Abstract
Background.
The National Outbreak Reporting System (NORS) captures data on foodborne, waterborne, and enteric illness outbreaks in the United States. This study describes enteric illness outbreaks reported during 11 years of surveillance.
Methods.
We extracted finalized reports from NORS for outbreaks occurring during 2009–2019. Outbreaks were included if caused by an enteric etiology or if any patients reported diarrhea, vomiting, bloody stools, or unspecified acute gastroenteritis.
Results.
A total of 38 395 outbreaks met inclusion criteria, increasing from 1932 in 2009 to 3889 in 2019. Outbreaks were most commonly transmitted through person-to-person contact (n = 23 812; 62%) and contaminated food (n = 9234; 24%). Norovirus was the most commonly reported etiology, reported in 22 820 (59%) outbreaks, followed by Salmonella (n = 2449; 6%) and Shigella (n = 1171; 3%). Norovirus outbreaks were significantly larger, with a median of 22 illnesses per outbreak, than outbreaks caused by the other most common outbreak etiologies (P < .0001, all comparisons). Hospitalization rates were higher in outbreaks caused by Salmonella and Escherichia coli outbreaks (20.9% and 22.8%, respectively) than those caused by norovirus (2%). Case fatality rate was highest in E. coli outbreaks (0.5%) and lowest in Shigella and Campylobacter outbreaks (0.02%).
Conclusions.
Norovirus caused the most outbreaks and outbreak-associated illness, hospitalizations, and deaths. However, persons in E. coli and Salmonella outbreaks were more likely to be hospitalized or die. Outbreak surveillance through NORS provides the relative contributions of each mode of transmission and etiology for reported enteric illness outbreaks, which can guide targeted interventions.
Keywords: outbreaks, enteric illness, surveillance, norovirus, Salmonella
Acute enteric illness or gastroenteritis, most commonly characterized by diarrhea and/or vomiting, has been estimated to cause approximately 179 million illnesses in the United States annually [1]. Norovirus is the leading cause of enteric illness outbreaks worldwide [2, 3]. However, enteric illnesses can be caused by many different viral, bacterial, and parasitic pathogens as well as by toxins and chemicals.
Surveillance for foodborne and waterborne disease outbreaks, including enteric illness outbreaks, has been a core function of the US Centers for Disease Control and Prevention (CDC) since the 1970s [4, 5]. In 2009, CDC expanded and streamlined its outbreak surveillance by establishing the National Outbreak Reporting System (NORS), a platform that combined foodborne and waterborne disease outbreak reporting and added reporting of enteric illness outbreaks transmitted by contact with environmental sources, infected persons or animals, or indeterminate or unknown modes of transmission [6]. State, local, and territorial public health agencies can voluntarily report these outbreaks to CDC via a web-based platform.
Data captured in NORS provide more information on enteric illness outbreaks than foodborne and waterborne disease outbreak surveillance alone by including outbreaks transmitted through other modes. The goal of this study is to describe the distribution of specific etiologies (ie, pathogens, toxins, chemicals), modes of transmission, and other characteristics of enteric illness outbreaks in the United States since establishment of NORS in 2009.
METHODS
Finalized outbreak reports were extracted on 24 February 2021 from NORS for outbreaks with first illness onset occurring during 2009–2019. Outbreaks in NORS are defined as 2 or more cases of similar illness associated with a common exposure. NORS captures outbreaks with exposures in US states, the District of Columbia, and US territories and Freely Associated States (referred to hereafter as “reporting sites”). Annual population estimates from 2009–2019 were available from the US Census Bureau for the 50 US states, the District of Columbia, and Puerto Rico and were used to calculate average annual outbreak reporting rates, expressed per 1 million population [7, 8]. Outbreaks in which exposure occurred in more than 1 reporting site (“multisite outbreaks”) were assigned as an outbreak to each site involved for the purpose of rate calculations. Primary mode of transmission was determined by each reporting site. Outbreak and case definitions might not be consistent across all sites. Medians were compared using the Wilcoxon rank-sum test. All analyses were conducted in SAS version 9.4 (SAS Institute).
The setting of exposure was the setting where patients were exposed to the outbreak etiology; for foodborne outbreaks, this was the location(s) of food preparation. Four of the most commonly reported symptoms (diarrhea, vomiting, bloody stools, and fever) were used to assess the symptom profile of patients. Not all symptoms or patient outcomes were reported for all outbreaks, and data were not available for all patients on specific symptoms or outcomes. Therefore, symptoms and outcomes are presented as the percentage of patients reporting that symptom or outcome among all patients with that information available.
Outbreak reports with only a reported nonenteric etiology were excluded from analysis (see Supplementary Methods and Supplementary Table 1). If no specific etiology was reported for an outbreak, the report was included if any patients reported symptoms of diarrhea, vomiting, bloody stools, or unspecified acute gastroenteritis. Outbreaks with no reported etiology and no symptom information were excluded from analysis. Outbreak etiologies were considered laboratory-confirmed if defined as such by the reporting site or if 2 or more laboratory-confirmed cases were identified. Unless otherwise specified, all figures and analyses include both laboratory-confirmed and suspected etiologies.
RESULTS
Reporting Trends
A total of 40 865 outbreaks were reported through NORS with a first illness onset during 2009–2019. Of these, 571 (1.4%) were outbreaks with a reported nonenteric etiology, primarily Legionella (n = 429), chlorine/chlorine gas (n = 37), and hepatitis A (n = 33) (Supplementary Table 1). An additional 1899 (4.6%) outbreak reports did not include an outbreak etiology and no patients reported vomiting, diarrhea, bloody stools, or unspecified gastroenteritis. The final analysis dataset included 38 395 enteric illness outbreaks.
The number of enteric illness outbreaks reported each year ranged from 1942 in 2009 to a peak of 4145 outbreaks in 2016 (Figure 1). The annual number of reported outbreaks was lowest during 2009–2011 and then stabilized, with a median of 3909 outbreaks reported annually during 2012–2019. Outbreaks were most commonly transmitted through direct person-to-person contact (n = 23 812; 62%), followed by foodborne transmission (n = 9234; 24%), unknown mode of transmission (n = 4313; 11%), waterborne transmission (n = 466; 1%), animal contact (n = 462; 1%), and environmental contamination (n = 108; 0.3%).
