Significance
Our study unravels the intricate meiotic processes, spotlighting the significance of Ring Finger E3 ligases. Specifically, we introduce RNF212B as a crucial player in prophase I progression in mice, essential for loading other Ring Finger ligases like RNF212 and HEI10. Understanding the regulatory mechanisms behind crossover (CO) designation and maturation is vital, as COs play a pivotal role in keeping bivalents together (chiasmata), critical for accurate chromosome segregation during meiosis. Dysregulation of these events is implicated in miscarriage, infertility, and birth defects in humans. Our findings contribute valuable insights into mammalian meiosis, into a conserved pathway from yeast to mammals that holds physiological significance for understanding the biology of COs and its role in shaping meiotic recombination landscape in eukaryotes.
Keywords: meiosis, crossing over, mouse, recombination, fertility
Abstract
Meiosis, a reductional cell division, relies on precise initiation, maturation, and resolution of crossovers (COs) during prophase I to ensure the accurate segregation of homologous chromosomes during metaphase I. This process is regulated by the interplay of RING-E3 ligases such as RNF212 and HEI10 in mammals. In this study, we functionally characterized a recently identified RING-E3 ligase, RNF212B. RNF212B colocalizes and interacts with RNF212, forming foci along chromosomes from zygonema onward in a synapsis-dependent and DSB-independent manner. These consolidate into larger foci at maturing COs, colocalizing with HEI10, CNTD1, and MLH1 by late pachynema. Genetically, RNF212B foci formation depends on Rnf212 but not on Msh4, Hei10, and Cntd1, while the unloading of RNF212B at the end of pachynema is dependent on Hei10 and Cntd1. Mice lacking RNF212B, or expressing an inactive RNF212B protein, exhibit modest synapsis defects, a reduction in the localization of pro-CO factors (MSH4, TEX11, RPA, MZIP2) and absence of late CO-intermediates (MLH1). This loss of most COs by diakinesis results in mostly univalent chromosomes. Double mutants for Rnf212b and Rnf212 exhibit an identical phenotype to that of Rnf212b single mutants, while double heterozygous demonstrate a dosage-dependent reduction in CO number, indicating a functional interplay between paralogs. SUMOylome analysis of testes from Rnf212b mutants and pull-down analysis of Sumo- and Ubiquitin-tagged HeLa cells, suggest that RNF212B is an E3-ligase with Ubiquitin activity, serving as a crucial factor for CO maturation. Thus, RNF212 and RNF212B play vital, yet overlapping roles, in ensuring CO homeostasis through their distinct E3 ligase activities.
During meiosis, a single round of DNA replication followed by two successive rounds of chromosome segregation leads to the formation of haploid gametes from diploid progenitors (1). The reductional division of homologous (maternal and paternal) chromosomes during the first meiotic division (MI) depends on a series of meiosis-specific processes that occur during prophase I. These include chromosome synapsis and recombination, both mediated by a proteinaceous structure, the synaptonemal complex [SC, (2)]. Recombination between homologous chromosomes is initiated by DNA double-strand break (DSB) induction, which results in the formation of noncrossovers (NCOs) and crossovers (COs), the latter which are essential precursors of the chiasmata that will maintain homolog interactions through until the first meiotic division.
DSBs are formed via the action of the topoisomerase, SPO11, and its associated cofactors. Through the course of prophase I, these DSBs undergo subsequent processing via the action of multiple DNA repair pathways whose action and assembly at electron-dense recombination nodules (RN) is dependent on the concurrent formation and integrity of the SC. Importantly, in mice, though approximately 250 DSBs form in preleptonema of prophase I, fewer than 10% resolve as COs in pachynema, while the remaining DSBs resolve as NCOs beginning in zygonema (3). The appropriate distribution of COs across the genome, and their stabilization by Rec8-containing cohesin complexes (4, 5), is critical for accurate segregation of homologous chromosomes at the MI in readiness for the subsequent equational division at MII. The importance of these physical linkages is underscored by the high rates of meiotic errors observed in women (6) and are the leading cause of spontaneous abortion, miscarriage, and birth defects in humans (7, 8).
How the appropriate frequency and distribution of COs is achieved during mammalian prophase I remains one of the major questions in the field and the subject of intense investigation. In zygonema of prophase I, the MutS heterodimer of MSH4/MSH5 (known as MutSγ) localizes to a subset of DSB repair intermediates along with the SUMO E3 ligase, RING-Finger protein-212 (RNF212), collectively licensing these DSB repair intermediates for CO formation (9–11). However, in mammals, approximately 150 MutSγ sites are identified at this stage, and only 23 to 26 of these will go on to load the MLH1-MLH3 heterodimer (known as MutLγ), which is considered to be the ultimate marker of the major class of CO events in mid-to-late pachynema of prophase I (12–15). The mechanism by which MutLγ designates this subset of MutSγ sites to become these “Class I” COs is unclear, but the interplay between two key pro-CO factors, RNF212 and the ubiquitin E3 ligase (and RING-finger protein), Human Enhancer of Invasion-10 (HEI10), has been implicated in this process (16, 17). Along these lines, recent studies across meiotic species have revealed a conserved role for RING-finger domain-containing proteins in crossover designation. Zip3, ZHP-3, Vilya, and RNF212 in Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, and Mus musculus, respectively, are all critical for appropriate levels of crossovers in their respective organisms (3, 9, 18, 19). At least two of these (Zip3, ZHP-3) interact with MutSγ, while HEI10 in plants interacts with MutLγ but not with MutSγ (20–22).
RNF212 and HEI10 are members of the SUMOylation and the ubiquitin–proteasome system (UPS), respectively, and are responsible for the conjugation of ubiquitin or SUMO chains to proteins in a process involving an activating enzyme (E1), a conjugating enzyme (E2), and finally an E3 ligase (23, 24). While addition of SUMO leads to altered protein function, localization, or stability, addition of ubiquitin results predominantly in targeting of proteins to the proteasome for degradation (25). The targets of the proteasome are misfolded proteins or short-lived proteins (26, 27) such as cyclins or more recently components of the meiotic CO machinery (16, 28, 29). Thus, SUMOylation, ubiquitylation, and proteasome activity are all required for proper homolog synapsis and recombination, as well as normal RN dynamics (16)
Variations in the frequency of COs across the genome among humans and other mammals have been well documented (30–32), yet the functional significance of these differences remains poorly understood (33). Notably, specific loci associated with key players in the meiotic process have been identified, including the histone methyltransferase PRDM9 (34), the meiotic cohesin RAD21L (30, 35), the E3 ligases RNF212 and HEI10 (30, 32), and the SC central element (CE) protein, SIX6OS1 (36, 37). Recent genome-wide association studies have highlighted RNF212B as a potential contributor to heritable variation in recombination rates in cattle (38).
In this study, we explore the biological role of RNF212B in mammalian meiosis. Our findings reveal RNF212B as a crucial player involved in CO licensing and designation in a DSB-independent and synapsis-dependent manner ultimately influencing mouse fertility. Proteome studies in testis and Hela cells indicate that RNF212B does not rewire the SUMOylome but functions instead as an E3 ligase with Ubiquitin activity which is crucial for its function as a pro-CO factor, since mutation of the catalytic domain of the protein phenocopies the null allele of Rnf212b. Importantly, RNF212B, despite its temporal, spatial, physical overlap with its paralog RNF212, is a pro-CO factor which is essential for normal prophase I progression and for ensuring appropriate CO designation in mammalian meiosis.
Results
RNF212B Is a Previously Uncharacterized Putative E3 Ligase.
In a GWAS of comparative heritability of the global recombination rate between male and female cattle (38), seven causative QTLs were identified including variants in known meiotic genes, including RNF212 and MLH3, as well as uncharacterized genes, exemplified by RNF212B. RNF212B, a poorly annotated paralog of RNF212 in vertebrates, exhibited a moderate similarity of 39.7% with its paralog. Despite its poorly characterized nature, RNF212B demonstrated high conservation, with an 83% sequence similarity between humans and mice (SI Appendix, Fig. S1A). RNF212B is transcribed at low and high levels in the kidney and testis, respectively. (https://www.gtexportal.com, https://www.biogps.org).
Similar to RNF212, RNF212B contains a RING finger domain that binds two zinc ions in a distinctive “cross-brace” arrangement through a defined motif of cysteine and histidine residues. Based on the distribution of the cysteines (C) and histidines (H), RNF212B belongs to the CH3C4 type of zinc finger proteins (SI Appendix, Fig. S1B). Notably, despite its low sequence conservation, the yeast RING finger protein ZIP3 has been demonstrated to exhibit SUMO ligase activity both in vitro and in vivo, earning its designation as a functional ortholog of RNF212 (9). These findings lead to the inference of SUMO ligase activity for RNF212, as opposed to ubiquitin, although this determination has yet to be investigated experimentally (see below).
To explore cell type-specific expression of the meiotic RNF212B, we performed single-cell RNA-sequencing of mouse testicular germ cells. This allowed us to plot average expression for Rnf212b across all cell clusters, and revealing overlapping transcriptional profiles for Rnf212b with Hei10, Rnf212, and with Msh4, Msh5, Mlh1, Mlh3, and Cntd1, suggesting its restriction to the germ lineage similar to these other prophase I genes (SI Appendix, Fig. S2).
DSB-Independent and Synapsis-Dependent Localization of RNF212B to the Chromosome Axes.
