Abstract
To gain insight into human foamy virus (HFV; also called spumaretrovirus)-induced alterations of cellular genes, the expression profiles of defined genes in HFV-infected primary human cells were analyzed by cDNA array assays. Several distinct cellular genes activated by HFV infection were identified; the identities of the cellular genes were confirmed by RNA blot analyses. Compared with mock-infected controls, the concentrations of cellular Kip2, Egr-1, COUP-TF1, insulin-like growth factor II (IGF-II), and EphB3 mRNAs were significantly increased in HFV-infected cells and showed a gene-specific and time-dependent induction. Immunoblot analyses with antibodies against some of the cellular gene products revealed increased levels of the corresponding proteins. To investigate mechanisms of HFV-induced alterations in cellular gene expression, the capacity of known HFV genes to increase expression of defined cellular genes was analyzed by transient expression experiments. Plasmids that encode the HFV Bel1 transcriptional transactivator were necessary and sufficient to strongly increase expression of p57Kip2, IGF-II, and EphB3 genes in 293T cells. Potential mechanisms and consequences of activation of cellular genes during HFV infection and Bel1 transactivation of the Kip2 gene are discussed.
Studies on the replication and gene expression of different foamy viruses (FVs) have deepened our understanding of the molecular biology of these nonconventional and complex retroviruses (for reviews, see references 1, 4, 13, and 15). FVs have been developed into viral vectors for defined targeted gene delivery applications (22). Features favoring FV-based vectors are the lack of an overt disease association of naturally occurring FV infections and the low prevalence in humans, which has been primarily attributed to the rarity of interspecies transmission from FV-positive nonhuman primates to humans (7). Simian FVs have the capacity to establish a persistent and apparently apathogenic infection in humans (7).
It has been reported that human FV (HFV) Env proteins are responsible for the characteristic cytopathic effects that result in formation of syncytia and cell lysis (1, 18). At present, it is unknown why FVs are apathogenic in the natural host whereas HFV-transgenic mice develop severe encephalopathy (1). The mechanisms responsible for induction and maintenance of HFV persistence are unclear. There is evidence, however, that cells that survive a lytic infection carry functionally inactivated FV genomes characterized by a deletion within the gene that encodes Bel1 (Tas), the transcriptional transactivator of FVs. This is achieved by reverse transcription of almost full-length genomic RNA spliced only at the intron of the accessory Bet protein of unknown function (13) that results in bel1-deleted proviruses (23). In contrast, some cell types allow a noncytopathic replication characterized by continuous production of genetically intact virus (30).
A better understanding of the multiple mutual interactions of FVs with cellular genes and gene products of infected cells, and especially regulatory factors in infected hosts, is essential for gaining insight into the molecular biology of FVs, their potential disease association or apathogenicity, and future utilization of FV-based retroviral vectors for gene therapy. Thus, this report is aimed at the identification of cellular genes that are specifically activated in cells infected with HFV. One of the cellular genes that was identified was analyzed in more detail and shown to be activated by the HFV Bel1 transcriptional transactivator.
MATERIALS AND METHODS
Plasmids and transfection.
Plasmids pBKCMV (Stratagene), pUC18, pCMVβ-gal, pBCbel (16), pHSRVΔMN (18), pbel1s (28), and pKip2 (27) were transfected into 293T cells by the coprecipitation method of Chen and Okayama (2). Bet expression plasmid pBCbet was derived from pBCbel by replacing bel1 sequences with PCR-derived bet cDNA sequences by using standard techniques (24).
Cell culture.
Cultivation of 293T, human embryonic lung (HEL299), and FAB cells and virus propagation were done as described elsewhere (18). To synchronize HEL299 cells, cultures were arrested in G0 by culturing in Dulbecco modified Eagle medium (DMEM) without fetal calf serum (FCS) for 3 days and subsequently released by the addition of 20% FCS in DMEM. To monitor cell cycle progression, cells were stained with propidium iodide (50 μg/ml, final concentration) and analyzed in a FACSort (Becton Dickinson, Hamburg, Germany) using the CellFit software.
RNA extraction, cDNA array and Northern blots.
