Abstract
Objectives
Electroacupuncture (EA) has been demonstrated to aid stroke recovery. However, few investigations have focused on identifying the potent molecular targets of EA by comparing EA stimulation between naïve and disease models. Therefore, this study was undertaken to identify the potent molecular therapeutic mechanisms underlying EA stimulation in ischemic stroke through a comparison of mRNA sequencing data obtained from EA-treated naïve control and ischemic stroke mouse models.
Methods
Using both naïve control and middle cerebral artery occlusion (MCAO) mouse models, EA stimulation was administered at two acupoints, Baihui (GV20) and Dazhui (GV14), at a frequency of 2 Hz. Comprehensive assessments were conducted, including behavioral evaluations, RNA sequencing to identify differentially expressed genes (DEGs), functional enrichment analysis, protein-protein interaction (PPI) network analysis, and quantitative real-time PCR.
Results
EA stimulation ameliorated the ischemic insult-induced motor dysfunction in mice with ischemic stroke. Comparative analysis between control vs. MCAO, control vs. control + EA, and MCAO vs. MCAO + EA revealed 4,407, 101, and 82 DEGs, respectively. Of these, 30, 7, and 1 were common across the respective groups. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses revealed upregulated DEGs associated with the regulation of inflammatory immune response in the MCAO vs. MCAO + EA comparison. Conversely, downregulated DEGs in the control vs. control + EA comparison were linked to neuronal development. PPI analysis revealed major clustering related to the regulation of cytokines, such as Cxcl9, Pcp2, Ccl11, and Cxcl13, in the common DEGs of MCAO vs. MCAO + EA, with Esp8l1 identified as the only common downregulated DEG in both EA-treated naïve and ischemic models.
Conclusion
These findings underscore the diverse potent mechanisms of EA stimulation between naïve and ischemic stroke mice, albeit with few overlaps. However, the potent mechanisms underlying EA treatment in ischemic stroke models were associated with the regulation of inflammatory processes involving cytokines.
Keywords: electroacupuncture, stroke, transcriptome analysis, inflammation, cytokine
INTRODUCTION
Ischemic stroke is characterized by severe hypoxemic damage to neurons due to the reduced supply of oxygen to the brain. Ischemic stroke can result in neuronal cell death and motor deficits [1]. Current approaches for managing ischemic stroke typically involve pharmacological thrombolysis, surgery, and rehabilitation efforts [2-4]. However, these approaches do not achieve complete functional recovery during the post-ischemic phase [5]. Therefore, it is necessary to identify alternative or complementary therapies that can enhance functional recovery. One such therapy is electroacupuncture (EA), which combines electrical stimulation with traditional acupuncture to promote the recovery of sensory and motor functions in patients with stroke [6, 7]. EA works by regulating the production of cytokines, neurotransmitters, and neurotrophic factors to alter various biological processes, including neuro-regeneration, angiogenesis, and suppression of inflammatory responses [7-9]. Despite considerable investigation into the therapeutic mechanisms of EA, a deeper and systematic analysis of the expression patterns of related mRNA is necessary.
RNA sequencing or microarray analysis of mRNA expression has emerged as a tool for elucidating biological pathways and identifying biomarkers relevant to the diagnosis and prognosis of various disorders [10, 11], including ischemic stroke [12-14]. Several researchers have employed mRNA sequencing to explore the therapeutic mechanisms underlying EA stimulation in disease models; however, not much research has been published regarding the molecular targets of EA investigated through a comparative analysis of EA stimulation between both naïve and disease models.
Even under normal physiological conditions, EA stimulation induces distinct patterns of neuronal excitability based on specific acupoints and frequencies [15]. Moreover, evidence of a temporary breakdown in the blood-brain barrier (BBB) observed in both healthy and middle cerebral artery occlusion (MCAO) models suggests that EA stimulation increases BBB permeability [16]. This study was conducted to compare the mRNA expression profiles between untreated naïve control, EA-treated naïve control, untreated ischemic stroke, and EA-treated ischemic stroke mouse models. The underlying hypothesis posited that genes commonly altered after EA stimulation would clarify the specific therapeutic mechanisms of EA. Therefore, the primary objective of this study was to explore the potential therapeutic mechanisms of EA stimulation in ischemic stroke through comparative mRNA sequencing analyses between EA-treated naïve and ischemic stroke mouse models.
MATERIALS AND METHODS
1. Animals
Six-week-old C57BL/6J mice were procured from DooYeol Biotech (Seoul, Korea). Male mice were procured to avoid the effects of estrogen. The mice were randomly allocated to four groups, each comprising five mice, as follows: naïve control, EA-treated control (control + EA), MCAO, and EA-treated MCAO (MCAO + EA). Based on behavioral assessments, MCAO mice that responded to EA treatment were subjected to subsequent RNA sequencing. All experimental procedures were approved by the Pusan National University Animal Ethics Committee (Approval Number PUN-2017-1448).