Figure 1.
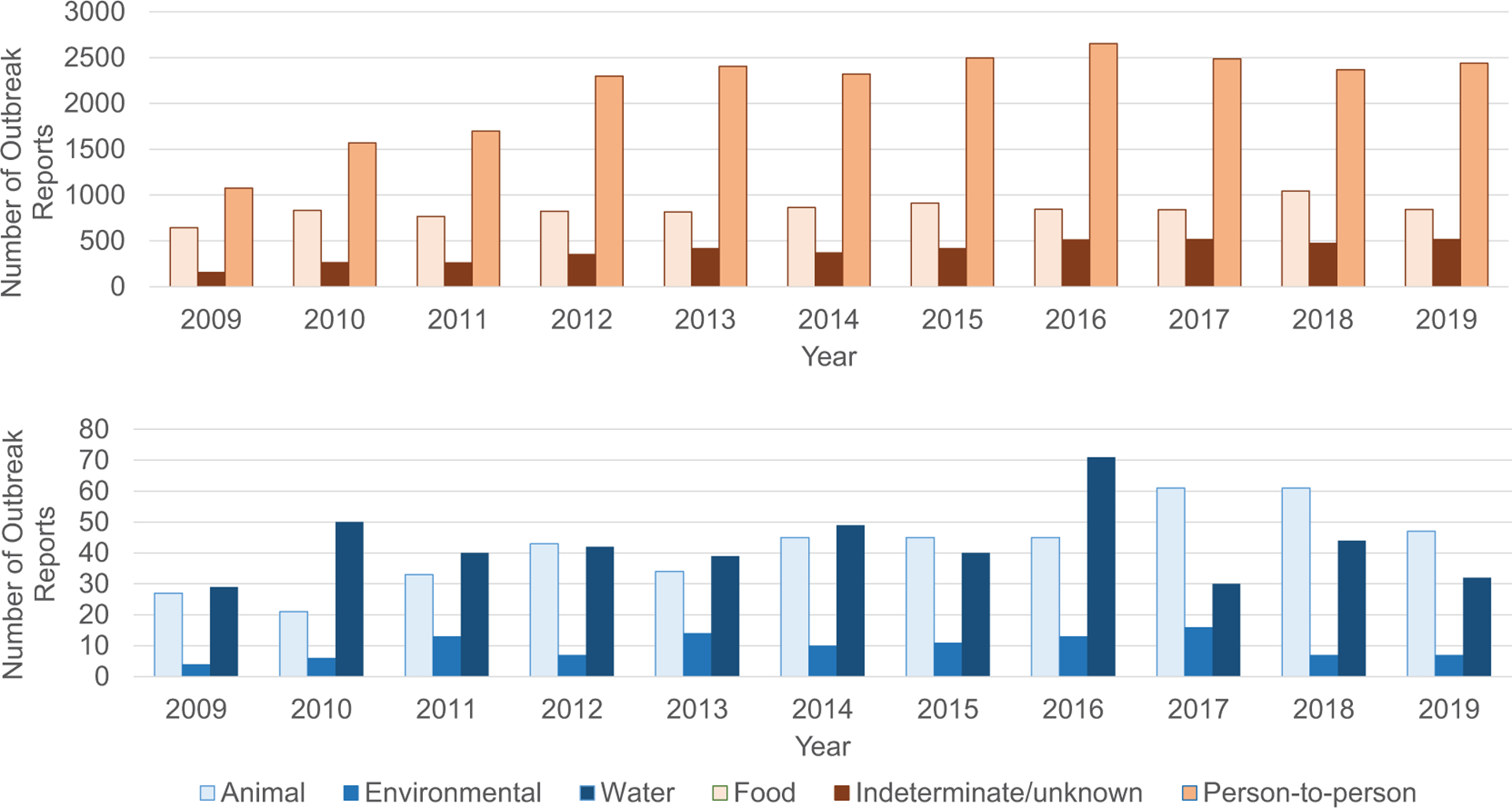
National Outbreak Reporting System outbreak reports by year and mode of transmission, 2009–2019 (N = 38 395).
Outbreak exposures occurred in all 50 US states; Washington, DC; Puerto Rico; and American Samoa. A total of 37 828 (99%) outbreaks resulted from exposures that occurred in a single reporting site. The 567 (1%) multisite outbreaks were most commonly associated with foodborne transmission (n = 386; 68%) or animal contact (n = 117; 21%).
A median of 15.8 outbreaks were reported in each reporting site per million population per year (range: 2.9 [New Jersey] to 60.0 [Rhode Island]) (Figure 2). Reporting varied by mode of transmission. A median of 52 sites reported foodborne outbreaks annually during both 2009–2011 and 2012–2019 (data not shown). The number of sites reporting at least 1 person-to-person outbreak annually increased from a median of 43 sites during 2009–2011 to 47.5 sites during 2012–2019 (P < .05), but this was still significantly lower than the median of 52 sites reporting foodborne outbreaks (P < .01).
Figure 2.
