Abstract
Infection with hepatitis B virus (HBV) was detected by serological testing for HBV surface antigen and by PCR assay for HBV DNA in serum samples from two common chimpanzees (Pan troglodytes subsp. verus) born in West Africa. The complete genome sequences obtained by nucleotide sequencing of overlapping DNA fragments amplified by PCR were compared with HBV variants recovered from other primates and with human genotypes A to F. Both chimpanzee sequences were 3,182 nucleotides in length, and the surface gene sequence predicted the existence of a, d, and w serological determinants. Neither sequence contained stop codons in the precore region. On phylogenetic analysis, the HBV variants infecting the chimpanzees clustered together with a third chimpanzee HBV isolate independently obtained from an infected captive animal (A. J. Zuckerman, A. Thornton, C. R. Howard, K. N. Tsiquaye, D. M. Jones, and M. R. Brambell, Lancet ii:652–654, 1978), with an overall sequence similarity of >94%. This provides strong evidence for a chimpanzee-specific genotype of HBV which circulates in nature. These findings add to the recent evidence for infection in the wild of other Old and New World primates (gibbon, orangutan, and woolly monkey) with species-specific variants of HBV. There is no evidence for close phylogenetic clustering of variants found so far in primates with any of the established HBV genotypes from humans. With the new evidence for the widespread distribution of HBV in primates, hypotheses for the origins of human infection are reviewed.
Hepatitis B virus (HBV) chronically infects approximately 5% of the human population. The toll of approximately one million deaths from chronic liver disease and hepatocellular carcinoma attributable to HBV infection per year (14) indicates the extent of the global health problem posed by this virus. HBV is transmitted by sexual contact and by parenteral exposure, although it is thought that mother-to-child perinatal transmission and the establishment of a lifelong highly infectious carrier state are responsible for the observed high rates of endemicity in high-prevalence regions such as South and East Asia and sub-Saharan Africa and among indigenous peoples in Central and South America.
HBV is classified in the family Hepadnaviridae and contains a partly double-stranded DNA genome of approximately 3,200 bases. HBV replicates via an RNA intermediate antigenome sequence, encoding a potentially error-prone polymerase enzyme with both reverse transcriptase and DNA polymerase activities. HBV variants from different geographical regions show genetic heterogeneity and are currently classified into six genotypes differing from each other by nucleotide sequence distances of approximately 10 to 13%. While genotypes A and D have global distributions, genotypes B and C are found predominantly in East and Southeast Asia, genotype E is found in West Africa, and genotype F is found among indigenous peoples in Central and South America (1, 11). Apart from humans, a range of much more genetically divergent hepadnaviruses infect the New World rodents woodchucks (Marmota monax), ground squirrels (Spermophilus beecheyi), and arctic ground squirrels (S. parryii) and a range of bird species (ducks, geese, and grey herons).
The origins of HBV infection in humans that have led to its current worldwide distribution have remained controversial. It is unclear whether nonhuman primates can be infected with HBV in the wild, as studies to date have used samples from captive animals exposed to both human and potentially other primate sources of infection (9, 12, 15, 17). In this study, we have genetically characterized HBV variants infecting chimpanzees (Pan troglodytes subsp. verus) born in the wild in West Africa and demonstrated that they share a genotype with that of a captive chimpanzee previously considered to have been infected from human sources (15). The existence of a chimpanzee-specific genotype, combined with the evidence of infection in other primate species, contributes to our understanding of the origins of HBV infection in humans, although a hypothesis consistent with all of the current experimental observations remains elusive.
MATERIALS AND METHODS
Samples.
Four samples from chimpanzees positive for HBV surface antigen (HBsAg) were available for the study. Three HBsAg-positive samples were collected from wild-born chimpanzees (Chimps 1 to 3) orphaned as a consequence of the bush meat trade and housed at a rescue center in Cameroon. Importation of samples from the center to the United Kingdom followed CITES regulations. Chimp 4 originated from West Africa, was smuggled to Spain for use in the beach chimp trade, confiscated by the Spanish authorities, and subsequently rehomed in an ape rescue center in the United Kingdom.
