Abstract
Luteoviruses avoid degradation in the hemolymph of their aphid vector by interacting with a GroEL homolog from the aphid's primary endosymbiotic bacterium (Buchnera sp.). Mutational analysis of GroEL from the primary endosymbiont of Myzus persicae (MpB GroEL) revealed that the amino acids mediating binding of Potato leafroll virus (PLRV; Luteoviridae) are located within residues 9 to 19 and 427 to 457 of the N-terminal and C-terminal regions, respectively, of the discontinuous equatorial domain. Virus overlay assays with a series of overlapping synthetic decameric peptides and their derivatives demonstrated that R13, K15, L17, and R18 of the N-terminal region and R441 and R445 of the C-terminal region of the equatorial domain of GroEL are critical for PLRV binding. Replacement of R441 and R445 by alanine in full-length MpB GroEL and in MpB GroEL deletion mutants reduced but did not abolish PLRV binding. Alanine substitution of either R13 or K15 eliminated the PLRV-binding capacity of the other and those of L17 and R18. In the predicted tertiary structure of GroEL, the determinants mediating virus binding are juxtaposed in the equatorial plain.
Potato leafroll virus (PLRV; Luteoviridae), a positive-stranded RNA virus, mainly replicates in the phloem tissue of its plant hosts and is transmitted by aphids in a persistent and circulative manner (29, 33). Based on ultrastructural studies of luteoviruses in vector and nonvector aphids, it has been postulated that virions are transcellularly transported through epithelial-cell linings in the gut and salivary gland (12). The hemolymph of an aphid acts as a reservoir in which acquired virus particles are retained in an infective form without replication for the life span of the aphid. A GroEL homolog synthesized by the primary bacterial endosymbiont (Buchnera sp.) of aphids is abundantly present in the hemolymph and plays a crucial role in determining the persistent nature of luteoviruses in the aphid's body fluid (32, 34). GroEL homologs are highly conserved and belong to the chaperonin-60 family of proteins, which are generally involved in the intracellular folding and assembly of nonnative proteins in an ATP-dependent manner (9). Crystallography of Escherichia coli GroEL has demonstrated that the protein forms a cylinder-shaped homooligomer of 14 subunits arranged in two heptameric rings stacked back to back. Buchnera GroEL 14-mers have been immunodetected in the hemolymph of aphids (32). In vitro ligand assays have shown that luteovirus particles display a strong affinity for native GroEL molecules as well as for the 60-kDa GroEL subunit (11, 17, 32, 34). Binding to Buchnera GroEL is mediated by the N-terminal part of the readthrough domain (RTD) of the minor capsid protein of a luteovirus (11, 32), which is produced as a result of translational readthrough of the major capsid protein. The luteovirus RTD is present on the surface of a virus particle (4). Furthermore, in vivo studies have shown that luteovirus mutants devoid of the RTD could not be sustained for a long period of time in the aphid hemolymph (32), indicating that the GroEL-RTD association protects the virus from rapid degradation in the aphid. Recently, it was suggested that a GroEL homolog of endosymbiotic origin exerted a protective function on Tomato yellow leaf curl virus (TYLCV; Geminiviridae) during its passage through the hemolymph of the whitefly Bemisia tabaci (25). Association with GroEL homologs of endosymbiotic origin may therefore be a common evolutionary adaptation shared by circulatively transmitted (plant) viruses.
Usually, hydrophobic residues of the apical domains, located on both sides of the GroEL cylinder, mediate the binding of nonnative proteins in the bacterial cytosol (3, 10). However, mutational analysis experiments of the gene encoding Buchnera GroEL of Myzus persicae (MpB GroEL) revealed that the determinants required for PLRV binding are located in the equatorial domain (17). The equatorial domain forms the waist of the GroEL 14-mer and holds the cylinder together (3). It is made up of two regions at the N terminus and C terminus that are not contiguous in the amino acid sequence but are in spatial proximity after folding of the GroEL polypeptide (3). This study identifies amino acid residues of GroEL that are critical in the binding of PLRV to the GroEL monomer by deletion mutant analysis and pepscan-assisted site-directed mutagenesis.
MATERIALS AND METHODS
Synthesis and cloning of Buchnera GroEL mutants.
pGEX-2T constructs for the expression of truncated mutants of MpB GroEL were generated by PCR using primers which contained additional restriction sites (BamHI, EcoRI, or HindIII sites) for cloning purposes (Table 1). PCR amplification was performed in 50 μl of 10 mM Tris-HCl (pH 8.3) containing 0.4 mM (total) deoxynucleoside triphosphates, 3 mM MgCl2, 50 mM KCl, 10 ng of template DNA (pCR[Buchnera GroEL] [12]), 0.25 μM each primer, and 2.5 U of Taq polymerase (Boehringer Mannheim). Mixtures were incubated for 2 min at 94°C, followed by 35 cycles of 1 min at 94°C, 1 min at 50°C, and 2 min at 72°C, with a final incubation of 10 min at 72°C. All PCR products were first cloned into pCRII (TA Cloning Kit; Invitrogen) digested with BamHI or BamHI/EcoRI and subsequently religated into the BamHI or BamHI/EcoRI sites of pGEX-2T. MpB GroEL[1-57/134-408] was synthesized with primer set F10 and R7 using pGEX MpB GroEL[1-408] as a template. The resulting fragment was digested with HindIII and self-ligated. Deletion mutants MpB GroEL[122-408/475-548], -[122-548], -[122-474], and -[122-408/475-548] have been described previously (17).
TABLE 1.
