Abstract
Borna disease virus is the prototype of a new family, Bornaviridae, within the order Mononegavirales, that is characterized by nuclear transcription, splicing, low level replication, and neurotropism. The products of five open reading frames predicted from the genomic sequence have been confirmed; however, expression of the sixth, corresponding to the putative viral polymerase (L), has not been demonstrated. Here, we describe expression and characterization of a 190-kDa protein proposed to represent L. Expression of this protein from the third transcription unit of the viral genome is dependent on a splicing event that fuses a small upstream open reading frame in frame with the larger downstream continuous open reading frame. The protein is detected by serum antibodies from infected rats and is present in the nucleus, where it colocalizes with the phosphoprotein. L is also shown to be phosphorylated by cellular kinases and to interact with the viral phosphoprotein in coimmunoprecipitation studies. These findings are consistent with the identity of the 190-kDa protein as the viral polymerase and provide insights and describe reagents that will be useful for Bornavirus molecular biology and pathobiology.
Borna disease virus (BDV) is a nonsegmented, negative-strand RNA virus that establishes persistent central nervous system infection and causes behavioral disturbances in warm-blooded animals (12, 16). Notable features of its molecular biology include replication and transcription in the nucleus (1, 3, 5), overlap of open reading frames (ORFs) and transcription units (2, 17), RNA splicing (7, 19), differential use of transcription termination sites and translation codons (17, 19), and requirements for phosphorylation by kinases with limited distribution within the central nervous system (20). The antigenome contains three transcription units and six ORFs (2, 6). The products of five ORFs have been described, including, from the 5′ end to the 3′ end on the antigenome, the nucleoprotein (N) (13), X protein (X) (26), phosphoprotein (P) (25), atypical glycosylated matrix protein (gp18) (9), and type 1 membrane glycoprotein (G) (8, 18). The sixth ORF occupies two-thirds of the antigenome, contains motifs conserved amongst polymerases of negative-strand RNA viruses, and is postulated to encode the BDV polymerase (L) (2, 6); however, the product of this ORF is not reported. Expression and characterization of the BDV polymerase were pursued with the objective of obtaining a more detailed understanding of BDV molecular biology.
L expression requires splicing and suppression of termination.
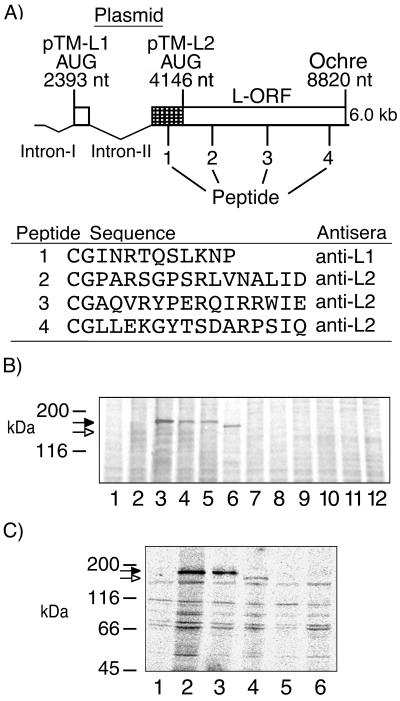
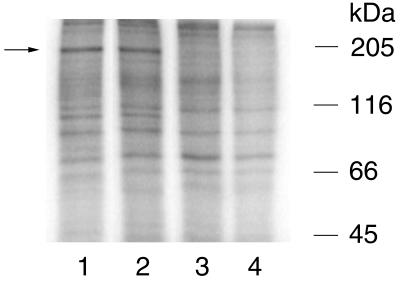
The third transcription unit of BDV strain V initiates at nucleotide (nt) 1889 and terminates at either termination site 3 (T3) (nt 4505) or T4 (nt 8855). Transcripts terminated at T3 may be spliced and translated into either or both gp18 and G (18, 19). Transcripts which read through T3 and terminate at T4 encode a continuous L ORF with five potential translation initiation sites. The AUG corresponding to strain V nt 4146 was proposed to initiate translation of L, resulting in expression of a 170-kDa protein (6); however, between the G ORF translation termination site and this AUG, the predicted amino acid sequence of strain V and strain He/80 is fully conserved, indicating that this region is likely to be translated (strain V, nt 3704 to 4146) (Fig. 1A). Recognition of splicing in BDV provided a mechanism for expression of the conserved sequence (nt 3704 to 4146): splicing of intron