Abstract
Transformation via Agrobacterium tumefaciens (Agrobacterium) is the predominant method used to introduce exogenous DNA into plant genomes1,2. Transfer DNA (T-DNA) originating from Agrobacterium can be integrated as a single copy or in complex concatenated forms3,4, but the mechanisms affecting final T-DNA structure remain unknown. In this study, we demonstrate that inclusion of retrotransposon (RT)-derived sequences in T-DNA can increase T-DNA copy number by more than 50-fold in Arabidopsis thaliana (Arabidopsis). These additional T-DNA copies are organized into large concatemers, an effect primarily induced by the long terminal repeats (LTRs) of RTs that can be replicated using non-LTR DNA repeats. We found that T-DNA concatenation is dependent on the activity of the DNA repair proteins MRE11, RAD17, and ATR. Finally, we show that T-DNA concatenation can be used to increase the frequency of targeted mutagenesis and gene targeting. Overall, this work uncovers molecular determinants that modulate T-DNA copy number in Arabidopsis and demonstrates the utility of inducing T-DNA concatenation for plant gene editing.
T-DNA integration into plant genomes via Agrobacterium-mediated transformation is used for a myriad of applications in plant biology, including the introduction of gene editing components and sequences conferring traits for crop improvement. In some instances, single copy insertions of T-DNA are desirable as they are expected to produce more consistent phenotypes between independent transgenic lines. However, it has long been recognized that multicopy T-DNA insertions in the form of concatemers are frequent outcomes of plant transformation3,5–7. A better understanding of these complex T-DNA structures could potentially be advantageous for specific applications that would benefit from increased transgene copy number. For example, DNA repair templates required for gene targeting can be delivered via T-DNA, and in vivo levels of these templates have been shown to correlate positively with the frequency of homology-directed repair8. While much progress has been made in understanding the mechanisms responsible for chromosomal integration of T-DNA in plants9,10, relatively little is known regarding how T-DNA copy number may be regulated.
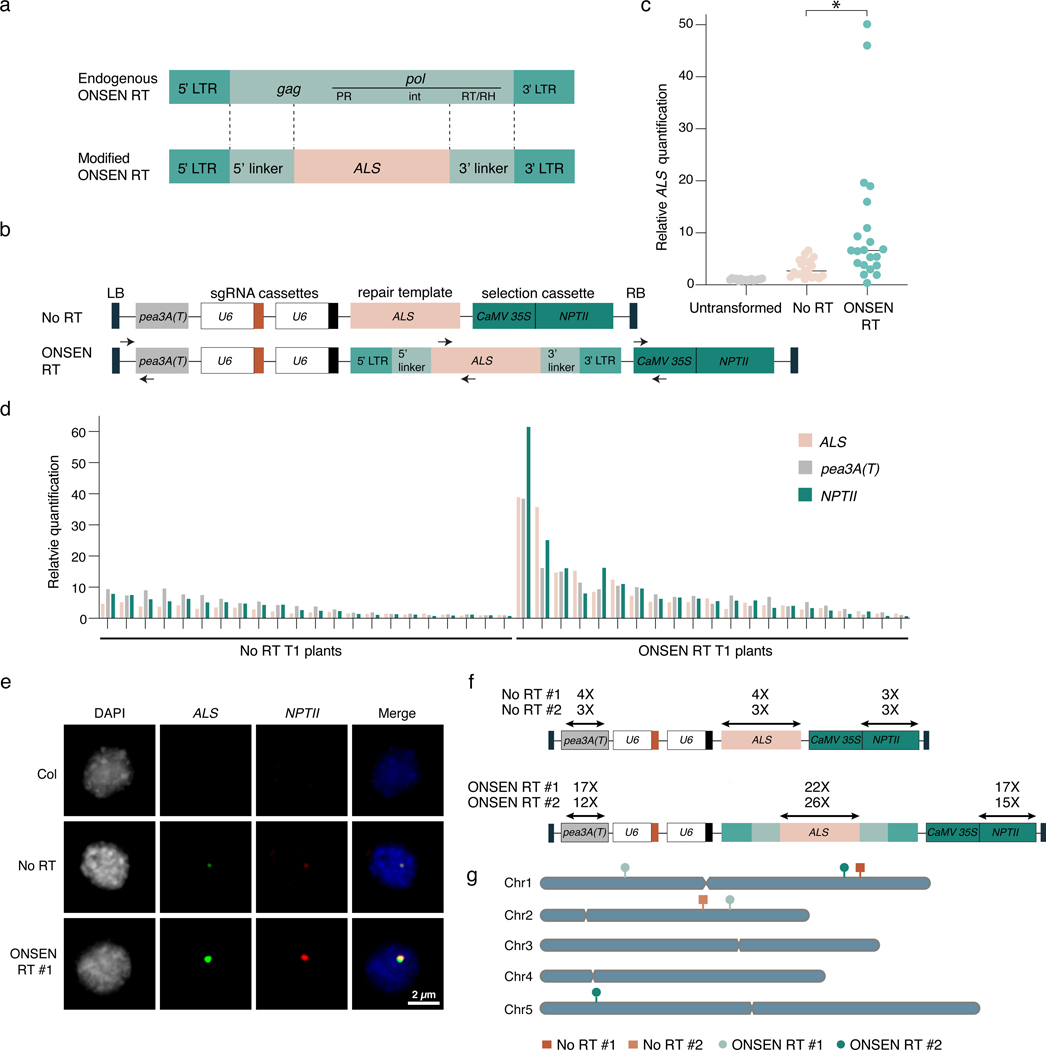
As a strategy to increase T-DNA copy number in Arabidopsis, we explored the possibility that including retrotransposon (RT)-derived sequences within a T-DNA may induce transgene amplification. In plants, copy number of RTs and other repetitive sequences are preferentially increased during genome replication over protein-coding genes in the absence of the histone mark H3.1K27me111, a process that depends on DNA repair12. In addition, RTs contain sequence features that can interfere with replication, transcription, and DNA repair, which can lead to locus-specific genomic amplification13. To assess if RT-derived DNA sequences increase T-DNA copy number, we designed a T-DNA vector based on the well-studied ONSEN family of long terminal repeat (LTR) RTs14,15. We first replaced part of the gag and pol genes of an ONSEN RT (At1g11265) with DNA from the Arabidopsis ACETOLACTATE SYNTHASE (ALS) locus, with the aim of using this ALS fragment 1) to measure T-DNA copy number relative to the Arabidopsis genome and 2) to serve as a repair template in gene targeting assays (Fig. 1a). The resulting ONSEN RT was then subcloned into the T-DNA region of a binary vector modified from a previously described gene targeting study (see Methods section)16. Identical plasmids, except for the presence of ONSEN sequences surrounding ALS, were then used to transform Arabidopsis plants (Col-0 ecotype) via Agrobacterium using the floral dip method (Fig. 1b)17. Genomic DNA was extracted from individual first-generation transformed (T1) plants and ALS quantification was performed by quantitative PCR (DNA-qPCR) using a primer set that can amplify both endogenous and T-DNA-associated ALS sequences. We found that T1 plants transformed with the plasmid containing ONSEN sequence (ONSEN RT) had, on average, higher ALS copy numbers compared to untransformed plants or T1 plants transformed with the control plasmid lacking ONSEN (No RT) (Fig. 1c). Individual T1 plants transformed with ONSEN RT showed wide variation in ALS levels, with copy number sometimes reaching a 50-fold increase relative to untransformed (UT) plants. In contrast, T1 plants transformed with the plasmid lacking ONSEN rarely showed more than 5-fold increase over untransformed plants (Fig. 1c). To determine if other parts of the T-DNA were being amplified, we quantified the relative levels of the pea3A terminator (pea3A(T)) and the kanamycin-resistance NPTII cassette present at the 5’ and 3’ regions of the T-DNA, respectively (Fig. 1b). We detected similar increases in copy numbers for ALS, pea3A(T), and the NPTII cassette in individual T1 plants transformed with ONSEN RT (Fig. 1d), indicating that the entire T-DNA, and not solely ONSEN-embedded ALS, was amplified.
Figure 1. T-DNAs featuring retrotransposon sequences are concatenated at higher levels in Arabidopsis.
a. Structure of the endogenous ONSEN RT and the modified RT used in the ONSEN RT-based vector. LTR: long terminal repeat; gag: gag-like protein; PR: protease; int: integrase; RT/RH: reverse transcriptase/RNase H. b. Schematic representation of the No RT (adapted from 16) and ONSEN RT T-DNAs. Arrows indicate the primers used for real time quantification shown in panels c and d. LB: left border; RB: right border. c. DNA-qPCR of ALS in Col-0 (untransformed), No RT, and ONSEN RT T1 plants. Each dot represents an individual plant (n = 16 for Col-0 and n = 21 for No RT and ONSEN RT individual T1 plants). Horizontal bars indicate the median. *P = 0.0007 (two-tailed Mann-Whitney U test). d. DNA-qPCR of ALS, pea3A(T) and NPTII. e. FISH in leaf nuclei using probes for ALS (green) and NPTII (red). Nuclei were stained with DAPI. f. Transgene copy number determined by whole genome sequencing analysis. g. T-DNA insertion sites in No RT and ONSEN RT T1 plants.