Oligonucleotides used for the construction of MpB GroEL deletion mutants
| Oligonucleotide | Sequence (5′→3′)a | Corresponding positionsb |
|---|---|---|
| F1 | ccggatccATGGCCGCTAAAGATGTA | 1–6 |
| F2 | ggatccatgAAAGCTGTTATTAGTGCTG | 122–127 |
| F6 | ggtgaagcttAACTATGGTTATAATGCAGC | 475–480 |
| F7 | ccggatccGGTAATGAAGCCCGAATT | 9–14 |
| F8 | ccggatccGGAGTTAATGTATTAGCAG | 19–24 |
| F9 | gaggatccGTAACTTTAGGTCCAAAAG | 29–34 |
| F10 | ttccaagcttTTATCTGTACCATGTTCAG | 134–139 |
| R1 | acggatccTTACATCATTCCaCCC | 545–548 |
| R2 | gaattcttaACCTTTTCCATCTTTTACG | 470–474 |
| R3 | caataagcttTTCAACAGCTGCACCAGT | 404–408 |
| R4 | gaattcttaTTCAACAGCTGCACGAG | 404–408 |
| R7 | caataagcttGATTTCACGAGCTACTGA | 53–57 |
| R8 | gaattcttaATTGGAAACAATTTGACGTAA | 452–457 |
| R9 | gaattcttaAGATGTTTTTCCAGCCACTC | 422–427 |
Uppercase letters indicate Buchnera groEL sequences, and lowercase letters indicate sequences which are not part of the gene. Restriction sites (EcoRI, BamHI, and HindIII) are boldfaced, start codons are double underlined, and termination codons are single underlined.
Numbering refers to the corresponding positions of the amino acid residues of MpB GroEL.
Single amino acid mutations were made using the QuickChange Site-Directed Mutagenesis Kit (Stratagene). Primer design and PCR were performed according to the manufacturer's recommendations. Constructs pGEX MpB GroEL[1-408] and pGEX MpB GroEL[122-548] were used as templates for generating point mutations at the N terminus and C terminus, respectively. To obtain full-length constructs of MpB GroEL containing the point mutations at the N terminus, the C-terminal XbaI/EcoRI restriction fragment of pGEX MpBGroEL[122-548] was cloned into the XbaI and EcoRI sites of pGEX MpBGroEL[1-408]R13A, pGEX MpBGroEL[1-408]K15A, or pGEX MpB GroEL[1-408]L17A/R18A. The XbaI restriction site is located in the region encoding the apical domain of MpB GroEL (17). To generate a full-length construct of MpB GroEL containing mutations at positions R13, R441, and R445, the C-terminal XbaI/EcoRI restriction fragment of pGEX MpBGroEL[122-548]R441A/R445A was cloned into the XbaI and EcoRI sites of pGEX MpBGroEL[1-408]R13A. The full-length constructs of MpB GroEL containing the C-terminal point mutations at positions R441 and R445 were obtained by ligation of the N-terminal BamHI/XbaI fragment of pGEX MpB GroEL[1-408] to the C-terminal XbaI/EcoRI fragment of pGEX MpBGroEL[122-548]R441A/R445A. The ligation product was cloned into the BamHI/EcoRI sites of pGEX-2T. All constructs were verified by nucleotide sequence analysis.
Expression and isolation of Buchnera GroEL mutants.
The pGEX constructs containing GroEL sequences were introduced into E. coli DH5α (Stratagene). For expression, overnight cultures were diluted 1:10 in Luria broth (LB) containing ampicillin (100 μg/ml) and grown at 37°C for 3 h. Subsequently, 1 mM isopropyl-β-d-thiogalactoside (IPTG) was added to induce protein synthesis, and cultures were allowed to grow at room temperature. After 7 h, cells were pelleted at 4,000 × g for 10 min and resuspended in 50 mM Tris-HCl (pH 7.5) containing 10 mM MgCl2. Cells were lysed by one cycle of freeze-thawing and sonication. After centrifugation, the glutathione S-transferase (GST) fusion proteins were affinity purified from the supernatant using glutathione-Sepharose (Pharmacia) according to the manufacturer's recommendations. To remove the GST moiety, fusion proteins were incubated with thrombin for 3 h at 10°C. Cleaved products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blot analysis with anti-MpB GroEL immunoglobulin G (IgG) (Plant Research International, Wageningen, The Netherlands). To ensure that similar quantities of deletion mutants were tested for their virus-binding capacities (described below), they were diluted to yield bands of similar intensities as assessed by amido black staining after electroblotting. Each mutant was named after the positions of the first and last amino acids bordering the included fragment.
Virus overlay assay.
PLRV (35) was maintained on Physalis floridana as previously described and purified from leaf material by a modified enzyme-assisted procedure (31). Virus overlay assays (far-Western assays) were performed as described before (17, 34). Similar amounts of MpB GroEL polypeptides were separated by SDS-PAGE. After electrophoresis, gels were conditioned in 10 mM 3-[cyclohexylamino]-1-propanesulfonic acid (pH 11.0) containing 10% methanol for 1 h, and proteins were electrotransferred onto nitrocellulose. All experiments were performed in duplicate. One protein blot was incubated overnight with purified PLRV (10 μg per ml), after which immunodetection with anti-PLRV IgG and alkaline phosphatase-conjugated anti-rabbit IgG was carried out (34). The other blot was stained with amido black to confirm whether similar amounts of the proteins were transferred to the membrane.
Pepscan analysis.
Decameric peptides were synthesized on cellulose membranes using 9-fluorenylmethoxy carbonyl (Fmoc)-amino acid active esters according to the manufacturer's instructions (Genosys Biotechnologies). Subsequently, membranes were incubated with blocking buffer (Genosys Biotechnologies) for 16 h at room temperature, followed by 10 μg of purified PLRV/ml in blocking buffer for 16 h at room temperature. Bound virus particles were detected with anti-PLRV IgG (Plant Research International), followed by goat anti-rabbit IgG-alkaline phosphatase conjugate (Sigma) at a concentration of 1 μg/ml in blocking buffer for 3 h at room temperature. Duplicate membranes, treated in the same manner as above but without the virus, served as negative controls.
Quantification of virus binding.
Virus binding in the far-Western assays and pepscan analyses was quantified using the Comparative Quantification module of Molecular Analyst software (Bio-Rad, Hercules, Calif.), which calculates the pixel intensities of the immunostained bands.
Comparative protein modeling.
