Highlights
-
•
Root exudates play an important role in regulating microbial communities and C.
-
•
Three common groups of root exudates have contrasting effects on soil C and Fe.
-
•
Sugar's application strongly reduced microbial diversity.
-
•
Carboxylic acids increased actinobacteria and facilitated C mobilization.
-
•
Amino acids increased proteobacteria that tended to prevent the release of soil C.
Keywords: Microbial community assembly, NanoSIMS imaging, Priming effects, Root exudate chemistry, Soil organic carbon, Rhizosphere processes
Abstract
Plant roots are one of the major mediators that allocate carbon captured from the atmosphere to soils as rhizodeposits, including root exudates. Although rhizodeposition regulates both microbial activity and the biogeochemical cycling of nutrients, the effects of particular exudate species on soil carbon fluxes and key rhizosphere microorganisms remain unclear. By combining high-throughput sequencing, q-PCR, and NanoSIMS analyses, we characterized the bacterial community structure, quantified total bacteria depending on root exudate chemistry, and analyzed the consequences on the mobility of mineral-protected carbon. Using well-controlled incubation experiments, we showed that the three most abundant groups of root exudates (amino acids, carboxylic acids, and sugars) have contrasting effects on the release of dissolved organic carbon (DOC) and bioavailable Fe in an Ultisol through the disruption of organo-mineral associations and the alteration of bacterial communities, thus priming organic matter decomposition in the rhizosphere. High resolution (down to 50 nm) NanoSIMS images of mineral particles indicated that iron and silicon co-localized significantly more organic carbon following amino acid inputs than treatments without exudates or with carboxylic acids. The application of sugar strongly reduced microbial diversity without impacting soil carbon mobilization. Carboxylic acids increased the prevalence of Actinobacteria and facilitated carbon mobilization, whereas amino acid addition increased the abundances of Proteobacteria that prevented DOC release. In summary, root exudate functions are defined by their chemical composition that regulates bacterial community composition and, consequently, the biogeochemical cycling of carbon in the rhizosphere.
Graphical abstract
1. Introduction
Roots exude considerable amounts (11–40%) of photosynthetically fixed carbon, creating a gradient of a variety of chemicals in the rhizosphere [1,2]. These complex cocktails of phyto-metabolites, including sugars, amino acids, organic acids, and secondary metabolites [3], exert various effects on bacterial community composition and microbiome assembly. Consequently, there are impacts on bacterial ecological functionality within the rhizosphere. For example, sugars have little [4] or marked [5] selection effects on soil microbiomes [3,6], owing to being more bioenergetically favorable [7]. In contrast, due to strong metal-complexing abilities and of limited bioenergetic use to microbes [7], organic acids, especially those involved in the tricarboxylic acid cycle, have strong selection effects [8] and can recruit plant growth promoting rhizobacteria (PGPRs) into the rhizosphere [9]. Proline, a proteinogenic amino acid, can be utilized as both a carbon and nitrogen (N) source by rhizosphere associated Pseudomonas [10]. Pseudomonas, a model root colonizer, has been identified as an essential species both for rhizosphere fitness and in evading host immune responses [11]. Root exudate fluxes are very difficult to determine and have been reported to vary from 0.1 to 36 mg g dry root−1 h−1, depending on the collection method and plant species [12]. Even with these previously identified insights, the specific effects of distinct root exudates on bacterial communities and their accompanied biogeochemical cycling remain largely undescribed.
Recently, the role of root exudate inputs in carbon cycling has attracted increasing attention [13]. Roots excrete multiple soluble organic compounds, which induces a rhizosphere “priming effect” towards the acceleration of soil organic carbon (SOC) turnover [7,14]. This “priming effect” is defined as strong, generally short-term changes in the turnover of native SOC induced by comparatively moderate treatments of the soil [14] and has been found to be strongly dependent on the chemical composition or functional groups of the root excreted compounds [3]. For example, oxalic acid induced a much stronger “priming effect” than glucose and acetic acid, which further promoted carbon loss by liberating soil mineral-protected organic carbon [7]. Therefore, the “priming effect” is not a purely biotic phenomena but may include synergistic effects between microorganisms and minerals [7]. In addition, root exudate-driven release of iron (Fe) and the subsequent formation of reactive Fe minerals was found to impart a direct control on soil carbon storage and loss [15]. It has been estimated that globally, one-quarter of the SOC pool (∼600 Gt, 1 Gt = 1 billion tons) has been retained by interactions with reactive Fe minerals (mainly amorphous or short-range-ordered Fe (oxyhydr)oxides) [16]. During the “priming effect”, bacterial communities, especially of fast-growing r-strategists with the ability to utilize various root-derived carbon substrates [17], are reported to trigger apparent “priming effect”. In contrast, such organisms, which are predominantly K-strategists that are more competitive in stable environments with limited resources, increase SOC decomposition and thus cause real “priming effect” [18]. However, we still lack a holistic understanding of the linkages between the composition of root exudates, bacterial community composition, and the biogeochemical cycling of carbon and other nutrients. Deciphering these linkages is essential to both better understand the function of the rhizosphere and to promote a better understanding of plant-microbial-soil tripartite interactions.
The objectives of this study were to (i) assess the impact of root exudate composition on soil bacterial community assembly; (ii) examine the subsequent effects of exudate chemistry on the mobilization of carbon and nutrients in soils; and (iii) identify the specific bacterial groups that prime SOC decomposition and lead to the release of dissolved organic carbon (DOC) and bioactive elements (i.e., Fe and silicon (Si)). To accomplish these objectives, a series of well-controlled incubation experiments were conducted in order to elucidate the role of the three most abundant groups of root exuded compounds (amino acids, carboxylic acids, and sugars) [19], in governing SOC dynamics in the rhizosphere originating from an Ultisol soil. Both 16S rRNA gene-based Illumina MiSeq high-throughput sequencing (HTS) and real-time quantitative PCR (q-PCR) were used to characterize the bacterial community and to quantify total soil bacteria, respectively. Scanning electron microscopy (SEM) and nanoscale secondary ion mass spectrometry (NanoSIMS) were integrated to identify the localization of carbon and other bioactive elements (Fe and Si) as well as the effects of root exudate chemistry on the mobility of mineral-protected SOC. Our systematic study provides both direct evidence of the contrasting effects of most abundant organic substances within root-exudates on bacterial communities and an improved understanding of the biogeochemical cycling of SOC and other bioactive elements.
2. Materials and methods
2.1. Soil preparation
Soil samples were collected from an agricultural field in Nanjing, China, in October 2018, where a typical subtropical monsoon climate prevails, with a mean annual temperature and precipitation of 18 °C and 1416 mm, respectively. The soil was classified as an Ultisol that is widely distributed throughout the subtropical areas in South China. After sampling, visible gravel, roots and other plant materials were removed and the soil sample was sieved. After sieving, the <2 mm soil fraction was used for chemical characterization and the incubation experiments. Related soil chemical properties measured included pH (7.64), total N (TN, 1.97 g kg–1), total carbon (TC, 2.49 g kg–1), available potassium (155 mg kg–1), total potassium (5.33 g kg–1), available phosphorous (106 mg kg–1), total phosphorous (0.54 g kg–1) and electric conductivity (309 μS cm–1).
2.2. Soil incubation experiment
Soil (15 g dry weight equivalent) was placed into each well of a 6-well plate. To activate soil microorganisms and remove germinating seeds from the naturally occurring seed bank, plates were pre-incubated in a growth chamber at 30 °C for 1 week. Following the pre-incubation, each well received 1.5 mL of artificial exudate solution twice a week for a total of 8.5 weeks (8 treatments). The experimental exudate solutions consisted of 18 representative compounds from root exudates selected based on the composition and relative abundance of typical root exudates (Table S1) as well as our previous study [20]. These compounds are closely related to that in Arabidopsis (Table S1) and then classified into three common groups: amino acids (A), short-chain carboxylic acids (C), and sugars (S). The concentration of the added compounds in soils was 2.3 ± 0.6 mg kg−1.
Three exudate solutions (A, C, and S) were prepared, each group containing the selected compounds in equal dosages at a final total concentration of 10 mM. Solution A contained 1.67 mM of isoleucine, leucine, methionine, proline, tryptophan, and ornithine. Solution C contained 1.67 mM of oxaloacetic acid, citric acid, aconitic acid, ketoglutaric acid, succinic acid, and malic acid, while solution S contained 1.67 mM of maltose, ribose, glucose, sucrose, fructose, and xylose (Table S2).
Incubation experiments were conducted with eight treatments in total, including one control soil in which only autoclaved water was added. The remaining seven treatments consisted of treatments A, C, and S which represented soils that received only solutions A, C, and S, respectively, treatments AC, AS, and CS which received two corresponding groups of solutions at equal volumes of each solution, and treatment ACS which received all three groups of solutions with equal volumes. Each treatment consisted of 18 replicates over 3 plates. All plates were randomly placed during the incubation period. During the incubation, soil humidity was determined by weighing twice a week and adjusted to 70% using sterile deionized water.
