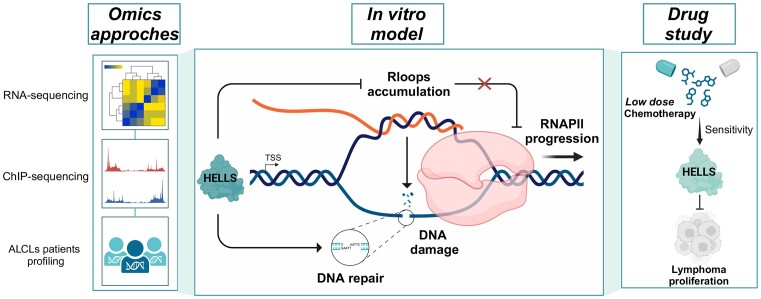
Abstract
Chromatin modifiers are emerging as major determinants of many types of cancers, including Anaplastic Large Cell Lymphomas (ALCL), a family of highly heterogeneous T-cell lymphomas for which therapeutic options are still limited. HELLS is a multifunctional chromatin remodeling protein that affects genomic instability by participating in the DNA damage response. Although the transcriptional function of HELLS has been suggested, no clues on how HELLS controls transcription are currently available. In this study, by integrating different multi-omics and functional approaches, we characterized the transcriptional landscape of HELLS in ALCL. We explored the clinical impact of its transcriptional program in a large cohort of 44 patients with ALCL. We demonstrated that HELLS, loaded at the level of intronic regions of target promoters, facilitates RNA Polymerase II (RNAPII) progression along the gene bodies by reducing the persistence of co-transcriptional R-loops and promoting DNA damage resolution. Importantly, selective knockdown of HELLS sensitizes ALCL cells to different chemotherapeutic agents, showing a synergistic effect. Collectively, our work unveils the role of HELLS in acting as a gatekeeper of ALCL genome stability providing a rationale for drug design.
Graphical Abstract
Graphical Abstract.
Introduction
ALK− Anaplastic Large Cell Lymphoma (ALCL) is one of the most aggressive and heterogeneous subtypes of T-cell lymphomas (TCLs). As relatively uncurable lymphoma with a poor prognosis (1,2), there is an urgent need to identify potential novel therapeutic targets. Maintaining a high transcription rate is a mandatory requirement for aggressive cancers. This serves to both keep up with uncontrolled cell proliferation and to guarantee the robust expression of cancer-essential genes (3). However, massive transcription imposes severe DNA-topological tensions leading to the generation of DNA–RNA structures (R-loops) and DNA double-strand breaks (DSBs) culminating in genome instability (4). Being at the nexus of all chromatin organization and function, chromatin remodeling complexes are emerging as pivotal players in cancer, including ALCLs (2).
HELLS, also known as LSH, is a chromatin remodeling protein related to SNF2/helicase family members, essential for the physiological proliferation of peripheral B- and T- lymphocytes (5).
HELLS is mutated in immunodeficiency, centromeric instability, facial anomalies (ICF) syndrome, a rare autosomal recessive disorder characterized by DNA hypomethylation in pericentromeric repeats and subtelomeres and by centromeric instability (6).
In ALCLs (7,8), as well as in other types of cancer (5), HELLS expression has a prognostic value, as its overexpression is associated with aggressive clinicopathological features and poorer patient outcomes. HELLS is involved in DSBs repair (5) and plays a critical role in maintaining chromatin structure by promoting the deposition of histone modifications, which can affect the accessibility of DNA to transcription factors (9). In addition to these canonical functions, a potential role for HELLS in controlling transcription has been proposed (10). In line with this hypothesis, we recently demonstrated that HELLS is an essential gene in ALCLs where it controls the activation of a precise transcriptional program required to sustain the proliferation, structural plasticity, and progression of ALCLs (7).
In this study, we explored this function in detail and provided, for the first time, a potential mechanism through which HELLS regulates lymphoma gene expression. We demonstrated that by binding to intronic regions of target genes, HELLS promotes transcription by easing unscheduled obstacles and favoring RNA Polymerase II (RNAPII) elongation. Thus, HELLS promotes the activation of a subset of ALCLs-specific support genes.
Materials and methods
Patients’ specimen
Clinicopathological characteristics of formalin-fixed paraffin-embedded (FFPE) retrospective cohorts of ALCL have been previously described (11). The study was approved by the Institutional Human Ethics Review Boards of the Ethical Committee AVEN and AUSL-IRCCS di Reggio Emilia (287/2018/OSS/IRCCSRE and 785/2022/TESS/IRCCSRE), and written informed consent was obtained from patients in accordance with the Declaration of Helsinki.
Gene expression profiling (GEP) by Nanostring
Total RNA was extracted by Maxwell® RSC RNA FFPE kit (Promega) starting from five slides of 5 μm FFPE tissue. RNA quantity and quality were assessed by NanoDrop2000 (Thermo Fisher Scientific). For samples that reached the quality standards (A260/A280 ≥ 1.7 and A260/A230 ≥ 1.8), we evaluated the gene expression profile (GEP) by the nCounter platform (NanoString Technologies) using a custom panel. This panel includes a total of 48 transcripts: 15 housekeeping genes, HELLS and 32 HELLS-direct genes (HDGs) (Supplementary Table S1). The set of 32 HDGs was chosen focusing on the cytoskeleton, DNA metabolism, and JAK/STAT signaling pathways deregulated after HELLS KD.
Molecular classification of ALCL samples has been performed by applying the three gene model (12) and considering the lack of ALK expression as previously described (11). Analysis of detected gene counts was performed by nSolver Analysis Software 4.0 (NanoString Technologies) as previously described (11).
Chromatin immunoprecipitation (ChIP) and ChIP-sequencing
ChIP-qRT-PCR experiments were performed using SimpleChIP Enzymatic Chromatin IP Kit (#9003Magnetic Beads, Cell Signaling Technology). Briefly, TLBR-2 HELLSKD cells (4 × 106/IP) were crosslinked for 10 min with 1% formaldehyde, lysed, and sonicated for 10 cycles (30 min ON, 30 min OFF) using Bioruptor Pico Sonicator (Diagenode, Denville, NJ, USA) to obtain 100–200 bp chromatin fragments. Chromatin was precipitated overnight using Dynabeads Protein G magnetic beads (Thermo Fisher Scientific), Phospho-Rpb1 CTD (Ser2) (1:50, E1Z3G, Rabbit mAb #13499, Cell Signaling Technology), Phospho-Rpb1 CTD (Ser5) (1:50, D9N5I, Rabbit mAb #13523, Cell Signaling Technology), 2.5 μg of RNAPII (#14958, Rabbit Monoclonal, Cell Signaling Technology), 3 μg of phosphor-histone H2A.X (Ser139) (#80312,Mouse mAb,Cell Signaling Technology) or IgG-isotype control (#66362, Cell Signaling Technology and sc-2025, Santa Cruz Biotechnology). A fraction equal to 0.25% of total chromatin was used as input. For Chip, each RT-qPCR value was normalized over the appropriate input control and reported as a % of input in graphs. The list of primers used is provided in Supplementary Table S2.
For ChIP-seq, TLBR-2 HELLSKD cells (40 × 106/IP) were crosslinked for 15 min with 1% formaldehyde, lysed and sonicated for 20 cycles (30 min ON, 30 min OFF) using Bioruptor Pico Sonicator (Diagenode, Denville, NJ, USA) to obtain 100–200 bp chromatin fragments. Chromatin was precipitated overnight using Dynabeads Protein G magnetic beads (Thermo Fisher Scientific) and 3.5 μg of HELLS (orb178580, Biorbyt), 1.5 μg of H3K4me3 (Ab 213223, Rabbit Polyclonal, Abcam), 2.5 μg of RNAPII (#14958, Rabbit Monoclonal, Cell Signaling Technology), or IgG-isotype control (#66362, Cell Signaling Technology). A fraction equal to 0.25% of total chromatin was used as input. Samples were quantified with Qubit (Thermo Fisher Scientific), and the quality was evaluated by Bioanalyzer (Agilent Technologies). Library for sequencing was obtained following the ThruPLEX DNA-Seq Kit (Takara Bio USA) using 3–10 ng ChIP DNA as starting material. Triplicates were sequenced on Illumina NextSeq500 high-output cartridge (single-stranded, read length 75 bp–1 × 75).
Library preparation and RNA-sequencing
RNA seq libraries were obtained starting from 100 ng of total RNA following Illumina Stranded TotalRNA Prep Ligation with Ribo-zero Plus protocol. Sequencing was performed using Illumina NEXTSeq high-output cartridge (double-stranded, read length 75 bp–2 × 75).
