Abstract
The field of psychoneuroimmunology (PNI) has grown substantially in both relevance and prominence over the past 40 years. Notwithstanding its impressive trajectory, a majority of PNI studies are still based on a relatively small number of analytes. To advance this work, we suggest that PNI, and health research in general, can benefit greatly from adopting a multi-omics approach, which involves integrating data across multiple biological levels (e.g., the genome, proteome, transcriptome, metabolome, lipidome, and microbiome/metagenome) to more comprehensively profile biological functions and relate these profiles to clinical and behavioral outcomes. To assist investigators in this endeavor, we provide an overview of multi-omics research, highlight recent landmark multi-omics studies investigating human health and disease risk, and discuss how multi-omics can be applied to better elucidate links between psychological, nervous system, and immune system activity. In doing so, we describe how to design high-quality multi-omics studies, decide which biological samples (e.g., blood, stool, urine, saliva, solid tissue) are most relevant, incorporate behavioral and wearable sensing data into multi-omics research, and understand key data quality, integration, analysis, and interpretation issues. PNI researchers are addressing some of the most interesting and important questions at the intersection of psychology, neuroscience, and immunology. Applying a multi-omics approach to this work will greatly expand the horizon of what is possible in PNI and has the potential to revolutionize our understanding of mind–body medicine.
Keywords: Psychoneuroimmunology, Multi-omics, Precision Medicine, Health, Disease
The last 40 years of psychoneuroimmunology (PNI) research has yielded exciting discoveries into how psychological, central nervous system, and immune system processes interact, and how these interactions in turn affect human health and behavior (Ader, 2006; Daruna, 2012; Slavich et al., 2020a). Compared to the vast amount of biological data that can now be collected, however, most PNI studies published today still only include a few select biological markers (e.g., of immune or genome function). Moreover, few PNI studies integrate data across more than one or two biological systems or levels of analysis. As a result, there remains a sizable schism between the great complexity of the human mind and body, and what is currently represented in the PNI literature.
Around twenty years ago, new biological profiling technologies began emerging that enabled researchers to quantify tens of thousands of markers from a standard blood sample. At first, these technologies were prohibitively expensive, and knowledge of these approaches was limited to a few research groups. Fast forward to today, however, and we are reaching a point where researchers can conduct highly integrative, well-powered studies using a variety of core labs. The cost of multi-omics analyses is also decreasing, such that in the near future, well-powered studies will be possible to conduct within the amount of support provided by standard funding schemes (e.g., NIH R01). As these technologies continue to become more accessible, biological profiling will be more commonly used across research disciplines. In this article, we address a particular issue in this broader context, which is how PNI can benefit from integrating data that span different biological data types, including, but not limited to, the genome, proteome, transcriptome, metabolome, lipidome, and metagenome. This approach is referred to as multi-omics, and it is incredibly powerful. Multi-omics is also a great match for PNI researchers, who, by nature, are already highly integrative, innovative, interdisciplinary, and collaborative.
To fully describe the potential of integrating multi-omics approaches into PNI studies, we first introduce multi-omics at a broad level, highlighting the most commonly studied types of omics and what researchers have discovered so far using these approaches in relation to health and disease (see Fig. 1, Panel A). Second, we discuss several key considerations when designing studies that incorporate multi-omics approaches, including the importance of collecting diverse samples of participants, the benefits of repeated measures study designs, how to decide between different types of biological samples, how to incorporate behavioral and psychological data into studies using wearable sensing technology and psychological evaluations, and how to approach data modeling and computation (see Fig. 1, Panel B). In doing so, we aim to enhance PNI researchers’ understanding and appreciation of the power of multi-omics approaches and to supercharge the next generation of PNI studies investigating how myriad biological systems in the human body work in concert to shape human health, well-being, and behavior.
Fig. 1.
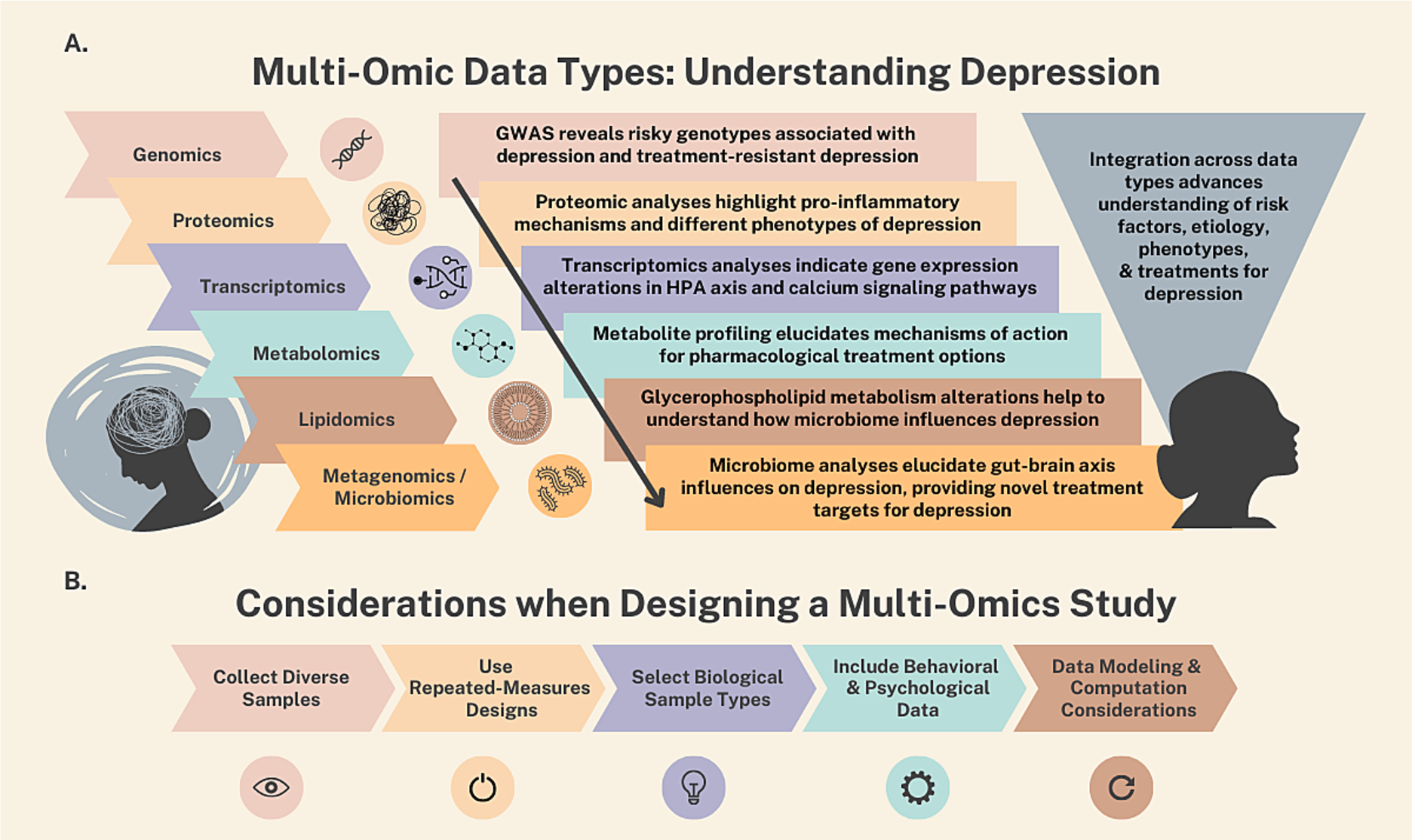
Overview of multi-omics approaches. (A) Multi-omics data types: understanding depression. Combining information across multi-omics data types to better understand risk factors, etiology, phenotypes, and treatment options in major depressive disorder. (B) Considerations when designing a multi-omics study.
1. Multi-omics
Multi-omics analysis refers to the assessment and quantification of thousands of molecules from biological samples (e.g., blood, cell, and tissue samples) that span multiple biological levels. At its core, multi-omics analysis is a scientific approach that uses multiple targeted and untargeted assays, alongside the use of multiplexed assays, to analyze biological samples. The addition of the term “omics” indicates a global or unbiased assessment of a set of molecules (Hasin et al., 2017). For example, genomics analyses can categorize entire genomes, or the complete set of genes present in an organism. Each type of molecule within an organism can be classified within one, or sometimes multiple, different omics, depending on the levels of analysis considered.
1.1. Genomics
In recent years, genome-wide association studies (GWAS) of thousands of individuals have enabled researchers to discover key genetic markers and differences in minor allele frequencies that influence the development and prognosis of disease, as well as how people with these genetic variants respond to treatment (Hasin et al., 2017). For example, a GWAS comparing the genomes of individuals with type II diabetes mellitus to control participants identified 13 new type II diabetes mellitus-associated loci (Scott et al., 2017). Discoveries such as these have led researchers one step closer to understanding the causal pathways through which type II diabetes mellitus develops. Unlike GWAS, studies using a multi-omics approach use more than just one set of omics. That is, instead of relying on genomic data alone, multi-omics studies combine information from at least two omics analyses (e.g., genomics and transcriptomics, proteomics, lipidomics, metabolomics, and/or metagenomics/microbiomics).
1.2. Transcriptomics
Although combining data from multiple biological systems is what makes multi-omics approaches so powerful, each type of omics analysis is also powerful on its own. Transcriptomics, for instance, includes genome-wide assessments of RNA levels. By investigating associations between both coding and non-coding RNA levels and disease outcomes, researchers have advanced our understanding of how the transcription of non-coding RNA influences disease processes. For example, Gupta and colleagues (2010) found that large intervening non-coding RNA, termed HOTAIR, regulates metastatic progression in breast cancer, providing new targets for treatment and diagnostics (Gupta et al., 2010). In the context of PNI, transcriptomics enables researchers to investigate cellular heterogeneity of the brain by characterizing complex cellular changes of tens of thousands of cells. A study of 48 individuals with Alzheimer’s disease that examined more than 80,000 single-cell transcriptomes revealed new insights into the disease pathology in specific brain cells (Mathys et al., 2019). Specifically, the authors observed that different brain cell types exhibit unique transcriptional changes in Alzheimer’s disease, and that these perturbations differed between males and females, highlighting the need for cell- and sex-specific therapeutic strategies. Similar single-cell transcriptomic analysis in mental health disorders, such as depression, have the potential to identify distinct molecular signatures and sex-specific transcriptional alterations, offering new insights for targeted therapies and personalized treatment strategies.
Combining genomics and transcriptomics can be especially powerful. As an example, by investigating both the genomics and transcriptomics of microglia cells, researchers have discovered that microglia cells exhibit functions beyond their previously known immune responses, such as synaptic modulation and neurotrophic support (Wes et al., 2016). This discovery would not have been possible by investigating genomics or transcriptomics alone. Furthermore, because the social environment influences gene expression, combining genomics and transcriptomics data can elucidate the mechanisms through which social-environmental factors, such as life stress and adversity, impact health and promote disease risk (Slavich, 2020b, 2022; Slavich et al., 2023a, Slavich et al., 2023b).
1.3. Proteomics
In contrast to transcriptomics, proteomics is used to quantify different proteins and peptide levels, alongside interactions between these proteins and peptides, and includes many analytes familiar to PNI research programs (e.g., proteins that influence cytokine expression). Proteomics studies also hold great potential for finding early biomarkers for psychiatric disorders (Nakayasu et al., 2021). A targeted proteomic analysis of individuals with Major Depressive Disorder (MDD), for example, identified 41 proteins in patient plasma that were strongly associated with the phenotype of cognitive function (Schubert et al., 2018). Although this study provided insights into cognitive deficits in MDD, it has not yet directly impacted treatment strategies for MDD or cognitive function. Nevertheless, the results demonstrate a new approach to discovering biomarkers in psychiatry by integrating transcriptomic and proteomic platforms. The study also identified new potential targets for therapeutic intervention, including the proteins CCND3, TXND5, and TRI26, which could lead to more effective treatments for cognitive deficits in MDD in the future. These findings require further investigation before they can be translated into clinically meaningful treatment approaches, especially within larger cohorts, underlining the need for additional multi-omics study in the field.