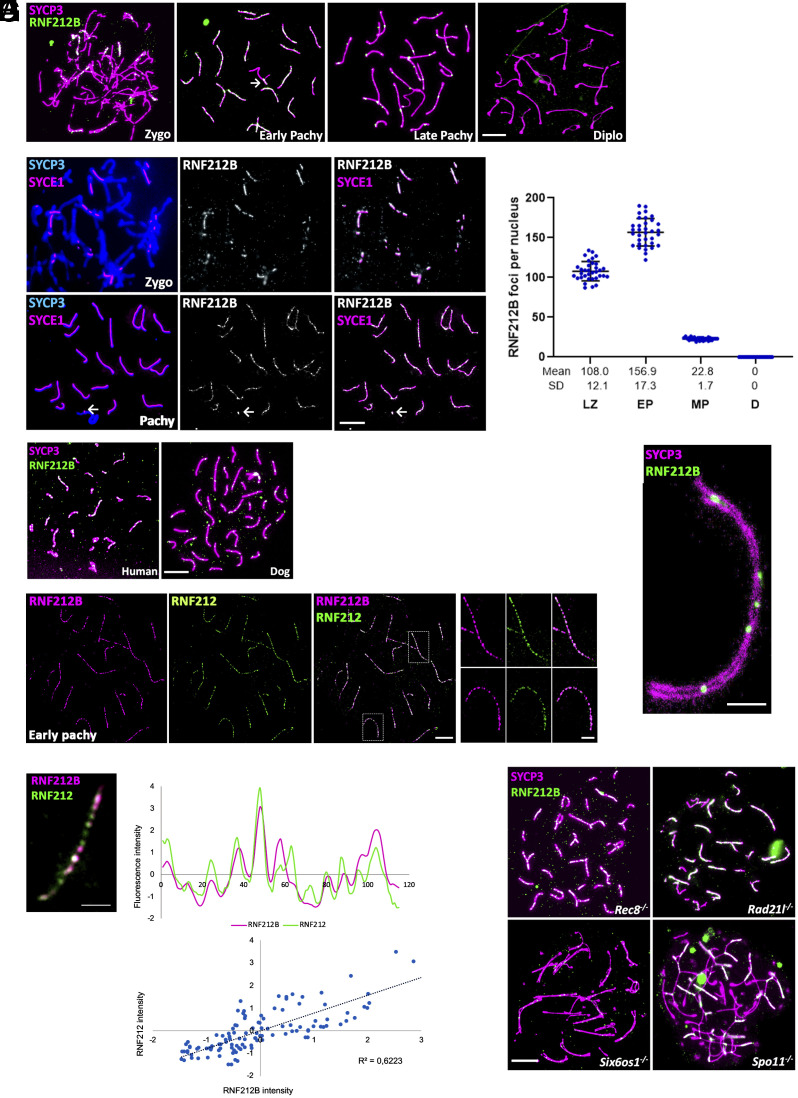
We next sought to delineate the chromosomal localization of RNF212B during prophase I. We analyzed mouse spermatocyte spreads prepared from adult males and stained with a custom-made antibody against RNF212B (Fig. 1A, specificity was tested by IF and western blot) and the structural protein of the lateral/axial elements (LE/AE) of the SC, SYCP3, or the central element protein, SYCE1 (a marker of chromosome synapsis; Fig. 1B). RNF2121B was first detected in early zygonema, labeling numerous foci (Fig. 1A) that overlapped with SYCE1 (Fig. 1B). As synapsis progressed, a notable increase in the number of RNF212B foci was observed along the synapsed LEs (Fig. 1 A–C). On the XY bivalent, the synapsed pseudoautosomal region (PAR) was also labeled (Fig. 1 A and B). From late zygonema to early pachynema there was a reduction in the number of foci down to a few per chromosome at late pachynema (Fig. 1A), similar to the numbers of MutLγ foci that mark the final distribution of Class I CO events. At early diplonema, when homologous start to desynapse, the RNF212B foci had disappeared. (Fig. 1 A and C). This localization was also shown by double immunolabeling of RNF212B and SYCP3 on paraffin-embedded testis sections (SI Appendix, Fig. S3A).
Fig. 1.
RNF212B colocalizes with RNF212, its loading to the chromosome axis is dependent on synapsis and is independent of Spo11-induced DSB. (A) Colabeling of RNF212B and SYCP3 from Zygonema to Diplonema. (B) Colabeling of spermatocytes spreads with SYCP3, central element protein SYCE1, and RNF212B. Arrow in (A) and (B) indicate the positive labeling of the PAR. (C) Plot quantitation of the number of RNF212B foci. LZ (Late Zygonema), EP (early Pachynema), MP (mid pachynema), and D (Diplonema). (D) Colabeling of RNF212B and SYCP3 in human and dog spermatocytes showing conserved pattern of localization onto the chromosome axes. (E) STED microscopy images of RNF212 and RNF212B colocalization in pachytene spermatocytes. (F) Double immunostaining of RNF212B and RNF212. Immunofluorescence signal levels were measured on synapsed chromosome axes. Upper plot represents normalized signal intensity profiles of RNF212B (magenta) with RNF212 (green). Lower plot shows regression analysis of the correlation between each pair corresponding to chromosome axes. Each of the dots represents the RNF212B fluorescence intensity in axes “x” and RNF212 fluorescence intensity in axe “y” for a given point along the AE. (G) STED images of pachynema chromosome immunostained for SYCP3 and RNF212B. (H) Double immunolabeling of RNF212B and SYCP3 in arrested spermatocytes of Rec8−/−, Rad21l−/−, Six6os1−/−, and Spo11−/−. Bar in panels (A), (B), (D), and (H) 10 μm. Bars in panels (G) and (F) represents 2.5 μm. All experiments have been carried out in at least three mice and 15 cells per mouse.
To explore the dynamics of RNF212B accumulation through prophase I in female germ cells, oocyte spreads were prepared from fetal ovaries taken from 16.5 to 18.5 days postcoitum (dpc). The loading of RNF212B initiates the formation of foci during zygonema, concurrent with synapsis. The number of foci gradually increases during early pachynema (SI Appendix, Fig. S3 B and C). RNF212B starts to unload progressively during diplonema, although some foci persist at synapsed lateral elements (LEs) at late diplonema (SI Appendix, Fig. S3 B and C). The number of foci in oocytes exceeds those observed in spermatocytes.
Given the high conservation of the RNF2121B paralogs within mammals (~80% mouse vs. dog, or human vs. mouse in average) we also detected RNF212B foci on synapsed LEs in spermatocytes from humans and dogs (Fig. 1D), including intense labeling at the XY PAR, underscoring the evolutionarily conserved role of RNF212B in meiosis across mammalian species.
We next performed dual immunofluorescence staining for RNF212B and RNF212, coupled with superresolution imaging (STED), exhibiting partial overlapping along the chromosome axes (regression coefficient r = 0.6; n = 20 cells) (Fig. 1 E and F). In addition, we used STED to clarify the specific localization of the RNF212B foci relative to the tripartite structure of the SC. RNF212B foci localized to the central region of the SC suggesting that RNF212B loading requires synapsis (Fig. 1G). Thus, we evaluated RNF212B distribution in the absence of synapsis using three different genetic models. Mutant mice lacking the cohesins RAD21L and REC8 exhibit partial synapsis between nonhomologous chromosomes and self-synapsis between sister chromatids, respectively (35, 39–41), while loss of SIX6OS1 results in a failure to form the CE and a consequent failure of synapsis (36). In spermatocytes from Rec8−/− and Rad21l−/− males, RNF212B foci localized to aberrant synapsed regions (Fig. 1H). However, in Six6os1−/− nuclei, despite RNF212B expression (SI Appendix, Fig. S4), RNF212B foci were not observed instead a very faint dispersed staining was discerned along those LEs that showed alignment (Fig. 1H). Thus, RNF212B loading and/or persistence is dependent on synapsis.
Next, we investigated the dependency between RNF212B focus formation and the initiation of double-strand breaks (DSBs) by SPO11, the pivotal event in the meiotic recombination cascade. To explore this, we analyzed RNF212B localization in a Spo11 null mutant, which intriguingly also exhibits pseudosynapsis of nonhomologous chromosome axes at a pachytene-like arrest (42). Notably, RNF212B was observed to associate with the pseudosynapsed axes even in the absence of SPO11-induced DSBs (Fig. 1H). These findings collectively suggest the existence of an unexplored DSB-independent and synapsis-dependent pathway for the recruitment of meiotic recombination proteins such as RNF212B to chromosome axes.
RNF212B Colocalization and Loading Dependency on Recombination Machinery.
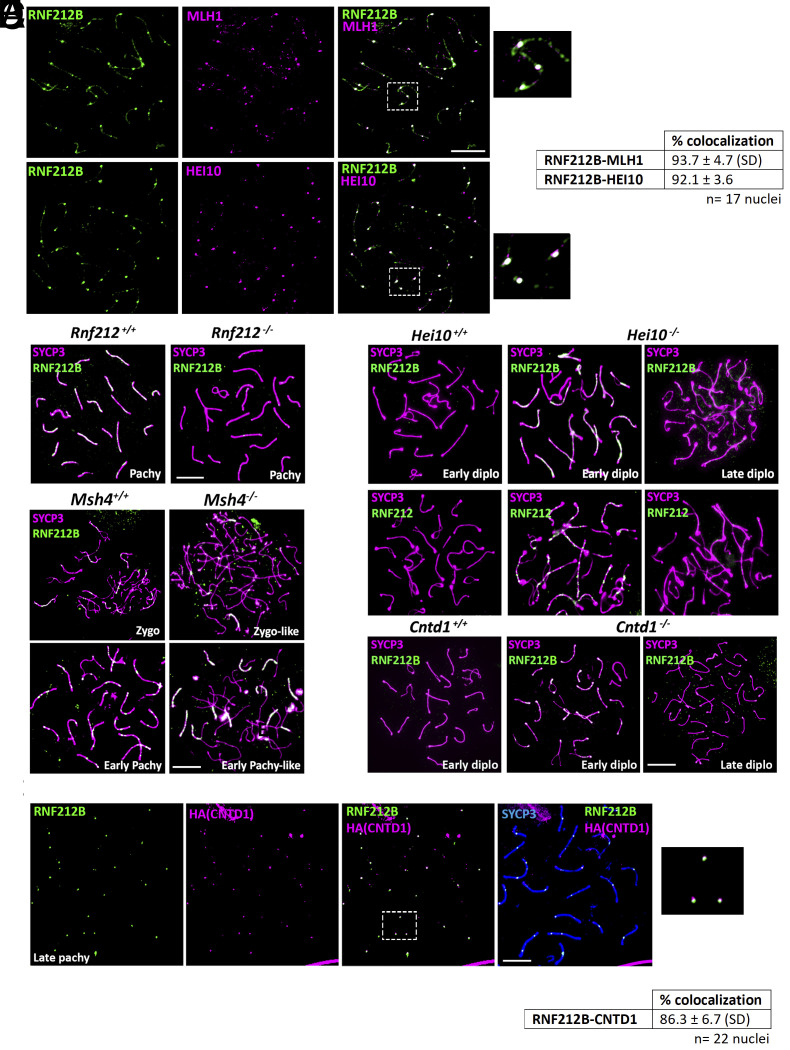
MLH1-MLH3 (MutLγ), HEI10, and the cyclin N-terminal domain-containing-1 protein (CNTD1) are terminal markers of the major class of CO events beginning in early pachynema (3). To assess CO site-associated RNF212B localization, we visualized RNF212B localization together with these markers. RNF212B foci colocalize with MLH1 in mouse spermatocytes and oocytes (Fig. 2A and SI Appendix, Fig. S3D). Similarly, HEI10 also colocalizes with RNF212B at pachynema (Fig. 2A). Altogether, RNF212B foci and RNF212 share similar dynamic spatiotemporal colocalization, marking most, if not all, of COs at pachynema and suggesting a role for RNF212B alongside RNF212 in CO designation/maturation.
Fig. 2.
RNF212B colocalizes with CO-associated proteins and its loading is dependent on RNF212, and its unloading on HEI10 and CNTD1. (A) Double immunolabeling of RNF212B with MLH1 and HEI10 in pachynema spermatocytes. Right panel show magnification of the indicated region. The quantitation of the colocalization of RNF212B with MLH1 and HEI10 are shown in right table. (B) Double immunolabeling of RNF212B and SYCP3 in Rnf212−/−in pachynema spermatocytes showing absence of foci in comparison with the numerous foci of the wild-type control. (C) Double immunolabeling of RNF212B and SYCP3 in Msh4+/+ and Msh4−/−spermatocytes at zygonema and early pachynema, showing persistence of RNF212B loading in the absence of MSH4. (D) Double immunolabeling of SYCP3 with RNF212B and RNF212 in Hei10−/− early diplonema spermatocytes showing numerous foci at the synapsed LEs in comparison with the wild-type diplonemas which lack foci and in Hei10−/− late diplonemas when foci have completely disappeared. (E) Colabeling of RNF212B, anti-HA (CNTD1), and SYCP3 showing colocalization. The Right panel shows magnification of the indicated region. The quantitation of the colocalization of RNF212B with CNTD1 is shown in the lower table. (F) Double immunolabeling of RNF212B and SYCP3 in Cntd1−/−early diplonema spermatocytes showing numerous foci at the synapsed LEs in comparison with the wild-type diplonema spermatocytes which lack foci and in Cntd1−/− late diplonema when foci have completely disappear. Bar in panels 10 μm. All experiments have been carried out in at least three mice and 15 cells per mouse.
