Abstract
MicroRNAs (miRNAs) are short endogenous non-coding RNAs that regulate gene expression at the post-transcriptional level in a broad range of eukaryotic species. In animals, it is estimated that more than 60% of mammalian genes are targets of miRNAs, with miRNAs regulating cellular processes such as differentiation and proliferation. In plants, miRNAs regulate gene expression and play essential roles in diverse biological processes, including growth, development, and stress responses. Arabidopsis mutants with defective miRNA biogenesis are embryo lethal, and abnormal expression of miRNAs can cause severe developmental phenotypes. It is therefore crucial that the homeostasis of miRNAs is tightly regulated. In this review, we summarize the key mechanisms of plant miRNA biogenesis and stabilization. We provide an update on nuclear proteins with functions in miRNA biogenesis and proteins linking miRNA biogenesis to environmental triggers.
Keywords: microRNAs, Biogenesis, Processing, AGO1, pri-miRNA
1. Introduction
microRNAs (miRNAs) are ∼20-to-24 nucleotide (nt) long endogenous non-coding RNAs that play an essential role in regulating gene expression in plants and animals. In 1993, lin-4, the first miRNA ever characterized, was identified as a regulator of developmental timing in Caenorhabditis elegans, with partial antisense complementarity to the lin-14 3′UTR [1,2]. In 2000, the second miRNA, let-7, was discovered in C. elegans through a similar approach [3]. lin-4 and let-7 are endogenous short RNAs that are 21 nt and 22 nt long, respectively. Both are partially complementary to the 3′ untranslated regions of their target protein-coding genes and cause degradation and translational inhibition of the messenger RNAs (mRNAs) [1,3]. Phylogenetic analysis of the let-7 RNA sequence revealed its conservation among bilaterian animals, such as humans and the fly Drosophila melanogaster, suggesting that similar gene expression regulatory mechanisms could be adopted by diverse species [4]. In 2001, in a multimodal approach involving bioinformatics and cDNA cloning, many small endogenous RNAs with shared features to lin-4 and let-7 were reported in Multiple species, including C. elegans, Drosophila and humans, and named miRNAs [5], [6], [7]. Arabidopsis miRNAs were reported in 2002 [8], [9], [10]. With the application of high-throughput sequencing in the past decades, the number of miRNA species has dramatically expanded. According to miRBase (v.22.1), the number of mature miRNAs is more than 2600 in humans, 430 in C. elegans, 460 in D. melanogaster, 420 in Arabidopsis thaliana, 320 in Zea mays, and 730 in Oryza sativa, and these numbers continue to grow. One caveat, however, is that the numbers may be inflated due to the mis-annotation of endogenous small interfering RNAs (siRNAs) as miRNAs in some cases in plant species [11]. In addition to eukaryotic species, miRNAs have also been found in viruses [12].
In plants, miRNAs are selectively incorporated into an ARGONAUTE (AGO) protein to form a miRNA-induced silencing complex (miRISC) [13,14]. The majority of plant miRNAs are preferentially loaded into AGO1, which prefers miRNAs with a 5′ uridine (U) [15]. miRISC then binds to the target messenger RNAs that contain sequences complementary to the miRNA, thus inducing degradation or translational repression of the target transcripts. In addition to regulating gene expression post-transcriptionally, certain 22-nt plant miRNAs are also capable of triggering the biogenesis of secondary siRNAs from protein-coding genes or long non-coding RNAs in a phased pattern. siRNAs produced through this pathway are referred to as phasiRNAs [16]. miRNA-mediated post-transcriptional gene silencing regulates critical biological processes in plants, including growth, development, hormone signaling, and biotic and abiotic stress responses, thus it is vital to tightly control the abundance of miRNAs. The steady-state level of a miRNA reflects the balance between biogenesis, stabilization, and turnover.
2. Plant miRNA biogenesis and regulation
Tens to hundreds of miRNAs have been identified in various plant species, and the vast majority of them are encoded by MIR genes located in intergenic regions [17]. In Arabidopsis the 420 MIR genes belong to ∼200 families (miRbase, v22.1), with most families being represented by a single member. Unlike their counterparts in metazoans, plant MIR genes are rarely clustered in the genome [17].
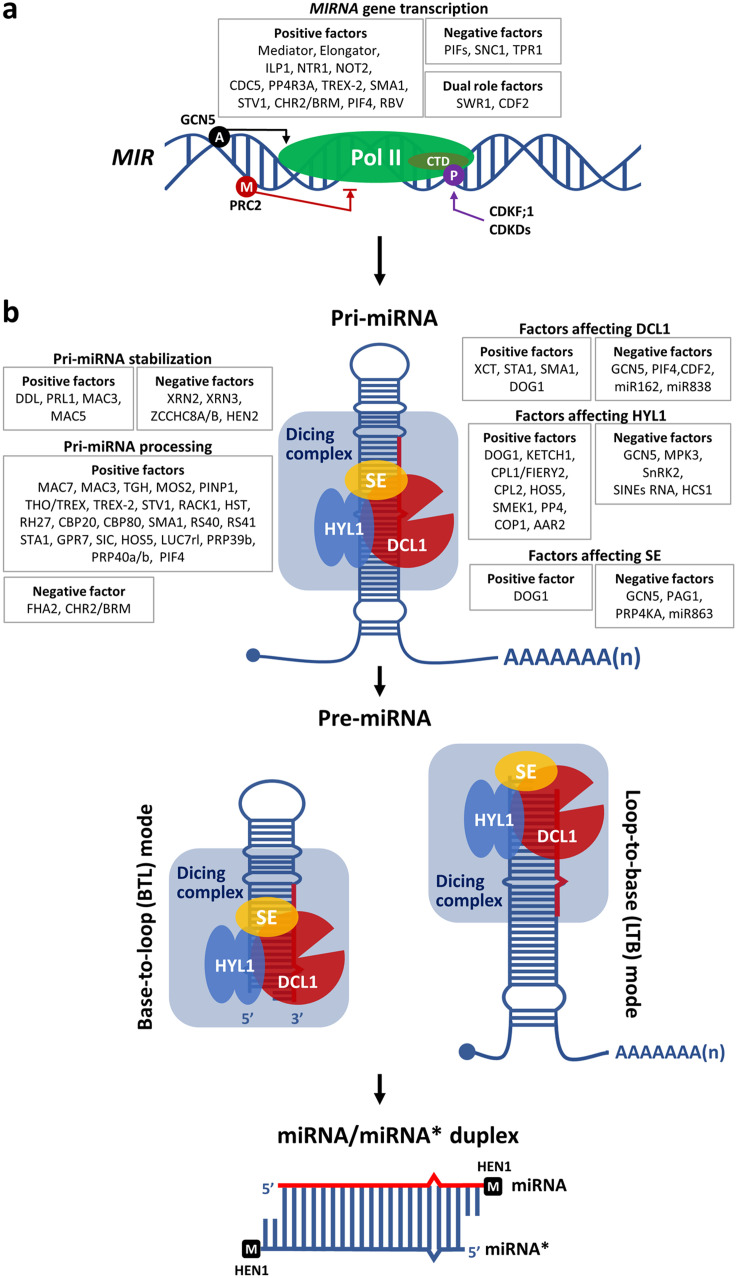
