Abstract
Amprenavir (Agenerase, 141-W94, VX-478) is a human immunodeficiency virus type 1 (HIV-1) protease inhibitor (PRI) recently approved for the treatment of HIV-1 infection in the United States. A major cause of treatment failure is the development of resistance to PRIs. One potential use for amprenavir is as salvage therapy for patients for whom treatment that includes one (or more) of the other four currently approved PRIs—saquinavir, indinavir, ritonavir, and nelfinavir—has failed. We evaluated the cross-resistance to amprenavir of viruses that evolved during treatment with the two most commonly prescribed PRIs, nelfinavir and indinavir. Unexpectedly, a dramatic increase in susceptibility (2.5- to 12.5-fold) was observed with 20 of 312 (6.4%) patient viruses analyzed. The most pronounced increases in susceptibility were strongly associated with an N88S mutation in protease. All viruses that carried the N88S mutation were hypersensitive to amprenavir. Site-directed mutagenesis studies confirmed the causal role of N88S in determining amprenavir hypersensitivity. The presence of the N88S mutation and associated amprenavir hypersensitivity may be useful in predicting an improved clinical response to amprenavir salvage therapy.
Since the discovery of human immunodeficiency virus (HIV) in the early 1980s, an intense drug discovery effort has resulted in the approval of 14 antiviral drugs, with many more in development (3). The approved drugs inhibit HIV replication by interfering with the enzymatic activities of either protease (PR) or reverse transcriptase (RT). The lack of proofreading functions that is inherent in RT, coupled with error-prone replication at a high rate, allows HIV to mutate readily. This high mutation frequency contributes to the ability of HIV to evade successful long-term therapy, resulting in viral load rebound (4).
Multi-drug-resistant HIV variants pose an increasing problem for the care of infected patients (2, 5, 6, 10). Determination of the genotypes of such viruses frequently reveals complex patterns of mutations in PR and RT, thus complicating predictions of sensitivity or resistance to antiviral drugs. Accurate measurements of phenotypic drug susceptibility can be used to determine patterns of cross-resistance to existing drugs. This type of analysis is an increasingly important tool for evaluating investigational or newly approved drugs. The most recently approved PR inhibitor (PRI), amprenavir, is a promising candidate for salvage therapies, in part because available data suggest good activity against viruses resistant to other PRIs (12, 13, 21).
Utilizing a recently described phenotypic drug-susceptibility assay (14, 16a), we determined the drug susceptibilities of viruses present in infected individuals whose combination therapy regimens, including nelfinavir or indinavir, the two most commonly prescribed PRIs, were failing (viral loads of >500 copies per ml). A measure of relative susceptibility is obtained by comparing the 50% inhibitory concentration (IC50) for the patient virus to that for a reference virus, derived from the NL4-3 infectious HIV type 1 (HIV-1) DNA clone (1). Values obtained from single measurements that are less than or more than 2.5-fold those of the reference exceed the normal variation of the assay and are considered indicative of altered susceptibility to a drug. Hypersensitivity is defined as a reduction in the IC50 of ≥2.5-fold (i.e., a susceptibility value of ≤0.4) relative to the NL4-3 reference strain. A significant proportion of viruses (20 of 312, or 6.4%) from patients that had been treated with nelfinavir and/or indinavir exhibited hypersensitivity to amprenavir (Table 1).
TABLE 1.