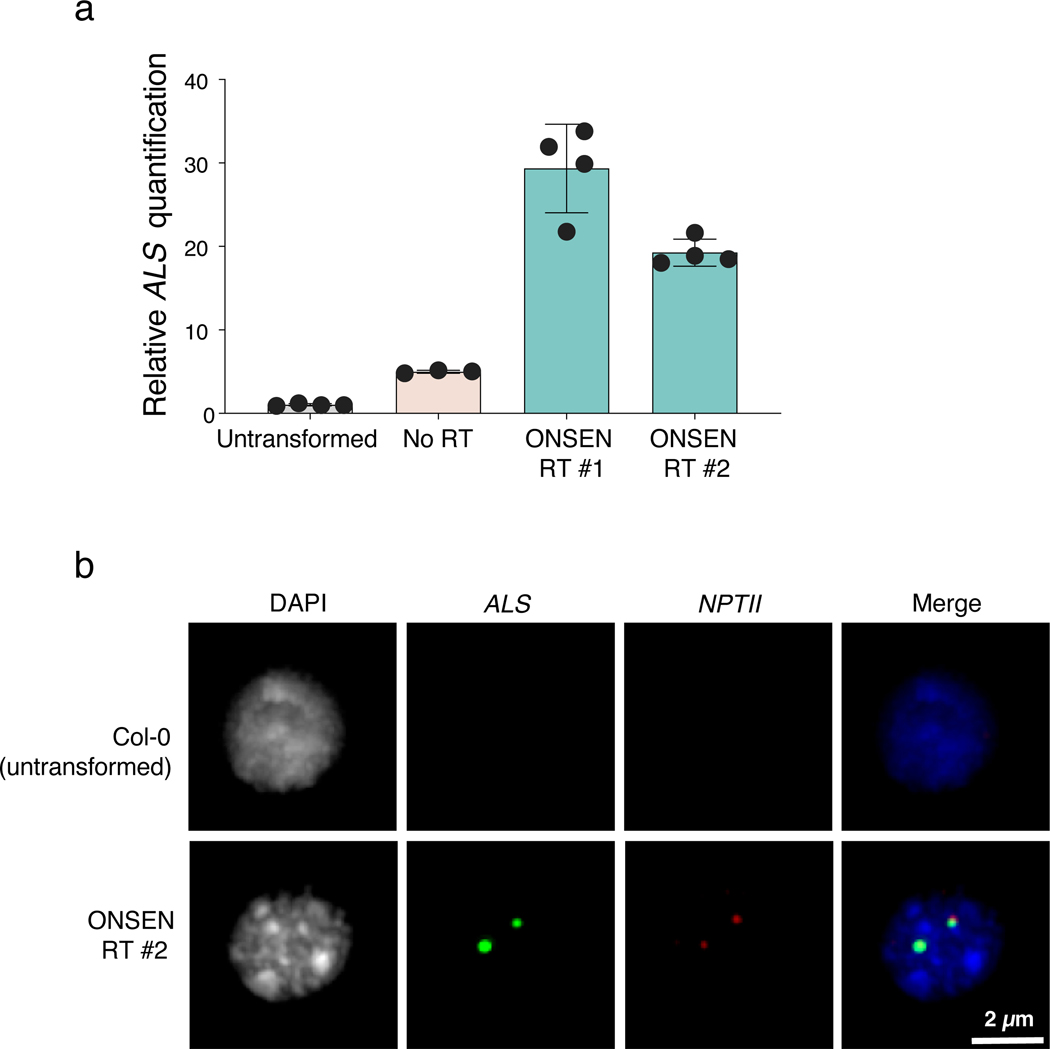
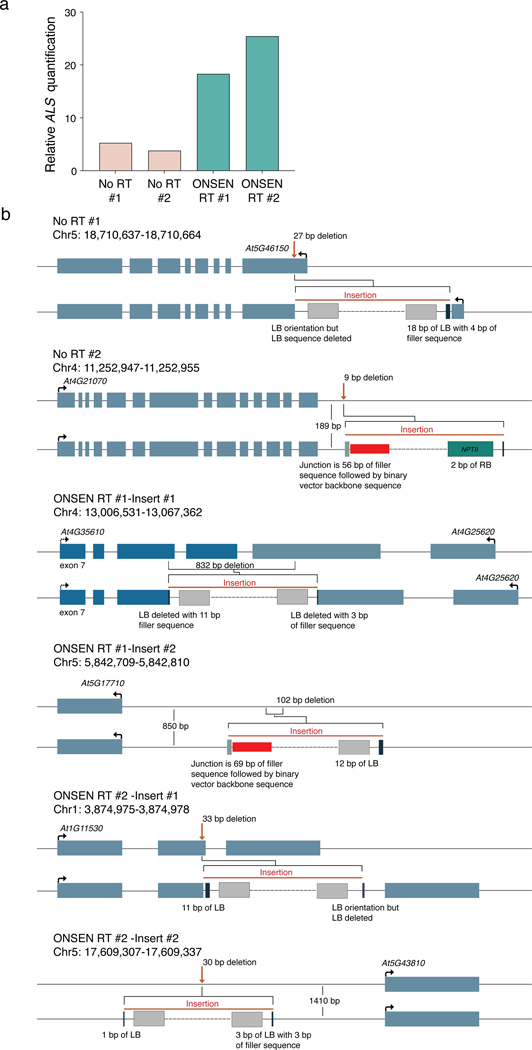
To determine if the ONSEN-mediated T-DNA copy increase is due to concatenation or multiple independent insertions at different loci, we performed DNA fluorescence in situ hybridization (FISH) experiments with probes designed to detect ALS and NPTII. In nuclei of T1 plants transformed with ONSEN RT and confirmed by DNA-qPCR to contain large numbers of T-DNA copies (>10-fold vs untransformed plants) (Extended Data Fig. 1a), we observed bright and overlapping signals for ALS and NPTII (Fig. 1e and Extended Data Fig. 1b). We typically detected 1–2 FISH signals per nucleus, which is in line with the average number of T-DNA integrations resulting from Arabidopsis floral transformation18–20. By contrast, FISH signals in nuclei from untransformed plants or T1 plants transformed using the plasmid lacking ONSEN were either undetectable or much weaker (Fig. 1e and Extended Data Fig. 1b). The fact that overlapping and high-intensity FISH signals can be observed for ALS and NPTII strongly suggests that ONSEN-induced T-DNA copy number gains are the result of concatenation rather than an increase in T-DNA insertion sites. To validate this, we sequenced the genome of a Col-0 control and four T1 plants (two plants transformed with each type of T-DNA plasmid) characterized by different levels of T-DNA copies as assessed by DNA-qPCR (Extended Data Fig. 2a). Analysis of the sequencing data confirmed the increase in copy number of the T-DNA and showed that these plants contained only one or two T-DNA insertion sites (Fig. 1f–g and Extended Data Fig. 2b), thus supporting that the additional T-DNA copies are due to concatenation.
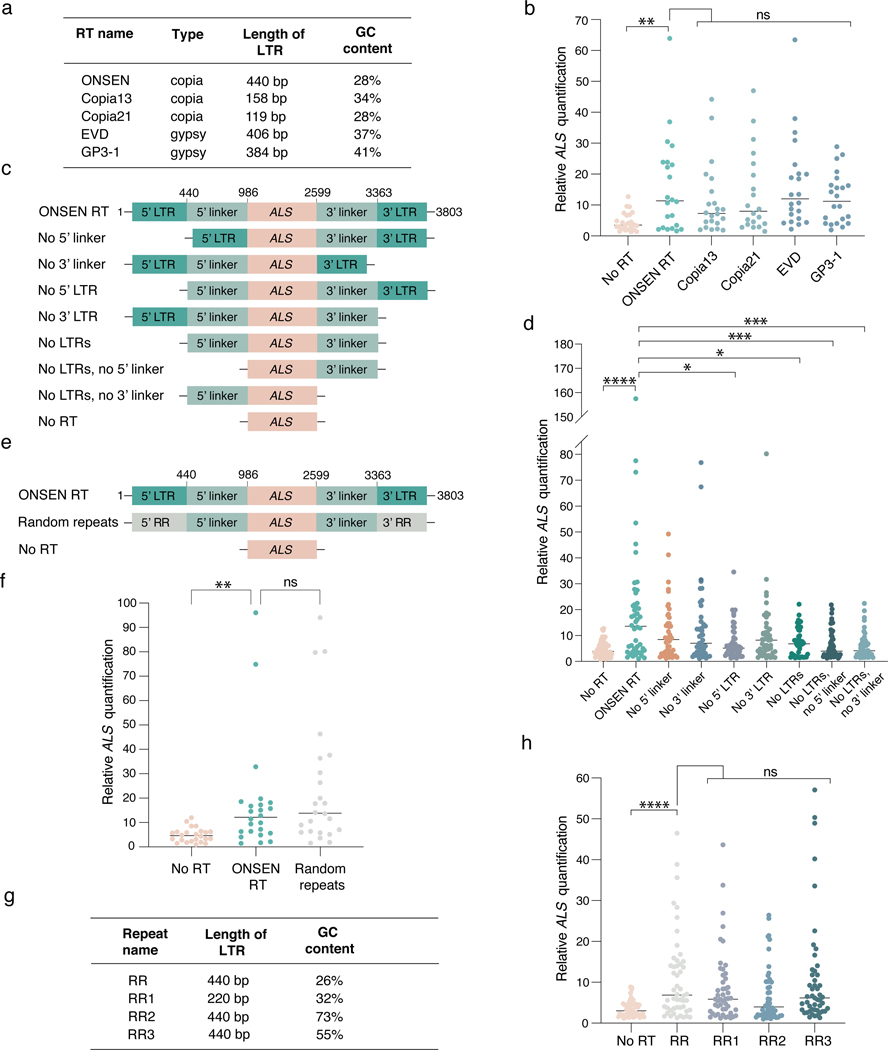
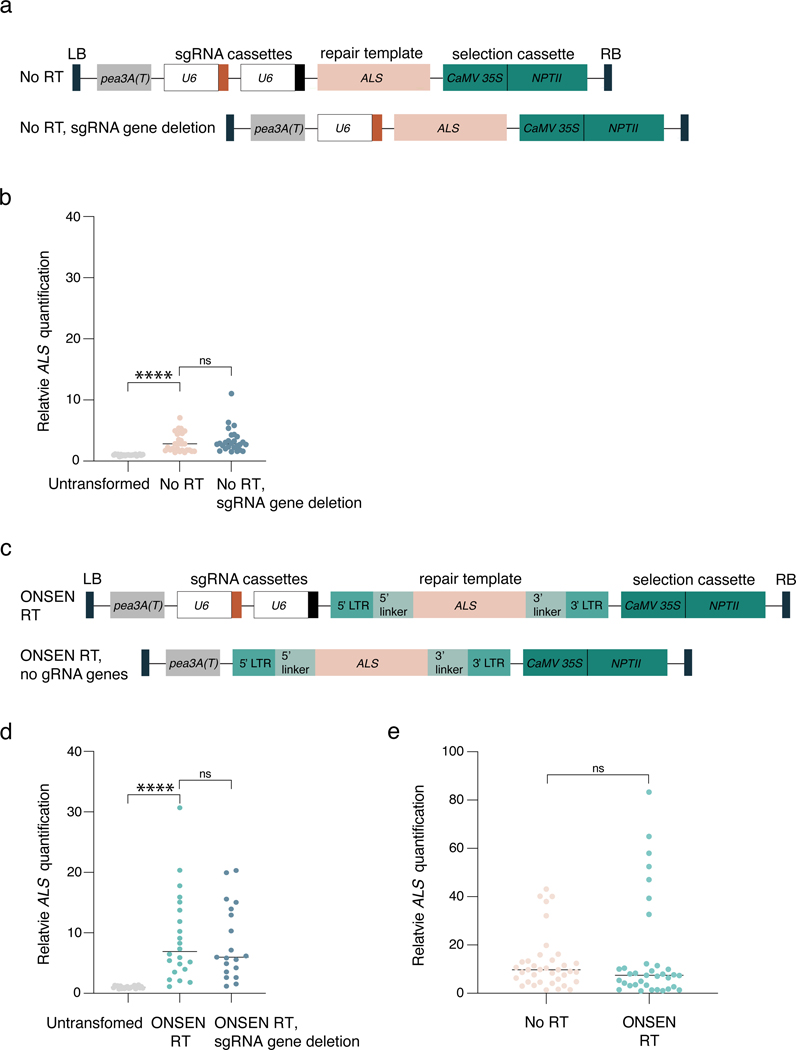
Next, we investigated the mechanism(s) involved in ONSEN-mediated T-DNA concatenation. First, we assessed if T-DNA concatenation could be conferred by sequences from RTs other than ONSEN. We tested four different Arabidopsis LTR RTs by replacing part of their gag-pol sequence with the same ALS fragment present in ONSEN RT (Fig. 2a and Fig. 1a), and observed a similar effect of increased T-DNA copies (Fig. 2b). We then investigated which region(s) of the ONSEN RT are involved in T-DNA concatenation by generating a series of binary plasmids containing various deletions of the ONSEN RT plasmid (Fig. 2c). Our results show that the LTRs had the most impact on concatenation levels (Fig. 2d). We hypothesized that the effect of the LTRs on T-DNA concatenation is mainly caused by their repetitive nature, which we verified by assessing T-DNA levels induced by a binary plasmid with random repeated sequences (RR) of the same length and GC content as the ONSEN LTRs (Fig. 2e–f). To validate this result, we tested three additional sets of artificial DNA repeats composed of different lengths (220 bp vs 440 bp) and GC content (26%, 55% and 73%). Our results showed that all DNA repeats could increase T-DNA copy number, although high GC levels (73%) led to a lower increase (RR vs RR2, P < 0.07; just outside of the statistical cutoff of P < 0.05) (Fig. 2g–h). Interestingly, not all DNA direct repeats within a T-DNA contribute to concatenation levels, as removal of one or two sgRNA genes containing the same Arabidopsis U6–26 promoter (387 bp) had no effect on T-DNA copy number (Extended Data Fig. 3a–d). Thus, multiple pairs of DNA repeats, and/or a linker sequence of defined length separating individual repeats, may be required to induce T-DNA concatenation. Finally, we tested if DNA repeats also contribute to increasing transgene copy number in plants transformed using biolistic particle delivery (BPD). Our results from BPD-transformed tobacco plants did not indicate any effect of the ONSEN repeats (Extended Data Fig. 3e), thus suggesting differences in the mechanisms involved in exogenous DNA integration.
