Abstract
Plasmid vectors containing Japanese encephalitis virus (JEV) premembrane (prM) and envelope (E) genes were constructed that expressed prM and E proteins under the control of a cytomegalovirus immediate-early gene promoter. COS-1 cells transformed with this plasmid vector (JE-4B clone) secreted JEV-specific extracellular particles (EPs) into the culture media. Groups of outbred ICR mice were given one or two doses of recombinant plasmid DNA or two doses of the commercial vaccine JEVAX. All mice that received one or two doses of DNA vaccine maintained JEV-specific antibodies 18 months after initial immunization. JEVAX induced 100% seroconversion in 3-week-old mice; however, none of the 3-day-old mice had enzyme-linked immunosorbent assay titers higher than 1:400. Female mice immunized with this DNA vaccine developed plaque reduction neutralization antibody titers of between 1:20 and 1:160 and provided 45 to 100% passive protection to their progeny following intraperitoneal challenge with 5,000 PFU of virulent JEV strain SA14. Seven-week-old adult mice that had received a single dose of JEV DNA vaccine when 3 days of age were completely protected from a 50,000-PFU JEV intraperitoneal challenge. These results demonstrate that a recombinant plasmid DNA which produced JEV EPs in vitro is an effective vaccine.
Japanese encephalitis (JE) is a mosquito-borne viral disease of major public health importance in Asia. More than 35,000 cases and 10,000 deaths are reported annually (52). Japanese encephalitis virus (JEV) is a member of the genus Flavivirus in the family Flaviviridae. More than 70 species in the Flavivirus genus have been genetically and serologically classified (29). Other important human pathogenic flaviviruses include yellow fever, dengue type 1 to 4 (DEN1 to DEN4), tick-borne encephalitis (TBE), and St. Louis encephalitis (SLE) viruses. Vaccination has been an effective mechanism for prevention of flavivirus infection in humans and domestic animals. Three JEV vaccines are in widespread production and use (52). These are inactivated virus from infected mouse brain, inactivated virus from primary hamster kidney cells, and a live attenuated SA14-14-2 vaccine. Only inactivated JEV vaccine, JEVAX, produced in mouse brain is distributed commercially and available internationally (52). Inactivated, mouse brain-derived whole virus vaccine is costly to prepare and carries the risk of allergic reaction to murine encephalitogenic basic proteins or gelatin stabilizer (45; M. M. Andersen, and T. Ronne, Letter, Lancet 337:1044, 1991). Since 1989, an unusual number of systemic reactions characterized by generalized urticaria and/or angioedema following JEVAX immunization have been reported from Australia, Canada, and Denmark (36). A major problem associated with use of the inactivated mouse brain vaccine is the failure to stimulate long-term immunity (39). Multiple immunization is recommended to provide adequate protection (28, 39). The attenuated JEV vaccine, SA14-14-2, is undergoing clinical trials (31). However, because of regulatory issues this vaccine has not found wide acceptance outside the People's Republic of China (11).
Several experimental recombinant virus, attenuated virus, and subunit JEV vaccines have been reported. Recombinant baculovirus vector that contained the JEV envelope (E) protein gene has been used to infect insect cells and produce E protein that has been studied as a biosynthetic immunogen (33). Recombinant vaccinia viruses expressing the JEV genes extending from premembrane (prM) to NS2B proteins have been the most promising candidate vaccines. These candidate vaccines produced extracellular virus-like particles (EPs) in infected cell culture that induced high titers of neutralizing and hemagglutination-inhibiting antibodies and protective immunity in mice (19–21, 47, 54). Recombinant vaccinia viruses expressing the same JEV genes based on the attenuated vaccinia virus strain, NYVAC-JEV, or canarypox, ALVAC-JEV, were tested in phase I human trials (18). In this trial, only 1 in 10 ALVAC-JEV recipients developed detectable viral neutralizing antibody, and vaccinia virus-preimmune recipients had a significantly lower humoral immune response.
Inoculation of animals with purified plasmid vectors (DNA) by the intramuscular (i.m.) or intradermal route leads to expression of the recombinant vector-encoded protein in transfected cells, resulting in stimulation of a protein-specific immune response. Plasmid DNA vaccines provide an alternative to attenuated, inactivated, or virus-vectored subunit vaccines. Flavivirus DNA vaccines for Murray Valley encephalitis, DEN2, JE, SLE, and TBE (Central European encephalitis and Russian spring summer encephalitis) viruses have been developed and tested in the mouse model (4, 17, 24, 30, 38, 49). All of these plasmid DNA constructs contained similar transcriptional regulatory elements and a flavivirus gene cassette. Vaccination of mice with these plasmid DNA vaccines induced a virus-specific antibody response, as detected by enzyme-linked immunosorbent assay (ELISA). However, production of neutralizing antibody leading to 100% protection of vaccinated animals from virus challenge was observed only after multiple immunizations or delivery of DNA to the epidermis by particle bombardment (4, 24, 49). In this study, we constructed a JEV prM and E gene cassette that incorporates an extended signal peptide sequence at the NH2 terminus of the prM gene and Kozak's sequence, an optimal translation enhancing element surrounding the AUG site. JEV protein expression was characterized using six different recombinant vectors containing the same insert. The humoral immune response and protection from virulent JEV challenge following immunization with the recombinant plasmid DNAs were compared to findings for the human vaccine, JEVAX, licensed by the U.S. Food and Drug Administration, in outbred ICR mice.
MATERIALS AND METHODS
Cell culture and virus strain.
COS-1, COS-7, and SV-T2 cells (1650-CRL, 1651-CRL, and 163.1-CCL; American Type Culture Collection) were grown at 37°C in Dulbecco's modified Eagle medium (Gibco Laboratories, Grand Island, N.Y.) supplemented with 10% heat-inactivated fetal bovine serum (HyClone Laboratories, Inc., Logan, Utah), 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 7.5% NaHCO3 (30 ml/liter), penicillin (100 U/ml), streptomycin (100 μg/ml). COS-1 and COS-7 cells were derived from simian virus 40 (SV40) transformed CV1 cells which have an African green monkey kidney cell origin. SV-T2 cells were derived from SV40-transformed mouse fibroblasts. Vero cells were grown under the same conditions except that 5% fetal calf serum without nonessential amino acid was used. C6/36 cells (13) were grown at 28°C in the same medium used for the COS-1 cells. The SA14 strain of JEV, propagated by intracranial inoculation into suckling mouse brain, was used for animal challenges and plaque reduction neutralization tests (PRNT). The SA14 virus used in ELISA and Western blot experiments was propagated in C6/36 cells and purified by ultracentrifugation on 30% glycerol–45% potassium tartrate gradients (37).
Construction of plasmids expressing JEV prM and E gene proteins.
Genomic RNA was extracted from 150 μl of SA14 mouse brain JEV by using a QIAamp viral RNA kit (Qiagen, Santa Clarita, Calif.). RNA was adsorbed on a silica membrane, eluted in 80 μl of diethyl pyrocarbonate (Sigma Chemical Co., St. Louis, Mo.)-treated water, and used as a template for amplification of JEV prM and E genes. Primer sequences were obtained from the published data (35). A single cDNA fragment containing genomic nucleotides (nt) 389 to 2478 was amplified by reverse transcriptase-mediated PCR (RT-PCR). Restriction enzyme sites for KpnI and XbaI and Kozak's sequence for an optimal translation initiation (25, 26) were engineered at the 5′ terminus of the cDNA by amplimer 14DV389. An in-frame translation termination codon, followed by a NotI restriction site, was introduced at the 3′ terminus of the cDNA by amplimer c14DV2453 (Fig. 1). A single-tube RT-PCR was performed using a Titan RT-PCR Kit (Roche Molecular Biochemical, Indianapolis, Ind.). The RT-PCR product was purified using a QIAquick PCR purification kit (Qiagen), and the DNA was eluted with 50 μl of 1 mM Tris-HCl (pH 7.5).
