Abstract
Primary features of the flavivirus Kunjin (KUN) subgenomic replicons include continuous noncytopathic replication in host cell cytoplasm and the ability to be encapsidated into secreted virus-like particles (VLPs). Previously we reported preparation of RNA-based KUN replicon vectors and expression of heterologous genes (HG) in cell culture after RNA transfection or after infection with recombinant KUN VLPs (A. N. Varnavski and A. A. Khromykh, Virology 255:366–375, 1999). In this study we describe the development of the next generation of KUN replicon vectors, which allow synthesis of replicon RNA in vivo from corresponding plasmid DNAs. These DNA-based vectors were able to direct stable expression of β-galactosidase (β-Gal) in several mammalian cell lines, and expression remained high (∼150 pg per cell) throughout cell passaging. The applicability of these vectors in vivo was demonstrated by β-Gal expression in the mouse lung epithelium for at least 8 weeks after intranasal inoculation and induction of anti-β-Gal antibody response after intramuscular inoculation of the β-Gal-encoding KUN replicon DNA. The noncytopathic nature of DNA-based KUN replicon vectors combined with high-level and stability of HG expression in a broad range of host cells should prove them to be useful in a variety of applications in vitro and in vivo.
Development of gene expression vectors based on subgenomic replicons of positive-strand RNA viruses has gained much attention over the last decade (11, 32). Genomes of the alphaviruses Semliki Forest virus (SFV) (7, 29), Sindbis (SIN) virus (1, 10, 15) and Venezuelan equine encephalitis virus (12, 37), as well as the poliovirus genome (34, 36), have all been used. An important characteristic feature of these systems is the ability of replicon RNA to self-replicate, thereby amplifying the input template in the host cell. This amplification in turn leads to increased production of encoded proteins. Replicon RNAs can be delivered into host cells by direct transfection with RNA transcripts produced in vitro from corresponding plasmid DNAs (20, 47) or by infection with virus-like particles (VLPs) containing encapsidated replicon RNAs (10, 12, 29, 37). Alternatively, they can be transcribed from transfected replicon-encoded plasmid DNAs utilizing cellular RNA polymerase II transcription machinery (1, 7, 14, 15). It was shown that replicon-based DNA vectors produced higher levels of encoded heterologous proteins than conventional plasmid DNA expression vectors and also elicited greatly enhanced immune responses (7, 19).
Applications for most of the alphavirus and poliovirus replicon vectors have been limited to only short-term transient expression due to the cytopathic effects (CPE) induced by vector replication in mammalian cells (18). To address this problem, noncytopathic SIN virus replicon-based vectors containing the puromycin resistance gene were developed by isolation of SIN replicon mutants adapted to puromycin selection in BHK cells (1, 17). However, the use of these vectors is restricted by a number of limitations, such as a narrow host range, relatively low levels of heterologous gene (HG) expression, and some instability of expression in cell populations during passaging (1).
We have been developing a gene expression system based on subgenomic replicons of another RNA virus, the flavivirus Kunjin (KUN), containing deletions in the structural region of the genome (25). In contrast to the alphavirus and poliovirus replicons, as well as full-length KUN RNA, replication of KUN replicons in mammalian cell cultures did not produce any apparent CPE (25). Recently we reported the construction and use of RNA-based KUN replicon vectors for HG expression in cell culture after the direct transfection of in vitro-synthesized recombinant KUN replicon RNAs or after infection with recombinant KUN VLPs (43). In this study we describe the development of DNA-based KUN replicon vectors and demonstrate their ability to direct high-level prolonged HG expression in a range of cell lines and in vivo. Moreover, we show the induction of antibody response against a KUN vector-encoded HG after immunization of mice with the corresponding KUN replicon DNA construct. These noncytopathic DNA-based KUN replicon expression vectors should be useful for a variety of applications both in vitro and in vivo.
MATERIALS AND METHODS
Cells.
BHK21 (baby hamster kidney), Vero (green African monkey kidney), HepG2 (human hepatocarcinoma), HeLa (human cervical epitheloid carcinoma), A172 (human glioblastoma), and 293 (transformed human embryonal kidney) cells were grown in Dulbecco's minimal essential medium (Gibco BRL) supplemented with 10% fetal bovine serum. HEp-2 (human larynx epidermoid carcinoma) cells were grown in RPMI 1640 medium supplemented with 15 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) and 5% fetal bovine serum.
Plasmid construction.
All the molecular constructs were prepared by using standard molecular biology techniques (3), and their sequences were confirmed by restriction digest analysis and/or sequencing. All the PCR amplifications for subsequent cloning were performed with high-fidelity Pfu DNA polymerase (Stratagene). The sequences of primers used in preparation of KUN replicon vectors and constructs are shown in Table 1.
TABLE 1.
Primers used in preparation of KUN replicon vectors and constructs
| Primer | Sequence (5′→3′)a | Restriction site used |
|---|---|---|
| Ub_F | ggttctagatgcatacgcgttaCAGATATTCGTGAAGACT | XbaI |
| Ub_R | ggtactagtacgtaaggcgcGCCACCACGGAGACG | SpeI |
| NS5dGDD_F | CTG GTT AAC TGT GTG GTA AAG CCC TT | |
| 3′UTRHDV | GAGAACACAGGATCTGGGTCGGCATGGCATCT | |
| SV40Pa_R | ggcctcgaGCAATTGTTGTTGTTAACTT | XhoI |
| CMV_F | gcgcttaaGACATTGATTATTGACTAGTTA | |
| CMV5′UTR | CGTTTAGTGAACCGAGTAGTTCGCCTGTGTGA | |
| FMDV2A_R | gtgacgcgtcggccGGGCCCTGGGTTGGA | EagI |
| PacPst_F | cgccctgcagccACCGAGTACAAGCCCA | PstI |
| 2AMluNsi_R | ctggatgcatacgcgtcGGGCCCTGGGTTG | NsiI |
| NsiIRES_F | ggtatgcatagCGGGATCAATT | NsiI |
| dNS1H3_R | ccgaagcttgTCCAGTATCAGCATGCA | SphI |
| NsiLacZ_F | ccggatgcatggGTCGTCTTACAGCGTCGT | NsiI |
| NsiLacZ_R | cgcgatgcatgTTTTTGACACCAGACTAACT | NsiI |
Nucleotides in capital letters represent the authentic KUN or cloned heterologous sequences (defined in the text), nucleotides in lowercase letters show artificial sequences, and nucleotides in boldface show introduced restriction sites used in cloning.
(i) RNA-based KUN replicon constructs.
