Abstract
Adamantinoma-like Ewing Sarcoma (ALES) has traditionally been considered a variant of Ewing Sarcoma since it generally harbors EWSR1::FLI1 fusions despite diffuse positivity for keratins and p40. However, it has become increasingly recognized that different tumors can have identical translocations, including shared fusions between carcinomas and sarcomas, raising questions as to whether ALES might represent a separate entity. Using methylation profiling, we further explore the relationship between Ewing sarcoma and ALES. The archives of multiple institutions were searched for candidate cases of ALES. DNA methylation profiling was performed and results were compared to corresponding data from conventional Ewing sarcoma. 12 cases of ALES (5 previously reported) were identified arising in 10 males and 2 females (20 to 72 years of age, median 41.5). Cases included tumors arising in the parotid gland (3), sinonasal cavity (2), submandibular gland (2), thyroid gland (1), neck (1), gingiva (1), hypopharynx (1) and mandible (1). Histologic review consistently showed sheets and nests of basaloid cells within a fibromyxoid or hyalinized stroma. All tumors were positive for at least one keratin and CD99 expression, while all 10 cases tested were positive for p63 or p40; S100 protein expression was noted in 2 cases. Cases harbored either EWSR1::FLI1 fusions (n=6), FUS::FLI1 fusions (n=1) and/or EWSR1 rearrangements (n=6). Methylation profiling was successful in 11/12 cases evaluated. Unsupervised clustering and dimensionality reduction (UMAP) of DNA methylation data revealed a distinct methylation cluster for all 11 cases, including the tumor with the FUS::FLI1 fusion, which clearly segregated from conventional Ewing sarcoma. Follow-up (n=11, 1–154 months) revealed that 4 patients experienced recurrence and 6 developed metastatic disease. ALES demonstrates a distinct methylation signature from conventional Ewing sarcoma. This finding adds to the distinctive immunoprofile of ALES, suggesting that these two tumors should be considered distinct entities rather than histologic extremes of the same disease.
Keywords: Ewing, sarcoma, adamantinoma, EWSR1
Introduction
Adamantinoma-like Ewing sarcoma (ALES) is currently considered a histologic variant of Ewing sarcoma in the current editions of the Soft Tissue and Bone as well as the Head and Neck WHO Classification scheme.(1, 2) It is defined by the presence of squamous differentiation including diffuse keratin expression, positivity for squamous markers p63 and p40, and even occasional keratin pearl formation. Although initially described as arising in the long bones by Bridge in 1999, ALES is now recognized to most commonly occur in the head and neck region (3). Over the past several years as more cases have been described, it has become increasingly clear that ALES arising in the head and neck is histologically and immunophenotypically distinct from conventional Ewing sarcoma even though they harbor the same EWSR1/FUS::FLI1 gene fusion. Historically, the presence of this gene fusion has been regarded as pathognomonic for Ewing sarcoma, leading to initial classification of ALES within this category. However, as many other fusions have been recognized to occur across multiple diverse tumor types and even lineages, the dictum that any specific fusion can only occur in a single tumor has become less convincing. Indeed, controversy has recently emerged regarding the classification of ALES as a subtype of Ewing sarcoma at all, with some observers regarding them as a distinctive category of carcinoma. Using methylation profiling, we sought to explore further the relationship between Ewing sarcoma and head and neck ALES.
Materials and Methods
Patient Cohort and Data Collection
This study was approved by the Institutional Review Boards at all participating institutions. Cases of ALES arising in the head and neck and confirmed by either EWSR1 rearrangement and/or EWSR1/FUS::FLI1 gene fusion were collected. A case of high-grade carcinoma with features of intrathyroidal thymic carcinoma (previously known as carcinoma showing thymic-like differentiation, or CASTLE) was included in the analysis as well. This latter case lacked EWSR1/FUS rearrangement but strongly mimicked ALES morphologically. DNA methylation and copy number profiling were performed and results were compared to corresponding data from conventional Ewing sarcoma. Clinical and immunohistochemical data were documented.
Array-based methylome profiling
DNA methylation analysis of all ALES was performed on FFPE tissue with at least 40% of tumor content. Methylome profiling was carried out using the Infinium Human Epic Array (850k) platform (Illumina) according to the manufacturer’s instructions. For comparison, the methylome profiles of 15 adamantinomas of the tibia, 55 (head and neck) squamous cell carcinomas, and 74 Ewing sarcomas were collected (including publicly available datasets) and investigated.(4) (5)
Methylation array process
Raw data were processed as described earlier.(6) Briefly, raw intensity data files (IDATs) from the Methylation Epic (=850k) BeadChips were processed with the R-package ‘minfi’. Epic arrays were converted to a virtual 450K array for joint normalization and processing of data from both platforms. Probes associated with known SNPs, non-CpGs and sex chromosomes were not taken into account for the evaluation. Moreover, samples with a mean detection p-value >0.05 were discarded. The ‘preprocessQuantile’ function was used before generating the dimension reduction visualization whereas the ‘preprocessIllumina’ was preferred before deriving copy number profiles. Finally, batch effect corrections were applied to the beta-values in order to remove any bias related to the sample type (FFPE / KRYO), the array type (450K / 850K) or the protocol (use or not of the FFPE restoration kit) using the R-package ‘ChAMP’.
Unsupervised methylation-based clustering
The set of probes was then restricted to the top 25’000 most differentially methylated (based on the standard deviation) determine on 15’500 reference datasets mostly derived from the cancer genome atlas (TCGA) and gene expression omnibus (GEO) as previously described.(7) Uniform manifold approximation and projection (UMAP) was performed on the results of a principal component analysis (40 PCs) calculated via the singular value decomposition of the beta methylation matrix. The R-package ‘uwot’ used for generating the graph can be found at (https://github.com/jlmelville/uwot).