1.4. Metabolomics
Metabolomics analyses quantify all the small molecules that are the product of cellular metabolisms, such as amino and fatty acids (Hasin et al., 2017). Metabolomics research in MDD has revealed the differential expression of several metabolites associated with depression, including those involved in cell signaling, inflammation, hormonal activation, and sleep regulation, among others (Costa et al., 2022). Key pathways implicated include the kynurenine and acylcarnitine metabolic pathways, with changes in these metabolites often correlating with the severity of depressive symptoms and treatment responses. For example, one study suggested that treatments like ketamine or esketamine may lead to decreased tryptophan metabolites and increased glutamic acid levels, shedding light on the role of the glutamatergic system in MDD (Rotroff et al., 2016). Despite these promising findings, a metabolic biomarker has yet to be fully translated into clinical practice for diagnosing or treating MDD.
1.5. Lipidomics
Although metabolomics can include analysis of lipids, lipidomics is emerging as a distinct multi-omics specialty due to the complex nature of lipids. Lipidomics is used to assess the cellular lipids, or hydrophobic metabolites, present in a cell or organism, and has proven valuable in understanding mechanisms of many diseases, including metabolic syndrome and neurological disorders (e.g., Ban et al., 2014; Mazereeuw et al., 2013). For example, lipidomics analyses have been used to discover biomarkers of treatment efficacy for neurodegenerative diseases such as Alzheimer’s disease, providing a better understanding of the mechanistic pathways through which treatment with benfotiamine impacts Alzheimer’s pathology (Bhawal et al., 2021). Beyond elucidating mechanisms through which pharmaceuticals impact health and disease, lipidomics has also been used to discover biological mechanisms through which other treatment modalities impact health. In one study, researchers used lipidomics to investigate the mechanisms through which acupuncture treatment impacts inflammatory cytokine release in a mouse model of depression (Jung et al., 2021), in turn advancing our understanding of how traditional treatment modalities such as acupuncture influence both inflammation and depression.
1.6. Immunomics
Beyond classifying omics exclusively by specific molecule types, immunomics is an emerging approach used to better understand the immune system as a whole by examining how different parts of the immune system work together. Currently, immunomics mainly uses single-cell-level transcriptomics of immune cells to investigate immunological-related research questions, but including other analytes and omics (e.g., cytokines from whole blood, metabolomics) can help answer system-wide questions (Bonaguro et al., 2022).
1.7. Metagenomics/Microbiomics
The gut microbiome consists of the microorganisms living in the gastrointestinal tract and is an important part of the gut-brain axis, a series of immunoregulatory, neuroendocrine, and vagus nerve pathways that facilitate bidirectional communication between the gut and the brain. These interactions are important throughout the lifespan, and growing evidence points to an important role of the microbiome in neurodevelopment (Lynch et al, 2023). Multi-omics approaches have been used to help identify mechanisms through which gut-brain interactions are associated with mental health disorders such as MDD (Amin et al, 2023; Xie et al., 2023; Zhao et al, 2022), bipolar depression (Li et al., 2022), and schizophrenia (Fan et al, 2022). Multi-omics studies have also been used to identify psychobiotics that could potentially be used to improve depressive mood (Hao et al, 2023). Indeed, preclinical and small clinical studies have suggested that prebiotics and probiotics may have a beneficial effect on mood and anxiety, although larger, more well-controlled studies are needed (Dinan et al, 2013; El Dib et al, 2021; Noonan et al, 2020).
1.8. The potential of multi-omics approaches
Multi-omics approaches can provide PNI researchers, and health researchers in general, with a large and dynamic set of new tools that will facilitate the discovery of new taxonomies of health and disease. So far, multi-omics studies in humans have revealed dynamic changes in molecular components and biological pathways across healthy and diseased conditions, alongside various medical risks, including type II diabetes mellitus (Chen et al., 2012; Schüssler-Fiorenza Rose et al., 2019). Multi-omics can also demonstrate how psychosocial or clinical interventions, focused on behaviors such as diet, can affect systems sequentially. For example, recent multi-omics studies have shown that dietary fiber supplementation can alter the microbiome, which in turn alters the metabolome to improve lipid profiles in hypercholesterolemic individuals (Lancaster et al., 2022). Additionally, dietary changes that influence the microbiome can have systemic effects also measured by multi-omics. These can affect immune function through the microbiome, inflammation as measured by cytokines, microbiome gut diversity, and the genetic function of the microbiome (Wastyk et al., 2021). These omics measurements can then be correlated with clinical data, the gold standard for assessing health and disease states, and with cytokine levels, which measure inflammatory activity, to gain a more holistic picture of how systems biology, measured using multi-omics, is related to human health and behavior.
Although many analyses that include only one data type are limited to interpretations of correlational associations, mostly reflecting reactive rather than causative processes, the integration of different omics data types can be used to identify potential causative changes that lead to disease, which can then be tested in further molecular studies (Hasin et al., 2017). Furthermore, finding the same biological mechanisms in multiple omics layers substantially increases confidence in findings (Lancaster et al., 2020). Given the transformative potential of incorporating multi-omics approaches into PNI research programs, we next provide an overview of how to design a multi-omics study, highlighting key decisions researchers must make along the way.
2. Designing a multi-omics study
Many factors important to consider when designing multi-omics studies are similar to those that should be weighed when designing PNI studies more broadly. For example, who will your participants be? How many participants do you need to collect data from and how many samples per participant do you need to collect? What types of biological samples should you collect? Once you have collected the samples, which analytes should you assay them for and how many analytes should you assay in total? Beyond biological samples, what other data should you collect from participants? Once you have completed your biological assays, how do you analyze and interpret the results? Although the answers to these questions are often specific to the specific research question(s) at hand, some approaches are more likely to yield success than others when conducting multi-omics research. We review these points in greater detail below.
2.1. Participant diversity is critical
When considering using a multi-omics approach, it is critical to accurately measure and consider the ways in which marginalization, diversity, and underrepresentation impact multi-omics outcomes and measurement. Indeed, failing to prioritize the collection of diverse samples when using new scientific approaches can actually cause health disparities across populations (e.g., by race, gender, sexual orientation) that were not previously present. This risk is especially high when investigating biological factors related to health and disease risk.
Jatoi and colleagues (2022) described a notable example of this problem. Namely, prior to the 1980s, breast cancer mortality risk was lower for Black vs. White women. In the 1980s, however, this dynamic changed starkly, and breast cancer mortality risk is now higher for Black as compared to White women (Siegel et al., 2019).
Why might this be the case? It appears that advancements in mammogram screening and endocrine treatments for hormone receptor positive breast cancer may be the cause. Although these advancements were developed to reduce breast cancer mortality risk—and do achieve this goal—they do not provide the same benefit for White and Black women due to the differential prevalence of hormone receptor positive and hormone receptor negative forms of breast cancer, in addition to differences in access to genetic and mammogram screening. Therefore, although these medical advancements reduce the rates of breast cancer mortality risk for both Black and White women, these advancements provide a far greater benefit to White women and have thus created sizable disparities in breast cancer mortality risk across these groups, with Black women now faring far worse than White women. Here, medical advancements created a health disparity that did not previously exist.
As health technology advances, PNI scholars and practitioners have the power to prevent these unfavorable outcomes from occurring. The first, and perhaps most important way to reduce such unintended consequences is to ensure that all research using multi-omics approaches is conducted in diverse populations from the beginning. That is, sample diversity should not be an afterthought, but rather should be incorporated into initial study designs. To date, 96% of all GWAS have used European samples, and the data gathered from these studies is already being used to identify variants associated with disease profiles (Popejoy & Fullerton, 2016). Given the heterogeneity of the samples collected, however, the diverse global population will not likely experience better health as a result of these discoveries. Moreover, the clinical benefits associated with identifying genetic variants that affect disease risk in diverse populations will lag far behind those for European samples, which has the potential to create or exacerbate health disparities over time in a manner similar to the breast cancer example described above. It is thus critical that researchers prioritize multi-omics research using diverse samples so as to not create or perpetuate disparities in health or healthcare.
Second, researchers must consider how social adversity, marginalization, and oppression impact the biological systems measured. It is well documented that many health-relevant omics of interest, as well as social-environmental factors that portent poor health outcomes, differ across populations (e.g., Gillespie et al., 2022; Hamlat et al., 2022; Mayer et al., 2023; Toussaint et al., 2022). For example, there is a large body of research documenting higher rates of inflammation-related diseases in marginalized populations, and many have attributed this phenomenon to the greater exposure to discrimination faced by diverse populations (Diamond et al., 2021; Simons et al., 2021; Slavich et al., 2023b). These differences suggest that although we need to pay attention to biological differences across racial groups that can impact treatment (e.g., Black women’s higher rates of types of breast cancer that do not respond to the endocrine treatments rolled out in the 1980s; Jatoi et al., 2022), we also need to consider how psychosocial stressors, social interactions, and individuals’ interactions with broader societal processes impact these biological processes and, in turn, disease progression and treatment success.
Finally, researchers must consider how social marginalization and structural oppression impact diverse populations’ access to, knowledge of, and comfort with new technological advancements. Given that marginalized populations have poorer access to health insurance and quality healthcare (Williams & Rucker, 2000), whenever possible, researchers should seek to ensure that medical advancements and discoveries are easily accessible and widely publicized in a variety of languages, and that they clearly communicate the benefits of these discoveries. Doing so will enable diverse populations to advocate for receiving the best and most up-to-date health care available, which is critical for increasing the likelihood that cutting-edge discoveries made using multi-omics approaches will translate into better health for all.
These recommendations are by no means exhaustive. Rather, they are intended to provide an overview of several of the most important points researchers should consider as they begin incorporating multi-omics approaches into studies (for additional recommendations, see Fatumo et al., 2022). Importantly, there are also numerous measures that researchers can use to measure social determinants of health and health disparities, such as identity specific measures of discrimination and inclusion (e.g., racial discrimination vs. sexual orientation-based discrimination). For additional information on how to assess key variables related to social determinants of health disparities such as biological sex, gender, race, sexual orientation, healthcare access, discrimination, and health literacy, among other measures, we recommend reviewing the NIH PhenX Social Determinants of Health Core Toolkit (Hamilton et al., 2011).
2.2. The power of repeated measures designs
After selecting the appropriate participant population for the research question at hand, researchers must next design their study protocol. Although this holds true in many areas of research, the power of repeated measures and longitudinal multi-omics designs, as compared to single time-point designs, cannot be overstated. Indeed, sampling multiple time points enables researchers to examine disease progression, starting (ideally) from a healthy state and transitioning to a disease state, as well as genotype-environment-phenotype dynamics within an individual—or set of individuals—over time.
Understanding the trajectories of multi-omics analytes can also provide a better indication of casualty and increased confidence in interpreting correlations between measurements. In this context, the number of participants you can sample is less meaningful when using a multi-omics approach than is the number of samples per participant that you are able to collect. In one recent study, for example, researchers collected multi-omics data for one individual over 14 months using whole genome sequencing paired with periodic measurements of transcriptomics, proteomics, metabolomics, and other biomarkers, and discovered that the participant was at an increased risk for type II diabetes mellitus. Later in the study, after a rhinovirus infection, the individual developed type II diabetes mellitus. With subsequent lifestyle modifications, however, the participant’s glucose levels eventually reversed to pre-type II diabetes mellitus levels, enabling researchers to observe a signaling network rewiring during these transitions (Chen et al., 2012; Kellogg et al., 2018). These processes were mapped to provide an in-depth picture of the transition from health to disease and to elucidate how lifestyle changes affect disease pathophysiology.
Larger, more traditional longitudinal multi-omics studies have also proven valuable in characterizing the biology underlying health and disease. For example, a 109-person cohort followed for up to eight years underwent quarterly longitudinal profiling that included genomics, transcriptomics, proteomics, immunomics, metabolomics, and microbiomics (i.e., gut microbe diversity). So far, the study has yielded at least 67 clinically actionable discoveries that can translate into better outcomes across several disorders (Schüssler-Fiorenza Rose et al., 2019).