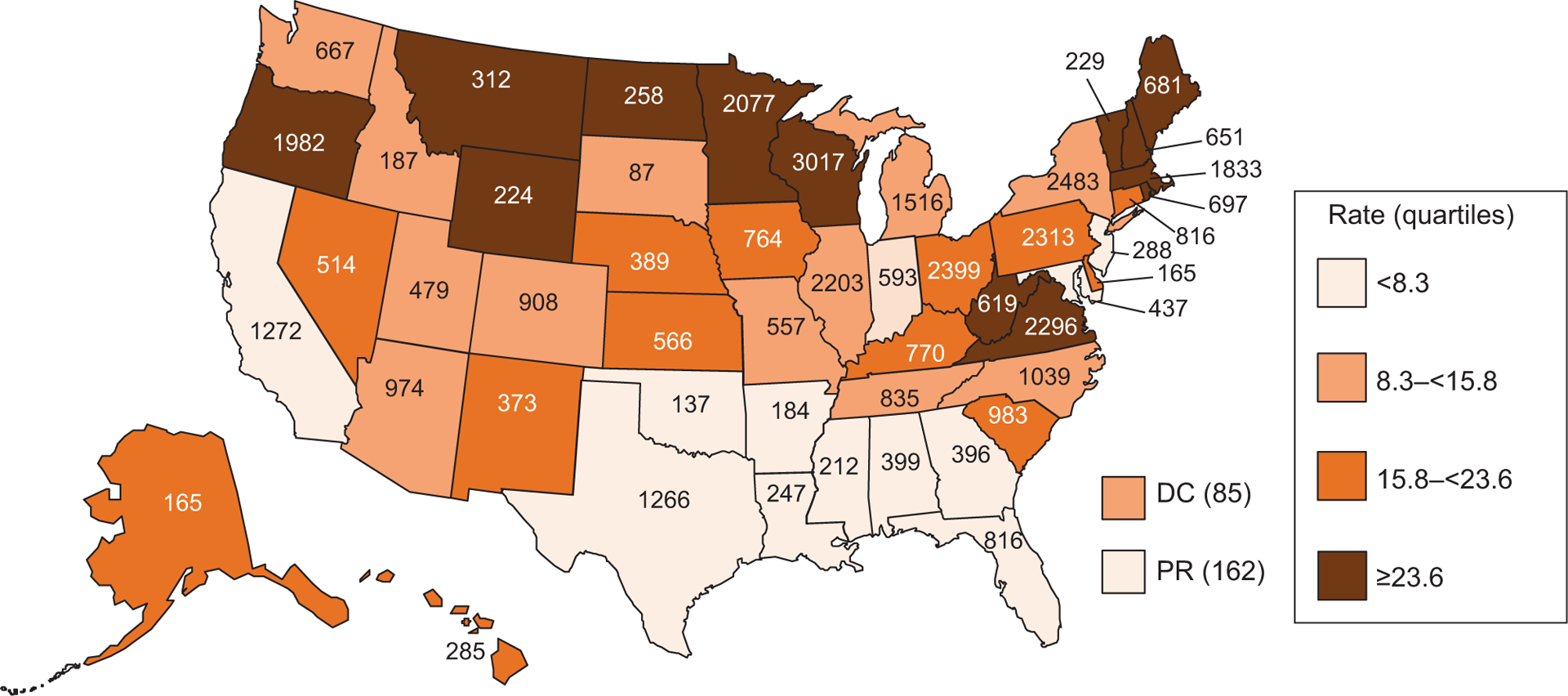
Rate* and number† of enteric illness outbreaks by state, 2009–2019 (N = 38 395). *Average annual incidence of outbreaks per state, per million population, on the basis of US Census Bureau population estimates, 2009–2019. †567 outbreaks in which exposure occurred in more than 1 state were assigned as an outbreak to each state involved. Abbreviations: DC, Washington, District of Columbia; PR, Puerto Rico.
Outbreaks by Etiology
Norovirus was the most commonly reported sole suspected or laboratory-confirmed outbreak etiology, reported in 22 820 (59%) of 38 395 outbreaks (Table 1). Other commonly reported outbreak etiologies were as follows: Salmonella in 2249 (6%) outbreaks, Shigella in 1171 (3%) outbreaks, Escherichia coli in 797 (2%) outbreaks, and Campylobacter in 634 (2%) outbreaks. A total of 16 319 outbreaks had at least 1 laboratory-confirmed etiology. Norovirus was the sole laboratory-confirmed etiology reported in 10 057 (62%) outbreaks, followed by 2183 (13%) laboratory-confirmed Salmonella outbreaks and 951 (6%) laboratory-confirmed Shigella outbreaks. A total of 7343 (19%) outbreak reports did not include either a suspected or laboratory-confirmed etiology.
Table 1.
Most Common Suspected and Laboratory-Confirmed Enteric Outbreak Etiologies, 2009–2019
| Suspected or Confirmed Etiology | Laboratory-Confirmed and Suspected (n = 38 395), n (%) | Laboratory-Confirmed Only (n = 16 319), n (%) |
|---|---|---|
| Norovirus | 22 820 (59) | 10 057 (62) |
| Salmonella | 2449 (6) | 2183 (13) |
| Shigella | 1171 (3) | 951 (6) |
| Escherichia coli | 797 (2) | 689 (4) |
| Campylobacter | 634 (2) | 424 (3) |
| Cryptosporidium | 512 (1) | 415 (3) |
| Clostridium | 461 (1) | 229 (1) |
| Scombroid toxin (histamine) | 173 (0.5) | 157 (1) |
| Ciguatoxin | 172 (0.5) | 157 (1) |
| Giardia | 170 (0.4) | 147 (1) |
| All other single etiologies | 1233 (3) | 760 (5) |
| Multiple etiologies | 460 (1) | 150 (1) |
| None reported | 7343 (19) | N/A |
Abbreviation: N/A, not applicable.
By mode of transmission, norovirus was the most commonly reported single laboratory-confirmed or suspected etiology in person-to-person (n = 17 927; 75%), foodborne (n = 3084; 33%), unknown mode (n = 1706; 40%), and environmental contamination (n = 57; 53%) outbreaks (Table 2). Cryptosporidium was the most commonly reported etiology in waterborne enteric illness outbreaks (n = 200; 43%). Salmonella was the most commonly reported etiology in animal contact outbreaks of enteric illness (n = 213; 46%).
Table 2.