Chimp 2 was HBsAg positive 2 years after arrival at the center in Cameroon but was HBsAg negative on retesting 2 years later. It has shown no signs of clinical hepatitis during its stay at the center. Chimp 4 has been HBsAg positive (Public Health Laboratory Service, Colindale, United Kingdom) since initial testing after arrival in the United Kingdom and remains so 7 years later.
Amplification and nucleotide sequencing.
Each HBsAg-positive sample was assayed by PCR for HBV DNA sequences. Nucleic acid was extracted from 200-μl volumes of serum by the Nuclisens extractor (Organon-Teknika, Boxtel, The Netherlands). Three microliters of extracted DNA was then amplified in a PCR mixture containing Access reagents (Promega, Chilworth, Southampton, United Kingdom) using primers S1 (5′-CATCAGGAYTCCTAGGACCCCT-3′; positions 171 to 192 numbered from the EcoRI site in the HBVADW sequence; GenBank accession no. V00866) and S5 (GAGGCATAGCAGCAGGATGMAGAGG; positions 406 to 430; all primers were produced by Oswel DNA, Southampton, United Kingdom). PCR conditions for all first-round amplification steps were 1 cycle of 94°C for 2 min; 30 cycles of 94°C for 30 s, 55°C for 21 s, and 72°C for 1.5 min; and 1 cycle of 72°C for 6 min. Amplified sequences in 1 μl of PCR product were amplified further in a second-round PCR using internal (nested) primers S3 (5′-CGTGTTACAGGCGGKGTKTTTCTTGT-3′; positions 196 to 221) and S6 (ATGATAAAACGCCGCAGACACATC; positions 379 to 402) and reagents described previously (7). The following combinations of outer and inner nested primers were used to amplify the rest of the HBV genome in overlapping fragments: 21 (GACTTCTCTCARTTTTCYAGGGG; positions 265 to 287) and 24 (AGTAAAYTGAGCCARGAGAAACGG; positions 661 to 684), followed by 22 and 23 (ACGGRCTRAGGCCCACTCCCATAG; positions 641 to 664); 13 (CAAYCCNYTGGGATTCTTYCCC; positions 2892 to 2913) and 16 (CCCCTRGAAAAYTGAGAGAAGTC; positions 265 to 287), followed by 14 (AATCCMGATTGGGACTTCAAYMC; positions 2953 to 2975) and 19 (CKGAACTGGAGCCACCARCAG; positions 69 to 89); 13 and 16, followed by 15 (GTCCACCACGAGTCTAGAYTCTK; positions 245 to 267) and 18 (CATCCTCAGGCCATGMAGTGG; positions 3181 to 3201); 21 and 24, followed by 17 (GTCYTGGCCAAAATTCGCAGTCC; positions 300 to 322) and 23; 22 (GATGTRTCTGCGGCGTTTTATCAT; positions 379 to 402) and 12 (AGACAMAAGAAAATTGGTAAYAG; positions 800 to 822), followed by 11 and 12; 2 (ACTGTTCAAGCCTCCAAGCT; positions 1862 to 1881) and 1 (GATAGGGGCATTTGGTGGTC; positions 2301 to 2320), followed by 3 and 4; 11 (AAWTGCACWTGTATTCCCATCCC; positions 592 to 615) and 27 (CCARCCAGTGGGDGTTGCRTC; positions 1192 to 1112), followed by 25 (TGGTATTGGGGGCCAARTCTG; positions 752 to 772) and 26 (AGCAAACACTTGGCACAGVCC; positions 1171 to 1191); 28 (TCGCCAACTTAYAAGGCCTTT; positions 1102 to 1122) and 31 (GCAGAGGTGAAGCGAAGTGCA; positions 1583 to 1603), followed by 29 (CCTTTACCCCGTTGCYCGGCA; positions 1153 to 73) and 30 (CACGGWCCGGCAGATGAGAAG; positions 1583 to 1603); 32 (ACGGGGCGCACCTCTCTTTA; positions 1522 to 1541) and 3 (AGTGCGAATCCACACTC; positions 2271 to 2287), followed by 33 (CGCGGWCTCCCCGTCTGTGC; positions 1542 to 1561) and 34 (CAATGYTCNGGAGACTCTAA; positions 2027 to 2046); 4 (GAGCTWCTGTGGAGTTACTCTC; positions 1932 