Abstract
Background:
Hospital refrigerators as essential food storage can be important source of food contamination. We aimed to investigate the frequency and antibiotic susceptibility of the pathogenic bacteria in three hospital refrigerators in Tehran.
Methods:
This study was performed on 254 samples, collected from 60 refrigerators of the various wards of three hospitals, A, B, and C, in Tehran, Iran from 2020 to 2021. Following isolation and identification of isolates, the antibiotic susceptibility pattern was determined. PCR-based assays were used to screen the presence of antibiotic resistance genes of resistant isolates.
Results:
From 254 collected samples, 236 samples (92.9%) were contaminated. Most strains were isolated from refrigerators with poorly cleaned, temperatures above 8 °C in non-critical wards. Most bacteria belonging to Enterobacteriaceae (68.8%), followed by Staphylococcus (11.9%), and Enterococcus (10.6%), while the frequency of non-Enterobacteriaceae isolates was 8.9%. The highest antibiotic resistant bacteria were in extended spectrum beta-lactamase (ESBL) 9.7%, vancomycin-resistant enterococci (VRE) 5.3%, methicillin-resistant S. epidermidis (MRSE) 0.4%, and methicillin-resistant S. aureus (MRSA) 0.4%, respectively. The blaOXA-48, blaCTX, and bclaTEM genes were found only in 10% of Enterobacteriaceae isolates. The blaOXA-51 gene was found in all non-Enterobacteriaceae isolates. The vanA and mecA genes were detected in antibiotic-resistant Enterococcus and Staphylococcus.
Conclusion:
Our findings suggests major concern about cross-contamination and the emergence of antibiotic-resistant isolates as a potential health threat with hospital refrigerators origin. More attention to hospital refrigerators cleaning is necessary to prevent foodborne diseases and nosocomial infections.
Keywords: Foodborne diseases, Antibiotic resistance, Hospital refrigerators, Food safety
Introduction
Improper cleanliness of surfaces, equipments, and places for food processing and storage may potentially accelerate the distribution of pathogenic bacteria, cross-contamination, and the prevalence of food-borne diseases (1, 2). Cross-contamination is the pollution of food from other sources. A good cleaning with appropriate disinfectants can help to prevent foodborne illness from occurring through cross-contamination (1). In addition, foods can be sources of the prominent pathogens of nosocomial infections (3). Therefore, this is a very important issue that needs special attention in refrigerators of various wards of hospitals, which can be a possible source of the prevalence of foodborne nosocomial infections in hospitalized high-risk patients (4). Taking antibiotics in such patients increases the chance of developing antibiotic resistance (5).
Currently, the worldwide prevalence of multi-drug resistant (MDR) bacteria are well-recognized to be one of the most common health issues (5, 6). The methicillin-resistant Staphylococcus (MRSA, MRSE), vancomycin-resistant Enterococcus (VRE), carbapenem-resistant Klebsiella, and extended-spectrum beta - lactamases (ESBLs)-producing Enterobacteriaceae are the most important MDR bacteria leading to increased morbidity and mortality of hospitalized patients (7, 8). Mechanisms of resistance to antibiotics may be related to the combination of the enzymatic degradation, changes in membrane permeability, and modification of target proteins (9). The resistance genes play a vital role in the virulence and antimicrobial resistance (10). The presence of antimicrobial-resistant microorganisms or antibiotic resistance genes in foods, it is a potential major public health challenge (11).
Although there are some reports on the bacterial contaminations in the surfaces, equipment, and environment of hospitals focusing on the antibiotic resistance (12, 13), less attention has been paid to food contaminated by antimicrobial-resistant bacteria in the hospital refrigerators.
The present study, for first time, aimed to investigate the presence, frequency, and distribution of the foodborne antibiotic-resistant bacteria in the hospital’s refrigerators in Tehran.
Materials and Methods
Bacterial isolates
Random sampling of 60 refrigerators in the wards of three hospitals with more than 400 beds (A, B, and C located in Tehran, Iran 2020 to 2021) was performed once a week for ten consecutive months. The sampling part of hospitals was critical and non-critical wards (ICU, CCU, and surgeries). Wet swabs were taken from the walls, floors, and the inner surface of the refrigerator doors. Samples were enriched for 24 h at 37 °C in tryptic soy broth (TSB) medium and then cultured in MacConkey, blood agar, and xylose lysine deoxycholate (XLD) agar followed by incubation for 24 h at 37 °C. After observing growth, the strains were then purified by re-culturing. The doubtful colonies were identified according to the Gram staining, and standard biochemical tests such as catalase, oxidase, coagulase, mannitol, glucose and lactose fermentation, hemolysis, indole, motility, citrate, urease, amino acid decarboxylase (lysine, ornithine, arginine), and MR-VP test (14). For further analyses, isolates were cultured in Luria-Bertani (LB) broth medium and were incubated at 37 °C for 24 h and their cultures were preserved at −70 °C supplemented with 20% glycerol.