Primary Mode of Transmission and Setting of Exposure by Confirmed or Suspected Outbreak Etiology, 2009–2019
| Norovirus, n (%) | Salmonella, n (%) | Shigella, n (%) | Escherichia coli, n (%) | Campylobacter, n (%) | All Other Single Etiologies, n (%) | Multiple Etiologies, n (%) | No Reported Etiology, n (%) | All Outbreaks, n (%) | |
|---|---|---|---|---|---|---|---|---|---|
| Total | 22 820 | 2449 | 1171 | 797 | 634 | 2721 | 460 | 7343 | 38 395 |
| By mode of transmission | |||||||||
| Person-to-person | 17 927 (79) | 253 (10) | 921 (79) | 165 (21) | 49 (8) | 543 (20) | 210 (46) | 3744 (51) | 23 812 (62) |
| Food | 3084 (14) | 1512 (62) | 62 (5) | 357 (45) | 314 (50) | 1511 (56) | 154 (33) | 2240 (31) | 9234 (24) |
| Water | 46 (0.2) | 3 (0.1) | 23 (2) | 28 (4) | 16 (3) | 301 (11) | 24 (5) | 25 (0.3) | 466 (1) |
| Animal contact | 0 (0) | 213 (9) | 0 (0) | 58 (7) | 62 (10) | 113 (4) | 15 (3) | 1 (0.01) | 462 (1) |
| Environmental contamination | 57 (0.3) | 15 (0.6) | 8 (0.7) | 9 (1) | 6 (1) | 4 (0.2) | 1 (0.2) | 8 (0.1) | 108 (0.3) |
| Indeterminate/unknown | 1706 (7) | 453 (19) | 157 (13) | 180 (23) | 187 (30) | 249 (9) | 56 (12) | 1325 (18) | 4313 (11) |
| By setting | |||||||||
| LTCFs | 12 193 (53) | 63 (3) | 10 (1) | 10 (1) | 16 (3) | 242 (9) | 95 (21) | 3080 (42) | 15 709 (41) |
| Restaurant | 2372 (10) | 559 (23) | 37 (3) | 115 (14) | 123 (19) | 666 (24) | 81 (18) | 1626 (22) | 5579 (15) |
| School/daycare | 2502 (11) | 150 (6) | 757 (65) | 145 (18) | 24 (4) | 187 (7) | 113 (25) | 668 (9) | 4546 (12) |
| Private home | 261 (1) | 482 (20) | 99 (8) | 152 (19) | 164 (26) | 448 (16) | 29 (6) | 134 (2) | 1769 (5) |
| Other single settings | 2441 (11) | 464 (19) | 140 (12) | 211 (26) | 186 (29) | 713 (26) | 88 (19) | 850 (12) | 5093 (13) |
| Multiple settings | 201 (1) | 211 (9) | 6 (1) | 48 (6) | 18 (3) | 152 (6) | 20 (4) | 132 (2) | 788 (2) |
| None reported | 2850 (12) | 520 (21) | 122 (10) | 116 (15) | 103 (16) | 313 (12) | 34 (7) | 853 (12) | 4911 (13) |
N = 38 395.
Abbreviation: LTCF, long-term care facility.
Outbreaks caused by the 4 most commonly reported bacterial pathogens occurred more often during May–October: Salmonella (n = 1544; 63%), Shigella (n = 677; 58%), E. coli (n = 577; 72%), and Campylobacter (n = 422; 67%) (Figure 3). Most suspected or laboratory-confirmed norovirus outbreaks (n = 18 824; 82%) and outbreaks with no reported etiology (n = 5096; 69%) occurred during November–April (Figure 3).
Figure 3.
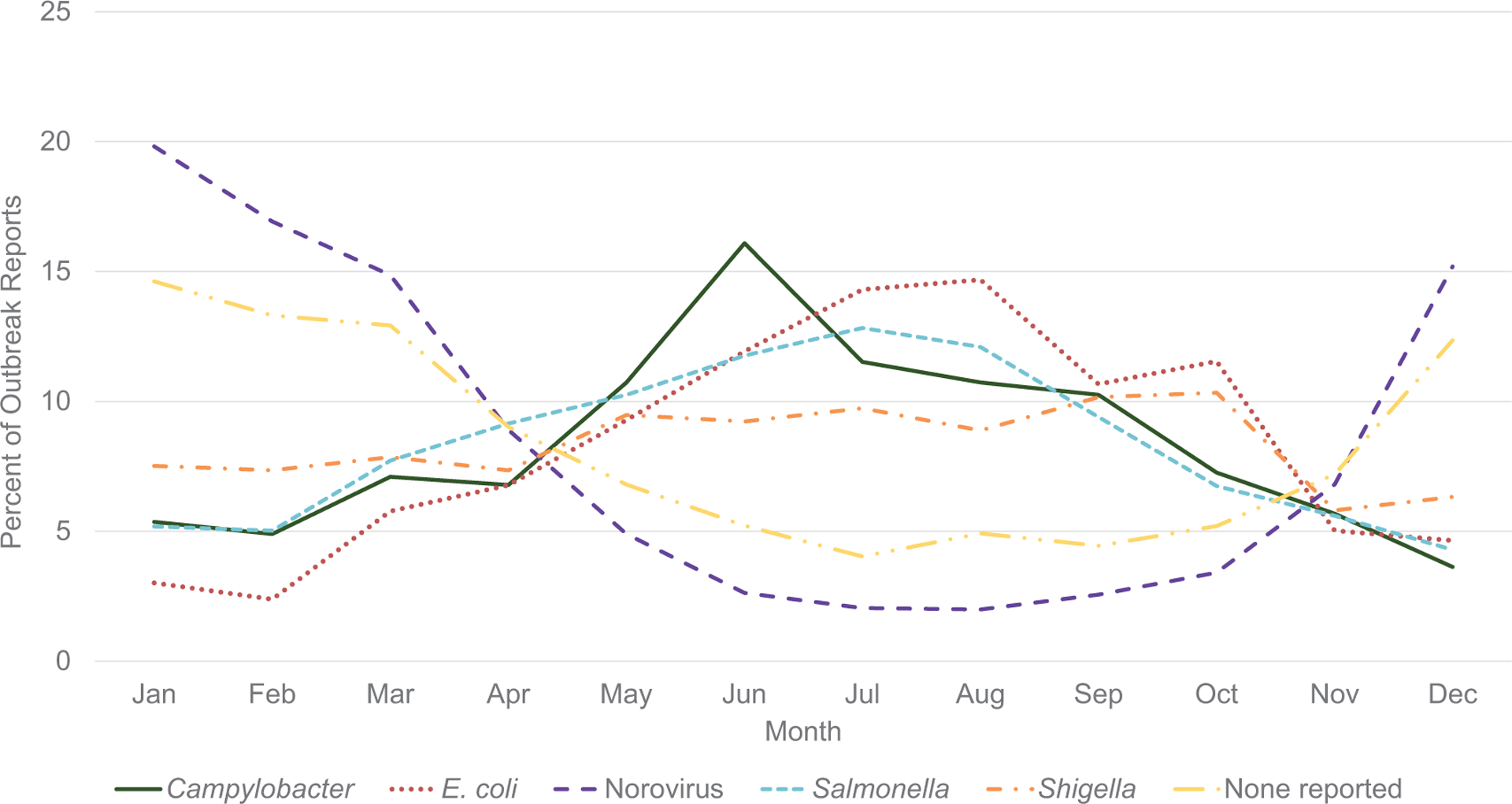
Outbreak reports by month of first illness for selected confirmed or suspected etiologies, 2009–2019 (N = 35 214).