PRI susceptibilities and PR genotypes of 20 patient-derived HIV-1 viruses displaying hypersensitivity to amprenavir
| Patient no. | Relative susceptibilitya
|
PR mutationb
|
|||||
|---|---|---|---|---|---|---|---|
| SQV | IDV | RTV | NFV | AMP | Resistance associated | Polymorphism(s) | |
| 1 | 0.73 | 2.11 | 1.72 | 8.92 | 0.08 | K20T, M36I, L63Q, N88S | K14R, I15V, E35D, R41K, I62V |
| 2 | 0.26 | 6.16 | 1.50 | 21.06 | 0.09 | M46L, L63P, N88S | I13I/V, E35D, I64V, I72V |
| 3 | 1.55 | 3.15 | 1.22 | 11.06 | 0.10 | L63P, V77I, N88S | I62V |
| 4 | 1.20 | 1.49 | 3.38 | 15.87 | 0.15 | M36I, L63P, A71A/T, N88S | I13V, E35D, I62V |
| 5 | 1.88 | 6.31 | 1.49 | 29.95 | 0.15 | K20M, M36V, M46I, L63P, N88S | I13V, N37A, I62V, I93L |
| 6 | 1.41 | 5.47 | 1.85 | 16.76 | 0.16 | M46I, L63P, V77I, N88S | I93I/L |
| 7 | 1.28 | 7.61 | 3.36 | 24.67 | 0.16 | M46I, L63P, N88S | I13V, K14R, N37D, I93L |
| 8 | 1.80 | 7.56 | 1.95 | 18.61 | 0.20 | M46I, L63P, V77I, N88S | I13V, R41K, I93L |
| 9 | 1.81 | 1.15 | 3.70 | 5.71 | 0.23 | L10I, K20K/R, M36I, I54V, L63H, L90L/M | I13V, I62V, I64V, I72V, N83D |
| 10 | 2.05 | 5.58 | 1.59 | 15.18 | 0.24 | L10I, M46I, L63P, A71T, V77I, N88S | E35D, N37S |
| 11 | 1.22 | 4.55 | 2.55 | 9.55 | 0.28 | L63P, V77I, N88N/S | R41K, I62V, I93I/L |
| 12 | 0.12 | 0.77 | 3.81 | 1.24 | 0.29 | L10L/F, M36M/L, M46M/I, L63S, V77I, V82A/T | I64V, I72T |
| 13 | 0.38 | 1.06 | 8.12 | 1.65 | 0.30 | K20R, M36I, I54V, V82A | R41K, I64V, H69Y |
| 14 | 8.99 | 13.59 | 6.29 | 63.05 | 0.30 | K20T, L63P, A71V, N88S, L90M | I15V, R41K, K45R, R57K, I72M |
| 15 | 0.54 | 1.27 | 0.83 | 2.59 | 0.32 | K20K/M, L63P, A71A/T, N88N/D/S | I13V, I15V, E35D, N37D |
| 16 | 0.62 | 1.12 | 4.36 | 3.56 | 0.32 | M36I, I54I/V, L63P, A71A/V, V82V/A | E35D, I62V, H69Q |
| 17 | 0.53 | 0.60 | 0.57 | 0.45 | 0.33 | M36M/I, L63Q, V77V/I, N88N/S | K14R, I15V, N37N/S, R41K, I62V |
| 18 | 0.42 | 1.28 | 0.63 | 27.62 | 0.36 | D30N, M36I, L63H, A71V | I13V, E35D, N37D, R57K, I62V, I64I/V, I93L |
| 19 | 0.32 | 0.69 | 0.35 | 2.99 | 0.37 | D30D/N, M36V | E35D, N37S, I64V |
| 20 | 1.16 | 1.37 | 1.15 | 29.13 | 0.37 | D30N, L33I, M36M/I, L63P, N88D | T12A/V, I13V, R57K, I93L |
Values represent the fold change in IC50 relative to the reference, representing the mean of the two determinations. Reproducibility studies have demonstrated the variability of PRI drug susceptibility results to be less than twofold with replicate testing of samples. Mean relative susceptibility values (the IC50 for virus from the patient divided by the IC50 for the reference) of >2, indicative of reduced susceptibility, or of <0.5, indicative of increased susceptibility, are highlighted in bold type. Values are rounded to two decimal places. SQV, saquinavir; IDV, indinavir; RTV, ritonavir; NFV, nelfinavir; AMP, amprenavir.
Mixed virus populations exhibiting more than one amino acid at given positions are indicated by slashes.
Table 1 lists drug susceptibility data for 20 patient-derived viruses, in order of decreasing susceptibility to amprenavir. All but one of the 20 viruses (patient 17) exhibited decreased susceptibility to at least one PRI, most frequently nelfinavir. The PR genotypes of the viruses displaying hypersensitivity to amprenavir were determined by DNA sequencing using conventional dideoxynucleotide chain terminating sequencing methods (Perkin-Elmer Biosystems, Foster City, Calif.). Deduced PR amino acid sequences were compared to that of NL4-3 (Table 1) and to a list of mutations associated with PRI resistance (19). Thirteen of the 20 patient virus populations (65%) and notably all of the viruses displaying the most pronounced increases in susceptibility (≤0.2 relative susceptibility, which corresponds to a ≥5-fold increase in susceptibility) had an asparagine (N) to serine (S) substitution in PR at amino acid position 88. N88S was always observed in combination with mutations at various other positions, including amino acids 20, 36, 46, 63, and 77. No viruses from drug-naïve patients were found to carry this substitution (2, 10; our unpublished data). While other changes at amino acid 88, most commonly N88D, occurred in viruses isolated from patients for whom treatment with PRIs was failing, these alterations are not correlated with a significant increase in susceptibility to amprenavir (data not shown). Mutations D30N and V82A, each found in 3 of the 20 patient-derived viruses listed in Table 1, were less frequently associated with amprenavir hypersensitivity. However, the increases in amprenavir susceptibility were less dramatic than those with N88S. These substitutions were also observed in viruses that did not exhibit hypersensitivity to amprenavir (data not shown), implying less direct roles for D30N and V82A in increased amprenavir susceptibility.