In light of the spatial colocalization of the RNF212 and RNF212B paralogs, the proposed interplay between RNF212 and HEI10 in the context of CO designation (16), and the colocalization with components of the MutSγ and MutLγ heterodimers, we evaluated the genetic requirements of RNF212B in relation to loss of these key genes in mutant mouse models (17, 43). In Rnf212−/− and Hei10−/− spermatocytes, RNs containing MutLγ fail to load in pachynema, resulting in disrupted crossing over and metaphase I arrest with an absence of most chiasmata (univalents are observed instead of bivalents; (16, 43). In the current study, we also show by immunolabeling the absence of RNF212B in Rnf212−/− spermatocytes, indicating that the loading and/or stabilization of RNF212B is dependent on RNF212 (Fig. 2B). To distinguish between these possibilities, we analyzed the expression of RNF212B in testis tissue extracts using western blotting, demonstrating RNF212B expression despite the absence of foci at the chromosome axis (SI Appendix, Fig. S4).
In contrast to RNF212, loss of the MutSγ complex had no impact on RNF212B loading onto synapsed chromosome cores in zygonema and early pachynema (Fig. 2C). Similarly, Hei10−/− spermatocytes show persistence and increased frequency of RNF212B and RNF212 foci from pachynema to mid-diplonema along the synapsed chromosome axes, ultimately disappearing in late diplonema (Fig. 2D). Collectively, the localization pattern of RNF212B parallels that of, and is dependent upon, RNF212 (9, 16), further substantiating its synapsis-dependent loading onto the central element of the SC. Additionally, like RNF212, loss of HEI10 results in delayed unloading of RNF212B at diplonema, implicating HEI10 in the turnover of these two RING-finger proteins.
Recent studies have identified Cyclin N-terminal domain-containing-1 (CNTD1), an ortholog of C. elegans COSA-1 (44), as a regulator of CO designation (45, 46). Like HEI10, CNTD1 is critical for unloading excess MutSγ and RNF212 to allow for the final stages of MutLγ loading in pachynema (45). We investigated RNF212B colocalization with CNTD1 using a FLAG and HA dual-tagged allele of mouse Cntd1 (45). CNTD1 loads in pachynema, and colocalized with RNF212B at a frequency of 1 to 2 foci per chromosome, similar to our previous colocalization studies with CNTD1 and other late RN components (Fig. 2E). Moreover, in Cntd1−/− males, we observe a dramatic upregulation and persistence of RNF212B foci in late pachynema and extending into mid-diplonema (Fig. 2F). Collectively, these results indicate that CO-associated proteins, HEI10 and CNTD1, play a crucial role in the turnover of these two RING-finger proteins with known proCO functions establishing a conditional regulation.
RNF212, RNF212B, and HEI10 Act Cooperatively through Direct Interactions.
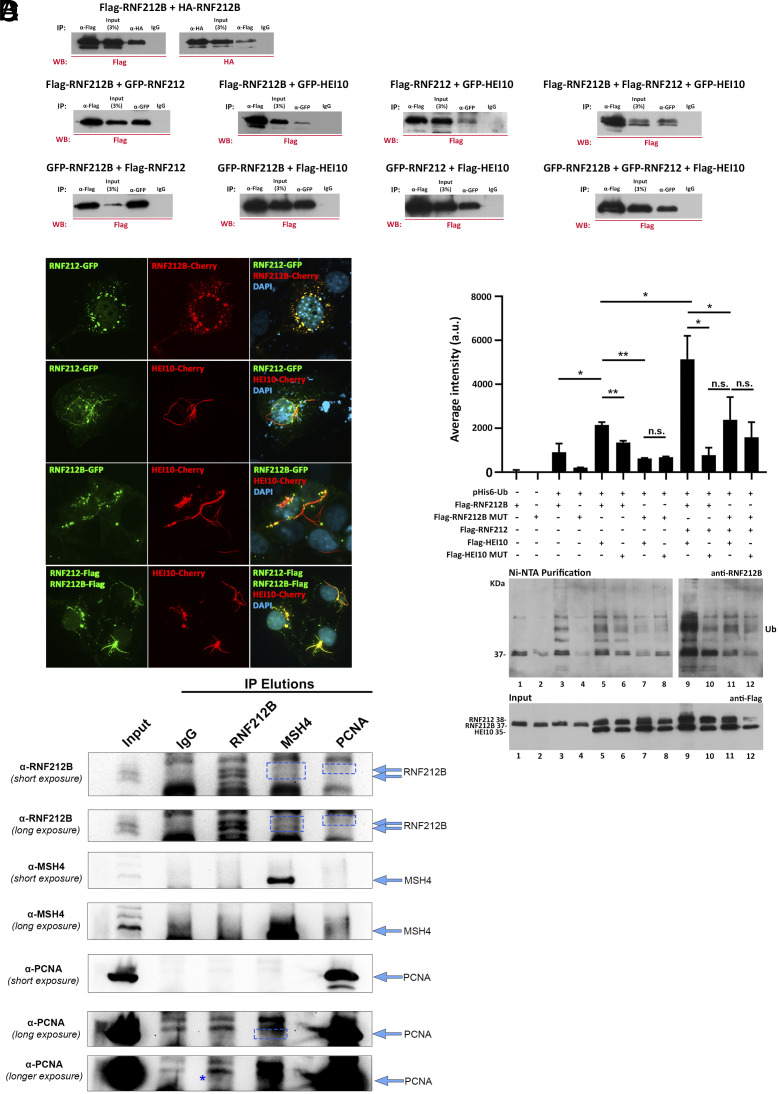
The dimerization of many RING-finger type E3 ligases has been structurally characterized and demonstrated to be essential for their ligase activity (47, 48). To investigate a potential interaction between RNF212 and its paralog RNF212B, we cotransfected both genes, each tagged differently, into HEK293T cells. Our analysis revealed a positive coimmunoprecipitation between RNF212B and itself as well as with RNF212 and HEI10 (Fig. 3A). The specificity of the interaction was validated using the unrelated E3 ligases TRIM37 and RNF34 (SI Appendix, Fig. S5A). We further confirmed the interactions of RNF212B, RNF212 through immunofluorescence analysis of transfected COS7 cells. The results show strong and specific colocalization in cytoplasmic aggregates (Fig. 3B and SI Appendix, Fig. S5B). Collectively, the observed protein–protein complex of RNF212 and RNF212B, coupled with their co-occurrence in time and space along the synapsed chromosome axes of mammalian meiocytes (Fig. 1E), strongly suggests the formation of a proteinaceous heterocomplex.
Fig. 3.
RNF212B interactions and Ubiquitylation activity. (A) Co immunoprecipitation of RNF212B with RNF212B, RNF212, and HEI10. HEK293T cells were cotransfected with plasmids encoding RNF212, RNF212B, and HEI10 tagged with FLAG or GFP. Protein complexes were immunoprecipitated overnight with an antibody anti-FLAG, anti-GFP, or IgGs (as a negative control) and analyzed by immunoblotting. Positive interaction of RNF212B with RNF212 and HEI10 as well as RNF212B (self-interaction) is shown. Additionally, a positive interaction between RNF212 and HEI10 is observed. (B) COS7 cells immunofluorescence shows colocalization of transfected RNF212B with RNF212 and HEI10. COS7 cells were transfected to express RNF212B (fused to either Cherry, Flag, or GFP) along with RNF212 (fused with Flag or GFP) and/or HEI10 (Cherry-tagged). Nuclear and punctuate cytoplasmic RNF212B signal perfectly colocalizes with RNF212 signal (Top). Transfected HEI10 forms a ring fiber-like pattern that when cotransfected with RNF212 and/or RNF212B recruits them to this ring structures altering their localization pattern. (Scale bars: 10 μm.) (C) RNF212B has autoubiquitynilation activity. HEK293T cells were cotransfected with Flag-tagged plasmids encoding RNF212, RNF212B (WT or mutant), and/or HEI10 (WT or mutant) in the presence or absence of a plasmid encoding 6xHis-Ubiquitin. Protein complexes were purified using Ni-NTA, favoring the purification of proteins containing 6xHis sequence. Then, ubiquitylation was analyzed by immunoblotting using antibodies anti-RNF212B and anti-Flag (Input). The results show RNF212B autoubiquitinylation. As observed in the statistics of the quantification of average intensity for each well, the strongest ubiquitinylation was exhibited when RNF212B was cotransfected along with RNF212 and HEI10 and was abolished when a RNF212B dead mutant was transfected. Quantification of the results of three independent experiments. Two-tailed Welch’s t test analysis: *P < 0.05. **P < 0.005. (D) IPs in isolated germ cells with the antibodies indicated (Top) against RNF212B, MSH4, and PCNA, and western blot analysis of the eluted proteins with the same antibodies (Left) detecting specific bands for RNF212B in the MSH4 and PCNA elution, and PCNA in the RNF212B and MSH4 elution (boxes and asterisk). Arrows indicate sizes for bands of interest in each blot.
Given the observed genetic interaction and cytological localization of the three E3 ligases, we further investigated their interaction in HEK293T and Cos7 cells. The results unequivocally demonstrate an interaction of HEI10 with both RNF212B and RNF212, accompanied by colocalization upon cotransfection in Cos7 cells (Fig. 3B). Furthermore, when expressed from a transgene in Cos7 cells, HEI10 forms ring structures that resemble polycomplexes of coiled-coil proteins of the central element of the SC (36), and these structures recruit RNF212 and RNF212B (Fig. 3B). These findings indicate a direct interaction among the three RING-finger E3 ligases, at least in a heterologous cell system.
Subsequently, we conducted a ubiquitin assay in transfected HEK293T cells using Rnf212, Rnf212b, and Hei10 and a 6XHis-ubiquitin expression vector. The results reveal consistently low basal levels of autoubiquitylation when each of the three E3 ligases (HEI10, RNF212B, RNF212) is transfected independently, which sharply escalates upon simultaneous transfection (Fig. 3C). Importantly, this synergistic activity is abolished when the RNF212B dead mutant is transfected alongside the RNF212 and HEI10 wild-type E3 ligases (Fig. 3C). Altogether, these results indicate that the three E3 ligases can interact and that this interaction promotes the autoubiquitylation of RNF212B in an autocatalytic-dependent manner.
RNF212B Interacts with Key Members of the Recombination Machinery in Mouse Spermatocytes.