Total RNA was harvested using the acidic guanidium-phenol-chloroform method (3). For cDNA array analysis, RNA was digested with RNase-free DNase I and selected for poly(A)+ mRNA [Dynabeads oligo(dT)25; DYNAL, Hamburg, Germany]. Poly(A)+ RNAs (1 μg of each) and [α-32P]dATP (10 μCi/μl; Amersham, Braunschweig, Germany) were used for generating complex, radioactively labeled cDNA probes. Samples were purified, diluted to 4 × 105 cpm/ml in hybridization solution, and hybridized to human cDNA expression array filters (Human Broad-Coverage; Clontech, Heidelberg, Germany) according to the manufacturer's instructions. After stringent washing, filters were analyzed by autoradiography and phosphorimaging (Molecular Dynamics, Krefeld, Germany). For Northern blot analysis, 10-μg aliquots of total RNA were separated on 1% agarose–morpholine propanesulfonic acid (MOPS) gels (20 mM MOPS, 5 mM sodium acetate, 1 mM EDTA [pH 7.0]), stained with ethidium bromide (EtBr), and transferred to Hybond N+ blotting membranes (Amersham). Hybridization probes directed against defined cellular genes were derived from inserts of IMAG clones IMAGp998P321787, COUP-TF1 (21), IMAGp998M21287, insulin-like growth factor II (IGF-II) (20), IMAGp998L15690, Kip2 (11), IMAGp998P17408, Egr-1 (14), IMAGp998G021963, EphB3 (32), and pGAPDH (expressing glyceraldehyde-3-phosphate dehydrogenase [GAPDH]). Probes were labeled with [α-32P]dCTP (10 μCi/μl) by random priming (Megaprime; Amersham). Hybridization was carried out at 68°C for at least 20 h in hybridization buffer (50 mM sodium phosphate, 1% sodium dodecyl sulfate [SDS], 1× Denhart's solution, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1 mg of tRNA per ml). Blots were washed with 2× SSC–0.1% SDS and analyzed by autoradiography and phosphorimaging.
Immunoblotting.
Cellular proteins were harvested by cell lysis in 1% SDS. Equal amounts of total protein as measured using the DC protein assay (Bio-Rad, Munich, Germany) were separated by SDS-polyacrylamide gel electrophoresis, blotted, and detected with enhanced chemoluminescence (Amersham) as described elsewhere (16). For detection of cellular proteins, sera against p57Kip2 (PharMingen, Hamburg, Germany), Egr-1 (Santa Cruz Biotechnology, Santa Cruz, Calif.), and IGF-II (Upstate Biotechnology Inc., Lake Placid, N.Y.) were used. Goat anti-mouse immunoglobulin G-peroxidase (Jackson ImmunoResearch, Hamburg, Germany) was used as secondary antibody.
RESULTS
Analysis of HFV-induced changes in cellular gene expression by cDNA microarray assays.
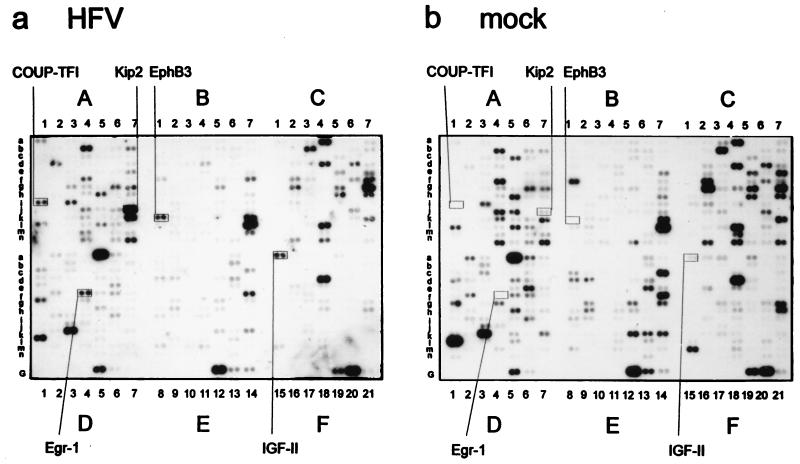
To analyze whether cellular genes were transcriptionally up- or down-regulated by HFV infection, we chose to use a commercially available cDNA microarray consisting of cDNA fragments of 588 defined human genes. HEL299 cells were synchronized by serum starvation and 16 h after release of the block infected with HFV (multiplicity of infection of 20) or mock infected with supernatants from uninfected cells. RNA harvested 72 h postinfection (p.i.) was used for mRNA selection. At this stage, syncytia had been formed in HFV-infected but not mock-infected cells. Poly(A)+ mRNAs (1 μg of each) from HFV- and mock-infected cells were used to generate random radioactively labeled cDNAs as hybridization probes for two identical cDNA array membranes. Filters were stringently washed and visualized by autoradiography (Fig. 1) or phosphorimaging. Cellular RNAs that showed strongly increased expression levels in HFV-infected cells are boxed in Fig. 1. Upon normalization of signals to GAPDH mRNA standards, the expression levels of cellular genes COUP-TF1, IGF-II, Kip2, Egr-1, and EphB3 were significantly increased in HFV-infected cells compared to noninfected controls (Table 1).
FIG. 1.
Autoradiograms of cDNA array membranes hybridized to labeled complex cDNA probes derived from HFV-infected (a) and mock-infected (b) HEL299 cells 72 h p.i. The locations (boxes) and names of strongly induced genes in HFV- and mock-infected HEL299 cells are indicated. Lines G and 1 to 21 represent negative and positive hybridization controls specified by the manufacturer.
TABLE 1.