FIG. 1.
Map of the JEV genomic structure (top) and the DNA sequence of oligonucleotides used in RT-PCR to construct the transcription unit for the expression of prM-E protein coding regions (bottom). Potential transmembrane helices of viral polyprotein are indicated by blackened areas.
All vector constructions and analyses were carried out using standard techniques (46). RT-PCR-amplified cDNA was digested with enzymes KpnI and NotI and inserted into the KpnI-NotI site of eukaryotic expression plasmid vector pCDNA3 (Invitrogen, Carlsbad, Calif.). Electroportion-competent Escherichia coli XL1-Blue cells (Stratagene, La Jolla, Calif.) were transformed by electroporation (Gene Pulser; Bio-Rad Laboratories, Hercules, Calif.) and plated on Luria broth (LB) agar plates that contained carbenicillin (100 μg/ml; Sigma). Clones were picked and inoculated into 3 ml of LB containing carbenicillin (100 μg/ml). Plasmid DNA was extracted from a 14-h LB culture by using a QIAprep Spin Miniprep kit (Qiagen). Automated DNA sequencing was performed as recommended on an ABI Prism 377 DNA sequencer (Perkin-Elmer/Applied Biosystems, Foster City, Calif.). Both strands of the cDNA were sequenced and compared to the published SA14 virus sequence (35).
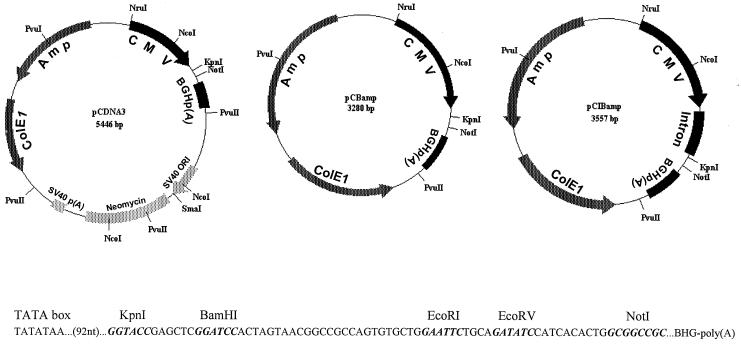
The pCDNA3 fragment from nt 1289 to nt 3455, which contained the f1-encoded eukaryotic origin of replication (ori), SV40 ori, neomycin coding region, and SV40 poly(A) elements, was deleted by PvuII digestion and then self-ligated to generate plasmid pCBamp. The pCIBamp vector, which contained a chimeric intron insertion at the NcoI-KpnI site of the pCB vector, was constructed by excising the intron sequence from pCI (Promega, Madison, Wis.) by digestion with NcoI and KpnI. The resulting 566-bp fragment was cloned into NcoI-KpnI-digested pCBamp to replace its 289-bp fragment. Figure 2 shows a schematic drawing of plasmids pCDNA3, pCBamp, and pCIBamp.
FIG. 2.
Schematic representations of plasmid vectors pCDNA3, pCBamp, and pCIBamp. These plasmids include the CMV promoter/enhancer element, BGH poly(A) signal and transcription termination sequence [BHGp(A)], ampicillin resistance gene (Amp), and ColE1 ori for selection and maintenance in E. coli. The f1 ori for single-stranded rescue in E. coli cells, SV40 ori, neomycin coding region, and SV40 poly(A) [SV40 p(A)] sequences were deleted from pCDNA3 to generate pCBamp. An intron sequence was inserted in the NcoI-KpnI site of pCBamp to generate pCIBamp. The multiple cloning site for the insertion of JEV genes, located between the TATA box of the CMV promoter/enhancer and BHG poly(A) site, is shown.
The DNA fragment containing the JEV coding region in the recombinant plasmid pCDJE2-7, derived from the pCDNA3 vector, was excised by NotI and KpnI or XbaI digestion and cloned into the KpnI-NotI sites of pCB, pCIB, pCEP4 (Invitrogen), and pREP4 (Invitrogen) and into the SpeI-NotI site of the pRc/RSV (Invitrogen) expression vector to create pCBJE1-14, pCIBJES14, pCEJE, pREJE, and pRCJE, respectively. Both strands of the cDNA from each plasmid vector were sequenced, and recombinant clones with a correct nucleotide sequence were identified. Plasmid DNA for in vitro transformation or mouse immunization was purified by anion-exchange chromatography using an EndoFree Plasmid Maxi kit (Qiagen).
IFA.
Expression of JEV-specific gene products by the various recombinant expression plasmids was evaluated by indirect immunofluorescence antibody assay (IFA) in the transient expression system using COS-1, COS-7, and SV-T2 cells. For transformation, cells were grown to 75% confluence in 150-cm2 culture flasks, trypsinized, and resuspended in 4°C phosphate-buffered saline (PBS) to a final density of 1 × 107 to 2 × 107 cells/ml. Five hundred microliters of cell suspension was then electroporated with 10 μg of plasmid DNA, using a Bio-Rad Gene Pulser II set at 250 V and 960 μF. Cells were diluted with 25 ml of fresh medium after electroporation and seeded into one 75-cm2 flask. Forty-eight hours after transformation, the medium was removed, and the cells were trysinized and resuspended in 5 ml of PBS with 3% normal goat serum. Ten-microliter aliquots of the cell suspension were then spotted onto slides, air dried, and fixed with acetone at 4°C for 10 min. Immunofluorescent mapping of the E protein-specific epitopes was performed using a panel of murine monoclonal antibodies (MAbs) (15, 42, 55) and JEV-specific hyperimmune mouse ascitic fluid (HIAF). All antibodies were tested at 1:400 dilution in PBS.
Selection of an in vitro-transformed stable cell line constitutively expressing JEV-specific gene products.
COS-1 cells transformed with 10 μg of pCDJE2-7 DNA by electroporation were incubated in nonselective culture medium for 24 h and then treated with neomycin (G418; 0.5 mg/ml; Sigma). G418-resistant colonies, which became visible after 2 to 3 weeks, were cloned by limited dilution in G418-containing medium. Expression of the JEV proteins was determined by IFA using JEV HIAF. One IFA-positive (JE-4B) and one IFA-negative (JE-5A) clone were selected for further analysis and maintained in medium containing 200 μg of G418 per ml. These stably transformed cells secreted antigen in the form of EPs (A. Hunt and G. J. Chang, unpublished data).
Antigen capture ELISA for detection of E protein secreted into culture fluid.
The antigen capture ELISA, a modification of the procedure described by Guirakhoo et al. (8), was used to detect E protein from transiently transformed cells or JE-4B culture fluid. Flavivirus group-reactive MAb 4G2 was used to capture the JEV antigens (7). The 4G2-captured antigen was detected using horseradish peroxidase-conjugated MAb 6B6C-1 by incubation for 1 h at 37°C. Enzyme activity on the solid phase was detected with 3,3′,5,5′-tetramethylbenzidine ELISA substrate (Life Technologies, Grand Island, N.Y.); the reaction was stopped with the addition of 2 M H2SO4, and the optical density was measured at 450 nm.
Mouse experiments.
Three-day-old mixed-sex or 3-week-old female ICR outbred mice were vaccinated i.m. with 50 or 100 μg of plasmid DNA at a concentration of 1 μg/μl in PBS or subcutaneously (s.c.) with 1/10 or 1/5 of the adult human dose of JEVAX (manufactured by the Research Foundation for Microbial Disease of Osaka University and distributed by Connaught Laboratories, Swiftwater, Pa.). The chloramphenicol acetyltransferase (CAT) protein expression plasmid pCDNA3/CAT (Invitrogen) was used as the vaccination control. Selected groups of mice were boosted 3 weeks later with an additional dose of plasmid vaccine or JEVAX. Mice were bled from the retro-orbital sinus; serum samples were evaluated for JEV antibody by ELISA and Western blotting using purified JEV and by PRNT.