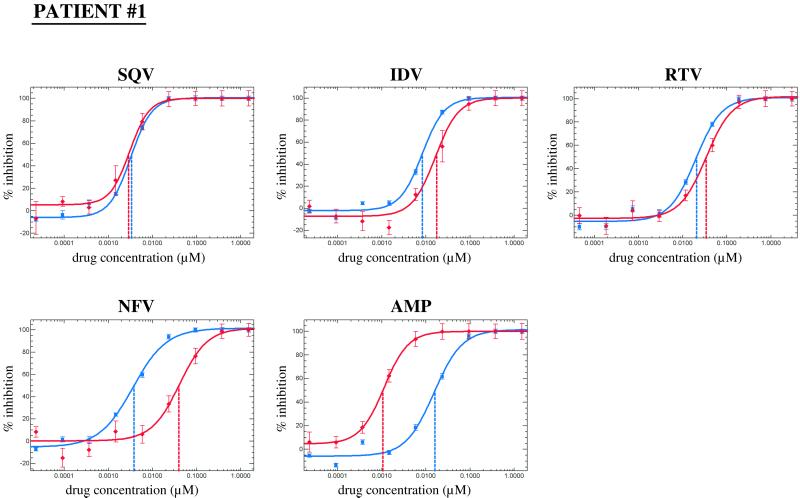
A representative PRI susceptibility profile for virus from patient 1 is shown in Fig. 1. The shift of the amprenavir susceptibility curve to lower drug concentrations, i.e., to the left of the reference curve, reflects increased susceptibility. The nelfinavir curve is shifted towards higher drug concentrations, i.e., to the right, indicating reduced susceptibility. Susceptibilities to saquinavir, indinavir, and ritonavir did not vary significantly (less than 2.5-fold) from those of the reference strain.
FIG. 1.
PRI susceptibility profile of virus from patient 1. Susceptibility to PRI drugs was assessed by comparing drug susceptibility curves obtained using the vector containing PR and RT coding sequences from the patient-derived virus (red lines) with a reference construct containing PR and RT from pNL4-3 (blue lines). The IC50 is determined from the curve and indicated by vertical lines. The IC50s of saquinavir (SQV), indinavir (IDV), ritonavir (RTV), nelfinavir (NFV), and amprenavir (AMP) for the reference control were 3.4, 8.3, 20.6, 3.8, and 16.0 nM, respectively.
Based on the available treatment histories of the patients harboring viruses with hypersensitivity to amprenavir, this phenotype appears to be associated with the failure of nelfinavir- or indinavir-containing treatment regimens. N88S has been identified previously in viruses isolated after in vitro passage in the presence of an investigational PRI, SC-55389A (20, 22), and in viruses isolated from patients treated with nelfinavir (16). Recently, viruses which emerged early after in vitro selection in the presence of an investigational PRI, BMS 232632, were found to contain the N88S mutation (4a). Intriguingly, N88S was found to arise in only two of the three laboratory strains used, suggesting that different genetic backgrounds could predispose viruses to alternative resistance pathways of decreased PRI susceptibility. These data raise the possibility that new PRIs may select more frequently for the N88S substitution than do nelfinavir or indinavir.
