Abstract
Adaptive evolution navigates a balance between chance and determinism. The stochastic processes of mutation and drift generate phenotypic variation; however, once mutations reach an appreciable frequency in the population, their fate is governed by the deterministic action of selection, enriching for favorable genotypes and purging the less-favorable ones. The net result is that replicate populations will traverse similar—but not identical—pathways to higher fitness. This parallelism in evolutionary outcomes can be leveraged to identify the genes and pathways under selection. However, distinguishing between beneficial and neutral mutations is challenging because many beneficial mutations will be lost due to drift and clonal interference, and many neutral (and even deleterious) mutations will fix by hitchhiking. Here, we review the best practices that our laboratory uses to identify genetic targets of selection from next-generation sequencing data of evolved yeast populations. The general principles for identifying the mutations driving adaptation will apply more broadly.
Introduction
In his 1989 book, Wonderful Life, Stephen Jay Gould proposed a thought experiment in which the reader is invited to “rewind the tape of life” and watch evolution play out again. The purpose of this exercise was to ask a seemingly unanswerable question: how likely was it that evolution would turn out this way? Or more broadly, how reproducible is the process of adaptive evolution? Sometimes nature provides partial answers to these questions. It has long been observed that populations adapting in parallel to the same environment will tend to find similar solutions to the same selective pressures (Grant et al. 2004, Protas et al. 2006, Jones et al. 2012). These “natural experiments” suggest a degree of predictability in evolutionary outcomes. Natural experiments, however, are not perfect replicates. Details of the environments will differ, the number of replicates is constrained, and experimental parameters typically cannot be adjusted.
Around the time that Wonderful Life was released, Richard Lenski—then at the University of California Irvine—initiated a landmark experiment using twelve replicate cultures of E. coli to address the question of reproducibility (Lenski et al. 1991, Elena and Lenski 2003). Remarkably, this experiment has now surpassed 75,000 generations and is still ongoing, now in the laboratory of Jeff Barrick at UT Austin (see (Lenski 2023)). Early observations from this experiment—now known simply as the LTEE for Long-Term Evolution Experiment—strongly supported parallelism at the phenotypic level such as increases in cell size (Elena et al. 1996) and loss of the ability to catabolize D-ribose (Cooper et al. 2001). This parallelism extended to the genetic level: mutations in pbpA operon resulted in reduction of penicillin-binding proteins 2, which improves fitness, while also conferred changes in cell morphology (Philippe et al. 2009). Independent deletions in the ribose operon (rbs genes) caused a phenotypic change from Rbs+ to Rbs- and confer a fitness advantage under the experimental conditions (Cooper et al. 2001).
Experimental evolution has been performed on a number of bacterial (Velicer et al. 1998, Barrett et al. 2005), viral (Bull et al. 1997, Wichman et al. 1999), and eukaryotic microorganisms (Paquin and Adams 1983, Ferea et al. 1999, Cowen et al. 2000, McDonald et al. 2009), as well as metazoan systems such as Drosophila (Rose et al. 1992, Burke et al. 2010). Some experiments predating the LTEE and some following by example. Despite the differences in the model systems, striking parallelism is observed at the phenotypic and genotypic levels. For instance, sterility often arises in the yeast Saccharomyces cerevisiae due to loss of the signaling through the mating pathway, which provides a fitness advantage to yeast in asexual populations (Lang et al. 2009, Rojas Echenique et al. 2019).
With the rise of high-throughput sequencing technology, it is possible to identify cases of parallel evolution directly from the genome, by identifying common targets of selection across a range of conditions. Therefore, in addition to testing evolutionary theory, laboratory evolution experiments are performed to compare adaptation strategies between conditions or strain backgrounds, to quantify the rate of adaptation, to optimize growth in a novel condition, and for functional genomics, identifying genes and pathways that respond to selective pressures. With beneficial mutations in hand, one can ask a new set of questions regarding the distributions of fitness effects, the degree of dominance, pleiotropy, and epistasis of mutations that underly adaptative evolution.
In this review article, we present the experimental design and fundamental parameters of evolution that our laboratory implements to identify targets of selection, with a focus on the yeast, Saccharomyces cerevisiae. We start by giving a guideline of experimental setups, considerations, and a broadly bioinformatic pipeline. Next, we introduce a statistical analysis approach that has allowed us to filter putative drivers of adaptation. As the use of experimental evolution continues to expand, more standardized protocols are needed. We hope that these practices will narrow the search of candidate targets of selection in experimental evolution.
Considerations when setting up a laboratory evolution experiment
Variations in experimental parameters often depend on the objective of the experiment. Here, our objective is to uncover the mutations that are driving adaptation by sequencing the genomes of evolved clones and identifying genes and pathways that are mutated across replicate populations more often than expected by chance. Below we outline five considerations when setting up a laboratory evolution experiment and how these choices affect the ability to identify common targets of selection.
(1). Experimental replicates.
The most important parameter is the number of replicate populations, simply stated: more is better. To increase throughput, many labs—including our own—perform laboratory evolution in 96-well plates and use liquid-handling robotics to automate serial transfer. This experimental throughput strategy allows us to scale up the number of evolving populations in a relatively easy way (Kerr et al. 2006, Lang and Murray 2011, Kryazhimskiy et al. 2014, Fisher et al. 2018, Marad et al. 2018, Johnson et al. 2021). Specifically, we perform daily dilution and dispensing of solutions into round-bottom 96-well plates using the Biomek FX Liquid Handler equipped with a 96 multichannel pod. This setup lends itself to performing evolution experiments where the total number of populations maintained is a multiple of 96. The ideal number of populations needed to be able to identify common targets of selection will depend on the distribution of beneficial mutation rate and the distribution of fitness effects in the experimental conditions, parameters that are typically not known at the start of the experiment. In practice, we find that 48 replicate populations are sufficient to identify beneficial mutations by overrepresentation.
(2). Population size and propagation regime.
Selection can act on fitness effects greater than 1/Ne, where Ne is the effective population size. In addition, a larger population will have more beneficial lineages present and competing within the population (Gerrish and Lenski 1998). In a serial dilution regimen, the population size increases exponentially between bottlenecks. This can be mitigated by imposing more-frequent dilutions (Van den Bergh et al. 2018) or by using a scalable continuous culture system (Miller et al. 2013). Practical constraints of using 96-well plates limit the maximum population size in our experiments. Effective population size can be increased by performing smaller dilutions more frequently. For example, we directly compared adaptation at two population sizes by diluting 1:32 every 12 hours or 1:1,024 every 24 hours. Both propagation regimes result in 10 generations of growth per day at Ne ~106 and Ne ~105, respectively (Lang and Murray 2011). For the sake of convenience and because the dynamics of adaptation were quite similar in these two conditions (Lang et al. 2013), our standard practice is to dilute each culture 1:1,024 (210) every 24 hours. Practically, we achieve this by doing serial 1:32 dilutions of 4 μl into 124 μl of fresh medium. A strength of laboratory evolution experiments —particularly in microorganisms—is the ability to maintain a “frozen fossil record” for each population. This collection allows one to revive ancestral populations to measure phenotypes, sequence their genomes, or replay the evolution of a population. In practice, freezing down once every week (70 generations) works well.