Antimicrobial susceptibility testing
Routine laboratory antibiotics (UK MAST company) including amikacin (AMK: 30 μg), ampicillin (AMP: 30μg), aztreonam (ATM: 30 μg), ciprofloxacin (CIP: 5 μg), ceftriaxone (CRO: 30 μg), ceftazidime (CAZ: 30 μg), cefoxitin (FOX: 30 μg), gentamicin (GEN: 10 μg), imipenem (IMP: 10 μg), lioezolid (LNZ: 10 μg), meropenem (MEM: 10 μg), tetracycline (TE: 10 μg), tobramycin (TN: 10 μg), and vancomycin (VAN: 30 μg) were used for antibiogram testing. Antibiotic susceptibility of antibiotics was determined by disk agar diffusion method by using Müller-Hinton-agar (MHA) medium according to Clinical and Laboratory Standards Institute (CLSI) guidelines and the results were reported as sensitive (S), intermediate (I), and resistance (R) (15).
For vancomycin, its susceptibility/resistance pattern was firstly reported using VAN disc; ≥17 mm as sensitive, 15–16 mm intermediate and ≤ 14 mm as resistant. Moreover, agar dilution method using BHI agar was also done as per CLSI (Supplement M100; 2021) guidelines. E. faecalis ATCC 29212 (susceptible) and E. faecalis ATCC 51299 (resistant) were used as control strains. Presence of >1 colony indicated presumptive vancomycin resistance. Interpretive criteria were defined as per CLSI: MIC ≤ 4 sensitive, 8–16 intermediate and ≥ 32 as resistant.
DNA Extraction
DNA extraction from bacteria was performed by the boiling method according to the described method (16) and its quality was quantified with a nanodrop.
Detection of antibiotic resistance genes by PCR
The antibiotic resistance genes including blaTEM, blaCTX-M, blaSHV (ESBL) and blaOXA-48 (carbapenemases) in Enterobacteriaceae, blaOXA-23, blaOXA-24, and blaOXA-51 (class D carbapenemases) in non-Enterobacteriaceae (Acinetobacter), vanA (VRE) in Enterococcus, and mecA (MRSA, MRSE) in Staphylococcus were assayed by PCR using specific oligonucleotide primers (Table 1). Amplification was performed using a BioRad MJ MiniTM PCR system with pervious reported protocols and the specific annealing temperature for each gene (Table 1) and the band pattern was analyzed by a gel documentation system (7, 8, 17). Positive control and negative control were included in each PCR run. Positive controls for ESBL genes and vanA were respectively E. coli ATCC 35218 and E. faecalis ATCC 51299. For blaOXA-48, class D carbapenemases, and mecA, a carbapenemase-resistant Klebsiella, Acinetobacter, and an MRSA isolate, characterized previously in our laboratory by sequencing served as positive control.
Table 1:
The list of primers, annealing temperatures, and expected amplicon sizes for molecular detection of antibiotic resistance genes of isolates.
| Group of Resistance | Type of bacteria | Gene | Primers (5′–3′) | Annealing | Product size (bp) | Ref. |
|---|---|---|---|---|---|---|
| ESBL | Enterobacteriaceae | bla TEM | F:TCCGCTCATGAGACAATAACC | 57 °C | 931 | (17) |
| R:TTGGTCTGACAGTTACCAATGC | ||||||
| bla CTX-M | F: CTTCCAGAATAAGGGAATCCC | 909 | ||||
| R: CCGTTTCCGCTATTACAAAC | ||||||
| bla SHV | F: TGGTTATGCGTTATATTCGCC | 64 °C | 868 | |||
| R:GGTTAGCGTTGCCAGTGCT | ||||||
| Carbapenemases | Enterobacteriaceae (Klebsiella) | bla OXA-48 | F: GCGTGGTTAAGGATGAACAC | 30 °C | 438 | (18) |
| R: CATCAAGTTCAACCCAACCG | ||||||
| Non-Enterobacteriaceae (Acinetobacter) | bla OXA-23 | F:GATCGGATTGGAGAACCAGA | 52 °C | 501 | (19) | |
| R:ATTTCTGACCGCATTTCCAT | ||||||
| bla OXA-24 | F:GGTTAGTTGGCCCCCTTAAA | 246 | ||||
| R:AGTTGAGCGAAAAGGGGATT | ||||||
| bla OXA-51 | F:TAATGCTTTGATCGGCCTTG | 353 | ||||
| R:TGGATTGCACTTCATCTTGG | ||||||
| VRE | Enterococcus | vanA | F:GGGAAAACGACAATTGC | 50 °C | 732 | (20) |
| R:GTACAATGCGGCCGTTA | ||||||
| MRSA, MRSE | Staphylococcus | mecA | F:GTAGAAATGACTGAACGTCCGATAA | 310 | (21) | |
| R:CCAATTCCACATTGTTTCGGTCTAA |
Statistical analysis
The results were expressed as absolute frequencies and percentages. For the statistical analyses, the statistical software SPSS version 21.0 for Windows (IBM Corp., Armonk, NY, USA) was used. Analyses were performed with using oneway analysis of variance (ANOVA) and Tukey’s test to compare the differences between the means (P<0.05). Chi-Square test was used for the significant relationship between the groups. The curves were plotted using Excel software version 2016 (Microsoft Corporation, USA).
