Abstract
The chemokine receptors CXCR4 and CCR5 are the principal coreceptors for infection of X4 and R5 human immunodeficiency virus type 1 (HIV-1) isolates, respectively. Here we report on the unexpected observation that the removal of the N-linked glycosylation sites in CXCR4 potentially allows the protein to serve as a universal coreceptor for both X4 and R5 laboratory-adapted and primary HIV-1 strains. We hypothesize that this alteration unmasks existing common extracellular structures reflecting a conserved three-dimensional similarity of important elements of CXCR4 and CCR5 that are involved in HIV envelope glycoprotein (Env) interaction. These results may have far-reaching implications for the differential recognition of cell type-dependent glycosylated CXCR4 by HIV-1 isolates and their evolution in vivo. They also suggest a possible explanation for the various observations of restricted virus entry in some cell types and further our understanding of the framework of elements that represent the Env-coreceptor contact sites.
Coreceptor molecules belonging or related to the chemokine receptor family of seven-transmembrane-domain G-protein-coupled receptors are required along with CD4 for human immunodeficiency virus (HIV-1) envelope glycoprotein (Env)-mediated membrane fusion and virus entry (reviewed in references 8, 12, 27, 36, 38, and 64). Although some 15 related coreceptor molecules have been shown by one or more laboratories to function in the fusion or entry of at least one HIV isolate, it is now well recognized that the principal HIV-1 coreceptors remain the initially discovered CXC chemokine receptor CXCR4 and the CC-chemokine receptor CCR5 (7, 21). Previously, it was hypothesized that the very first step in HIV-1 entry involves the formation of a trimolecular complex between the viral Env, CD4, and a coreceptor molecule (18, 33, 52), and both functional and biochemical evidence to support this hypothesis has been reported (35, 59, 84, 88, 90). Defining the elements of the coreceptor molecules involved in these interactions is of critical importance for our understanding of the virus entry mechanism. To date, multiple studies, largely employing the use of genetic chimeras, have provided an extensive framework of important structural information on several coreceptors, which has been reviewed in detail (10, 12, 34, 38, 64). Although not in complete agreement with each other, these studies have clearly shown that multiple extracellular domains are required for coreceptor function, which involves cooperativity between particular elements of the N-terminal domain and one or more of the three extracellular loops. However, there remains the possibility that many of the chimeric receptors produced in these studies are functional due to compensatory conditions brought about by distal regions of the background receptor, and this could result in regions important for coreceptor function being overlooked (64).
We recently studied a large battery of CXCR4 mutants (23). Those results indicated that negatively charged glutamic acid residues in the N terminus and the aspartic acid residue D97 in extracellular loop 1 (ecl-1) are important for CXCR4 function as a coreceptor for X4 isolates. We and others have speculated that the role of the negatively charged CXCR4 residues is related to their electrostatic interactions with positively charged Env residues in the V3 loop region of the previously classified syncytium-inducing X4 Envs (32, 50). However, other consequences of the site-directed mutagenesis, specifically the effect of removal of N-linked glycosylation sites (resulting in what will henceforth be referred to as non-N-linked glycosylated CXCR4) on the coreceptor function of CXCR4, have not been completely explored. Here we report the first observation of an unexpected and dramatic effect of N-linked glycosylation site removal in an AIDS-related molecule: the expansion of both laboratory-adapted and primary R5 HIV-1 isolates.
We found that CXCR4 N-linked glycosylation results in the loss of coreceptor activity for those HIV-1 isolates which are dominant during the early stages of HIV-1 disease as well as in virus transmission to an uninfected individual (so-called non-syncytium-forming, macrophage-tropic R5 isolates). Thus, alleviation of the N-linked glycosylation of CXCR4 can allow the protein to serve as a universal coreceptor for some R5 and X4 HIV-1 Envs. Taken together with other reports, our data suggest that there is an existing underlying conserved framework of elements between CXCR4 and CCR5 and that removal of masking glycosylation moieties reveals these structures. Thus, differential glycosylation of CXCR4 may modulate the ability of particular HIV-1 isolates to infect various cell types. The possible existence of CXCR4 glycoforms with altered coreceptor activities, perhaps as the result of cell type-dependent glycosylation differences, may have far-reaching implications for the differential recognition of CXCR4 by HIV-1 isolates, for their evolution in vivo, and for the mechanism of HIV-1 entry into cells, as well as for future development of novel drugs and vaccines.
MATERIALS AND METHODS
Cells and culture conditions.
Human HeLa cells and simian BSC-1 cells were obtained from the American Type Culture Collection, Manassas, Va., while human glioblastoma cell line U373-MG and the U373-MG-CD4+ derivative cell line were provided by Adam P. Geballe, Fred Hutchinson Cancer Research Center, Seattle, Wash. (53). Cell cultures were maintained at 37°C in a humidified 5% CO2 atmosphere. HeLa and BSC-1 cell monolayers were maintained in Dulbecco's modified Eagle's medium (Quality Biologicals, Gaithersburg, Md.) supplemented with 10% bovine calf serum (BCS), 2 mM l-glutamine, and antibiotics (DMEM-10). U373 cell monolayers were maintained in the same way except that 15% BCS was used (DMEM-15). U373-MG-CD4+ cell monolayers were also supplemented with 200 μg of G418 (Calbiochem, La Jolla, Calif.)/ml.
Plasmids and recombinant vaccinia viruses.
For Env expression, we employed a battery of plasmids and recombinant vaccinia viruses encoding the env genes from several R5, X4, and R5X4 HIV-1 isolates. The following recombinant vaccinia viruses expressing gp160 from different HIV-1 isolates (whose names are in parentheses) were used: vCB-28 (JR-FL), vCB-32 (SF162), vCB-34 (SF2), vCB-39 (ADA), vCB-41 (LAV), vCB-43 (Ba-L), vCB-52 (CM235), vCB-53 (CM243) (17), and vDC-1 (89.6 [28]) gp160 linked to a strong synthetic vaccinia virus early-late promoter (pSC59 [25]). Purified vaccinia virus stocks were used at a multiplicity of infection of 10 PFU/cell. Plasmids encoding functional gp160, from a variety of HIV-1 primary isolates, linked to the T7 promoter were obtained from the National Institutes of Health AIDS Research and Reagent Program (Rockville, Md.). They include 93BR019.10 (clade F/B), 92UG975.10 (clade G), 93BR029.2 (clade F), 92TH022.4 (clade E), MA301965.26 (clade C), 92BR025.9 (clade C), 91US005.11 (clade B), 92BR020.4 (clade B), 92UG037.8 (clade A), and 92RW020.5 (clade A). For CD4 expression, we used recombinant vaccinia virus vCB-3 (19). Bacteriophage T7 RNA polymerase was produced by infection with vTF1-1 (containing the P11 natural late vaccinia virus promoter) (2). The Escherichia coli lacZ gene linked to the T7 promoter was introduced into cells by infection with vaccinia virus recombinant vCB21R-LacZ, which was described previously (3). For coreceptor expression, we employed recombinant vaccinia viruses or one of two alternative plasmid expression protocols. Vaccinia virus vHC-1 (also vvCCR5-1107), encoding CCR5, was described previously (90). Vaccinia viruses encoding wild-type and mutant CXCR4 (vHC-3 [wild-type CXCR4], vHC-5 [N176A CXCR4], vHC-6 [C-terminal CXCR4 deletion], vHC-7 [N11A N176A CXCR4], and vHC-8 [N11A D187A CXCR4]) were prepared by subcloning the appropriate cDNA into the SmaI site of pMC1107 (22). The recombinant viruses were then obtained by standard techniques employing (Ecogpt) selection (20). For cell fusion assays, we either infected cells with the appropriate vaccinia virus encoding a chemokine receptor linked to the 7.5k vaccinia virus promoter or transfected cell monolayers with plasmids containing coreceptor genes linked to a strong synthetic vaccinia virus early-late promoter (pSC59) (25) followed by infection 2 h later with the Western Reserve (WR) wild-type strain of vaccinia virus, and transfection of monolayers was performed with DOTAP {N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate}. For virus infection assays, cells were transfected with coreceptor genes linked to the cytomegalovirus promoter in pCDNA3 (Invitrogen, Carlsbad, Calif.) and transfection was performed by the DEAE-dextran procedure, as described below.