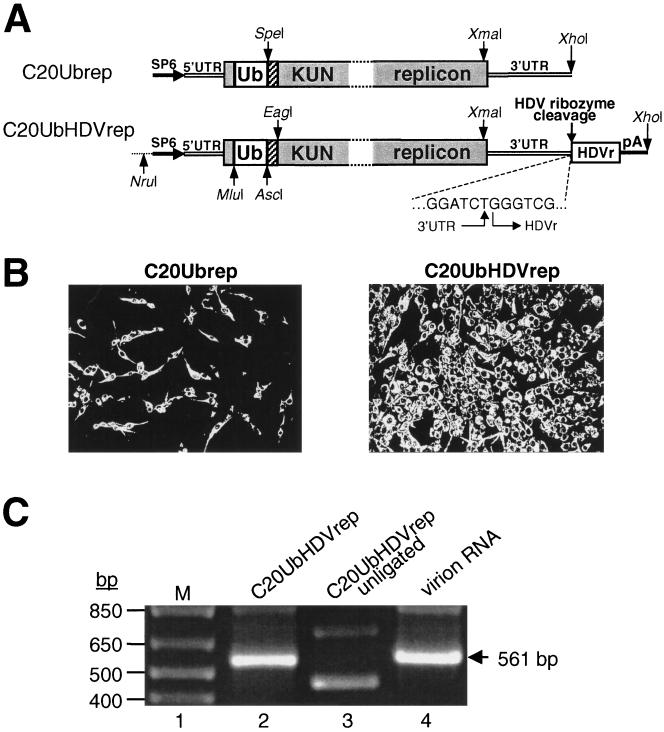
The C20Ubrep plasmid was prepared by cloning the mouse Ubiquitin (Ub) gene sequence (16), PCR amplified from the plasmid pRB269 (5) by using Ub_F and Ub_R primers with incorporated XbaI and SpeI restriction sites, respectively (Table 1), into the SpeI site of the previously reported C20DX2Arep plasmid (43) (Fig. 1A). To construct the C20UbHDVrep plasmid, the hepatitis delta virus antigenomic ribozyme (HDVr) sequence (35) followed by the simian virus 40 (SV40) polyadenylation signal (pA) was inserted immediately downstream of the last nucleotide of the KUN replicon sequence. The fragment containing the last 1,331 nucleotides of the KUN replicon sequence followed by the HDVr/SV40-pA cassette was produced in a fusion PCR (23) using NS5dGDD_F, 3′UTRHDV, and SV40pA_R primers (Table 1) and two plasmid templates, pTMSV5A (obtained from Tom Macnaughton, Sir Albert Sakzewski Virus Research Centre, Brisbane, Australia) and C20DXrep (26). The resulting PCR product was digested with XmaI (5′ end; KUN NS5 sequence) and XhoI (3′ end) and cloned into the XmaI-XhoI-digested C20Ubrep DNA, producing the C20UbHDVrep construct (see Fig. 1A).
FIG. 1.
Incorporation of the HDVr sequence into KUN replicons. (A) RNA-based KUN replicon constructs. Filled boxes represent translated regions of the KUN replicon, as for the previously described C20DX2Arep plasmid (43). SP6, the SP6 RNA polymerase promoter. 5′UTR and 3′UTR represent the KUN 5′ and 3′ untranslated regions, respectively. Ub, mouse Ubiquitin gene (16). Hatched boxes indicate the FMDV-2A sequence (40). HDVr, the HDVr sequence (35) with its cleavage site indicated by an arrow. pA, the SV40-derived polyadenylation signal sequence. Restriction sites used in construct preparation are as shown. Sequence of the 3′ UTR-HDVr junction, confirmed by sequencing, is indicated. (B) IF analysis with KUN anti-NS3 antibodies of BHK21 cells 24 h after transfection with equal amounts of C20Ubrep or C20UbHDVrep in vitro-transcribed RNAs. In vitro transcription and transfection of KUN replicon RNA, as well as anti-NS3 IF analysis, were performed as described previously (24, 25). (C) RT-PCR analysis of the KUN RNA transcripts. The RNA templates were either in vitro transcribed from the C20UbHDVrep plasmid DNA (as described in reference 24 with omission of the synthetic cap analogue; lanes 2 and 3) or extracted from purified KUN virus grown in Vero cells and decapped (24; lane 4). The RNA templates were purified by phenol-chloroform extraction and ethanol precipitation and were self-ligated with T4 RNA ligase (Pharmacia) at 17°C for 12 h in the reaction buffer (50 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 10 mM ATP, 60 μg of bovine serum albumin/ml) (lanes 2 and 4). The resulting circular RNAs were then RT-PCR amplified across the 3′ end-5′ end junction by using One-Step RT-PCR System (Gibco BRL), essentially as described by the manufacturer. The primers for RT-PCR were forward primer 5′-GCTGCGAAGTGATCCATGTAA-3′ (representing nucleotides 10582 to 10603 of the KUN 3′UTR sequence; 24) and reverse primer 5′-GGGCCCTCCTGGTTTCTT-3′ (complementary to nucleotides 119 to 102 of the KUN core-5′UTR region; 24). The size of the HDVr-pA sequence was 279 bp, and the expected sizes of RT-PCR products were 561 bp and/or 840 bp, depending on whether the HDVr cleavage occurred or not. Lane 3, the product of RT-PCR from unligated in vitro-transcribed C20UbHDVrep RNA (negative control). The ∼450- and ∼700-bp fragments in lane 3 represent nonspecific amplification products. Lane 4 (see above), a positive control for the experiment; lane 1, 1-kb Plus DNA molecular size marker (Gibco BRL).
(ii) DNA-based KUN replicon constructs.
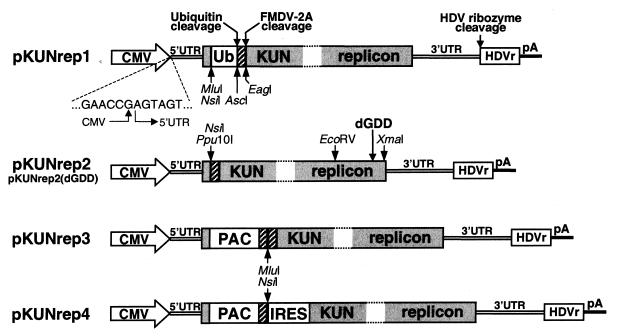
The pKUNrep1 plasmid was prepared by replacing the SP6 promoter in the RNA-based construct C20UbHDVrep with the cytomegalovirus (CMV)-derived immediate-early enhancer/promoter region (8). The fragment containing the CMV sequence followed by the 5′ end of the KUN replicon sequence was produced in a fusion PCR (23) using CMV_F, CMV5′UTR, and FMDV2A_R primers (Table 1) and the plasmid templates pCI (Promega) and C20Ubrep. The resulting PCR product was digested with EagI (3′ end) and cloned into the C20UbHDVrep plasmid digested with the NruI and EagI endonucleases, producing the pKUNrep1 vector (see Fig. 1A and 2). The pKUNrep2 vector was prepared by deletion of the Ub gene from the pKUNrep1 vector, i.e., by digestion with the AscI and MluI endonucleases and subsequent religation (Fig. 2). The pKUNrep2(dGDD) construct with the deletion of the RNA polymerase motif GDD in the NS5 gene (designated as dGDD) was prepared by replacing the EcoRV-XmaI restriction fragment in the pKUNrep2 plasmid with the EcoRV-XmaI fragment derived from the FLdGDD plasmid (26). The pKUNrep3 vector was constructed by cloning the puromycin N-acetyltransferase (PAC; 42) gene followed by the foot-and-mouth disease virus autoprotease 2A (FMDV-2A; 40) sequence into the NsiI site of the pKUNrep2 vector (Fig. 2). The cloned PAC-FMDV-2A fragment was PCR amplified from the C20/GFPpac/2Arep plasmid (to be described elsewhere) by using PacPst_F and 2AMluNsi_R primers with incorporated PstI and NsiI restriction sites, respectively (Table 1). The pKUNrep4 vector was prepared by substituting the second FMDV-2A sequence in the pKUNrep3 vector with the encephalomyelocarditis virus internal ribosome entry site (IRES) sequence, PCR amplified from the C20DX/CAT/IRESrep plasmid (43) by using NsiIRES_F and dNS1H3_R primers with incorporated NsiI and SphI restriction sites, respectively (Table 1). The Escherichia coli β-galactosidase (β-Gal) gene, which was PCR amplified from the C20DX/β-gal/2Arep construct (43) by using NsiLacZ_F and NsiLacZ_R primers with incorporated NsiI restriction sites (Table 1), was cloned as a reporter gene into the NsiI sites of the pKUNrep2, pKUNrep2(dGDD), pKUNrep3, and pKUNrep4 vectors, producing the pKUNβrep2, pKUNβrep2(dGDD), pKUNβrep3, and pKUNβrep4 constructs, respectively.
FIG. 2.
DNA-based KUN replicon vectors. Most of the designations are as in Fig. 1. CMV, the eukaryotic CMV-derived immediate-early enhancer/promoter region (8). Ub- and FMDV-2A-mediated cleavages are indicated by arrows. Sequence of the CMV-5′UTR junction, confirmed by sequencing, is shown. dGDD with an arrow (in pKUNrep2) indicates the position of the deletion of the RNA polymerase motif GDD (26). PAC, the PAC gene (42). IRES, the sequence of the encephalomyelocarditis virus IRES. Restriction sites used in construct preparation are as shown.
