol 6000 and 0.5 M NaCl. After centrifugation at 16,000 × g for 30 min, the phage pellets were resuspended in sterile phosphate-buffered saline (PBS), giving a 100-fold concentration of phage.
Phage titer.
Due to decreased infectivity of the g3p-truncated phage, the titers were determined by using denaturing agarose gels. Free RNA and double-stranded DNA was removed through incubation with RNase A and DpnI for 2 h at 37°C. Virions were disassembled by incubation in SDS-containing buffer (1% SDS, 1× TAE, 5% glycerol, 0.25% bromophenol blue) at 70°C for 20 min. The phage DNA content was then quantified by 0.7% agarose gel electrophoresis. After electrophoresis, the gels were stained in ethidium bromide for 1 h and were destained in MilliQ-water for 1 h to overnight. Wild-type phage DNA with known titer was used to determine the actual amount of single-stranded DNA and thereby determine the titer of the different phage stocks.
Infection experiments.
A logarithmic-phase culture of the suitable bacterial strain in TB medium was used in all infection assays. The number of infecting phages was determined through titration on diverse strains. When using the g3p mutant phage, the proper complementation of the phage with wild-type g3p was crucial in order to visualize PFU. Even if the g3p on the surface of the mutated phage was of various lengths, i.e., N1L1, N2L2, and CT, the packed phage DNA was deleted of the complete gene 3 (R408d3). Strain K1762 that was used for the visualization of plaques allows the proper complementation of g3p through the presence of the vectors pJARA112 and pJARA131. Other strains that were transformed with the pJARA112 and pJARA131 vectors were XL1-Blue, BL21DE3F−, K17tolADE3F−, and K17tolADE3F+. The transformed strains were assayed for infectivity by using R408wt phage. This was in order to see that no leakage of g3p occurred in the absence of filamentous bacteriophage, thereby abolishing the possibility of proper infection through the downregulation of the receptors.
Phages were allowed to infect for 20 min at 37°C in the presence or absence of 50 mM CaCl2. Thereafter, the mixed bacteria and phage samples were plated in soft agar on TB plates without antibiotic. The plates were incubated overnight at 37°C.
The blocking experiments using the N1L1 phage were performed in the presence or absence of the RNA phage MS2. The K1762 cells were preincubated for 10 min with 50 μl of MS2 (stock, 1012 phage/ml) before the addition of different amounts of N1L1 phage.
Inhibition analysis.
Infection of F+TolA+ bacteria (strain K1762) with either N1L1 phage or wild-type phage (VCSM13) was performed in the presence of the different protein constructs N1, N1L1, N2, and N1L1N2, respectively. Different amounts of these proteins were used to assay their inhibitory effects on the infection process. The infections were performed by using 100 μl of a log-phase culture (OD600 of 0.4 to 0.6) of K1762. The bacteria were preincubated with 10 μl of g3p fragments at 37°C for 15 min under moderate shaking (70 rpm). Thereafter, 10-μl samples of different dilutions of phage was added to the mixture. After another 10 min at 37°C and 70 rpm, the samples were plated in soft agar on TB plates without antibiotic.
Pili preparation and enzyme-linked immunosorbent assay.
To elucidate the pili-binding properties of the N1L1 phage, functional F pili were prepared from Top 10F′ bacteria (1). Briefly, the bacteria were grown to high density on rich agar and were subsequently scraped off and stirred gently in ice-cold sterile SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 2 h. Cells were removed through repetitive slow-speed centrifugation steps and were washed in ice-cold SSC buffer. The supernatants were mixed with 10% polyethylene glycol 6000 and 0.5 M NaCl in order to precipitate the pili. After a second precipitation of the pili, the preparation was resuspended in PBS to a concentration of 1 mg of total protein/ml.
The pili preparation was coated onto polystyrene microtiter plates overnight at room temperature, using 100 ng/well. Coated plates were blocked with 0.1% bovine serum albumin in PBS for 1 h at room temperature. After washing, the plates were incubated with dilutions of both the N1L1 phage and the VCSM13 wild-type phage in 100 μl of PBS. The plates were incubated with the phage for 2 h at 37°C. Bound phage was detected by using horseradish peroxidase-conjugated anti-M13 antiserum from Pharmacia (Uppsala, Sweden) diluted 1/5,000 in PBS. Orthophenylene amine and hydrogen peroxide (Sigma, Lund, Sweden) were added to each well, as chromogen and substrate, respectively. The reaction was stopped after 15 min by adding 150 μl of 1 M sulfuric acid, and the absorbance was measured at 490 nm.