2.3. Sample collecting, sequencing, and data processing
After a 8.5-week incubation, soil samples from two wells of each treatment were randomly pooled into one ‘sample’ and stored at −80 °C. A total of nine samples were obtained for each treatment. Total soil DNA was extracted from 0.5 g of soil using the Power Lyzer PowerSoil DNA Isolation Kit (Qiagen, Germany) following the manufacturer's protocol. A 1% agarose gel and a NanoDrop 2000 spectrophotometer were used to evaluate DNA quality and quantity (Thermo Scientific, Waltham, MA, USA).
For taxonomical profiling, PCR products that targeted the V4 region of the bacterial 16S rRNA gene were synthesized with the primers 515F: GTGYCAGCMGCCGCGGTAA and 806R: GGACTACNVGGGTWTCTAAT) [21] to yield an amplicon of 292 bp. The PCR amplification was performed using a BioRad S1000 (Bio-Rad Laboratory, CA) with conditions previously reported [22]. Products were run on a 1% agarose gel and those with clear bands between 290 and 310 bp were combined for sequencing. PCR products were mixed in equidensity ratios and then purified with an EZNA Gel Extraction Kit (Omega, USA). Sequencing libraries were generated using the NEBNext® Ultra™ DNA Library Prep Kit for Illumina® (New England Biolabs, USA) following the manufacturer's recommendations. Index codes were added and library quality was assessed with a Qubit® 2.0 Fluorometer (Thermo Scientific) and an Agilent BiCAnalyzer 2100 system. Finally, the library was sequenced on an Illumina MiSeq sequencing platform using PE250 chemistry at the Genomics Core of Michigan State University. Reads were quality filtered to obtain high-quality reads according to Trimmomatic (V0.33, http://www.usadellab.org/cms/?page=trimmomatic) and sequences were assigned to each sample based on its unique barcode. These reads were then processed using the pipelines detailed below.
For sequencing data analyses, usearch (V. 10.1) and vsearch (V. 0.6.3) were used for the primary processing. First, “vsearch –fastq_mergepairs” script was used for pair end sequence merge; “vsearch –fastx_filter” script was used for primers removal; “vsearch –derep_fulllength” script was used to identify unique sequences; “usearch -cluster_otus” script was used for the clustering of operational taxonomic units (OTUs) at 97% similarity. “vsearch –usearch_global” script was used to create the OTU table. Annotation was performed using the “vsearch –sintax” script and the RDP taxonomic database.
In total, 2634,804 high-quality sequences were obtained across a total of 72 soil samples from eight treatments consisting of nine replicates each. The average read count of each sample was 36,594 (standard deviation (SD) 14,578). All sequences were clustered into 10,146 OTUs. The number of OTUs, generally ranged between 1178 and 3235 per sample with an average of 2119 (SD 431). The majority of OTUs belonged to the phyla TM7 (39.71% with SD 0.17), Proteobacteria (23.77% with SD 0.1), Cyanobacteria (8.95% with SD 0.06), Chloroflexi (5.53% with SD 0.03), Bacteroidetes (5.34% with SD 0.02), Acidobacteria (5.04% with SD 0.03), Actinobacteria (3.48% with SD 0.01), Gemmatimonadetes (2.71% with SD 0.02), Verrucomicrobia (2.17% with SD 0.01) and Firmicutes (1.56% with SD 0.01).
In order to further explore the response of microbial communities to soil DOC, we divided all the soils into three groups based on their relative DOC content, as compared to the control soil. This included group 1 (Up) with the soil DOC content significantly higher than that in Control soil; group 2 (Down) with soil DOC content significantly lower than that in Control soil; and group 3 (Nose) with no significant difference in soil DOC content between the treated soil and the Control soil. Random forests (RF) analyses [23] were then utilized to construct models using relative abundances as a candidate feature, which has previously been shown to outperform other modeling approaches for environmental bacterial datasets [24]. Random forest classifier model training was performed using the R function randomForest (package: randomForest; 1000 trees, other parameters set by default). Classifier performance evaluation was established through fivefold cross-validation within the training set. Cross-validation was performed by the rfcv function for selecting appropriate features. The varImpPlot function was used to illustrate the importance of features in the classification and the curves were visualized by using the ggplot2 package in R v.3.6.1.
2.4. Quantification of soil bacteria by q‐PCR
Real-time q-PCR was performed to quantify total soil bacterial abundances using an Applied Biosystems StepOne Plus (Applied Biosystems, CA, USA) qPCR machine and SYBR Premix Ex TaqTM (TliRnaseH Plus) (TaKaRa Biotechnology Co., Ltd). Reaction conditions conformed to a previously published protocol [25] using primers 347F: 5′-GGAGGCAGCAGTRRGGAAT-3′ and 531R: 5′-CTNYGTMTTACCGCGGCTGC-3′, yielding a 182 bp product. Quantitative PCR was performed for the nine biological replicates using three technical replicates. Amplification specificity was verified by melting-curve analysis and agarose gel electrophoresis.
2.5. NanoSIMS analyses
Three contrasting samples from Control, A, and C were chosen for NanoSIMS analysis in duplicate. The soil was first suspended in ultrapure water at a ratio of 1:5 (w/v), shaken for 8 h at 25 °C, and centrifuged for 6 min at 2500 g. Aliquots of the supernatant suspensions containing water-dispersible soil colloids were dispersed in ethanol and dropped onto Au foil, air-dried and gold-coated. Imaging with SEM (Zeiss Sigma 500) was performed with a 20 kV accelerating potential. Sample analyses using NanoSIMS-50 L (Cameca, Gennevilliers, France) were carried out at the School of Earth System Science, Tianjin University, China.
Prior to analysis, the gold coating layer (∼10 nm) and possible contamination of the sample surface were pre-sputtered using a high primary beam current [26]. During the pre-sputtering, reactive Cs+ ions were implanted into the sample to enhance secondary ion yields. Secondary ions of 12C−, 28Si−, and 56Fe16O− were collected simultaneously by electron multipliers with an electronic dead time of 44 ns. The presence of the 12C− ion was interpreted as organic carbon, while 28Si− and 56Fe16O− ions were corresponded to silicates and Fe minerals, respectively [26]. The charging effect resulting from nonconductive mineral particles was compensated through an electron flood gun. Composite multi-element images were constructed using Image J (version 1.45) with the OpenMIMS plugin (http://www.nrims.hms.harvard.edu/NRIMS_ImageJ.php).
2.6. Soil chemical analyses
Soil pH (Fig. S1) was determined using a glass electrode pH meter (PHS-3C, INESA Scientific Instrument Co., Ltd., Shanghai, China) in a 1:5 soil water (w/v) suspension [26]. To determine soil DOC, 5 g soil and 45 g ultrapure water were mixed in a 250 ml flask and shaken for 8 h at 200 r min–1 [26]. Then, soil DOC was filtered with 0.45 µm filter membrane, and the concentration of DOC in the <0.45 µm filtrate was measured with a Shimadzu TOC analyzer (TOC-L Shimadzu, Japan). Concentrations of available Fe (aFe) and available Si (aSi) [26] in the same filtrate were measured with ICP-AES (NexION 300XX, PerkinElmer, USA).
2.7. Statistical analyses
For the statistical analysis and visualization of microbial communities, we checked and removed one sample from group A, due to a significantly lower number of OTUs, as compared with other samples within the group. Alpha diversity measures of Richness, Shannon diversity, and Pielou's evenness were calculated after the OTU table was re-sampled with the minimum read number (12,170) among all treatments and a boxplot was used for data visualization. The relative abundance of each OTU was standardized by the normalize_table.py script with Qiime (1.9.1) [27] and Bray-Curtis similarity matrices were prepared with the beta_diversity.py script. Principal coordinate analysis (PCoA) plots were generated from Bray-Curtis similarity matrices. A nonparametric t-test was used to determine the significance. For beta diversity, PERMANOVA (Adonis, transformed data by Bray-Curtis, permutation = 999) was used to determine the similarities between the composition of bacterial communities among treatments. Common and specific OTUs were calculated with an R script and visualized with a flower plot [28]. Linear regression was calculated and plotted with the package “ggpubr”. The concentrations of aFe, aSi, and DOC were used as explanatory variables for redundancy analyses (RDA) computed on Bray-Curtis distances using the vegan package [29]. Network analysis based on “pearson” correlations and their properties were calculated with package “igraph” [30]. To explore the direct and indirect relationships among soil chemical properties (i.e., DOC, aFe, and aSi), bacterial community composition and the applied compounds, directed graphs of the partial least squares path model (PLS-PM) analysis were calculated with the package “plspm” [31]. R software was used for all statistical analyses. A threshold of α = 0.05 was considered as statistically significant, except nodes, and p values were adjusted with a “fdr” correction in cases of multiple tests [32].