2. MCAO models and tissue preparation
C57BL/6J mice were subjected to surgical procedures involving the insertion of a silicon-coated 7-0 monofilament (Docol Corporation, Sharon, MA, USA) into the middle cerebral artery via the internal carotid artery. Following a 40-minute occlusion period, reperfusion was initiated by withdrawing the filament. Cerebral blood flow in each region was continuously monitored using the PeriFlux Laser Doppler System 5000 (Perimed, Stockholm, Sweden), with probes affixed to the skulls. Anesthesia was induced using a calibrated vaporizer (Midmark NIP 3000, Orchard, OH, USA) to deliver a mixture of 20% O2 and 80% N2O, supplemented with 2% isoflurane (200 mL/min O2, 800 mL/min N2O; VSP corporation, Choongwae, Seoul, Korea) during surgery.
3. EA stimulation
Two stainless-steel needles (0.18 mm in diameter and 30 mm in length) connected to a Grass S88 electrostimulator (Grass Instrument Co., West Warwick, RI, USA) were inserted to a depth of approximately 2 mm at specific acupoints corresponding to Baihui (GV20, located at the midpoint of the line connecting the apexes of both ears on the parietal bone) and Dazhui (GV14, positioned along the posterior midline and in the depression below the spinous process of the seventh cervical vertebra) in human anatomy. EA stimulation (output voltage 2 V, frequency 2 Hz) was provided for 20 min. Three days after MCAO, EA treatment was administered once daily over five days for gene expression profiling. Mice in the non-EA group were administered only isoflurane anesthesia for 20 min.
4. Behavioral assessment
The behavior of mice was assessed by blinded observers and independent researchers. For the rotarod test, mice were placed on a rotating rod, and the rotational speed of the rod was increased steadily from 4 rpm to 20 rpm. Total latency was assessed over three trials, calculated by averaging three recordings for each mouse. For the pole test, mice were positioned on a tape-enclosed pole (measuring 50 cm in height and 0.5 cm in diameter) oriented with their heads upward. Total latency, encompassing the time required for a complete turn (T-turn) at the top and descent to the bottom, was recorded across five trials, calculated by averaging three recordings for each mouse. For the wire grip test, mice were placed on a wire mesh (0.3 cm) positioned 45 cm above ground level. The results were calculated as the average score of five trials as follows: 0 for mice falling from the wire; 1 for mice gripping the wire using the contralateral forelimb; 2 for mice using both forelimbs; and 3 for mice using both forelimbs and tail.
5. RNA extraction and sequencing
Total RNA was extracted according to the manufacturer’s instructions. Subsequently, the quantity and quality of RNA samples were evaluated using the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) and Agilent Bioanalyzer 2100 (Agilent Technologies, USA). Libraries were prepared using specific kits, following the manufacturer’s instructions. Purified libraries were quantified using Qubit 2.0 and Agilent 2100 Bioanalyzer. Clusters were created using the cBot library and sequenced on the Illumina HiSeq 2500 platform. The sequencing was performed by the designated company.
6. RNA sequencing analysis
Fast quality control assessed the quality of RNA sequencing reads. Trimming procedures were performed using seqtk to eliminate known Illumina TruSeq adapter sequences, poor-quality reads, and ribosomal RNA reads. Subsequently, the trimmed reads were mapped to the UCSC mm10 reference database using Hisat2.2.1. Gene expression was quantified using HTseq-count 0.11.3 to enumerate each gene from the trimmed reads. Differential gene expression analysis was performed using DESeq2, with significance defined by an absolute log2-fold change value and a p-value of < 0.05 for each comparison group. A heatmap of differentially expressed genes (DEGs) was generated using the R package heatmap. To elucidate the functional implications of DEGs, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analyses were conducted using the R software. Database for Annotation, Visualization, and Integrated Discovery (DAVID, http://david.ncifcrf.gov/) was used for the functional analysis of DEGs based on GO terms and enrichment analysis of KEGG pathways. Cytoscape software (version 3.8.1) was used for protein–protein interaction (PPI) network analysis using an interaction threshold of mid-confidence 0.4.