The biogenesis of plant miRNAs is composed of three major steps. A MIR gene is first transcribed by Pol II into a long pri-miRNA, which folds into a stem-loop structure (Fig.1a) [18]. The pri-miRNA is then recognized and processed by the dicing complex into a miRNA/miRNA* duplex with 2-nt overhangs at the 3′ ends [8,19,20]. Next, the miRNA/miRNA* duplex undergoes 2′-O-methylation at the 3′ ends by HUA ENHANCER1 (HEN1) (Fig.1b) [21]. The nuclear endonuclease DCL1 [8,19,20], the double-stranded RNA binding protein HYPONASTIC LEAVES 1 (HYL1) [22,23], also known as double-stranded RNA-binding protein1 (DRB1), and the zinc finger protein SERRATE (SE) [24,25] form the dicing complex. In this section, we will review the miRNA biogenesis pathway and summarize recent findings.
Fig. 1.
Mechanisms for the biogenesis of plant miRNAs. (a) MIR gene transcription. RNA polymerase II (Pol II)-mediated MIR gene transcription is regulated positively and/or negatively by protein factors or other mechanisms. Phosphorylation of Pol II at the C-terminal domain (CTD) of its largest subunit by CDKF;1 and CDKDs promotes MIR gene transcription. Phosphorylation is indicated by “P” in a purple circle. Histone acetylation by GCN5 and methylation by PRC2 at MIR loci are indicated by “A” in a black circle and “M” in a red circle, respectively. (b) Pri-miRNA processing in the base-to-loop (BTL) and loop-to-base (LTB) mode. The capped and poly-A tailed pri-miRNA folds into a stem-loop structure that harbors mismatches, and is cleaved by the dicing complex into precursor miRNA (pre-miRNA). The pre-miRNA is further cleaved by the dicing complex to release an approximately 21-nt long miRNA/miRNA* duplex with a 2-nt 3′ overhang on each strand. The duplex is 2′-O-methylated at the 3′ ends by HEN1 (the methyl group is marked with “M” in a black square). The miRNA strand is highlighted in red.
2.1. MIR gene transcription
The transcription of MIR genes into pri-miRNAs is by Pol II, thus the pri-miRNA transcripts are capped and poly-A tailed [18,26,27]. While most MIR genes that reside in the non-coding regions between genes are independently transcribed, others are found within introns of protein-coding or non-coding genes, such as MIR400 and MIR402, and are co-transcribed with their host genes by Pol II and regulated by RNA splicing [28,29].
As with protein-coding genes, the transcription of MIR genes is regulated by both cis- and trans-regulatory modules, and general regulatory mechanisms that control protein-coding gene transcription might also apply to MIR gene transcription. For instance, general cis- regulators, such as the TATA box, are located upstream of the transcription start sites of MIR genes [18]. Binding sites for the transcription factors MYC2, ARF, SORLREP3, and LFY are overrepresented in MIR promoters [30]. In addition, phosphorylation of the C-terminal domain (CTD) of the largest subunit of Pol II governed by CYCLIN-DEPENDENT KINASE F;1 (CDKF;1) and CYCLIN-DEPENDENT KINASE Ds (CDKDs) promotes pri-miRNA capping and polyadenylation [31]. A panel of trans-regulatory modules promotes the transcription of MIR genes by regulating Pol II occupancy at the MIR loci. Trans-regulatory module mutants show reduced levels of pri-miRNAs, including those of the Mediator complex subunit MED20a [32], the Elongator complex members ELP2 and ELP5 [33], two intron-lariat spliceosome (ILS) complex disassembly factors POLYPLOIDY1–1D (ILP1) and NTC-Related protein 1 (NTR1) [34], the transcription factor NEGATIVE ON TATA LESS 2 (NOT2) [35], the MOS4-ASSOCIATED COMPLEX (MAC) subunit CELL DIVISION CYCLE 5 (CDC5) [36], the chromatin-associated protein REGULATORY SUBUNIT 3A (PP4R3A) of the PROTEIN PHOSPHATASE4 (PP4) complex [37], the Three PRIME REPAIR EXONUCLEASE 2 (TREX-2) complex core components THO/HRP1 PHENOTYPE 1 (THP1) and SUPPRESSOR OF ACTIN 3A (SAC3A) [38], the splicing factor SMALL1 (SMA1) [39], the ribosomal protein SHORT VALVE 1 (STV1) [40], and a WD40 domain protein REDUCTION IN BLEACHED VEIN AREA (RBV) [41].
It is important to note that many MIR gene transcriptional regulators also interact with the dicing complex members, indicating they might have dual roles and/or promote the co-transcriptional processing of the pri-miRNAs. For example, NOT2, TREX-2 complex, ILP1, and NTR1 are found to interact with both Pol II and the dicing complex [34,35,38]. PP4 and the MAC subunit CDC5 are associated with MIR promoters, Pol II, and the dicing complex [36,37].