(3). The number of generations.
How long do you need to run an evolution experiment to detect parallel evolution? More is not always better and the optimum generation number will depend on mutation rate, population size, and the size of mutation targets. The first beneficial mutations will arise early in the evolution (Gerstein et al. 2011, Venkataram et al. 2016, Blundell et al. 2019), whereas other beneficial mutations will emerge after thousands of generations (Wiser et al. 2013, Johnson et al. 2021). Although we cannot control this stochasticity, we can get an idea of the population dynamics by knowing the initial fitness of the ancestor (Barrick et al. 2010, Kryazhimskiy et al. 2014, Jerison et al. 2017, Rojas Echenique et al. 2019, Johnson et al. 2021), and the strength of the selective pressure (Barrick and Lenski 2013, Bailey et al. 2015, Cisneros-Mayoral et al. 2022). Parallelism has been observed in both short-and long-term experiments (Good et al. 2017). In general, more replicate populations and more generations will improve the ability to detect parallel evolution (Bailey et al. 2017). The extent to which populations have changed relative to the each other and to their ancestor can be assessed by quantifying changes in fitness. This can be done by measuring strain-specific growth rate (Dykhuizen et al. 1990; Nilsson et al. 2006; Lindsey et al. 2013), pairwise competitions (Lang et al. 2011, Wiser and Lenski 2015, Payen and Dunham 2016), or by tracking changes in barcode frequencies (Venkataram et al. 2016; Kinsler et at. 2020).
Over very long evolutionary times, as seen in the LTEE populations, may no longer be evolving from the same relevant genetic background or even to the same environment (Quandt et al. 2015, Bajić et al. 2018). Statistical analysis of mutations appearing before and after Generation 17,500 in the LTEE shows a shift from strong parallelism, where all twelve populations are sampling from the same set of beneficial mutations to a situation where more available beneficial mutations are specific to each population, contingent on the identify of fixed mutations that are unique to its history (Good et al. 2017). It is expected that the populations will eventually diverge, following their own idiosyncratic evolutionary trajectories; however, in yeast, long-term evolution experiments out to 10,000 generations still show a strong signature of parallelism at the genetic level (Johnson et al. 2021). The discussion that follows is based on populations of yeast that have evolved for several thousand generations—long enough that many selective sweeps have occurred but not too long that the initial assumptions (replicate populations adapting to the same selective environment) are no longer met.
(4). Choice of strain background.
For laboratory evolution experiments, it is common to use laboratory strains (e.g. S288C, W303, and BY) that have well-annotated reference genomes and are well-suited to propagation under controllable laboratory conditions. These backgrounds contain ancestral signatures of lab domestication that can cause fitness differences. However, the accessibility and convenient phenotypes of these strains make experimental findings comparable between laboratories. Nevertheless, all lab strains may differ by spontaneous mutations, sequencing the ancestor prior to the evolution experiment will aid in identifying background-specific mutations, structural variants, and heterozygosity that are not included in the genomic reference.
Beyond strain background, the yeast Saccharomyces cerevisiae has two mating types and can be propagated as a haploid or diploid. This choice will impact both the dynamics of adaptation and the targets of selection. Recessive beneficial mutations have no selective benefit when they first appear as heterozygous in diploid population. Therefore, these mutations are only immediately beneficial in haploids, leading to faster adaptation compared to diploids (Gerstein et al. 2011, Marad et al. 2018, Sharp et al. 2018, Johnson et al. 2021). Despite the slower rate of adaptation in diploids, haploid populations will often undergo autoduplication events leading to increased ploidy (Venkataram et al. 2016, Fisher et al. 2018, Tung et al. 2021). Diploid populations, on the other hand, acquire beneficial mutations that are at least partially dominant, and often overdominant (Fisher et al. 2021, Aggeli et al. 2022). If heterozygosity exists in the starting strain, loss-of-heterozygosity (LOH) events are likely to dominate the mutational spectra (Gerstein et al. 2014, Smukowski Heil et al. 2017, James et al. 2019) due to higher rates of LOH compared to point mutation (Dutta et al. 2021).
Previous studies have initiated evolution experiments using strains harboring engineered or evolved mutations in order to assess evolutionary outcomes from different initial fitnesses, to identify compensatory mutations in compromised strains, or to alter mutation rates (Thompson et al. 2006, Harcombe et al. 2009, McDonald et al. 2012, Kryazhimskiy et al. 2014, Szamecz et al. 2014, Laan et al. 2015, Cooper 2018, Helsen et al. 2020, LaBar et al. 2020, Vignogna et al. 2022). In the early days of experimental evolution, it was assumed that the dynamics of adaptation are dominated by rare beneficial mutations that occasionally survive drift and increase in frequency until they fix. Indeed, early observations seemed to support this view (Atwood et al. 1951, Paquin and Adams 1983). Given the assumption that evolution was limited by the supply of mutations, the question often comes up as to whether the experimentalist should “speed things up” by using strains with an elevated mutation rate (Taddei et al. 1997, Arjan G et al. 1999). Over time, the field came to realize that mutations are not limiting—that for even a modestly-sized population, mutation rate is high enough that multiple beneficial mutations will be spreading through the population simultaneously (Joseph and Hall 2004, Perfeito et al. 2007). Using mutator strains, therefore, has no practical benefit in terms of speeding up evolution experiments and comes at a great cost, and can potentially decrease statistical power to identify common targets of selection by flooding the genome with hitchhiker mutations. It is also important to recognize that mutators often have a very different mutational spectrum compared to non-mutators (Foster et al. 2015, Foster et al. 2018, Sharp et al. 2018), which could in turn shift the distribution of beneficial mutations (Sane et al. 2022, Tuffaha et al. 2022) or, in the case of mutators that destabilize homopolymeric runs and microsatellites, present challenges for sequence aligners (Lang et al. 2013).
(5). Choice of environment.
Just as mutator strains were once thought to be a necessary—or at least a pragmatic—choice for experimental evolution, it was once conventional wisdom that some kind of stress needed to be applied to see evolution, to knock the organism off of its fitness peak. After all, yeast and E. coli have been workhorse model systems in Genetics and Molecular Biology for over a century—surely they are well-adapted to standard laboratory medium. However, standard laboratory procedures and best practices initiate new experiments from frozen collections, and when strains propagate, they are routinely subjected to single-cell bottlenecks, mitigating the effect of selection. Laboratory strains (or any strain for that matter) are far from optimized for growth in laboratory conditions. So even under a “simple” and “optimal” environment, there is some degree of stress imposed that leads to an adaptive response (Hallsworth 2018). However, in most evolution experiments, a standard laboratory medium is used because it supports robust growth, and is therefore a good starting point for laboratory evolution. Regimens such as different concentrations or types of nutrients, temperature, shaking, competition, pathogens, and so on, can be varied as the experimenter chooses to introduce higher cellular stress (Kawecki et al. 2012). The key point to consider is the ability to maintain growth conditions consistently over the duration of the experiment. For example, we use a separate incubator for our evolution experiments to avoid the constant opening and closing of shared incubators. We produce medium in large batches and keep it separate at 4°C to reduce the chance of contamination. Because our dilutions are performed in non-sterile conditions, we need to add ampicillin (100 mg/ml) and tetracycline (25 mg/ml) to our growth medium to prevent bacterial contamination.