3. Results
3.1. Changes in dissolved organic carbon, available Fe and Si, and mineral protected carbon under root exudate inputs
In order to investigate the effects of specific classes of root exudates on the mobilization of soil carbon and selected bioactive elements, we evaluated the yields of soil DOC, aFe and aSi that corresponded to the addition of various organic compound classes. Compared to the Control soil, addition of short-chain carboxylic acids (treatment C) led to higher (ANOVA; LSD method for multiple comparisons) DOC and aFe in the soils. In contrast, addition of amino acids (treatment A) decreased both DOC and aFe (Figs. 1 and S2). The sugar input (treatment S) led to an intermediate content of both DOC and aFe. All three organic compound classes resulted in a lower aSi content, as compared to the Control soil.
Fig. 1.
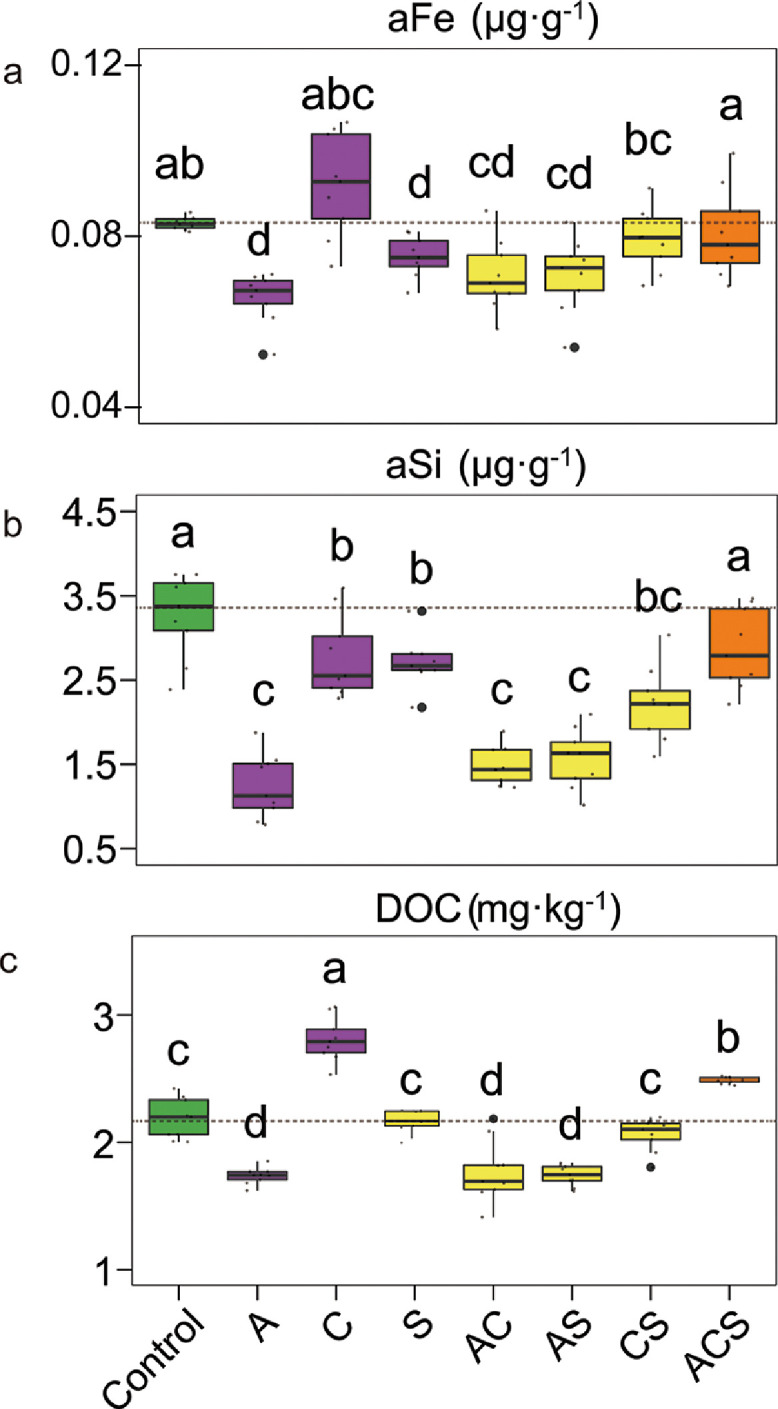
Effects of root exudate chemistry on available Fe (aFe) (a), available Si (aSi) (b), and dissolved organic carbon (DOC) (c) in soil depending on the addition of various groups of root exudates. A, C, S represent soils received only amino acids (A), carboxylic acids (C), and sugars (S), respectively; AC, AS, and CS indicate soils received two corresponding organic compound-classes with equal final molar concentrations for each compound; ACS denotes soil received all three organic compounds with the same final molar concentration for each compound. The horizontal bars within boxes represent the median. The tops and bottoms of boxes represent 75th and 25th quartiles, respectively. Letters represent significant differences between groups (nonparametric t-test).
The combination of root exudate classes (treatment ACS) led to higher contents of DOC, aFe, and aSi than all soils treated with only two root exudate components: AC, AS, and CS. Soils with AC and AS additions exhibited a lower amount of DOC, aFe, and aSi than those amended with CS (Fig. 1). Lower DOC, aFe, and aSi contents in the soils with AC, AS, and CS inputs may be related to a lower soil pH (Fig. S1) that altered microbial activities and the surface charges of soil colloids and possibly influenced DOC molecular weight and the solubility and chemical speciation of Fe. As such, the mobilization of carbon, aFe, and aSi may have been impacted. Significant positive correlations between DOC and aFe (R = 0.60, p < 0.001) and aSi (R = 0.78, p < 0.001) support this supposition (Fig. S3).
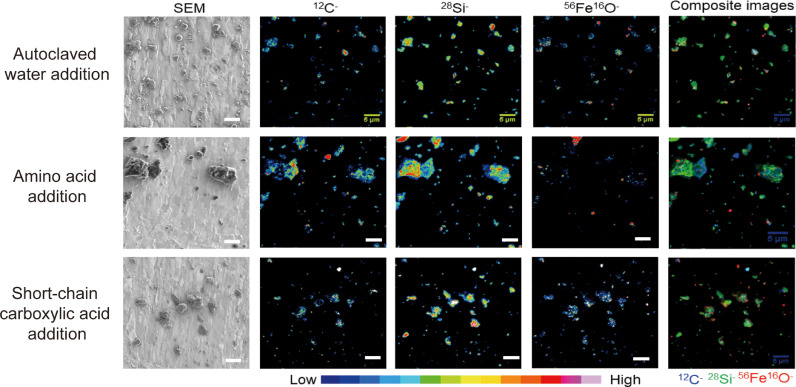
Correlative SEM and NanoSIMS images allowed for the visualization of the effects of root exudate components on organo-mineral associations and mineral-protected carbon within water dispersible soil colloids (Figs. 2 and S4–S7). For this analysis, only three contrasting samples (Fig. 1) were chosen for NanoSIMS analysis in duplicate, owing to its high cost. The particle size of the examined soil samples ranged from submicron to ∼50 µm (Fig. 2). NanoSIMS images of 12C−, 56Fe16O−, and 28Si− ion masses illustrated the elemental distribution and spatial heterogeneity in the water dispersible soil colloids (Figs. 2 and S6). Composite NanoSIMS images clearly demonstrated that both mineral elements (56Fe16O− and 28Si−) and organic carbon (12C−) are highly heterogeneous but colocalized in colloidal soil particles at the submicron scale (Figs. 2 and S5–S7). In the soil particles after amino acid addition, Fe and Si were co-localized with much more organic carbon than both the Control soil and after carbohydrate addition (Figs. 2 and S5–S7). This is consistent with the results from the bulk soil characterization (Fig. 1) after amino acid addition which was found to lead to lower DOC, aFe, and aSi contents. Lastly, soil with carbohydrate addition contained higher colloidal Fe than that with no addition or amino acid addition (Fig. 2), which likely contributed to the overall higher aFe content (Fig. 1).
Fig. 2.
Correlative SEM and NanoSIMS images of water dispersible soil colloids. SEM, scanning electron microscopy. NanoSIMS, nanoscale secondary ion mass spectrometry. Composite images showing the co-localization of C (12C−, blue), Fe (56Fe16O−, red), and Si (28Si−, green) in samples. Note that the color intensity on the images corresponds to the relative contents of individual elements (bottom bare scale), but cannot be used to compare the contents one element with another. Scale bar = 5 µm.