Mutagenesis.
CCR5 and CXCR4 mutations were induced by using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) in accordance with the manufacturer's instructions. Two mutagenic polyacrylamide gel electrophoresis (PAGE)-purified oligonucleotides were used per mutation. The identities of all CXCR4 mutant constructs were confirmed by DNA sequencing.
Cell-cell fusion assays.
Fusion between Env-expressing and receptor-expressing cells was measured by a reporter gene assay in which the cytoplasm of one cell population contained vaccinia virus-encoded T7 RNA polymerase and the cytoplasm of the other contained the E. coli lacZ gene linked to the T7 promoter; β-galactosidase (β-Gal) was synthesized in fused cells (66). Vaccinia virus-encoded proteins were produced by incubating infected cells at 31°C overnight (9). Cell-cell fusion reactions were conducted with the various cell mixtures in 96-well plates at 37°C. Typically, the ratio of CD4-expressing to Env-expressing cells was 1:1 (2 × 105 total cells per well, 0.2-ml total volume). Cytosine arabinoside (40 μg/ml) was added to the fusion reaction mixture to reduce nonspecific β-Gal production (9). For quantitative analyses, Nonidet P-40 was added (0.5% final) at 2.5 h and aliquots of the lysates were assayed for β-Gal at ambient temperature with the substrate chlorophenol red–β-d-galactopyranoside (Roche Molecular Biochemicals). Fusion results were calculated and expressed as rates of β-Gal activity (change in optical density at 570 nm per minute × 1,000) (66).
HIV-1 infection studies.
U373-MG-CD4+ target cells were prepared in 48-well plates and transfected with the desired coreceptor-encoding plasmid by the DEAE-dextran method. Briefly, 0.2 μg of DNA mixed in 110 μl of DMEM-2.5 with 100 μM chloroquine diphosphate and 1.1 μl of a DEAE-dextran stock (10 mg/ml) in phosphate-buffered saline (PBS) was added to each well of semiconfluent cells. After 4 h, the medium was replaced with PBS–10% dimethyl sulfoxide for 2 min. Monolayers were then washed with PBS and incubated overnight in DMEM-15. Viral infection assays were performed with a luciferase reporter HIV-1 Env pseudotyping system (29). Viral stocks were prepared, as previously described, by transfecting 293T cells with plasmids encoding the luciferase virus backbone (pNL-Luc-ER) and Env from HIV strain JR-FL (67) or NL4-3 (LAV) (1, 71). The resulting supernatant was clarified by centrifugation for 10 min at 2,000 rpm in a Sorvall RT-7 centrifuge (RTH-750 rotor) and stored at 4°C. Monolayers were infected with 100 μl of virus preparation containing 8 μg of DEAE-dextran/ml. After 2 h, 0.5 ml of DMEM-15 was added to each well. Cells were lysed at 72 h postinfection by resuspension in 105 μl of cell lysis buffer (Promega, Madison, Wis.), and 50 μl of the resulting lysate was assayed for luciferase activity, using an equal volume of luciferase substrate (Promega).
Western blot analysis.
BSC-1 cell monolayers were infected overnight with vaccinia virus encoding wild-type or mutant CXCR4 at multiplicity of infection of 10. Western blot detection of CXCR4 was performed essentially as described previously (49) but with some modifications. Cells were extracted with 0.5% Triton X-100 in 20 mM Tris-HCl, pH 8.0, containing 100 mM NaCl, and the nuclei were removed by centrifugation. Extracts from 5 × 104 cells (total) were loaded per well onto a sodium dodecyl sulfate (SDS)–10% polyacrylamide gel; samples were incubated for 30 min at 37°C and not boiled, since boiling often induces aggregation of seven-transmembrane-domain proteins. Following electrophoresis, proteins were transferred to nitrocellulose paper, the blot was probed with 4G10, an anti-CXCR4 murine monoclonal antibody (MAb) (C. C. Broder and E. A. Berger, unpublished observations). The blot was then incubated with horseradish peroxidase-conjugated rabbit anti-mouse immunoglobulin G and developed with a Pierce (Rockford, Ill.) SuperSignal chemiluminescence kit.
Cell surface staining.
Coreceptor expression levels were determined by fluorescent-antibody cell surface staining. Appropriate cells were transfected or infected and incubated overnight as described above. Cells were then kept on ice; 106 cells were washed twice with PBS and once with PBS containing 2.5% BCS, incubated in PBS containing 2.5% BCS and 4 μg of 2D7 MAb (for CCR5) or 2 μg of 12G5 or 4G10 MAb (for CXCR4)/100 μl for 1 h washed three times with PBS, incubated in PBS containing 2.5% goat serum and 10 of phycoerythrin-labeled goat anti-mouse immunoglobulin G per 100 μl for 45 min, washed three times with PBS, and fixed with PBS–2% paraformaldehyde. Fluorescence was measured with a Coulter (Miami, Fla.) EPIC XL flow cytometer.
Molecular modeling.
Theoretical three-dimensional structures of the HIV-1 coreceptors CXCR4 and CCR5 were based on the physically determined structures of both bacteriorhodopsin and rhodopsin (79, 85, 86), as well as analysis of the amino acid sequences of related G-protein-coupled receptors (34, 36). A molecular-modeling software package (Insight II 98.0; Molecular Simulations, Inc., San Diego, Calif.) was used to add a hypothetical three-branched N-linked carbohydrate structure with a molecular mass of 6 kDa (based on the analysis of the CXCR4 non-N-linked glycosylated mutants) to the N terminus of the CXCR4 molecule.
RESULTS
R5 isolate use of altered CXCR4 molecules.
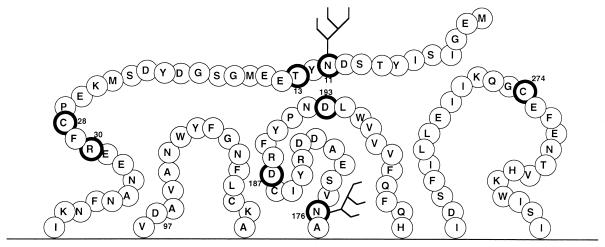
Coreceptor genes were expressed by using either a vaccinia virus promoter system or the cytomegalovirus promoter in a plasmid transfection protocol, depending on the particular assay employed. An alanine-scanning mutagenesis strategy (51) was performed to identify residues involved in CXCR4 coreceptor activity (23). Shown in Fig. 1 is a representation of the extracellular domains of CXCR4 with the locations of several point mutations highlighted: N11A, C28A, and R30A in the N terminus; N176A, D187A, and D193A in ecl-2; and C274A in ecl-3. The mutation of these residues individually by substitution with alanine was noted to enhance the ability of CXCR4 to serve as a coreceptor for otherwise R5-dependent HIV-1 isolates, while full function was retained for X4 and R5X4 isolates. Only the cysteine mutations had moderately reduced X4 Env coreceptor activity (23). A combination of both substitutions for cysteine residues (i.e., C28A C274A) yielded a mutant CXCR4 with an enhanced coreceptor activity for R5 Envs, better than that resulting from substitution for either one alone. Figure 1 also shows the locations of the two potential N-linked glycosylation sites, located in the N terminus and in ecl-2. Using a protocol involving 12G5 or 4G10 MAbs, cell surface staining, and fluorescence-activated cell sorter analysis, all CXCR4 mutants used in the present study had surface expression levels quite comparable to that of wild-type CXCR4 (85 to 115%) (reference 23 and data not shown). Similar N-linked glycosylation site-deleted mutant CXCR4 receptor cell surface expression findings were reported by others (69). Further, the non-N-linked glycosylated CXCR4 mutants in this study reacted equally well with a panel of six additional conformation-dependent anti-CXCR4 MAbs supplied by R&D Systems (D. J. Chabot, F. Baribaud, R. W. Doms, and C. C. Broder, unpublished observations).
FIG. 1.
Diagram of CXCR4 extracellular domains. Residues that when converted to alanine enhance fusion with R5 HIV-1 Env glycoproteins are highlighted by enclosure within dark black circles. Locations of the potential N-linked glycosylation sites are indicated.
Removal of N-linked glycosylation sites expands CXCR4 coreceptor activity.