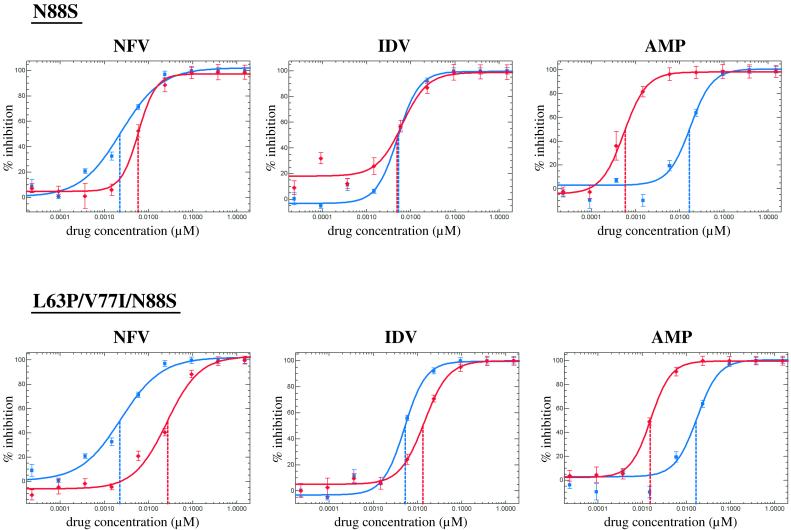
To delineate the role of individual mutations in PRI susceptibility, site-specific oligonucleotide-directed mutagenesis was used to generate a series of virus mutants. Mutations were introduced into the PR coding region of the NL4-3-derived reference vector using a PCR “megaprimer” method (18). The presence of the desired mutation(s) and absence of other substitutions was confirmed by DNA sequencing. Oligonucleotide RsrII (5′-ACTTTCGGACCGTCCATTCCTGGCTTTAATTTTACTGGTACAG-3′) was used to introduce a unique RsrII restriction site (underlined) approximately 50 nucleotides downstream of the PR coding region into our drug-sensitive reference vector. The three point mutations introduced (bold) do not change the coding sequence of RT. The resulting construct allows an exchange of the PR coding region using a 590-bp ApaI-RsrII fragment, since both restriction sites are unique. All subsequent mutants were constructed by exchanging ApaI-RsrII fragments. The following oligonucleotides were used to introduce specific point mutations (bold type): K20T (5′-GGGGGGCAATTAACGGAAGCTCTATTAG-3′; coding strand), M36I (5′-GTATTAGAAGAAATAAATTTGCCAGGAAG-3′; coding strand), M46L (5′-GATGGAAACCAAAATTGATAGGGGGAATTG-3′; coding strand), L63P (5′-GTATGATCAGATACCCATAGAAATCTGC-3′; coding strand), N88S (5′-CTGAGTCAACAGACTTCTTCCAATTATG-3′; noncoding strand). Phenotypic analysis was performed on a total of 12 mutants (Table 2). Representative susceptibility curves for nelfinavir, indinavir, and amprenavir for mutant viruses carrying N88S and L63P/V77I/N88S are shown in Fig. 2. The N88S mutant and all other viable mutants that carried N88S were hypersensitive to amprenavir, with relative susceptibility values ranging from 0.04 to 0.14. Mutants containing N88S also displayed reduced susceptibility to nelfinavir (2.39- to 12.89-fold). Mutant viruses containing L63P or L63P/V77I alone were not hypersensitive to amprenavir, thus confirming that the N88S mutation is sufficient to determine this phenotype. Clearly, M46L, L63P, and V77I each play a role in decreased susceptibility to indinavir and nelfinavir when present in combination with N88S. The L63P/V77I double mutant exhibited a small, 2.49-fold reduction in nelfinavir susceptibility. The K20T virus displayed increased susceptibility to all five PRIs, with values ranging from 0.37 to 0.47. No change in susceptibility to any of the nine approved RT inhibitors was found (data not shown).
TABLE 2.
Susceptibility to PRIs and relative luciferase activity of HIV-1 mutants
| Site-directed PR mutation(s) | Relative susceptibilitya
|
Relative luciferase activityb | ||||
|---|---|---|---|---|---|---|
| SQV | IDV | RTV | NFV | AMP | ||
| N88S | 0.47 | 1.56 | 0.36 | 2.39 | 0.04 | 1.0 |
| L63P, N88S | 1.44 | 2.56 | 0.77 | 5.10 | 0.11 | 20.7 |
| M46L, L63P, N88S | 1.15 | 2.30 | 0.85 | 8.18 | 0.12 | 28.0 |
| L63P, V77I, N88S | 1.24 | 3.09 | 1.39 | 12.89 | 0.08 | 29.3 |
| M46L, L63P, V77I, N88S | 1.45 | 2.97 | 1.33 | 12.24 | 0.14 | 53.2 |
| K20T | 0.37 | 0.47 | 0.47 | 0.43 | 0.38 | 10.9 |
| L63P | 1.04 | 1.12 | 1.27 | 1.43 | 1.06 | 163.9 |
| L63P, V77I | 1.24 | 1.72 | 1.73 | 2.49 | 0.91 | 75.6 |
| K20T, N88S | Not viable | <0.01 | ||||
| K20T, L63P, N88S | Not viable | <0.01 | ||||
| K20T, M36I, L63P, N88S | Not viable | <0.01 | ||||
| K20T, M46L, L63P, N88S | Not viable | <0.01 | ||||
Multiple clones that contained the indicated mutations were tested for PRI susceptibility individually (two to five independent clones each), and the mean relative susceptibilities were determined. See Table 1, footnote a, for PRI abbreviations.
