Abstract
Background:
We aimed to investigate miR-21-5p inhibition effect on lncRNA-XIST expression and apoptosis status of MCF-7 cells.
Methods:
The MCF-7 cells were cultured and transfected by the anti-miR-21-5p oligonucleotide and expression of miR-21-5p, lncRNA-XIST, apoptosis-associated genes (bax and p53) and one miR-21-5p-unrelated lncRNA (BC200) was assessed by RT-qPCR. Furthermore, cell viability checked by MTT assay and apoptosis and cell cycle in transfected cells were detected by flow cytometry. Also, bioinformatics analysis on the transcriptome data confirmed that the lncRNA XIST might have a critical role in breast cancer (BC) cell apoptosis through ceRNAs mechanism and possible regulatory interactions with miR-21-5p.
Results:
Expression of miR-21-5p and lncRNA-XIST was significantly down- and up-regulated respectively (P<0.05). However, there was no significant change in lncRNA-BC200 expression. Also, the expression of bax and p53 upraised significantly (P<0.05). In transfected cells, MTT and flow cytometry assays reported a highly significant decrease and increase in viability and apoptosis respectively.
Conclusion:
Inhibition of miR-21-5p resulted in significant upregulation of lncRNA-XIST and apoptosis-associated genes bax and p53, which led to the induction of apoptosis in MCF-7 cells. Therefore, more investigations may provide a valuable target for studies on molecular therapies for BC.
Keywords: Non-coding RNA, X-inactive specific transcript, Apoptosis, Breast cancer
Introduction
Being the most common cancer in females, breast cancer (BC), is one of the main global health concerns and a major cause of women deaths worldwide. Although the development and advancement of screening methods and therapies have improved survival rates, Global Cancer Observatory reports over two million new BC cases and more than 500 thousand deaths annually (1).
BC is a heterogeneous disease, which can be classified into different molecular subtypes with diverse molecular mechanisms. The risk for BC is a combination of a variety of factors such as older age, genetic mutations and polymorphisms, family history of BC and lifestyle (2) etc. In the same way, prognosis and diagnosis and response to therapies vary for different BC cases. Therefore, there are still many unaddressed questions about the molecular mechanisms of BC and the efficiency of the treatments. Consequently, research on underlying cellular and molecular mechanisms of BC and providing enough knowledge for new therapeutic approaches is urgently needed.
Due to their crucial regulatory roles, recently non-coding RNAs (ncRNAs) have gained a lot of attention in cancer research. In this regard, there is a major focus on the applicabilities of microRNAs (miRNAs) and long ncRNAs (lncRNAs) in BC diagnosis and treatment. Indeed, miRNAs have been proposed as master regulators of metastasis, tumor suppressors and oncogenic factors in BC with a range of promising opportunities for therapeutic and prognostic approaches. lncRNAs contribute in a wide range of molecular processes of BC such as regulation of transcriptional and post-transcriptional events, the plasticity of cancer stem cells and tumor development (3).
One of the most studied miRNAs in cancer that targets multiple genes involved in BC-signaling pathways is miR-21-5p. It interacts with BC suppressor genes and helping tumor progress represents an oncomiR activity. Accordingly, its downregulation has been suggested as a potential prevention and therapy approach for BC (4). X-inactive specific transcript (XIST), which is one of the first discovered lncRNAs in mammals with a master regulatory role in females’ X chromosome transcriptional inactivation (5), is dysregulated in numerous types of human cancers and implicates in many aspects of carcinogenesis. The lncRNAXIST expression was found to be significantly downregulated in BC tumor samples and model cell lines. In fact, it is shown that lncRNA-XIST has a suppressor role in BC by downregulating AKT activation (6).
The crosstalk and interaction of lncRNA-XIST with several types of miRNAs have been studied in several cellular pathways of cancer. Zhang et al reported reciprocal effects of lncRNA-XIST and miR-21-5p on each other’s expression, which results in inhibition of osteosarcoma (7).
Considering the above-mentioned arguments, there is an undeniable gap in current knowledge about the miR-21-5p and lncRNA-XIST interaction in BC. Consequently, we aimed to do a preliminary analysis for determining miR-21-5p inhibition effects on lncRNA-XIST expression and apoptosis in MCF-7 BC cells. In order to strengthen our results, we also examined miR-21-5p inhibition effect on lncRNA-BC200 expression. There is no experimental or predicted evidence for relatedness between lncRNA-BC200 and miR-21-5p, however, it has a high expression in invasive BC tumors (8).
Material and Methods
Bioinformatics analysis
To survey interacrtion between miR-21 and XIST, before other analysises, online databases like DAINA-LncBase were applied. Analysis of co-expressed genes is one of the ways to detect lncRNAs function in the cell. Therefore, to investigate the function of our lncRNAs and biological processes related to them, we extracted co-expressed mRNAs (Spearman correlation coefficient > 0.5) with the lncRNAs based on the breast cancer RNA-Seq data using co-lncRNA web tool (9). Given the cell lines used in this study, we selected TCGA breast cancer progesterone and estrogen receptor data in the co-lncRNA tool. To investigate the pathways and processes related to our lncRNAs, Enrichment analysis was applied to the extracted mRNAs based on Gene Ontology (GO) Biological Process (BP) data (10)and KEGG pathways (11) using co-lncRNA. The Benjamin and Hochberg method (12) was applied to correct the p-values and the results with corrected p-value>0.05 were ignored. Some co-expressed mRNAs with XIST physically interact with each other at the protein level, were investigated on the base of data extracted from the STRING database (13) and visualized with Cytoscape v. 3.6.1 (14). Based on competing endogenous RNAs (ceRNAs) hypothesis, the lncRNAs regulate the co-expressed mRNAs through shared miRNAs as mediators (15, 16). Therefore, to investigate the regulatory relationship between XIST and its co-expressed mRNAs based on the ceRNAs hypothesis, the TarBase database (17) was used to extract miRNA-mRNA data. Also, the lncBase v2 database (18) was used to identify those miRNAs that targeted previously extracted mRNAs.