Figure 2. The repetitive nature of LTRs promotes T-DNA concatenation.
a. Classification and characteristics of retrotransposons used to make RT-based T-DNAs. b. DNA-qPCR of ALS in plants transformed using various RT-based plasmids. Each dot represents an individual T1 plant (n = 22). Horizontal bars indicate the median. PNoRT = 0.00639, ns = not significantly different (Kruskal-Wallis ANOVA followed by Dunn’s test). c. Schematic representation of ONSEN deletion constructs used in panel d. d. DNA-qPCR of ALS in plants transformed with ONSEN RT deletion constructs. Each dot represents an individual T1 plant (n = 48). Horizontal bars indicate the median. PNo RT = 0.00001, PNo5’ LTR = 0.00290, PNo LTRs = 0.02991, PNo LTRs, No 5’ linker = 0.00095, PNo LTRs, No 3’ linker = 0.00035 (Kruskal-Wallis ANOVA followed by Dunn’s test). e. Schematic representation of the random repeat construct used in panel f. f. DNA-qPCR of ALS in plants transformed with ONSEN RT and the random repeat construct. Each dot represents an individual T1 plant (n = 24). Horizontal bars indicate the median. PNoRT = 0.00182, ns = not significantly different (Kruskal-Wallis ANOVA followed by Dunn’s test). g. Characteristics of artificial repeat constructs used in panel h. h. DNA-qPCR of ALS in plants transformed using various random repeat-based plasmids. Each dot represents an individual T1 plant (n = 48). Horizontal bars indicate the median. PNoRT = 0.00003, ns = not significantly different (Kruskal-Wallis ANOVA followed by Dunn’s test). ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
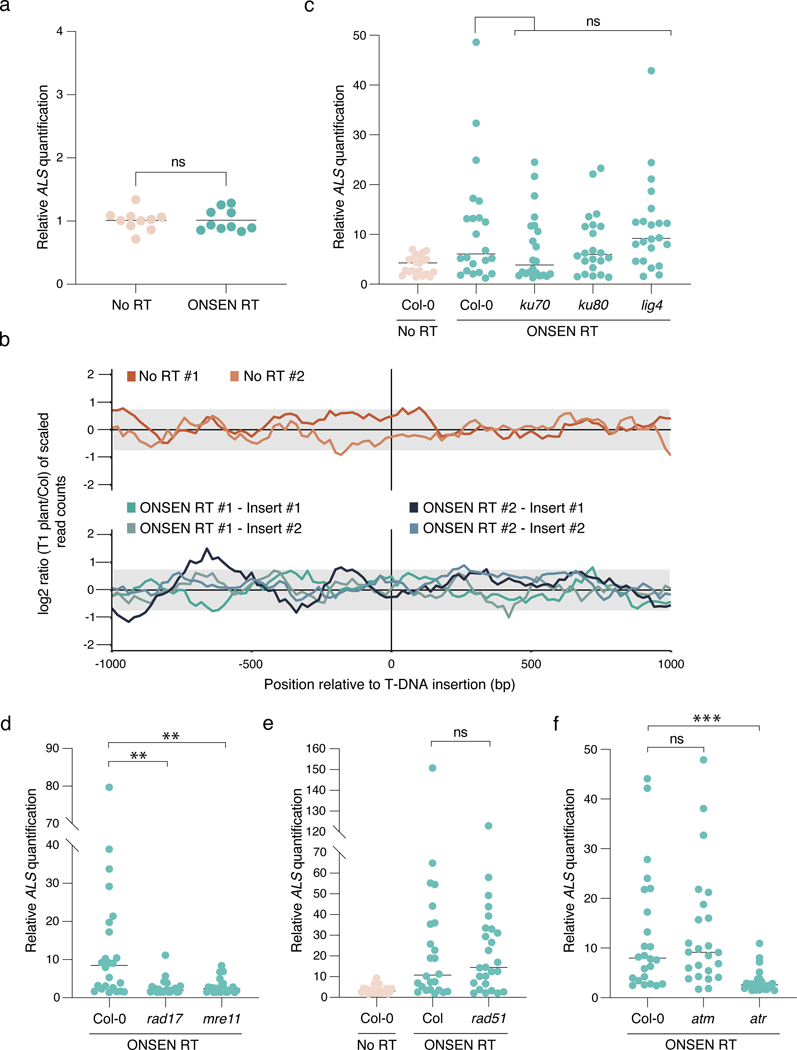
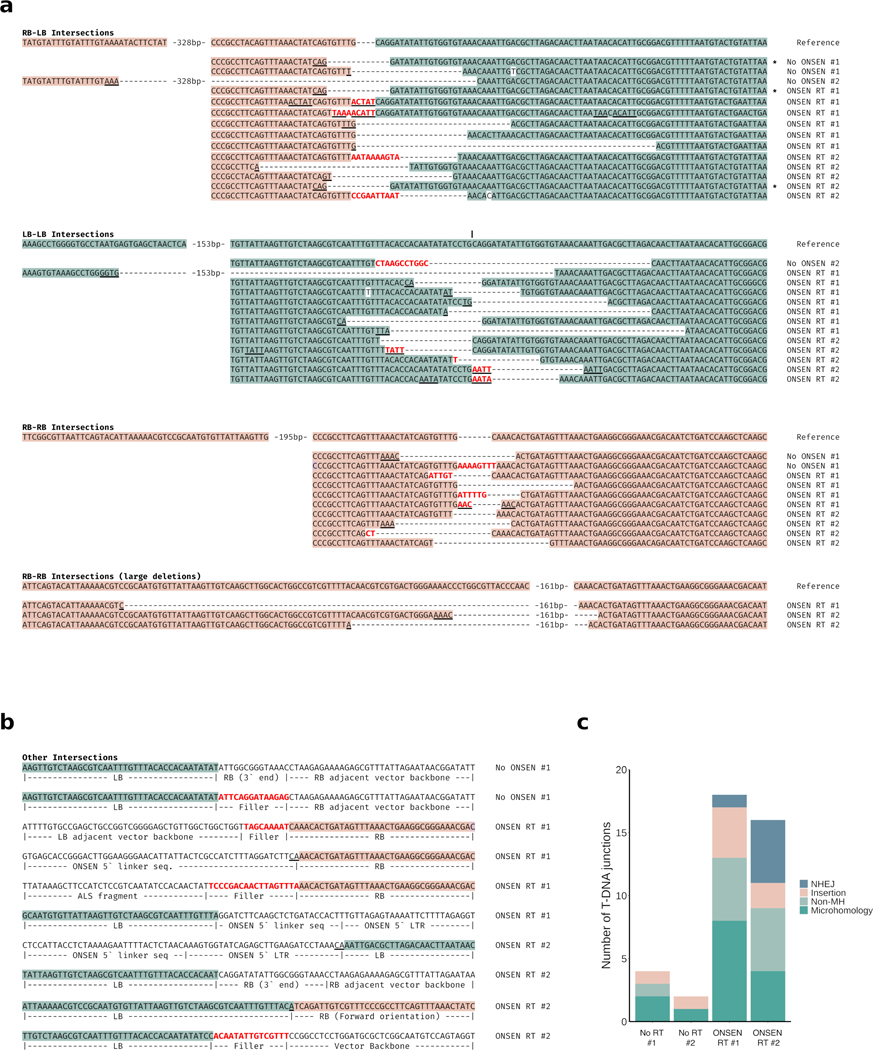
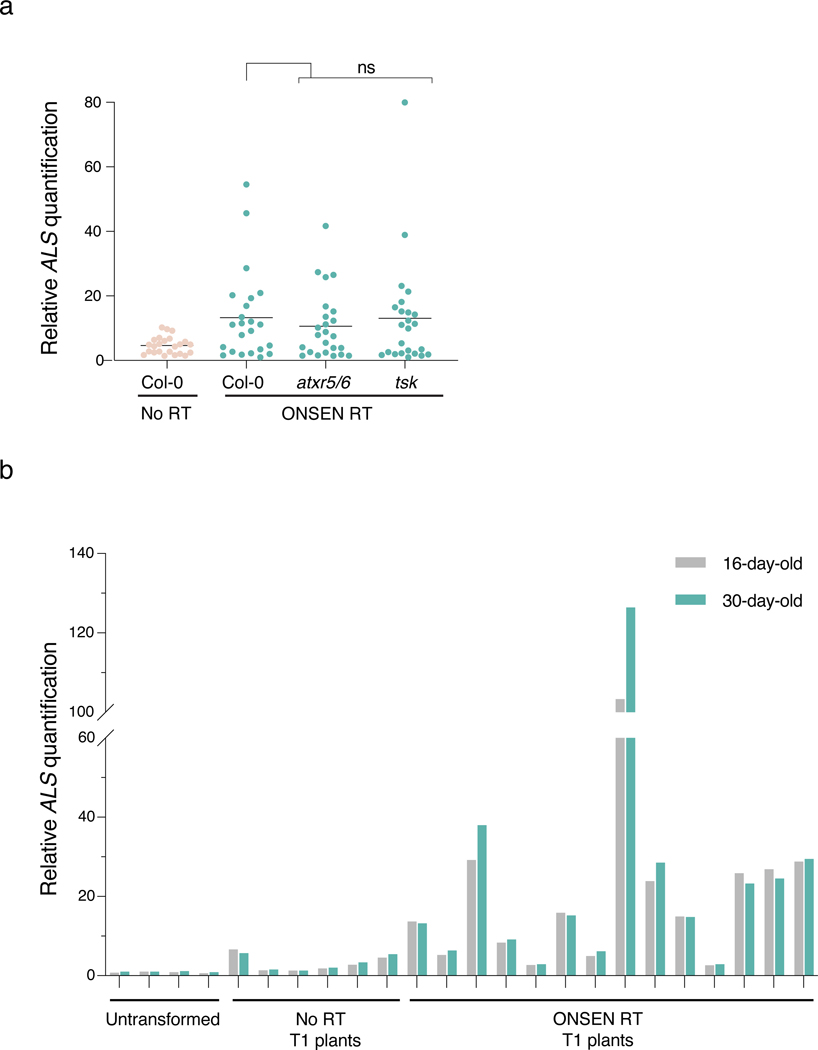
T-DNA concatenation may originate in Agrobacterium or arise in plants before, during or after T-DNA integration in the genome. Our results indicate that T-DNA levels are similar in cultured Agrobacterium strains carrying plasmids with or without RT sequences (Fig. 3a), arguing that T-DNA concatenation takes place within the plant. Furthermore, while whole-genome sequencing analysis revealed substantial increases in T-DNA copy number (Fig. 1f), the copy number of the genomic regions flanking these T-DNA insertion sites did not differ from the levels observed in the untransformed Col-0 genome (Fig. 3b). This suggests that concatenation occurs at the time of, or before, T-DNA integration. In Arabidopsis, two DNA repair pathways contribute to T-DNA integration: theta-mediated end joining (TMEJ) and non-homologous end joining (NHEJ)9,10. In the absence of homologous recombination (HR), TMEJ acts as a backup DNA repair pathway that is error-prone and responsible for the presence of large (>5 kb) tandem genomic amplifications21. Similarly, Arabidopsis mutants lacking the histone mark H3.1K27me1 (e.g., atxr5 atxr6 mutant22,23) are characterized by amplification of heterochromatin in a manner dependent on TMEJ and the replication fork-repair factor TONSOKU (TSK)12. To test if DNA repeat-mediated T-DNA concatenation is caused by specific DNA repair pathways, we transformed our binary plasmids (with and without ONSEN) into several mutant backgrounds. First, we tested NHEJ by transforming ku70, ku80 and lig4 mutants, but we did not detect a significant effect on T-DNA concatenation levels (Fig. 3c). To assess TMEJ, we could not directly verify the involvement of DNA polymerase theta (the main component of this repair pathway; encoded by the POLQ/TEBICHI gene), as we failed to obtain transformed plants using floral dip in the absence of this protein, as previously observed10,24, even when using the hypomorphic polq mutant teb-325. Therefore, we assessed mutant backgrounds of TMEJ-associated RAD17 and MRE119,26, and observed strong suppression of T-DNA concatenation (Fig. 3d). RAD17 and MRE11 also contribute to HR-mediated repair27–29, but, using a rad51 mutant background, we found that HR is not involved in inducing T-DNA concatenation (Fig. 3e). In support of the involvement of TMEJ in T-DNA concatenation, we identified mutational signatures consistent with this repair pathway (e.g., microhomology-associated deletions and small filler sequence insertions21) in sequencing reads spanning T-DNA copy junctions (Extended Data Fig. 4). Further investigations of DNA repair proteins revealed that the DNA damage-induced kinase ATR, but not ATM, is required to increase T-DNA copy number (Fig. 3f). Finally, in contrast to heterochromatin amplification in the absence of H3.1K27me1, T-DNA concatenation is not affected by mutations in ATXR5/ATXR6 or TSK, thus suggesting that the mechanism is not dependent on DNA replication (Extended Data Fig. 5a). In accordance, T-DNA levels are relatively constant in tissues of T1 plants separated by large number of replication cycles (Extended Data Fig. 5b). In sum, our results suggest a key role for TMEJ in inducing T-DNA concatenation.
