Abstract
Factors accounting for long-term nonprogression may include infection with an attenuated strain of human immunodeficiency virus type 1 (HIV-1), genetic polymorphisms in the host, and virus-specific immune responses. In this study, we examined eight individuals with nonprogressing or slowly progressing HIV-1 infection, none of whom were homozygous for host-specific polymorphisms (CCR5-Δ32, CCR2-64I, and SDF-1-3′A) which have been associated with slower disease progression. HIV-1 was recovered from seven of the eight, and recovered virus was used for sequencing the full-length HIV-1 genome; full-length HIV-1 genome sequences from the eighth were determined following amplification of viral sequences directly from peripheral blood mononuclear cells (PBMC). Longitudinal studies of one individual with HIV-1 that consistently exhibited a slow/low growth phenotype revealed a single amino acid deletion in a conserved region of the gp41 transmembrane protein that was not seen in any of 131 envelope sequences in the Los Alamos HIV-1 sequence database. Genetic analysis also revealed that five of the eight individuals harbored HIV-1 with unusual 1- or 2-amino-acid deletions in the Gag sequence compared to subgroup B Gag consensus sequences. These deletions in Gag have either never been observed previously or are extremely rare in the database. Three individuals had deletions in Nef, and one had a 4-amino-acid insertion in Vpu. The unusual polymorphisms in Gag, Env, and Nef described here were also found in stored PBMC samples taken 3 to 11 years prior to, or in one case 4 years subsequent to, the time of sampling for the original sequencing. In all, seven of the eight individuals exhibited one or more unusual polymorphisms; a total of 13 unusual polymorphisms were documented in these seven individuals. These polymorphisms may have been present from the time of initial infection or may have appeared in response to immune surveillance or other selective pressures. Our results indicate that unusual, difficult-to-revert polymorphisms in HIV-1 can be found associated with slow progression or nonprogression in a majority of such cases.
Disease induction following human immunodeficiency virus type 1 (HIV-1) infection, or the ability to control the infection in rare cases, is likely to be regulated by the balance of at least three factors: genetic susceptibility of the host, the ability of the host to elicit effective immune responses, and the sequence of the incoming virus.
Twenty-five to 30% of long-term nonprogressors (LTNPs) who have remained AIDS free for more than 15 years in the absence of antiretroviral drugs express specific mutant forms of the HIV-1 second receptors CCR5 and CCR2 (11, 23, 52). Individuals who are homozygous for a 32-bp deletion (Δ32) in CCR5, which encodes the most frequently used second receptor for HIV-1, are resistant to infection by HIV-1 (11, 36). HIV-1-infected individuals who are heterozygous for CCR5Δ32 (−/+) show a slightly slower disease progression than CCR5+/+ homozygotes (11, 23, 36, 49). A V64I polymorphism in the CCR2 gene, which is linked to a point mutation in the regulatory region of the CCR5 gene, and a common polymorphism in the 3′-untranslated region of the stromal derived factor gene (SDF-1) are also associated with delayed disease progression (3, 5, 6, 57). SDF-1 is the natural ligand for CXCR4, the second receptor for syncytium-inducing strains of HIV-1. Intracellular factors may also contribute to the inherent susceptibility of host cells to support HIV replication (13, 22, 35).
HIV-1-infected individuals who are LTNPs also appear in general to have more effective HIV-1-specific immune responses. Perhaps most remarkable is the HIV-specific CD4 helper cell activity seen in nonprogressors controlling their HIV infection (47). This vigorous helper cell response contrasts with the absent or low response typically observed in progressors (47). Nonprogressors also generally exhibit more vigorous cytotoxic T-lymphocyte (CTL) activity than typical progressors (18, 21). The ability to mount effective immune responses may be influenced by the extent of heterozygosity at major histocompatibility complex class I (MHC-I) loci or by the specific MHC-I genotype or both (6). The ability to mount effective immune responses may of course be related in some cases to the virulence of the infecting strain of HIV-1 and/or the inherent susceptibility of the host.
One patient from central Massachusetts (27) and nine from Australia (10) are infected with Nef-deleted forms of HIV-1. Infection with this replication-competent, attenuated form of HIV-1 is clearly responsible for slow progression or nonprogression in these individuals (10, 27). This is particularly evident for eight of the individuals in the Australian cohort, who were all infected by a single blood donor, who all have the Nef-defective form of the virus, and who all have been slow progressors or nonprogressors (33, 43). An additional LTNP with consistently deficient forms of Nef has been described (48). One LTNP whose rev gene exhibited deficient activity over more than 4.5 years of observation has been described (25), another with a consistently defective p17 sequence (5) and a nonprogressing infant-mother pair containing defects clustered in the C terminus of Vpr have been documented (54).
In this report we describe full-length HIV-1 sequences from eight nonprogressors living in New England. Additional examples of uniform, nonrevertible inactivating deletions in auxiliary genes were not discovered. However, unusual, difficult-to-revert polymorphisms were noted in the HIV-1 present in seven of these eight individuals.
MATERIALS AND METHODS
LTNP HIV-1 cultures.
Peripheral blood mononuclear cells (PBMC; 5 × 106) from LTNPs were cocultivated with an equal number of phytohemagglutinin-activated PBMC from seronegative donors. These cultures were monitored for p24 production using an HIV-1 core antigen kit (Coulter, Hialeah, Fla.). If HIV-1 replication was not detected in these cultures, CD8 cells were depleted using immunomagnetic beads (Dynabeads; Dynal Inc, Great Neck, N.Y.) prior to cocultivation (14). Using these protocols, HIV-1 was recovered from seven of the eight LTNPs described in this report.
Isolation of cellular DNA.
Infected cultured cells (5 × 106) or LTNP PBMC were lysed in 0.5 ml of lysis buffer (10 mM Tris [pH 8.2], 0.4 M NaCl, 2 mM EDTA [pH 8.2]) that was supplemented with 33 μl of 10% sodium dodecyl sulfate (SDS) and 10 μl of proteinase K (10 mg/ml) for 1 h at 56°C. After lysis, 160 μl of saturated NaCl was added and the tube was inverted to mix the reagents. The mixture was then centrifuged at 14,000 rpm in a microcentrifuge for 10 min. The clear supernatant was removed and placed in a fresh tube, and 700 μl of isopropanol was added. The mixture was inverted and centrifuged for 10 min at 14,000 rpm. The supernatant was then removed, and the pellet was washed with 70% ethanol and then air dried for 1 h.
Amplification of LTNP HIV-1 sequences.
One microgram of cellular DNA served as the template for PCR amplifications. Primers for PCR that annealed to highly conserved regions of HIV-1 were chosen. These primers were designed such that their sequences were rare in both the documented human genome and HIV-1 sequences according to the Right Primer analysis program (Biodisk, San Francisco, Calif.). In addition, they were designed to have annealing temperatures of approximately 70°C as determined by the Oligo primer analysis program (National Biosciences, Plymouth, Minn.). Stringent PCR conditions that had been previously shown to optimize the amplification of large, low-copy-number templates were chosen (3, 8, 58). A 200-μl reaction volume in a 0.5-ml thin-walled PCR tube (Perkin-Elmer Cetus, Norwalk, Conn.) was used. The volume included 2 U of rTthXL (Perkin-Elmer Cetus), 200 mM deoxynucleoside triphosphates, and a 0.2 μM concentration of each primer. This PCR mixture was preheated for 60 s at 80°C before 1.0 mM magnesium acetate was added. The sample was then inserted into an Omnigene PCR cycler (Hybaid, Franklin, Mass.) that was preheated to 80°C. Seventy-five cycles/round of PCR was used to amplify LTNP HIV-1 sequences. Each cycle consisted of a 93°C denaturation step followed by a rapid-cooling step to 70°C and a slow-cooling step to 65°C. The 65°C annealing and polymerization step was carried out for 1 min for every 1 kbp of amplified HIV-1 sequence. The complete HIV-1 sequences from the seven LTNPs for which virus was successfully isolated were obtained by amplification of 11 overlapping fragments of approximately 1 kbp each using a single round of PCR for each fragment. HIV-1 from 161J has not been isolated, and thus overlapping fragments of approximately 500 bp were amplified using two rounds of PCR with nested primer sets to complete the HIV-1 sequences. Nested PCR was also employed for amplification of HIV-1 sequences from PBMC samples of LTNPs from whom virus was recovered. Overlapping PCR fragments were treated with T4 DNA polymerase (New England Biolabs, Beverly, Mass.) and inserted into a SmaI-digested pUC18 vector (Promega, Madison, Wis.). The DNA sequence of the inserted fragment was then determined with M13 primers using an ABI 377 DNA sequencer (Perkin-Elmer Cetus). Sequence data for LTNP HIV-1 presented in this report represent a consensus of two independent clones from each of two independent PCR amplifications. Nested PCR was used to amplify HIV-1 DNA from the LTNP designated LTNP 5 and the infectious clone NL 4-3 in two approximate halves which could be combined at a common restriction site for transfection into cells. The internal oligonucleotide for the amplification of each half genome contained an A-to-G change from the consensus sequence which introduces an XmaI site (CCCGGG) into a highly conserved region (nucleotide 4402 for NL 4-3) (1) without changing the pol amino acid coding sequence. 5′ PCR fragments included bp 297 to 4402, and 3′ fragments included bp 4402 to 9607 (base pairs are based on NL 4-3 sequences).
Phylogenetic analysis of LTNP sequences.