2.2. Developmental stages affect the transcription of MIR genes
While some transcription factors have a general impact on the transcription of MIR genes, others positively and/or negatively regulate individual MIR genes at a specific developmental stage. Two B3 domain family transcription factors FUSCA3 (FUS3) and ABSCISIC ACID INSENSETIVE3 (ABI3) directly promote MIR156 transcription during the embryo-to-seedling transition and early stages of seed development, respectively [42,43]. Unlike FUS3, which functions primarily as a transcriptional activator, ABI3 represses MIR156D during later seed development and downregulates MIR160B, which regulates AUXIN RESPONSE FACTOR (ARF)10 and ARF16 that participate in seed dormancy [43]. Besides FUS3 and ABI3, the expression levels of MIR156A are stabilized through a feedback loop - the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) 9 and SPL10 genes are both targets of miR156 and encode transcriptional activators of MIR156A [44].
Some miRNAs can act upstream to regulate the expression of other MIR genes. For example, SPL9 and SPL10, targets of miR156, are found to be enriched at the MIR172B locus to promote its transcription [44], [45], [46]. miR172b is critical for the juvenile-to-adult transition in Arabidopsis and maize, and during the transition, miR156 levels decline while miR172 levels increase [44]. As with MIR156, the transcription of MIR172 is subject to multiple layers of regulation. The SANT-domain-containing protein POWERDRESS (PWR) was found to enhance the transcription of only three of the five MIR172 members, MIR172A, B, and C, by increasing Pol II occupancy at the promoters [47]. The transcription factor APETALA2 (AP2) plays a complex role in MIR172 regulation. AP2 was found to be enriched at two MIR gene loci, MIR156 and MIR172 [48]. AP2 positively regulates MIR156 expression, which indirectly dampens the expression of MIR172 [48]. Meanwhile, together with the transcriptional co-repressors LEUNIG (LUG) and SEUSS (SEU), AP2 represses the expression of MIR172 [49]. As a regulator of MIR172, AP2 is also subject to the regulation by its cognate miRNA to form a feedback loop [48,49]. The AP2 protein is probably largely restricted to the outer floral whorls as its mRNA translation is repressed in the inner whorls by miR172 [50]. In another case, during leaf polarity development, class II homeodomain leucine zipper (HD-ZIPs) transcriptional factors HAT3 and ATHB4 interact with the class III HD-ZIP protein REVOLUTA (REV) to directly repress the transcription of MIR165/166 genes via a conserved cis-element in their promoters while REV is a target of miR165/166 [51,52].
2.3. Epigenetic marks affect MIR gene transcription
Histone modification epigenetically regulates gene expression, and studies have shown that such mechanisms also affect MIR gene transcription. For instance, GENERAL CONTROL NON-REPRESSED PROTEIN 5 (GCN5) promotes the expression of a subset of MIR genes through the addition of acetylation marks on H3K14 at the MIR loci [53]. By increasing H3K27me3 marks at the MIR156A/MIR156C loci, POLYCOMB REPRESSIVE COMPLEX 2 (PRC2) subunits CURLY LEAF (CLF) and SWINGER (SWN) redundantly mediate their transcriptional repression during vegetative phase change [54]. CHROMOSOME REMODELING2 (CHR2), also known as BRAHMA (BRM), is the ATPase subunit of the large switch/sucrose non-fermentable (SWI/SNF) complex, and a defect in CHR2/BRM reduces MIR gene transcription [55]. In addition, chromatin modification can affect MIR genes differently, for example the SWR1 chromatin remodeling complex (SWR1-C) alters the nucleosome occupancy at MIR gene promoters to regulate their expression positively or negatively [56]. It remains unclear how some transcription regulators distinguish one group of MIR genes from another.
2.4. Stabilization of pri-miRNAs
During transcription, pri-miRNAs are protected by 5′ capping and 3′ poly(A) tailing from nuclear RNA exosome-mediated degradation [57,58]. Additional regulation has also been reported to mediate pri-miRNA stability - mutations in specific proteins cause reduced accumulation of pri- and mature miRNA without compromising MIR gene transcription. Proteins in this group include the RNA binding protein DAWDLE (DDL) [59] and MAC subunits PLEIOTROPIC REGULATORY LOCUS1 (PRL1) [60], MAC3 [61] and MAC5A [62], with all four proteins showing a direct interaction with pri-miRNA transcripts and the dicing complex. MAC5A interacts with the stem-loop of pri-miRNAs and SE [62]. The reduced accumulation of pri-miRNAs in mac5a is partially rescued by introducing xrn2 or xrn3, mutations in two nuclear 5′-to-3′ exoribonucleases, suggesting that MAC5A binds to the pri-miRNA stem-loop to prevent it from XRN2- and XRN3-mediated degradation [62]. Interestingly, an se mutation also suppresses the molecular defect of mac5a, indicating that the reduced accumulation of pri-miRNAs in mac5a is SE-dependent [62]. The direct interaction between SE and XRN2, identified in the same study, suggests a possible link between SE and pri-miRNA degradation, in addition to its role in pri-miRNA processing [62]. In line with this hypothesis, a recent study found that SE interacts with the NUCLEAR EXOSOME TARGETING (NEXT) complex subunits ZCCHC8A and ZCCHC8B [57]. In addition, in the absence of HEN2, another NEXT complex subunit, a subset of pri-miRNAs is increased in abundance while mature miRNA levels are not affected [57]. Whereas, in another study, it was found that loss of SUPPRESSORS OF THE PAS2 1 (SOP1), which is also a component of the nuclear exosome and co-localizes with HEN2, does not affect pri-miRNA accumulation [58,63]. Furthermore, both hen2 and sop1 partially increase the accumulation of pri-miRNAs in the hyl1 mutant, and subsequently restore the levels of a subset of the corresponding miRNAs [63]. Together, these studies showed that HEN2-mediated pri-miRNA degradation is independent of HYL1, while HYL1 might antagonize SOP1-mediated pri-miRNA degradation. In another study, HYL1 was found to negatively impact pri-miRNA stability. AAR2, a homolog of a U5 snRNP assembly factor involved in pre-mRNA splicing in yeast and humans, interacts with HYL1 in the cytoplasm, nucleus, and dicing bodies. Mutations in AAR2 lead to reduced pri-miRNA levels and higher pri-miRNA decay rates in the se-1 mutant but not in the hyl1–2 mutant, suggesting that pri-miRNA degradation caused by loss of AAR2 function requires HYL1 [64]. Collectively, these discoveries imply a complex role of HYL1 and SE in pri-miRNA stabilization and processing and show that pri-miRNAs are subject to exosome-mediated degradation.