Figure 3. T-DNA concatenation is dependent on DNA repair.
a. DNA-qPCR of ALS in No RT- and ONSEN RT-transformed Agrobacterium. Each dot represents a liquid culture initiated from an independent colony (n = 20). Horizontal bars indicate the mean. ns = not significantly different (two-tailed Mann-Whitney U test). b. log2 ratio between the number of reads corresponding to the genomic regions surrounding the T-DNA insertion sites compared to Col-0 in 20 bp bins. The grey box represents the interval where 95% of copy number values from the Arabidopsis genome are present. c-f. DNA-qPCR of ALS in mutant backgrounds of the (c) NHEJ, (d) TMEJ, and (e) HR pathways, and of (f) DNA damage-induced kinases. Each dot represents an individual T1 plant (n = 24 for rad17, mre11, atm and atr, n = 22 for ku70, ku80 and lig4 and n = 26 for rad51). Horizontal bars indicate the median. Prad17 = 0.00233, Pmer11 = 0.00438 (Kruskal-Wallis ANOVA followed by Dunn’s test), Prad51 = 0.00233 (two-tailed Mann-Whitney U test), Patr = 0.00011 (Kruskal-Wallis ANOVA followed by Dunn’s test), ns = not significantly different. ns, P > 0.05; **, P < 0.01; ***, P < 0.001.
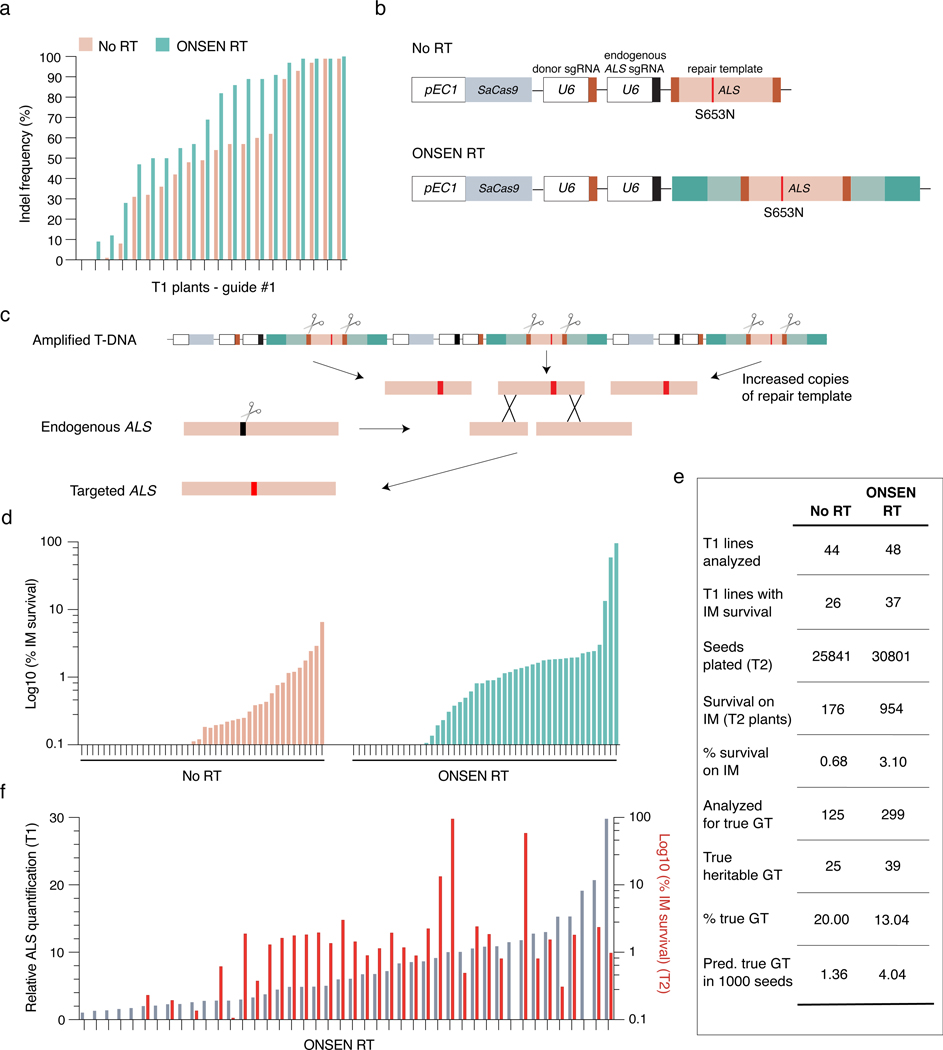
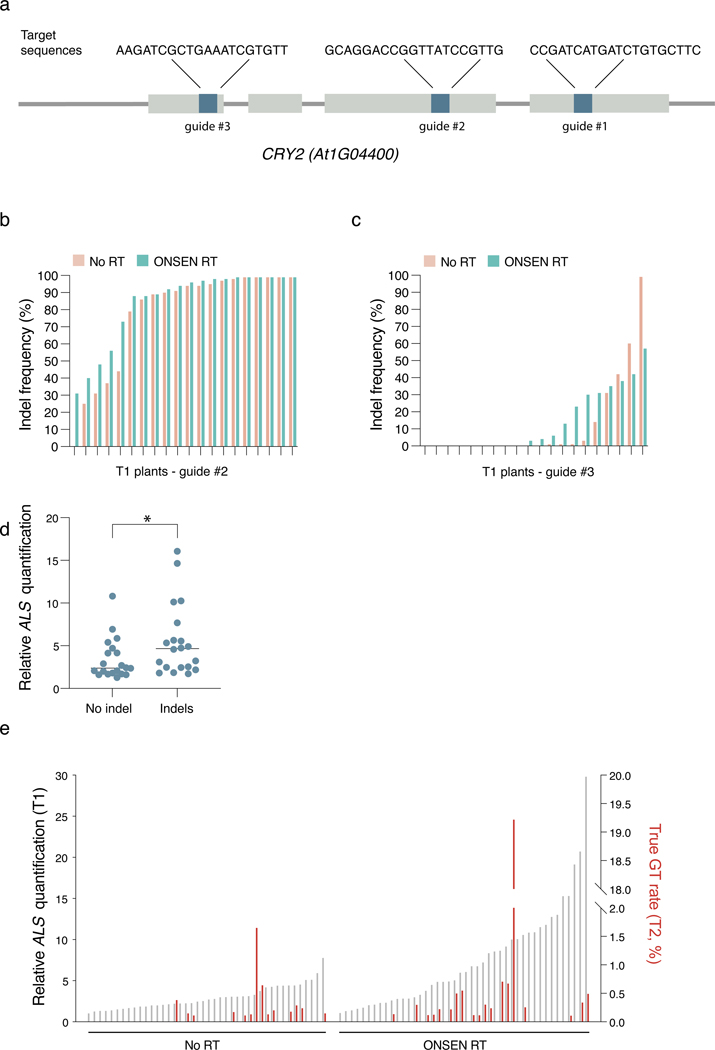
DNA repeat-mediated T-DNA concatenation allows us to increase the number of T-DNA copies in Arabidopsis transformants. Higher copy numbers of T-DNA has the potential to impact many biotechnological applications in plants. To provide a proof-of-concept of the utility of inducing T-DNA concatenation, we measured the efficiency of targeted mutagenesis by CRISPR/Cas9 using RT-derived plasmids. We designed and tested three different sgRNAs that target CRYPTOCHROME 2 (CRY2) (Extended Data Fig. 6a). Our data indicates that all three sgRNAs induced a slight increase in the mutation rates (although not statistically significant) when present on plasmids containing RT sequences (Fig. 4a and Extended Data Fig. 6b–c). In addition, individuals with CRISPR/Cas9-mediated indels displayed higher levels of T-DNA copies than plants with no detectable indels (Extended Data Fig. 6d). Taken together, these results demonstrate the benefits of inducing T-DNA concatenation in Arabidopsis to increase targeted mutagenesis rates.
Figure 4. T-DNA concatenation increases gene editing efficiency.
a. INDEL frequency of CRY2 PCR products amplified from DNA extracted from leaves of individual T1 plants transformed with either No RT or ONSEN RT constructs containing CRY2 sgRNA #1. b. Schematic representations of constructs used for gene targeting of ALS. SaCas9: Staphylococcus aureus Cas9. c. IPGT approach with ONSEN RT-induced T-DNA concatenation. d. Percentage of IM-resistant T2 plants from individual T1 parents transformed with No RT or ONSEN RT constructs. e. Summary of gene targeting events using No RT and ONSEN RT constructs. f. DNA-qPCR of ALS in ONSEN RT T1 plants (gray) in relation to percentage of IM-resistance for associated T2 plants (red).
Increasing T-DNA concatenation levels may also improve methods used to perform gene targeting in plants. For example, in planta gene targeting (IPGT) relies on the chromosomal integration of a T-DNA containing Cas9, two sgRNAs genes, and a repair template (Fig. 4b)30. One sgRNA is used to create a double-stranded DNA break to initiate DNA repair at a target locus, while the other directs Cas9 to the T-DNA to excise the repair template, which facilitates homology-directed repair (HDR). For single-copy T-DNA integrations, IPGT must rely on only one repair template copy per diploid cell in T1 plants. By inducing T-DNA concatenation, more copies of the repair template can be made available for HDR in each cell, which may result in higher gene targeting levels (Fig. 4c). To test this hypothesis, we modified a previously described system targeting the endogenous ALS locus of Arabidopsis16. Mutating serine 653 to asparagine (S653N) in ALS confers resistance to the herbicide imazapyr (IM), thus providing a visual assay to detect and quantify gene targeting events16. We designed an IPGT plasmid based on the ALS-IM system that contained the ONSEN RT sequences. Col-0 plants were transformed with the RT-based IPGT plasmid (ONSEN RT) or a standard IPGT plasmid (No RT) (Fig. 4b), and gene targeting levels were measured in T2 seed populations (from individual T1 parents) grown on IM-containing plates. Our results show that more T1 lines produced IM-resistant seedlings when transformed with the ONSEN RT plasmid (37/48 or 77.1%) compared to the control plasmid (26/44 or 59.1%) (Fig. 4d–e), and that, in general, higher T-DNA copy numbers in T1 plants produced more IM-resistant T2 seedlings (Fig 4e–f). Comparing all T2 seedlings analyzed, we detected a higher percentage of IM-resistant seedlings when they were transformed with the RT plasmid (3.10% versus 0.68%) (Fig. 4e). In these experiments, plants can gain resistance to IM via gene targeting, or through a process known as ectopic gene targeting (EGT). EGT occurs when part of the ALS genomic sequence is copied onto the T-DNA to generate a functional ALS S653N gene, which is subsequently integrated randomly into the genome16. Using the ALS-IM system, EGT can easily be differentiated from true gene targeting events by PCR16, and our results indicate that the frequency of true gene targeting events is approximately three times higher when the ONSEN RT plasmid is used (Fig. 4e, Extended Data Fig. 6e). Overall, these results indicate that T-DNA-dependent gene targeting systems in plants can be improved by inducing T-DNA concatenation.