Increased expression of cellular genes in HFV-infected HEL299 cells determined by cDNA array assays
| Cellular gene | Fold induction in HFV-infected HEL299 cellsa
|
|
|---|---|---|
| 36 h p.i. | 72 h p.i. | |
| COUP-TF1 | 0.5 | 14.0 |
| Kip2 | 1.5 | 20.0 |
| EphB3 | 2.6 | 77.0 |
| Egr1 | 1.3 | 12.0 |
| IGF-II | 1.2 | 8.5 |
cDNA arrays were hybridized to labeled probes from HFV- and mock-infected cells. Signals were quantitated by phosphorimaging and normalized to GAPDH standards. Induction is expressed as relative increase of HFV-infected compared with mock-infected HEL299 cells.
In independent experiments, HFV- and mock-infected cells were harvested 36 h p.i. At this time, increases of less than fivefold for any of the cDNAs were detectable in cellular gene expression when hybridization patterns from HFV- and mock-infected cells were compared. Table 1 shows values only for cellular genes that were identified as up-regulated 72 h p.i.
The number of genes identified is likely to be lower than the number of genes actually affected by HFV infection, since we focused on late events during HFV infection and since a fraction of cells remained uninfected early after addition of HFV. Similarly, a background of unaffected cells will mask decreases in RNA levels taking place in infected cells only, which likely explains why we did not unambiguously identify HFV-induced down-regulation of cellular genes.
Detection of HFV-induced changes in cellular gene expression by Northern blot analyses.
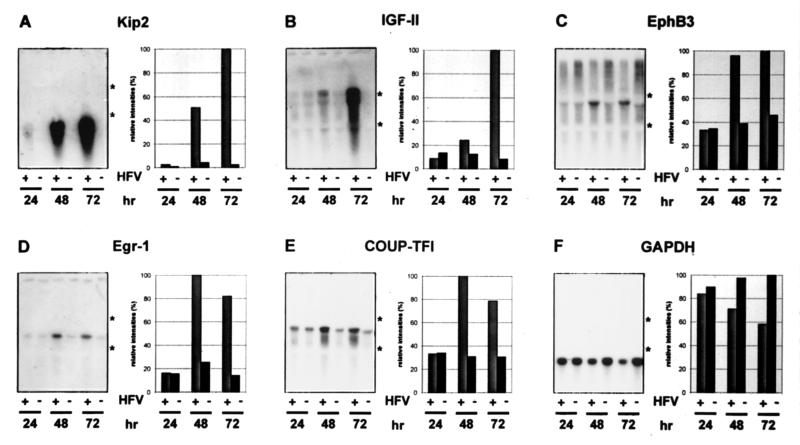
To confirm the cDNA array data on the induction of cellular genes after HFV infection, RNA blot analyses were performed. To avoid unnecessary handling of samples during selection of poly(A)+ mRNA, total RNAs from nonsynchronized HFV- and mock-infected HEL299 cells were harvested 24, 48, and 72 h p.i. and directly used for Northern blotting. For blotting on nitrocellulose membranes, RNAs (10 μg of each) were separated on denaturing gels and probed with cloned DNAs complementary to cellular Kip2, IGF-II, EphB3, Egr-1, COUP-TFI, and GAPDH mRNAs (Fig. 2). Gels had been loaded with equal amounts of RNAs as estimated from similar intensities of rRNA bands after EtBr staining (not shown). Autoradiograms revealed significant increases in RNA levels of these genes in HFV-infected cells except for the housekeeping gene GAPDH, which was reduced after HFV infection. Quantification of the data is presented graphically next to each blot in Fig. 2. Whereas at 24 h p.i. almost no changes in cellular gene expression were detectable, minor (IGF-II), intermediate (COUP-TFI, Egr-1, and EphB3), and strong increases in HFV-infected cells (Kip2) were clearly detectable 48 h after infection consistent with the data from cDNA array analyses. RNA levels for IGF-II and Kip2 increased even further during incubation, whereas other mRNAs did not show additional increments.
FIG. 2.
Northern blot analysis of HFV-infected (+) or mock-infected (−) HEL299 cells harvested 24, 48, and 72 h p.i. Total RNAs (10 μg of each) were separated on 1% agarose–MOPS gels and blotted onto nitrocellulose filters. The filters were probed with 32P-labeled cDNA fragments directed against Kip2 (A), IGF-II (B), EphB3 (C), Egr-1 (D), and COUP-TFI (E). After autoradiography and phosphorimaging, the filters were reprobed with GAPDH (F). Positions of the 28S and 18S rRNAs observed after Etbr staining are indicated with asterisks. Individual mRNA signals were quantified using a phosphorimager and are shown next to the autoradiograms. The signals for each RNA spot were normalized to the corresponding GAPDH signals and are shown relative to the strongest signal on each blot.
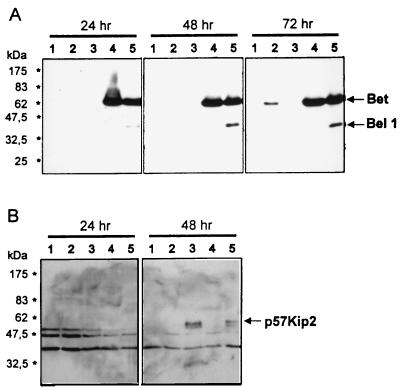
Detection of HFV-induced changes in HEL299 gene expression by immunoblot analyses.