to 1953) and 40 (GTTTGGAARTAATGATTAAC; positions 2726 to 2746), followed by 35 (ACCATACNGCACTCAGGCAAG; positions 2055 to 2075) and 39 (CTGGATAATAAGGTTTAAT; positions 2700 to 2718); and 41 (CGTCGCMGAAGATCCAATCT; positions 2423 to 2443) and 19, followed by 42 (GTATYCCTTGGACTCATAAGG; positions 2461 to 2481) and 43 (CCACTGCATGGCCTGAGGATG; positions 3181 to 3201). Amplified DNA was directly sequenced using a commercially available cycle sequencing method (Amersham, Amersham, United Kingdom).
Sequence analysis.
Nucleotide sequences were edited and assembled using the SIMMONIC sequence analysis package. Each amplicon was sequenced in both directions, and most of the nucleotide sequence data was derived from two independent readings. The two complete chimpanzee-derived HBV sequences were aligned with a data set of 145 complete HBV genome sequences available from GenBank (a list is available from the authors), which excluded sequences containing insertions/deletions, and sequences shown previously to be intergenotype recombinants (HBVDNA [accession no. X68292] and HPBADW1 [accession no. D00329]; reference 4). For phylogenetic analysis, genotype C was reduced to 34 representative sequences, as they were heavily overrepresented in the original data set, leaving 90 complete hepadnavirus genomes with an alignment length of 3,244 bases. The sequence of woolly monkey HBV (WMHBV) (9) was used as an outgroup to root the phylogenetic tree. In a second phylogenetic analysis, the S open reading frame (ORF) sequences were extracted from a sample of 22 hepadnaviruses (see below) and combined with those of seven recently published S ORF sequences obtained from wild-caught orangutans (GenBank accession no. Y17559 to Y17565; reference 16) to produce a data set of 29 sequences of 1,212 bp.
Phylogenetic trees were constructed using a maximum-likelihood (ML) method incorporating the general reversible Markov model of nucleotide substitution and a gamma distribution of rate variation among sites (assumed to contain eight different rate categories), the latter of which has been shown to have a major effect on HBV phylogenies (2). Parameter values were estimated from the data during tree reconstruction and are available from the authors on request. The Kishino-Hasegawa test (8) was used to compare, using ML, different hypotheses of phylogenetic relationship among the human and primate hepadnaviruses, although for ease of computation, this analysis was based on a sample of 22 complete genome sequences, including all of those from nonhuman primates and 3 from each human genotype, apart from genotype E, for which only two complete genomes are available. Finally, to assess the robustness of the phylogenetic groupings obtained, a bootstrap neighbor-joining (NJ) analysis with 1,000 replications was performed using the same model of nucleotide substitution as in the ML analysis. All of the phylogenetic analyses described in this paper were performed using the PAUP* package (version 4; reference 13).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been submitted to the GenBank database and assigned accession no. AF242585 and AF242586.
RESULTS
Nucleotide sequences of HBV variants from chimpanzees.