DNA transfection and selection of stably expressing cell cultures.
Plasmid DNAs were transfected with FuGENE 6 transfection reagent (Boehringer Mannheim) essentially as described by the manufacturer. For HG expression and stable cell line selection, ∼0.8 μg of DNA was used with 2 μl of FuGENE 6 to transfect ∼1.3 × 105 cells in 16-mm-diameter wells (of a 24-well cell culture plate). Cells transfected with the plasmids containing the PAC gene, a selection marker conferring resistance to the antibiotic puromycin, were incubated in the appropriate growth medium for ∼48 h after transfection, followed by their selection with puromycin (Sigma) added to the medium at 1 to 5 μg/ml.
Metabolic labeling, RIP, and Northern blot hybridization.
Metabolic labeling of BHK21 cells in 35-mm-diameter dishes with [35S]methionine-cysteine at ∼30 h after transfection with ∼2 μg of pKUNrep2 or pKUNrep2(dGDD) plasmid DNAs was performed essentially as described previously (27). Actinomycin D (ACD) was added, where indicated, for 1 h prior to radiolabeling at 10 μg/ml and during the 1-h labeling at 3 μg/ml. Radioimmunoprecipitation (RIP) analysis of the labeled cell lysates with KUN anti-NS3 antibodies was performed as previously described (27). Northern blot hybridization of 10 μg of total RNA isolated from BHK21 cells at ∼36 h after transfection with pKUNrep2 or pKUNrep2(dGDD) plasmids was performed with the [32P]dCTP-labeled probe representing the 3′-terminal 761 nucleotides of the KUN cDNA (24) as described previously (25, 27).
In situ β-Gal reaction staining and β-Gal assay.
Expression of β-Gal in transfected cells was detected by staining cell monolayers with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) at 30 to 40 h after transfection with corresponding β-Gal-encoding constructs or by determination of β-Gal activity in lysates of these cells by using the β-Galactosidase Enzyme Assay System (Promega, Madison, Wis.) essentially as described by the manufacturer.
In vivo DNA administration and antibody titration.
Six- to 9-week-old female BALB/c mice anesthetized by intraperitoneal injection with ketamine-xylazine were inoculated intranasally with ∼5 μg of the pKUNβrep2 DNA complexed with 15 μl of FuGENE 6 transfection reagent (Boehringer Mannheim) in a total volume of 100 μl. Primary and booster intramuscular (i.m.) immunizations were performed by injection of 25 μg of pKUNrep2, pKUNβrep2, pSCAβ, or pCMVβ DNA in phosphate-buffered saline (PBS) in a total volume of 80 μl into the quadricep muscles of 6- to 9-week-old female BALB/c mice. Booster immunizations were performed 5 weeks later.
The immunized mice were bled 4, 8, and 11 weeks postpriming, and anti-β-Gal immunoglobulin G (IgG) responses were determined by enzyme-linked immunosorbent assay (ELISA) using purified recombinant β-Gal protein (Promega) as follows. Wells of microtiter plates were incubated with 0.2 μg of β-Gal/ml in 50 μl of coating buffer (15 mM Na2CO3, 35 mM NaHCO3, pH 9.6) overnight at 4°C, blocked with 50 μl of PBS containing 1% skim milk powder and 1% sucrose (PBS/MP/S) for 6 to 7 h at 4°C, and incubated with 50 μl of twofold serial dilutions of the sample sera in a 1/5 dilution of PBS/MP/S supplemented with 0.05% Tween 20 (PBS/MP/S/T) overnight at 4°C. After three washes of the wells with distilled water, they were incubated with 50 μl of a 1/1,000 dilution of horseradish peroxidase-conjugated goat anti-mouse IgG solution (Protos Immunoresearch, Burlingame, Calif.) for 30 min at room temperature. After three washes in water, bound conjugate was developed by incubation with 50 μl of K-blue TMB substrate (Graphic Scientific). The reaction was stopped by the addition of 50 μl of 2 M H2SO4, and readings of the optical density at 450 nm of the reaction products were determined.
Mouse lung sections.
One, 2, 4, and 8 weeks after intranasal delivery of the pKUNβrep2 DNA, mice were euthanatized with CO2 and their lungs were removed, rinsed in PBS, and fixed in 4% paraformaldehyde for 2 to 4 h at room temperature. Whole lungs were stained with X-Gal (see above), post-fixed in formalin, and paraffin embedded, and ∼5-μm sections were prepared, mounted, and photographed.
RESULTS
Ub-containing KUN replicon vectors.
The Ub sequence (16) was originally introduced into KUN replicon vectors (see Fig. 1A and 2) for two purposes: firstly, to improve the induction of cytotoxic T-lymphocytes (CTL) against encoded HG product, and secondly, to facilitate the formation of a precise amino terminus of the HG product. Ub is specifically recognized by a cellular protease complex and cleaved at its carboxy terminus (5). Thus, cloning of an HG upstream of the Ub sequence (at a unique MluI or NsiI site; Fig. 1A) will result in production of a C20-HG-Ub fusion product containing the first 20 amino acid residues encoded by the KUN open reading frame (C20) at its amino terminus and those encoded by Ub at its carboxy terminus. This (ubiquitinated) C20-HG-Ub product will presumably be more efficiently targeted to the cellular proteasome-mediated major histocompatibility complex class I presentation pathway similar to that described by Rodriguez et al. (38). The insertion of an HG between the Ub and FMDV-2A sequences (at a unique AscI site; Fig. 1A) will result in synthesis of an HG-FMDV-2A fusion product with a precise HG sequence-defined amino terminus (as described in references 4 and 5). A correct processing of encoded HG products from the Ub-containing KUN replicon vectors was confirmed by RIP analysis (data not shown), and the immunogenic potentials of these vectors will be addressed elsewhere. For the purpose of this study, Ub-containing vectors were used as intermediates for construction of other vectors (Fig. 2).
Specific restriction of KUN replicon RNA by an encoded HDVr.
As a first step in the construction of DNA-based KUN replicon vectors, the HDVr-pA cassette containing the HDV-derived ribozyme and the SV40-derived transcription termination signal (see Materials and Methods) was inserted immediately downstream of the last nucleotide of the RNA-based KUN replicon vector C20Ubrep, producing the C20UbHDVrep construct (Fig. 1A). The number of cells positively transfected with in vitro-transcribed C20UbHDVrep RNA was significantly (∼fivefold) higher than that detected after transfection with the same amount of C20Ubrep RNA, as judged by immunofluorescent (IF) analysis with KUN anti-NS3 antibodies (Fig. 1B). This is presumably indicative of higher infectivity and/or replication efficiency of the C20UbHDVrep-derived RNA, since we previously demonstrated that the number of NS3-positive cells directly correlated with the amount of KUN RNA accumulated in the replicon RNA-transfected cells (25, 26).
The increased efficiency of the C20UbHDVrep RNA is most likely due to the HDVr cleavage-mediated formation of the authentic KUN RNA 3′ terminus, which has been shown to be important for efficient initiation of RNA synthesis by positive-strand RNA viruses (9). Other studies have found that the extent of self-cleavage by HDVr depended on a specific RNA conformation (46). To examine the catalytic activity of the KUN replicon-encoded HDVr, KUN replicon RNA was in vitro transcribed from the C20UbHDVrep DNA template, self-ligated, and used for reverse transcription (RT)-PCR amplification across the 3′ end-5′ end junction (Fig. 1C). The RT-PCR product amplified from the ligated C20UbHDVrep RNA (Fig. 1C, lane 2) contained a 561-bp fragment corresponding in size to the HDVr-cleaved RNA and identical to the fragment amplified from the decapped and self-ligated virion RNA (24) (Fig. 1C, lane 4). No RT-PCR fragment of the same size was detected in the reaction with the unligated C20UbHDVrep RNA template (Fig. 1C, lane 3), thus demonstrating the specificity of the RT-PCR (see the legend to Fig. 1C). Sequencing of the corresponding RT-PCR products confirmed the authenticity of the KUN RNA 3′ end formed by the HDVr-mediated cleavage during the RNA preparation and/or ligation. Overall, this experiment demonstrated the catalytic activity of HDVr encoded in the KUN replicon construct, but it did not define the extent of HDVr cleavage nor whether this cleavage occurred during RNA preparation in vitro and/or after RNA transfection into cells. It is also possible that the C20UbHDVrep-derived RNA with uncleaved HDVr is more stable and therefore may be more infectious.