Differential methylation analysis
Raw data were preprocessed as described above although the dataset was restricted to ALES and Ewing sarcomas evaluated by 850K arrays and for which the mean detection p-value was below 1%. The R-package ‘DMRcate’ was loaded and the function ‘cpg.annotate’ was used on the M-values matrix with the default parameters. Differentially methylated regions were called using the function ‘dmrcate’ for a minimum of five differentially methylated CpGs with a maximal distance between two CpGs of 500 base pairs. Finally, each region was labelled using the R-package ‘annotatr’ in order to identify the proximal promoters of protein-coding genes.
Results
Clinical cohort
12 cases of ALES (5 previously reported) were identified arising in 10 males and 2 females (20 to 72 years of age, median 41.5). (8–10) Cases included tumors arising in the parotid gland (3), sinonasal tract (2), submandibular gland (2), thyroid gland (1), neck (1), gingiva (1), hypopharynx, and mandible (1) (Table 1).
Table 1.
Clinicopathologic features of case of ALES
| Site | Age/Sex | Immunoprofile | Genetics | Treatment for primary | Additional treatment | Outcome | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cytokeratins | p63/p40 | S100 | CD99 | FEV::ETS | EWSR1/FUS rearrangement | Recurrence (mo) | Metastasis (site, mo) | Status | Length of follow-up (mo) | |||||
| 1* | Nasal cavity | 51/F | AE1/3+ CAM5.2+ CK5/6+ |
+/ | + | + | EWSR1::FLI1 | chemotherapy (VDC/IE) and radiation at diagnosis | Recurrence treated with surgery and chemotherapy with temozolamide/iriniotecan |
Y (138) | N | AWOD | 154 | |
| 2 | Neck | 59/M | pankeratin+ | /+ | EWSR1+ | N | N | AWOD | 61 | |||||
| 3 | Lung metastasis from submandibular primary | 26/M | AE1/3+ CAM5.2+ |
/ | - | + | EWSR1::FLI1 | Resection (regional lymph nodes) | VATS resection for lung metastasis and chemotherapy (VDC/IE x 17 cycles) | N | Y (lung, 4) | AWD | 40 | |
| 4* | Parotid | 72/M | AE1/3+ | /+ | - | + | EWSR1+ | Surgery, additional treatment pending | N | N | AWOD | 1 | ||
| 5 | Thyroid gland | 23/M | AE1/3+ | /+ | - | + | EWSR1+ | Surgery, radiation, and chemotherapy (VDC/IE) | Chemo after recurrence (irenotecan, temozolamide and vincristine) | N | Y (spine, 0; femur, 12) | Dead of complications of chemotherapy (Klebsiella pneumoniae pneumonia, sepsis, and ARDS) (15) | 15 | |
| 6* | Sinonasal | 37/F | pankeratin+ | /+ | + | + |
EWSR1+
FLI1+ |
Surgery | Recurrence with surgery, radiation, chemotherapy (docetaxel, carboplatin, capecitabine, methotrexate) | Y (24) | Y (dura, 46) | DWD (52) | 52 | |
| 7 | Gingiva | 20/M | CK5/6+ | +/ | - | weak | FUS::FLI1 | Mandibulectomy with neck dissection followed by concurrent radiation and chemotherapy (VDC/IE) | Metastasis treated with radiation | N | Y (scapula, 10) | AWOD | 16 | |
| 8* | Parotid | 32/M | AE1/3+ | /+ | - | + | EWSR1::FLI1 | EWSR1+ | surgery, radiation and chemotherapy (VDC/IE) | N | N | AWOD | 72 | |
| 9* | Parotid | 46/M | AE1/3+ | /+ | - | + | EWSR1+ | surgery, radiation, VDC/IE | N | N | AWOD | 58 | ||
| 10 | Submandibular gland | 69/M | AE1/3+ CAM5.2+ CK5/6+ |
/ | - | weak | EWSR1::FLI1 | Submandibular gland excision with neck dissection | Recurrence treated with chemotherapy (doxorubicin, ifosfamide and vincristine with mesna and growth factor support) and radiation | Y (3) | Y (lung, 17) | AWD | 17 | |
| 11 | Neck recurrence from mandible primary | 24/M | CK5/6+ 34BE12+ CK7- CK20- |
+/ | - | + | EWSR1::FLI1 | Radiation for mandibular primary | Resection and radiation for neck recurrence; Adjuvant cisplatin and proton for 2nd neck recurrence; After lung metastasis VAC ifosfamide/etoposide and Nivolumab | Y (22) | Y (regional nodes, 36; lung 61) | DOD (63) | 63 | |
| 12 | Hypopharynx | 61/M | AE1/3+ | /+ | - | + | EWSR1:FLI1 | Recent diagnosis | Recent diagnosis | Recent diagnosis | Recent diagnosis | 0 | ||
, previously reported; NP, not performed; +, positive; -, negative; chemo, chemotherapy; AWOD, alive without disease; NED, no evidence of disease; DOC, died of complications from chemotherapy; DWD, died with disease; DOD, died of disease; N/A, not available
Pathologic features
Histologic examination of all cases revealed sheet-like growth of cohesive epithelioid to ovoid-shaped cells arranged in variably sized cords, nests, and islands embedded in a desmoplastic stroma (Figure 1A). Peripheral palisading was prominent imparting a basaloid appearance to the tumors (Figure 1B). High power review showed a monomorphic population of cells with uniform nuclei and variably prominent nucleoli (Figure 1C). The gingival primary contained cells with relatively abundant pale pink cytoplasm (Figure 2A), while the tumor cells in the remaining cases had high N:C ratios and scant eosinophilic cytoplasm. A review of the post-chemotherapy specimen revealed prominent neuronal maturation (Figure 2B). There were no other significant morphologic differences in our cohort. Mitotic figures and apoptotic cells were common. Comedo-type necrosis within the tumor nests was present in a subset of cases. One case showed single cells with abundant intensely eosinophilic cytoplasm (Figure 2C), suggestive of early keratinization (Case 4), and one case (Case 1) had deposition of brightly eosinophilic matrix (Figure 2D). One case showed focal keratin pearl formation, but no other evidence of keratinization was observed across the cohort. No dysplasia or in situ carcinoma was appreciated in cases with evaluable epithelium.