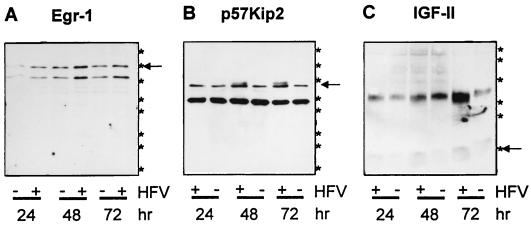
Next, the expression of HFV-induced cellular genes was studied kinetically by means of defined antisera. Randomly growing HEL299 cells were HFV or mock infected and harvested 24, 48, and 72 h p.i. Aliquots of protein (10 μg of each) were analyzed by immunoblotting with sera directed against the cellular proteins identified. Temporal expression profiles of Egr-1, p57Kip2, and IGF-II in HFV- and mock-infected HEL299 cells are shown in Fig. 3. Whereas Egr-1 (arrow in Fig. 3A) was significantly increased after 48 and 72 h in HFV-positive cells, p57Kip2 (arrow in Fig. 3B) was detectable in Hel299 cells only 48 and 72 h after HFV infection and was virtually absent 24 h p.i. and in mock-infected samples. In HFV-infected cells, increased levels of mature cell-associated 6.5-kDa IGF-II (arrow in Fig. 3C) and high-molecular-mass precursors were detectable. Sera against EphB3 and COUP-TFI did not yield specific bands or were not available.
FIG. 3.
Immunoblots of 10 μg of total protein from whole cell lysates obtained from HFV-infected (+) or mock-infected (−) HEL299 cells harvested 24, 48, and 72 h p.i. Antisera against the 85-kDa Egr-1 protein (arrow in panel A), the p57Kip2 protein (arrow in panel B), and the mature 7.5-kDa IGF-II (arrow in panel C) and IGF-II precursor forms of about 25 kDa were used. Asterisks at the right margin correspond to the following apparent molecular masses from marker proteins separated in parallel (from bottom to top), 6.5, 16.5, 25.0, 32.5, 47.5, 62.0, 83.0, and 175.0 kDa; 83.0- and 175.0-kDa bands are not shown in panel C).
Induction of cellular genes by the HFV Bel1 transactivator.
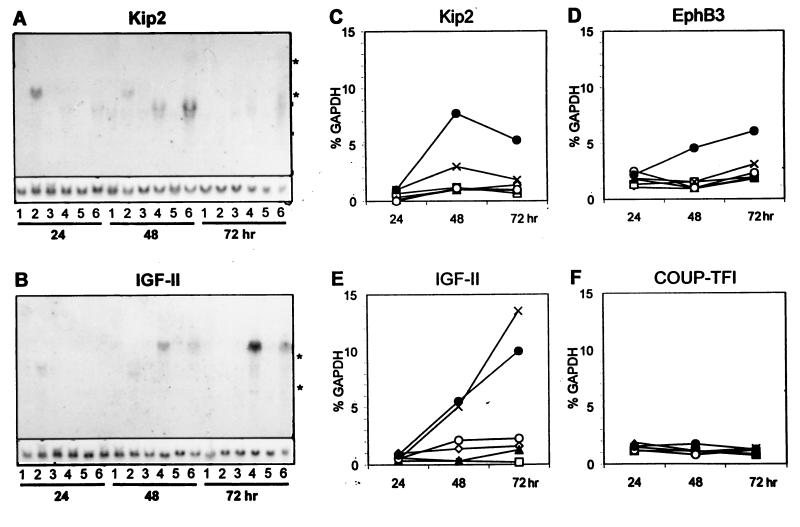
We then studied whether a defined HFV gene is responsible for induction of the identified cellular genes to confirm the specificity of the results described above. Cytomegalovirus immediate-early (CMV IE) promoter-based expression vectors coding for HFV Bel1 (pbel1s [Fig. 4, lanes 4]), Bet (pBCbet [lanes 5]), and Bel1 and Bet (pBCbel [lanes 6]), each containing simian virus 40 origin sequences, were used together with a replication-deficient, env-deleted HFV provirus directing HFV gene expression from the viral promoters (pHSRVΔMN [lanes 3]). As control vectors devoid of HFV sequences, plasmids pUC18 (lanes 1) and pBKCMV (which contains the CMV IE promoter and the simian virus 40 origin [lanes 2]) were used for high-efficiency transfection of randomly growing 293T human kidney cells. 293T cells transfected with the different Bel1 and Bet expression plasmids and the controls were harvested 24, 48, and 72 h after transfection, and total RNA was analyzed as described above. Hybridization probes were specific for p57Kip2 (Fig. 4A), IGF-II (Fig. 4B), EphB3, COUP-TFI, and Egr-1 (not shown). Graphic presentation of the data is shown in Fig. 4C to F. Kip2, IGF-II, and EphB3 mRNAs were clearly induced with time by the Bel1/Bet plasmid pBCbel (lanes 6). Remarkably, Kip2 RNAs and proteins (see below) showed a peak 48 h after transfection and slightly reduced but still elevated levels at 72 h p.i. IGF-II showed strongest induction in cells that expressed Bel1 but not Bet (plasmid pbel1s [lanes 4]). Other plasmids, including an HFV proviral genome deficient in env (pHSRVΔMN), did not show significant increase in the expression of any of the analyzed genes above background (Fig. 4 and 5). Gels had been loaded with equal RNA quantities, as shown by reprobing the blots against GAPDH mRNAs. COUP-TFI mRNA levels did not change relative to the time of harvest or transfected DNA, whereas Egr-1 mRNAs were not detectable at all.