The subtype of each LTNP HIV-1 sequence was determined by comparison to HIV-1 subtype reference sequences using the Los Alamos subtyping program (B. Korber, www.ncbi.nih.gov, Los Alamos National Laboratory, 1999). Phylogenetic trees based on neighbor-joining analyses were produced to determine the genetic distance between LTNP and reference gag and nef sequences using the PAUP phylogenetic analysis software (D. L. Swofford, Phylogenetic analysis using parsimony [and other methods], 4th ed., Sinauer Associates, Sunderland, Mass., 1998). The reference sequences in these analyses, indicated by (in order) code, subgroup, strain, identification number, and accession number(s) are as follows: A1A, U455, 328902, M230; A2, A, 92UG037.1, U51190; B1, B, OYI, 328440, M26727; B2, B, RF, 328565, M17451, M12508, USA; C1, C, ETH2220, 1353860, U46016; C2, C, 92BR025.8, 2194183, U529523; D1, D, NDK, 328154, M27323; D2, D, Z2Z6, 329377, M22639; F1, F, 93BR020.1, 3114544, AF005494; H1, H, 90CR056.1, 3114562, AF005496; AEI, A/E (E in most of env, A in gag and pol, and a mixture of A and E in regulatory genes), CM240, 1537050, U54771; G1, G, SE6165, 3403225, AF061642; E1, MP38; E2, MP59.
Plasmid construction.
NL 4-3/LTNP Gag chimeras were engineered by insertion of an ApaI-BclI-digested 320-base fragment that contained the coding sequence for LTNP p6gag-pol into ApaI-BclI-digested p83-2 plasmid which contained NL 4-3 sequences. In order to engineer simian immunodeficiency virus/HIVnef (SHIVnef) recombinants, SIVmac nef sequences were deleted from the 3′ extent of the SIVmac env gene to 120 bases 5′ of the NF-κB binding site in SIVmac239 (45) and HIV-1 sequences derived from the LTNP 1486D were inserted as previously described (2). Thorough DNA sequence analysis verified that all recombinant clones selected for study contained exactly the desired sequences.
HIV-1 and SHIVnef replication assays.
All stocks of NL 4-3 or recombinants described in this report were generated by DEAE-dextran transfection of the cell line CEMx174 (41). NL 4-3 or recombinant constructs were transfected into CEMx174 cells, and virus in the cell-free supernatant was harvested at or near the peak of virus production as previously described (15). Transfected or infected CEMx174 cells were grown in RPMI 1640 (Gibco BRL, Grand Island, N.Y.) that was supplemented with 10% fetal calf serum (Gibco BRL). The levels of p27 viral protein that were produced from transfections or infections or that were contained within viral stocks were quantified using a SIV core antigen kit (Coulter).
Analysis of Vpr incorporation.
Virus from NL 4-3 or HIV-1Δvpr or recombinants with LTNP p6gag sequences that contained 500 ng of p24 antigen were centrifuged at 13,000 × g for 2 h at 4°C. The supernatant was removed, and the virus pellet was resuspended in Laemmli sample buffer prior to electrophoresis through an SDS–12% polyacrylamide gel electrophoresis gel. The proteins were electroblotted onto an Immobilon-P membrane (Millipore, Bedford, Mass.) which was then blocked with 5% skim milk in phosphate-buffered saline–0.05% Tween 20 (PBST). The blot was incubated overnight at 4°C in blocking solution that contained the anti-HIV-1 Vpr-specific polyclonal antibody (NIH AIDS Research and Reference Reagent Program) at a dilution of 1:2,000. Primary antibodies were removed by washing the membranes three times for 30 min with PBST at room temperature. The dilution of the secondary antibody and the detection of Vpr were performed according to the protocol of the enhanced chemiluminescence system (Amersham, Chicago, Ill.).
Vif functional assay.
The Vif genes of LTNP 5 virus clones 2 and 3 were isolated as XbaI-XhoI fragments using PCR and inserted into the HIV-1HXB-3 Vif expression vector pgVIF. Each Vif expression vector as well as the negative control (pgΔvif) was cotransfected into H9 cells with the vif-deficient proviral vector pIIIB/Δvif. At 24 h posttransfection, viruses were harvested from the culture supernatants, normalized according to reverse transcriptase activity, and used in single-cycle infections of the indicator cell line C8166/HIV-CAT. After an additional 30 h, the levels of chloramphenicol acetyltransferase (CAT) expression were measured from whole-cell lysates and used to determine relative levels of infectivity. Whole-cell lysates of transfected H9 cultures were also evaluated for Vif accumulation by Western blotting using a Vif-specific monoclonal antibody as previously described (51).
Experimental infection of rhesus monkeys.
Virus diluted to contain 50 ng of p27 antigen was inoculated intravenously into juvenile rhesus monkeys (Macaca mulatta) (2). At various time points postinoculation, blood samples were collected as previously described (15).
Determination of viral RNA and infectious cell loads.
Cell-associated virus loads in infected monkeys were determined by quantitative cocultivation of PBMC with CEMx174 cells (12). PBMC were purified, counted in a hemocytometer, and cocultured with CEMx174 cells in various numbers. On day 21, the presence of SIV p27 antigen was determined and the numbers of PBMC needed to recover SIV were calculated. The results described in this report represent averages of duplicate determinations. Virion-associated SIV RNA in plasma samples was quantified using a reverse transcription-PCR assay on an Applied Biosystems Prism 7700 sequence detection system (Perkin-Elmer Cetus) (53).
Determination of the percentages of CD4+ cells in blood of SHIVnef-infected animals.
Whole blood was drawn from SHIV nef-inoculated animals at various times postinoculation and stained with OKT4, a fluorescein isothiocyanate-conjugated murine monoclonal antibody that reacts with rhesus macaque CD4. The stained samples were analyzed using a FACSscan flow cytometer (Becton Dickinson, San Jose, Calif.).
CCR5, CCR2, and SDF-1 genetic analysis.
Genomic DNA was extracted from patient samples using a DNA/RNA extraction kit (Amersham Life Science, Piscataway, N.J.). CCR5, CCR2, and SDF-1 genetic polymorphisms were amplified using previously described oligonucleotides and procedures (11, 52, 57).
RESULTS
Introduction to patient population.
The patients presented here are HIV-1-infected individuals from the Massachusetts area who have remained AIDS free with low viral loads without antiretroviral intervention (Table 1). Seven of the eight (LTNP 1, LTNP 2, LTNP 3, LTNP 5, LTNP 6, LTNP 7, and 161J) are individuals with hemophilia who were likely infected by contaminated blood products. 1486D is a homosexual male. Of the eight LTNPs, seven continue to be monitored for CD4 concentrations and viral load (Table 1) (17). LTNP 5 died in June 1997 from complications of hepatitis C infection. At the time of his last visit, LTNP 5 exhibited a CD4 concentration of 464/ml and maintained low but detectable viral RNA loads (Table 1). In recent evaluations, CD4 concentrations from LTNP 1 have declined although viral RNA loads remained below the level of detection (Table 1) (19). This individual has maintained a large deletion in Nef and 3′ LTR sequences over the entire course of infection, and no reversion of Nef sequence was observed during the period when CD4 concentrations have declined (19, 27). Except for the recent treatment of LTNP 1, none of the LTNPs in this report have received any antiretroviral therapy for their HIV-1 infection despite infection in most cases for over 15 years (Table 1). 161J continues to maintain detectable CD4 helper T-cell responses previously shown to be associated with nonprogression (47). Detectable proliferation responses to p24 antigen have been observed in LTNPs 1, 2, and 3 but not in LTNPs 6 and 7 (17). LTNP 6 was previously reported to contain a polymorphism in Sp1 binding site III which resulted in suboptimal Tat-mediated transactivation (28), consistent with the independent analysis described in this report. LTNP 6 isolates were also shown to be defective in a single cell killing assay in comparison to isolates from progressors (44). LTNP 2, LTNP 3, and LTNP 6 Nef sequences have been previously determined and were shown to be functional in a CD4 down-regulation assay (38).
TABLE 1.
Clinical history of LTNPs
| LTNP | Date of first seropositivity detectiona (mo-day-yr or yr) | Date of last visitb (mo-day-yr or mo-yr) | Absolute CD4 T-cell counts/ml at last visit | Plasma viral RNA level at last visit (copies/ml) |
|---|---|---|---|---|
| LTNP 1 | 12-21-83 | 2-11-99 | 257c | <50 |
| LTNP 2 | 3-7-84 | 9-16-98 | 589 | 58 |
| LTNP 3 | 12-7-83 | 4-21-99 | 530 | 180 |
| LTNP 5 | 2-1-84 | 3-2-95 | 464 | 644 |
| LTNP 6 | 5-16-84 | 3-24-99 | 1,122 | 5,000 |
| LTNP 7 | 2-29-84 | 1-6-99 | 606 | 97 |
| 161J | 1979 | 6-15-99 | 843 | <50 |
| 1486D | 1991 | 12-98 | 630 | 8,360 |
LTNP 1, LTNP 2, LTNP 3, LTNP 6, and LTNP 7 were all seropositive at their first visit. LTNP 5, 161J, and 1486D seroconverted between regularly scheduled visits.
The date shown is the date of the last visit. LTNP 5 died from complications of hepatitis C infection in June 1997. All other patients are healthy as of their last visit.
CD4 T-cell counts in LTNP 1, who is infected with Nef-deleted HIV-1, began declining around 1996 to 1997 despite consistently undetectable viral loads (19).
Analysis of HIV-1 second-receptor genotypes.
Specific mutations in the genes for HIV-1 second receptors CCR5 (Δ32) and CCR2 (64I) and for the chemokine SDF-1 (3′A) that is the ligand for the CXCR4 second receptor have been associated with slower progression to AIDS (11, 23, 30, 39, 49, 52, 57). We investigated the extent to which these polymorphisms were present in the eight LTNPs studied in this report. LTNP 5 and LTNP 7 carried +/Δ32CCR5 and +/64ICCR2 protective genotypes, respectively, and the other six patients contained more common, nonprotective genotypes (Table 2). No patient was homozygous for CCR5-Δ32, for CCR2-64I, or for SDF-1-3′A (Table 2).
TABLE 2.