Figure 1.
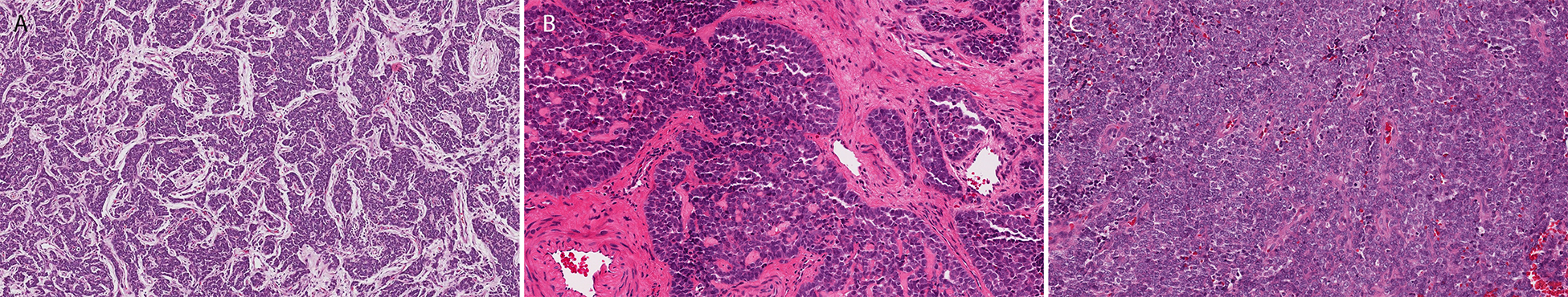
Histologic sections of ALES showed relatively uniform features including a nested architecture (A), desmoplastic stroma (B), and peripheral palisading (D).
Figure 2.
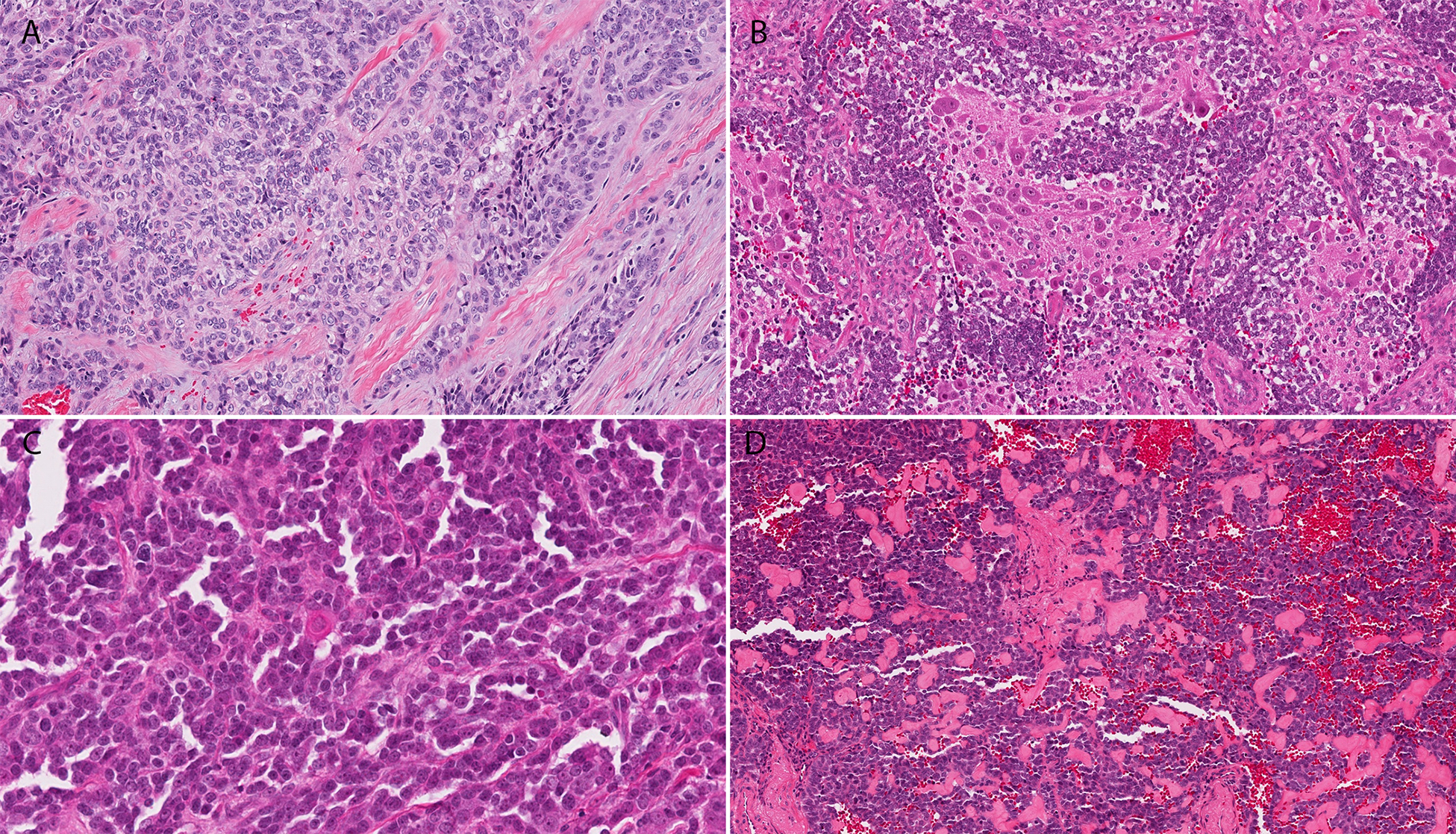
The cytoplasm of the gingival ALES was more abundant and pale pink (A), while the case which was reviewed post-chemotherapy showed large ganglion-like cells consistent with neuronal maturation (B). Case 4 showed single cell keratinization (C), while case 1 contained deposition of eosinophilic matrix between tumor nests (D).