Norovirus outbreaks were most often transmitted by person-to-person contact (n = 17 927; 79%) and foodborne transmission (n = 3084; 14%) (Table 2). Just over half (n = 12 193; 53%) of reported norovirus outbreaks occurred in long-term care facilities (LTCFs). An additional 2502 (11%) norovirus outbreaks occurred in schools and daycare centers, and 2372 (10%) outbreaks occurred in restaurants. Similarly, outbreaks without a reported etiology were most commonly transmitted by person-to-person contact (n = 3744; 51%) and by food (n = 2240; 31%). These outbreaks were reported primarily in LTCFs (n = 3080; 42%) and restaurants (n = 1626; 22%). Shigella outbreaks were also most commonly transmitted by person-to-person contact (n = 921; 79%) (Table 2). Most Shigella outbreaks (n = 757; 65%) occurred in schools and daycare centers, and 99 (8%) outbreaks occurred in private homes.
Food was the most commonly reported mode of transmission for Salmonella (n = 1512; 61%), E. coli (n = 357; 45%), and Campylobacter (n = 314; 50%) outbreaks, followed by unknown mode of transmission (n = 453 [19%], n = 180 [23%], and n = 187 [30%], respectively) (Table 2). Salmonella outbreaks were most commonly reported in restaurants (n = 559; 23%) and private homes (n = 482; 20%). Escherichia coli outbreaks were most commonly reported in private homes (n = 152; 19%) and schools and daycare centers (n = 145; 18%). Similarly, Campylobacter outbreaks were most often reported in private homes (n = 164; 26%) and restaurant settings (n = 123; 19%). Multisite outbreaks were most commonly caused by Salmonella in 361 (64%) outbreaks, followed by E. coli in 103 (18%) outbreaks.
Over 1 million enteric illnesses were reported from all outbreaks (median: 100 563 illnesses per year; range: 55 787–109 014) (Table 3). Of these, 739 220 (72%) illnesses were reported in norovirus outbreaks. The median number of illnesses per norovirus outbreak was 22 (interquartile range [IQR]: 11–41). This was significantly higher than the median number of cases in outbreaks due to Salmonella (7; IQR: 3–17), Shigella (6; IQR: 3–14), E. coli (5; IQR: 3–11), and Campylobacter (4; IQR: 2–7) (P < .0001, all comparisons). An additional 138 765 illnesses were reported in outbreaks without a reported etiology, with a median of 11 (IQR: 5–24) illnesses per outbreak.
Table 3.
Outbreak-Associated Illnesses and Outcomes by Confirmed or Suspected Etiology, 2009–2019
| Suspected or Confirmed Etiology | Total Illnesses | Median Illnesses (IQR) | Hospitalizations (of Those With Information) | Hospitalization Rate | Deaths (of Those With Information) | Case Fatality Rate |
|---|---|---|---|---|---|---|
| Norovirus | 739 220 | 22 (11–41)* | 10 384/532 329 | 2.0 | 894/563 502 | 0.16 |
| Salmonella | 51 383 | 7 (3–17) | 8038/38 469 | 20.9 | 81/40 050 | 0.20 |
| Shigella | 20 187 | 6 (3–14) | 861/14 661 | 5.9 | 4/16 166 | 0.02 |
| Escherichia coli | 9192 | 5 (3–11) | 1828/8030 | 22.8 | 38/8089 | 0.47 |
| Campylobacter | 5528 | 4 (2–7) | 372/4315 | 8.6 | 1/4476 | 0.02 |
| All other single etiologies | 43 032 | 5 (3–14) | 1990/34 699 | 5.7 | 187/36 388 | 0.51 |
| Multiple etiologies | 17 984 | 15 (7–38) | 339/13 943 | 2.4 | 16/13 383 | 0.12 |
| None reported | 138 765 | 11 (5–24) | 1850/117 473 | 1.6 | 121/120 144 | 0.10 |
| All outbreaks | 1 025 291 | 16 (7–33) | 25 662/763 919 | 3.4 | 1342/802 148 | 0.17 |
N = 38 395.
P < .0001 for comparisons to median illnesses of Salmonella, Shigella, E. coli, and Campylobacter outbreaks.
The highest case fatality rates (CFRs) and hospitalization rates were reported in E. coli outbreaks, with 38 deaths (CFR: 0.47%) and 1828 hospitalizations (hospitalization rate: 22.8%), followed by Salmonella outbreaks, with 81 deaths (CFR: 0.20%) and 8038 hospitalizations (hospitalization rate: 20.9%) (Table 3). The highest absolute number of deaths and hospitalizations occurred in norovirus outbreaks, with 894 deaths (CFR: 0.16%) and 10 384 hospitalizations (hospitalization rate: 2.0%).
Diarrhea was the most commonly reported symptom in outbreaks attributed to the most common etiologies and in outbreaks with no etiology reported (Figure 4). Vomiting was nearly as frequent in norovirus outbreaks as in outbreaks with no etiology reported, with 78% of patients in both types of outbreaks reporting diarrhea, and 70% and 67% of patients, respectively, reporting vomiting. Conversely, only 21% of patients in norovirus outbreaks and 22% of patients in outbreaks without an etiology reported a fever, and 1% in both types of outbreaks reported bloody stools. Fever and bloody stools were more commonly reported in outbreaks attributed to the 4 most common bacterial etiologies. Forty-three percent of patients in E. coli outbreaks reported bloody stools, and more than half of Salmonella (62%), Shigella (55%), and Campylobacter (63%) patients reported a fever.
Figure 4.
