Abstract
Coxsackievirus B3 (CVB3) is a common factor in human myocarditis. Apoptotic events are present in CVB3-induced disease, but it is unclear how CVB3 is involved in apoptosis and which viral proteins may induce the apoptotic pathway. In this report we demonstrate that the human and murine proapoptotic protein Siva specifically interact with the CVB3 capsid protein VP2. Furthermore, the transcription of Siva is strongly induced in tissue of CVB3-infected mice and is present in the same area which is positively stained for apoptosis, CD27, and CD70. It has been proposed that Siva is involved in the CD27/CD70-transduced apoptosis. Therefore, we suggest a molecular mechanism through which apoptotic events contributes to CVB3-caused pathogenesis.
Coxsackievirus B3, a member of the picornavirus family, is an important human pathogen. Most CVB3-caused disease are mild, but some acute infections are severe and lethal. Clinically, coxsackievirus infections are known to be associated with different forms of subacute, acute, and chronic myocarditis (45, 50). CVB3 may cause cardiac arrhythmias and acute heart failure; chronic forms of this disease may supervene, leading to dilated cardiomyopathy, requiring heart transplantation, or to death. The pathogenesis of coxsackievirus infection has been studied extensively in different murine models, demonstrating that the outcome of the disease is determined by complex interactions among several variables, such as virus genotype (11, 25), mouse strains used (11, 24), and the sex (23, 24, 26), age (32), and immune status (18, 27, 37, 49) of the host. In addition, the molecular biology of CVB3 is well documented, especially in view of the availability of sequence data (9, 34, 35, 39, 46) and infectious cDNA molecules (30, 35) as well as the characterization of the genome organization (36) and RNA structures (52). Despite the accumulation of molecular data, so far there are no virus-specific preventive or therapeutic procedures available to protect humans against coxsackievirus-induced heart diseases. In addition, the mechanisms how CVB3 causes acute or chronic myocarditis are not well characterized (6).
One detail of CVB3-induced pathogenesis is apoptosis. For example, apoptotic processes are present in myocardial tissue of patients with dilated cardiomyopathy (42), and depending on the mouse strain and the virus variant used, apoptotic cells are detectable in inflammatory lesions as well as myocardial tissue outside inflamed areas (12, 16, 18, 22, 25). However, it is not established which cell type undergoes apoptosis. Furthermore, CVB3 infection of HeLa cells induces caspase-3 activation, but this may not be responsible for the characteristic cytopathic effect produced by coxsackieviruses (8). It is not clear how CVB3 is involved in apoptotic processes and which viral proteins may interact with host cell proteins. To study these possible protein-protein interactions in more detail, we used the yeast two-hybrid system. A HeLa cDNA expression library was screened for proteins that interact with structural and functional proteins of CVB3. We identified the interaction between the CVB3 capsid protein VP2 and the proapoptotic protein Siva, which is involved in the CD27/CD70-transduced apoptotic pathway (44). CD27, a member of the tumor necrosis factor receptor superfamily, is known to be expressed in T and B cells. Two basic functions of CD27 signaling are known so far. First, the binding of CD27 to CD70, a protein which is also expressed in T and B cells, can provide costimulatory signals in lymphocyte proliferation and immunoglobulin production (19, 44). For example, it was demonstrated that the cytoplasmic tail of CD27 directly associates with Traf-2 and signals to the Jun N-terminal kinase activation in primary murine lymph node T cells (17). On the other hand, CD27 was shown to be involved in apoptotic processes (44). The cytoplasmic tail of CD27 lacks the death domain, but Siva, which has a death domain-like region, can bind to this part of CD27 under in vitro conditions. Overexpression of Siva in different cell lines induces apoptosis in the absence of CD70. Therefore, only the association of CD27 with the intracellular Siva can result in the induction of apoptotic events. Furthermore, under in vivo conditions Siva was also found to be present in a number of nonlymphatic tissues (44). Using a rat model of acute ischemic injury, it was demonstrated (43) that the rat equivalent of Siva is produced within the kidney cells after injury and could be the mediator of apoptosis via an interaction of CD27 which is expressed in the kidney as well.
Using the murine model of CVB3 infection, we demonstrate that this virus infection induced the transcription of the murine equivalent of Siva (muSiva) in pancreas and heart tissue. The interaction between VP2 and muSiva was confirmed using a yeast two-hybrid approach. Interestingly, transcription of muSiva as well as CD27-, CD70-, active caspase-3-, and TUNEL (terminal deoxynucleotidyltransferase [TdT]-mediated dUTP-biotin nick end labeling) assay-positive cells were present in the same area of tissue in CVB3-infected mice. These findings indicate a newly discovered mechanism by which apoptosis may contribute to coxsackievirus-dependent pathogenesis.
MATERIALS AND METHODS
Mice.
Inbred BALB/c (H-2dd) mice were obtained from the Friedrich Schiller University breeding colony. Adult males 7 to 9 weeks of age were used in this study. Experimental groups consisted of a minimum of four mice, and experiments were repeated at least twice and usually three or four times.
Viruses and cell lines.
The CVB3 (Nancy) variant used is a cDNA-generated virus obtained after transfection of HeLa cells with plasmid pCVB3M2, originally obtained by R. Zell (38). The virus was propagated in HeLa cells and then purified and quantified as described previously (18).
Constructions of plasmids.