Cell culture
The human breast cancer cell line MCF-7 was obtained from the National Cell Bank of Iran (NCBI, Pasteur Institute of Iran). The cells were grown and maintained using Dulbecco’s Modified Eagle Medium (Sigma, United States) supplemented with 10% heat-inactivated FBS (Gibco, Beijing, China), 100 μg/ml penicillin, 100 μg/ml streptomycin, 2 mM L-Glutamine (Invitrogen, United States) at 37°C under a 5% humidified CO2 atmosphere. All experiments were performed on cultures that were 70% confluent and cells were harvested using the trypsin-EDTA treatment.
Cell transfection
The sequence of anti-miR-21-5p was: 5′UCAACAUCAGUCUGAUAAGCUA3′ and 5′ end was marked with 6-FAM due to its fluorescent property to detect whether it has entered to the cells. The oligonucleotide of anti-miR-21-5p was purified by high-pressure liquid chromatography. The MCF-7 cells were seeded in 6-well plates and transfected with anti-miR-21-5p (Bioneer, Korea) using lipofectamine™2000 (Invitrogen, USA) according to the manufacturer’s instructions. Briefly, 176 μl of media was mixed separately with 12μl of lipofectamine in a microtube along with 10 μl of anti-miR-21-5p (50pmol/μl) and then incubated in dark at room temperature for 20 min to form a complex. After that, the medium was removed from wells and cells were washed using phosphate-buffered saline (PBS)1X and serum-free medium(SFM) in triplicate. Then, 1800 μl of medium containing 2.5% serum was added to the wells and after that lipofectamine and anti-miR-21-5p complex was added gently. The plate was incubated at 37 °C and 5% CO2 and after 8 h, the medium was removed and cells were washed with SFM. Then, the culture medium containing FBS 10% was added to the wells and incubated for 48 h at 37°C and 5% CO2 as described previously (7).
RNA extraction
Total RNA was extracted using Invitrogen™ TRI-zol™ Reagent (Carlsbad,CA,USA) and SanPrep Column microRNA Mini-Preps Kit (BIOBASIC,Canada) was used to extract ncRNAs following manufacturers’ protocol. To avoid genomic contamination, the RNA samples were treated with DNase.
Quantitative real-time PCR analysis
The cDNA was synthesized using PrimeScriptTM RT Master Mix (Takara Biotechnology, Dalian, China). The cDNA samples were amplified using WizPureTM qPCR Master (SYBR) Kit and Corbett Rotor gene 6000 real-time PCR cycler, with an initial denaturation temperature 95 °C for 5 min followed by 40 cycles of 95 °C for 10 s, 60 °C for 20 s, and 72 °C for 30 s. The melting temperature was set for 60 to 95°C. The relative quantification expression of target genes was calculated by the 2−ΔΔCT method. All RT-qPCR reactions were performed in triplicate. U6 and GAPDH expressions were used as internal controls for ncRNAs and other studied genes respectively. In addition, to forward and reverse primers (Table 1), the stem-loop primer was used for miR-21-5p with this sequence: CAATTAGACTACACCTGTCCGGTAGTGTGGCC CTGCGTCCTGTAGTCTAATTGTCAACA
Table 1:
The primers sequence for the RT-real time PCR
| Target genes | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| XIST | AGCTCCTCGGACAGCTGTAA | CTCCAGATAGCTGGCAACC |
| BC200 | GCCTGTAATCCCAGCTCTCA | GTTGCTTTGAGGGAAGTTACGCT |
| GAPDH | TCGTGGAAGGACTCATGACC | CCATCACGCCACAGTTTCCC |
| Bax | GATGCGTCCACCAAGAAG | AGTTGAAGTTGCCGTCAG |
| p53 | ACCTATGGAAACTACTTCCTGAAA | ACCATCGCTATCTGAGCAGC |
| mir-21 | GTGCAGGGTCCGAGGT | AGACTGCACCTGTCCGG |
| U6 | TGCGGGTGCTCGCTTCGGCAGC | CCAGTGCAGGGTCCGAGGT |
Cell viability assay
To investigate the inhibitory effect of anti-miR-21-5p on the viability of the MCF-7 cells, cell viabillity assay was used. The MTT stock prepared in PBS (0.5 mg/ml, pH 7.2) and 20 μl of it was added to each well and cells were incubated for 4 h at 37 °C with 5% CO2. Then, 150 μl of dimethyl sulfoxide was added to each well and 5 min incubation was repeated. Then, the absorbance at 570 nm was measured using a plate reader (BioTek, Epoch). The cell viabillity Assay was performed in triplicate and the percentage of surviving cells in transfected and untreated control cells was compared.
Apoptosis assay
The apoptotic transfected cell rates were analysed by Flow cytometry assay BD FACS Calibur (BD Biosciences, San Jose, CA, USA) with Annexin VFITC assay. Briefly, cells 48h after transfection were collected and according to instructions of apoptotic detection kits were prepared for staining. Untreated cells were used as control. Using FlowJo software, data were analyzed in terms of two-dimensional curves, and results were presented as the percentage of apoptotic cells.
Cell cycle assay
Transfected and control cells were collected and washed with PBS and fixed using cold 70% ethanol. After another PBS washing and spinning at 850 g in a centrifuge, 1ml of PI Master Mix containing 40 μl of PI, 10 μl of RNase, and 950 μl of PBS was added and incubated for 30 min. The measurements of forward scatter and side scatter were done by flow cytometry instrument, BD FACS Calibur (BD Biosciences, San Jose, CA, USA). Pulse processing was used to exclude cell doublets from the analysis. The data were analyzed by FlowJo software.