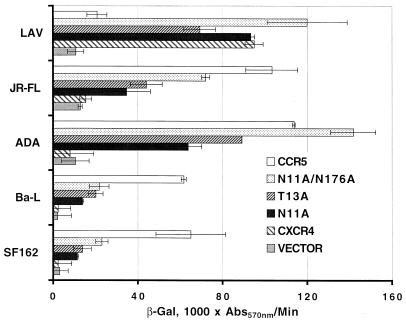
The vaccinia virus-based β-Gal cell fusion assay was performed to examine the non-N-linked glycosylated CXCR4 mutants in an experiment in which human U373 target cells expressing CD4 and infected with the vCB-21R-LacZ reporter virus were transfected with plasmids encoding mutant or wild-type CXCR4. Env-expressing HeLa effector cells were produced by infection with the appropriate Env-encoding vaccinia virus and a vaccinia virus encoding T7 RNA polymerase (9, 17, 66). One of the most potent single mutations that allowed CXCR4 to serve as an R5 isolate coreceptor was N11A (Fig. 2). This alteration potentially eliminated an N-terminal N-linked glycosylation structure. Site-directed mutagenic removal of the asparagine residue of an N-linked glycosylation site motif is a more definitive means of glycosylation site identification than enzymatic removal of the carbohydrate moiety, and more importantly for the present study, it permits a functional examination of effects of glycosylation. We confirmed by mutagenesis that the N11A phenotype was likely due to the elimination of a glycosylation site by disruption of the consensus glycosylation sequence (N-X-S/T) by an alternative mutation (T13A). In Env-mediated cell fusion assays with a panel of prototypic R5 Envs (Fig. 2), the T13A CXCR4 mutant exhibited coreceptor activity nearly equivalent to that of the N11A mutant. This result strongly indicates that it was the carbohydrate modification at asparagine 11, rather than the asparagine amino acid itself, that was mediating the CXCR4 mutant phenotype of enhanced R5 coreceptor activity. We also examined the other potential N-linked glycosylation site in CXCR4, located in ecl-2, using the N176A mutant and found that it also had some enhanced coreceptor activity for R5 Env-mediated fusion, although the level of activity was significantly lower than that of the N-terminal site mutant. Therefore, we chose to not examine an additional S178A mutant but rather focus on the N-terminal glycosylation site and examine a double mutant both functionally and biochemically. The double non-N-linked glycosylated CXCR4 (N11A N176A) was constructed, and this non-N-linked glycosylated CXCR4 exhibited a coreceptor activity for several R5 HIV-1 isolate Envs, including JR-FL, ADA, Ba-L, and SF162, that was further enhanced over that of the single N11A construct (Fig. 2). The data shown in Fig. 2 are the actual rates of reporter activity, with background levels of both vector alone, wild-type CXCR4, and the CCR5 activity shown for comparison. The N11A N176A mutant CXCR4 retained full coreceptor activity for LAV Env (Fig. 2) and for several other X4 or R5X4 Envs (data not shown), in agreement with earlier reports (16, 69). The expanded tropism of the non-N-linked glycosylated CXCR4 was quite significant, with activities ranging from 35 to 125% of the level of coreceptor activity observed with CCR5 in the same experiment (Fig. 2). Similar results were achieved when these CXCR4 mutants were expressed along with CD4 in mouse 3T3 cells, rabbit RK13 cells, and monkey BSC-1 cells (data not shown). These N-linked glycosylations could potentially block interactions between CXCR4 and certain regions within a particular Env, or they might alter the conformation of CXCR4 in a way that prevents such interactions.
FIG. 2.
Coreceptor function of non-N-linked glycosylated CXCR4 mutants in cell fusion assays with R5 HIV-1 Envs. U373 target cells were transfected with a plasmid encoding the wild-type or a mutant coreceptor linked to the vaccinia virus promoter and infected with vCB21R-LacZ and vCB-3 (CD4). HeLa effector cells were infected with a vaccinia virus encoding an HIV-1 Env and with a vaccinia virus encoding T7 polymerase (vTF1-1). Cell mixtures (duplicates) were incubated at 37°C for 2.5 h. Fusion was assessed by measurement of β-Gal in detergent lysates of cells. The rates of β-Gal activity shown were obtained from separate samples in the same experiment. Error bars indicate the standard deviations of the mean values obtained from duplicate fusion assays. This experiment was performed three times, and the data from a representative experiment are shown in the figure. Abs570nm, absorbance at 570 nm.
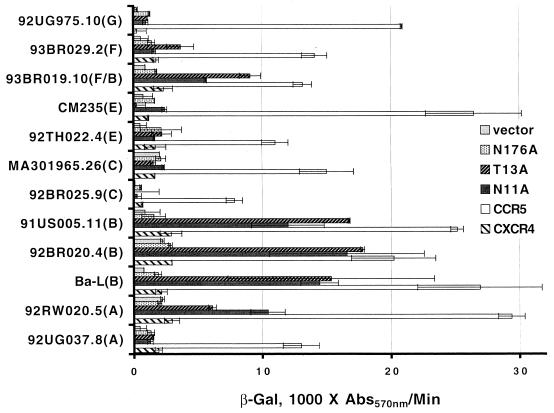
In light of these surprising observations, we examined the Env-mediated fusion activities of HIV-1 primary isolates and those of alternate clades. Shown in Fig. 3 are the results obtained in testing a battery of primary R5 isolate Envs with the individual non-N-linked glycosylated CXCR4 mutants in comparison to wild-type CXCR4 and CCR5 in the cell fusion assay. Two primary clade B isolates (91US005.11 and 92BR020.4), a clade F/B mosaic (93BR019.10-all gp120 F), and a clade A (92RW020.5) R5 Env could utilize the N11A CXCR4 coreceptor. The overall reporter gene signal levels were lower in this experiment than in the experiment shown in Fig. 2 because both the coreceptor genes and the Env genes (most driven by a T7 promoter system) were transfected as plasmids in this assay. For this reason, a plasmid encoding the Ba-L Env is included for comparison. In other experiments (data not shown), the double non-N-linked glycosylated CXCR4 mutant yielded a slightly more elevated level of coreceptor activity than the single N11A mutant for the same panel of Envs tested in the study shown in Fig. 3, but the N-terminal site clearly had the greatest influence, and the control mutation (T13A) imparted this expanded coreceptor activity as well (Fig. 3). The fact that some R5 Envs from alternate clades, like the two clade C Envs examined, were not able to use the non-N-linked glycosylated CXCR4 is not surprising because clade C X4 isolates rarely occur (14, 70). This may indicate that the clade C R5 Env-coreceptor interaction is more distinct than that of R5 Envs from other clades, like A, B, and F; on the other hand, we predict that a clade D R5 isolate would have been able to utilize the non-N-linked CXCR4 mutant coreceptor had there been a cloned gp160 available for testing.
FIG. 3.
Coreceptor function of non-N-linked glycosylated CXCR4 mutants in cell fusions assays with primary isolate R5 HIV-1 Envs. U373 target cells were transfected with a plasmid encoding the wild-type or a mutant coreceptor linked to the vaccinia virus promoter and infected with vCB21R-LacZ and vCB-3 (CD4). HeLa effector cells were transfected with a plasmid encoding Env (clades are indicated in parentheses) linked to a T7 promoter and infected with a vaccinia virus encoding T7 polymerase (vTF1-1). Duplicate cell mixtures were incubated at 37°C for 3 h. Fusion was assessed by measurement of β-Gal in detergent lysates of cells. The rates of β-Gal activity shown were obtained from separate samples in the same experiment. Error bars indicate the standard deviations of the mean values obtained for duplicate fusion assays. Data from a representative experiment are shown in the figure. Abs570nm, absorbance at 570 nm.