The experiments were carried out based on guidelines of the Research Ethical Committee of Islamic Azad University-Science and Research Branch.
Results
The frequency of isolates
Among 254 samples collected from the refrigerators of three hospitals, 236 (92.9%) samples showed bacterial growth while no bacterial growth were observed in 18 (7.1%) samples. Chi-Square test showed a significant relationship between the apparent cleanliness of refrigerators and the growth of bacteria (P<0.05). Most isolated bacteria were found at temperatures above 8°C (70/74; 94.6%) and the lowest bacteria were obsreved at temperatures below 2 °C (11/13; 84.6%). The isolated bacteria from the floor of refrigerators (91/93; 97.8%) was significantly higher than their door (84/88; 95.5%) and inner walls (61/73; 83.6%) (P=0.001). In addition, 90.8% (119/131) of samples of critical wards were positive, while this rate was 95.1% (117/123) for non-critical wards. In addition, 91.9% (204/222) of samples collected from weekly cleaned refrigerators were showed the bacterial contamination.
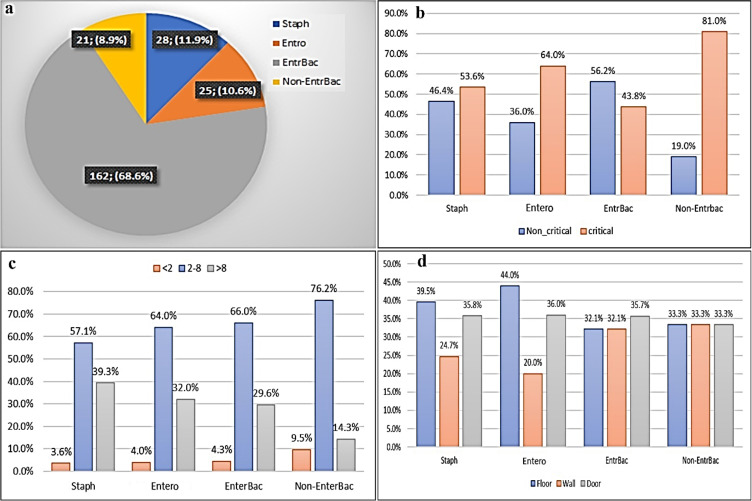
The isolates were divided into four bacterial groups: Staphylococcus, Enterococcus, Enterobacteriaceae and non-Enterobacteriaceae. The highest isolates belonged to Enterobacteriaceae and the non-Enterobacteriaceae was the lowest isolates (Fig. 1a). The Chi-Square analysis showed a significant association between the frequency of isolated bacteria with the wards of the hospital (P<0.05). Most isolates of Staphylococcus, Enterococcus, and non-Enterobacteriaceae groups were obtained from the critical wards, while most isolates of Enterobacteriaceae were isolated from the non-critical wards (Fig. 1b). The most isolates in each bacterial group were isolated from refrigerators with temperatures 2–8 °C (Fig. 1c). In all temperature ranges (<2, 2–8, and >8), the highest frequency isolates belonged to the Enterobacteriaceae (162/236; 68.6%). The highest frequency of Staphylococcus and Enterococcus isolates were obtained from the floors, door, and inner walls of refrigerators, respectively. Most of the Enterobacteriaceae isolates were identified in the refrigerator’s door, while the frequency of the non-Enterobacteriaceae isolates was equal in different parts of the refrigerators (Fig. 1d). Among the Enterobacteriaceae isolates, E. coli, K. pneumoniae, and P. aeruginosa were the frequent isolates, whereas the Salmonella spp were not found in any of the refrigerators.