RESULTS
Display of different domains of g3p on the phage surface.
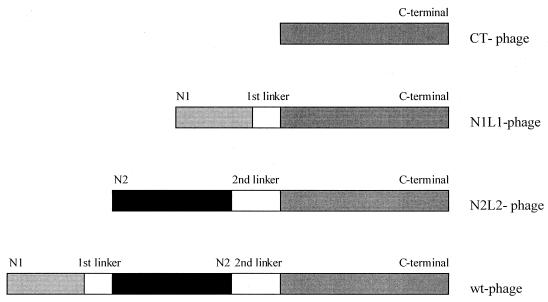
In order to evaluate the function of the different domains of g3p, including the glycine-rich linker regions, different phage constructs were designed. It was important to maintain the normal configuration of the g3p molecule as intact as possible. Three distinct g3p mutants were engineered and were expressed on the phage surface (Fig. 1). The CT phage displays residues 257 to 406 of the g3p molecule. The N1L1 phage displays the N1 domain (residues 1 to 67), together with the first glycine-rich linker region (L1) (residues 68 to 86) and the CT domain (residues 257 to 406). The N2L2 phage expresses the N2 domain (residues 87 to 217), the second glycine-rich linker region (L2), and the CT domain (residues 218 to 406). The titers obtained with these different phage constructs were lower than what is normally obtained with wild-type phage. This is in part due to the use of a low-copy-number plasmid for the engineered g3p molecule. Furthermore, the expression of the g3p domains separately might have a more toxic effect on the host cell than the expression of wild-type g3p (17, 25, 30). The phage stocks complemented with different g3p mutants did not contain any wild-type g3p, as determined by Western blot analysis (data not shown). This was a crucial control, since the phage stocks were propagated by using R408d3 as a helper phage, displaying wild-type g3p on their surface.
FIG. 1.
A schematic representation of the diverse phage constructs. The linker regions indicated in the figure are the naturally occurring first (residues 68 to 86) and second (residues 218 to 256) glycine-rich linkers. The engineered g3p genes are cloned into an expression vector (pAM18chlr) and are complementing the g3p-deleted filamentous R408 phage. wt, wild type.
Mutant N1L1 phage efficiently infects E. coli.
The infection levels of the engineered phage together with wild-type phage were analyzed utilizing different bacterial strains. It was important to determine the infection frequencies on E. coli that were positive or negative for the primary receptor (F pilus) and/or the coreceptor (TolQRA). For that purpose, the E. coli strains K1762 (F+TolA+), BL21DE3 (F−TolA+), K17tolADE3F+ (F+TolA−), and K17tolADE3F+ (F−TolA−) were used (Table 1).
TABLE 1.
Infectivity of different R408 phage constructsa
| Phage | Infectivity of:
|
|||
|---|---|---|---|---|
| F+TolA+ | F−TolA+ | F+TolA− | F−TolA− | |
| N1L1 | 2.6 × 109 (1.0 × 1010) | 1.0 × 102 (1.0 × 1010) | 0 (3.0 × 108) | 0 (3.0 × 108) |
| N2L2 | 0 (3.5 × 108) | 0 (3.5 × 108) | 0 (3.5 × 108) | 0 (3.5 × 108) |
| Wild type | 3.0 × 1012 (5.0 × 1012) | 3.0 × 104 (5.0 × 1012) | 1.5 × 101 (5.0 × 1012) | 1.0 × 101 (5.0 × 1012) |
PFU obtained when infecting bacteria that express F pilus and/or TolA. The number in parentheses represents the total amount of input phage used.
The bacteriophage displaying the N1 domain fused to the first glycine-rich linker region and the CT domain, i.e., N1L1 phage, infects cells almost at the same frequency as the wild-type phage. At an input ratio of 100 phage per cell, 26% of the N1L1 phage could infect bacteria expressing both the F pilus and the TolA protein (compared to 60% of the wild-type phage). The N1L1 phage was dependent on F pilus for infection, since without the F pilus, no infection was detected with 1.0 × 1010 phage. For infection to occur, an interaction was required between the N1 domain and its corresponding coreceptor complex, TolQRA. This was evident since infection of TolA− bacterial cells with N1L1 phage resulted in no plaques.