Statistical analysis
The data were analyzed statistically by Graph pad prism V6.0 software (Graph Pad Software Inc., San Diego, CA, USA) and were presented as mean ± SD (Standard deviation). Statistical significance was determined using t-test and P- values of * P < 0.05, ** P< 0.01, *** P < 0.001 were considered.
Results
Bioinformatics analysis
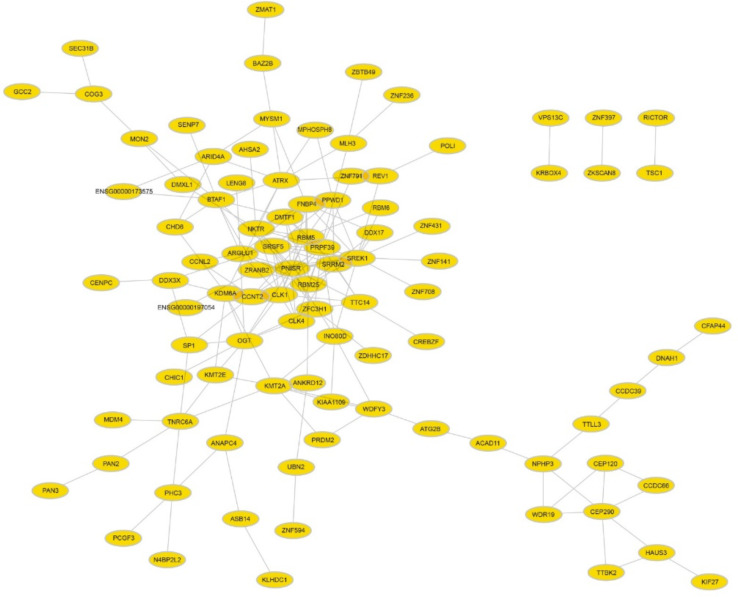
Obtained results from databases like DIANA-LncBase showed direct interaction between miR-21 and XIST (Fig. 1a). Moreover, the results from TCGA RNA-Seq data revealed the function of our lncRNAs and the processes and pathways related to them and extract the co-expressed mRNAs with XIST. Totally, 179 and 172 mRNAs were obtained from estrogen and progesterone RNASeq data respectively (Supplementary Files 1&2). These mRNAs are co-expressed with XIST, and 95 of them physically interact with each other at the protein level, which is shown in (Fig. 1b).
Fig.1a:
Direct interaction between miR-21 and XIST
Fig. 1b:
The protein interaction network of the mRNAs co-expressed with XIST (the data extracted from the STRING database and visualized with Cytoscape v. 3.6.1
However, cancer analysis of the other mRNAs which can potentially act as ceRNAs with XIST shows that there are some other apoptotic genes that may be overexpressed after overexpression of this lncRNA and drive the cancer cell to apoptosis. Among obtained mRNAs, there were some important genes such as bax, bad, and bak that participate in apoptosis processes (Supplementary File 3).
Cell transfection
Anti-miR-21-5p movement into the cells was confirmed by a fluorescent microscope due to 6-FAM in the 5′ end of anti-miR, which produces fluorescent dots (Fig. 2a and b).
Fig. 2a:
MCF-7 cells before transfection with anti-miR-21-5p
Fig. 2b:
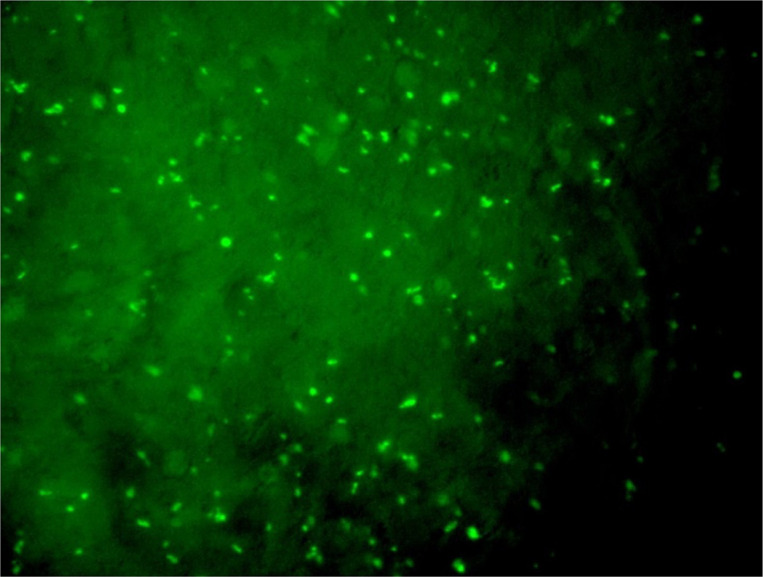
MCF-7 cells after transfection with anti-miR-21-5p. Bright fluorescent dots are due to 6-FAM in the 5′ end of anti-miR-5p, which confirms the oligonucleotide entrance into the cells
Quantitative real-time PCR analysis
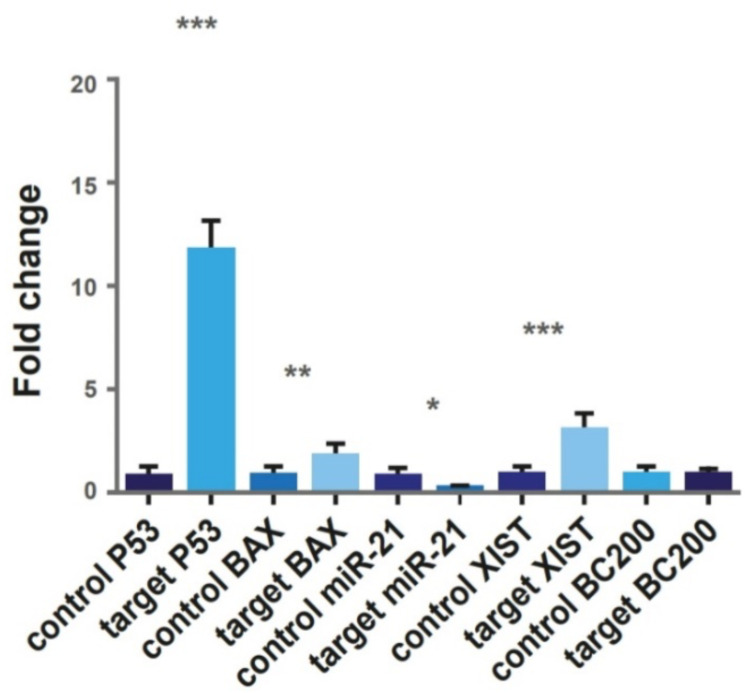
In transfected cells, expression of miR-21-5p decreased by 0.3 fold. The lncRNA-BC200 expression was not significantly altered but XIST expression was upregulated significantly 3.9 fold. In addition, upregulation of apoptosis biomarkers, Bax and P53 was detected as 2.23 and 14.62 folds respectively. Expression changes in all genes except lncRNA-BC200 was statistically significant (Fig. 2c).