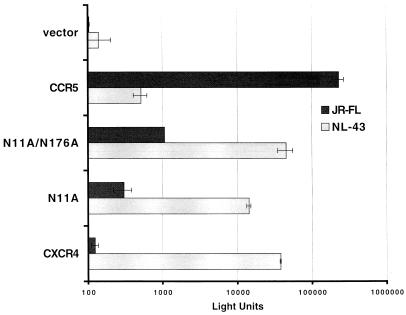
The HIV-1 Env-mediated cell fusion assay presents a proven and reliable model of HIV-1 Env-mediated fusion and receptor function (4, 17, 26, 41, 42, 49, 62, 76). However, an examination of the activities of these CXCR4 mutants in an alternate assay for virus entry was also performed. The data in Fig. 4 show that the non-N-linked glycosylated CXCR4 molecule can also support infection by a CCR5-dependent virus in an R5 JR-FL-pseudotyped luciferase reporter system. Both the single (N11A) and double (N11A N176A) non-N-linked glycosylated CXCR4 mutants supported this R5 Env-mediated virus infection, and the double mutant yielded a higher level of coreceptor activity. The background signals obtained with plasmid vector alone and with wild-type CXCR4 and CCR5 are shown for comparison. In additional experiments, the level of signal obtained with the single N176A CXCR4 mutant appeared no higher than that of wild-type CXCR4 activity in the pseudovirus assay. We have consistently observed that levels of R5 virus entry signal for cells expressing non-N-linked glycosylated CXCR4 in the luciferase pseudovirus assay are often lower than those of the cell fusion assay, and this was also observed with the charged-to-alanine (D187A) CXCR4 mutant, another R5-enhancing alteration also found by another group (23, 87). On the one hand, the pseudovirus assay is dependent on Env-mediated fusion as well as reverse transcription, pre-integration complex formation, and nuclear translocation, while our recombinant Env cell-cell fusion system was devised to eliminate the dependency on these postentry events in measuring Env fusion activity (17). These differences between the two systems may be inherent. Alternatively, the differences in the relative signal levels of the two assays might reflect postbinding roles of CCR5 for R5 HIV-1 isolates. Whether there are indeed other mechanisms at work in the entry process of HIV that can account for the efficiency differences described here, such as an event during the postentry phase of virus replication, as demonstrated with simian immunodeficiency virus (24), remains to be shown. Nevertheless, the results obtained by both of these assay systems support the hypothesis that non-N-linked glycosylated CXCR4 can serve as a coreceptor for R5 Envs.
FIG. 4.
Coreceptor function of non-N-linked glycosylated CXCR4 mutants in virus infection assays with an R5 HIV-1 Env. U373 CD4+ cells were transfected with a plasmid (pCDNA3) encoding the wild-type or a mutant coreceptor. Wells of cells (triplicate) were infected with the indicated HIV-1 Env-luciferase reporter virus. Infection was assessed at day 4 by measuring the amounts of luciferase activity in cell lysates. Error bars indicate the standard deviations of the mean values obtained from triplicate wells. This experiment was performed three times, and the data from a representative experiment are shown in the figure.
Identification of CXCR4 N-linked glycosylations.
Previously we examined recombinant vaccinia virus-expressed CXCR4 by Western blot analysis with a polyclonal rabbit antiserum raised against the N terminus of the molecule (49). In those studies, the predominant molecular species of CXCR4 had a molecular mass of approximately 47 to 48 kDa. Also apparent was a second, less-intense band with an apparent molecular mass of ∼94 to 97 kDa, double that of the predominant species. We have consistently observed this monomer/dimer pattern for CXCR4 as well as CCR5 (data not shown), and the ratios of the two bands can vary depending on the SDS-PAGE conditions. Virtually identical results were obtained by Doms and colleagues with an hemagglutinin epitope-tagged CXCR4-vaccinia virus construct (11). The predicted molecular mass of CXCR4, 39.7 kDa, indicated that a posttranslational N-linked glycosylation event likely had occurred on the protein. The first clear evidence that CXCR4 was indeed N glycosylated on at least one of these sites was provided by experiments using endoglycosidase F treatment of recombinant vaccinia virus-expressed CXCR4 followed by SDS-PAGE and Western blotting (11). To expand on these observations and precisely identify the sites of N-linked glycosylation—and, more importantly, correlate these findings to functional activity—we performed a biochemical analysis by SDS-PAGE and Western blotting of the non-N-linked glycosylated CXCR4 mutants.
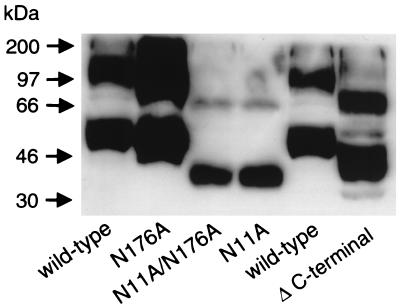
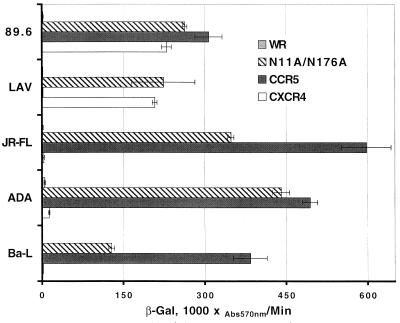
Western blot detection of CXCR4 following SDS-PAGE separation has been notoriously difficult. We and others have found analysis by SDS-PAGE and Western blotting of wild-type CXCR4 or mutants that are expressed transiently by plasmid-transfected cells unsatisfactory. This is likely due to a combination of low-level expression and low-affinity antibody. In order to obtain unambiguous results, we constructed several new recombinant vaccinia viruses encoding wild-type or one of several mutant CXCR4s. We analyzed lysates of cells infected with these vaccinia viruses to biochemically characterize the CXCR4 N glycosylations by Western blotting with MAb 4G10, raised against the CXCR4 N terminus. Binding of 4G10 to CXCR4 also appeared to be unaffected by the N-terminal glycosylation (23) (Fig. 5 and data not shown). Using this approach, we determined that the N11A mutant had a significantly lower apparent molecular mass (monomer, ∼41 to 42 kDa) than wild-type CXCR4 and was very close to the predicted size of unmodified CXCR4 (∼40 kDa). There was no detectable size difference between the single N176A mutant and wild-type CXCR4 or between the double N11A N176A mutant and the single N11A CXCR4 mutant. The blot was purposefully overexposed to show the two-band pattern in the N11A and N11A N176A lanes. A shorter autoradiographic exposure period revealed the same broad band at both positions for the other samples (data not shown), as was noted in prior work with wild-type and epitope-tagged CXCR4 (11, 49). Thus, if there is any N-linked glycosylation present at N176A, it is too small of a modification to be measurable in this assay. The non-N-linked glycosylated CXCR4 monomer was even smaller than that of an additional CXCR4 mutant that was used as a relative molecular mass marker in this experiment (a C-terminal 42-amino-acid [5-kDa] deletion construct with a molecular mass of ∼42 to 43 kDa). Also, the apparent dimer bands were equally shifted downward in all cases (Fig. 5). Taken together, our data indicate that the principal site of N-linked glycosylation of CXCR4 is in the N terminus and consists of an approximated 5- to 6-kDa carbohydrate moiety. Using these CXCR4-encoding recombinant vaccinia viruses to express CXCR4, we observed identical molecular mass patterns of monomeric and dimeric CXCR4 in a variety of cells, including primary human macrophages as well as the mouse 3T3 and human HeLa, U373, and HOS cell lines (data not shown). The double non-N-linked glycosylated CXCR4 mutant-encoding vaccinia virus was also examined functionally (Fig. 6), and although there might have been a slight decrease in relative expression efficiencies in whole-cell lysates (Fig. 5), this mutant was clearly quite efficient in supporting Env-mediated fusion by several prototypic R5 isolates in comparison to vaccinia virus-expressed CCR5. The non-N-linked glycosylated mutants were fully functional for prototypic X4 (LAV) and R5X4 (89.6) Envs as well (Fig. 6).
FIG. 5.
Biochemical analysis of non-N-linked glycosylated mutant CXCR4s: Western blot of wild-type CXCR4 and non-N-linked glycosylated CXCR4 mutants expressed by recombinant vaccinia viruses. Δ C-terminal, C-terminal 42-amino-acid deletion construct with a molecular mass of ∼43 kDa. Lysates were prepared from BSC-1 cells infected with a vaccinia virus encoding the indicated CXCR4 gene and analyzed by SDS-PAGE followed by Western blotting with a mouse MAb to CXCR4 (4G10).
FIG. 6.