Immunohistochemistry
All tumors were positive for at least one keratin, with specific expression of high molecular weight keratin CK5/6 in all 4 cases tested. Three (of 3) cases were positive for p63 and 7 (of 7) were positive for p40 (Figure 3A-B). Of the two cases not tested for either p63 or p40, one was positive for CK5/6 and one represented a metastasis of a known submandibular primary. S100 protein expression was noted in 2 cases (Table 1). Membranous CD99 expression was present in all cases tested (11/11) (Figure 3C).
Figure 3.
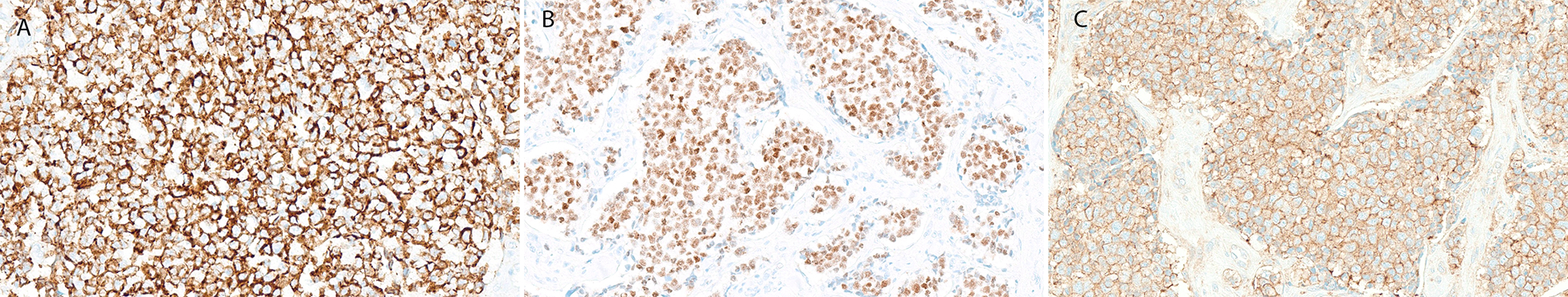
Cases of ALES in our cohort had uniform immumoprofiles with consistent expression of high molecular weight cytokeratin (A), p40 (C), and membranous CD99 (D).
Molecular genetic findings
Cases harbored either EWSR1::FLI1 fusion (n=6), FUS::FLI1 fusion (n=1) and/or EWSR1 rearrangement (n=6) (Table 1). One case with EWSR1 rearrangement also had FLI1 rearrangement. RNA sequencing did not detect any fusions in the case of high-grade carcinoma with thymic-like features.
Epigenetic findings
All samples (n=12) were subjected to DNA methylation profiling, yielding interpretable results in 11 cases. Using an unsupervised clustering approach (UMAP, Uniform Manifold Approximation and Projection), the methylomes of ALES were compared to the methylome profiles of conventional Ewing sarcomas and adamantinomas of the tibia. Furthermore, a series of tumors known to harbor gene-fusions involving EWSR1 was also used for comparison, including publicly available reference sets.(4, 5) As observed in Figure 4, methylation-based clustering allowed to clearly distinguish ALES samples from all other tumor subtypes included in our study. Although morphologically suggestive of ALES, the high-grade carcinoma with features of CASTLE clustered with conventional head and neck squamous cell carcinomas (Figure 4). Two of the most differentially methylated promoters between ALES and Ewing sarcoma are the FLI1 promoter (mean difference of methylation = 0.30; adjusted p-value = 1.64 × 1015) and the DDR2 promoter (mean difference of methylation = 0.42; Adjusted p-value = 8.67 × 1062) (Figure 5). Six of the eleven cases of ALES had interpretable copy number profiles. The highly variable amount of rearrangements observed in Ewing sarcoma were also found in ALES. A subset showed flat copy number profiles whereas numerous aberrations were observed in others (Supplementary Figure 1).
Figure 4.