DNA-based KUN replicon vectors.
To enable RNA polymerase II-mediated in vivo transcription of the KUN replicon-encoding plasmid DNA, the KUN replicon sequence was placed under the control of the CMV-derived promoter (8) (Fig. 2). To ensure formation of precise 5′ termini in KUN replicon transcripts, KUN DNA-based constructs were engineered so that the last nucleotide of the CMV promoter was immediately followed by the first nucleotide of the KUN sequence (see Materials and Methods).
The pKUNrep1 vector is a DNA-based equivalent of the C20UbHDVrep construct (Fig. 1A). The pKUNrep2 vector differs from pKUNrep1 only by the absence of the Ub sequence (Fig. 2). Both pKUNrep1 and pKUNrep2 were designed for transient gene expression. The DNA-based vectors pKUNrep3 and pKUNrep4, which encode a PAC selection marker (42) which confers resistance to the antibiotic puromycin, were designed for the generation of cell populations stably expressing the encoded proteins (see Materials and Methods). An HG cloned into a unique MluI or NsiI site of the pKUNrep3 vector was initially translated as a part of a single polyprotein. The subsequent cleavages by two flanking FMDV-2A autoproteases (Fig. 2) should result in the release of the HG fusion product containing the FMDV-2A peptide (∼19 amino acid residues; 40) at its carboxy terminus. In the dicistronic pKUNrep4 vector, on the other hand, translation of the HG is separated from translation of the rest of the polyprotein by an IRES (Fig. 2), allowing production of the HG product with a precise carboxy terminus determined by its termination codon. The amino termini of HG products from both pKUNrep3 and pKUNrep4 vectors, generated by the FMDV-2A cleavage (39, 40), will contain only two to five additional vector-derived amino acids, depending on the choice of the cloning site.
Evidence of self-amplification of in vivo-transcribed KUN replicon RNA.
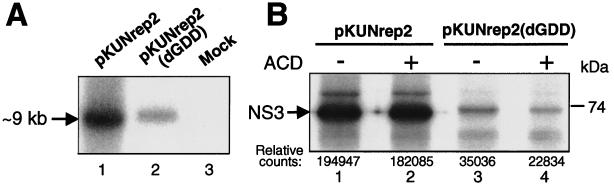
To demonstrate self-amplification of the in vivo-transcribed KUN replicon RNAs, as well as to estimate the effect of this amplification on the level of gene expression, a comparison of RNA and protein syntheses from the DNA-based KUN replicon vector pKUNrep2 and from the same vector but with a deletion of the RNA polymerase motif GDD in pKUNrep2(dGDD) (Fig. 2) was performed. The GDD deletion was previously shown to completely abolish KUN RNA replication (26). Therefore, the KUNrep2(dGDD) RNA could only be produced via transcription from the transfected DNA by cellular RNA polymerase II, resembling RNA synthesis from conventional plasmid DNA expression vectors.
Initially, we compared the relative amounts of KUN replicon RNA produced in BHK21 cells 36 h after transfection with equal amounts of the pKUNrep2 (RNA replication competent) or pKUNrep2(dGDD) (RNA replication defective) DNAs using Northern blot hybridization analysis. The amount of KUN replicon RNA produced from the former was significantly (∼fivefold) higher than that produced from the latter (Fig. 3A). We then examined the expression of one of the vector proteins (NS3) in BHK21 cells transfected with the pKUNrep2 or pKUNrep2(dGDD) constructs, using RIP analysis with KUN anti-NS3 antibodies (Fig. 3B). Similarly to the synthesis of replicon RNA, a significantly higher (∼sixfold) amount of NS3 was produced in the pKUNrep2-transfected cells compared to that produced in the pKUNrep2(dGDD)-transfected cells, as determined by quantitative phosphorimager analysis (Fig. 3B, compare lanes 1 and 3). Importantly, NS3 expression in cells transfected with the pKUNrep2 DNA for 30 h was not apparently reduced in the presence of ACD, an inhibitor of DNA-dependent RNA synthesis (Fig. 3B, compare lanes 1 and 2), while that for pKUNrep2(dGDD) was (Fig. 3B, compare lanes 3 and 4). The small amount of labeled NS3 in lane 4 was presumably translated from replicon RNA transcribed prior to addition of ACD.
FIG. 3.
Comparison of RNA and protein syntheses from the RNA replication-competent and RNA replication-defective KUN replicon DNA vectors. (A) Northern blot hybridization analysis of total RNA isolated from BHK21 cells 36 h after transfection with equal amounts of pKUNrep2 (lane 1) or pKUNrep2(dGDD) (lane 2) plasmid DNAs, or untransfected cells (lane 3). The probe was a [32P]dCTP-labeled cDNA fragment representing the last 761 nucleotides of the KUN genome (24). (B) RIP analysis with KUN anti-NS3 antibodies of BHK21 cells radiolabeled for 1 h at 30 h after transfection with pKUNrep2 (lanes 1 and 2) or pKUNrep2(dGDD) (lanes 3 and 4) plasmid DNAs. Shown are RIP samples from cells labeled in the absence (−) (lanes 1 and 3) or presence (+) (lanes 2 and 4) of ACD. The specificity of KUN anti-NS3 antibodies in RIP and IF analyses was demonstrated previously (45). Relative phosphorimager counts of the radiolabeled NS3 bands in corresponding RIP samples are shown with the background level deducted. DNA transfection, Northern blot hybridization, protein labeling, ACD treatment, and RIP procedures were performed as described in Materials and Methods.
Comparative analysis of transient HG expression kinetics by KUN replicon and other DNA-based vectors.
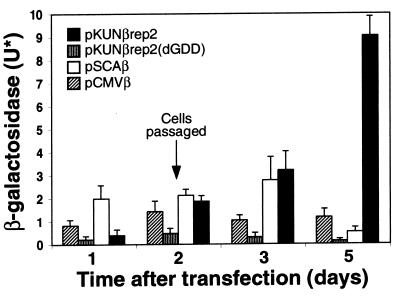
The β-Gal gene was used as a reporter HG for comparative analysis of protein expression levels from the DNA-based KUN replicon vector and other DNA-based vectors. The KUN replicon constructs pKUNβrep2 and pKUNβrep2(dGDD) were prepared by cloning β-Gal into pKUNrep2 and pKUNrep2(dGDD) vectors, respectively (see Materials and Methods). The levels of β-Gal expression in BHK21 cells, transfected with the same amounts of pKUNβrep2, pKUNβrep2(dGDD), pSCAβ (SFV replicon-based construct; 14) and pCMVβ (a conventional plasmid DNA expression construct; Clontech) plasmid DNAs, were compared at different times posttransfection. Transfection efficiencies, determined by counting X-Gal-staining-positive cells of duplicate parallel transfection samples, were ∼40% for all DNA constructs, except for pCMVβ DNA (∼50 to 60%), probably due to its relatively small size (7.2 kb versus 16.5 kb for pKUNβrep2 and 14.5 kb for pSCAβ DNAs).
