List of abbreviations
- AFP
Alpha‐Fetoprotein
- ANOVA
Analysis of Variance
- BCLC
Barcelona‐Clinic Liver Cancer
- CCK‐8
Cell Counting Kit‐8
- CI
confidence interval
- DEGs
differentially expressed genes
- EHHADH
enoyl‐CoA, hydratase/3‐hydroxyacyl CoA dehydrogenase
- GO
gene ontology
- HCC
hepatocellular carcinoma
- M stage
distant metastasis according to the International Union Against Cancer
- HMGA1
high‐mobility group AT‐hook 1
- HR
hazard ratio
- mimic‐NC
control‐mimic
- miR‐1290
microRNA‐1290
- miRNAs
microRNAs
- NT
normal healthy liver tissues
- N stage
lymph node metastasis according to the International Union Against Cancer
- OR
odds ratio
- SHC
steatohepatitis cirrhosis
- T stage
tumor depth according to the International Union Against Cancer
- TCGA
The Cancer Genome Atlas
- UTR
untranslated region
Hepatocellular carcinoma (HCC) is the second most common cancer and the third leading cause of cancer‐related death worldwide [1]. Recently, HCC incidence and mortality rates have further increased due to changes in the global social environment and dietary habits [2]. HCC is treated by surgical resection or combination chemotherapy, but the overall survival rate of HCC patients has not improved, and the recurrence rate is still high due to strong invasiveness and resistance to chemotherapy [3]. Therefore, it is necessary to understand the pathogenesis of HCC and identify novel diagnostic biomarkers and therapeutic targets for the treatment of HCC. Recently, microRNA‐1290 (miR‐1290) has been reported to regulate the progression of many types of malignant cancers, such as colorectal cancer [4], lung cancer [5], and HCC [6]. However, research on the therapeutic targets of miR‐1290 is still needed. Thus, in this study, we will use HCC patient samples to assess the potential of miR‐1290 as both a diagnostic marker and a therapeutic target, and we will demonstrate its oncogenic properties through functional and mechanistic studies. The methods and materials were described in Supplementary Materials andMethods.
To discover HCC development‐specific microRNAs (miRNAs), we performed NanoString‐based miRNA expression profiling in 5 steatohepatitis cirrhosis (SHC) tissues and 5 early‐stage HCC tissues. Eleven miRNAs were discovered to be differentially expressed between SHC and HCC tissues (Figure 1A). Among them, miR‐1290 was upregulated in HCC compared to SHC and was selected based on its cell proliferation and differentiation function in cancers [7, 8]. In a subsequent validation step, the miR‐1290 was found to be significantly elevated in HCC tissues compared to either SHC (P = 0.043) or normal healthy liver tissues (NT) (P = 0.019) (Figure 1B). In addition, miR‐1290 expression was significantly upregulated as the TNM stage increased in an independent cohort of 111 HCC patients (Figure 1C; ANOVA test P = 0.002). To demonstrate the clinical relevance of miR‐1290 expression in patients with HCC, we determined the association between miR‐1290 expression and various clinicopathological variables in a cohort of 111 HCC patients (Supplementary Table S1). The expression of miR‐1290 was associated with factors reflecting disease progression, such as T stage (P < 0.001), M stage (P = 0.011), TNM stage (P = 0.003), and BCLC stage (P < 0.001). Moreover, the 5‐year overall survival rate was significantly lower in high miR‐1290‐expressing HCC patients (log‐rank P < 0.001) (Figure 1D). Next, the Univariate Cox proportional hazards analysis (Supplementary Table S2) revealed that T stage (HR 4.60; 95% CI 2.76 to 7.66; P < 0.001), N stage (HR 3.21; 95% CI 1.52 to 6.78; P = 0.002), M stage (HR 5.66; 95% CI 2.73 to 11.72; P < 0.001), BCLC stage (HR 5.93; 95% CI 3.64 to 9.65; P < 0.001), AFP expression (HR 2.80; 95% CI 1.73 to 4.53; P < 0.001), and high expression of miR‐1290 (HR 3.16; 95% CI 1.95 to 5.14; P < 0.001) were associated with a poor prognosis. In the multivariate analysis, T stage (HR 2.26; 95% CI 1.12 to 4.57; P = 0.024), BCLC stage (HR 2.59; 95% CI 1.21 to 5.55; P = 0.014), and high expression of miR‐1290 (HR 2.05; 95% CI 1.15 to 3.67; P = 0.016) were significantly associated with poor survival, while AFP expression was not associated with prognosis. Since miR‐1290 expression was associated with M stage, we also examined the association of miR‐1290 expression with HCC metastasis. Univariate logistic regression analysis (Supplementary Table S3) revealed that T stage (OR 8.84; 95% CI 1.08 to 72.13; P = 0.042) and high expression of miR‐1290 (OR 5.09; 95% CI 1.32 to 19.58; P = 0.018) were significantly associated with distant metastasis. In the multivariate analysis, only high expression of miR‐1290 (OR 12.90; 95% CI 1.17 to 142.08; P = 0.037) was independently able to predict metastasis in HCC patients. Furthermore, we validated the expression of exosomal‐miR‐1290 in sera from patients with SHC and HCC. The levels of exosomal‐miR‐1290 showed a significant gradual increase as the disease progressed (ANOVA test, P < 0.001) (Figure 1E). Taken together, these data provide strong evidence that miR‐1290 could be a useful biomarker for predicting HCC progression as well as distant metastasis.