Mice vaccinated at 3 days of age were challenged intraperitoneally (i.p.) 7 weeks postvaccination with JEV strain SA14 (50,000 PFU/100 μl) and observed for 3 weeks. To evaluate passive protection by maternal antibody, pups were obtained from mating of nonimmunized males with immunized females 9 weeks following their vaccination with plasmid DNA at 3 weeks of age. Pups were challenged by the i.p. route 3 to 15 days after birth with SA14 virus (5,000 PFU/100 μl) and observed daily for 3 weeks. Postchallenge serum was collected from survivors and tested for reactivity with JEV antigens by ELISA and Western blotting.
Serological tests.
Postvaccination and postchallenge serum samples were tested for the ability to bind to purified JEV by ELISA, neutralize JEV infectivity by PRNT, or recognize JEV proteins by Western blotting (12, 41, 48). The PRNT assay was performed by incubating ∼200 PFU of SA14 virus in 100 μl of Dulbecco's modified Eagle medium containing 5% bovine serum albumin and 20 mM HEPES buffer (pH 8.0) with serial twofold dilutions of serum specimens, started at 1:10, in 100 μl of the same buffer in 96-well trays at 4°C overnight. Serum specimens were heat inactivated at 56°C for 30 min before use. Duplicate 100-μl aliquots were assayed for infective virus by plaque formation on Vero cell monolayers. The percent plaque reduction was calculated relative to virus controls without serum. Titers were expressed as the reciprocal of serum dilutions yielding a 90% reduction in plaque number (PRNT90).
RESULTS
Effect of the promoter and poly(A) signal on the efficiency of JEV prM and E protein expression.
Four eukaryotic cell expression plasmids that contained the JEV coding region extending from genomic nt 390 to nt 2478 were constructed. This region of the genome encoded the prM and E genes. The Kozak sequence for the eukaryotic translation initiation site (underlined) of −9 to +4, GCCGCCGCCATGG, at the 5′ terminus (2, 25, 26, 27) and the in-frame translation termination sequence at the 3′ terminus of cDNA were incorporated directly into cDNA by RT-PCR using viral RNA as a template. Transcription of the JEV genes in plasmid pCDJE2-7 was controlled by the human cytomegalovirus (CMV) early IA gene promoter/enhancer. The resulting mRNA is terminated and stabilized by a bovine growth hormone (BGH) transcription terminator and a poly(A) signal, respectively. The transcriptional control elements in pREJE were replaced by the Rous sarcoma virus (RSV) long terminal repeat promoter and SV40 poly(A). The pCEJE and pRCJE plasmids contain CMV plus SV40 poly(A) and RSV plus BGH poly(A), respectively (Table 1).
TABLE 1.
Transient expression of JEV prM and E proteins by various recombinant plasmids in two transformed cell lines
|
|
Recombinant plasmid | IFA intensity/% positivea
|
|||||
|---|---|---|---|---|---|---|---|
| Name | Promoter | Intron | Poly(A) | Ori | COS-1 | COS-7 | |
| pCDNA3 | CMV | No | BGH | SV40 | pCDJE2-7 | 3+/40 | 3+/35 |
| pCBamp | CMV | No | BGH | No | pCBJE1-14 | 3+/45 | ND |
| pCIBamp | CMV | Yes | BGH | No | pCIBJES14 | 3+/39 | ND |
| pCEP4 | CMV | No | SV40 | OriP | pCEJE | 2+/4 | 2+/3 |
| pREP4 | RSV | No | SV40 | OriP | pREJE | 1+/3 | 1+/2 |
| pRc/RSV | RSV | No | BGH | SV40 | pRCJE | 1+/3 | 1+/3 |
| pCDNA3 | CMV | No | BGH | SV40 | pCDNA3/CAT | − | − |
Various cell lines were transformed with pCDNA3/CAT (negative control), pCDJE2-7, pCBJE1-14, pCIBJES14, pCEJE, pREJE, or pRCJE. Cells were trypsinized 48 h later and tested by IFA with JEV HIAF. Data are presented as the intensity (scale of 1+ to 4+) and percentage of IFA-positive cells. pCDNA3/CAT-transformed cells were used as the negative control. ND, not determined. −, negative.
To determine the influence of the promoter and poly(A) elements on JEV prM and E protein expression, recombinant plasmids pCDJE2-7, pCEJE, pRCJE, and pREJE were initially tested for the ability to express JEV prM and E proteins following transformation of various mammalian cells. COS-1, COS-7, and SV-T2 cells were transiently transformed with equal amounts of pCDJE2-7, pCEJE, pRCJE, or pREJE plasmid DNA. The SV-T2 cell line was excluded from further testing after preliminary results showed that less than 1% of pCDJE2-7-transformed SV-T2 cells were expressing JEV antigen.
JEV antigens were expressed in COS-1 and COS-7 cells transformed by all four recombinant plasmids, thus confirming that the CMV or RSV promoter and BGH or SV40 poly(A) elements were functionally active. However, the percentage of transformed cells and the level of JEV antigens expressed, as determined by the number of IFA-positive cells and IFA intensity, respectively, differed significantly (Table 1). A significantly higher percentage of pCDJE2-7-transformed COS-1 cells expressed JEV proteins with greater IFA intensity at a level equal to that observed with JEV-infected cells. Cells transformed with the pCEJE, pREJE, or pRCJE vector, on the other hand, showed a lower percentage of antigen-expressing cells as well as a lower IFA intensity. Vectors containing the CMV promoter and BGH poly(A) were selected for further analysis (Fig. 2).
To determine whether the enhanced expression of JEV proteins by the pCDJE2-7 vector was influenced by the SV40 ori, we constructed the pCBJE1-14 vector in which a 2,166-bp fragment containing the f1 ori, SV40 ori, neomycin coding region, and SV40 poly(A) elements was deleted. A chimeric intron was then inserted into pCBJE1-14 to generate pCIBJES14. Plasmid pCIBJES14 was used to determine whether the expression of JEV proteins could be enhanced by an intron sequence. Following transformation, both pCBJE1-14 and pCIBJES14 vectors resulted in cells expressing levels of JEV proteins similar to that observed with the pCDJE2-7 vector (Table 1). These results indicated that expression of the JEV proteins was influenced only by the transcriptional regulatory elements encoded in the recombinant plasmid. Neither the SV40 ori nor the intron sequence enhanced JEV protein expression in the cells used.
Epitope mapping of E protein expressed by a stably transformed cell line constitutively expressing JEV-specific gene products.
Authenticity of the JEV E protein expressed by the JE-4B clone was demonstrated by epitope mapping by IFA using a panel of JEV E-specific murine MAbs. JEV HIAF and one irrelevant mouse ascitic fluid were used as positive and negative antibody controls, respectively. Four JEV-specific, six flavivirus subgroup-specific, and two flavivirus group-reactive MAbs reacted similarly with the 4B clone and with JEV-infected COS-1 cells (Table 2).
TABLE 2.