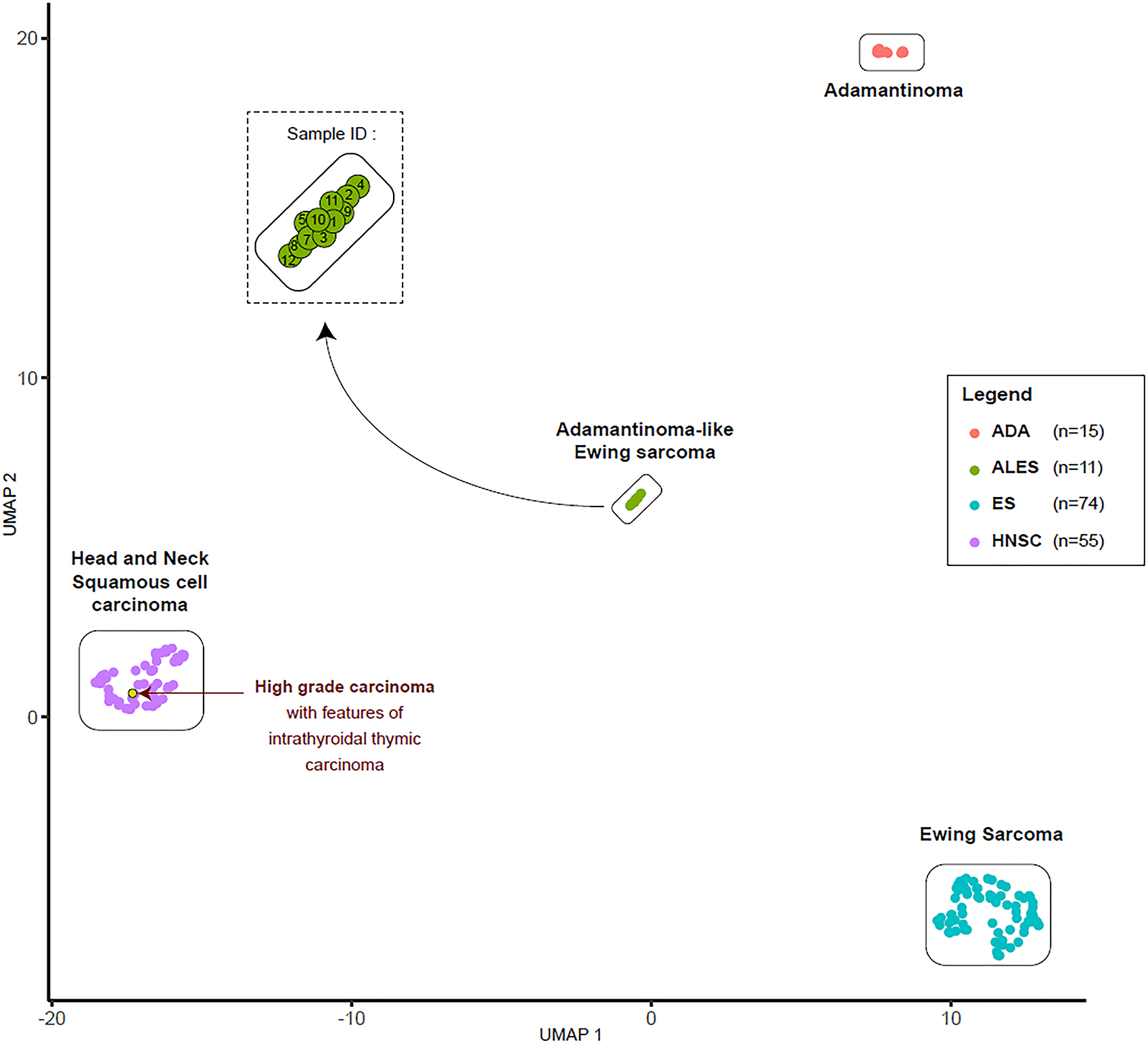
DNA methylation-based clustering of ALES and potential differential diagnoses, after non-linear dimensionality reduction (UMAP).
Figure 5.
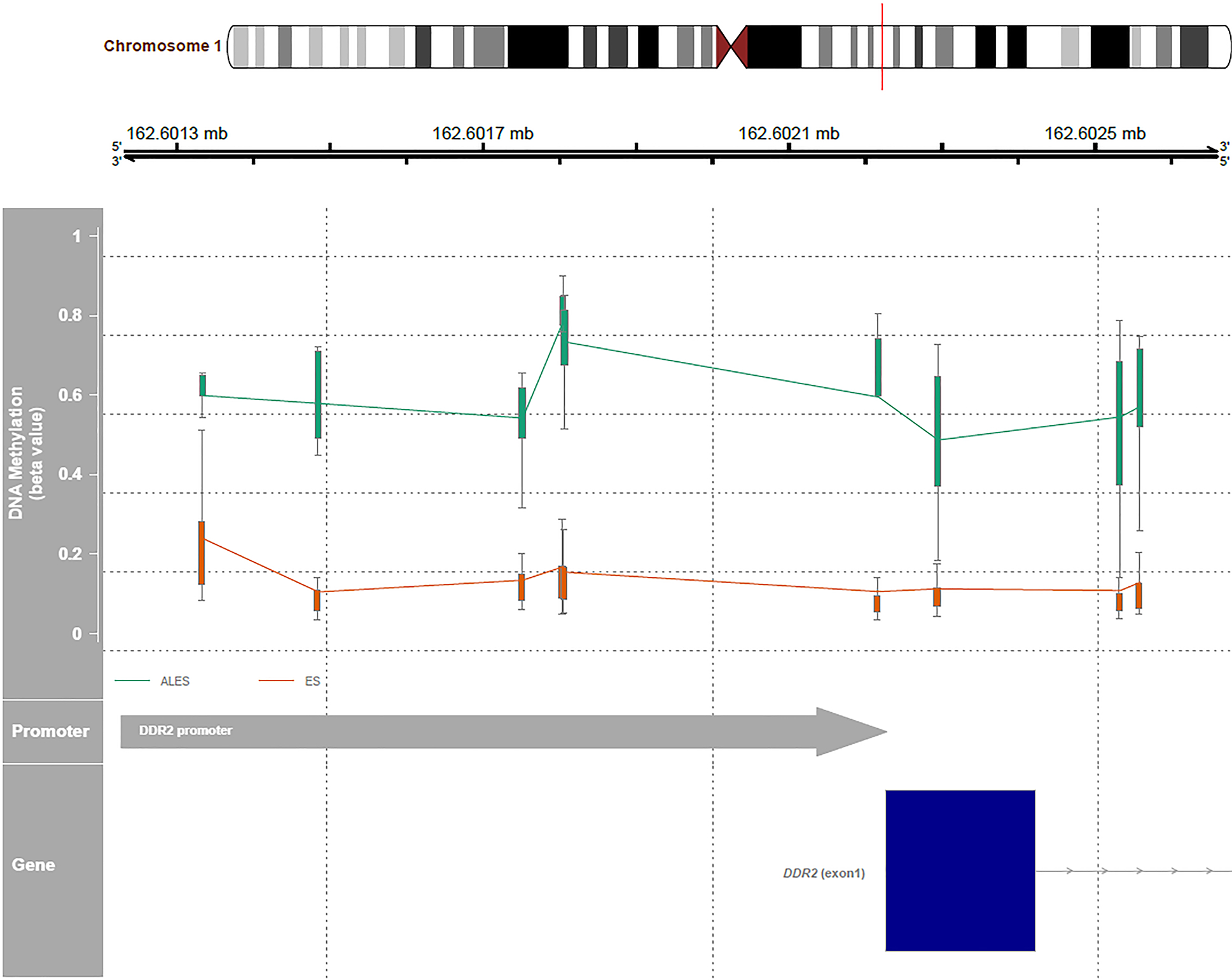
Comparison of methylation of the promoter of the DDR2 gene between ALES and conventional Ewing sarcoma.