FIGURE 1.
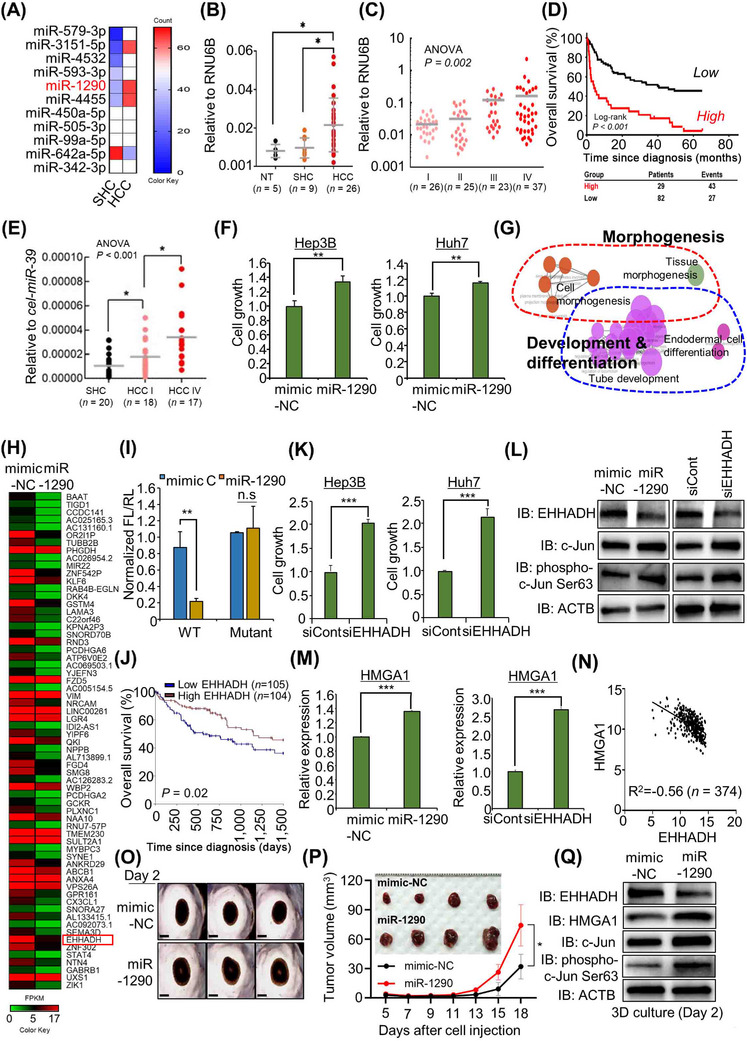
miR‐1290 is a potential therapeutic and diagnostic marker for HCC. (A) Eleven differentially expressed HCC‐specific microRNAs between SHC and HCC tissues in NanoString analysis (white; middle‐expression, red; high‐expression, blue; low‐expression). (B) Expression status of miR‐1290 in NT, SHC tissues, and HCC tissues. The gray horizontal bars represent mean expression levels. The RNU6B is used as a reference in miR‐1290 expression analysis; *P < 0.05, student t‐tests. (C) Expression level of miR‐1290 in different TNM stages (I‐IV) of HCC (n = 111) in the 8th editions of the AJCC TNM staging system. The RNU6B is used as a reference in miR‐1290 expression analysis; P = 0.002, ANOVA. (D) Kaplan‒Meier overall survival analysis based on miR‐1290 expression in HCC patients (n = 111; P < 0.001, log‐rank test). (E) Expression status of exosomal‐miR‐1290 in sera from SHC patients, TNM stage I of HCC patients, and TNM stage IV of HCC patients. The gray horizontal bars represent mean expression levels. The synthetic cel‐miR‐39 is added for spike‐in during RNA extraction in serum samples, which is used as a reference in exosomal‐miR‐1290 expression analysis; *P < 0.05, student t‐test. (F) CCK‐8 solution was added to the culture medium, and the cells were incubated for 5 min at 37°C. Cell growth was assessed by detecting the absorbance with a microplate reader (450 nm). The mean ± SD of three independent experiments are shown, and P‐values were calculated using Student's t‐tests (**P < 0.01). (G) GO pathway term enrichment networks. GO pathway term networks in the miR‐1290‐mimic and control groups were functionally grouped by ClueGO. Terms in the functionally grouped networks were cut off at P‐values > 0.05. (H) Heatmap of RNA‐seq data in the control and miR‐1290 mimic groups. (I) Construction of a dual luciferase reporter vector including normal seed match sequences (WT) or mutant sequences (MUT) of the miR‐1290 binding site in the EHHADH 3’ UTR (upper). Luciferase reporter assay of the 3’ UTR of EHHADH in HEK293T cells transfected with miR‐1290 mimic and the luciferase reporter vector. The mean ± SD of three independent experiments are shown, and P‐values were calculated using Student's t‐tests (** P < 0.01, n.s. not significant). (J) Kaplan‒Meier plot of overall survival for HCC patients in the TCGA cohort. The survival rate of the high EHHADH group was significantly increased compared with that of the low EHHADH group (P = 0.02). (K) CCK‐8 solution was added to the culture medium, and