3. Dicing complex assembly and pri-miRNA processing
Maturation of miRNAs requires the precise processing of pri-miRNAs. Pri-miRNAs vary largely in stem-loop shape and length. The dicing complex relies on structural features of pri-miRNAs to distinguish them from other dsRNA species and define the position for the initial cut. In most cases, DCL1 processes the pri-miRNA in a base-to-loop mode, with the dicing complex recognizing a 15–17 bp dsRNA stem below the miRNA/miRNA* and above an internal loop to perform the initial cut that defines one end of the miRNA [65]. In other cases, such as pri-miR159a and pri-miR319a, the first cut is generated at the distal region of the pri-miRNA in a loop-to-base mode, where DCL1 recognizes a 15–17 bp dsRNA stem above the miRNA/miRNA* duplex and below a small terminal loop to perform the initial cut [66,67]. In either case, after the initial cut, DCL1 acts as a molecular ruler and generates a second cut ∼21 nt away from the first cut to release the miRNA/miRNA* [14]. While most miRNA precursors are substrates of DCL1, a few young miRNAs including miR822 and miR839 are processed by DCL4 in Arabidopsis [68]. In addition, a class of 24-nt miRNAs requires DCL3 for biogenesis in rice [69].
3.1. Secondary structures of pri-miRNAs
The secondary structures of pri-miRNAs are not all alike, with differing features such as GC content, branched terminal loops, intron position, and flexible internal base-pairing having been shown to affect the efficiency of DCL1-mediated pri-miRNA processing. GC bias and signatures play a role in determining the precision of pri-miRNA processing and the abundance of miRNAs [70]. G/C enrichment at positions 8–9, 18–19 and A/U enrichment at positions 5, 7 and 10 within miRNA regions positively correlate with miRNA abundance, with one possible explanation being that these signatures influence the structure of pri-miRNAs to promote HYL1 recruitment and binding [70].
Alternative secondary structures of miRNA precursors, such as a terminal branched loop, have been observed to affect the processing of several pri-miRNAs. Pri-miR165 and pri-miR166 possess a terminal branched loop, which is recognized by the dicing complex and leads to bi-directional processing [71]. The canonical base-to-loop mode promotes miRNA production whereas the non-canonical loop-to-base mode leads to unproductive cuts that diminish miRNA biogenesis [71]. For pri-miR157c, the terminal branched loop followed by an ∼18 bp dsRNA segment can be recognized by the dicing complex to generate mature miRNAs, albeit the loop-to-base mode of processing is relatively slow [72].
Many pri-miRNAs possess introns, and certain introns residing in the stem-loop region can disrupt the secondary structure of pri-miRNAs and thus must be removed by splicing, such as those in pri-miR842 and pri-miR846 [73]. In contrast, pri-miR161, pri-miR163 and pri-miR172b possess introns downstream of the stem-loop that play a positive role in miRNA maturation, possibly due to the 5ʹ splice site of these introns being recognized by U1 snRNP, which in turn recruits these pri-miRNAs to the dicing complex for processing [29,74,75].
While most miRNA precursors generate only one miRNA, some can produce multiple miRNAs due to their unique sequence features. The miR168/miR168* duplex undergoes flexible internal base-pairing enabling three alternative structural configurations, which promote the production of isomeric miRNAs (isomiRs) [76]. isomiR168s vary in length and 5′ terminal nucleotides hence leading to distinct AGO sorting outcomes [76]. In particular, a 22-nt miR168 isoform is enriched in AGO10 instead of AGO1, and AGO10-miR168 promotes the repression of AGO1 by triggering the production of secondary siRNAs from the AGO1 mRNA [76]. In addition, certain miRNA precursors can be sequentially processed by DCL1 multiple times to produce several small RNA duplexes [77].
Protein factors are capable of regulating pri-miRNA processing by altering the pri-miRNA secondary structure. In addition to promoting MIR gene transcription, CHR2/BRM directly binds to pri-miRNAs and alters their secondary structure thereby inhibiting processing [55]. This inhibition requires the interaction of CHR2/BRM with SE [55]. Interestingly, the association with SE is not necessary for CHR2/BRM to promote MIR gene transcription [55]. Extensive cytidylation and uridylation on the 3′ termini of pre-miRNAs processed through the base-to-loop mode was reported, with the nucleotidyl transferases HEN1 SUPPRESSOR1 (HESO1), NTP6 and NTP7 contributing to a portion of the cytidine addition and HESO1 being responsible for the majority of uridine addition [78]. This untemplated tailing can restore trimmed pre-miRNAs to their intact length thereby promoting processing [78]. pri-miRNA structures can also be altered by post-transcriptional modification. The mRNA adenosine methylase (MTA) introduces N6-methyladenosine (m6A) in pri-miRNAs [79]. A deficiency in MTA leads to less structured stem-loop regions, which hampers the association of HYL1 with these precursors and results in decreased miRNA levels [79]. MTA also interacts with Pol II and TOUGH (TGH), which are required in the early stages of miRNA biogenesis, suggesting that MTA might also act at early steps [79].