We next asked whether RNF212B interacts, either directly or indirectly, with components of the CO machinery to direct CO designation and maturation. In particular, we investigated RNF212B interactions with the MutSγ complex and with PCNA, a sliding clamp complex that is critical for activation of MutL heterodimers in somatic cells, and which has been implicated in MutLγ activation in mammalian meiosis (45, 49, 50). To investigate RNF212B interactions with MutSγ (via anti-MSH4 antibodies) and other pro-CO regulators in vivo, we performed IPs in isolated germ cell suspension, minimizing somatic cell contamination and enriching lysates with germ cell proteins, including spermatocytes. IPs with antibodies against RNF212B, MSH4, and PCNA showed robust purification of each respective antigen. We then probed each IP elution and detected specific bands for RNF212B in the MSH4 and PCNA elution, and PCNA in the RNF212B and MSH4 elution. The relatively faint bands as evidenced by long exposure times suggest that these interactions may be low affinity, transient, and/or indirect. However, we also found that RNF212B was insoluble in typical IP buffer conditions, so we used a modified buffer with relatively high salt (500 mM) and detergent (1% Triton) concentrations, which likely affected the efficiency of the IPs (Fig. 3D and SI Appendix, Fig. S6A). We further analyzed these interactions by IP coupled to mass spectrometry (IP-MS). Among the immunoprecitpated proteins related with DNA repair and recombination, we identified RNF212B (positive control), RNF212 (its interactor), PCNA, the three components of the PCNA clamp loader (RFC1, RFC2, and RFC4), and the Muts protein MSH2 and MSH6 (Dataset S1). In somatic cells, PCNA and RFC work cooperatively to activate MutL heterodimeric complexes (51). The interaction with the MSH4/MSH5 and PCNA obtained by IP-WB in germ cells was also addressed in HEK293 cells transfected with Rnf212b and Msh4, Msh5, or Pcna. We obtained co-IP of RNF212B with MSH5 but not with MSH4 and PCNA, suggesting either that the interactions are through transient and intricate multiprotein complexes, or that they require the physiological milieu of the SC and chromatin of the spermatocyte (SI Appendix, Fig. S6B). The absence of the heteroduplex MSH4/MSH5 in the IP-MS suggests a transient and/or indirect interaction in low abundance zygotene cells that are poorly represented by whole testis protein extracts. Taken together, our IP-MS and IP-WB results suggest that RNF212B acts together with RNF212 in a complex in mammalian spermatocytes and that RNF212B further interacts with components of the CO designation machinery to activate MutLγ.
Rnf212b-Knockout Male and Female Mice Are Sterile and Show Synapsis Defects Specifically between the Sex Chromosomes.
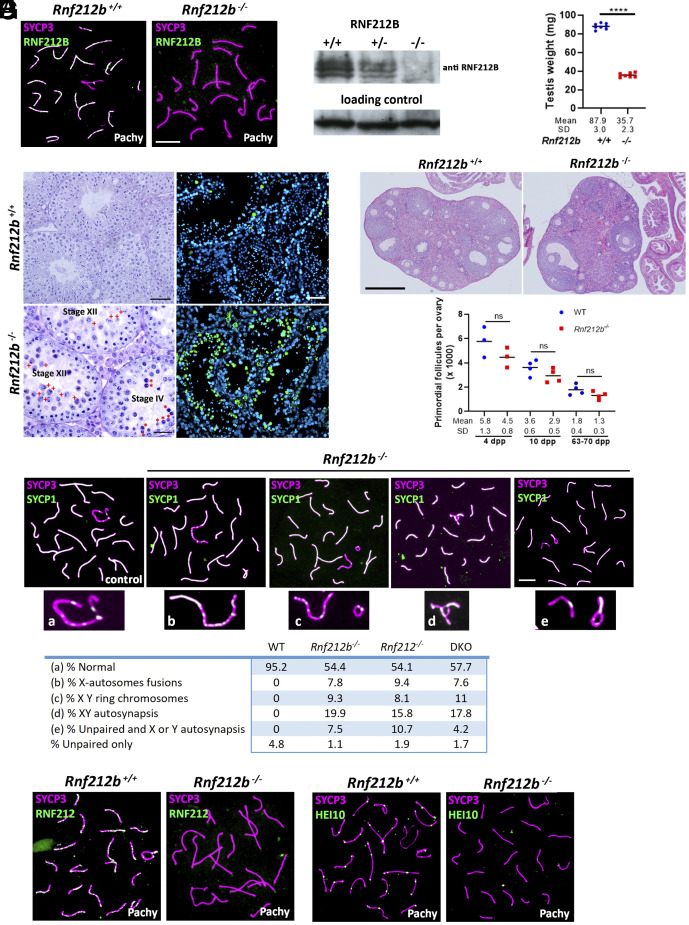
To further investigate the function of RNF212B, mice lacking the Zn-RING domain were generated (SI Appendix, Fig. S7A). The Rnf212bwt/edited founder with the selected mutation was crossed with C57/Bl6 and the heterozygous interbred (SI Appendix, Fig. S7B and Materials and Methods). Rnf212bedited/edited mice did not show RNF212B foci at their chromosomes and no protein band was observed in the western blot analysis of testis extracts from Rnf212b−/− mutant mice (Fig. 4 A and B). This result indicates that the mutation represents a null allele (herein Rnf212b−/−) and further validates the specificity of the polyclonal antibodies developed during our studies. Homozygous mutant mice were obtained at Mendelian frequencies and were apparently healthy. Both male and female Rnf212b−/− mice were infertile after crossing for a year with both male and female wild types (4 crosses each). Mutant mice show reduced testes (70% reduction; Fig. 4C) and normal ovaries (Fig. 4E), respectively. Histological analysis of Rnf212b−/− spermatocytes revealed an accumulation of tubules at stage XII of the seminiferous epithelial cycle with massive apoptosis of spermatocytes that failed to carry out the first meiotic division, and absence of spermatocytes progressing beyond this stage. This observation suggests a metaphase I arrest, corresponding to stage XII (Fig. 4D). Additionally, we observed pachytene spermatocytes entering into apoptosis, characterized by large densely staining cells toward the center of the lumen, at a stage corresponding to stage IV of the seminiferous epithelial cycle (Fig. 4D). We confirmed these spermatocytes were dying via apoptosis in Rnf212b−/−males by TUNEL assay (Fig. 4D).
Fig. 4.
Characterization of Rnf212b-deficient mice. (A) Double immunolabeling of RNF212B and SYCP3 showing absence of fluorescent signal in the Rnf212b−/−and foci labeling in the control spermatocytes. Bar 10 μm. (B) Western blot analysis of whole testis extracts showing two bands in the wild type and heterozygous that are absent in the Rnf212b−/−. (C) Plot representation of the reduction in the weight of the testis from Rnf212b−/−in comparison with the wild-type (WT). (D) Left panel, histological section of a testis from Rf212b+/+ and Rf212b−/−mice showing abnormal accumulation of tubules at stage XII of the seminiferous epithelial cycle with abnormal and apoptotic metaphase I cells (red crosses) and at stage IV of the seminiferous epithelial cycle having apoptotic pachynemas (red asterisks). Bar 50 μm in Rnf212b+/+and 25 μm in Rf212b−/−. Right panel, immunofluorescence detection of apoptotic cells by TUNEL staining showing an increase of apoptotic cells in Rf212b−/−seminiferous tubules. Bar 50 μm. (E) Upper panel, histological staining of representative sections of ovaries from an Rnf212b+/+and an Rnf212b−/−adult mice, showing oocytes and follicles at all stages of follicular development. Bar 200 μm. Lower panel, representative plot of primordial follicles quantitation from Rnf212b−/−and wild-type ovaries showing a reduced trend in primordial follicles numbers in the mutant ovaries, which is not statistically different. (F) Double immunolabeling of SYCP3 and SYCP1 showing aberrant synapsis and structural abnormalities in the XY chromosome pair including X-autosome fusions (b), ring chromosomes (c), XY autosynapsis (d), and unpaired and autosynapsis (e). The quantitation numerical values of these aberrant configurations in the XY bivalent in Rnf212b−/−, Rnf212−/−, Rnf212b−/− Rnf212−/−(dKO), and wild-type spermatocytes (WT) are shown in the lower table. (G) Double immunolabeling of SYCP3 with RNF212 and with HEI10 showing absence of fluorescent foci in Rnf212b−/− in comparison with the wild-type controls. Bar in (E–G) 10 μm. All experiments have been carried out in at least three mice and 100 cells per mouse in (F), and 15 cells in (G).
Ovaries from adult female mice were normal in size and showed follicles at different stages of folliculogenesis and several corpora lutea (Fig. 4E). Furthermore, ovaries from mutant females at 4, 10, and 63 to 70 dpp showed a slight, but not significant, reduction in the number of primordial follicles when compared with WT (Fig. 4E). Taken together, these observations are consistent with progression through prophase I in females as other recombinant mutants (9, 14, 17).
Despite previous reports suggesting normal SC formation in Rnf212-deficient meiocytes (9, 16), worm zhp-3 mutants exhibit defects in SC assembly (18). Consequently, we investigated synapsis and SC assembly in the absence of Rnf212b by colocalization of SYCP1 and SYCP3 through prophase I. During zygonema, the SYCP3-labeled axial elements initiate synapsis, as demonstrated by SYCP1 staining of the transverse filaments of the SC. Full synapsis is attained at pachynema, revealing complete overlap between SYCP1 and SYCP3 along the autosomes, mirroring to the wild-type spermatocytes. The synapsis of the XY chromosome pair is markedly sensitive to synaptic alterations (52) due to the PAR-restricted domain of synapsis between these chromosomes PAR. Rnf212b−/− mice displayed pachytene spermatocytes with structural abnormalities involving the X and Y chromosomes including a variety of phenomena such as ring chromosomes, fusions between X and an autosome, and autosynapsis of the X chromosome, among others (Fig. 4F). Such defects were never observed in wild-type (WT) mice. Oocyte chromosome spreads also revealed, albeit to a lesser extent, nonhomologous synapsis involving several autosomes (SI Appendix, Fig. S8A). Subsequent staining against the phosphorylated form of histone H2AX (γH2AX) which, in pachynema marks the heterochromatin-rich XY-containing Sex Body domain, revealed abnormal γH2AX staining in a fraction of the cells with structural abnormalities in Rnf212b−/− males. Specifically, we observed faint γH2AX signal that extended beyond the Sex Body into the autosomal domain, contrasting with the compact restricted labeling of γH2AX to the XY-containing Sex Body domain of control pachytene spermatocytes (SI Appendix, Fig. S8B). Interestingly, similar XY synapsis errors were observed in our Rnf212-deficient mice (53) (Fig. 4F and SI Appendix, Fig. S8 A and B), an observation that was overlooked or not observed in a previous study of a different Rnf212-deficient mouse line (9, 16). Finally, and given the interplay between the E3 ligases, we also evaluated the genetic interactions and loading dependency of RNF212 and Hei10 with RNF212B. The results show that RNF212 and HEI10 foci are not loaded in Rnf212b-deficient spermatocytes, despite the expression of both RNF212 and HEI10 proteins in testis tissue extracts from this mutant (Fig. 4G and SI Appendix, Fig. S8C).
RNF212B Is Not Required for Early DSB Induction and Repair Events but Is Essential for CO and Chiasmata Formation.
To evaluate DSB induction and early processing, we used antibodies against the phosphorylated form of histone H2AX (γH2AX) and the RecA homolog, DMC1. In early prophase I, prior to Sex Body formation, γH2AX marks sites across the genome that have undergone DSB induction and thus can identify defects in SPO11-mediated initiation of recombination. As expected by the temporal pattern of RNF212B localization, Rnf212b−/− males showed no significant differences from WT mice both for DSB induction in leptonema, as well as for the accumulation DMC1 in zygonema. Importantly, DMC1 disappears in pachynema, indicating the normal progression of early DSB processing events (SI Appendix, Fig. S9).