The four HBsAg-positive samples from Chimps 1 to 4 were assayed for HBV DNA by nested PCR using primer pairs S1-S5 and S3-S6. Samples from Chimps 2 and 4 were positive. From these samples, complete genome sequences were obtained by assembly of the nucleotide sequences of overlapping fragments amplified with nested primers (see Materials and Methods). The genome sequences obtained, from Chimps 2 and 4, were circular, and both were 3,182 nucleotides in length. For comparison, the sequence lengths of representative members of the six human HBV genotypes were 3,221 (A; HBVADW2), 3,215 (B; HPBADW2), 3,215 (C; HPBCG), 3,182 (D; HBVGEN1), 3,212 (E; HHVBE4), and 3,215 (F; ADW4A) nucleotides. The chimpanzee sequences were the same length as the HPBVCG sequence obtained from an infected chimpanzee in the London zoo (15) and the HBU46935 sequence from a white-handed gibbon (12) but longer than the HBV sequence from a captive woolly monkey (WMHBV; accession no. AF046996; 3,179 bp; reference 9). The length and number of ORFs encoding the core, surface, polymerase, and X genes were identical to those of HPBVCG and some human HBV genotypes.
Neither the Chimp 2 nor the Chimp 4 sequence contained a precore stop codon, in contrast to the HPBVCG sequence. In the surface gene, the presence of lysine residues at positions 122 and 160 predicted the existence of d and w serological determinants, while isoleucine and alanine at positions 134 and 159 in the Chimp 4 sequence predicted w2. While the subtype adw2 predicted for the Chimp 4 sequence is shared with the HPBVCG sequence, the Chimp 2 sequence contains the less common amino acid residues leucine and valine at positions 134 and 159, making subtype prediction more problematic.
Pairwise comparison of the chimpanzee sequences with other HBV variants (Table 1) showed the Chimp 2 and 4 sequences to be most closely related to the HPBVCG sequence (2.4 to 5.8% sequence divergence) and distinct from human genotypes (8.9 to 13.1% divergence) or the primate sequences HBU46935 (gibbon; 9 and 9.8% divergence) and WMHBV (22.1 and 22.2% divergence). The pairwise distance between Chimps 2 and 4 (6% divergence) was greater than the mean pairwise distances between members of the same human genotype (1.2 to 4.5% divergence between representative sequences of genotypes A to F used for Table 1).
TABLE 1.
Sequence divergencea between new chimpanzee variants and other HBV variants
| Source of genome sequence | Chimp 4 | HPBVCG | Gibbon | Human HBV genotypesb:
|
WMHBV | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | |||||
| Chimp 2 | 0.0598 | 0.0582 | 0.0900 | 0.0996 | 0.1065 | 0.1047 | 0.1032 | 0.0891 | 0.1261 | 0.2210 |
| Chimp 4 | 0.0239 | 0.0976 | 0.0987 | 0.1054 | 0.1069 | 0.1064 | 0.0936 | 0.1309 | 0.2223 | |
| HPBVCG | 0.0905 | 0.0956 | 0.1021 | 0.1007 | 0.1004 | 0.0900 | 0.1243 | 0.2229 | ||
| Gibbon | 0.1023 | 0.1072 | 0.1007 | 0.1109 | 0.1036 | 0.1294 | 0.2235 | |||
| A | 0.0895 | 0.0892 | 0.0970 | 0.0992 | 0.1355 | 0.2229 | ||||
| B | 0.0913 | 0.1047 | 0.1095 | 0.1376 | 0.2170 | |||||
| C | 0.1038 | 0.1044 | 0.1354 | 0.2168 | ||||||
| D | 0.0752 | 0.1345 | 0.2191 | |||||||
| E | 0.1330 | 0.2194 | ||||||||
| F | 0.2325 | |||||||||
Uncorrected nucleotide distances measured between complete genome sequences.
Mean pairwise distances among five representative sequences from genotypes A to D, two of genotype E, and three of genotype F.
Phylogenetic analysis of hepadnavirus genomes.