II (nt 2410 to 3703) results in fusion of 17 nt to the 5′ end of the conserved sequence at nt 3704 and provides an in-frame AUG (nt 2393) that allows utilization of the entire ORF, encoding a protein predicted to be 190 kDa (Fig. 1A) (19). Consistent with this model, a protein with an apparent molecular mass of 190 kDa was precipitated by anti-L1 antisera directed against a peptide within the conserved sequence (Fig. 1A) from lysates of strain V-infected COS-7 cells (Fig. 1B, lane 4) and from lysates of BSR-T7 cells (BHK-21 cells stably transfected with a T7 RNA polymerase expression plasmid) following Lipofectin (GIBCO-BRL) transfection with pTM-L1 (Fig. 1B, lane 3). pTM-L1 expresses the entire proposed strain V L ORF, nt 2393 to 2410 fused to nt 3704 to 8822, from an internal ribosome entry site-containing T7 promoter plasmid (14). Similar results were obtained with pB-L, a T7 expression plasmid which lacks the internal ribosome entry site. pCD-L, a cytomegalovirus (CMV) promoter plasmid, failed to produce L mRNA and/or protein. The 190-kDa protein was also precipitated from lysates of infected COS-7 cells by anti-L2 antisera directed against peptides distributed throughout the remainder of the L ORF (Fig. 1A and Fig. 1B, lane 5). Neither anti-L1 nor anti-L2 antisera precipitated proteins of similar molecular weights from lysates of noninfected cells (Fig. 1B, lanes 1 and 8) or mock-transfected cells (Fig. 1B, lanes 2 and 7). Anti-L2 antisera did not precipitate a smaller protein of 170 kDa from infected cell lysates (Fig. 1B, lane 5), consistent with the size of the protein precipitated from pTM-L2 (nt 4146 to 8822)-transfected BSR-T7 cells (Fig. 1B, lane 6). Anti-L1 antisera failed to precipitate a protein lacking sequence upstream of nt 4146 from lysates of pTM-L2-transfected BSR-T7 cells (Fig. 1C, lane 5), confirming the presence of the conserved sequence (nt 3704 to 4146) in the protein precipitated from infected cell lysates.
FIG. 1.
Detection of L in infected and transfected cells. (A) Representation of BDV third transcription unit, indicating inserts of L-T7 expression plasmids pTM-L1 and pTM-L2. The relative positions of the peptides used to elicit murine antisera to L are indicated, and the sequences are shown underneath. The cross-hatched area indicates a region of conserved sequence predicted to be incorporated into L (2). (B) Immunoprecipitation of 190-kDa wild-type or recombinant L from COS-7 cells or from BSR-T7 cells, respectively, by using anti-L1 antisera (lanes 1 to 4), anti-L2 antisera (lanes 5 to 8), or preimmune mouse sera (lanes 9 to 12) with protein G-Sepharose (Pharmacia). Metabolically labeled lysates (labeled with [35S]methionine) were obtained from noninfected COS-7 cells (lanes 1 and 8), mock (vector)-transfected BSR-T7 cells (lanes 2, 7, and 12), pTM-L1-transfected BSR-T7 cells (lanes 3 and 10), infected COS-7 cells (lanes 4, 5, and 9), or pTM-L2-transfected BSR-T7 cells (lanes 6 and 11). (C) Immunoprecipitation of lysates from pTM-L1 (lanes 1 to 3) or pTM-L2 (lanes 4 to 6) transiently transfected BSR-T7 cells by using preimmune mouse sera (lanes 1 and 6), anti-L2 antisera (reactive with sequence common to pTM-L1 and pTM-L2) (lanes 2 and 4), or anti-L1 antisera (reactive within the conserved sequence [nt 3704 to 4146] of L unique to pTM-L1) (lanes 3 and 5). The filled arrows indicate the position of L initiating at the AUG at nt 2393; the open arrows indicate the position of L initiating at the AUG at nt 4146. Immunoprecipitation analysis was done by SDS-7.5% PAGE.
Infected rats have antibodies reactive with L.