Sometimes slight changes in the environment can affect evolution in subtle and surprising ways (Worthan et al. 2023). For example, antibiotics added to yeast growth medium to prevent bacterial contamination have no detectable effect on our ancestral yeast strain. Nevertheless, some evolved genotypes have lower fitness if the antibiotics are removed (Aggeli et al. 2022). Another example is the choice to shake the cultures during cell growth. Although we typically use non-shaking conditions for practical reasons, this static environment creates a problem of accessing nutrients as the cells settled. Mutations in the ergosterol pathway allow cells to stick to the sides of the well, and because these mutations are deleterious in a homogeneous culture, they create stable subpopulations, occupying different spatial niches and maintained by frequency-dependent selection (Frenkel et al. 2015). A similar phenomenon occurs in non-shaken bacterial cultures (Rainey and Travisano 1998). As a final example, citrate is often added to the medium as either a buffer or a chelating agent. In a remarkable series of papers, the Lenski and Barrick groups dissected the complex series of events that allowed one population in the LTEE to evolve the ability to use citrate as a sole carbon source (Blount et al. 2012, Quandt et al. 2014, Quandt et al. 2015).
Identifying unique mutations by whole-genome sequencing
Computer programmers are wont to use the phrase “Garbage In, Garbage Out” as a warning that inferences are only as good as the data used to derive them. Accordingly, our ability to identify common targets of selection based on statistical overrepresentation necessitates that we first employ robust methods for identifying mutations in our evolved populations. One strategy is to sequence the whole population at different time points. This can give a bigger picture of the evolutionary dynamics of adapting lineages, but it makes it difficult to estimate which mutations co-occur in the same lineage within a population (Lang et al. 2013, Good et al. 2017). A point of consideration is that this strategy will depend on the sequencing depth, where only mutations that are present in at least 1% of individuals can be detected with 100-fold coverage. A second strategy is to sequence endpoint clones from each population (McDonald 2019). This strategy identifies fixed mutations but can miss much of the genetic variation present in the whole population. Sequencing a single clone will also pick up low-frequency mutations that happen to be in the selected clone. For this reason, our standard procedure for S. cerevisiae is to sequence at least two clones from a given population, which allows us to distinguish between high-frequency mutations (shared between the clones) and low-frequency mutations (unique to a single clone). However, for species where multiple subpopulations coexist (Good et al. 2017, Behringer et al. 2018, Harris et al. 2021), two clones will not be sufficient and the best approach may be a combination of clone and whole-population sequencing.
Though many sequencing platforms and computational tools exist for aligning reads and calling variants, our lab uses almost exclusively the Illumina platform and analyzes reads using BWA for alignment and FreeBayes for variant calling. Methods to reduce the cost of Illumina library preparation (Baym et al. 2015) and the ability to multiplex a large number of samples, make Illumina our preferred platform for whole-genome sequencing. All downstream computation analysis is performed on a High-Performance Computing Cluster. We remove adapters and low-quality bases using Trimmomatic (Bolger et al. 2014). We align our reads to the appropriate reference genome using BWA-MEM (Li and Durbin 2009). The resulting sequence alignment (SAM) files are large and, therefore, are immediately converted to binary format (BAM) using SAMtools (Li et al. 2009). Next, we use the FreeBayes (Garrison and Marth 2012) variant detector to call SNVs and indels in the evolved genome in a Variant Call format (VCF). Though our laboratory uses BWA/FreeBayes, other aligners (e.g. Bowtie, STAR, Segemehl) and variant callers (e.g. GATK, Breseq) are routinely used for analysis of clone and population-level sequencing (e.g. (Zhu et al. 2014, Behringer et al. 2018, Johnson et al. 2021)).
VCF files typically contain hundreds (and sometimes over 1,000) putative variants, many of which are spurious calls due to sequencing or alignment error, often in low complexity regions of the genome or in repeated sequences such as tRNAs or paralogous gene pairs. These spurious calls often appear in many replicate populations—an attribute that can be used to remove them from the dataset. Observing parallelism at the nucleotide level is much less common than at the level of genes (Tenaillon et al. 2012, Bailey et al. 2015). The few bona fide examples of parallelism at the level of nucleotide change involve reversion of auxotrophies (Johnson et al. 2021), frameshift mutations at short homopolymeric runs within genes (Lang et al. 2013), and alteration-of-function mutations near the active sites of proteins in the LTEE (Good et al. 2017, Maddamsetti et al. 2017). When the same variant is called in multiple populations, it is important to consider other explanations such as standing genetic variation generated during the initial overnight culture before the evolution or cross-contamination during library preparation or during the isolation of single clones.
To remove spurious calls, we use vcf-isec to first identify mutations that are common to most of the VCF files and to remove the common calls that are from each population (Danecek et al. 2011). If the sequenced strain differs from the reference genome, those differences will also be removed at this step. We next annotate the functional effect for each mutation using SnpEff (Cingolani et al. 2012). This step requires a GFF annotation file identifying the gene boundaries, which is typically only available for a subset of laboratory strains. The final, and the most laborious step, is manually validating variant calls in Integrative Genome Viewer (Robinson et al. 2011, Thorvaldsdóttir et al. 2013). Despite all the filters applied, many dubious variants manage to pass the filters. It is a good practice to always visually confirm variant calls and heterozygosity, if applicable. The inference of the zygosity will depend on the parameters that the variant caller uses and the read qualities. For consistent thresholds, we recommend to annotate homozygous variants with a read depth of ~15 and an alternative allele frequency of ~0.9 (Fisher et al. 2018, Marad et al. 2018, Martínez et al. 2022).
Identification of common targets of selection
Putative genetic targets of selection can be identified as genes that are mutated more frequently than expected by chance across replicate populations. Different statistical methods have been used to predict genetic parallelism in experimental populations. The log-likelihood ratio test (G-test) is one of the approaches widely used in experimental evolution, principally for E. coli, where there is a large number of mutations (Tenaillon et al. 2016, Behringer et al. 2018). For smaller number of mutations, other studies have used Bray-Curtis Similarity (Turner et al. 2018) and Dice’s Coefficient of Similarity (Deatherage et al. 2017). In principle, prediction of candidate target of selection is easiest if one assumes under the null model that all genes are equally likely to be hit by mutation (Shoemaker and Lennon 2022). For example, Lang et al. 2013 identified 723 coding-sequence mutations. If we distribute these randomly over the 5,799 genes in the yeast genome, we expect, according to the Poisson distribution, only two genes to have been mutated in three or more populations. The data, however, show 24 genes mutated in at least three populations, far in excess of the null expectation. Further increases in statistical power can be gained by restricting the statistical analysis to nonsynonymous mutations. Although codon usage bias selection implies that natural selection acts on synonymous variation in natural populations (Hershberg and Petrov 2008), they are rarely identified as beneficial mutations in experimental evolution, with a few notable exceptions (Bailey et al. 2021). Ignoring synonymous mutations removes a large background of hitchhiker mutations and produces more robust predictions of true targets of selection (Martínez et al. 2022).