The baits for the two-hybrid screening were constructed as follows. Sequences specific for the capsid proteins VP1 (851 bp), VP2 (788 bp), and the protease 2A (440 bp) were amplified by PCR from the CVB3 cDNA (35). The upstream primer contains an NdeI site. The downstream primers contain a BamHI site 3′ to the stop codon. VP1, VP2, and 2A samples were amplified at 94°C (5 min) for 1 cycle followed by 30 cycles at 94°C (1 min), 58°C (1 min), and 72°C (2 min), and a final cycle at 72°C for 5 min in a total volume of 100 μl. The PCR products were digested with NdeI and BamHI, gel purified, and inserted between the appropriate sites of pAS2-1 (Clontech Laboratories Inc., Palo Alto, Calif.) to produce pAS2-1/VP1, pAS2-1/VP2, and pAS2-1/2A. In a similar way, the coding sequences of PV1-VP2 and TMEV-VP2 were PCR amplified using the cDNA of the relevant virus as a template and cloned into pAS2-1. The fusion of the entire human Siva (huSiva) protein to the Gal4 DNA-binding and activation domains (Gal4BD and Gal4AD) was achieved by PCR amplification of the identified prey plasmid lacking the first seven residues of huSiva, using an upstream extended primer containing the missing codons and a cleavage site for EcoRI. In the downstream primer, the stop codon of huSiva was followed by an XhoI site. Digested PCR fragments were ligated to the EcoRI/XhoI sites of pGADGH (Clontech) or to the EcoRI/SalI sites of pAS2-1, yielding Gal4AD-huSiva and Gal4BD-huSiva, respectively. The same primers were applied for the generation of a glutathione S-transferase (GST)–huSiva fusion using the vector pGEX-4T-1 (Pharmacia, Freiburg, Germany). The in-frame fusions of all PCR-amplified fragments were confirmed by sequencing.
Yeast strains, transformation, and two-hybrid analyses.
The Saccharomyces cerevisiae strain used for the two-hybrid studies was Y190 (MATa ura3-52 his3-200 lys2-801 ade2-101 trp1-901 leu2-3,-112 gal4Δ gal80Δ cyhr2 LYS2::GAL1UAS-HIS3TATA-HIS3 URA3::GAL1UAS-GAL1TATA-lacZ; Clontech). Transformation of yeast cells was carried out by the method of Klebe et al. (33). Yeast transformants were selected and cultivated on SD synthetic medium (2% glucose and 0.67% yeast nitrogen base without amino acids) supplemented with the appropriate nutrients. S. cerevisiae Y190 expressing each of the analyzed viral baits fused to the Gal4BD was transformed with a Gal4AD-tagged HeLa cell cDNA library (Clontech), and the cotransformants were initially selected for growth on medium lacking histidine. To enhance the stringency of a two-hybrid interaction, the medium was particularly supplemented with 40 mM 3-amino-1,2,4-triazole. Growing yeast colonies were subsequently analyzed for β-galactosidase expression using a colony lift filter assay with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as a substrate as specified by Breeden and Nasmyth (7). The library plasmids of yeast cells expressing both reporter genes were rescued by transformation of total yeast DNA into Escherichia coli HB101. Transformants were selected on M9 minimal medium lacking leucine. To ensure the identification of the correct cDNA preys, isolated plasmids were retransformed into yeast strain Y190 containing the appropriate bait proteins, and the cotransformants were again tested for β-galactosidase activity.
GST pull-down experiments.
GST-huSiva was maintained and expressed in E. coli BL21 as instructed by the manufacturer (Pharmacia). The fusion protein was purified from crude bacterial cell extracts with glutathione-Sepharose. Proteins of noninfected and CVB3-infected HeLa cells were isolated by detaching the cells with 10 mM EDTA. Cells were centrifuged at 250 × g, resuspended in NTE buffer (10 mM Tris-HCl [pH 7.4], 100 mM NaCl, 0.5% NP-40), and immediately vortexed for 30 s. Cell debris were removed by centrifugation at 12,000 × g at 4°C for 20 min. Protein concentration of the clear supernatant was determined by the Bradford protein assay (Bio-Rad Laboratories, Hercules, Calif.). The GST-pull down assays were performed by the protocol of MacDonald et al. (40), with slight modification. Briefly, 30 μg of the HeLa cell crude extract was incubated with 2 μg of GST-huSiva or 2 μg of GST alone in a total volume of 200 μl of binding buffer (20 mM Tris-HCl [pH 7.6], 100 mM NaCl, 0.1% NP-40, 0.1 nM phenylmethylsulfonylfluoride, 1 mM dithiothreitol, pepstatin [50 μg/ml], aprotinin [2 μg/ml], leupeptin [2 μg/ml]). Subsequently, 50 μl of a 50% slurry of glutathione-Sepharose, equilibrated with the binding buffer, was added. The mixture was incubated for 1 h at 4°C under slight shaking. Associated proteins were pelleted by centrifugation and washed five times in 10 volumes of binding buffer. Finally the pellet was resuspended in 10 μl of sodium dodecyl sulfate sample buffer and analyzed for VP2 content by Western blotting. For this, the samples were loaded on a 10 to 20% Tris-glycine gradient gel. After electrophoresis, proteins were electroblotted on a nitrocellulose membrane. Detection of proteins was performed with the ProtoBlot II AP system (Promega Corp., Madison, Wis.). As a primary antibody, a polyclonal anti-CVB3/VP4-2 rabbit antibody (dilution 1:500) was applied, and color reaction was obtained by using the Promega ProtoBlot II AP detection system.
Preparation and staining of routine histology.
Aseptically removed pancreas and heart tissue was fixed with for at least 24 h with 4% formaline and mounted in paraffin, and 6-μm sections were cut and stained with hematoxylin-eosin.
Reverse transcription-PCR (RT-PCR).
Total RNA was isolated from pancreas and heart tissues of infected and noninfected BALB/c mice according to the acid guanidinium thiocyanate phenol chloroform method described in detail by Chomczynski and Sacchi (10). Following ultraspeed homogenization, RNA was extracted using 4 M guanidinium thiocyanate–25 mM sodium citrate–0.5% sarcosyl–100 mM mercapthoethanol (pH 7.0). DNA and protein contaminations were removed by phenol-chloroform treatment. After ethanol precipitation, RNA pellets were dissolved in diethyl pyrocarbonate-treated water and incubated with DNase I (Boehringer, Mannheim, Germany) for 15 min at room temperature (RT) to digest remaining DNA. The DNase I was inactivated by adding 10 mM EDTA and heating to 65°C for 10 min. Reverse transcription was performed as follows, using 5 μg of total RNA. Random hexamer primers were allowed to anneal for 10 min at 70°C. Samples were heated to 42°C for 50 min using 200 U of Superscript II reverse transcriptase (Life Technologies Inc., Rockeville, Md.) in buffer containing 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1 mM dithiothreitol, and 2.5 mM MgCl2. The reaction was terminated by heating to 90°C for 5 min. The remaining RNA was digested with E. coli RNase H (Life Technologies). Primers were designed to correspond to the 5′ and 3′ ends of murine Siva, VP2, and β-actin cDNA sequences.