Although many longitudinal studies are conducted over months or years, it is possible to collect multiple samples over the course of a few days, or even across one hour, to produce an intensive, repeated measures dataset. For example, a recent acute exercise study used a repeated measures multi-omics analysis to collect five intravenous blood samples over one hour from 36 participants exercising on a treadmill. A time-series analysis identified key biological processes of peak VO2, and a prediction model found molecules that predicted peak VO2 using baseline multi-omics profiling, which could be used to develop a new resting biomarker for aerobic fitness (Contrepois et al., 2020). High intensity study designs such as these have many advantages over more traditional longitudinal designs, as assessment windows that are longer than the naturally occurring timeline for causal effects can attenuate effect sizes, leading to imprecise estimates of the magnitude of effects (Dormann & Griffin, 2015; Dwyer, 1983). For a more thorough review of the benefits of intensive longitudinal data collection in PNI, see Moriarity & Slavich (2023).
2.2.1. Balancing costs with study goals
The associated expenses of both multi-omics and longitudinal research raises concerns about the feasibility of longitudinal and repeated measures multi-omics studies. To this effect, we recommend carefully considering the primary goals of a study to balance the costs of the study with the breadth of biological and temporal assessment required to address the main research aims. For example, if descriptive characterization of the biological features of a disease state is the goal, cross-sectional data with a more diverse multi-omics panel and a large number of participants would be appropriate. Conversely, if the goal is to identify stress-induced changes in multiple domains of biological functioning, it would be better to select a smaller panel of analytes based on prior research and theory to be able to afford repeated measures in a smaller sample of participants. Still some studies will benefit from large sample sizes and longitudinal profiling, such as those characterizing biological transitions to disease states with the goal of understanding biological subtypes and the mechanistic pathways that define these disease subtypes.
Relatedly, regarding power, it is important to consider whether the study hypotheses are about between-person differences or within-person variability. This will determine whether power analyses should focus on the number of participants, number of observations per participant, or both. Determining the appropriate sample size can be challenging in multi-omics studies that assess many features simultaneously and depends in part on the goal of the study (e.g., differential features, longitudinal change, predictive modeling/machine learning; Guo et al., 2010; Krassowski et al., 2020). See Box 1 for a few tools that can help researchers determine a study’s appropriate sample size.
Box 1. Multi-omics data analysis resources and tools.
| Description | Name/Key Reference |
|---|---|
| Platforms that contain common analytic tools for both supervised and unsupervised data analytic approaches | • MixOmics (Cao et al., 2016) • MetaboAnalyst (Pang et al., 2022) • 3omics (Kuo et al., 2013) • PaintOmics (Liu et al., 2022) • OmicsNet2.0 (Zhou et al., 2022) • Mergeomics2.0 (Ding et al., 2021) |
| Tools for determining appropriate sample size | • MultiPower/MultiML (Tarazona et al., 2020) • PowerTools (Acharjee et al., 2020) |
| Reviews of analysis techniques used in multi-omics studies of psychiatric disorders | • Amasi-Hartoonian et al., 2022 • Sathyanarayanan et al., 2023 |
| Review of unsupervised multi-omics data integration methods | • Vahabi & Michailidis, 2022 |
2.3. Biological samples and assay selection
Determining which multi-omics assays and analyses to use depends on the study goal and budget. For a large-scale multi-omics study, assay costs can range widely from $200-$3,450 per sample, depending on the quality of the assays selected and number of analytes the researcher seeks to quantify (see Table 1). Although genomics costs are expected to decline in the near future, whole genome sequencing is still relatively expensive; on the other hand, the high cost may be worth it, as genomics studies hold great potential to reveal underlying mechanisms of disease. Proteomics also tend to be an expensive choice, especially for an untargeted assay; in contrast, transcriptomics has a relatively low cost, as it involves a fraction of the sequencing length required for genomics analyses and is showing promise in identifying potential biomarkers of disease (Slavich et al., 2023b). A recent study using multi-omics analysis to study genetic components of psychiatric disorders, for example, compared gene expression from brain derived tissue to that from whole blood and found that transcriptomics data derived from whole blood could be used as a starting point for identifying neurological and psychiatric outcomes, including attention-deficit hyperactivity disorder, Alzheimer’s disease, bipolar disorder, depression, intelligence, insomnia, neuroticism, and schizophrenia (Korologou-Linden et al., 2021).
Table 1.
Summary of molecules present in different sample types for multi-omics assays and the approximate assay costs.
| Omics | Assesses | Sample Types | Approx. Number of Analytes | Approx. Cost per Sample |
|---|---|---|---|---|
|
| ||||
| Transcriptomics | Genome-wide RNA levels | Tissue, whole blood, PBMCs, urine, stool | 12,000+ | $250-$400 |
| Genomics | Complete set of genes in an organism | Whole blood, saliva, buccal swabs | Complete genome | $500 |
| Proteomics | Proteins and peptide levels | Whole blood, plasma, serum, DBS, urine, stool | 100–3,000 | $100-$1,200 |
| Lipidomics | Cellular lipids | Tissue, whole blood, plasma, serum, DBS, urine, stool | ~1,000 | $100 |
| Metabolomics | Cellular metabolites (i.e., amino acids and fatty acids) | Tissue, whole blood, plasma, serum, DBS urine, stool, | ~700 | $150 |
| Metagenomics/microbiomics | Community of microorganisms in a sample (i.e., microbes) | Tissue, skin, urine, stool | 25,000–100,000 | $100-$1,200 |
Note. Prices are for research-grade assays, conducted at scale, as of March 2023. Approx. = approximate; PBMCs = peripheral blood mononuclear cells; DBS = dried blood spot.
Determining the correct biological sample type for a study design and analysis of choice requires considering the key analytes of interest, feasibility of collecting each type of sample, burden each type of sample collection places on the participant, research question(s) at hand, and cost of collection, storage, and processing of these sample types. Typically, multi-omics studies rely on blood, stool, urine, saliva, buccal swabs, or solid tissue samples, and can be paired with continuous physiological measurements by wearable devices (see Table 1). Solid tissue samples are typically only used to answer very specific research questions and usually require a clinician to collect. For example, to examine the association between environmental pollution exposure and facial skin conditions, skin tissue can be collected and used to quantify the presence and levels of specific skin metabolites (Misra et al., 2021).
Other types of biological samples are easier to obtain, less invasive, and provide the opportunity to assess a greater range of analytes. For example, collecting saliva, stool, and urine using at-home collection kits is relatively low-burden for participants. Moreover, there are a growing number of vendors that supply these collection kits, providing researchers with a simplified and streamlined process for collecting and processing these samples. Generally, saliva samples can be an excellent choice when conducting research on children and neonates, as saliva is fairly easy to collect, and dozens of biomarkers across genomics, transcriptomics, proteomics, metabolomics, and microbiomics have already been identified in pediatric diseases (Pappa et al., 2019). Saliva sampling can also be a good option in stress studies, as salivary cortisol is a biomarker for stress and considered the “gold standard” in clinical research assessing acute stress reactivity (Pappa et al., 2019).
Urine samples, on the other hand, are particularly useful for determining the metabolic effects of kidney disorders, and when a researcher is interested in daily levels of metabolites (for recommendations, see González-Domínguez et al., 2020). In contrast, stool samples can help elucidate associations between microbes, their host, and diet in cardiometabolic disorders alongside gastrointestinal diseases. For example, stool genomics can reveal the microbial composition of the identification of disease-specific genes, and transcriptomics was previously used as a proxy for functional output comparison in inflammatory bowel disease patients (Sauceda et al., 2022). Finally, the metagenome is a bulk analysis of the entire bacterial DNA inside a stool sample, usually containing hundreds of bacteria. The bacteria within stool is known as the microbiome and is exquisitely interwoven with the host and responsive to changes in diet and other factors. The microbiome can thus be analyzed to understand the systems biology effects of microbiome changes due to (for example) dietary interventions (Lancaster et al., 2022) and stress (Kim et al., 2021; Sichko et al., 2021) that affect the gut-brain axis and health.
Although there are cases when other sample types are preferred, blood-based samples are the most commonly collected in multi-omics research. There are a few different methods of blood collection that may be appropriate depending on the study budget, study needs, and a researcher’s access to clinical resources. Collecting venipuncture blood using standard blood draw procedures requires access to a phlebotomist and for the participant to be physically present at the research lab or clinic, which often causes venipuncture blood collection to be an expensive option for researchers and places a high burden on research participants. Obtaining blood from a vein has many benefits, especially the ability for researchers to collect large volumes of blood (i.e., up to 50 ml), with different anticoagulants to aid in the processing and storage of blood into serum and plasma samples that can be used for multiple immunoassays. The large volume of blood that can be collected during venipuncture aids in the detection of most analytes available, allows for repeated analysis if needed, and in general requires less processing and validation than do other, less invasive methods of obtaining blood (Dasgupta & Krasowski, 2019; Koh et al., 2022). However, these samples must be processed relatively quickly after the blood is collected to avoid the degradation of many analytes, and samples must be stored at sub-zero temperatures.
One common, less invasive method for obtaining blood samples is using dried blood spot sampling, which can be obtained using a finger prick method or an at-home collection kit. Although these methods typically only provide researchers with a small amount of blood (usually less than 100 μl), there are many benefits to using this method of blood collection. For example, using commercially available devices, such as the TASSO M20 device, participants can collect their own blood sample at home and return it by mail at room temperature. The main advantages of such a sampling method are the relatively low cost and convenient sampling for participants. However, such methods require higher expertise in processing, and due to the low blood volume of samples, there are a lower number of detectable analytes in dried blood samples compared to venipuncture blood samples (Lei & Prow, 2019). A recent multi-omics study using the volumetric absorptive microsampling method (i.e., finger prick method, collecting 10 μl of blood) showed a high correlation in the measurement of thousands of metabolites, lipids, cytokines, and proteins, when compared to venipuncture blood sampling method (Shen et al., 2023). Although all of the blood sampling methods mentioned above enable the multi-omics study of proteomics, metabolomics, lipidomics, and immunomics, whole blood or peripheral blood mononuclear cells are typically needed to assess genomics and transcriptomics, and not all blood sample types produce the same results (Gautam et al., 2019).
2.4. Behavioralomics: Incorporating behavioral data using sensing technology
Just as multi-omics sampling captures biological signatures of participants, high-frequency wearable technology (“wearables”) with optical sensors and behavioral tracking provide detailed indicators of participants’ individual physiological processes and fluctuating health states. Incorporating wearable technology into multi-omics studies can provide researchers with access to a multitude of digital biomarkers and behavioral metrics that enable a comprehensive examination of the whole physiome and behaviors, in addition to biological processes. In doing so, researchers can examine additional physiological and behavioral factors that contribute to inter- and intra-individual differences in multi-omics data. Accelerometers and gyroscopes in wearables that assess positioning and movement are useful in monitoring behaviors that may be mediated by the immune system, since immune system shifts—whether the result of mental or physical illness—initiate behavioral changes as well (i.e., “sickness behavior”; Dantzer, 2009; Maes et al., 2012; Slavich et al., 2020a; Slavich and Auerbach, 2018).
One recent study that examined patients with myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) and healthy controls using wearables found that time spent upright was significantly associated with ME/CFS presence and severity. Specifically, the researchers found that whereas healthy participants spent more than 30% of their time upright, moderately ill ME/CFS participants spent between 20 and 30% of the day upright, and severely ill participants spent less than 20% of the day upright (Palombo et al., 2020). Another study tracked physical activity using a wearable accelerometer and found that physical activity was associated with improved neutrophil migration in elderly participants, suggesting it may be a mechanism through which exercise is associated with better immunity (Bartlett et al., 2016). Indicators of autonomic system activity assessed using commonly used wearable devices (e.g., heart rate, nocturnal heart rate variability, respiratory rate) have also been associated with measures reflecting mental health disorders such as depression and anxiety, as well as stress and well-being more generally (Coutts et al., 2020; Ritsert et al., 2022). In summary, wearables and other remote sensing technology can thus be used alongside other multi-omics data to create personalized prediction models of early disease development, highly dynamic health profiling and disease-state profiling (Kellogg et al., 2018; Shen et al., 2023), and even evaluate therapeutic outcomes (Antikainen et al., 2022).