Thus far, we have assumed that all genes are equally likely to acquire a mutation by chance. Given the known variation in mutation rate across the genome (Lang and Murray 2011), this assumption is almost certainly violated. Without knowing the per gene mutation rate, we can attempt to correct for this by weighing our probabilities by gene size (with larger genes being more likely to acquire mutations by chance). We estimate the probability that the observed mutation will occur in each established genomic region (assuming a constant mutation rate): the expected number of mutations for each coding region (), weighted for the coding sequence length () of the gene across the total length of the coding regions, in a given total number of mutations () is:
The probability of observing evolved mutations in each gene is:
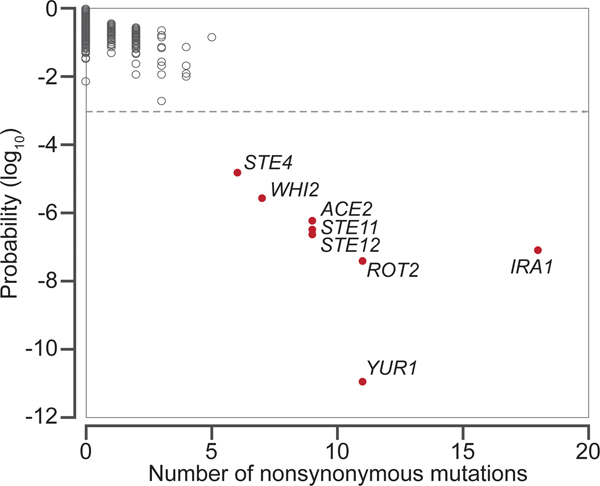
The Benjamin-Hochberg post hoc adjustment correction can be applied to correct for multiple hypothesis testing. Common targets of selection, then, must satisfy a significance threshold rather than a minimal number of observed mutations (Figure 1).
Figure 1. Identifying common targets of selection.
The observed number of nonsynonymous mutations in each of the ~5800 genes in the yeast genome and the probability that the observed number of mutations in each gene occurred by chance, after controlling for gene length. The data shown are from Lang et al. 2013.
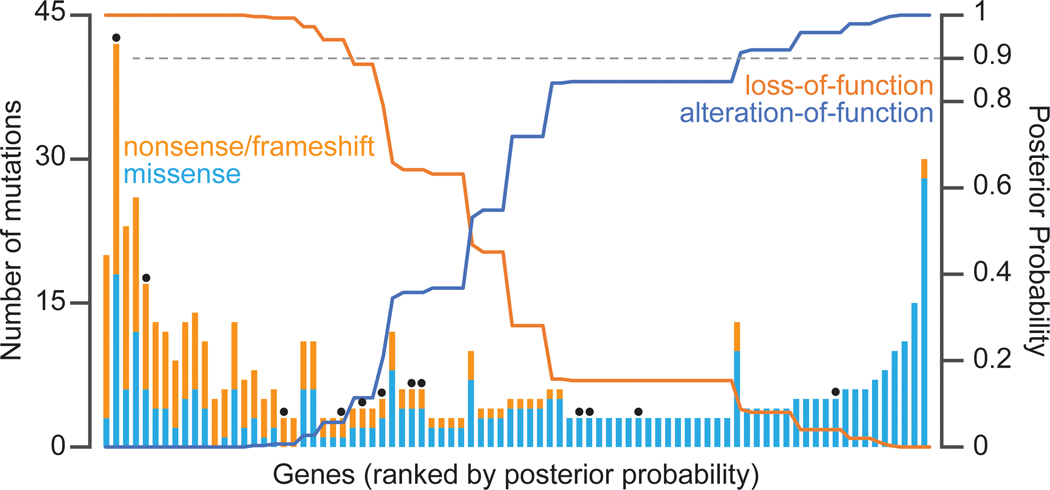
Beyond identifying parallel evolution at gene level, Gene-Ontology (GO) term enrichment can be used to identify biological pathways in which mutations are overrepresented. This identifies putative targets of selection based on their functional association, even when individual genes are not significant on their own. Based on the types of mutations we observe, we can infer how selection is acting on the target genes and pathways. For example, in S. cerevisiae, genes in the Ras pathway appear to acquire missense, nonsense, and frameshift mutations indiscriminately, suggesting that selection is acting on loss-of-function. In contrast, genes in cell-wall assembly acquire missense mutations almost exclusively, inferring that selection is acting on alteration or attenuation, not loss, of function. Similarly, in E. coli, mutations in the NAD biosynthesis/regulator gene, nadR, to be disruptive, whereas mutations in the DNA topoisomerase gene, topA, are single-base substitution mutations that modify enzymatic activity (Deatherage and Barrick 2021). Due to their large target size, loss-of-function mutations are easiest to identify and, at first glance, appear to dominate laboratory evolution experiments (Bailey and Bataillon 2016). However, over long-time scales, parallelism at the level of amino acid substitution becomes apparent, often at protein interfaces (Maddamsetti et al. 2017). A similar trend can be seen by aggregating data across multiple laboratory evolution experiments using a statistical framework to estimate the log-likelihood and posterior probabilities that selection is acting on loss-of-function or alteration-of-function given the observed mutational spectrum for any given gene (Figure 2) (Vignogna et al. 2022).
Figure 2. Laboratory evolution selects for both loss-of-function and alteration-of-function mutations.
The number and type of mutations observed in common targets of selection in experimental evolution (left y-axis), and the posterior probability that evolution selects for loss or alteration of function (right y-axis). Black dots are genes absent from the deletion collection (Giaever et al. 2002). These data were aggregated from seven yeast evolution experiments. We used a Bayesian approach to determine the posterior probabilities that selection acts on loss or alteration of function. Note that for genes where selection is inferred to be selecting for alteration-of-function, nonsense and frameshift mutations are occasionally observed. Prior probabilities for loss or alteration of function and conditional probabilities for mutational spectra were determined from experimental data as described in Vignogna et al. 2022.
Adaptive evolution is not just driven by point mutation, but also by structural variants, including deletions, insertions, and aneuploidies (Gorkovskiy and Verstrepen 2021). Like point mutations, copy number variation (CNVs) can also present strong signatures of parallelism. Chromosome-scale mutations may occur at rates orders of magnitude higher than base substitutions (Zhang et al. 2013). Therefore, contrary to single nucleotide polymorphism (SNPs), the chance of seeing CNVs that are in the exact same region is more often than SNPs. To identify CNVs from Illumina coverage we use Control-FREEC (Boeva et al. 2012) and we confirm CNVs by visually inspecting coverage plots. A more thorough guide for identifying copy number variants, loss of heterozygosity events, and other karyotype changes is presented elsewhere (Smukowski Heil 2023, Spealman et al. 2023).