Immunohistochemistry.
Immunohistochemical studies were carried out with cryomicrotome sections. Aseptically removed pancreas and heart tissue was quickly frozen in frozen specimen embedding medium (Cryomatrix; Life Technologies). For lymphocyte characterization, 10-μm sections were obtained, air dried for 2 h, fixed for 3 min with acetone at RT, washed with Hanks solution, and treated with 0.04% H2O2 to block cellular peroxidase activity. Thereafter, sections were incubated separately with an avidin solution (30 min, RT), biotin solution (30 min, RT), and a 2% nonfat dry milk solution (30 min, RT) to block nonspecific binding. Between each procedure, sections were washed three times with Hanks solution (3 min, RT). All incubation and washing procedures to detect active caspase-3 were performed in the presence of 0.05% saponin. Primary antibodies were applied for 14 h at 4°C. These consisted of rabbit anti-active caspase-3 antibodies (clone 67341A; PharMingen, San Diego, Calif.), Armenian hamster anti-CD27 antibodies (clone LG.3A10; PharMingen), and rat anti-CD70 antibodies (clone FR70; PharMingen). After a 12-min wash with Hanks solution, secondary antibodies (biotin-conjugated anti-rabbit [PharMingen]; biotin-conjugated mouse anti-hamster [PharMingen]; biotin-conjugated goat anti-rat [Jackson ImmunoResearch Laboratories, West Grove, Pa.]) were applied for 30 min at RT. A color reaction was obtained after washing of the slides for 12 min (RT) and sequential treatment with streptavidin-horseradish peroxidase conjugate and ACE Red peroxidase substrate kit (Camon Labor-Service GmbH, Wiesbaden, Germany). The sections were counterstained with Mayer's hemalaun solution.
In situ hybridization.
Digoxigenin-labeled DNA probes for in situ detection of viral RNA and RNA of muSiva were synthesized from plasmid pCMV/VP1 or pAS2-1/muSiva by PCR using digoxigenin-labeled nucleotides. Formalin-fixed, paraffin-embedded sections (6 μm) obtained from pancreas and heart tissue were rehydrated by sequential incubation with xylene, 100, 95, 70, and 50% ethanol, and phosphate-buffered saline (PBS) for 5 min each and treated with protease VIII solution (50 μg/ml; Sigma Biosciences, St. Louis, Mo.) for 1 h at 37°C. After dehydration by sequential incubation with 70, 95, and 100% ethanol, sections were air dried, denatured at 90°C for 8 min, and hybridized at 37°C for 18 h. Thereafter, slides were washed twice at 37°C for 10 min. Background binding was blocked with blocking solution (Kreatech Inc., Amsterdam, The Netherlands), and the sections were incubated with antidigoxigenin antibody solution (Boehringer) for 1 h at RT. A color reaction was obtained after washing of the slides three times with PBS at RT and treatment with nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate solution (Boehringer). The sections were counterstained with eosin or hematoxylin solution.
Apoptosis assay.
Apoptotic cells in pancreas and heart tissue of CVB3-infected mice were detected by using a TACS Blue Label (TBL) in situ apoptosis detection kit (Genzyme Corp., Cambridge, Mass.) as described in the Genzyme manual. Briefly, formalin-fixed paraffin sections (6 μm) were obtained from pancreas and heart tissue and rehydrated by sequential incubation with xylene, 100, 95, and 70% ethanol, distilled H2O, and PBS for 5 min each. After protease K digestion for 15 min at RT and quenching of endogenous peroxidase using 2% H2O2 solution for 5 min at RT, tissue sections were incubated with the enzyme TdT, reaction buffer with Co2+ cations, and digoxigenin-labeled nucleotides. As a negative control, the TdT enzyme was omitted from the protocol. Single positive cells were detected after color reaction with TBL-streptavidin-horseradish peroxidase detection solution. The slides were counterstained with eosin.
RESULTS
Coxsackievirus protein VP2 interacts with the proapoptotic protein huSiva.
To identify human proteins that interact with CVB3 proteins, we used a yeast two-hybrid system based on the yeast Gal4 transactivator (15). As viral baits we selected the CVB3 proteins VP1, VP2, and protease 2A. The full-length cDNAs of these proteins fused to the yeast Gal4BD were applied to screen a Gal4AD-tagged HeLa cell cDNA library. Our initial selection criteria for positive interactors were the expression of the reporter genes HIS3 and lacZ. Out of approximately 2.2 × 105 analyzed yeast cotransformants, we identified two candidate clones expressing proteins which may bind to VP2. No reporter-positive clones were detected using the baits VP1 and 2A. The cotransformation of Gal4BD-2A-containing plasmids with the Gal4AD library plasmids resulted in a low transformation frequency. Additionally, the obtained transformants grew very slowly on selective media, which may indicate a toxic effect of 2A expression on yeast cells as described earlier for the 2A protease of poliovirus type 1 (3).