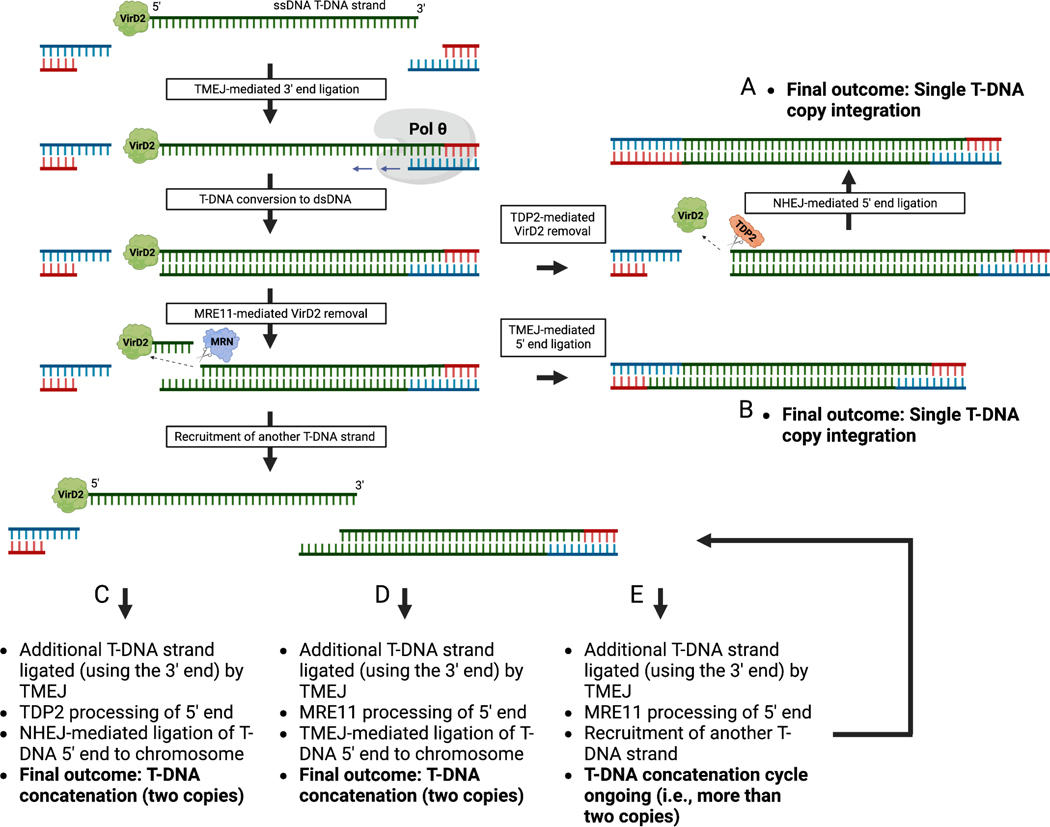
In summary, this study reveals that T-DNA composition and specific DNA repair proteins in plants play important roles in regulating T-DNA copy number at individual insertion sites. Our data supports that concatenation occurs before and/or at the time of T-DNA integration. The current model for T-DNA integration stipulates that T-DNA strand capture at the 3’ end is mediated exclusively by TMEJ, while ligation of the T-DNA 5’ end is redundantly catalyzed by TMEJ and NHEJ9. Specific to the capture of the 5’ end of the T-DNA is the requirement to remove the Agrobacterium protein VirD2, which is covalently attached to the T-DNA right border sequence31,32. NHEJ partners with TDP2 to accomplish this step, while TMEJ relies on MRE119. In contrast to TDP2, which severs the chemical bond between VirD2 and the 5’ end nucleotide of the T-DNA, MRE11 has been proposed to nick the protein-linked T-DNA strand internally9,33, which would generate a staggered end (Extended Data Fig. 7). It is possible that the differential processing of the T-DNA 5’ end by MRE11 generate a T-DNA repair intermediate that is more prone to recruit additional T-DNA strands to induce concatenation. Variation in T-DNA composition such as presence of DNA repeats may influence the number of available T-DNA strands and/or their accessibility for ligation, thus potentially increasing concatenation levels. Although further work will be required to fully elucidate the mechanism of T-DNA concatenation, our study demonstrates the benefits of modulating T-DNA copy number at single genomic sites for specific methods (e.g., gene editing) in plants.
Methods
Plant materials
Arabidopsis plants were grown under cool-white fluorescent lights (~100 μmol m−2 s−1) in long-day conditions (16 h light/8 h dark, 22°C). The T-DNA insertion mutants atxr5/6 (At5g09790 / At5g24330, SALK_130607 / SAIL_240_H0123), tsk/bru1–4 (At3g18730, SALK_03420737), rad51 (At5G20850, GK_134A0138), ku70–2 (At1g16970, SALK_123114c39), ku80–7 (At1g48050, SALK_11292139), lig4–4 (At5g57160, SALK_04402740), rad17–2 (At5g66130, SALK_00938441), mre11 (At5g54260, SALK_02845042), atm (At3g48190, SALK_040423C43), and atr (At5g40820, SALK_032841C44) are in the Col-0 genetic background. They were obtained from the Arabidopsis Biological Resource Center (Columbus, Ohio).
Cloning
All plasmids used in this study are derived from the DSB/DSB PcUbi4–2 and DSB/DSB AtEC1.1/1.2 vectors previously described16. These vectors encode a Staphylococcus aureus CRISPR/Cas9 system. The derivative plasmids lacking the ubiquitin promoter and Cas9 sequence were made using the DSB/DSB PcUbi4–2 plasmid, which was digested using AscI and EcoRI, blunted with T4 DNA Polymerase (New England Biolabs, Ipswitch, MA), and re-ligated using T4 DNA Ligase (NEB). All RT-based plasmids (lacking Cas9) were generated by inserting an RT-ALS cassette (described below) in place of the ALS only cassette using the AatII and PacI restriction sites.
To make the ONSEN RT cassette, ONSEN (At1g11265, 4956 bp) was amplified from Col-0 (−109 bp from the beginning of 5’LTR, to +114 bp from the end of the 3’LTR) and cloned into pCR2.1-TOPO (Invitrogen, Waltham, MA). An AscI site was then created in ONSEN at nucleotide position 987–994 (relative to 5’ LTR), replacing GTCACCGT with GGCGCGCC. The ALS repair template and the surrounding sgRNA binding sites were amplified from DSB/DSB AtEC1.1/1.2 and inserted into the pCR2.1-TOPO-ONSEN vector using the AscI and BsrGI sites to generate pCR2.1-TOPO-ONSEN-ALS. The resulting ONSEN-ALS cassette was transferred to the binary plasmid (lacking Cas9) using the AatII and PacI restriction sites. For the binary plasmids expressing the other RTs, the LTRs and linker sequences of Copia13 (At2g13940), Copia21 (At5g44925), EVD (At5g17125) and GP3–1 (At3g11970) were mapped in the Arabidopsis genome and then synthesized at GenScript (Piscataway, NJ). The length of the 5’ linker (546 bp) and 3’ linker sequences (734 bp) synthesized for each RT was based on the length of the linker sequences in the ONSEN RT vector. A multicloning sequence that includes a NotI restriction site was inserted between the 5’ and 3’ linker sequences of each RT. The ALS repair template in DSB/DSB AtEC1.1/1.2 was removed from the vector using NotI and inserted in the NotI cloning site of the four synthesized RTs. Finally, the RT-ALS cassettes were transferred to the binary plasmid (lacking Cas9) using AatII and PacI.
The series of truncated ONSEN RT plasmids were created from a synthesized (GenScript) ONSEN-ALS cassette cloned into pUC57. The sequence of this synthesized ONSEN-ALS cassette is identical to the ONSEN-ALS cassette described in the previous paragraph, except for the insertion of restriction sites (each one cutting only at a single location) right before and after the different sections of ONSEN-ALS: 5’ LTR, 5’ linker, ALS, 3’ linker, and 3’ LTR. To remove a specific section of the synthesized ONSEN-ALS cassette, two restriction enzymes targeting the borders of that section were used to digest pUC57-ONSEN-ALS. The digested plasmid was then blunted using Quick Blunting kit (NEB) and re-ligated using T4 DNA Ligase (NEB) to generate the deletion. Finally, all modified ONSEN-ALS cassettes were cloned into the binary plasmid (lacking Cas9) using AatII and PacI.
The binary plasmids lacking either one or both sgRNA genes were generated by digesting the No RT plasmid with XmaI and PacI (one sgRNA deleted) or the ONSEN RT plasmid with MluI and PacI (two sgRNAs deleted), blunting (Quick Blunting kit, NEB), and re-ligating using T4 DNA Ligase (NEB). The random repeat sequence (RR) was generated using the online tool Random DNA Sequence Generator online tool (The Maduro Lab, UC Riverside), synthesized at GenScript, and cloned into pUC57-ONSEN-ALS, replacing the 5’ and 3’ LTRs. The three additional artificial repeat elements (RR1, RR2, and RR3) used to test the effect of length and GC content were cloned from bacterial derived vector backbone sequences. RR1 was cloned from the backbone of pBluescript II SK(+) (GenBank: X52328.1), while repeats RR2 and RR3 were cloned from the backbone of pLN18 (a cosmid vector derived from pLAFR3). All elements were inserted into pUC57-ONSEN-ALS using the corresponding restriction sites and binary vector as described above.
The binary plasmids (No RT and ONSEN RT) used for transformation into the Arabidopsis mutant backgrounds were modified to replace the kanamycin resistance gene (NPTII) with an hygromycin resistance gene. Briefly, the NPTII gene cassette was removed from the No RT and ONSEN RT plasmids using the restriction enzymes AatII and PmeI. In parallel, the NPTII gene cassette (including promoter and terminator) was cloned into pAGM1311 and modified using HiFi DNA Assembly Cloning Kit (NEB, Ipswitch, MA) to replace the complete coding sequence of NPTII with the coding sequence of the hygromycin resistance gene from pMDC745. The modified cassette was then reinserted into the No RT and ONSEN RT plasmids.
For the gene editing experiments involving the detection of mutations at the CRY2 locus, the DSB/DSB PcUbi4–2 plasmid was first modified by replacing the original ALS cassette with an ALS cassette lacking the sgRNA binding sites using AatII and PacI. The sgRNA binding sites were eliminated to prevent cutting of the T-DNA locus by the Cas9-sgRNA complex, which could affect T-DNA quantification. To build the equivalent ONSEN RT plasmid, an ALS cassette without the sgRNA binding sites was inserted in the pCR2.1-TOPO-ONSEN vector using the AscI and BsrGI sites, and the resulting ONSEN-ALS cassette was then cloned into the DSB/DSB PcUbi4–2 plasmid at the AatII and PacI sites. Finally, the sgRNA gene targeting the ALS endogenous locus was replaced by a sgRNA gene targeting CRY2. This was done by digesting the ONSEN RT plasmid with XmaI and PacI and subcloning the fragment (containing the endogenous ALS gRNA gene) into a pENTR/D-Topo vector (Thermo fisher Scientific, Waltham, MA). The resulting plasmid was PCR amplified with a primer pair to change the ALS sgRNA spacer sequence to one of three different CRY2 sequences (sgRNA#1, 5’-AAGATCGCTGAAATCGTGTT-3’; sgRNA#2, 5’-GCAGGACCGGTTATCCGTTG-3’; and sgRNA#3, 5’-CCGATCATGATCTGTGCTTC-3’). The amplified PCR products were then ligated with T4 DNA Ligase (NEB) and the CRY2 sgRNA genes were subcloned into the modified DSB/DSB PcUbi4–2 plasmids (i.e., lacking sgRNA binding sites) with XmaI and PacI.