The mean relative luciferase activity (expressed as a percentage of that of the reference) is defined as relative light units produced by the PR mutant divided by relative light units produced by the reference virus, in the absence of a drug.
FIG. 2.
NFV, IDV, and AMP susceptibility curves for two representative mutants with site-directed mutations, N88S and L63P/V77I/N88S. Red lines represent respective mutant constructs; blue lines represent the reference construct. For drug abbreviations, see the legend for Fig. 1.
In the absence of drug, the N88S mutant produced significantly less luciferase activity than the parental reference virus (∼1%). A virus containing both mutations K20T and N88S could not be analyzed, since it did not produce sufficient luciferase activity. The introduction of additional substitutions at position 63, positions 36 and 63, or positions 46 and 63 to K20T/N88S mutants did not restore luciferase activity. To ensure that the K20T/N88S mutants did not carry other mutations that might result in inactive constructs, the ApaI-RsrII restriction fragment covering all of the HIV PR of the K20T/N88S mutant was exchanged with the equivalent fragment of the M46L/L63P/V77I/N88S mutant. Luciferase activity was restored, and phenotypic data were consistent with the results presented in Table 2 (data not shown). Thus, the vector backbone used to generate all mutants harboring K20T/N88S is functional.
Viruses derived from patients 1 and 14 carried both K20T and N88S substitutions and generated modest amounts of luciferase activity, although the activity was markedly less than that of the control (data not shown). One attractive model that may explain these observations is that these mutants are severely fitness impaired. The luciferase activity measured in the absence of a drug may be interpreted as an indicator of how well the recombinant virus pool is able to replicate. Luciferase activity may serve as a potential measure of replicative fitness for the following reasons: (i) given equal transfection efficiencies with the same test vector DNA pool, relative luciferase activity is highly reproducible; (ii) relative luciferase activity values obtained with different viral constructs vary; and (iii) most, though not all, virus mutants generate lower relative luciferase activity values than the drug-sensitive reference virus (our unpublished observations). This is in accordance with the general idea that the fitness of most drug-resistant HIV variants is less than that of wild-type HIV. Recombinant viruses containing N88S have been reported to display delayed growth kinetics compared to parental wild-type HIV-1 (20, 22), adding support to this concept. If this is correct, it is likely that additional mutations, generally referred to as polymorphisms (e.g., I15V and R41K), enhance the fitness of patient 1- or 14-derived virus compared to that of the K20T/N88S site-directed mutant. Experiments to shed light on these issues and to explore the suitability of this single-replication-cycle assay to evaluate viral fitness are currently under way.
It is relevant to determine how the amprenavir concentrations used in the phenotypic assay correlate with plasma drug levels in patients undergoing therapy. The IC50s and IC95s for the NL4-3 reference strain and the amprenavir-hypersensitive patient strains were compared to the estimated trough concentrations of amprenavir in plasma (17). Amprenavir has an IC50 of 8 to 16 nM and an IC95 about 10 times higher (100 to 139 nM) for the NL4-3 reference strain. Similar IC50s and IC95s for wild-type strains have been reported by other investigators using different assays (9, 13, 15, 21). Values for N88S mutant viruses ranged from 0.9 to 5.8 nM (IC50) and 7 to 70 nM (IC95). The concentration of amprenavir in plasma 12 h after a dose of 1,200 mg is approximately 0.65 μg/ml, or 1,300 nM (17). Since 90% of circulating amprenavir is bound to plasma proteins (11), the effective minimum concentration is 130 nM. Thus, the range in IC95s for N88S-containing viruses is below the amprenavir trough level, whereas that for wild-type viruses sometimes exceeds it. The decreases in amprenavir IC50 and IC95 observed for viruses carrying the N88S mutation in PR therefore appear to be potentially clinically relevant, since such viruses are less likely to escape the suppressive effects of the drug than are wild-type viruses with higher IC95 values. When other variables that negatively affect drug trough levels are considered, such as alterations in levels of amprenavir binding proteins (in particular α1-acid glycoprotein) or brief interruptions in drug dosing, the effect of N88S on the viral response to amprenavir could be even more important.