The phage displaying the N2 domain in conjunction with the second glycine-rich linker region and the CT domain, i.e., N2L2 phage, could not infect any of the tested bacterial strains. Thus, when 3.5 × 108 N2L2 phage were used, neither the F pilus nor the TolA molecule rendered the bacteria susceptible to the N2L2 phage. The titer of the N2L2 phage was too low to detect any receptor-independent background infection. As expected, in a similar manner was the wild-type infection dependent on the presence of both F pilus and TolA, and the lack of either receptor reduced the infection >108 times.
Infection of N1L1 phage is dependent on F pilus expression.
It has been shown that the filamentous phage is interacting with the TolQRA complex in the outer membrane and that it is the N1 domain itself that mediates this interaction (27, 31). Since the N1L1 phage, in our experiments, seemed to be dependent on the presence of the F pilus, it was important to elucidate if the engineered phage physically interacted with the primary receptor (Table 1). The fact that an extracellular level of divalent cations was required for infection with RNA phage (8, 22) was used to set up an assay where the F pilus was blocked by the RNA phage MS2. Thus, no translocation of MS2 RNA took place in the absence of cations. Preincubation of the F+TolA+ bacterial cells (K1762) with MS2 phage rendered the cells completely insensitive towards N1L1 phage infection (Table 2). This assay demonstrated that the expression of F pilus is necessary for the N1L1 phage to translocate into the cytoplasm of the bacterial cell. The inhibitory effect of MS2 phage was also shown for wild-type infection where the frequency was reduced in the same manner as the N1L1 phage infection (data not shown). The absence of detectable plaque in this assay was not due to lysis of the indicator cells, since control experiments demonstrated that the viability of the cells was not affected by the addition of MS2 at the multiplicity of infection used for blocking experiments.
TABLE 2.
Inhibiting effect of MS2 phage on N1L1 phage infectiona
| Amt of N1L1 phage | Infection
|
|
|---|---|---|
| Without MS2 phage | With MS2 phage | |
| 5.0 × 107 | 5.2 × 105 | 0 |
| 5.0 × 106 | 5.8 × 104 | 0 |
| 5.0 × 105 | 1.1 × 103 | 0 |
| 5.0 × 104 | 5.2 × 101 | 0 |
PFU obtained when using different amounts of N1L1 phage infecting K1762 bacterial strain. Experiments were performed in the presence or absence of 50 μl of MS2 phage with a titer of 1012 phage/ml.
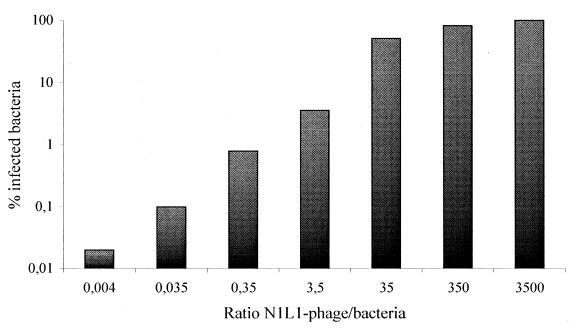
Earlier reports have indicated that only 1.5 × 10−4% of the phage lacking the N2 domain together with the glycine-rich linker regions (L1/L2) was capable of infecting F+TolA+ bacterial cells (27). To investigate if all bacterial cells were susceptible to N1L1 phage infection, the effect of the phage-to-bacteria ratio was tested over a wider range (Fig. 2). The results indicated that at a ratio of 350 N1L1 phage per bacterium, it was possible to infect all bacterial cells.
FIG. 2.
All bacteria studied could be infected with the N1L1 phage construct. Different amounts of N1L1 phage were used to evaluate if 100% of the bacterial cells could be infected by using this mutated g3p phage. At an input ratio of 350 phages per bacterium, it proved possible to achieve maximal infection.
The effect of CaCl2 on N1L1 and N2L2 phage infection.