LTNP CCR5, CCR2, and SDF-1 genotypesa
| LTNP | Genotypeb of:
|
||
|---|---|---|---|
| CCR5 | CCR2 | SDF-1 | |
| LTNP 1 | +/+ | +/+ | +/3′A |
| LTNP 2 | +/+ | +/+ | +/+ |
| LTNP 3 | +/+ | +/+ | +/+ |
| LTNP 5 | +/Δ32 | +/+ | +/+ |
| LTNP 6 | +/+ | +/+ | +/+ |
| LTNP 7 | +/+ | +/64I | +/3′A |
| 161J | +/+ | +/+ | +/3′A |
| 1486D | +/+ | +/+ | +/3′A |
Patient samples were analyzed for HIV-1 second-receptor (CCR5 and CCR2) and chemokine (SDF-1) polymorphisms associated with slower progression.
+, most common allele for each gene; Δ32, 64I, and 3′A, CCR5, CCR2, and SDF-1 polymorphisms, respectively, associated with slower 64I and 3′A progression in the homozygous state; and Δ32 and 64I in the heterozygous state.
HIV-1 sequences of LTNPs.
Replication-competent HIV-1 was successfully isolated from seven of the eight LTNPs described in this report by cocultivation of uninfected donor PBMC with PBMC samples obtained in 1994 for LTNP 1, LTNP 2, LTNP 3, LTNP 5, LTNP 6, and LTNP 7 as well as in 1995 for 1486D (Table 3) (17). DNA from cells infected with these isolates served as the template for the amplification of HIV-1 sequences. HIV-1 was not successfully isolated from 161J (Table 3); we used DNA from a PBMC sample obtained in 1994 from 161J to serve as template for PCR.
TABLE 3.
Summary of LTNP HIV-1 genotypic and phenotypic characteristics
| LTNP | Virus isolationa | In vitro growthb | Unusual polymorphismsc | Frequency (%) of polymorphism in the databased |
|---|---|---|---|---|
| LTNP 1 | Yes | Good | ΔNef (>100 aa) | 0 |
| LTNP 2 | Yes | Good | None | |
| LTNP 3 | Yes | Good | Δ112p17gag | 0 |
| LTNP 5 | Yes | Poor | Δ23TMenv | 0 |
| LTNP 6 | Yes | Good | 4-base insertion 324U3LTR | 0 |
| 4-aa insertion Vpu | 4 | |||
| Δ9-11TMenv | 2 | |||
| Δ49-50nef | 9 | |||
| 1-aa deletion in Nef C terminus | 0 | |||
| LTNP 7 | Yes | Good | Δ466-467p6gag-pol | 0 |
| 161J | No | NA | Δ483-484p6gag-pol | 0 |
| Δ48-49Nef | 0 | |||
| 1486D | Yes | Good | Δ370p2gag | 2 |
| Δ465-466p6gag-pol | 0 | |||
| Δ48-49Nef | 0 |
For virus isolation LTNP PBMC were cocultivated with either bulk or CD8-depleted, PHA-activated PBMC from seronegative donors.
Good production of >100 ng of p24 antigen/ml in cocultivation assays. LTNP 5 isolates produced a maximum of 5 ng of p24 antigen/ml (poor growth). NA, not applicable.
Includes those that are difficult to revert and those that were consistently observed in cultured virus and in patient PBMC.
Frequency of polymorphism in the Los Alamos sequence database.
The laboratories in which cellular DNA was purified and HIV-1 sequences were amplified for this study are not otherwise used for HIV-1 studies. All of the sequences from the eight LTNPs described in this report were uniquely different from one another and uniquely different from sequences of laboratory strains of HIV-1 in the Los Alamos HIV-1 sequence database (B. Korber, http://hiv-web.lanl.gov, Los Alamos National Laboratory, 1999). Furthermore, as described below, the examination of unusual HIV-1 polymorphisms in PBMC taken at different times affirmed the authenticity of each sequence. Thus, the sequences reported here are authentic and cannot be due to contamination.
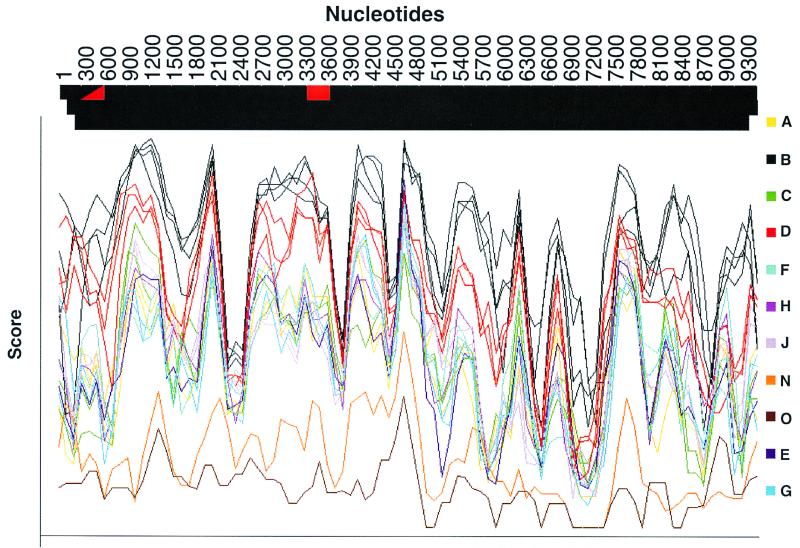
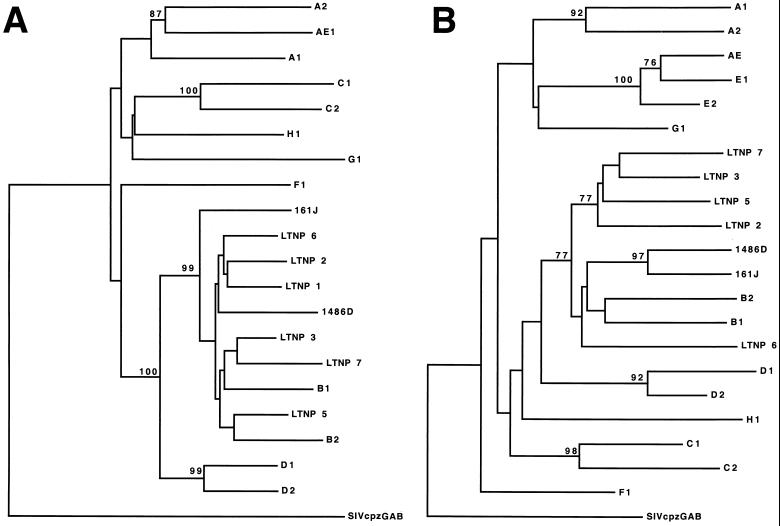
All eight LTNPs described here lived, and were likely infected, in the United States, where group M, subtype B, isolates vastly predominate. Full-length sequences from all eight LTNPs were analyzed using the Los Alamos subtyping program and subtype reference sequences (B. Korber, www.ncbi.nih.gov). Nucleotide sequences of all eight LTNPs had the highest relatedness to subgroup B sequences across the entire length of the HIV-1 genome. The relatedness of 1486D sequences to sequences of different subgroups is shown as an example in Fig. 1. In addition, dendrograms based on alignments of LTNP Gag and Nef amino acid sequences were generated (Fig. 2). Neighbor-joining analyses demonstrated that the LTNP Gag (Fig. 2A) (confidence value of 99) and Nef sequences (Fig. 2B) (confidence value of 77) were most closely related to subgroup B sequences. Trees using parsimony analysis revealed identical intersubtype relationships among the LTNP and reference Gag and Nef sequences.
FIG. 1.
Subtyping of the HIV-1 sequence of LTNP 1486D. 1486D sequences from bp 297 to 9607 (numbers based on NL 4-3 sequences) were analyzed using the Los Alamos subtyping program and subtyping reference sequences. The score value indicates the relative relatedness of 1486D sequences at different loci throughout the genome. The color of the bar at the top indicates the subtype that 1486D sequences are most closely related to at loci throughout the HIV-1 genome.
FIG. 2.
Dendrograms of reference sequences of HIV-1 isolates of different subtypes and LTNP sequences for Gag (A) and Nef (B). These analyses were performed using PAUP. Bootstrap confidence values were assigned when a minimum value of 70 was achieved for a particular branch. The dendrogram was rooted to the SIVcpzGAB sequences. The identification of the subtype isolates which are symbolized here is described in Material and Methods.