All of the N88S viruses we analyzed had increased susceptibility to amprenavir. N88S mutant viruses emerge in patients undergoing nelfinavir and/or indinavir treatment, and this mutation has been linked to decreased susceptibility to both drugs. This finding is reminiscent of the increase in susceptibility to zidovudine and adefovir caused by the M184V mutation in RT, which confers high-level resistance to 3TC (8; H. Tian et al., Abstr. 2nd Int. Workshop HIV Drug Resistance Treatment Strategies, abstr. 30, 1998; M. Miller et al., Abstr. 2nd Int. Workshop HIV Drug Resistance Treatment Strategies, abstr. 34, 1998). However, M184V severely decreases susceptibility to 3TC, while the N88S substitution, in combination with other mutations, causes a relatively moderate decrease in susceptibility to nelfinavir and indinavir. N88S alone marginally affects susceptibility to nelfinavir (Table 2). Therefore, in contrast to M184V, which is readily selected by 3TC treatment, N88S is less likely to arise as a primary resistance mutation in response to nelfinavir or indinavir therapy. This notion is supported by the finding that viruses containing the N88S substitution alone have not been isolated from patients, suggesting that N88S may be selected primarily in viruses that already have other mutations in PR. Alternatively, N88S mutants may arise early but require additional changes in order to overcome fitness constraints before becoming a predominant quasispecies.
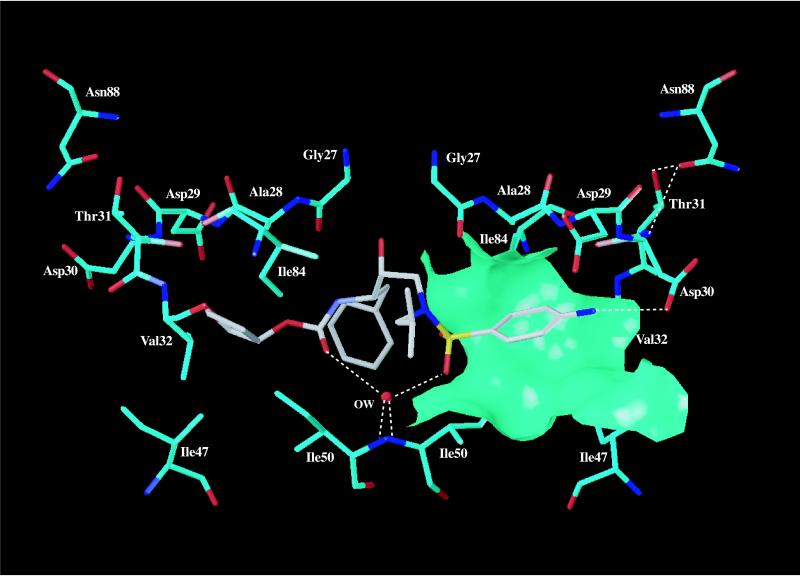
Based on HIV-1 PR crystal structure data, residue 88 is not directly involved in substrate or inhibitor binding (7). In wild-type PR structures, the side chain of N88 forms hydrogen bonds with the main-chain amide of T74, the main-chain carbonyl of T31, and the side-chain Oγ1 hydroxyl of T31 (Fig. 3). Mutation of N88 to a serine residue introduces a smaller side chain. In the published structure of the HIV-1 PR dimer complexed with amprenavir (Protein Data Bank entry 1HPV [7]; www.rcsb.org/pdb/), the D30 side chain of one PR subunit forms a hydrogen bond with the bound drug, and V32 forms part of a hydrophobic patch in the drug-binding pocket. The N88S mutation could lead to the formation of a hydrogen bond between S88 Oγ and the main-chain amide of T31, potentially pulling the D30-T31 region towards the site of mutation. This potential change in the binding pocket could allow the aromatic ring of amprenavir to slide further in toward the protein surface, improving its hydrophobic interactions with amino acid residues such as A28, V32, I47, I50, and I84 (Fig. 3). Overall, these changes could improve the amprenavir binding, which would be consistent with the observed hypersensitivity of N88S HIV-1 PR to amprenavir. Biochemical characterization will be required to elucidate the underlying mechanism of drug susceptibility.
FIG. 3.
Postulated interactions of amprenavir bound to HIV-1 protease. Amprenavir (in gray) is shown interacting with key amino acid residues (in cyan) of HIV-1 PR in the crystal structure of HIV-1 PR complexed with amprenavir (7). Nitrogen and oxygen atoms are colored blue and red, respectively, and OW represents a key conserved water molecule in the pocket. The molecular surface is calculated using the side-chain atoms of A28, V32, I47, I50, and I84 and Cα of A28. The figure was drawn using the program SYBYL, version 6.3 (Tripos, Inc.).
The intricate drug resistance phenotypes displayed by the N88S (in PR) and M184V (in RT) mutants may be paradigms for other potentially clinically relevant manifestations. The complexities of the resistance phenotypes could not have been accurately predicted by genotypic analysis alone. In light of the increasing number of patients undergoing highly active antiretroviral therapy, these results suggest that phenotypic testing is helpful in unraveling the potential synergistic interplay of multiple mutations in determining drug efficacy in vivo. The detection of drug hypersensitivity suggests a strategy for therapy management that can be tested in the context of a clinical trial.