3.2. Responses of soil microbiota to root exudate component inputs
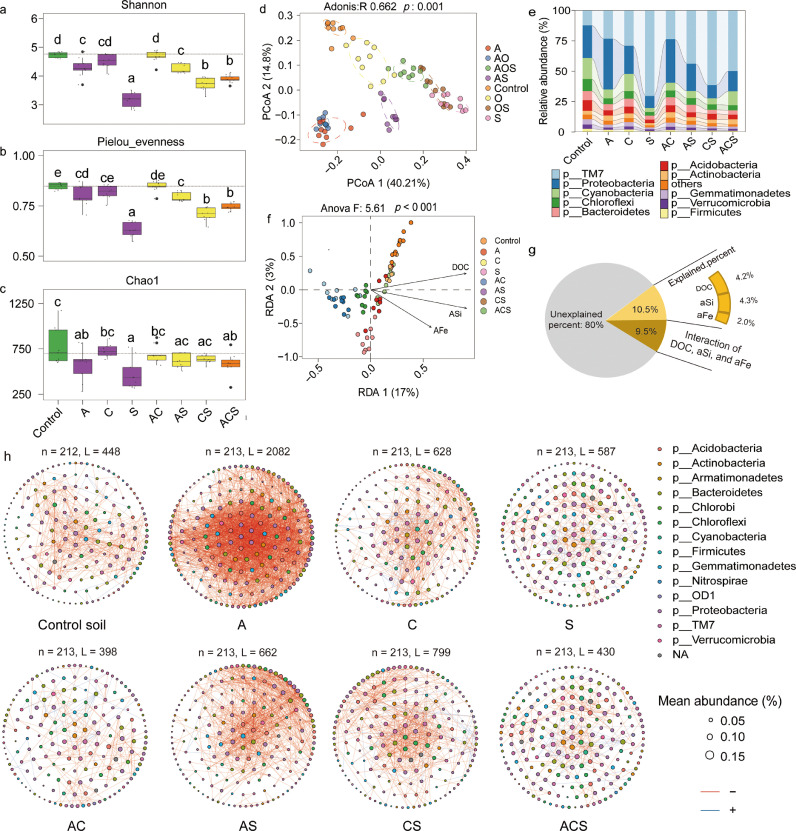
To assess the influence of the various root exudate classes on the soil microbiota, HTS was used to reveal the composition of the bacterial community. We first examined differences in diversity with the addition of single components (A, C, and S) as well as the Control. Alpha diversity measures, consisting of Richness, Shannon diversity, and Pielou's evenness were all decreased with the addition of each class of exudate compounds. Sugar (S) addition caused the most drastic reduction (Fig. S8a). Ordination using PCoA, based on Bray-Curtis distances, illustrated a significant (Adonis, p = 0.001, R = 0.64, permutational multivariate analysis of variance (PERMANOVA)) difference among the four treatments (Fig. S8b). The carboxylic acid (C) treated soil harbored the highest number of unique OTUs (250), followed by the Control soil (142), and soils with the addition of S (105) and A (89) (Fig. S8c). Based on the alluvial diagram, the relative abundances of TM7 and Proteobacteria were highest in sugar-infused soils and amino acids-infused soils, respectively (Fig. S8d).
We also explored the synergistic effects with the addition of three compound classes grouped together. Compared to the Control soil, the addition of all compounds did not significantly increase alpha diversity. Bacterial diversity was found to be decreased in all soils that received sugar, including the S, CS, and ACS treatments (Fig. 3a–c). Ordination using PCoA based on Bray-Curtis distances illustrated a significant (Adonis, p = 0.001, R = 0.69, PERMANOVA) difference among all treatments (Fig. 3d). A higher abundance of Proteobacteria was identified in treatment A, while a larger population of TM7 was associated with S (Fig. 3e). Cyanobacterial abundances were associated with treatment C and the Control, as compared to others (Fig. 3e). Bray-Curtis distances between each treatment and the Control were highest for soils with sugar addition, while soils amended with carboxylic acids were more closely related to the control (Fig. S9). When sugars or carboxylic acids were added with other classes, Bray-Curtis distances were different from those in soils amended with only S or C (Fig. S9). These results indicate that among all treatments, sugar addition strongly reduced microbial diversity and led to the highest dissimilarity in bacterial community composition, as compared to the Control. In contrast, the addition of carboxylic acids-addition resulted in the lowest distance between soil bacterial communities, as compared to the Control.
Fig. 3.
General descriptions of the soil bacterial communities. (a) - (c) illustrate the alpha diversity of soil bacterial communities. The Chao1, Shannon and Pielou indices were calculated with all OTUs. The horizontal bars within boxes represent the median. The tops and bottoms of boxes represent 75th and 25th quartiles, respectively. All outliers were plotted as individual points. Letters represent significant differences between groups (nonparametric t-test). (d) the principal coordinates analysis (PCoA) with Bray-Curtis dissimilarity performed on the taxonomic profile (at the OTU level) of bacterial communities. R- and p-values were evaluated via Adonis test. (e) The relative abundance (%) of the major phyla present in the bacterial communities. (f) The effects of soil chemical properties on the microbial communities using Redundancy analysis (RDA) analysis. (g) Pie charts showed the relative contribution of soil chemical properties to microbial communities and a permutation test result of each soil chemical property. (h) Co-occurrence networks of the abundance OTU (> 1%). n, node. L, line. Edges represent significant Spearman correlations (ρ > |0.8|, p < 0.05). Light blue lines represent a significant negative correlation and light red lines represent a significant positive.
To better reveal the effects of root exudate compositions on soil microbiome, abundant OTUs (number of 213; relative abundance > 0.1%) were selected for co-occurrence network analysis. Bacterial interactions of conditioned soils were illustrated with the Erdős-Rényi random network which exhibited various patterns (Fig. 3h, Table S9). Overall, the inputs of different root exudate compositions to soils did not alter network size (i.e., node number), while they markedly affected network connections (i.e., link number). Specifically, soils with amino acid addition contained more connections (link number: 2082) than other soils. In contrast, amino acid plus carboxylic acids-treated soils possessed the least number of connections (link number: 398). Among network connections, positive associations (positive correlations between links) were markedly larger than negative associations (negative correlations between links) (Table S9). Soils with carboxylic acids exhibited more clusters (34) and a larger negative correlation ratio (30%) than soils with other substances. The smallest number of clusters and a negative correlation ratio (16%) was associated in the soil with amino acid addition. A higher number of negative correlations was identified in the soils amended with a mixture of amino acids and sugars, such as treatments AS (34%) and ACS (33%), compared to soils with either amino acids or sugar alone. The trends of network connections (i.e., link number) were similar to those of clustering connectivity (i.e., the extent to which nodes are clustered) and centralization degree (Table S9).
Redundancy analyses (RDA) were used to illustrate the relationship between soil microbial community composition and DOC, aFe, and aSi in soils with the addition of one individual compound class. DOC, aFe and aSi were all found to impact bacterial community composition with a total of 26% explained (p < 0.001), including 16.7% from single factors and 9.3% from their interactions (Fig. S8e). Individually, soil DOC, aFe, and aSi explained 7.2%, 3.7%, and 5.8% of the total variation, respectively (Fig. S8f). Similar results were observed using RDA analysis on all samples. For example, the DOC, aFe, and aSi together explained 20% of the variation of the soil bacterial community, including 10.5% from single factors and 9.5% from interactions (Fig. 3g), with low contribution from the single factors of DOC (4.2%), aFe (1%), and aSi (4.3%). There was only a 3.6% contribution by pH to the overall variation (Fig. S10). These results suggested a minor effect of the pH and released DOC on soil bacterial community composition.
3.3. Linkage between bacterial groups and organic carbon mobility
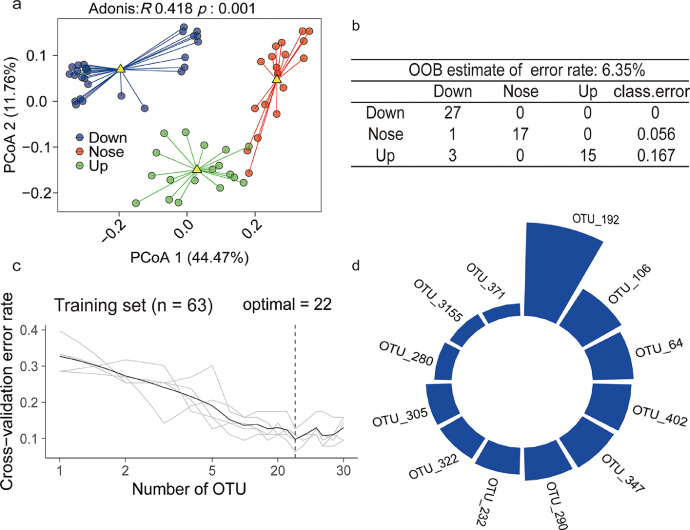
In order to evaluate the relationship between microbial community composition and soil carbon mobility, we divided all soils into three groups based on their relative DOC contents, as compared to the Control soils. This included group 1 (Up) with the soil DOC content significantly higher than that in Control soil; group 2 (Down) with soil DOC content significantly lower than that in Control; and group 3 (Nose) without a significant difference in DOC content between the treated soil and the Control. The PCoA, based on Bray-Curtis distances, indicated that these three groups significantly (Adonis, p = 0.001, R = 0.20, PERMANOVA) separated the composition of soil bacterial communities (Fig. 4a).