Limitations to the recurrence-based approach
The statistical approaches described above identify candidate genes and pathways underlying adaptation. Verifying these predictions requires measuring the fitness effects of putative beneficial mutations by reconstructing them in the ancestral background (Chou et al. 2011, Khan et al. 2011) or by using bulk-segregant approaches that separate the effects of individual mutations (Buskirk et al. 2017, Aggeli et al. 2022). CRISPR-Cas9 methods in yeast have made allele replacement feasible at large scale (DiCarlo et al. 2013, Shen et al. 2017, Sadhu et al. 2018, Sharon et al. 2018) and barcode-based fitness assays allow fitness to be quantified in bulk (Venkataram et al. 2016, Jagdish and Ba 2022, Kinsler et al. 2023).
The success of beneficial mutations is dependent on their fitness effects. Mutations that have a ~1.5% effect reach a higher frequency and are more likely to fix compared to mutations that occur at exactly the same rate but have only a ~0.5% effect (Lang et al. 2011). Though the underlying distribution of fitness effects in any condition is unknown, both theory and experiment suggest that it is skewed with a large number of small-effect mutations and fewer mutations of large effect (Eyre-Walker and Keightley 2007). Experimental evolution samples from the middle of the fitness distribution (Hegreness et al. 2006). At one end of the distribution, drift and clonal interference impede the spread of weakly beneficial mutations, making it unlikely for them to appear in replicate populations (Barrick and Lenski 2013). At the same time, some large-effect beneficial mutations will be too rare to be sampled in multiple replicate populations, for example where only specific point mutations confer a fitness advantage.
Beyond weak and/or rare beneficial mutations, strict recurrence-based methods are also prone to miss genetic interactions: mutations that are beneficial only when another mutation is present in the same background. In principle, the approach above can be extended to identify pairs of genes (rather than individual genes) in which mutations co-occur more often than expected by chance. Applying this approach to two large laboratory evolution experiments in yeast showed that genetic interactions significantly affect evolution at the gene level; however, the power to identify specific pairs of interacting genes is limited (Fisher et al. 2019, Johnson et al. 2021).
Overview
The combination of experimental evolution and whole-genome sequencing has become a powerful tool for functional genomics (Cooper 2018). Though it is not often described as such, experimental evolution is a versatile genetic screen, revealing the genes and pathways under selection in any environment. With population sizes ~105, evolution can in principle act on differences as small as 0.001%, far below the ability to resolve by any phenotypic assay. With a genome size of 107 bp, a mutation rate of 10−10 per bp per generation, and continuous selective pressure exerted over 103 generations, each population will sample hundreds of thousands of coding-sequence mutations. Furthermore, evolution can identify complex compensatory interactions involving multiple mutations. Establishing best practices for performing laboratory evolution experiments, standardizing methods for calling mutations, and developing statistical and experimental methods for identifying and validating targets of selection is crucial as our field continues to evolve.
Acknowledgements
We thank Dimitra Aggeli for comments on the manuscript. This study was supported by the National Institutes of Health: R01GM127420.
References
- Aggeli D, Marad DA, Liu X, Buskirk SW, Levy SF and Lang GI (2022). “Overdominant and partially dominant mutations drive clonal adaptation in diploid Saccharomyces cerevisiae.” Genetics 221(2): iyac061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjan G J, M. d., C. W. Zeyl, P. J. Gerrish, J. L. Blanchardand Lenski RE(1999). “Diminishing returns from mutation supply rate in asexual populations.” Science 283(5400): 404–406. [DOI] [PubMed] [Google Scholar]
- Atwood KC, Schneider LK and Ryan FJ (1951). “Periodic selection in Escherichia coli.” Proceedings of the National Academy of Sciences 37(3): 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SF, Alonso Morales LA and Kassen R. (2021). “Effects of synonymous mutations beyond codon bias: The evidence for adaptive synonymous substitutions from microbial evolution experiments.” Genome biology and evolution 13(9): evab141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SF and Bataillon T. (2016). “Can the experimental evolution programme help us elucidate the genetic basis of adaptation in nature?” Molecular ecology 25(1): 203–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SF, Blanquart F, Bataillon T. and Kassen R. (2017). “What drives parallel evolution? How population size and mutational variation contribute to repeated evolution.” BioEssays 39(1): 1–9. [DOI] [PubMed] [Google Scholar]
- Bailey SF, Rodrigue N. and Kassen R. (2015). “The Effect of Selection Environment on the Probability of Parallel Evolution.” Molecular Biology and Evolution 32(6): 1436–1448. [DOI] [PubMed] [Google Scholar]
- Bajić D, Vila JC, Blount ZD and Sánchez A. (2018). “On the deformability of an empirical fitness landscape by microbial evolution.” Proceedings of the National Academy of Sciences 115(44): 11286–11291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RD, MacLean RC and Bell G. (2005). “Experimental evolution of Pseudomonas fluorescens in simple and complex environments.” The American Naturalist 166(4): 470–480. [DOI] [PubMed] [Google Scholar]
- Barrick JE, Kauth MR, Strelioff CC and Lenski RE (2010). “Escherichia coli rpoB mutants have increased evolvability in proportion to their fitness defects.” Molecular biology and evolution 27(6): 1338–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE and Lenski RE (2013). “Genome dynamics during experimental evolution.” Nature Reviews Genetics 14(12): 827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM and Kishony R. (2015). “Inexpensive multiplexed library preparation for megabase-sized genomes.” PloS one 10(5): e0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer MG, Choi BI, Miller SF, Doak TG, Karty JA, Guo W. and Lynch M. (2018). “Escherichia coli cultures maintain stable subpopulation structure during long-term evolution.” Proceedings of the National Academy of Sciences 115(20): E4642–E4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount ZD, Barrick JE, Davidson CJ and Lenski RE (2012). “Genomic analysis of a key innovation in an experimental Escherichia coli population.” Nature 489(7417): 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell JR, Schwartz K, Francois D, Fisher DS, Sherlock G. and Levy SF (2019). “The dynamics of adaptive genetic diversity during the early stages of clonal evolution.” Nature ecology & evolution 3(2): 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeva V, Popova T, Bleakley K, Chiche P, Cappo J, Schleiermacher G, Janoueix-Lerosey I, Delattre O. and Barillot E. (2012). “Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data.” Bioinformatics 28(3): 423–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M. and Usadel B. (2014). “Trimmomatic: a flexible trimmer for Illumina sequence data.” Bioinformatics 30(15): 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull J, Badgett M, Wichman HA, Huelsenbeck JP, Hillis DM, Gulati A, Ho C. and Molineux I. (1997). “Exceptional convergent evolution in a virus.” Genetics 147(4): 1497–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke MK, Dunham JP, Shahrestani P, Thornton KR, Rose MR and Long AD (2010). “Genome-wide analysis of a long-term evolution experiment with Drosophila.” Nature 467(7315): 587–590. [DOI] [PubMed] [Google Scholar]