We conclude from our studies that amprenavir may have increased clinical efficacy against N88S viruses. Previous clinical studies found that 3TC-zidovudine combination therapy was more beneficial than zidovudine monotherapy for patients harboring M184V viruses (8). By analogy, dual PRI therapy combining amprenavir with nelfinavir, indinavir, or new drugs such as BMS-232632 could delay the emergence of viruses resistant to both PRIs. Some such combinations have already been reported to be well tolerated (J. Eron et al., Abstr. 5th Conf. Retroviruses Opportunistic Infect., abstr. 6, 1998).
Acknowledgments
We thank the following individuals for patient-derived samples: Richard Haubrich and the CCTG 575 study team, Jon Condra and Emilio Emini, Martin Markowitz, James Kahn, and Patrick Joseph. We also thank Ron Swanstrom and Terri Smith for an intermediate vector construct, members of the ViroLogic Clinical Reference Laboratory for PhenoSense assays, Pam Johnson for data management, Shannon Utter and Nicole Whitehurst for genotyping, and Jeannette Whitcomb, Terri Wrin, Wei Huang, and Lai-Mun Gong for discussion, communication of unpublished results, and technical assistance. Finally, we are grateful to the following individuals for critical reviews of the manuscript: David Ho and Martin Markowitz, Steve Hughes, Ron Swanstrom, and Nicholas Hellmann.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boden D, Hurley A, Zhang L, Cao Y, Guo Y, Duran M, Tsay J, Ip J, Farthing C, Limoli K, Parkin N, Markowitz M. HIV-1 drug resistance in newly infected individuals. JAMA. 1999;282:1135–1141. doi: 10.1001/jama.282.12.1135. [DOI] [PubMed] [Google Scholar]
- 3.Carpenter C C J, Fischl M A, Hammer S M, Hirsch M S, Jacobsen D M, Katzenstein D A, Montaner J S G, Richman D D, Saag M S, Schooley R T, Thompson M A, Vella S, Yeni P G, Volberding P A. Antiretroviral therapy for HIV infection in 1998. Updated recommendations of the International AIDS Society-USA panel. JAMA. 1998;280:78–86. doi: 10.1001/jama.280.1.78. [DOI] [PubMed] [Google Scholar]
- 4.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 4a.Gong, Y.-F., B. S. Robinson, R. E. Rose, C. Deminie, T. P. Spicer, D. Stock, R. J. Colonno, and P.-F. Lin. In vitro resistance profile of the human immunodeficiency virus type 1 protease inhibitor BMS-232632. Antimicrob. Agents Chemother., in press. [DOI] [PMC free article] [PubMed]
- 5.Hecht F M, Grant R M, Petropoulos C J, Dillon B, Chesney M A, Tian H, Hellmann N S, Bandrapalli N I, Digilio L, Branson B, Kahn J O. Sexual transmission of an HIV-1 variant resistant to multiple reverse-transcriptase and protease inhibitors. N Engl J Med. 1998;339:307–311. doi: 10.1056/NEJM199807303390504. [DOI] [PubMed] [Google Scholar]
- 6.Hirsch M S, Conway B, D'Aquila R T, Johnson V A, Brun-Vezient F, Clotet B, Demeter L M, Hammer S M, Jacobsen D M, Kuritzkes D R, Loveday C, Mellors J W, Vella S, Richman D D. Antiretroviral drug resistance testing in adults with HIV infection: implications for clinical management. JAMA. 1998;279:1984–1991. doi: 10.1001/jama.279.24.1984. [DOI] [PubMed] [Google Scholar]
- 7.Kim E E, Baker C T, Dwyer M D, Murcko M A, Rao B G, Tung R D, Navia M A. Crystal structure of HIV-1 protease in complex with VX-478, a potent and orally bioavailable inhibitor of the enzyme. J Am Chem Soc. 1995;117:1181–1182. [Google Scholar]
- 8.Larder B A, Kemp S D, Harrigan P R. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science. 1995;269:696–699. doi: 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- 9.Lazdins J, Mestan J, Goutte G, Walker M, Bold G, Capraro H, Klimkait T. In vitro effect of α1-acid glycoprotein on the anti-human immunodeficiency virus (HIV) activity of the protease inhibitor CGP 61755: a comparative study with other relevant HIV protease inhibitors. J Infect Dis. 1997;175:1063–1070. doi: 10.1086/520352. [DOI] [PubMed] [Google Scholar]