Sequencing data processing
Sequencing data quality was assessed using the FastQC v0.11.8 software (www.bioinformatics.babraham.ac.uk/projects/fastqc/), low-quality reads were discarded, and where residual adapters were present, they were removed using Trimmomatic (v.0.39) software. For ChIP-seq and ATAC-seq, filtered reads were aligned to the human reference genome (GRCh38/hg38 assembly) by using Bowtie2 version 2.3.5.1. Picard tool (http://broadinstitute.github.io/picard) and samtools 1.9v (http://samtools.sourceforge.net/) were used to remove duplicates and unmapped reads and to retain uniquely aligned reads for downstream analyses. For ChIP-seq, peak calling was performed using MACS2 (v.2.1.3.3). Significant peaks (q < 0.05) were merged, and high-confidence peaks enriched in at least two of three replicates were retained for the analysis. For the HELLS ChIP-seq experiment cut-off analysis was performed and ap-score <2.3 was set to call significant peaks. ChIPseeker R package was used for assigning peaks to the nearest genes, according to GRCh38/hg38 annotation, using a transcription start site (TSS) window of ±3 kb. Differential binding analysis was computed using the DiffBind R package (v2.13.1) (13,14). Differentially bound sites were retrieved by applying the edgeR method, considering a false discovery rate (FDR) of 10% and setting the date of the experiment (confounder) as a blocking factor for batch effect correction.
For ATAC-seq, peak calling was performed for each condition on replicas merged tracks using MACS2 (v.2.1.3.3.) with customize options (–nomodel –shift –100 –extsize 200 –bdg). Significant peaks (q-value < 0.05) were retained for subsequent analysis and assigned to the nearest genes (ChIPseeker R package), according to GRCh38/hg38 annotation, using a transcription start site (TSS) window of 2 kb (0.5 kb upstream and 1.5 kb downstream). Differential Enrichment analysis between the two conditions was performed with DESeq2 R package (v.1.42.0.), using the matrix of peaks read counts obtained with deepTools v.3.5.1 as input. (multiBamSummary BED-file mode). The shrunken log2 fold changes (15) were used for comparisons.
For RNA-seq, filtered paired-end reads were aligned to the human reference transcriptome (GRCh38, Gencode release 30 using STAR version 2.7), and gene abundances were estimated with the RSEM algorithm (v1.3.1). Differential analysis was performed with R package DESeq2, considering a false discovery rate (FDR) of 10% and excluding genes with low read counts. Significantly deregulated genes underwent enrichment analysis, performed through the enriched package on Gene Ontology biological processes using a significance threshold of 0.05 on the P value adjusted for multiple testing using the Benjamini–Hochberg correction. ChIP-seq and RNA-seq R analyses have been carried out on R version 3.6.3. ATAC-seq R analysis was performed on R version 4.3.2.
DRIP-qPCR
The DRIP assay was performed according to previous literature (16). After 48 h of doxycycline treatment, 8 × 106 TLBR-2 control and HELLS-KD cells were resuspended in 1.6 ml of TE with 0.5% of SDS and 5 μl of Proteinase K (Roche Life Sciences), then the cells were incubated overnight at 37°C. The genomic DNA was extracted with phenol/chloroform in MaXtract High-Density phase lock gel tubes (Qiagen) and precipitated with ethanol/sodium acetate. After washing with 70% ethanol, the genomic DNA was resuspended in TE. DNA was digested with HindIII, BsrGI, XbaI, EcoRI and SspI at 37°C overnight and was purified by phenol/chloroform and ethanol method. Digested genomic DNA was treated with or without 4 μl RNase H (Cat#: M0297S, NEB) for 6 h at 37°C. Then, 4.4 μg DNA was bound with 10 μg of S9.6 antibody (MABE1095, Millipore) and 50 μl Dynabeads Protein A (Thermo Fisher) in 1 × DRIP binding buffer (10 mM NaPO4 pH7.0, 140 mM NaCl, 0.05% Triton X-100) overnight at 4°C. After washing three times in 1 × DRIP binding buffer, the bound immunocomplexes were eluted in Elution Buffer (50 mM Tris pH 8.0, 10 mM EDTA, 0.5% SDS, Proteinase K) at 55°C with rotation. The eluted samples were then subjected to nucleic acid purification using phenol/chloroform in MaXtract High-Density phase lock gel tubes (Qiagen). The immunoprecipitated nucleic acid and input DNA were analyzed by qPCR using SsoFast™ EvaGreen® Supermix (Biorad) with a CFX-Connect Real-Time PCR machine (Biorad).
Proximity ligation assay (PLA)
To perform the proximity ligation assay, the Duolink® In Situ Red Mouse/Rabbit kit (DUO92101) from Sigma-Aldrich was employed. Briefly, cells were spotted on glass slides using Cytospin (Thermo Scientific), fixed with ice-cold methanol for 10 min at −20°C, permeabilized with acetone for 1 min at −20°C, and washed twice for 5 min with 4× SSC (pH 7). Dots were blocked in 5% BSA/0.1% Tween-20/4× SSC (pH 7) for 1 h at RT. Samples were incubated with Phospho-Rpb1 CTD (Ser2) antibody (E1Z3G, Rabbit mAb #13499, 1:200 Cell Signaling Technology) and anti-DNA–RNA Hybrid primary antibody (clone S9.6, MABE 1095, 1:200, Merck) (either or together) in all conditions (control versus doxycycline) at 4°C overnight. The next day, coverslips were washed two times with Wash Buffer A. According to the manufacturer's instructions, PLA probes (both MINUS and PLUS) were then diluted in the antibody diluent and placed on the slides in a humidification chamber for 1 h at 37°C. Ligation was later performed by adding ligase into the ligation solution buffer and incubating the slides in a pre-heated humidity chamber for 30 min at +37°C. After the amplification and probing processes (100 min at 37°C), slides were later washed two times with Buffer B for 10 min and one time with 0,01% Wash Buffer B and prepared for Imaging. DAPI was used to stain nuclei. Cells were imaged by placing the slides on the stage of a Nikon Eclipse (Ni) epifluorescence microscope using 60× and excited using the appropriate laser line. Images were acquired using a 16-bit resolution with ImageJ.
RNase H1 overexpression
ppyCAG_RNaseH1_WT (Addgene plasmid # 111906) and ppyCAG_RNaseH1_D210N (Addgene plasmid # 111904) were a gift from Xiang-Dong Fu. HEK-293T cells were plated into chamber slides (60 000) and transfected with plasmids (1 μg) by using 1 μl of Lipofectamine 2000 (Invitrogen # 11668019). After 6 h, the medium was changed, and half well was treated with doxycycline (100 ng/ml) for 48 h.
DNA repair assay
pimEJ5GFP and EJ2GFP-puro constructs were gifts from Jeremy Stark (Addgene plasmids # 44026, 44025). pDRGFP was a gift from Maria Jasin (Addgene plasmid # 26475). Each plasmid was pre-digested with I-SceI restriction enzyme and the digestion products following purification were run on a 1% agarose gel to confirm that each reporter had been digested. After induction of HELLSKD, HEK-293T cells were plated into a 96-well plate (20,000 cells per well) in triplicates and transfected with 0.15 ug SceI-linearized reporter plasmids by using 0.3 μl of Lipofectamine 2000 (Invitrogen # 11668019) per well, and the medium was changed 6 h later. Two hours later efficiency of the GFP gene repair in the reporter plasmids was evaluated by the IncuCyte®, following the manufacturer's instructions. The DNA repair efficiency was estimated as the ratio of GFP+ cells/total cells. All the analyses were performed with the IncuCyte® Live-Cell Analysis Systems (Model S3; Sartorius AG, Goettingen, Germany). Data were acquired using 10× and 20× objective lenses in phase contrast.
