Significance
Extracellular vesicles (EVs) serve as one mechanism by which cells communicate with one another to coordinate behaviors, exchange small molecules, and alter their environment. During infection, many parasitic organisms, like Trichomonas vaginalis, are known to manipulate their host through the shedding of EVs and their subsequent internalization by host cells. Here, we explored the question of how parasite uptake of parasite-derived EVs affects communication between T. vaginalis strains and show that parasites up-regulate and directly transfer proteins that aid in adherence to host cells via EVs. This study broadens our understanding of the role of parasite EVs in infection and provides valuable insights that may be exploited to design strategies to use EVs in treatment and vaccination against this parasite.
Keywords: extracellular vesicles, host:pathogen interactions, Trichomonas vaginalis, dSTORM
Abstract
Trichomonas vaginalis, a common sexually transmitted parasite that colonizes the human urogenital tract, secretes extracellular vesicles (TvEVs) that are taken up by human cells and are speculated to be taken up by parasites as well. While the crosstalk between TvEVs and human cells has led to insight into host:parasite interactions, roles for TvEVs in infection have largely been one-sided, with little known about the effect of TvEV uptake by T. vaginalis. Approximately 11% of infections are found to be coinfections of multiple T. vaginalis strains. Clinical isolates often differ in their adherence to and cytolysis of host cells, underscoring the importance of understanding the effects of TvEV uptake within the parasite population. To address this question, our lab tested the ability of a less adherent strain of T. vaginalis, G3, to take up fluorescently labeled TvEVs derived from both itself (G3-EVs) and TvEVs from a more adherent strain of the parasite (B7RC2-EVs). Here, we showed that TvEVs generated from the more adherent strain are internalized more efficiently compared to the less adherent strain. Additionally, preincubation of G3 parasites with B7RC2-EVs increases parasite aggregation and adherence to host cells. Transcriptomics revealed that TvEVs up-regulate expression of predicted parasite membrane proteins and identified an adherence factor, heteropolysaccharide binding protein (HPB2). Finally, using comparative proteomics and superresolution microscopy, we demonstrated direct transfer of an adherence factor, cadherin-like protein, from TvEVs to the recipient parasite’s surface. This work identifies TvEVs as a mediator of parasite:parasite communication that may impact pathogenesis during mixed infections.
Communication between cells plays a vital role in how organisms respond to and manipulate their environment. The secretion of extracellular vesicles (EVs), a heterogenous set of small membranous structures which are shed from the plasma membrane and contain proteins, nucleic acids, and other small molecules, are now acknowledged as one mechanism that cells use to communicate with each other (1). Additionally, there is now a growing appreciation for the role of EVs during parasitic infection (2). EVs have been demonstrated to play a role during infection in a diverse set of eukaryotic parasites ranging from the Apicomplexan parasites (Plasmodium falciparum, Cryptosporidium parvum, and Toxoplasma gondii) (3–11), Kinetoplastids (Leishmania donovani and Trypanosoma brucei) (12–15), Amoebozoa (16), Diplomonads (17), and parasitic worms (Dicrocoelium dendriticum, Heligmosomoides polygyrus, Litomosoides sigmodontis, and Schistosoma japonicum) (18–25).
Trichomonas vaginalis is a sexually transmitted parasite and the causative agent of trichomoniasis, the most common nonviral sexually transmitted disease worldwide (26, 27). T. vaginalis causes an estimated ¼ billion infections annually and in the United States is the most common parasitic infection, with an annual incidence of about 5 million cases (26, 27). T. vaginalis infection, while commonly asymptomatic, can cause a number of clinical pathologies in women and men such as vaginitis, urethritis, prostatitis, preterm delivery, and infertility (28). There is also evidence that T. vaginalis is a cofactor for HIV spread and is associated with increased risk of urogenital cancers (29–37).
In the past decade, our lab and others have demonstrated that T. vaginalis secretes EVs (TvEVs) that can be internalized by human epithelial cells derived from both the prostate and female reproductive tract (FRT) via caveolin-dependent endocytic pathways and that these TvEVs modulate the ability of the parasite to adhere to host cells, an important step in establishing infection (38, 39). Studying the crosstalk between TvEVs and human epithelial cells has led to insight into host:parasite interactions. However, little is known about the effect of TvEV uptake within the parasite population.
Recent work has shown that TvEVs isolated from more adherent strains of the parasite increased the number of cytoneme-like membrane protrusions on parasites, and that these protrusions are associated with increased aggregation and adherence to host cells (40). This work also showed that cytoneme and filipodia formation can be induced by paracrine signaling between strains of T. vaginalis mediated by TvEVs. The ability of TvEVs to mediate communication within a parasite population is important as approximately 11% of clinical isolates worldwide are found to be coinfections with multiple strains (41). It is also known that clinical isolates of T. vaginalis differ greatly in their adherence to and cytolysis of human epithelial cells (42, 43). These observations underscore the importance of determining whether TvEVs are internalized by T. vaginalis within a mixed infection setting, and whether the potential exchange of virulence factors between parasites via TvEVs, impacts infection outcomes.
Here, we assessed the ability of T. vaginalis to internalize TvEVs from both a less adherent strain (G3) and a more adherent strain (B7RC2) and explored what effect this form of parasite:parasite communication has on host cell adherence, parasite aggregation, and parasite gene expression (43–45). We show that T. vaginalis is capable of taking up TvEVs and that TvEVs derived from a more adherent strain (B7RC2-EVs) are taken up more efficiently than TvEVs from a less adherent strain (G3-EVs). Additionally, using adherence and aggregation assays we determined that preincubation with B7RC2-EVs increases both parasite adherence to host cells and parasite aggregation. Transcriptomic analysis of T. vaginalis treated with TvEVs revealed that TvEVs up-regulate the expression of predicted parasite membrane proteins and known adherence factors. Additionally, we showed that overexpression of two TvEV-up-regulated genes increased adherence of parasites to human cells, and identified an adherence factor, heteropolysaccharide binding protein, HPB2. Using proteomics to compare the protein composition of TvEVs from more adherent and less adherent parasites, we identified a parasite cadherin-like protein (CLP), that is more abundant in B7RC2-EVs. Finally, using direct stochastic optical reconstruction microscopy (dSTORM), we showed that CLP can be directly transferred between parasites via TvEVs, leading to an increase in parasite adherence to host cells.
Results
Trichomonas-Derived EVs Are Internalized by T. vaginalis.
We have previously shown that T. vaginalis secretes EVs that are internalized by human epithelial cells and that internalization of EVs increases parasite adherence to host cells in a strain-specific manner (38, 39). Approximately 11% of T. vaginalis clinical isolates result from coinfections with multiple parasite strains where cell-to-cell communication between diverse strains of T. vaginalis may play a role in pathogenesis (41). Additionally, multiple labs have published indirect evidence suggesting that T. vaginalis is capable of EV uptake (38, 40). This led us to ask whether T. vaginalis could internalize its own EVs. To determine whether potential protein contamination from parasite growth media (Diamond’s) would affect our downstream analysis, we isolated TvEVs from either T. vaginalis parasites grown in serum-free Diamond’s media or isolated from equivalent volumes of Diamond’s media alone (mock). We found that Diamond’s media alone (mock) did not yield protein concentrations above our limit of detection in a Bicinchoninic acid assay (BCA) (SI Appendix, Fig. S1 A–C).
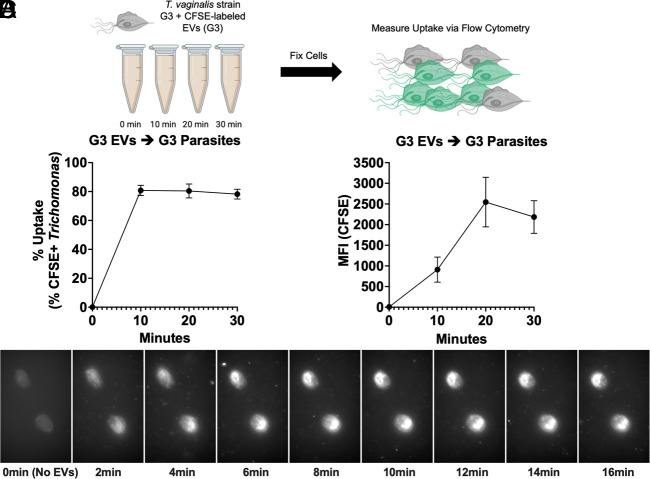
To determine whether T. vaginalis could internalize its own EVs, we extracted EVs from T. vaginalis strain G3 (G3-EVs), fluorescently labeled them with carboxyfluorescein succinimidyl ester (CFSE), and then incubated G3-EVs with G3 parasites. Internalization of G3-EVs (10 µg/mL) by G3 parasites was measured over 0 to 30 min using flow cytometry (Fig. 1A and SI Appendix, Fig. S2) (39). We found that internalization of G3-EVs was detectable as early as 10 min at 10 µg/mL (Fig. 1B). TvEV uptake also continues over time as indicated by the increase in mean fluorescent intensity (MFI) from 10 to 30 min (Fig. 1C). Additionally, uptake of mock prepared (Diamond’s media alone) only resulted in minimal CFSE+ parasites (>5%) (SI Appendix, Fig. S1 D and E). As an additional proof of principle, we performed live cell imaging of T. vaginalis strain G3 incubated with ExoGlowTM labeled G3-EVs (50 µg/mL). We imaged uptake every 2 min over the course of 16 min. With this method, we observed TvEV uptake by the parasite as early as 2 min, which continued over the 16-min time course, as indicated by the accumulation of ExoGlowTM signal in the parasite (Fig. 1D and Movie S1). Together, these data directly show that T. vaginalis is capable of internalizing TvEVs.
