Abstract
The Us9 gene is conserved among most alphaherpesviruses. In pseudorabies virus (PRV), the Us9 protein is a 98-amino-acid, type II membrane protein found in the virion envelope. It localizes to the trans-Golgi network (TGN) region in infected and transfected cells and is maintained in this compartment by endocytosis from the plasma membrane. Viruses with Us9 deleted have no observable defects in tissue culture yet have reduced virulence and restricted spread to retinorecipient neurons in the rodent brain. In this report, we demonstrate that Us9-promoted transneuronal spread in vivo is dependent on a conserved acidic motif previously shown to be essential for the maintenance of Us9 in the TGN region and recycling from the plasma membrane. Mutant viruses with the acidic motif deleted have an anterograde spread defect indistinguishable from that of Us9 null viruses. Transneuronal spread, however, is not dependent on a dileucine endocytosis motif in the Us9 cytoplasmic tail. Through alanine scanning mutagenesis of the acidic motif, we have identified two conserved tyrosine residues that are essential for Us9-mediated spread as well as two serine residues, comprising putative consensus casein kinase II sites, that modulate the rate of PRV transneuronal spread in vivo.
Pseudorabies virus (PRV) is a member of the neurotropic Alphaherpesvirus subfamily which also includes the human pathogens herpes simplex virus types 1 and 2 (HSV-1 and -2) and varicella-zoster virus (VZV) and the animal pathogens bovine herpesvirus type I (BHV-1), equine herpesvirus type I (EHV-1), and feline herpesvirus type I (FHV-1). PRV establishes a latent infection in its natural host, the adult swine, but causes a lethal encephalitis in young piglets and in a wide variety of mammals and birds (5, 27, 45). PRV displays a striking neurotropism for both the central and peripheral nervous systems, which it enters by primary infection of cells lining mucosal or epithelial surfaces (18, 19). Once PRV has invaded the nervous system, it is capable of spreading between chains of synaptically connected neurons in pathways consistent with known neuronal connections (22, 26, 33). The spread of PRV between neurons is confined to sites of synaptic contact and can occur in both the anterograde and retrograde directions (reviewed in references 9 and 17).
In attempts to decipher the molecular mechanism of directional spread of alphaherpesviruses in the nervous system, our laboratory has studied the spread of both wild-type and mutant PRV strains in the rodent nervous system following either intravitreal or intracerebral infection of the brain (7, 10, 12, 13, 25, 36, 38, 44). Examination of viral strains containing defined mutations in the unique short region of the viral genome has allowed for the identification of three gene products which play key roles in PRV transneuronal spread in the anterograde direction (spread in the direction of the nerve impulse) following either infection of the retina or direct infection of the brain. These genes encode the envelope membrane proteins gI, gE, and Us9 (7, 12, 44). The absence of any of these gene products, either individually or in combination, leads to a restricted spread of the virus in the rodent nervous system. For instance, following direct infection of the retina, viruses with either gI, gE, or Us9 deleted replicate efficiently in retinal ganglion cells and their target neurons that are involved in circadian rhythms but are defective in anterograde spread to second-order neurons in centers involved in visual perception and reflex movement of the eyes (7, 12, 44). However, retrograde spread of virus from the eye to the brain is not compromised by the loss of gE, gI, or Us9 (7, 12, 44). Similarly, when these mutant strains are injected directly into the prefrontal cortex of living animals, there is a notable decrease in anterograde spread of virus to the cortical projection targets while wild-type levels of retrograde spread are maintained (7, 10). In addition, viruses with gI, gE, or Us9 deleted show decreased virulence as measured by time to death and appearance of symptoms following intravitreal infection (7, 12, 44).
The Us9 protein is a phosphorylated membrane protein present in the lipid envelope of PRV particles in a tail-anchored type II membrane topology (6). In both infected and transfected cells, Us9 exhibits a steady-state residence in a cellular compartment in or near the trans-Golgi network (TGN) (8). Maintenance of Us9 in the TGN region is a dynamic process involving retrieval of molecules from the cell surface and is dependent on at least two motifs in the cytoplasmic tail (8). These motifs include an acidic cluster containing putative tyrosine and casein kinase II phosphorylation sites and a dileucine endocytosis signal. Deletion of the acidic domain relocalizes a Us9–enhanced green fluorescent protein (EGFP) fusion protein from the TGN region to the plasma membrane while playing no role in Us9 virion incorporation (8). Conversely, mutation of the dileucine endocytosis signal did not completely inhibit Us9–EGFP endocytosis but rather reduced the protein's overall rate of internalization.
Despite the conservation of the acidic cluster and the dileucine-based internalization motifs in the cytosolic tail of the Us9-homologous proteins (discussed in reference 8), the function of Us9 internalization in the viral life cycle remains to be elucidated. To address this issue, we tested the abilities of PRV strains containing defined mutations in either the acidic domain or the dileucine endocytosis motif to promote spread of the virus both in vitro and in vivo. In this study, we found that viral mutants lacking the highly conserved Us9 acidic motif required for endocytosis and TGN localization exhibited restricted infection in the rat nervous system that is indistinguishable from that of a Us9 null virus. In contrast, mutants lacking the Us9 dileucine motif required for efficient endocytosis from the plasma membrane exhibited wild-type transneuronal spread. Neither of these motifs, however, was required for efficient cell-to-cell spread in tissue culture or for targeting of Us9 to the viral envelope. Through alanine scanning mutagenesis of the acidic motif, we found that Us9-mediated transneuronal spread in vivo is dependent on two highly conserved tyrosine residues whereas the rate of spread of the virus is dependent on the phosphorylation status of two conserved serine residues.
MATERIALS AND METHODS
Virus strains and cells.
PK15 (pig kidney epithelial) and MDBK (Madin-Darby bovine kidney) cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). All PRV strains used in this study (see Fig. 1) were propagated on PK15 cells in DMEM containing 2% FBS as previously described (44). The wild-type strain PRV Becker (PRV Be), the isogenic Us9 null strains (PRV 160 and PRV 161), and the revertants (PRV 160R and PRV 161R) have been described previously (4, 7). PRV 162 is an isogenic strain of PRV Be encoding a mutant Us9 protein in which the nucleotide sequence encoding amino acids 46 to 55 has been removed (8). PRV 91 contains a deletion removing the gE open reading frame and has been described previously (12). Revertants of selected viruses have been described by Brideau et al. (7).
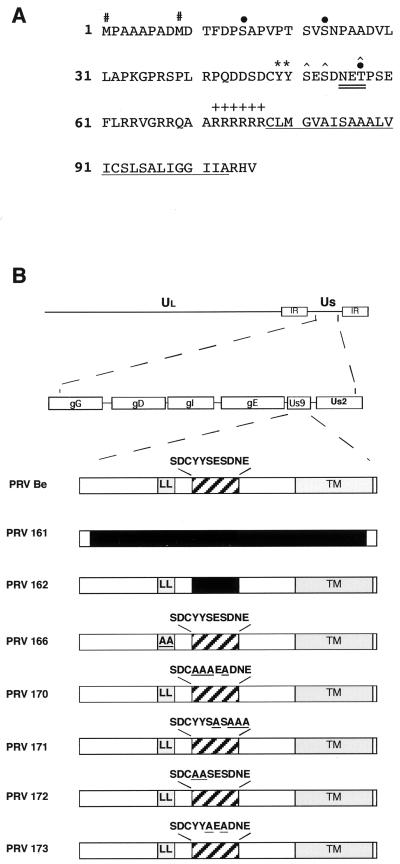
FIG. 1.
(A) Amino acid sequence of the Us9 open reading frame product. The two in-frame methionine residues (#) and the potential tyrosine kinase phosphorylation sites (∗) are indicated. The consensus casein kinase I sites [(S/T)X2–3(S/T)X] (31) each are each indicated by a solid circle above the S or T boldfaced here, and the consensus casein kinase II sites [X(S/T)XX(D/E)] are each marked with a caret above the S or T (PROSITE pattern). The potential N-linked glycosylation [NX(S/T)] sequence is double underlined. The putative transmembrane domain is underlined, and the surrounding basic residues are indicated by plus signs. (B) PRV genome and maps of viruses used in this study. The PRV genome is shown on the first line, and the unique short region (Us) is expanded on the second line. The Us9 proteins from the various viruses used in this study are diagrammed below. The dileucine endocytosis motif (LL) and the transmembrane domain (TM) are indicated by shaded boxes. The acidic domain is indicated by a hatched box with the amino acid sequence given above. All amino acid substitutions are underlined, and deletions are indicated by solid boxes. PRV Be is the wild-type strain used in this study. PRV 161 contains a 258-bp deletion in the Us9 open reading frame. PRV 162 contains a 10-amino-acid deletion removing a conserved acid motif in the Us9 cytoplasmic tail. PRV 166 contains a leucine-to-alanine substitution at amino acids 30 and 31 in the Us9 protein. PRV 170 (Y49–50A S51A S53A), PRV 171 (E52A D54A N55A E56A), PRV 172 (Y49–50A), and PRV 173 (S51A S53A) contain alanine substitutions in the conserved acidic domain in the Us9 cytoplasmic tail (see the text for details).
PK15–BB14 cells expressing a Us9–EGFP fusion protein were grown in DMEM supplemented with 10% FBS and 1 mg of G418 (Gibco/BRL) per ml (6). PK15–AB62, PK15–AB63, PK15–AB64, and PK15–AB65 cells stably express the Us9 (Y49A Y50A [Y49–50A] S51A S53A)–EGFP, Us9(E52A D54A N55A E56A)–EGFP, Us9(Y49–50A)–EGFP, and Us9(S51A S53A)–EGFP fusion proteins, respectively (Table 1). To construct these cell lines, 40% confluent dishes of PK15 cells were transfected by the calcium phosphate method (23) with 10 μg of plasmid pAB62, pAB63, pAB64, or pAB65 (see below for description of plasmids). Two days after transfection, the cells were split 1:20 and replated into selection medium containing 1 mg of G418/ml. G418-positive cells were pooled and grown to confluence.
TABLE 1.
Plasmids, cell lines, and viruses used in this study
| Us9 mutation(s) | EGFP fusion plasmid | Cell line | Virus |
|---|---|---|---|
| Wild type | pBB14 | PK15–BB14 | PRV Be |
| del 46–55 | pAB35a | PK15–AB35 | PRV 162 |
| L30–31A | pAB37a | PK15–AB37 | PRV 166 |
| Y49–50A S51A S53A | pAB62 | PK15–AB62 | PRV 170 |
| E52A D54A N55A E56A | pAB63 | PK15–AB63 | PRV 171 |
| Y49–50A | pAB64 | PK15–AB64 | PRV 172 |
| S51A S53A | pAB65 | PK15–AB65 | PRV 173 |
Plasmids pAB35 and pAB37 and the corresponding cell lines, PK15–AB35 and PK15–AB37, have been described previously (8).
Antisera.