Fig. 4.
Differences in three bacterial communities and their characteristic OTUs. (a) The principal coordinates analysis (PCoA) with Bray-Curtis dissimilarity performed on the taxonomic profile (at the OTU level) of three groups. R- and p-values were evaluated via Adonis test. (b) The number of correct (59) and incorrect (4) predictions of DOC content, based on a random forest classification of the OTUs belonged to Actinobacteria. (c) Ten-fold cross-validation error as a function of the number of OTUs used to differentiate the Groups with different DOC contents. (d) The top 22 bacterial OTUs identified by applying RandomForest classification.
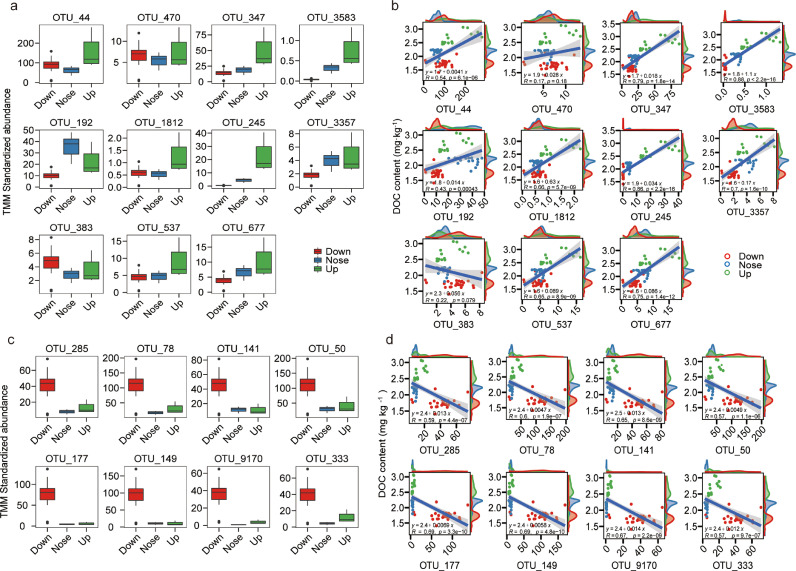
The Edger method was used to identify OTUs that were enriched in the Up group, in which 11 OTUs (0.6% of total reads) were identified and significantly (FDR < 0.05) correlated with DOC (Fig. 5a, Table S4). Five of these OTUs belonged to Actinobacteria and were classified as Nocardioidaceae (OTU_470), Mycobacteriaceae (OTU_347), Pseudonocardiaceae (OTU_3538), Nocardioidaceae (OTU_1812) and unknown (OTU_192; order: Actinomycetales) at the family level (Fig. 5a, Table S4). Linear corrections showed that the relative abundances of nine OTUs increased (p < 0.001) with soil DOC content (Fig. 5b). Eight OTUs were enriched in the Down group, which consisted of six groups belonging to the Deltaproteobacteria and Gammaproteobacteria (Fig. 5c). Linear correlations showed that the relative abundances of these eight OTUs decreased (p < 0.001) with an increase in DOC (Fig. 5d).
Fig. 5.
Response of bacterial groups to organic carbon mobilization. (a) The trimmed mean of M-values (TMM). The corrected relative abundance of OTUs belongs to Actinobacteria increased in the soil, where DOC content was significantly higher than control. (b) Linear regression analyses were performed for DOC and the TMM standardized abundance of OTUs belong to Actinobacteria. (c) The TMM corrected relative abundance of OTUs belongs to Actinobacteria increased in the soil, in which DOC content was significantly lower than control. (d) Linear regression analyses were performed for DOC content and the TMM standardized abundance of OTUs belong to Bacteriodetes and Gammaproteobacteria.
Random forest models, based on the OTU level, were used to classify the three DOC groups (Up, Down, Nose) in order to identify microbes responsible for soil DOC release. For these final classifier models an average accuracy of 85% was obtained (Table S6). This accuracy was lower for half of the samples from the Up group that could not be adequately classified (Table S6). Linear regression analyses indicated that 16S rRNA gene copies and alpha diversity indices were both independent of DOC content (Fig. S11). Random forest model analyses were then performed with OTUs from each phylum, which resulted in the highest average accuracy (92.8%) associated with OTUs belonging to Actinobacteria (Fig. 4b, Tables S7–S8). Ten-fold cross-validation with five repeats illustrated the importance of bacterial OTUs as potential indicators. The cross-validation error curve stabilized at the top 22 most relevant OTUs with three OTUs: OTU192 (o: Actinomycetales), OTU470 (f: Nocardioidaceae), and OTU347 (g: Mycobacterium) identified as the most important groups enriched with soil DOC (random forest model, Fig. 4b–d).
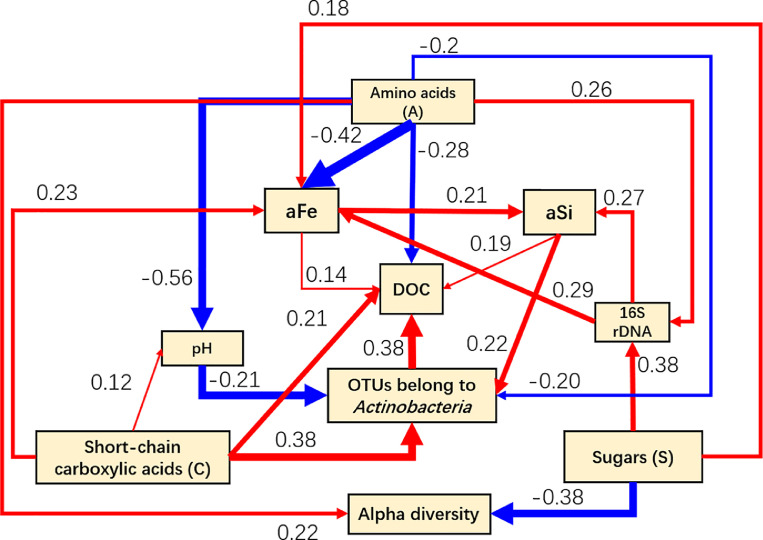
3.4. Partial least squares path modeling (PLSPM) showing the effects of bacterial communities on carbon mobilization
The complex relationships between Actinobacteria, aFe, aSi, and DOC were profiled in order to identify the most important properties that were associated with the responses to soil carbon. To achieve this, we conducted PLSPM analyses with the factors consisting of: applied compound classes, 16S rRNA gene copies, alpha diversity measures (Richness, Shannon, and Pielou index), identified OTUs (Actinobacteria), pH, aFe, and aSi (Fig. 6). Additions of root exudates were found to impact microbial community composition, aFe, and aSi. The model best fit was achieved with a GOF of 0.63. Root exudate inputs influenced DOC content directly (path coefficient of A= −0.28 and C = 0.21) or indirectly by its effect on soil properties (mainly soil pH, aFe, and Actinobacteria). Actinobacteria attributes (0.38) exerted a greater impact on soil DOC than on aFe (0.14) and aSi (0.19). Addition of exudate compounds to soil influenced aFe or aSi contents indirectly by shaping the total bacterial community abundance (copies of 16S rRNA; A: 0.26; S: 0.38).
Fig. 6.
Partial Least Squares Path Modeling (PLSPM). Red and blue lines represent positive and negative effects, respectively. Numbers on the lines in the PLSPM model are the ‘total effects’ values.