FIG. 4.
Northern blot analyses of 293T cells transfected with plasmids pUC18 (lanes 1), pBKCMV (lanes 2), pHSRVΔMN (lanes 3), pbel1s (lanes 4), pBCbet (lanes 5), and pBCbel (lanes 6). Total RNAs (10 μg of each) harvested 24, 48, and 72 h after transfection were separated, blotted, hybridized against 32P-labeled Kip2 (A) and IGF-II (B) cDNA probes, and analyzed by autoradiography. Blots reprobed with a GAPDH-specific probe are shown in the lower part of panels A and B. 28S and 18S rRNAs are marked by asterisks. In cells transfected with control DNA pBKCMV, unspecific signals slightly above the 18S rRNA were observed. (C to F) Quantification of RNA blots probed with Kip2-, EphB3-, IGF-II-, and COUP-TF1-specific probes. Signals of each blot were normalized to the corresponding GAPDH signals. Symbols in the graphs represent plasmids pUC18 (✧), pBKCMV (□), pHSRVΔMN (▴), pbel1s (X), pBCbet (○), and pBCbel (●).
FIG. 5.
Immunoblot analyses of HFV Bel1 and Bet (A) and p57Kip2 protein expression (B) in transfected 293T cells. Whole cell lysates (10 μg of each) from 293T cells harvested 24, 48, and 72 h after transfection with pUC18 (lanes 1), pHSRVΔMN (lanes 2), pbel1s (lanes 3), pBCbet (lanes 4), and pBCbel (lanes 5) were analyzed. The positions of molecular mass markers and of Bel1, Bet, and p57Kip2 are shown at the margins. For technical reasons, the 72-h value in panel B is not shown.
To directly correlate HFV Bel1 transactivator expression with increases in cellular gene expression, Bel1 expression was monitored in parallel using a Bel1/Bet-specific antiserum. Whereas Bet was strongly expressed from the CMV IE promoter in plasmids pBCbel and pBCbet between 24 and 72 h after transfection, Bet expression was delayed in HFV promoter-dependent deletion clone pHSRVΔMN (Fig. 5A). Bel1 was consistently detectable in pBCbel-transfected cells and only after longer exposure (not shown) in pbel1s-transfected cells but was virtually absent in extracts from other cells, including those transfected with pHSRVΔMN. This Bel1 expression profile corresponds to the temporal patterns of HFV-mediated induction of cellular genes. Thus, it appears that the concentration and/or duration of Bel1 expression in transfected cells is a crucial parameter of the induction of distinct cellular genes.
Detection of Bel1-induced p57Kip2 expression by immunoblotting.
To study whether the Bel1-mediated increase of distinct cellular transcripts is correlated with a concomitant increase in the corresponding protein level, 293T cells transfected with different Bel1/Bet expression plasmids and pUC18 control DNA were analyzed. Protein samples harvested in regular aliquots were analyzed by immunoblot analysis with p57Kip2 and IGF-II monoclonal antibodies (MAbs). As shown in Fig. 5B, protein bands specific for p57Kip2 were detectable in 293T cells only 48 h and 72 h (data not shown) after transfection with plasmid pBCbel or pbel1s (lanes 3 and 5). Neither the Bet expression clone nor the pHSRV13 deletion clone pHSRVΔMN or control plasmid pUC18 resulted in detectable levels of p57Kip2 protein (Fig. 5B, lanes 3 and 5). IGF-II precursors and mature IGF-II were not clearly detectable in transfected 293T cells. The results prove that the Bel1 transactivator alters cellular gene expression, resulting in significantly changed concentrations of essential cellular regulatory proteins.
HFV Bel1-mediated activation of the human Kip2 promoter.
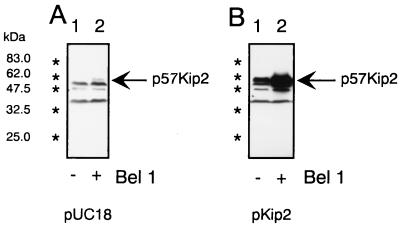
Plasmid clone pKip2, which directs human Kip2 expression from the human Kip2 promoter (27), was used for cotransfections with the Bel1 expression plasmid pbel1s and pUC18 control DNA (Fig. 6). 293T cells were transfected with 5 μg of pUC (Fig. 6A) or pKip2 (Fig. 6B) DNA in the absence or presence of the Bel1 expression plasmid pbel1s. Proteins were harvested 48 h after transfection, and equal amounts of proteins were used for immunoblotting with the Kip2-specific MAbs. Whereas p57Kip2 expression from the cellular Kip2 gene was undetectable in the absence of Bel1, p57Kip2 was clearly visible in Bel1-expressing 293T cells (Fig. 6A). Similarly, p57Kip2 expression in cells transfected with plasmid pKip2 (Fig. 6B) was strongly and reproducibly increased by HFV Bel1 (lane 2). This result strongly indicates that HFV Bel1 directly transactivates the authentic human Kip2 promoter, enhancing the expression of p57Kip2 proteins. However, alternative mechanisms could be also employed as discussed below.