The rabbit polyvalent gE antiserum was kindly provided by K. Bienkowska-Szewcyzk (University of Gdansk). Rb133 is a rabbit polyvalent antiserum made against acetone-inactivated PRV Be and recognizes all of the major envelope glycoproteins (12). The polyvalent goat PRV gB (Ab 284) antiserum has been described previously (32). The rabbit polyvalent antiserum directed against GFP was purchased from Clontech.
The rabbit polyvalent Us9 antiserum has been described previously (6). Mouse monoclonal antiserum 5F10 was raised against the PRV Us9 protein. A GST-Us9 fusion protein was expressed in DH5α cells and isolated as inclusion bodies essentially as described previously (6). The fusion protein was dialyzed against phosphate-buffered saline (PBS) and used to immunize BALB/c mice following standard procedures.
Construction of PRV 165 and PRV 166.
PRV 165 is a Us9 null virus containing the EGFP gene under the control of the cytomegalovirus (CMV) immediate-early (IE) promoter in the Us9 locus. The transfer plasmid for the construction of PRV 165 was made by the insertion of a cassette expressing EGFP under the control of the CMV IE promoter into the AflII site of plasmid pAB25, in which the Us9 nucleotide sequences are deleted (7). Specifically, pAB25 was linearized with AflII, treated with calf intestinal phosphatase (New England Biolabs), and ligated to an AflII-PvuII fragment containing the CMV IE-EGFP cassette (Clontech). The ligation reaction product was then treated with Klenow fragment (New England Biolabs) to fill in the remaining available AflII end, and the entire plasmid was self-ligated to create plasmid pAB29. The CMV IE-EGFP cassette in pAB29 is inserted in a 3′-to-5′ orientation and is, therefore, transcribed from the minus strand. PRV 165 was then constructed by cotransfection of plasmid pAB29 and PRV Be viral DNA. Recombinant viruses were plaque purified based on their green fluorescence under UV illumination.
PRV 166 is an isogenic strain of PRV Be encoding a mutant Us9 protein in which the leucine residues at positions 30 and 31 (L30–31) were replaced with alanine residues. To construct PRV 166, amino acids 30 and 31 of the Us9 open reading frame product were changed to alanine residues by oligonucleotide mutagenesis on plasmid pRS3 containing the SphI-MluI region of the PRV BamHI7 fragment as described previously (8). A 424-bp EagI and BbsI fragment of the mutagenized plasmid was then used to replace the corresponding fragment in plasmid pGS166, which contains a SphI fragment spanning the region from the gE tail to the middle of the Us2 gene (36). The nucleotide sequence of the resulting plasmid, pAB40, was verified by DNA sequencing with Sequenase (United States Biochemical). Subcloning the EagI/BbsI fragment out of the site-directed-mutagenesis plasmid allowed us to reduce the amount of mutagenized DNA used to make the recombinant virus. Plasmid pAB40 was then cotransfected into PK15 cells with viral DNA from PRV 165, a Us9 null virus which expresses EGFP under the control of the CMV IE promoter (see above). The cells and media were collected from the cotransfection when a complete cytopathic effect was evident. The virus stock was then replated on PK15 cells, and recombinant viruses were identified by the loss of EGFP fluorescence under UV illumination and subjected to four rounds of plaque purification.
Construction of alanine substitution mutant viruses.
PRV 170 (Y49–50A S51A S53A), PRV 171 (E52A D54A N55A E56A), PRV 172 (Y49–50A), and PRV 173 (S51A S53A) are all isogenic strains of PRV Be containing alanine substitutions in the conserved acidic motif in the Us9 cytoplasmic tail (see Fig. 1). To construct PRV 170, the nucleotide sequences encoding tyrosine residues at positions 49 and 50 (Y49–50) and serines at positions 51 and 53 (S51 and S53) in the Us9 gene were changed to sequences encoding alanine residues by oligonucleotide mutagenesis on plasmid pRS3. The oligonucleotide used to introduce the alanine substitutions also introduced a unique SacII restriction enzyme site. The PRV 170 transfer plasmid (pAB51) was then constructed, replacing the 443-bp StyI fragment of pGS166 with the corresponding StyI fragment of the mutagenized DNA. For PRV 171, an oligonucleotide was used to change the glutamic acid residues at positions 52 and 56 (E52 and E56), the aspartic acid residue at position 54 (D54), and the asparagine residue at position 55 (N55) to alanine residues. This oligonucleotide also introduced a unique SacII site in the Us9 gene. For PRV 172, oligonucleotide mutagenesis was used to replace the tyrosine residues at positions 49 and 50 with alanine residues. For PRV 173, the oligonucleotide used replaced the serine residues at positions 51 and 53 with alanine residues. The transfer plasmids for PRV 171, PRV 172, and PRV 173 were constructed by replacing the EagI/BbsI fragment of pGS166 with the corresponding fragments of the mutagenized DNAs. These final transfer vectors were named pAB53 (E52A D54A N55A E56A), pAB54 (Y49–50A), and pAB56 (S51A S53A), respectively. The correct nucleotide sequences of all transfer plasmids were verified by DNA sequencing. Plasmids pAB51, pAB53, pAB54, and pAB56 were then cotransfected with PRV 165 viral DNA, and recombinant viruses were isolated as described above for PRV 166.
Genotype verification of recombinant viruses.
Southern blot analysis was used to verify the genotypes of the viruses described above. The genotype of PRV 165 was verified by digestion of viral DNA with the restriction endonuclease SphI, as insertion of the CMV IE-EGFP cassette introduced an additional SphI site in the BamHI7 fragment of the Us region. Double digestion of PRV 165 with SphI and BamHI resulted in the cleavage of a 2,640-bp fragment into a 657- and a 1,983-bp fragment. In the case of PRV 166, substitution of the nucleotides encoding leucine residues at positions 30 and 31 in the Us9 open reading frame with nucleotides encoding alanine residues introduced a unique SacII restriction enzyme site in the Us9 gene. Digestion of the BamHI7 fragment of PRV 166 viral DNA with SacII cleaved a 1,351-bp SacII fragment into two fragments of 604 and 747 bp. Likewise, the alanine substitutions in both PRV 170 (Y49–50A S51A S53A) and PRV 171 (E52A D54A N55A E56A) created a unique SacII site in the Us9 gene. For PRV 170, digestion of viral DNA with SacII and BamHI cleaved the wild-type 1,351-bp fragment into a 799- and a 552-bp fragment. Similarly, the 1,351-bp BamHI/SacII fragment in PRV 171 is cleaved into an 814- and a 537-bp fragment. In the case of PRV 172 (Y49–50A) and PRV 173 (S51A S53A), no new restriction sites were introduced with the alanine substitutions that could easily distinguish the recombinant viruses from wild-type virus. Thus, to verify the genotypes of PRV 172 and PRV 173, the Us9 gene was PCR amplified from the corresponding viral DNAs and the nucleotide sequences of the PCR products were confirmed by DNA sequencing (Princeton University Sequencing Facility).
Plasmids.
Plasmid pBB14 encodes a Us9–EGFP fusion protein under the control of the CMV IE promoter and has been described previously (6). Plasmids pAB62, pAB63, pAB64, and pAB65 each contain a hybrid Us9 gene in which the EGFP open reading frame was fused to the carboxy terminus of Us9(Y49–50A S51A S53A), Us9(E52A D54A N55A E56A), Us9(Y49–50A), or Us9(S51A S53A), respectively (Table 1). These plasmids were created by PCR mutagenesis with pfu DNA polymerase (Stratagene) and plasmids pAB51, pAB53, pAB54, and pAB56 (see above) as templates. The forward PCR primer introduced an EcoRI site upstream of the first methionine of Us9, and the reverse PCR primer replaced the Us9 stop codon (TAG) with a codon for an arginine residue. The reverse primer also introduced a BamHI site after the mutated stop codon to allow for an in-frame fusion with EGFP. The PCR products were digested with EcoRI and BamHI and cloned into pEGFP-N1 (Clontech). DNA sequencing (Princeton University Sequencing Facility) verified the nucleotide sequences of the resulting plasmids.
Immunoprecipitations.
For steady-state analysis of viral proteins, monolayers of PRV-infected PK15 cells (multiplicity of infection [MOI], 10) were labeled with [35S]cysteine-methionine (170 μCi/ml; New England Nuclear) beginning at 5 h after infection as described previously (44). For phosphate labeling, [33P]orthophosphate (120 μCi/ml; Amersham) was added to PK15-infected cells at 5 h postinfection. After 16 h of infection, the cell lysates were prepared and immunoprecipitated with rabbit polyclonal Us9 antiserum. All immunoprecipitated samples were suspended in Laemmli sample buffer, heated to 90°C for 2.5 min, and loaded onto a sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gel.
For pulse-chase analysis, infected PK15 monolayers were incubated in cysteine- and methionine-free medium for 30 min beginning at 5.5 h postinfection. This starvation period was followed by a 7-min pulse with 125 μCi of [35S]cysteine-methionine. The label was then removed, and the cells were rinsed in PBS, followed by the addition of a nonradioactive medium containing cysteine and methionine. Cell lysates were prepared immediately after the 7-min pulse (0 min) or at 15, 30, 60, 90, and 120 min after the pulse. All samples were immunoprecipitated with Us9 polyvalent antiserum under denaturing conditions.
Isolation of virions.
Viral particles from the medium of PK15 cells infected for 15 h with PRV Be, PRV 170, PRV 171, PRV 172, or PRV 173 (MOI, 10) were isolated by centrifugation through sucrose as described previously (6). The purified virions were separated on an SDS–12.5% polyacrylamide gel, transferred to nitrocellulose, and Western blotted with a Us9 monoclonal (5F10) or gE polyclonal antiserum as described previously (6).
Plaque size determination.
Monolayers of MDBK cells in 60-mm dishes were infected with virus to yield approximately 200 plaques per dish. At 48 h after infection, the plaques were visualized by a black plaque immunoreactivity assay with gB antiserum (1:200) as described previously (7). The diameters of 48 random immunoreactive plaques were measured on an Olympus inverted microscope with an ocular reticule.
Immunofluorescence microscopy.
PK15 cells grown on glass coverslips to approximately 50% confluency were transfected with 8 μg of plasmid pBB14, pAB35, pAB62, pAB63, pAB64, or pAB65. At 48 h after transfection, the cells were rinsed with PBS, fixed with 2% paraformaldehyde, and mounted onto glass coverslips with VectaShield (Vector Laboratories). The intracellular localization of Us9–EGFP, Us9(Y49–50A S51A S53A)–EGFP, Us9(E52A D54A N55A E56A)–EGFP, Us9(Y49–50A)–EGFP, and Us9(S51A S53A)–EGFP was analyzed by fluorescence microscopy.
Antibody uptake assays.