To produce the ONSEN RT plasmid for gene targeting, the ONSEN RT cassette (containing ALS and the sgRNA binding sites) was amplified from the pCR2.1-TOPO-ONSEN-ALS vector and inserted into DSB/DSB AtEC1.1/1.2 using the AatII and PacI sites, replacing the ALS only cassette. The original DSB/DSB AtEC1.1/1.2 plasmid (No RT plasmid) served as a control.
Arabidopsis transformation
Arabidopsis plants were transformed by using the floral dip method46. Briefly, one day prior to floral dip transformation, 300 μL of a stationary Agrobacterium (strain GV310) liquid culture was used to inoculate 200 mL of LB containing 100 mg/L gentamycin, 100 mg/L spectinomycin. The culture was incubated with shaking overnight at 28°C. The bacterial culture was spun down at 3,220 x g for 25 min and resuspended in 200 mL of transformation solution (5% sucrose and 0.02% Silwet L-77). Arabidopsis flowers were dipped into the bacterial solution, gently agitated for 10 seconds, then stored horizontally in a tray with a blackout lid overnight in a long-day growth chamber. T1 plants were selected on ½ MS plates containing 1% sucrose, carbenicillin (200 μg/mL) and either kanamycin (100 μg/mL) or hygromycin (25 μg/mL). Herbicide-resistant seedlings were transferred to soil after 7–10 days on plates. Transformed DNA repair mutants were all homozygous except for rad51 and mre11, due to sterility38,47. We therefore transformed rad51 and mre11 heterozygous plants and analyzed homozygous T1 mutants.
Tobacco transformation
Leaf pieces of in vitro grown plants of Nicotiana tobaccum “Glurk” were used as initial explant for the particle bombardment. The procedure was performed with a Biolistic PDS-1000/He particle delivery system (Bio-Rad) using a protocol previously described48. In summary, 0.6 μm gold microcarriers (Bio-Rad, Hercules, CA, USA) were defrosted and water-bath sonicated for 1 min. Plasmid DNA (final concentration of 1 μg/μL) was added to the gold microcarriers and vortexed for 1 min. 50 μL of 2.5 M calcium chloride and 20 μL of 0.1 M spermidine were then added onto the inside of the cap of the tube and mixed by pipetting up and down 2–3 times. The cap of the tube was closed, and the tube was tapped down to let all the components mix at the bottom. The mixture was centrifuged for 5 s at top speed, and the supernatant was removed. The pellet was resuspended in 150 μL of 100% ethanol vortexed for 1 min. The components were centrifuged for 5 s at top speed, and the supernatant was removed. The pellet was then resuspended in 40 μL of 100% ethanol. The macrocarriers were loaded on the macrocarrier holders. The gold-plasmid complex in the 100% ethanol solution was slowly loaded onto the center of the macrocarrier. The gold-plasmid DNA complex was then allowed to air dry on the macrocarriers for 5–10 min. The settings used for particle delivery were as follows: 2.5 cm distance gap between the rupture disk and macrocarrier, 9 cm target distance between the stopping screen and the target plate, 0.8 cm distance between the macrocarrier and the stopping screen, 28–29” Hg vacuum, 5.0 vacuum flow rate, and 4.5 vacuum vent rate.
Leaf pieces of tobacco plants were placed on tobacco pre-culture media, as previously described49, containing 4.3 g/L of Murashige and Skoog (MS) salts; 1mL/L of Gamborg’s vitamins 1000x; 1mL/L of Benzylaminopurine (BAP 1mg/mL); 1mL/L of Napthalene acetic acid (NAA 1mg/mL); 1ml/L of p-Chloro-phenoxy acetic acid (pCPA 8mg/mL); 30g/L of sucrose and 6.5g/L of Difco Agar, and pH adjusted to 5.7. Plates containing the leaf pieces were bombarded, removed from the delivery system, wrapped with PVC film, and placed under light at 25°C for 4 days. Leaf pieces were then transferred to selection/regeneration media containing 4.3 g/L of Murashige and Skoog (MS) salts; 1mL/L of Gamborg’s vitamins 1000x; 1mL/L of Benzylaminopurine (BAP 1mg/mL); 0.1mL/L of Napthalene acetic acid (NAA 1mg/mL); 30g/L of sucrose; 75mg/L of kanamycin and 6.5g/L of Difco Agar, and pH adjusted to 5.7. The plates were kept under light at 25°C and subcultured every two weeks until shoot formation. All plants were rooted under selection media. Leaves were collected for genomic DNA extraction 1.5 months after regeneration (total timing since bombardment: 3 months).
Plant DNA extraction
Leaves from plants that were grown for two weeks on soil (unless otherwise indicated) were homogenized in 500 μL DNA extraction buffer (200 mM Tris-HCl pH 8.0, 250 mM NaCl, 25 mM EDTA, and 1% SDS) and 50 μL phenol:chloroform:isoamyl alcohol (25:24:1). Each sample was centrifuged for 7 min at 16,000 x g, and 300 μL of the aqueous layer was transferred to a 1.5 mL tube containing 300 μL isopropanol. Samples were vortexed, incubated at room temperature for 5 min, and then spun down at 16,000 x g for 10 min. The supernatant was removed and the pellets were washed with 400 μL 70% ethanol. After centrifugation at 16,000 x g for 5 min, the ethanol was removed and the pellets were dissolved in 100 μL of water.
Agrobacterium DNA extraction
GV310-transformed colonies were grown in liquid culture overnight at 28°C. 300 μL of each culture was transferred to a 1.5 ml tube and centrifuged for 1 min at 16,000 x g. The resulting pellet was resuspended in 250 μL of a lysozyme solution (200 mM CaCl2 with 1% lysozyme). The samples were incubated at 42°C for 5 min and 750 μL of 96% ethanol was added, followed by centrifugation for 10 min at 16,000 x g. The supernatant was removed, and the pellet was air dried for 10 min and resuspended in 100 μL of water.
DNA-qPCR
Real-time PCR was carried out using a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA) with a KAPA SYBR FAST qPCR Master Mix (2×) Kit (Kapa Biosystems, Wilmington, MA). Relative quantities were determined by the 2(-delta delta Ct ) method50 using Actin7 (At5g09810), the gene coding for hypothetical protein WP_046033610.1 (NCBI accession number), and the L25 gene as the normalizers for DNA extracted from Arabidopsis, Agrobacterium, and tobacco, respectively. Relative quantities of ALS in Arabidopsis plants were calculated using Col-0 DNA as the calibrator. For relative quantification of pea3A(T) and the NPTII cassette in Arabidopsis and of ALS in Agrobacterium, a DNA sample from a “No RT” transformant was used as the calibrator. Graphpad prism 9 was used to analyze the data.
Fluorescence in situ probe design
Primary FISH probes were designed with a procedure previously described51, with some modifications. First, a pool of primary targeting sequences were designed with OligoArray2.152, with the following parameters: sequence length 30 nt; minimum melting temperature 66°C; maximum melting temperature 100°C; secondary structure melting temperature limit 76°C; cross-hybridization melting temperature limit 72°C; minimum GC content 30%; maximum GC content 90%; avoiding 6 or more consecutive A, T, G or C’s; allowing at most 20-nt overlap between adjacent target sequences. The primary targeting sequences were compared against the TAIR10 genome to ensure specificity. Then, a 30 nt secondary probe binding sequence (reverse complement of the dye-labeled secondary probe sequence) was appended to the 3’ end of each primary targeting sequence to generate the full-length primary probes (Integrated DNA Technologies, Coralville, IA). To detect the ALS and the NPTII genes, 39 and 29 primary probes were designed, respectively (Supplementary Table 1). The secondary probe sequences were adapted from a previous report51. The secondary probes for ALS and NPTII were conjugated to 5’ Alex Fluor 488 and ATTO 590, respectively (IDT).
Fluorescence in situ hybridization
Leaves from 3-week-old plants were fixed in cold 4% formaldehyde in Tris buffer (10 mM Tris-HCl pH 7.5, 10 mM NaEDTA, 100 mM NaCl) for 20 min, and then washed twice in Tris buffer. The leaves were chopped with a razor blade in 500 μl LB01 buffer (15 mM Tris-HCl pH7.5, 2 mM NaEDTA, 0.5 mM spermine-4HCl, 80 mM KCl, 20 mM NaCl and 0.1% Triton X-100), and the resulting slurry was filtered through a 30 μM mesh (Sysmex Partec, Gorlitz, Germany). The filtered solution was mixed 1:1 with sorting buffer (100 mM Tris-HCl pH 7.5, 50 mM KCl, 2mM MgCl2, 0.05% Tween-20 and 5% sucrose), spread onto a coverslip, and dried. Cold methanol was added to the coverslips for 3 min, followed by TBS-Tx (20 mM Tris pH 7.5, 100 mM NaCl, 0.1% Triton X-100). 0.1 mg/ml of RNase A in 2x SSC (0.3M NaCl, 30 nM sodium citrate, pH 7.0) was added onto the coverslips and incubated at 37°C for 45 min, followed by two washes in 2x SSC. Pre-hybridization buffer (2X SSC, 50% Formamide and 0.1% Tween 20) was added for 30 min at room temperature. Probes were added to the hybridization buffer at a concentration of 1 uM, which was applied to each coverslip. The coverslips were then incubated at 80°C for 3 min and at 37°C overnight in a humid chamber. Coverslips were washed twice with SSC-0.1% Tween at 60°C for 15 min, and once for 15 min at room temperature. Secondary probes at a concentration of 40 nM in buffer (2x SSC + 40% formamide) were added to coverslips. The coverslips were incubated at room temperature for 30 min, washed with secondary wash buffer (2x SSC + 40% formamide), and washed twice with 2x SSC. Coverslips were mounted on microscope slides with Vectashield containing DAPI (Vector Laboratories, Burlingame, CA). Nuclei were imaged under a Nikon Eclipse Ni-E microscope with a 100X CFI PlanApo Lamda objective (Nikon, Minato City, Tokyo, Japan) and an Andor Clara camera. Z-series optical sections of each nucleus were obtained at 0.3 μm steps. Images were generated using the NIS-Elements software, and deconvolved by FIJI using the DeconvolutionLab plugin53,54. The nuclei selected for imaging were ≤ 55 μm3 to enrich for 2C nuclei55. The nuclear volume was measured using FIJI with the 3D ImageJ suite54,56. FISH experiments were performed > 3 times with similar results.
Library construction, sequencing and bioinformatic analyses
DNA sequencing libraries were prepared at the Yale Center for Genome Analysis. Genomic DNA was sonicated to a mean fragment size of 350 bp using a Covaris E220 instrument (Covaris, Woburn, MA) and libraries were generated using the xGen Prism library prep kit for NGS (Integrated DNA Technologies, Coralville, IA). Paired-end 150 bp sequencing was performed on an Illumina NovaSeq 6000 using the S4 XP workflow (Illumina, San Diego, CA). Raw FASTQ files were pre-processed and trimmed using the fastp tool57 (--length_required 20 --average_qual 20 --detect_adapter_for_pe -w 10). Subsequently, the command line program grep (Free Software Foundation, Boston, MA) was used to search FASTQ files using the 20 bp sequences inside the T-DNA neighboring the LB and RB sites (and their reverse complement) as a query. The filtered sequences were then processed using BioPython58 to isolate flanking sequence tags (FSTs) adjacent to the LB and RB. FSTs were then used as BLAST queries59 to identify regions of the genome where T-DNA sequences were inserted. Further supporting reads were obtained by mapping the FASTQ files to the genome (TAIR1060) using BWA-MEM61 and isolating reads for which only a single read of a mate pair maps to the genome at the identified point of insertion, as the other read will map to the T-DNA insertion. Assembling the T-DNA insertion junctions with the genome was then done using the MAFFT multiple sequence aligner62, and a reference sequence was manually created. Insert sites were confirmed using PCR and Sanger sequencing.