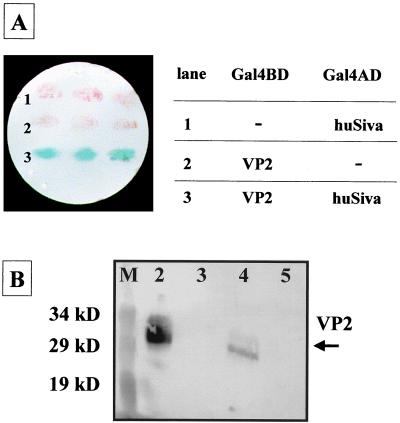
One of the identified preys interacting with VP2 encoded huSiva. Recently, it has been shown that huSiva is involved in mediating CD27/CD70-transduced apoptotic processes (44). Nucleotide sequencing revealed that the encoded fusion protein of the original two-hybrid clone lacked the first seven residues of huSiva. To reduce the probability of a false positive interaction, which often occurs in two-hybrid screens (4), the entire huSiva sequence was fused to the Gal4AD and analyzed for interaction with Gal4BD-VP2. Yeast cells containing both fusion proteins induced lacZ gene expression similarly to cells expressing Gal4BD-VP2 together with Gal4AD–amino-terminally truncated huSiva (Fig. 1A). The reporter gene was not activated in cells expressing Gal4BD-VP2 or Gal4AD-Siva alone. These data indicated that the first seven amino acids of huSiva were not required for the interaction with VP2.
FIG. 1.
Yeast two-hybrid filter lift assay demonstrating the interaction between VP2 of CVB3 and huSiva. Plasmids carrying Gal4BD-VP2 and Gal4AD-huSiva or the Gal4BD and Gal4AD alone were transformed into yeast reporter strain Y190. The resulting cells were analyzed for β-galactosidase activity (A). Only the interaction of Gal4BD-VP2 and Gal4AD-huSiva reconstituted an active Gal4 transcription factor, demonstrated by blue-stained yeast colonies detected after 2 h of incubation with X-Gal (lane 3). GST pull-down experiments confirmed the VP2-huSiva interaction in vitro (B). Thirty-microgram of crude extract proteins of CVB3-infected HeLa cells was incubated with 2 μg of GST (lane 3) or 2 μg of GST-huSiva (lane 4). Protein complexes were precipitated with glutathione-Sepharose and analyzed for VP2 content as described in Materials and Methods. Lane 2, 30 μg of CVB3-infected HeLa cell crude extract proteins; lane 5, 30 μg of noninfected HeLa cell crude extract proteins. A prestained protein marker was used as a size standard (lane M).
The specificity of the two-hybrid interaction between CVB3 VP2 and huSiva was demonstrated by analyzing the ability of huSiva to bind to VP2 of two other picornaviruses, such as poliovirus type 1 and Theiler's murine encephalomyelitis virus, which are also involved in apoptotic processes (1, 29). None of these Gal4BD-tagged VP2 proteins were able to activate reporter gene expression in Gal4BD-huSiva-expressing yeast reporter strains, indicating that they did not interact with huSiva (data not shown). To confirm the two-hybrid in vivo binding of VP2 to huSiva, we performed in vitro assays using GST-huSiva and whole cell lysates of CVB3-infected HeLa cells. After the components were mixed, the huSiva-specific protein complexes were precipitated with glutathione-Sepharose and analyzed for VP2 content by Western blotting using VP2-specific polyclonal antibodies. Figure 1B shows that GST-huSiva efficiently precipitated endogenous VP2 from the CVB3-infected cell extracts. These data not only verified the VP2-huSiva interaction found in the yeast two-hybrid system by an in vitro interaction but also showed that huSiva selectively bound to VP2 even in the high background of other cellular and viral components, which were contained in the crude extracts.
Induction of muSiva transcription in different tissue of CVB3-infected mice.
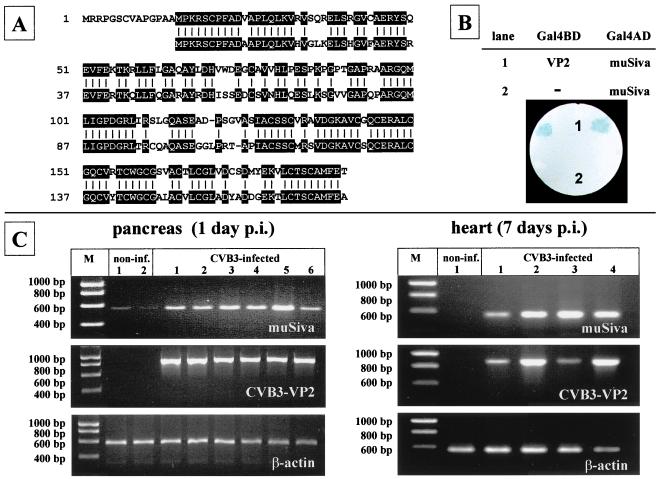
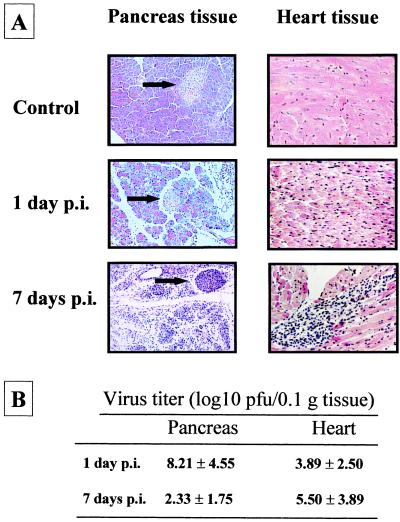
Comparison of the 189-amino-acid sequence of huSiva (44) with the 177-amino-acid sequence of muSiva (described by K. V. Prasad et al. [GenBank accession no. AF033115]) revealed an identity of 71% (Fig. 2A). The entire muSiva cDNA sequence was obtained by PCR from the cDNA of CVB3-infected pancreas tissue and fused to the DNA sequence coding for the yeast Gal4AD. The cotransformants of yeast strain Y190 expressing Gal4BD-VP2 and Gal4AD-muSiva were analyzed for lacZ activity by a filter lift assay. As shown in Fig. 2B, the blue color could be detected after 2 h of incubation with X-Gal, demonstrating that VP2 also binds to the muSiva. To study the VP2-Siva interaction under in vivo conditions, we used the mouse model of CVB3-induced disease. Because apoptotic events seem to be involved in CVB3-caused myocarditis, the induction of muSiva mRNA was analyzed in pancreas as well as heart tissue of CVB3-infected mice by RT-PCR 1 day and 7 days p.i., respectively. As demonstrated in Fig. 2C, CVB3 caused the induction of high levels of muSiva mRNA in different tissue of individual mice compared to noninfected mice. This indicates that the CVB3-caused induction of muSiva expression might be involved in apoptosis during coxsackievirus-depending pathogenesis. Upon intraperitoneal (i.p.) inoculation of 106 PFU, CVB3 replicated primarily in tissue of the exocrine pancreas. High levels of viral progenies were detectable at the first day of viral replication (Fig. 3B), causing massive tissue destruction 1 to 3 days postinfection (p.i.) as demonstrated in Fig. 3A. Thereafter, only the tissue of the endocrine pancreas (islets of Langerhans) was still present 5 to 7 days p.i. Via blood circulation CVB3 entered the heart tissue, causing myocytolysis and infiltration of mononuclear cells into the infected area (Fig. 3A).