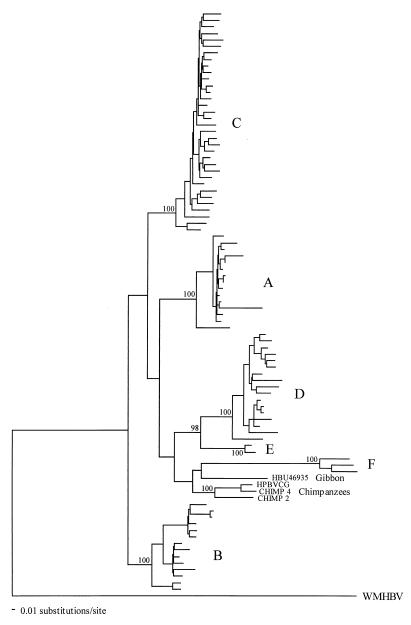
Evolutionary relationships among 90 representative complete hepadnavirus genome sequences rooted by that from the woolly monkey (WMHBV) were analyzed by ML analysis with NJ bootstraps (Fig. 1). Chimps 2 and 4 formed a strongly supported clade with the virus previously obtained from the London Zoo chimpanzee (100% bootstrap support), thereby constituting powerful evidence that all three animals are infected with a chimpanzee-specific virus. The distinctive nature of the six human genotypes was also apparent (all had 100% bootstrap support), although, with the exception of a D-E grouping, intergenotype relationships were uncertain in that none had strong bootstrap support, as was the case with the placement of the gibbon virus. It is therefore difficult to resolve the branching order of the human and primate viruses on these data, although it is noteworthy that the chimpanzee and gibbon viruses fell within, rather than outside, the human part of the tree.
FIG. 1.
ML phylogenetic analysis of complete genome sequences of Chimps 2 and 4, other primates, and available sequences of human genotypes A to F (genotype C is restricted to 34 representative sequences). The tree was rooted with the outgroup WMHBV. NJ bootstrap values of ≥70% are indicated on branches.
To determine the extent of phylogenetic resolution in these data in more statistically rigorous fashion, we undertook a series of likelihood significance tests, although, for ease of computation, this analysis was performed on a smaller sample of 22 representative viruses (see Materials and Methods). First, 10 model tree topologies were constructed based on the ML tree of all 90 sequences (Fig. 1) but with the WMHBV sequence occupying different positions, thereby generating topologies in which the chimpanzee and gibbon viruses, as well as each human genotype, were pictured as the first to diverge. Next, the likelihoods of these model topologies were estimated on the data and compared using the Kishino-Hasegawa test (Table 2). Although a tree with genotype B as the most divergent had the highest likelihood, as was also the case for the ML tree of all 90 complete genomes, the nine competing topologies had very similar likelihoods; only the trees in which genotype D or E diverged first were significantly worse than the best (B), although a tree with genotypes D and E placed together as the most divergent could not be rejected. This analysis reveals that there is little phylogenetic resolution at the base of this tree, perhaps because the WMHBV sequence is too divergent to serve as a viable outgroup. Finally, ML trees for this sample of 22 viruses were also constructed on the genome regions covered by single or double reading frames taken separately and with the most variable sites in the sequence removed. In no case did this produce a major change in branching order (the trees are not shown but are available from the authors on request).
TABLE 2.
Likelihoods of different tree topologies
| Treea | −Ln Lb | Diff −ln L | SD (diff) | t | Pc |
|---|---|---|---|---|---|
| A | 19,113.91571 | 8.21828 | 4.75478 | 1.7284 | 0.0840 |
| B | 19,105.69743 | Best | |||
| C | 19,108.31650 | 2.61907 | 5.75045 | 0.4555 | 0.6488 |
| D | 19,122.28125 | 16.58382 | 8.05901 | 2.0578 | 0.0397 |
| E | 19,122.49123 | 16.79380 | 8.08440 | 2.0773 | 0.0379 |
| D + E | 19,118.31469 | 12.61726 | 6.57211 | 1.9198 | 0.0550 |
| F | 19,113.65749 | 7.96006 | 8.69884 | 0.9151 | 0.3602 |
| Chimpanzees | 19,113.09801 | 7.40058 | 8.78119 | 0.8428 | 0.3994 |
| Gibbon | 19,113.15832 | 7.46089 | 8.75796 | 0.8519 | 0.3943 |
| Apesd | 19,113.15832 | 7.46089 | 8.75795 | 0.8519 | 0.3943 |
Most divergent clade on tree (excluding the WMHBV outgroup).