Fig. 1:
The total frequency of bacteria groups (Staphylococcus, Enterococcus, Enterobacteriaceae. and non-Enterobacteriaceae) isolated from hospitals refrigerators (a), the frequency of bacteria groups in hospitals refrigerators based on ward type (b), temperature (c), and different parts (floor, wall, door) of refrigerators (d)
Determination of antibiotic susceptibility
The antibiotic susceptibility test (Table 2) showed that 18.2% (43/236) of isolates were antibiotic-resistant and 81.8% (193/236) of the isolated bacteria were sensitive to antibiotics. Among the antibiotic-resistant isolates, the highest percentage of resistance belonged to ESBL group followed by VRE. The lowest resistance was also related to MRSE and MRSA groups (Table 3).
Table 2:
Antimicrobial resistance profile of isolates
| Antibiotic | No. (%) of antibiotic resistance isolates | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enterobacteriaceae (n=162) | Non-Enterobacteriaceae (n=21) | Staphylococcus (n=28) | Enterococcus (n=25) | |||||||||
| S | I | R | S | I | R | S | I | R | S | I | R | |
| Amikacin | 162 (100) | 0 | 0 | 21 (100) | 0 | 0 | - | - | - | - | - | - |
| Ampicillin | 79 (48.8) | 83 (51.2) | 0 | 17 (80.9) | 4 (19.1) | 0 | - | - | - | - | - | - |
| Aztreonam | 159 (98.2) | 1 (0.6) | 2 (1.2) | 16 (76.2) | 0 | 5 (23.9) | - | - | - | - | - | - |
| Ciprofloxacin | 159 (98.2) | 2 (1.2) | 1 (0.6) | 16 (76.2) | 0 | 5 (23.9) | - | - | - | - | - | - |
| Ceftriaxone | 161 (99.4) | 0 | 1 (0.6) | 20 (95.2) | 0 | 1 (4.8) | - | - | - | - | - | - |
| Ceftazidime | 156 (96.3) | 0 | 6 (3.7) | 15 (71.4) | 0 | 6 (28.6) | - | - | - | - | - | - |
| Imipenem | 157 (96.9) | 0 | 5 (3.1) | 15 (71.4) | 0 | 6 (28.6) | - | - | - | - | - | - |
| Meropenem | 158 (97.5) | 0 | 4 (2.5) | 15 (71.4) | 0 | 6 (28.6) | - | - | - | - | - | - |
| Tetracycline | 162 (100) | 0 | 0 | 17 (80.9) | 0 | 4 (19.1) | 28 (100) | 0 | 0 | 25 (100) | 0 | 0 |
| Gentamycin 10 | 162 (100) | 0 | 0 | 17 (80.9) | 0 | 4 (19.1) | 28 (100) | 0 | 0 | 25 (100) | 0 | 0 |
| Gentamycin 120 | 162 (100) | 0 | 0 | - | - | - | - | - | - | 25 (100) | 0 | 0 |
| Linezolid | 162 (100) | 0 | 0 | - | - | - | - | - | - | 24 (96) | 0 | 1 (4) |
| Vancomycin | - | - | - | - | - | - | - | - | - | 13* (52) | 0 | 12** (48) |
| Cefoxitin | - | - | - | - | - | - | 26 (92.8) | 0 | 2 (7.2) | - | - | - |
S: sensitive, I: intermediate, R: resistance
The vancomycin date repored based on MIC results:
* MIC ≤ 4, sensitive;
** MIC ≥ 32, resistant
Table 3:
Antibiotic resistance profile of isolates
| Antibiotic-resistance group | Total frequency (%) | Bacterial group | |||
|---|---|---|---|---|---|
|
| |||||
| Enterobacteriaceae | Non-Enterobacteriaceae | Staphylococcus | Enterococcus | ||