PK15 cells stably expressing pBB14, pAB62, pAB63, pAB64, or pAB65 were grown on glass coverslips. The slips were washed once in 37°C medium prior to being transferred to a clean 35-mm dish. The coverslips were then incubated in prewarmed medium (25 μl) containing polyvalent GFP antiserum (diluted 1:75) and placed in a 37°C incubator. After 60 min of exposure to the exogenous antibodies, the cells were fixed with 2% paraformaldehyde and the localization of the GFP antiserum was detected by indirect immunofluorescence microscopy with an Alexa 568-conjugated goat immunoglobulin anti-rabbit immunoglobulin G (IgG) secondary antibody. Optical sections were taken through the center of the cell with a Nikon MRC600 confocal microscope.
Intraocular injections.
Adult male Sprague-Dawley rats weighing 210 to 290 g were used in this assay. The photoperiod was standardized to 14 h of light and 10 h of darkness (light on at 0600 h). Food and water were freely available throughout the experiments. Experimental protocols were approved by the Animal Welfare Committee at Princeton University and were consistent with the regulations of the American Association for Accreditation of Laboratory Animal Care and those in the Animal Welfare Act (Public Law 99-198). During the course of the experiment, all animals were confined to a biosafety level 2 laboratory and experiments were conducted with the specific safeguards noted by Enquist and Card (16). Animals were deeply anesthetized by intraperitoneal or intramuscular injection of ketamine and xylazine prior to all experimental manipulations.
Infection of the retina was performed as described previously (13). Briefly, 2.5 μl of virus (approximately 2 × 108 to 6 × 108 PFU/ml) was injected into the vitreous humor of the left eye of anesthetized animals using a Hamilton microliter syringe. The inoculum was injected slowly over a period of 15 min, and the needle was left in the eye for an additional 5 min in an effort to reduce leakage of virus into the orbit. The animals were sacrificed and exsanguinated when symptoms of infection were overt. The brains were then removed and sliced into 35-μm-thick coronal sections as previously described (16). The detection of PRV antigen in the brain sections by immunohistochemistry with Rb133 antisera has been described previously (13). Us9-immunoreactive polypeptides were detected in the brain sections with a Us9-specific polyvalent antiserum diluted 1:700.
RESULTS
Construction of viral mutants.
The cytoplasmic tail of PRV Us9 contains at least two motifs important for the intracellular localization and trafficking of the protein. These include a conserved acidic domain and a dileucine endocytosis signal (8). Based on a previous report (21), we also noted that the acidic motif contains, in addition to two conserved putative tyrosine phosphorylation sites, a consensus casein kinase I and a casein kinase II phosphorylation site. However, reevaluation of this region using the PROSITE program has revealed that the serine residues within the region that we have termed the acidic domain (amino acids 46 to 55) actually comprise two consensus casein kinase II sites (S51 and S53). The putative casein kinase I sites lies just downstream of the acidic domain at threonine 57 (Fig. 1A).
To determine the role of Us9 internalization in the context of viral infection, we created viruses containing defined mutations in either the acidic domain or the dileucine-based endocytosis motif. PRV 162 (del 46–55) contains a 10-amino-acid deletion in the Us9 open reading frame removing the conserved acidic cluster (Fig. 1B) and has been described previously (8). PRV 166 (L30–31A) is isogenic to PRV Be except that it expresses a Us9 protein in which the dileucine endocytosis motif has been mutated to alanine residues. The construction of PRV 166 is described in Materials and Methods.
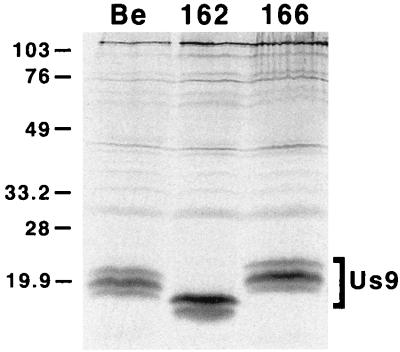
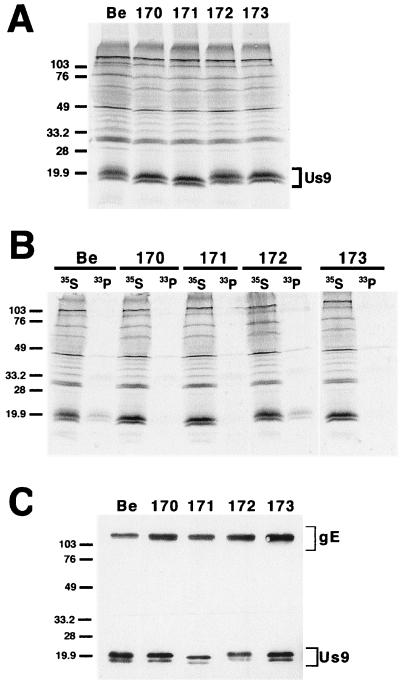
Expression of the Us9 protein in PRV 162 (del 46–55)- and PRV 166 (L30–31A)-infected cells was examined by immunoprecipitation with a Us9-specific polyvalent antiserum (Fig. 2). As reported previously (6), the Us9 antiserum precipitated three polypeptides migrating at approximately 17, 18, and 20 kDa from [35S]cysteine-methionine-labeled PRV Be-infected cells. In PRV 162-infected cells, two Us9 immunoreactive polypeptides were precipitated which migrated slightly faster than wild-type Us9 due to the deletion of amino acids 46 to 55 (8). We suggest that the loss of the slowest-migrating Us9 polypeptide in PRV 162-infected cells is due to the removal of the putative tyrosine and casein kinase II phosphorylation sites present in this region. Like PRV Be, PRV 166 expressed three Us9-immunoreactive polypeptides with the middle form being the most predominant. The amino acid substitutions in PRV 166 led to a slight decrease in the electrophoretic mobility of the Us9 protein. Additional experiments showed that PRV Be, PRV 162, and PRV 166 all produce identical species of the gE, gI, and gC envelope proteins as determined by Western blot analysis (data not shown). Moreover, the mutations in the Us9 cytosolic tail in PRV 162 and PRV 166 had no effect on the incorporation of gE or gC, or of Us9 itself, into the virion envelope (data not shown).
FIG. 2.
Expression of Us9 proteins. PK15 cells were infected with either PRV Be (wild type), PRV 162 (del 46–55), or PRV 166 (L30–31A) at an MOI of 10. The infected monolayers were radiolabeled for 11 h beginning at 5 h postinfection. At 16 h postinfection, cellular lysates were prepared and immunoprecipitated with Us9 polyvalent antiserum. All immunoprecipitated products were analyzed by electrophoresis on an SDS–12.5% polyacrylamide gel followed by autoradiography. Positions of molecular mass markers are indicated on the left in kilodaltons.
Plaque sizes on MDBK cells.
Viruses containing a mutation in the tyrosine-based endocytosis motif in the gE cytoplasmic tail form slightly smaller plaques than those formed by wild-type PRV Be (36), thereby suggesting a role for gE endocytosis in the spread of virus between adjacent cells in tissue culture. In contrast, our results indicated that Us9 internalization plays no role in PRV cell-to-cell spread in monolayers of MDBK cells in tissue culture (Table 2). The plaques formed from viruses that had the acidic motif deleted (PRV 162) or contained alanine substitutions in the dileucine endocytosis signal (PRV 166) were identical to plaques formed by PRV Be. These results are consistent with the observation that Us9 null viruses form wild-type-sized plaques in this assay (7). As reported previously, the diameters of plaques made by a gE null virus (PRV 91) were approximately 70% of the diameters of wild-type plaques (36).
TABLE 2.
Plaque sizes on MDBK cells
| Virus | Size (mm)a | SD (mm) | % of wild type | Pb |
|---|---|---|---|---|
| PRV Be | 0.78 | 0.11 | 100 | |
| PRV 91 | 0.55 | 0.06 | 70 | <0.005 |
| PRV 162 | 0.79 | 0.11 | 101 | 0.947 |
| PRV 166 | 0.81 | 0.09 | 103 | 0.169 |
An average of 48 plaques were measured 48 h after infection.
P with respect to PRV Be was determined by Student's t test.
Pulse-chase analysis of Us9 protein processing and stability.
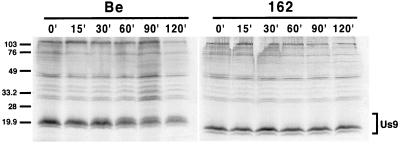
Pulse-chase analysis of Us9 was performed on PRV 162 (del 46–55)-infected cells to determine if the 10-amino-acid deletion in this virus affected the overall stability and processing of Us9. In this experiment, PRV Be- or PRV 162-infected PK15 cells were pulsed at 6 h postinfection with [35S]methionine-cysteine for 7 min, rinsed with PBS, and chased with cold medium for the times indicated prior to immunoprecipitation with Us9 polyvalent antiserum. The results in Fig. 3 show that immediately following (0 min) and up to 15 min after the 7-min pulse, only two forms of Us9, the 17- and 18-kDa polypeptides, were detected in PRV Be-infected cells. The slowest-migrating Us9 form, the 20-kDa polypeptide, did not appear until approximately 30 min of chase, with maximum conversion of Us9 to this form around 60 min after the pulse. All three Us9 polypeptides remained present up to 120 min after the chase. Pulse-chase analysis of PRV 162-infected cells revealed that, like wild-type Us9, the PRV 162-expressed Us9 protein was initially detected as two polypeptides. However, in contrast to wild-type Us9, the Us9 protein expressed from PRV 162 was not converted to the slowest-migrating Us9 form even after 120 min of pulse. This is consistent with the observation that only two Us9 immunoreactive proteins were precipitated from PRV 162-infected cells under steady-state conditions (Fig. 2) (8). These results also show that the stability of PRV 162-expressed Us9 was not compromised by the 10-amino-acid deletion, as both PRV 162 Us9 polypeptides can be detected up to 120 min postchase without any obvious decrease in signal intensity. Immunoprecipitations of both the PRV Be- and PRV 162-infected lysates for the envelope protein gC controlled for the extent of infection and total protein loaded per lane (data not shown).
FIG. 3.
Pulse-chase analysis of Us9. PK15 cells were infected with either PRV Be (wild type) or PRV 162 (del 46–55) at an MOI of 10. The cells were pulse-labeled with 125 μCi of [35S]methionine-cysteine for 7 min, rinsed with PBS, and chased for the times indicated. Cellular lysates were immunoprecipitated with a Us9 polyvalent antiserum and fractionated on a 12.5% polyacrylamide gel. Molecular mass markers (kilodaltons) are indicated on the left.
Role of Us9 intracellular trafficking in transneuronal spread.