Results
HELLS binds to intragenic regions to selectively control transcription of a subset of ALK−ALCL supporting genes
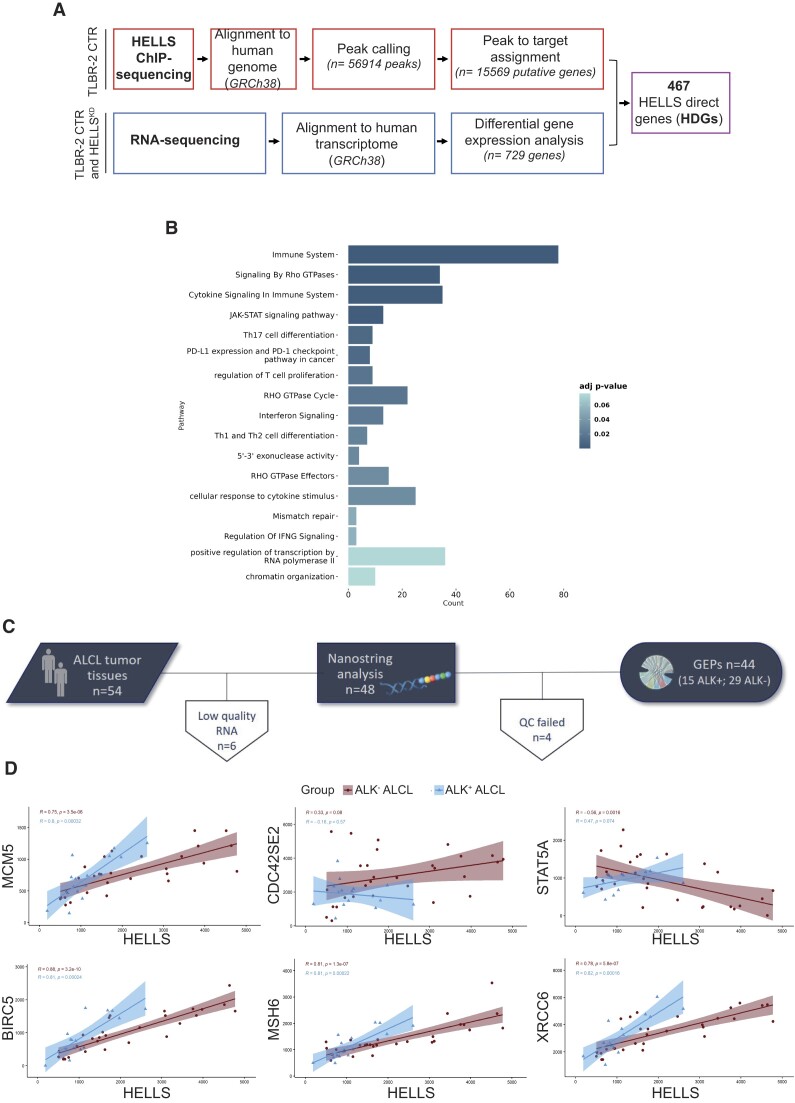
To explore the transcriptional function of HELLS in ALCLs, we performed HELLS Chromatin immunoprecipitation sequencing (ChIP‐seq) analysis in the TLBR-2 cell line, the principal cell line used in our earliest studies that elucidated the role of HELLS in lymphomagenesis (7,8). We identified 5.6 × 104 HELLS-specific peaks (Figure 1A) in line with the peak density previously reported for other chromatin remodeling factors (17). Genome-wide peaks of HELLS are enriched in distal regions (47.10%) (Supplementary Figure S1A) mirroring its previously described remodeling activity (18,19).
Figure 1.
Transcriptional landscape of HELLS in T-cell Lymphomas. (A) Schematic workflow for the identification of HELLS direct genes (HDGs). (B) GO enrichment bar plot of the most significantly enriched pathways (adjusted P-value < 0.05) for HDGs. Colors indicate the adjusted P-values and the size of bars is proportional to the ratio of differentially expressed genes on the total genes of the given pathway. (C) Outline of the study workflow for validating HELLS signature in the FFPE retrospective cohorts of ALCL by the nCounter platform. (D) Correlation plots of HELLS and a selection of HDGs in ALK− ALCL and ALK+ ALCL patients. For each gene pair, the expression HELLS is on the x-axis, while the expression HDGs is on the y-axis. Blue and red areas represent 95% confidence intervals.
We intersected these data with the list of HELLS-dependent genes that we previously identified by performing RNA sequencing (RNA-seq) in TLBR-2 HELLSKD and control (7). Out of the 729 genes significantly affected by HELLS knockdown (KD), 467 genes (64% of the total) were directly bound by HELLS based on ChIP-seq analysis (Figure 1A, Supplementary Figure S1B). We termed these genes HELLS-direct genes (HDGs). By using ChIP-qPCR in TLBR-2 control and HELLSKD cells, we validated HELLS binding on a representative set of genes confirming our ChIP-seq results (Supplementary Figure S1C, D). Gene-set enrichment analysis of HDGs revealed diverse biological processes associated with deregulated genes including T-cell proliferation, the JAK/STAT signaling pathway, chromatin organization, cytokine signaling in the immune system, DNA repair and the interferon γ signaling pathway (Figure 1B). Using RT-qPCR, we validated a representative set of downregulated genes confirming the RNA-seq results upon HELLS KD (Supplementary Figure S1E-F). To confirm the functional association between HELLS and its HDGs in ALCL patients, we evaluated the expression of a set of HDGs (n = 32 genes), in a retrospective cohort of 44 patients with ALCLs (n = 15 ALK+ ALCL and n = 29 ALK− ALCL cases) (Figure 1C). These genes were selected among those belonging to cytoskeleton regulation, JAK/STAT signaling pathway, and DNA metabolic regulation categories as major determinants of ALK−(2,7,8). Consistent with our previous observation (8), HELLS results significantly deregulated between the two ALCL subtypes showing higher expression in the ALK−ALCL one (Supplementary Figure S1G). Gene expression correlation analysis showed that HELLS was significantly associated with its target genes showing a positive correlation with 22 genes and a negative correlation with 10 genes in ALK−ALCL primary samples. BIRC5, MSH6, and MCM5 resulted in the most significantly correlated genes (Figure 1D, Supplementary Figure S1H). Collectively, these data demonstrate that HELLS can bind to specific genomic regions and regulate genes associated with T-cell lymphoma aggressiveness.
Inactivation of HELLS alters the profile of RNAPII along the genome
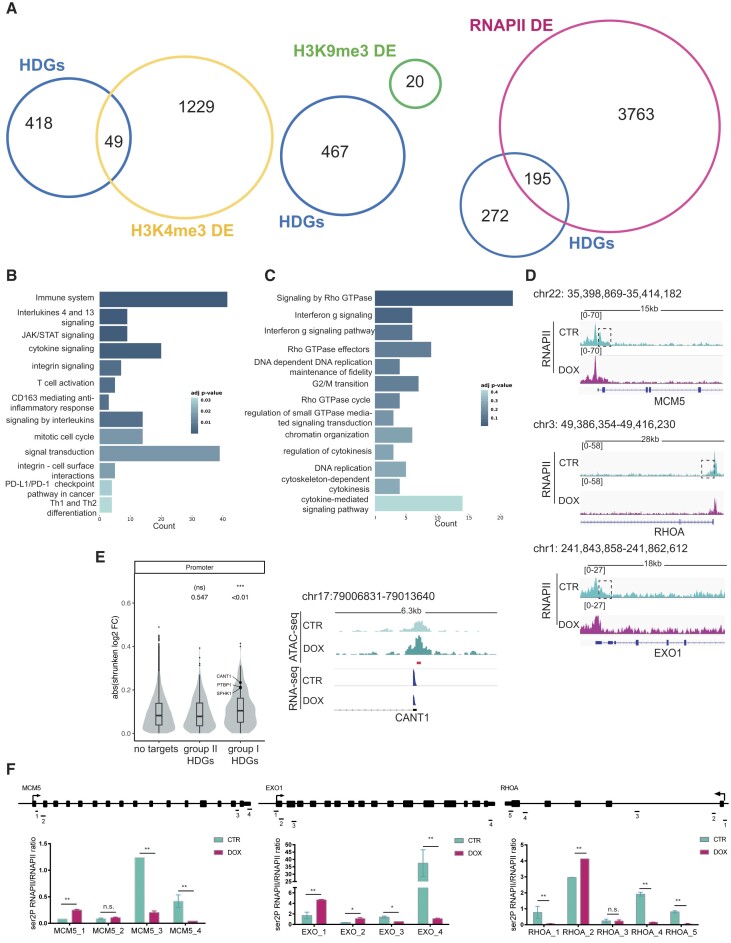
Analysis of the HELLS binding profile at the HDGs site showed that 67% of HELLS binding sites had intragenic localization being associated with promoters (24.38%), introns (39.50%) and exons (2.73%) (Supplementary Figure S2A). Interestingly in these genes, HELLS binding is more enriched in promoter regions rather than distal regions suggesting an active role in transcription regulation (Supplementary Figure S2A). To assess the contribution of HELLS to HDGs transcription, we explored the effects of HELLS KD on chromatin organization. Distribution of histone 3 trimethyl lysine 4 (H3K4me3) and histone 3 trimethyl lysine 9 (H3K9me3) -as HELLS associated histone marks (20,21)- and RNA‐polymerase II (RNAPII) was investigated by ChIP‐seq in TLBR-2 HELLSKD and control cells. Peak annotation at different genomic regions is shown in Supplementary Figure S2B. 13.6% (n = 3278) of the total H3K4me3 peaks identified (n = 24 080) in TLBR-2 control cells were enriched for HELLS binding (Supplementary Table S3). Upon HELLS depletion, 1571 sites—corresponding to 1278 associated genes—were differentially enriched for H3K4me3. In total, 3.75% (n = 49) of the predicted genes were HDGs (Figure 2A).