| MRSA | 1 (0.42) | 0 | 0 | 1 (3.57) | 0 |
| MRSE | 1 (0.42) | 0 | 0 | 1 (3.57) | 0 |
| VRE | 12 (5.10) | 0 | 0 | 0 | 12* (48) |
| ESBL | 10 (4.23) | 10 (6.18) | 0 | 0 | 0 |
| Carbapenemases | 13 (5.52) | 13 (61.90) | 0 | 0 | |
| Non-resistance | 193 (81.77) | 148 (91.36) | 6 (28.58) | 26 (92.85) | 13 (52) |
| Other antibiotic-resistance group | 6 (2.54) | 4 (2.46) | 2 (9.52) | 0 | 0 |
| Total | 236 (100) | 162 (100) | 21 (100) | 28 (100) | 25 (100) |
* MIC ≥ 32, resistant
Antibiotic resistance genes analysis
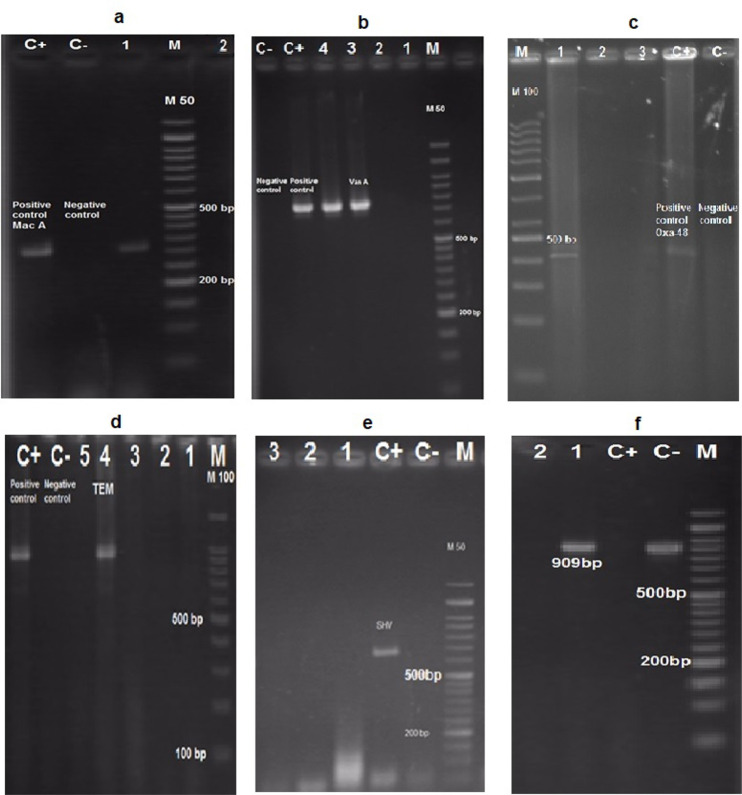
The antibiotic-resistant bacteria were selected to investigate the presence of resistant genes. The PCR reaction confirmed the presence of resistance genes of blaTEM, blaCTX-M, and blaOXA-48 in Enterobacteriaceae group, blaOXA-23 and blaOXA51 in non-Enterobacteriaceae, vanA in Enterococcus, and mecA in Staphylococcus in compared with positive controls (Fig.2). The frequency of antibiotic resistance genes revealed that the blaTEM, blaCTX-M, and blaOXA-48 genes in 1 of 10 resistant Enterobacteriaceae, while the blaSHV gene was not found in them. The presence of blaOXA-48 gene confirmed the carbapenem-resistant Klebsiella. The blaOXA-51 and blaOXA-23 genes were respectively identified in 13 and 2 resistant non-Enterobacteriaceae isolates, while the blaOXA-24 gene was not found in any of the isolates. The high frequency of blaOXA-51 genes indicated that the prevalence of carbapenemase encoding Acinetobacter. The result also showed that one of 2 isolates of the MRSA/MRSE Staphylococcus group carried the mecA gene and 4 isolates of the VRE Enterococcus group (out of 12 cases) were encoded the vanA gene. The presence of MRSA and VRE isolates containing these genes in the hospital refrigerators.
Fig. 2:
Gel electrophoresis for characterization of resistance genes of mecA (a), vanA (b), blaOXA-48 (c), blaTEM (d), blaSHV (e), and blaCTX-M (f). C+: positive control; C−: negative control
Discussion
The role of hospital refrigerators in the food contamination and the prevalence of foodborne illness or nosocomial infections are important. This investigation provided detailed evidence about the presence, frequency, and distribution of the foodborne antibiotic-resistant pathogenic bacteria in the refrigerators of three hospitals in Tehran. Among 254 samples, 236 (92.9%) samples were shown bacterial contamination, and only in 18 (7.1%) samples, no bacterial growth was observed, which was close to the results of related studies performed on the bacterial and fungal contaminations in the home and restaurant refrigerators (22, 23). Most frequent isolates belonged to Enterobacteriaceae, Staphylococcus, Enterococcus, and non-Enterobacteriaceae, respectively.