- 10.Little S, Daar E, D'Aquila R, Keiser P, Connick E, Whitcomb J, Hellmann N, Petropoulos C, Pitt J, Rosenberg E, Koup R, Richman D. Reduced antiretroviral drug susceptibility among patients with primary HIV infection. JAMA. 1999;282:1142–1149. doi: 10.1001/jama.282.12.1142. [DOI] [PubMed] [Google Scholar]
- 11.Livingston D, Pazhanisamy S, Porter D, Partaledis J, Tung R, Painter G. Weak binding of VX-478 to human plasma proteins and implications for anti-human immunodeficiency virus therapy. J Infect Dis. 1995;172:1238–1245. doi: 10.1093/infdis/172.5.1238. [DOI] [PubMed] [Google Scholar]
- 12.Murphy R, Gulick R, DeGruttola V, D'Aquila R, Eron J, Sommadossi J, Currier J, Smeaton L, Frank I, Caliendo A, Gerber J, Tung R, Kuritzkes D. Treatment with amprenavir alone or amprenavir with zidovudine and lamivudine in adults with human immunodeficiency virus infection. J Infect Dis. 1999;179:808–816. doi: 10.1086/314668. [DOI] [PubMed] [Google Scholar]
- 13.Palmer S, Shafer R W, Merigan T C. Highly drug-resistant HIV-1 clinical isolates are cross-resistant to many antiretroviral compounds in current clinical development. AIDS. 1999;13:661–667. doi: 10.1097/00002030-199904160-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkin N, Lie Y, Hellmann N, Markowitz M, Bonhoeffer S, Ho D, Petropoulos C. Phenotypic changes in drug susceptibility associated with failure of human immunodeficiency virus type 1 (HIV-1) triple combination therapy. J Infect Dis. 1999;180:865–870. doi: 10.1086/314928. [DOI] [PubMed] [Google Scholar]
- 15.Partaledis J A, Yamaguchi K, Tisdale M, Blair E E, Falcione C, Maschera B, Myers R E, Pazhanisamy S, Futer O, Cullinan A B, Stuver C M, Byrn R A, Livingston D J. In vitro selection and characterization of human immunodeficiency virus type 1 (HIV-1) isolates with reduced sensitivity to hydroxyethylamino sulfonamide inhibitors of HIV-1 aspartyl protease. J Virol. 1995;69:5228–5235. doi: 10.1128/jvi.69.9.5228-5235.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patick A K, Duran M, Cao Y, Shugarts D, Keller M R, Mazabel E, Knowles M, Chapman S, Kuritzkes D R, Markowitz M. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob Agents Chemother. 1998;42:2637–2644. doi: 10.1128/aac.42.10.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Petropoulos C J, Parkin N T, Limoli K L, Lie Y S, Wrin M T, Huang W, Tian H, Smith D, Winslow G A, Capon D, Whitcomb J. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000;44:920–928. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadler B M, Hanson C D, Chittick G E, Symonds W T, Roskell N S. Safety and pharmacokinetics of amprenavir (141W94), a human immunodeficiency virus (HIV) type 1 protease inhibitor, following oral administration of single doses to HIV-infected adults. Antimicrob Agents Chemother. 1999;43:1686–1692. doi: 10.1128/aac.43.7.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkar G, Sommer S S. The “megaprimer” method of site-directed mutagenesis. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 19.Schinazi R F, Larder B A, Mellors J W. Mutations in retroviral genes associated with drug resistance. Int Antivir News. 1999;7:46–69. [Google Scholar]
- 20.Smidt M L, Potts K E, Tucker S P, Blystone L, Stiebel T R, Jr, Stallings W C, McDonald J J, Pillay D, Richman D D, Bryant M L. A mutation in human immunodeficiency virus type 1 protease at position 88, located outside the active site, confers resistance to the hydroxyethylurea inhibitor SC-55389A. Antimicrob Agents Chemother. 1997;41:515–522. doi: 10.1128/aac.41.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.St. Clair M H, Millard J, Rooney J, Tisdale M, Parry N, Sadler B M, Blum M R, Painter G. In vitro antiviral activity of 141W94 (VX-478) in combination with other antiretroviral agents. Antivir Res. 1996;29:53–56. doi: 10.1016/0166-3542(95)00916-7. [DOI] [PubMed] [Google Scholar]
- 22.Tucker S P, Stiebel T R, Jr, Potts K E, Smidt M L, Bryant M L. Estimate of the frequency of human immunodeficiency virus type 1 protease inhibitor resistance within unselected virus populations in vitro. Antimicrob Agents Chemother. 1998;42:478–480. doi: 10.1128/aac.42.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]