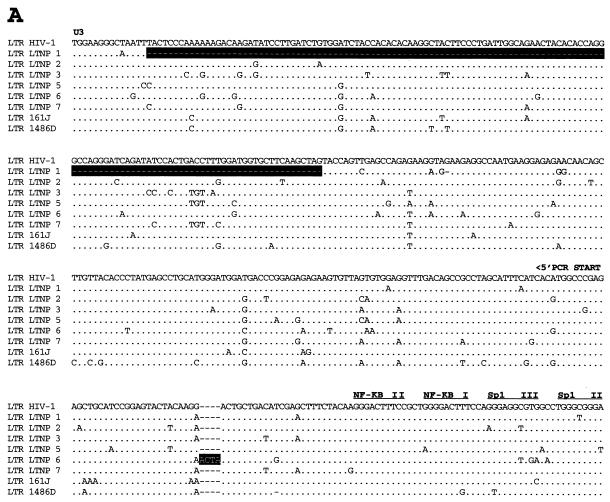
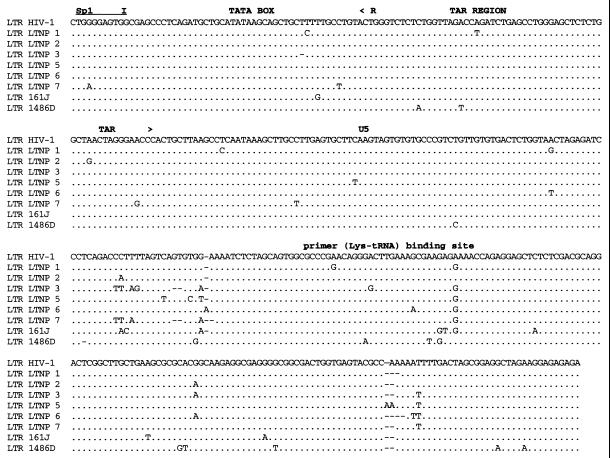
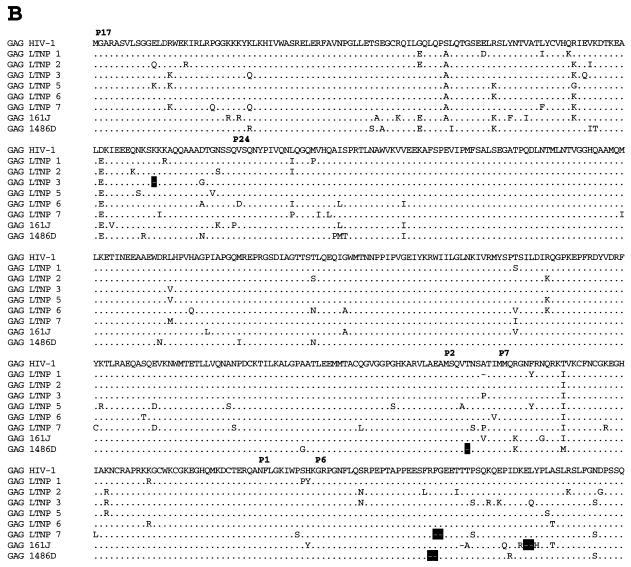
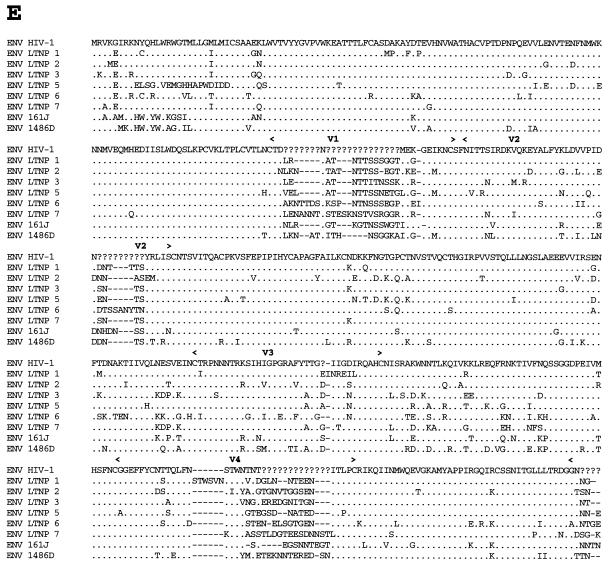
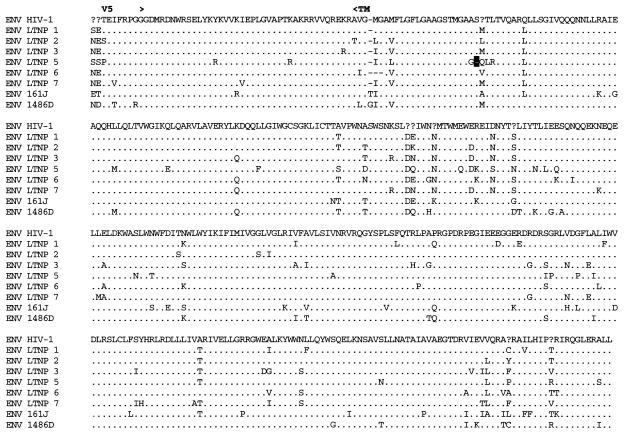
Analysis of Gag sequences revealed that five of the eight LTNPs contained 1- or 2-amino-acid (aa) deletions in Gag compared to subgroup B consensus sequences (Fig. 3B and Table 3). HIV-1 from LTNP 3 had a deletion in p17gag (1 aa); HIV-1 from LTNP 1 had a deletion in p2gag (1 aa); HIV-1 from LTNP 7 had a deletion in the p6gag-pol overlap region (2 aa); HIV-1 from 1486D had deletions in both p2gag (1 aa) and p6gag-pol (2 aa); and 161J had two separate deletions in p6gag-pol (1 and 2 aa) (Fig. 3B and Table 3). None of the Gag deletions were independently observed in different subjects. To ensure that these deletions were representative of sequences maintained in these subjects, DNA was isolated from LTNP PBMC obtained at distinct times and used as the template for PCR. These experiments revealed that the polymorphisms in Gag sequences described here were consistently observed for PBMC samples obtained in 1985 and 1994 for LTNP 3, 1983 and 1994 for LTNP 1 and LTNP 7, 1992 and 1995 for 1486D, and 1994 and 1998 for 161J (data not shown). The Los Alamos HIV-1 sequence database (B. Korber, http://hiv-web.lanl.gov) containing Gag sequences from 93 group M, group O, and SIVcpz isolates was examined for the Gag polymorphisms observed in our LTNP sequences. The p17gag Δ112 polymorphism contained in HIV-1 from LTNP 3 was not observed in any of the isolates in the database. The 2-aa p6gag polymorphisms contained in HIV-1 from 161J (Δ483-484), LTNP 7 (Δ466-467), and 1486D (Δ465-466) were also not observed in any of the isolates in the database, although Δ467 was observed for four subgroup A sequences (B. Korber, http://hiv-web.lanl.gov). The p2gag Δ370 polymorphism contained in HIV-1 from 1486D was observed in two database sequences: one from subgroup B (GA.OYI) and one from subgroup F (FI.FIN9363). We independently derived Gag sequences from four Massachusetts subjects who had progressed to AIDS. In contrast to what was observed in the LTNP Gag sequences, we observed no unusual polymorphisms in the Gag sequences from these four progressors (data not shown). In addition, we scanned 90 group M Gag sequences in the Los Alamos database and did not find a single example of a deletion unique to a single individual. Thus, the polymorphisms observed in four of the LTNPs are unique among determined HIV-1 Gag sequences and their occurrence appears to be nonrandom in nature.
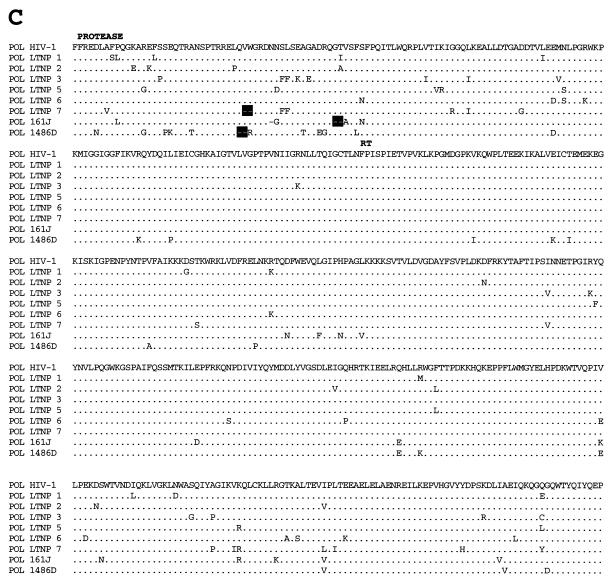
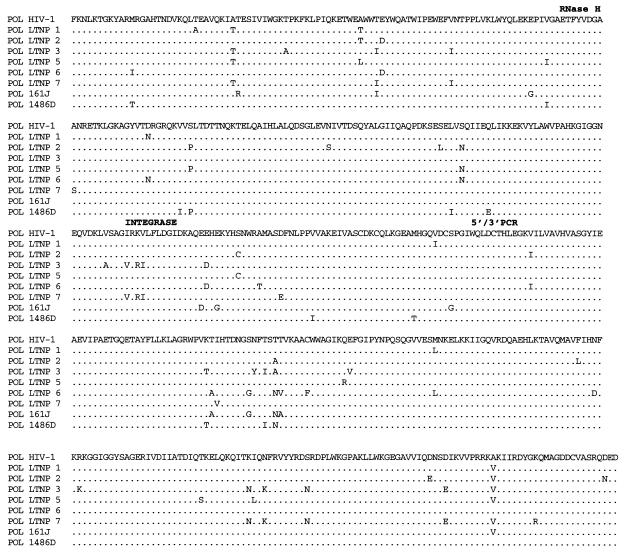
FIG. 3.
Alignments of LTNP sequences. Nucleotide (A) and amino acid (B to J) sequences of the indicated HIV-1 genetic elements and proteins were aligned to the group M, subgroup B, consensus sequences (HIV-1). Periods indicate conservation between LTNP and consensus B sequences. A dash indicates that a base or amino acid is not contained in a particular sequence. Unusual polymorphisms consistently observed in HIV-1 sequences from LTNPs are highlighted in black. The data presented represent consensus sequences from two clones from each of two independent PCRs.