Follow-up
Follow-up available on 11 patients (1–154 months, median 52 months) revealed that 4 patients experienced recurrence (at intervals of 3 to 138 months) and 6 patients developed metastatic disease to site including to lung, bone, dura and regional lymph nodes (at intervals of 0 to 46 months) (Table 1). At last follow-up (n=11), 6 patients were alive without disease, 2 were alive with disease, and 1 each died with disease, died of disease, and died due to complications from chemotherapy.
Treatment data were available in 9 patients in our cohort (Table 1). (8–10) 5 patients received a Ewing sarcoma chemotherapy regimen which consisted of vincristine, doxorubicin, and cyclophosphamide alternating with ifosfamide and etoposide (VDC/IE) at the time at diagnosis. 4 patients are alive without disease (15 to 154 months); the fifth died from complications of chemotherapy while undergoing treatment for recurrence. 2 of the 4 patients who received either surgery or radiation only at the time of diagnosis died with disease or of disease, respectively. A third received VDC/IE at the time of lung metastasis and is currently alive with disease.
Discussion
In 2008, Weinreb and colleagues described an extraskeletal malignancy with epithelial differentiation and EWSR::FL11 fusion arising in the lateral neck.(11) In their description, the authors noted that the cytology of the tumor cells was akin to an ‘atypical’ Ewing sarcoma, and areas with dyskeratotic cells and squamous perils were identified. In this publication it was not entirely clear if this lesion was identical to the ‘adamantinoma-like Ewing sarcoma’ of long bones first recognized in the 1970s and later genetically characterized by Bridge in 1999.(3, 12–15) Over the past several years, increasing numbers of similar cases have been reported in the head and neck literature using the “ALES” terminology. Concordantly, there has been growing skepticism whether this entity is truly a subtype of conventional Ewing sarcoma given its predominance at head and neck sites, epithelial differentiation, and consistent expression of high molecular weight keratins coupled with the recognition that gene fusions are not specific to a single diagnostic entity. In fact, there have been two thyroid tumors with features of ALES harboring EWSR1::FLI1 fusions labeled as ‘Carcinomas of the thyroid with Ewing Family Tumor Elements.’(16, 17) In this study, we sought to study a cohort of molecularly confirmed ALES of the head and neck to better understand their relationship to Ewing sarcoma.
Morphologic examination of our cohort of ALES revealed relatively uniform histologic features distinct from Ewing sarcoma. In one of the earliest reports of ALES by Meister and colleagues, “peripheral grouping of cells” was recognized, and this feature seems to be one of this entity’s most characteristic morphologic findings. (12) Tumors in our series consistently showed peripheral palisading rimming variably sized tumor nests. Although the tumor population exhibits the monotony of Ewing sarcoma, the neoplastic cells in our cases appeared to be more ovoid/epithelioid than round and showed vague nuclear streaming. Only two cases in our series showed any keratin production as manifested by single cell keratinization or rare keratin pearls, but more diffuse squamous nests and eddies have been well described in cases of ALES. This finding is not appreciated in conventional Ewing sarcoma.(9, 18, 19) Finally, striking desmoplasia and deposition of eosinophilic basement membrane-like material, unusual for Ewing sarcoma, was noted in our cases. Conversely, one novel point of overlap between these tumors was the neuronal maturation seen in a single post-chemotherapy ALES specimen- a finding that has previously been reported in Ewing sarcoma.(20, 21)
The immunoprofile in our series was consistent, showing robust expression of pankeratin, including high molecular weight keratin, p63 and/or p40 expression, and membranous CD99. Although keratin expression has been well-documented in a subset of Ewing sarcoma, diffuse expression of high molecular weight keratin seems to be restricted to the ‘adamantinoma-like’ variant.(22) Likewise, p63 and p40 staining is unusual in sarcomas, and when present in Ewing sarcoma, the staining pattern is focal or weak.(23–25) In the keratin-positive content of ALES, the presence of p63 and p40 expression supports true squamous differentiation.
Methylome profiling has recently been shown to correlate with cellular differentiation and to reliably distinguish tumor types with overlapping histologic features in the brain. Similar findings have been described in selected soft tissue and bone tumors, suggesting this new approach to be particularly helpful in better defining the relationship between tumors including those with different histotypes but shared genetics.(26, 27) Unsupervised clustering and dimensionality reduction (UMAP) of DNA methylation data of our ALES cohort revealed a distinct methylation cluster for all 10 cases with informative data, including the tumor with FUS::FLI1 fusion and that with neuronal maturation post-therapy. These cases clearly segregated from conventional Ewing sarcoma. Notably, a case diagnosed as high-grade carcinoma with thymic-like features clustered separately from the ALES cohort. Since all ALES cases formed a distinct cluster, methylome profiling could serve as an alternative for molecular confirmation in cases where RNA fusion testing fails. The current Heidelberg classifier does not contain data on ALES cases; for that reason, an unsupervised methylation based clustering as used here would be superior to a supervised approach (Heidelberg) when investigating novel tumor entities. Since methylation profiling is not yet routinely used in the work-up of non-CNS tumors, increased turn-around time and lack of a robust methylation classifier may be prohibitive; however, as this methodology becomes more established, methylation studies may become more cost effective than current molecular work-ups.