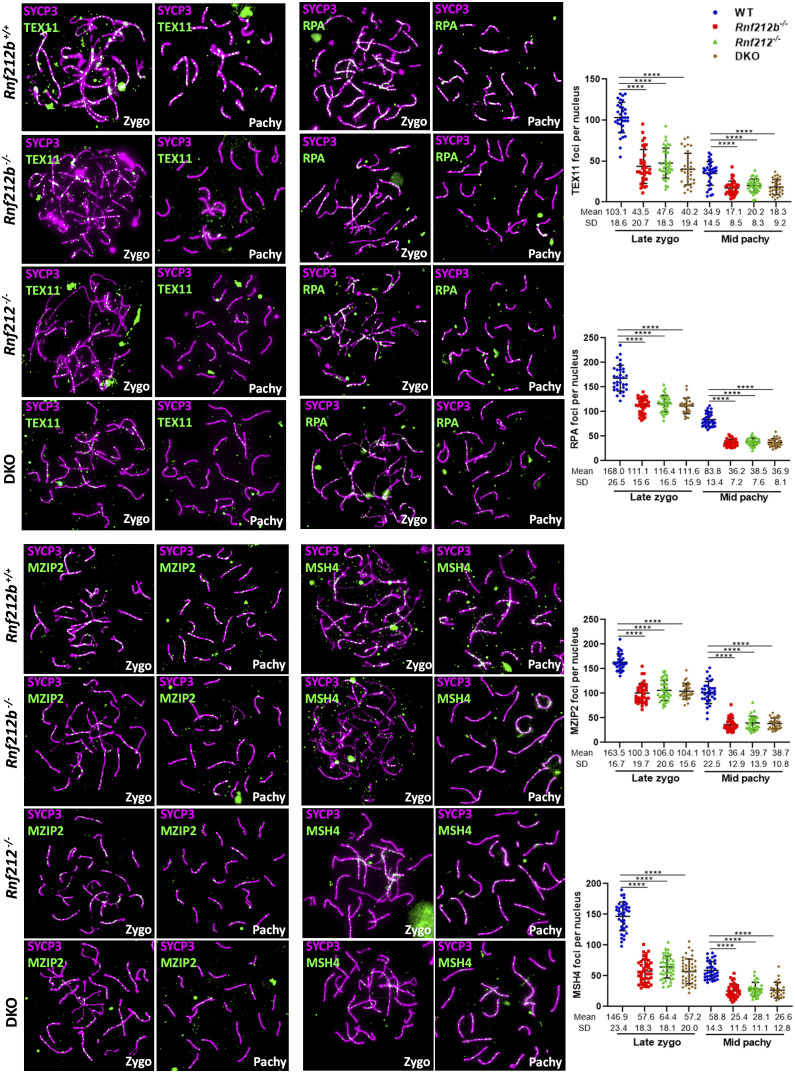
We next conducted a comprehensive assessment of DSB repair processing on chromosome spreads from Rnf212b−/− spermatocytes compared to WT spermatocytes. We utilized a range of antibodies against pro-CO factors, including the orthologs of yeast ZMM proteins (54), TEX11 (yeast Zip4), MZIP1 (yeast Zip2), MSH4/MSH5, and RPA. Although the initial formation of TEX11, MZIP2, MSH4, and RPA (55) foci appeared normal (SI Appendix, Fig. S10), their localization was significantly reduced in Rnf212b−/− (as well as in Rnf212−/−) from late zygonema onward (Fig. 5), indicating a pronounced deficiency in CO licensing. This reduction was also observed in oocytes (SI Appendix, Fig. S11A).
Fig. 5.
Localization of pro-CO factors is decreased in wild-type, Rnf212b−/−, Rnf212−/−, and Rnf212b−/− Rnf212−/− (DKO) spermatocytes. Double immunolabeling of SYCP3 and TEX11, RPA, MZIP2, or MSH4. In late zygonema and mid pachynema, foci were significantly reduced in Rnf212b−/−, Rnf212−/−, and Rnf212b−/− Rnf212−/− (DKO) in comparison with the wild-type control. Plots in the Right of the panels represent the quantitation of the values. Two-tailed Welch’s t test analysis: ****P < 0.0001. Bar in all panels, 10 μm. All experiments have been carried out in at least three mice and 15 cells per mouse.
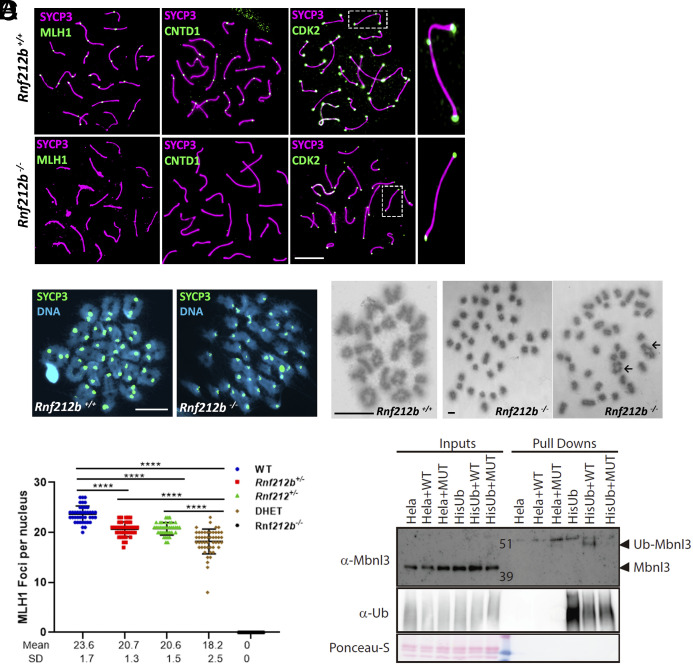
To assess CO designation and maturation, we performed immunofluorescence localization for CO-associated proteins: MLH1, CNTD1, CDK2, and HEI10. In Rnf212b−/− spermatocytes, we found no pachytene accumulation of any of these markers, while Rnf212b+/+ spermatocytes showed the usual 1-2 foci per chromosome (Figs. 4G and 6A). Telomere-associated CDK2 localization, a pattern that is not associated with DSB repair, was normal in Rnf212b−/− spermatocytes. The failure to load MLH1, CNTD1, interstitial CDK2, and HEI10 indicates that crossing over through the major CO pathway is severely disrupted in the absence of RNF212B (Figs. 4G and 6A). Subsequently, we studied the presence of chiasmata, the physical manifestation of crossing over. Using the standard air drying technique (56), most of the metaphase I/diakinesis from mutant spermatocytes displayed univalents with only a fraction forming chiasmata (0.55 chiasma/metaphase I), in contrast with wild-type spermatocytes (24.5 chiasmata/metaphase I) (Fig. 6B). Similarly, pachytene oocytes from Rnf212b−/− females lack MLH1 foci and metaphase I chromosomes have no chiasmata (SI Appendix, Fig. S11 B and C).
Fig. 6.
Major CO pathway disrupted in the absence of RNF212B, and MBNL3 ubiquitination by RNF212B. (A) Double immunolabeling of SYCP3 with MLH1, CNTD1, and CDK2 in Rnf212b−/−and wild-type mouse spermatocytes (Rnf212b+/+). Right magnified panels show the presence (Up) and absence (Low) of an interstitial CDK2 foci at a CO are shown. (B) Metaphase I cells from chromosome spreads (stained with SYCP3 and DAPI, Left panel) and from Methanol/Acetic chromosome preparations stained with Giemsa (Right) show the presence of univalents (Rnf212b−/−) and a few bivalents (black arrows) in comparison with all bivalents (Rnf212b+/+). (C) Plot quantitation of the number of MLH1 foci in pachynema spermatocytes from Rnf212b+/+(WT), Rnf212b+/−, and Rnf212+/−single mutants, Rnf212+/− Rnf212b+/− (DHet) and Rnf212b−/−. Welch’s t test analysis: ****P < 0.0001. (D) Ubiquitination assay performed on Hela cells to verify MBNL3 ubiquitination by RNF212B activity. Hela cells stably expressing 10xHis-Ub were transfected with the indicated expression vectors. 10xHis-ubiquitin conjugated proteins were purified from cell lysates using Ni-NTA beads. Purified proteins (Pull-Downs) and Lysate samples (Inputs) were analyzed by western blotting using specific Ubiquitin antibody and MBNL3 antibody to detect its ubiquitinated form. We observe a 10 KDa higher band corresponding to the ubiquitinated form of MBNL3 that is only detectable when transfecting HELA-Ub with RNF212B-WT expression vector. No band is detected when transfecting RNF212B dead mutant form (MUT). Bar in all panels, 10 μm. All experiments have been carried out in at least three mice and 12 cells per mouse in (A) and (C), and 5 cells in (B).
Our results presented herein have demonstrated that RNF212B and RNF212 interact and colocalize, and their genetic depletion shows similar meiotic phenotypes. These observations suggest a potential collaborative and codependent function. To assess their genetic interaction, we generated double heterozygous Rnf212b+/−, Rnf212+/− and double Rnf212B−/−, Rnf212−/−mice. Unexpectedly, double heterozygous mice showed a more severe phenotype in the number of nascent COs with fewer MLH1 foci (18.20 ± 2.47) relative to the single heterozygote mutants (20.62 ± 1.47 in Rnf212b+/− and 20.74 ± 1.25 in Rnf212+/−) (Fig. 6C and SI Appendix, Fig. S11D), indicating a dosage-sensitive and cumulative effect of Rnf212 and Rnf212b on CO maturation. In fact, 37.5% of the double heterozygous mice failed to have offspring after mating for 3 mo (n = 16 mice). However, the phenotype of the double mutant for Rnf212 and Rnf212b, ultimately leading also to a reduction of TEX11, MZIP2, MSH4, and RPA foci, was indistinguishable from the corresponding single null mice attending also to the presence of aberrant XY chromosomes and aberrant É£H2AX staining (Figs. 4F and 5 and SI Appendix, Fig. S8B).
In Vivo Evidence Supports a Critical Role for RNF212B Ubiquitin E3 Ligase Activity in Meiotic Prophase I.
Although RNF212 has been implicated in the maturation of COs events through its proposed role as a SUMO E3 ligase (16, 43), formal proof of this enzymatic activity is lacking. Given the promiscuity of the in vitro assays for Ub or SUMO activity, we endeavored to gain insight into which proteins are SUMOylated in the testis, by comparing the SUMOylome of young WT and mutant Rnf212b mice together with Hei10 deficient mice as an additional control with a similar meiotic arrest phenotype but with a different molecular target. We utilized the sole methodology capable of purifying and identifying lysine residues modified by endogenous SUMO2/3 (57), combined with selective high-resolution mass spectrometry (58). From quintuplicate analyses, we identified 1,115 unique SUMO2/3-modified peptides, corresponding to 857 unique SUMO2/3 modification sites (Dataset S2), across 477 SUMO target proteins (Dataset S2); representing the most comprehensive mouse testis SUMOylome to date. Numerically, 48.2% of SUMO sites adhered to the KxE consensus motif. When also accounting for the biological abundance of each SUMOylation site, 64.2% of total SUMO modified proteins resided on KxE motifs. The SUMOylomes were highly reproducible, with an average Pearson correlation of 0.84, and the most abundant SUMO target proteins were virtually all nuclear proteins and possessed canonical SUMO functions such as transcriptional regulation, chromosome organization, and modulation of DNA repair. Intriguingly, we did not observe a notable rewiring of the global SUMOylome in either Rnf212b- or Hei10-knockout mice, suggesting either that RNF212B has no SUMO E3 activity, or has activity that is targeted very specifically to proteins that have yet to be determined (Dataset S2).