Figure 2.
Loss of HELLS alters RNAPII dynamics. (A) The Venn diagrams show the overlap, the number of genes identified in each condition, and the number of HELLS direct genes (HDGs) that are differentially enriched (DE) for the trimethylated histone H3 on Lysine 4 (H3K4me3), trimethylated histone H3 on Lysine 9 (H3K9me3) and RNA-Polymerase II (RNAPII) in TLBR-2 HELLSKD cells relative to control. (B) GO enrichment bar plots of the most significantly enriched pathways (adjusted P-value < 0.05) for HELLS direct genes (HDGs) belonging to group I (n = 195). Colors indicate the adjusted P-values and the size of the bars is proportional to the ratio of differentially expressed genes to the total of genes of the given pathway. (C) GO enrichment bar plots of the most significantly enriched pathways (adjusted P-value < 0.05) for group II of HDGs (n = 272). Colors indicate the adjusted P-values, and the size of bars is proportional to the ratio of differentially expressed genes to the total of genes of the given pathway. (D) RNAPII enrichment profile of representative HELLS direct genes in TLBR-2 HELLSKD (DOX) and control (CTR) cells. (E) The violin plot shows changes in chromatin accessibility—after HELLS KD in TLBR-2 cells—between genes that are not bound by HELLS (no targets) and groups I and II of HDGs. On top, P-values associated with beta coefficients estimated by linear regression model with log2 shrunken fold-change as the response variable, and gene groups as the independent variable (left panel). Example of an ATAC-seq and RNA-seq IGV browser view in a representative HDG in TLBR-2 control and HELLSKD. The y-axis represents the normalized tag densities relative to hg19 genomic coordinates. The red bar represents the HELLS peak (based on ChIP-seq analysis) (right panel). (F) ChIP-RT-qPCR detection of ser2P-RNAPII normalized on total RNAPII levels for HDGs in TLBR-2 cell line. Data are representative of five independent experiments and are shown as the mean ± SEM. Two-tailed t-test. *P < 0.05; **P < 0.01.
In contrast, 86% (n = 463) of identified H3K9me3 peaks in TLBR-2 control cells were also enriched for HELLS (Supplementary Table S3). Only 36 genomic regions—corresponding to 20 genes—were significantly differentially enriched for this histone modification after HELLS KD. However, none of these 20 genes belong to HDGs (Figure 2A). Notably, significant perturbations in the H3K9me3 binding profiles were observed in HELLS-bound pericentromeric genes (Supplementary Figure S2C) consistent with the hypothesis that the methylase activity of HELLS is not directly involved in the regulation of gene expression (22).
A total of 76005 RNAPII binding peaks were identified in the TLBR-2 control, of which 3280 (4,3%) were also enriched for HELLS (Supplementary Table S3). 6581 sites—corresponding to 3958 associated genes—were differentially bound by RNAPII in TLBR-2 HELLSKD cells compared to control cells. Intersecting this list of genes with HDGs, about 40% (n = 195, group I) of HDGs showed a significant alteration in RNAPII occupancy upon HELLS depletion (Figure 2A). At these HDGs, RNAPII binding perturbation was consistent with the expression changes observed upon HELLS KD (Supplementary Figure S2D). GO analysis of these HDGs showed enrichment in T-cell mediated immunity, cytokines signaling, cell cycle and the JAK/STAT signaling pathway (Figure 2B). This observation indicates a previously underscored potential function of HELLS in mediating the signaling of immune-related pathways.
Conversely, on the remaining 60% of HDGs (n = 272, group II)—which were enriched for selective biological functions, including cytoskeleton regulation and DNA metabolic processes (Figure 2C)—the loss of HELLS did not affect the recruitment of RNAPII but it caused a selective loss of RNAPII signal in the regions immediately downstream of the transcription start site (TSS) (Figure 2D). These observations seemed to indicate that, depending on targets, HELLS may foster transcription through two alternative mechanisms: by promoting RNAPII loading (group I) and by promoting RNAPII elongation along the gene body (group II). To test this hypothesis, we evaluated changes in accessibility by performing transposase-accessible chromatin using sequencing (ATAC-seq), identifying a total of 124 440 peaks, of which 2166 were associated with HDGs (Supplementary Figure S2E). We did not notice differences in ATAC-seq peak distribution at genome-wide level between TLBR-2 HELLSKD and control cell conditions and no individual differential enrichment applying a standard cut-off on significance (79.7% of overlap, FDR < 0.05). Nevertheless, we observed an overall change in genome accessibility of promoter peaks with functional HELLS loss only for HDGs belonging to group I (Figure 2E and Supplementary Figure S2F). This confirms that HELLS helps the recruitment of transcriptional machinery to sustain open chromatin at immune-related HDG promoters.
In parallel, on HDGs belonging to group II, we monitored by ChIP-qPCR the levels of RNAPII phosphorylation of serine 5 (ser5P-RNAPII) and serine 2 (ser2P-RNAPII) residues, which are hallmarks of early transcription or active elongation, respectively (23). While little or no effects were observed on the levels of ser5P-RNAPII between control and HELLS KD cells (Supplementary Figure S2G-I), levels of ser2P-RNAPII were slightly upregulated in the proximity of TSS and then reduced along the gene body in the absence of HELLS (Figure 2F, Supplementary Figure S2H, J) indicative of RNAPII arrest. This data suggests that, in the absence of HELLS, RNAPII is elongation-proficient, but upon unscheduled obstacles, it becomes unable to sustain transcription.
Collectively, these data indicate a bimodal function of HELLS on RNAPII regulation.