7. Quantitative real-time PCR
Total RNA was extracted from the striatum using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA concentration and quality were assessed using the NanoDrop spectrophotometer (nd-1000; Thermo Fisher Scientific, Waltham, MA, USA), with an acceptable 260/280 nm ratio of 1.8-2.0. The TOPscriptTM cDNA Synthesis Kit (EZ005S; Enzynomics, Daejeon, Korea) was used to reverse-transcribe 2 μg of RNA from each sample into cDNA in a reaction volume of 20 μL. PCR amplification was performed using 2 μg of cDNA and 30 pmol of gene-specific primers in SYBR Green Master Mix (RR420A; Takara Bio, Kusatsu, Japan). The primer sets used were as follows:
Cxcl9 Forward: CCTAGTGATAAGGAATGCACGATG
Cxcl9 Reverse: CTAGGCAGGTTTGATCTCCGTTC
Pcp2 Forward: GGAAGGCTTCTTCAACCTGCTG
Pcp2 Reverse: CTGGGTGTTGACCAGCATATCC
Ccl11 Forward: TCCATCCCAACTTCCTGCTGCT
Ccl11 Reverse: CTCTTTGCCCAACCTGGTCTTG
Cxcl3 Forward: CATAGATCGGATTCAAGTTACGCC
Cxcl13 Reverse: GTAACCATTTGGCACGAGGATTC
Esp8l1 Forward: TCCTCTCCAGAGCTGTTGCACT
Esp8l1 Reverse: TCACCAGGAGTGACATTGTCGC
Gapdh Forward: GGGTGTGAACCACGAGAAAT
Gapdh Reverse: CCTTCCACAATGCCAAAGTT
8. Statistical analysis
All data were generated, and statistical analyses were performed using Sigmaplot version 12.5 (SystatSoftware, San Jose, CA, USA). A graph was plotted as individual data points along with mean ± standard error of the mean. Statistical analyses were conducted using one-way analysis of variance and Tukey’s post-hoc test. Results were deemed statistically significant at a threshold of p < 0.05.
RESULTS
1. Therapeutic effects of EA stimulation on motor function recovery post-ischemic stroke
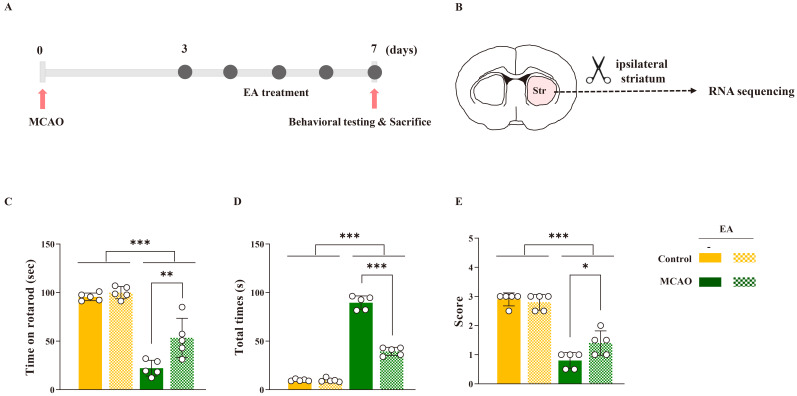
Mice were administered EA treatment once daily over five days, commencing 3 days post-MCAO. Behavioral assessments were performed following the final EA session (Fig. 1A). Comparative analysis revealed significant impairment in motor function in the MCAO group compared to that in the control group (rotarod test: F[3,16] = 57.733, p < 0.001; pole test: F[3,16] = 396.587, p < 0.001; wire grip test: F[3,16] = 39.635, p < 0.001) (Fig. 1C-E). EA stimulation significantly restored motor function in the MCAO group; however, no significant effects were observed in the naïve control group. These results underscore the efficacy of EA treatment in reducing functional motor impairments post-ischemic stroke.
Figure 1.
Therapeutic effects of EA stimulation in ischemic stroke. (A) Experimental timeline of electroacupuncture treatment and (B) schematic images of RNA sequencing. Quantification of motor function results of (C) rotarod, (D) pole, and (E) wire grip tests. Data are expressed as means ± standard error of the mean. *p < 0.05, **p < 0.01, and ***p < 0.001 vs. each group using one-way analysis of variance with Tukey’s test. n = 5 mice/group.
2. Identification of DEGs
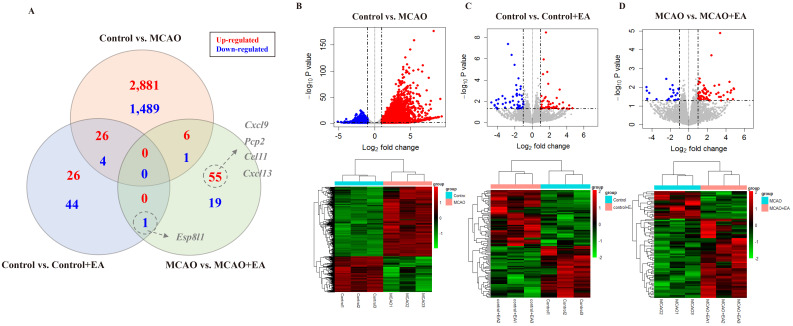
Following behavioral assessments, the ipsilateral striatum was extracted from each mouse for RNA sequencing (Fig. 1B). The DEGs observed in each group are shown in Fig. 2. Comparing the control and MCAO groups, 4,407 DEGs (|FC| > 2, p < 0.05) were identified, comprising 2,913 upregulated and 1,494 downregulated genes. Comparing the control and control + EA groups, 101 DEGs (|FC| > 2, p < 0.05) were identified, comprising 52 upregulated and 49 downregulated genes. Comparing the MCAO and MCAO + EA groups, 82 DEGs (|FC| > 2, p < 0.05) were identified, comprising 61 upregulated and 21 downregulated genes (Fig. 2A-D). Significantly more DEGs (4,407) were noted when comparing the control and MCAO groups than in other pairwise group comparisons (control vs. control + EA: 101 genes, MCAO vs. MCAO + EA: 82 genes). In addition, the number of DEGs in each pairwise comparison was notably smaller than the total number of DEGs, particularly evident in the control vs. control + EA and MCAO vs. MCAO + EA comparisons (30 genes between control vs. control + EA and control vs. MCAO, 7 genes between control vs. MCAO and MCAO vs. MCAO + EA, and only one gene between control vs. control + EA and MCAO vs. MCAO + EA). These findings suggest that EA treatment in both naïve control and ischemic stroke models altered the expression of multiple genes involving diverse pathways.