Epitope mapping of E protein expressed by JE-4B, a pCDJE2-7 stably transformed clone of COS-1 cells, with JEV-reactive antibodiesa
| MAb or antiserum | Biological activity of MAb
|
IFA intensity of cells
|
||
|---|---|---|---|---|
| Specificity | Biological function | JEV infected | 4B | |
| MAbs | ||||
| MC3 | JEV specific | 2+ | 2+ | |
| 2F2 | JEV specific | HI, N | 4+ | 4+ |
| 112 | JEV specific | 4+ | 4+ | |
| 503 | JEV specific | N | 4+ | 3+ |
| 109 | Subgroup | HI | 2+ | 1+ |
| N.04 | Subgroup | HI, N | 3+ | 4+ |
| 201 | Subgroup | 1+ | 1+ | |
| 203 | Subgroup | 4+ | 3+ | |
| 204 | Subgroup | 2+ | 2+ | |
| 301 | Subgroup | HI | 2+ | 2+ |
| 504 | Flavivirus | 4+ | 4+ | |
| 6B6C-1 | Flavivirus | 2+ | 2+ | |
| 3B4C-4 | VEE | − | − | |
| HIAF | ||||
| Anti-JEV | 4+ | 3+ | ||
| Anti-WEE | − | − | ||
| PBS | − | − | ||
VEE, Venezuelan equine encephalomyelitis virus; WEE, Western equine encephalomyelitis virus. −, negative.
Detection of JEV E protein secreted by the JE-4B COS-1 cell line.
An antigen capture ELISA, employing flavivirus group-reactive, anti-E MAbs 4G2 and 6B6C-1, was used to detect JEV E proteins that were secreted into the culture fluid by the COS-1 cell clone JE-4B. Antigen could be detected in the culture fluid the first day following seeding of the cells with maximum ELISA titers that ranged from 1:16 to 1:32.
Comparison of immune responses in mice vaccinated with pCDJE2-7 genetic vaccine and JEVAX.
Plasmid pCDJE2-7 was used as a nucleic acid vaccine to induce an antibody response in mice by immunizing groups of five 3-week-old female ICR outbred mice. Mice were bled at 3, 6, 9, 23, 40, and 60 weeks after immunization, and antibody titers were determined by ELISA or by PRNT. As expected, sera from animals in the pCDNA3/CAT control group did not contain JEV antibody. All animals immunized with pCDJE2-7 and JEVAX seroconverted by 3 weeks after the first vaccination (Table 3). The antibody titers were similar irrespective of the number of doses of pCDJE2-7 or JEVAX given. Mouse serum samples collected 9 weeks after immunization were also tested by Western blotting using purified JEV. Serum specimens from DNA-vaccinated mice, which had reactivity similar to that of JEV HIAF, detected E and prM proteins (Fig. 3). However, mouse serum from JEVAX-immunized mice reacted only with E protein. Comparable ELISA antibody titers were maintained in DNA-vaccinated groups for up to 60 weeks, at which time the experiment was terminated. Only one of four mice in the JEVAX group remained JEV antibody positive at 60 weeks postinoculation. These results demonstrated that one dose of JEV-specific nucleic acid vaccine was more effective in maintaining JEV antibody levels in mice than the commercially available vaccine JEVAX.
TABLE 3.
Persistence of the immune response in mice (five per group) immunized with pCDJE2-7 or JEVAX
| Inoculationa | ELISA titer (log10)
|
PRNT90 titer
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 3b | 6 | 9 | 23 | 40 | 60c | 3 | 6 | 9 | |
| pCDJE2-7 | |||||||||
| 1 dose | 2.6–3.2 | 3.8–5.0 | 3.8–4.4 | >3.2 | >3.2 | 2.4, 2.4, 3.8, 4.4 | <20 | 20 | 40–160 |
| 2 doses | 2.6–3.8 | 4.4 | 3.8–4.4 | >3.2 | >3.2 | 2.6, 3.8, 3.8 | <20 | 20–40 | 40–160 |
| JEVAX, 2 doses | 2.6–3.8 | 4.4–5.0 | 3.8–5.6 | >3.2 | >3.2 | <2, <2, <2, 4.4 | <20 | 20–40 | 20–160 |
| pCDNA3/CAT, 2 doses | <100 | <100 | <100 | NDd | ND | ND | <20 | <20 | <20 |
Three-week-old mice were inoculated i.m. with one or two 100-μg doses of plasmid DNA or twice s.c. with one-fifth of the human dose of JEVAX.
Weeks postimmunization.
Individual serum titers.
ND, not determined.
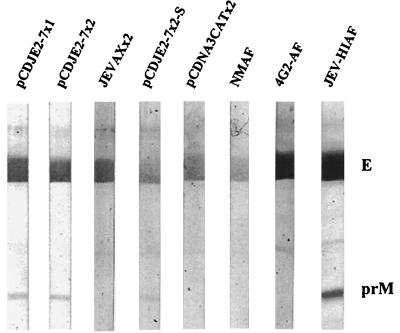
FIG. 3.
JEV-specific reactivity of prechallenge and postchallenge serum samples obtained from mice immunized with DNA vaccine or JEVAX. Serum specimens collected from the mice used in the experiments represented in Tables 3 and 4 were randomly selected and tested at 1:1,000 dilution by Western blot analysis using purified JEV as the antigen. pCDJE2-7x2-S was the serum from one of the mice challenged at 4 days of age (Table 4). NMAF, 4G2-AF, and JEV HIAF were the mouse ascitic fluids included as normal mouse, E-specific, and JEV hyperimmune controls, respectively.
Comparison of various nucleic acid vaccine constructs and JEVAX for ability to induce JEV-reactive antibody in different age groups of mice.
Similar amounts of JEV protein were expressed by COS-1 cells transformed by either pCDJE2-7, pCBJE1-14, or pCIBJES14. JEV antibody induction by these nucleic acid constructs was compared to results for JEVAX in two different age groups of mice. Three-day-old mixed-sex or 3-week-old female ICR outbred mice, 10 per group, were vaccinated i.m. with 50 or 100 μg of plasmid DNA or s.c. with 1/10 or 1/5 of the adult human dose of JEVAX, respectively. Serum specimens were collected at 7 weeks after immunization and tested at 1:400 or 1:1,600 by ELISA. Ninety to 100% of all 3-week-old mice that received pCBJE1-14, pCDJE2-7, pCIBJES14, or JEVAX had antibody titers of ≥1:1,600. However, a significant difference in antibody response was observed in 3-day-old groups that received various vaccines. None of the 3-day-old JEVAX-vaccinated mice had antibody titers higher than 1:400. All 3-day-old mice vaccinated with pCBJE1-14 had antibody titers higher than 1:1,600. Seroconversion of 100% was observed at 1:400 in 3-day-old mice that received pCDJE2-7 or pCIBJES14, but only 60% of both mouse groups were positive at 1:1,600. pCBJE1-14 was the most effective of three DNA constructs tested. The minimum dose of this DNA construct capable of providing 100% seroconversion (1:400 by ELISA) by i.m. immunization in 3-week-old mice was determined to be 25 μg (data not shown).
Protective immunity conferred by the nucleic acid vaccine.
Mice immunized at 3 days of age were challenged by the i.p. route at 7 weeks postvaccination with the SA14 strain of JEV (50,000 PFU/100 μl) and observed for 3 weeks. One hundred percent of the animals that received various nucleic acid vaccine constructs were protected. In contrast, only 40 and 30% of mice that received JEVAX and pCDNA3/CAT, respectively, survived virus challenge (Fig. 4). These results suggested that the DNA vaccine could be effective as a neonatal vaccine. In contrast, JEVAX was not as effective in neonatal animals.
FIG. 4.
Postchallenge survival rates of mice (10 per group) that were immunized with pCDJE2-7, pCBJE1-14, pCIBJES14, pcDNA3/CAT, or JEVAX at 3 days of age and challenged i.p. with 50,000 PFU of JEV (SA14) 7 weeks postimmunization. A P value of 0.003 was obtained by Fisher's exact test when the survival rate of the JEV DNA-immunized groups was compared with that of the pcDNA3/CAT or JEVAX group.
Passive protection of neonatal mice correlated with the maternal antibody titer.