HELLS prevents R-loops accumulation to promote the transcription
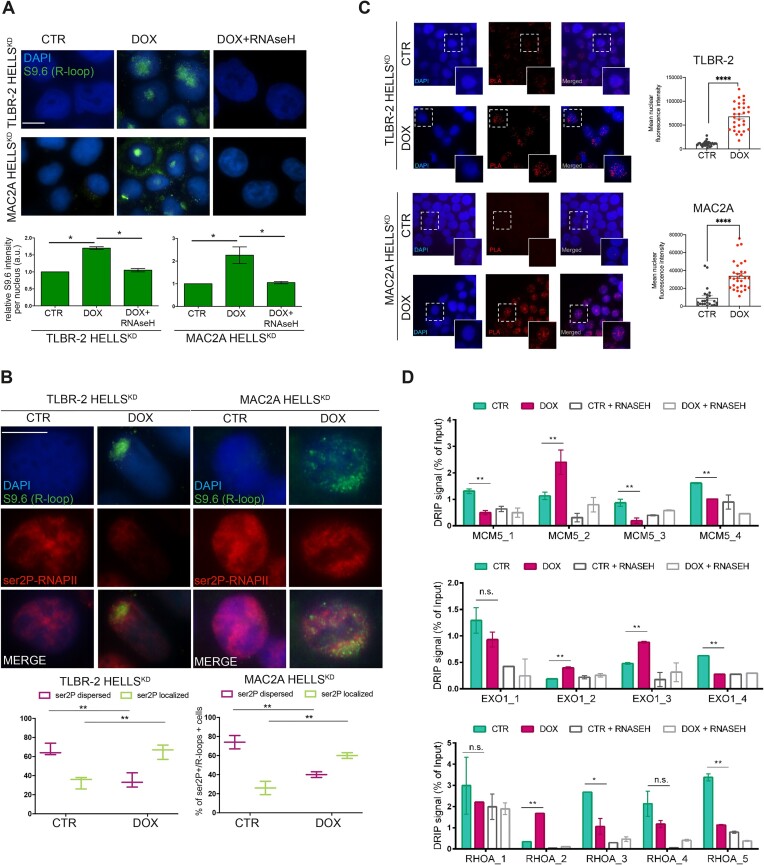
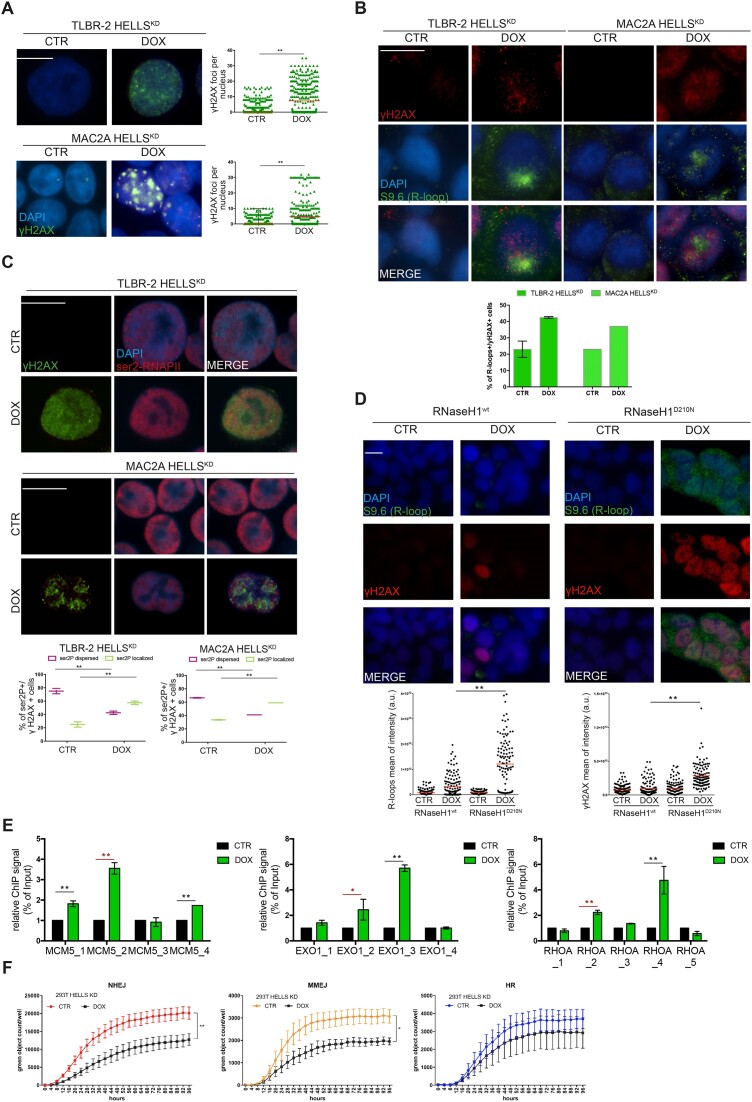
Stacked RNAPII during elongation drives DNA–RNA hybrids (R-loops) formation and it is a primary source of genome instability (24). We hypothesized that HELLS could promote RNAPII elongation by relieving R-loops, thereby decreasing potential obstacles to the transcription machinery. We investigated the effect of HELLS depletion on R-loops accumulation by performing immunofluorescences using the S9.6 antibody to detect R-loops in control and HELLSKD cells. After the depletion of HELLS, we observed a significant increase in the nuclear S9.6 signal in TLBR-2 and MAC2A HELLSKD cells compared to control cells (Figure 3A, Supplementary Figure S3A). The signals detected by the S9.6 antibody were significantly reduced by RNase H treatment, confirming its specificity for DNA-RNA hybrids (Figure 3A). Notably, the increased S9.6 signal was accompanied by a different pattern of nuclear ser2P-RNAPII with clusters adjacent to R-loop structures suggesting that R-loops are associated with stalled transcription (Figure 3B, Supplementary Figure S3B). In situ proximity ligation assay (PLA) using S9.6 and ser2P-RNAPII antibodies confirmed the interaction of the elongating form of RNAPII with R-loops (Figure 3C). To strengthen these results, we performed DNA-RNA hybrid immunoprecipitation (DRIP) using the S9.6 antibody followed by qPCR (DRIP-qPCR) on the same subset of the genes in which RNAPII elongation is defective. DRIP-qPCR showed an increase in the abundance of R-loops nearby or in the same sites where productive elongation is perturbed by HELLS (Figure 3D, Supplementary Figure S3C). To confirm the specificity of DNA-RNA hybrid enrichment, we pretreated whole genomic extracts with RNase H to specifically degrade R-loops before performing DRIP. This resulted in the loss of the qPCR signal, indicating that the signal enrichment was due to the accumulation of R-loops after TSS is specific (Figure 3D, Supplementary Figure S3C).
Figure 3.
Loss of HELLS leads to co-transcriptional R-loops accumulation. (A) Representative immunofluorescences showing the presence of R-loops in HELLSKD cells in the absence (CTR) or presence (DOX) of doxycycline to induce shRNA against HELLS (48 h) and after treatment with RNase H1. Cells were stained with S9.6 antibody and DAPI. The white scale bar represents 10 μm. For specific quantification of nuclear S9.6 staining, regions of interest were overlaid on the DAPI signal and selectively quantified to exclude cytoplasmic S9.6 signal and nucleolar S9.6 signal (n = 3 independent experiments). Relative bar plots indicate the intensity of S9.6-stained cells per nucleus (n = 1000 cells). Each data represents the mean ± SEM (n = 3). Two-tailed t-test. *P < 0.05; **P < 0.01 relative to CTR. (B) Immunofluorescences show the distribution of ser2P-RNAPII and R-loops in HELLSKD cells. Cells were stained with ser2P-RNAPII, S9.6 antibodies, and DAPI. The white scale bar represents 10 μm. Relative box plots indicate the distribution of ser2P-RNAPII of S9.6 positive cells (n = 500 cells). Each data represents the mean ± SEM (n = 3). Two-tailed t-test. *P < 0.05; **P < 0.01 relative to CTR. (C) Representative in situ proximity ligation assay (PLA) showing the interaction between R-loops and ser2P-RNAPII in TLBR-2 and MAC2A cell models, comparing CTR versus DOX conditions. Each red spot represents a single interaction between the two targets. DNA was stained with DAPI (in blue). Quantification of dots per cell is represented in the graph on the right by analyzing the mean nuclear fluorescence intensity as described in Materials and methods. Statistical analyses were performed with a t-test and specified with asterisks (**P < 0.01, ***P < 0.001, ****P < 0.0001). Data are represented as mean ± SEM. (D) Representative DRIP‐qPCR signal values at the MCM5, EXO1 and RHOA loci in TLBR-2 control and HELLSKD cells w/o pretreatment with RNase H. Each data represents the mean ± SEM. Two-tailed t-test. *P < 0.05; **P < 0.01 relative to CTR.
Together, these data indicate that impaired productive elongation accounts for the effect of the absence of HELLS in reducing the formation of R-loops.
DNA damage accumulates in actively transcribed genes in HELLS depleted cells
HELLS acts as platform to recruit proteins involved in DNA damage ensuring an efficient double-strand break (DSBs) repair (25).
We investigated whether the defects in RNAPII dynamics and concomitant R-loops accumulation caused by HELLS depletion could also be associated with increased DNA damage. We initially assayed the effect of the KD of HELLS through the detection of phosphorylated H2AX (γ-H2AX) as a general readout of DNA damage accumulation. We performed immunofluorescence staining and we observed an overall increase in the intensity of the nuclear γH2AX signal and a significant increase in the formation of γH2AX foci in TLBR-2 and MAC2A cell lines after HELLS KD (Figure 4A). Notably, the increase in γH2AX foci was found in S9.6 positive cells (Figure 4B) and was associated with a distinctive pattern of ser2P-RNAPII (Figure 4C), suggesting that the accumulation of DSBs caused by loss of HELLS is a direct result of R-loops formation. To address this question, we overexpressed RNase H1 to resolve hybrids that form while depleting HELLS. Unfortunately, ALK−ALCL cells did not tolerate the overexpression of RNase H1. For this reason, we used 293T cells in which HELLS depletion affects the DNA damage accumulation (Supplementary Figure S3D). In these assays, we transiently expressed the wild-type RNase H1 (Supplementary Figure S3E) and we quantified the intensity γH2AX and the presence of R-loops. The catalytically inactive RNase H1 mutant (D210N) was also overexpressed as control. Although the doxycycline (DOX) partially impaired the overexpression of RNase H1 (Supplementary Figure S3F), the remaining low level of exogenous RNase H1 was sufficient to significantly reduce the accumulation of R-loops leading to the attenuation of γH2AX signal in HELLS-depleted cells compared to cells overexpressing RNase H1D210N (Figure 4D, Supplementary Figure S3G). We concluded that R-loops accumulation concurs to DNA damage events.