Figure 2.
Significant DEGs. (A) Venn diagram showing significant gene expression profiles. (B-D) Upper panel: Volcano plots displaying significant gene expression profiles. Red and blue points represent upregulated and downregulated differentially expressed genes (DEGs), respectively. Low panel: Heat map and hierarchical clustering of DEGs. Higher and lower gene expressions are indicated by a deeper red color and a deeper green color, respectively. n = 3 mice/group.
3. GO enrichment analysis of DEGs
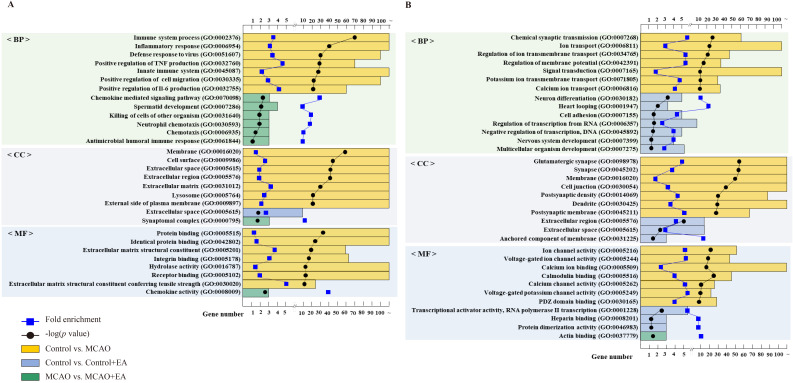
For GO functional analysis of upregulated DEGs using DAVID, the 2,913 upregulated DEGs identified in the control vs. MCAO comparison were significantly enriched in 1,302 GO terms. Among these, 928 were related to biological processes (BP), 155 to cellular components (CC), and 219 to molecular function (MF) (p < 0.05). In the ontology analysis, the regulation of inflammatory and immune responses constituted most of the enriched GO categories, encompassing immune system process (GO:0002376), inflammatory response (GO:0006954), positive regulation of tumor necrosis factor production (GO:0032760), and positive regulation of interleukin (IL)-6 production (GO:0032755). In the control vs. control + EA comparison, the 52 upregulated DEGs were significantly enriched in one GO term related to CC (p < 0.05), specifically the extracellular space (GO:0005615). In the MCAO vs. MCAO + EA comparison, the 61 upregulated DEGs were significantly enriched in eight GO terms, six of which were related to BP, one to CC, and one to MF (p < 0.05). The ontology results revealed that inflammatory cytokines predominantly produced by and involved in the regulation of inflammatory and immune responses constituted most of the enriched GO categories such as chemokine-mediated signaling pathways (GO:0070098), neutrophil chemotaxis (GO:0030593), and chemotaxis (GO:0006935). The GO terms associated with the highest enrichment factors for upregulated DEGs are shown in Fig. 3A.
Figure 3.
GO functional enrichment analysis of DEGs. Bar chart displaying the top seven significant biological processes, cellular components, and molecular functions of (A) upregulated and (B) downregulated differentially expressed genes. Bar plot represents the number of genes in each group; blue square dot represents fold enrichment; black circle dot represents the adjusted p-value. n = 3 mice/group.
In the control vs. MCAO comparison, 1,494 downregulated DEGs were significantly enriched in 533 GO terms. Among these, 293 were associated with BP, 121 with CC, and 119 with MF (p < 0.05). The ontology results revealed synaptic regulation based on action potential and neurotransmitter as the predominantly enriched GO categories, including chemical synaptic transmission (GO:0007268), ion transport (GO:0006811), synapse organization (GO:0045202), and calcium channel activity (GO:0005262). In the control vs. control + EA comparison, GO enrichment analysis identified 13 GO terms enriched among 49 downregulated DEGs. Among these, seven were related to BP, three to CC, and three to MF (p < 0.05). The ontology results identified the regulation of synaptic plasticity based on neuronal development-mediated gene transcription as the predominantly enriched GO categories, including neuronal differentiation (GO:0030182), cell adhesion (GO:0007155), and nervous system development (GO:0007399). In the MCAO vs. MCAO + EA comparison, GO enrichment analysis revealed only one GO term associated with MF among 21 downregulated DEGs (p < 0.05). Specifically, the enriched MF was actin binding (GO:00037779). The GO terms associated with the highest enrichment factors for downregulated DEGs are presented in Fig. 3B.