In addition to the Gag deletions, small deletions and insertions were also observed in other LTNP HIV-1 sequences that have not previously been seen in any subgroup B sequence in the database (B. Korber, http://hiv-web.lanl.gov). LTNP 5 contained a single amino acid deletion in the transmembrane (gp 41) protein encoded by the env gene (Fig. 3E and Table 3). This mutation was consistently contained in PBMC samples obtained from LTNP 5 in 1987 and 1994 and was not observed in the sequences of the 131 isolates in the Los Alamos HIV-1 sequence database (B. Korber, http://hiv-web.lanl.gov). LTNP 6 HIV-1 cultures from virus obtained in 1984 and 1995 had a 3-aa deletion in gp41 that was previously observed in only three subgroup C isolates (C.DJ.DJ259A, C.DJ.DJ373A, and C.ET.ETH2220). Cultured virus from LTNP 6 from 1984 and 1995 also retained a 4-bp insertion in nef and 3′-LTR sequences (Fig. 3A) that resulted in truncation of LTNP 6 Nef by 1 aa short of the consensus C-terminal cysteine (Fig. 3J). This polymorphism has not been previously noted in any of the 256 Nef sequences in the Los Alamos HIV-1 sequence database (B. Korber, http://hiv-web.lanl.gov). HIV-1 from LTNP 6 obtained in 1984 and 1995 also had a 2-aa deletion in Nef (Δ49-50; Fig. 3J and Table 3) which appears in about 9% of subgroup B Nef sequences and represents the consensus for subgroup E Nef. LTNP 6 isolates also contained a 4-aa insertion near the N terminus of Vpu (Fig. 3G; Table 3) which is similar to a 4-aa insertion contained in four subgroup B isolates in the database. The 4 aa insertion was not detected in LTNP 6 PBMC samples obtained in 1984 but was detected in samples obtained in 1987 and at later times (Fig. 4). Cultured LTNP 1 virus maintained a large deletion in the 3′-LTR and Nef sequences (Fig. 3A and Table 3), similar to what has been observed previously (27). The PBMC sample obtained from 161J in 1994 revealed a 2-aa deletion in Nef (Δ48-49) (Fig. 3J and Table 3) that was also retained in PBMC obtained in 1998 from 161J. Cultured 1486D HIV-1 from 1995 maintained Δ48-49 Nef and also maintained a 1-aa insertion in Nef (61Q; Fig. 3J and Table 3). These polymorphisms were retained in 1486D 1995 PBMC (Fig. 5). However, the 61Q insertion was not observed in 1486D 1992 PBMC, suggesting that the addition of this amino acid took place in the intervening period (1992 to 1995) between samplings. As described above, Δ49-50 is maintained in about 9% of subgroup B isolates and represents the subgroup E Nef consensus sequence. However, Δ48-49 and the 61Q insertion have not been previously observed (B. Korber, http://hiv-web.lanl.gov).
FIG. 4.
Alignment of LTNP 6 Vpu sequences. LTNP 6 amino acid sequences from the time points indicated (e.g., P84, PBMC sample obtained in 1984) were aligned to the group M, subgroup B, consensus sequences (HIV-1). V, sequences were obtained from isolated HIV-1. Each sequence represents a consensus of two clones from each of two independent PCRs. Periods indicate conservation between LTNP 6 and the consensus B sequences. A dash indicates that an amino acid is not contained in a particular sequence. The 4-aa insertion observed in Vpu sequences from LTNP 6 is highlighted in black.
FIG. 5.
Amino acid sequences of 1486D Nef after infection into rhesus monkeys with SHIVnef recombinants. A SHIVnef recombinant containing 1486D Nef sequences from PBMC obtained in 1992 (P92) was inoculated into Mm 32-97 and Mm 33-97, and a recombinant containing 1486D Nef sequences obtained in 1995 was inoculated into Mm 34-97 and Mm 35-97. The sequences in the inoculum and sequences obtained from the infected animal 44 weeks postinoculation were aligned to the group M, subgroup B, consensus (HIV-1) sequences. Each sequence represents a consensus of two clones from each of two independent PCRs. Periods indicate conservation between 1486D and the consensus B sequences. A dash indicates that an amino acid is not contained in a particular sequence. The deletion and insertion polymorphisms observed in Nef sequences from 1486D are highlighted in black.
HLA haplotypes.
Specific sequences in Gag and Nef serve as MHC-I-restricted epitopes for CTL recognition (20, 31, 34). Sequence variations in these proteins can result in escape from CTL recognition. Six of the eight LTNPs described here had deletions in Gag and/or Nef. In many cases, these deletions were located in close proximity to each other: 1486D p2gag Δ370 versus LTNP 1 p2gag Δ373, 1486D p6gag Δ465-466 versus LTNP 7 p6gag Δ466-467 and 161J p6gag Δ471, and 1486D and 161J Δ48-49 Nef versus LTNP 6 Δ49-50 Nef (Fig. 3 and Table 4). We investigated whether common HLA types correlated with clustered polymorphisms among the LTNPs. 1486D and LTNP 1 shared the HLA C6 allele in addition to p2gag deletions that were only 3 aa apart (Fig. 3B and Table 4). Conversely, 1486D, LTNP 7, and 161J, who had p6gag deletions at similar locations, did not have any common HLA alleles. Similarly, 1486D, 161J, and LTNP 6, who had Nef deletions at similar locations, also did not have any common HLA alleles (Table 4).
TABLE 4.
Comparison of LTNP Gag and Nef deletions and HLA types
| LTNP | Gag deletionsa | Nef deletionsa | HLA typeb |
|---|---|---|---|
| LTNP 1 | 373 | >100 aa | A2, B13, B62, C2, C5 |
| LTNP 2 | 112 | None | A2, B44, C2, C5 |
| LTNP 6 | None | 49–50 | A1, A2, B44, B57, C5, C6 |
| LTNP 7 | 466–467 | None | A3, A31, B38, B64, C5, C12 |
| 161J | 471, 483–484 | 48–49 | A2, A3, B7, B60, C3 |
| 1486D | 370, 465–466 | 48–49 | A1, A24, B57, B65, C4, C6 |
Amino acid(s) consistently absent in LTNP sequences.
Designation of a single A, B, or C allele indicates that a patient is homozygous at that locus.
Incorporation of HIV-1 Vpr by LTNP p6gag.
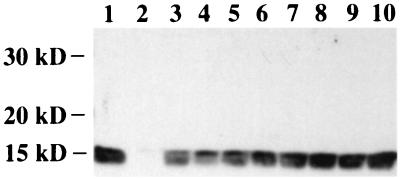
p6gag facilitates the incorporation of Vpr into HIV-1 particles (29, 37). Three of the eight LTNPs in this study harbored HIV-1 that had deletions in p6gag. We tested the ability of LTNP p6gag sequences to facilitate the incorporation of Vpr into recombinant HIV-1. ApaI-BclI restriction fragments derived from amplified LTNP sequences were inserted into p83-2, a vector which contained 5′ half sequences of the HIV-1 strain NL 4-3 (1). Recombinant virus containing 500 ng of p24 as well as NL 4-3 and ΔVpr controls were subjected to SDS-PAGE and assayed for Vpr incorporation by Western blotting with polyclonal anti-Vpr sera (Fig. 6). A Vpr-specific band was detected from purified NL 4-3 (Fig. 6, lane 1) that was not detected from purified ΔVpr virus (Fig. 6, lane 2). A Vpr-specific band was also detected from all eight purified recombinant viruses that expressed LTNP p6gag sequences at levels similar to that of the NL 4-3 control (Fig. 6, lanes 3 to 10). Thus, all LTNPs contained p6gag sequences that facilitated the efficient incorporation of Vpr into virions.
FIG. 6.
LTNP p6gag-mediated Vpr incorporation. Recombinant HIV-1 containing LTNP p6gag sequences were purified by centrifugation. Virally associated proteins were separated by SDS-PAGE and electroblotted onto a membrane filter. HIV-1 Vpr was detected using a Vpr-specific polyclonal antiserum. Lane 1, NL 4-3 parental virus; lane 2, NL 4-3ΔVpr; lanes 3 to 10, recombinant HIV-1 containing LTNP p6gag sequences: lane 3, LTNP 2; lane 4, LTNP 1; lane 5, LTNP 3; lane 6, LTNP 5; lane 7, LTNP 6; lane 8, LTNP 7; lane 9, 161J; lane 10, 1486D.
Infection of monkeys with SHIVnef containing 1486D Nef sequences.
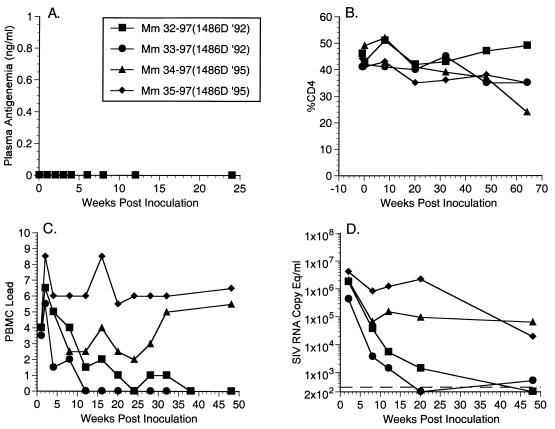
We have previously demonstrated that HIV-1 Nef can substitute for SIVmac Nef in vivo to produce a pathogenic infection in rhesus monkeys (2). 1486D Nef sequences isolated in 1992 (Δ48-49) and 1995 (Δ48-49 and insertion 61Q; Fig. 5) were evaluated for their performance in rhesus monkeys in the context of SHIVnef. We created SHIVnef recombinants by engineering the corresponding nef sequences into SIVΔnefXESAB, resulting in SHIVnef-1486D92 and SHIVnef-1486D95, respectively. Two juvenile rhesus monkeys were inoculated intravenously with either SHIVnef-1486D92 (Mm 32-97 and Mm 33-97) or SHIVnef-1486D95 (Mm 34-97 and Mm 35-97) containing 50 ng of p27 antigen. We monitored plasma antigenemia, CD4 counts, numbers of infectious cells in PBMC, and viral RNA loads with blood samples obtained at intervals following experimental infection of these monkeys. These analyses revealed that Mm 32-97 and Mm 33-97 did not maintain consistently measurable numbers of infectious cells in PBMC or RNA loads in a readily measurable range (Fig. 7C and D). Conversely, Mm 34-97 and Mm 35-97 displayed persistently high numbers of infectious cells in PBMC and high RNA loads. We investigated if Nef sequences in Mm 32-97 and Mm 33-97 were under selective pressure to acquire the 61Q insertion. At week 44 postinoculation we did not observe this polymorphism in Mm 32-97 and Mm 33-97. The 61Q polymorphism was retained in Mm 34-97 and Mm 35-97 at the same time point (Fig. 5). We also examined if the Δ48-49 deletion was stable in the 1486D Nef sequences and found that this polymorphism had been maintained in all four animals at 44 weeks postinoculation (Fig. 5).
FIG. 7.