Figure 4.
HELLS manages DNA damage repair. (A) Quantification and representative IF images of γH2AX intensities in HELLSKD cells. The white scale bar represents 10 μm. Data presented as mean ± SEM (n = 3 separate experiments, 500 cells analyzed in each experiment). Quantification and representative IF images of γH2AX and nuclear S9.6 intensities (B) and ser2P-RNAPII and nuclear S9.6 intensities (C) in HELLSKD cells. The white scale bar represents 10 μm. Data presented as mean ± SEM (n = 3 separate experiments, 500 cells analyzed in each experiment). (D) Quantification and representative IF images of R-loops and γH2AX intensities in 293T HELLSKD cells overexpressing wild-type (WT) and D210N mutant RNase H1. The white scale bar represents 10 μm. Data is representative of n = 3 independent experiments (>150 cells analyzed in each experiment). Red bars represent the mean of S9.6 or γH2AX intensities of the indicated cell populations. (E) ChIP-RT-qPCR detection of γH2AX levels for representative HDGs in TLBR-2 cell line. Data are represented as relative % of input over CTR and are presented as mean ± SEM (n = 2 independent experiments). Two-tailed t-test. *P < 0.05; **P < 0.01 (in red the regions affected by R-loops accumulation and ser2P-RNAPII stall). (F) The effects of HELLS-KD on NHEJ, MMEJ and HR efficiency were examined in 293T HELLSKD cells using the GFP reporter assay.
Next, we asked if DNA damage occurs in the proximity of sites affected by R-loops accumulation and thus by the stall of RNAPII. To address this question, we monitored by ChIP-qPCR the levels of γH2AX along HDGs. Figure 4E and Supplementary Figure S3H show that the accumulation of DNA damage affects the same site in which R-loops accumulate and the terminal transcription sites suggesting that most of the DNA damage events occur in transcribed regions. Although these data are in disagree with the previously described role of HELLS in promoting DNA strand repair mainly in not transcribed and fragile regions (25), these results are in accord with the ability of ATP-remodeling complexes to prevent DNA damage at R-loops sites (26,27).
We finally investigated which pathway mediates the ability of HELLS to repair DNA damage using reporter-based non-homologous end joining (NHEJ), microhomology-mediated end joining (MMEJ), and homologous recombination (HR) assays. Although all three tested pathways were active under basal conditions, the loss of HELLS resulted in decreased NHEJ and MMEJ efficiencies (approximately 37% and 36%, respectively, over time) without significant effects on the HR system (Figure 4F, Supplementary Figure 3I). Importantly, these data are in accord with the previous findings showing that HELLS-deficient and HELLS-mutated cells harbor delay in protein accumulation at DNA damage sites resulting in defects in NHEJ (28,29). This is also consistent with previous observations suggesting that DNA resection deficiency due to RNAPII inhibition and R-loops accumulation favors the NHEJ pathway (30)
Collectively, these results strongly indicate that the efficient management and resolution of R-loops and associated DNA damage events concur to guarantee HELLS-dependent genome stability in lymphomas.
Depletion of HELLS sensitizes T-cell lymphoma cells to chemotherapeutic drugs
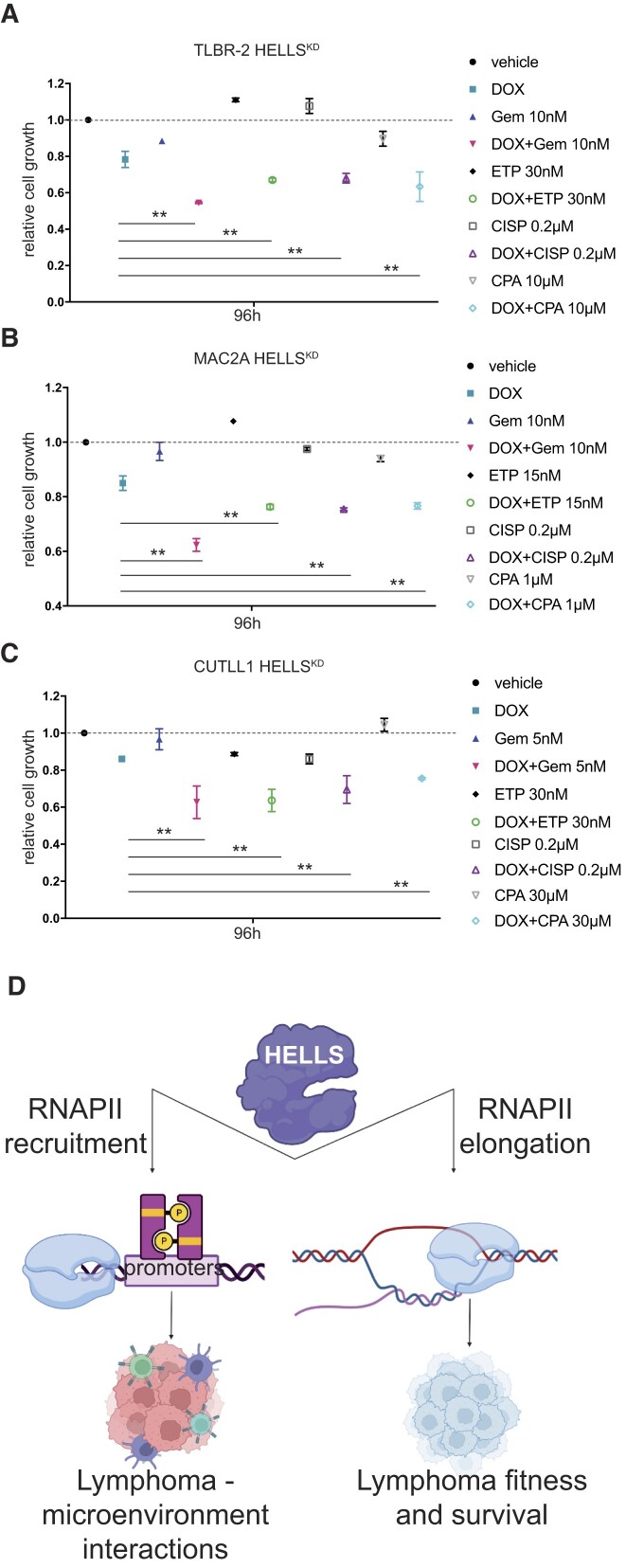
Our data indicated that HELLS is a major keeper of ALCL genome organization and function, explaining the dependency of ALCL cells on this protein. Therefore, we explored the potential implications of targeting HELLS in T-cell lymphomas. We treated HELLSKD and control cells with low doses of several chemotherapeutic drugs currently used in the clinic (including gemcitabine [GEM], cisplatin [CISP], etoposide [ETP] and cyclophosphamide [CPA]) and evaluated cellular proliferation (Figure 5A–C, Supplementary Figure S4A–C). We also included an additional non-ALCL cell line, CUTLL1, in which HELLS depletion impaired proliferation and DNA damage accumulation (Supplementary Figure S4D–G). In all cell lines tested, HELLS depletion was found to synergize with all compounds, causing significant synthetic lethality with drugs, compared to the use of drugs alone or control cells (Figure 5A–C). Notably, the most significant synergistic effects of HELLS depletion were observed upon treatment with gemcitabine (GEM) and etoposide (ETP) (Figure 5A–C, Supplementary Figure S4H), which preferentially relied on NHEJ-mediated repair to modulate their cytotoxic effects.
Figure 5.
Loss of HELLS sensitizes to low doses of drugs. Graphs showing the relative cell growth of TLBR-2 HELLSKD (A), MAC2A HELLSKD (B) and CUTLL1 HELLSKD (C) cells in the absence (CTR) and in presence (DOX) of doxycycline and treated with different chemotherapeutic drugs (Gem: gemcitabine, ETP: etoposide, CISP: cisplatin, CPA: cyclophosphamide) for 96 h. (D) Graphical model of HELLS function in T-cell lymphomas. Created with BioRender.com
Collectively, these data provide a strong rationale for the selective inhibition of HELLS as a new strategy to improve chemotherapy efficacy in TCLs.
Discussion
Previous studies have suggested the role of HELLS in genome maintenance (5), but it was unclear how the remodeling properties of HELLS could be relevant for gene expression regulation in cancers. Using functional genomics approaches, we identified a direct transcriptional function of HELLS and we defined the transcriptional landscape of HELLS and its unique gene signature in the aggressive ALK−ALCL subtype. Among the genes directly controlled by HELLS, we identified many targets linked to ALCL pathogenesis, such as BATF3, STAT5A, RHOA, PD-L1, IL6 and JAK2.