4. KEGG mapper
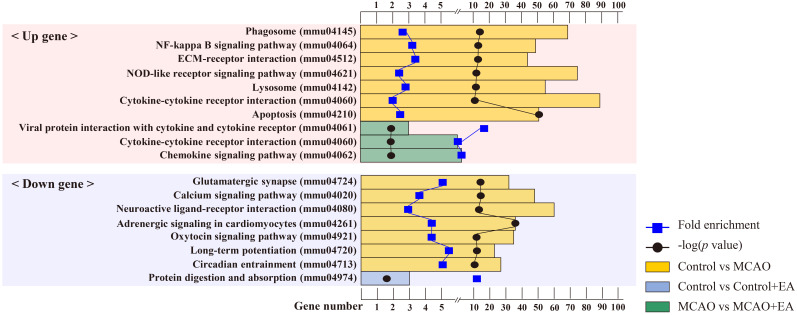
In the control vs. MCAO comparison, KEGG enrichment analysis revealed 211 pathway terms in common between the 2,913 upregulated DEGs and 1,494 downregulated DEGs. Among these, 10 were significant (p < 0.05). Analysis of the 2,913 upregulated DEGs revealed major categories related to inflammatory response and apoptosis mediated by cytokines occupying the top ranks. Notable pathways included phagosome (mmu04145), nuclear factor (NF)-kappa B signaling pathway (mmu04064), cytokine-cytokine receptor interaction (mmu04060), and apoptosis (mmu04210). Similarly, analysis of the 1,494 downregulated DEGs revealed that the top-ranked categories were primarily involved in synaptic plasticity regulation. Key pathways included glutamatergic synapse (mmu04724), calcium signaling pathway (mmu04020), neuroactive ligand-receptor interaction (mmu04080), and long-term potentiation (mmu04720). However, in the control vs. control + EA comparison, KEGG enrichment analysis of the common downregulated DEGs revealed only one key pathway, encompassing protein digestion and absorption (mmu04974). In the MCAO vs. MCAO + EA comparison, three pathway terms were analyzed for the 61 common upregulated DEGs. Enriched KEGG categories included inflammatory cytokine responses, encompassing cytokine-cytokine receptor interaction (mmu04060), and chemokine signaling pathway (mmu04062). The enriched pathways are shown in Fig. 4.
Figure 4.
KEGG pathway enrichment analysis of DEGs. Bar chart showing the top seven significant pathways of differentially expressed genes. Bar plot represents the number of genes in each group; blue square dot represents fold enrichment; black circle dot represents the adjusted p-value. n = 3 mice/group.
5. PPI network
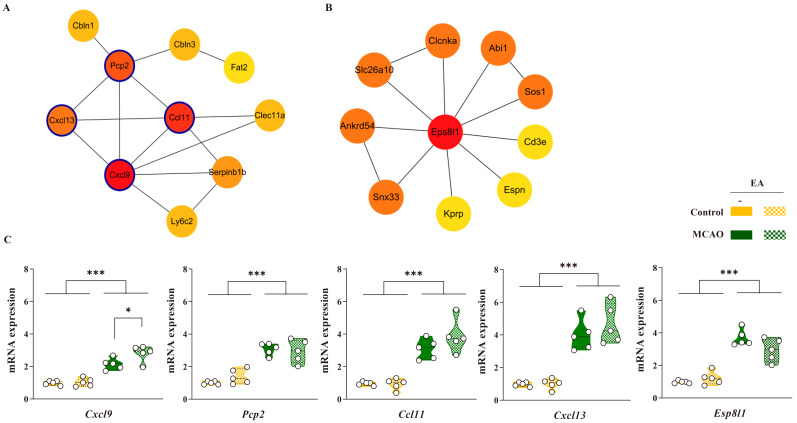
In the MCAO vs. MCAO + EA comparison, significant common DEGs were used to construct a PPI network, which was based on data from the STRING database. A total of 61 upregulated DEGs (|FC| > 2, p < 0.05) were identified. The resulting PPI network comprised 49 nodes and 18 interactions (PPI enrichment: 1.01E-06; enrichment clustering coefficient: 0.222). One major cluster was identified, mainly involving Cxcl9, Pcp2, and Ccl11, with Cxcl13 serving as a central hub. The top nodes ranked by maximal clique centrality (MCC) are shown in Fig. 5A. Additionally, 49 downregulated DEGs (|FC| > 2, p < 0.05) were identified in the MCAO vs. MCAO + EA comparison. The PPI network comprised 17 nodes and 8 interactions (PPI enrichment: 0.0026; enrichment clustering coefficient: 0.294). Despite interactions between these genes, the genes did not form any major clusters (data not shown).
Figure 5.