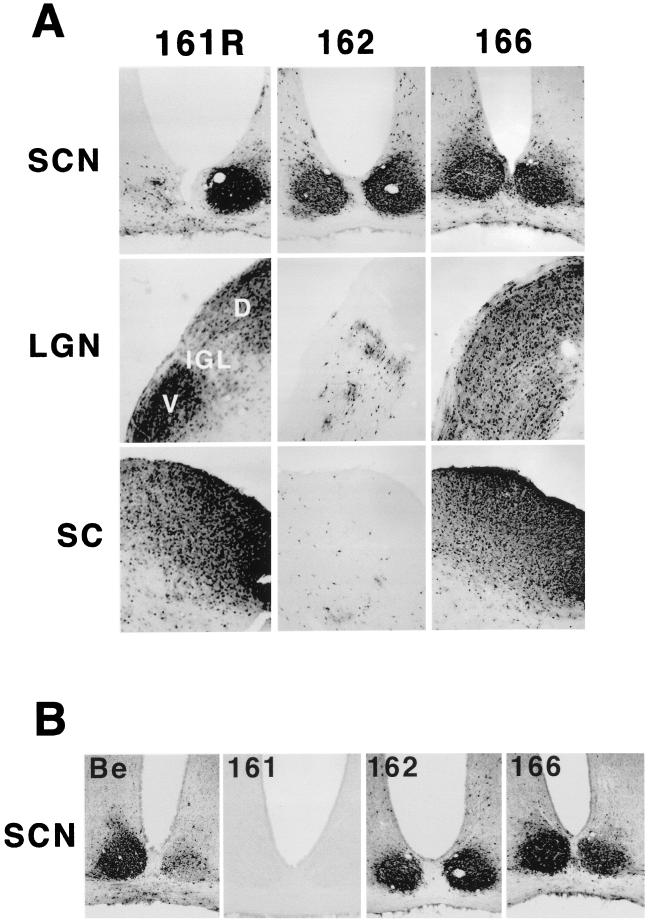
Intraocular injection of PRV Be into the vitreous humor of the rat eye leads to infection of the retinal ganglion cells lining the retina and subsequent anterograde transsynaptic spread of virus to all regions of the brain that receive retinal input (11, 13). When the wild-type strain PRV Be is examined in this system, viral antigen is found in the visual centers such as the superior colliculus (SC) and the lateral geniculate nuclei (LGN; dorsal and ventral aspects). Viral antigen evident of a robust infection is also detected in the circadian-rhythm centers, which include the suprachiasmatic nucleus (SCN) and the intergeniculate leaflet (IGL). Spread of the virus, however, to both the dorsal aspect of the lateral geniculate complex (dorsal geniculate nuclei [DGN]) and the SC is dependent on the envelope proteins gE, gI, and Us9 (7, 12, 44). Deletion of any of these envelope proteins from the wild-type strain PRV Be leads to a failure of the virus to spread to the DGN and the SC, while wild-type levels of spread to the SCN and IGL are maintained.
We tested the ability of viruses defective in Us9 internalization to undergo anterograde spread in the rat nervous system. As detailed in Materials and Methods, we injected approximately 1 × 106 PFU of PRV 162 (del 46–55) and 6.8 × 105 PFU of PRV 166 (L30–31A) into the vitreous humor of a rodent eye. After survival times of up to 80 h for PRV 162 and up to 75 h for PRV 166, the animals were sacrificed, the brains were sliced into 35-μm-thick coronal sections, and the localization of viral antigen was detected with an immunohistochemistry reaction using antiserum directed against total PRV particles (Rb133). When the brains of animals infected with PRV 161R (revertant of the Us9 null virus PRV 161) were analyzed, viral antigen was detected in all retinorecipient areas: the SCN, the SC, and all aspects of the LGN (Fig. 4A). The pattern of infection in the brains of animals infected with PRV 166 (L30–31A) was identical to that of PRV 161R, with viral antigen detected in all areas examined. In contrast, PRV 162 (del 46–55) displayed a restricted pattern of anterograde spread in which viral antigen was detected in both the SCN and IGL but was absent from the DGN and the SC. Retrograde spread, however, was not affected by the loss of the Us9 acidic domain or by mutagenesis of the dileucine endocytosis motif (data not shown).
FIG. 4.
Anterograde spread of Us9 trafficking mutants in the rodent visual system. Approximately 1 × 106 PFU of PRV 162 (del 46–55) and 6.8 × 105 PFU of PRV 166 (L30–31A) was injected into the vitreous humor of Sprague-Dawley male rats. (A) Thirty-five-micrometer-thick coronal sections of the infected brains were examined for total PRV antigen with polyvalent antiserum Rb133, which recognizes all of the major envelope glycoproteins. (B) SCNs of PRV-infected animals stained for the Us9 protein with a polyvalent Us9-specific antiserum. Representative sections are shown for each virus. The data for the wild-type strain PRV Be, the Us9 null virus PRV 161, and the revertant virus PRV 161R have been reported previously (7) and are included here only for comparison. Due to sectioning of the PRV 161R-infected brain at an oblique angle, the image in this figure shows viral antigen in only one SCN. D, dorsal, V, ventral.
As the anterograde spread defect of PRV 162 is identical to that of a Us9 null virus (7), we wanted to ensure that the Us9 protein was actually expressed in the brains of animals infected with this virus. To this end, we examined the brain tissue of PRV 162-infected rats for the Us9 protein by immunohistochemistry with Us9 polyvalent antiserum (Fig. 4B). PRV Be (wild type)-, PRV 161 (Us9 null)-, and PRV 166 (L30–31A)-infected tissues were also stained with the Us9 antiserum as controls. It can be clearly seen in Fig. 4B that the SCNs of PRV 162 (del 46–55)-infected animals contained amounts of Us9 antigen equivalent to those in PRV Be- and PRV 166 (L30–31A)-infected animals. This experiment also showed that Us9 immunoreactive polypeptides were present in the IGL of PRV 162-infected animals but not in either the DGN or SC (data not shown). Furthermore, as anticipated, no Us9 immunoreactive proteins were detected in the brains of animals infected with the Us9 null virus PRV 161.
Alanine scanning mutagenesis of the acidic domain.
The data presented in Fig. 4 indicate the importance of the conserved 10-amino-acid acidic domain in Us9-promoted anterograde spread in the rat nervous system. Not only is this region highly acidic in nature, but it also contains potential tyrosine and casein kinase II consensus phosphorylation sites. To determine which residues in the conserved acidic domain are important in promoting Us9-mediated anterograde spread in vivo, we performed alanine scanning mutagenesis of amino acids 46 to 56 in the Us9 open reading frame. Rather than mutating all the amino acids in this region individually to alanine residues, we specifically targeted the phosphorylation motifs. In PRV 170, we mutated both the putative tyrosine and casein kinase II phosphorylation sites by replacing tyrosine residues 49 and 50 and serine residues 51 and 53, respectively, with alanine residues (see Fig. 1). Based on the predicted consensus sequence for casein kinase I (31), mutation of S53 may also affect casein kinase I phosphorylation of the threonine residue at position 57. In PRV 171, the acidic residues in this region, which are necessary components of the casein kinase I and II recognition sequences, were mutated by alanine substitution of glutamic acid residues 52 and 56 (E52 and E56) and aspartic acid residue 54 (D54). The asparagine residue at position 55 was also changed to an alanine residue in this virus. Strain PRV 172 contains alanine substitutions in the potential tyrosine phosphorylation sites at positions 49 and 50. Lastly, PRV 173 contains alanine substitutions in the putative casein kinase II phosphorylation sites at serines 51 and 53. As noted above, these mutations may also affect the phosphorylation of T57 by casein kinase I. All the alanine-scanning-mutagenesis mutant viruses were made by oligonucleotide mutagenesis as described in Materials and Methods.
After confirmation of the genotypes of these viruses by both Southern blot analysis and PCR, we examined the steady-state expression levels of the Us9 protein expressed by these viral mutants by immunoprecipitation (Fig. 5A). Although all the viruses examined produced approximately equivalent amounts of the Us9 protein, the number of Us9 polypeptides varied among the viruses. For instance, three Us9 immunoreactive polypeptides were precipitated from both [35S]cysteine-methionine-labeled PRV Be- and PRV 172 (Y49–50A)-infected PK15 cells. In contrast, only two Us9 polypeptides were precipitated from PRV 170 (Y49–50A S51A S53A)-, PRV 171 (E52A D54A N55A E56A)-, and PRV 173 (S51A S53A)-infected cells. This is similar to results observed for PRV 162 (del 46–55) (see Fig. 2). The Us9 amino acid substitutions in the mutant viruses also had varying effects on the electrophoretic mobilities of the Us9 polypeptides, with the replacement of the acidic residues in PRV 171 having the most obvious effect. Western blot analysis on infected cell lysates revealed that PRV Be, PRV 170, PRV 171, PRV 172, and PRV 173 all produced identical species of both the gE and gI proteins (data not shown).
FIG. 5.
Analysis of the alanine scanning Us9 proteins. (A) Steady-state expression of alanine scanning Us9 proteins. PK15 cells infected with PRV Be (wild type), PRV 170 (Y49–50A S51A S53A), PRV 171 (E52A D54A N55A E56A), PRV 172 (Y49–50A), or PRV 173 (S51A S53A) were radiolabeled overnight with [35S]methionine-cysteine. Cellular extracts were prepared at 16 h postinfection and immunoprecipitated with Us9 polyvalent antiserum. (B) Analysis of Us9 phosphoforms. PRV Be (wild type)-, PRV 170 (Y49–50A S51A S53A)-, PRV 171 (E52A D54A N55A E56A)-, PRV 172 (Y49–50A)-, and PRV 173 (S51A S53A)-infected cells were radiolabeled overnight in the presence of either [35S]methionine-cysteine or [33P]orthophosphate. Cellular extracts were prepared at 16 h postinfection and subjected to immunoprecipitation with Us9 polyvalent antiserum. All of the immunoprecipitated products were analyzed by electrophoresis on an SDS–12.5% polyacrylamide gel followed by autoradiography (exposure time, 1.5 days). (C) Incorporation of Us9 into viral particles. Monolayers of PK15 cells were infected at an MOI of 10 with either PRV Be (wild type), PRV 170 (Y49–50A S51A S53A), PRV 171 (E52A D54A N55A E56A), PRV 172 (Y49–50A), or PRV 173 (S51A S53A) for 15 h. Cellular extracts were prepared, and virions were isolated from the medium by centrifugation through a 30% sucrose cushion. The purified virion extracts were fractionated on an SDS–12.5% polyacrylamide gel and analyzed by Western blotting with gE polyvalent and Us9 monoclonal (5F10) antisera. Molecular mass markers (kilodaltons) are indicated on the left in all three panels.
Analysis of Us9 phosphoforms.