3.2. Assembly of the dicing complex
Although many proteins have been found to assist with pri-miRNA processing, the two main dicing complex components promoting the functions of DCL1 are HYL1 and SE, with these three components forming the dicing complex and being colocalized in nuclear loci termed ‘dicing bodies’ (D-bodies) [80,81]. SE and HYL1 interact with each other, and both interact with DCL1 [80]. HYL1 functions as a dimer to bind the stem region of pri-miRNAs and partner with SE, a C2H2 zinc finger protein that also binds pri-miRNAs, to ensure the precise cleavage of miRNA precursors by DCL1 [82], [83], [84], [85], [86]. A lack of HYL1 or SE causes reduced accumulation of mature miRNAs globally and increases levels of pri-miRNAs [83], [84], [85], [86].
Dicing complex formation is regulated by multiple factors. MAC subunits have been shown to be involved in dicing complex formation, including MAC7 and MAC3, which are associated with the dicing complex and promote the recruitment of HYL1 [61,87]. Interestingly, the RNA binding protein MODIFIEROFSNC1,2 (MOS2) [88] and the DEAH-box helicase PSR1-INTERACTING PROTEIN 1 (PINP1) [89] promote the assembly of the dicing complex without interacting with it. In the absence of MOS2, the association between pri-miRNA and HYL1 is reduced and localization of HYL1 in the d-bodies is compromised [88]. A recent study shows that d-bodies are phase-separated condensates, and the phase separation property of SE drives the formation of D-body [90]. Truncation of the intrinsically disordered regions (IDRs) of SE abrogates D-body formation and results in reduced miRNA processing [90]. Moreover, the TREX-2 complex interacts with SE and promotes D-body formation in addition to its role in MIR gene transcription [38]. DEAD-box RNA helicase 6 (RH6), RH8, and RH12 have recently been identified as D-body components that interact with SE and promote the phase separation of SE and the formation of D-bodies [91].
3.3. Regulation of pri-miRNA processing
Recruitment of pri-miRNAs by the dicing complex is facilitated by MOS2 [88], the RNA binding protein TGH [92], the ribosomal protein STV1 [40], and the THO/TREX complex [93]. TGH is a component of the dicing complex, it binds pri- and pre-miRNAs in vivo, and is required for the interaction between HYL1 and pri-miRNAs [92]. In addition, a lack of TGH reduced the activity of DCL1 in vitro [92]. STV1 binds pri-miRNAs and facilitates the recruitment of pri-miRNAs to the dicing complex [40]. The THO/TREX complex promotes the association of pri-miRNAs with HYL1, however, it does not appear to interact with any miRNA biogenesis pathway factors [93].
It has long been proposed that pri-miRNA processing occurs in D-bodies as many processing complex components, such as DCL1 and HYL1, are co-localized there [80]. However, considering that certain dicing complex cofactors, such as MOS2 [88], are not associated with the D-bodies, miRNA processing might not exclusively take place in D-bodies. Several studies show that dicing complex components are recruited to MIR loci during MIR transcription, suggesting that the processing of the pri-miRNAs can occur co-transcriptionally like in animals [94], alternatively, this allows for an initial assembly of the dicing complex. First, DCL1 is associated with the chromatin regions of MIR genes [33]. In addition, the Elongator complex [33], CDC5 [36], NOT2 [35], and TREX-2 [38] interact with both Pol II and the dicing complex and might act to recruit the dicing complex to MIR loci during MIR transcription. In accordance with this assumption, mutants of Elongator and NOT2 show impaired DCL1 localization in D-bodies or defects in D-body formation [33,35]. In addition, TREX-2 interacts with SE and promotes D-body formation [38]. It is unclear whether D-body formation is coupled with dicing complex component's recruitment to the MIR loci. HASTY (HST), the plant homolog of animal EXPORTIN 5 (EXP5/XPO5), has been shown to facilitates miRNA biogenesis independently of its proposed, yet unproven, role in miRNA nuclear export in Arabidopsis. HST could act as a scaffold to stabilize the DCL1-MED37 complex which in turn enhances the recruitment of DCL1 to MIR loci [95].
A recent study provides cogent evidence that pri-miRNAs can be processed co-transcriptionally, with the evidence being that pri-miRNAs can be detected at their MIR loci and that pri-miRNA processing intermediates are associated with Pol II [96]. This study also reported that the entire processing steps occur co-transcriptionally for loop-to-base (LTB) pri-miRNAs, while the base-to-loop (BTL) pri-miRNAs undergo the first processing step co-transcriptionally with the resulting pre-miRNA being further processed in the nucleoplasm [96]. R-loop, a DNA-RNA hybrid structure formed during transcription, appears to promote co-transcriptional processing of pri-miRNAs when within close distance to and upstream of the hairpin [96].
The activities of the dicing complex are modulated by specific factors. For example, RECEPTOR FOR ACTIVATED C KINASE1 (RACK1) promotes pri-miRNA processing through interaction with SE [97]. As it also interacts with AGO1, RACK 1 might bridge pri-miRNA processing and AGO1 loading or play a separate role downstream of pri-miRNA processing [97]. Some factors regulate the dicing complex in a tissue-specific manner. The DEAD-box RNA HELICASE 27 (RH27), which is expressed in embryos, shoot apical meristems and root apical meristems, is associated with pri-miRNAs and interacts with HYL1, SE and DDL to promote pri-miRNA processing [98].
3.4. Roles of splicing in pri-miRNA processing
A number of proteins modulating both pri-miRNA processing and splicing have been identified. For example, the dicing complex core components HYL1 [99] and SE [100,101], CAP-BINDING PROTEIN 20 (CBP20) and CBP80 [100,102], the splicing factor SMA1 [39], HIGH OSMOTIC STRESS GENE EXPRESSION 5 (HOS5), ARGININE/SERINE-RICH SPLICING FACTOR 40 (RS40) and RS41 [103], the pre-mRNA processing factor 6 homolog STABILIZED1 (STA1) [104], GLYCINE-RICH RBP 7 (GRP7) [105], SICKLE (SIC) [106], THO2 of the THO/TREX complex [93], the U1 snRNP subunit LETHAL UNLESS CBC 7 RL (LUC7rl), and the PRE-MRNA-PROCESSING PROTEIN (PRP)39b, PRP40a, PRP40b [29] are positive regulators of pri-miRNA processing and are involved in splicing. In addition, the MAC complex has been shown to associate with the spliceosome [107]. The fact that many proteins influence both pri-miRNA processing and splicing does not necessarily implicate a role of splicing in pri-miRNA processing, as the proteins may have independent functions in the two processes. However, as discussed in Section 3.1, splicing is necessary to form the proper structures of some pri-miRNAs to enable processing. Furthermore, intronic lariat RNAs, the by-products of pre-mRNA splicing, inhibit pri-miRNA processing by acting as a decoy to sequester the dicing complex to prevent its binding to pri-miRNAs [108]. The two intron-lariat spliceosome (ILS) complex subunits ILP1 and NTR interact with DCL1 and SE and facilitate the degradation of lariat RNAs, which may in turn promote miRNA processing [34,108].