A structural model of Buchnera GroEL was generated using the automated homology modeling server SWISS-MODEL (13) running at Glaxo Wellcome Experimental Research (Geneva, Switzerland) and using the three-dimensional (3D) structure model of E. coli GroEL (3) as a modeling template. The predicted 3D template was displayed using the Swiss-PdbViewer 3.5 (13), and the generated graphic was further enhanced using the POV-Ray ray tracing package 3.1 (POV-Team, Williamstown, Australia).
RESULTS
Identification of amino acids in the N-terminal part of the equatorial domain of MpB GroEL mediating virus binding.
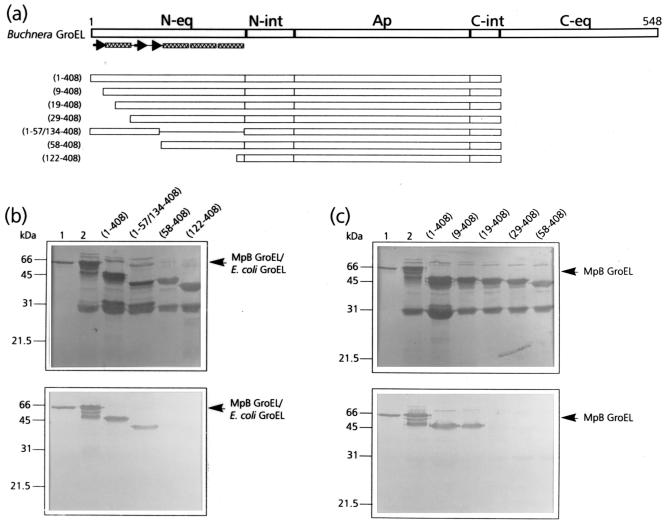
To determine which region of the N-terminal part of the equatorial domain of MpB GroEL harbors components with a PLRV-binding capacity, a series of deletion mutants was synthesized based on the predicted secondary structure of GroEL (Fig. 1a). All deletion mutants, which were devoid of the C-terminal equatorial domain, were expressed in E. coli, and after removal of the GST moiety, similar amounts of the recombinant polyproteins were tested for their capacities to bind purified PLRV in a virus overlay assay. It was shown that deletion of the distal 57 amino acids of the N-terminal equatorial domain abolished PLRV binding; PLRV bound as readily to MpB GroEL(1-408) as it did to recombinant GroEL, but no binding to MpB GroEL(58-408) was detected (Fig. 1b and c). MpB GroEL(1-57/134-408), which lacks the proximal part of the equatorial domain, was still recognized by PLRV, although binding was reduced by 45%. By homology to GroEL of E. coli, the distal half of the N terminus of the equatorial domain of MpB GroEL is characterized by four structural elements: three β-sheets and an α-helix (Fig. 1a). To further map the PLRV-binding site, additional deletion mutants designed according to these secondary structures were expressed and tested for virus binding in the overlay assay (Fig. 1c). This revealed that residues in the first β-sheet were not critical for binding, since PLRV bound as readily to MpB GroEL(9-408) as to MpB GroEL(1-408). However, no virus binding to MpB GroEL(19-408) or MpB GroEL(29-408) was detected, indicating that the region containing the α-helical structure, and more particularly the residues between amino acids 9 and 19, is required for the interaction with PLRV (Fig. 1c).
FIG. 1.
Mapping of the N-terminal PLRV-binding site by virus overlay assays of deletion derivatives of MpB GroEL. (a) Schematic representation of MpB GroEL deletion mutants. The numbers in parentheses correspond to the positions of amino acid residues of MpB GroEL (17) and mark the borders of the deletion mutants. N-eq, N-terminal region of the equatorial domain; N-int, N-terminal region of the intermediate domain; Ap, apical domain; C-int, C-terminal region of the intermediate domain; C-eq, C-terminal region of the equatorial domain. Secondary structural elements are indicated by boxed sine waves (α-helices) and arrows (β-strands). (b) Virus overlay assay showing that the first 57 amino acid residues are involved in PLRV binding (bottom) and amido black-stained blot (top). (c) Virus overlay assay showing that residues 10 to 18 are involved in virus binding (bottom) and amido black-stained blot (top). Lanes 1, wild-type MpB GroEL isolated from M. persicae; lanes 2, recombinant MpB GroEL. All other lanes contain the indicated deletion mutants of MpB GroEL as depicted in panel a. The positions of MpB GroEL and E. coli GroEL are indicated.
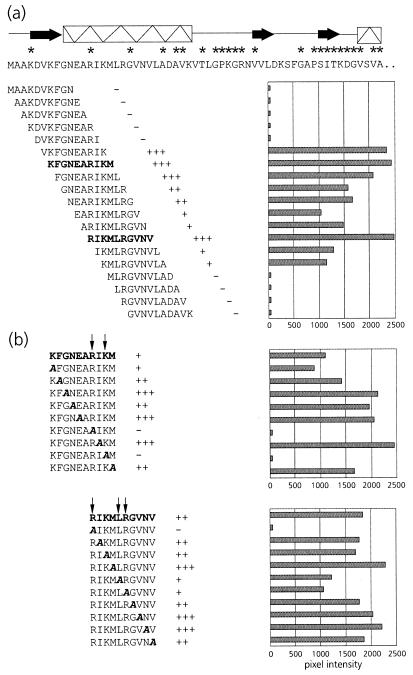
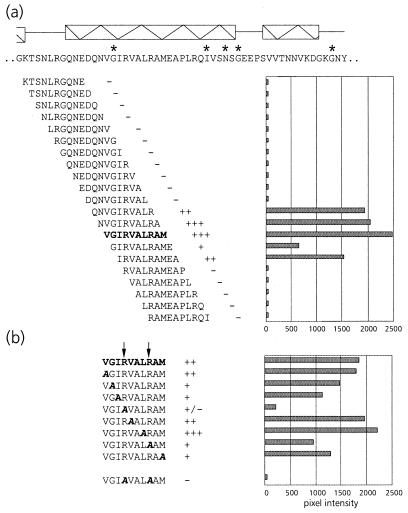
To assist site-directed mutagenesis, a pepscan analysis of the N terminus of the equatorial domain was performed. This showed that PLRV exhibited affinity only for peptides in the region between amino acid residues 5 and 24 (Fig. 2a), thus corroborating the results of the mutational analysis. Alanine scanning (Fig. 2b) using the two peptides, KFGNEARIKM and RIKMLRGVNV, that most strongly interacted with PLRV identified the arginine at position 13 (R13) as the key residue in maintaining affinity for the virus. Both peptides failed to bind PLRV when R13 was replaced by alanine. Although alanine replacement of K15 eliminated PLRV binding of the former peptide, it did not eliminate PLRV binding of the latter. Alanine replacement of L17 and R18 only slightly reduced the PLRV affinity of peptide RIKMLRGVNV.