When comparing the methylation of promoters between ALES and conventional Ewing sarcoma, the DDR2 promoter appeared to be one of the most differentially methylated genes. DDR2 (discoidin domain receptor tyrosine kinase 2) is a receptor tyrosine kinase that uses collagen as its ligand and plays a role in cell adhesion, proliferation and extracellular matrix remodeling.(28) Differential expression analysis and proteomic evaluation have yet to be performed, but such studies might help to clarify the role of methylation of this gene promoter.
Reports of ALES of long bones treated with traditional Ewing sarcoma neoadjuvant chemotherapy regimens have shown poor response with significant amounts of viable tumor remaining in the resection specimens.(3, 19, 29) Despite these findings, patients overall seem to fare relatively well, remaining free of disease, with the caveat that follow-up is limited.(19, 29) Similarly, patients with head and neck ALES appear to have better outcomes when treated with Ewing sarcoma regimens either at initial diagnosis or the time of recurrence/metastasis, even though data regarding histologic treatment effect is unavailable.(10, 30) Although there is some data that copy number profiles correlate with adverse behavior in Ewing sarcoma, copy number profiles did not differentiate ALES from Ewing sarcoma or predict behavior in our cohort.(31, 32) This may be a consequence of the limited number of interpretable copy number profiles in our cohort of ALES which prevented us from drawing a robust conclusion. Additional investigations would be required to assess the relevance of this approach for the distinction of these two tumor types.
In summary, ALES arising in the head and neck show distinct epigenetic features when compared to conventional Ewing sarcoma, complementing their unique morphologic and immunophenotypic profiles. These findings suggest that although ALES shares the same genetic alteration with Ewing sarcoma, it might be considered a separate entity possibly representing an unusual carcinoma rather than a subtype of the latter. It is still unclear whether ALES of the long bones and head and neck represent the same entity. Given limited treatment and follow-up data and increasing differences with Ewing sarcoma, additional studies are necessary to determine the best treatment protocol for these tumors when they arise in the head and neck.
Supplementary Material
Acknowledgements:
The authors have no acknowledgements to declare.
Funding:
A subset of this project was funded by the Anatomic Pathology Department at Cleveland Clinic. BA, VA and DB were supported by the Gertrude von Meissner-Stiftung, the Stiftung für Krebskranke Kinder beider Basel (SKKK), and the Basel Bone Tumor Reference Center Foundation. BX was supported by the Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30CA008748.
Footnotes
Conflict of interest: The authors have no relevant conflicts of interest to disclose.
Ethics approval/consent to participate: Approval for this study was granted by the Institutional Review Boards of all participating institutions.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data availability statement:
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
- 1.WHO Classification of Tumours Editorial Board. Soft tissue and bone tumours. 5th ed. Lyon (France): International Agency for Research on Cancer. [Google Scholar]
- 2.WHO Classification of Tumours Editorial Board. Head and neck tumours. 5th ed. Lyon (France): International Agency for Research on Cancer. [Google Scholar]
- 3.Bridge JA, Fidler ME, Neff JR, Degenhardt J, Wang M, Walker C, et al. Adamantinoma-like Ewing’s sarcoma: genomic confirmation, phenotypic drift. Am J Surg Pathol. 1999;23(2):159–65. [DOI] [PubMed] [Google Scholar]
- 4.Koelsche C, Schrimpf D, Stichel D, Sill M, Sahm F, Reuss DE, et al. Sarcoma classification by DNA methylation profiling. Nat Commun. 2021;12(1):498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyskjaer I, De Noon S, Tirabosco R, Rocha AM, Lindsay D, Amary F, et al. DNA methylation-based profiling of bone and soft tissue tumours: a validation study of the ‘DKFZ Sarcoma Classifier’. J Pathol Clin Res. 2021;7(4):350–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ameline B, Nathrath M, Nord KH, de Flon FH, Bovee J, Krieg AH, et al. Methylation and copy number profiling: emerging tools to differentiate osteoblastoma from malignant mimics? Mod Pathol. 2022;35(9):1204–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haefliger S, Tzankov A, Frank S, Bihl M, Vallejo A, Stebler J, et al. NUT midline carcinomas and their differentials by a single molecular profiling method: a new promising diagnostic strategy illustrated by a case report. Virchows Arch. 2021;478(5):1007–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M, Hoschar AP, Budd GT, Chao ST, Scharpf J. Primary Ewing’s sarcoma of the ethmoid sinus with intracranial and orbital extension: case report and literature review. Am J Otolaryngol. 2013;34(5):563–8. [DOI] [PubMed] [Google Scholar]
- 9.Bishop JA, Alaggio R, Zhang L, Seethala RR, Antonescu CR. Adamantinoma-like Ewing family tumors of the head and neck: a pitfall in the differential diagnosis of basaloid and myoepithelial carcinomas. Am J Surg Pathol. 2015;39(9):1267–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rooper LM, Jo VY, Antonescu CR, Nose V, Westra WH, Seethala RR, et al. Adamantinoma-like Ewing Sarcoma of the Salivary Glands: A Newly Recognized Mimicker of Basaloid Salivary Carcinomas. Am J Surg Pathol. 2019;43(2):187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinreb I, Goldstein D, Perez-Ordonez B. Primary extraskeletal Ewing family tumor with complex epithelial differentiation: a unique case arising in the lateral neck presenting with Horner syndrome. Am J Surg Pathol. 2008;32(11):1742–8. [DOI] [PubMed] [Google Scholar]