PPI network analysis of DEGs. The top genes were selected using MCODE in Cytoscape for the (A) upregulated differentially expressed genes (DEGs) in the middle cerebral artery occlusion (MCAO) vs. MCAO + EA comparison. (B) Common downregulated DEGs between control vs. control + EA and MCAO vs. MCAO + EA comparisons. (C) Bar charts of gene expression in the ipsilateral-striatum using qPCR. The expression of these genes was significantly changed by EA stimulation with MCAO. Data are expressed as means ± standard error of the mean. *p < 0.05 and ***p < 0.001 vs. each group using one-way analysis of variance with Tukey’s test. n = 5 mice/group.
To identify the molecular mechanisms of EA stimulation, a PPI network was constructed using significant common DEGs between control vs. control + EA and MCAO vs. MCAO + EA groups. One common downregulated DEG (|FC| > 2, p < 0.05) was identified, namely Eps8l1. The PPI network constructed around Eps8l1 comprised 11 nodes and 13 interactions (PPI enrichment: 1.0E-16; enrichment clustering coefficient: 0.915), with no major clustering patterns observed. Eps8l1-related genes were ranked based on MCC and organized, as shown in Fig. 5B.
Based on significant DEGs identified through PPI analysis, genes demonstrating significant changes in response to EA treatment were selected, including Cxcl9, Pcp2, Ccl11, Cxcl13, and Eps8l1. qPCR analysis revealed that the expression of Cxcl9, Pcp2, Ccl11, and Cxcl13, which are related to cytokine-mediated inflammatory and immune responses, was significantly increased in the MCAO group compared to that in the naïve control group. Esp8l1 expression was also significantly altered in the MCAO group. In addition, the expression of these genes increased in response to EA stimulation in both naïve control and MCAO models, with Cxcl9 exhibiting significant changes in the MCAO model. These findings suggest that the potential therapeutic mechanisms of EA treatment involve the regulation of inflammatory and immune responses.
DISCUSSION
In this study, mRNA expression profiles in the ipsilateral striatum of EA-treated naïve control and MCAO mice were analyzed. DEGs were identified in each pairwise group comparison. Subsequent investigations included GO enrichment analysis, as well as PPI and PPI module analyses, aimed at exploring the therapeutic mechanisms of EA stimulation in the context of ischemic stroke. Transcriptome analysis revealed that in the comparison between control and MCAO groups, upregulated DEGs were associated with the regulation of inflammatory and immune responses. Previous studies on ischemia have highlighted the activation of neuroimmune processes, including cytokine-mediated inflammatory responses, cell migration, defense responses, and wound healing [17, 18]. The downregulated DEGs were related to synaptic regulation based on action potential and neurotransmitters. Synaptic regulation plays a crucial role in the development of brain networks following brain injury, and improving plasticity is recognized as a therapeutic strategy for stroke management [19, 20].
However, the number of common genes between EA-treated naïve control and ischemic stroke mice was considerably lower than that in the comparison between control and MCAO groups. GO and KEGG analyses revealed that the upregulated DEGs in the comparison between MCAO and MCAO + EA groups were primarily associated with the regulation of inflammatory immune responses. These findings suggest that the therapeutic effects of EA stimulation are limited to specific molecular targets involved in the regulation of inflammatory immune responses implicated in the pathogenesis of ischemic stroke.
To identify the molecular mechanisms underlying the effects of EA treatment in ischemic stroke models, we focused on DEGs in the pairwise comparison between MCAO and MCAO + EA groups. Transcriptome analysis revealed a significant enrichment of cytokine responses among the upregulated DEGs. Subsequent PPI analysis of the common DEGs in the comparison between MCAO and MCAO + EA groups revealed major clustering genes, including Cxcl9, Pcp2, Ccl11, and Cxcl13. Most of these genes were significantly associated with the interaction of cytokine and cytokine receptors. Cxcl9 plays an important role in inflammation and immune responses, as well as microglia and macrophage activation [21-23]. The Cxcl family, expressed by microglia, macrophages, and astrocytes, releases various neurotrophic factors and promotes recovery from ischemic stroke [24, 25]. Our results suggest that EA stimulation regulates inflammatory and immune responses by influencing levels of cytokines.
To elucidate the common molecular mechanisms underlying the effects of EA stimulation under naïve control and ischemic conditions, common genes between the control vs. control + EA and MCAO vs. MCAO + EA groups were analyzed. Eps8l1 was the only common downregulated DEG. Eps8l1 is a subtype of Eps8 and is recognized as a key regulator of various signaling pathways. Eps8 is important for TLR4-MyD88 interaction, which is necessary for macrophage phagocytosis during lipopolysaccharide stimulation [26]. TLR4 is commonly implicated in the pathogenesis of stroke, and its activation via damage-associated molecular patterns is necessary for immunological responses to promote the release of inflammatory cytokines in the brain [27, 28]. In addition, Eps8 controls the angiopoietin 1-AKT pathway, modulating the barrier function of spinal cord microvascular endothelial cells. Eps8 expressed in mouse hippocampal neurons regulates neuronal morphogenesis and structural plasticity [29]. The results of this study highlighted Eps8l1 as a major gene affected by EA stimulation, potentially serving as a major factor in regulating phagocytosis, angiogenesis, and morphogenesis through its involvement in various signaling pathways.