- Buskirk SW, Peace RE and Lang GI (2017). “Hitchhiking and epistasis give rise to cohort dynamics in adapting populations.” Proceedings of the National Academy of Sciences 114(31): 8330–8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H-H, Chiu H-C, Delaney NF, Segrè D. and Marx CJ (2011). “Diminishing returns epistasis among beneficial mutations decelerates adaptation.” Science 332(6034): 1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X. and Ruden DM (2012). “A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3.” Fly 6(2): 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisneros-Mayoral S, Graña-Miraglia L, Pérez-Morales D, Peña-Miller R. and Fuentes-Hernández A. (2022). “Evolutionary history and strength of selection determine the rate of antibiotic resistance adaptation.” Molecular biology and evolution 39(9): msac185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper VS (2018). “Experimental Evolution as a High-Throughput Screen for Genetic Adaptations.” mSphere 3(3): e00121–00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper VS, Schneider D, Blot M. and Lenski RE (2001). “Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B.” Journal of Bacteriology 183(9): 2834–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen LE, Sanglard D, Calabrese D, Sirjusingh C, Anderson JB and Kohn LM (2000). “Evolution of drug resistance in experimental populations of Candida albicans.” Journal of bacteriology 182(6): 1515–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT and Sherry ST (2011). “The variant call format and VCFtools.” Bioinformatics 27(15): 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage DE and Barrick JE (2021). “High-throughput characterization of mutations in genes that drive clonal evolution using multiplex adaptome capture sequencing.” Cell Systems 12(12): 1187–1200. e1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage DE, Kepner JL, Bennett AF, Lenski RE and Barrick JE (2017). “Specificity of genome evolution in experimental populations of Escherichia coli evolved at different temperatures.” Proceedings of the National Academy of Sciences 114(10): E1904–E1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Norville JE, Mali P, Rios X, Aach J. and Church GM (2013). “Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems.” Nucleic Acids Res 41(7): 4336–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Dutreux F. and Schacherer J. (2021). “Loss of heterozygosity results in rapid but variable genome homogenization across yeast genetic backgrounds.” Elife 10: e70339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena SF, Cooper VS and Lenski RE (1996). “Punctuated evolution caused by selection of rare beneficial mutations.” Science 272(5269): 1802–1804. [DOI] [PubMed] [Google Scholar]
- Elena SF and Lenski RE (2003). “Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation.” Nature Reviews Genetics 4(6): 457–469. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A. and Keightley PD (2007). “The distribution of fitness effects of new mutations.” Nature Reviews Genetics 8(8): 610–618. [DOI] [PubMed] [Google Scholar]
- Ferea TL, Botstein D, Brown PO and Rosenzweig RF (1999). “Systematic changes in gene expression patterns following adaptive evolution in yeast.” Proceedings of the National Academy of Sciences 96(17): 9721–9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher KJ, Buskirk SW, Vignogna RC, Marad DA and Lang GI (2018). “Adaptive genome duplication affects patterns of molecular evolution in Saccharomyces cerevisiae.” PLoS genetics 14(5): e1007396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher KJ, Kryazhimskiy S. and Lang GI (2019). “Detecting genetic interactions using parallel evolution in experimental populations.” Philosophical Transactions of the Royal Society B 374(1777): 20180237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher KJ, Vignogna RC and Lang GI (2021). “Overdominant mutations restrict adaptive loss of heterozygosity at linked loci.” Genome biology and evolution 13(8): evab181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL, Lee H, Popodi E, Townes JP and Tang H. (2015). “Determinants of spontaneous mutation in the bacterium Escherichia coli as revealed by whole-genome sequencing.” Proceedings of the National Academy of Sciences 112(44): E5990–E5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL, Niccum BA, Popodi E, Townes JP, Lee H, MohammedIsmail W. and Tang H. (2018). “Determinants of Base-Pair Substitution Patterns Revealed by Whole-Genome Sequencing of DNA Mismatch Repair Defective Escherichia coli.” Genetics 209(4): 1029–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel EM, McDonald MJ, Van Dyken JD, Kosheleva K, Lang GI and Desai MM (2015). “Crowded growth leads to the spontaneous evolution of semistable coexistence in laboratory yeast populations.” Proceedings of the National Academy of Sciences 112(36): 11306–11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E. and Marth G. (2012). “Haplotype-based variant detection from short-read sequencing.” arXiv preprint arXiv:1207.3907. [Google Scholar]
- Gerrish PJ and Lenski RE (1998). “The fate of competing beneficial mutations in an asexual population.” Genetica 102–103(1–6): 127–144. [PubMed] [Google Scholar]
- Gerstein A, Cleathero L, Mandegar M. and Otto S. (2011). “Haploids adapt faster than diploids across a range of environments.” Journal of evolutionary biology 24(3): 531–540. [DOI] [PubMed] [Google Scholar]
- Gerstein A, Kuzmin A. and Otto S. (2014). “Loss-of-heterozygosity facilitates passage through Haldane’s sieve for Saccharomyces cerevisiae undergoing adaptation.” Nature communications 5(1): 1–9. [DOI] [PubMed] [Google Scholar]
- Good BH, McDonald MJ, Barrick JE, Lenski RE and Desai MM (2017). “The dynamics of molecular evolution over 60,000 generations.” Nature 551(7678): 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorkovskiy A. and Verstrepen KJ (2021). “The Role of Structural Variation in Adaptation and Evolution of Yeast and Other Fungi.” Genes 12(5): 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PR, Grant BR, Markert JA, Keller LF and Petren K. (2004). “Convergent evolution of Darwin’s finches caused by introgressive hybridization and selection.” Evolution 58(7): 1588–1599. [DOI] [PubMed] [Google Scholar]
- Hallsworth JE (2018). “Stress-free microbes lack vitality.” Fungal biology 122(6): 379–385. [DOI] [PubMed] [Google Scholar]
- Harcombe W, Springman R. and Bull J. (2009). “Compensatory evolution for a gene deletion is not limited to its immediate functional network.” BMC Evolutionary Biology 9(1): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KB, Flynn KM and Cooper VS (2021). “Polygenic adaptation and clonal interference enable sustained diversity in experimental Pseudomonas aeruginosa populations.” Molecular biology and evolution 38(12): 5359–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegreness M, Shoresh N, Hartl D. and Kishony R. (2006). “An Equivalence Principle for the Incorporation of Favorable Mutations in Asexual Populations.” Science 311(5767): 1615–1617. [DOI] [PubMed] [Google Scholar]
- Helsen J, Voordeckers K, Vanderwaeren L, Santermans T, Tsontaki M, Verstrepen KJ and Jelier R. (2020). “Gene loss predictably drives evolutionary adaptation.” Molecular biology and evolution 37(10): 2989–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R. and Petrov DA (2008). “Selection on codon bias.” Annual review of genetics 42: 287–299. [DOI] [PubMed] [Google Scholar]