Fig. 1.
Trichomonas-derived EVs are taken up by T. vaginalis. (A) Schematic of TvEV uptake assay. T. vaginalis strain G3 was exposed to 10 µg/mL of CFSE-labeled G3-EVs for 0, 10, 20, and 30 min and TvEV uptake was measured via flow cytometry. Created with BioRender.com. (B and C) Percent uptake and MFI of CFSE-positive parasites plotted over time. Percent uptake was measured by the percent of CFSE-positive parasites in the entire population and the amount of TvEV uptake was quantified by the increase in MFI plotted over time. Dots = mean ± SD. N = 3 replicates/experiment, three experiments total. (D) Individual images from 2-min intervals of time-lapse video showing uptake of ExoGlowTM labeled G3-EVs by T. vaginalis.
EVs Derived from a More Adherent Strain of T. vaginalis Are Taken Up More Efficiently than EVs Derived from a Less Adherent Strain.
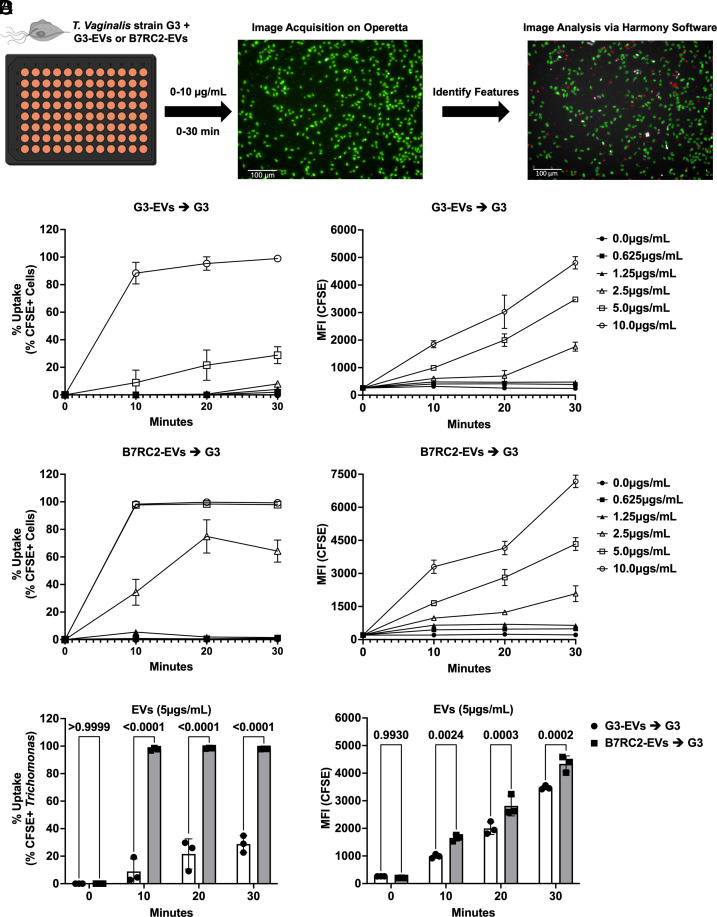
We and others have demonstrated that T. vaginalis strains differ greatly in their adherence to and cytolysis of host cells (40, 42, 43). For example, strain B7RC2 is approximately 20-fold more adherent to benign prostatic hyperplasia (BPH-1) cells compared to strain G3 (43). These phenotypic differences between strains caused us to wonder whether there might be differences in TvEV composition that could alter their uptake by T. vaginalis. In keeping with this line of thought, a number of TvEV proteomes from different strains have been published and reveal varying levels of overlap in TvEV protein content (38, 40, 46, 47). To test whether TvEVs isolated from different strains were taken up more efficiently than others, we compared the uptake of TvEVs produced by G3 parasites (G3-EVs) with that of TvEVs produced by B7RC2 parasites (B7RC2-EVs). We fluorescently labeled B7RC2-EVs or G3-EVs with CFSE and exposed a recipient strain of T. vaginalis (T. vaginalis strain G3) to various concentrations of TvEVs ranging from 0 to 10 µg/mL and measured TvEV internalization over time (0 to 30 min) using an automated imaging system, the Operetta® CLSTM platform and Harmony software (Fig. 2A) (48). We observed that G3-EVs were taken up less efficiently than B7RC2-EVs at multiple concentrations and time points (Fig. 2 B and C). Specifically, we observed a statistically significant decrease in the uptake of G3-EVs compared to B7RC2-EVs at 2.5 µg/mL and 5 µg/mL, over 10 to 30 min as measured by both the percent of the parasite population that were positive for TvEV uptake (% Uptake) and the amount of TvEVs that had been internalized (MFI) (Fig. 2D and SI Appendix, Fig. S3 A and B).
Fig. 2.
B7RC2-EVs are taken up more efficiently than G3-EVs. (A) Schematic of TvEV uptake assay. T. vaginalis strain G3 was exposed to CFSE-labeled B7RC2-EVs or G3-EVs ranging from 0 to 10 µg/mL for 0, 10, 20, and 30 min. Entire wells were imaged on the Operetta® CLSTM platform and analyzed using Harmony™ software. Created with BioRender.com. (B and C) Percent uptake and MFI of CFSE-positive parasites plotted over time. Percent uptake was measured by the percent of CFSE-positive parasites in the entire population and the amount of TvEV uptake was quantified by the increase in MFI plotted over time for either G3-EVs (B) or B7RC2-EVs (C). Dots = mean ± SD. N = 3 wells/experiment, three experiments total. (D) Quantification of % uptake and total MFI between G3-EVs and B7RC2-EVs at 5 µg/mL. Bars, mean ± SD. N = 3 wells/experiment, three experiments total. Numbers above bars indicate P-values for two-way ANOVA, Dunnett’s multiple comparisons test compared to G3-EVs.
Together, these results demonstrate that T. vaginalis is capable of taking up TvEVs isolated from multiple strains of the parasite, demonstrating a method of parasite:parasite communication between strains. Additionally, we observed that TvEVs from the more adherent parasite strain, B7RC2-EVs, are taken up more efficiently, in a time and concentration-dependent manner, compared to those from a less adherent strain, G3-EVs, suggesting a role for strain diversity in the quality of TvEVs and their effect on parasite biology.
Uptake of EVs from a More Adherent Strain of T. vaginalis Increases Host Cell Adherence of a Less Adherent Parasite Strain.
We have previously shown that preincubation of host cells with TvEVs from B7RC2 increases adherence of G3 parasites to host cells (38). Given the ability of T. vaginalis to directly take up TvEVs derived from different strains of the parasite (Fig. 2 B and C), we decided to directly test whether preincubation of G3 parasites with B7RC2-EVs also increased parasite adherence to host cells. We also examined whether treatment with TvEVs from more adherent parasites (B7CR2-EVs) versus those from less adherent parasite (G3-EVs) would modulate differences in adherence.
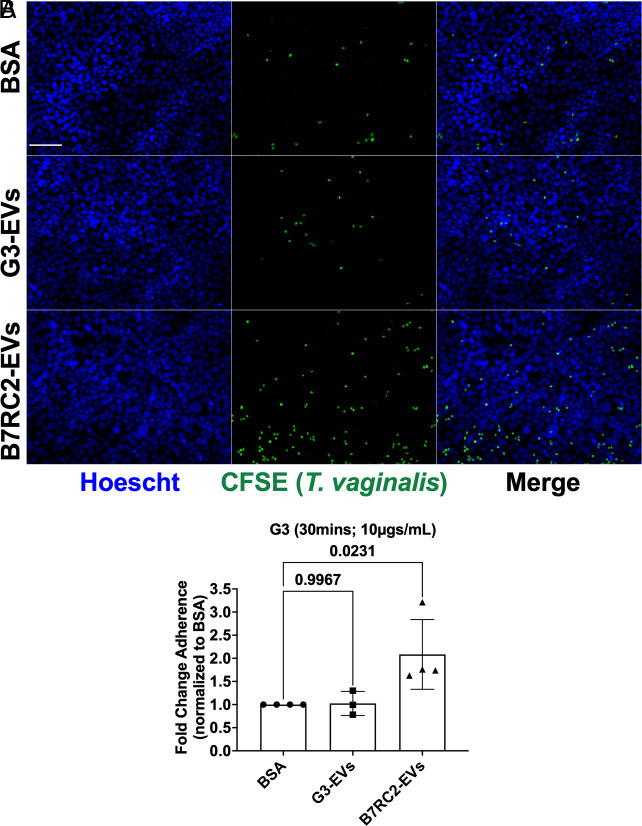
To this end, we pretreated less adherent G3 parasites with either G3-EVs, B7RC2-EVs, or bovine serum albumin (BSA) as a negative control, and then quantified parasite adherence to the prostate-derived cell line BPH-1 (49, 50). We found that compared to BSA control, there was no difference in parasite adherence to host cells when G3 parasites were pretreated with their own TvEVs (Fig. 3 A and B). However, when G3 parasites were pretreated with B7RC2-EVs there was a twofold increase in host cell adherence compared to BSA controls (Fig. 3 A and B). This shows that internalization of TvEVs by T. vaginalis can alter the adherence properties of parasites in a strain-dependent manner. It also suggests that the cargo of B7RC2-EVs and G3-EVs may differ, as pretreatment of G3-EVs did not alter adherence of T. vaginalis strain G3.