Female 3-week-old ICR mice were vaccinated with one or two doses of pCDJE2-7 plasmid DNA (100 μg/100 μl) or twice with one-fifth of the adult human dose of JEVAX. For evaluation of passive protection by maternal antibody, pups were obtained from matings of experimental females with nonimmunized male mice. Pups were challenged by the i.p. route at 3 to 5 or 13 to 15 days after birth with SA14 virus (5,000 PFU/100 μl). Survival rates and average survival time correlated with the maternal neutralizing antibody titers (Table 4). One hundred percent of pups nursed by mothers with a PRNT of 1:80 survived viral infection regardless of the type of vaccine received by the mothers. None of the pups from mothers which received pCDNA3/CAT plasmid DNA survived (Table 4). Partial protection (45% [5 of 11 pups] to 67% [8 of 12 pups]) was observed in older pups that were nursed by the mothers which had serum PRNT titers of 1:20 and 1:40, respectively. However, none of the 3-day-old pups survived virus challenge when the mothers had a serum PRNT titer of 1:20 or 1:40. Maternally transferred antibody can only be detected in the circulation of the young mouse up to 40 days after birth. An appreciable level of maternally derived antibody is maintained in the circulation of the young mouse 24 days or more postpartum (1). JEV ELISA antibody detected in the serum of 97% (29 of 30) of the postchallenge pups at 12 weeks after virus challenge was unlikely to be residual maternally transferred antibody. The presence of JEV antibody in the surviving pups challenged at 3 to 4 or 13 to 15 days of age strongly suggested that maternal antibody did not provide sterilizing immunity to the pups. It also indicated that 3- to 4- or 13- to 15-day-old mice could mount an immune reaction to a live-virus challenge. Partial protection in older pups could be explained by the opportunity to accumulate a large quantity of passive antibody due to the length of nursing time before challenge. One randomly selected postchallenge serum sample also reacted with prM and E proteins by Western blotting (Fig. 3).
TABLE 4.
Ability of maternal antibody from JEV nucleic acid-vaccinated female mice to protect their pups from fatal JE
| Vaccinated mothera
|
JEV-challenged pups
|
||||
|---|---|---|---|---|---|
| Vaccine | PRNT90 | Age (days) | No. of survivors/total in litter | Avg survival time (days) | ELISAb |
| 1 × pCDJE2-7 | 40 | 4 | 0/11 | 5.27 | |
| 2 × pCDJE2-7 | 80 | 4 | 12/12 | NAc | 12/12 |
| 2 × JEVAX | 20 | 3 | 0/16 | 4.75 | |
| 2 × pCDNA3/CAT | <10 | 5 | 0/14 | 4.00 | |
| 1 × pCDJE2-7 | 20 | 15 | 5/11 | 10.0 | 5/5 |
| 2 × pCDJE2-7 | 40 | 14 | 8/12 | 13.75 | 7/8 |
| 2 × JEVAX | 80 | 13 | 5/5 | NA | 5/5 |
| 2 × pCDNA3/CAT | <10 | 14 | 0/14 | 6.14 | |
Mice were inoculated i.m. with one or two 100-μg doses of pCDJE2-7 DNA or twice s.c. with one-fifth of the adult human dose of JEVAX. Serum samples were collected 9 weeks postvaccination for PRNT testing prior to mating with nonimmune male.
Number of JEV ELISA antibody-positive animals (titer ≥ 1:400)/number of survivors. Serum specimens were collected for testing 12 weeks after challenge.
NA, not applicable.
DISCUSSION
The flavivirus virion contains a capsid protein (C), a membrane protein (M), and an E protein. The prM MAbs, exhibiting weak or undetectable neutralizing activity in vitro, can provide passive protection following DEN2 virus challenge (16). However, the E protein plays a dominant role in generating neutralizing antibodies and providing protective immunity in the host. Passive transfer of JEV E-specific neutralizing MAbs has been shown to protect recipients from JEV-induced fatal encephalitis (3, 16, 32, 55). Antigenic and structural analysis using various panels of MAbs has shown that most of the E protein epitopes that elicit virus-neutralizing antibodies are conformationally dependent (9, 40). Coexpression of both proteins as type I transmembrane proteins is essential to maintain proper E conformation and prevent the E protein from undergoing irreversible, low-pH-catalyzed conformational changes (8–10, 19, 50). A 2-kb genomic region, from the internal signal peptide at the carboxyl terminus of C to the transmembrane domain at the carboxyl terminus of the E gene, is essential for expressing authentic proteins. These authentic prM and E proteins are able to self-assemble into virus-like particles in cells infected by either recombinant vaccinia virus or alphavirus vector or in cells transformed by recombinant plasmid DNA (4, 19, 22, 48; Hunt and Chang, unpublished data).
A gene cassette including the elements listed above was amplified from SA14 virus by RT-PCR in the present study. Optimal sequence composition surrounding the translation initiation site (−9 to +4) was incorporated into the 14DV398 amplifying primer (2, 26, 27) (Fig. 1). Recombinant plasmids containing the CMV early gene promoter/enhancer and the BHG poly(A) terminator as transcription regulatory elements expressed JEV proteins with the highest efficiency in three different cell lines. Protein expression and the serological response of mice immunized with DNA vaccine were not influenced by the presence or absence of the SV40 ori or an intron sequence in recombinant plasmids. Virus-specific proteins, secreted into culture medium, could be detected by antigen capture ELISA as early as 48 h after plasmid transformation (data not shown). The authenticity of the E protein produced by the pCDJE2-7 stably transformed cell line, JE-4B, was demonstrated by MAb epitope mapping.
Vaccine potential and characteristics of various eukaryotic plasmids that express flavivirus prM and E proteins are summarized in Tables 5 and 6. All constructs listed had the same transcriptional control elements and similar viral gene cassettes. DEN2 plasmid, which contains prM and 91% of E, is the only exception (Table 6). The JEV DNA vaccine reported in this study is the only construct that stimulated complete protective immunity in mice by a single dose of vaccine given by the i.m. route (Table 5). Sequences surrounding the translation initiation site and the composition of the signal peptide preceding the prM protein are the two major differences among the constructs that may contribute to increasing the vaccine potential of our construct (Table 6). Conserved features of the sequences which flank vertebrate translation initiation sites include a strong preference for purine at the −3 position; a higher frequency of G at positions −9, −6, −3, and +4; and a preference for A or C at positions −5, −4, −2, and −1 (2). Instead of the sequence used in previous publications, the sequence used in our construct was −9 · GCCGCCGCCATGG, which fits the general criteria listed above. Although less than 1% of eukaryotic mRNA sequences exhibit this sequence, the experimental data have suggested that this sequence provides exceptionally high levels of translation potential (2, 26).
TABLE 5.
Vaccine potential of various eukaryotic plasmids that express flavivirus prM and E proteinsa
| Virus | In vitro secretion of EPs | Immunization
|
Protection from virus challenge | Reference | ||
|---|---|---|---|---|---|---|
| Dosage | Route/method | Neutralizing antibodyb | ||||
| JE | Yes | 25–100 μg × 1 | i.m./needle | Yes (1:20–1:16090%) | 100% | This report |
| ND | 100 μg × 2 | i.m./needle | No | Partial | 30 | |
| ND | 10–100 μg × 2 | i.m. or i.d./needle | Yes (1:10–1:2090%) | 100% | 24 | |
| MVE | Yes | 100 μg × 4 | i.m./needle | ND | Partial | 4 |
| Yes | 1–2 μg × 2–4 | i.d./gene gun | Yes (80–32050%) | 100% | 4 | |
| SLE | ND | 100 μg × 2 | i.m./needle | No | Partial | 38 |
| CEE | ND | 1 μg × 1–2 | i.d./gene gun | Yes (1:100–1:1,60080%) | 100% | 49 |
| RSSE | ND | 1 μg × 1–2 | i.d./gene gun | ND | 100% | 49 |
| DEN2 | ND | 200 μg × 3 | i.d./needle | Yes (1:10–1:32050%) | None | 17 |
MVE, Murray Valley encephalitis; CEE, Central European encephalitis; RSSE, Russian spring-summer encephalitis; i.d., intradermal; ND, not done.