Functional analysis of vaccinia virus-expressed double non-N-linked glycosylated CXCR4: coreceptor function of recombinant vaccinia virus-encoded non-N-linked glycosylated mutant CXCR4 in cell fusion assays with clade B Envs. Target cells were infected with vCB-21R-LacZ, vCB-3 (CD4), and either WR, vHC-7 (N11A/N176A), vHC-1 (CCR5), or vHC-3 (CXCR4). HeLa HIV-1 Env-expressing effector cells were infected with a vaccinia virus encoding T7 polymerase (vTF1-1) and the indicated Env. Cell mixtures (duplicates) were incubated at 37°C for 2.5 h. Fusion was assessed by measurement of β-Gal in detergent lysates of cells. The rates of β-Gal activity shown were obtained from separate samples in the same experiment. Error bars indicate the standard deviations of the mean values obtained from duplicate fusion assays. This experiment has been performed multiple times, and the data from a representative experiment are shown in the figure. Abs570nm, absorbance at 570 nm.
DISCUSSION
The HIV-1 coreceptors have complex membrane topologies consisting of an N-terminal domain, three extracellular and intracellular loops, and a cytoplasmic C-terminal tail. Their exact three-dimensional structure is presently unknown. However, theoretical models of CXCR4 and CCR5 based on the physically determined structures of both bacteriorhodopsin and rhodopsin (79, 85, 86) as well as analysis of the amino acid sequences of related receptors have been constructed (34, 36; S. Durell, personal communication). Most recently, a model for CCR5 derived by a similar methodology was presented in which further constraints were placed on the theoretical modeling procedure by incorporation of MAb binding data (61). These models suggest that the coreceptors are barrel shaped, with the extracellular loops and N terminus brought closer together by two extracellular disulfide bonds; perhaps more significantly, a clear distinction of electrostatic potentials in the extracellular elements is evident, with CXCR4 possessing a more negative surface charge. Indeed, data suggesting that both pairs of cysteines are likely involved in disulfide bond formation have been provided (15, 23), and these observations aid in grasping the many reports that indicate multiple extracellular regions in CCR5 (5, 13, 39, 43, 46, 47, 54, 62, 68, 72, 75, 76, 89) as well as CXCR4 (16, 23, 40, 62, 69, 83) coreceptor function and that coreceptor-Env interactions are quite complex. An additional complexity to this interaction comes from the assessment of the Env regions involved in coreceptor use and goes well beyond the notion of just an electrostatic interaction between the V3 loop and a coreceptor (see recent reviews in references 8 and 55).
In this study we showed that the removal of N-linked glycosylation sites of CXCR4 allows this molecule to serve as a functional coreceptor for otherwise-restricted R5 isolate Envs. Unlike CXCR4, the other principal coreceptor, CCR5, does not appear to possess any N-linked glycosylation modifications, although there is a potential site in the receptor's ecl-3 (48). Rather, it was concluded that only O-linked glycosylation modifications to CCR5 were evident. Further, it was reported that sulfation of the N terminus of the CCR5 coreceptor is important for function, with sulfated tyrosines contributing to the binding of CCR5 natural ligands as well as gp120-CD4 complexes (48). Along this line, the removal of a carbohydrate moiety from CXCR4 likely results in an even more-negative surface charge on the molecule, although at this time we cannot exclude the possibility that the N-linked carbohydrate structure is further modified in a way that affects the overall charge (e.g., sialic acid addition). Although our results show some additive biological effect of double N-linked glycosylation site removal, they clearly show that only one site (i.e., at the N terminus) predominates in its biological effect; additionally, we showed biochemically that only the N-terminal site imparts a modification measurable by SDS-PAGE, and since N-linked carbohydrate structures are invariably large, this strongly indicates that only this site is used in the BSC-1 (Fig. 5), murine 3T3, human HOS, HeLa, and U373-MG cell lines and in primary human macrophages (data not shown). Our attempts to measure these biological effects of N-linked glycosylation site removal by other means, including tunicamycin treatment of cells, were unsuccessful, apparently due to cellular toxicity (data not shown). Indeed, the finding that prevention of N-linked glycosylation would allow CXCR4 to function as a coreceptor for R5 Envs while full function for X4 Envs was retained was quite unexpected.
We hypothesize that removal of the carbohydrate moieties in CXCR4 results in its enhanced coreceptor activity with CCR5-dependent R5 Envs by unmasking existing structures capable of interacting with these Envs. It may be that CCR5-restricted Envs are adapted to recognizing a coreceptor with a non-N-linked glycosylated N terminus whereas CXCR4-restricted Envs can accommodate such a configuration. Taken together, our data further support the notion that despite differences in the primary sequences of their extracellular regions, there is perhaps an underlying conserved similarity between the three-dimensional structures of CXCR4 and CCR5 and that subtle alterations in CXCR4 (i.e., removal of carbohydrate), or mutation in Env to accommodate CXCR4 N glycosylation, allows R5 Envs to utilize it as a coreceptor. We also note that among the set of CXCR4 mutations that result in enhanced coreceptor activity for R5 Envs is the disruption of what is likely a disulfide bond between the N terminus and ecl-3 (23). A possible explanation for the underlying mechanism of this prior observation, in light of the present data, is that this alteration relaxes the barrel shape of CXCR4 and thereby repositions the existing N-terminal glycosylation moiety, allowing better exposure of contact sites for R5 Env interaction.
We have also consistently observed the apparent monomer/dimer pattern of CXCR4 and CCR5 (Fig. 5 and data not shown). The first suggestion of an oligomeric feature of an HIV coreceptor was shown by an immunoprecipitation assay using metabolically labeled CCR5; a monomer/dimer pattern was reported in a low-SDS environment (6). More recently, a similar SDS-PAGE pattern has been reported for CXCR4 and CCR5 (74); the chemokine ligands were shown to induce a monomer-to-dimer transition. Further studies need to be performed to determine the nature and significance of these observations. Our data are derived from SDS-PAGE analysis under reducing conditions, but samples were not boiled; this suggests that strong hydrophobic interactions are involved. We feel that the high-molecular-mass species we observed is not an alternative or more heavily glycosylated CXCR4 molecule, since we still observed a dimer in the double non-N-linked glycosylated mutant, although one might speculate that the N-terminal carbohydrate moiety plays a role in stabilizing the dimer, based on the data shown in Fig. 5. The fact that the anti-CXCR4 MAb 12G5 had been demonstrated to differentially inhibit HIV-1 infection in both a cell type- and virus strain-dependent manner (63) had first prompted the suggestion that the CXCR4 molecule itself is differentially processed, as in macrophages, resulting in it being utilized differently by various isolates (37). It has been suggested that a high-molecular-mass species of CXCR4 that is defective in coreceptor activity is present in human macrophages (60) and that posttranslational glycosylation could account for this observation; however, we noted that recombinant-expressed wild-type and non-N-linked glycosylated CXCR4 in human macrophages yielded molecular mass patterns identical to those shown in Fig. 5. Indeed, if multimeric-complex formation between the oligomeric HIV-1 Env and its cellular receptors is required for fusion pore formation (65) and subsequent virus entry, then the very existence of CD4-independent strains of HIV-1, HIV-2 (56, 58, 73), and simian immunodeficiency virus (44) supports the notion of oligomeric coreceptors.
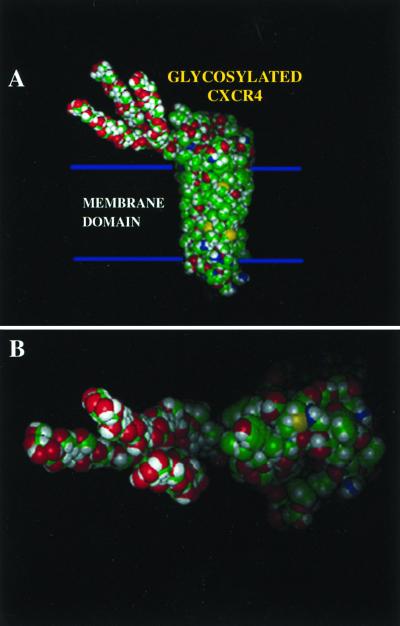
A revised schematic model of CXCR4, in which we have incorporated a hypothetical three-branched 6-kDa carbohydrate structure on the molecule's N-terminal domain based on our estimated molecular mass differences, is shown in Fig. 7. In viewing this model, it becomes readily apparent how such a structure could potentially mask elements of the coreceptor's extracellular domains. Although theoretical, we feel it is quite relevant to present this model in the context of this report because it dramatically shows how readily such a structure could cover underlying elements of not only the coreceptor's N terminus but the extracellular loops as well, a feature rarely appreciated in stick figure diagrams. As more MAbs specific for the CXCR4 coreceptor become available to complement the growing number of available mutant coreceptor molecules, further detailed mapping and modeling constraints will be possible to help refine these theoretical three-dimensional models.