Plasma antigenemia, CD4 percentage, PBMC load, and RNA copy (equivalents per milliliter) measurements for animals infected with SHIVnef recombinants containing 1486D Nef sequences. (A) Plasma antigenemia in SHIVnef-infected rhesus monkeys. p27 concentrations in plasma were determined at the time points indicated. The limit of detection is approximately 0.05 ng/ml. The week 0 sample is a preinfection sample taken immediately before inoculation with SHIVnef. (B) CD4 percentages in SHIVnef-infected rhesus monkeys. Whole blood was drawn from SHIVnef-inoculated animals at various times postinoculation and stained with OKT4, a fluorescein isothiocyanate-conjugated murine monoclonal antibody that was raised against rhesus macaque CD4 (American Type Culture Collection). The stained samples were analyzed using a FACSscan flow cytometer (Becton Dickinson). (C) Frequencies of infectious cells in PBMC of SHIVnef-infected rhesus macaques. Viral loads were graded on a scale from 0 to 10 indicating the number of PBMC needed to recover SIV. A value of 0 denotes that no virus was recovered using 106 cells, 1 denotes successful virus recovery from 106 cells, and 2 to 10 denote successful virus recovery from 333,333, 111,111, 37,037, 12,345, 4,115, 1,371, 457, 152, or 51 cells, respectively. (D) Plasma SIV RNA levels at the indicated weeks postinoculation for animals infected with SHIVnef recombinants. The dashed line indicates the threshold sensitivity of the assay, 300 copy eq/ml. A value of 0 was assumed for week 0.
Determinants for poor growth of HIV-1 from LTNP 5.
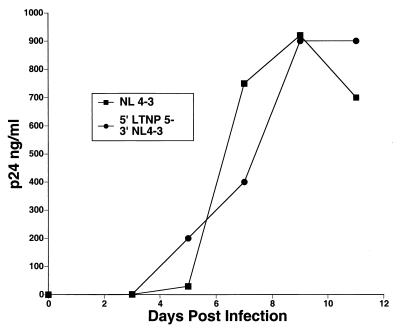
Of the seven LTNP HIV-1 isolates that were successfully cultured, six grew well in infected PBMC cultures (Table 3). Only LTNP 5 HIV-1 grew poorly (Table 3), reaching a maximum p24 concentration of 5 ng/ml in PBMC obtained from several seronegative donors (17). We investigated potential determinants of this slow- or low-growth phenotype. Chimeric LTNP 5/NL 4-3 HIV-1 was created to assess the effect of LTR, Gag, and Pol sequences on LTNP 5 replication. LTNP 5 and NL 4-3 sequences from bp 297 to 4402 (5′) and NL 4-3 sequences from bp 4402 to 9607 (3′) (Fig. 3) were amplified separately. An XmaI (CCCGGG) site was introduced at the junction of the PCR fragments (bp 4402) as described in Materials and Methods. XmaI-digested 5′ LTNP 5 and 5′ NL 4-3 fragments were cotransfected with 3′ NL 4-3 fragments into permissive CEMx174 cells. Supernatants were collected starting at day 3 posttransfection and assayed for p27 antigen concentration. Production of both wild-type NL 4-3 and recombinant 5′ LTNP 5–3′ NL 4-3 peaked at day 9 posttransfection with similar peak yields (Fig. 8). Thus, the 5′ sequences in LTNP 5 to bp 4402 seem suitably competent for viral replication.
FIG. 8.
Growth of recombinant HIV-1 containing 5′ LTNP 5 sequences. LTNP 5 and NL 4-3 sequences from bp 297 to 4402 (5′) and NL 4-3 sequences from bp 4402 to 9607 (3′) were amplified separately. An XmaI (CCCGGG) site was introduced at the junction of the PCR fragments (bp 4402) as described in Materials and Methods. XmaI-digested 5′ LTNP 5 and 5′ NL 4-3 fragments were cotransfected with 3′ NL 4-3 fragments into permissive CEMx174 cells. Supernatants were collected and assayed for p27 antigen concentration.
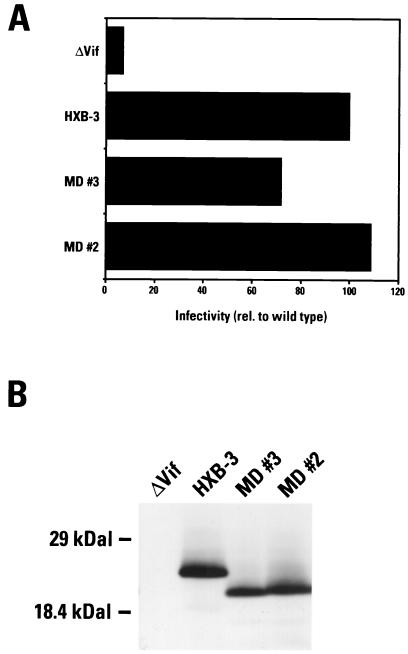
Previous studies have shown that vpu, vpr, and nef can be deleted from HIV-1 with little or no effect on replication in lectin-stimulated PBMC cultures (16, 26). Therefore, vpu, vpr, and nef are unlikely to be responsible for the poor growth of HIV-1 from LTNP 5. Among additional LTNP 5 sequences contained in the 3′ LTNP 5 sequences that could contribute to slow growth, no unusual, difficult-to-revert mutations were observed in integrase, vif, tat, rev, and U3 LTR sequences (Fig. 3 and Table 3). Among these sequences, LTNP 5 vif sequences displayed the greatest divergence from the subgroup B consensus as well as from the LTNP vif sequences presented here (Fig. 1D). LTNP 5 vif allele 2 and LTNP 5 vif allele sequences as well as HBX-3 vif-positive control sequences were inserted into an expression vector and were cotransfected with a vif-deficient proviral vector pIIIB/Δvif into H9 cells. Western blots of whole-cell lysates from transfected H9 cultures indicated that the vectors containing LTNP 5 vif alleles 2 and 3 and HBX-3 vif sequences expressed comparable levels of Vif (Fig. 10B). HIV-1 produced from these cells was used in single-cycle challenges of the indicator cell line C8166/HIV-CAT to assess Vif function. Cells transfected with LTNP 5 vif alleles 2 and 3 and HXB-3 vif produced similar levels of CAT (Fig. 10A) in this assay, indicative of the wild-type-like activity of these LTNP 5 vif genes.
FIG. 10.
Vif functional assay. (A) LTNP 5 (MD) vif sequences 2 and 3 as well as HXB-3 vif were cotransfected into H9 cells with a vif-deficient proviral vector. Harvested viruses were used to challenge the indicator cell line C8166/HIV-CAT. Levels of CAT activity indicate levels of Vif-mediated infectivity. These data represent the means of four independent experiments. (B) Whole-cell lysates of transfected H9 cells were separated by SDS-PAGE and electroblotted onto a membrane filter. HIV-1 Vif was detected using a Vif-specific monoclonal antibody.
Our analysis of LTNP 5 isolate sequences revealed a difficult-to-revert polymorphism in LTNP 5 env sequences (Fig. 3E and Table 3) that was also contained in the 3′ LTNP 5 PCR fragments. A single amino acid deletion in the fusion peptide region of the transmembrane domain (Fig. 3E and Table 3) was consistently retained in LTNP 5 virus isolates and in PBMC samples obtained in 1987 and 1994. This polymorphism is not observed in any of the 131 group M, group O, or SIVcpz Env sequences in the Los Alamos database (B. Korberg, http: //hiv-web.lanl.gov) and represents the only deletion or insertion contained in LTNP 5 HIV-1 sequences (Fig. 3 and Table 3).
DISCUSSION
HIV-1 gene sequences from LTNPs have been investigated extensively in order to determine if inactivating polymorphisms are associated with nonprogression. Nucleotide and amino acid substitutions have been associated with defective p17 and Rev activities (5, 25). Among the auxiliary genes, several examples of large deletions in nef have been documented (10, 27) but have not been observed in LTNP vpu or vpr sequences. Thus, the types of polymorphisms described here are unusual in that they are difficult to revert and in many cases without precedent in the Los Alamos HIV-1 sequence data base (B. Korber, http://hiv-web.lanl.gov).
Seven of the eight LTNPs contained a total of 13 unusual, difficult-to-revert polymorphisms in HIV-1 sequences. The rate of misincorporation during the process of reverse transcription has been estimated at approximately 3 × 10−4 bases or approximately one misincorporation per genome transcribed (4, 40, 46). Amino acid polymorphisms that are detrimental to the virus rapidly revert (26, 32). Conversely, the elimination of insertion or deletion polymorphisms requires slippage or stuttering of reverse transcriptase, which is a much rarer occurrence. A 4-aa deletion in SIV Nef has been repaired by duplication of adjacent sequences and subsequent reversion to wild-type sequences (56). However, reversion of this deletion is rare and is not observed in most infected monkeys. In all cases presented here, with the exception of the 4-aa insertion in LTNP 6 Vpu, difficult-to-revert polymorphisms were consistently present in historical samples from the LTNPs.
Five of the eight LTNPs in this study had deletions of 1 or 2 aa in p17gag, p2gag, and/or p6gag (Fig. 3B and Table 3). Four had deletions that did not appear in 93 group M, group O, or SIVcpz isolates in the Los Alamos HIV-1 sequence database (B. Korber, http://hiv-web.lanl.gov). In contrast, we did not observe any difficult-to-revert polymorphisms in the Gag sequences isolated from four individuals that progressed to AIDS and we did not observe a single example of a unique deletion in Gag among 90 group M sequences in the Los Alamos database. These results suggest that the deletions in Gag that we observed occurred in a nonrandom fashion in our group of LTNPs. HIV-1 isolated from the LTNPs containing these polymorphisms was able to replicate in culture (Table 3) (17), indicating that they do not dramatically debilitate the virus. However, they could possibly have subtle effects on Gag function that may lower replicative capacity or result in attenuation. Three of the four unique Gag deletions occurred in p6 sequences (161J Δ483-484, LTNP 7 Δ466-467, and 1486D Δ465-466). Vpr incorporation into virions was not detectably affected by the p6 deletions (Fig. 6).