- Jagdish T. and Ba ANN (2022). “Microbial experimental evolution in a massively multiplexed and high-throughput era.” Current Opinion in Genetics & Development 75: 101943. [DOI] [PubMed] [Google Scholar]
- James TY, Michelotti LA, Glasco AD, Clemons RA, Powers RA, James ES, Simmons DR, Bai F. and Ge S. (2019). “Adaptation by loss of heterozygosity in Saccharomyces cerevisiae clones under divergent selection.” Genetics 213(2): 665–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerison ER, Kryazhimskiy S, Mitchell JK, Bloom JS, Kruglyak L. and Desai MM (2017). “Genetic variation in adaptability and pleiotropy in budding yeast.” Elife 6: e27167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MS, Gopalakrishnan S, Goyal J, Dillingham ME, Bakerlee CW, Humphrey PT, Jagdish T, Jerison ER, Kosheleva K. and Lawrence KR (2021). “Phenotypic and molecular evolution across 10,000 generations in laboratory budding yeast populations.” Elife 10: e63910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC and White S. (2012). “The genomic basis of adaptive evolution in threespine sticklebacks.” Nature 484(7392): 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SB and Hall DW (2004). “Spontaneous mutations in diploid Saccharomyces cerevisiae: more beneficial than expected.” Genetics 168(4): 1817–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawecki TJ, Lenski RE, Ebert D, Hollis B, Olivieri I. and Whitlock MC (2012). “Experimental evolution.” Trends in ecology & evolution 27(10): 547–560. [DOI] [PubMed] [Google Scholar]
- Kerr B, Neuhauser C, Bohannan BJ and Dean AM (2006). “Local migration promotes competitive restraint in a host–pathogen’tragedy of the commons’.” Nature 442(7098): 75–78. [DOI] [PubMed] [Google Scholar]
- Khan AI, Dinh DM, Schneider D, Lenski RE and Cooper TF (2011). “Negative epistasis between beneficial mutations in an evolving bacterial population.” Science 332(6034): 1193–1196. [DOI] [PubMed] [Google Scholar]
- Kinsler G, Schmidlin K, Newell D, Eder R, Apodaca S, Petrov D. and Geiler-Samerotte K. (2023). “Extreme sensitivity of fitness to environmental conditions; lessons from #1BigBatch.” Journal of Molecular Evolution (This Issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S, Rice DP, Jerison ER and Desai MM (2014). “Global epistasis makes adaptation predictable despite sequence-level stochasticity.” Science 344(6191): 1519–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan L, Koschwanez JH and Murray AW (2015). “Evolutionary adaptation after crippling cell polarization follows reproducible trajectories.” Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBar T, Hsieh Y-YP, Fumasoni M. and Murray AW (2020). “Evolutionary repair experiments as a window to the molecular diversity of life.” Current Biology 30(10): R565–R574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Botstein D. and Desai MM (2011). “Genetic variation and the fate of beneficial mutations in asexual populations.” Genetics 188(3): 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI and Murray AW (2011). “Mutation rates across budding yeast chromosome VI are correlated with replication timing.” Genome biology and evolution 3: 799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Murray AW and Botstein D. (2009). “The cost of gene expression underlies a fitness trade-off in yeast.” Proceedings of the National Academy of Sciences 106(14): 5755–5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Parsons L. and Gammie AE (2013). “Mutation rates, spectra, and genome-wide distribution of spontaneous mutations in mismatch repair deficient yeast.” G3 (Bethesda) 3(9): 1453–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Rice DP, Hickman MJ, Sodergren E, Weinstock GM, Botstein D. and Desai MM (2013). “Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations.” Nature 500(7464): 571–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski RE (2023). “Revisiting the Design of the Long-Term Evolution Experiment with Escherichia coli.” Journal of Molecular Evolution (This Issue). [DOI] [PubMed] [Google Scholar]
- Lenski RE, Rose MR, Simpson SC and Tadler SC (1991). “Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations.” The American Naturalist 138(6): 1315–1341. [Google Scholar]
- Li H. and Durbin R. (2009). “Fast and accurate short read alignment with Burrows–Wheeler transform.” bioinformatics 25(14): 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. and Subgroup GPDP (2009). “The Sequence alignment/map (SAM) format and SAMtools.” Bioinformatics 25(16): 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddamsetti R, Hatcher PJ, Green AG, Williams BL, Marks DS and Lenski RE (2017). “Core genes evolve rapidly in the long-term evolution experiment with Escherichia coli.” Genome biology and evolution 9(4): 1072–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marad DA, Buskirk SW and Lang GI (2018). “Altered access to beneficial mutations slows adaptation and biases fixed mutations in diploids.” Nature ecology & evolution 2(5): 882–889. [DOI] [PubMed] [Google Scholar]
- Martínez AA, Conboy A, Buskirk SW, Marad DA and Lang GI (2022). “Long-term adaptation to galactose as a sole carbon source selects for mutations in nutrient signaling pathways.” bioRxiv: 2022.2005.2017.492354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MJ (2019). “Microbial experimental evolution–a proving ground for evolutionary theory and a tool for discovery.” EMBO reports 20(8): e46992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MJ, Gehrig SM, Meintjes PL, Zhang X-X and Rainey PB (2009). “Adaptive divergence in experimental populations of Pseudomonas fluorescens. IV. Genetic constraints guide evolutionary trajectories in a parallel adaptive radiation.” Genetics 183(3): 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MJ, Hsieh Y-Y, Yu Y-H, Chang S-L and Leu J-Y (2012). “The evolution of low mutation rates in experimental mutator populations of Saccharomyces cerevisiae.” Current Biology 22(13): 1235–1240. [DOI] [PubMed] [Google Scholar]
- Miller AW, Befort C, Kerr EO and Dunham MJ (2013). “Design and use of multiplexed chemostat arrays.” JoVE (Journal of Visualized Experiments)(72): e50262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin C. and Adams J. (1983). “Frequency of fixation of adaptive mutations is higher in evolving diploid than haploid yeast populations.” Nature 302(5908): 495–500. [DOI] [PubMed] [Google Scholar]
- Payen C. and Dunham MJ (2016). Experimental evolution and resequencing analysis of yeast. Yeast Functional Genomics, Springer: 361–374. [DOI] [PubMed] [Google Scholar]
- Perfeito L, Fernandes L, Mota C. and Gordo I. (2007). “Adaptive mutations in bacteria: high rate and small effects.” science 317(5839): 813–815. [DOI] [PubMed] [Google Scholar]
- Philippe N, Pelosi L, Lenski RE and Schneider D. (2009). “Evolution of penicillin-binding protein 2 concentration and cell shape during a long-term experiment with Escherichia coli.” Journal of Bacteriology 191(3): 909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, Zon LI, Borowsky R. and Tabin CJ (2006). “Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism.” Nature genetics 38(1): 107–111. [DOI] [PubMed] [Google Scholar]