We demonstrated that, in a small fraction of HDGs (group I), HELLS controls RNAPII positioning and recruitment at target promoters. At these sites, the loss of HELLS causes a displacement of RNAPII associated with concomitant changes in the level of H3K4me3 enrichment and with changes in chromatin accessibility, suggesting an effect on transcription initiation. Surprisingly, we noticed these genes belonged to biological processes involved in the dysregulation of circuitries that control T-cell-dependent host interactions. We speculate that the aberrant expression of HELLS may create a different immune response and contribute, at least in part, to a more aggressive tumor phenotype observed in this malignancy.
For the largest fraction of HDGs (group II), HELLS is required to guarantee the progression of RNAPII during elongation on a selected subset of genes central to ALCL cell self-maintenance. On these genes, HELLS, by binding to intragenic regions, eases RNAPII progression along the gene body by resolving co-transcriptional DNA-RNA hybrid structures formed by aberrant transcription. Reduced formation of R-loops appears to be a mechanism widely used by HELLS to control the transcription (19). This highlights the importance of HELLS-dependent R-loop homeostasis in transcriptional regulation, as unbalanced levels alter transcriptional dynamics and promote genome instability.
By performing ChIP experiments, we found that the same sites in which R-loops accumulate are prone to accumulate DNA damage defects. RNase H1 overexpression rescues DNA damage in HELLSKD cells suggesting that co-transcriptional R-loops contribute to a high level of DNA damage observed in HELLS-depleted cells. This complex HELLS activity has two major consequences. On the one hand, HELLS ensures robust transcription of vital genes supporting ALCL fitness and survival; on the other hand, it prevents excessive accumulation of DNA damage avoiding irreversible genomic crashes and consequential cell death. The ability to resolve transcriptional conflicts is emerging as a new mechanism by which the SWI/SNF complex protects genome integrity (27).
Taken together these data indicate that: (i) HELLS works with a bi-modal function in controlling gene expression by regulating both the loading and the elongation of RNAPII; (ii) the different work modalities have different biological consequences. For genes that are vital for the cancer cell itself, HELLS controls the rate of elongation of RNAPII, whereas, for genes that control cancer cell interaction with the environment, HELLS fine-tunes their transcription activation (Figure 5D).
HELLS-depleted cells showed defects in NHEJ and MMEJ activity. The same defects in NHEJ repair have been observed in lymphoblastoid cells derived from ICF patients (29) and in HELLS-deficient B-lymphocytes with impaired class switch recombination (28). These findings suggest a predominant role of HELLS in NHEJ regulation in lymphocytes.
At the crossroads between transcription and DNA repair, HELLS is an attractive target for the development of novel anticancer treatments for hematological malignancies. Using in vitro drug assays, we demonstrated synergistic activity between the loss of HELLS and low doses of chemotherapeutic drugs. These findings suggest that the clinical response of TCLs to standard chemotherapeutic regimens can be significantly improved by concurrent changes in chromatin accessibility.
In summary, in this study, we consolidated the transcriptional role of HELLS in T-cell lymphomas, providing evidence that this chromatin remodeling protein acts as a functional multitasking unit capable of coordinating transcriptional progression, thereby supervising genomic stability.
We believe that our study could promote the development of selective HELLS inhibitors in ALCLs to enhance their sensitivity to chemotherapeutic drugs and expand the therapeutic portfolio for patients with T-cell lymphoma.
Supplementary Material
Acknowledgements
The authors are grateful to Marina Grassi, Eleonora Dufrusine, Laura Moretti, Riccardo Fuoco and Cristian Ascione for their technical help and to all the members of the laboratory for helpful discussions.
Author contributions: A.T., S.M., G.G., S.M. and N.P. performed experiments and analyzed data, B.D. performed gene expression and Nanostring analysis, V.M., F.T., E.S., E.V., S.M. and E.S. performed RNA-seq, ChIP-seq, ATAC-seq and bioinformatics analyses. M.Z., I.T., S.A., F.S. and G.M. provided tissue samples and lymphoma diagnosis. A.N. provided important experimental and analytic support. A.C. interpreted the results and helped to discuss the results. V.F. designed the project, interpreted the results, and wrote the manuscript.
All the authors read and approved the final version of the manuscripts.
Contributor Information
Annalisa Tameni, Laboratory of Translational Research, Azienda USL-IRCCS di Reggio Emilia, Viale Risorgimento 80, 42123, Reggio Emilia, Italy.
Selene Mallia, Laboratory of Translational Research, Azienda USL-IRCCS di Reggio Emilia, Viale Risorgimento 80, 42123, Reggio Emilia, Italy.
Veronica Manicardi, Laboratory of Translational Research, Azienda USL-IRCCS di Reggio Emilia, Viale Risorgimento 80, 42123, Reggio Emilia, Italy.
Benedetta Donati, Laboratory of Translational Research, Azienda USL-IRCCS di Reggio Emilia, Viale Risorgimento 80, 42123, Reggio Emilia, Italy.
Federica Torricelli, Laboratory of Translational Research, Azienda USL-IRCCS di Reggio Emilia, Viale Risorgimento 80, 42123, Reggio Emilia, Italy.
Emanuele Vitale, Laboratory of Translational Research, Azienda USL-IRCCS di Reggio Emilia, Viale Risorgimento 80, 42123, Reggio Emilia, Italy; Clinical and Experimental Medicine Ph.D. Program, University of Modena and Reggio Emilia, Modena 41125, Italy.
Elisa Salviato, Laboratory of Translational Research, Azienda USL-IRCCS di Reggio Emilia, Viale Risorgimento 80, 42123, Reggio Emilia, Italy.
Giulia Gambarelli, Laboratory of Translational Research, Azienda USL-IRCCS di Reggio Emilia, Viale Risorgimento 80, 42123, Reggio Emilia, Italy.
Silvia Muccioli, Laboratory of Translational Research, Azienda USL-IRCCS di Reggio Emilia, Viale Risorgimento 80, 42123, Reggio Emilia, Italy.
Magda Zanelli, Pathology Unit, Department of Oncology, Azienda Unità Sanitaria Locale – IRCCS di Reggio Emilia, Reggio Emilia, 42123, Italy.
Stefano Ascani, Pathology Unit, Azienda Ospedaliera Santa Maria di Terni, University of Perugia, 05100 Terni, Italy.
Giovanni Martino, Pathology Unit, Azienda Ospedaliera Santa Maria di Terni, University of Perugia, 05100 Terni, Italy; Institute of Hematology and CREO, University of Perugia, Perugia 06129, Italy.
Francesca Sanguedolce, Pathology Unit, Policlinico Riuniti, University of Foggia, 71122 Foggia, Italy.
Elisabetta Sauta, IRCCS Humanitas Clinical and Research Center, via Manzoni 56, 20089 Rozzano, Milan, Italy.
Ione Tamagnini, Pathology Unit, Department of Oncology, Azienda Unità Sanitaria Locale – IRCCS di Reggio Emilia, Reggio Emilia, 42123, Italy.
Noemi Puccio, Laboratory of Translational Research, Azienda USL-IRCCS di Reggio Emilia, Viale Risorgimento 80, 42123, Reggio Emilia, Italy.
Antonino Neri, Scientific Directorate, Azienda USL-IRCCS di Reggio Emilia, Viale Umberto I 50, 42123, Reggio Emilia, Italy.
Alessia Ciarrocchi, Laboratory of Translational Research, Azienda USL-IRCCS di Reggio Emilia, Viale Risorgimento 80, 42123, Reggio Emilia, Italy.
Valentina Fragliasso, Laboratory of Translational Research, Azienda USL-IRCCS di Reggio Emilia, Viale Risorgimento 80, 42123, Reggio Emilia, Italy.
Data availability
All data generated and/or analyzed in this study are included in this article and its Online Supplementary Appendix. Gene expression profile data are available in the Gene Expression Omnibus (GEO) repository (accession number: GSE217426).
Raw RNA-seq data in fastq.gz format are available in the ArrayExpress repository, dataset E-MTAB-9918 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-9918).
ChIP-seq raw data in fastq.gz format are available in the ArrayExpress repository, dataset E-MTAB-12935.