Plaque reduction neutralization titer followed by percentage reduction endpoint used in the test.
TABLE 6.
Characteristics of various eukaryotic plasmids expressing flavivirus prM and E proteins
| Virusa | Plasmid | Sequence surrounding translation initiation site | Amino acids preceding prM proteinb | SP potential (C score)c | Reference |
|---|---|---|---|---|---|
| JE | pCDJE2-7 | −9•GCCGCCGCCATGG•+4 | MGRKQNKRGGNEGSIMWLASLAVVIACAGA /MKL | Yes (0.921) | This report |
| pJME | −9•GGCTCAATCATGG•+4 | MWLASLAVVIACAGA /MKL | Yes (0.578) | 30 | |
| pCDNA3JEME | −9•GAATTCACCATGG•+4 | MNEGSIMWLASLAVVIACAGA /MKL | Yes (0.921) | 24 | |
| MVE | pCDNA3.prM-E | −9•TGATTTCAAATGT•+4 | MSKKRGGSETSVLMVIFMLIGFAAA /LKL | Yes (0.819) | 4 |
| SLE | pSLE1 | ? | ?LDTINRRPSKKRGGTRSLLGLAALIGLASS /LQL | Yes (0.709) | 38 |
| DEN2 | p1012D2ME | ? | ?AGMIIMLIPTVMA /FHL | Yes (0.646) | 17 |
| TBE | SV-PEwt | −9•GCGGCCGCCATGG•+4 | MVGLQKRGKRRSATDWMSWLLVITLLGMTLA /ATV | Yes (1.000) | 48 |
| RSSE | pWRG7077 | −9•GTAGACAGGATGG•+4 | MGWLLVVVLLGVTLA /ATV | Yes (0.762) | 50 |
| CEE | pWRG7077 | −9•ACGGACAGGATGG•+4 | MSWLLVITLLGMTIA /ATV | Yes (0.609) | 50 |
Abbreviations are as given in Table 5, footnote a.
Single amino acid code. Positively charged amino acid is indicated by bold letter. Signal peptidase cleavage site is indicated by /.
Cleavage potential of signal peptide (SP) predicted by SignalP-NN at http://www.cbs.dtu.dk/services (34).
Signal peptides determine translocation and orientation of inserted protein, hence the topology of prM and E. Signal peptide differences in our plasmid construct may account for the efficient translocation and correct topology, thus increasing prM and E secretion. A machine-learning program using neural networks trained on eukaryotes (SignalP-NN at http://www.cbs.dtu.dk/services/) was applied to test the efficiency of the prM signal peptide sequence in the different plasmid constructs (34) (Table 6). The most probable location and orientation of transmembrane helices in the prM-E protein were then determined by a hidden Markov model-trained computer program (6 [TMHMM at http://www.cbs.dtu.dk/services/]). SignalP-NN searches correctly predicted the signal peptidase cleavage site of all constructs. However, a considerable difference in cleavage potential (C score, between 0.578 and 1.000) was observed (Table 6). Cleavage potential differences may be influenced by the amino acid composition and length of the h region in various constructs (44).
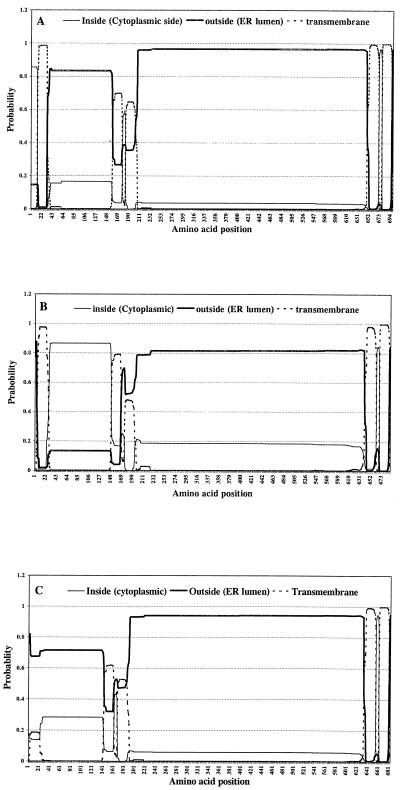
The TMHMM program correctly predicted five transmembrane helices encoded in the prM-E protein. Significant difference in the probable orientation of the first transmembrane helix was observed in three JEV constructs (Fig. 5). In our pCDJE2-7 construct, the first 12 amino acids of the n region form a short loop in the cytoplasmic side that causes the following h region (transmembrane helix) to be inserted in a tail orientation. Secretion of JEV protein could be detected by antigen capture ELISA in pCDJE2-7 transient expression studies in which less than 5% of the cells were positive by IFA (data not shown). Thus, there is a high probability that prM and E proteins expressed by pCDJE2-7 would be expressed in the correct orientation, as type I transmembrane proteins (Fig. 5A). There is also a high probability that the prM protein of pcDNA3JEME could be expressed as a type II membrane protein with its transmembrane h region inserted in a head orientation because of the absence of positively charged amino acids in its n region (Fig. 5B). Efficient protein synthesis in conjunction with correct topology of expressed prM and E (Fig. 5A) would most likely enhance EP formation and secretion in transformed cells.
FIG. 5.
Graphic representation, generated by the TMHMM program, indicating probable orientations of five transmembrane helices in the prM-E protein expressed by pCDJE2-7 (A), pcDNA3JEME (B), and pJME (C). ER, endoplasmic reticulum.
Another characteristic that could explain the excellent vaccine potential of our JEV construct is its ability to produce EPs which have a virus-like polymeric structure that enhances antigenic stability and provides a high-density presentation to antigen-presenting cells, such as macrophages, dendritic cells, and Langerhans cells (5). When DNA is given by the i.m. route, the majority of antigen is expressed by non-antigen-presenting muscle cells. The efficacy of a DNA vaccine is therefore dependent on transfection of antigen-presenting cells or to reprocessing of antigen derived from other cells. Muscle cells transfected by our construct could conceivably synthesize and secrete EPs, which are highly immunogenic and have been shown to elicit good cellular and humoral responses (22, 23).
Genetic JEV vaccine that induced a completely protective immunity in neonatal mice and a maternally transferable protective immunity in young adult mice by a single i.m. immunization was demonstrated in this study. Additional studies are planned to address the effectiveness of a DNA vaccine in overcoming the potential influence of maternally transferred flavivirus antibodies on the induction of JEV antibody in neonatal mice.
Immunization of pigs is a theoretical means of interrupting transmission and amplification of JEV and thereby preventing human infections (43). The JEV DNA vaccine could also be used as a veterinary vaccine in pregnant sows to prevent JEV-induced stillbirth and abortion (51, 53). Maternally transferred antibody could also interrupt piglets as the JEV-amplifying host and thus reduce human infection.
ACKNOWLEDGMENTS
We thank K. Yasui and M.-J. Zhang for providing JEV MAbs and J. Roehrig and B. Miller for useful discussion and advice. We thank D. Holmes, C. Lin, N. Frank, and T. Springfield for superb technical assistance and animal care.