3.5. Regulation of the dicing complex components DCL1, HYL1 and SE
The expression and function of the dicing complex core components DCL, HYL1 and SE are regulated at transcriptional, post-transcriptional, and post-translational levels.
The transcription of DCL1 is promoted by XAP5 CIRCADIAN TIMEKEEPER (XCT) [109] and STA1 [104]. The splicing factor SMA1 facilitates the splicing of the ninth intron of DCL1 pre-mRNA to promote miRNA processing [39]. DELAY OF GERMINATION1 (DOG1) promotes the expression of DCL1, HYL1, SE, TGH, and CDC5 [110], while the histone acetyltransferase GCN5 indirectly represses the expression of DCL1, HYL1, and SE [53], and the AGO1 gene encoding the miRNA effector [111], to negatively regulate miRNA maturation. In addition, the levels of DCL1 mRNA are subject to the negative feedback regulation by miR162 [112] and miR838, the latter residing in intron 14 of DCL1 pre-mRNA [68]. Furthermore, SE RNA is targeted and negatively regulated by miR863 during bacterial infection [113].
At the post-transcriptional level, KARYOPHERIN ENABLING THE TRANSPORT OF THE CYTOPLASMIC HYL1 (KETCH1), an importin beta protein, enhances miRNA processing by promoting the translocation of HYL1 from the cytoplasm to the nucleus [114]. In addition, pri-miRNA-like SHORT INTERSPERSED ELEMENTS (SINEs) RNAs can act as a decoy and sequester HYL1 to reduce its participation in pri-miRNA processing [115]. A recent study shows that the 20S core proteasome α subunit G1 (PAG1) promotes pri-miRNA processing by recruiting the SE protein for degradation in a ubiquitin-independent manner [116]. Presumably the degraded SE is disordered and non-functional [116]. In the pag1 mutant, SE is distributed in both the cytoplasm and the nucleus, while it is predominantly detected in the nucleus in the wild type [116].
The activity of the dicing complex is regulated by phosphorylation. The phosphorylation of DCL1 is required for its interaction with DDL, with this interaction being necessary for pri-miRNA processing [117,118]. HYL1 activity is also regulated by its phosphorylation states with phosphorylation inhibiting HYL1 function [119,120]. C-TERMINAL DOMAIN PHOSPHATASE-LIKE 1 (CPL1)/FIERY2 and CPL2 dephosphorylate HYL1, with SE serving as a bridge for the interaction between HYL1 and CPL1 [121]. HOS5, also known as REGULATOR OF CBF GENE EXPRESSION 3 (RCF3) and SHINY1, interacts with CPL1 and CPL2 to assist HYL1 dephosphorylation in a tissue specific manner [122]. In addition, SUPPRESSOR OF MEK 1 (SMEK1) partners with PROTEIN PHOSPHATASE 4 (PP4) to dephosphorylate and stabilize HYL1, thus enhancing pri-miRNA processing [123]. Furthermore, AAR2, a splicing factor, associates with HYL1 and is required for its dephosphorylation and D-body localization [64]. MITOGEN-ACTIVATED PROTEIN KINASE 3 (MPK3) and SNF1-RELATED PROTEIN KINASE 2 (SnRK2) can phosphorylate HYL1 in vitro, and both kinases interact with HYL1 in vivo [119,120]. Besides, SnRK2 also associates with and phosphorylates SE in vitro, although the physiological role of this modification remains unclear [120]. PRE-MRNA PROCESSING 4 KINASE A (PRP4KA) interacts with SE and phosphorylates at least five residues of SE in vitro and in vivo [124]. An se mutant can be rescued by the expression of hypo-phosphorylated SE variants in vivo, but not its hyper-phosphorylated counterparts [124]. In addition, compared to the hyper-phosphorylated SE variants, the hypo-phosphorylated SE variants displayed stronger binding affinity toward HYL1 and were more resistant to degradation by the 20S proteasome [124].
3.6. Regulation of miRNA biogenesis by environmental cues
As plants are sessile organisms, effective responses to environmental changes are essential. MIR gene expression and the activity and subcellular localization of the dicing complex components are regulated by environmental cues in a tissue- and developmental stage-specific manner.