L, likelihood of tree.
Probability of getting a more extreme t value under the null hypothesis of no difference between the two trees (two-tailed test). Values significant at P < 0.05 are underlined.
Chimpanzees and gibbon.
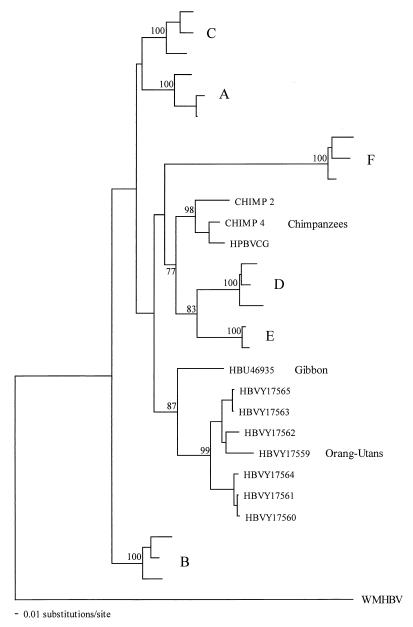
In a second phylogenetic analysis, we constructed trees on the S gene sequences from our sample of 22 viruses plus the 7 viruses recently obtained from orangutans by using ML analysis with NJ bootstraps (Fig. 2). Intriguingly, the orangutan sequences were not only monophyletic, suggesting that they were species specific, but also clustered with the virus taken from the gibbon, an animal that occupies a similar geographic range in Southeast Asia. There is also some (weak) support for a phylogenetic relationship between the chimpanzee viruses and genotypes D and E, although this is not supported at the level of the whole genome. The remainder of the tree is similar to that based on the 90 whole-genome sequences, apart from the slight movement shown by the genotype A sequences.
FIG. 2.
ML phylogenetic analysis of the HBsAg gene sequences of HBV recovered from primates (including those from orangutans) and representative sequences of human genotypes A to F. The tree was rooted with the WMHBV sequence. NJ bootstrap values of ≥70% are indicated on branches.
DISCUSSION
This study provides evidence for infection of chimpanzees (P. troglodytes subsp. verus) born in the wild. Despite the different geographical origins and handling histories of the two study chimpanzees, the nucleotide sequences of HBV amplified from Chimps 2 and 4 were similar to each other but phylogenetically distinct from those of humans or other primate species. HBV infection appeared to be relatively common among chimpanzees in West Africa; in the refuge center in Cameroon, HBsAg screening of other wild-born chimpanzees has identified a further 8 positive samples among 26 tested (J.C.M.L., unpublished data). Further PCR testing and sequence analysis are required to confirm the serology results and to investigate further the distribution of the chimpanzee genotype reported in this study.
The existence of a chimpanzee-specific genotype of HBV is supported by the close similarity of this genotype to that of HBV recovered from a captive chimpanzee (HPBVCG [strain LSH]; references 15 and 17), a finding which clarifies the origin of HBV infection in the London Zoo outbreak. The original investigators found HBV infection in three offspring of a male and a female that had been captured in Africa. However, neither in the original description nor in the subsequent report of the sequence of strain LSH were the authors able to determine whether HBV infection was acquired by either the parents or the offspring in the London Zoo, by the parents after capture in Africa (possibly by inoculation of human immunoglobulin preparations), or in the wild from other chimpanzees. The genetic relatedness of strain LSH to human genotypes has led both these (15) and subsequent investigators (3, 9) to conclude that the chimpanzee infection arose from human contact. The discovery of the same HBV genotype in chimpanzees sampled independently in this study provides convincing evidence against this hypothesis.