Fig. 2c:
Expression of molecules of P53, Bax, miR-21-5p, lncRNA-XIST and lncRNA-BC200 in untreated control cells and transfected target cells. After transfection, miR-21 showed significantly decreased expression, while, other genes showed significant upregulation. The change in the expression level of lncRNA-BC200 was not significant
Cell viability assay
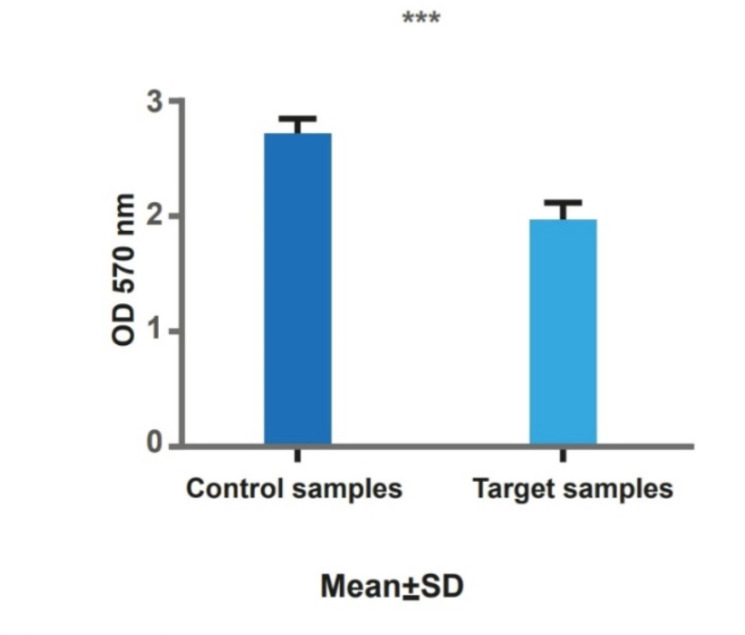
Results showed a significant decrease in the viability of transfected cells (Fig. 2d).
Fig.2d:
It is shown that there is a significant decrease in cell viability in transfected cells (target) in comparison with controls. * P<0.05, ** P<0.01, *** P<0.001
Apoptosis assay
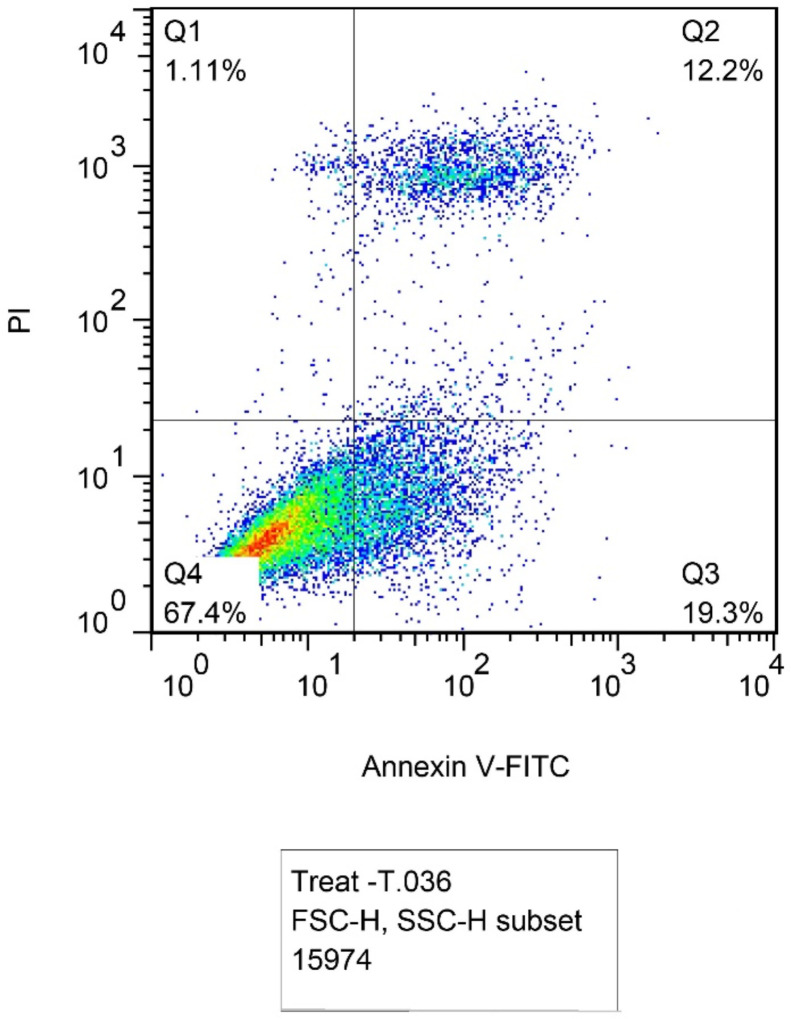
Flow cytometry method with Annexin V-FITC and PI staining showed apoptosis induction. Fig. 3a and b represent the plots including four regions: Q1 for necrotic cells (Anx−, PI+), Q2 for late apoptotic cells (Anx+, PI+), Q3 for early apoptotic cells (Anx+, PI−), and Q4 for normal cells (Anx−, PI−). In transfected cells, the percentage of apoptotic cells was significantly enhanced compared to un-transfected cells. As shown in the plots, in the Q2 region of late apoptosis, the presence of PI has increased in the nucleus and phosphatidylserines in the inner leaflet of the plasma membrane transferred to the outer leaflet. In total, the percentage of late apoptotic cells increased from 5.58% in untransfected cells to 12.2% in transfected cells and the population of early apoptotic cells increased from 1.94% to 19.3% of cells.