We primarily examined the relationship between bacterial contamination and the appearance of the refrigerators in different wards of the hospitals. The lowest bacterial contamination was observed in the cleaned refrigerators (16.4%), that demonstrate that the regular and weekly cleaning of the refrigerators by means of appropriate detergents and disinfectants, can effectively reduce their bacterial contamination. The low temperatures, similar to the refrigerators, is another vital factor in the prevention of the growth of pathogenic bacteria in foods. When the appliance is full, the temperature may increase to a higher range via inhibition of air circulation, which can potentially affect the microbial contamination of the refrigerator (24, 25). The current findings were consistent with this principle.
In the current study, the highest isolated bacteria belonged to the floor and door. The preservation of different materials on the floors, and not cleaning the refrigerators at short intervals can be the reasons for high contamination of the floors (26). The higher bacterial contamination (90.8%) in the critical wards (ICU and CCU) in this study may reflected the improper cleaning of refrigerators located in those parts. The high prevalence of Enterobacteriaceae in non-critical wards may be attributed to the human origin that can possibly create foods cross-contamination by coliform bacteria due to poor hygiene. In addition, the resistance of these bacteria to detergents and antibiotics can be another factor. Here, among the common foodborne bacteria, E. coli, and S. aureus were the frequent isolates, whereas Salmonella spp were not found in any of the refrigerators, which was similar to the previous studies (27–29).
Antibiotic-resistant bacteria are a major cause of hospital-acquired infections and a serious public health concern (6). A high percentage of MRSA and ESBL resistant bacteria (~65%) were reported by Taheri in Arak hospital (22). Ekrami et al reported MRSA isolates in 60% of isolates (29). In another study, the frequency of MDR-MRSA was found at 91.6% in hospital’s kitchens samples and 18.7% in ICU (30). The results of these studies were higher than the results of the current study. In the current study due to our samples only collected from hospital’s refrigerators, the frequency of antibiotic-resistant bacteria were low, whereas in the previous studies samples have collected from different sources in the hospital.
The antibiotic resistance genes are the basis of the high prevalence of the antibiotic-resistant bacterial. The frequency of blaTEM, blaCTX-M, and blaOXA-48 genes (10%) in isolated Enterobacteriaceae was similar to the Tamma et al results, which observed blaCTX-M gene in 11% of gram-negative bacteria (31). The both blaOXA-51 and blaO-XA-23 genes were observed in 2 isolates, suggesting the carbapenemase-resistant isolates (32, 33). The co-existence of multiple resistance genes was probably due to the presence of multiple resistance plasmids in the isolates, which can enhance antimicrobial resistance (34).
The co-resistance to vancomycin and methicillin emphasizes that a concern regarding the emergence of MDR isolates. Vancomycin and methicillin resistance can be conferred by the vanA and mecA genes (35). In a study conducted by Hussein et al, out of 109 MRSA isolates, 55 (50.4%) isolates carried the mecA gene (36). In other studies, the frequency of vanA gene was found 30% (37) and 47.4% (38). The frequency of mecA and vanA genes in these studies were higher than the present study (mecA:0.4% and vanA:5.3%). Antibiotic resistance is often achieved by horizontal transfer of resistance genes. Plasmid-mediated transfer of antibiotic resistance play a crucial role in the transfer of resistance genes between different species (39). The simultaneous presence of VRE and MRSA/MRSE isolates can possibly transmit vanA and mecA genes to other Staphylococcus or Enterococcus and their resistance (8, 40).
This study was limited by its relatively small sample size. The study collection comprised only strains isolated within one year, collected from only the refrigerators of a few hospitals in Tehran, the capital of Iran and may not necessarily be representative of the epidemiology of foodborne pathogenic bacteria in other areas of Iran. The antibiotic-resistant isolates from the refrigerator varies from one hospital to another, depending on the local pattern of antibiotic resistance, infection control activities, and the level of public health. For this reason, it is necessary to perform further studies on antibiotic-resistant bacteria isolated from refrigerators of hospitals in various regions of Iran.
Conclusion
Although the strains isolated in our study did not have high resistance to the antibiotics tested in this study, there were only a few resistant strains with refrigerators origin with a potential to acquire other resistance. The utilization of broad-spectrum antibiotics by patients admitted to the hospital can be one of the effective factors in increasing the resistance of foodborne bacteria in the hospital environment. Therefore, it is recommended that the proper and timely cleaning of hospital refrigerators to prevent the prevalence of foodborne antibiotic-resistant bacteria and nosocomial infections.
Journalism Ethics considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors. Since this study did not include any human or animal sample,the institutional research board did not require to obtain an ethical code.
Acknowledgements
The authors wish to thank Dr. Shayan Ziaee and Leyla Pourgholi for the technical assistance and molecular experiments.