To investigate the E3 ligase activity of RNF212B in an in vivo cell system, we conducted an unbiased pull-down assay in HeLa cells that stably expressed either 10XHis-ubiquitin or 10xHis-SUMO. Following infection with RNF212B or an inactive RNF212B mutant coupled with proteasome inhibition, pulldowns were performed using Ni-NTA beads, and the enriched proteins were identified using mass spectrometry (Datasets S3 and S4). While the SUMO analysis did not yield any definitive candidates, the ubiquitin assay identified a clear candidate, the splicing factor MBNL3, as a potential RNF212B-dependent ubiquitylated protein. To validate these results further, purified proteins (from pull-downs) and lysate samples (inputs) were analyzed using western blotting with specific antibodies against ubiquitin and MBNL3 to detect its ubiquitinated form. We observed a higher 10 KDa band corresponding to the ubiquitinated form of MBNL3, which was only detectable upon infection wild-type RNF212B in HeLa cells stably expressing HisUb (Fig. 6D). No band was detected when infecting with the catalytically inactive mutant form of RNF212B (MUT). This observation confirms that the ubiquitination activity of RNF212B on MBNL3 in the in vivo HelA cell system is dependent on the catalytic activity of the E3 ligase (Fig. 6D and SI Appendix, Fig. S12).
Finally, we investigated whether the catalytic E3 ligase activity of RNF212B is crucial for its biological function within the proteinaceous complex or if it functions solely as a scaffold. To address this, we generated mutant mice with a histidine-to-alanine (H24A) substitution in the catalytic tetrad, resulting in a RING E3 ligase dead mutant protein (SI Appendix, Fig. S7C) (59). As histidine substitutions in the catalytic domain can impact proper folding (59), we initially analyzed RNF212B loading dynamics, which appeared normal in the Rnf212bH24A/H24A mutants (SI Appendix, Fig. S13A). Cytological examination of Rnf212bH24A/H24A spermatocytes recapitulated the phenotype observed in Rnf212b−/− mice, including synapsis and aberrant XY chromosomes, univalents at metaphase I (0.64 chiasma/metaphase I), and decreased loading of TEX11 and MSH4 foci (SI Appendix, Fig. S13 B–D). Collectively, these findings strongly suggest that the E3 ligase activity of RNF212B is indispensable for its essential pro-CO function.
Discussion
The establishment and resolution of COs between homologous chromosomes occurs in a tightly regulated fashion during prophase I and is vital ensuring their proper segregation at metaphase I. Of the 250 DSBs that form in early prophase I in the mouse, approximately 90% are repaired as NCOs beginning in zygonema. The remaining 10% of DSB events are processed largely through the class I CO pathway involving the MutSγ and MutLγ heterodimers (10, 12, 14, 60, 61) (SI Appendix, Fig. S14). In this process, MutSγ loading on to a defined proportion of DSB repair intermediates marks the process of CO licensing, followed by a further reduction of intermediates which then load the MutLγ complex, together with HEI10, CNTD1, and CDK2, in the process of CO designation (3). Together, these canonical markers of Class I COs facilitate final CO maturation. The regulatory mechanisms governing the selective licensing and designation of DSBs for CO resolution remain poorly understood across many meiotic species but clearly involve an array of auxiliary factors Pro-CO factors with roles in cell cycle regulation, protein–protein interactions, and posttranslational modifications of key CO proteins (17, 43–46, 62).
In the current study, we have further refined the Pro-CO family by the characterization of a RING-finger E3 ligase named RNF212B which is indispensable for CO formation. The genetic depletion of RNF212B in mice results in loss of CO designation that ultimately leads to loss of most COs, similar to other Class I regulators and Pro-CO proteins, including MLH1, MLH3, CNTD1, RNF212, and HEI10, (14, 17, 46, 61), resulting in the loss of most chiasmata. Consistent with this, we observe reduced localization of earlier CO mediators, including MSH4, TEX11, and RPA. Moreover, the observed biochemical and genetic interactions between RNF212B and other Pro-CO factors, including RNF212 and HEI10, suggest that RNF212B functions either directly or indirectly with these other E3 ligases in the process of CO designation. Importantly, our results also suggest that the function of RNF212B in CO designation is dependent on its catalytic activity. These findings are supported in part by recent studies by Ito and colleagues who have generated another knockout allele of Rnf212b (63) and by the recent identification of an infertile man with a pathogenic variant in RNF212B (64).
Meiotic recombination and chromosome synapsis during SC assembly are well known to be functionally coupled processes through mechanisms that are poorly understood in mammals. After DSBs are generated by SPO11 and its accessories factors, the meiotic recombination machinery components (MSH4, MSH5, RPA) of early RNs assemble in a DSB-dependent manner (42, 53). Counterintuitively, however, intermediate and late RNs, as defined by their RNF212, MLH1, CNTD1, and HEI10 accumulation, are loaded onto the axes independently of DSBs but in a synapsis- and CE-dependent manner (16, 43, 65). The observed synapsis-dependent loading of RNF212B onto the SC occurs not only on the normally synapsed axes of zygotene and pachytene spermatocytes but also in mutants exhibiting synapsis defects between nonhomologous chromosomes or between sister chromatids. Thus, RNF212B can localize to SCs independent of recombination and even when SC formation is aberrant. However, it remains unknown what subunit of the CE mediates this RNF212B loading. Of note, the spontaneous ring structures obtained in somatic cells overexpressing Hei10, which recruit RNF212 and RNF212B, resemble C. elegans polycomplexes of SC proteins that exhibit liquid-like behaviors (66, 67), suggesting a similar liquid crystal-like mechanism for RN maturation in the mouse (35, 68, 69).
Given the similar meiotic phenotypes we describe herein for null mutants of Rnf212 and Rnf212b, along with the observed interaction between both RING-finger proteins, one would predict that phenotype of double mutant mice would be indistinguishable from single mutant mice (70). However, the observed phenotype in Rnf212 and Rnf212b double heterozygous mutants, suggests the existence of a critical threshold of activity below of which functional activity is severely compromised. The identification of two essential pro-COs RING-finger E3 ligases acting at the level of CO designation has also been reported for the worm ZHP-3 and ZHP-4 (44, 71). Vertebrates show a 1:1 ortholog relation between Rnf212b and Rnf212, indicating evolutionary conservation of the process as observed in the foci distribution of RNF212B in human and dog spermatocytes. Thus, a set of RING-finger ligases evolutionary conserved from yeast and worms to mammals regulate CO homeostasis. Excitingly in the mouse, there appears to be unique interdependence of the three pro-CO Ring Finger E3 ligases: the loading of RNF212 and RNF212B are codependent, while the loading of HEI10 is dependent on both RNF212 and RNF212B. Conversely, the timely unloading of RNF212 and RNF212B is dependent on HEI10.
Despite completing the most comprehensive mouse testis SUMOylome to date, we find no global alterations in the SUMOylome in Rnf212b and Hei10 mutant males. Furthermore, we observe only Ubiquitin, not SUMO, activity of RNF212B in somatic cells, strongly suggesting that the function of RNF212B in CO maturation does not involve widespread SUMO E3 ligase activity. In addition, given that genetic depletion of RNF212B also leads to a concomitant functional depletion of RNF212 (and vice versa), we would expect to see a loss of widespread RNF212-associated SUMO activity in our Rnf212b-deficient mice, but this is not the case. Thus, our conclusions do not support a model in which RNF212 drives widespread SUMOylation including key regulators of CO designation and maturation, as proposed previously (16). It is possible that the loss of RNF212B leads to increased activity of other SUMO E3 ligases, but these proteins were not revealed in our mass spectrometry analysis and can only be speculative for now. Furthermore, we cannot rule out functions of RNF212 or RNF212B outside of the nucleus or independent of each other or in somatic tissues.
Taken together, we have refined the Pro-CO family by the functional characterization of a RING-finger E3 ligase, RNF212B, that is indispensable for CO maturation (SI Appendix, Fig. S9). Together with HEI10 and RNF212, this brings to three the number of E3 ligases implicated in CO control. Our in vitro evidence points to a Ubiquitin E3 ligase activity for RNF212B, while previous studies have ascribed a similar function to HEI10 (3, 16, 43, 72). Verification of the in vivo activities of all these Pro-CO factors will shed further light on the mechanisms of posttranslation modifications orchestrated in a stepwise fashion to drive CO designation and maturation. We envisage a model in which each RING-finger ligase will have its own set of target substrates that must be modified in a specific sequence either to promote crossing over, or to divert the DSB repair intermediate toward a NCO fate. For example, an attractive hypothesis is one that involves RNF212B-mediated modification of one of the late RN/Class I CO mediators. Such posttranslational modifications, which are not restricted to SUMOylation or ubiquitylation, but might also include phosphorylation, have previously been identified for early RN components such as yeast Msh4 (73), but not yet for components of the late RN/CO Designation machinery. Thus, we propose that a primary function of RNF212B is to ubiquitylate, and thus activate/recruit key components of the CO Designation machinery either directly or indirectly, leading to activation of the MutLγ complex. Our IP-MS and IP-WB results suggest that PCNA might be one potential regulator of this process: In somatic cells, PCNA is critical for canonical MMR activity involving various heterodimers of the MutL homologs in a manner that is dependent on mono- and poly-ubiquitylation at lysine 164 (51). Furthermore, loss of this ubiquitylation activity in meiotic cells causes a phenotype that is similar to that of the Rnf212B mutant presented herein (16). However, while PCNA is just one possible link between RNF212B and the CO Designation machinery, the general model we propose could indeed apply to other key mediators of CO designation. Such a model makes it easy to visualize how subtle variations in RNF212B structure can alter the CO frequency (genome-wide) and distribution, leading to outcomes such as infertility, aneuploidy, and birth defects. In essence, our elucidation of this intricate interplay advances our understanding of the conserved pathway of meiotic dynamics through the regulation of CO designation and maturation.
Materials and Methods
Generation of CRISPR/Cas9-Edited Mice.
For developing the Rnf212b−/− loss of function mice, two sgRNAs targeting the exon 2 were used. For developing the Rnf212bH24A/H24A model, a single sgRNA1 targeting exon 4 was used with a template ssODN that contains the mutation (CAT>GCC, that results in the change of His>Ala). The crRNAs, the tracrRNAs, and the ssODNs were produced by chemical synthesis by IDT. The crRNA and tracrRNA were annealed to obtain the mature sgRNA and complexed with the Cas9 protein and ssODN before being microinjected into B6/CBA F2 zygotes. Edited founders were identified by PCR amplification and Sanger sequencing. Selected Rnf212b founders, carrying the desired alleles, were crossed with wild-type C57BL/6 J mice to generate the edited homozygotes Rnf212b−/− mice. Hemizygous Rnf212bH24A/H24A mice were directly processed for the analysis. Rnf212−/−, Hei10−/−, Msh4−/−, Cntd1−/− mice were previously described (10, 45, 53). (see SI Appendix for the sequence of ODNs).
Immunocytology and Antibodies.