Estimation of transgene sequence copy number was achieved by mapping reads using BWA-MEM to a panel of 7,535 genes, along with three regions of the T-DNA transgene. The selected genes are reported in the PLAZA 5.0 database63 to originate from single gene families in the genome (omitting plastid genes and At3g48560 from which the ALS sequence of the transgene derives). PCR duplicates were removed using Picard (http://broadinstitute.github.io/picard/) and the read counts from these alignments were extracted using SAMtools idxstats function64 and normalized to Bins Per Million mapped reads (BPM; [Reads Per Kilobases]/[sum(Reads Per Kilobases)*1ê6]). The mean BPM of the selected genes were taken as the normalized copy number of a single copy gene in the genome and the relative fold change, in BPM, for regions of the T-DNA transgene was calculated using this as a reference.
Measurement of coverage around mapped T-DNA insertion sites was done using the bamCompare function of the deepTools package65 which scales by read count and returns the log2 ratio of two alignments for a genome split into equally sized bins. The bin size was set to 20bp and the genome alignment of each line was compared with a wild-type Col-0 sample sequenced at the same time.
Internal T-DNA junctions were assembled in a similar manner to the identification of insertion junctions with the genome, but using reads with FSTs that match the binary vector used to transform the plants. Once assembled, the FASTQ files were searched again with consensus intersection sequences (+/− 15 bp from either the point of intersection or edge of filler sequence) and only intersections with two or more independent read pairs supporting it were retained.
Gene editing
To assess the efficiency of targeted mutagenesis of CRY2, genomic DNA was extracted from individual 2-week-old T1 plants and used to amplify the CRY2 gene. The resulting PCR products were sequenced and analyzed for INDEL frequency by Inference of CRISPR Edits (ICE) analysis (Synthego Performance Analysis, ICE Analysis. 2019. V3.0. Synthego).
To measure IPGT rates, T2 seeds from individual T1 lines (44 and 48 lines for No RT and ONSEN RT, respectively) were plated on ½ MS plates containing 5 μM Imazapyr (IM) and 1% sucrose. Seeds were stratified for 3 days at 5°C. The seed count for each plate was determined by using the ImageJ ‘analyze particles’ function after binary processing. The seeds were allowed to germinate and grow for one week in long day conditions and the imazapyr resistant seedling were then counted manually. Relative quantification of ALS in T1 plants was assessed by DNA-qPCR as described above. T1 lines (two No RT and four ONSEN RT) were eliminated from further analysis due to technical failure of qPCR.
The true gene targeting events were characterized as previously described16, with some modifications. Briefly, the endogenous ALS locus was amplified with primers specific for regions outside of the repair template and the PCR product was Sanger sequenced. The codon corresponding to amino acid 653 and the gRNA binding site were analyzed for a gene targeting event using Sequencher 5.4.6 (Gene Codes Corporation, Ann Arbor, MI). Samples with different types of editing events were re-analyzed using Synthego ICE or subcloning and further Sanger sequencing. Plants were considered as having undergone true gene targeting when samples displayed, at a minimum, ~50% GT-edited sequences.
Statistics and Reproducibility
Statistical analyses were performed using GraphPad Prism version 9.4.0 for macOS (GraphPad Software, San Diego, California USA, www.graphpad.com). The sample sizes, statistical tests, and P values are indicated in the figure legends. All experiments in this study were performed at least three times with similar results.
Primers
All primers used in this study are in Supplementary Table 1.
Data availability
The data generated from this study are included within the main figures, extended data and supplementary information. Raw sequencing data are deposited at the NCBI SRA (BioProject: PRJNA892619). Analysis of the data made use of the TAIR10 Arabidopsis genome (https://www.arabidopsis.org/download/index-auto.jsp%3Fdir%3D%252Fdownload_files%252FGenes%252FTAIR10_genome_release) and the Orthologous gene family list sourced from the Dicots PLAZA 5.0 database (https://ftp.psb.ugent.be/pub/plaza/plaza_public_dicots_05/GeneFamilies/genefamily_data.ORTHOFAM.csv.gz). There are no restrictions on data availability.
Extended Data
Extended Data Figure 1. DNA FISH analysis.
a. DNA q-PCR of ALS in Col-0 (untransformed) and T1 plants used for the FISH experiment (panel b and Fig. 1e). Dots represent DNA samples from individual leaves from the same T1 plant (n = 4 for Col-0, ONSEN RT #1 and ONSEN RT #2, n = 3 for No RT). Horizontal bars indicate the mean. SD is shown. b. FISH in leaf nuclei targeting the T-DNA sequence. Nuclei were stained with DAPI and probes for ALS (green) and NPTII (red). FISH experiment was performed >3 times with similar results.
Extended Data Figure 2. Whole genome sequencing analysis of T-DNA insertions.
a. DNA- qPCR of ALS in the No RT and ONSEN RT T1 plants used for whole genome sequencing (panel b and Fig. 1f–g). b. Locus diagrams for the identified T-DNA insertions. The coordinates for each insertion are based on the TAIR10 annotation and correspond to the Arabidopsis genomic borders surrounding each identified T-DNA. For each insertion, top lines represent unaltered genomic sequence with annotated genes. Red arrows represent insertion points. The bottom lines show the borders of the insertion in more detail, with the identified binary vector (non-T-DNA region) or T-DNA components shown. Dashed lines represent contiguous T-DNA-associated cassettes. Red bars indicate binary vector sequence (non-T-DNA), dark blue LB bars to light gray Pea3A terminator sequence bars indicate the 5’ end of the T-DNA construct (though it may be in 5’ or 3’ orientation in the plant genome). Darker gray bars adjacent to the red bars are filler sequences, and the teal bar represents NPTII sequence.
Extended Data Figure 3. Repetitive sgRNA genes do not contribute to T-DNA concatenation.
a. Schematic representation of the sgRNA gene deletion from the No RT vector. b. DNA-qPCR of ALS in Col-0 (untransformed) and in T1 plants transformed with the No RT plasmid and the No RT plasmid with a gRNA gene deleted. Each dot represents and individual plant (n = 21 for Col-0, n = 26 individual T1 plants for No RT and No RT, sgRNA gene deletions). Horizontal bars indicate the median. PCol-0 = 0.00000002, ns = not significantly different (Kruskal-Wallis ANOVA followed by Dunn’s test). c. Schematic representation of the sgRNA gene deletion from the ONSEN RT vector. d. DNA-qPCR of ALS in Col-0 (untransformed) and in T1 plants transformed with the ONSEN RT plasmid and the ONSEN RT plasmid with both gRNA genes deleted. Each dot represents and individual plant (n = 22). Horizontal bars indicate the median. PCol-0 = 0.00000004, ns = not significantly different (Kruskal-Wallis ANOVA followed by Dunn’s test). e. DNA-qPCR of ALS in tobacco plants transformed by biolistic bombardment with particles coated with No RT or ONSEN RT plasmid DNA. Each dot represents an individual plant. Horizontal bars indicate the median. ns = not significantly different (two-tailed Mann-Whitney U test). ns, P > 0.05; ****, P < 0.0001.
Extended Data Figure 4. T-DNA junctions support the involvement of TMEJ in T-DNA concatenation.
a. Multiple sequence alignments of junctions between two T-DNA sequences grouped by orientation of the two sequences (RB-LB, LB-LB or RB-RB). Reference sequences were constructed assuming T-DNA molecules began and ended immediately upstream of the endonuclease recognition sequence (5′-CAGGATATATT-3′)60,61. Filler sequences are in red and sequences consistent with microhomology-associated deletions are underlined. If filler sequences have a similar sequence nearby, they (and the nearby sequence) are also underlined. Asterisks indicate identical junctions occurring in independent plants. b. Depiction of T-DNA junctions with another T-DNA or binary vector sequence not immediately internally adjacent to the LB or RB. c. Classification of RB-LB, LB-LB and RB-RB sequences for each T1 line following a procedure previously described62. NHEJ (<4 bp deletions and <5bp insertions), insertions (≥5bp with any deletion), non-microhomology (Non-MH; ≥4bp deletion or <5bp insertions with microhomologies <2), and microhomology (MH; ≥4bp deletion with microhomologies ≥2). The latter three are associated with DNA polymerase theta.
Extended Data Figure 5. T-DNA concatenation is not DNA replication-dependent.
a. DNA-qPCR of ALS in Col-0, atxr5/6, and tsk plants transformed with the ONSEN RT construct. Each dot represents an individual T1 plant (n = 24). Horizontal bars indicate the median. ns = not significantly different (Kruskal-Wallis ANOVA followed by Dunn’s test). b. DNA-qPCR of ALS in DNA extracted from leaves of T1 plants at 16 days and 30 days after germination.
Extended Data Figure 6. T-DNA concatenation increases the efficiency of targeted mutagenesis and gene targeting.
a. Gene structure of CRY2. Gray bars represent exons, and blue bars represent regions targeted by sgRNAs. b-c. INDEL frequency of CRY2 PCR products amplified from DNA extracted from leaves of individual T1 plants transformed with either No RT or ONSEN RT constructs. Constructs carried either (b) sgRNA #2 or (c) sgRNA #3. d. DNA-qPCR of ALS in T1 plants with no detectable CRY2 indels (No indels) or detectable indels (Indels) using CRY2 guide #3. Each dot represents an individual T1 plant (n = 20). Horizontal bars indicate the median. *P = 0.0350 (two-tailed Mann-Whitney U test) e. DNA-qPCR of ALS in No RT and ONSEN RT T1 plants (gray) in relation to the percentage of true (non-ectopic) gene targeting rates in the T2 generation (red).
Extended Data Figure 7. Model of T-DNA concatenation.
The model builds on the T-DNA integration model from Kralemann et al., 20226. Chromosomal capture of a T-DNA strand 3’ end is mediated by TMEJ. After conversion to a double-stranded T-DNA intermediate, capture of the T-DNA 5’ end is accomplished by removal of the Agrobacterium protein VirD2 by TDP2 or MRE11. TDP2-mediated removal of VirD2 creates blunt-ended DNA at the T-DNA 5’ end that is ligated to the chromosome by NHEJ. In contrast, MRE11, acting as part of the MRN complex (MRE11-RAD50-NBS1; loaded on DNA by RAD1725), removes VirD2 by cutting the T-DNA internally, generating a staggered end at the T-DNA 5’ end. TDP2/NHEJ activity leads to a single T-DNA copy integration (outcome A), while MRE11/TMEJ activity leads to multiple outcomes (B-E), with the simplest one being chromosomal capture of the T-DNA 5’ end (outcome B). Alternatively, the staggered T-DNA 5’ end can facilitate recruitment of additional T-DNA strands for ligation, leading to concatenation. Capture of the 5’ end of an additional T-DNA strand by TDP2/NHEJ instead of MRE11/TMEJ is more likely to terminate the concatenation cycle. In this model, T-DNA features like DNA repeats may increase concatenation levels by increasing the number of available T-DNA strands for integration, and/or their accessibility. ssDNA: single-stranded DNA. dsDNA: double-stranded DNA. Created with BioRender.com.
Supplementary Material
Acknowledgments
We thank all current and former members of the Jacob lab, especially Franziska Langhammer and Gonzalo Villarino, for reagents, discussions, and advice, and Christopher Bolick and his staff at Yale for help with plant growth and maintenance. This project was supported by grant #R35GM128661 from the National Institutes of Health (Y.J.) and an NIH Director’s New Innovator Award (DP2GM137414) (S.W.). We want to thank Dr. Holger Puchta from the Karlsruhe Institute of Technology for his generous gifts of the gene targeting binary plasmids used in this work. Finally, we want to acknowledge Marco Molina and Mayra Molina (Multi-Crop Transformation Facility, Texas A&M University) for generating the transgenic tobacco plants used in this study.