Footnotes
Conflict of interest
The authors declare that there is no conflict of interests.
References
- 1.Pieniz S, Rodrigues D, Arndt R, et al. (2019). Molecular identification and microbiological evaluation of isolates from equipments and food contact surfaces in a hospital food and nutrition unit. Braz J Biol, 79:191–200. [DOI] [PubMed] [Google Scholar]
- 2.Kirchner M, Goulter RM, Chapman BJ, et al. (2021). Cross-contamination on atypical surfaces and venues in food service environments. J Food Prot, 84(7):1239–51. [DOI] [PubMed] [Google Scholar]
- 3.Akova M. (2016). Epidemiology of antimicrobial resistance in bloodstream infections. Virulence, 7(3):252–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuster-Valls N, Hernández-Herrero M, Marínde-Mateo M, et al. (2008). Effect of different environmental conditions on the bacteria survival on stainless steel surfaces. Food Control, 19(3):308–14. [Google Scholar]
- 5.Beceiro A, Tomás M, Bou G. (2013). Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev, 26(2):185–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pourgholi L, Farhadinia H, Hosseindokht M, et al. (2022). Analysis of carbapenemases genes of carbapenem-resistant Klebsiella pneumoniae isolated from Tehran heart center. Iran J Microbiol, 14(1):38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chellat MF, Raguž L, Riedl R. (2016). Targeting antibiotic resistance. Angew Chem Int Ed Engl, 55(23):6600–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheraghi S, Pourgholi L, Shafaati M, et al. (2017). Analysis of virulence genes and accessory gene regulator (agr) types among methicillin-resistant Staphylococcus aureus strains in Iran. J Glob Antimicrob Resist, 10:315–20. [DOI] [PubMed] [Google Scholar]
- 9.Sultan I, Rahman S, Jan AT, et al. (2018). Antibiotics, resistome and resistance mechanisms: a bacterial perspective. Front Microbiol, 9:2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan Y, Zeng J, Li L, et al. (2020). Coexistence of antibiotic resistance genes and virulence factors deciphered by large-scale complete genome analysis. mSystems, 5(3):e00821–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verraes C, Van Boxstael S, Van Meervenne E, et al. (2013). Antimicrobial resistance in the food chain: a review. Int J Environ Res Public Health, 10(7):2643–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chowdhury AMA, Uddin KN. (2022). Analysis of the occurrence of antibiotic resistant bacteria in the hospital’s effluent and its receiving environment. Microbiol Insights, 15:11786361221078211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mirón-Rubio M. (2021). Treatment of infections caused by multi-resistant microorganisms in hospital at home units. Rev Esp Quimioter, 34 Suppl 1(Suppl1):18–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray PR, Rosenthal KS, Pfaller MA. Medical microbiology e-book: Elsevier Health Sciences; 2020. [Google Scholar]
- 15.Humphries R, Bobenchik AM, Hindler JA, et al. (2021). Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100. J Clin Microbiol, 59(12):e00213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chelab RL. (2019). Rapid and Inexpensive DNA extraction protocol from gram negative and gram positive bacteria. Indian J Public Health Res Dev, 10(5):542–44. [Google Scholar]
- 17.Soriano-Moreno DR, Yareta J, Rojas-Cosi AF, et al. (2021). Hospital effluents as a reservoir of beta-lactamase-and carbapenemase-producing Enterobacteriaceae. Rev Peru Med Exp Salud Publica, 38(2):302–307. [DOI] [PubMed] [Google Scholar]
- 18.Hatrongjit R, Kerdsin A, Akeda Y, et al. (2018). Detection of plasmid-mediated colistin-resistant and carbapenem-resistant genes by multiplex PCR. MethodsX, 5:532–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowings M, Ehlers MM, Dreyer AW, et al. (2015). High prevalence of oxacillinases in clinical multidrug-resistant Acinetobacter baumannii isolates from the Tshwane region, South Africa–an update. BMC Infect Dis, 15:521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tatsing Foka FE, Ateba CN. (2019). Detection of virulence genes in multidrug resistant Enterococci isolated from feedlots dairy and beef cattle: Implications for human health and food safety. Biomed Res Int, 2019:5921840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahrami N, Motamedi H, Tofighi SER, et al. (2019). SCCmec typing and Panton-valentine leukocidin occurrence in methicillin resistant Staphylococcus aureus (MRSA) isolates from clinical samples of Ahvaz, southwest of Iran. Microbiologia Medica, 34(2):43–48. [Google Scholar]