To examine the phosphorylation status of the Us9 proteins expressed by the alanine scanning mutant viruses, infected PK15 cells were labeled overnight with [33P]orthophosphate prior to immunoprecipitation with Us9 antiserum (Fig. 5B). As a control, Us9 polypeptides were immunoprecipitated from [35S]cysteine-methionine-labeled infected cells in parallel. As noted previously (6), the wild-type Us9 protein was present in PRV Be-infected cells as at least three phosphoforms when labeled with [33P]orthophosphate (Fig. 5B). Likewise, the Us9 protein produced by PRV 172-infected cells was present as multiple phosphoforms despite mutagenesis of the putative tyrosine phosphorylation sites, thereby suggesting that the tyrosine residues do not serve as major sites of Us9 phosphorylation. The low level of 33P incorporated into the Us9 protein in both PRV Be- and PRV 172-infected cells may indicate that only a subset of the various Us9 polypeptides are modified by the addition of phosphate. In contrast, either no Us9 phosphorylated polypeptides or very small amounts were precipitated from [33P]orthophosphate-labeled PK15 cells infected with PRV 170 (Y49–50A S51A S53A), PRV 171 (E52A D54A N55A E56A), or PRV 173 (S51A S53A). These results indicate that, in contrast to the putative tyrosine phosphorylation sites, the consensus casein kinase II sites at positions S51 and S53 are major sites of Us9 phosphorylation. This is consistent with the observation that recombinant Us9 is efficiently phosphorylated by purified casein kinase II in vitro (C. Hengartner and L. W. Enquist, unpublished data). Increased exposure of the gel to film (9 days) revealed that the Us9 polypeptides expressed by PRV 170, PRV 171, and PRV 173 were in fact modified by some degree of phosphorylation, although at a much lower level than the Us9 protein expressed from either PRV Be or PRV 172. This is consistent with what we observed previously with PRV 162 (del 46–55)-produced Us9 (8). Immunoprecipitations of the same lysates for the envelope protein gB indicated that all lysates contained equivalent amounts of total protein and incorporated label (data not shown).
Virion incorporation of alanine mutants.
We next determined if the Us9 molecules produced by PRV 170, PRV 171, PRV 172, and PRV 173 were incorporated into viral particles. As shown in Fig. 5C, the Us9 proteins produced by the alanine scanning mutant viruses were incorporated into viral particles as efficiently as wild-type Us9. In all viruses examined, only two Us9 polypeptides were found in purified virions. This is consistent with what we observed previously for PRV Be (6). Overall, these results suggest that neither the conserved tyrosine residues nor the consensus casein kinase II phosphorylation sites are needed to target Us9 to the virion envelope. Western blot analysis with gE antiserum on the purified virion extracts showed that the Us9 mutations had no effect on the targeting of gE to the virion envelope (Fig. 5C). The absence of the immature form of gE (93 kDa) indicated that the virions are free of intracellular membrane contamination.
Intracellular localization and internalization of mutant Us9 constructs.
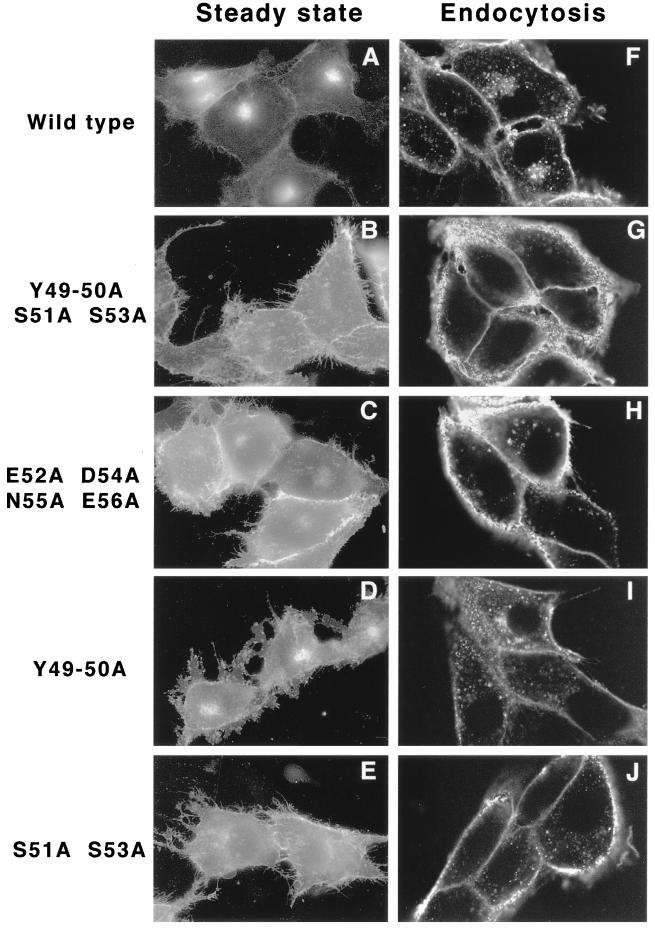
The phosphorylation status of the acidic internalization motif has been shown to be important for the intracellular trafficking of both the endoprotease furin (15, 34, 35, 44) and the human CMV (hCMV) gB membrane protein (20). Consequently, we were interested in determining what role, if any, phosphorylation of Us9's acidic motif plays in the localization of Us9 in the cell. In this same experiment, we also tested the significance of the conserved tyrosine residues in Us9 steady-state localization. Plasmids encoding Us9(Y49–50A S51A S53A)–EGFP, Us9(E52A D54A N55A E56A)–EGFP, Us9(Y49–50A)–EGFP, and Us9(S51A S53A)–EGFP gene fusions were transfected into PK15 cells (see Table 1). Fusion of the GFP to the carboxy terminus of the alanine scanning Us9 proteins not only aided in the visualization of the various proteins in transfected cells but was also necessary for subsequent endocytosis assays (see below). The localization of the mutant Us9–EGFP fusion proteins was compared to that of wild-type Us9–EGFP after 48 h of transfection. The results of these experiments are shown in Fig. 6A through E. Wild-type Us9–EGFP was found predominantly in the TGN region, with a few molecules in cytoplasmic vesicles and on the plasma membrane (Fig. 6A). In cells transfected with plasmids encoding Us9(Y49–50A S51A S53A)–EGFP (Fig. 6B), Us9(E52A D54A N55A E56A)–EGFP (Fig. 6C), Us9(Y49–50A)–EGFP (Fig. 6D), or Us9(S51A S53A)–EGFP (Fig. 6E), GFP fluorescence was detected both on the plasma membrane and in the TGN region. However, the plasma membranes in cells transfected with the Us9–EGFP alanine mutant constructs appeared to stain more intensely than those in cells transfected with wild-type Us9–EGFP (Fig. 6; compare panels B, C, D, and E with panel A). This is especially evident when one examines the amount of GFP fluorescence found in the cellular extensions. Although the relative levels of Us9–EGFP molecules retained in the TGN regions of cells transfected with the various alanine mutants have not been quantitatively analyzed, immunofluorescence analysis suggests that mutation of tyrosine residues 49 and 50 had the least effect on Us9–EGFP localization to the TGN (Fig. 6D). Overall, the results presented in Fig. 6A through E indicate that both the conserved serine residues and the conserved tyrosine residues, although to a lesser extent, are important in the localization of Us9–EGFP to the TGN region.
FIG. 6.
(A through E) Transient transfection of Us9 alanine scanning mutant constructs. PK15 cells grown on glass coverslips were transfected by the calcium phosphate method with plasmids encoding Us9–EGFP (A), Us9(Y49–50A S51A S53A)–EGFP (B), Us9(E52A D54A N55A E56A)–EGFP (C), Us9(Y49–50A)–EGFP (D), and Us9(S51A S53A)–EGFP (E). At 48 h posttransfection, the localization of the various Us9–EGFP fusion proteins was detected by fluorescence microscopy. (F through J) Internalization of alanine scanning Us9 mutant proteins from the cell surface. PK15 cells stably expressing wild-type Us9–EGFP (F), Us9(Y49–50A S51A S53A)–EGFP (G), Us9(E52A D54A N55A E56A)–EGFP (H), Us9(Y49–50A)–EGFP (I), and Us9(S51A S53A)–EGFP (J) fusion proteins were plated on glass coverslips. The coverslips were incubated at 37°C in medium containing a 1:75 dilution of polyclonal GFP-specific antiserum. After 1 h, the cells were fixed and permeabilized, and the localization of the GFP-specific antibodies was detected by indirect immunofluorescence followed by confocal microscopy. Only the red fluorescence of the Alexa 568-conjugated goat anti-rabbit IgG secondary antibody is shown.
We next performed antibody uptake experiments to test if the increased plasma membrane staining of the alanine mutants was due to a defect in internalization. In these experiments, we examined the ability of Us9–EGFP molecules (EGFP fused to the carboxy terminus of Us9) to bind and to internalize exogenously added GFP-specific antibodies. Specifically, these experiments were performed on PK15 cells stably expressing the Us9–EGFP, Us9(Y49–50A S51A S53A)–EGFP, Us9(E52A D54A N55A E56A)–EGFP, Us9(Y49–50A)–EGFP, and Us9(S51A S53A)–EGFP fusion proteins. When Us9–EGFP-expressing cells were allowed to internalize the GFP-specific antiserum for 1 h, the GFP antibodies were detected both in a perinuclear compartment and in cytoplasmic vesicles (Fig. 6F). Staining was also detected on the plasma membranes of these cells. As a control, no staining was detected when the cells were incubated with exogenous Us9-specific antiserum (data not shown). In contrast, when PK15 cells expressing Us9(Y49–50A S51A S53A)–EGFP were incubated with exogenous GFP antibodies, the plasma membrane stained brightly and very few to no Us9 molecules could be detected in the interior of the cell (Fig. 6G). Moreover, when the antibody uptake assay was performed on cells expressing either Us9(E52A D54A N55A E56A)–EGFP (Fig. 6H) or Us9(S51A S53A)–EGFP (Fig. 6J), some vesicles containing the fusion proteins could be observed in the interior of the cell. Whereas some of the internalized Us9(E52A D54A N55A E56A)–EGFP molecules appeared to cluster near the nucleus, Us9(S51A S53A)–EGFP internalized molecules seemed scattered throughout the cytoplasm (compare Fig. 6H and J). In contrast to observations with the other Us9–EGFP alanine mutant fusion proteins, a considerable amount of the polyclonal GFP antibodies was internalized when the assay was performed on PK15 cells stably expressing Us9(Y49–50)–EGFP (Fig. 6I). Like internalized wild-type Us9–EGFP molecules (Fig. 6F), internalized Us9(Y49–50A)–EGFP molecules accumulated in a perinuclear staining pattern. However, the accumulation of internalized Us9(Y49–50A)–EGFP molecules in the TGN region was not as common or as prevalent as that observed for wild-type Us9–EGFP. In all, these experiments suggest that all of the alanine scanning Us9–EGFP fusion proteins are defective, although to varying extents, in internalizing and recycling Us9–EGFP molecules back to the TGN region. These results also indicate the importance of the phosphorylation status of the Us9 cytoplasmic tail in Us9 endocytosis and TGN localization.
Spread of the Us9 alanine scanning viral mutants in vivo.