Whether other primate species are infected with HBV in the wild has been controversial. Norder et al. (12) obtained the sequence of an HBV variant (HBU46935) infecting a chimpanzee after inoculation with serum from a captive white-handed gibbon (Hybolates lar), although whether the original source of HBV was from the wild has remained controversial. However, the recent unpublished description of six complete genome sequences of HBV that originated from gibbons (S. Grethe et al., accession no. AJ131568 to AJ131574) and group closely with HBU46935 (data not shown) suggests infection of gibbons in the wild, although clarification of this hypothesis awaits publication of the data.
HBV has also been recovered from a captive woolly monkey (Lagothrix lagotricha; reference 9). The sequence of WMHBV is the most divergent of all primate HBV variants, and unlike other primate HBVs, there was some evidence for a restriction in host range; an inoculum from an infected woolly monkey failed to efficiently infect a chimpanzee (9). Very recently, HBV infection was detected in a number of originally wild-caught orangutans (16). Although sequences from the surface gene appeared to show some relationship to the HBU46935 gibbon virus, that all seven grouped together on the tree also implies that orangutans have a specific variant of HBV. In summary, there is growing evidence from this and previous studies for widespread infection of primates with HBV and for the existence of species-specific genotypes. Furthermore, the three genotypes recovered from Old World primate species show a level of sequence divergence from each other (9 to 10%) similar to that among human HBV genotypes A to E.
The finding of HBV infecting native chimpanzees has a number of implications for theories of the origins of human HBV infection. A hypothesis for a relatively recent origin proposed that HBV spread from the Americas into Europe and elsewhere in the Old World after contact between Europeans and indigenous peoples around 400 years ago (3). However, the finding that chimpanzees, as well as orangutans (16), can be infected with HBV in the wild makes the proposed recent spread of HBV from the New World extremely unlikely.
It has been alternatively proposed that HBV infection was present in anatomically modern humans as they migrated from Africa approximately 150,000 to 100,000 years ago (10, 11), and the different genotypes infecting humans evolved since this dispersal. The problem with the hypothesis is that it also does not explain the origin of the various nonhuman primate viruses which, with the exception of the variant found in woolly monkeys, are interspersed among the human genotypes in the phylogenetic tree.
Finally, it could be argued that the variants found in chimpanzees in this study and previously in gibbons, orangutans, and New World primate woolly monkeys are viruses that coevolved with their primate hosts over periods of 10 to 35 million years. The numerous genotypes found in humans would therefore originate through multiple zoonotic transmission episodes from several nonhuman primate species infected with different species-specific genotypes. Such a scenario is not unprecedented; human immunodeficiency virus type 1 infection in humans originated through at least three separate cross-species transmissions from different subspecies of chimpanzees (6), while human infection with human immunodeficiency virus type 2 in West Africa arose independently several times through contact with sooty mangabeys (5). The first difficulty here is that the phylogenetic tree of the various primate HBV variants in no way reflects the phylogeny of the host species, as would be expected for cospeciation. Second, if human genotypes A to F originated in primates, then the actual species involved in transmission to humans remain unidentified.
At this stage, the problems associated with each of three hypotheses for the origin of HBV prevent a definitive conclusion. First, as indicated previously (2), resolution of this issue requires more extensive HBsAg screening and sequence analysis of HBV infecting wild-caught primates from each of their three principal geographical ranges. Second, indigenous human populations in areas of high endemicity for HBV infection (such as sub-Saharan Africa) are poorly sampled and evidence for the existence of other genotypes should be sought. Combined human and primate studies may succeed in identifying the immediate sources of each of the six human genotypes and the circumstances under which HBV subsequently came to be globally distributed.
ACKNOWLEDGMENTS
Eddie Holmes is supported by a fellowship from the Royal Society. Peter Simmonds is supported by a fellowship from the Darwin Trust. Deborah MacDonald was funded by a studentship from the Roseanne Campbell Research Fund.