4. Discussion
4.1. Effects of root exudate compounds on bacterial communities and soil carbon cycling
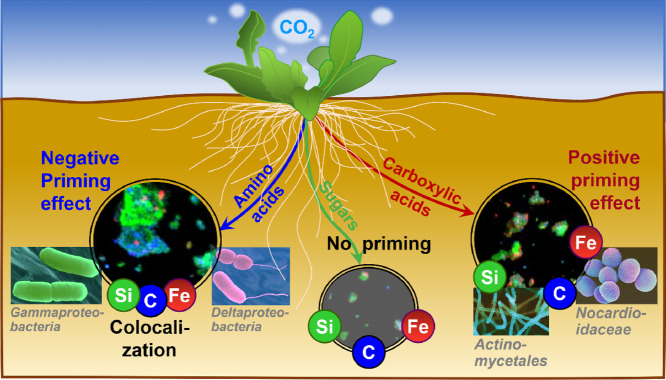
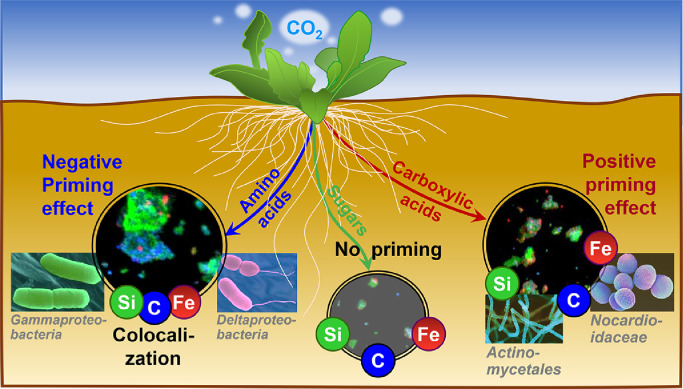
Multiple lines of existing evidence have shown that root exudates can alter i) the composition of bacterial communities [33] and ii) soil carbon cycling [14]. However, specific linkages among distinct category of root exudates, bacterial community composition, and soil carbon cycling remains largely unknown. Using a series of well-controlled incubation experiments combined with high-throughput sequencing and q-PCR, our results indicated that three groups of root exudates, including amino acids, short-chain carboxylic acids, and sugars, exerted distinct effects on the primed release of DOC and available Fe, and that the altered bacterial communities were the driving force behind these processes in the rhizosphere (Fig. 7). Specifically, the addition of short-chain carboxylic acids (C) had a negative priming effect on the release of DOC and available Fe, whereas the addition of amino acids (A) increased both. In contrast, sugars (S) induced an inconspicuous priming effect. Both DOC and available Fe were found to be positively correlated, indicating that carbon loss was accompanied by aFe release (Fig. S3). This is supported by earlier evidence where an enhanced release of organic acids within the rhizosphere increased the solubility of poorly available Fe pools [34] and where carboxylic acids from root exudates were found to release soil carbon by counteracting mineral protection [7]. However, the effects of other groups of root exudates, e.g., amino acids and sugars, on mineral-protected carbon within soil remain poorly understood. In contrast to the positive effect of carboxylic acids on soil organic carbon mobilization, amino acids exerted a negative effect, whereas sugars exhibited no effect (Fig. 1).
Fig. 7.
Conceptual model illustrating the role of root exudate chemistry in driving microbial community assembly and soil organic carbon mobilization in the rhizosphere.
Due to the complicated effects of phyto-metabolites on the rhizosphere, it is currently necessary to select the most abundant root-exuded compounds in order to approximate the rhizosphere environment [7]. However, it should be noted that the three selected categories of organic compounds used in the current study are only a fraction of the total root exudates that are dynamic across spatiotemporal scales and much more diverse than these simplified categories. In addition, the composition of root exudates has been reported to be closely related to plant species and their growth stages [35,36], thus imparting another variable. Thus, the changes caused by the addition of amino acids, carboxylic acids, and sugars found here may be distinct from those associated with roots from different plants and also likely impacted by soil edaphic properties.
4.2. Linking compound class‐specific priming effects to soil carbon mobilization
Plant-derived carbon inputs such as root exudates are closely tied to the “priming effect” [37]. However, root exudates can result in a positive or negative priming effect on soil carbon storage [38], varying from −70% to 380% [39]. Our incubation experiments showed the highest and the lowest DOC content in soil with the addition of carboxylic acids and amino acids, respectively (Fig. 1), attesting to a compound class-specific priming on soil carbon decomposition. These priming effects may result from the competition for energy and nutrient acquisition between the microorganisms specialized in the decomposition of fresh or polymerized organic matter [40]. Indeed, the response of DOC and aFe to the addition of root-exudates was closely related to the bioavailability/reactivity and energy source of the root exudates for microorganisms (Fig. 1). For example, short-chain carboxylic acids are principally metabolic by-products of microorganism metabolism. The addition of carboxylic acids did not seem to provide much metabolizable energy for bacteria, but rather forces them to utilize metabolizable soil organic matter which resulted in the release of DOC. In contrast, the addition of sugars provided a more bioavailable or metabolizable energy source which reduced the decomposition of soil organic matter and thus the release of soil DOC. In contrast, the addition of amino acids, although also highly bioavailable to microorganisms, resulted in a significant decrease in soil pH (Fig. S1), which likely altered or depressed microbial activity or metabolic pathways, resulting in a lower soil DOC release. Root exudates have been shown to lead to the activation of some bacterial populations which leads to additional DOC decomposition, possibly through the production of reactive oxygen species [41], [42], [43], [44]. In addition, DOC was usually found to be positively correlated with available Fe content and negatively correlated to soil pH [45]. However, our PLSPM analyses showed that soil pH had no direct or indirect effects on DOC, aFe, and aSi (Fig. 6), which may be related to a strong buffer ability of soil matrix [46]. Overall, the addition of amino acids may indirectly decrease soil DOC content by lowering the soil pH and decreasing available Fe, while the opposite was associated with the addition of carboxylic acids.
4.3. The underlying biological mechanisms of distinct category of root exudates on soil carbon mobilization
Combined with bulk analysis, spatial localization, and high-throughput sequencing, our findings demonstrate that in soils with carboxylic acids addition, higher bacterial richness index (Fig. 3) is one of the driving forces that promotes the release of mineral-protected carbon (12C−) (Fig. 2) by mobilizing more DOC and Fe (Fig. 1). This result may be related to the fact that soil microorganisms have a high demand for soluble low molecular weight carbon and N [47]. The major advantages of the NanoSIMS instrument over other microscopic techniques lie in [48]: (1) elemental mapping can be done with better lateral resolution, (2) the low depth penetration (∼10–20 nm) of the NanoSIMS primary beam allows thin surface layers to be examined, and (3) highly accurate isotope detection allows the operator to track isotopic labeling organics onto distinct minerals in an intact micro-environment and thus enables process-level studies. However, the two major disadvantages of NanoSIMS are that (1) it only detects very small scale images and thus much more replicate images are needed to acquire reliable conclusions; and (2) the mass difference between species must be smaller than 22 (i.e., Mmax/Mmin<=22) [49]. Bacterial communities in soils are always limited by carbon [37] and play an essential role in the turnover of organic carbon and other nutrients (e.g., N, P, Fe, Si, and Ca) [50]. Our results are directly supported by an earlier study where oxalic acid, a carboxylic acid, derived from root exudates was shown to accelerate soil carbon mineralization in the rhizosphere [7], though the underlying biological mechanisms were un-resolved in that study.
The impact of sugar addition on soil microbiomes are inconsistent [4,5], which may be related to distinct soil types that harbor various microbiomes [4,5] and substrate pulses [5]. Compared to the Control soil, addition of all root exudates (especially sugars) reduced microbial diversity (Fig. 3a). This diversity reduction is consistent with a previous report [51], strongly indicating that decreases in microbial diversity in the rhizosphere is a common phenomenon, at least temporarily across temporal scales. The distinct effects of the different applied root exudate classes on reduced microbial diversity, as compared to the Control (Fig. S9), may be attributed to the fact that sugars can be utilized by the majority of microorganisms which depresses their utilization of soil organic matter. In contrast, carboxylic acids can only be assimilated by some bacterial groups [52], forcing the remaining microbes to decompose soil organic matter. Similarly, sugars (glucose, sucrose, and fructose) were found to decrease bacterial community richness in a greater degree than carboxylic acids (quinic, lactic, and maleic acids) [53]. Therefore, a strongly reduced microbial diversity with sugar addition is likely explained by their availability to a wide range of microorganisms. We also identified a greater number of connections (edge number: 2082) within bacterial networks with sugar addition (Fig. 3). A similar result identified a higher network complexity after the addition of sugars in a Fusarium wilt pathogen-infested soil [54].
It is clear that the application of amino acids increased the relative abundance of Proteobacteria and reduced soil carbon mobilization partly through the associated decrease in soil pH (Fig. 4). Conversely, the application of carboxylic acids increased the release of DOC and Fe directly, through the breakage of organo-mineral associations (Fig. 2), and indirectly, via stimulation Actinobacteria (Fig. 6). Five Actinobacterial OTUs, classified as Mycobacteriaceae, Pseudonocardiaceae, and Nocardioidaceae, increased with DOC content (Fig. 5a, Table S4). Actinobacteria are known to have the ability to release soil carbon or increase carbon bioavailability across various soils [55], reinforcing the idea that Actinobacteria may be associated with the solubilization of organo-mineral associations. In addition, higher soil DOC has been concurrently identified with more abundant Actinobacteria [56] with reduced carbon availability associated with decreased Actinobacteria abundance [55]. Along these lines, within the rhizosphere of wild oat, DOC content was found to be increased with Actinobacteria abundance [57]. Due to their ability to conserve metabolism and lie dormant under dry conditions, Actinobacteria persist well in drought-impacted soils [55] and have been used as predictive markers of disease-suppressive soils at the continental scale [58]. In addition to Actinobacteria, the abundance of other several OTUs, e.g., Bacteroidetes (OTU_50, OTU_177) and Gammaproteobacteria (OTU_149, OTU_333), decreased with increasing DOC content (Fig. 5d; Table S5), suggesting a negative rhizosphere priming effect.