Fig. 3a:
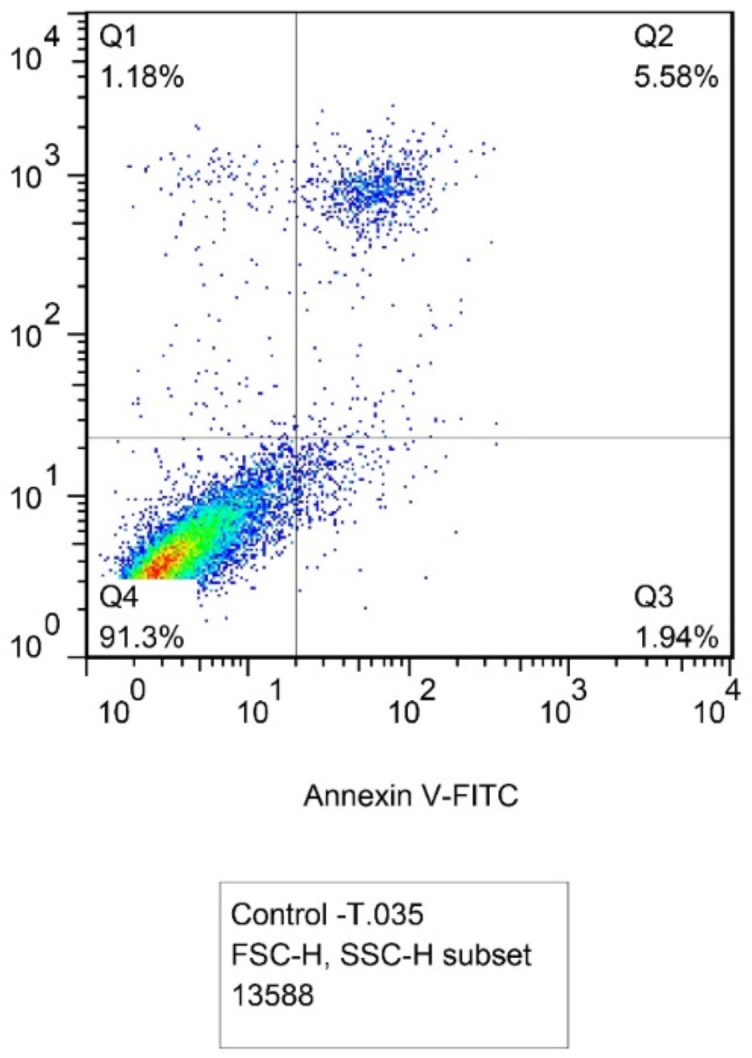
Un-transfected MCF-7 cells population with 5.58% of late apoptotic cells in Q2 region and 1. 94% of early apoptotic cells in Q3
Fig.3b:
Transfected cells with 12.2% of late apoptotic cells and 19.3% of early apoptotic cells
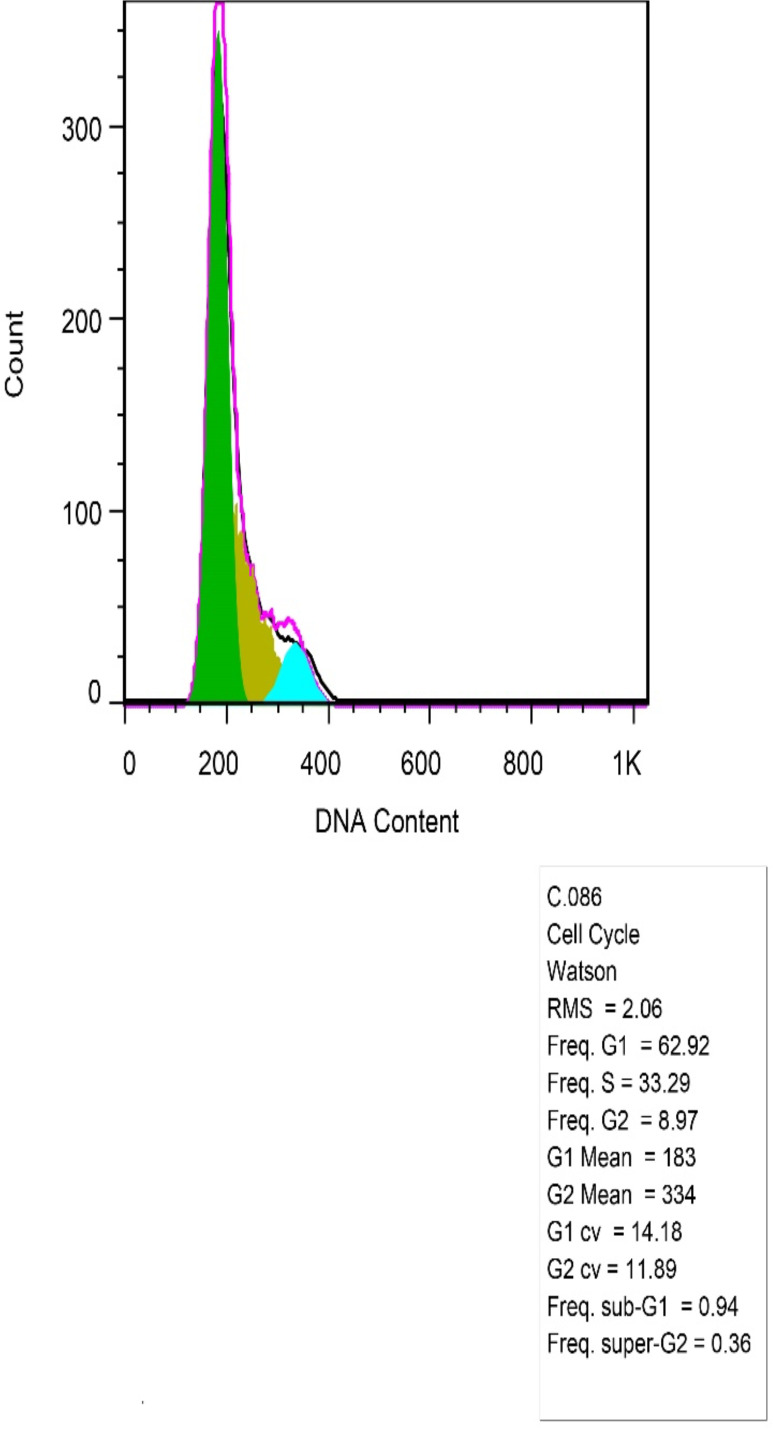