FIG. 2.
CVB3-caused induction of muSiva. Sequence comparison between the human (first row) and the murine (second row) proapoptotic protein Siva reveals a 71% identity (A). The interaction between VP2 of CVB3 and muSiva was confirmed using the yeast two-hybrid system (B), demonstrated by the blue-colored yeast colonies coexpressing Gal4BD-VP2 and Gal4AD-muSiva (lane 1). Male BALB/c mice were infected with CVB3 i.p. RNA was isolated from pancreas and heart tissue 1 or 7 days p.i. Transcription of muSiva, CVB3-VP2, and β-actin in tissue of individual noninfected or CVB3-infected mice was analyzed by RT-PCR, demonstrating the induction of high levels of muSiva mRNA only in tissue of CVB3-infected mice (C).
FIG. 3.
Coxsackievirus-induced pathology in pancreas and heart tissue of BALB/c mice. BALB/c mice were infected with CVB3 i.p. (A) Pancreas tissue and heart tissue was isolated from infected animals 1 day and 7 days p.i., respectively, and from noninfected animals. After hematoxylin-eosin staining, virus-caused tissue damage in the pancreas was obvious, demonstrated by massive destruction of the exocrine pancreas up to 7 days p.i. (original magnification, ×500). Only the islets of Langerhans remained unaffected (arrows). In the heart tissue, CVB3 infection caused massive inflammation accompanied by infiltration of mononuclear cells 7 days p.i. (original magnification, ×500). (B) At the indicated time points, eight mice were sacrificed and virus titers were measured by plaque formation assays. Average titers and standard deviations are shown as log10 values of PFU/0.1 g of tissue.
Presence of apoptotic, CD27-positive, and CD70-positive cells in different tissues of CVB3-infected mice.
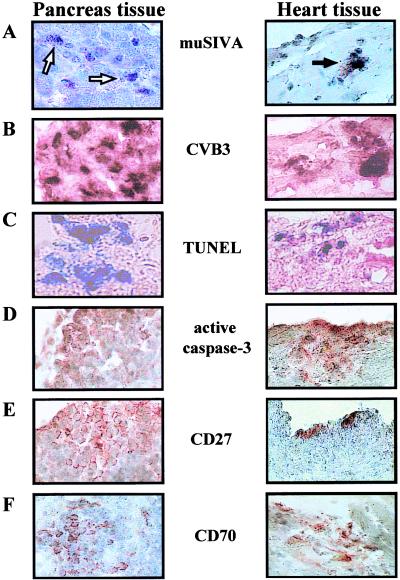
During CVB3 infection many host cell functions are altered, including the induction or suppression of several genes encoding the information of structural and nonstructural cellular proteins (51). These experiments indicate that CVB3 infections are dynamic molecular processes in which timely interactions between viral and host proteins determine the outcome for both the virus and the host cells. To analyze whether the CVB3-caused induction of muSiva transcription (Fig. 2B) and the presence of high amounts of infectious virus particles (Fig. 3B) were accompanied by apoptosis, the TUNEL assay and immunohistochemistry to detect active caspase-3 protein were applied, using pancreas and heart tissue 1 and 7 days p.i. As shown in Fig. 4C and D, TUNEL assay- and active caspase-3-positive cells were easily detectable in pancreas and heart tissue in which viral RNA (Fig. 4B) as well as muSiva RNA (Fig. 4A) were present. Furthermore, CVB3 infections activated also the expression of CD27 and CD70 in cells which were localized in the infected area (Fig. 4E and F), indicating that the CD27/CD70 apoptotic pathway seemed to be induced in CVB3-caused pathogenesis. Tissue sections of noninfected animals were negative relating to in situ hybridization of muSiva and CVB3 as well as TUNEL assay and immunohistochemistry of active caspase-3, CD27, and CD70 (data not shown).
FIG. 4.
Detection of muSiva, apoptotic cells, and CD27- and CD70-positive cells in CVB3-infected tissue. BALB/c mice were infected with CVB3 i.p. Pancreas tissue and heart tissue were isolated at 1 day and 7 days after infection, respectively, and used to perform in situ hybridization studies (A and B), TUNEL assays (C), and immunohistochemistry stainings (D to F). In the area of CVB3-infected tissue (B), transcriptional activity of muSiva (A, arrows) as well as TUNEL assay (C)- and active caspase-3 (D)-positive cells were detectable. CVB3-caused inflammation also induced the accumulation of CD27- and CD70-positive cells in both tissue (E and F). Original magnification, ×1575.
DISCUSSION
In this report, we demonstrate that the structural protein VP2 of CVB3 interacts specifically with the proapoptotic protein Siva. This observation was obtained using the yeast two-hybrid system which was successfully applied for the identification of other virus-host cell protein interactions. For example, this technique was used to demonstrate that the IE2 protein of cytomegalovirus interacts with several different human ribonucleoproteins (48) and that the core protein of hepatitis C virus is able to bind to the intracellular domain of the lympho-toxin β receptor, influencing hepatitis C virus-caused pathology (20).