- 12.Meister P, Konrad E, Hubner G. Malignant tumor of humerus with features of “adamantinoma” and Ewing’s sarcoma. Pathol Res Pract. 1979;166(1):112–22. [DOI] [PubMed] [Google Scholar]
- 13.Lipper S, Kahn LB. Case report 235. Ewing-like adamantinoma of the left radial head and neck. Skeletal Radiol. 1983;10(1):61–6. [DOI] [PubMed] [Google Scholar]
- 14.Ishida T, Kikuchi F, Oka T, Machinami R, Kojima T, Iijima T, et al. Case report 727. Juxtacortical adamantinoma of humerus (simulating Ewing tumor). Skeletal Radiol. 1992;21(3):205–9. [DOI] [PubMed] [Google Scholar]
- 15.van Haelst UJ, de Haas van Dorsser AH. A perplexing malignant bone tumor. Highly malignant so-called adamantinoma or non-typical Ewing’s sarcoma. Virchows Arch A Pathol Anat Histol. 1975;365(1):63–74. [DOI] [PubMed] [Google Scholar]
- 16.Eloy C, Cameselle-Teijeiro J, Vieira J, Teixeira MR, Cruz J, Sobrinho-Simoes M. Carcinoma of the thyroid with Ewing/PNET family tumor elements: a tumor of unknown histogenesis. Int J Surg Pathol. 2014;22(6):579–81. [DOI] [PubMed] [Google Scholar]
- 17.Eloy C, Oliveira M, Vieira J, Teixeira MR, Cruz J, Sobrinho-Simoes M. Carcinoma of the thyroid with ewing family tumor elements and favorable prognosis: report of a second case. Int J Surg Pathol. 2014;22(3):260–5. [DOI] [PubMed] [Google Scholar]
- 18.Bal M, Shah A, Rekhi B, Mittal N, Rane SU, Rabade K, et al. Adamantinoma-Like Ewing Sarcoma of the Head and Neck: A Case-Series of a Rare and Challenging Diagnosis. Head Neck Pathol. 2022;16(3):679–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rekhi B, Kothari R, Shah S, Shetty O, Shah MC. A unique case of adamantinoma-like Ewing sarcoma in the calcaneus, exhibiting prominent squamous differentiation and displaying EWSR1 gene rearrangement. Skeletal Radiol. 2022;51(1):209–17. [DOI] [PubMed] [Google Scholar]
- 20.Salet MC, Vogels R, Brons P, Schreuder B, Flucke U. Maturation toward neuronal tissue in a Ewing sarcoma of bone after chemotherapy. Diagn Pathol. 2016;11(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ushigome S, Shimoda T, Nikaido T, Nakamori K, Miyazawa Y, Shishikura A, et al. Primitive neuroectodermal tumors of bone and soft tissue. With reference to histologic differentiation in primary or metastatic foci. Acta Pathol Jpn. 1992;42(7):483–93. [PubMed] [Google Scholar]
- 22.Folpe AL, Goldblum JR, Rubin BP, Shehata BM, Liu W, Dei Tos AP, et al. Morphologic and immunophenotypic diversity in Ewing family tumors: a study of 66 genetically confirmed cases. Am J Surg Pathol. 2005;29(8):1025–33. [PubMed] [Google Scholar]
- 23.Jo VY, Fletcher CD. p63 immunohistochemical staining is limited in soft tissue tumors. Am J Clin Pathol. 2011;136(5):762–6. [DOI] [PubMed] [Google Scholar]
- 24.Rekhi B, Shetty O, Vora T, Gulia A, Bajpai J, Laskar S. Clinicopathologic, immunohistochemical, molecular cytogenetic profile with treatment and outcomes of 34 cases of Ewing sarcoma with epithelial differentiation, including 6 cases with “Adamantinoma-like” features, diagnosed at a single institution, India. Ann Diagn Pathol. 2020;49:151625. [DOI] [PubMed] [Google Scholar]
- 25.Bishop JA, Montgomery EA, Westra WH. Use of p40 and p63 immunohistochemistry and human papillomavirus testing as ancillary tools for the recognition of head and neck sarcomatoid carcinoma and its distinction from benign and malignant mesenchymal processes. Am J Surg Pathol. 2014;38(2):257–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dermawan JK, Vanoli F, Herviou L, Sung YS, Zhang L, Singer S, et al. Comprehensive genomic profiling of EWSR1/FUS::CREB translocation-associated tumors uncovers prognostically significant recurrent genetic alterations and methylation-transcriptional correlates. Mod Pathol. 2022;35(8):1055–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elkamhawy A, Lu Q, Nada H, Woo J, Quan G, Lee K. The Journey of DDR1 and DDR2 Kinase Inhibitors as Rising Stars in the Fight Against Cancer. Int J Mol Sci. 2021;22(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujii H, Honoki K, Enomoto Y, Kasai T, Kido A, Amano I, et al. Adamantinoma-like Ewing’s sarcoma with EWS-FLI1 fusion gene: a case report. Virchows Arch. 2006;449(5):579–84. [DOI] [PubMed] [Google Scholar]
- 30.Wei CH, Thompson LDR, Lee K, Chow W, Liang Y. Outcome for Neoadjuvant Treatment of Parotid Gland Adamantinoma-Like Ewing Sarcoma: Case Report and Review of Literatures. Int J Surg Pathol. 2022;30(7):776–83. [DOI] [PubMed] [Google Scholar]
- 31.Roberts P, Burchill SA, Brownhill S, Cullinane CJ, Johnston C, Griffiths MJ, et al. Ploidy and karyotype complexity are powerful prognostic indicators in the Ewing’s sarcoma family of tumors: a study by the United Kingdom Cancer Cytogenetics and the Children’s Cancer and Leukaemia Group. Genes Chromosomes Cancer. 2008;47(3):207–20. [DOI] [PubMed] [Google Scholar]
- 32.Shulman DS, Whittle SB, Surdez D, Bailey KM, de Alava E, Yustein JT, et al. An international working group consensus report for the prioritization of molecular biomarkers for Ewing sarcoma. NPJ Precis Oncol. 2022;6(1):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.