3.6.1. Regulation of MIR gene transcription and pri-miRNA processing by environmental cues
A growing number of MIR gene transcription and processing factors have been reported as being subject to the regulation by environmental changes. For example, in Arabidopsis, red light treatment and white light treatment of etiolated seedlings can alter the expression of a group of miRNAs. During shade avoidance response, the level of PHYTOCHROME-INTERACTING FACTORS (PIFs) is highly induced, and the PIFs bind to the promoters of multiple MIR156 genes and repress their expression [125]. Another light signaling transcription factor ELONGATED HYPOCOTYL 5 (HY5) directly regulates the expression of several MIR genes [126]. Partnering with HY5-HOMOLOG (HYH), HY5 mediates light-induced expression of HEN1, which protects miRNAs from degradation [127]. The transcription of MIR395, MIR399, and MIR398 genes is specifically induced upon sulfur, phosphate and copper deprivation, respectively [128], [129], [130], [131], [132], [133]. In addition to copper deprivation, miR398 has also been reported to respond to heat [134,135]. Tocopherols (vitamin E) facilitate the accumulation of 3′-phosphoadenosine 5′-phosphate (PAP), which inhibits the activity of nuclear exoribonucleases XRN2 and XRN3 to promote miRNA biogenesis, possibly by protecting pri-miRNAs from being degraded [135]. The induction of tocopherols and PAP by heat is required for the increased accumulation of miR398 [135]. MiR402, an intronic miRNA, is induced by heat stress [136]. In contrast to miR402, pri-miR400 is retained in the host RNA upon heat stress, which results in reduced accumulation of miR400 [28]. At cold temperatures, the expression of STA1 is highly induced, which facilitates pri-miRNA splicing and promotes DCL1 transcription [104,137]. The sic mutant is hypersensitive to cold and salt stresses [106]. SIC participates in pre-mRNA splicing, colocalizes with HYL1 and positively regulates pri-miRNA processing [106]. The cycling DOF transcription factor CDF2, which participates in photoperiodic flowering, binds to the promoter of some MIR genes and activates or represses their transcription [138,139]. CDF2 also interacts with DCL1 and suppresses pri-miRNA processing [139]. Under salt stress, the transcription of MIR163 and MIR829 genes is increased, while the transcription of MIR161 and MIR173 genes is repressed by AGO1 [111]. Furthermore, pathogen responses can affect miRNA biogenesis. The nuclear localized disease resistance protein SNC1 partners with its interacting protein TPR1 to represses the transcription of MIR genes [140].
3.6.2. Regulation of the dicing complex by environmental cues
Among the abiotic and biotic stimuli affecting pri-miRNA processing, light plays an influential role. In the light, CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), an E3 ubiquitin ligase, translocates from the nucleus to the cytoplasm, where it stabilizes HYL1 by preventing its degradation by HYL1-CLEAVAGE SUBTILASE 1 (HCS1), which would otherwise degrade HYL1 by cleaving its N-terminal region [141,142]. Consistent with this observation, during de-etiolation, pri-miRNAs and the dicing complex components DCL1, HYL1, and SE were observed to accumulate to higher levels [143]. However, the accumulation of most miRNAs did not change; this discrepancy suggests an unknown suppressor hampers pri-miRNA processing during de-etiolation [143]. A recent study shows that FORHEAD-ASSOCIATED DOMAIN 2 (FHA2), a light stabilized protein, might be the suppressor. FHA2 associates with DCL1, HYL1, and SE to suppress their pri-miRNA processing activity [144]. In the dark, the levels of HYL1 are reduced, and the ratio between active (dephosphorylated) and inactive (phosphorylated) HYL1 is also strongly diminished during extended periods of darkness or shade [145]. Interestingly, although inactive phosphorylated HYL1 is stabilized in the nucleus in the dark, HYL1 can be quickly activated (dephosphorylated) on plants’ exposure to light [145]. During a dark to red light transition, PHYTOCHROME-INTERACTING FACTOR 4 (PIF4), a basic helix-loop-helix (bHLH) transcription factor, interacts with and destabilizes DCL1 in a ubiquitin-proteasome (UPS) independent pathway [146]. PIF4 also interacts with HYL1, and acts as a transcription activator of a group of miRNA genes [146].
Besides light, ABA treatment and other environmental stresses could activate SnRK2s, thereby phosphorylating HYL1 and SE. SnRK2s-mediated phosphorylation promotes HYL1 stability during ABA treatment [120]. In contrast to SnRK2s, MPK3-mediated phosphorylation promotes HYL1 degradation [119]. ABA treatments induce SMEK1 to antagonize HYL1 degradation mediated by MPK3 [123]. In addition, the expression levels of HOS5/RCF3/SHINY are reduced by salt and ABA treatments [103,147]. Furthermore, multiple MAC subunits such as CDC5, PRL1, and MAC7, which promote miRNA biogenesis, are responsive to different environmental cues [36,60,87].
4. miRNA stabilization and RISC assembly
The miRNA/miRNA* duplexes are 2′-O-methylated by the methyltransferase HEN1 at the 3′ terminus of each strand to protect miRNAs from degradation [21,148]. Mutations in HEN1 lead to reduced levels of nearly all miRNAs [21,148,149]. Recent studies show that HEN1 interacts with DCL1 and HYL1 but not SE, suggesting HEN1 might replace SE in the dicing complex after pri-miRNA processing [150]. Next, the methylated miRNA/miRNA* duplex is loaded into an AGO to form a miRISC. Upon loading, AGO unwinds the miRNA/miRNA* duplex and selectively retains one strand (guide strand or miRNA), which directs AGOs for target gene inhibition, whereas the other strand (passenger strand or miRNA*) is ejected [13].
4.1. miRNA strand selection
The thermodynamic differences in the base-pairing stabilities and/or the identity of the first nucleotide of the miRNA are key features for strand selection and AGO sorting. Usually, the strand with lower 5′ thermostability in the duplex is selected as the guide strand and retained by AGOs [151,152]. In Arabidopsis, most miRNA guide strands start with a 5′ uridine and are preferentially incorporated into AGO1, the major miRNA effector of the AGO family [15]. Small RNAs that start with a 5′ adenosine are predominantly associated with AGO2 and AGO4, whereas those starting with a 5′ cytosine are mainly associated with AGO5 [15]. In addition, the miRNA duplex structure has also been shown to affect their sorting. For instance, AGO1 prefers miRNA duplexes with central mismatches, while AGO2 prefers duplexes with no middle mismatches [153]. Arabidopsis AGO7 associates specifically with miR390, which triggers TAS3 trans-acting siRNA biogenesis [154,155]. The 5′ adenosine of the miR390 guide strand and three nucleotides in the middle of the miR390/miR390* duplex are crucial for the specific interaction with AGO7 [154,155].
miRNAs can be loaded into more than one AGO protein. Indeed, in Arabidopsis, miR165/166 is associated with both AGO1 and AGO10 [156,157]. While most miRNA* strands are degraded, some are loaded into AGO proteins and function as the guide strand in gene repression. For instance, miR393b* starts with a 5′ adenosine, and is enriched in AGO2, while miR393 starts with a 5′ uridine, and is mainly loaded into AGO1 [158]. In 2013, the miRBase database ceased using the miR/miR* nomenclature, instead, miR-5p and miR-3p are assigned to small RNA sequences derived from the 5′ and 3′ arm of the miRNA precursor, respectively [159]. Furthermore, protein cofactors such as HYL1 [151], CPL1 [121], HOS5, RS40, and RS41 [103] have also been reported to assist with strand selection.