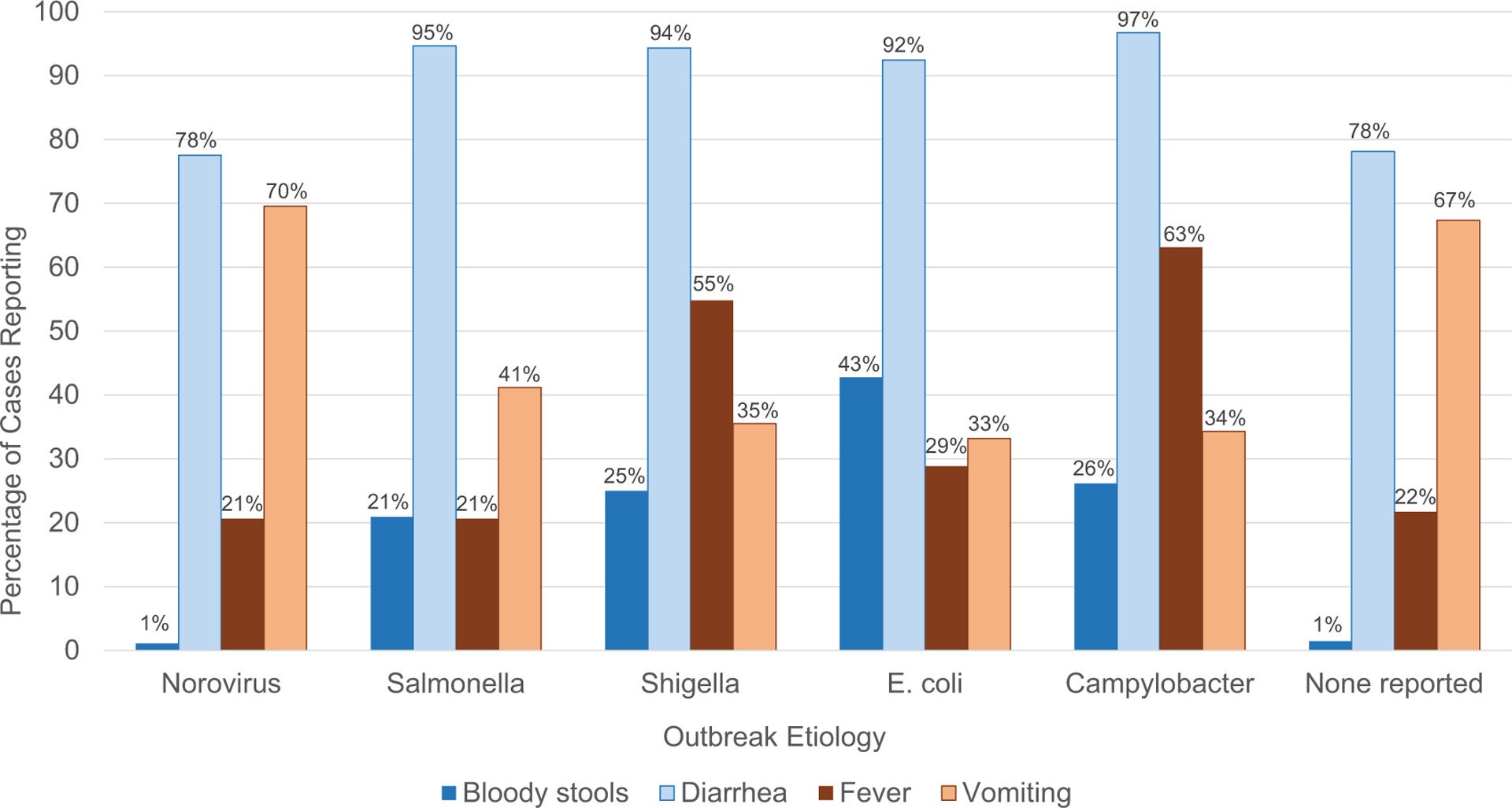
Symptoms reported by case-patients in outbreaks of selected confirmed or suspected etiologies, 2009–2019.
DISCUSSION
Outbreak reports form the basis for attribution estimations when considering policies for prevention, and robust outbreak surveillance is vital in guiding appropriate, targeted interventions [9, 10]. The CDC conducted foodborne and waterborne disease outbreak surveillance prior to creating NORS, but these 2 modes of transmission do not account for all enteric illness outbreaks. The number of enteric illness outbreaks reported through NORS more than doubled from 2009 to 2019, largely due to inclusion of person-to-person transmission starting in 2009. Person-to-person contact was the primary mode of transmission in nearly two-thirds of enteric illness outbreaks, and it is likely that sites needed additional time to implement reporting of these outbreaks. The increased reporting over time may reflect this additional time needed rather than a true increase in the number of outbreaks occurring.
Outbreaks were reported to NORS with exposures occurring in all 50 US states; Washington, DC; Puerto Rico; and American Samoa. However, reporting rates varied widely by reporting site, with the highest reporting rate more than 20 times higher than the lowest reporting rate. The 3 most populous US states—California, Texas, and Florida—all had outbreak reporting rates in the lowest quartile. Reporting also varied by mode, with fewer sites reporting person-to-person outbreaks than foodborne outbreaks even during recent years of surveillance. Differences in reporting rates by site and mode may thus be a function of resource availability and prioritization for outbreak investigation rather than a true reflection of outbreak incidence.
Norovirus was the most common suspected or laboratory-confirmed outbreak etiology, identified in over half (59%) of all reported enteric illness outbreaks. Norovirus spreads easily by many modes of transmission, due to a low infectious dose and ability to survive for long periods on surfaces [11]. Typically, norovirus causes a self-limited illness characterized by diarrhea and vomiting, although it can cause severe outcomes in the elderly and immunocompromised [12]. Norovirus outbreaks in this study exhibited a strong winter seasonality and were most often reported in LTCFs, consistent with other studies [13, 14]. Norovirus is also the leading cause of foodborne enteric illness outbreaks, identified here in 44% of foodborne outbreaks with a reported etiology, consistent with previous studies [15]. Because norovirus outbreaks are so prevalent and cause significantly larger outbreaks than other common etiologies, norovirus is also the leading cause of enteric illness outbreak–associated hospitalizations and deaths, despite being associated with comparatively low case fatality and hospitalization rates. Outbreaks with no reported etiology also exhibited a strong winter seasonality, were most often spread by person-to-person transmission, and were most often reported in LTCFs, similar to norovirus and suggesting norovirus as a possible etiology. Salmonella was identified as a leading cause of foodborne enteric outbreaks, second only to norovirus, in this study as in other studies [15]. Salmonella outbreaks were responsible for nearly two-thirds of multisite outbreaks. Salmonella has long been recognized as the predominant cause of multisite outbreaks, which have been on the rise as food distribution, consumption habits, and laboratory testing methods have evolved [15, 16]. PulseNet, a laboratory-based subtyping network, and the use of whole-genome sequencing (WGS) aid in the detection of more clusters of illness more quickly, linking cases to one another, and traceback to a source [17, 18]. Multisite outbreaks generally cause more illnesses than single-site outbreaks and, when combined with the high hospitalization rate of Salmonella outbreaks (20.9% in this study), can contribute disproportionately to more illnesses and hospitalizations [16].