Calcium ions will alter the structure of the bacterial membrane so that the accessibility of the coreceptor complex TolQRA will be increased (29). The exposed coreceptor will facilitate an F-pilus-independent infection through the direct interaction between the N1 domain and the TolA molecule. Therefore, increased efficiency of infection utilizing CaCl2 has only been observed with N2-deleted g3p mutants, while no apparent effect has been detected by using either N1-deleted g3p mutants or wild-type phages (13, 27). The g3p mutant phage constructs, together with wild-type phage, were tested for normal F+TolA+ infection in the presence or absence of 50 mM CaCl2 (Table 3). The infections of wild-type and N2L2 phage were not affected by the addition of Ca2+ ions. Furthermore, a 70% decrease in the frequency of infection was observed when CaCl2 was present during the N1L1 phage infection, which should be compared to CaCl2-dependent enhancement of the infection of a N1 mutant phage lacking the L1 region (27).
TABLE 3.
The effect of CaCl2 on the deleted-phage infectionsa
| Phage | Infection
|
|
|---|---|---|
| Without CaCl2 | With 50 mM CaCl2 | |
| N1L1 | 2.6 × 109 (1.0 × 1010) | 8.6 × 108 (1.0 × 1010) |
| N2L2 | 0 (3.5 × 108) | 0 (3.5 × 108) |
| Wild type | 3.0 × 1012 (5.0 × 1012) | 2.1 × 1012 (5.0 × 1012) |
PFU obtained when infecting the K1762 bacterial strain. The effect of 50 mM CaCl2 on the infection rate was evaluated. The number in parentheses represents the total amount of input phage used.
The F-pilus-binding capacity of the N1L1 phage is due to the first glycine-rich linker region.
To determine if the observed interaction between the F pilus and the N1L1 phage was mediated, at least in part, by the L1 region (residues 68 to 86), a phage (N1 phage) that lacked residues 68 to 243 of g3p was assayed for infection. The fd-D13 phage contained only the N1 domain and was analyzed for its ability to infect F+ and F− bacterial cells (TolA+) in the presence or absence of 50 mM CaCl2. The results obtained were in agreement with the data presented earlier (27). The infection frequency of fd-D13 increased when adding CaCl2, but the expression of F pilus on the host cell had no profound effect on this enhancement, indicating an F-pilus-independent infection mechanism. However, the high infection rate observed with the N1L1 phage was not achieved by using only the N1 phage (fd-D13 phage), demonstrating a direct interaction of the L1 region with F pilus, which was of functional importance (data not shown).
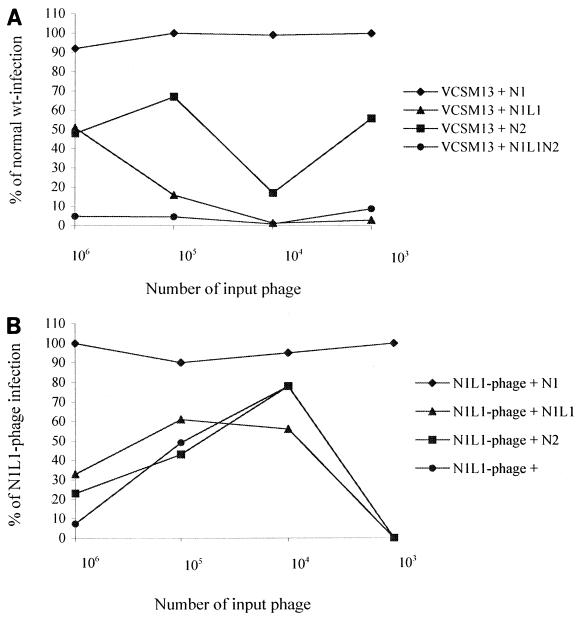
Inhibition of N1L1 and wild-type phage infection by different g3p domains.
To determine the involvement of the different domains of g3p and the first glycine-rich linker region (L1) during both normal and N1L1 phage infection, these domains were expressed as single proteins and were tested for inhibition properties during the infection of F+TolA+ bacteria (Fig. 3A and B). Figure 3A shows the effect of the N1, N1L1, N2, and N1L1N2 regions on wild-type infection. It was obvious that the N1 domain itself has no effect on the wild-type infection, although it has been shown by others that the N1 fragment can have a weak effect at much higher concentrations than those used in these experiments (13). The presence of N2 domain has an inhibitory effect on the wild-type infection, giving an infection that ranges from 20 to 68% of the normal value, while the presence of N1L1 or N1L1N2 almost completely abrogated the infection.