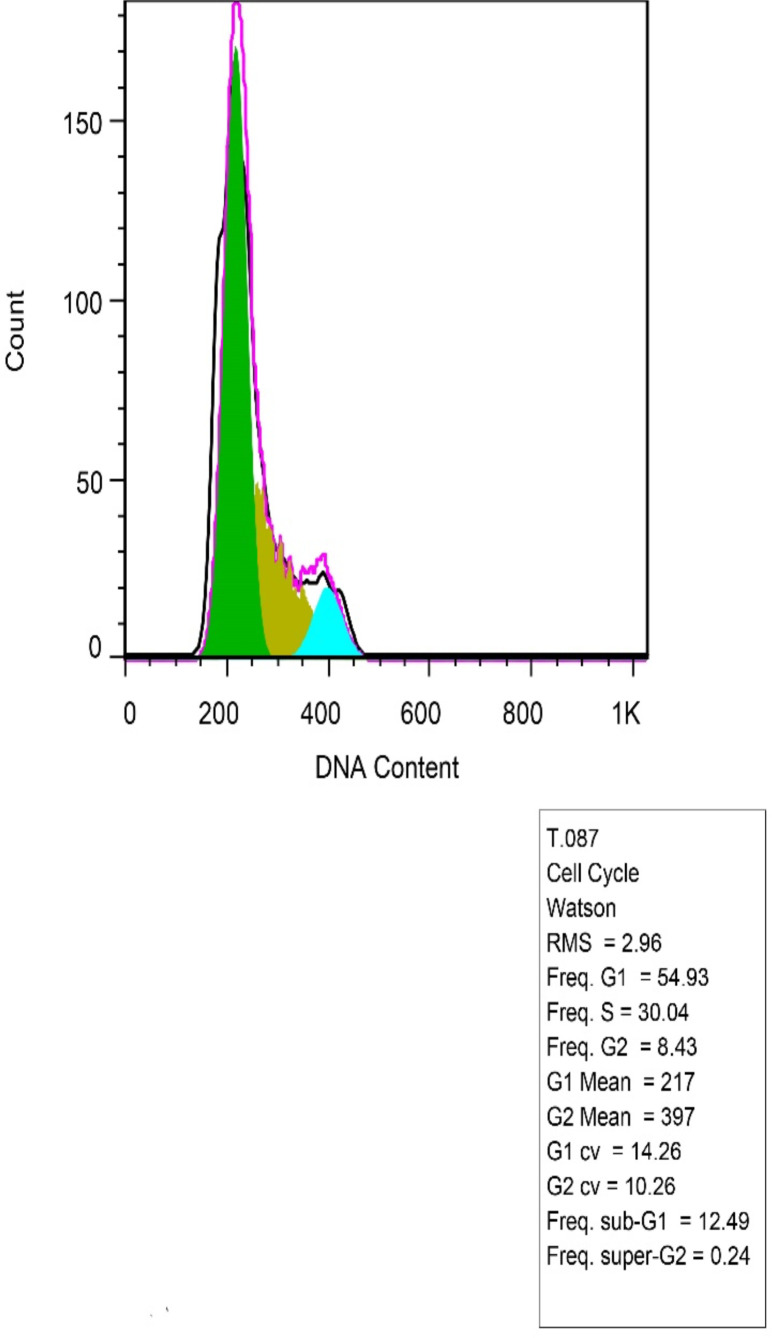
Cell cycle assay
This assay showed that the percentage of apoptotic sub-G1 cells increased from 0.94 in untransfected cells to 12.49 in transfected cells (Fig. 3c and d). There weren’t any remarkable changes in other cell cycle phases.