Using wearable monitors also enables researchers to make use of multi-omics data more precisely. For example, many biological processes, such as cortisol production and energy metabolism, have circadian rhythms that play a role in health as well as the development of diseases (Patke et al., 2020). However, individuals vary in their circadian rhythms, which are affected by behaviors such as sleep onset and wake time. Similarly, physical activity exerts a multitude of physiological processes that, if left unmeasured, introduce unnecessary variability in models (Nieman et al., 2019). Researchers conducting studies involving multi-omics can accurately account for these differences in daily rhythms and behaviors, and contextualize within-person changes and between-person differences, by using wearable technology that can track these health behaviors with relative accuracy without requiring users to keep an accurate sleep or physical activity diary (de Zambotti et al., 2019).
In addition, because of the potential for participant burden and the high cost of multi-omics assessments, it is often not feasible to include high-frequency multi-omics data collection in studies. In studies using multi-omics data at lower frequencies, it may be possible to use wearable data to estimate multi-omics data between blood collection(s). Indeed, data derived from wearables, such as heart rate features, have demonstrated utility in predicting molecular changes. In one study, for example, researchers matching high-frequency multi-omics data collection with wearable data found that wearable-derived data—in particular, heart rate data (e.g., heart rate variability, range, and maximum heart rate)—were associated with 447 of the 2,223 molecules examined, most of which were lipids and cytokines (Shen et al., 2023). Although the models yielded thus far are in a nascent stage, including wearable data to expand what is possible in lower-frequency multi-omics studies has a multitude of benefits.
2.5. Survey and psychological data collection
Whenever biological data are collected, it is vital to also consider what other data researchers can collect that will help them understand and interpret biological associations and outcomes. At a basic level, demographic information including sex, race, age, and socioeconomic measures are essential. Beyond these variables, collecting health-relevant information–such as disease status, recent illnesses, medication use, and mental health conditions–is useful when seeking to understand unexpected data patterns. Combining historical health data, such as what is available in electronic health records, with multi-omics approaches is another way to provide insight into unexpected data patterns and the progression of disease states.
In repeated measures designs with biological data collection, it is also beneficial to collect measures of psychological and behavioral variables of interest (e.g., current levels of stress, social support, hunger, mood; for publicly available measures, see Hamilton et al., 2011) concurrently with biological sample collection. Furthermore, correlating real-time psychological data collection, such as Ecological Momentary Assessments (EMAs), with real-time information from wearable devices has proven valuable in discovering digital biomarkers of distress in MDD patients; they can also enable real-time interventions, called Ecological Momentary Interventions, which can be used to relieve anxiety- and stress-related symptoms as they occur (Lautman and Lev-Ari, 2022; see also Byrne et al., 2021).
Given the clarity that can be gleaned from understanding both biological and psychological factors that influence health and behavior, future empirical work should incorporate a more holistic approach, sampling both biological and physiological factors together with psychological data collection. Multi-omics approaches to these questions can undoubtedly contribute to this holistic goal. By utilizing recent advancements in minimally invasive blood sampling, multi-omics approaches give researchers the ability to measure biological factors longitudinally in conjunction with minimally taxing longitudinal psychological assessments such as EMAs, which enables researchers to capture the transient changes that are often of interest in PNI studies, alongside genotype-environment-phenotype effects within an individual. This approach will also enable greater opportunities for intervention. If researchers can identify the multiple processes (e.g., biological, psychological, environmental) that contribute to disease risk and progression, for example, they can use this information to develop more effective personalized interventions aimed at reducing risk in multiple domains as opposed to attempting to intervene on a single domain (e.g., biology) while ignoring the influence of others (e.g., psychology). Although the recommendation to collect psychological data alongside of biological data may seem obvious to PNI researchers, this is not standard practice in all labs conducting multi-omics research, and it is one area in which PNI researchers could provide valuable expertise to researchers conducting studies that use multi-omics approaches to study health and disease.
2.6. Multi-omics data analysis
Analyzing and integrating multi-omics data is complex. Each individual type of omics analysis presents its own unique set of challenges, and the task of integrating multiple and vastly different types of data, such as omics data (including genomics, proteomics, metabolomics, etc.), EMAs (which capture experiences in real-time), wearable data (generated by devices that collect physiological metrics), and psychological data (from behavioral and cognitive assessments) presents new challenges as well (Krassowski et al., 2020; Mirza et al., 2019; Hasin et al., 2017). These data types are not only diverse in their nature but also their scale, quality, and format, leading to complexities in handling and interpreting them in a harmonious way.
The best way to approach this challenge is to form multi-disciplinary teams composed of experts who specialize in each of the various data types. A lone researcher attempting to master each type of data is unlikely to yield high-quality results. By working together, interdisciplinary teams are able to balance needs across different data types, ensuring accurate collection, preprocessing, integration, and interpretation of the data. The collaboration of diverse researchers increases not only the quality of data collected, but also promotes insightful and meaningful interpretations of results through the discussion of connections between the different data types (e.g., Schüssler-Fiorenza Rose et al., 2019; Shen et al., 2023).
In general, methods used in multi-omics studies are intended to investigate associations within a high-dimensional space, usually with limited samples, while considering the inherent noise unique to each omics and the number of features from each omics. For example, in a typical workflow, thousands of transcripts can be generated as compared to only hundreds of metabolites, such that the information in the transcriptome can hide more pertinent information contained in the metabolome. In this context, we next provide a brief overview of the steps and challenges involved in multi-omics data analysis and some promising approaches to tackle them. We also stress that broad recommendations are necessary but not sufficient to ensure appropriate use of multi-omics data.
2.6.1. Identifying analysis goals
The first and most important step is to clearly define the question and goals of the analysis. Common goals of multi-omics analyses include identifying disease-associated molecular patterns, identifying and classifying disease subtypes, identifying biomarkers for diagnosis and prognosis, understanding biological mechanisms of disease, and generating temporal predictions of disease onset, recurrence, and survival (Subramanian et al., 2020; Athieniti & Spyrou 2023). Each of these goals requires different types of tools (for a repository of software packages for multi-omics analyses, see: https://github.com/mikelove/awesome-multi-omics).
2.6.2. Data preprocessing
The second step of multi-omics data analysis is to perform data preprocessing. Data preprocessing includes ensuring the quality and handling the variability of the data, and presents one of the greatest challenges in multi-omics data analysis. Multi-omics data can be affected by technical and biological factors, such as batch effects, sample heterogeneity, and measurement error, which can introduce noise and biases in results. This noise is not evenly distributed across omics, and variability in this distribution of errors must be taken into account during study design and evaluation. As mentioned above, each individual omics investigated will yield a varying number of features, and these will be associated with their own amount of noise (Tarazona et al., 2020). Therefore, it is critical to perform quality control and preprocessing steps to ensure the consistency and reliability of the data. If possible, analyze all biological samples in one batch, to avoid batch effects (i.e., systematic differences due to technical and other factors in sample processing) that can obscure true biological associations (Čuklina et al., 2021). When this is not possible, researchers can minimize the chance of batch effects confounding biological associations through careful experimental design, using strategies such as randomization and blocking (Čuklina et al., 2020).
One step researchers can take to facilitate batch correction is to include biological and technical replicates across batches. Although determining the optimal ways to perform batch correction in multi-omics studies remains an empirical question, a number of tools and methodologies have been developed to perform batch correction, including normalization (Čuklina et al., 2020). Normalization methods include loess, quantile, systematic error removal using random forest (Fan et al., 2019), and tunable median polish of ratio (Dammer et al., 2023), among many others. Normalization is followed by batch correction procedures such as surrogate variable estimation (Leek et al., 2012), empirical bayes methods (ComBat/ComBat-seq; Johnson et al., 2007; Zhang et al., 2020), and others, including a recently developed method, MultiBaC, that performs batch correction across omics datasets under specified conditions (Ugidos et al., 2022; Ugidos et al., 2020). For analysis methods that include modeling, “batch” can be included as a covariate in analyses, but this is not possible with all methods. It is always important to evaluate the efficacy of the batch correction method chosen, as over corrections can obscure meaningful results, while inappropriately accounting for batch effects can produce spurious results. Additional data pre-processing steps depend on planned analyses, but common steps are data transformations and standardization to enable the integration of data that come from different distributions and scales (see Graw et al., 2021).
2.6.3. Data integration
There are multiple approaches to integrating omics data, and omics and non-omics data. For example, in multistage analysis, associations are first found between data types and then between the phenotype(s) of interest (Ritchie et al., 2015). Simultaneous analysis of multiple data types is divided into three main categories: (a) concatenation-based or early integration combines multiple omics datasets into a single dataset, and this combined data set is then analyzed; (b) transformation-based integration, where each data type is transformed into an intermediate form such as a graph or kernel and then integrated, which can occur jointly or independently; and (c) model-based or late integration, where each omics dataset is analyzed separately and then the results are combined (Kaur et al., 2021; Ritchie et al., 2015). A fourth strategy is hierarchical integration, where known regulatory associations between omics layers are taken into account when integrating datasets (Picard et al., 2021).
A similar set of strategies has been proposed for integrating omics data with non-omics data, including joint modeling (early integration), independent modeling (late integration), and a conditional approach in which a clinical model is defined first with non-omics variables and then omics variable selection is performed by considering a model that contains or adjusts for the non-omics variables (López de Maturana et al., 2019). Analysis tools for multi-omics data analysis include univariate and multivariate statistical methods, machine learning methods that include both supervised and unsupervised methods, and deep learning. For an overview of analysis techniques used in multi-omics studies of psychiatric disorders, see Amasi-Hartoonian et al. (2022) and Sathyanarayanan et al. (2023).
2.6.4. Data analysis methods
The goals of the analysis are extremely important for selecting which methods of analysis are most appropriate. Unsupervised clustering is agnostic to the phenotype or outcome and is used both for dimensionality reduction, understanding the underlying structure of the data, and downstream analyses. Dimensionality reduction is one approach for addressing the high dimensional nature of multi-omics data and reducing model complexity. Common clustering techniques used for dimensionality reduction are principal components analysis, multi-dimensional scaling, and T-distributed Stochastic Neighbor Embedding. Newer techniques aimed specifically at multi-omics data include multi-omics factor analysis (Argelaguet et al., 2018) and Multiple Co-Inertia analysis (Meng et al., 2014). Clustering is also commonly used to identify disease subtypes or disease trajectory subtypes of populations. Common clustering techniques include hierarchical, k-means, self-organizing maps, and gaussian mixture models, among others (Eicher et al., 2020), and fuzzy c-means clustering is often used to cluster longitudinal trajectories.
Researchers seeking to understand biological mechanisms underlying health and disease may use statistical methods to identify associations of omics analytes with outcomes of interest and then follow this analytic step with integrated pathway analysis, which can be further integrated across omes. Statistical tools include univariate methods, both parametric and non-parametric, and multivariate methods. When using univariate methods, it is important to conduct multiple hypothesis testing correction given the large number of analytes. Common methods include Bonferroni (Bland & Altman, 1995) which is the most conservative, the Benjamini-Hochberg False Discovery Rate (FDR; Benjamini & Hochberg, 1995), and the optimized FDR approach of Storey and Tibshirani (2003) that produces q-values, a measure of the FDR that would occur by accepting the given test and all other tests with a similar p-value. For example, Misiewicz and colleagues (2019) used limma eBayes (a linear model with empirical Bayes variance estimation) to identify differentially expressed genes and proteins affected by chronic stress, and then compared stress-resilient, stress-susceptible, and same-strain control mice. The researchers first performed pathway analysis on the individual omics level and then used an integrative analysis to identify the most widely stress-affected pathways. They found that pathways related to mitochondrial function and transcriptional control were significantly enriched but in opposite directions for stress-resilient vs. stress-resistant mice (Misiewicz et al., 2019). Other methods for understanding biological associations are correlation networks, partial correlation networks, Weighted Gene Coexpression Networks, and Topological Analysis of networks (Eicher et al 2020).