FIG. 3.
Inhibition of wild-type and N1L1 phage infection with the different domains of g3p. (A) The inhibiting capacity of the different domains of g3p on the infection of F+TolA+ bacteria with VCSM13 wild-type phage was evaluated. In all experiments different amounts of phage were utilized (103 to 106 phage), together with a constant number (3 × 107) of bacteria. The final concentration in each experiment of the g3p fragments were 8.0 μM N1, 3.4 μM N1L1, 2.6 μM N2, and 1.9 μM N1L1N2. The results are displayed as percent infection versus the value of wild-type infection, i.e., in the absence of the different g3p fragments. (B) The inhibition of N1L1 phage infection was evaluated in the same manner as the VCSM13 infection. The number of phage and bacteria utilized were the same as above. Furthermore, the final concentrations of the g3p fragments were the same in the two experiments, and the results are displayed as percent infection versus the value of N1L1 phage infection, i.e., in the absence of the different g3p fragments.
The N1L1 phage infection was then evaluated in the presence of the same protein preparations (Fig. 3B). All of the proteins, except the N1 domain itself, had inhibitory effects on the infection. The inhibition of the N1L1 phage infection was similar using the N1L1, the N2, or the N1L1N2 fragment (Fig. 3B), although the dose response differed when compared to inhibition of wild-type phage (Fig. 3A). The reduction in infection to 0% at an input of 103 phage depends on the fact that an average of 1% of the N1L1 phage are infectious (at a phage-to-bacterium ratio of 10 to 1), and thereby are not detectable at these low phage titers.
Finally, the F-pilus-binding properties of the N1L1 phage were directly assayed by enzyme-linked immunosorbent assay. Pili were coated onto plastic microtiter plates, and serial dilutions of wild-type and N1L1 phage were added. An OD490 value of 0.45 (10× background) was obtained by using 107 wild-type phage while the same amount of N1L1 phage gave an OD490 of 0.2 (5× background), indicating a significant but weaker binding (data not shown).
DISCUSSION
One of the minor coat proteins, i.e., g3p, is responsible for the interaction between the filamentous phage and the F+ bacterial cell, as well as for the subsequent incorporation into the cell (12). The N-terminal part of g3p can be divided into two distinct domains, N1 and N2, that are separated by a glycine-rich linker region (1, 30). N2 mediates the interaction with the first bacterial receptor, the F pilus (1). The first domain (N1) is believed to interact with the second receptor, the TolQRA protein complex, spanning the periplasmic space and the inner membrane (5, 9, 27, 31). It has previously been shown that phage with only the N1 and the CT domains on its surface will infect at a drastically reduced level. Furthermore, in the absence of N1, but presence of N2, the infection is almost abolished (13, 27). So far, no proper function has been demonstrated for the two glycine-rich linker regions present in g3p. In this study, we have observed that it is possible to delete both the N2 domain and the second glycine-rich linker region (L2) and still maintain an almost intact level of infection. Thus, we demonstrated that the maintenance of the first glycine-rich linker region (L1) (residues 68 to 86) in conjunction with the N1 domain and the CT part on the surface of the bacteriophage, will result in a level of infection close to what is obtained when using wild-type phage.
The g3p-engineered phages were constructed (Fig. 1), and in order to ensure a normal configuration of the adsorption protein, the glycine-rich linker regions were kept intact. For the same reason, extra care was taken not to include any supplementary amino acids during the construction and cloning. The N1L1 phage proved to be highly infectious, in contrast to already reported observations (27). The crucial difference from the earlier constructs is the maintenance of amino acid residues 68 to 86 in our N1L1 phage. We observed an average of 1%, and a maximum of 26%, of the N1L1 phage to be infective (at a phage-to-bacterium ratio of 10:1). This can be compared to the 1.5 × 10−4% result obtained when a phage comprised of only the N1 domain and the CT part, and lacking the first glycine-rich linker region (L1), was used (13, 27). The latter result was observed in the presence of CaCl2, which is known to enhance DNA translocation. We also found that the infection of N1L1 phage was dependent on the presence of the F pilus. Without the F pilus, the infection decreased by 2 × 107 times. As expected, in the absence of TolA expression, the infection rate was entirely abrogated. The N2L2 phage turned out to completely lack the ability to infect. These results were in agreement with data obtained when using a TolA− bacterial strain, since without the proper coreceptor or the interacting N1 domain, any detectable infection was obtained.