CVB3 infections are usually accompanied by dramatic changes of the cellular metabolism and the release of newly synthesized virus particles. For a better understanding of this disease, several mouse model systems demonstrating the complexity of CVB3-caused pathogenesis have been established. Upon CVB3 binding to the coxsackievirus and adenovirus receptor, the viral RNA enters the cytoplasm (5). There it is translated into a single polyprotein which is proteolytically processed by virus-specific proteases into structural and nonstructural proteins. The virus-encoded RNA-dependent RNA polymerase transcribes negative-strand RNA, which is the template for multiple rounds of virus genome synthesis. During this viral replication several host cellular processes are altered, inducing host cellular protein synthesis shutoff; e.g., the virus-specific protease 2A cleaves the eucaryotic initiation factor 4 gamma-1 and -2, stimulating the translation of uncapped mRNA like the CVB3 genome (14, 41). In addition, recently it has been shown that 2A of CVB3 can also inactivate both the poly(A)-binding protein (31) as well as the cytoskeletal protein dystrophin (2). Furthermore, 2B of CVB3 can modify plasma membrane and endo-plasmic reticulum permeability (13), thus inducing an increased level of cytosolic-free calcium (28, 47). Using in vitro conditions, CVB3 infection results in tyrosine phosphorylation of two cellular proteins, increasing viral progeny production (21). With the help of the differential mRNA display technique, it was demonstrated that in heart tissue of CVB3-infected mice several genes were up- as well as downregulated in comparison to cells of noninfected animals. Among these genes, mRNA levels of the mouse Nip21 were decreased. The human equivalent of Nip21, Nip2, may interacts with the Bcl-2 protein to promote cell survival. Downregulation of Nip21 by CVB3 infection may therefore increase myocyte cell death (51).
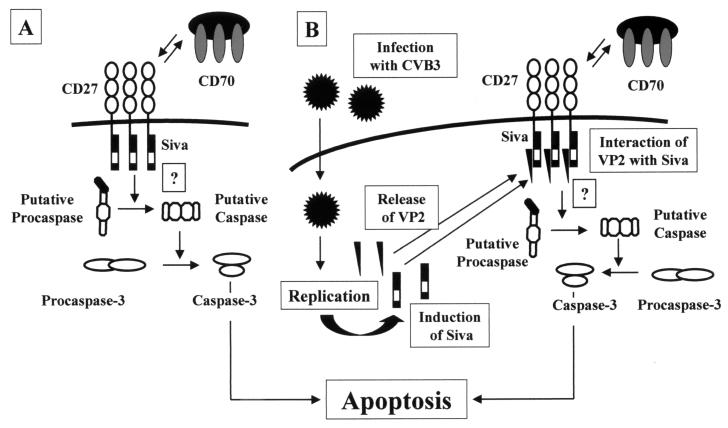
In our murine model of CVB3 infection, we were able to demonstrate that the transcription of muSiva was increased in pancreas and heart tissue in the presence of infectious virus particles. A molecular model that illustrates the role of Siva in CD27/CD70-caused apoptosis is shown in Fig. 5A. With or without binding of CD70 to CD27 on the surface of the cellular membrane, Siva interacts with the cytoplasmic tail of CD27, providing death domain-like structures. The further events of this apoptotic pathway are unknown so far. Due to the fact that CD27 belongs to the tumor necrosis factor receptor superfamily, it is quite possible that a protein which is similar to FADD—a protein which is necessary for the Fas/Fas ligand-caused apoptosis—binds to Siva and induces the activation of a putative caspase. Therefore, this activation might be responsible for the activation of effector caspases (e.g., caspase-3) and finally for the induction of programmed cell death. In CVB3-infected cells, the expression of Siva is induced (Fig. 5B) and both CD27- and CD70-positive cells are present in the same area of the infected tissue. Siva binds to the cytoplasmatic tail of CD27; thereafter, VP2 of CVB3 may take the role of a putative FADD-like protein, inducing apoptosis by caspase activation as shown in Fig. 4D by detecting the active form of caspase-3. One other possibility is that the direct binding between Siva and VP2 in the cytoplasm may attract caspases and activate the death pathway in the absence of CD70 ligation and CD27 complex formation. In addition, in yeast Siva forms homodimeric complexes (data not shown), but whether this observation has a physiological function in apoptotic pathways is not clear.
FIG. 5.
Model for the putative induction of apoptosis in CD27/Siva-mediated pathways (A) and possible role of VP2 in CD27/Siva-mediated apoptotic events after CVB3 infection (B).
Given that coxsackievirus infections can cause pancreatitis as well as acute or dilated cardiomyopathy and apoptotic events are present in virus-infected tissue, our results indicate a molecular mechanism by which the regulation of cell death proteins may be an important early event of CVB3 infection before and after inflammation.
ACKNOWLEDGMENTS
We thank H.-P. Saluz for helpful discussions during the preparation of this article.