Testes and ovaries (E16.5-18.5 embryos) were processed for spreading using a conventional “dry-down” technique. Affinity-purified Abs against RNF212B recombinant protein (Escherichia coli) were produced by Proteogenix. Meiocytes from males and females were incubated with primary antibodies for immunofluorescence (see list in SI Appendix, Materials and Methods). The secondary antibodies used conjugated with fluorochromes were goat α-mouse, donkey α-rabbit, goat α-rat, and goat Fab α-rabbit. Slides were visualized using a microscope (Axioplan2; Carl Zeiss, Inc.) with 63× objectives. Images were taken with an ORCA-ER (Hamamatsu) and processed with OPENLAB 4.0.3 and Photoshop (Adobe).
Stimulated Emission Depletion Microscopy.
Stimulated emission depletion (STED) microscopy (SP8, Leica) was used to generate the superresolution images. Fluorescence signals were measured along the 19 autosomal AEs of pachytene cells using the “Plot profile” tool of ImageJ. Signal intensities were standardized, acquiring values between −1 and + 1, and the overlay profiles of RNF212B and other RNF212 were plotted. Regression analysis for the pair of proteins was performed to determine the correlation between their profiles.
Oocyte Quantification.
Histologically fixed ovaries were serially sliced into 5 μm thick sections and stained with hematoxylin and eosin. For each mouse, the number of oocytes was counted in every fifth section of one ovary, and the sum of oocytes from all counted sections was considered the total number of oocytes per ovary.
Generation of Plasmids.
Full-length cDNAs encoding RNF212, RNF212B, and HEI10 were RT-PCR amplified from testis RNA and cloned into pcDNA3 2xFlag, pcDNA3 2xHA, pmCherry-C1, and pEGFP-C1 mammalian expression vectors under the CMV promoter. Mutant constructs of the HEI10, RNF212B, and RNF212 bearing the two Cys>Ala mutations in the Zn domain (MUT), were carried out by PCR mutagenesis (see SI Appendix for additional information).
Immunoprecipitation and Western Blotting.
HEK293T cells were transiently transfected and whole cell extracts were cleared with protein G Sepharose beads (GE Healthcare) and incubated with the corresponding antibodies. Immunocomplexes were isolated by adsorption to protein G-Sepharose beads o/n. After washing, the proteins were eluted from the beads with SDS. Proteins were detected by western blotting with the indicated antibodies. Antibodies were detected by using Immobilon Western Chemiluminescent HRP Substrate from Millipore.
Statistics.
In order to compare counts between genotypes, we used the Welch’s t test (unequal variances t test). We applied a two-sided test in all the cases. Asterisks denote statistical significance: *P-value < 0.05, **P-value < 0.01, ***P-value < 0.001 and ****P-value < 0.0001.
Ethics Statement.
Mice were housed in a temperature-controlled facility (specific pathogen free) using individually ventilated cages, standard diet, and a 12 h light/dark cycle, according to EU laws. Mouse protocols have been approved by the Ethics Committee for Animal Experimentation of the University of Salamanca (USAL).
Human and Dog Samples.
The human sample was obtained from CREA fertility clinic (Valencia, Spain) and was deidentified prior to its use in the study to maintain patient confidentiality and compliance with ethical guidelines. The dog sample was obtained from Cabrerizos veterinary clinic (Salamanca, Spain).
Hela Transduction and His-Pull Downs.
HELA cells stably expressing either 10xHis-ubiquitin or 10xHis-SUMO (15 plates) were infected with FLAG-RNF212b plasmid (empty vector, WT, or MUT). Forty-eight hours postinfection, cells were scraped in ice-cold PBS and an input sample was taken for immunoblotting analysis. The rest was lysed Guanidinium lysis buffer for subsequent His-Pull down experiments (69). (see SI Appendix for details). For electrophoresis and immunoblotting, samples were separated on Novex 4 to 12% gradient gels (Thermo Fisher Scientific) using NuPAGE® MOPS SDS running buffer and transferred onto Nitrocellulose membranes (see SI Appendix for details). Trypsin digestion of His-Ubiquitin and His-SUMO2/3 purified conjugates were carried out as follows: Eluted proteins were supplemented with ABC to 50 mM. Subsequently, samples were reduced with 1 mM dithiothreitol (DTT) for 30 min and alkylated with 5 mM chloroacetamide (CAA) for 30 min. After an additional reduction with 5 mM DTT for 30 min at RT, conjugates were diluted with 50 mM ABC and digested with 500 ng of sequencing-grade modified trypsin (Promega) o/n in dark.
Mass Spectrometry Sample Preparation for Pull Down Analysis.
Digested peptides were acidified by adding 2% TriFlourAcetic (TFA) acid. Subsequently, peptides were desalted and concentrated (74). Stage-tips were activated by passing through methanol and after elution in acetonitrile/formic acid samples were and stored at −20 °C prior to mass spectrometry analysis. All the experiments were analyzed by on‐line nanoHPLC MS/MS with a system consisting of an EASY-nLC 1200 system (Thermo) coupled to an Orbitrap Fusion Lumos Trybrid mass spectrometer (Thermo). Samples were measured in a 2-column setup. The mass spectrometer was operated in data-dependent MS/MS mode (see SI Appendix for details).
Mass Spectrometry Data Analysis in Pull-Down Analysis.
All raw data were analyzed using MaxQuant (version 1.6.14) as previously described (75). We performed the search against an in silico digested UniProt reference proteome for. All peptides were used for protein quantification. Output from MaxQuant Data were processed in MS Excel for further filtering and visualization. For the statistical analysis, output from the analysis in MaxQuant was further processed in the Perseus computational platform version 1.6.14 (76).
Preparation of Germ Cell Lysates, Immunoprecipitation, and Western Blotting.
Germ cell suspension was prepared from 12-wk-old C57BL6/J males (SI Appendix). Pelleted cells were resuspended in Lysis Buffer, sonicated, and cleared by centrifuging. For the germ cell suspension IP, 2.95 mg protein lysate was used for each IP. Input gel sample corresponds to ~5% of the total input for each IP. For each IP, 10 µg of each primary antibody and normal rabbit IgG was added and incubated overnight at 4 °C. Protein A Dynabead slurry was added to antibody/lysate reaction and incubated. Beads were then washed and bound protein was eluted at 65 °C. One-half of each IP gel sample (elutions and input) was used for western blot analysis (see SI Appendix for details).
SUMOylome Analysis from Mouse Samples.
We performed the sumoylome on an n = 5 analysis of the samples. To maximize the yield, we pooled 8 testes per replicate for wild type (labeled as “C” in the data), eight testes per replicate for Rnf212b KO (labeled “RKO”), and 5 testes per replicate for Hei10 KO (labeled “HKO”). Wild-type controls were 17-19 dpp to avoid spermatocytes as Rnf212b and Hei10 lack. Testes were stored at −80 °C. Homogenization of testes and subsequent purification of peptides modified by endogenous SUMO2/3 was performed as described previously (57) (see SI Appendix for details).
Peptide Preparation for Endogenous SUMO-IP.
Samples were digested with Endoproteinase Lys-C (Wako) in a 1:200 enzyme-to-protein ratio (w/w). Digested samples (room temperature) were diluted with ammonium bicarbonate (ABC) to dilute guanidine, redigested with Endoproteinase Lys-C, and after acidification with TFA, were centrifuged. Peptides from the clarified were purified using C8 Sep-Pak cartridges (Waters). SUMOylated peptides were eluted and frozen overnight at −80 °C. Deep-frozen samples were lyophilized to dryness for 96 h.
StageTip Purification and High-pH Fractionation of SUMO-IP Samples.
Preparation of StageTips and high-pH fractionation of SUMO-IP samples on StageTip was previously described (57, 77). Quad-layer StageTips were prepared using four punch-outs of C18 material (Sigma-Aldrich, Empore™ SPE Disks, C18, 47 mm). Samples were loaded onto equilibrated StageTips, washed twice with 150 μL 50 mM ammonium, and eluted as six fractions using increased concentrations from 4 to 25% ACN in 50 mM ammonium. All fractions were dried and peptides dissolved using in 0.1% formic acid (see SI Appendix for details).
Statistical Processing of SUMOylome MS Data.
To further minimize false-positive discovery, additional manual filtering was performed at the peptide level (SI Appendix). Only proteins containing at least one SUMO site were considered as SUMO target proteins, and other putative SUMO target proteins were discarded. Statistical handling and quantification of SUMO site and SUMO target protein data was performed using Perseus software (versions 1.5.5.3 and 1.6.14.0) (76) (see SI Appendix for description).The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (78) partner repository with the dataset identifiers PXD050239, PXD047332, and PXD047019 (79–81).
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Acknowledgments
This research was funded by the Ministerio de Ciencia e Innovación (PID2020–120326RB-I00 and PID2023-152857NB-I00) and Junta de Castilla y León (Unidad de Investigación Consolidada 066, CSI148P20 and CSI017P23) and R.G.-P. is funded by Dutch Cancer society, KWF Kankerbestrijding, Young Investigator Grant (11367/2017-2), and by Consejería de Economía, Conocimiento, Empresas y Universidad, Junta de Andalucía, Programa EMERGIA (EMERGIA20_00276). P.E.C., I.D.W., and R.B. are supported by funding from the Eunice Kennedy Shriver National Institute of Child Health and Development (HD041012 to P.E.C., HD106630 to I.D.W.). R.B. is supported by a graduate fellowship from The Natural Sciences and Engineering Research Council of Canada (PGSD-577969). R.S.-U. and Y.B.C. are funded by Ministerio de Ciencia (FPU) and L.G.-H. is funded by Max planck and JCyL (beginning of the project). M.L.N. lab was supported by the Novo Nordisk Foundation (NNF14CC0001). The proteomics technology applied was part of a project that has received funding from the European Union’s Horizon 2020 research and innovation program (EPIC-XS-823839). We are indebted to Attila Toth (Universitat Dresden, Germany), Neil Hunter (UC Davis, California, USA), Chao Yu (University of Gothenburg, Sweden), Christer Hoog (Karolinska Inst., Sweden), and Jeremy Wang (University of Pennsylvania) for providing Abs against CNTD1, RNF212, MZIP2, SYCE1, and TEX11 respectively. We thank J.M. Pereda (Centro de Investigación del Cáncer, Spain) for structural Zn-ring domain 3D modelling.
Author contributions
P.E.C., A.M.P., and E.L. designed research; Y.B.C., R.S.-U., L.G.-H., D.S.-L., N.F.-M., R.B., I.D.W., M.S.-M., I.A.H., M.L.N., R.G.-P., P.E.C., and E.L. performed research; J.L.B., M.L.N., and P.E.C. contributed new reagents/analytic tools; L.G.-H., S.T., D.d.R., A.M.P., and E.L. analyzed data; and P.E.C., A.M.P., and E.L. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Alberto M. Pendas, Email: amp@usal.es.
Elena Llano, Email: ellano@usal.es.
Data, Materials, and Software Availability
Proteomics data have been deposited in ProteomeXchange (PDX047332 and PXD047019) (80, 81). All other data are included in the manuscript and/or supporting information.