Fig. 3c:
As depicted control cells did not show any significant changes in cellular phases of G1, S, G2 (from left to right)
Fig. 3d:
Transfected cells, was shown apoptosis more than control cells and there was a significant difference between two samples in the phase before G1 (sub-G1)
Discussion
Although it has revealed interactions of different miRNAs like miR-429 with lncRNA-XIST in molecular pathways of cancer including BC (19), the miR-21-5p crosstalk with XIST in BC has not addressed. Based on our bioinformatics investigation, among all of LncRNAs in relation to miR-21-5p, XIST with high score was considered. As displayed in Fig. 1a, miR-21 and XIST are in interaction together. To study experimentally, MCF-7 cells treated by anti-miR-21 to inhibit miR-21 (Fig. 2a and b) resulted in lncRNA-XIST up regulation that increased apoptotic factors such as P53 and Bax (Fig. 2c). P53 and XIST have reciprocal interaction and their expression have effect on each other (20). Induced apoptosis was confirmed by apoptosis and cell cycle flow cytometry. Then significant difference was identified between the transfected and control cells in early (Q3) and late (Q2) in apoptosis and the frequency of apoptotic sub-G1 cells in cell cycle assays respectively (Fig. 3). While no significant change was observed in the expression level of BC200, the other analyzed lncRNA (Fig. 2c). In recent years LncRNAs and microRNAs as key molecules obtained much consideration. Already, role of XIST as tumor suppressor and miR-21 as oncogene in BC has been proved. The expression level of lncRNA-XIST is down-regulated in cervical, ovarian, and BC cell lines and it has already been proposed that decreased lncRNA-XIST levels may potentially be considered as a therapeutic target and a biomarker for BC diagnosis (5, 6) and the lncRNA XIST/miR-92b-3p/MTF1 regulatory axis for the progression of breast cancer. The XIST lncRNA could inhibit cancer proliferation and EMT and promote apoptosis in breast cancer (21). LncRNA NEAT1 as a significantly dysregulated RNA in the breast cancer patients has significant interactions with XIST and hsa-miR-612 (22).
One of the well-known oncomiRs is miR-21-5p, which participates in migration, invasion, and apoptosis of ovarian, cervical and BC cancer cells (23). It targets many important tumor suppressor genes and promotes molecular and cellular processes of BC development, which provides a potential prevention and therapy approach for BC (24). MiR-21-5p has been demonstrated in relation to other LncRNAs and inhibiting apoptosis in BC. CASC7 lncRNA as tumor suppressor is in interaction with miR-21-5p in breast cancer (25). Over-expression of activity of GAS5 resulted in down regulation of miR-21, which increased both radio sensitivity and the activation of the apoptosis pathway, including bax and caspase 3 (26). One of the consequences of miR-21-5p up-regulation in cancerous cells is their resistance against apoptosis. Indeed, it is well known that miR-21 exerts its oncogenic actions mainly through the inhibition of apoptosis. Therefore, miR-21-5p may affect the regulation of specific lncRNAs such as XIST. For instance, it inhibits apoptosis of tongue squamous cell carcinomas partly via TPM1 silencing (27). In addition, down-regulation of miR-21-5p induces glioma cell apoptosis through caspase 9 and 3 activations, which are possibly mediated by TIMP3. Consistent with these findings, TIMP3 levels are inversely correlated with miR-21 in BC and over-expression of TPM1 in the MCF-7 cell line suppresses cell growth (28).
Furthermore, oncogenic function of miR-21 in relation to other molecules has been declared. Shi et al. reported that miR-21-5p overexpression protects glioblastoma cells from temozolomide-induced apoptosis by down-regulation of pro-apoptotic bax gene and up-regulation of anti-apoptotic bcl-2 gene. Previously, they showed that transfection of MCF-7 cells with anti-miR-21-5p oligonucleotides results in suppression of both cell growths in vitro and tumor growth in the xenograft mouse model by increasing of apoptosis and reducing of cell proliferation owing to down-regulation of the bcl-2 gene (29). In the same way, Frankel et al. reported a link between miR-21 and tumor suppressor proteins p53 and PDCD4 in MCF-7 cells and showed that inhibition of mir-21 causes reduced growth of MCF-7 cells (30). Therefore, interaction between miR-21 and XIST is another target that can be considered as therapy for BC.
Based on our bioinformatic analysis on the transcriptome data, lncRNA-XIST was found to co-expressed with several mRNAs in BC. This indicates that miR-21-5p may function as a mediator between XIST and these mRNAs and lncRNA XIST can influence those identified mRNAs through the ceRNAs mechanism. Furthermore, we found miRNAs other than miR-21-5p, which target lncRNA-XIST and some transcripts including Bax, Bad and Bak mRNAs, which are the most known pro-apoptotic factors. So, overexpression of XIST may lead to apoptosis through regulating these genes via the ceRNAs mechanism. Since these genes are not co-expressed with XIST based on our expression data analysis, this result should be investigated more precisely in future studies. As mentioned above, overexpressing XIST led to apoptosis in cancer cells and also in our investigations, we could upregulated it via down-regulation of miR-21 which induced apoptosis. Investigating of the apoptotic effect of XIST on the cancer cells may be important to control cancer cell apoptosis but how this process occurs? In our bioinformatics studies, we could not find any co-expressed apoptotic mRNAs with XIST in the analyzed expression data and should be more investigated in future studies. Meanwhile, due to our limitations, we can not absolutely claim and confirm our main goal and it entails further studies and experiments of pro and anti-apoptotic proteins in protein level. Besides, it can be proposed that interactions among other mRNAs with miR-21 and XIST experimentally are investigated in the future.