To accomplish the high rate of N1L1-mediated phage infection, it seemed necessary to have expression of the F pilus. To further elucidate the meaning of the primary receptor during this infection, experiments were performed where a shaft-binding RNA phage was utilized for its F-pilus-binding properties. Adding the RNA phage MS2 to pilus-expressing bacteria completely blocked the N1L1 phage infection as well as the VCSM13 infection. These results argue against the existence of any other primary receptor on the bacterial surface, apart from the F pilus, that might mediate the infection of the N1L1 phage.
Furthermore, we showed that the addition of CaCl2 had no enhancing effect on the N1L1 phage infection. The presence of CaCl2 during infection is believed to have a profound effect on the bacterial membrane, making the TolQRA coreceptor more accessible for interaction with the N1 domain (13, 27, 29). This alteration in the membrane structure could explain the increased F-pilus-independent infection observed by others. The fact that CaCl2 has no positive effect on the infection frequency of the N1L1 phage further strengthens the hypothesis that the primary receptor of this phage is distinct from the TolQRA complex.
Inhibition analyses were performed to determine the involvement of the individual domains of g3p in the N1L1 phage and the wild-type phage infection of F+TolA+ bacteria. Expressed and purified N1 (residues 1 to 67), N1L1 (residues 1 to 86), N2 (residues 87 to 217), and N1L1N2 (residues 1 to 217) proteins were used in an attempt to block the above infections. Surprisingly, both the N1L1 phage and the wild-type phage infections were affected in the same manner. The N1 domain demonstrated no inhibitory characteristics, in contrast to other observations where the wild-type infection was impaired when utilizing a higher concentration (10−5 M) of the g3p fragment than in the present assay (13). The other proteins, N1L1, N2, and N1L1N2, proved to inhibit the infections well. The inhibitory role of the N2 domain in normal wild-type phage infection is reasonable. However, more remarkable is the observed involvement of the N2 domain during N1L1 phage infection, although this observation might be explained as a steric phenomenon, i.e., the N2 fragment will block the N1L1 phage from interacting with the F pilus.
Consequently, the present evidence suggests that an interaction takes place between the F pilus and the N1L1 phage, i.e., the phage displaying a truncated version of g3p comprised only of the N1 domain, the first glycine-rich linker region (L1), and the CT part. The consensus has been that it is the N2 domain alone that physically interacts with the tip of the F pilus, thereby mediating the transport of the bacteriophage through the outer membrane by an unknown mechanism (30). We suggest, based on the results presented here, that there might exist a second point of interaction between the F pilus and the bacteriophage. Recently, others have shown that the N1 and N2 domains are physically bound to each other in the absence of F pilus (11, 17). It was implied that once the N2 domain binds to the tip of the pilus, its affinity for the N1 domain decreases, leaving this part of g3p free to interact with its secondary receptor. After depolymerization of the pilus, the released N1 domain can interact with the CT part of TolA localized on the inside of the outer membrane (7, 9, 27). Our observation of a second F-pilus-interacting domain (L1) could reflect the fact that amino acid residues 68 to 86 of g3p might constitute the pilus-binding epitope together with residues in the N2 domain. A more speculative interpretation is that the N1 domain, together with the glycine-rich linker region, interacts independently with the F pilus. This event might take place in the vicinity of the tip, since the N1 domain is physically hindered from extruding far away from the already-bound N2 domain. The results of the inhibition analyses using the N1L1 protein could support this speculation through the efficient blocking of the infection of both N1L1 and wild-type phage.
This is the first observation indicating an active role for one of the g3p glycine-rich linker regions in the infection process, although earlier observations have demonstrated a number of L1-dependent effects on the outer membrane of E. coli (3). These include deoxycholate sensitivity, leakage of periplasmic β-lactamase, impaired F pili expression, and alterations in the tolerance to certain colicins.