FIG. 6.
Immunoblot analyses of p57Kip protein expression after HFV Bel1 transfection into 293T cells. Cell lysates (38 μg of each) harvested 48 h after cotransfection of pUC18 (A) and human Kip2 promoter-based p57Kip2 expression plasmid pKip2 (B) with and without HFV Bel1 expression plasmid pbel1s were used for immunoblotting and reacted with human p57Kip2-specific MAbs. Positions of the p57Kip2 protein bands are marked with arrows; positions of molecular size markers are shown at the left.
DISCUSSION
Here we report for the first time that HFV infection alters the expression of defined cellular genes, thereby likely changing or modifying cellular pathways in which these genes play important roles. Whereas the human cellular genes Kip2, Egr-1, EphB3, COUP-TF1, and IGF-II were strongly induced late during HFV infection of HEL299 cells, Kip2, IGF-II, and EphB3 were induced in Bel1-expressing 293T cells, too. In contrast, transcription factor COUP-TF1 expression was not affected by Bel1, and Egr-1 mRNAs were undetectable in 293T cells. This implicates cell-type-specific differences in HFV-mediated up-regulation of distinct cellular genes. It is likely that Egr-1 and COUP-TF1 were induced independently of Bel1 in infected HEL299 cells, for instance, by HFV-induced processes such as cell stress and cytopathogenicity. Since 293T cells did not show HFV-induced cytopathogenicity, signals required for up-regulation of transcriptional activators Egr-1 and COUP-TF1 may be absent in these cells. As a consequence, it is unlikely that COUP-TF1 and Egr-1 were responsible for increased p57Kip2, IGF-II, and EphB3 mRNA and protein levels in 293T cells. Taken together, different mechanisms for altering cellular gene expression during HFV infection obviously exist: defined cellular genes are activated by the Bel1 transactivator (Kip2, IGF-II, and EphB3), whereas others, such as Egr-1 and COUP-TF1, may be induced as a consequence of lytic FV replication. It is likely that Bel1 functions indirectly or in concert with presently undefined but limiting cellular factors, explaining the delayed activation of these genes in HFV-infected and Bel1-transfected cells. The biological functions and consequences of induction of cellular genes by HFV are unknown. A situation where growth-promoting IGF-II (20) and growth-inhibiting p57Kip2 (11) cellular functions are simultaneously activated by HFV might appear paradoxical. Intracellular overexpression of p57Kip2 leads to growth arrest of cells (11), and this may allow efficient replication of HFV, a function similar to the Vpr-mediated cell cycle delay in human immunodeficiency virus type 1 replication (5). On the other hand, the HFV-enhanced secretion of IGF-II may act locally and/or systemically to increase the rate of replication of those cells on which HFV replication depends. In line with this model, 293T cells expressing Bel1 showed increased populations of S and G2/M-phase cells (data not shown).
As to the increased levels of p57Kip2 and Egr-1 in HFV-infected cells, it is noteworthy that the normal function of p57Kip2 is to act as a cell cycle-dependent kinase inhibitor and inducer of differentiation (11, 29, 31) and the pleiotropic growth-suppressive effects of Egr-1 (14) are in agreement with the nononcogenicity of HFV. In contrast, the mechanistically distinct interactions of the human T-cell leukemia virus type 1 (HTLV-1) Tax transactivator with other defined negative regulators of cell cycle progression of the INK4 family result in the inactivation or reduced expression of cyclin-dependent kinase inhibitors, and these events are considered to contribute to the oncogenic potential of HTLV-1 Tax (25, 26). In summary, the different complex human retroviruses have the capacity to target the control and regulation of the cell cycle of infected host cells by fundamentally different mechanisms which result in distinct cellular responses.
The transcriptional transactivator Bel1 is necessary and sufficient for specifically increasing levels of mRNAs and protein of the cellular genes Kip2, IGF-II, and EphB3. The experiments indicate that a critical threshold level of Bel1 has to be present in the cells, as induction of IGF-II, p57Kip2, and EphB3 is correlated to Bel1 expression levels by the different effector plasmids, independently confirming the specificity of the activations (Fig. 5). The Bel1 concentration required for induction of cellular genes was physiological, since these genes were also induced in wild-type HFV-infected cells. The requirement for critical Bel1 concentration is comparable to that during HFV gene expression in which the HFV internal promoter is preferentially activated by Bel1 prior to activation of the 5′ long terminal repeat promoter (16, 17).