Fig. 3.
Preincubation of T. vaginalis strain G3 with B7RC2-EVs increases adherence to host cells. (A) Representative fluorescence microscopy image of host cell adherence assay. T. vaginalis strain G3 was labeled with CFSE, then pretreated with BSA, G3-EVs, or B7RC2-EVs at 10 µg/mL for 30 min. Images depict CFSE-labeled T. vaginalis strain G3 (Green) and BPH-1 nuclei stained with Hoechst (Blue). (Scale bar, 100 µm.) (B) Quantification of parasite adherence to BPH-1. Data are depicted as fold change in adherence compared to BSA control. Bars, mean ± SD. N = 3 to 6 wells/experiment, four experiments total. Numbers above bars indicate P-values for one-way ANOVA, Dunnett’s multiple comparisons test compared to BSA control.
Uptake of EVs Secreted by More Adherent T. vaginalis Strains Increases Parasite Aggregation.
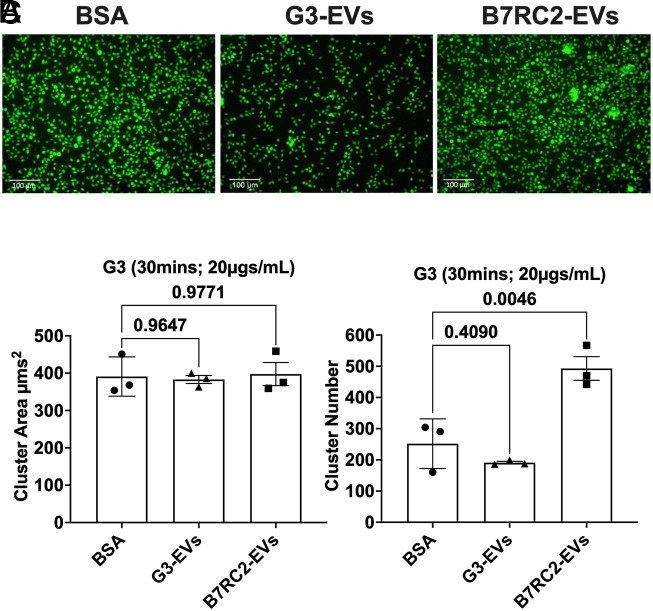
T. vaginalis is known to form aggregates on both ectocervical and prostate-derived cell lines (51). Furthermore, we and others have shown that parasite aggregation correlates with increased host cell adherence (51, 52). Given our observation that B7RC2-EVs, but not G3-EVs increase adherence of T. vaginalis to BPH-1, we tested whether increased adherence might be partially attributed to parasite aggregation promoted by TvEV uptake. We fluorescently labeled T. vaginalis strain G3 with CFSE and incubated the labeled parasites with either G3-EVs, B7RC2-EVs, or BSA as a negative control for 30 min and allowed them to form aggregates. We then quantified parasite aggregation using the Operetta® CLSTM platform (48) where an aggregate was defined as a cluster of parasites with an area greater than 250 µm2. We observed the formation of parasite aggregates as indicated by “clusters” of CFSE-positive parasites (Fig. 4A). We then quantified the number of parasite clusters and cluster area using Harmony software. We observed no difference in overall cluster size between BSA, G3-EV, and B7RC2-EV treated parasites (Fig. 4B); however, we did observe a statistically significant increase in parasite cluster number when G3 parasites were treated with B7RC2-EVs compared to pretreatment with either G3-EVs or the BSA control (Fig. 4C). These data show that more adherent TvEVs (B7RC2-EVs), but not TvEVs from less adherent parasites (G3-EVs), promote an increase in parasite aggregation.
Fig. 4.
Preincubation of T. vaginalis strain G3 with B7RC2-EVs increases parasite aggregation. (A) Representative fluorescence microscopy image of parasite aggregation assay. T. vaginalis strain G3 was labeled with CFSE, then pretreated with BSA, G3-EVs, or B7RC2-EVs at 20 µg/mL for 30 min. Images depict CFSE-labeled T. vaginalis strain G3 (Green). (B) Quantification of parasite aggregate area. Bars, mean ± SD. N = 3 wells/experiment, three experiments total. (C) Quantification of parasite aggregate number. Bars, mean ± SD. N = 3 wells/experiment, three experiments total. (B and C) Numbers above bars indicate P-values for one-way ANOVA, Dunnett’s multiple comparisons test compared to BSA control.
Transcriptomic Analysis of T. vaginalis Strain G3 Treated with B7RC2-EVs Reveals Upregulation of Previously Characterized and Uncharacterized Adherence Factors.
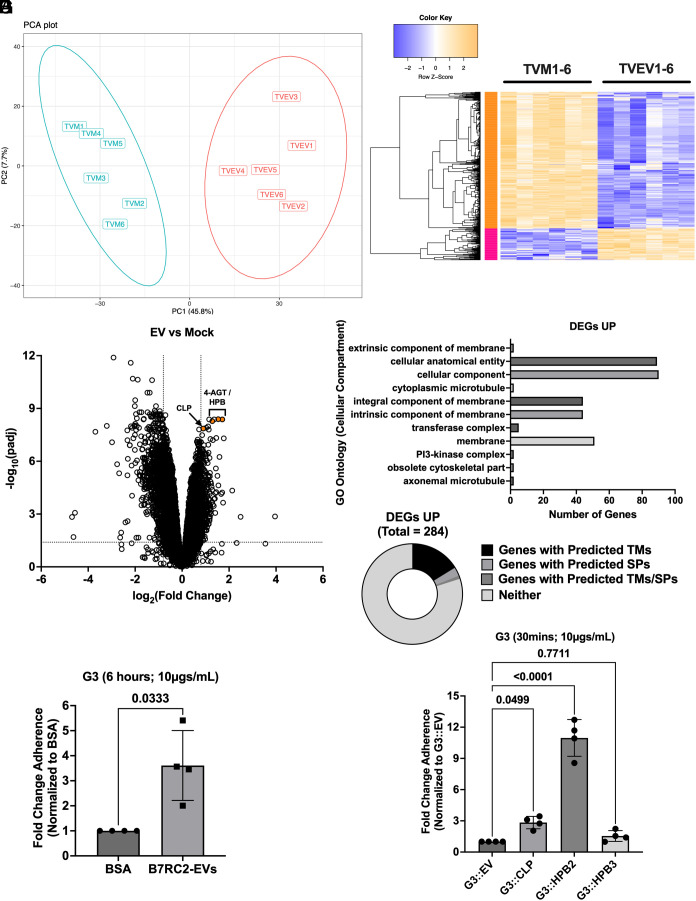
We next sought to identify what effect B7RC2-EVs had on T. vaginalis gene expression as a way of identifying potential mediators of the adherence and aggregation phenotypes observed. To achieve this goal, we either mock treated or treated T. vaginalis strain G3 with 50 µg/mL of B7RC2-EVs for 6 h and then extracted total RNA for RNA-seq. Sample alignment and quality was assessed using MultiQC (SI Appendix, Fig. S4) (53). Transcriptional profiles of the 6 Mock (TVM) and 6 TvEV-treated (TVEV) samples were distinguishable by Principle Component Analysis (Fig. 5A), with 1,086 genes [281 up-regulated and 805 down-regulated; Log2(FC) > 0.8 or Log2(FC) < −0.8; padj < 0.05] identified as differentially expressed between the two conditions (Fig. 5B and Dataset S1). The larger number of down-regulated genes suggests that B7RC2-EVs have an overall suppressive effect on G3 gene expression. Consistent with this finding, a large number (>20) of the most strongly down-regulated genes are predicted RNA polymerase II transcription regulator recruiting proteins (Dataset S2). Additionally, we performed GO enrichment analysis (54, 55) for predicted molecular function, cell compartment, and biological process of the 805 down-regulated genes (Datasets S3–S5). The top 15 most significantly enriched GO pathways for predicted molecular function included multiple annotations for RNA polymerase II activity as well as transcription regulator activity and nucleotide binding (SI Appendix, Fig. S5 and Dataset S3). GO enrichment analysis of the 805 down-regulated DEGs for predicted biological process included multiple annotations for RNA metabolism and regulation of gene expression (SI Appendix, Fig. S5 and Dataset S4). GO enrichment analysis for predicted cell compartment identified the nucleus as enriched (SI Appendix, Fig. S5 and Dataset S5).