This work was partly supported by grant HE 2910/2-1 MU 1395/1-1 from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Agol V I, Belov G A, Bienz K, Egger D, Kolesnikova M S, Raikhlin N T, Romanova L I, Smirnova E A, Tolskaya E A. Two types of death of poliovirus-infected cells: caspase involvement in the apoptosis but not cytopathic effect. Virology. 1998;252:343–353. doi: 10.1006/viro.1998.9438. [DOI] [PubMed] [Google Scholar]
- 2.Badorff C, Lee G H, Lamphear B J, Martone M E, Campbell K P, Rhoads R E, Knowlton K U. Enteroviral protease 2A cleaves dystrophin: evidence of cytoskeletal disruption in an acquired cardiomyopathy. Nat Med. 1999;5:320–326. doi: 10.1038/6543. [DOI] [PubMed] [Google Scholar]
- 3.Barco A, Carrasco L. Poliovirus 2Apro expression inhibits growth of yeast cells. FEBS Lett. 1995;371:4–8. doi: 10.1016/0014-5793(95)00841-v. [DOI] [PubMed] [Google Scholar]
- 4.Bartel P, Chien C T, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. BioTechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- 5.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 6.Bowles N E, Towbin J A. Molecular aspects of myocarditis. Curr Opin Cardiol. 1998;13:179–184. [PubMed] [Google Scholar]
- 7.Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- 8.Carthy C M, Granville D J, Watson K A, Anderson D R, Wilson J E, Yang D, Hunt D W, McManus B M. Caspase activation and specific cleavage of substrates after coxsackievirus B3-induced cytopathic effect in HeLa cells. J Virol. 1998;72:7669–7675. doi: 10.1128/jvi.72.9.7669-7675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman N M, Tu Z, Tracy S, Gauntt C J. An infectious cDNA copy of the genome of a non-cardiovirulent coxsackievirus B3 strain: its complete sequence analysis and comparison to the genomes of cardiovirulent coxsackieviruses. Arch Virol. 1994;135:115–130. doi: 10.1007/BF01309769. [DOI] [PubMed] [Google Scholar]
- 10.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 11.Chow L H, Beisel K W, McManus B M. Enteroviral infection of mice with severe combined immunodeficiency. Evidence for direct viral pathogenesis of myocardial injury. Lab Investig. 1992;66:24–31. [PubMed] [Google Scholar]
- 12.Colston J T, Chandrasekar B, Freeman G L. Expression of apoptosis-related proteins in experimental coxsackievirus myocarditis. Cardiovasc Res. 1998;38:158–168. doi: 10.1016/s0008-6363(97)00323-4. [DOI] [PubMed] [Google Scholar]
- 13.Doedens J R, Kirkegaard K. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 1995;14:894–907. doi: 10.1002/j.1460-2075.1995.tb07071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Etchison D, Milburn S C, Edery I, Sonenberg N, Hershey J W. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- 15.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 16.Gebhard J R, Perry C M, Harkins S, Lane T, Mena I, Asensio V C, Campbell I L, Whitton J L. Coxsackievirus B3-induced myocarditis: perforin exacerbates disease, but plays no detectable role in virus clearance. Am J Pathol. 1998;153:417–428. doi: 10.1016/S0002-9440(10)65585-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gravestein L A, Amsen D, Boes M, Calvo C R, Kruisbeek A M, Borst J. The TNF receptor family member CD27 signals to Jun N-terminal kinase via Traf-2. Eur J Immunol. 1998;28:2208–2216. doi: 10.1002/(SICI)1521-4141(199807)28:07<2208::AID-IMMU2208>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 18.Henke A, Huber S, Stelzner A, Whitton J L. The role of CD8+ T lymphocytes in coxsackievirus B3-induced myocarditis. J Virol. 1995;69:6720–6728. doi: 10.1128/jvi.69.11.6720-6728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hintzen R Q, Lens S M, Koopman G, Pals S T, Spits H, van Lier R A. CD70 represents the human ligand for CD27. Int Immunol. 1994;6:477–480. doi: 10.1093/intimm/6.3.477. [DOI] [PubMed] [Google Scholar]
- 20.Hsieh T Y, Matsumoto M, Chou H C, Schneider R, Hwang S B, Lee A S, Lai M M. Hepatitis C virus core protein interacts with heterogeneous nuclear ribonucleoprotein K. J Biol Chem. 1998;273:17651–17659. doi: 10.1074/jbc.273.28.17651. [DOI] [PubMed] [Google Scholar]
- 21.Huber M, Selinka H C, Kandolf R. Tyrosine phosphorylation events during coxsackievirus B3 replication. J Virol. 1997;71:595–600. doi: 10.1128/jvi.71.1.595-600.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber S A. Coxsackievirus-induced myocarditis is dependent on distinct immunopathogenic responses in different strains of mice. Lab Investig. 1997;76:691–701. [PubMed] [Google Scholar]
- 23.Huber S A, Kupperman J, Newell M K. Hormonal regulation of CD4+ T-cell responses in coxsackievirus B3-induced myocarditis in mice. J Virol. 1999;73:4689–95. doi: 10.1128/jvi.73.6.4689-4695.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber S A, Lodge P A. Coxsackievirus B-3 myocarditis in Balb/c mice. Evidence for autoimmunity to myocyte antigens. Am J Pathol. 1984;116:21–29. [PMC free article] [PubMed] [Google Scholar]
- 25.Huber S A, Mortensen A, Moulton G. Modulation of cytokine expression by CD4+ T cells during coxsackievirus B3 infections of BALB/c mice initiated by cells expressing the γδ+ T-cell receptor. J Virol. 1996;70:3039–3044. doi: 10.1128/jvi.70.5.3039-3044.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huber S A, Pfaeffle B. Differential Th1 and Th2 cell responses in male and female BALB/c mice infected with coxsackievirus group B type 3. J Virol. 1994;68:5126–5132. doi: 10.1128/jvi.68.8.5126-5132.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huber S A, Stone J E, Wagner D H, Jr, Kupperman J, Pfeiffer L, David C, O'Brien R L, Davis G S, Newell M K. γδ+ T cells regulate major histocompatibility complex class II(IA and IE)-dependent susceptibility to coxsackievirus B3-induced autoimmune myocarditis. J Virol. 1999;73:5630–5636. doi: 10.1128/jvi.73.7.5630-5636.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irurzun A, Perez L, Carrasco L. Enhancement of phospholipase activity during poliovirus infection. J Gen Virol. 1993;74:1063–1071. doi: 10.1099/0022-1317-74-6-1063. [DOI] [PubMed] [Google Scholar]