FIG. 7.
Schematic model of the HIV-1 coreceptor CXCR4 with a representation of the amino-terminal N-linked glycosylation moiety. The carbohydrate is drawn as a simple three-branch structure with a molecular mass of ∼6 kDa based on the measured molecular mass shift observed during SDS-PAGE analysis under reducing conditions. Green represents carbon, red represents oxygen, yellow represents nitrogen, and white represents hydrogen. (A) Side view; (B) top view.
Our results concerning the CXCR4 N-terminal glycosylation also suggest that just a posttranslational modification of CXCR4 could prevent an R5 isolate from exhibiting a dualtropic phenotype. The CCR5 coreceptor does not have an N-terminal N-linked glycosylation site, and, interestingly, the rarely employed CCR2 coreceptor, whose primary sequence is most homologous to that of CCR5, contains an N-terminal N-linked glycosylation site. However, removal of this site did not expand CCR2 coreceptor activity; only a fourfold increase, to 10% of the activity of CCR5 with a single R5 Env, ADA (data not shown), occurred, while several other isolates yield 35 to 125% of the level of CCR5 coreceptor activity with the non-N-linked glycosylated CXCR4 (Fig. 2). A larger study with more R5 isolates appears warranted in order to fully assess and validate the breadth of these present findings. Taken together, our data provide further evidence suggesting that there is significant homology on a three-dimensional level for Env interaction sites of the CXCR4 and CCR5 coreceptors, perhaps even more so than that between CCR5 and CCR2.
It is R5 HIV-1 strains that are largely responsible for virus transmission, and individuals who lack CCR5 due to a natural mutation in the gene (ccr5Δ32 allele) are resistant to HIV-1 infection (31, 57, 78). HIV-1 X4 isolates tend to emerge much later in the infection course, and the tropism switch from R5 to X4 viruses correlates with progression of the infection to symptomatic AIDS (30, 81, 82). Part of the nature of this tropism switch is clearly related to genetic changes in the env nucleotide sequence, but not all infected individuals who progress to AIDS develop X4 isolates, and the precise nature of the driving force behind the in vivo evolution of HIV is not wholly understood. Our findings now add further complexity to this area because they demonstrate the possibility that early-stage primary R5 isolates could infect target cells via CD4-CXCR4 receptors under circumstances of differential glycosylation of CXCR4 without an accompanying genetic change in Env. Although clearly speculative, the existence of non-N-linked glycosylated CXCR4 or CXCR4 glycoforms in an individual, as result of genetic or environmental influences or even the infection process itself, that allow for R5 Env recognition could have broad-reaching implications for the HIV infection and pathogenesis process, as well as for outcomes in the host. Indeed, glycoforms of a protein may be cell type or even cell cycle dependent (77). Whether there are alternate glycoforms of CXCR4 in vivo remains to be determined by experiments that would certainly be highly challenging considering the present difficulty in detecting endogenous CXCR4 by blotting techniques.
Finally, unlike the CCR5-CD4 interaction, the association of CXCR4 with CD4 appears to be greatly enhanced by gp120; the coimmunoprecipitation of CXCR4 and CD4 is largely gp120 dependent (35, 59, 90). We are now examining whether these CXCR4 mutants, which have the capacity to serve as universal HIV-1 coreceptors, have an enhanced affinity for CD4, similar to the CCR5-CD4 interaction, or whether they exhibit their effect through an enhanced Env-coreceptor interaction. A more universal coreceptor has better potential in the development of novel therapeutic agents targeted against HIV-infected cells, such as a cytocidal pseudovirus (45, 80) with CD4 and non-N-linked glycosylated CXCR4 on its surface, by targeting a wider range of HIV-1 isolate-infected cells. The continued characterization of the interactions between HIV-1 Env and its coreceptors will aid in our understanding of the virus infection process and in devising methods to prevent it.
ACKNOWLEDGMENTS
We thank Agnes Jones-Trower for assistance in densitometer scanning and molecular mass determinations, Tzanko Stantchev for help with flow cytometry, and Joseph Isaac for viruses and cells. We are grateful to Frantisek Bizik, Stuart Durell, and Pradman Qasba for help with the molecular modeling of CXCR4 structures. A number of reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
This study was supported by NIH grants R29AI414110 and R01AI43885 and USUHS grant RO73FG to C.C.B.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander W A, Moss B, Fuerst T R. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alkhatib G, Broder C C, Berger E A. Cell type-specific fusion cofactors determine human immunodeficiency virus type 1 tropism for T-cell lines versus primary macrophages. J Virol. 1996;70:5487–5494. doi: 10.1128/jvi.70.8.5487-5494.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 5.Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M A. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 6.Benkirane M, Jin D Y, Chun R F, Koup R A, Jeang K T. Mechanism of transdominant inhibition of CCR5-mediated HIV-1 infection by ccr5Δ32. J Biol Chem. 1997;272:30603–30606. doi: 10.1074/jbc.272.49.30603. [DOI] [PubMed] [Google Scholar]
- 7.Berger E A, Doms R W, Fenyö E M, Korber B T, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 8.Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 9.Berger E A, Nussbaum O, Broder C C. HIV envelope glycoprotein/CD4 interactions: studies using recombinant vaccinia virus vectors. In: Karn J, editor. HIV: a practical approach. Vol. 2. Oxford, United Kingdom: Oxford University Press; 1995. pp. 123–145. [Google Scholar]
- 10.Berson J F, Doms R W. Structure-function studies of the HIV-1 coreceptors. Semin Immunol. 1998;10:237–248. doi: 10.1006/smim.1998.0130. [DOI] [PubMed] [Google Scholar]
- 11.Berson J F, Long D, Doranz B J, Rucker J, Jirik F R, Doms R W. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bieniasz P D, Cullen B R. Chemokine receptors and human immunodeficiency virus infection. Front Biosci. 1998;3:D44–D58. doi: 10.2741/a265. [DOI] [PubMed] [Google Scholar]
- 13.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S, Caron M G, Cullen B R. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bjorndal A, Sonnerborg A, Tscherning C, Albert J, Fenyö E M. Phenotypic characteristics of human immunodeficiency virus type 1 subtype C isolates of Ethiopian AIDS patients. AIDS Res Hum Retrovir. 1999;15:647–653. doi: 10.1089/088922299310944. [DOI] [PubMed] [Google Scholar]
- 15.Blanpain C, Lee B, Vakili J, Doranz B J, Govaerts C, Migeotte I, Sharron M, Dupriez V, Vassart G, Doms R W, Parmentier M. Extracellular cysteines of CCR5 are required for chemokine binding, but dispensable for HIV-1 coreceptor activity. J Biol Chem. 1999;274:18902–18908. doi: 10.1074/jbc.274.27.18902. [DOI] [PubMed] [Google Scholar]
- 16.Brelot A, Heveker N, Pleskoff O, Sol N, Alizon M. Role of the first and third extracellular domains of CXCR-4 in human immunodeficiency virus coreceptor activity. J Virol. 1997;71:4744–4751. doi: 10.1128/jvi.71.6.4744-4751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broder C C, Berger E A. Fusogenic selectivity of the envelope glycoprotein is a major determinant of human immunodeficiency virus type 1 tropism for CD4+ T-cell lines vs. primary macrophages. Proc Natl Acad Sci USA. 1995;92:9004–9008. doi: 10.1073/pnas.92.19.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broder C C, Dimitrov D S. HIV and the 7-transmembrane domain receptors. Pathobiology. 1996;64:171–179. doi: 10.1159/000164032. [DOI] [PubMed] [Google Scholar]