FIG. 2.
(a) Schematic representation of PLRV-binding activities of synthetic decameric peptides corresponding to amino acid residues 1 to 57 of the N-terminal region of the equatorial domain of MpB GroEL. The results of the first 19 peptides are shown; no PLRV binding to any of the subsequent peptides in this region was detected. Secondary structural elements are indicated by thick arrows (β-strands) and boxed sine waves (α-helices). Conserved sequences in GroEL/Hsp60 sequences are indicated by asterisks (14). (b) Alanine scanning of the two decameric peptides with the strongest binding capacities (boldfaced). The affinity of PLRV for the peptides has been quantified using Molecular Analyst software (histograms) and is interpreted as follows: +++, high affinity; ++, intermediate affinity; +, low affinity; −, no PLRV binding detected. Arrows indicate residues critical for binding PLRV.
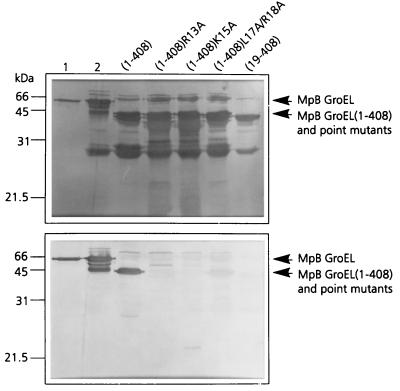
To verify the importance of R13, K15, L17, and R18 in PLRV binding, these residues were replaced by alanines in the context of MpB GroEL(1-408), resulting in MpB GroEL(1-408)R13A, MpB GroEL(1-408)K15A, and MpB GroEL(1-408)L17A/R18A. When the respective polypeptides were tested in the virus overlay assay, it was shown that MpB GroEL(1-408)R13A and MpB GroEL(1-408)K15A lost their virus-binding capacities completely, whereas PLRV binding to MpB GroEL(1-408)L17A/R18A was reduced by 70% compared to binding to the recombinant full-length product (Fig. 3). The site-directed mutagenesis results are entirely consistent with those obtained from the pepscan analysis (Fig. 2): alanine replacement of R13 or K15 eliminated the other's virus-binding capacity as well as those of L17 and R18.
FIG. 3.
Virus overlay assay (bottom panel) of alanine replacement mutants of MpB GroEL(1-408) to identify individual amino acids involved in PLRV binding. Lane 1, wild-type MpB GroEL isolated from M. persicae; lane 2, recombinant MpB GroEL. All other lanes contain wild-type MpB GroEL(1-408) and point mutants of MpB GroEL(1-408) as indicated. The positions of full-length MpB GroEL, MpB GroEL(1-408), and alanine replacement mutants of MpB GroEL(1-408) are indicated by arrows. (Top panel) Amido black-stained blot.
Identification of amino acids involved in the C-terminal binding site of MpB GroEL.
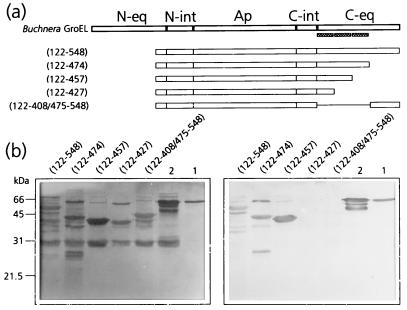
In a recent report it was shown that MpB GroEL(122-474) bound PLRV, whereas MpB GroEL(122-408/475-548) did not (17), indicating that the determinants implicated in PLRV binding are located between residues 408 and 475 of the C-terminal equatorial domain of MpB GroEL. To identify the amino acids responsible for PLRV binding, additional mutants, all of which were devoid of the N-terminal equatorial domain, were created based on the predicted secondary structure of MpB GroEL (Fig. 4a). Virus overlay assays revealed that MpB GroEL(122-427) did not bind PLRV, whereas MpB GroEL(122-457) and MpB GroEL(122-474) both did (Fig. 4b), although virus binding to the latter mutant was reduced by about 35%. It was therefore concluded that the amino acid(s) critical for binding PLRV is present in the region between residues 427 and 457.
FIG. 4.
Mapping of the C-terminal PLRV-binding site by virus overlay assays of deletion derivatives of MpB GroEL. (a) Schematic representation of MpB GroEL deletion mutants. The numbers in parentheses correspond to the positions of amino acid residues of MpB GroEL (17) and mark the borders of the deletion mutants. N-eq, N-terminal region of the equatorial domain; N-int, N-terminal region of the intermediate domain; Ap, apical domain; C-int, C-terminal region of the intermediate domain; C-eq, C-terminal region of the equatorial domain. Secondary structural elements are indicated by boxed sine waves (α-helices). (b) Virus overlay assay (right) showing that the putative α-helix between residues 427 and 457 is part of the PLRV-binding site. Lanes 1, wild-type MpB GroEL isolated from M. persicae; lanes 2, recombinant MpB GroEL. All other lanes contain the indicated deletion mutants of MpB GroEL as depicted in panel a. (Left) Amido black-stained blot.
Pepscan analysis of this region showed that PLRV displayed a strong affinity for the decameric peptide with the sequence VGIRVALRAM (Fig. 5a). The substitutions R441A and R445A diminished the PLRV-binding capacity of this decameric peptide by 90 and 50%, respectively (Fig. 5b). When both arginines were simultaneously replaced, no binding of PLRV was detected (Fig. 5b).