Taken together, our findings provide a systematic illustration that functional groups of root exudates have distinct effects on the biogeochemical cycling of carbon and mineral elements through re-structuring the composition of soil bacterial communities (Fig. 7). These findings have several important implications. First, the rhizosphere represents a critical hotspot for biogeochemical cycling of carbon and other bioactive elements within terrestrial ecosystems [3,14,37]. The active microorganisms within these rhizosphere hotspots whose metabolisms are driven by root exudates are ∼2–20 times higher in abundance than in the bulk soil [59]. Therefore, knowledge on the effects of molecular composition of root exudates on bacterial communities is essential to improve our understanding of the patterns of microbial community assembly and functions in the rhizosphere and the biogeochemical cycling of bioactive elements, such as carbon, N, P, Fe, and others. Second, given that climate change and elevated CO2 increase the root exudation of organic compounds into soils [60], there is an urgent need to further understand changes in microbial community composition, the effects of root exudate species and composition on plant-microbial interactions under future scenarios. Lastly, plant growth-promoting rhizobacteria (PGPR) have multiple beneficial effects on plant growth [61]. Thus, understanding the underlying mechanisms of root exudate chemistry that recruit and shape PGPR populations is critical, especially in the context of sustainable ecological agriculture, as it is central to the breeding of root-microbial partnerships for plant health and productivity [62].
5. Conclusion
Combining high-throughput sequencing, q-PCR, and NanoSIMS analyses, we characterized the effects of three groups of root exudate compound classes on bacterial community composition and abundance and analyzed consequences for mineral-protected carbon within the soil. The three common groups of root exudates examined imparted contrasting effects on the release of DOC and bioavailable Fe in an Ultisol by disrupting organo-mineral associations and influencing the soil bacterial communities. In the mineral particles, Fe and Si colocalized much more mineral-associated organic carbon after amino acid inputs than those without exudates or with carboxylic acids. Among these root exudates, sugars strongly reduced microbial diversity and led to the largest difference in bacterial community composition, as compared to the control soil. In contrast, the addition of carboxylic acids resulted in the smallest impact on soil bacterial community composition. Furthermore, carboxylic acids increased the abundance of Actinobacteria and facilitated organic carbon mobilization, whereas amino acids increased Proteobacteria that prevented DOC release. Overall, this unique combination of physical, chemical and molecular-biology state-of-the-art approaches allowed for deep insights into the mechanisms that underlie priming effects and the biogeochemical cycling of soil organic carbon, Fe, and Si in the rhizosphere.
Data availability
All the raw sequencing data were deposited (PRJCA001214) in the Genome Sequence Archive in the BIG Data Center and the accession codes CRA001372 for sequencing data of bacterial 16S rRNA gene. All scripts for computational analysis and corresponding raw data are available at https://github.com/taowenmicro/2021_Root-Exudate_TaoWen.etal.
Declaration of Competing Interest
The authors declare that they have no conflicts of interest in this work.
Acknowledgments
This work is supported by National Natural Science Foundation of China (Grants No. 31902107 and 41977271), Natural Science Foundation of Jiangsu Province (Grant No. BK20211577), the Innovative Research Team Development Plan of the Ministry of Education of China (Grant No. IRT_17R56). J. Y. was supported by Qing Lan Project of Jiangsu Province. Y.K. thanks for the support by the RUDN University Strategic Academic Leadership Program. T.W. thanks the WeChat subscription ID “meta-Genome” and “Micro-Bioinformatics and microflora” for the analysis methods.
Biographies

Tao Wen is a Ph.D. candidate at the Research Center for Organic-based Fertilizers, Nanjing Agricultural University. His-major research interests are the meta-analysis of Environmental Microbiome data, the workflow of microbiome data development, and machine learning analysis.

Jun Yuan is an associate professor at the Research Center for Organic-based Fertilizers, Organic Solid Waste Utilization, Nanjing Agricultural University. His-major research interests are plant-microbe interactions via root exudates, meta-analysis of environmental microbiome data, and multiple-omics analysis of environmental microbiome.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fmre.2021.12.016.
Appendix. Supplementary materials
References
- 1.Lynch J.M., Whipps J.M. Substrate flow in the rhizosphere. Plant Soil. 1990;129:1–10. [Google Scholar]
- 2.Haichar F.E., Santaella C., Heulin T., Achouak W. Root exudates mediated interactions belowground. Soil Biol. Biochem. 2014;77:69–80. [Google Scholar]
- 3.Zhalnina K., Louie K.B., Hao Z., et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018;3:470–480. doi: 10.1038/s41564-018-0129-3. [DOI] [PubMed] [Google Scholar]
- 4.Kamilova F., Kravchenko L.V., Shaposhnikov A.I., et al. Organic acids, sugars, and l-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol. Plant Microbe Interact. 2006;19:250–256. doi: 10.1094/MPMI-19-0250. [DOI] [PubMed] [Google Scholar]
- 5.Mau R.L., Liu C.M., Aziz M., et al. Linking soil bacterial biodiversity and soil carbon stability. ISME J. 2015;9:1477–1480. doi: 10.1038/ismej.2014.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haichar F.e.Z., Marol C., Berge O., et al. Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2008;2:1221–1230. doi: 10.1038/ismej.2008.80. [DOI] [PubMed] [Google Scholar]
- 7.Keiluweit M., Bougoure J.J., Nico P.S., et al. Mineral protection of soil carbon counteracted by root exudates. Nat. Clim. Change. 2015;5:588–595. [Google Scholar]
- 8.Chaparro J.M., Badri D.V., Bakker M.G., et al. Root exudation of phytochemicals in arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS ONE. 2013;8:e55731. doi: 10.1371/journal.pone.0055731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan J., Zhang N., Huang Q., et al. Organic acids from root exudates of banana help root colonization of PGPR strain Bacillus amyloliquefaciens NJN-6. Sci. Rep. 2015;5:13438. doi: 10.1038/srep13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vílchez S., Molina L., Ramos C., et al. Proline catabolism by Pseudomonas putida: cloning, characterization, and expression of the put genes in the presence of root exudates. J. Bacteriol. 2000;182:91–99. doi: 10.1128/jb.182.1.91-99.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lugtenberg B.J.J., Dekkers L., Bloemberg G.V. Molucular determinants of rhizosphere colonization by Pseudomonas. Annu. Rev. Phytopath. 2001;39:461–490. doi: 10.1146/annurev.phyto.39.1.461. [DOI] [PubMed] [Google Scholar]
- 12.Williams A., Langridge H., Straathof A.L., et al. Comparing root exudate collection techniques: an improved hybrid method. Soil Biol. Biochem. 2021;161 doi: 10.1016/j.soilbio.2021.108391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malik A.A., Martiny J.B.H., Brodie E.L., et al. Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change. ISME J. 2020;14:1–9. doi: 10.1038/s41396-019-0510-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuzyakov Y., Friedel J.K., Stahr K. Review of mechanisms and quantification of priming effects. Soil Biol. Biochem. 2000;32:1485–1498. [Google Scholar]
- 15.Arredondo M.G., Lawrence C.R., Schulz M.S., et al. Root-driven weathering impacts on mineral-organic associations in deep soils over pedogenic time scales. Geochim. Cosmochim. Ac. 2019;263:68–84. [Google Scholar]
- 16.Kramer M.G., Chadwick O.A. Climate-driven thresholds in reactive mineral retention of soil carbon at the global scale. Nat. Clim. Change. 2018;8:1104–1108. [Google Scholar]
- 17.Philippot L., Raaijmakers J.M., Lemanceau P., et al. Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013;11:789–799. doi: 10.1038/nrmicro3109. [DOI] [PubMed] [Google Scholar]
- 18.Kuzyakov Y. Priming effects: interactions between living and dead organic matter. Soil Biol. Biochem. 2010;42:1363–1371. [Google Scholar]
- 19.Neumann G., Roemheld V. CRC Press; 2007. The Rhizosphere: Biochemistry and Organic Substances At the Soil-Plant Interface. [Google Scholar]