ATAC-seq raw data in fastq.gz format are available in the ArrayExpress repository, datase data E-MTAB-13851.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
A.T. was supported by Fondazione AIRC per la Ricerca sul Cancro (AIRC); V.M. was supported by Fondazione GRADE Onlus; E.V. was supported by the Fondazione Guido Berlucchi; Bando per la Valorizzazione della Ricerca Istituzionale 2021—fondi 5 per Mille 2020 (to V.F.); Fondazione AIACE (to V.F.); Fondazione AIRC per la Ricerca sul Cancro (AIRC) [MFAG 2023 ID 28974 project – P.I. Fragliasso Valentina and ID IG24365 project - P.I. Antonino Neri]; Italian Ministry of Health-Ricerca Corrente Annual Program 2025 (in part). Funding for open access charge: Fondazione AIRC per la Ricerca sul Cancro.
Conflict of interest statement. None declared.
References
- 1. Alaggio R., Amador C., Anagnostopoulos I., Attygalle A.D., Araujo I.B.D.O., Berti E., Bhagat G., Borges A.M., Boyer D., Calaminici M. et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: lymphoid Neoplasms. Leukemia. 2022; 36:1720–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fiore D., Cappelli L.V., Broccoli A., Zinzani P.L., Chan W.C., Inghirami G. Peripheral T cell lymphomas: from the bench to the clinic. Nat. Rev. Cancer. 2020; 20:323–342. [DOI] [PubMed] [Google Scholar]
- 3. Bradner J.E., Hnisz D., Young R.A. Transcriptional addiction in cancer. Cell. 2017; 168:629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Crossley M.P., Bocek M., Cimprich K.A. R-loops as cellular regulators and genomic threats. Mol. Cell. 2019; 73:398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen X., Li Y., Rubio K., Deng B., Li Y., Tang Q., Mao C., Liu S., Xiao D., Barreto G. et al. Lymphoid-specific helicase in epigenetics, DNA repair and cancer. Br. J. Cancer. 2022; 126:165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thijssen P.E., Ito Y., Grillo G., Wang J., Velasco G., Nitta H., Unoki M., Yoshihara M., Suyama M., Sun Y. et al. Mutations in CDCA7 and HELLS cause immunodeficiency–centromeric instability–facial anomalies syndrome. Nat. Commun. 2015; 6:7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tameni A., Sauta E., Mularoni V., Torricelli F., Manzotti G., Inghirami G., Bellazzi R., Fragliasso V., Ciarrocchi A. The DNA-helicase HELLS drives ALK− ALCL proliferation by the transcriptional control of a cytokinesis-related program. Cell Death. Dis. 2021; 12:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fragliasso V., Verma A., Manzotti G., Tameni A., Bareja R., Heavican T.B., Iqbal J., Wang R., Fiore D., Mularoni V. et al. The novel lncRNA BlackMamba controls the neoplastic phenotype of ALK− anaplastic large cell lymphoma by regulating the DNA helicase HELLS. Leukemia. 2020; 34:2964–2980. [DOI] [PubMed] [Google Scholar]
- 9. Ni K., Ren J., Xu X., He Y., Finney R., Braun S.M.G., Hathaway N.A., Crabtree G.R., Muegge K. LSH mediates gene repression through macroH2A deposition. Nat. Commun. 2020; 11:5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Law C., Wei L., Tsang F.H., Chan C.Y., Xu I.M., Lai R.K., Ho D.W., Lee J.M., Wong C.C., Ng I.O. et al. HELLS regulates chromatin remodeling and epigenetic silencing of multiple tumor suppressor genes in Human hepatocellular carcinoma. Hepatology. 2019; 69:2013–2030. [DOI] [PubMed] [Google Scholar]
- 11. Mularoni V., Donati B., Tameni A., Manicardi V., Reggiani F., Sauta E., Zanelli M., Tigano M., Vitale E., Torricelli F. et al. Long non-coding RNA mitophagy and ALK- anaplastic lymphoma associated transcript: a novel regulator of mitophagy in T cell lymphoma. Haematol. 2023; 108:3333–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Agnelli L., Mereu E., Pellegrino E., Limongi T., Kwee I., Bergaggio E., Ponzoni M., Zamò A., Iqbal J., Piccaluga P.P. et al. Identification of a 3-gene model as a powerful diagnostic tool for the recognition of ALK-negative anaplastic large-cell lymphoma. Blood. 2012; 120:1274–1281. [DOI] [PubMed] [Google Scholar]
- 13. Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R. et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012; 481:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stark R., Brown G. DiffBind. 2017; 10.18129/B9.BIOC.DIFFBIND. [DOI]
- 15. Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sanz L.A., Chédin F. High-resolution, strand-specific R-loop mapping via S9.6-based DNA–RNA immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2019; 14:1734–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grossi E., Raimondi I., Goñi E., González J., Marchese F.P., Chapaprieta V., Martín-Subero J.I., Guo S., Huarte M. A lncRNA-SWI/SNF complex crosstalk controls transcriptional activation at specific promoter regions. Nat. Commun. 2020; 11:936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cousu C., Mulot E., De Smet A., Formichetti S., Lecoeuche D., Ren J., Muegge K., Boulard M., Weill J.-C., Reynaud C.-A. et al. Germinal center output is sustained by HELLS-dependent DNA-methylation-maintenance in B cells. Nat. Commun. 2023; 14:5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Unoki M., Sharif J., Saito Y., Velasco G., Francastel C., Koseki H., Sasaki H. CDCA7 and HELLS suppress DNA:RNA hybrid-associated DNA damage at pericentromeric repeats. Sci. Rep. 2020; 10:17865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dunican D.S., Cruickshanks H.A., Suzuki M., Semple C.A., Davey T., Arceci R.J., Greally J., Adams I.R., Meehan R.R. Lsh regulates LTR retrotransposon repression independently of Dnmt3b function. Genome Biol. 2013; 14:R146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu X., Ni K., He Y., Ren J., Sun C., Liu Y., Aladjem M.I., Burkett S., Finney R., Ding X. et al. The epigenetic regulator LSH maintains fork protection and genomic stability via MacroH2A deposition and RAD51 filament formation. Nat. Commun. 2021; 12:3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. De Dieuleveult M., Bizet M., Colin L., Calonne E., Bachman M., Li C., Stancheva I., Miotto B., Fuks F., Deplus R. The chromatin remodelling protein LSH/HELLS regulates the amount and distribution of DNA hydroxymethylation in the genome. Epigenetics. 2022; 17:422–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol. Cell. 2009; 36:541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Niehrs C., Luke B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 2020; 21:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peixoto E., Khan A., Lewis Z.A., Contreras-Galindo R., Czaja W. The chromatin remodeler HELLS: a new regulator in DNA repair, genome maintenance, and cancer. Int. J. Mol. Sci. 2022; 23:9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sessa G., Gómez-González B., Silva S., Pérez-Calero C., Beaurepere R., Barroso S., Martineau S., Martin C., Ehlén Å., Martínez J.S. et al. BRCA2 promotes DNA-RNA hybrid resolution by DDX5 helicase at DNA breaks to facilitate their repair‡. EMBO J. 2021; 40:e106018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bayona-Feliu A., Barroso S., Muñoz S., Aguilera A. The SWI/SNF chromatin remodeling complex helps resolve R-loop-mediated transcription–replication conflicts. Nat. Genet. 2021; 53:1050–1063. [DOI] [PubMed] [Google Scholar]
- 28. He Y., Ren J., Xu X., Ni K., Schwader A., Finney R., Wang C., Sun L., Klarmann K., Keller J. et al. Lsh/HELLS is required for B lymphocyte development and immunoglobulin class switch recombination. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:20100–20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Unoki M., Funabiki H., Velasco G., Francastel C., Sasaki H. CDCA7 and HELLS mutations undermine nonhomologous end joining in centromeric instability syndrome. J. Clin. Invest. 2018; 129:78–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gómez-Cabello D., Pappas G., Aguilar-Morante D., Dinant C., Bartek J. CtIP-dependent nascent RNA expression flanking DNA breaks guides the choice of DNA repair pathway. Nat. Commun. 2022; 13:5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and/or analyzed in this study are included in this article and its Online Supplementary Appendix. Gene expression profile data are available in the Gene Expression Omnibus (GEO) repository (accession number: GSE217426).
Raw RNA-seq data in fastq.gz format are available in the ArrayExpress repository, dataset E-MTAB-9918 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-9918).
ChIP-seq raw data in fastq.gz format are available in the ArrayExpress repository, dataset E-MTAB-12935.
ATAC-seq raw data in fastq.gz format are available in the ArrayExpress repository, datase data E-MTAB-13851.