At present it is unknown how Bel1 increases the expression of cellular genes. It is well known that Bel1 activates both HFV promoters by direct binding to short defined but promiscuous DNA target sequences of the FV promoters (9, 10, 33) and that for full transactivation levels, interactions of Bel1 with unknown cellular proteins seem to be required (10). Since the potent Bel1 transactivator has the capacity to bind different DNA target sequences, it is likely that Bel1-mediated activation of cellular genes proceeds by a similar mechanism. One question is whether HFV Bel1 binds to and directly transactivates the promoters of IGF-II, p57Kip2, and EphB3 or whether Bel1-mediated activation of these genes is indirectly mediated by activation of unknown cellular transcription factors needed to activate expression of IGF-II, p57Kip2, and EphB3. The transactivator Tax has been reported to form Tax-DNA-CREB complexes that contribute to full transactivation levels in vitro (12), a result comparable with findings for Bel1, which is postulated to interact with and recruit cellular transcription factors and/or coactivators to the promoters of the target genes (10).
The results presented in Fig. 6 may indicate that Bel1 acts directly on the promoter of the Kip2 gene. Transcriptional regulation and activation of cellular genes is context dependent and relies on molecular interactions of defined cellular transcription factors (6, 8, 19). It is likely that Bel1 interacts with defined cellular partner molecules of comparable higher-order oligomeric complexes for similarly enhancing and activating expression of target genes. Alternatively, Bel1 may increase the processing, stability, and/or transport of cellular genes. The mechanism on how Bel1 achieves transactivation of gene expression is the subject of our current studies.
ACKNOWLEDGMENTS
This work was supported by DFG and BEO-BMFT grants to M.L. and R.M.F.
We thank Bernhard Korn for IMAG clones, Peter Hexel for help with the FACSort instrument, Jennifer Reed for critically reading the manuscript, and Harald zur Hausen for support.
REFERENCES
- 1.Aguzzi A, Marino S, Tschopp R, Rethwilm A. Regulation of expression and pathogenic potential of human foamy virus in vitro and in transgenic mice. Curr Top Microbiol Immunol. 1996;206:243–273. doi: 10.1007/978-3-642-85208-4_13. [DOI] [PubMed] [Google Scholar]
- 2.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 4.Coffin J M. Retroviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. New York, N.Y: Raven Press; 1996. pp. 1767–1847. [Google Scholar]
- 5.Emerman M, Malim M H. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science. 1998;280:1880–1884. doi: 10.1126/science.280.5371.1880. [DOI] [PubMed] [Google Scholar]
- 6.Fry C J, Farnham P J. Context-dependent transcriptional regulation. J Biol Chem. 1999;274:29583–29586. doi: 10.1074/jbc.274.42.29583. [DOI] [PubMed] [Google Scholar]
- 7.Heneine W, Switzer W M, Sandstrom P, Brown J, Vedapuri S, Schable C A, Khan A S, Lerche N W, Schweizer M, Neumann-Haefelin D, Chapman L E, Folks T. Identification of a human population infected with simian foamy viruses. Nat Med. 1998;4:403–407. doi: 10.1038/nm0498-403. [DOI] [PubMed] [Google Scholar]
- 8.Kadonaga J T, Courey A J, Ladika J, Tjian R. Distinct regions of Sp1 modulate DNA binding and transcriptional activation. Science. 1988;242:1566–1570. doi: 10.1126/science.3059495. [DOI] [PubMed] [Google Scholar]
- 9.Kang Y, Cullen B R. Derivation and functional characterization of a consensus DNA binding sequence for the tas transcriptional activator of simian foamy virus type 1. J Virol. 1998;72:5502–5509. doi: 10.1128/jvi.72.7.5502-5509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang Y, Blair W S, Cullen B R. Identification and functional characterization of a high-affinity Bel-1 DNA binding site located in the human foamy virus internal promoter. J Virol. 1998;72:504–511. doi: 10.1128/jvi.72.1.504-511.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee M-H, Reynisdottir I, Massague J. Cloning of p57KIP2, a cyclin-dependent kinase inhibitor with unique domain structure and tissue distribution. Genes Dev. 1995;9:639–649. doi: 10.1101/gad.9.6.639. [DOI] [PubMed] [Google Scholar]
- 12.Lenzmeier B, Baird A, Dervan E E, Nyborg J K. The tax protein-DNA interaction is essential for HTLV-I transactivation in vitro. J Mol Biol. 1999;291:731–744. doi: 10.1006/jmbi.1999.2969. [DOI] [PubMed] [Google Scholar]