the cells were incubated for 5 min at 37°C. Cell growth was assessed by detecting the absorbance with a microplate reader (450 nm). The mean ± SD of three independent experiments are shown, and P‐values were calculated using Student's t‐tests (***P < 0.001). (L) Western blot analysis after transfection with miR‐1290 (left) and siEHHADH (right) using anti‐EHHADH, anti‐c‐JUN, anti‐phospho‐c‐Jun (Ser63), and anti‐ACTB antibodies. ACTB was used as the internal control in Hep3B cells. (M) qRT‒PCR analysis of HMGA1 expression after transfection with miR‐1290 (left) and siEHHADH (right). The mean ± SD of three independent experiments are shown, and P‐values were calculated using Student's t‐tests (***P < 0.001). (N) Scatter plots of HMGA1 and EHHADH in the TCGA portal. Each dotted line indicates a linear regression line of the expression of HMGA1 and EHHADH. P‐values and correlation coefficients (r) between two genes were obtained using the Pearson correlation method. (O) The 3D spheroid formation assay. Hep3B cells transfected with miR‐1290 or mimic‐NC were loaded onto ULA plates and incubated until 48 h when the spheroids were photographed under a microscope, scale bar, 500 µm. (P) 3 × 106 Huh7 cells with mimic‐NC or miR‐1290 mimic were subcutaneously injected into NOG mice. After cell injection, the tumor volume and body weight of mice were measured every 2‐3 days. The mean ± SEM from four independent experiments is shown, with P‐values calculated using Two‐way ANOVA (*P < 0.05) (below graph). Eighteen days after cell injection, all tumors were isolated from mice (upper picture). (Q) Western blot analysis after miR‐1290 and mimic‐NC transfection using anti‐EHHADH, anti‐HMGA1, anti‐c‐JUN, anti‐phospho‐c‐Jun (Ser63), and anti‐ACTB antibodies. ACTB was used as the internal control in Hep3B spheroids.
Abbreviations: ACTB, β‐actin; ANOVA, Analysis of Variance; AT‐hook 1; c‐JUN, Jun Proto‐Oncogene protein; CCK‐8, Cell Counting Kit‐8; EHHADH, enoyl‐CoA, hydratase/3‐hydroxyacyl CoA dehydrogenase; GO, gene ontology; HCC, hepatocellular carcinoma; HMGA1, high‐mobility group AT‐hook 1; M stage, distant metastasis according to the International Union Against Cancer; miR‐1290, microRNA‐1290; mimic‐NC, control‐mimic; NT, normal healthy liver tissues; N stage, lymph node metastasis according to the International Union Against Cancer; SD, Standard Deviation; SHC, steatohepatitis cirrhosis; T stage, tumor depth according to the International Union Against Cancer; TCGA, The Cancer Genome Atlas; ULA, ultralow attachment; UTR, untranslated region.