- 22.Taheri N, Abtahi H, Amozande-Nobaveh A, et al. (2014). The antibiotic resistant determinant of pathogenic bacteria isolated from medical equipment and hospital environment in Valiasr hospital, Arak, 2013. J Maz Univ Med Sci, 24(114):60–73. [Google Scholar]
- 23.Rahimi SM, Ebrahimi M, Barikbin B, et al. (2019). Evaluation of bacterial and fungal contamination of kitchens of Birjand University of Medical Sciences. BMC Res Notes, 12(1):703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belman-Flores JM, Pardo-Cely D, Gómez-Martínez MA, et al. (2019). Thermal and energy evaluation of a domestic refrigerator under the influence of the thermal load. Energies, 12(3):400. [Google Scholar]
- 25.Lalitha C. (2019). Contamination of refrigerator is a threat for infections. International Journal of Advance Research, Ideas and Innovations in Technology, 5(2):1514–17. [Google Scholar]
- 26.Kumar MR, Rishu B, Osborne J. (2012). Isolation of various bacterial pathogens from domestic refrigerators. Asian J Pharm Clin Res, 5(3):151–53. [Google Scholar]
- 27.Mori M, Sakagami Y, Tanaka M, et al. (2020). Analysis of the relationship of microbial contamination with temperature and cleaning frequency and method of domestic refrigerators in Japan. J Food Prot, 83(7):1234–40. [DOI] [PubMed] [Google Scholar]
- 28.Andritsos ND, Stasinou V, Tserolas D, et al. (2021). Temperature distribution and hygienic status of domestic refrigerators in Lemnos island, Greece. Food Control, 127:108121. [Google Scholar]
- 29.Ekrami A, Kayedani A, Jahangir M, et al. (2011). Isolation of common aerobic bacterial pathogens from the environment of seven hospitals, Ahvaz, Iran. Jundishapur J Microbiol, 4(2):75–82. [Google Scholar]
- 30.Gholammostafaei F, Alebouyeh M, Jabari F, et al. (2014). Prevalence of antibiotic resistant bacteria isolated from foodstuff in kitchen of a hospital in Tehran. J Ilam Univ Med Sci, 22(2):1–9. [Google Scholar]
- 31.Tamma PD, Smith TT, Adebayo A, et al. (2021). Prevalence of blaCTX-M genes in gram-negative bloodstream isolates across 66 hospitals in the United States. J Clin Microbiol, 59(6):e00127–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selasi GN, Nicholas A, Jeon H, et al. (2015). Genetic basis of antimicrobial resistance and clonal dynamics of carbapenem-resistant Acinetobacter baumannii sequence type 191 in a Korean hospital. Infect Genet Evol, 36:1–7. [DOI] [PubMed] [Google Scholar]
- 33.Hasan B, Perveen K, Olsen B, et al. (2014). Emergence of carbapenem-resistant Acinetobacter baumannii in hospitals in Pakistan. J Med Microbiol, 63(Pt 1):50–55. [DOI] [PubMed] [Google Scholar]
- 34.Rastegar-Lari A, Mohammadi-Barzelighi H, Arjomandzadegan M, et al. (2015). Distribution of class I integron among isolates of Acinetobacter baumannii recoverd from burn patients. J Med Bacteriol, 2(1–2):1–11. [Google Scholar]
- 35.Périchon B, Courvalin P. (2009). VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother, 53(11):4580–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hussein N, Salih RS, Rasheed NA. (2019). Prevalence of methicillin-resistant Staphylococcus aureus in hospitals and community in Duhok, Kurdistan region of Iraq. Int J Infect, 6(2):e89636. [Google Scholar]
- 37.Mirzaei B, Babaei R, Asiabar APD, et al. (2015). Detection of both vanA & vanB genes in vanA phenotypes of Enterococci by Taq Man RT-PCR. Braz J Microbiol, 46:161–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khairy RM, Mahmoud MS, Esmail MAM, et al. (2019). First detection of vanB phenotypevanA genotype vancomycin-resistant enterococci in Egypt. J Infect Dev Ctries, 13(9):837–42. [DOI] [PubMed] [Google Scholar]
- 39.Lerminiaux NA, Cameron AD. (2019). Horizontal transfer of antibiotic resistance genes in clinical environments. Can J Microbiol, 65(1):34–44. [DOI] [PubMed] [Google Scholar]
- 40.Heinze K, Kabeto M, Martin ET, et al. (2019). Predictors of methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococci co-colonization among nursing facility patients. Am J Infect Control, 47(4):415–20. [DOI] [PMC free article] [PubMed] [Google Scholar]