- 13.Linial M. Foamy viruses are unconventional retroviruses. J Virol. 1999;73:1747–1755. doi: 10.1128/jvi.73.3.1747-1755.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C, Calogero A, Ragona G, Adamson E, Mercola D. EGR-1, the reluctant suppression factor: EGR-1 is known to function in the regulation of growth, differentiation, and also has significant tumor suppressor activity and a mechanism involving the induction of TGF-beta1 is postulated to account for this suppressor activity. Crit Rev Oncol. 1996;7:101–125. [PubMed] [Google Scholar]
- 15.Löchelt M, Flügel R M. The molecular biology of primate spumaviruses. In: Levy J A, editor. The Retroviridae. Vol. 4. New York, N.Y: Plenum Press; 1995. pp. 239–292. [Google Scholar]
- 16.Löchelt M, Flügel R M, Aboud M. The human foamy virus internal promoter directs the expression of the functional Bel 1 transactivator and Bet protein early after infection. J Virol. 1994;68:638–645. doi: 10.1128/jvi.68.2.638-645.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Löchelt M, Muranyi W, Flügel R M. Human foamy virus genome pocesses an internal, bel-1 dependent and functional promoter. Proc Natl Acad Sci USA. 1993;90:7317–7321. doi: 10.1073/pnas.90.15.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Löchelt M, Yu S F, Linial M L, Flügel R M. The human foamy virus internal promoter is required for efficient gene expression and infectivity. Virology. 1995;206:601–610. doi: 10.1016/s0042-6822(95)80077-8. [DOI] [PubMed] [Google Scholar]
- 19.Naar A M, Beaurang P A, Robinson K M, Oliner J D, Avizonis D, Scheek S, Zwicker J, Kadonaga J T, Tjian R. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev. 1998;12:3020–3031. doi: 10.1101/gad.12.19.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Dell S D, Day I N M. Insulin-like growth factor II (IGF-II) Int J Biochem Cell Biol. 1998;30:767–771. doi: 10.1016/s1357-2725(98)00048-x. [DOI] [PubMed] [Google Scholar]
- 21.Qiu Y, Krishnan V, Pereira F A, Tsai S Y, Tsai M-J. Chicken ovalbumin upstream promoter-transcription factors and their regulation. J Steroid Biochem Mol Biol. 1996;56:81–85. doi: 10.1016/0960-0760(95)00225-1. [DOI] [PubMed] [Google Scholar]
- 22.Russell D W, Miller A D. Foamy virus vectors. J Virol. 1996;70:217–222. doi: 10.1128/jvi.70.1.217-222.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saib A, Koken M H M, van der Spek P, Peries J, de The H. Involvement of a spliced and defective human foamy virus in the establishment of chronic infection. J Virol. 1995;69:5261–5268. doi: 10.1128/jvi.69.9.5261-5268.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 25.Suzuki T, Kitao S, Matsushime H, Yoshida M. HTLV-1 Tax protein interacts with cyclin-dependent kinase inhibitor p16INK4A and counteracts its inhibitory activity towards CDK4. EMBO J. 1996;15:1607–1614. [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki T, Narita T, Uchida-Toita M, Yoshida M. Down-regulation of the INK4 family of cyclin-dependent kinase inhibitors by tax protein of HTLV-1 through two distinct mechanisms. Virology. 1999;259:384–391. doi: 10.1006/viro.1999.9760. [DOI] [PubMed] [Google Scholar]
- 27.Tokino T, Urano T, Furuhata T, Matsushima M, Miyatsu T, Sasaki S, Nakamura Y. Characterization of the human p57KIP2 gene: alternative splicing, insertion/deletion polymorphisms in VNTR sequences in the coding region, and mutational analysis. Hum Genet. 1996;97:625–631. doi: 10.1007/BF02281873. [DOI] [PubMed] [Google Scholar]
- 28.Venkatesh L K, Yang C, Theodorakis P A, Chinnadurai G. Functional dissection of the human spumaretrovirus transactivator identifies distinct classes of dominant-negative mutants. J Virol. 1993;67:161–169. doi: 10.1128/jvi.67.1.161-169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan Y, Frisen J, Lee M H, Massague J, Barbacid M. Ablation of the CDK inhibitor p57Kip2 results in increased apoptosis and delayed differentiation during mouse development. Genes Dev. 1997;11:973–983. doi: 10.1101/gad.11.8.973. [DOI] [PubMed] [Google Scholar]
- 30.Yu S F, Stone J, Linial M L. Productive persistent infection of hematopoietic cells by human foamy virus. J Virol. 1996;70:1250–1254. doi: 10.1128/jvi.70.2.1250-1254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang P, Wong C, Liu D, Finegold M, Harper J W, Elledge S J. p21(CIP1) and p57(KIP2) control muscle differentiation at the myogenin step. Genes Dev. 1999;13:213–224. doi: 10.1101/gad.13.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou R. The Eph family receptors and ligands. Pharmacol Ther. 1998;77:151–181. doi: 10.1016/s0163-7258(97)00112-5. [DOI] [PubMed] [Google Scholar]
- 33.Zou J X, Luciw P A. The transcriptional transactivator of simian foamy virus 1 binds to a DNA target element in the viral internal promoter. Proc Natl Acad Sci USA. 1996;93:326–330. doi: 10.1073/pnas.93.1.326. [DOI] [PMC free article] [PubMed] [Google Scholar]