Because of a delay of ∼16 h in detectable β-Gal expression from the pKUNβrep2 DNA, the comparative expression results are shown commencing at 24 h (1 day) posttransfection (Fig. 4). To account for the noncytopathic nature and persistence of replication of KUN replicon vectors, the transfected cells were allowed to propagate continuously during the entire experiment by passaging them into larger plates 2 days after the initial transfection (see legend of Fig. 4). The level of β-Gal expression in continuously propagating BHK21 cells transfected with the KUN replicon DNA construct pKUNβrep2 steadily increased with time after transfection. Five days after the initial transfection, this level was ∼18-fold higher than that after transfection with the SFV DNA construct pSCAβ and ∼7- to 10-fold higher than that generated by the conventional plasmid DNA construct pCMVβ (Fig. 4). Most of the pKUNβrep2-transfected cells appeared healthy and formed β-Gal-expressing cell colonies, thus demonstrating transfer of the replicating KUNβrep2 RNA into daughter cells during cell division. In contrast, β-Gal expression from the pSCAβ DNA was significantly reduced by day 5 posttransfection, presumably due to the vector-induced death of the expressing cells. Notably, similar expression kinetics for pSCAβ construct was reported by DiCiommo and Bremner (14). In addition, β-Gal expression from the pCMVβ and pKUNβrep2(dGDD) construct, both noncytopathic and RNA replication defective, did not increase from day 2 to day 5 posttransfection (Fig. 4), demonstrating that the corresponding ∼fivefold increase in the β-Gal level detected in pKUNβrep2 DNA-transfected cells was indeed due to the replicon RNA self-amplification rather than only to propagation of the β-Gal-expressing cells.
FIG. 4.
Comparative analyses of transient β-Gal expression from the KUN replicon [pKUNβrep2, pKUNβrep2(dGDD); see Materials and Methods], pSCAβ (SFV replicon-based; 14), and pCMVβ (conventional plasmid; Clontech) DNA constructs. BHK21 cell monolayers (∼1.3 × 105 cells in 16-mm-diameter wells of 24-well plates) were transfected with ∼0.8 μg of corresponding DNAs and incubated until reaching confluency (∼2 days posttransfection). The cells were then trypsinized and transferred into larger plates to allow continuous cell division. Forty percent of the trypsinized cells were transferred into 35-mm-diameter plates, allowed to propagate, and assayed for β-Gal activity at day 3 after initial transfection. Fifty percent of the trypsinized cells were transferred into 60-mm-diameter plates, allowed to propagate, and assayed for β-Gal activity at day 5 after initial transfection. U*, the amounts of biochemically active β-Gal protein, in units of the enzymatic activity, produced in the total volume of each cell lysate collected from 16-mm-diameter wells (days 1 and 2), 35-mm-diameter plates (day 3), and 60-mm-diameter plates (day 5). The values are the means ± standard deviation for triplicate experiments.
Transient and stable HG expression from DNA-based KUN replicon vectors in different cell lines and in vivo.
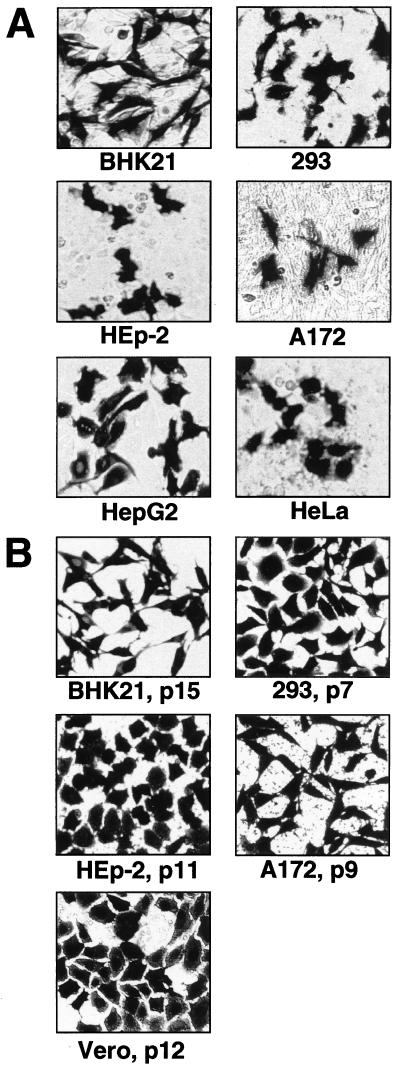
Transient expression of the reporter gene β-Gal from transfected pKUNβrep2 DNA was demonstrated in a number of cell lines of different origin (Fig. 5A). To examine the efficacy of puromycin-selectable DNA-based KUN replicon vectors pKUNrep3 and pKUNrep4 for the generation of cell cultures stably expressing HGs, the corresponding β-Gal-recombinant constructs pKUNβrep3 and pKUNβrep4 were prepared (see Materials and Methods). Several cell lines stably expressing β-Gal after transfection with pKUNβrep3 or pKUNβrep4 plasmid DNAs and subsequent puromycin selection were successfully established (Fig. 5B and Table 2). Significantly, no β-Gal expression-negative cells were detected and the expression levels remained high throughout passaging (see Table 2). Maximum production of β-Gal in selected BHK21 cells was ∼200 pg/cell in the first two or three passages, which was similar to the amount of β-Gal per expressing cell produced during transient expression from pKUNβrep2 DNA by 48 h after transfection. Importantly, the expression levels decreased by only ∼20 to 25% after extensive cell passaging (Table 2), indicating the highly stable nature of HG expression directed by the DNA-based KUN replicon vectors. We have previously observed similar results on the stability of HG expression from the selectable RNA-based KUN replicon vector C20DX2ArepNeo (43). The level of β-Gal expression from pKUNβrep4 construct was slightly lower than that obtained from pKUNβrep3 construct in selected BHK21 cells (see Table 2).
FIG. 5.
Expression of β-Gal in different cell lines using DNA-based KUN replicon vectors. (A) Transient β-Gal expression in indicated cell lines, detected by X-Gal staining 30 to 40 h after transfection with the pKUNβrep2 DNA. (B) Stable β-Gal expression in different cell lines at indicated passages after transfection with the pKUNβrep3 (BHK21, Vero, 293, HEp-2 cells) or pKUNβrep4 (A172 cells) DNAs and subsequent puromycin selection. DNA transfection and puromycin selection of cell cultures were performed as described in Materials and Methods.
TABLE 2.
Expression of β-Gal in cells using DNA-based KUN replicon vectors
| DNA construct (expression type) | Cell type | Cell passage | Amt of β-Gal (pg/cell)a |
|---|---|---|---|
| pKUNβrep2 (transient) | BHK21 | 186 ± 27 | |
| pKUNβrep3 (stable) | BHK21 | 2 | 194 ± 9 |
| BHK21 | 3 | 198 ± 14 | |
| BHK21 | 13 | 153 ± 4 | |
| BHK21 | 22 | 156 ± 7 | |
| Vero | 10 | 153 ± 6 | |
| HEp-2 | 15 | 154 ± 22 | |
| 293 | 20 | 122 ± 4 | |
| pKUNβrep4 (stable) | BHK21 | 6 | 130 ± 6 |
| A172 | 8 | 140 ± 12 |
Amounts of β-Gal were determined by using the β-Galactosidase Enzyme Assay System (Promega) by comparison with β-Gal concentration standards. To calculate β-Gal amounts per expressing cell, the amounts of β-Gal detected in the total volume of cell lysates were divided by the number of expression-positive cells at 48 h posttransfection in transient-expression experiments or by the total number of cells in stably expressing (100% β-Gal positive) cultures at indicated passages. The values are the means ± standard deviations for triplicate experiments.