Fig. 5.
B7RC2-EVs up-regulate expression of parasite proteins involved in host cell adherence. (A) Principal components analysis showing distinct clustering of mock treated parasites (TVM; n = 6) and B7RC2-EV treated parasites (TVEV; n = 6) from RNA-seq data generated in this study. (B) Heatmap of 1,086 genes [281 up-regulated and 805 down-regulated; Log2(FC) > 0.8 or Log2(FC) < −0.8; padj < 0.05] identified as differentially expressed between the mock and B7RC2-EV treated parasites. (C) Volcano plot depicting differences in gene expression between B7RC2-EVs and mock treated parasites. The x axis corresponds to the log2 fold change in gene expression and the y axis indicates the adjusted P value. Genes with a −log10 P value of 1.3 or greater (P value of ≤ 0.05) and a log2 fold change −0.8 ≤ or ≥ 0.8 were deemed significantly differentially expressed. (D) GO enrichment analysis 281 up-regulated DEGs for predicted cellular compartment enrichment. (E) Pie chart of percentage of 281 up-regulated DEGs with predicted transmembrane domain or signal peptides as annotated in TrichDB (www.trichdb.org). (F) Quantification of parasite adherence to BPH-1. Data are depicted as fold change in adherence compared to BSA control. Bars, mean ± SD. N = 6 wells/experiment, four experiments total. Numbers above bars indicate P-values for Welch’s t test compared to BSA control. (G) Quantification of parasite adherence to BPH-1. Data are depicted as fold change in adherence compared to empty vector (G3::EV) control. Bars, mean ± SD. N = 6 wells/experiment, four experiments total. Numbers above bars indicate P-values for one-way ANOVA, Dunnett’s multiple comparisons test compared to G3::EV control.
Interestingly, while B7RC2-EVs had an overall suppressive effect on parasite gene expression, four of the top most significantly up-regulated DEGs in response to B7RC2-EVs were genes previously identified as playing a role in either parasite adherence to human cells or TvEV uptake by human cells (SI Appendix, Table S1, Dataset S6, and Fig. 5C) (39, 52). Three of these genes include predicted heteropolysaccharide binding proteins (HPB) [TVAGG3_0163930 (HPB1), TVAGG3_0232100 (HPB2), and TVAGG3_0969590 (HPB3)] previously annotated as 4-α-glucanotransferase (4-AGT) genes (TVAG_154680, TVAG_157940, and TVAG_222040) that our lab had identified as playing a role in TvEV internalization by human epithelial cells (39). These three genes (referred to by their current annotation HPB from here on) were also found in a number of previously published T. vaginalis EV and surface membrane proteomes (38, 56–58). We also observed upregulation of a previously described parasite surface protein, cadherin-like protein (CLP; TVAGG3_0583720 previous annotation TVAG_393390) that was shown to mediate parasite aggregation, host cell adherence, and host cell cytolysis (52). Additionally, GO enrichment analysis of the 281 up-regulated DEGs for predicted cellular compartment identified cell membrane proteins as enriched (Fig. 5D). Consistent with membrane proteins being up-regulated by B7RC2-EVs, 16% of up-regulated DEGs are predicted to contain transmembrane domains (Fig. 5E). Interestingly, our prior data indicated that as little as 30 min of exposure to B7RC2-EVs was sufficient to increase adherence of recipient parasites to host cells (Fig. 3), while our transcriptomic data suggest that gene expression changes at 6 h post exposure to B7RC2-EVs may play a role as well. To identify whether longer incubation (6 h) with B7RC2-EVs would further increase adherence over the initial 30-min data, we performed adherence assays with G3 parasites incubated with B7RC2-EVs for 6 h. We observed a 3.5-fold increase in adherence when parasites were incubated with B7RC2-EVs for 6 h compared to BSA controls (Fig. 5F) which was higher than the previous adherence assays performed at 30 min (twofold increase over BSA controls) (Fig. 3B).
Taken together, these results indicate that gene expression changes due to B7RC2-EVs may alter the surface membrane composition of recipient parasites to facilitate parasite adhesion to host cells and parasite aggregation.
To directly test whether the up-regulated genes identified in our transcriptomic data were involved in mediating adherence to host cells, we overexpressed CLP (TVAGG3_0583720) and two of the three predicted HPB genes (TVAGG3_0232100 and TVAGG3_0969590), which will be referred to as HPB2 and HPB3 respectively. We ectopically expressed a second copy of each gene in T. vaginalis strain G3 using our standard T. vaginalis expression vector, pMasterNeo (pMNeo), where each gene is under the control of a T. vaginalis α-succinyl coenzyme A (CoA) synthetase promoter and fused to two C-terminal hemagglutinin (HA) tags (59). We confirmed expression of the tagged proteins via IFA (SI Appendix, Fig. S6A) and quantified expression levels of each construct via MFI (SI Appendix, Fig. S6 B and C). IFA analysis showed similar levels of expression between each construct, while an empty vector control (G3::EV) showed little to no detectable levels of HA staining. Overexpression of CLP increased adherence of G3 parasites to BPH-1 cells by ~2.8-fold on average compared to empty vector control, consistent with previous overexpression studies performed in T. vaginalis strain RU393 (ATCC 50142) (Fig. 5G) (52). Additionally, while overexpression of HPB3 showed no statistically significant difference in adherence to BPH-1 compared to empty vector control, overexpression of HPB2 resulted in an ~11-fold increase in adherence compared to empty vector control (Fig. 5G). Taken together, these results indicate that B7RC2-EVs up-regulate the expression of genes involved in parasite adherence to host cells and identify HPB2 as a parasite adherence factor.
Comparative Proteomics Identifies CLP as a Mediator of Host Cell Adherence That Can Be Transferred between T. vaginalis Parasites via the Secretion and Uptake of EVs.
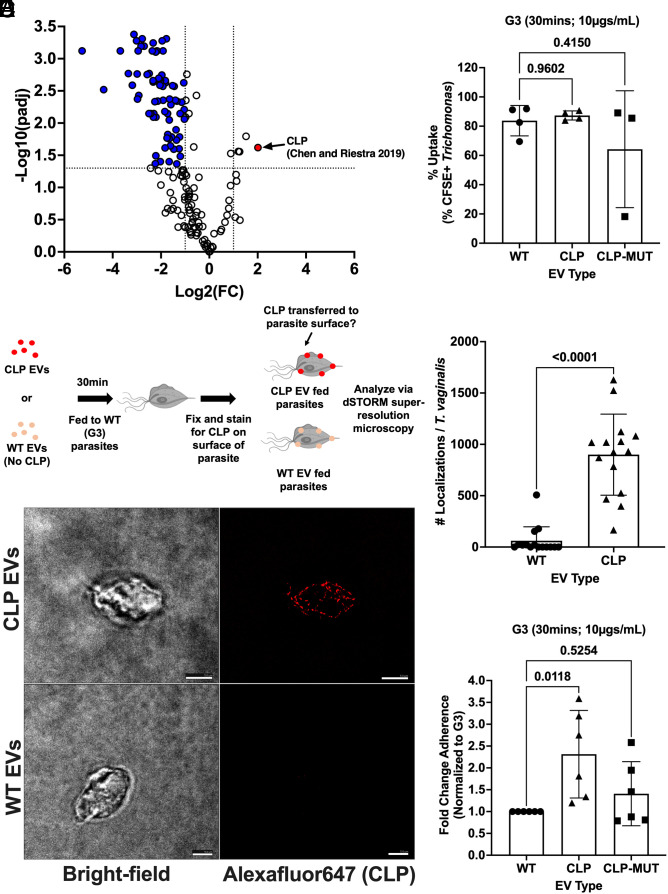
Given that the increase in adherence to host cells and parasite aggregation was specific to B7RC2-EVs, but not G3-EVs, we reasoned that the cargo of TvEVs derived from more adherent and less adherent parasites must differ. Furthermore, while our transcriptomic data indicate that B7RC2-EVs mediates increased adherence to host cells and parasite aggregation by up-regulating membrane proteins including known (CLP) and uncharacterized (HPB2) adherence factors, we observe increased adherence within as little as 30 min after parasite uptake of B7RC2-EVs, suggesting that proteins present in the TvEVs themselves are potentially transferred to the membrane of the recipient strain to aid in adherence. To this end, we decided to couple our transcriptomic data with proteomics. To identify potential TvEV proteins driving these phenotypes, we used available proteomic data in the lab to profile and quantify differences in the protein content of TvEVs from a nonadherent strain of T. vaginalis (NYH209-EVs) (43, 60) and B7RC2-EVs. We identified a total of 303 proteins packaged within T. vaginalis–derived EVs. Of the 303 proteins identified, many were predicted homologues of known exosome proteins such as tetraspanins, small Rab GTPases, small Ras GTPases, and VSPs (Dataset S10) (1). We identified a total of 77 differentially abundant proteins [Log2(FC) > 1 or Log2(FC) < −1; padj < 0.05] with four proteins found more abundantly in the B7RC2-EVs compared to NYH209-EVs and 73 proteins found more abundantly in NYH209-EVs compared to B7RC2-EVs (Fig. 6A and Dataset S11). Interestingly, the most differentially abundant protein identified in the B7RC2-EVs compared to the NYH209-EVs was CLP, which was also up-regulated in our transcriptomic data (Fig. 6A and SI Appendix, Table S1).