- Quandt EM, Deatherage DE, Ellington AD, Georgiou G. and Barrick JE (2014). “Recursive genomewide recombination and sequencing reveals a key refinement step in the evolution of a metabolic innovation in Escherichia coli.” Proceedings of the National Academy of Sciences 111(6): 2217–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt EM, Gollihar J, Blount ZD, Ellington AD, Georgiou G. and Barrick JE (2015). “Fine-tuning citrate synthase flux potentiates and refines metabolic innovation in the Lenski evolution experiment.” Elife 4: e09696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey PB and Travisano M. (1998). “Adaptive radiation in a heterogeneous environment.” Nature 394(6688): 69–72. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G. and Mesirov JP (2011). “Integrative genomics viewer.” Nature Biotechnology 29(1): 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas Echenique JI, Kryazhimskiy S, Nguyen Ba AN and Desai MM (2019). “Modular epistasis and the compensatory evolution of gene deletion mutants.” PLoS genetics 15(2): e1007958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MR, Vu LN, Park SU and Graves JL Jr (1992). “Selection on stress resistance increases longevity in Drosophila melanogaster.” Experimental gerontology 27(2): 241–250. [DOI] [PubMed] [Google Scholar]
- Sadhu MJ, Bloom JS, Day L, Siegel JJ, Kosuri S. and Kruglyak L. (2018). “Highly parallel genome variant engineering with CRISPR-Cas9.” Nat Genet 50(4): 510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sane M, Diwan GD, Bhat BA, Wahl LM and Agashe D. (2022). “Shifts in mutation spectra enhance access to beneficial mutations.” bioRxiv: 2020.2009.2005.284158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon E, Chen SA, Khosla NM, Smith JD, Pritchard JK and Fraser HB (2018). “Functional Genetic Variants Revealed by Massively Parallel Precise Genome Editing.” Cell 175(2): 544–557 e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp NP, Sandell L, James CG and Otto SP (2018). “The genome-wide rate and spectrum of spontaneous mutations differ between haploid and diploid yeast.” Proceedings of the National Academy of Sciences 115(22): E5046–E5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JP, Zhao D, Sasik R, Luebeck J, Birmingham A, Bojorquez-Gomez A, Licon K, Klepper K, Pekin D, Beckett AN, Sanchez KS, Thomas A, Kuo CC, Du D, Roguev A, Lewis NE, Chang AN, Kreisberg JF, Krogan N, Qi L, Ideker T. and Mali P. (2017). “Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions.” Nat Methods 14(6): 573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker WR and Lennon JT (2022). “Predicting Parallelism and Quantifying Divergence in Microbial Evolution Experiments.” Msphere 7(1): e00672–00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smukowski Heil CS (2023). “Loss of heterozygosity and its importance in genome evolution.” Journal of Molecular Evolution (This Issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smukowski Heil CS, DeSevo CG, Pai DA, Tucker CM, Hoang ML and Dunham MJ (2017). “Loss of heterozygosity drives adaptation in hybrid yeast.” Molecular biology and evolution 34(7): 1596–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spealman P, De T, Chuong J. and Gresham D. (2023). “Best Practices in Microbial Experimental Evolution: Using reporters and long read sequencing to identify copy number variation in experimental evolution.” Journal of Molecular Evolution (This Issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamecz B, Boross G, Kalapis D, Kovács K, Fekete G, Farkas Z, Lázár V, Hrtyan M, Kemmeren P. and Groot Koerkamp MJ (2014). “The genomic landscape of compensatory evolution.” PLoS biology 12(8): e1001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei F, Radman M, Maynard-Smith J, Toupance B, Gouyon P-H and Godelle B. (1997). “Role of mutator alleles in adaptive evolution.” Nature 387(6634): 700–702. [DOI] [PubMed] [Google Scholar]
- Tenaillon O, Barrick JE, Ribeck N, Deatherage DE, Blanchard JL, Dasgupta A, Wu GC, Wielgoss S, Cruveiller S. and Médigue C. (2016). “Tempo and mode of genome evolution in a 50,000-generation experiment.” Nature 536(7615): 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O, Rodríguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD and Gaut BS (2012). “The Molecular Diversity of Adaptive Convergence.” Science 335(6067): 457–461. [DOI] [PubMed] [Google Scholar]
- Thompson DA, Desai MM and Murray AW (2006). “Ploidy controls the success of mutators and nature of mutations during budding yeast evolution.” Current Biology 16(16): 1581–1590. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT and Mesirov JP (2013). “Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration.” Briefings in bioinformatics 14(2): 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuffaha M, Varakunan S, Castellano D, Gutenkunst RN and Wahl LM (2022). “Shifts in mutation bias promote mutators by altering the distribution of fitness effects.” bioRxiv: 2022.2009.2027.509708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung S, Bakerlee CW, Phillips AM, Nguyen Ba AN and Desai MM (2021). “The genetic basis of differential autodiploidization in evolving yeast populations.” G3 11(8): jkab192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CB, Marshall CW and Cooper VS (2018). “Parallel genetic adaptation across environments differing in mode of growth or resource availability.” Evolution letters 2(4): 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bergh B, Swings T, Fauvart M. and Michiels J. (2018). “Experimental design, population dynamics, and diversity in microbial experimental evolution.” Microbiology and Molecular Biology Reviews 82(3): e00008–00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velicer GJ, Kroos L. and Lenski RE (1998). “Loss of social behaviors by Myxococcus xanthus during evolution in an unstructured habitat.” Proceedings of the National Academy of Sciences 95(21): 12376–12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataram S, Dunn B, Li Y, Agarwala A, Chang J, Ebel ER, Geiler-Samerotte K, Herissant L, Blundell JR, Levy SF, Fisher DS, Sherlock G. and Petrov DA (2016). “Development of a Comprehensive Genotype-to-Fitness Map of Adaptation-Driving Mutations in Yeast.” Cell 166(6): 1585–1596 e1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignogna RC, Allocca M, Monticelli M, Norris JW, Steet R, Perlstein EO, Andreotti G. and Lang GI (2022). “Evolutionary rescue of phosphomannomutase deficiency in yeast models of human disease.” eLife 11: e79346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichman H, Badgett M, Scott L, Boulianne C. and Bull J. (1999). “Different trajectories of parallel evolution during viral adaptation.” Science 285(5426): 422–424. [DOI] [PubMed] [Google Scholar]
- Wiser MJ and Lenski RE (2015). “A comparison of methods to measure fitness in Escherichia coli.” PloS one 10(5): e0126210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiser MJ, Ribeck N. and Lenski RE (2013). “Long-term dynamics of adaptation in asexual populations.” Science 342(6164): 1364–1367. [DOI] [PubMed] [Google Scholar]
- Worthan SB, McCarthy RDP and Behringer MG (2023). “Case Studies in the Assessment of Microbial Fitness: Seemingly Subtle Changes Can Have Major Effects on Phenotypic Outcomes.” Journal of Molecular Evolution (This Issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zeidler AF, Song W, Puccia CM, Malc E, Greenwell PW, Mieczkowski PA, Petes TD and Argueso JL (2013). “Gene copy-number variation in haploid and diploid strains of the yeast Saccharomyces cerevisiae.” Genetics 193(3): 785–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YO, Siegal ML, Hall DW and Petrov DA (2014). “Precise estimates of mutation rate and spectrum in yeast.” Proceedings of the National Academy of Sciences 111(22): E2310–E2318. [DOI] [PMC free article] [PubMed] [Google Scholar]