The study results revealed that EA stimulation affected the expression of different genes and regulated different molecular pathways in EA-treated naïve control and ischemic stroke models, but there was limited overlap between the various genes involved. The therapeutic effects of EA were mostly related to the modulation of inflammatory cytokines. However, results of the transcriptome analysis need to be further validated through additional animal experiments.
CONCLUSION
In this study, the molecular pathways underlying the therapeutic effect of EA stimulation in EA-treated naïve control and ischemic stroke models were identified. EA stimulation regulates pathways controlling the production of inflammatory cytokines and cell death processes in ischemic stroke conditions. Therefore, EA could be considered to modulate inflammatory cytokines and cell death pathways to restore motor function after ischemic stroke.
ACKNOWLEDGEMENTS
This work was supported by a 2-Year Research Grant of Pusan National University.
Footnotes
CONFLICTS OF INTEREST
Hwa Kyoung Shin has been an editorial board member of Journal of Pharmacopuncture since 2022 but has no role in the decision to publish this article. No other potential conflicts of interest relevant to this aricle were reported.
REFERENCES
- 1.Herpich F, Rincon F. Management of acute ischemic stroke. Crit Care Med. 2020;48(11):1654–63. doi: 10.1097/CCM.0000000000004597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rabinstein AA. Update on treatment of acute ischemic stroke. Continuum (Minneap Minn) 2020;26(2):268–86. doi: 10.1212/CON.0000000000000840. [DOI] [PubMed] [Google Scholar]
- 3.Jolugbo P, Ariens RAS. Thrombus composition and efficacy of thrombolysis and thrombectomy in acute ischemic stroke. Stroke. 2021;52(3):1131–42. doi: 10.1161/STROKEAHA.120.032810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell BCV, Khatri P. Stroke. Lancet. 2020;396(10244):129–42. doi: 10.1016/S0140-6736(20)31179-X. [DOI] [PubMed] [Google Scholar]
- 5.Toni D, Risitano A, Gentile L. A revolution in stroke therapy: reperfusion therapy effective even if late. Eur Heart J Suppl. 2020;22(Suppl E):E157–61. doi: 10.1093/eurheartj/suaa083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao H, Lu Y, Wang Y, Han X, Zhang Y, Han B, et al. Electroacupunture contributes to recovery of neurological deficits in experimental stroke by activating astrocytes. Restor Neurol Neurosci. 2018;36(3):301–12. doi: 10.3233/RNN-170722. [DOI] [PubMed] [Google Scholar]
- 7.Liu L, Zhang Q, Xie HY, Gua WJ, Bao CR, Wang NH, et al. Differences in post-ischemic motor recovery and angiogenesis of MCAO rats following electroacupuncture at different acupoints. Curr Neurovasc Res. 2020;17(1):71–8. doi: 10.2174/1567202617666191223151553. [DOI] [PubMed] [Google Scholar]
- 8.Shin HK, Lee SW, Choi BT. Modulation of neurogenesis via neurotrophic factors in acupuncture treatments for neurological diseases. Biochem Pharmacol. 2017;141:132–42. doi: 10.1016/j.bcp.2017.04.029. [DOI] [PubMed] [Google Scholar]
- 9.Geng Y, Chen Y, Sun W, Gu Y, Zhang Y, Li M, et al. Electroacupuncture ameliorates cerebral I/R-induced inflammation through DOR-BDNF/TrkB pathway. Evid Based Complement Alternat Med. 2020;2020:3495836. doi: 10.1155/2020/3495836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurvits L, Lättekivi F, Reimann E, Kadastik-Eerme L, Kasterpalu KM, Kõks S, et al. Transcriptomic profiles in Parkinson's disease. Exp Biol Med (Maywood) 2021;246(5):584–95. doi: 10.1177/1535370220967325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen YL, Wang K, Xie F, Zhuo ZL, Liu C, Yang Y, et al. Novel biomarkers identified in triple-negative breast cancer through RNA-sequencing. Clin Chim Acta. 2022;531:302–8. doi: 10.1016/j.cca.2022.04.990. [DOI] [PubMed] [Google Scholar]
- 12.Xie Q, Zhang X, Peng S, Sun J, Chen X, Deng Y, et al. Identification of novel biomarkers in ischemic stroke: a genome-wide integrated analysis. BMC Med Genet. 2020;21(1):66. doi: 10.1186/s12881-020-00994-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bao J, Zhou S, Pan S, Zhang Y. Molecular mechanism exploration of ischemic stroke by integrating mRNA and miRNA expression profiles. Clin Lab. 2018;64(4):559–68. doi: 10.7754/Clin.Lab.2017.171044. [DOI] [PubMed] [Google Scholar]