- 20.Yuan J., Zhao J., Wen T., et al. Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome. 2018;6:156. doi: 10.1186/s40168-018-0537-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walters W., Hyde E.R., Berg-Lyons D., et al. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems. 2016;1:e00009–e00015. doi: 10.1128/mSystems.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan J., Wen T., Zhang H., et al. Predicting disease occurrence with high accuracy based on soil macroecological patterns of Fusarium wilt. ISME J. 2020;14:2936–2950. doi: 10.1038/s41396-020-0720-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liaw A., Wiener M. Classification and regression by randomForest. R. News. 2002;2:18–22. [Google Scholar]
- 24.Smith M., Rocha A.M., Smillie C., et al. Natural bacterial communities serve as quantitative geochemical biosensors. MBio. 2015;6 doi: 10.1128/mBio.00326-15. e00326-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao M., Jun Y., Zongzhuan S., et al. Predominance of soil vs root effect in rhizosphere microbiota reassembly. FEMS Microbiol. Ecol. 2019;95:fiz139. doi: 10.1093/femsec/fiz139. [DOI] [PubMed] [Google Scholar]
- 26.Yu G., Xiao J., Hu S., et al. Mineral availability as a key regulator of soil carbon storage. Environ. Sci. Technol. 2017;51:4960–4969. doi: 10.1021/acs.est.7b00305. [DOI] [PubMed] [Google Scholar]
- 27.Sun R., Zhang X.X., Guo X., et al. Bacterial diversity in soils subjected to long-term chemical fertilization can be more stably maintained with the addition of livestock manure than wheat straw. Soil Biol. Biochem. 2015;88:9–18. [Google Scholar]
- 28.Caporaso J.G., Kuczynski J., Stombaugh J., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugawara M., Epstein B., Badgley B.D., et al. Comparative genomics of the core and accessory genomes of 48 Sinorhizobium strains comprising five genospecies. Genome Biol. 2013;14:1–20. doi: 10.1186/gb-2013-14-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dixon P. VEGAN, a package of R functions for community ecology. J. Veget. Sci. 2003;14:927–930. [Google Scholar]
- 31.Csardi G., Nepusz T. The igraph software package for complex network research. InterJournal Complex Syst. 2006;1695:1–9. [Google Scholar]
- 32.Russolillo Giorgio. Non-metric partial least squares. Electron. J. Statist. 2012;6:1641–1669. [Google Scholar]
- 33.Henry S., Texier S., Hallet S., et al. Disentangling the rhizosphere effect on nitrate reducers and denitrifiers: insight into the role of root exudates. Environ. Microbiol. 2008;10:3082–3092. doi: 10.1111/j.1462-2920.2008.01599.x. [DOI] [PubMed] [Google Scholar]
- 34.Mimmo T., Del Buono D., Terzano R., et al. Rhizospheric organic compounds in the soil–microorganism–plant system: their role in iron availability. Europ. J. Soil Sci. 2014;65:629–642. [Google Scholar]
- 35.Zhao M., Zhao J., Yuan J., et al. Root exudates drive soil-microbe-nutrient feedbacks in response to plant growth. Plant Cell Environ. 2021;44:613–628. doi: 10.1111/pce.13928. [DOI] [PubMed] [Google Scholar]
- 36.Liao Q., Liu H., Lu C., et al. Root exudates enhance the PAH degradation and degrading gene abundance in soils. Sci. Tot. Environ. 2021;764 doi: 10.1016/j.scitotenv.2020.144436. [DOI] [PubMed] [Google Scholar]
- 37.Kuzyakov Y. Priming effects: interactions between living and dead organic matter. Soil Biol. Biochem. 2010;42:1363–1371. [Google Scholar]
- 38.Hamer U., Marschner B. Priming effects in different soil types induced by fructose, alanine, oxalic acid and catechol additions. Soil Biol. Biochem. 2005;37:445–454. [Google Scholar]
- 39.Cheng W., Parton W.J., Gonzalez-Meler M.A., et al. Synthesis and modeling perspectives of rhizosphere priming. New Phytolog. 2014;201:31–44. doi: 10.1111/nph.12440. [DOI] [PubMed] [Google Scholar]
- 40.Fontaine S., Mariotti A., Abbadie L. The priming effect of organic matter: a question of microbial competition? Soil Biol. Biochem. 2003;35:837–843. [Google Scholar]
- 41.el Z. Haichar F., Marol C., Berge O., et al. Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2008;2:1221–1230. doi: 10.1038/ismej.2008.80. [DOI] [PubMed] [Google Scholar]
- 42.Kleber M., Bourg I.C., Coward E.K., et al. Dynamic interactions at the mineral–organic matter interface. Nat. Rev. Earth Environ. 2021;2:402–421. [Google Scholar]
- 43.Yu G.-.H., Kuzyakov Y. Fenton chemistry and reactive oxygen species in soil: abiotic mechanisms of biotic processes, controls and consequences for carbon and nutrient cycling. Earth Sci. Rev. 2021;214 [Google Scholar]
- 44.Chi Z.-.L., Yu G.-.H. Nanozyme-mediated elemental biogeochemical cycling and environmental effects. Sci. China Earth Sci. 2021;64:1015–1025. [Google Scholar]
- 45.Marschner H., Römheld V. In vivo measurement of root-induced pH changes at the soil-root interface: effect of plant species and nitrogen source. Zeitschrift für Pflanzenphysiologie. 1983;111:241–251. [Google Scholar]
- 46.Yu G.-.H., Kuzyakov Y., Luo Y., et al. Molybdenum bioavailability and asymbiotic nitrogen fixation in soils are raised by iron (oxyhydr)oxide-mediated free radical production. Environ. Sci. Technol. 2021;55:14979–14989. doi: 10.1021/acs.est.1c04240. [DOI] [PubMed] [Google Scholar]
- 47.Coody P.N., Sommers L.E., Nelson D.W. Kinetics of glucose uptake by soil microorganisms. Soil Biol. Biochem. 1986;18:283–289. [Google Scholar]
- 48.Mueller C.W., Weber P.K., Kilburn M.R., et al. Advances in the Analysis of Biogeochemical Interfaces: nanoSIMS to Investigate Soil Microenvironments. Adv. Agronomy. 2013;121:1–46. [Google Scholar]
- 49.Yang W., Hu S., Zhang J., et al. NanoSIMS analytical technique and its applications in earth sciences. Sci. China Earth Sci. 2015;10:1758–1767. [Google Scholar]
- 50.De Nobili M., Contin M., Mondini C., et al. Soil microbial biomass is triggered into activity by trace amounts of substrate. Soil Biol. Biochem. 2001;33:1163–1170. [Google Scholar]
- 51.Shi S., Nuccio E., Herman D.J., et al. Successional trajectories of rhizosphere bacterial communities over consecutive seasons. MBio. 2015;6 doi: 10.1128/mBio.00746-15. e00746-00715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Landi L., Valori F., Ascher J., et al. Root exudate effects on the bacterial communities, CO2 evolution, nitrogen transformations and ATP content of rhizosphere and bulk soils. Soil Biol. Biochem. 2006;38:509–516. [Google Scholar]
- 53.Shi S., Richardson A.E., O'Callaghan M., et al. Effects of selected root exudate components on soil bacterial communities. FEMS Microbiol. Ecol. 2011;77:600–610. doi: 10.1111/j.1574-6941.2011.01150.x. [DOI] [PubMed] [Google Scholar]
- 54.Ren G., Meng T., Ma Y. Sugars altered fungal community composition and caused high network complexity in a Fusarium wilt pathogen-infested soil. Biol. Fert. Soils. 2020;56:395–409. [Google Scholar]
- 55.Jansson J.K., Hofmockel K.S. Soil microbiomes and climate change. Nat. Rev. Microbiol. 2020;18:35–46. doi: 10.1038/s41579-019-0265-7. [DOI] [PubMed] [Google Scholar]
- 56.Liu X., Lamb E.G., Zhang S. Nitrogen addition impacts on soil microbial stoichiometry are driven by changes in plant resource stoichiometry not by the composition of main microbial groups in an alpine meadow. Biol. Fert. Soils. 2020;56:261–271. [Google Scholar]
- 57.DeAngelis K.M., Brodie E.L., DeSantis T.Z., et al. Selective progressive response of soil microbial community to wild oat roots. ISME J. 2009;3:168–178. doi: 10.1038/ismej.2008.103. [DOI] [PubMed] [Google Scholar]
- 58.Trivedi P., Leach J.E., Tringe S.G., et al. Plant–microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 2020;18:607–621. doi: 10.1038/s41579-020-0412-1. [DOI] [PubMed] [Google Scholar]
- 59.Kuzyakov Y., Blagodatskaya E. Microbial hotspots and hot moments in soil: concept & review. Soil Biol. Biochem. 2015;83:184–199. [Google Scholar]
- 60.Usyskin-Tonne A., Hadar Y., Yermiyahu U., et al. Elevated CO2 and nitrate levels increase wheat root-associated bacterial abundance and impact rhizosphere microbial community composition and function. ISME J. 2020;15:1073–1084. doi: 10.1038/s41396-020-00831-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lugtenberg B., Kamilova F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 62.Beattie G.A. Metabolic coupling on roots. Nat. Microbiol. 2018;3:396–397. doi: 10.1038/s41564-018-0139-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the raw sequencing data were deposited (PRJCA001214) in the Genome Sequence Archive in the BIG Data Center and the accession codes CRA001372 for sequencing data of bacterial 16S rRNA gene. All scripts for computational analysis and corresponding raw data are available at https://github.com/taowenmicro/2021_Root-Exudate_TaoWen.etal.