When the goal of a study is to predict disease or discover biomarkers of disease, researchers frequently rely on a large range of statistical and machine learning methods. Unlike pathway analysis, which requires a large number of associated analytes to perform, biomarker studies often aim to identify a parsimonious number of analytes. Both univariate and multivariate statistical techniques can be used. Univariate techniques include regularized regression such as the least absolute shrinkage and selection operator (LASSO; Tibshirani, 1996), and elastic net, which often performs better than LASSO when the number of features exceeds the number of subjects and the dataset contains groups of correlated variables (Friedman et al., 2010). Regularization shrinks the coefficients of less important features and can be used for feature selection when the coefficients of unimportant features are set to zero. Multivariate statistical methods, in turn, include sparse Partial Least Squares (Cao et al., 2008) and sparse canonical correlation analysis (Witten & Tibshirani, 2009; Witten et al., 2009) and variations of these methods. These are latent variable models that seek to maximize the covariance (i.e., sPLS) or correlation (i.e., sCCA) between two sets of data (Mihalik et al., 2022).
Other machine learning techniques include Random Forests which is a tree-based algorithm, and Support Vector Machines, which can handle nonlinear data as well as causal discovery feature selection methods (Saxe et al., 2017). Deep learning methods, which use neural networks and can capture nonlinear and hierarchical features, have powerful predictive capacities but often require much larger sample sizes than are typical of multi-omics studies (Kang et al., 2022). When sample sizes are small, additional attention must be paid to the method used for evaluating model performance. Instead of using the traditional training and test set method or simple k-fold cross-validation, methods such as nested cross validation and bootstrap bias corrected validation are more appropriate, as these more reliably estimate performance in small samples and make much better use of the data (Tsamardinos, 2022; Vabalas et al., 2019).
As an example of what can be achieved when appropriate data analytic approaches to multi-omics data are used, Ghaemi et al. (2019) developed a model to estimate gestational age during pregnancy. The researchers considered multiple factors including appropriate cross validation strategies to minimize data leakage (i.e., using different portions of the data to train a model and then test the hypotheses), regularization, and stack generalization to combine results across omics. These steps enabled the researchers to overcome challenges inherent to multi-omics data analysis, to develop a promising machine learning model of gestational age with limited samples, and to set the stage for future research aimed at examining deviations associated with pregnancy-related complications (i.e., preeclampsia, gestational diabetes, preterm birth). In sum, although multi-omics data analysis is complicated, there are a number of recently developed platforms that contain common analytic tools for supervised and unsupervised analysis that reduce some barriers related to conducting multi-omics data analysis (see Box 1).
2.7. The future of multi-omics in PNI: Challenges and next steps
The breadth and depth of biological information collected with multi-omics assessments is well-poised to increase our fundamental understanding of biological systems and how they interact with psychological and environmental factors. With each promise of multi-omics approaches, however, there also arises challenges that researchers must face to ensure that they are using these approaches appropriately. For example, it is impossible to understate the necessity of considering the biological plausibility of inferences when interpreting statistical output. This is important for all research but especially important for multi-omics research, which is typically exploratory and data-driven. Access to multi-omics data and a plan to use an unsupervised approach does not preclude the need for careful thought about biologically important variables. That is, if a revealed pattern of results is not biologically plausible, alternative modifications/models must be considered.
It is also important to recognize the limitations of current biological knowledge. This can affect pathway analysis which relies on existing databases, as these databases are constantly evolving as new knowledge becomes available. Databases are much better developed for some omics compared to others. For example, transcriptomics databases are more established than metabolomics databases, where there are still large amounts of unknowns and metabolite identification is a challenge (Wieder et al., 2021).
As mentioned above, the cost of collecting, storing, and analyzing high-quality multi-omics data often limits the sample sizes that researchers are able to collect (e.g., given typical research budgets). We advocate for a balanced approach that appreciates the exploratory potential that multi-omics data provide for highlighting potential biomarkers or mechanisms of health and disease, and that carefully considers where it is possible to restrict the scope of data collection to preserve funding (i.e., number of analytes vs. number of participants vs. number of repeated measures). Another consideration is the depth of sequencing needed for the study, which affects the cost of genomic analysis. For example, whereas deep, whole metagenome sequencing enables researchers to identify rare microbes and can identify single nucleotide variants in individual microbiome species, it can be cost prohibitive for large longitudinal studies and the taxonomic resolution of shallow shotgun sequencing may be adequate for the questions being asked, as a viable and cost-effective alternative to 16S sequencing (La Reau et al., 2023). Central to this consideration should be the ability to test the primary research question(s) of interest with adequate power.
To the extent that resource scarcity presents a problem, multi-lab collaborations might help to pool together enough resources to conduct meaningful analyses. Another approach is to bank samples for future additional omics analysis. In general, as more and more multi-omics studies demonstrate the value of this type of research, funding agencies which traditionally have prioritized hypothesis-driven research are recognizing the value of data-driven research for biological discovery. For example, although large, exploratory multi-omics studies are cost and resource expensive to conduct, there is value in discovering the pathways, mechanisms, and phenotypes of disease and diseases processes, as opposed to conducting multiple one-off studies over the course of decades to identify select parts of the overall picture.
For newer omics technologies, challenges associated with measurement accuracy are ongoing. Specifically, different collection, assay, and analytical techniques can yield different results and/or lack standardization, resulting in inconsistency between different laboratories (Katz et al., 2022; Raffield et al., 2020). It will take time before best practices are established and consensus reached for optimal data compatibility and comparison. In the meantime, researchers need to be acutely aware of the limitations of any new technologies and work with expert collaborators to optimize reproducibility and understand the limitations of the methods used.
For PNI researchers, the potential to advance our understanding of immunology from a systems perspective is a key next step toward understanding associations between neural processes, immune function, psychological factors, and health, and multi-omics approaches can help to identify novel treatment targets and potentially guide the delivery of just-in-time adaptive interventions. However, most PNI research still assesses a few select proteins or uses simplified composite scores created from standardizing and averaging a limited panel of proteins to assess immune system activities (Moriarity et al., 2021). In this context, guided by theoretical insights and biological plausibility, multi-omics data and approaches can provide a more comprehensive, multi-system resource to explore the complexity of immunology and guide future data collection and analytic strategies. Similarly, multi-omics data approaches provide a more powerful platform to conduct foundational descriptive research on the physiometrics (i.e., key measurement properties of biological variables; Moriarity & Alloy, 2021; Segerstrom & Smith, 2012) of both individual facets (e.g., proteins) and broader immunological systems/processes (e.g., markers involved in the acute phase reaction). PNI researchers who specialize in physiometrics or psychometrics may also be a valuable resource for researchers seeking to improve the reliability, validity, and standardization of multi-omics assessments and analysis practices.
3. Conclusion
In conclusion, there are a number of broad recommendations that we believe will help realize the full potential of multi-omics in PNI and health research. First, work in teams and, ideally, in teams that include experts from different omics and academic backgrounds, as doing multi-omics research well requires a lot of conceptual, technical, statistical, and computational expertise. Do not feel as though you or your lab needs to master every part of the process; instead, connect with others at your institution or another institution who have other expertise (i.e., computer science, machine learning) and resources (i.e., Mass Spectrometry, diverse sample access, supplemental funding) to get the job done. Second, pool financial and technical resources, because multi-omics research is both expensive and resource intensive. Third, do not forget the P in PNI. Muti-omics research will be most useful and informative when it is linked to psychological, behavioral, and/or clinical processes, so make sure to assess those constructs with the same thoughtfulness and care you apply to the biological ones. Finally, do not forget the upside. Many research programs are focused on disease risk reduction, which is important, but researching the psychobiological basis of constructs such as belonging, resilience, and thriving is equally, if not more, important, as this work has the potential to reduce pre-clinical and clinical disease processes before they take hold and cause human suffering (Allen et al., 2021; Slavich et al., 2022).
We are now at the point where the technologies of tomorrow have arrived. The future of PNI and health research is here, and our ability to understand how complex, multi-faceted systems work together to influence human health and disease risk on a high-resolution, within-person basis, is within our reach. Given the highly integrative and collaborative nature of our field, we believe no researchers are better poised to take full advantage of multi-omics than those in PNI.
Funding
Preparation of this article was supported by grant #OPR21101 from the California Governor’s Office of Planning and Research/California Initiative to Advance Precision Medicine and a National Institutes of Health (NIH) Grant K08 ES028825. Additionally, DPM was supported by National Research Service Award F32 MH130149. These organizations had no role in planning, writing, editing, or reviewing this article, or in deciding to submit this article for publication.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Data availability
No data were used for the research described in this article.
References
- Acharjee A, Larkman J, Xu Y, Cardoso VR, Gkoutos GV, 2020. A random forest based biomarker discovery and power analysis framework for diagnostics research. BMC Medical Genomics 13 (1), 1–14. 10.1186/s12920-020-00826-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ader R (Ed.), 2006. Psychoneuroimmunology, Two-volume Set, fourth ed. Academic Press. [Google Scholar]