- 29.Jelachich M L, Lipton H L. Restricted Theiler's murine encephalomyelitis virus infection in murine macrophages induces apoptosis. J Gen Virol. 1999;80:1701–1705. doi: 10.1099/0022-1317-80-7-1701. [DOI] [PubMed] [Google Scholar]
- 30.Kandolf R, Hofschneider P H. Molecular cloning of the genome of a cardiotropic coxsackie B3 virus: full-length reverse-transcribed recombinant cDNA generates infectious virus in mammalian cells. Proc Natl Acad Sci USA. 1985;82:4818–4822. doi: 10.1073/pnas.82.14.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerekatte V, Keiper B D, Badorff C, Cai A, Knowlton K U, Rhoads R E. Cleavage of poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J Virol. 1999;73:709–717. doi: 10.1128/jvi.73.1.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khatib R, Chason J L, Silberberg B K, Lerner A M. Age-dependent pathogenicity of group B coxsackieviruses in Swiss-Webster mice: infectivity for myocardium and pancreas. J Infect Dis. 1980;141:394–403. doi: 10.1093/infdis/141.3.394. [DOI] [PubMed] [Google Scholar]
- 33.Klebe R J, Harriss J V, Sharp Z D, Douglas M G. A general method for polyethylene-glycol-induced genetic transformation of bacteria and yeast. Gene. 1983;25:333–341. doi: 10.1016/0378-1119(83)90238-x. [DOI] [PubMed] [Google Scholar]
- 34.Klump W M, Bergmann I, Muller B C, Ameis D, Kandolf R. Complete nucleotide sequence of infectious coxsackievirus B3 cDNA: two initial 5′ uridine residues are regained during plus-strand RNA synthesis. J Virol. 1990;64:1573–83. doi: 10.1128/jvi.64.4.1573-1583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knowlton K U, Jeon E S, Berkley N, Wessely R, Huber S. A mutation in the puff region of VP2 attenuates the myocarditic phenotype of an infectious cDNA of the Woodruff variant of coxsackievirus B3. J Virol. 1996;70:7811–7818. doi: 10.1128/jvi.70.11.7811-7818.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kräusslich H G, Nicklin M J, Lee C K, Wimmer E. Polyprotein processing in picornavirus replication. Biochemie. 1988;70:119–130. doi: 10.1016/0300-9084(88)90166-6. [DOI] [PubMed] [Google Scholar]
- 37.Leipner C, Borchers M, Merkle I, Stelzner A. Coxsackievirus B3-induced myocarditis in MHC class II-deficient mice. J Hum Virol. 1999;2:102–114. [PubMed] [Google Scholar]
- 38.Lindberg A M, Crowell R L, Zell R, Kandolf R, Pettersson U. Mapping of the RD phenotype of the Nancy strain of coxsackievirus B3. Virus Res. 1992;24:187–196. doi: 10.1016/0168-1702(92)90006-u. [DOI] [PubMed] [Google Scholar]
- 39.Lindberg A M, Stalhandske P O, Pettersson U. Genome of coxsackievirus B3. Virology. 1987;156:50–63. doi: 10.1016/0042-6822(87)90435-1. [DOI] [PubMed] [Google Scholar]
- 40.MacDonald P N, Sherman D R, Dowd D R, Jefcoat S C, Jr, DeLisle R K. The vitamin D receptor interacts with general transcription factor IIB. J Biol Chem. 1995;270:4748–4752. doi: 10.1074/jbc.270.9.4748. [DOI] [PubMed] [Google Scholar]
- 41.Ohlmann T, Rau M, Pain V M, Morley S J. The C-terminal domain of eucaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996;15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- 42.Olivetti G, Abbi R, Quaini F, Kajstura J, Cheng W, Nitahara J A, Quaini E, Di Loreto C, Beltrami C A, Krajewski S, Reed J C, Anversa P. Apoptosis in the failing human heart. N Engl J Med. 1997;336:1131–1141. doi: 10.1056/NEJM199704173361603. [DOI] [PubMed] [Google Scholar]
- 43.Padanilam B J, Lewington A J, Hammerman M R. Expression of CD27 and ischemia/reperfusion-induced expression of its ligand Siva in rat kidneys. Kidney Int. 1998;54:1967–1975. doi: 10.1046/j.1523-1755.1998.00197.x. [DOI] [PubMed] [Google Scholar]
- 44.Prasad K V, Ao Z, Yoon Y, Wu M X, Rizk M, Jacquot S, Schlossman S F. CD27, a member of the tumor necrosis factor receptor family, induces apoptosis and binds to Siva, a proapoptotic protein. Proc Natl Acad Sci USA. 1997;94:6346–6351. doi: 10.1073/pnas.94.12.6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reyes M P, Lerner A M. Coxsackievirus myocarditis—with special reference to acute and chronic effects. Prog Cardiovasc Dis. 1985;27:373–394. doi: 10.1016/0033-0620(85)90001-5. [DOI] [PubMed] [Google Scholar]
- 46.Tracy S, Chapman N M, Tu Z. Coxsackievirus B3 from an infectious cDNA copy of the genome is cardiovirulent in mice. Arch Virol. 1992;122:399–409. doi: 10.1007/BF01317202. [DOI] [PubMed] [Google Scholar]
- 47.van Kuppeveld F J, Hoenderop J G, Smeets R L, Willems P H, Dijkman H B, Galama J M, Melchers W J. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 1997;16:3519–3532. doi: 10.1093/emboj/16.12.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y F, Chen S C, Wu F Y, Wu C W. The interaction between human cytomegalovirus immediate-early gene 2 (IE2) protein and heterogeneous ribonucleoprotein A1. Biochem Biophys Res Commun. 1997;232:590–594. doi: 10.1006/bbrc.1997.6334. [DOI] [PubMed] [Google Scholar]
- 49.Woodruff J F. The influence of quantitated post-weaning undernutrition on coxsackievirus B3 infection of adult mice. II. Alteration of host defense mechanisms. J Infect Dis. 1970;121:164–181. doi: 10.1093/infdis/121.2.164. [DOI] [PubMed] [Google Scholar]
- 50.Woodruff J F. Viral myocarditis. A review. Am J Pathol. 1980;101:425–484. [PMC free article] [PubMed] [Google Scholar]
- 51.Yang D, Yu J, Luo Z, Carthy C M, Wilson J E, Liu Z, McManus B M. Viral myocarditis: identification of five differentially expressed genes in coxsackie-virus B3-infected mouse heart. Circ Res. 1999;84:704–712. doi: 10.1161/01.res.84.6.704. [DOI] [PubMed] [Google Scholar]
- 52.Zell R, Stelzner A. Application of genome sequence information to the classification of bovine enteroviruses: the importance of 5′- and 3′-nontranslated regions. Virus Res. 1997;51:213–229. doi: 10.1016/s0168-1702(97)00096-8. [DOI] [PubMed] [Google Scholar]