Conclusion
Here we showed in our bioinformatics and experiments studies that down-regulation of miR-21-5p in MCF-7 cells was concurrent with the up-regulation of lncRNA-XIST, which resulted in up regulated pro-apoptotic genes bax, and p53 and induction of apoptosis. Therefore, miR-21-5p and lncRNA-XIST relation may provide valuable targets for more studies on molecular therapies and diagnosis of breast cancer.
Journalism Ethics considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Acknowledgements
We are grateful to Prof. Majid Sadeghizadeh for critical reviewing and editing the paper and his kindly scientific advice and to Dr. Azam Bolhassani for providing lipofectamine as a gift.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.WHO , 2018: World Health Organization International Agency for Research on Cancer. The Global Cancer Observatory. 2018 statistics.
- 2.Ma X, Liu C, Xu X, et al. (2020). Biomarker expression analysis in different age groups revealed age was a risk factor for breast cancer. J Cell Physiol,235(5):4268–4278. [DOI] [PubMed] [Google Scholar]
- 3.Esmatabadi MJD, Motamedrad M, Sadeghizadeh M. (2018). Down-regulation of lncRNA, GAS5 decreases chemotherapeutic effect of dendrosomal curcumin (DNC) in breast cancer cells. Phytomedicine, 42:56–65. [DOI] [PubMed] [Google Scholar]
- 4.Esmatabadi MJD, Farhangi B, Montazeri M, et al. (2017). Up-regulation of miR-21 decreases chemotherapeutic effect of dendrosomal curcumin in breast cancer cells. Iran J Basic Med Sci,20(4):350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loda A, Heard E. (2019). Xist RNA in action: Past, present, and future. PLoS Genet, 15(9):e1008333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang YS, Chang CC, Lee SS, et al. (2016). Xist reduction in breast cancer upregulates AKT phosphorylation via HDAC3-mediated repression of PHLPP1 expression. Oncotarget,7(28):43256–43266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang R, Xia T. (2017). Long non-coding RNA XIST regulates PDCD4 expression by interacting with miR-21-5p and inhibits osteosarcoma cell growth and metastasis. Int J Oncol, 51(5):1460–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iacoangeli A, Lin Y, Morley EJ, et al. (2004). BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis, 25(11):2125–33. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Z, Bai J, Wu A, et al. (2015). Co-LncRNA: investigating the lncRNA combinatorial effects in GO annotations and KEGG pathways based on human RNA-Seq data. Database (Oxford), 2015:bav082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Gene Ontology Consortium (2019). The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res,47(D1):D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanehisa M, Goto S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res,28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benjamini Y, Hochberg Y. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Meth, 57(1):289–300. [Google Scholar]
- 13.Szklarczyk D, Gable AL, Lyon D, et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res,47(D1):D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shannon P, Markiel A, Ozier O, et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res,13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kouhsar M, Azimzadeh Jamalkandi S, Moeini A, et al. (2019). Detection of novel biomarkers for early detection of Non-Muscle-Invasive Bladder Cancer using Competing Endogenous RNA network analysis. Sci Rep, 9:8434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Bryan S, Dong S, Mathis JM, et al. (2017). The roles of oncogenic miRNAs and their therapeutic importance in breast cancer. Eur J Cancer,72:1–11. [DOI] [PubMed] [Google Scholar]
- 17.Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, et al. (2018). DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res,46(D1):D239–D245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. (2016). DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res,44(D1):D231–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Z, Li Z, Ma K, et al. (2017). Long Non-coding RNA XIST Promotes Glioma Tumorigenicity and Angiogenesis by Acting as a Molecular Sponge of miR-429. J Cancer, 8(19):4106–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parfenyev S, Singh A, Fedorova O, et al. (2021). Interplay between p53 and non-coding RNAs in the regulation of EMT in breast cancer. Cell Death Dis,12(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang B, Zhu H, Feng W, et al. (2022). Database Mining Detected a Cuproptosis-Related Prognostic Signature and a Related Regulatory Axis in Breast Cancer. Dis Markers, 2022:9004830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azadeh M, Salehzadeh A, Ghaedi K, et al. (2022). NEAT1 can be a diagnostic biomarker in the breast cancer and gastric cancer patients by targeting XIST, hsa-miR-612, and MTRNR2L8: integrated RNA targetome interaction and experimental expression analysis. Genes Environ, 44(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Si ML, Zhu S, Wu H, et al. (2007). miR-21-mediated tumor growth. Oncogene,26(19):2799–803. [DOI] [PubMed] [Google Scholar]
- 24.Yan LX, Liu YH, Xiang JW, et al. (2016). PIK3R1 targeting by miR-21 suppresses tumor cell migration and invasion by reducing PI3K/AKT signaling and reversing EMT, and predicts clinical outcome of breast cancer. Int J Oncol,48(2):471–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang G, Duan P, Liu F, et al. (2021). Long non-coding RNA CASC7 suppresses malignant behaviors of breast cancer by regulating miR-21-5p/FASLG axis. Bioengineered,12(2):11555–11566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma Y, Yu L, Yan W, et al. (2022). lncRNA GAS5 Sensitizes Breast Cancer Cells to Ionizing Radiation by Inhibiting DNA Repair. Biomed Res Int, 2022:1987519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Huang H, Sun L, et al. (2009). MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res,15(12):3998–4008. [DOI] [PubMed] [Google Scholar]
- 28.Buscaglia LE, Li Y. (2011). Apoptosis and the target genes of microRNA-21. Chin J Cancer, 30(6):371–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi L, Chen J, Yang J, et al. (2010). MiR-21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity. Brain Res,1352:255–64. [DOI] [PubMed] [Google Scholar]
- 30.Frankel LB, Christoffersen NR, Jacobsen A, et al. (2008). Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem, 283(2):1026–33. [DOI] [PubMed] [Google Scholar]