REFERENCES
- 1.Appleby P, Catty D. Transmission of immunoglobulin to foetal and neonatal mice. J Reprod Immunol. 1983;5:203–213. doi: 10.1016/0165-0378(83)90236-x. [DOI] [PubMed] [Google Scholar]
- 2.Cavener D R, Ray S C. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991;19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cecilia D, Gadkari D A, Kedarnath N, Ghosh S N. Epitope mapping of Japanese encephalitis virus envelope protein using monoclonal antibodies against an Indian strain. J Gen Virol. 1988;69:2741–2747. doi: 10.1099/0022-1317-69-11-2741. [DOI] [PubMed] [Google Scholar]
- 4.Colombage G, Hall R, Pavy M, Lobigs M. DNA-based and alphavirus-vectored immunisation with prM and E proteins elicits long-lived and protective immunity against the flavivirus, Murray Valley encephalitis virus. Virology. 1998;250:151–163. doi: 10.1006/viro.1998.9357. [DOI] [PubMed] [Google Scholar]
- 5.Condon C, Watkins S C, Celluzzi C M, Thompson K, Falo L D., Jr DNA-based immunization by in vivo transfection of dendritic cells. Nat Med. 1996;2:1122–1128. doi: 10.1038/nm1096-1122. [DOI] [PubMed] [Google Scholar]
- 6.Erik L L, von Heijne S G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. In: Glasgow J, Littlejohn T, Major F, Lathrop R, Sankoff D, Sensen C, editors. Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. Menlo Park, Calif: AAAI Press; 1998. pp. 175–182. [PubMed] [Google Scholar]
- 7.Gentry M K, Henchal E A, McCown J M, Brandt W E, Dalrymple J M. Identification of distinct antigenic determinants on dengue-2 virus using monoclonal antibodies. Am J Trop Med Hyg. 1982;31:548–555. doi: 10.4269/ajtmh.1982.31.548. [DOI] [PubMed] [Google Scholar]
- 8.Guirakhoo F, Bolin R A, Roehrig J T. The Murray Valley encephalitis virus prM protein confers acid resistance to virus particles and alters the expression of epitopes within the R2 domain of E glycoprotein. Virology. 1992;191:921–931. doi: 10.1016/0042-6822(92)90267-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heinz F X, Berger R, Tuma W, Kunz C. A topological and functional model of epitopes on the structural glycoprotein of tick-borne encephalitis virus defined by monoclonal antibodies. Virology. 1983;126:525–537. doi: 10.1016/s0042-6822(83)80010-5. [DOI] [PubMed] [Google Scholar]
- 10.Heinz F X, Stiasny K, Puschner-Auer G, Holzmann H, Allison S L, Mandl C W, Kunz C. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology. 1994;198:109–117. doi: 10.1006/viro.1994.1013. [DOI] [PubMed] [Google Scholar]
- 11.Hennessy S, Liu Z, Tsai T F, Strom B L, Wan C M, Liu H L, Wu T X, Yu H J, Liu Q M, Karabatsos N, Bilker W B, Halstead S B. Effectiveness of live-attenuated Japanese encephalitis vaccine (SA14-14-2): a case-control study. Lancet. 1996;347:1583–1586. doi: 10.1016/s0140-6736(96)91075-2. [DOI] [PubMed] [Google Scholar]
- 12.Hunt A R, Calisher C H. Relationships of bunyamwera group viruses by neutralization. Am J Trop Med Hyg. 1979;28:740–749. [PubMed] [Google Scholar]
- 13.Igarashi A. Isolation of a Singh's Aedes albopictus cell clone sensitive to dengue and Chikungunya viruses. J Gen Virol. 1978;40:531–544. doi: 10.1099/0022-1317-40-3-531. [DOI] [PubMed] [Google Scholar]
- 14.Kaufman B M, Summers P L, Dubois D R, Cohen W H, Gentry M K, Timchak R L, Burke D S, Eckels K H. Monoclonal antibodies for dengue virus prM glycoprotein protect mice against lethal dengue infection. Am J Trop Med Hyg. 1989;41:576–580. doi: 10.4269/ajtmh.1989.41.576. [DOI] [PubMed] [Google Scholar]
- 15.Kimura-Kuroda J, Yasui K. Topographical analysis of antigenic determinants on envelope glycoprotein V3 (E) of Japanese encephalitis virus, using monoclonal antibodies. J Virol. 1983;45:124–132. doi: 10.1128/jvi.45.1.124-132.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura-Kuroda J, Yasui K. Protection of mice against Japanese encephalitis virus by passive administration with monoclonal antibodies. J Immunol. 1988;141:3606–3610. [PubMed] [Google Scholar]
- 17.Kochel T, Wu S J, Raviprakash K, Hobart P, Hoffman S, Porter K, Hayes C. Inoculation of plasmids expressing the dengue-2 envelope gene elicit neutralizing antibodies in mice. Vaccine. 1997;15:547–552. doi: 10.1016/s0264-410x(97)00215-6. [DOI] [PubMed] [Google Scholar]
- 18.Konishi E, Kurane I, Mason P W, Shope R E, Kanesa-Thasan N, Smucny J J, Hoke C H, Jr, Ennis F A. Induction of Japanese encephalitis virus-specific cytotoxic T lymphocytes in humans by poxvirus-based JE vaccine candidates. Vaccine. 1998;16:842–849. doi: 10.1016/s0264-410x(97)00265-x. [DOI] [PubMed] [Google Scholar]
- 19.Konishi E, Mason P W. Proper maturation of the Japanese encephalitis virus envelope glycoprotein requires cosynthesis with the premembrane protein. J Virol. 1993;67:1672–1675. doi: 10.1128/jvi.67.3.1672-1675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konishi E, Pincus S, Fonseca B A, Shope R E, Paoletti E, Mason P W. Comparison of protective immunity elicited by recombinant vaccinia viruses that synthesize E or NS1 of Japanese encephalitis virus. Virology. 1991;185:401–410. doi: 10.1016/0042-6822(91)90788-d. [DOI] [PubMed] [Google Scholar]
- 21.Konishi E, Pincus S, Paoletti E, Laegreid W W, Shope R E, Mason P W. A highly attenuated host range-restricted vaccinia virus strain, NYVAC, encoding the prM, E, and NS1 genes of Japanese encephalitis virus prevents JEV viremia in swine. Virology. 1992;190:454–458. doi: 10.1016/0042-6822(92)91233-k. [DOI] [PubMed] [Google Scholar]
- 22.Konishi E, Pincus S, Paoletti E, Shope R E, Burrage T, Mason P W. Mice immunized with a subviral particle containing the Japanese encephalitis virus prM/M and E proteins are protected from lethal JEV infection. Virology. 1992;188:714–720. doi: 10.1016/0042-6822(92)90526-u. [DOI] [PubMed] [Google Scholar]
- 23.Konishi E, Win K S, Kurane I, Mason P W, Shope R E, Ennis F A. Particulate vaccine candidate for Japanese encephalitis induces long-lasting virus-specific memory T lymphocytes in mice. Vaccine. 1997;15:281–286. doi: 10.1016/s0264-410x(96)00180-6. [DOI] [PubMed] [Google Scholar]
- 24.Konishi E, Yamaoka M, Khin-Sane-Win, Kurane I, Mason P W. Induction of protective immunity against Japanese encephalitis in mice by immunization with a plasmid encoding Japanese encephalitis virus premembrane and envelope genes. J Virol. 1998;72:4925–4930. doi: 10.1128/jvi.72.6.4925-4930.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984;12:857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 27.Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–2492. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ku C C, King C C, Lin C Y, Hsu H C, Chen L Y, Yueh Y Y, Chang G J. Homologous and heterologous neutralization antibody responses after immunization with Japanese encephalitis vaccine among Taiwan children. J Med Virol. 1994;44:122–131. doi: 10.1002/jmv.1890440204. [DOI] [PubMed] [Google Scholar]