Furthermore, the data suggests that the interaction between the N1 domain and the TolA molecule is necessary for infection to occur, while the interaction between the F pilus and the N2 only enhances the rate of infection through the increased recruitment of phage. Others have speculated on the existence of an ancestral filamentous phage without an N2 domain, circumventing the requirement for an interaction with the primary receptor (27). This forefather of today's bacteriophage was suggested to be less efficient in infecting bacteria through the direct interaction of the N1 domain and the TolQRA coreceptor. It is tempting to speculate that the N1 in conjunction with the first glycine residues actually would have provided the phage with a powerful tool to ensure proper infection. Later in evolution, the efficiency might have been enhanced even further through the development of the N2 domain.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Swedish Council for Engineering Sciences and the SSF Functional Genomic Program.
REFERENCES
- 1.Armstrong J, Perham R N, Walker J E. Domain structure of bacteriophage fd adsorption protein. FEBS Lett. 1981;135:167–172. doi: 10.1016/0014-5793(81)80969-6. [DOI] [PubMed] [Google Scholar]
- 2.Beck E, Zink B. Nucleotide sequence and genome organisation of filamentous bacteriophages f1 and fd. Gene. 1981;16:35–58. doi: 10.1016/0378-1119(81)90059-7. [DOI] [PubMed] [Google Scholar]
- 3.Boeke J D, Model P. A procaryotic membrane anchor sequence: carboxyl terminus of bacteriophage f1 gene III protein retains it in the membrane. Proc Natl Acad Sci USA. 1982;79:5200–5204. doi: 10.1073/pnas.79.17.5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeke J D, Model P, Zinder N D. Effects of bacteriophage f1 gene III protein on the host cell membrane. Mol Gen Genet. 1982;186:185–192. doi: 10.1007/BF00331849. [DOI] [PubMed] [Google Scholar]
- 5.Burke J M, Novotny C P, Fives-Taylor P. Defective F pili and other characteristics of Flac and Hfr Escherichia coli mutants resistant to bacteriophage R17. J Bacteriol. 1979;140:525–531. doi: 10.1128/jb.140.2.525-531.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caro L G, Schnös M. The attachment of the male-specific bacteriophage f1 to sensitive strains of Escherichia coli. Proc Natl Acad Sci USA. 1966;56:126–132. doi: 10.1073/pnas.56.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Click E M, Webster R E. Filamentous phage infection: required interactions with the TolA protein. J Bacteriol. 1997;179:6464–6471. doi: 10.1128/jb.179.20.6464-6471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Date T. Kinetic studies of the interaction between MS2 phage and F pilus of Escherichia coli. Eur J Biochem. 1979;96:167–175. doi: 10.1111/j.1432-1033.1979.tb13026.x. [DOI] [PubMed] [Google Scholar]
- 9.Derouiche R, Gavioli M, Benedetti H, Prilipov A, Lazdunski C, Lloubes R. TolA central domain interacts with Escherichia coli porins. EMBO J. 1996;15:6408–6415. [PMC free article] [PubMed] [Google Scholar]
- 10.Grant R, Lin T-C, Konigsberg W, Webster R E. Structure of the filamentous bacteriophage f1. J Biol Chem. 1981;256:539–546. [PubMed] [Google Scholar]
- 11.Holliger P, Riechmann L. A conserved infection pathway for filamentous bacteriophages is suggested by the structure of the membrane penetration domain of the minor coat protein g3p from phage fd. Structure. 1997;5:265–275. doi: 10.1016/s0969-2126(97)00184-6. [DOI] [PubMed] [Google Scholar]
- 12.Jacobson A. Role of F pili in the penetration of bacteriophage f1. J Virol. 1972;10:835–843. doi: 10.1128/jvi.10.4.835-843.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krebber C, Spada S, Desplancq D, Krebber A, Ge L, Plückthun A. Selectively-infective phage (SIP): a mechanistic dissection of a novel in vivo selection for protein-ligand interactions. J Mol Biol. 1997;268:607–618. doi: 10.1006/jmbi.1997.0981. [DOI] [PubMed] [Google Scholar]