FIG. 5.
(a) Schematic representation of the virus overlay assays of decameric peptides corresponding to amino acid residues 423 to 476 of the C-terminal region of the equatorial domain of MpB GroEL. The results of the first 21 peptides are shown; no PLRV binding to any of the other peptides in this region was detected. Secondary structural elements are indicated by boxed sine waves (α-helices). Conserved sequences in GroEL/Hsp60 sequences are indicated by asterisks. (b) Alanine scanning of the decameric peptide with the strongest binding capacity as indicated in panel a (in boldface). The affinity of PLRV for the peptides has been quantified using Molecular Analyst software (histograms) and is interpreted as follows: +++, high affinity; ++, intermediate affinity; +, low affinity; −, no PLRV binding detected. Arrows indicate residues critical for binding PLRV.
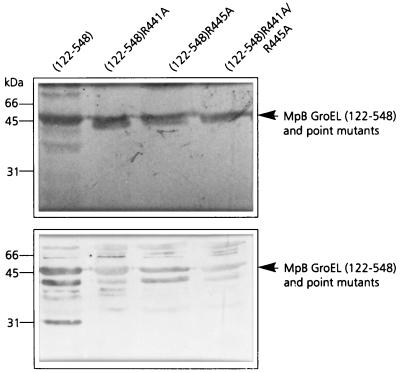
Alanine substitution of R441 and R445 in MpB GroEL(122-548) confirmed the results of the pepscan analysis, since PLRV binding by the mutants MpB GroEL(122-548)R441A and MpB GroEL(122-548)R445A was only 20% of that of the recombinant full-length GroEL product (Fig. 6). Unlike in the pepscan analysis, changing both arginine residues to alanine simultaneously (MpB GroEL(122-548)R441A/R445A) did not abolish virus binding but reduced it by approximately 80%. This suggests that the context of R441 and R445 (residues between position 427 and 457) also contributes to the PLRV-binding capacity of the N-terminally truncated GroEL mutant.
FIG. 6.
Virus overlay assay (bottom panel) of alanine replacement mutants of MpB GroEL(122-548) to identify individual amino acids involved in PLRV binding. Lanes contain wild-type MpB GroEL(122-548) and point mutants of MpB GroEL(122-548). The positions of MpB GroEL(122-548), and alanine replacement mutants of MpBGroEL(122-548) are indicated by arrowheads. (Top panel) Amido black-stained blot.
As was shown previously, PLRV also interacted with endogenous E. coli GroEL, copurified with some of the GST fusion products (17). Consequently, some lanes contain a 60-kDa polypeptide that binds PLRV (Fig. 4b and 6). Comparison of the amino acid sequences of GroEL from M. persicae and E. coli revealed that all amino acids identified in this study as implicated in virus binding are conserved except for R441, which is a lysine in GroEL of E. coli. Site-directed mutagenesis showed that the modifications R441K and R445K in MpB GroEL(122-548) did not change its virus-binding properties (data not shown).
Alanine substitutions in full-length MpB GroEL.
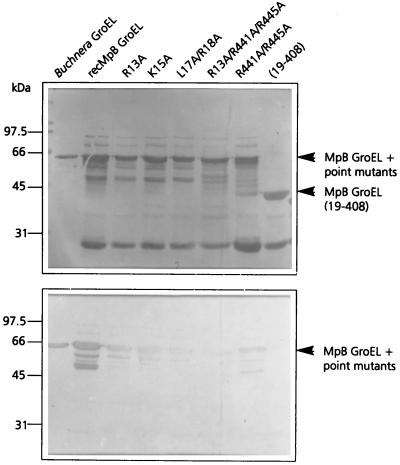
To verify whether the amino acid residues of the equatorial domain of MpB GroEL, which mediate PLRV binding to truncated GroEL polypeptides, exert similar effects in full-length GroEL, single and simultaneous alanine substitutions were made (Fig. 7). In the N-terminal equatorial domain of full-length MpB GroEL, the same set of alanine substitutions was made as in truncated GroEL (Fig. 3): R13A, K15A, and L17A R18A. All three substitutions strongly reduced the ability to bind purified PLRV, by 52% (R13A), 67% (K15A), and 72% (L17A R18A) in the virus overlay assay in comparison with that of recombinant MpB GroEL (Fig. 7). Moreover, R13, K15, and L17–R18 strongly affected each other's capacity to bind the virus, which is consistent with the results obtained using MpB GroEL deletion mutants (Fig. 3). It was also shown that alanine substitutions in the N terminus of the equatorial domain did not completely eliminate virus binding (Fig. 7), which is most likely due to PLRV binding by the unaffected C terminus. However, it should be noted that the residual binding capacity of the amino acids in the wild-type terminus differs from what one would expect if all residues contributed equally to the virus-binding capacity of the molecule. A similar observation was made for alanine substitutions in the C terminus (R441A/R445A in Fig. 7). Only when mutations were made in both termini (R13A/R441A/R445A in Fig. 7) did full-length MpB GroEL lose virtually all of its capacity to bind purified PLRV.
FIG. 7.
Virus overlay assay (bottom panel) of recombinant MpB GroEL, alanine replacement mutants of MpB GroEL, and MpB GroEL(19-408). (Top panel) Amido black-stained blot. The positions of MpB GroEL, MpB GroEL point mutants, and MpB GroEL(19-408) are indicated by arrowheads.