Fig. 6.
CLP can be transferred between T. vaginalis parasites via TvEVs resulting in increased parasite adherence to host cells. (A) Volcano plot depicting differences in protein abundance between B7RC2-EVs and NYH209-EVs. The x axis corresponds to the log2 fold change in protein abundance and the y axis indicates the adjusted P value. Proteins with a −log10 P value of 1.3 or greater (P value of ≤ 0.05) and a log2 fold change −1 ≤ or ≥ 1 were deemed differentially abundant. (B) Percent uptake of CFSE-labeled TvEVs from indicated strains by T. vaginalis strain G3. Percent uptake was measured by the percent of CFSE-positive parasites in the entire population. Bars, mean ± SD. N = 3 replicates/experiment, 3 to 4 experiments total. (C) Top, graphical depiction of CLP-EV transfer experiment and subsequent dSTORM imaging. Created with BioRender.com. Bottom, representative IFA for GFP-tagged CLP of parasites fed either WT-EVs or CLP-EVs using dSTORM. (Scale bar, 5 µm.) (D) Quantification of the number of localizations for WT-EV and CLP-EV treated parasites. Bars, mean ± SD. N = 5 parasites/experiment, 15 parasites total were imaged and quantified. (E) Quantification of parasite adherence to BPH-1. Data are depicted as fold change in adherence compared to G3 control. Bars, mean ± SD. N = 6 wells/experiment, five experiments total. (B and E) Numbers above bars indicate P-values for one-way ANOVA, Dunnett’s multiple comparisons test compared to WT control. ns = not significant. (D) Numbers above bars indicate P-values for the Mann–Whitney test compared to WT control.
Given that CLP was identified in both our transcriptomic and proteomic data, we decided to directly test whether CLP was packaged into TvEVs and was able to be transferred between strains. To confirm that CLP was packaged in TvEVs, as indicated by our TvEV proteomics, we overexpressed an N-terminally GFP-tagged version of CLP or a calcium-binding site mutant version of CLP (CLP-MUT) using our standard pMNeo vector (52, 59). We confirmed the presence of the CLP or CLP-MUT gene using PCR (SI Appendix, Fig. S7A) and then performed western blot analysis to show that the CLP and CLP-MUT proteins were expressed by the parasite and loaded into the TvEVs (SI Appendix, Fig. S7 B and C). To ensure that any differences we saw in downstream adherence assays were not due to alterations in TvEV uptake caused by the overexpression of CLP or CLP-MUT, we performed our standard TvEV uptake assay (39). There were no statistically significant differences in TvEV uptake between wild type (WT) EVs, CLP EVs, or CLP-MUT EVs (Fig. 6B). To determine whether CLP could be directly transferred from one parasite to the surface of another via TvEV uptake by a recipient strain, we performed dSTORM (61, 62) on G3 parasites treated with either TvEVs from wild type (G3) parasites or TvEVs from parasites overexpressing an N-terminally GFP-tagged version of CLP (SI Appendix, Fig. S7 and Fig. 6C) (52). Parasites treated with CLP EVs showed consistent surface staining for the GFP-tagged CLP via dSTORM (Fig. 6C) and had a higher total number of localization events compared to parasites treated with WT EVs (Fig. 6D) consistent with the transfer of CLP to the recipient parasite surface. Finally, we pretreated wild type G3 parasites with either WT EVs, CLP EVs, or CLP-MUT EVs, and then performed an adherence assay and quantified parasite adherence to BPH-1 cells. We found that compared to WT EVs, incubation of wild type G3 parasites with CLP EVs increased host cell adherence by ~2.5-fold (Fig. 6E). However, disruption of CLP’s calcium-binding domain ablated this effect, consistent with previous reports (Fig. 6E) (52). Taken together, these data show that T. vaginalis delivers an adherence factor, CLP, to neighboring parasites via secreted TvEVs, resulting in increased parasite adhesion to host cells. These data establish a mechanism by which diverse strains of T. vaginalis can communicate with one another to alter parasite:host interactions.
Discussion
The data presented here show that TvEV uptake by T. vaginalis constitutes a form of parasite:parasite communication that may have implications for mixed infections. TvEV uptake mediates parasite adherence to host cells by both the upregulation of gene expression and the direct transfer of parasite adherence factors. We showed that TvEV uptake efficiency varies between strains and that TvEVs produced from more adherent strains are taken up more efficiently than those produced by less adherent strains. We also showed that T. vaginalis uptake of TvEVs from a more adherent strain of T. vaginalis increases parasite aggregation and parasite adherence. Transcriptomics revealed that TvEVs mediate increased gene expression of parasite membrane proteins and established T. vaginalis adherence factors. Additionally, TvEVs up-regulate the expression of three predicted heteropolysaccharide binding proteins, one of which (HPB2), we confirmed as an adherence factor through overexpression studies. Finally, we also showed using dSTORM that at least one parasite adherence factor, CLP, can be transferred from one strain of T. vaginalis to the surface of another via TvEVs, resulting in increased adherence to host cells.
The observation that TvEVs can alter host cell adherence of recipient parasites in a strain-specific manner correlating with adherence of the T. vaginalis strain from which the TvEVs were isolated has been shown by our group and others (38, 40). Additionally, there is strong evidence that parasite-shed EVs play a role in parasite: parasite communication in Plasmodium, Trypanosoma, Entamoeba, Giardia, and Leishmania spp. where they alter parasite development, mediate transfer of resistance factors, small RNAs, and affect social motility behaviors (3, 4, 12–17). Despite these observations, direct internalization of parasite derived EVs by T. vaginalis was assumed, but not directly shown. Here, we present direct evidence using a combination of flow cytometry, immunofluorescence microscopy, and live cell imaging that T. vaginalis is capable of TvEV internalization. This establishes a mechanism of parasite:parasite communication, whereby molecular cargo (parasite proteins, small RNAs, and other molecules) can be transferred between T. vaginalis strains. Epidemiological data estimate that approximately 11% of T. vaginalis clinical infections result from coinfections with multiple parasite strains (41) raising the question of how TvEV-mediated parasite:parasite interactions might alter infection outcomes. Recently, our group established a mouse male urogenital tract model of T. vaginalis infection and showed that preincubation of the parasite with TvEVs from B7RC2, a strain that is highly adherent to prostate epithelial cells in vitro, prior to mouse infection resulted in increased parasite burden and survival in vivo (43, 63). Together, the data from our mouse male urogenital tract model of infection and the data in this paper provide evidence that TvEVs play a role in altering infection outcomes in mixed infection settings.
Our experiments showing direct internalization of TvEVs by T. vaginalis also revealed that TvEVs derived from B7RC2, a more adherent strain of the parasite, were taken up more efficiently than ones derived from a less adherent strain, G3, indicating that TvEVs themselves differ qualitatively in a way that alters the kinetics of uptake. It is likely that this difference in uptake efficiency is driven by differences in either the membrane composition of the TvEVs themselves, surface proteins that decorate TvEVs, or other molecular modifications such as differential glycosylation of membrane proteins or lipids. Additionally, our group has shown that human host cell uptake of TvEVs relies on caveolin lipid raft mediated endocytosis (CLRDE), but not clathrin-mediated endocytosis (CME) (39). Interestingly, T. vaginalis lacks any known homologues to caveolins, as such CLRDE pathways likely do not exist in this parasite. CLDRE pathways themselves appear to have arisen relatively late in eukaryotic evolution and are specific to the metazoans but are absent from the fungi, plants, and nonmetazoan parasites (64, 65). Future comparative studies between B7RC2-EVs and G3-EVs may prove useful in characterizing the molecular features of TvEVs that drive increased uptake by T. vaginalis and may help define the mechanisms of TvEV uptake by the parasite by identifying the surface receptors and endocytic pathways involved.
It has recently been shown that Trichomonas vaginalis virus (TVV) is released from small EVs shed by the parasite and can alter the molecular cargo carried by TvEVs (47). Hence, we determined whether TVV is present in B7RC2, the parasite strain used for TvEV analyses here. Using RT-PCR to detect TVV1-4 on RNA isolated from B7RC2, this parasite strain was found to be negative for all four of the previously identified TVV genomes (SI Appendix, Fig. S8). These data eliminate a possible role for TVV in the effects seen in this study. However, the observation that T. vaginalis is capable of transferring TvEVs between strains may indicate that one mechanism of TVV spread among parasites may be through internalization of TvEVs.