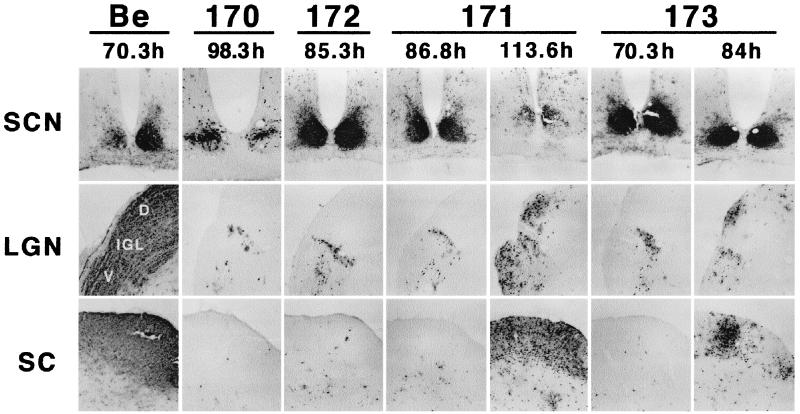
We next determined the abilities of the various mutant viruses containing alanine substitutions in the conserved acidic motif to spread in the anterograde direction following intraocular infection. These experiments were performed essentially as described above for PRV 162 (del 46–55) and PRV 166 (L30–31A) (see Fig. 4). As shown in Fig. 7, both PRV 170 (Y49–50A S51A S53A) and PRV 172 (Y49–50A) displayed a restricted-spread phenotype, even with survival times of up to 98 and 87 h, respectively (Table 3). The brains of these animals had large amounts of viral antigen in both the SCN and the IGL, whereas the SC and the DGN were essentially void of antigen. The restricted-spread phenotype of PRV 170 and PRV 172 is identical to that of either a Us9 null virus (7) or a virus with the entire acidic motif deleted (PRV 162 [Fig. 4]). In contrast, the extent of viral spread in the brains of animals infected with either PRV 171 (E52A D54A N55A E56A) or PRV 173 (S51A S53A) was dependent on how long the animal survived infection (Fig. 7; Table 3). For instance, an animal that survived 87 h of infection with PRV 171 had a restricted-spread pattern, whereas in an animal that survived 113 h of infection, viral antigen was detected in all areas receiving retinal input. Similarly, viral antigen was detected in the SC and DGN of a PRV 173-infected animal that lived up to 84 h postinfection but not in an animal that died after 74 h of infection. It should be noted, however, that even though viral antigen was detected in the DGN and SC of PRV 171- and PRV 173-infected animals with long survival times, the amounts of antigen in these areas were not nearly as great as those seen in PRV Be-infected brains. Furthermore, all the Us9 alanine scanning mutant viruses showed reduced virulence as measured by time to death and severity of symptoms, and this did not correlate with the extent of anterograde spread in the brain (see Table 3).
FIG. 7.
Localization of Us9 alanine scanning mutant viruses in the brain. Male Sprague-Dawley rats were subjected to intravitreous injection with approximately 5 × 105 PFU of PRV 170 (Y49–50A S51A S53A), PRV 171 (E52A D54A N55A E56A), PRV 172 (Y49–50A), or PRV 173 (S51A S53A). The localization of viral antigen in 35-μm-thick coronal sections was detected with an antigen against whole-virus particles (Rb133). Each vertical set of three panels shows sections from one animal, and the time to which the animal survived after infection is indicated. Representative samples are shown for each virus. Images of brain tissues from animals infected with PRV Be (1.2 × 106 PFU) are included for comparison (7).
TABLE 3.
Summary of intravitreal injections of Us9 alanine scanning mutant viruses
| Animal no. | Virus | Genotype | Time to death (h) | Spreada | Symptomb | Plaque sizec | Endocytosisd |
|---|---|---|---|---|---|---|---|
| 99-AB17 | PRV Bee | Wild type | 70.3 | +++ | Wild type | +++ | +++ |
| 99-AB18 | PRV Be | Wild type | 63.6 | +++ | Wild type | +++ | +++ |
| 99-AB10 | PRV 162 | del 46–55 | 79.5 | − | Attenuated | +++ | −f |
| 99-AB11 | PRV 162 | del 46–55 | 75.8 | − | Attenuated | +++ | − |
| 99-AB12 | PRV 162 | del 46–55 | 73.0 | − | Attenuated | +++ | − |
| 99-AB15 | PRV 166 | L30–31A | 74.8 | +++ | Wild type | +++ | ++f |
| 99-AB16 | PRV 166 | L30–31A | 64.4 | +++ | Wild type | +++ | ++ |
| 99-AB25 | PRV 170 | Y49–50A S51A S53A | 86.6 | − | Attenuated | +++g | − |
| 99-AB26 | PRV 170 | Y49–50A S51A S53A | 68.7 | − | Attenuated | +++ | − |
| 99-AB33 | PRV 170 | Y49–50A S51A S53A | 98.3 | − | Attenuated | +++ | − |
| 99-AB34 | PRV 170 | Y49–50A S51A S53A | 96.8 | − | Attenuated | ||
| 99-AB27 | PRV 171 | E52A D54A N55A E56A | 113.6 | ++ | Attenuated | +++g | + |
| 99-AB28 | PRV 171 | E52A D54A N55A E56A | 86.8 | − | Attenuated | +++ | + |
| 99-AB35 | PRV 171 | E52A D54A N55A E56A | 77.9 | − | Attenuated | +++ | + |
| 99-AB36 | PRV 171 | E52A D54A N55A E56A | 68.2 | − | Attenuated | +++ | + |
| 99-AB29 | PRV 172 | Y49–50A | 85.3 | − | Attenuated | +++g | ++ |
| 99-AB30 | PRV 172 | Y49–50A | 73.5 | − | Attenuated | +++ | ++ |
| 99-AB37 | PRV 172 | Y49–50A | 87.3 | − | Attenuated | +++ | ++ |
| 99-AB38 | PRV 172 | Y49–50A | 78.3 | − | Attenuated | ||
| 99-AB31 | PRV 173 | S51A S53A | 70.3 | − | Attenuated | +++g | + |
| 99-AB32 | PRV 173 | S51A S53A | 84.0 | + | Attenuated | +++ | + |
| 99-AB39 | PRV 173 | S51A S53A | 71.5 | − | Attenuated | +++ | + |
| 99-AB40 | PRV 173 | S51A S53A | 74.0 | − | Attenuated | +++ | + |
Relative amount of PRV viral antigen found in the SC and DGN compared to that found after PRV Be infection. +++, amount found after PRV Be infection.
Wild-type versus attenuated symptoms have been described previously (7).
Relative sizes of plaques measured 48 h after infection compared to sizes of plaques formed by PRV Be (+++).
Endocytosis was examined by antibody uptake assays and indirect immunofluorescence internalization assays (data not shown) on PK15 cells stably expressing the various mutant Us9-EGFP fusion proteins. The relative amount of the mutant Us9–EGFP fusion proteins internalized is compared to that of wild-type Us9–EGFP (+++).
The data for animals infected with PRV Be have been reported previously and are included for comparison (7).
The endocytosis phenotypes for PRV 162 and PRV 166 have been described previously (8).
Data not shown.
DISCUSSION
The cytoplasmic tails of several herpesvirus envelope proteins contain motifs that are involved in endocytosis of the proteins from the cell surface in both the infected and the transfected cell. For instance, functional internalization motifs have been identified in the cytoplasmic tails of the PRV gE protein (36, 37), the VZV gE and gI proteins (1, 2, 28, 29, 46), the HSV-1 gE protein (3), and the HCMV gB protein (20, 39, 40). Moreover, we have demonstrated that the 68-amino-acid Us9 cytoplasmic tail contains two functional endocytosis motifs: a conserved acidic cluster containing putative phosphorylation sites and a dileucine endocytosis motif (8). Despite these observations, the functional significance of endocytosis in the herpesvirus life cycle remains unknown. Tirabassi and Enquist have recently shown that internalization of the envelope protein gE during viral infection is not necessary for insertion of gE into the viral envelope, for transneuronal anterograde spread of virus in the rat brain, or for PRV pathogenesis in vivo (36, 37). In fact, the only phenotype associated with gE endocytosis mutants thus far is a slight defect in cell-to-cell spread in tissue culture. In this report, we present experiments designed to test the role of Us9 intracellular trafficking and endocytosis in the viral life cycle. Specifically, we tested the abilities of viruses containing defined mutations in the Us9 internalization domains to be targeted to the virion envelope, to form plaques in tissue culture, and to promote virulence and spread of the virus in the rodent central nervous system following direct infection of the retina.
Analysis of Us9 internalization viral mutants in tissue culture.
We have previously identified two motifs in the Us9 cytoplasmic tail that are important in the steady-state intracellular localization and endocytosis of the protein. These include a 10-amino-acid acidic motif containing both putative tyrosine and casein kinase II phosphorylation sites and a dileucine-based internalization signal. Deletion of the acidic motif from the Us9 cytoplasmic tail results in the loss of Us9 TGN localization and an inhibition of internalization (8). Mutagenesis of the dileucine internalization motif causes an increase in Us9 molecules on the plasma membrane due to a defect in the overall rate of endocytosis (8). Further analysis of the acidic domain, which we present in this report, revealed that the appropriate intracellular localization of the Us9 protein is dependent on the tyrosine residues at positions 49 and 50 in this region as well as the serine residues at positions 51 and 53 and the surrounding acidic residues. Alanine substitutions of these residues resulted in an increase in the number of Us9–EGFP molecules on the plasma membrane compared to that of wild-type Us9–EGFP. Antibody uptake experiments indicated that the increase in plasma membrane staining was due, in part, to a defect in Us9 internalization and transport back to the TGN. Mutagenesis of both tyrosine residues 49 and 50 together with alanine substitutions of serine residues 51 and 53 [Us9(Y49–50A S51A S53A)–EGFP] led to a complete defect in Us9–EGFP internalization, while mutagenesis of the putative casein kinase II sites alone [Us9(E52A D54A N55A E56A)–EGFP and Us9(S51A S53A)–EGFP] led to a partial defect in endocytosis. In contrast, Us9–EGFP molecules containing alanine replacements of the conserved tyrosine residues only [Us9(Y49–50A)–EGFP] were able to internalize and be targeted to the TGN, although less efficiently than wild-type Us9–EGFP. These results are in agreement with published reports indicating the importance of casein kinase II phosphorylation for the intracellular localization and trafficking of furin (15, 24, 42), hCMV gB (20), and VZV gE membrane proteins (2).