Shigella was also identified as a leading cause of enteric illness outbreaks in this study. Similar to norovirus outbreaks, Shigella outbreaks also mostly spread through direct person-to-person contact but were more often identified in school and daycare settings. Shigella is a well-recognized cause of enteric illness outbreaks in children [19, 20]. While Shigella is considered a year-round enteric pathogen, these findings align with other studies that have found a slight increase in cases in summer months [21, 22]. Shigella often causes a mild, self-limiting diarrheal illness, as illustrated by the very few Shigella outbreak–related deaths reported to NORS [23]. Previous studies have shown that patients with bloody diarrhea and concurrent fever, symptoms commonly reported in Shigella outbreaks, are more likely to seek medical care than those without, potentially contributing to the high hospitalization rate seen in this study [24].
The other leading bacterial enteric pathogens—Salmonella, E. coli, and Campylobacter—were all most commonly associated with foodborne outbreaks. Outbreaks of these pathogens were identified year-round, but this study found a marked increase in the summer months, as has been noted elsewhere [21, 22]. While more than half of norovirus and Shigella outbreaks occurred in a single identified setting (LTCFs and schools or daycare centers, respectively), outbreaks caused by Salmonella, Campylobacter, and E. coli did not have a predominant single setting. This may be due in part to upstream contamination of food products that are then prepared and consumed in a variety of settings, rather than point-source contamination of food by an infected foodworker, as is often the case in foodborne norovirus outbreaks [25].
Although Salmonella outbreaks contributed the most absolute numbers of deaths and hospitalizations after norovirus, the highest case fatality and hospitalization rates in this study were attributed to E. coli outbreaks. While many strains of E. coli are relatively harmless, other strains such as Shiga toxin–producing E. coli are a well-known cause of severe and often bloody diarrhea and can lead to serious outcomes like hemolytic uremic syndrome and death [26]. Campylobacter infection typically causes a mild illness but can lead to severe complications and is the leading bacterial cause of diarrheal illness in the United States [27]. In this study, only 1 death was attributed to a Campylobacter outbreak, but nearly 9% of patients were hospitalized. Like Shigella and other bacterial pathogens, multidrug resistance of Salmonella and Campylobacter is of significant concern and may, in turn, lead to more severe outcomes [28, 29]. Prevention efforts to reduce the overall burden of Salmonella, Campylobacter, and E. coli outbreaks must address multidrug resistance, multiple modes of transmission, contamination throughout the food chain, and the diversity of settings in which these outbreaks have been reported.
There are multiple limitations to this study. NORS is a voluntary reporting system that relies on state, local, and territorial public health agencies to detect, investigate, and report outbreaks. Therefore, data captured by NORS may not be generalizable to all outbreaks or illness associated with the etiologies reported here. Persons with an enteric illness often do not seek medical care, and among those who do, few provide a stool specimen for etiologic testing [24]. Variability in reporting by site and mode of transmission may introduce bias in which outbreaks are reported via NORS. NORS also collects data on foodborne and waterborne outbreaks of nonenteric illness; our inclusion criteria may have excluded some enteric illness outbreaks that did not include either an etiology or symptom information in the outbreak report, or else included outbreaks of nonenteric illness due to a pathogen that can cause both enteric and nonenteric illness. Nearly 60% of outbreak reports did not have a laboratory-confirmed etiology, and 19% did not report any suspected or confirmed etiology.
This study reviewed 11 years of data on enteric illness outbreaks for all reported etiologies and modes of transmission, building on the knowledge gained through decades of foodborne and waterborne outbreak surveillance. Norovirus is the leading cause of enteric illness outbreaks as well as outbreak-related deaths and hospitalizations in the United States. However, outbreaks due to common bacterial etiologies like Shigella and Salmonella continue to contribute significantly to morbidity and mortality. The continued efforts of local public health agencies to investigate and report enteric illness outbreaks to CDC and collect and test specimens are essential in understanding the national burden of enteric illness outbreaks. Expanded enteric illness outbreak reporting through NORS has provided a more comprehensive picture of the relative contribution of each outbreak etiology and mode of transmission and the characteristics of those outbreaks. Robust NORS data can, in turn, help track trends over time and guide targeted interventions by identifying settings, sources, and patients most at risk for enteric illness.
Supplementary Material
Acknowledgments.
The authors thank Caroline Pilewski for her early efforts on this analysis and Bryn Pape for her efforts in etiologic classification.
Financial support.
The National Outbreak Reporting System is funded by the Centers for Disease Control and Prevention.
Footnotes
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Disclaimer. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Scallan E, Griffin PM, Angulo FJ, Tauxe RV, Hoekstra RM. Foodborne illness acquired in the United States—unspecified agents. Emerg Infect Dis 2011; 17:16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yen C, Wikswo ME, Lopman BA, Vinje J, Parashar UD, Hall AJ. Impact of an emergent norovirus variant in 2009 on norovirus outbreak activity in the United States. Clin Infect Dis 2011; 53:568–71. [DOI] [PubMed] [Google Scholar]
- 3.Kirk MD, Pires SM, Black RE, et al. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 2015; 12:e1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Systems for tracking waterborne disease and outbreaks. Available at: https://www.cdc.gov/healthywater/surveil-lance/tracking-systems.html. Accessed 27 March 2021.