Next, to assess the function of miR‐1290 in HCC cell growth, we used the control‐mimic (NC) and miR‐1290 mimic. After treatment with miR‐1290 mimic and NC, we performed CCK‐8 assays and observed an increase in cell growth in Hep3B and Huh7 cell lines (Figure 1F). In gene ontology (GO) analysis with RNA‐seq results, Figure 1G showed that morphogenesis‐, development‐ and differentiation‐related GO terms were enriched by miR‐1290 mimic treatment. To identify the direct target of miR‐1290, we focused on the downregulated genes after miR‐1290 mimic treatment compared to mimic‐NC treatment, and we selected the 247 downregulated genes (cut off >2‐fold) in RNA‐seq results (Figure 1H). By comparing the TargetScan results of miR‐1290, we finally selected the enoyl‐CoA, hydratase/3‐hydroxyacyl CoA dehydrogenase (EHHADH) [9] as a direct target of miR‐1290. In a luciferase assay using wild‐type (wt) and mutant (mut) miR‐1290 binding sites in the 3’ untranslated region (UTR) of EHHADH, we observed that mutation of the EHHADH 3’ UTR did not change the luciferase activity compared to wt (Figure 1I). In prognosis analysis using the RNA‐seq result from The Cancer Genome Atlas (TCGA) portal, low expression of EHHADH in HCC indicated a poor prognosis (Figure 1J). Also, EHHADH downregulation by siEHHADH showed induction of cell growth in HCC cell lines (Figure 1K). Thus, our results clearly suggest that EHHADH could be a direct target for miR‐1290 in HCC.
GO analysis showed enrichment in the term positive regulation of JUN kinase activity after treatment with the miR‐1290 mimic (Supplementary Figure S1A). Additionally, a phosphor‐array after treatment with miR‐1290 mimic or siEHHADH clearly showed that the serine 63 phosphorylation of c‐JUN was increased compared to that in the mimic‐NC or siCont (siControl RNA) group (Supplementary Figure S1B). Thus, we suggest that the induction of cell growth by miR‐1290 is related to the activation of c‐JUN via downregulation of EHHADH. To confirm the induction of phosphorylated c‐JUN, we found the induction of serine 63 phosphorylation by miR‐1290 mimic or siEHHADH compared to mimic‐NC or siCont, respectively (Figure 1L). In addition, to assess how the miR‐1290‐EHHADH axis activated c‐JUN, we identified a down‐target using RNA‐seq results after treatment with miR‐1290 mimic or siEHHADH, and finally, we selected high‐mobility group AT‐hook 1 (HMGA1) [10] and observed HMGA1 upregulation in qRT‐PCR results (Figure 1M). Furthermore, correlation analysis using HCC TCGA results (n = 374) showed a negative correlation (R2 = ‐0.56) between EHHADH and HMGA1 expression (Figure 1N). To verify the relationship between EHHADH and HMGA1 in cell growth, we performed recovery analysis after cotreatment with siEHHADH and siHMGA1 and found that a single treatment with siEHHADH induced an increase in c‐JUN phosphorylation, while cotreatment group decreased c‐JUN phosphorylation (Supplementary Figure S2A andB). In the 3D spheroid model and in vivo xenograft mouse model, we observed an increase in the size of the 3D spheroids and tumors following treatment with the miR‐1290 mimic (Figure 1O and 1P, Supplementary Figure S3). Furthermore, the phosphorylation of c‐JUN was significantly promoted by miR‐1290 mimic treatment in 3D spheroid models (Figure 1Q). Thus, we verified that c‐JUN is activated by the miR‐1290‐EHHADH‐HMGA1 axis using a 3D culture model.
This study demonstrates the potential role of miR‐1290 as a biomarker for predicting HCC progression and distant metastasis. We discovered HCC‐specific upregulation of miR‐1290 by comparing cirrhosis and HCC tissue samples. Through a validation step using several large and independent HCC patient cohorts, we determined that miR‐1290 was upregulated in HCC compared with normal liver, and its level increased in a TNM stage‐dependent manner along with HCC progression. In addition, one of the unique strengths of our study is that we successfully elucidated the potential role of miR‐1290 expression as a novel biomarker for HCC. Patients with high miR‐1290 expression had a poor survival rate in the 5‐year follow‐up analysis. Notably, high miR‐1290 expression in HCC was a significant indicator of HCC prognosis and metastasis and performed better than AFP.
In conclusion, using HCC and normal clinical samples, we identified overexpression of miR‐1290 and suggested EHHADH as a direct target of miR‐1290 via a luciferase assay. Additionally, the negative regulation of EHHADH by miR‐1290 induced c‐JUN activation via HMGA1 upregulation. Thus, we suggest that the miR‐1290‐EHHADH‐c‐JUN activation axis plays an important role in HCC cell proliferation and that miR‐1290 is a novel marker of prognosis and metastasis risk for HCC (Supplementary Figure S4).