Footnotes
Competing Interests Statement
A patent application (Y.J, L.D. and W.Y. as the inventors), related to the use of repetitive sequences and genetic mutations to regulate T-DNA copy number and improve gene editing, has been filed. The remaining authors declare no competing interests.
References
- 1.Gelvin SB Integration of Agrobacterium T-DNA into the Plant Genome. Annu Rev Genet 51, 195–217, doi: 10.1146/annurev-genet-120215-035320 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Gelvin SB Plant DNA Repair and Agrobacterium T-DNA Integration. Int J Mol Sci 22, doi: 10.3390/ijms22168458 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jupe F. et al. The complex architecture and epigenomic impact of plant T-DNA insertions. PLoS Genet 15, e1007819, doi: 10.1371/journal.pgen.1007819 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pucker B, Kleinbolting N. & Weisshaar B. Large scale genomic rearrangements in selected Arabidopsis thaliana T-DNA lines are caused by T-DNA insertion mutagenesis. BMC Genomics 22, 599, doi: 10.1186/s12864-021-07877-8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chilton MD et al. Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell 11, 263–271, doi: 10.1016/0092-8674(77)90043-5 (1977). [DOI] [PubMed] [Google Scholar]
- 6.Galbiati M, Moreno MA, Nadzan G, Zourelidou M. & Dellaporta SL Large-scale T-DNA mutagenesis in Arabidopsis for functional genomic analysis. Funct Integr Genomics 1, 25–34, doi: 10.1007/s101420000007 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Zambryski P. et al. Tumor DNA structure in plant cells transformed by A. tumefaciens. Science 209, 1385–1391, doi: 10.1126/science.6251546 (1980). [DOI] [PubMed] [Google Scholar]
- 8.Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA & Voytas DF DNA replicons for plant genome engineering. Plant Cell 26, 151–163, doi: 10.1105/tpc.113.119792 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kralemann LEM et al. Distinct mechanisms for genomic attachment of the 5’ and 3’ ends of Agrobacterium T-DNA in plants. Nat Plants 8, 526–534, doi: 10.1038/s41477-022-01147-5 (2022). [DOI] [PubMed] [Google Scholar]
- 10.van Kregten M. et al. T-DNA integration in plants results from polymerase-theta-mediated DNA repair. Nat Plants 2, 16164, doi: 10.1038/nplants.2016.164 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Jacob Y. et al. Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature 466, 987–991, doi: 10.1038/nature09290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davarinejad H. et al. The histone H3.1 variant regulates TONSOKU-mediated DNA repair during replication. Science 375, 1281–1286, doi: 10.1126/science.abm5320 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaratiegui M. Cross-Regulation between Transposable Elements and Host DNA Replication. Viruses 9, doi: 10.3390/v9030057 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavrak VV et al. How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet 10, e1004115, doi: 10.1371/journal.pgen.1004115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito H. et al. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472, 115–119, doi: 10.1038/nature09861 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Wolter F, Klemm J. & Puchta H. Efficient in planta gene targeting in Arabidopsis using egg cell-specific expression of the Cas9 nuclease of Staphylococcus aureus. Plant J 94, 735–746, doi: 10.1111/tpj.13893 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Bechtold N, Ellis J, and Pelletier G. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. . Comp. Rend. L’Acad. des Sci. Serie III 316, 1194–1199 (1993). [Google Scholar]
- 18.Alonso JM et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657, doi: 10.1126/science.1086391 (2003). [DOI] [PubMed] [Google Scholar]
- 19.McElver J. et al. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159, 1751–1763, doi: 10.1093/genetics/159.4.1751 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sessions A. et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994, doi: 10.1105/tpc.004630 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamp JA, van Schendel R, Dilweg IW & Tijsterman M. BRCA1-associated structural variations are a consequence of polymerase theta-mediated end-joining. Nat Commun 11, 3615, doi: 10.1038/s41467-020-17455-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacob Y. et al. Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science 343, 1249–1253, doi: 10.1126/science.1248357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacob Y. et al. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat Struct Mol Biol 16, 763–768, doi: 10.1038/nsmb.1611 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishizawa-Yokoi A. et al. Agrobacterium T-DNA integration in somatic cells does not require the activity of DNA polymerase theta. New Phytol 229, 2859–2872, doi: 10.1111/nph.17032 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Inagaki S. et al. Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell 18, 879–892, doi: 10.1105/tpc.105.036798 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hussmann JA et al. Mapping the genetic landscape of DNA double-strand break repair. Cell 184, 5653–5669 e5625, doi: 10.1016/j.cell.2021.10.002 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Budzowska M. et al. Mutation of the mouse Rad17 gene leads to embryonic lethality and reveals a role in DNA damage-dependent recombination. Embo J 23, 3548–3558, doi: 10.1038/sj.emboj.7600353 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q. et al. Rad17 recruits the MRE11-RAD50-NBS1 complex to regulate the cellular response to DNA double-strand breaks. Embo J 33, 862–877, doi: 10.1002/embj.201386064 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams GJ, Lees-Miller SP & Tainer JA Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair (Amst) 9, 1299–1306, doi: 10.1016/j.dnarep.2010.10.001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fauser F. et al. In planta gene targeting. Proc Natl Acad Sci U S A 109, 7535–7540, doi: 10.1073/pnas.1202191109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheiffele P, Pansegrau W. & Lanka E. Initiation of Agrobacterium tumefaciens T-DNA processing. Purified proteins VirD1 and VirD2 catalyze site- and strand-specific cleavage of superhelical T-border DNA in vitro. J Biol Chem 270, 1269–1276, doi: 10.1074/jbc.270.3.1269 (1995). [DOI] [PubMed] [Google Scholar]
- 32.Ward ER & Barnes WM VirD2 protein of Agrobacterium tumefaciens very tightly linked to the 5′ end of T-strand DNA. Science 242 (1988). [Google Scholar]
- 33.Neale MJ, Pan J. & Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436, 1053–1057, doi: 10.1038/nature03872 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jasper F, Koncz C, Schell J. & Steinbiss HH Agrobacterium T-strand production in vitro: sequence-specific cleavage and 5’ protection of single-stranded DNA templates by purified VirD2 protein. Proc Natl Acad Sci U S A 91, 694–698, doi: 10.1073/pnas.91.2.694 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleinboelting N. et al. The Structural Features of Thousands of T-DNA Insertion Sites Are Consistent with a Double-Strand Break Repair-Based Insertion Mechanism. Mol Plant 8, 1651–1664, doi: 10.1016/j.molp.2015.08.011 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Feng W. et al. Marker-free quantification of repair pathway utilization at Cas9-induced double-strand breaks. Nucleic Acids Res 49, 5095–5105, doi: 10.1093/nar/gkab299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brzezinka K, Altmann S. & Baurle I. BRUSHY1/TONSOKU/MGOUN3 is required for heat stress memory. Plant Cell Environ 42, 771–781, doi: 10.1111/pce.13365 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Li W. et al. The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci U S A 101, 10596–10601, doi: 10.1073/pnas.0404110101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valuchova S. et al. Protection of Arabidopsis Blunt-Ended Telomeres Is Mediated by a Physical Association with the Ku Heterodimer. Plant Cell 29, 1533–1545, doi: 10.1105/tpc.17.00064 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heacock ML, Idol RA, Friesner JD, Britt AB & Shippen DE Telomere dynamics and fusion of critically shortened telomeres in plants lacking DNA ligase IV. Nucleic Acids Res 35, 6490–6500, doi: 10.1093/nar/gkm472 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heitzeberg F. et al. The Rad17 homologue of Arabidopsis is involved in the regulation of DNA damage repair and homologous recombination. Plant J 38, 954–968, doi: 10.1111/j.1365-313X.2004.02097.x (2004). [DOI] [PubMed] [Google Scholar]
- 42.Samanic I, Simunic J, Riha K. & Puizina J. Evidence for distinct functions of MRE11 in Arabidopsis meiosis. PLoS One 8, e78760, doi: 10.1371/journal.pone.0078760 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng W. et al. Large-scale heterochromatin remodeling linked to overreplication-associated DNA damage. Proc Natl Acad Sci U S A 114, 406–411, doi: 10.1073/pnas.1619774114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Culligan K, Tissier A. & Britt A. ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16, 1091–1104, doi: 10.1105/tpc.018903 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Curtis MD & Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133, 462–469, doi: 10.1104/pp.103.027979 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clough SJ & Bent AF Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16, 735–743, doi: 10.1046/j.1365-313x.1998.00343.x (1998). [DOI] [PubMed] [Google Scholar]
- 47.Puizina J, Siroky J, Mokros P, Schweizer D. & Riha K. Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16, 1968–1978, doi: 10.1105/tpc.104.022749 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molina-Risco M. et al. Optimizing Agrobacterium-Mediated Transformation and CRISPR-Cas9 Gene Editing in the tropical japonica Rice Variety Presidio. Int J Mol Sci 22, doi: 10.3390/ijms222010909 (2021). [DOI] [Google Scholar]
- 49.Clemente T. Nicotiana (Nicotiana tobaccum, Nicotiana benthamiana). Methods Mol Biol 343, 143–154, doi: 10.1385/1-59745-130-4:143 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Livak KJ & Schmittgen TD Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408, doi: 10.1006/meth.2001.1262 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Wang S. et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science 353, 598–602, doi: 10.1126/science.aaf8084 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rouillard JM, Zuker M. & Gulari E. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res 31, 3057–3062, doi: 10.1093/nar/gkg426 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sage D. et al. DeconvolutionLab2: An open-source software for deconvolution microscopy. Methods 115, 28–41, doi: 10.1016/j.ymeth.2016.12.015 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Schindelin J. et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682, doi: 10.1038/nmeth.2019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jovtchev G, Schubert V, Meister A, Barow M. & Schubert I. Nuclear DNA content and nuclear and cell volume are positively correlated in angiosperms. Cytogenet Genome Res 114, 77–82, doi: 10.1159/000091932 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Ollion J, Cochennec J, Loll F, Escude C. & Boudier T. TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics 29, 1840–1841, doi: 10.1093/bioinformatics/btt276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen S, Zhou Y, Chen Y. & Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890, doi: 10.1093/bioinformatics/bty560 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cock PJ et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423, doi: 10.1093/bioinformatics/btp163 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ Basic local alignment search tool. J Mol Biol 215, 403–410, doi: 10.1016/S0022-2836(05)80360-2 (1990). [DOI] [PubMed] [Google Scholar]
- 60.Lamesch P. et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40, D1202–1210, doi: 10.1093/nar/gkr1090 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997 (2013). [Google Scholar]
- 62.Katoh K. & Standley DM MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30, 772–780, doi: 10.1093/molbev/mst010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Bel M. et al. PLAZA 5.0: extending the scope and power of comparative and functional genomics in plants. Nucleic Acids Res 50, D1468–D1474, doi: 10.1093/nar/gkab1024 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Danecek P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, doi: 10.1093/gigascience/giab008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramirez F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–165, doi: 10.1093/nar/gkw257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated from this study are included within the main figures, extended data and supplementary information. Raw sequencing data are deposited at the NCBI SRA (BioProject: PRJNA892619). Analysis of the data made use of the TAIR10 Arabidopsis genome (https://www.arabidopsis.org/download/index-auto.jsp%3Fdir%3D%252Fdownload_files%252FGenes%252FTAIR10_genome_release) and the Orthologous gene family list sourced from the Dicots PLAZA 5.0 database (https://ftp.psb.ugent.be/pub/plaza/plaza_public_dicots_05/GeneFamilies/genefamily_data.ORTHOFAM.csv.gz). There are no restrictions on data availability.