In this report, however, we showed that neither the maintenance of Us9 in the TGN region nor endocytosis from the plasma membrane is necessary for efficient viral replication or growth of PRV in tissue culture. Both PRV 162, containing a 10-amino-acid deletion of the conserved acidic motif, and PRV 166, containing an alanine substitution of the dileucine endocytosis signal, formed wild-type-sized plaques on monolayers of MDBK cells. Similarly, all viruses containing alanine substitutions in either the conserved tyrosine residues or the consensus casein kinase II phosphorylation sites showed no defect in cell-to-cell spread in tissue culture as measured by plaque size (data not shown). Moreover, all Us9 mutant viruses examined in this study grew to titers equivalent to those of wild-type virus and showed no defect in the expression or localization of the gE, gI, or gC envelope proteins (data not shown). Furthermore, examination of purified viral particles by Western blot analysis revealed that the targeting of Us9 to the virion envelope is not dependent on either the TGN localization or the endocytosis of Us9. All viral mutants containing alanine substitutions of the acidic domain were efficiently incorporated into viral particles. Likewise, viruses with the entire acidic cluster deleted (PRV 162) (8) or containing alanine substitutions in the dileucine signal (PRV 166) (data not shown) incorporate wild-type levels of Us9 into their envelopes. It is interesting that with all Us9 mutants examined to date, only two of the three Us9 polypeptides were detected in the envelopes of purified viral particles at all times. This observation suggests the existence of a selection process and indicates that not all Us9 forms are suitable for incorporation into virions.
Phosphorylation of the conserved acidic cluster.
Previous experiments have revealed that the Us9 protein expressed in PRV Be-infected cells is present as at least three phosphoforms (6), which we postulated is due to the addition of phosphate residues to the conserved tyrosine residues and/or casein kinase II sites in the Us9 cytoplasmic tail. In support of this prediction, removal of the 10-amino-acid acidic cluster from the Us9 open reading frame results in a reduction in the number of steady-state Us9 forms expressed in the infected cell (8). However, the results presented in this study indicate that the conserved tyrosine residues at positions 49 and 50 of the Us9 cytoplasmic tail are not major sites of Us9 phosphorylation. Replacement of these tyrosine residues with alanine residues had no effect on the number of Us9 phosphoforms in the infected cell. In contrast, mutagenesis of the serine residues comprising the consensus casein kinase II phosphorylation sites, as well as the surrounding acidic residues, resulted in a loss of all predominant Us9 phosphoforms, indicating that S51 and S53 are, in fact, major sites of Us9 phosphorylation. These results are consistent with unpublished observations by Hengartner and Enquist showing that recombinant casein kinase II can phosphorylate Us9 in vitro. However, the low level of Us9 phosphorylation detected in cells infected with PRV 162 (8), PRV 170, PRV 171, or PRV 173 after increased exposure of the immunoblot to film (data not shown) suggests the presence of minor Us9 species with serine- or threonine-linked phosphorylation outside of the acidic motif. Serine and threonine residues account for 14% of the amino acids in the Us9 open reading frame (30, 41) which may be available for phosphorylation by other cellular kinases or by the virally encoded serine-threonine protein kinase, Us3 (14).
The acidic cluster is essential for Us9-mediated anterograde spread in vivo.
We recently reported that the ability of PRV to spread to all neuronal connections in the anterograde direction in the rat nervous system is dependent, in part, on the Us9 protein (7). Following direct infection of the retina, viruses with Us9 deleted are defective in anterograde spread to the visual centers controlling reflex movements of the eye but displaying wild-type spread to the circadian-rhythm centers. This restricted-spread phenotype is indistinguishable from that observed for viruses with gE or gI deleted or for the attenuated strain PRV Bartha (12, 13, 44). In this report, we found that the dileucine endocytosis motif plays no role in Us9-promoted anterograde transneuronal spread. PRV 166, containing an alanine substitution of the dileucine internalization motif, was indistinguishable from wild-type virus in spreading to all retinorecipient areas following intraocular injection. This virus also displayed wild-type virulence (data not shown). These results indicate that the dileucine motif, whose mutation leads only to a partial defect in Us9 internalization, plays no detectable role in Us9-mediated spread or virulence. In contrast, we found that a virus containing a defined mutation removing the 10-amino-acid acidic motif from the Us9 cytoplasmic tail (PRV 162) was defective in spread in the anterograde direction to secondary neurons of the visual centers in a manner indistinguishable from that of a Us9 null virus. We also found that removal of the acidic domain led to a decrease in the virulence of the virus as measured by severity of symptoms and time to death (data not shown). Immunohistochemistry analysis of PRV 162-infected brains with a Us9-specific antiserum confirmed that the Us9 protein was abundantly expressed in these animals and that the restricted-spread pattern observed was not due to the inefficient expression or degradation of Us9. These results are also in agreement with those of the pulse-chase analysis of Us9 in PRV 162-infected cells, which indicated that the 10-amino-acid deletion did not significantly alter the stability of Us9.
Moreover, we demonstrated a dependence of PRV transneuronal spread on a pair of conserved tyrosine residues (Y49 and Y50) within the acidic domain. Both viruses containing alanine substitutions at tyrosine 49 and tyrosine 50 (PRV 170 and PRV 172) exhibited restricted spread to the brain after eye infection. Interestingly, we also observed that the rate of viral spread to all retinorecipient areas was dependent on phosphorylation of serine residues 51 and 53. Animals infected with either PRV 171, containing alanine substitutions of the acidic residues surrounding the putative casein kinase II sites, or PRV 173, containing alanine replacement of serine residues 51 and 53, displayed wild-type anterograde spread when the animals lived for extended times. In contrast, if animals succumbed to the virus at earlier times after retinal infection, viral antigen was restricted to the circadian-rhythm centers. Although these results suggest that the Us9 phosphorylation by casein kinase II is important for the efficiency of Us9-promoted transneuronal spread, we have yet to show if S51 and/or S53 is in fact a casein kinase II substrate in vivo. Moreover, at this point, it remains unclear how the phosphorylation status of Us9 determines the efficiency of Us9-promoted spread. A similar dependence of the length of survival of an infected animal on the rate of transneuronal spread has been recently shown by Tirabassi and Enquist for viral mutants lacking the cytoplasmic tail of the envelope protein gI (35). It should be noted that, regardless of the extent of viral spread in the brain, all the alanine scanning viral mutants examined in this study displayed a reduction in virulence as measured by time to death and severity of symptoms. The role of Us9 in virulence is not yet understood.
To ensure that there were no additional mutations in Us9 or in the adjacent gE and Us2 genes, we sequenced the Us9 region in all PRV strains examined in this study (data not shown). Revertants of selected mutants were constructed (e.g., PRV 160R and PRV 161R) and proved to have restored wild-type phenotypes. These results, as well as confirmation of the genotypes by Southern blot analysis, indicate that the phenotypes observed were in fact due to the desired Us9 mutations.
Role of Us9 endocytosis in promoting PRV anterograde spread.
We have previously proposed that the role of the Us9 protein in mediating PRV anterograde spread between synaptically connected neurons is to transport either newly enveloped viral particles and/or glycoprotein-containing vesicles down the axon to the site of viral egress (7). In this model, we also suggested that Us9 internalization back to the TGN region or targeting to specific vesicles may be critical for Us9-mediated transneuronal spread. In support of these speculations, the results presented in this report show that viruses defective in Us9 internalization and TGN retention, such as those with the acidic cluster deleted (PRV 162) or containing alanine substitutions of both the conserved tyrosine (Y49 and Y50) and serine (S51 and S53) residues (PRV 170), have an anterograde spread defect indistinguishable from that of a Us9 null virus (Table 3). Based on the observations made in this report on the trafficking of Us9 in nonpolarized epithelial cells, we further extend this model by proposing that phosphorylation of the Us9 cytoplasmic tail by casein kinase II modulates Us9 internalization from the neuronal-cell surface and retrieval to the TGN region. Moreover, we suggest that in the absence of casein kinase II phosphorylation, Us9 internalization and intracellular targeting in the neuron would be inefficient, thereby leading to an overall reduction in Us9-promoted spread. This speculation is supported by the data obtained for PRV 171 (E52A D54A N55A E56A) and PRV 173 (S51A S53A). These viruses were partially defective in Us9 endocytosis and TGN localization in nonpolarized tissue culture cells, and they were also inefficient in promoting anterograde transneuronal spread in vivo (see Table 3). Lastly, the observations made with PRV 166 (L30–31A) and PRV 172 (Y49–50A) indicate additional functions of the conserved tyrosine residues other than promoting Us9 internalization. Although both PRV 162 and PRV 172 had similar Us9 internalization rate defects in tissue culture, these two viruses had distinctly different anterograde spread phenotypes in vivo (see Table 3). For instance, whereas mutation of the dileucine endocytosis motif (PRV 166) had no effect on Us9-promoted anterograde spread, the two conserved tyrosine residues (PRV 172) were essential for transneuronal spread. Tyrosine residues 49 and 50 of the Us9 cytoplasmic tail may serve as a critical domain for the interaction of Us9 with itself, other viral glycoproteins, or the cellular transport machinery. Experiments are currently in progress to test these ideas.
Clearly, the experiments presented in this report indicate a link between Us9 endocytosis and Us9-mediated transneuronal spread between synaptically connected neurons in vivo. As the endocytosis of Us9 has been examined only in nonpolarized tissue culture cells, additional experiments need to be conducted to examine the steady-state localization and intracellular trafficking of Us9 in neurons.
ACKNOWLEDGMENTS
We thank K. Bienkowska-Szewcyzk for the generous gift of gE polyvalent antiserum. We acknowledge P. Card for insightful discussions regarding the analysis of the infected animal tissues. Many thanks to J. Schwarzbauer for the use of her fluorescence microscope and imaging system and to members of the Enquist lab for their continuous support.
This work was supported by NINDS grant 1RO133506 to L.W.E.