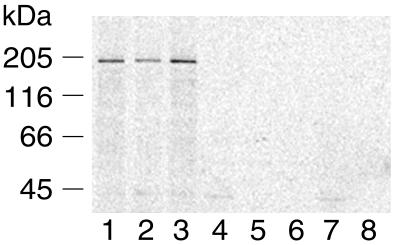
As an independent test for expression of L in vivo, Lewis rats with clinical signs of Borna disease were bled 2 and 4 months after intracranial infection and assayed for the presence of serum antibodies to recombinant L. Sera obtained from two infected rats precipitated L from metabolically [35S]methionine-labeled BSR-T7 cells that were transiently transfected with pTM-L1 (Fig. 2, lanes 2 and 3). Signal was higher in samples precipitated using 4-months-postinfection sera, suggesting a higher titer of antibodies to L. Sera from noninfected rats did not precipitate L (Fig. 2, lane 4).
FIG. 2.
Sera from BDV strain V-infected rats recognizes recombinant 190-kDa L. Metabolically labeled lysates obtained from pTM-L1-transfected BSR-T7 cells (lanes 1 to 4) or mock (vector)-transfected BSR-T7 cells (lanes 5 to 8) were immunoprecipitated using anti-L1 antisera (lanes 1 and 5), rat sera 2 months (lanes 2 and 6) or 4 months (lanes 3 and 7) post-BDV infection, or noninfected rat sera (lanes 4 and 8). Immunoprecipitation analysis was done by SDS-7.5% PAGE.
L localizes to the nucleus of infected or L-transfected cells.
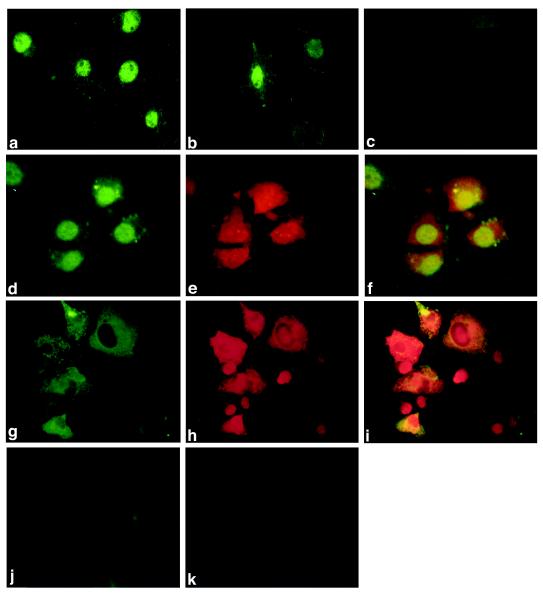
BDV transcribes its genome in the nucleus (1, 3, 5); thus, if L represented the viral polymerase, it would be anticipated to be present in the nucleus. Potential nuclear localization sequences within the L ORF have been proposed (6); however, other BDV proteins also contain nuclear localization sequences (10, 15, 21, 23) and might facilitate nuclear localization of L through protein-protein interaction-mediated transport. The distribution of wild-type and recombinant L was examined in strain V-infected COS-7 (Fig. 3a) and BSR-T7 (Fig. 3d) cells and in noninfected BSR-T7 cells transiently transfected with pTM-L1 (Fig. 3b). In all three systems, L was predominantly nuclear, as determined by immunofluorescence analysis with anti-L1 antisera, indicating that the protein is imported into the nucleus in the absence of other viral proteins.
FIG. 3.
Localization of L and of L with P by indirect immunofluorescence in infected COS-7 or BSR-T7 cells or in noninfected, transfected BSR-T7 cells. (a) Infected COS-7 cells, anti-L1 antisera; (b) pTM-L1-transfected BSR-T7 cells, anti-L1 antisera; (c) noninfected COS-7 cells, anti-L1 antisera; (d) infected BSR-T7 cells, anti-L1 antisera; (e) infected BSR-T7 cells, anti-P antisera; (f) infected BSR-T7 cells, anti-L1 and anti-P antisera; (g) pTM-L1- and pcDNA-P-transfected BSR-T7 cells, anti-L1 antisera; (h) pTM-L1- and pcDNA-P-transfected BSR-T7 cells, anti-P antisera; (i) pTM-L1- and pcDNA-P-transfected BSR-T7 cells, anti-L1 and anti-L2 antisera; (j) noninfected, nontransfected BSR-T7 cells, anti-L1 antisera; (k) noninfected, nontransfected BSR-T7 cells, anti-P antisera.