Previous work by us and others established that preincubation of T. vaginalis with TvEVs derived from highly adherent strains of the parasite resulted in increased adherence to vaginal and prostate epithelial cells and increased parasite aggregation (40, 43). However, the specific mechanisms underlying these adherence phenotypes were unclear. Using transcriptomics, we showed that TvEVs up-regulate the expression of predicted parasite membrane proteins and known parasite adherence factors. We also identified three predicted heteropolysaccharide binding proteins (HPB1-3) as up-regulated by B7RC2-EVs and showed that overexpression of HPB2 increased adherence of G3 parasites to BPH-1 cells. The exact mechanism that mediates HPB2’s ability to increase adherence remains unknown. However, the three HPB genes up-regulated by TvEVs [TVAGG3_0163930 (HPB1), TVAGG3_0232100 (HPB2), and TVAGG3_0969590 (HPB3)] are i) 88% similar to each other by amino acid identity, ii) contain 2 N-terminal Carbohydrate-Binding Module 20 (CBM20) domains, and iii) are also predicted to contain a C-terminal glycosyl hydrolase domain often found in 4-α-glucanotransferases, which have been described as playing a role in glycogen and starch metabolism in bacteria, plants, and several unicellular eukaryotes (66). Additionally, previous research by our group has shown that pretreatment of human cells with recombinant CBM20, a 248-amino acid polypeptide that contains the CBM20 domains from HPB2, was capable of blocking uptake of TvEVs by 80% (39). It is possible that the CBM20 domain of HPB2 is directly involved in interactions with the host cell surface. Alternatively, the C-terminal glycosyl hydrolase domain may play a role in modifying sugar moieties on the surface of the parasite which have also been shown to mediate host:parasite interactions (67). Additionally, our transcriptomic data also revealed upregulation of other previously identified adherence factors such as BspA proteins (68) and potential pore forming and hemolytic factors that may aid in cytolysis and hemolysis (69).
Transfer of membrane-bound proteins from one cell to another has been observed in both prokaryotic and eukaryotic systems (70, 71). Our observation that B7RC2-EVs increased host cell adherence within as little as 30 min after parasite uptake suggested that direct transfer of TvEV proteins to the surface membrane of the recipient parasite was responsible for the increased adherence. Our proteomic data comparing TvEVs from a lowly adherent strain of T. vaginalis (NYH209) and a highly adherent strain (B7RC2) revealed a promising TvEV transferred candidate, cadherin-like protein (CLP), which was found to be significantly more abundant in TvEVs derived from the more adherent strain, B7RC2. CLP had been previously identified and characterized by our group as a parasite transmembrane protein that mediates parasite aggregation, host cell adherence, and host cell cytolysis (52). Our proteomics data suggested that CLP was also packaged in the TvEVs and thus capable of being transferred from one parasite cell to another. Using TvEVs isolated from parasites that expressed a tagged version of CLP, we confirmed that CLP was packaged into TvEVs, and using dSTORM, we demonstrated that CLP residing in TvEVs was transferred to the surface of recipient strains of T. vaginalis. Transmission of CLP via TvEVs to recipient parasites also resulted in increased adherence to host cells, establishing a mechanism by which one strain of T. vaginalis can redecorate the surface of another to aid in host:parasite interactions. The full extent to which TvEV proteins localize to the plasma membrane of recipient parasites and the mechanisms driving transfer to the recipient parasite’s surface remains to be explored.
Collectively the research reported here furthers our understanding of Trichomonas:Trichomonas interactions by establishing TvEVs as a mechanism for parasite:parasite communication via alteration of parasite gene expression and the direct transfer of TvEV proteins to the T. vaginalis surface. This work likewise establishes a mechanism by which different strains of T. vaginalis may aid each other in the context of a mixed infection. A better understanding of how TvEV transfer between parasites affects infection will reveal additional physiological roles of these vesicles and provide valuable insight to guide the use of TvEVs in future therapeutic and vaccination strategies.
Materials and Methods
Parasite Maintenance.
T. vaginalis strains B7RC2 and G3 were cultured in Diamond’s modified Trypticase-yeast extract-maltose (TYM) medium supplemented with 10% horse serum (Sigma-Aldrich), 10 U/mL penicillin, and 10 μg/mL streptomycin (Gibco), 180 μM ferrous ammonium sulfate, and 2 μM sulfosalicylic acid (72). Parasites were grown at 37 °C and passaged daily for no more than 2 wk at a time.
Human Cell Culture.
Human benign prostate hyperplasia 1 (BPH-1) epithelial cells were cultured at 37 °C with 5% CO2 in RPMI 1640, L-glutamine, and HEPES media (Gibco) supplemented with 10 U/mL penicillin, 10 μg/mL streptomycin, and 10% fetal bovine serum (FBS; Gibco) as previously described (50).
Generation of Transgenic Parasites.
For exogenous expression of CLP, HPB2, and HPB3 in poorly adherent T. vaginalis G3 parasites, genes were PCR amplified from B7RC2 genomic DNA using primers with overhangs homologous to the pMasterNeo plasmid and assembled via In-fusion cloning using In-Fusion® Snap Assembly Starter Bundle (Takara; Cat. #638945). Detailed methods for nucleofection, selection, and verification of transgenic parasites are described in SI Appendix, Supplemental Methods.
EV Isolation.
T. vaginalis was grown to confluency (1 × 106 cells/mL) in 50 mL of complete Diamond’s media. Parasites were spun down at 3,200 rpm for 10 min and washed with 50 mL serum-free Diamond’s media twice. Cells were transferred to 50 mL serum-free Diamond’s media and incubated at 37 °C for 4 h. Cells were spun down at 3,000 rpm at 4 °C for 20 min to remove cell debris. The supernatant was kept and mixed 2:1 with Total Exosome Isolation Reagent (Invitrogen #4478359). The supernatant mixture was incubated overnight at 4 °C. Precipitated TvEVs were recovered by standard centrifugation at 10,000 × g for 60 min at 4 °C. The exosome pellet was resuspended in sterile 1x PBS plus 1:100 Halt Protease Inhibitor Cocktail (Thermo Scientific #78428) and stored at −80 °C. The concentration of TvEV protein was determined using the Pierce BCA Kit (Thermo Scientific #23225). To generate the mock control used in our transcriptomic analyses, we used 50 mL serum-free Diamond’s alone (no parasites) and isolated as described above.
EV Uptake Assay.
EVs were labeled with CFSE (BioLegend) as previously described (39). Detailed methods for uptake of extracellular vehicles in parasites are described in SI Appendix, Supplemental Methods.
Live Cell Imaging of TvEV Uptake.
G3-EVs were labeled with the ExoGlowTM-Membrane EV Labeling Kit [System Biosciences Innovation (SBI); Cat. # EXOGM600A-1] per manufacturer protocol for 24 h prior to use in live cell imaging. T. vaginalis strain G3 (5*105 parasites) were incubated with 50 µg/mL of ExoGlowTM labeled G3-EVs and imaged on a Nanoimager (Onibio) at 100x magnification at 37 °C. 30 frames were captured every 2 min for 16 min total.
Adherence Assay.
Adherence of T. vaginalis to BPH-1 cells was performed as described previously (52) with modification for use in 96-well plates on the Operetta® CLSTM platform. Detailed methods for adherence assays are described in SI Appendix, Supplemental Methods.
Aggregation Assay.
Aggregation assay was performed as previously described (52) with modifications for use in 96-well plates on the Operetta® CLSTM platform. Detailed methods for aggregation assays are described in SI Appendix, Supplemental Methods.
Transcriptomic Analysis.
T. vaginalis strain G3 (1 × 106 parasites per condition per replicate) was treated with 50 µg/mL of B7RC2-EVs or mock treated for 6 h in serum-free Diamonds media at 37 °C prior to total RNA isolation. Total RNA was isolated using the Direct-zolTM RNA Miniprep Kit (Zymo Research; Cat. #R2071) according to the manufacturer’s instructions. Total RNA was sent to Novogene for RNA quality control (QC) analysis, library prep, and sequencing. RNA QC was analyzed, and quantification of RNA preparations and libraries was carried out using an Agilent 5400. Samples were sequenced on an Illumina NovoSeq 6000 to produce 150–base pair paired end reads with a mean sequencing depth of 20 million reads per sample. After read mapping with Kallisto (73), version 0.46.2, TxImport (74) was used to read Kallisto outputs into the R environment. Annotation data from TrichDB (https://trichdb.org/common/downloads/release-64/TvaginalisG32022/) was used to summarize data from transcript-level to gene-level. All subsequent analyses were carried out using the statistical computing environment R version 4.3.0 in RStudio version 1.1.456, and Bioconductor version 3.8. Briefly, transcript quantification data were summarized to genes using the tximport package and normalized using the trimmed mean of M values (TMM) method in edgeR (75). Genes with <1 CPM in n + 1 of the samples, where n is the size of the smallest group of replicates, were filtered out. Normalized filtered data were variance-stabilized using the voom function in limma (76), and differentially expressed genes were identified with linear modeling using limma (FDR ≤ 0.05; absolute logFC ≥ 0.8) after correcting for multiple testing using the Benjamini–Hochberg procedure. GO analysis was carried out on TrichDB (https://trichdb.org/trichdb/app/search/transcript/GenesByGoTerm).