AUTHOR CONTRIBUTIONS
Conception and design: Mi‐Young Son, Dae‐Soo Kim, Keun Hur, Hyun‐Soo Cho. Development of methodology: Jinkwon Lee, Tae‐Su Han, Eunsun Jung, Kwangho Kim, Tae Young Ryu, In Hwan Tae, Yunsang Kang, Taesang Son, Kiyoon Kwon, Yuna Roh. Analysis and interpretation of data and Clinical data: Gyeonghwa Kim, Byungheon Lee, Yu Rim Lee, Soo Young Park, Won Young Tak. Writing and review of manuscript: Mi‐Young Son, Dae‐Soo Kim, Keun Hur, Hyun‐Soo Cho. Study supervision: Mi‐Young Son, Dae‐Soo Kim, Keun Hur, Hyun‐Soo Cho.
CONFLICT OF INTEREST STATEMENT
The authors have declared that no competing interest exists.
FUNDING INFORMATION
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science, ICT and Future Planning (2018M3A9H3023077/ 2021M3A9H3016046, 2020R1C1C100743, 2019R1A2C1083892, 2021R1A5A2021614, RS‐2023‐00225239), the Korean Fund for Regenerative Medicine (KFRM) grant funded by the Korea government(the Ministry of Science and ICT, the Ministry of Health & Welfare, 21A0404L1) and by the KRIBB Research Initiative Program. The funders had no role in the study design, data collection or analysis, decision to publish, or manuscript preparation.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study protocol was approved by the ethics committee of our institution (Kyungpook National University Hospital) approved the study (#KNUH‐2014‐04‐056‐001), and all patients provided written informed consent prior to sample collection. All animal experiments were approved by the Committee on Animal Experimentation of the Korea Research Institute of Bioscience and Biotechnology (KRIBB‐AEC‐23188).
CONSENT FOR PUBLICATION
Not applicable.
Supporting information
Supporting information
ACKNOWLEDGMENTS
Not applicable.
Jinkwon Lee, Gyeonghwa Kim, and Tae‐Su Han contributed equally to this work.
Contributor Information
Dae‐Soo Kim, Email: kds2465@kribb.re.kr.
Mi‐Young Son, Email: myson@kribb.re.kr.
Keun Hur, Email: KeunHur@knu.ac.kr.
Hyun‐Soo Cho, Email: chohs@kribb.re.kr.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author Hyun‐Soo Cho (email: chohs@kribb.re.kr) upon reasonable request.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–49. [DOI] [PubMed] [Google Scholar]
- 2. Gao X, Wang X, Zhang S. Bioinformatics identification of crucial genes and pathways associated with hepatocellular carcinoma. Biosci Rep. 2018;38(6):BSR20181441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen W, Xiao W, Zhang K, Yin X, Lai J, Liang L, et al. Activation of c‐Jun predicts a poor response to sorafenib in hepatocellular carcinoma: Preliminary Clinical Evidence Sci Rep. 2016;6:22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu X, Xu X, Pan B, He B, Chen X, Zeng K, et al. Circulating miR‐1290 and miR‐320d as Novel Diagnostic Biomarkers of Human Colorectal Cancer. J Cancer. 2019;10(1):43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu Y, Wei J, Zhang W, Xie M, Wang X, Xu J. Serum Exosomal miR‐1290 is a Potential Biomarker for Lung Adenocarcinoma. Onco Targets Ther. 2020;13:7809–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Q, Wang G, Niu L, Zhao S, Li J, Zhang Z, et al. Exosomal MiR‐1290 Promotes Angiogenesis of Hepatocellular Carcinoma via Targeting SMEK1. J Oncol. 2021;2021:6617700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghafouri‐Fard S, Khoshbakht T, Hussen BM, Taheri M, Samadian M. A Review on the Role of miR‐1290 in Cell Proliferation, Apoptosis and Invasion. Front Mol Biosci. 2021;8:763338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kalhori MR, Soleimani M, Arefian E, Alizadeh AM, Mansouri K, Echeverria J. The potential role of miR‐1290 in cancer progression, diagnosis, prognosis, and treatment: An oncomiR or onco‐suppressor microRNA? J Cell Biochem. 2022;123(3):506–31. [DOI] [PubMed] [Google Scholar]
- 9. Okamura S, Yoshino H, Kuroshima K, Tsuruda M, Osako Y, Sakaguchi T, et al. EHHADH contributes to cisplatin resistance through regulation by tumor‐suppressive microRNAs in bladder cancer. BMC Cancer. 2021;21(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vignali R, Marracci S. HMGA Genes and Proteins in Development and Evolution. Int J Mol Sci. 2020;21(2):654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author Hyun‐Soo Cho (email: chohs@kribb.re.kr) upon reasonable request.