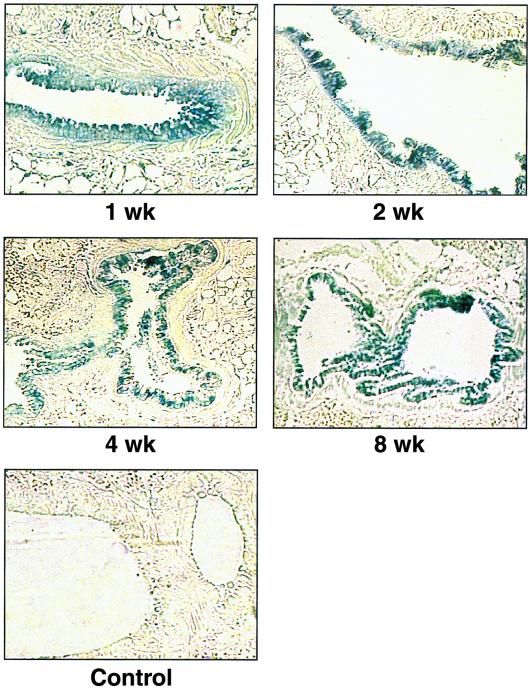
The potential of KUN replicon-based vectors for delivery and expression of HGs in vivo was evaluated by intranasal administration of pKUNβrep2 DNA into BALB/c mice (see Materials and Methods). Expression of β-Gal was detected in epithelial cells lining the lung airways at 1, 2, 4, and 8 weeks after inoculation (Fig. 6), clearly demonstrating the ability of KUN replicon DNA to direct prolonged expression of HGs in vivo.
FIG. 6.
β-Gal expression in mouse lung epithelial cells at indicated times after intranasal inoculation with the DNA-based KUN replicon construct pKUNβrep2. Control panel shows a lung section of a mouse inoculated with the pKUNrep2 DNA. DNA administration and preparation of lung sections were performed as described in Materials and Methods.
Induction of anti-β-Gal-specific antibodies in mice immunized with β-Gal-encoding KUN replicon DNA.
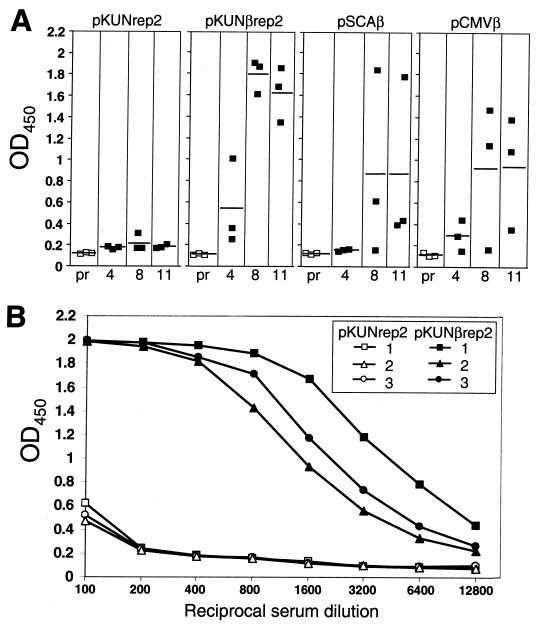
The ability of the KUN replicon-based plasmid DNA to induce specific antibodies against the encoded β-Gal protein after i.m. immunization was examined in BALB/c mice. Anti-β-Gal antibody levels induced by pKUNβrep2 DNA were compared with those induced by SFV replicon-based and conventional plasmid DNA-based β-Gal-encoding vectors. The anti-β-Gal antibody responses in mice immunized twice i.m. with 25 μg of either pKUNβrep2, pSCAβ, pCMVβ, or control pKUNrep2 DNAs were examined by ELISA (per Materials and Methods). The antibody levels were low or undetectable at 4 weeks postpriming with all the DNA constructs but increased significantly at 8 and 11 weeks postpriming (3 and 6 weeks postbooster, respectively) in all mice immunized with pKUNβrep2 DNA, but only in one out of three and in two out of three mice immunized with pSCAβ and pCMVβ DNAs, respectively (Fig. 7A). Figure 7B shows ELISA titration results for the 8-week sera from the individual mice immunized with the pKUNβrep2 or pKUNrep2 DNAs. Interestingly, no detectable antibody responses against the KUN vector-encoded NS5 protein were induced in mice immunized with pKUNrep2 or pKUNβrep2 DNAs (data not shown).
FIG. 7.
Induction of β-Gal-specific antibodies in mice after i.m. immunization with β-Gal-encoding plasmid DNA constructs. (A) β-Gal-specific IgG responses in a 1/800 dilution of sera from BALB/c mice 4, 8, and 11 weeks after i.m. immunization with pKUNrep2, pKUNβrep2, pSCAβ, and pCMVβ plasmid DNA constructs (as per Materials and Methods). ELISA readings for individual mouse sera are shown by open squares (preimmune sera [pr]) and filled squares (immunized sera; 4, 8, and 11 weeks postpriming). Horizontal lines show the averages for each group. (B) β-Gal-specific IgG responses induced in individual sera of BALB/c mice 8 weeks post i.m. immunization (3 weeks postboost) with the pKUNβrep2 and control pKUNrep2 plasmid DNAs. OD450, optical density at 450 nm.
DISCUSSION
We have described the construction and potential applications of the DNA-based KUN replicon vectors, which allow efficient in vivo synthesis of KUN replicon RNAs from the corresponding plasmid DNAs. This is a further step in the development of the KUN replicon-based gene expression system (25, 27, 43) and, to our knowledge, is the first demonstration of the production of a replicating flavivirus RNA from plasmid DNA in vivo. The use of DNA-based vectors eliminates expensive and cumbersome RNA preparation and handling steps and allows utilization of conventional protocols for delivery of plasmid DNA.
KUN replicon sequence in the DNA-based vectors was placed under transcriptional control of the CMV-derived promoter and SV40-derived transcription termination signal (Fig. 2), allowing KUN replicon RNAs to be transcribed by cellular RNA polymerase II. The importance of precise 5′ and 3′ termini for infectivity and/or replication efficiency was reported for different positive-strand RNA virus genomes (6, 9). Therefore, the CMV promoter sequence was placed immediately upstream of the first nucleotide of the KUN sequence, thus ensuring the formation of authentic KUN 5′ ends during KUN replicon RNA transcription. To facilitate as well the formation of authentic KUN 3′ ends, the HDVr sequence (35) was incorporated immediately downstream of the KUN replicon sequence (Fig. 1 and 2), as reported for other viral RNA transcripts produced from DNA-based constructs (15, 21, 22, 33). The catalytic activity of HDVr was demonstrated for the KUN replicon RNA in vitro transcribed from the C20UbHDVrep DNA (see Results and Fig. 1).
Efficient self-replication of in vivo-transcribed KUN replicon RNAs was demonstrated by comparison of RNA and protein syntheses from the KUN replicon DNA constructs and their corresponding RNA-replication-defective mutants with a deleted GDD motif. Thus, the levels of RNA and protein syntheses from the RNA replication-competent vector pKUNrep2 were five- to sixfold higher than those from the nonreplicating vector pKUNrep2(dGDD) (Fig. 3). Also, the total amount of β-Gal produced in continuously propagating cells from the pKUNβrep2 DNA increased by ∼20-fold from day 1 to day 5 after transfection, compared to no apparent increase in β-Gal expression in cells transfected with the RNA replication-defective pKUNβrep2(dGDD) DNA construct during the same time period (Fig. 4). Apparently, noncytopathic replication of KUN replicon RNA allowed the transfected cells to divide normally and to transfer replicon RNA into daughter cells where it continued its self-amplification, resulting in a significant increase in total HG expression. Although replicon RNA produced from the SFV DNA-based construct pSCAβ is also capable of self-amplification, its replication and corresponding increase in HG expression is limited to only ∼3 days due to the induction of strong CPE in the host cells. On the other hand, the expression from noncytopathic pCMVβ and pKUNβrep2(dGDD) constructs, producing nonreplicating mRNAs by nuclear transcription, did not increase after day 2 posttransfection (Fig. 4), thus reflecting the enhancing effect of cytoplasmic self-amplification (of replicon-based vectors) on levels of gene expression.