Proteomic Analysis.
B7RC2-EVs and NYH209-EVs used for proteomic analysis were isolated and protein extracted as previously described (38). Raw data were processed using MaxQuant (77) and MSstats (78) on the Galaxy server (https://usegalaxy.org/). Searches were performed using the T. vaginalis database from TrichDB (https://trichdb.org/trichdb/app/record/dataset/DS_47fb0ba7f2). Briefly, the MS/MS database search was performed using default settings, with a 20 ppm mass tolerance for the main search. Trypsin was selected as the protease, with up to two missed cleavages allowed. Results were filtered by a 0.05 false discovery rate at both protein and peptide levels. The minimum length of acceptable identified peptides was set at seven amino acids. Proteins with a −log10 P-adjusted value of 1.3 or greater (P-adjusted value of ≤ 0.05) and a log2 fold change −1 ≤ or ≥ 1 were deemed differentially abundant.
Data Collection for dSTORM.
Prior to dSTORM data acquisition, a single frame snapshot of wide-field image was taken for each excitation wavelength for comparison purposes and a single frame snapshot of bright-field image was taken as well. A maximum of 5,000 frames were collected using an exposure time of 20 ms. A photoswitching buffer of 100 mM mercaptoethylamine in PBS was mounted on top of the samples during data acquisition. Detailed methods for immunofluorescent staining are described in SI Appendix, Supplemental Methods.
RT-PCR Screen of TVV 1-4 in B7RC2 Parasites.
Total RNA was isolated from 2*106 parasites from T. vaginalis strains NYH209, LSU160, and B7RC2 using the Direct-zolTM RNA Miniprep Kit (Zymo Research; Cat. #R2071) according to the manufacturer’s instructions. RT-PCR screening on 200 ng of total RNA was carried out using SuperScript™ III One-Step RT-PCR System with Platinum™ Taq High Fidelity DNA Polymerase (Invitrogen; Cat. #12574035) with species-selective screening primers (SI Appendix, Table S2), designed from previously reported TVV sequences (79).
Statistical Analysis.
Graphs were generated and statistical analyses were performed using Prism 10.0.2 software. All experiments were performed at least three independent times, and statistical analyses were conducted on the composite data unless reported otherwise. Unless otherwise specified, the data were analyzed using a one-way ANOVA with Dunnett’s multiple-comparison test to the control.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Dataset S09 (XLSX)
Dataset S10 (XLSX)
Dataset S11 (XLSX)
ExoglowTM Green-labeled TvEVs are internalized by T. vaginalis. Time-lapse video showing the uptake of fluorescently labeled G3-EVs by T. vaginalis strain G3.
Acknowledgments
We want to thank our colleagues in the Johnson lab and at University of California Los Angeles for their helpful discussions and critiques. This research was supported by NIH grants R01AI103182 and R01AI148475 (to P.J.J.). J.A.K., P.M.M., S.E., and K.M. received support from NIH Ruth L. Kirschstein National Research Service award T32AI007323. A.M.R. received support from Howard Hughes Medical Institute Gilliam Fellowship.
Author contributions
J.A.K., P.M.M., K.M., and P.J.J. designed research; J.A.K., P.M.M., S.E., and K.M. performed research; J.A.K., K.M., A.K.R., and A.M.R. contributed new reagents/analytic tools; J.A.K., P.M.M., S.E., K.M., and P.J.J. analyzed data; and J.A.K. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: R.H., University of Newcastle upon Tyne; and U.S., Stanford University.
Data, Materials, and Software Availability
Raw sequence data are available on the Gene Expression Omnibus (GEO; accession # GSE253522) (80). All other data are included in the manuscript and/or supporting information.
Supporting Information
References
- 1.Doyle L. M., Wang M. Z., Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells 8, 727 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szempruch A. J., Dennison L., Kieft R., Harrington J. M., Hajduk S. L., Sending a message: Extracellular vesicles of pathogenic protozoan parasites. Nat. Rev. Microbiol. 14, 669–675 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Regev-Rudzki N., et al. , Cell-cell communication between malaria-infected red blood cells via exosome-like vesicles. Cell 153, 1120–1133 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Mantel P.-Y., et al. , Malaria-infected erythrocyte-derived microvesicles mediate cellular communication within the parasite population and with the host immune system. Cell Host Microbe 13, 521–534 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kioko M., et al. , Extracellular vesicles could be a putative posttranscriptional regulatory mechanism that shapes intracellular RNA levels in Plasmodium falciparum. Nat. Commun. 14, 6447 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aline F., Bout D., Amigorena S., Roingeard P., Dimier-Poisson I., Toxoplasma gondii antigen-pulsed-dendritic cell-derived exosomes induce a protective immune response against T. gondii infection. Infect. Immun. 72, 4127–4137 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wowk P. F., et al. , Proteomic profiling of extracellular vesicles secreted from Toxoplasma gondii. Proteomics 17, 1600477 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Silva V. O., et al. , Extracellular vesicles isolated from Toxoplasma gondii induce host immune response. Parasite Immunol. 40, e12571 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Quiarim T. M., et al. , Characterization of extracellular vesicles isolated from types I, II and III strains of Toxoplasma gondii. Acta Trop. 219, 105915 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Gómez-Chávez F., Murrieta-Coxca J. M., Caballero-Ortega H., Morales-Prieto D. M., Markert U. R., Host-pathogen interactions mediated by extracellular vesicles in Toxoplasma gondii infection during pregnancy. J. Reprod. Immunol. 158, 103957 (2023). [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., et al. , Induction of inflammatory responses in splenocytes by exosomes released from intestinal epithelial cells following cryptosporidium parvum infection. Infect. Immun. 87, e00705-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douanne N., et al. , Leishmania parasites exchange drug-resistance genes through extracellular vesicles. Cell Rep. 40, 111121 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Silva M. R., et al. , Extracellular vesicles shed by Trypanosoma cruzi are linked to small RNA pathways, life cycle regulation, and susceptibility to infection of mammalian cells. Parasitol. Res. 113, 285–304 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Szempruch A. J., et al. , Extracellular vesicles from Trypanosoma brucei mediate virulence factor transfer and cause host anemia. Cell 164, 246–257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eliaz D., et al. , Exosome secretion affects social motility in Trypanosoma brucei. PLoS Pathog. 13, e1006245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma M., et al. , Characterization of extracellular vesicles from entamoeba histolytica identifies roles in intercellular communication that regulates parasite growth and development. Infect. Immun. 88, e00349-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Natali L., et al. , The exosome-like vesicles of Giardia assemblages A, B, and E are involved in the delivering of distinct small RNA from parasite to parasite. Int. J. Mol. Sci. 24, 9559 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernal D., et al. , Surface analysis of Dicrocoelium dendriticum. The molecular characterization of exosomes reveals the presence of miRNAs. J. Proteomics 105, 232–241 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Sánchez-López C. M., et al. , Extracellular vesicles from the trematodes Fasciola hepatica and Dicrocoelium dendriticum trigger different responses in human THP-1 macrophages. J. Extracell. Vesicles 12, e12317 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buck A. H., et al. , Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun. 5, 5488 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L., et al. , Exosome-like vesicles derived by Schistosoma japonicum adult worms mediates M1 type immune- activity of macrophage. Parasitol. Res. 114, 1865–1873 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Zhu S., et al. , Release of extracellular vesicles containing small RNAs from the eggs of Schistosoma japonicum. Parasit. Vectors 9, 574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J., et al. , Isolation and characterization of extracellular vesicles from adult Schistosoma japonicum. J. Vis. Exp. 135, 57514 (2018), 10.3791/57514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J., et al. , Schistosoma japonicum extracellular vesicle miRNA cargo regulates host macrophage functions facilitating parasitism. PLoS Pathog. 15, e1007817 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L., et al. , Sja-miR-71a in Schistosome egg-derived extracellular vesicles suppresses liver fibrosis caused by schistosomiasis via targeting semaphorin 4D. J. Extracell. Vesicles 9, 1785738 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwebke J. R., Burgess D., Trichomoniasis. Clin. Microbiol. Rev. 17, 794–803 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Secor W. E., Meites E., Starr M. C., Workowski K. A., Neglected parasitic infections in the United States: Trichomoniasis. Am. J. Trop. Med. Hyg. 90, 800–804 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fichorova R. N., Impact of T. vaginalis infection on innate immune responses and reproductive outcome. J. Reprod. Immunol. 83, 185–189 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gram I. T., Macaluso M., Churchill J., Stalsberg H., Trichomonas vaginalis (TV) and human papillomavirus (HPV) infection and the incidence of cervical intraepithelial neoplasia (CIN) grade III. Cancer Causes Control 3, 231–236 (1992). [DOI] [PubMed] [Google Scholar]
- 30.Zhang Z.-F., et al. , Trichomonas vaginalis and cervical cancer. Ann. Epidemiol. 5, 325–332 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z. F., et al. , Trichomonas vaginalis and cervical cancer. A prospective study in China. Ann. Epidemiol. 5, 325–332 (1995). [DOI] [PubMed] [Google Scholar]