L interacts with the BDV P protein.
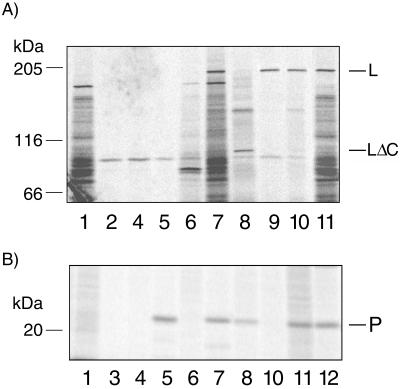
Polymerases of rhabdoviruses and paramyxoviruses interact with viral phosphoproteins to form the active polymerase complex (24). Previous studies have shown that BDV P interacts with BDV N and BDV X (22). To assess whether BDV P might also interact with L, coimmunoprecipitation and colocalization experiments were performed. Colocalization experiments were performed in infected COS-7 cells (data not shown) or BSR-T7 cells (Fig. 3d to f) or in BSR-T7 cells transiently transfected for coexpression of L and P (Fig. 3g to i). Whereas in infected cells both proteins colocalized in the nucleus, in transfected cells both proteins colocalized predominantly in the cytoplasm. This shift in localization may result from the overexpression of L-P complexes in the absence of other viral factors within transfected cells. Coimmunoprecipitation experiments were performed with metabolically labeled lysates (labeled with [35S]methionine) from infected COS-7 cells or BSR-T7 cells transfected with pTM-L1 (L expression) and/or pcDNA-P (P expression), and polyclonal rabbit antisera to P or anti-peptide antisera (anti-L1 or anti-L2). Anti-P antisera coprecipitated L with P from lysates of infected cells (Fig. 4, lanes 11) or from lysates of cells transfected for expression of both L and P (Fig. 4, lanes 7). A carboxyl-terminal deletion mutant of L (deletion of 941 carboxyl-terminal amino acids; pTM-LΔC) also coprecipitated with P (Fig. 4, lanes 8); however, L was not coprecipitated from lysates of cells transfected for expression of L and a phosphorylation-deficient mutant of P (pcDNA-Pmut), in which all known phosphorylation sites (20) have been mutated to alanine (Fig. 4, lanes 5). In concert, these results indicate that the interaction of P with L involves phosphorylation of P and implicate the amino-terminal half of L in this interaction. The inability of anti-L1 (Fig. 4, lanes 6 and 10) and anti-L2 (data not shown) to coprecipitate P together with L is consistent with a model where the major epitopes detected by anti-L1 and anti-L2 are obscured by binding of L to P; however, sites of L-P interaction have not yet been mapped.
FIG. 4.
Coimmunoprecipitation of L with P. Metabolically labeled lysates obtained from infected COS-7 cells or noninfected BSR-T7 cells transfected with pTM-L1 (L expression) or pcDNA-P (P expression) were immunoprecipitated by anti-L1 or anti-P antisera. Samples were split and analyzed by SDS-7.5% PAGE for L (A) and by SDS-10% PAGE for P (B). Lanes: 1, infected COS-7 cells, preimmune sera; 2, pTM-L1 transfected BSR-T7 cells, anti-P antisera; 3, pcDNA-P transfected BSR-T7 cells, anti-L1 antisera; 4, pTM-L1 and pcDNA-P cotransfected BSR-T7 cells, preimmune sera; 5, pTM-L1 and pcDNA-Pmut cotransfected BSR-T7 cells, anti-P antisera; 6, pTM-L1 and pcDNA-P cotransfected BSR-T7 cells, anti-L1 antisera; 7, pTM-L1 and pcDNA-P cotransfected BSR-T7 cells, anti-P antisera; 8, pTM-LΔc and pcDNA-P cotransfected BSR-T7 cells, anti-P antisera; 9, pTM-L1 transfected BSR-T7 cells, anti-L1 antisera; 10, infected COS-7 cells, anti-L1 antisera; 11, infected COS-7 cells, anti-P antisera; 12, pcDNA-P-transfected BSR-T7 cells, anti-P antisera.
L is phosphorylated.