- 29.Kuno G, Chang G J, Tsuchiya K R, Karabatsos N, Cropp C B. Phylogeny of the genus Flavivirus. J Virol. 1998;72:73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin Y L, Chen L K, Liao C L, Yeh C T, Ma S H, Chen J L, Huang Y L, Chen S S, Chiang H Y. DNA immunization with Japanese encephalitis virus nonstructural protein NS1 elicits protective immunity in mice. J Virol. 1998;72:191–200. doi: 10.1128/jvi.72.1.191-200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z L, Hennessy S, Strom B L, Tsai T F, Wan C M, Tang S C, Xiang C F, Bilker W B, Pan X P, Yao Y J, Xu Z W, Halstead S B. Short-term safety of live attenuated Japanese encephalitis vaccine (SA14-14-2): results of a randomized trial with 26,239 subjects. J Infect Dis. 1997;176:1366–1369. doi: 10.1086/517323. [DOI] [PubMed] [Google Scholar]
- 32.Mason P W, Dalrymple J M, Gentry M K, McCown J M, Hoke C H, Burke D S, Fournier M J, Mason T L. Molecular characterization of a neutralizing domain of the Japanese encephalitis virus structural glycoprotein. J Gen Virol. 1989;70:2037–2049. doi: 10.1099/0022-1317-70-8-2037. [DOI] [PubMed] [Google Scholar]
- 33.McCown J, Cochran M, Putnak R, Feighny R, Burrous J, Henchal E, Hoke C. Protection of mice against lethal Japanese encephalitis with a recombinant baculovirus vaccine. Am J Trop Med Hyg. 1990;42:491–499. doi: 10.4269/ajtmh.1990.42.491. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst. 1997;8:581–599. doi: 10.1142/s0129065797000537. [DOI] [PubMed] [Google Scholar]
- 35.Nitayaphan S, Grant J A, Chang G J, Trent D W. Nucleotide sequence of the virulent SA-14 strain of Japanese encephalitis virus and its attenuated vaccine derivative, SA-14-14-2. Virology. 1990;177:541–552. doi: 10.1016/0042-6822(90)90519-w. [DOI] [PubMed] [Google Scholar]
- 36.Nothdurft H D, Jelinek T, Marschang A, Maiwald H, Kapaun A, Loscher T. Adverse reactions to Japanese encephalitis vaccine in travellers. J Infect. 1996;32:119–122. doi: 10.1016/s0163-4453(96)91281-5. [DOI] [PubMed] [Google Scholar]
- 37.Obijeski J F, Bishop D H, Palmer E L, Murphy F A. Segmented genome and nucleocapsid of La Crosse virus. J Virol. 1976;20:664–675. doi: 10.1128/jvi.20.3.664-675.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillpotts R J, Venugopal K, Brooks T. Immunisation with DNA polynucleotides protects mice against lethal challenge with St. Louis encephalitis virus. Arch Virol. 1996;141:743–749. doi: 10.1007/BF01718332. [DOI] [PubMed] [Google Scholar]
- 39.Poland J D, Cropp C B, Craven R B, Monath T P. Evaluation of the potency and safety of inactivated Japanese encephalitis vaccine in US inhabitants. J Infect Dis. 1990;161:878–882. doi: 10.1093/infdis/161.5.878. [DOI] [PubMed] [Google Scholar]
- 40.Roehrig J T, Bolin R A, Kelly R G. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology. 1998;246:317–328. doi: 10.1006/viro.1998.9200. [DOI] [PubMed] [Google Scholar]
- 41.Roehrig J T, Hunt A R, Johnson A J, Hawkes R A. Synthetic peptides derived from the deduced amino acid sequence of the E-glycoprotein of Murray Valley encephalitis virus elicit antiviral antibody. Virology. 1989;171:49–60. doi: 10.1016/0042-6822(89)90509-6. [DOI] [PubMed] [Google Scholar]
- 42.Roehrig J T, Mathews J H, Trent D W. Identification of epitopes on the E glycoprotein of Saint Louis encephalitis virus using monoclonal antibodies. Virology. 1983;128:118–126. doi: 10.1016/0042-6822(83)90323-9. [DOI] [PubMed] [Google Scholar]
- 43.Rosen L. The natural history of Japanese encephalitis virus. Annu Rev Microbiol. 1986;40:395–414. doi: 10.1146/annurev.mi.40.100186.002143. [DOI] [PubMed] [Google Scholar]
- 44.Sakaguchi M, Tomiyoshi R, Kuroiwa T, Mihara K, Omura T. Functions of signal and signal-anchor sequences are determined by the balance between the hydrophobic segment and the N-terminal charge. Proc Natl Acad Sci USA. 1992;89:16–19. doi: 10.1073/pnas.89.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakaguchi M, Yoshida M, Kuroda W, Harayama O, Matsunaga Y, Inouye S. Systemic immediate-type reactions to gelatin included in Japanese encephalitis vaccines. Vaccine. 1997;15:121–122. doi: 10.1016/s0264-410x(96)00170-3. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Sato T, Takamura C, Yasuda A, Miyamoto M, Kamogawa K, Yasui K. High-level expression of the Japanese encephalitis virus E protein by recombinant vaccinia virus and enhancement of its extracellular release by the NS3 gene product. Virology. 1993;192:483–490. doi: 10.1006/viro.1993.1064. [DOI] [PubMed] [Google Scholar]
- 48.Schalich J, Allison S L, Stiasny K, Mandl C W, Kunz C, Heinz F X. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J Virol. 1996;70:4549–4557. doi: 10.1128/jvi.70.7.4549-4557.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmaljohn C, Vanderzanden L, Bray M, Custer D, Meyer B, Li D, Rossi C, Fuller D, Fuller J, Haynes J, Huggins J. Naked DNA vaccines expressing the prM and E genes of Russian spring summer encephalitis virus and Central European encephalitis virus protect mice from homologous and heterologous challenge. J Virol. 1997;71:9563–9569. doi: 10.1128/jvi.71.12.9563-9569.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stocks C E, Lobigs M. Signal peptidase cleavage at the flavivirus C-prM junction: dependence on the viral NS2B-3 protease for efficient processing requires determinants in C, the signal peptide, and prM. J Virol. 1998;72:2141–2149. doi: 10.1128/jvi.72.3.2141-2149.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takashima I, Watanabe T, Ouchi N, Hashimoto N. Ecological studies of Japanese encephalitis virus in Hokkaido: interepidemic outbreaks of swine abortion and evidence for the virus to overwinter locally. Am J Trop Med Hyg. 1988;38:420–427. doi: 10.4269/ajtmh.1988.38.420. [DOI] [PubMed] [Google Scholar]
- 52.Tsai T F, Chang G J, Yu Y X. Japanese encephalitis vaccines. In: Stanley A P, Orenstein W A, editors. Vaccines. 3rd ed. Philadelphia, Pa: W. B. Saunders; 1999. pp. 672–710. [Google Scholar]
- 53.Ueba N, Kimura T, Nakajima S, Kurimura T, Kitaura T. Field experiments on live attenuated Japanese encephalitis virus vaccine for swine. Biken J. 1978;21:95–103. [PubMed] [Google Scholar]
- 54.Yasuda A, Kimura-Kuroda J, Ogimoto M, Miyamoto M, Sata T, Sato T, Takamura C, Kurata T, Kojima A, Yasui K. Induction of protective immunity in animals vaccinated with recombinant vaccinia viruses that express PreM and E glycoproteins of Japanese encephalitis virus. J Virol. 1990;64:2788–2795. doi: 10.1128/jvi.64.6.2788-2795.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang M J, Wang M J, Jiang S Z, Ma W Y. Passive protection of mice, goats, and monkeys against Japanese encephalitis with monoclonal antibodies. J Med Virol. 1989;29:133–138. doi: 10.1002/jmv.1890290211. [DOI] [PubMed] [Google Scholar]