Stable expression of β-Gal reporter was examined in different cell cultures transfected with pKUNβrep3 or pKUNβrep4 DNAs and subsequently selected with puromycin (Table 2). The slight variation in the levels of β-Gal expression from pKUNβrep3 DNA in distinct cell lines was probably due to differences in replication efficiency of KUN replicon RNA and/or cellular conditions at the time of the analysis. On the other hand, in the selected BHK21 cells, β-Gal expression from the pKUNβrep4 construct was ∼20 to 30% lower than that from the pKUNβrep3 construct. This difference could result from a less efficient replication of the pKUNβrep4-derived RNA possibly due to a less efficient initiation of translation of KUN nonstructural proteins from the IRES compared with that from the native KUN 5′ end. Replication of KUN replicon RNA may also be inhibited by an extended IRES RNA secondary structure. Clearly, further investigations including direct comparison of the replication efficiencies of the RNAs produced from pKUNrep3 and pKUNrep4 vectors are required to make definite conclusions.
The levels of gene expression from the DNA-based KUN replicon vectors were comparable with those achieved by the most efficient cytopathic SFV replicon vectors in transient expression experiments (7) and significantly higher than those reported for noncytopathic SIN replicon vectors (1). The selectable SIN replicon vectors were isolated by selection of PAC-encoding mutants adapted to replication in BHK21 cells in the presence of puromycin. These mutants replicated ∼50- to 100-fold less efficiently than the wild-type cytopathic SIN replicon RNA and produced only ∼1 pg of β-Gal per cell (1, 17). Pairing of the noncytopathic SIN puromycin-selectable replicon RNA with the G418-selectable SIN DI vectors (required double-antibiotic selection) increased the expression level to ∼30 pg per cell (1), which is still at least fivefold lower than the expression levels achieved by the single KUN replicon vector (∼150 pg/cell; Table 2). Importantly, the level of HG expression in KUN replicon-derived stable cell lines was reduced only slightly during the extensive cell passaging (Table 2), indicating high stability of KUN replicon-directed gene expression and suggesting a potential application of such KUN replicon-derived cell cultures for large-scale continuous production of heterologous proteins.
Another useful feature of KUN replicons is their ability to replicate efficiently in a broad range of host cells without producing any apparent CPE. Thus, using KUN replicon DNA-based vectors, we were able to express HGs (stably and/or transiently) in different cell lines, including BHK21, Vero, HeLa, HepG2, HEp-2, 293, and A172 (Fig. 5A and B), as well as L929 (mouse fibroblasts) and Jurkat (human lymphoma) cells (unpublished data). Noticeably, a broad host range was also observed for KUN replicon RNA- and VLP-based vectors (unpublished data). In addition, we demonstrated expression of the β-Gal reporter gene in mouse lung epithelial cells for at least 8 weeks following the intranasal inoculation of mice with the β-Gal-encoding KUN replicon DNA (Fig. 6). Although we did not compare the longevity of in vivo expression with other replicon-based vectors, the reported studies for alphavirus vectors have been limited to only 5 to 14 days (2, 15, 20). The ability to direct prolonged noncytopathic gene expression in different host cells is a unique property of KUN replicon vectors, presumably reflecting the inherent nature of their noncytopathicity (25, 26). Similarities in the levels of HG (β-Gal) expression after transient transfection with the KUN replicon construct and in selected stably expressing cells also indicate that noncytopathicity of KUN replicons is defined in their original sequence, rather than being acquired by adaptive mutations during the selection of replicon-expressing cells.
Prolonged high-level HG expression afforded by the KUN replicon vectors discussed above may be advantageous in the context of vaccine applications for maintaining long-lasting immunity. KUN β-Gal-encoding DNA construct pKUNβrep2 induced appreciable levels of anti-β-Gal antibodies for at least 11 weeks in all the immunized mice (Fig. 7A and B), and in a separate experiment we were able to detect antibodies to green fluorescent protein (GFP) over a year after immunization with GFP-recombinant KUN VLPs (unpublished results). The equimolar synthesis of encoded HG and KUN nonstructural proteins by the KUN replicon constructs raises a concern about the immune response against the vector-encoded proteins. Surprisingly, we could not detect antibodies when tested against one of the KUN proteins, NS5 (see Results). It is possible that in the context of noncytopathic expression, the tight association of the KUN nonstructural proteins in the membrane-bound RNA replication complex (30, 45) prevented their efficient presentation to the immune system. It is also possible that β-Gal expressed from the KUN replicon construct could be more immunogenic than NS5. Another potentially beneficial factor of using KUN replicon constructs as vaccines is the synthesis of double-stranded RNA during the KUN RNA replication, which is known to facilitate production of immune-enhancing cytokines (44) and to activate protein kinase-dependent antigen presentation pathways (13). In addition, flaviviruses, unlike alphaviruses, have been shown to up-regulate expression of major histocompatibility complex class I and class II molecules (28, 31, 41). Whether the KUN replicon-based vectors will also induce such up-regulation is not yet known, and we intend to address this, as well as other aspects of KUN replicon-directed immune responses, in future studies.
In conclusion, we have developed highly efficient DNA-based KUN replicon vectors allowing prolonged high-level expression of HGs in cells and in laboratory animals. We believe that these vectors should prove their usefulness in a variety of applications both in vitro and in vivo.
ACKNOWLEDGMENTS
We are grateful to Petra Sedlak for technical assistance, Tom Macnaughton for providing the pTMSV5A plasmid containing the HDVr-pA sequence, Rod Bremner for the pSCAβ plasmid, Donna West for assistance in mouse experiments, Christine Lee for preparation of lung sections, and Ed Westaway for critical review of the manuscript.
This work was supported by a grant from the National Health and Medical Research Council of Australia.
Footnotes
Publication 104 from Sir Albert Sakzewski Virus Research Centre.