The sequence of L contains at least 40 potential phosphorylation sites (11). Because phosphorylation is implicated in regulating the activity of catalytic enzymes (4), including polymerases of negative-strand RNA viruses, phosphorylation of BDV L was investigated. Metabolically labeled extracts (labeled with [32P]orthophosphate) of infected BSR-T7 cells and noninfected BSR-T7 cells transfected with pTM-L1 were immunoprecipitated with anti-L1 antisera and size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). A 190-kDa radiolabeled protein was present in both infected and transfected cells, consistent with phosphorylation of L by cellular kinases (Fig. 5, lanes 1 and 2).
FIG. 5.
Phosphorylation of wild-type and recombinant L. Metabolically labeled lysates obtained from infected BSR-T7 cells (lanes 1 and 4), pTM-L1-transfected BSR-T7 cells (lane 2), or mock (vector)-transfected BSR-T7 cells (lane 3) were immunoprecipitated by anti-L1 antisera (lanes 1 to 3) or preimmune mouse sera (lane 4). Immunoprecipitation analysis was done by SDS-7.5% PAGE.
The characteristics of the 190-kDa protein described here, including size, position on the viral antigenome, presence of motifs consistent with polymerases of negative-strand RNA viruses, nuclear localization, potential for phosphorylation, and colocalization and interaction with the BDV phosphoprotein provide indirect but strong support for the conclusion that it is the BDV polymerase. Although native L and its corresponding mRNA appear to be present only at low levels in infected cells, we have established methods for overexpression of L that afford unprecedented opportunities for pursuing functional assays and reverse genetic systems.
Acknowledgments
We are grateful to Xi Yu Jia for helpful discussions, to Bernard Moss for the gift of the plasmid pTM1, and to Karl-Klaus Conzelmann for helpful comments and the gift of the BSR-T7 cell line.
This work was supported by grant NS29425 from the National Institutes of Health.
REFERENCES
- 1.Briese T, de la Torre J C, Lewis A, Ludwig H, Lipkin W I. Borna disease virus, a negative-strand RNA virus, transcribes in the nucleus of infected cells. Proc Natl Acad Sci USA. 1992;89:11486–11489. doi: 10.1073/pnas.89.23.11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briese T, Schneemann A, Lewis A J, Park Y S, Kim S, Ludwig H, Lipkin W I. Genomic organization of Borna disease virus. Proc Natl Acad Sci USA. 1994;91:4362–4366. doi: 10.1073/pnas.91.10.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carbone K M, Moench T R, Lipkin W I. Borna disease virus replicates in astrocytes, Schwann cells and ependymal cells in persistently infected rats: location of viral genomic and messenger RNAs by in situ hybridization. J Neuropathol Exp Neurol. 1991;50:205–214. doi: 10.1097/00005072-199105000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Creighton T E. Proteins structures and molecular properties. 2nd ed. Heidelberg, Germany: W. H. Freeman and Company; 1993. [Google Scholar]
- 5.Cubitt B, de la Torre J C. Borna disease virus (BDV), a nonsegmented RNA virus, replicates in the nuclei of infected cells where infectious BDV ribonucleoproteins are present. J Virol. 1994;68:1371–1381. doi: 10.1128/jvi.68.3.1371-1381.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cubitt B, Oldstone C, de la Torre J C. Sequence and genome organization of Borna disease virus. J Virol. 1994;68:1382–1396. doi: 10.1128/jvi.68.3.1382-1396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cubitt B, Oldstone C, Valcarcel J, de la Torre J C. RNA splicing contributes to the generation of mature mRNAs of Borna disease virus, a non-segmented negative strand RNA virus. Virus Res. 1994;34:69–79. doi: 10.1016/0168-1702(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez-Dunia D, Cubitt B, Grasser F A, de la Torre J C. Characterization of Borna disease virus p56 protein, a surface glycoprotein involved in virus entry. J Virol. 1997;71:3208–3218. doi: 10.1128/jvi.71.4.3208-3218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kliche S, Briese T, Henschen A H, Stitz L, Lipkin W I. Characterization of a Borna disease virus glycoprotein, gp18. J Virol. 1994;68:6918–6923. doi: 10.1128/jvi.68.11.6918-6923.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi T, Shoya Y, Koda T, Takashima I, Lai P K, Ikuta K, Kakinuma M, Kishi M. Nuclear targeting activity associated with the amino terminal region of the Borna disease virus nucleoprotein. Virology. 1998;243:188–197. doi: 10.1006/viro.1998.9049. [DOI] [PubMed] [Google Scholar]