REFERENCES
- 1.Arauz-Ruiz P, Norder H, Visona K A, Magnius L O. Genotype F prevails in HBV infected patients of Hispanic origin in Central America and may carry the precore stop mutant. J Med Virol. 1997;51:305–312. [PubMed] [Google Scholar]
- 2.Bollyky P L, Holmes E C. Reconstructing the complex evolutionary history of hepatitis B virus. J Mol Evol. 1999;49:130–141. doi: 10.1007/pl00006526. [DOI] [PubMed] [Google Scholar]
- 3.Bollyky P L, Rambaut A, Grassly N, Carman W F, Holmes E C. Hepatitis B virus has a New World evolutionary origin. Hepatology. 1997;26:765. [Google Scholar]
- 4.Bollyky P L, Rambaut A, Harvey P H, Holmes E C. Recombination between sequences of hepatitis B virus from different genotypes. J Mol Evol. 1996;42:97–102. doi: 10.1007/BF02198834. [DOI] [PubMed] [Google Scholar]
- 5.Feng G, Yue L, White A T, Pappas P G, Barchue J, Greene B M, Sharp P M, Shaw G M, Hahn B H. Human infection by genetically diverse SIVsm-related HIV-2 in West Africa. Nature. 1992;358:495–499. doi: 10.1038/358495a0. [DOI] [PubMed] [Google Scholar]
- 6.Gao F, Bailes E, Robertson D L, Chen Y, Rodenburg C M, Michael S F, Cummins L B, Arthur L O, Peeters M, Shaw G M, Sharp P M, Hahn B H. Origins of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 7.Jarvis L M, Davidson F, Hanley J P, Yap P L, Ludlam C A, Simmonds P. Infection with hepatitis G virus among recipients of plasma products. Lancet. 1996;348:1352–1355. doi: 10.1016/s0140-6736(96)04041-x. [DOI] [PubMed] [Google Scholar]
- 8.Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data and the branching order in Hominoidea. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 9.Lanford R E, Chavez D, Brasky K M, Burns R B, 3rd, Rico-Hesse R. Isolation of a hepadnavirus from the woolly monkey, a New World primate. Proc Natl Acad Sci USA. 1998;95:5757–5761. doi: 10.1073/pnas.95.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magnius L O, Norder H. Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene. Intervirology. 1995;38:24–34. doi: 10.1159/000150411. [DOI] [PubMed] [Google Scholar]
- 11.Norder H, Courouce A M, Magnius L O. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- 12.Norder H, Ebert J W, Fields H A, Mushahwar I K, Magnius L O. Complete sequencing of a gibbon hepatitis B virus genome reveals a unique genotype distantly related to the chimpanzee hepatitis B virus. Virology. 1996;218:214–223. doi: 10.1006/viro.1996.0181. [DOI] [PubMed] [Google Scholar]
- 13.Swofford D L. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland, Mass: Sinauer Associates; 1998. [Google Scholar]
- 14.Thomas H C, Jacyna M R. Hepatitis B virus: pathogenesis and treatment of chronic infection. In: Zuckerman A J, Thomas H C, editors. Viral hepatitis. Edinburgh, United Kingdom: Churchill Livingstone; 1993. pp. 185–207. [Google Scholar]
- 15.Vaudin M, Wolstenholme A J, Tsiquaye K N, Zuckerman A J, Harrison T J. The complete nucleotide sequence of the genome of a hepatitis B virus isolated from a naturally infected chimpanzee. J Gen Virol. 1988;69:1383–1389. doi: 10.1099/0022-1317-69-6-1383. [DOI] [PubMed] [Google Scholar]
- 16.Warren K S, Heeney J L, Swan R A, Heriyanto, Verschoor E J. A new group of hepadnaviruses naturally infecting orangutans (Pongo pygmaeus) J Virol. 1999;73:7860–7865. doi: 10.1128/jvi.73.9.7860-7865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuckerman A J, Thornton A, Howard C R, Tsiquaye K N, Jones D M, Brambell M R. Hepatitis B outbreak among chimpanzees at the London Zoo. Lancet. 1978;ii:652–654. doi: 10.1016/s0140-6736(78)92761-7. [DOI] [PubMed] [Google Scholar]