- 32.Yap E. H., et al. , Serum antibodies to Trichomonas vaginalis in invasive cervical cancer patients. Genitourin. Med. 71, 402–404 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyle D. C. M., Smith J. R., Infection and cervical intraepithelial neoplasia. Int. J. Gynecol. Cancer 9, 177–186 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Yang S., et al. , Trichomonas vaginalis infection-associated risk of cervical cancer: A meta-analysis. Eur. J. Obstet. Gynecol. Reprod. Biol. 228, 166–173 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Sutcliffe S., et al. , Plasma antibodies against Trichomonas vaginalis and subsequent risk of prostate cancer. Cancer Epidemiol. Biomarkers Prev. 15, 939–945 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Stark J. R., et al. , Prospective study of Trichomonas vaginalis infection and prostate cancer incidence and mortality: Physicians’ health study. J. Natl. Cancer Inst. 101, 1406–1411 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Twu O., et al. , Trichomonas vaginalis homolog of macrophage migration inhibitory factor induces prostate cell growth, invasiveness, and inflammatory responses. Proc. Natl. Acad. Sci. U.S.A. 111, 8179–8184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Twu O., et al. , Trichomonas vaginalis exosomes deliver cargo to host cells and mediate host:parasite Interactions. PLoS Pathog. 9, e1003482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rai A. K., Johnson P. J., Trichomonas vaginalis extracellular vesicles are internalized by host cells using proteoglycans and caveolin-dependent endocytosis. Proc. Natl. Acad. Sci. U.S.A. 116, 21354–21360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salas N., et al. , Role of cytoneme structures and extracellular vesicles in Trichomonas vaginalis parasite-parasite communication. Elife 12, e86067 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conrad M. D., et al. , Extensive genetic diversity, unique population structure and evidence of genetic exchange in the sexually transmitted parasite Trichomonas vaginalis. PLoS Negl. Trop. Dis. 6, e1573 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krieger J. N., Ravdin J. I., Rein M. F., Contact-dependent cytopathogenic mechanisms of Trichomonas vaginalis. Infect. Immun. 50, 778–786 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lustig G., Ryan C. M., Secor W. E., Johnson P. J., Trichomonas vaginalis contact-dependent cytolysis of epithelial cells. Infect. Immun. 81, 1411–1419 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hollander D. H., Tysor J. S., Isolation of a stable clone of the ameboid-adherent (AA) variant of Trichomonas vaginalis. J. Parasitol. 73, 1074–1075 (1987). [PubMed] [Google Scholar]
- 45.Carlton J. M., et al. , Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315, 207–212 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Artuyants A., et al. , Extracellular vesicles produced by the protozoan parasite Trichomonas vaginalis contain a preferential cargo of tRNA-derived small RNAs. Int. J. for Parasitol. 50, 1145–1155 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Rada P., et al. , Double-stranded RNA viruses are released from Trichomonas vaginalis inside small extracellular vesicles and modulate the exosomal cargo. Front. Microbiol. 13, 893692 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Massey A. J., Multiparametric cell cycle analysis using the operetta high-content imager and harmony software with PhenoLOGIC. PLoS One 10, e0134306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayward S. W., et al. , Establishment and characterization of an immortalized but non-transformed human prostate epithelial cell line: BPH-1. In Vitro Cell Dev. Biol. Anim. 31, 14–24 (1995). [DOI] [PubMed] [Google Scholar]
- 50.Wu Q., et al. , Benign prostatic hyperplasia (BPH) epithelial cell line BPH-1 induces aromatase expression in prostatic stromal cells via prostaglandin E2. J. Endocrinol. 195, 89–94 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Coceres V. M., et al. , The C-terminal tail of tetraspanin proteins regulates their intracellular distribution in the parasite Trichomonas vaginalis. Cell Microbiol. 17, 1217–1229 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Chen Y.-P., Riestra A. M., Rai A. K., Johnson P. J., A novel cadherin-like protein mediates adherence to and killing of host cells by the parasite Trichomonas vaginalis. mBio 10, e00720-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ewels P., Magnusson M., Lundin S., Käller M., MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ashburner M., et al. , Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aleksander et al. ; Gene Ontology Consortium, The Gene Ontology knowledgebase in 2023. Genetics 224, iyad031 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Miguel N., et al. , Proteome analysis of the surface of Trichomonas vaginalis reveals novel proteins and strain-dependent differential expression. Mol. Cell Proteomics 9, 1554–1566 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riestra A. M., et al. , A Trichomonas vaginalis rhomboid protease and its substrate modulate parasite attachment and cytolysis of host cells. PLoS Pathog. 11, e1005294 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Molgora B. M., et al. , A novel Trichomonas vaginalis surface protein modulates parasite attachment via protein: Host cell proteoglycan interaction. mBio 12, e03374-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dyall S. D., et al. , Presence of a member of the mitochondrial carrier family in hydrogenosomes: Conservation of membrane-targeting pathways between hydrogenosomes and mitochondria. Mol. Cell Biol. 20, 2488–2497 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Müller M., Meingassner J. G., Miller W. A., Ledger W. J., Three metronidazole-resistant strains of Trichomonas vaginalis from the United States. Am. J. Obstet. Gynecol. 138, 808–812 (1980). [DOI] [PubMed] [Google Scholar]
- 61.Bates M., Huang B., Dempsey G. T., Zhuang X., Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science 317, 1749–1753 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van de Linde S., et al. , Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 6, 991–1009 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Molgora B. M., et al. , Trichomonas vaginalis adherence phenotypes and extracellular vesicles impact parasite survival in a novel in vivo model of pathogenesis. PLoS Negl. Trop. Dis. 17, e0011693 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Field M. C., Gabernet-Castello C., Dacks J. B., Reconstructing the evolution of the endocytic system: Insights from genomics and molecular cell biology. Adv. Exp. Med. Biol. 607, 84–96 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Kirkham M., et al. , Evolutionary analysis and molecular dissection of caveola biogenesis. J. Cell Sci. 121, 2075–2086 (2008). [DOI] [PubMed] [Google Scholar]
- 66.Takaha T., Smith S. M., The functions of 4-alpha-glucanotransferases and their use for the production of cyclic glucans. Biotechnol. Genet. Eng. Rev. 16, 257–280 (1999). [DOI] [PubMed] [Google Scholar]
- 67.Bastida-Corcuera F. D., Okumura C. Y., Colocoussi A., Johnson P. J., Trichomonas vaginalis lipophosphoglycan mutants have reduced adherence and cytotoxicity to human ectocervical cells. Eukaryot. Cell 4, 1951–1958 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Handrich M. R., Garg S. G., Sommerville E. W., Hirt R. P., Gould S. B., Characterization of the BspA and Pmp protein family of trichomonads. Parasit. Vectors 12, 406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Margarita V., et al. , Two different species of Mycoplasma endosymbionts can influence Trichomonas vaginalis pathophysiology. mBio 13, e0091822 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Niu X., Gupta K., Yang J. T., Shamblott M. J., Levchenko A., Physical transfer of membrane and cytoplasmic components as a general mechanism of cell-cell communication. J. Cell Sci. 122, 600–610 (2009). [DOI] [PubMed] [Google Scholar]
- 71.Ducret A., Fleuchot B., Bergam P., Mignot T., Direct live imaging of cell-cell protein transfer by transient outer membrane fusion in Myxococcus xanthus. Elife 2, e00868 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Diamond L. S., The establishment of various trichomonads of animals and man in axenic cultures. J. Parasitol. 43, 488–490 (1957). [PubMed] [Google Scholar]
- 73.Bray N. L., Pimentel H., Melsted P., Pachter L., Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Soneson C., Love M. I., Robinson M. D., Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Res. 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ritchie M. E., et al. , Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tyanova S., Temu T., Cox J., The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016). [DOI] [PubMed] [Google Scholar]
- 78.Kohler D., et al. , MSstats version 4.0: Statistical analyses of quantitative mass spectrometry-based proteomic experiments with chromatography-based quantification at scale. J. Proteome Res. 22, 1466–1482 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goodman R. P., et al. , Clinical isolates of Trichomonas vaginalis concurrently infected by strains of up to four Trichomonasvirus species (Family Totiviridae). J. Virol. 85, 4258–4270 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kochanowsky J. A., et al. , RNA sequencing Data from “Trichomonas vagainlis extracellular vesicles up-regulate and directly transfer adherence factors promoting host cell colonization.” Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE253522. Deposited 18 January 2024. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Dataset S09 (XLSX)
Dataset S10 (XLSX)
Dataset S11 (XLSX)
ExoglowTM Green-labeled TvEVs are internalized by T. vaginalis. Time-lapse video showing the uptake of fluorescently labeled G3-EVs by T. vaginalis strain G3.
Data Availability Statement
Raw sequence data are available on the Gene Expression Omnibus (GEO; accession # GSE253522) (80). All other data are included in the manuscript and/or supporting information.