- 19.Broder C C, Dimitrov D S, Blumenthal R, Berger E A. The block to HIV-1 envelope glycoprotein-mediated membrane fusion in animal cells expressing human CD4 can be overcome by a human cell component(s) Virology. 1993;193:483–491. doi: 10.1006/viro.1993.1151. [DOI] [PubMed] [Google Scholar]
- 20.Broder C C, Earl P L. Recombinant vaccinia viruses: design, generation and isolation. Mol Biotechnol. 1999;13:223–246. doi: 10.1385/MB:13:3:223. [DOI] [PubMed] [Google Scholar]
- 21.Broder C C, Jones-Trower A. Coreceptor use by primate lentiviruses. In: Korber B, Foley B, Leitner T, McCutchan F, Hahn B, Mellors J W, Myers G, Kuilen C, editors. Human retroviruses and AIDS. III. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1999. pp. 20–45. [Google Scholar]
- 22.Carroll M W. Expression, analysis and immunogenicity of human immunodeficiency type I envelope glycoprotein in vaccinia virus. Ph.D. dissertation. Manchester, United Kingdom: University of Manchester; 1993. [Google Scholar]
- 23.Chabot D J, Zhang P F, Quinnan G V, Broder C C. Mutagenesis of CXCR4 identifies important domains for human immunodeficiency virus type 1 X4 isolate envelope-mediated membrane fusion and virus entry and reveals cryptic coreceptor activity for R5 isolates. J Virol. 1999;73:6598–6609. doi: 10.1128/jvi.73.8.6598-6609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chackerian B, Long E M, Luciw P A, Overbaugh J. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakrabarti S, Sisler J R, Moss B. Compact, synthetic, vaccinia virus early/late promoter for protein expression. BioTechniques. 1997;23:1094–1097. doi: 10.2144/97236st07. [DOI] [PubMed] [Google Scholar]
- 26.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 27.Clapham P R, Reeves J D, Simmons G, Dejucq N, Hibbitts S, McKnight A. HIV coreceptors, cell tropism and inhibition by chemokine receptor ligands. Mol Membr Biol. 1999;16:49–55. doi: 10.1080/096876899294751. [DOI] [PubMed] [Google Scholar]
- 28.Collman R, Balliet J W, Gregory S A, Friedman H, Kolson D L, Nathanson N, Srinivasan A. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J Virol. 1992;66:7517–7521. doi: 10.1128/jvi.66.12.7517-7521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type 1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 30.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O'Brien S J Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, and ALIVE Study. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 32.De Jong J-J, De Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dimitrov D S. Fusin—a place for HIV-1 and T4 cells to meet. Nat Med. 1996;2:640–641. doi: 10.1038/nm0696-640. [DOI] [PubMed] [Google Scholar]
- 34.Dimitrov D S, Broder C C. HIV and membrane receptors. Austin, Tex: Landes Bioscience; 1997. [Google Scholar]
- 35.Dimitrov D S, Norwood D, Stantchev T S, Feng Y, Xiao X, Broder C C. A mechanism of resistance to HIV-1 entry: inefficient interactions of CXCR4 with CD4 and gp120 in macrophages. Virology. 1999;259:1–6. doi: 10.1006/viro.1999.9747. [DOI] [PubMed] [Google Scholar]
- 36.Dimitrov D S, Xiao X, Chabot D J, Broder C C. HIV coreceptors. J Membr Biol. 1998;166:75–90. doi: 10.1007/s002329900450. [DOI] [PubMed] [Google Scholar]
- 37.Dittmar M T, McKnight A, Simmons G, Clapham P R, Weiss R A, Simmons P. HIV-1 tropism and co-receptor use. Nature. 1997;385:495–496. doi: 10.1038/385495a0. [DOI] [PubMed] [Google Scholar]
- 38.Doms R W, Peiper S C. Unwelcomed guests with master keys: how HIV uses chemokine receptors for cellular entry. Virology. 1997;235:179–190. doi: 10.1006/viro.1997.8703. [DOI] [PubMed] [Google Scholar]
- 39.Doranz B J, Lu Z H, Rucker J, Zhang T Y, Sharron M, Cen Y H, Wang Z X, Guo H H, Du J G, Accavitti M A, Doms R W, Peiper S C. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doranz B J, Orsini M J, Turner J D, Hoffman T L, Berson J F, Hoxie J A, Peiper S C, Brass L F, Doms R W. Identification of CXCR4 domains that support coreceptor and chemokine receptor functions. J Virol. 1999;73:2752–2761. doi: 10.1128/jvi.73.4.2752-2761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 42.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 43.Edinger A L, Amedee A, Miller K, Doranz B J, Endres M, Sharron M, Samson M, Lu Z H, Clements J E, Murphey-Corb M, Peiper S C, Parmentier M, Broder C C, Doms R W. Differential utilization of CCR5 by macrophage and T cell tropic simian immunodeficiency virus strains. Proc Natl Acad Sci USA. 1997;94:4005–4010. doi: 10.1073/pnas.94.8.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edinger A L, Blanpain C, Kunstman K J, Wolinsky S M, Parmentier M, Doms R W. Functional dissection of CCR5 coreceptor function through the use of CD4-independent simian immunodeficiency virus strains. J Virol. 1999;73:4062–4073. doi: 10.1128/jvi.73.5.4062-4073.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Endres M J, Jaffer S, Haggarty B, Turner J D, Doranz B J, O'Brien P J, Kolson D L, Hoxie J A. Targeting of HIV- and SIV-infected cells by CD4-chemokine receptor pseudotypes. Science. 1997;278:1462–1464. doi: 10.1126/science.278.5342.1462. [DOI] [PubMed] [Google Scholar]
- 46.Farzan M, Choe H, Martin K A, Sun Y, Sidelko M, Mackay C R, Gerard N P, Sodroski J, Gerard C. HIV-1 entry and macrophage inflammatory protein-1β-mediated signaling are independent functions of the chemokine receptor CCR5. J Biol Chem. 1997;272:6854–6857. doi: 10.1074/jbc.272.11.6854. [DOI] [PubMed] [Google Scholar]
- 47.Farzan M, Choe H, Vaca L, Martin K, Sun Y, Desjardins E, Ruffing N, Wu L, Wyatt R, Gerard N, Gerard C, Sodroski J. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J Virol. 1998;72:1160–1164. doi: 10.1128/jvi.72.2.1160-1164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard N P, Gerard C, Sodroski J, Choe H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 49.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 50.Fouchier R A, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gibbs C S, Zoller M J. Identification of electrostatic interactions that determine the phosphorylation site specificity of the cAMP-dependent protein kinase. Biochemistry. 1991;30:5329–5334. doi: 10.1021/bi00236a001. [DOI] [PubMed] [Google Scholar]
- 52.Golding H, Dimitrov D S, Manischewitz J, Broder C C, Robinson J, Fabian S, Littman D R, Lapham C K. Phorbol ester-induced down modulation of tailless CD4 receptors requires prior binding of gp120 and suggests a role for accessory molecules. J Virol. 1995;69:6140–6148. doi: 10.1128/jvi.69.10.6140-6148.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harrington R D, Geballe A P. Cofactor requirement for human immunodeficiency virus type 1 entry into a CD4-expressing human cell line. J Virol. 1993;67:5939–5947. doi: 10.1128/jvi.67.10.5939-5947.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hill C M, Kwon D, Jones M, Davis C B, Marmon S, Daugherty B L, DeMartino J A, Springer M S, Unutmaz D, Littman D R. The amino terminus of human CCR5 is required for its function as a receptor for diverse human and simian immunodeficiency virus envelope glycoproteins. Virology. 1998;248:357–371. doi: 10.1006/viro.1998.9283. [DOI] [PubMed] [Google Scholar]
- 55.Hoffman T L, Doms R W. HIV-1 envelope determinants for cell tropism and chemokine receptor use. Mol Membr Biol. 1999;16:57–65. doi: 10.1080/096876899294760. [DOI] [PubMed] [Google Scholar]
- 56.Hoxie J A, LaBranche C C, Endres M J, Turner J D, Berson J F, Doms R W, Matthews T J. CD4-independent utilization of the CXCR4 chemokine receptor by HIV-1 and HIV-2. J Reprod Immunol. 1998;41:197–211. doi: 10.1016/s0165-0378(98)00059-x. [DOI] [PubMed] [Google Scholar]