Supporting Information
References
- 1.Zickler D., Kleckner N., Meiosis: Dances between homologs. Annu. Rev. Genet. 57, 1–63 (2023). [DOI] [PubMed] [Google Scholar]
- 2.Llano E., Pendas A. M., Synaptonemal complex in human biology and disease. Cells 12, 1718 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray S., Cohen P. E., Control of meiotic crossovers: From double-strand break formation to designation. Annu. Rev. Genet. 50, 175–210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoop-Myer C., Amon A., Meiosis: Rec8 is the reason for cohesion. Nat. Cell Biol. 1, E125–127 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Kudo N. R., et al. , Role of cleavage by separase of the Rec8 kleisin subunit of cohesin during mammalian meiosis I. J. Cell Sci. 122, 2686–2698 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jessberger R., Age-related aneuploidy through cohesion exhaustion. EMBO Rep. 13, 539–546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S., et al. , Inefficient crossover maturation underlies elevated aneuploidy in human female meiosis. Cell 168, 977–989.e917 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagaoka S. I., Hassold T. J., Hunt P. A., Human aneuploidy: Mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 13, 493–504 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reynolds A., et al. , RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat. Genet. 45, 269–278 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kneitz B., et al. , MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 14, 1085–1097 (2000). [PMC free article] [PubMed] [Google Scholar]
- 11.Edelmann W., et al. , Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 21, 123–127 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Kolas N. K., et al. , Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J. Cell Biol. 171, 447–458 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker S. M., et al. , Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13, 336–342 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Edelmann W., et al. , Meiotic pachytene arrest in MLH1-deficient mice. Cell 85, 1125–1134 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Moens P. B., et al. , The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J. Cell Sci. 115, 1611–1622 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Rao H. B., et al. , A SUMO-ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science 355, 403–407 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward J. O., et al. , Mutation in mouse hei10, an e3 ubiquitin ligase, disrupts meiotic crossing over. PLoS Genet 3, e139 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhalla N., Wynne D. J., Jantsch V., Dernburg A. F., ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 4, e1000235 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lake C. M., et al. , Vilya, a component of the recombination nodule, is required for meiotic double-strand break formation in Drosophila. Elife 4, e08287 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y., et al. , COSA-1 mediated pro-crossover complex formation promotes meiotic crossing over in C. elegans. Nucleic Acids Res. 52, 4375–4392 (2024), 10.1093/nar/gkae130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agarwal S., Roeder G. S., Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102, 245–255 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Wang T., et al. , HEI10 is subject to phase separation and mediates RPA1a degradation during meiotic interference-sensitive crossover formation. Proc. Natl. Acad. Sci. U.S.A. 120, e2310542120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sriramachandran A. M., Dohmen R. J., SUMO-targeted ubiquitin ligases. Biochim. Biophys. Acta 1843, 75–85 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Wilkinson K. A., Henley J. M., Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 428, 133–145 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins G. A., Goldberg A. L., The logic of the 26S proteasome. Cell 169, 792–806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo X., et al. , Site-specific proteasome phosphorylation controls cell proliferation and tumorigenesis. Nat. Cell Biol. 18, 202–212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belle A., Tanay A., Bitincka L., Shamir R., O’Shea E. K., Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. U.S.A. 103, 13004–13009 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glotzer M., Murray A. W., Kirschner M. W., Cyclin is degraded by the ubiquitin pathway. Nature 349, 132–138 (1991). [DOI] [PubMed] [Google Scholar]
- 29.Meyer H. J., Rape M., Enhanced protein degradation by branched ubiquitin chains. Cell 157, 910–921 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong A., et al. , Common and low-frequency variants associated with genome-wide recombination rate. Nat. Genet. 46, 11–16 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Kong A., et al. , Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467, 1099–1103 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Petit M., et al. , Variation in recombination rate and its genetic determinism in sheep populations. Genetics 207, 767–784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritz K. R., Noor M. A. F., Singh N. D., Variation in recombination rate: Adaptive or not? Trends Genet. TIG 33, 364–374 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Baudat F., et al. , PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327, 836–840 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herran Y., et al. , The cohesin subunit RAD21L functions in meiotic synapsis and exhibits sexual dimorphism in fertility. EMBO J. 30, 3091–3105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez H. L., et al. , C14ORF39/SIX6OS1 is a constituent of the synaptonemal complex and is essential for mouse fertility. Nat. Commun. 7, 13298 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halldorsson B. V., et al. , Characterizing mutagenic effects of recombination through a sequence-level genetic map. Science 363, eaau1043 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Kadri N. K., et al. , Coding and noncoding variants in HFM1, MLH3, MSH4, MSH5, RNF212, and RNF212B affect recombination rate in cattle. Genome Res. 26, 1323–1332 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bannister L. A., Reinholdt L. G., Munroe R. J., Schimenti J. C., Positional cloning and characterization of mouse mei8, a disrupted allelle of the meiotic cohesin Rec8. Genesis 40, 184–194 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Xu H., Beasley M. D., Warren W. D., van der Horst G. T., McKay M. J., Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev. Cell 8, 949–961 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Llano E., et al. , Meiotic cohesin complexes are essential for the formation of the axial element in mice. J. Cell Biol. 197, 877–885 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baudat F., Manova K., Yuen J. P., Jasin M., Keeney S., Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6, 989–998 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Qiao H., et al. , Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat. Genet. 46, 194–199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yokoo R., et al. , COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149, 75–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gray S., Santiago E. R., Chappie J. S., Cohen P. E., Cyclin N-terminal domain-containing-1 coordinates meiotic crossover formation with cell-cycle progression in a cyclin-independent manner. Cell Rep. 32, 107858 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holloway J. K., Sun X., Yokoo R., Villeneuve A. M., Cohen P. E., Mammalian CNTD1 is critical for meiotic crossover maturation and deselection of excess precrossover sites. J. Cell Biol. 205, 633–641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakatani Y., et al. , Regulation of ubiquitin transfer by XIAP, a dimeric RING E3 ligase. Biochem. J. 450, 629–638 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Chasapis C. T., Spyroulias G. A., RING finger E(3) ubiquitin ligases: Structure and drug discovery. Curr. Pharm. Design 15, 3716–3731 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Cannavo E., et al. , Regulation of the MLH1-MLH3 endonuclease in meiosis. Nature 586, 618–622 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Kulkarni D. S., et al. , PCNA activates the MutLgamma endonuclease to promote meiotic crossing over. Nature 586, 623–627 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boehm E. M., Gildenberg M. S., Washington M. T., The many roles of PCNA in eukaryotic DNA replication. Enzymes 39, 231–254 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferguson-Smith M. A., X-Y chromosomal interchange in the aetiology of true hermaphroditism and of XX Klinefelter’s syndrome. Lancet 2, 475–476 (1966). [DOI] [PubMed] [Google Scholar]
- 53.Felipe-Medina N., et al. , A missense in HSF2BP causing primary ovarian insufficiency affects meiotic recombination by its novel interactor C19ORF57/BRME1. Elife 9, e56996 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pyatnitskaya A., Borde V., De Muyt A., Crossing and zipping: Molecular duties of the ZMM proteins in meiosis. Chromosoma 128, 181–198 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Yang F., et al. , Meiotic failure in male mice lacking an X-linked factor. Genes Dev. 22, 682–691 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Evans E. P., Breckon G., Ford C. E., An air-drying method for meiotic preparations from mammalian testes. Cytogenetics 3, 289–294 (1964). [DOI] [PubMed] [Google Scholar]
- 57.Hendriks I. A., et al. , Site-specific characterization of endogenous SUMOylation across species and organs. Nat. Commun. 9, 2456 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hendriks I. A., Akimov V., Blagoev B., Nielsen M. L., MaxQuant.Live enables enhanced selectivity and identification of peptides modified by endogenous SUMO and ubiquitin. J. Proteome Res. 20, 2042–2055 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Garcia-Barcena C., Osinalde N., Ramirez J., Mayor U., How to inactivate human ubiquitin E3 ligases by mutation. Front Cell Dev. Biol. 8, 39 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Toledo M., et al. , A mutation in the endonuclease domain of mouse MLH3 reveals novel roles for MutLgamma during crossover formation in meiotic prophase I. PLoS Genet. 15, e1008177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lipkin S. M., et al. , Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 31, 385–390 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Haversat J., et al. , Robust designation of meiotic crossover sites by CDK-2 through phosphorylation of the MutSgamma complex. Proc. Natl. Acad. Sci. U.S.A. 119, e2117865119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ito M., et al. , Distinct and interdependent functions of three RING proteins regulate recombination during mammalian meiosis. bioRxiv [Preprint] (2023). 10.1101/2023.11.07.566091 (Accessed 10 November 2023). [DOI]
- 64.Gershoni M., et al. , A pathogenic variant in the uncharacterized RNF212B gene results in severe aneuploidy male infertility and repeated IVF failure. HGG Adv. 4, 100189 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pyatnitskaya A., Andreani J., Guérois R., De Muyt A., Borde V., The Zip4 protein directly couples meiotic crossover formation to synaptonemal complex assembly. Genes Dev. 36, 53–69 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Laghmach R., Potoyan D. A., Liquid-liquid phase separation driven compartmentalization of reactive nucleoplasm. Phys. Biol. 18, 015001 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rog O., Kohler S., Dernburg A. F., The synaptonemal complex has liquid crystalline properties and spatially regulates meiotic recombination factors. Elife 6, e21455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cloutier J. M., et al. , Histone H2AFX links meiotic chromosome asynapsis to prophase i oocyte loss in mammals. PLoS Genet. 11, e1005462 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rinaldi V. D., Bolcun-Filas E., Kogo H., Kurahashi H., Schimenti J. C., The DNA damage checkpoint eliminates mouse oocytes with chromosome synapsis failure. Mol. Cell 67, 1026–1036.e1022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuzmin E., et al. , Systematic analysis of complex genetic interactions. Science 360, eaa01729 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim J., Ercan S., Cohesin mediated loop extrusion from active enhancers form chromatin jets in C. elegans. bioRxiv [Preprint] (2023). 10.1101/2023.09.18.558239 (Accessed 19 March 2024). [DOI]
- 72.Toby G. G., Gherraby W., Coleman T. R., Golemis E. A., A novel RING finger protein, human enhancer of invasion 10, alters mitotic progression through regulation of cyclin B levels. Mol. Cell Biol. 23, 2109–2122 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He W., et al. , Regulated proteolysis of MutSgamma controls meiotic crossing over. Mol. Cell 78, 168–183.e165 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rappsilber J., Mann M., Ishihama Y., Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Tyanova S., Temu T., Cox J., The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016). [DOI] [PubMed] [Google Scholar]
- 76.Tyanova S., et al. , The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 77.Theurillat I., et al. , Extensive SUMO modification of repressive chromatin factors distinguishes pluripotent from somatic cells. Cell Rep. 32, 108146 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Perez-Riverol Y., et al. , The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.González-Prieto R., RNF212B E3 ligase is essential for crossover designation and maturation during male and female meiosis in the mouse. PRIDE. https://www.ebi.ac.uk/pride/archive/projects/PXD050239). Deposited 29 February 2024. [DOI] [PMC free article] [PubMed]
- 80.González-Prieto R., RNF212B E3 ligase is essential for crossover designation and maturation during male and female meiosis in the mouse. PRIDE. https://www.ebi.ac.uk/pride/archive/projects/PXD047332. Deposited 28 November 2023. [DOI] [PMC free article] [PubMed]
- 81.Hendriks I., RNF212B E3 ligase is essential for crossing over maturation and mouse fertility. PRIDE. https://www.ebi.ac.uk/pride/archive/projects/PXD047019. Deposited 16 November 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Data Availability Statement
Proteomics data have been deposited in ProteomeXchange (PDX047332 and PXD047019) (80, 81). All other data are included in the manuscript and/or supporting information.