DISCUSSION
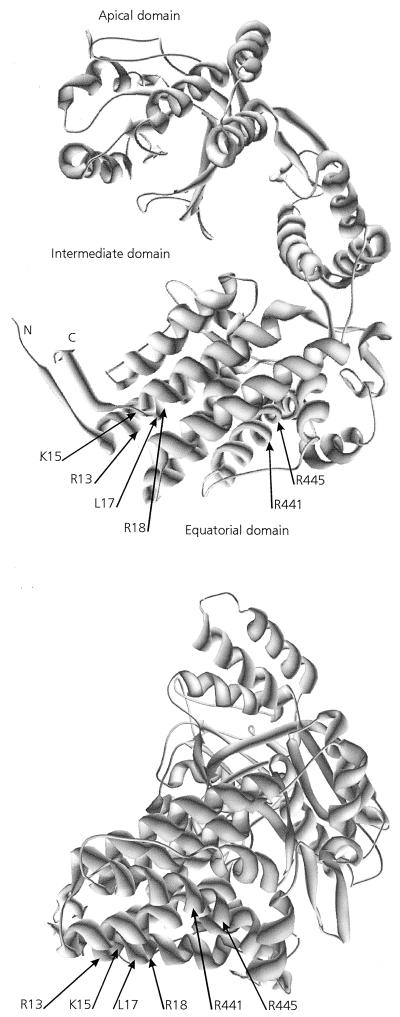
Our data reveal that only a limited number of amino acid residues markedly influence the affinity of MpB GroEL for PLRV. These residues are located between amino acid residues 9 and 19 and between amino acids 427 and 457 of the N-terminal and C-terminal regions, respectively, of the equatorial domain. This domain has previously been identified as the one harboring the PLRV binding site (17). Computer-generated structural predictions of the tertiary structure of GroEL of E. coli demonstrate that the N-terminal and C-terminal regions of the equatorial domain assemble to form the complete equatorial domain in the E. coli GroEL monomer (3). Buchnera GroEL shares >92% amino acid sequence identity, as well as structural and functional features, with E. coli GroEL (11, 17, 27). Also, the Buchnera GroEL model suggests that the N-terminal and C-terminal regions of the equatorial domain of MpB GroEL are in spatial proximity, as they assemble into a single equatorial domain (Fig. 8).
FIG. 8.
Model of a Buchnera GroEL subunit showing the positions of amino acid residues critical for PLRV binding. R13, K15, L17, and R18 belong to the N-terminal region of the equatorial domain, and R441 and R445 are in the C-terminal region of the equatorial domain. Two different rotation angles of the model are presented.
In the predicted tertiary structure, residues 9 to 19 of the N-terminal equatorial domain assemble into an α-helix (Fig. 1, 2, and 8). Virus-binding studies with decameric peptides and with full-length MpB GroEL and mutants thereof all demonstrated that amino acid residues R13, K15, L17, and R18 are critical for PLRV binding to the N-terminal equatorial domain. These residues are clustered on the hydrophilic side of the putative α-helix. The Buchnera GroEL model also predicts that residues R441 and R445 of the C-terminal PLRV-binding site of MpB GroEL (Fig. 4b) are part of a helical structure. Far-Western analysis further revealed that an additional determinant besides residues R441 and R445 is involved in composing the PLRV-binding site at the C-terminal region of the equatorial domain. This determinant may be a structural component rather than a single amino acid, since it was not identified in the pepscan analysis. It is not unlikely that replacement of R441 and R445 by the neutral alanine residues causes more changes in the physical and structural characteristics of the decameric peptide VGIRVALRAM than the alanine replacement of these residues in constructs of MpB GroEL, which may suggest that the complete α-helix between residues 431 and 459 is involved in PLRV binding. Since replacement of R441 and R445 by lysines did not change the PLRV-binding capacity of VGIRVALRAM or MpB GroEL mutants, it seems likely that the hydrophilic nature of the α-helix rather than the identities of single amino acids is important for PLRV binding. The α-helix harboring R441 and R445 is located toward the exterior of the GroEL 14-mer, whereas the α-helix containing R13, K15, L17, and R18 is located toward the cavity of the GroEL cylinder (3). It is not unusual for amino acids of the N-terminal and C-terminal parts of the equatorial domain to join so as to form a complex binding site. The ATP-binding site of E. coli GroEL is also composed of amino acids from both termini of the equatorial domain (2). Structure predictions of Buchnera GroEL show that all six residues are juxtaposed in the equatorial plain, which is potentially accessible from the outside the native molecule. However, it remains to be investigated to what extent the in vivo virus-binding activity of GroEL can be extrapolated from the in vitro binding data. Based on the fact that minor deletions in the termini of E. coli GroEL affect the formation of the 14-mers (see, e.g., references 5, 19, and 22), it is unlikely that the truncated Buchnera GroEL mutants used in this study are able to assemble into the native state. The role of the six identified residues in the folding and multimerization of Buchnera GroEL is yet to be determined.
The amino acid residues of MpB GroEL implicated in the binding of PLRV (an extracellular event) are mainly hydrophilic in nature, whereas residues involved in the binding of nonnative proteins in the cytosol of a bacterial cell are generally hydrophobic (3, 10). The involvement of hydrophilic residues in protein-protein interactions has been reported for other systems as well (7, 28). In the N-terminal part of the RTD of a luteovirus, both hydrophilic and hydrophobic regions are present. The region previously suggested to be involved in virus binding to GroEL is mainly hydrophobic and is highly conserved among luteoviruses (32). Moreover, this region, which is characterized by the conserved Ser-Tyr-Gly triplet, is enriched in Trp, Tyr, Arg, and Ile relative to the rest of the N-terminal part of the RTD. Analysis of hot spots in protein interfaces has shown that these residues are preferred over other amino acids in interactions between proteins in a heterodimer (1). It is therefore tempting to suggest that the interaction between GroEL and PLRV is of a hydrophilic-hydrophobic nature.