- Allen K-A, Kern ML, Rozek CS, McInerney DM, Slavich GM, 2021. Belonging: A review of conceptual issues, an integrative framework, and directions for future research. Aust. J. Psychol. 73, 87–102. 10.1080/00049530.2021.1883409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amasi-Hartoonian N, Pariante CM, Cattaneo A, Sforzini L, 2022. Understanding treatment-resistant depression using “omics” techniques: A systematic review. J. Affect. Disord. 318, 423–455. 10.1016/j.jad.2022.09.011. [DOI] [PubMed] [Google Scholar]
- Amin N, Liu J, Bonnechere B, MahmoudianDehkordi S, Arnold M, Batra R, Chiou Y-J, Fernandes M, Ikram MA, Kraaij R, Krumsiek J, Newby D, Nho K, Radjabzadeh D, Saykin AJ, Shi L, Sproviero W, Winchester L, Yang Y, Nevado-Holgado AJ, Kastenmüller G, Kaddurah-Daouk R, van Duijn CM, 2023. Interplay of Metabolome and Gut Microbiome in Individuals With Major Depressive Disorder vs Control Individuals. JAMA Psychiat 80 (6), 597. 10.1001/jamapsychiatry.2023.0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antikainen E, Njoum H, Kudelka J, Branco D, Rehman RZU, Macrae V, Davies K, Hildesheim H, Emmert K, Reilmann R, Janneke van der Woude C, Maetzler W, Ng W-F, O’Donnell P, Van Gassen G, Baribaud F, Pandis I, Manyakov NV, van Gils M, Ahmaniemi T, Chatterjee M, 2022. Assessing fatigue and sleep in chronic diseases using physiological signals from wearables: A pilot study. Front. Physiol 13 10.3389/fphys.2022.968185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argelaguet R, Velten B, Arnol D, Dietrich S, Zenz T, Marioni JC, Buettner F, Huber W, Stegle O, 2018. Multi-Omics Factor Analysis—a framework for unsupervised integration of multi-omics data sets. Mol. Syst. Biol. 14 (6) 10.15252/msb.20178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athieniti E, Spyrou GM, 2023. A guide to multi-omics data collection and integration for translational medicine. Comput. Struct. Biotechnol. J. 21, 134–149. 10.1016/j.csbj.2022.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban RH, Kamvissi V, Schulte KM, Bornstein SR, Rubino F, Graessler J, 2014. Lipidomic profiling at the interface of metabolic surgery and cardiovascular disease. Curr. Atheroscler. Rep. 16, 1–9. 10.1007/s11883-014-0455-8. [DOI] [PubMed] [Google Scholar]
- Bartlett DB, Fox O, McNulty CL, Greenwood HL, Murphy L, Sapey E, Goodman M, Crabtree N, Thøgersen-Ntoumani C, Fisher JP, Wagenmakers AJM, Lord JM, 2016. Habitual physical activity is associated with the maintenance of neutrophil migratory dynamics in healthy older adults. Brain Behav. Immun. 56, 12–20. 10.1016/j.bbi.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y, 1995. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. Roy. Stat. Soc.: Ser. B (Methodol.) 57 (1), 289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Bhawal R, Fu Q, Anderson ET, Gibson GE, Zhang S, 2021. Serum metabolomic and lipidomic profiling reveals novel biomarkers of efficacy for benfotiamine in Alzheimer’s disease. Int. J. Mol. Sci. 22 (24), 13188. 10.3390/ijms222413188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland JM, Altman DG, 1995. Multiple significance tests: The Bonferroni method. BMJ 310 (6973), 170. 10.1136/bmj.310.6973.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaguro L, Schulte-Schrepping J, Ulas T, Aschenbrenner AC, Beyer M, Schultze JL, 2022. A guide to systems-level immunomics. Nat. Immunol. 23 (10), 1412–1423. 10.1038/s41590-022-01309-9. [DOI] [PubMed] [Google Scholar]
- Byrne ML, Lind MN, Horn SR, Mills KL, Nelson BW, Barnes ML, Slavich GM, Allen NB, 2021. Using mobile sensing data to assess stress: Associations with perceived and lifetime stress, mental health, sleep, and inflammation. Digital Health 7, 1–11. 10.1177/20552076211037227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K-AL, Rohart F, Gonzalez I, & Dejean S, (2016). MixOmics: Omics Data Integration Project. R package version 6.1.1. https://CRAN.R-project.org/package=mixOmics 10.18129/B9.bioc.mixOmics. [Google Scholar]
- Cao K-A-L, Rossouw D, Robert-Granié C, Besse P, 2008. A Sparse PLS for Variable Selection when Integrating Omics Data. Stat. Appl. Genet. Mol. Biol. 7 (1) 10.2202/1544-6115.1390. [DOI] [PubMed] [Google Scholar]
- Chen R, Mias G, Li-Pook-Than J, Jiang L, Lam HK, Chen R, Miriami E, Karczewski K, Hariharan M, Dewey F, Cheng Y, Clark M, Im H, Habegger L, Balasubramanian S, O’Huallachain M, Dudley J, Hillenmeyer S, Haraksingh R, Sharon D, Euskirchen G, Lacroute P, Bettinger K, Boyle A, Kasowski M, Grubert F, Seki S, Garcia M, Whirl-Carrillo M, Gallardo M, Blasco M, Greenberg P, Snyder P, Klein T, Altman R, Butte AJ, Ashley E, Gerstein M, Nadeau K, Tang H, Snyder M, 2012. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 148 (6), 1293–1307. 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contrepois K, Wu S.i., Moneghetti KJ, Hornburg D, Ahadi S, Tsai M-S, Metwally AA, Wei E, Lee-McMullen B, Quijada JV, Chen S, Christle JW, Ellenberger M, Balliu B, Taylor S, Durrant MG, Knowles DA, Choudhry H, Ashland M, Bahmani A, Enslen B, Amsallem M, Kobayashi Y, Avina M, Perelman D, Schüssler-Fiorenza Rose SM, Zhou W, Ashley EA, Montgomery SB, Chaib H, Haddad F, Snyder MP, 2020. Molecular choreography of acute exercise. Cell 181 (5), 1112–1130.e16. 10.1016/j.cell.2020.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa LNG, Carneiro BA, Alves GS, Silva DHL, Guimaraes DF, Souza LS, Lins-Silva D, 2022. Metabolomics of major depressive disorder: A systematic review of clinical studies. Cureus 14 (3). 10.7759/cureus.23009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts LV, Plans D, Brown AW, Collomosse J, 2020. Deep learning with wearable based heart rate variability for prediction of mental and general health. J. Biomed. Inform. 112 (103610), 1–8. 10.1016/j.jbi.2020.103610. [DOI] [PubMed] [Google Scholar]
- Čuklina J, Pedrioli PGA, Aebersold R, 2020. Review of batch effects prevention, diagnostics, and correction approaches. In: Matthiesen R (Ed.), Mass Spectrometry Data Analysis in Proteomics. Springer, pp. 373–387. 10.1007/978-1-4939-9744-2_16. [DOI] [PubMed] [Google Scholar]
- Čuklina J, Lee CH, Williams EG, Sajic T, Collins BC, Rodríguez Martínez M, Sharma VS, Wendt F, Goetze S, Keele GR, Wollscheid B, Aebersold R, Pedrioli PGA, 2021. Diagnostics and correction of batch effects in large-scale proteomic studies: A tutorial. Mol. Syst. Biol. 17 (8), e10240. 10.15252/msb.202110240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammer EB, Seyfried NT, Johnson ECB, 2023. Batch correction and harmonization of –Omics datasets with a tunable median polish of ratio. Front. Syst. Biol. 3 10.3389/fsysb.2023.1092341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer R, 2009. Cytokine, sickness behavior, and depression. Immunol. Allergy Clin. North Am 29 (2), 247–264. 10.1016/j.iac.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daruna JH, 2012. Introduction to psychoneuroimmunology. Academic Press. 10.1016/B978-0-12-382049-5.00001-2. [DOI] [Google Scholar]
- Dasgupta A, Krasowski M, 2019. Therapeutic drug monitoring data: a concise guide. Academic Press. 10.1016/B978-0-12-815849-4.00001-3. [DOI] [Google Scholar]
- de Zambotti M, Cellini N, Goldstone A, Colrain IM, Baker FC, 2019. Wearable sleep technology in clinical and research settings. Med. Sci. Sports Exerc. 51 (7), 1538–1557. 10.1249/MSS.0000000000001947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond LM, Dehlin AJ, Alley J, 2021. Systemic inflammation as a driver of health disparities among sexually-diverse and gender-diverse individuals. Psychoneuroendocrinology 129 (105215), 1–18. 10.1016/j.psyneuen.2021.105215. [DOI] [PubMed] [Google Scholar]
- Dinan TG, Stanton C, Cryan JF, 2013. Psychobiotics: A Novel Class of Psychotropic. Biol. Psychiatry 74 (10), 720–726. 10.1016/j.biopsych.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Ding J, Blencowe M, Nghiem T, Ha S, Chen Y-W, Li G, Yang X, 2021. Mergeomics 2.0: A web server for multi-omics data integration to elucidate disease networks and predict therapeutics. Nucleic Acids Res. 49 (W1), W375–W387. 10.1093/nar/gkab405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann C, Griffin MA, 2015. Optimal time lags in panel studies. Psychol. Methods 20 (4), 489. 10.1037/met0000041. [DOI] [PubMed] [Google Scholar]
- Dwyer JH, 1983. Statistical models for the social and behavioral sciences. Oxford University Press, USA. [Google Scholar]
- Eicher T, Kinnebrew G, Patt A, Spencer K, Ying K, Ma Q, Machiraju R, Mathé EA, 2020. Metabolomics and Multi-Omics Integration: A Survey of Computational Methods and Resources. Metabolites 10 (5), 202. 10.3390/metabo10050202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Dib R, Periyasamy AG, de Barros JL, França CG, Senefonte FL, Vesentini G, Alves MGO, Rodrigues J.V.d.S., Gomaa H, Gomes Júnior JR, Costa LF, Von Ancken T.d.S., Toneli C, Suzumura EA, Kawakami CP, Faustino EG, Jorge EC, Almeida JD, Kapoor A, 2021. Probiotics for the treatment of depression and anxiety: A systematic review and meta-analysis of randomized controlled trials. Clinical Nutrition ESPEN 45, 75–90. 10.1016/j.clnesp.2021.07.027. [DOI] [PubMed] [Google Scholar]
- Fan Y, Gao Y, Ma Q, Yang Z, Zhao B, He X, Yang J, Yan B, Gao F, Qian L, Wang W, Zhu F, Ma X, 2022. Multi-Omics Analysis Reveals Aberrant Gut-Metabolome-Immune Network in Schizophrenia. Front. Immunol. 13 10.3389/fimmu.2022.812293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Kind T, Cajka T, Hazen SL, Tang WHW, Kaddurah-Daouk R, Irvin MR, Arnett DK, Barupal DK, Fiehn O, 2019. Systematic Error Removal Using Random Forest for Normalizing Large-Scale Untargeted Lipidomics Data. Anal. Chem. 91 (5), 3590–3596. 10.1021/acs.analchem.8b05592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatumo S, Chikowore T, Choudhury A, Ayub M, Martin AR, Kuchenbaecker K, 2022. A roadmap to increase diversity in genomic studies. Nat. Med. 28 (2), 243–250. 10.1038/s41591-021-01672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R, 2010. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw 33 (1), 1–22. 10.18637/jss.v033.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam A, Donohue D, Hoke A, Miller SA, Srinivasan S, Sowe B, Detwiler L, Lynch J, Levangie M, Hammamieh R, Jett M, Chalmers J, 2019. Investigating gene expression profiles of whole blood and peripheral blood mononuclear cells using multiple collection and processing methods. PLoS One 14 (12), e0225137. 10.1371/journal.pone.0225137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemi MS, DiGiulio DB, Contrepois K, Callahan B, Ngo TT, Lee-McMullen B, Aghaeepour N, 2019. Multiomics modeling of the immunome, transcriptome, microbiome, proteome and metabolome adaptations during human pregnancy. Bioinformatics 35 (1), 95–103. 10.1093/bioinformatics/bty537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie SL, Christian LM, Mackos AR, Nolan TS, Gondwe KW, Anderson CM, Hall MW, Williams KP, Slavich GM, 2022. Lifetime stressor exposure, systemic inflammation during pregnancy, and preterm birth among Black American women. Brain Behav. Immun 101, 266–274. 10.1016/j.bbi.2022.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Domínguez R, González-Domínguez Á, Sayago A, Fernández-Recamales Á, 2020. Recommendations and best practices for standardizing the pre-analytical processing of blood and urine samples in metabolomics. Metabolites 10(6), 229, 1–18. 10.3390/metabo10060229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graw S, Chappell K, Washam CL, Gies A, Bird J, Robeson MS, Byrum SD, 2021. Multi-omics data integration considerations and study design for biological systems and disease. Molecular Omics 17 (2), 170–185. 10.1039/D0MO00041H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Graber A, McBurney RN, Balasubramanian R, 2010. Sample size and statistical power considerations in high-dimensionality data settings: A comparative study of classification algorithms. BMC Bioinf. 11 (1), 447. 10.1186/1471-2105-11-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai M-C, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY, 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464 (7291), 1071–1076. 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton CM, Strader LC, Pratt JG, Maiese D, Hendershot T, Kwok RK, Hammond JA, Huggins W, Jackman D, Pan H, Nettles DS, Beaty TH, Farrer LA, Kraft P, Marazita ML, Ordovas JM, Pato CN, Spitz MR, Wagener D, Williams M, Junkins HA, Harlan WR, Ramos EM, Haines J, 2011. The PhenX Toolkit: get the most from your measures. Am. J. Epidemiol. 174 (3), 253–260. 10.1093/aje/kwr193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlat EJ, Laraia B, Bleil ME, Deardorff J, Tomiyama AJ, Mujahid M, Shields GS, Brownell K, Slavich GM, Epel ES, 2022. Effects of early life adversity on pubertal timing and tempo in black and white girls: The National Growth and Health Study. Psychosom. Med. 84, 297–305. 10.1097/PSY.0000000000001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z, Meng C, Li L, Feng S, Zhu Y, Yang J, Han L, Sun L, Lv W, Figeys D, Liu H, 2023. Positive mood-related gut microbiota in a long-term closed environment: A multiomics study based on the “Lunar Palace 365” experiment. Microbiome 11 (1), 88. 10.1186/s40168-023-01506-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin Y, Seldin M, Lusis A, 2017. Multi-omics approaches to disease. Genome Biol. 18 (1), 1–15. 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jatoi I, Sung H, Jemal A, 2022. The Emergence of the Racial Disparity in U.S. Breast-Cancer Mortality. N. Engl. J. Med. 386, 2349–2352. 10.1056/nejmp2200244. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A, 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8 (1), 118–127. 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Jung J, Lee SM, Lee M-J, Ryu J-S, Song J-H, Lee J-E, Kang G, Kwon OS, Park J-Y, 2021. Lipidomics reveals that acupuncture modulates the lipid metabolism and inflammatory interaction in a mouse model of depression. Brain Behav. Immun. 94, 424–436. 10.1016/j.bbi.2021.02.003. [DOI] [PubMed] [Google Scholar]