- 14.Kremser A, Rasched I. The adsorption protein of filamentous phage fd: assignment of its disulfide bridges and identification of the domain incorporated in the coat. Biochemistry. 1994;33:13954–13958. doi: 10.1021/bi00250a051. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Levengood S K, Webster R E. Nucleotide sequences of the tolA and tolB genes and localization of their products, components of a multistep translocation system in Escherichia coli. J Bacteriol. 1989;171:6600–6609. doi: 10.1128/jb.171.12.6600-6609.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lubkowski J, Hennecke F, Plückthun A, Wlodawer A. The structural basis of phage display elucidated by the crystal structure of the N-terminal domains of g3p. Nat Struct Biol. 1998;5:140–147. doi: 10.1038/nsb0298-140. [DOI] [PubMed] [Google Scholar]
- 18.Malmborg A-C, Söderlind E, Frost L, Borrebaeck C A K. Selective phage infection mediated by epitope expression on F pilus. J Mol Biol. 1997;273:544–551. doi: 10.1006/jmbi.1997.1332. [DOI] [PubMed] [Google Scholar]
- 19.Marwin D A. Filamentous phage structure, infection and assembly. Curr Opin Struct Biol. 1998;8:150–158. doi: 10.1016/s0959-440x(98)80032-8. [DOI] [PubMed] [Google Scholar]
- 20.Model P, Russel M. Filamentous bacteriophage. In: Calendar R, editor. The bacteriophages. New York, N.Y: Plenum Press; 1988. pp. 375–456. [Google Scholar]
- 21.Nelson F K, Friedman S M, Smith G P. Filamentous phage DNA cloning vectors: a noninfective mutant with a nonpolar deletion in gene III. Virology. 1981;108:338–350. doi: 10.1016/0042-6822(81)90442-6. [DOI] [PubMed] [Google Scholar]
- 22.Paranchych W. Stages in phage R17 infection: the role of divalent cations. Virology. 1966;28:90–99. doi: 10.1016/0042-6822(66)90309-6. [DOI] [PubMed] [Google Scholar]
- 23.Rakonjac J, Jovanovic G, Model P. Filamentous phage infection-mediated gene expression: construction and propagation of the gIII deletion mutant helper phage R408d3. Gene. 1997;198:99–103. doi: 10.1016/s0378-1119(97)00298-9. [DOI] [PubMed] [Google Scholar]
- 24.Rakonjac J, Model P. Roles of pIII in filamentous phage assembly. J Mol Biol. 1998;282:25–41. doi: 10.1006/jmbi.1998.2006. [DOI] [PubMed] [Google Scholar]
- 25.Rampf B, Bross P, Vocke T, Rasched I. Release of periplasmic proteins induced in E. coli by expression of an N-terminal proximal segment of the phage fd gene 3 protein. FEBS Lett. 1991;280:27–31. doi: 10.1016/0014-5793(91)80196-a. [DOI] [PubMed] [Google Scholar]
- 26.Rasched I, Oberer E. Ff coliphages: structural and functional relationships. Microbiol Rev. 1986;50:401–427. doi: 10.1128/mr.50.4.401-427.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riechmann L, Holliger P. The C-terminal domain of TolA is the coreceptor for filamentous phage infection of E. coli. Cell. 1997;90:351–360. doi: 10.1016/s0092-8674(00)80342-6. [DOI] [PubMed] [Google Scholar]
- 28.Russel M, Kidd S, Kelley M R. An improved filamentous helper phage for generating single-stranded plasmid DNA. Gene. 1986;45:333–338. doi: 10.1016/0378-1119(86)90032-6. [DOI] [PubMed] [Google Scholar]
- 29.Russel M, Whirlow H, Sun T-P, Webster R E. Low-frequency infection of F− bacteria by transducing particles of filamentous bacteriophages. J Bacteriol. 1988;170:5312–5316. doi: 10.1128/jb.170.11.5312-5316.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stengele I, Bross P, Garcés X, Giray J, Rasched I. Dissection of functional domains in phage fd adsorption protein. J Mol Biol. 1990;212:143–149. doi: 10.1016/0022-2836(90)90311-9. [DOI] [PubMed] [Google Scholar]
- 31.Sun T-P, Webster R E. fii, a bacterial locus required for filamentous phage infection and its relation to colicin-tolerant tolA and tolB. J Bacteriol. 1986;165:107–115. doi: 10.1128/jb.165.1.107-115.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]