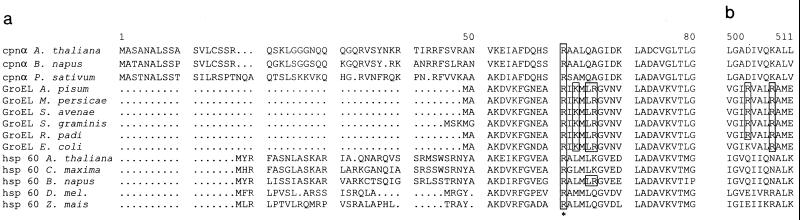
Chaperonins have been classified into two groups (14, 20). One contains chaperonins of bacterial origin (like GroEL) and of eukaryotic organelles such as the mitochondrial Hsp60 or the ribulose-1,5-biphosphate carboxylase-oxygenase binding protein from chloroplasts, all of which exhibit at least 50% sequence identity (14, 15). The second group contains chaperonins from thermophilic bacteria such as the 2-subunit TF55 from Sulfolobus shibatae or Sulfolobus solfataricus and the 9-subunit eukaryotic cytosolic TCP-1, which are 32 to 39% identical (21, 30). The two groups are weakly related but carry out similar functions, i.e., folding of proteins in the cell cytosol, and have structural similarities as well (6, 20, 23, 24, 26). The fact that PLRV binds to Buchnera GroEL from several aphid species (18) and to E. coli GroEL (17) indicates that the amino acid residues implicated in virus binding should be highly conserved among GroEL homologs. Alignment of amino acid sequences of Buchnera GroEL and E. coli GroEL indeed demonstrates that most amino acids involved in binding PLRV (R13, K15, L17, R18, and R445) are conserved between Buchnera GroEL and E. coli GroEL. The arginine at position 441 of Buchnera GroEL proteins is a lysine in the GroEL of E. coli (Fig. 9). But substitution of this residue by a lysine did not influence the PLRV-binding capacity of MpB GroEL(122-548) in virus overlay assays. Interestingly, R13 of MpB GroEL is conserved among all Hsp60 sequences (Fig. 9) and is also found in two subunits of TCP-1 (20, 21). Whether PLRV and other luteoviruses also exhibit an affinity for GroEL homologs of eukaryotic origin is yet to be determined.
FIG. 9.
Alignment of Hsp60/GroEL amino acid sequences of mitochondria, E. coli, and chloroplasts. Shown are chloroplast cpn α (subunit of chloroplast Hsp60) of Arabidopsis thaliana (accession no. P21238), Brassica napus (P34794), and Pisum sativum (P08926); mitochondrial hsp60 of A. thaliana (P29197), Cucurbita maxima (Q05046), B. napus (P35480), Drosophila melanogaster (O02649), and Zea mais (P29185); Buchnera GroEL sequences of Acyrthosiphon pisum (P25750), M. persicae (AF003957), Schizaphis graminum (Q59177), Sitobion avenae (U77379), and Rhopalosiphum padi (U77380); and GroEL of E. coli (reference 14). Sequences were aligned using the PILEUP program (Genetics Computer Group, Madison, Wis. [8]). (a) Alignment of the first 80 amino acids of the N-terminal regions of GroEL/Hsp60 equatorial domains. (b) Alignment of amino acids 500 to 511 of the C-terminal regions of GroEL/Hsp60 equatorial domains. Amino acids shown to be involved in PLRV-binding are boxed. The highly conserved R13 is indicated by an asterisk.
ACKNOWLEDGMENTS
This work was supported in part by Priority Program Crop Protection grant 45.014 from the Netherlands Organization for Scientific Research (NWO) and the Ministry of Agriculture, Nature Management, and Fisheries (LNV).
REFERENCES
- 1.Bogan A A, Thorn K S. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 2.Boisvert D C, Wang J, Otwinowski Z, Horwich A L, Sigler P. The 2.4 Å crystal structure of the bacterial chaperonin GroEL complexed with ATPγS. Nat Struct Biol. 1996;3:170–177. doi: 10.1038/nsb0296-170. [DOI] [PubMed] [Google Scholar]
- 3.Braig K, Otwinowski Z, Hegde R, Boisvert D C, Joachimiak A, Horwich A L, Sigler P. The crystal structure of the bacterial chaperonin GroEL at 2.8 Å. Nature. 1994;371:578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 4.Brault V, van den Heuvel J F J M, Verbeek M, Ziegler-Graff V, Reutenauer A, Herrbach E, Garaud J-C, Guilley H, Richards K, Jonard G. Aphid transmission of beet western yellows luteovirus requires the minor capsid read-through protein P74. EMBO J. 1995;14:650–659. doi: 10.1002/j.1460-2075.1995.tb07043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burnett B P, Horwich A L, Low K B. A carboxy-terminal deletion impairs the assembly of GroEL and confers a pleiotropic phenotype in Escherichia coli K-12. J Bacteriol. 1994;176:6980–6985. doi: 10.1128/jb.176.22.6980-6985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Creutz C E, Liou A, Snyder S L, Brownawell A, Willison K. Identification of the major chromaffin granule-binding protein, chromobindin A, as the cytosolic chaperonin CCT (chaperonin containing TCP-1) J Biol Chem. 1994;269:32035–32038. [PubMed] [Google Scholar]
- 7.Davis S J, Ikemizu S, Wild M K, van der Merwe P A. CD2 and the nature of protein interactions mediating cell-cell recognition. Immunol Rev. 1998;163:217–236. doi: 10.1111/j.1600-065x.1998.tb01199.x. [DOI] [PubMed] [Google Scholar]
- 8.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis R J, van der Vies S M. Molecular chaperones. Annu Rev Biochem. 1991;60:321–347. doi: 10.1146/annurev.bi.60.070191.001541. [DOI] [PubMed] [Google Scholar]
- 10.Fenton W A, Kashi Y, Furtak K, Horwich A L. Residues in chaperonin GroEL required for polypeptide binding and release. Nature. 1994;371:614–619. doi: 10.1038/371614a0. [DOI] [PubMed] [Google Scholar]
- 11.Filichkin S A, Brumfield S, Filichkin T P, Young M. In vitro interactions of the aphid endosymbiotic SymL chaperonin with barley yellow dwarf virus. J Virol. 1997;71:569–577. doi: 10.1128/jvi.71.1.569-577.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gildow F E. Virus-membrane interactions involved in circulative transmission of luteoviruses by aphids. Curr Top Vector Res. 1987;4:93–120. [Google Scholar]