- Kang M, Ko E, Mersha TB, 2022. A roadmap for multi-omics data integration using deep learning. Brief. Bioinform. 23 (1), bbab454. 10.1093/bib/bbab454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz DH, Robbins JM, Deng S, Tahir UA, Bick AG, Pampana A, Yu Z, Ngo D, Benson MD, Chen Z-Z, Cruz DE, Shen D, Gao Y, Bouchard C, Sarzynski MA, Correa A, Natarajan P, Wilson JG, Gerszten RE, 2022. Proteomic profiling platforms head to head: Leveraging genetics and clinical traits to compare aptamer- and antibody-based methods. Science. Advances 8 (33), eabm5164. 10.1126/sciadv.abm5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, Singh Y, Singh S, Singh RB, 2021. Gut microbiome-mediated epigenetic regulation of brain disorder and application of machine learning for multi-omics data analysis. Genome 64 (4), 355–371. 10.1139/gen-2020-0136. [DOI] [PubMed] [Google Scholar]
- Kellogg RA, Dunn J, Snyder MP, 2018. Personal omics for precision health. Circ. Res. 122, 1169–1171. 10.1161/CIRCRESAHA.117.310909. [DOI] [PubMed] [Google Scholar]
- Kim S, Zhang W, Pak V, Aqua JK, Hertzberg VS, Spahr CM, Slavich GM, Bai J, 2021. How stress, discrimination, acculturation and the gut microbiome affect depression, anxiety and sleep among Chinese and Korean immigrants in the USA: A cross-sectional pilot study protocol. BMJ Open 11 (7), e047281. 10.1136/bmjopen-2020-047281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh EJ, Kim SH, Hwang SY, 2022. Sample management: A primary critical starting point for successful omics studies. Mol. Cell. Toxicol. 18 (2), 141–148. 10.1007/s13273-021-00213-x. [DOI] [Google Scholar]
- Korologou-Linden R, Leyden GM, Relton CL, Richmond RC, Richardson TG, 2021. Multi-omics analyses of cognitive traits and psychiatric disorders highlights brain-dependent mechanisms. Hum. Mol. Genet. 00, 1–12. 10.1093/hmg/ddab016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krassowski M, Das V, Sahu SK, Misra BB, 2020. State of the field in multi-omics research: from computational needs to data mining and sharing. Front. Genet. 11 (610798), 1–17. 10.3389/fgene.2020.610798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo TC, Tian TF, Tseng YJ, 2013. 3Omics: a web-based systems biology tool for analysis, integration and visualization of human transcriptomic, proteomic and metabolomic data. BMC Syst. Biol. 7, 1–15. 10.1186/1752-0509-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Reau AJ, Strom NB, Filvaroff E, Mavrommatis K, Ward TL, Knights D, 2023. Shallow shotgun sequencing reduces technical variation in microbiome analysis. Sci. Rep. 13 (1) 10.1038/s41598-023-33489-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster SM, Sanghi A, Wu S.i., Snyder MP, 2020. A Customizable Analysis Flow in Integrative Multi-Omics. Biomolecules 10 (12), 1606. 10.3390/biom10121606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster SM, Lee-McMullen B, Abbott CW, Quijada JV, Hornburg D, Park H, Perelman D, Peterson DJ, Tang M, Robinson A, Ahadi S, Contrepois K, Hung C-J, Ashland M, McLaughlin T, Boonyanit A, Horning A, Sonnenburg JL, Snyder MP, 2022. Global, distinctive, and personal changes in molecular and microbial profiles by specific fibers in humans. Cell Host Microbe 30 (6), 848–862. e7. 10.1016/j.chom.2022.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautman Z, Lev-Ari S, 2022. The Use of Smart Devices for Mental Health Diagnosis and Care. Journal of. Clin. Med. 11(18), 5359, 1–2. 10.3390/jcm11185359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD, 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28 (6), 882–883. 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei BUW, Prow TW, 2019. A review of microsampling techniques and their social impact. Biomed. Microdevices 21(4), 81, 1–30. 10.1007/s10544-019-0412-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lai J, Zhang P, Ding J, Jiang J, Liu C, Huang H, Zhen H, Xi C, Sun Y, Wu L, Wang L, Gao X, Li Y, Fu Y, Jie Z, Li S, Zhang D, Chen Y, Zhu Y, Lu S, Lu J, Wang D, Zhou H, Yuan X, Li X, Pang L, Huang M, Yang H, Zhang W, Brix S, Kristiansen K, Song X, Nie C, Hu S, 2022. Multi-omics analyses of serum metabolome, gut microbiome and brain function reveal dysregulated microbiota-gut-brain axis in bipolar depression. Mol. Psychiatry 27 (10), 4123–4135. 10.1038/s41380-022-01569-9. [DOI] [PubMed] [Google Scholar]
- Liu T, Petek M, Martinez-Mira C, Balzano-Nogueira L, Ramšak Ž, McIntyre L, Gruden K, Tarazona S, Conesa A, 2022. PaintOmics 4: new tools for the integrative analysis of multi-omics datasets supported by multiple pathway databases. Nucleic Acids Res. 50 (W1), W551–W559. 10.1093/nar/gkac352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López De Maturana E, Alonso L, Alarcón P, Martín-Antoniano IA, Pineda S, Piorno L, Calle ML, Malats N, 2019. Challenges in the Integration of Omics and Non-Omics Data. Genes 10 (3), 238. 10.3390/genes10030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch CMK, Cowan CSM, Bastiaanssen TFS, Moloney GM, Theune N, van de Wouw M, Florensa Zanuy E, Ventura-Silva AP, Codagnone MG, Villalobos-Manríquez F, Segalla M, Koc F, Stanton C, Ross P, Dinan TG, Clarke G, Cryan JF, 2023. Critical windows of early-life microbiota disruption on behaviour, neuroimmune function, and neurodevelopment. Brain Behav. Immun 108, 309–327. 10.1016/j.bbi.2022.12.008. [DOI] [PubMed] [Google Scholar]
- Maes M, Berk M, Goehler L, Song C, Anderson G, Gałecki P, Leonard B, 2012. Depression and sickness behavior are Janus-faced responses to shared inflammatory pathways. BMC Med. 10 (66) 10.1186/1741-7015-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, Menon M, He L, Abdurrob F, Jiang X, Martorell AJ, Ransohoff RM, Hafler BP, Bennett DA, Kellis M, Tsai L-H, 2019. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570 (7761), 332–337. 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer SE, Guan J, Lin J, Hamlat E, Parker JE, Brownell K, Price C, Mujahid M, Tomiyama AJ, Slavich GM, Laraia BA, Epel ES, 2023. Intergenerational effects of maternal lifetime stressor exposure on offspring telomere length in Black and White women. Psychological Medicine 1–12. 10.1017/S0033291722003397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazereeuw G, Herrmann N, Bennett SAL, Swardfager W, Xu H, Valenzuela N, Fai S, Lanctôt KL, 2013. Platelet activating factors in depression and coronary artery disease: a potential biomarker related to inflammatory mechanisms and neurodegeneration. Neurosci. Biobehav. Rev. 37 (8), 1611–1621. 10.1016/j.neubiorev.2013.06.010. [DOI] [PubMed] [Google Scholar]
- Meng C, Kuster B, Culhane AC, Gholami AM, 2014. A multivariate approach to the integration of multi-omics datasets. BMC Bioinf. 15 (1), 162. 10.1186/1471-2105-15-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalik A, Chapman J, Adams RA, Winter NR, Ferreira FS, Shawe-Taylor J, Mourão-Miranda J, 2022. Canonical correlation analysis and partial least squares for identifying brain-behavior associations: A tutorial and a comparative study. Biol. Psych. Cognit. Neurosci. Neuroimag. 7 (11), 1055–1067. 10.1016/j.bpsc.2022.07.012. [DOI] [PubMed] [Google Scholar]
- Mirza B, Wang W, Wang J, Choi H, Chung NC, Ping P, 2019. Machine learning and integrative analysis of biomedical big data. Genes 10(2), 87, 1–30. 10.3390/genes10020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misiewicz Z, Iurato S, Kulesskaya N, Salminen L, Rodrigues L, Maccarrone G, Martins J, Czamara D, Laine MA, Sokolowska E, Trontti K, Rewerts C, Novak B, Volk N, Park DI, Jokitalo E, Paulin L, Auvinen P, Voikar V, Chen A, Erhardt A, Turck CW, Hovatta I, Flint J, 2019. Multi-omics analysis identifies mitochondrial pathways associated with anxiety-related behavior. PLoS Genet. 15 (9), e1008358. 10.1371/journal.pgen.1008358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra N, Clavaud C, Guinot F, Bourokba N, Nouveau S, Mezzache S, Palazzi P, Appenzeller BMR, Tenenhaus A, Leung MHY, Lee PKH, Bastien P, Aguilar L, Cavusoglu N, 2021. Multi-omics analysis to decipher the molecular link between chronic exposure to pollution and human skin dysfunction. Sci. Rep. 11 (1) 10.1038/s41598-021-97572-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarity DP, Alloy LB, 2021. Back to basics: The importance of measurement properties in biological psychiatry. Neurosci. Biobehav. Rev. 123, 72–82. 10.1016/j.neubiorev.2021.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarity DP, Ellman LM, Coe CL, Olino TM, Alloy LB, 2021. A physiometric investigation of inflammatory composites: Comparison of “a priori” aggregates, empirically-identified factors, and individual proteins. Brain Behav. Immun. Health 18 (100391), 1–10. 10.1016/j.bbih.2021.100391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarity DP, Slavich GM, 2023. The future is dynamic: A call for intensive longitudinal data in immunopsychiatry. Brain Behav. Immun. 112, 118–124. 10.1016/j.bbi.2023.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu ES, Gritsenko M, Piehowski PD, Gao Y, Orton DJ, Schepmoes AA, Fillmore TL, Frohnert BI, Rewers M, Krischer JP, Ansong C, Suchy-Dicey AM, Evans-Molina C, Qian W-J, Webb-Robertson B-J, Metz TO, 2021. Tutorial: Best practices and considerations for mass-spectrometry-based protein biomarker discovery and validation. Nat. Protoc. 16 (8), 3737–3760. 10.1038/s41596-021-00566-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman DC, Lila MA, Gillitt ND, 2019. Immunometabolism: a multi-omics approach to interpreting the influence of exercise and diet on the immune system. Annu. Rev. Food Sci. Technol. 10 (1), 341–363. 10.1146/annurev-food-032818-121316. [DOI] [PubMed] [Google Scholar]
- Noonan S, Zaveri M, Macaninch E, Martyn K, 2020. Food & mood: a review of supplementary prebiotic and probiotic interventions in the treatment of anxiety and depression in adults. BMJ Nutrit. Prevent. Health 3 (2), 351–362. 10.1136/bmjnph-2019-000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombo T, Campos A, Vernon SD, Roundy S, 2020. Accurate and objective determination of myalgic encephalomyelitis/chronic fatigue syndrome disease severity with a wearable sensor. J. Transl. Med. 18 (1), 1–9. 10.1186/s12967-020-02583-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z, Zhou G, Ewald J, Chang L, Hacariz O, Basu N, Xia J, 2022. Using MetaboAnalyst 5.0 for LC–HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 17 (8), 1735–1761. 10.1038/s41596-022-00710-w. [DOI] [PubMed] [Google Scholar]
- Pappa E, Kousvelari E, Vastardis H, 2019. Saliva in the “Omics” era: A promising tool in paediatrics. Oral Dis. 25 (1), 16–25. 10.1111/odi.12886. [DOI] [PubMed] [Google Scholar]