REFERENCES
- 1.Agapov E V, Frolov I, Lindenbach B D, Pragai B M, Schlesinger S, Rice C M. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc Natl Acad Sci USA. 1998;95:12989–12994. doi: 10.1073/pnas.95.22.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman-Hamamdzic S, Groseclose C, Ma J X, Hamamdzic D, Vrindavanam N S, Middaugh L D, Parratto N P, Sallee F R. Expression of beta-galactosidase in mouse brain: utilization of a novel nonreplicative Sindbis virus vector as a neuronal gene delivery system. Gene Ther. 1997;4:815–822. doi: 10.1038/sj.gt.3300458. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 4.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 5.Baker R T, Smith S A, Marano R, McKee J, Board P G. Protein expression using cotranslational fusion and cleavage of ubiquitin. Mutagenesis of the glutathione-binding site of human Pi class glutathione S-transferase. J Biol Chem. 1994;269:25381–25386. [PubMed] [Google Scholar]
- 6.Ball L A, Li Y. cis-acting requirements for the replication of flock house virus RNA 2. J Virol. 1993;67:3544–3551. doi: 10.1128/jvi.67.6.3544-3551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berglund P, Smerdou C, Fleeton M N, Tubulekas I, Liljestrom P. Enhancing immune responses using suicidal DNA vaccines. Nat Biotechnol. 1998;16:562–565. doi: 10.1038/nbt0698-562. [DOI] [PubMed] [Google Scholar]
- 8.Boshart M, Weber F, Jahn G, Dorsch-Hasler K, Fleckenstein B, Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- 9.Boyer J-C, Haenni A-L. Infectious transcripts and cDNA clones of RNA viruses. Virology. 1994;198:415–426. doi: 10.1006/viro.1994.1053. [DOI] [PubMed] [Google Scholar]
- 10.Bredenbeek P, Frolov I, Rice C M, Schlesinger S. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bredenbeek P, Rice C M. Animal RNA virus expression systems. Semin Virol. 1992;3:297–310. [Google Scholar]
- 12.Caley I J, Betts M R, Irlbeck D M, Davis N L, Swanstrom R, Frelinger J A, Johnston R E. Humoral, mucosal, and cellular immunity in response to a human immunodeficiency virus type 1 immunogen expressed by a Venezuelan equine encephalitis virus vaccine vector. J Virol. 1997;71:3031–3038. doi: 10.1128/jvi.71.4.3031-3038.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Der S D, Yang Y L, Weissmann C, Williams B R. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:3279–3283. doi: 10.1073/pnas.94.7.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiCiommo D P, Bremner R. Rapid, high level protein production using DNA-based Semliki Forest virus vectors. J Biol Chem. 1998;273:18060–18066. doi: 10.1074/jbc.273.29.18060. [DOI] [PubMed] [Google Scholar]
- 15.Dubensky T W, Jr, Driver D A, Polo J M, Belli B A, Latham E M, Ibanez C E, Chada S, Brumm D, Banks T A, Mento S J, Jolly D J, Chang S M W. Sindbis virus DNA-based expression vectors: utility for in vitro and in vivo gene transfer. J Virol. 1996;70:508–519. doi: 10.1128/jvi.70.1.508-519.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finch J S, Bonham K, Krieg P, Bowden G T. Murine polyubiquitin mRNA sequence. Nucleic Acids Res. 1990;18:1907. doi: 10.1093/nar/18.7.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frolov I, Agapov E, Hoffman T A, Jr, Pragai B M, Lippa M, Schlesinger S, Rice C M. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J Virol. 1999;73:3854–3865. doi: 10.1128/jvi.73.5.3854-3865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frolov I, Schlesinger S. Comparison of the effects of Sindbis virus and Sindbis virus replicons on host cell protein synthesis and cytopathogenicity in BHK cells. J Virol. 1994;68:1721–1727. doi: 10.1128/jvi.68.3.1721-1727.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hariharan M J, Driver D A, Townsend K, Brumm D, Polo J M, Belli B A, Catton D J, Hsu D, Mittelstaedt D, McCormack J K, Karavodin L, Dubensky T W, Jr, Chang S M W, Banks T A. DNA immunization against herpes simplex virus: enhanced efficacy using a Sindbis virus-based vector. J Virol. 1998;72:950–958. doi: 10.1128/jvi.72.2.950-958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johanning F M, Conry R M, LoBuglio A F, Wright M, Sumerel L A, Pike M J, Curiel D T. A sindbis virus mRNA polynucleotide vector achieves prolonged and high level heterologous gene expression in vivo. Nucleic Acids Res. 1995;23:1495–1501. doi: 10.1093/nar/23.9.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson K L, Ball L A. Replication of flock house virus RNAs from primary transcripts made in cells by RNA polymerase II. J Virol. 1997;71:3323–3327. doi: 10.1128/jvi.71.4.3323-3327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson K L, Ball L A. Induction and maintenance of autonomous flock house virus RNA1 replication. J Virol. 1999;73:7933–7942. doi: 10.1128/jvi.73.10.7933-7942.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karreman C. Fusion PCR, a one-step variant of the “Mega-primer” method of mutagenesis. BioTechniques. 1998;24:736–742. doi: 10.2144/98245bm08. [DOI] [PubMed] [Google Scholar]
- 24.Khromykh A A, Westaway E G. Completion of Kunjin virus RNA sequence and recovery of an infectious RNA transcribed from stably cloned full-length cDNA. J Virol. 1994;68:4580–4588. doi: 10.1128/jvi.68.7.4580-4588.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khromykh A A, Westaway E G. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J Virol. 1997;71:1497–1505. doi: 10.1128/jvi.71.2.1497-1505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khromykh A A, Kenney M T, Westaway E G. trans-complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J Virol. 1998;72:7270–7279. doi: 10.1128/jvi.72.9.7270-7279.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khromykh A A, Varnavski A N, Westaway E G. Encapsidation of the flavivirus Kunjin replicon RNA by using a complementation system providing Kunjin structural proteins in trans. J Virol. 1998;72:5967–5977. doi: 10.1128/jvi.72.7.5967-5977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King N J, Kesson A M. Interferon-independent increases in class I major histocompatibility complex antigen expression follow flavivirus infection. J Gen Virol. 1988;69:2535–2543. doi: 10.1099/0022-1317-69-10-2535. [DOI] [PubMed] [Google Scholar]
- 29.Liljestrom P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology. 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- 30.Mackenzie J M, Khromykh A A, Jones M K, Westaway E G. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology. 1998;245:203–215. doi: 10.1006/viro.1998.9156. [DOI] [PubMed] [Google Scholar]
- 31.Mullbacher A, Lobigs M. Up-regulation of MHC class I by flavivirus-induced peptide translocation into the endoplasmic reticulum. Immunity. 1995;3:207–214. doi: 10.1016/1074-7613(95)90090-x. [DOI] [PubMed] [Google Scholar]
- 32.Palese P. RNA virus vectors: where are we and where do we need to go? Proc Natl Acad Sci USA. 1998;95:12750–12752. doi: 10.1073/pnas.95.22.12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pattnaik A K, Ball L A, LeGrone A W, Wertz G W. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 34.Percy N, Barclay W S, Sullivan M, Almond J W. A poliovirus replicon containing the chloramphenicol acetyltransferase gene can be used to study the replication and encapsidation of poliovirus RNA. J Virol. 1992;66:5040–5046. doi: 10.1128/jvi.66.8.5040-5046.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perrotta A T, Been M D. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature (London) 1991;350:434–436. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
- 36.Porter D C, Ansardi D C, Choi W S, Morrow C D. Encapsidation of genetically engineered poliovirus minireplicons which express human immunodeficiency virus type 1 Gag and Pol proteins upon infection. J Virol. 1993;67:3712–3719. doi: 10.1128/jvi.67.7.3712-3719.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pushko P, Parker M, Ludwig G V, Davis N L, Johnston R E, Smith J F. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogenes in vivo. Virology. 1997;239:389–401. doi: 10.1006/viro.1997.8878. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez F, Zhang J, Whitton J L. DNA immunization: ubiquitination of a viral protein enhances cytotoxic T-lymphocyte induction and antiviral protection but abrogates antibody induction. J Virol. 1997;71:8497–8503. doi: 10.1128/jvi.71.11.8497-8503.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryan M D, Drew J. Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J. 1994;13:928–933. doi: 10.1002/j.1460-2075.1994.tb06337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan M D, King A M Q, Thomas G P. Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J Gen Virol. 1991;72:2727–2732. doi: 10.1099/0022-1317-72-11-2727. [DOI] [PubMed] [Google Scholar]
- 41.Shen J, To S S T-, Schrieber L, King N J C. Early E-selecin, VCAM-1, ICAM-1, and late major histocompatibility complex antigene induction on human endothelial cells by flavivirus and comodulation of adhesion molecule expression by immune cytokines. J Virol. 1997;71:9323–9332. doi: 10.1128/jvi.71.12.9323-9332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vara J A, Portela A, Ortin J, Jimenez A. Expression in mammalian cells of a gene from Streptomyces alboniger conferring puromycin resistance. Nucleic Acids Res. 1986;14:4617–4624. doi: 10.1093/nar/14.11.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varnavski A N, Khromykh A A. Noncytopathic flavivirus replicon RNA-based system for expression and delivery of heterologous genes. Virology. 1999;255:366–375. doi: 10.1006/viro.1998.9564. [DOI] [PubMed] [Google Scholar]
- 44.Vilcek J, Sen G C. Interferons and other cytokines. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 375–399. [Google Scholar]
- 45.Westaway E G, Mackenzie J M, Kenney M T, Jones M K, Khromykh A A. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu H-N, Lai M M C. RNA conformational requirements of self-cleavage of hepatitis delta virus RNA. Mol Cell Biol. 1990;10:5575–5579. doi: 10.1128/mcb.10.10.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ying H, Zaks T Z, Wang R-F, Irvine K R, Kammula U S, Marincola F M, Leitner W W, Pestifo N P. Cancer therapy using a self-replicating RNA vaccine. Nat Med. 1999;7:823–827. doi: 10.1038/10548. [DOI] [PMC free article] [PubMed] [Google Scholar]