The Nef sequences from the LTNPs belonged to group M, subgroup B (Fig. 2), as would be expected since these individuals live and were likely infected in the United States where subgroup B sequences predominate. The polymorphism seen in Nef sequences isolated from LTNP 6 (Δ49-50) (Fig. 3J) is found in about 9% of subgroup B sequences although it represents the subgroup E consensus, which is predominant in Asia. Conversely, the Nef polymorphism (Δ48-49) observed in 161J and 1486D isolates (Fig. 3J) was not observed in any of the 256 Nef sequences in the Los Alamos HIV-1 sequence database (B. Korber, http://hiv-web.lanl.gov). Monkeys infected with SHIVnef that contained 1486D sequences from 1992 (Δ48-49) maintained low viral loads in contrast to monkeys infected with 1486D sequences from 1995 (Δ48-49 and 61Q insertion), which maintained persistently high viral loads (Fig. 7). These results are consistent with 1486D 1992 Nef sequences being less than optimal for Nef function and with the 61Q insertion resulting in a more functional Nef. However, these results are based on only four animal infections and are not statistically significant (2).
The extreme slow/low growth phenotype observed for HIV-1 isolates from LTNP 5 (17) appears to map to the env region. LTNP 5 Env contains a polymorphism in the fusion peptide region of gp41 which was not observed in any of the 131 isolates in the Los Alamos HIV-1 sequence database (B. Korber, http://hiv-web.lanl.gov). The fusion peptide of gp41 inserts into target cell membranes, which is critical to the fusion process between cellular and viral membranes (7). The LTNP 5 Env polymorphism could affect this or other Env functions, resulting in the observed slow/low growth phenotype. Sequences responsible for a slow/low growth phenotype in other isolates have previously been mapped to Env amino acid substitutions in the V3 and C2-V4 regions (9, 24, 42, 50, 55) and thus could easily revert. The polymorphism seen in LTNP 5 Env is not an amino acid substitution, is maintained through long-term passage in cell culture, and is not likely to revert easily to a more rapid/high phenotype.
The origins of the unusual, difficult-to-revert polymorphisms observed in seven of our eight patients cannot be definitively determined with the samples available. As was clearly the case with eight Australians infected with Nef-deleted HIV-1 from a single blood donor (33), it is possible that many of the polymorphisms in our seven subjects were present in the infecting inoculum. Since the noted polymorphisms would be difficult to revert, they could have long-term attenuating effects on the virus. Alternatively, the unusual polymorphisms could have arisen in the infected hosts in response to selective pressures. Two types of selective pressures can be envisioned: the host immune response and polymorphisms in host cell proteins that must partner with HIV-1 proteins. With respect to selective pressures from host immune responses, strong CTL responses have been measured in all eight of these LTNPs. These strong cellular responses could put significant pressure on the virus to escape immune surveillance. However, deletions and insertions within ordinarily conserved sequences would be an unusual way for the virus to avoid immune recognition and would not be obviously advantageous over simple point mutations. Furthermore, we did not observe any consistent pattern of MHC-I haplotypes associated with similarly located deletions in the HIV-1 sequence. MHC-II-restricted CD4 helper cell activity could also conceivably be a selective force for driving sequence changes in the virus. With respect to possible polymorphisms in host cell proteins that must partner HIV-1 proteins, it is curious to note that heterozygosity for the SDF-1-3′A polymorphism seemed to be associated with similarly located Gag and Nef deletions in 1486D, 161J, and LTNP 7.
For the LTNPs in this study, we did not observe homozygous polymorphisms in second-receptor or chemokine genes (Table 2) that have been previously documented to be associated with nonprogression (11, 23, 39, 52, 57). We have observed unusual, difficult-to-revert polymorphisms in HIV-1 sequences from LTNPs that were not observed in the evaluated sequences from individuals who progressed to AIDS. Furthermore, most of these unusual polymorphisms are without precedent among observed HIV-1 sequences. Although we have not directly demonstrated that these deletions have contributed to their rate of disease progression, they appear to be associated with the unusual clinical status of these individuals in an apparently nonrandom fashion. Additional studies using monkeys and SIV or SHIV recombinants with deletions analogous to those observed in the LTNPs will be required to document any contribution to lowering viral load or attenuation of disease progression.
FIG. 9.
Alignment of LTNP 5 Vif sequences. The product of LTNP 5 vif allele representing the consensus (allele 2) and the product of an allele representing the most divergent sequences from the group M, subgroup B, consensus (allele 3) were aligned to subgroup B consensus sequences (HIV-1). Each sequence represents a consensus of two clones from each of two independent PCRs. A period indicates conservation between subgroup B and LTNP 5 sequences.
ACKNOWLEDGMENTS
We thank J. Lifson for plasma RNA measurements, Susan Czajak for technical assistance, P. Sehgal and E. Roberts for animal care, blood sampling, and clinical care, L. Denekamp for technical assistance and manuscript proofreading, and J. Newton for manuscript preparation.
This study was supported by PHS grants AI25328, AI38559, AI28568, AI39400, HL42257, and RR00168.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander L, Du Z, Howe A Y M, Czajak S, Desrosiers R C. Induction of AIDS in rhesus monkeys by a recombinant simian immunodeficiency virus expressing nef of human immunodeficiency virus type 1. J Virol. 1999;73:5814–5825. doi: 10.1128/jvi.73.7.5814-5825.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes W M. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bebenek K, Abbotts J, Roberts J D, Wilson S H, Kunkel T A. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989;264:16948–16956. [PubMed] [Google Scholar]
- 5.Binley J M, Jin X, Huang Y, Zhang L, Cao Y, Ho D D, Moore J P. Persistent antibody responses but declining cytotoxic T-lymphocyte responses to multiple human immunodeficiency virus type 1 antigens in a long-term nonprogressing individual with a defective p17 proviral sequence and no detectable viral RNA expression. J Virol. 1998;72:3472–3474. doi: 10.1128/jvi.72.4.3472-3474.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrington M, Nelson G W, Martin M P, Kissner T, Vlahov D, Goedert J J, Kaslow R, Buchbinder S, Hoots K, O'Brien S J. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 7.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 8.Cheng S, Fockler C, Barnes W M, Higuchi R. Effective amplification of long targets from cloned inserts and human genomic DNA. Proc Natl Acad Sci USA. 1994;91:5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng-Mayer C, Shioda T, Levy J A. Host range, replicative, and cytopathic properties of human immunodeficiency virus type 1 are determined by very few amino acid changes in tat and gp120. J Virol. 1991;65:6931–6941. doi: 10.1128/jvi.65.12.6931-6941.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Ellett A, Chatfield C, Lawson V A, Crowe S, Maerz A, Sonza S, Learmont J, Sullivan J S, Cunningham A, Dwyer D, Downton D, Mills J. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 11.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O'Brien S J. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CCR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. . (Erratum, 274:5290.) [DOI] [PubMed] [Google Scholar]
- 12.Desrosiers R C, Lifson J D, Gibson J S, Czajak S C, Howe A Y, Arthur L O, Johnson R P. Identification of highly attenuated mutants of simian immunodeficiency virus. J Virol. 1998;72:1431–1437. doi: 10.1128/jvi.72.2.1431-1437.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fauci A S. Host factors and the pathogenesis of HIV-induced disease. Nature. 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 14.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D D, Richman D D, Siliciano R F. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 15.Gibbs J S, Lackner A A, Lang S M, Simon M A, Sehgal P K, Daniel M D, Desrosiers R C. Progression to AIDS in the absence of a gene for vpr or vpx. J Virol. 1995;69:2378–2383. doi: 10.1128/jvi.69.4.2378-2383.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibbs J S, Regier D A, Desrosiers R C. Construction and in vitro properties of HIV-1 mutants with deletions in “nonessential” genes. AIDS Res Hum Retroviruses. 1994;10:343–350. doi: 10.1089/aid.1994.10.343. [DOI] [PubMed] [Google Scholar]
- 17.Greenough T C, Brettler D B, Kirchhoff F, Alexander L, Desrosiers R C, O'Brien S J, Somasundaran M, Luzuriaga K, Sullivan J L. Long-term nonprogressive infection with human immunodeficiency virus type 1 in a hemophilia cohort. J Infect Dis. 1999;180:1790–1802. doi: 10.1086/315128. [DOI] [PubMed] [Google Scholar]
- 18.Greenough T C, Brettler D B, Somasundaran M, Panicali D L, Sullivan J L. Human immunodeficiency virus type 1-specific cytotoxic T lymphocytes (CTL), virus load, and CD4 T cell loss: evidence supporting a protective role for CTL in vivo. J Infect Dis. 1997;176:118–125. doi: 10.1086/514013. [DOI] [PubMed] [Google Scholar]
- 19.Greenough T C, Sullivan J L, Desrosiers R C. Declining CD4 T-cell counts in a person infected with nef-deleted HIV-1. N Engl J Med. 1999;340:236–237. doi: 10.1056/NEJM199901213400314. [DOI] [PubMed] [Google Scholar]