- Patke A, Young MW, Axelrod S, 2020. Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 21, 67–84. 10.1038/s41580-019-0179-2. [DOI] [PubMed] [Google Scholar]
- Picard M, Scott-Boyer M-P, Bodein A, Périn O, Droit A, 2021. Integration strategies of multi-omics data for machine learning analysis. Comput. Struct. Biotechnol. J. 19, 3735–3746. 10.1016/j.csbj.2021.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popejoy AB, Fullerton SM, 2016. Genomics is failing on diversity. Nature 538 (7624), 161–164. 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffield LM, Dang H, Pratte KA, Jacobson S, Gillenwater LA, Ampleford E, Barjaktarevic I, Basta P, Clish CB, Comellas AP, Cornell E, Curtis JL, Doerschuk C, Durda P, Emson C, Freeman CM, Guo X, Hastie AT, Hawkins GA, Herrera J, Johnson WC, Labaki WW, Liu Y, Masters B, Miller M, Ortega VE, Papanicolaou G, Peters S, Taylor KD, Rich SS, Rotter JI, Auer P, Reiner AP, Tracy RP, Ngo D, Gerszten RE, O’Neal WK, Bowler RP, 2020. Comparison of proteomic assessment methods in multiple cohort studies. Proteomics 20 (12), 1900278. 10.1002/pmic.201900278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D, 2015. Methods of integrating data to uncover genotype–phenotype interactions. Nat Rev Genet 16 (2), 85–97. 10.1038/nrg3868. [DOI] [PubMed] [Google Scholar]
- Ritsert F, Elgendi M, Galli V, Menon C, 2022. Heart and breathing rate variations as biomarkers for anxiety detection. Bioengineering 9 (711), 1–9. 10.3390/bioengineering9110711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotroff DM, Corum DG, Motsinger-Reif A, Fiehn O, Bottrel N, Drevets WC, Singh J, Salvadore G, Kaddurah-Daouk R, 2016. Metabolomic signatures of drug response phenotypes for ketamine and esketamine in subjects with refractory major depressive disorder: new mechanistic insights for rapid acting antidepressants. Transl. Psychiatry 6 (9), e894–e. 10.1038/tp.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanarayanan A, Mueller TT, Ali Moni M, Schueler K, Baune BT, Lio P, Mehta D, Baune BT, Dierssen M, Ebert B, Fabbri C, Fusar-Poli P, Gennarelli M, Harmer C, Howes OD, Janzing JGE, Lio P, Maron E, Mehta D, Minelli A, Nonell L, Pisanu C, Potier M-C, Rybakowski F, Serretti A, Squassina A, Stacey D, van Westrhenen R, Xicota L, 2023. Multi-omics data integration methods and their applications in psychiatric disorders. Eur. Neuropsychopharmacol. 69, 26–46. 10.1016/j.euroneuro.2023.01.001. [DOI] [PubMed] [Google Scholar]
- Sauceda C, Bayne C, Sudqi K, Gonzalez A, Dulai PS, Knight R, Gonzalez DJ, Gonzalez CG, 2022. Stool multi-omics for the study of host-microbe interactions in inflammatory bowel disease. Gut Microbes 14 (1), 1–19. 10.1080/19490976.2022.2154092, 2154092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxe GN, Ma S, Ren J, Aliferis C, 2017. Machine learning methods to predict child posttraumatic stress: A proof of concept study. BMC Psychiatry 17 (1), 223. 10.1186/s12888-017-1384-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert KO, Stacey D, Arentz G, Clark SR, Air T, Hoffmann P, Baune BT, 2018. Targeted proteomic analysis of cognitive dysfunction in remitted major depressive disorder: Opportunities of multi-omics approaches towards predictive, preventive, and personalized psychiatry. J. Proteomics 188, 63–70. 10.1016/j.jprot.2018.02.023. [DOI] [PubMed] [Google Scholar]
- Schüssler-Fiorenza Rose SM, Contrepois K, Moneghetti KJ, Zhou W, Mishra T, Mataraso S, Dagan-Rosenfeld O, Ganz AB, Dunn J, Hornburg D, Rego S, Perelman D, Ahadi S, Sailani MR, Zhou Y, Leopold SR, Chen J, Ashland M, Christle JW, Avina M, Limcaoco P, Ruiz C, Tan M, Butte AJ, Weinstock GM, Slavich GM, Sodergren E, McLaughlin TL, Haddad F, Snyder MP, 2019. A longitudinal big data approach for precision health. Nat. Med. 25 (5), 792–804. 10.1038/s41591-019-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RA, Scott LJ, Mägi R, Marullo L, Gaulton KJ, Kaakinen M, Franks PW, 2017. An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes 66 (11), 2888–2902. 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segerstrom SC, Smith GT, 2012. Methods, Variance, and Error in Psychoneuroimmunology Research: The Good, the Bad, and the Ugly. Oxford University Press, pp. 421–432. [Google Scholar]
- Shen X, Kellogg R, Panyard DJ, Bararpour N, Castillo KE, Lee-McMullen B, Delfarah A, Ubellacker J, Ahadi S, Rosenberg-Hasson Y, Ganz A, Contrepois K, Michael B, Simms I, Wang C, Hornburg D, Snyder MP, 2023. Multi-omics microsampling for the profiling of lifestyle-associated changes in health. Nat. Biomed. Eng. 10.1038/s41551-022-00999-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sichko S, Bui TQ, Vinograd M, Shields GS, Saha K, Devkota S, Olvera-Alvarez HA, Carroll JE, Cole SW, Irwin MR, Slavich GM, 2021. Psychobiology of Stress and Adolescent Depression (PSY SAD) Study: Protocol overview for an fMRI-based multi-method investigation. Brain Behav. Immun. Health 17, 100334. 10.1016/j.bbih.2021.100334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A, 2019. Cancer statistics, 2019. CA A Cancer J. Clin 69 (1), 7–34. 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- Simons RL, Lei M-K, Klopack E, Zhang Y, Gibbons FX, Beach SRH, 2021. Racial discrimination, inflammation, and chronic illness among african american women at midlife: support for the weathering perspective. J. Racial Ethn. Health Disparities 8 (2), 339–349. 10.1007/s40615-020-00786-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavich GM, 2020a. Psychoneuroimmunology of stress and mental health. In: Harkness KL, Hayden EP (Eds.), The Oxford Handbook of Stress and Mental Health. Oxford University Press, New York, pp. 519–546. 10.1093/oxfordhb/9780190681777.013.24. [DOI] [Google Scholar]
- Slavich GM, 2020b. Social safety theory: A biologically based evolutionary perspective on life stress, health, and behavior. Annu. Rev. Clin. Psychol. 16, 265–295. 10.1146/annurev-clinpsy-032816-045159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavich GM, 2022. Social Safety Theory: Understanding social stress, disease risk, resilience, and behavior during the COVID-19 pandemic and beyond. Curr. Opin. Psychol. 45, 101299 10.1016/j.copsyc.2022.101299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavich GM, Auerbach RP, 2018. Stress and its sequelae: Depression, suicide, inflammation, and physical illness. In: Butcher JN, Hooley JM (Eds.), APA handbook of psychopathology: Vol. 1. Psychopathology: Understanding, assessing, and treating adult mental disorders. American Psychological Association, Washington, DC, pp. 375–402. 10.1037/0000064-016. [DOI] [Google Scholar]
- Slavich GM, Roos LG, Zaki J, 2022. Social belonging, compassion, and kindness: Key ingredients for fostering resilience, recovery, and growth from the COVID-19 pandemic. Anxiety Stress Coping 35, 1–8. 10.1080/10615806.2021.1950695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavich GM, Mengelkoch S, Cole SW, 2023a. Human social genomics: Concepts, mechanisms, and implications for health. Lifestyle Medicine 4(2), e75, 1–15. 10.1002/lim2.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavich GM, Roos LG, Mengelkoch S, Webb CA, Shattuck EC, Moriarity DP, Alley JC, 2023b. Social Safety Theory: Conceptual foundation, underlying mechanisms, and future directions. Health Psychol. Rev. 17, 5–59. 10.1080/17437199.2023.2171900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R, 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. 100 (16), 9440–9445. 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian I, Verma S, Kumar S, Jere A, Anamika K, 2020. Multi-omics data integration, interpretation, and its application, 1177932219899051 Bioinf. Biol. Insights 14, 1–24. 10.1177/1177932219899051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S, Balzano-Nogueira L, Gómez-Cabrero D, Schmidt A, Imhof A, Hankemeier T, Tegnér J, Westerhuis JA, Conesa A, 2020. Harmonization of quality metrics and power calculation in multi-omic studies. Nature. Communications 11 (1). 10.1038/s41467-020-16937-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R, 1996. Regression Shrinkage and Selection Via the Lasso. J. Roy. Stat. Soc.: Ser. B (Methodol.) 58 (1), 267–288. 10.1111/j.2517-6161.1996.tb02080.x. [DOI] [Google Scholar]
- Toussaint LL, Moriarity DP, Kamble S, Williams DR, Slavich GM, 2022. Inflammation and depression symptoms are most strongly associated for Black adults. Brain, Behavior, and Immunity –. Health 26, 100552. 10.1016/j.bbih.2022.100552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsamardinos I, 2022. Don’t lose samples to estimation. Patterns 3 (12). 10.1016/j.patter.2022.100612, 100612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugidos M, Tarazona S, Prats-Montalbán JM, Ferrer A, Conesa A, 2020. MultiBaC: A strategy to remove batch effects between different omic data types. Stat. Methods Med. Res. 29 (10), 2851–2864. 10.1177/0962280220907365. [DOI] [PubMed] [Google Scholar]
- Ugidos M, Nueda MJ, Prats-Montalbán JM, Ferrer A, Conesa A, Tarazona S, 2022. MultiBaC: An R package to remove batch effects in multi-omic experiments. Bioinformatics 38 (9), 2657–2658. 10.1093/bioinformatics/btac132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabalas A, Gowen E, Poliakoff E, Casson AJ, Hernandez-Lemus E, 2019. Machine learning algorithm validation with a limited sample size. PLoS One 14 (11), e0224365. 10.1371/journal.pone.0224365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahabi N, Michailidis G, 2022. Unsupervised multi-omics data integration methods: a comprehensive review. Front. Genet. 13, 854752 10.3389/fgene.2022.854752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wastyk HC, Fragiadakis GK, Perelman D, Dahan D, Merrill BD, Yu FB, Topf M, Gonzalez CG, Van Treuren W, Han S, Robinson JL, Elias JE, Sonnenburg ED, Gardner CD, Sonnenburg JL, 2021. Gut-microbiota-targeted diets modulate human immune status. Cell 184 (16), 4137–4153.e14. 10.1016/j.cell.2021.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wes PD, Holtman IR, Boddeke EWGM, Möller T, Eggen BJL, 2016. Next generation transcriptomics and genomics elucidate biological complexity of microglia in health and disease. Glia 64 (2), 197–213. 10.1002/glia.22866. [DOI] [PubMed] [Google Scholar]
- Wieder C, Frainay C, Poupin N, Rodríguez-Mier P, Vinson F, Cooke J, Lai RPJ, Bundy JG, Jourdan F, Ebbels T, Patil KR, 2021. Pathway analysis in metabolomics: Recommendations for the use of over-representation analysis. PLoS Comput. Biol. 17 (9), e1009105. 10.1371/journal.pcbi.1009105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DR, Rucker TD, 2000. Understanding and addressing racial disparities in health care. Health Care Financ. Rev. 21 (4), 75–90. [PMC free article] [PubMed] [Google Scholar]
- Witten DM, Tibshirani RJ, 2009. Extensions of sparse canonical correlation analysis with applications to genomic data. Stat. Appl. Genet. Mol. Biol. 8 (1), 1–27. 10.2202/1544-6115.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten DM, Tibshirani R, Hastie T, 2009. A penalized matrix decomposition, with applications to sparse principal components and canonical correlation analysis. Biostatistics 10 (3), 515–534. 10.1093/biostatistics/kxp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Zhong Q, Wu W, Chen J, 2023. Multi-omics data reveals the important role of glycerophospholipid metabolism in the crosstalk between gut and brain in depression. J. Transl. Med. 21 (1), 93. 10.1186/s12967-023-03942-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Parmigiani G, Johnson WE, 2020. ComBat-seq: Batch effect adjustment for RNA-seq count data. lqaa078 NAR Gen. Bioinform. 2 (3). 10.1093/nargab/lqaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Jin K, Jiang C, Pan F, Wu J, Luan H, Zhao Z, Chen J, Mou T, Wang Z, Lu J, Lu S, Hu S, Xu Y, Huang M, 2022. A pilot exploration of multi-omics research of gut microbiome in major depressive disorders. Transl. Psych. 12 (1), Article 1. 10.1038/s41398-021-01769-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Pang Z, Lu Y, Ewald J, Xia J, 2022. OmicsNet 2.0: A web-based platform for multi-omics integration and network visual analytics. Nucleic Acids Res. 50 (W1), W527–W533. 10.1093/nar/gkac376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data were used for the research described in this article.