REFERENCES
- 1.Alconada A, Bauer U, Baudoux L, Piette J, Hoflack B. Intracellular transport of the glycoproteins gE and gI of the varicella-zoster virus. J Biol Chem. 1998;273:13430–13436. doi: 10.1074/jbc.273.22.13430. [DOI] [PubMed] [Google Scholar]
- 2.Alconada A, Bauer U, Hoflack B. A tyrosine-based motif and a casein kinase II phosphorylation site regulate the intracellular trafficking of the varicella-zoster virus glycoprotein I, a protein localized in the trans-Golgi network. EMBO J. 1996;15:6096–6110. [PMC free article] [PubMed] [Google Scholar]
- 3.Alconada A, Bauer U, Sodeik B, Hoflack B. Intracellular traffic of herpes simplex virus glycoprotein gE: characterization of the sorting signals required for its trans-Golgi network localization. J Virol. 1999;73:377–387. doi: 10.1128/jvi.73.1.377-387.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker C H. Zur primaren Schadingung vegetativer Ganglien nach Infektion mit dem Herpes suis Virus bei verschiedenen Tierarten. Experientia. 1967;23:209. doi: 10.1007/BF02136291. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Porat T, Kaplan A S. Molecular biology of pseudorabies virus. In: Roizman B, editor. The herpesviruses. New York, N.Y: Plenum Publishing Corp.; 1985. pp. 105–173. [Google Scholar]
- 6.Brideau A D, Banfield B W, Enquist L W. The Us9 gene product of pseudorabies virus, an alphaherpesvirus, is a phosphorylated, tail-anchored type II membrane protein. J Virol. 1998;72:4560–4570. doi: 10.1128/jvi.72.6.4560-4570.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brideau A D, Card J P, Enquist L W. The role of pseudorabies virus Us9, a type II membrane protein, in infection of tissue culture cells and the rat nervous system. J Virol. 2000;74:834–845. doi: 10.1128/jvi.74.2.834-845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brideau A D, del Rio T, Wolffe E J, Enquist L W. Intracellular trafficking and localization of the pseudorabies virus Us9 type II envelope protein to host and viral membranes. J Virol. 1999;73:4372–4384. doi: 10.1128/jvi.73.5.4372-4384.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Card J P. Exploring brain circuitry with neurotropic viruses: new horizons in neuroanatomy. Anat Rec (New Anat) 1998;253:176–185. doi: 10.1002/(SICI)1097-0185(199812)253:6<176::AID-AR6>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 10.Card J P, Levitt P, Enquist L W. Different patterns of neuronal infection after intracerebral injection of two strains of pseudorabies virus. J Virol. 1998;72:4434–4441. doi: 10.1128/jvi.72.5.4434-4441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Card J P, Rinaman L, Schwaber J S, Miselis R R, Whealy M E, Robbins A K, Enquist L W. Neurotropic properties of pseudorabies virus: uptake and transneuronal passage in the rat central nervous system. J Neurosci. 1990;10:1974–1994. doi: 10.1523/JNEUROSCI.10-06-01974.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Card J P, Whealy M E, Robbins A K, Enquist L W. Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat visual system. J Virol. 1992;66:3032–3041. doi: 10.1128/jvi.66.5.3032-3041.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Card J P, Whealy M E, Robbins A K, Moore R Y, Enquist L W. Two alphaherpesvirus strains are transported differentially in the rodent visual system. Neuron. 1991;6:957–969. doi: 10.1016/0896-6273(91)90236-s. [DOI] [PubMed] [Google Scholar]
- 14.Daikoku T, Kurachi R, Tsurumi T, Nishiyama Y. Identification of a target protein of US3 protein kinase of herpes simplex virus type 2. J Gen Virol. 1994;75:2065–2068. doi: 10.1099/0022-1317-75-8-2065. [DOI] [PubMed] [Google Scholar]
- 15.Dittie A S, Thomas L, Thomas G, Tooze S A. Interaction of furin in immature secretory granules from neuroendocrine cells with the AP-1 adaptor complex is modulated by casein kinase II phosphorylation. EMBO J. 1997;16:4859–4870. doi: 10.1093/emboj/16.16.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enquist L W, Card J P. Pseudorabies virus: a tool for tracing neuronal connections. In: Lowenstein P R, Enquist L W, editors. Protocols for gene transfer in neuroscience: towards gene therapy of neurological disorders. New York, N.Y: John Wiley & Sons Ltd.; 1996. pp. 333–348. [Google Scholar]
- 17.Enquist L W, Husak P J, Banfield B W, Smith G A. Infection and spread of alphaherpesviruses in the nervous system. Adv Virus Res. 1999;51:237–347. doi: 10.1016/s0065-3527(08)60787-3. [DOI] [PubMed] [Google Scholar]
- 18.Field H J, Hill T J. The pathogenesis of pseudorabies in mice following peripheral inoculation. J Gen Virol. 1974;23:145–157. doi: 10.1099/0022-1317-23-2-145. [DOI] [PubMed] [Google Scholar]
- 19.Field H J, Hill T J. The pathogenesis of pseudorabies in mice: virus replication at the inoculation site and axonal uptake. J Gen Virol. 1975;26:145–148. doi: 10.1099/0022-1317-26-1-145. [DOI] [PubMed] [Google Scholar]
- 20.Fish K N, Soderberg-Naucler C, Nelson J A. Steady-state plasma membrane expression of a human cytomegalovirus gB is determined by the phosphorylation state of Ser900. J Virol. 1998;72:6657–6664. doi: 10.1128/jvi.72.8.6657-6664.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fletcher T, III, Gray W. DNA sequence and genetic organization of the unique short (Us) region of the simian varicella virus genome. Virology. 1993;193:762–773. doi: 10.1006/viro.1993.1185. [DOI] [PubMed] [Google Scholar]
- 22.Goodpasture E W, Teague O. Transmission of the virus of herpes along nerves in experimentally infected rabbits. J Med Res. 1923;44:139–184. [PMC free article] [PubMed] [Google Scholar]
- 23.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 24.Jones B G, Thomas L, Molloy S S, Thulin C D, Fry M D, Walsh K A, Thomas G. Intracellular trafficking of furin is modulated by the phosphorylation state of a casein kinase II site in its cytoplasmic tail. EMBO J. 1995;14:5869–5883. doi: 10.1002/j.1460-2075.1995.tb00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knapp A C, Enquist L W. Pseudorabies virus recombinants expressing functional virulence determinants gE and gI from bovine herpesvirus 1.1. J Virol. 1997;71:2731–2739. doi: 10.1128/jvi.71.4.2731-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kristensson K. Sorting signals and targeting of infectious agents through axons: an annotation to the 100 years' birth of the name “axon.”. Brain Res Bull. 1996;41:327–333. doi: 10.1016/s0361-9230(96)00255-9. [DOI] [PubMed] [Google Scholar]
- 27.Mettenleiter T C. Molecular biology of pseudorabies (Aujesky's disease) virus. Comp Immunol Microbiol Infect Dis. 1991;14:151–163. doi: 10.1016/0147-9571(91)90128-z. [DOI] [PubMed] [Google Scholar]
- 28.Olson J, Grose C. Endocytosis and recycling of varicella-zoster virus Fc receptor glycoprotein gE: internalization mediated by a YXXL motif in the cytoplasmic tail. J Virol. 1997;71:4042–4054. doi: 10.1128/jvi.71.5.4042-4054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olson J K, Grose C. Complex formation facilitates endocytosis of the varicella-zoster virus gE:gI Fc receptor. J Virol. 1998;72:1542–1551. doi: 10.1128/jvi.72.2.1542-1551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petrovskis E A, Post L E. A small open reading frame in pseudorabies virus and implications for evolutionary relationships between herpesviruses. Virology. 1987;159:193–195. doi: 10.1016/0042-6822(87)90368-0. [DOI] [PubMed] [Google Scholar]
- 31.Pinna L A, Ruzzene M. How do protein kinases recognize their substrates? Biochim Biophys Acta. 1996;1314:191–225. doi: 10.1016/s0167-4889(96)00083-3. [DOI] [PubMed] [Google Scholar]
- 32.Robbins A K, Dorney D J, Wathen M W, Whealy M E, Gold C, Watson R J, Holland L E, Weed S D, Levine M, Glorioso J C, Enquist L W. The pseudorabies virus gII gene is closely related to the gB glycoprotein gene of herpes simplex virus. J Virol. 1987;61:2691–2701. doi: 10.1128/jvi.61.9.2691-2701.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabin A B. Progression of different nasally instilled viruses along different nervous system pathways in the same host. Proc Soc Exp Biol Med. 1938;38:270–275. [Google Scholar]
- 34.Teuchert M, Schafer W, Berghofer S, Hoflack B, Klenk H D, Garten W. Sorting of furin at the trans-Golgi network. Interaction of the cytoplasmic tail sorting signals with AP-1 Golgi-specific assembly proteins. J Biol Chem. 1999;274:8199–8207. doi: 10.1074/jbc.274.12.8199. [DOI] [PubMed] [Google Scholar]
- 35.Tirabassi R S, Enquist L W. Role of the pseudorabies virus gI cytoplasmic domain in neuroinvasion, virulence, and posttranslational N-linked glycosylation. J Virol. 2000;74:3505–3516. doi: 10.1128/jvi.74.8.3505-3516.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tirabassi R S, Enquist L W. Mutation of the YXXL endocytosis motif in the cytoplasmic tail of pseudorabies virus gE. J Virol. 1999;73:2717–2728. doi: 10.1128/jvi.73.4.2717-2728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tirabassi R S, Enquist L W. Role of envelope protein gE endocytosis in the pseudorabies virus life cycle. J Virol. 1998;72:4571–4579. doi: 10.1128/jvi.72.6.4571-4579.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tirabassi R S, Townley R A, Eldridge M G, Enquist L W. Characterization of pseudorabies virus mutants expressing carboxy-terminal truncations of gE: evidence for envelope incorporation, virulence, and neurotropism domains. J Virol. 1997;71:6455–6464. doi: 10.1128/jvi.71.9.6455-6464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tugizov S, Maidji E, Xiao J, Pereira L. An acidic cluster in the cytosolic domain of human cytomegalovirus glycoprotein B is a signal for endocytosis from the plasma membrane. J Virol. 1999;73:8677–8688. doi: 10.1128/jvi.73.10.8677-8688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tugizov S, Maidji E, Xiao J, Zheng Z, Pereira L. Human cytomegalovirus glycoprotein B contains autonomous determinants for vectorial targeting to apical membranes of polarized epithelial cells. J Virol. 1998;72:7374–7386. doi: 10.1128/jvi.72.9.7374-7386.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Zijl M, van der Gulden H, de Wind N, Gielkens A, Berns A. Identification of two genes in the unique short region of pseudorabies virus: comparison with herpes simplex virus and varicella-zoster virus. J Gen Virol. 1990;71:1747–1755. doi: 10.1099/0022-1317-71-8-1747. [DOI] [PubMed] [Google Scholar]
- 42.Voorhees P, Deignan E, van Donselaar E, Humphrey J, Marks M S, Peters P J, Bonifacino J S. An acidic sequence within the cytoplasmic domain of furin functions as a determinant of trans-Golgi network localization and internalization from the cell surface. EMBO J. 1995;14:4961–4975. doi: 10.1002/j.1460-2075.1995.tb00179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wan L, Molloy S S, Thomas L, Liu G, Xiang Y, Rybak S L, Thomas G. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell. 1998;94:205–216. doi: 10.1016/s0092-8674(00)81420-8. [DOI] [PubMed] [Google Scholar]
- 44.Whealy M E, Card J P, Robbins A K, Dubin J R, Rziha H J, Enquist L W. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J Virol. 1993;67:3786–3797. doi: 10.1128/jvi.67.7.3786-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wittmann G, Rziha H-J. Aujeszky's disease (pseudorabies) in pigs. In: Wittmann G, editor. Herpesvirus diseases of cattle, horses and pigs. Boston, Mass: Kluwer; 1989. pp. 230–325. [Google Scholar]
- 46.Zhu Z, Hao Y, Gershon M D, Ambron R T, Gershon A A. Targeting of glycoprotein I (gE) of varicella-zoster virus to the trans-Golgi network by an AYRV sequence and an acidic amino acid-rich patch in the cytosolic domain of the molecule. J Virol. 1996;70:6563–6575. doi: 10.1128/jvi.70.10.6563-6575.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]