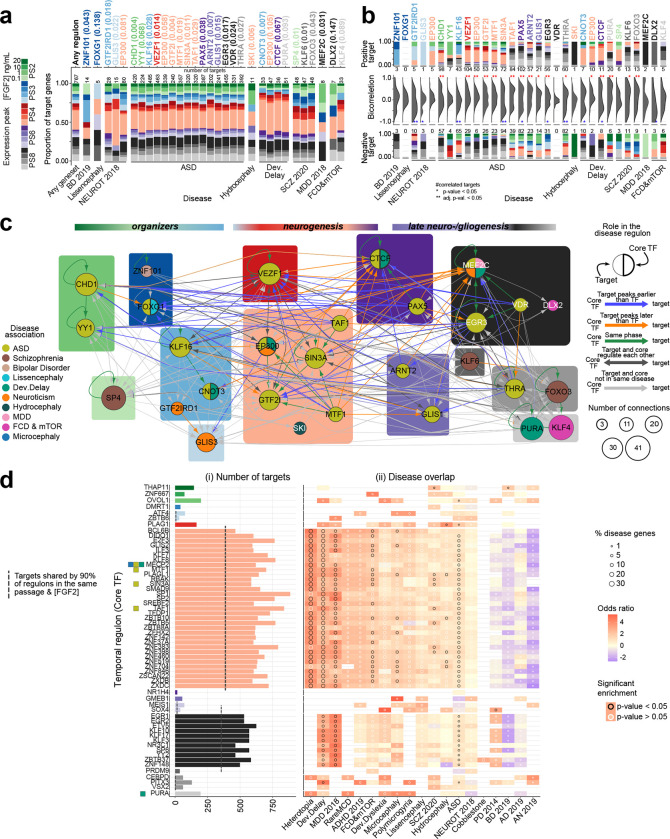
Fig. 3. Sequential gene regulatory networks across hNSC progression.
a) Proportion of target genes for each disease regulon with expression peak across passages and FGF2 conditions. Each core TF (top axis) is colored by its expression peak with p-value associated. The category “Any geneset/Any disease” includes all the genes of the disease regulons. Number of targets per regulon indicated on the top bar. b) Distribution of expression correlations between core TFs and their targets for each disease with positively and negatively correlated targets, and their number shown in the top and bottom summary bar plots, respectively. The color of the bars and TF labelling represent the expression peaks. Significantly high number of positively or negatively correlated targets are marked as ‘*’: p-value < 0.05, or ‘**’: corrected p-value < 0.05. c) Predicted gene regulatory network of the core TFs in the disease regulons with nodes representing genes colored by disease. For a TF found in multiple disease regulons, the thicker stroke indicates the disease in which it is a core TF; the node size indicates the connections to other core TFs. Background colors indicate gene expression peak in the in vitro hNSCs, same colors as in (a). Edge colors represent the expression peak relation between core TF and target, if they regulate each other, if associated with same or different disease. d) Temporal regulons ordered by the core TF peak expression across hNSC progression, same colors as in (a). Core TFs of disease regulons are indicated and colored by disease as in panel c. MECP2, although not a core TF in the disease regulons, is disease-associated. i) Number of targets for each regulon. ii) Overlap of temporal regulons with disease shown as odds ratio of the regulon gene enrichment in each disease (color of the grid), fraction of disease genes present in a regulon (dot size), and the significance of the enrichment (dot color).