- 11.Kreegipuu A, Blom N, Brunak S. PhosphoBase, a database of phosphorylation sites: release 2.0. Nucleic Acids Res. 1999;27:237–239. doi: 10.1093/nar/27.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ludwig H, Bode L, Gosztonyi G. Borna disease: a persistent virus infection of the central nervous system. Prog Med Virol. 1988;35:107–151. [PubMed] [Google Scholar]
- 13.McClure M A, Thibault K J, Hatalski C G, Lipkin W I. Sequence similarity between Borna disease virus p40 and a duplicated domain within the paramyxovirus and rhabdovirus polymerase proteins. J Virol. 1992;66:6572–6577. doi: 10.1128/jvi.66.11.6572-6577.1992. . (Erratum, 67:1746, 1993.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. Product review. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 15.Pyper J M, Gartner A E. Molecular basis for the differential subcellular localization of the 38- and 39-kilodalton structural proteins of Borna disease virus. J Virol. 1997;71:5133–5139. doi: 10.1128/jvi.71.7.5133-5139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rott R, Becht H. Natural and experimental Borna disease in animals. Curr Top Microbiol Immunol. 1995;190:17–30. doi: 10.1007/978-3-642-78618-1_2. [DOI] [PubMed] [Google Scholar]
- 17.Schneemann A, Schneider P A, Kim S, Lipkin W I. Identification of signal sequences that control transcription of borna disease virus, a nonsegmented, negative-strand RNA virus. J Virol. 1994;68:6514–6522. doi: 10.1128/jvi.68.10.6514-6522.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider P A, Hatalski C G, Lewis A J, Lipkin W I. Biochemical and functional analysis of the Borna disease virus G protein. J Virol. 1997;71:331–336. doi: 10.1128/jvi.71.1.331-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider P A, Schneemann A, Lipkin W I. RNA splicing in Borna disease virus, a nonsegmented, negative-strand RNA virus. J Virol. 1994;68:5007–5012. doi: 10.1128/jvi.68.8.5007-5012.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwemmle M, De B, Shi L, Banerjee A, Lipkin W I. Borna disease virus P-protein is phosphorylated by protein kinase C epsilon and casein kinase II. J Biol Chem. 1997;272:21818–21823. doi: 10.1074/jbc.272.35.21818. [DOI] [PubMed] [Google Scholar]
- 21.Schwemmle M, Jehle C, Shoemaker T, Lipkin W I. Characterization of the major nuclear localization signal of the Borna disease virus phosphoprotein. J Gen Virol. 1999;80:97–100. doi: 10.1099/0022-1317-80-1-97. [DOI] [PubMed] [Google Scholar]
- 22.Schwemmle M, Salvatore M, Shi L, Richt J, Lee C H, Lipkin W I. Interactions of the borna disease virus P, N, and X proteins and their functional implications. J Biol Chem. 1998;273:9007–9012. doi: 10.1074/jbc.273.15.9007. [DOI] [PubMed] [Google Scholar]
- 23.Shoya Y, Kobayashi T, Koda T, Ikuta K, Kakinuma M, Kishi M. Two proline-rich nuclear localization signals in the amino- and carboxyl-terminal regions of the Borna disease virus phosphoprotein. J Virol. 1998;72:9755–9762. doi: 10.1128/jvi.72.12.9755-9762.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takacs A M, Barik S, Das T, Banerjee A K. Phosphorylation of specific serine residues within the acidic domain of the phosphoprotein of vesicular stomatitis virus regulates transcription in vitro. J Virol. 1992;66:5842–5848. doi: 10.1128/jvi.66.10.5842-5848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thierer J, Riehle H, Grebenstein O, Binz T, Herzog S, Thiedemann N, Stitz L, Rott R, Lottspeich F, Niemann H. The 24K protein of Borna disease virus. J Gen Virol. 1992;73:413–416. doi: 10.1099/0022-1317-73-2-413. [DOI] [PubMed] [Google Scholar]
- 26.Wehner T, Ruppert A, Herden C, Frese K, Becht H, Richt J A. Detection of a novel Borna disease virus-encoded 10 kDa protein in infected cells and tissues. J Gen Virol. 1997;78:2459–2466. doi: 10.1099/0022-1317-78-10-2459. [DOI] [PubMed] [Google Scholar]