- 13.Guex N, Peitsch M C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 14.Gupta R S. Evolution of the chaperonin families (Hsp60, Hsp10 and Tcp-1) of protein and the origin of eukaryotic cells. Mol Microbiol. 1995;15:1–11. doi: 10.1111/j.1365-2958.1995.tb02216.x. [DOI] [PubMed] [Google Scholar]
- 15.Gupta R S, Picketts D J, Ahmad S. A novel ubiquitous protein ‘chaperonin’ supports the endosymbiotic origin of mitochondrion and plant protoplasts. Biochem Biophys Res Commun. 1989;163:780–787. doi: 10.1016/0006-291x(89)92290-0. [DOI] [PubMed] [Google Scholar]
- 16.Hemmingsen S M, Woolford C, van der Vies S M, Tilly K, Dennis D T, Georgopoulos C P, Hendrix R W, Ellis R J. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature. 1988;333:330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- 17.Hogenhout S A, van der Wilk F, Verbeek M, Goldbach R W, van den Heuvel J F J M. Potato leafroll virus binds to the equatorial domain of the aphid endosymbiotic GroEL homolog. J Virol. 1998;72:358–365. doi: 10.1128/jvi.72.1.358-365.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hogenhout S A, Verbeek M, Hans F, Houterman P M, Fortass M, van der Wilk F, Huttinga H, van den Heuvel J F J M. Molecular bases of the interactions between luteoviruses and aphids. Agronomie. 1996;16:167–173. [Google Scholar]
- 19.Horowitz A, Bochkareva E S, Kovalenko O, Girshovich A S. Mutation Ala2→Ser destabilizes intersubunit interactions in the molecular chaperone GroEL. J Mol Biol. 1993;231:58–64. doi: 10.1006/jmbi.1993.1256. [DOI] [PubMed] [Google Scholar]
- 20.Kim S, Willison K R, Horwich A L. Cytosolic chaperonin subunits have a conserved ATPase domain but diverged polypeptide-binding domains. Trends Biochem Sci. 1994;19:543–548. doi: 10.1016/0968-0004(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 21.Kubota H, Morita T, Nagata T, Takemoto Y, Nozaki M, Gachelin G, Matsushiro A. Nucleotide sequence of mouse Tcp-1a cDNA. Gene. 1991;105:269–273. doi: 10.1016/0378-1119(91)90162-5. [DOI] [PubMed] [Google Scholar]
- 22.Luo G-X, Horowitz P M. The stability of the molecular chaperonin cnp60 is affected by site-directed replacement of cysteine 518. J Biol Chem. 1994;269:32151–32154. [PubMed] [Google Scholar]
- 23.Marco S, Carrascosa J L, Valpuesta J M. Reversible interaction of beta-actin along the channel of the TCP-1 cytoplasmic chaperonin. Biophys J. 1994;67:364–368. doi: 10.1016/S0006-3495(94)80489-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melki R, Cowan N J. Facilitated folding of actins and tubulins occurs via a nucleotide-dependent interaction between cytoplasmic chaperonin and distinctive folding intermediates. Mol Cell Biol. 1994;14:2895–2904. doi: 10.1128/mcb.14.5.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morin S, Ghanim M, Zeidan M, Czosnek H, Verbeek M, van den Heuvel J F J M. A GroEL homologue from endosymbiotic bacteria of the whitefly Bemisia tabaci is implicated in the circulative transmission of tomato yellow leaf curl virus. Virology. 1999;256:75–84. doi: 10.1006/viro.1999.9631. [DOI] [PubMed] [Google Scholar]
- 26.Mummert E, Grimm R, Speth V, Eckerskorn C, Schiltz E, Gatenby A A, Schäfer E. A TCP1-related molecular chaperone from plants refolds phytochrome to its photoreversible form. Nature. 1993;363:644–648. doi: 10.1038/363644a0. [DOI] [PubMed] [Google Scholar]
- 27.Ohtaka C, Nakamura H, Ishikawa H. Structures of chaperonins from an intracellular symbiont and their functional expression in Escherichia coli groE mutants. J Bacteriol. 1992;174:1869–1874. doi: 10.1128/jb.174.6.1869-1874.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh J, Garber E, van Vlijmen H, Karpusas M, Hsu Y M, Zheng Z, Naismith J H, Thomas D. The role of polar interactions in the molecular recognition of CD40L with its receptor CD40. Protein Sci. 1998;7:1124–1135. doi: 10.1002/pro.5560070506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sylvester E S. Circulative and propagative virus transmission by aphids. Annu Rev Entomol. 1980;25:257–286. [Google Scholar]
- 30.Trent J D, Nimmesgern E, Wall J S, Hartl F U, Horwich A L. A molecular chaperone from a thermophilic archaebacterium is related to the eukaryotic protein t-complex polypeptide-1. Nature. 1991;354:490–493. doi: 10.1038/354490a0. [DOI] [PubMed] [Google Scholar]
- 31.van den Heuvel J F J M, Boerma T M, Peters D. Transmission of potato leafroll virus from plants and artificial diets by Myzus persicae. Phytopathology. 1991;81:150–154. [Google Scholar]
- 32.van den Heuvel J F J M, Bruyère A, Hogenhout S A, Ziegler-Graff V, Brault V, Verbeek M, van der Wilk F, Richards K. The N-terminal region of the luteovirus readthrough domain determines virus binding to Buchnera GroEL and is essential for virus persistence in the aphid. J Virol. 1997;71:7258–7265. doi: 10.1128/jvi.71.10.7258-7265.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van den Heuvel J F J M, de Blank C M, Peters D, van Lent J W M. Localization of potato leafroll virus in leaves of secondarily-infected potato plants. Eur J Plant Pathol. 1995;101:567–571. [Google Scholar]
- 34.van den Heuvel J F J M, Verbeek M, van der Wilk F. Endosymbiotic bacteria associated with circulative transmission of potato leafroll virus by Myzus persicae. J Gen Virol. 1994;75:2559–2565. doi: 10.1099/0022-1317-75-10-2559. [DOI] [PubMed] [Google Scholar]
- 35.van der Wilk F, Huisman M J, Cornelissen B J C, Huttinga H, Goldbach R. Nucleotide sequence and organization of potato leafroll virus genomic RNA. FEBS Lett. 1989;245:51–56. doi: 10.1016/0014-5793(89)80190-5. [DOI] [PubMed] [Google Scholar]