- 20.Hadida F, Parrot A, Kieny M P, Sadat-Sowti B, Mayaud C, Debre P, Autran B. Carboxyl-terminal and central regions of human immunodeficiency virus-1 NEF recognized by cytotoxic T lymphocytes from lymphoid organs. An in vitro limiting dilution analysis. J Clin Investig. 1992;89:53–60. doi: 10.1172/JCI115585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrer T, Harrer E, Kalams S A, Barbosa P, Trocha A, Johnson R P, Elbeik T, Feinberg M B, Buchbinder S P, Walker B D. Cytotoxic T lymphocytes in asymptomatic long-term nonprogressing HIV-1 infection. Breadth and specificity of the response and relation to in vivo viral quasispecies in a person with prolonged infection and low viral load. J Immunol. 1996;156:2616–2623. [PubMed] [Google Scholar]
- 22.Haynes B F, Pantaleo G, Fauci A S. Toward an understanding of the correlates of protective immunity to HIV infection. Science. 1996;271:324–328. doi: 10.1126/science.271.5247.324. [DOI] [PubMed] [Google Scholar]
- 23.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 24.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 25.Iversen A K N, Shpaer E G, Rodrigo A G, Hirsch M S, Walker B D, Sheppard H W, Merigan T C, Mullins J I. Persistence of attenuated rev genes in a human immunodeficiency virus type 1-infected asymptomatic individual. J Virol. 1995;69:5743–5753. doi: 10.1128/jvi.69.9.5743-5753.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kestler H W, III, Ringler D J, Mori K, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for the development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 27.Kirchhoff F, Greenough T C, Brettler D B, Sullivan J L, Desrosiers R C. Absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 28.Kirchhoff F, Greenough T C, Hamacher M, Sullivan J L, Desrosiers R C. Activity of human immunodeficiency virus type 1 promoter/TAR regions and tat1 genes derived from individuals with different rates of disease progression. Virology. 1997;232:319–331. doi: 10.1006/viro.1997.8586. [DOI] [PubMed] [Google Scholar]
- 29.Kondo E, Mammano F, Cohen E A, Gottlinger H G. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J Virol. 1995;69:2759–2764. doi: 10.1128/jvi.69.5.2759-2764.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kostrikis L G, Huang Y, Moore J P, Wolinsky S M, Zhang L, Guo Y, Deutsch L, Phair J, Neumann A U, Ho D D. A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nat Med. 1998;4:350–353. doi: 10.1038/nm0398-350. [DOI] [PubMed] [Google Scholar]
- 31.Koup R A, Pikora C A, Luzuriaga K, Brettler D B, Day E S, Mazzara G P, Sullivan J L. Limiting dilution analysis of cytotoxic T lymphocytes to human immunodeficiency virus gag antigens in infected persons: in vitro quantitation of effector cell populations with p17 and p24 specificities. J Exp Med. 1991;174:1593–1600. doi: 10.1084/jem.174.6.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lang S M, Weeger M, Stahl-Henning C, Coulibaly C, Hunsmann G, Müller J, Müller-Hermelink H, Fuchs D, Wachter H, Daniel M D, Desrosiers R C, Fleckenstein B. The importance of vpr for the infection of rhesus monkeys with simian immunodeficiency virus. J Virol. 1993;67:902–912. doi: 10.1128/jvi.67.2.902-912.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Learmont J C, Geczy A F, Mills J, Ashton L J, Raynes-Greenow C H, Garsia R J, Dyer W B, McIntyre L, Oelrichs R B, Rhodes D I, Deacon N J, Sullivan J S. Immunologic and virologic status after 14 to 18 years of infection with an attenuated strain of HIV-1. A report from the Sydney Blood Bank Cohort. N Engl J Med. 1999;340:1715–1722. doi: 10.1056/NEJM199906033402203. [DOI] [PubMed] [Google Scholar]
- 34.Lieberman J, Fabry J A, Fong D M, Parkerson G R., III Recognition of a small number of diverse epitopes dominates the cytotoxic T lymphocyte response to HIV type 1 in an infected individual. AIDS Res Hum Retroviruses. 1997;13:383–392. doi: 10.1089/aid.1997.13.383. [DOI] [PubMed] [Google Scholar]
- 35.Lifson J D, Nowak M A, Goldstein S, Rossio J L, Kinter A, Vasquez G, Wiltrout T A, Brown C, Schneider D, Wahl L, Lloyd A L, Williams J, Elkins W R, Fauci A S, Hirsch V M. The extent of early viral replication is a critical determinant of the natural history of simian immunodeficiency virus infection. J Virol. 1997;71:9508–9514. doi: 10.1128/jvi.71.12.9508-9514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 37.Lu Y L, Bennett R P, Wills J W, Gorelick R, Ratner L. A leucine triplet repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J Virol. 1995;69:6873–6879. doi: 10.1128/jvi.69.11.6873-6879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mariani R, Kirchhoff F, Greenough T C, Sullivan J L, Desrosiers R C, Skowronski J. High frequency of defective nef alleles in a long-term survivor with nonprogressive human immunodeficiency virus type 1 infection. J Virol. 1996;70:7752–7764. doi: 10.1128/jvi.70.11.7752-7764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin M P, Dean M, Smith M W, Winkler C, Gerrard B, Michael N L, Lee B, Doms R W, Margolick J, Buchbinder S, Goedert J J, O'Brien T R, Hilgartner M W, Vlahov D, O'Brien S J, Carrington M. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science. 1998;282:1907–1911. doi: 10.1126/science.282.5395.1907. [DOI] [PubMed] [Google Scholar]
- 40.Mendelman L V, Boosalis M S, Petruska J, Goodman M F. Nearest neighbor influences on DNA polymerase insertion fidelity. J Biol Chem. 1989;264:14415–14423. [PubMed] [Google Scholar]
- 41.Naidu Y M, Kestler III H W, Li Y, Butler C V, Silva D P, Schmidt D K, Troup C D, Sehgal P K, Sonigo P, Daniel M D, Desrosiers R C. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J Virol. 1988;62:4691–4696. doi: 10.1128/jvi.62.12.4691-4696.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Brien W A, Koyanagi Y, Namazie A, Zhao J Q, Diagne A, Idler K, Zack J A, Chen I S. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 43.Oelrichs R, Tsykin A, Rhodes D, Solomon A, Ellett A, McPhee D, Deacon N. Genomic sequence of HIV type 1 from four members of the Sydney Blood Bank Cohort of long-term nonprogressors. AIDS Res Hum Retroviruses. 1998;14:811–814. doi: 10.1089/aid.1998.14.811. [DOI] [PubMed] [Google Scholar]
- 44.Pal A, Greenough T C, Sullivan J L, Somasundaran M. In vitro characterization of adult primary human immunodeficiency virus type 1: demonstration of distinctive single-cell killing phenotypes in spite of similar levels of viral replication. J Infect Dis. 1997;176:933–940. doi: 10.1086/516537. [DOI] [PubMed] [Google Scholar]
- 45.Regier D A, Desrosiers R C. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res Hum Retroviruses. 1990;6:1221–1231. doi: 10.1089/aid.1990.6.1221. [DOI] [PubMed] [Google Scholar]
- 46.Ricchetti M, Buc H. Reverse transcriptases and genomic variability: the accuracy of DNA replication is enzyme specific and sequence dependent. EMBO J. 1990;9:1583–1593. doi: 10.1002/j.1460-2075.1990.tb08278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenberg E S, Billingsley J M, Caliendo A M, Boswell S L, Sax P E, Kalams S A, Walker B D. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science. 1997;278:1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- 48.Salvi R, Garbuglia A R, Di Caro A, Pulciani S, Montella F, Benedetto A. Grossly defective nef gene sequences in a human immunodeficiency virus type 1-seropositive long-term nonprogressor. J Virol. 1998;72:3646–3657. doi: 10.1128/jvi.72.5.3646-3657.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 50.Shioda T, Levy J A, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 51.Simon J H, Miller D L, Fouchier R A, Soares M A, Peden K W, Malim M H. The regulation of primate immunodeficiency virus infectivity by Vif is cell species restricted: a role for Vif in determining virus host range and cross-species transmission. EMBO J. 1998;17:1259–1267. doi: 10.1093/emboj/17.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith M W, Dean M, Carrington M, Winkler C, Huttley G A, Lomb D A, Goedert J J, O'Brien T R, Jacobson L P, Kaslow R, Buchbinder S, Vittinghoff E, Vlahov D, Hoots K, Hilgartner M W, O'Brien S J. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997;277:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 53.Suryanarayana K, Wiltrout T A, Vasquez G M, Hirsch V M, Lifson J D. Plasma SIV RNA viral load by determination by real-time quantification of product generation in reverse transcriptase-polymerase chain reaction. AIDS Res Hum Retroviruses. 1998;14:183–189. doi: 10.1089/aid.1998.14.183. [DOI] [PubMed] [Google Scholar]
- 54.Wang B, Ge Y C, Palasanthiran P, Xiang S H, Ziegler J, Dwyer D E, Randle C, Dowton D, Cunningham A, Saksena N K. Gene defects clustered at the C-terminus of the vpr gene of HIV-1 in long-term nonprogressing mother and child pair: in vivo evolution of vpr quasispecies in blood and plasma. Virology. 1996;223:224–232. doi: 10.1006/viro.1996.0471. [DOI] [PubMed] [Google Scholar]
- 55.Westervelt P, Trowbridge D B, Epstein L G, Blumberg B M, Li Y, Hahn B H, Shaw G M, Price R W, Ratner L. Macrophage tropism determinants of human immunodeficiency virus type 1 in vivo. J Virol. 1992;66:2577–2582. doi: 10.1128/jvi.66.4.2577-2582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whatmore A M, Cook N, Hall G A, Sharpe S, Rud E W, Cranage M P. Repair and evolution of nef in vivo modulates simian immunodeficiency virus virulence. J Virol. 1995;69:5117–5123. doi: 10.1128/jvi.69.8.5117-5123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winkler C, Modi W, Smith M W, Nelson G W, Wu X, Carrington M, Dean M, Honjo T, Tashiro K, Yabe D, Buchbinder S, Vittinghoff E, Goedert J J, O'Brien T R, Jacobson L P, Detels R, Donfield S, Willoughby A, Gomperts E, Vlahov D, Phair J, O'Brien S J. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC) Science. 1998;279:389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 58.Zimmermann K, Pischinger K, Mannhalter J W. Nested primer PCR detection limits of HIV-1 in the background of increasing numbers of lysed cells. BioTechniques. 1994;17:18–20. [PubMed] [Google Scholar]