- 57.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 58.Kolchinsky P, Mirzabekov T, Farzan M, Kiprilov E, Cayabyab M, Mooney L J, Choe H, Sodroski J. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J Virol. 1999;73:8120–8126. doi: 10.1128/jvi.73.10.8120-8126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Evidence for cell-surface association between fusin and the CD4-gp120 complex in human cell lines. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 60.Lapham C K, Zaitseva M B, Lee S, Romanstseva T, Golding H. Fusion of monocytes and macrophages with HIV-1 correlates with biochemical properties of CXCR4 and CCR5. Nat Med. 1999;5:303–308. doi: 10.1038/6523. [DOI] [PubMed] [Google Scholar]
- 61.Lee B, Sharron M, Blanpain C, Doranz B J, Vakili J, Setoh P, Berg E, Liu G, Guy H R, Durell S R, Parmentier M, Chang C N, Price K, Tsang M, Doms R W. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J Biol Chem. 1999;274:9617–9626. doi: 10.1074/jbc.274.14.9617. [DOI] [PubMed] [Google Scholar]
- 62.Lu Z, Berson J F, Chen Y, Turner J D, Zhang T, Sharron M, Jenks M H, Wang Z, Kim J, Rucker J, Hoxie J A, Peiper S C, Doms R W. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc Natl Acad Sci USA. 1997;94:6426–6431. doi: 10.1073/pnas.94.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McKnight A, Wilkinson D, Simmons G, Talbot S, Picard L, Ahuja M, Marsh M, Hoxie J A, Clapham P R. Inhibition of human immunodeficiency virus fusion by a monoclonal antibody to a coreceptor (CXCR4) is both cell type and virus strain dependent. J Virol. 1997;71:1692–1696. doi: 10.1128/jvi.71.2.1692-1696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moore J P, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 65.Munoz-Barroso I, Durell S, Sakaguchi K, Appella E, Blumenthal R. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J Cell Biol. 1998;140:315–323. doi: 10.1083/jcb.140.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nussbaum O, Broder C C, Berger E A. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O'Brien W A, Koyanagi Y, Namazie A, Zhao J Q, Diagne A, Idler K, Zack J A, Chen I S Y. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 68.Picard L, Simmons G, Power C A, Meyer A, Weiss R A, Clapham P R. Multiple extracellular domains of CCR-5 contribute to human immunodeficiency virus type 1 entry and fusion. J Virol. 1997;71:5003–5011. doi: 10.1128/jvi.71.7.5003-5011.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Picard L, Wilkinson D, McKnight A, Gray P, Hoxie J, Clapham P, Weiss R. Role of the amino-terminal extracellular domain of CXCR-4 in human immunodeficiency virus type 1 entry. Virology. 1997;231:105–111. doi: 10.1006/viro.1997.8506. [DOI] [PubMed] [Google Scholar]
- 70.Ping L H, Nelson J A, Hoffman I F, Schock J, Lamers S L, Goodman M, Vernazza P, Kazembe P, Maida M, Zimba D, Goodenow M M, Eron J J, Jr, Fiscus S A, Cohen M S, Swanstrom R. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J Virol. 1999;73:6271–6281. doi: 10.1128/jvi.73.8.6271-6281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quinnan G V, Jr, Zhang P F, Fu D W, Dong M, Margolick J B. Evolution of neutralizing antibody response against HIV type 1 virions and pseudovirions in multicenter AIDS cohort study participants. AIDS Res Hum Retrovir. 1998;14:939–949. doi: 10.1089/aid.1998.14.939. [DOI] [PubMed] [Google Scholar]
- 72.Rabut G E, Konner J A, Kajumo F, Moore J P, Dragic T. Alanine substitutions of polar and nonpolar residues in the amino-terminal domain of CCR5 differently impair entry of macrophage- and dualtropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:3464–3468. doi: 10.1128/jvi.72.4.3464-3468.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reeves J D, Hibbitts S, Simmons G, McKnight A, Azevedo-Pereira J M, Moniz-Pereira J, Clapham P R. Primary human immunodeficiency virus type 2 (HIV-2) isolates infect CD4-negative cells via CCR5 and CXCR4: comparison with HIV-1 and simian immunodeficiency virus and relevance to cell tropism in vivo. J Virol. 1999;73:7795–7804. doi: 10.1128/jvi.73.9.7795-7804.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rodriguez-Frade J M, Vila-Coro A J, Martín A, Nieto M, Sanchez-Madrid F, Proudfoot A E, Wells T N, Martínez A C, Mellado M. Similarities and differences in RANTES- and (AOP)-RANTES-triggered signals: implications for chemotaxis. J Cell Biol. 1999;144:755–765. doi: 10.1083/jcb.144.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ross T M, Bieniasz P D, Cullen B R. Multiple residues contribute to the inability of murine CCR-5 to function as a coreceptor for macrophage-tropic human immunodeficiency virus type 1 isolates. J Virol. 1998;72:1918–1924. doi: 10.1128/jvi.72.3.1918-1924.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rucker J, Samson M, Doranz B J, Libert F, Berson J F, Broder C C, Vassart G, Doms R W, Parmentier M. Regions in β-chemokine receptors CCR-5 and CCR-2b that determine HIV-1 cofactor specificity. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 77.Rudd P M, Joao H C, Coghill E, Fiten P, Saunders M R, Opdenakker G, Dwek R A. Glycoforms modify the dynamic stability and functional activity of an enzyme. Biochemistry. 1994;33:17–22. doi: 10.1021/bi00167a003. [DOI] [PubMed] [Google Scholar]
- 78.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C-M, Saragosti S, Lapoum-Eroulie C, Cogniaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection of Caucasian individuals bearing mutant alleles of the CCR5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 79.Schertler G F, Hargrave P A. Projection structure of frog rhodopsin in two crystal forms. Proc Natl Acad Sci USA. 1995;92:11578–11582. doi: 10.1073/pnas.92.25.11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schnell M J, Johnson J E, Buonocore L, Rose J K. Construction of a novel virus that targets HIV-1-infected cells and controls HIV-1 infection. Cell. 1997;90:849–857. doi: 10.1016/s0092-8674(00)80350-5. [DOI] [PubMed] [Google Scholar]
- 81.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, de Goede R E, van Steenwijk R P, Lange J M, Schattenkerk J K, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Strizki J M, Turner J D, Collman R G, Hoxie J, González-Scarano F. A monoclonal antibody (12G5) directed against CXCR-4 inhibits infection with the dual-tropic human immunodeficiency virus type 1 isolate HIV-189.6 but not the T-tropic isolate HIV-1HxB. J Virol. 1997;71:5678–5683. doi: 10.1128/jvi.71.7.5678-5683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 85.Unger V M, Hargrave P A, Baldwin J M, Schertler G F. Arrangement of rhodopsin transmembrane alpha-helices. Nature. 1997;389:203–206. doi: 10.1038/38316. [DOI] [PubMed] [Google Scholar]
- 86.Unger V M, Schertler G F. Low resolution structure of bovine rhodopsin determined by electron cryo-microscopy. Biophys J. 1995;68:1776–1786. doi: 10.1016/S0006-3495(95)80354-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang Z X, Berson J F, Zhang T Y, Cen Y H, Sun Y, Sharron M, Lu Z H, Peiper S C. CXCR4 sequences involved in coreceptor determination of human immunodeficiency virus type-1 tropism. Unmasking of activity with M-tropic Env glycoproteins. J Biol Chem. 1998;273:15007–15015. doi: 10.1074/jbc.273.24.15007. [DOI] [PubMed] [Google Scholar]
- 88.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 89.Wu L, LaRosa G, Kassam N, Gordon C J, Heath H, Ruffing N, Chen H, Humblias J, Samson M, Parmentier M, Moore J P, Mackay C R. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J Exp Med. 1997;186:1373–1381. doi: 10.1084/jem.186.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiao X, Wu L, Stantchev T S, Feng Y R, Ugolini S, Chen H, Shen Z, Riley J L, Broder C C, Sattentau Q J, Dimitrov D S. Constitutive cell surface association between CD4 and CCR5. Proc Natl Acad Sci USA. 1999;96:7496–7501. doi: 10.1073/pnas.96.13.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]