- 14.Akimniyazova A, Kamenova S, Duchshanova G, Aisina D, Goncharova A, et al. Kondybayeva А, author. Prediction of miRNA interaction with mRNA of stroke candidate genes. Neurol Sci. 2020;41(4):799–808. doi: 10.1007/s10072-019-04158-x. [DOI] [PubMed] [Google Scholar]
- 15.Fang JF, Du JY, Shao XM, Fang JQ, Liu Z. Effect of electroacupuncture on the NTS is modulated primarily by acupuncture point selection and stimulation frequency in normal rats. BMC Complement Altern Med. 2017;17(1):182. doi: 10.1186/s12906-017-1690-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Lin X, Zhou H, Chen Y, Xiao S, Jiao J, et al. Electroacupuncture: a new approach to open the blood-brain barrier in rats recovering from middle cerebral artery occlusion. Acupunct Med. 2018;36(6):377–85. doi: 10.1136/acupmed-2017-011496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S, Liu J, Wang Y, Deng L, Chen S, Wang X, et al. Differentially expressed genes induced by β-caryophyllene in a rat model of cerebral ischemia-reperfusion injury. Life Sci. 2021;273:119293. doi: 10.1016/j.lfs.2021.119293. [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Gong J, Chen H, Li X, Wang L, Qian X, et al. Ischemic stroke induces cardiac dysfunction and alters transcriptome profile in mice. BMC Genomics. 2021;22(1):641. doi: 10.1186/s12864-021-07938-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S, Lu Y, Ding D, Ma Z, Xing X, Hua X, et al. Fibroblast growth factor 2 contributes to the effect of salidroside on dendritic and synaptic plasticity after cerebral ischemia/reperfusion injury. Aging (Albany NY) 2020;12(11):10951–68. doi: 10.18632/aging.103308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ou Z, Zhao M, Xu Y, Wu Y, Qin L, Fang L, et al. Huangqi Guizhi Wuwu decoction promotes M2 microglia polarization and synaptic plasticity via Sirt1/NF-κB/NLRP3 pathway in MCAO rats. Aging (Albany NY) 2023;15(19):10031–56. doi: 10.18632/aging.204989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis SM, Collier LA, Messmer SJ, Pennypacker KR. The poststroke peripheral immune response is differentially regulated by leukemia inhibitory factor in aged male and female rodents. Oxid Med Cell Longev. 2020;2020:8880244. doi: 10.1155/2020/8880244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Shui X, Sun R, Wan L, Zhang B, Xiao B, et al. Microglial phenotypic transition: signaling pathways and influencing modulators involved in regulation in central nervous system diseases. Front Cell Neurosci. 2021;15:736310. doi: 10.3389/fncel.2021.736310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai W, Liu S, Hu M, Sun X, Qiu W, Zheng S, et al. Post-stroke DHA treatment protects against acute ischemic brain injury by skewing macrophage polarity toward the M2 phenotype. Transl Stroke Res. 2018;9(6):669–80. doi: 10.1007/s12975-018-0662-7. [DOI] [PubMed] [Google Scholar]
- 24.Gelderblom M, Weymar A, Bernreuther C, Velden J, Arunachalam P, Steinbach K, et al. Neutralization of the IL-17 axis diminishes neutrophil invasion and protects from ischemic stroke. Blood. 2012;120(18):3793–802. doi: 10.1182/blood-2012-02-412726. [DOI] [PubMed] [Google Scholar]
- 25.Kang Z, Altuntas CZ, Gulen MF, Liu C, Giltiay N, Qin H, et al. Astrocyte-restricted ablation of interleukin-17-induced Act1-mediated signaling ameliorates autoimmune encephalomyelitis. Immunity. 2010;32(3):414–25. doi: 10.1016/j.immuni.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen YJ, Hsieh MY, Chang MY, Chen HC, Jan MS, Maa MC, et al. Eps8 protein facilitates phagocytosis by increasing TLR4-MyD88 protein interaction in lipopolysaccharide-stimulated macrophages. J Biol Chem. 2012;287(22):18806–19. doi: 10.1074/jbc.M112.340935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caso JR, Pradillo JM, Hurtado O, Lorenzo P, Moro MA, Lizasoain I. Toll-like receptor 4 is involved in brain damage and inflammation after experimental stroke. Circulation. 2007;115(12):1599–608. doi: 10.1161/CIRCULATIONAHA.106.603431. [DOI] [PubMed] [Google Scholar]
- 28.Liesz A, Dalpke A, Mracsko E, Antoine DJ, Roth S, Zhou W, et al. DAMP signaling is a key pathway inducing immune modulation after brain injury. J Neurosci. 2015;35(2):583–98. doi: 10.1523/JNEUROSCI.2439-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang CC, Lin YS, Lee CC, Hsu KS. Cell type-specific expression of Eps8 in the mouse hippocampus. BMC Neurosci. 2014;15:26. doi: 10.1186/1471-2202-15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]