- 5.Centers for Disease Control and Prevention. Frequently asked questions about the Foodborne Disease Outbreak Surveillance System (FDOSS). Available at: https://www.cdc.gov/fdoss/faq.html. Accessed 27 March 2021.
- 6.Centers for Disease Control and Prevention. About NORS. Available at: https://www.cdc.gov/nors/about.html. Accessed 8 August 2019.
- 7.US Census Bureau. Table 1. Annual estimates of the resident population for the United States, regions, states, and Puerto Rico: April 1, 2010 to July 1, 2019 (NST-EST2019–01). Available at: https://www.census.gov/data/tables/time-series/demo/popest/2010s-state-total.html. Accessed 5 January 2021.
- 8.US Census Bureau. Table 1. Intercensal estimates of the resident population for the United States, regions, states, and Puerto Rico: April 1, 2000 to July 1, 2010. Available at: https://www.census.gov/data/tables/time-series/demo/popest/intercensal-2000-2010-state.html. Accessed 5 January 2021.
- 9.Interagency Food Safety Analytics Collaboration. Foodborne illness source attribution estimates for 2018 for Salmonella, Escherichia coli O157, Listeria monocytogenes, and Campylobacter using multi-year outbreak surveillance data, United States. Atlanta, GA, and Washington, DC: US Department of Health and Human Services, Centers for Disease Control and Prevention, Food and Drug Administration, US Department of Agriculture’s Food Safety and Inspection Service, 2020. [Google Scholar]
- 10.Kambhampati A, Shioda K, Gould LH, et al. A state-by-state assessment of food service regulations for prevention of norovirus outbreaks. J Food Prot 2016; 79:1527–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel MM, Hall AJ, Vinjé J, Parashar UD. Noroviruses: a comprehensive review. J Clin Virol 2009; 44:1–8. [DOI] [PubMed] [Google Scholar]
- 12.Cardemil CV, Parashar UD, Hall AJ. Norovirus infection in older adults: epidemiology, risk factors, and opportunities for prevention and control. Infect Dis Clin North Am 2017; 31:839–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grytdal S, Browne H, Collins N, et al. Trends in incidence of norovirus-associated acute gastroenteritis in four Veterans Affairs medical center populations in the United States, 2011–2015. Clin Infect Dis 2019; 70:40–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vega E, Barclay L, Gregoricus N, Shirley SH, Lee D, Vinje J. Genotypic and epidemiologic trends of norovirus outbreaks in the United States, 2009 to 2013. J Clin Microbiol 2014; 52:147–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gould LH, Walsh KA, Vieira AR, et al. Surveillance for foodborne disease outbreaks—United States, 1998–2008. MMWR Surveill Summ 2013; 62:1–34. [PubMed] [Google Scholar]
- 16.Crowe SJ, Mahon BE, Vieira AR, Gould LH. Vital signs: multistate foodborne outbreaks—United States, 2010–2014. MMWR Morb Mortal Wkly Rep 2015; 64:1221–5. [DOI] [PubMed] [Google Scholar]
- 17.Ribot EM, Hise KB. Future challenges for tracking foodborne diseases: PulseNet, a 20-year-old US surveillance system for foodborne diseases, is expanding both globally and technologically. EMBO Rep 2016; 17:1499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitney BM, McClure M, Hassan R, et al. A series of papaya-associated Salmonella illness outbreak investigations in 2017 and 2019—a focus on traceback, laboratory, and collaborative efforts. J Food Prot 2021; doi: 10.4315/JFP-21-082. [DOI] [PubMed] [Google Scholar]
- 19.Arvelo W, Hinkle CJ, Nguyen TA, et al. Transmission risk factors and treatment of pediatric shigellosis during a large daycare center-associated outbreak of multidrug resistant Shigella sonnei: implications for the management of shigellosis outbreaks among children. Pediatr Infect Dis J 2009; 28:976–80. [DOI] [PubMed] [Google Scholar]
- 20.Centers for Disease Control and Prevention. Outbreaks of multidrug-resistant Shigella sonnei gastroenteritis associated with day care centers—Kansas, Kentucky, and Missouri, 2005. MMWR Morb Mortal Wkly Rep 2006; 55:1068–71. [PubMed] [Google Scholar]
- 21.Williams VF, Stahlman S, Oh GT. Incidence of Shigella intestinal infections, active component, U.S. Armed Forces, 2007–2016. MSMR 2017; 24:11–5. [PubMed] [Google Scholar]
- 22.Naumova EN, Jagai JS, Matyas B, DeMaria A Jr, MacNeill IB, Griffiths JK. Seasonality in six enterically transmitted diseases and ambient temperature. Epidemiol Infect 2007; 135:281–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotloff KL, Riddle MS, Platts-Mills JA, Pavlinac P, Zaidi AKM. Shigellosis. Lancet 2018; 391:801–12. [DOI] [PubMed] [Google Scholar]
- 24.Scallan E, Jones TF, Cronquist A, et al. ; FoodNet Working Group. Factors associated with seeking medical care and submitting a stool sample in estimating the burden of foodborne illness. Foodborne Pathog Dis 2006; 3:432–8. [DOI] [PubMed] [Google Scholar]
- 25.Dewey-Mattia D, Manikonda K, Hall AJ, Wise ME, Crowe SJ. Surveillance for foodborne disease outbreaks—United States, 2009–2015. MMWR Surveill Summ 2018; 67:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brooks JT, Sowers EG, Wells JG, et al. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983–2002. J Infect Dis 2005; 192:1422–9. [DOI] [PubMed] [Google Scholar]
- 27.Tack DM, Ray L, Griffin PM, et al. Preliminary incidence and trends of infections with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2016–2019. MMWR Morb Mortal Wkly Rep 2020; 69:509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medalla F, Hoekstra RM, Whichard JM, et al. Increase in resistance to ceftriaxone and nonsusceptibility to ciprofloxacin and decrease in multidrug resistance among Salmonella strains, United States, 1996–2009. Foodborne Pathog Dis 2013; 10:302–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geissler AL, Bustos Carrillo F, Swanson K, et al. Increasing Campylobacter infections, outbreaks, and antimicrobial resistance in the United States, 2004–2012. Clin Infect Dis 2017; 65:1624–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