The assembly of AGO1-containing RISC requires the assistance of the molecular chaperone HEAT SHOCK PROTEIN 90 (HSP90), which binds AGO1 in an ATP-dependent manner and triggers a conformational change of AGO1 to allow sRNA duplex loading [160,161]. Following ATP hydrolysis, HSP90 dissociates from and likely prompts a second conformational change of AGO1, which results in the proper removal of the passenger strand [160,161]. The unwinding and removal of miRNA* strands from AGO1 do not require the endonuclease activity of AGO1 [160,162,163]. On the contrary, cleavage of the miR390* strand is essential for the maturation of the AGO7–miR390 complex [154]. The AGO1-miRISC assembly is also facilitated by CYCLOPHILIN 40/SQUINT (CYP40/SQN) and inhibited by PROTEIN PHOSPHATASE 5 (PP5) in an HSP90-dependent manner [164]. In addition, two importin-beta family proteins, ENHANCED MIRNA ACTIVITY 1 (EMA1) and TRANSPORTIN 1 (TRN1), interact with AGO1 and negatively and positively regulate AGO1-miRISC formation, respectively [165,166]. Furthermore, RBV, a nuclear WD40 domain protein, promotes the loading of miRNAs into AGO1, in addition to its role in promoting the transcription of MIR genes [41].
4.2. Regulation of AGO1 stability
AGO1 is subjected to post-transcriptional and post-translational regulation. At the post-transcriptional level, AGO1 transcripts are targeted by AGO1-bound miR168, forming a negative feedback regulatory loop [167]. At the post-translational level, the F-box protein F-BOX WITH WD-40 2 (FBW2) negatively regulates AGO1 protein levels, with the overexpression of FBW2 decreasing the accumulation of AGO1 protein but not AGO1 mRNA, whereas a lack of FBW2 increases AGO1 protein levels [168]. Several studies show that AGO1 is also targeted by viral F-box RNA silencing suppressors. Polerovirus-encoded F-box protein (P0) and Enamovirus-encoded P0 destabilize host AGO1 upon viral infection [169,170]. Although F-box proteins are known for tethering poly-ubiquitins to mark proteins for degradation through the ubiquitin-26S proteasome system (UPS), treatment with a proteasome inhibitor does not prevent AGO1 from FBW2-, Polerovirus P0-, and Enamovirus P0-mediated destabilization, suggesting AGO1 is not degraded via the UPS pathway in these cases [168], [169], [170]. Interestingly, AGO1 levels were significantly increased in P0-expressing transgenic plants treated with an autophagy inhibitor [171]. In addition, AGO1 co-localizes with AUTOPHAGY 8 (ATG8), a ubiquitin-like autophagosome membrane protein [171]. Taken together, the data imply that FBW2 and P0 mediate the degradation of ubiquitinated AGO1 through an autophagy-dependent pathway. Another study shows that AGO1 could also be degraded through the UPS pathway depending on a viral RNA silencing suppressor. The Potato virus X protein P25 interacts with and destabilizes AGO1 through the UPS pathway, as the UPS inhibitor MG132 significantly promotes the accumulation of AGO1 despite the presence of P25 [172]. A recent study shows that under UV exposure, CLF, a methyltransferase subunit of the PRC2, inhibits FBW2 expression thereby enhancing AGO1 stability [173].
4.3. The cell biology of RISC assembly
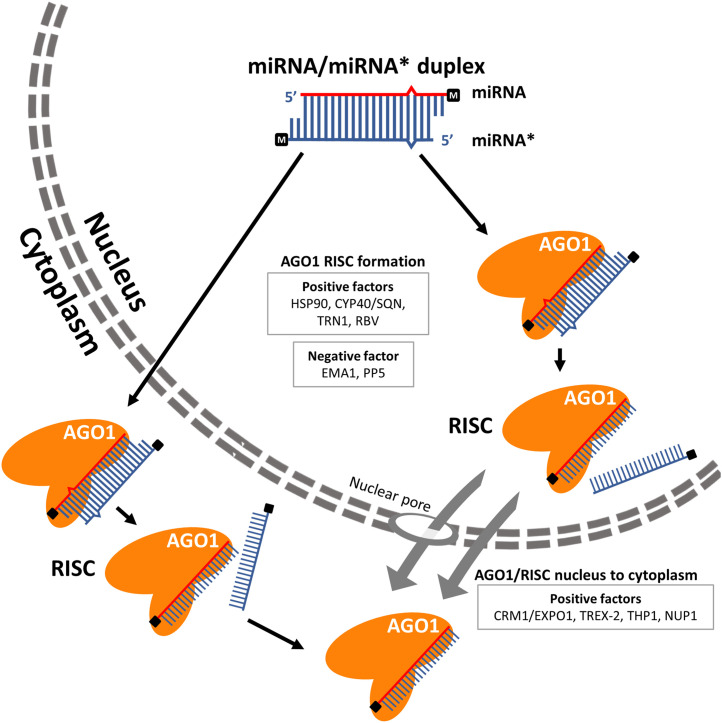
Animal pre-miRNAs are exported to the cytosol by EXPORTIN5 (EXP5/XPO5), a RanGTP-dependent dsRNA-binding protein, for final processing and RISC formation [174], [175], [176]. Unlike animals, plant miRNA maturation is thought to take place exclusively in the nucleus. It was proposed that plant miR/miR* duplexes are exported from the nucleus to the cytoplasm by HASTY (HST), the plant ortholog of EXP5/XPO5, and loaded into AGO1 in the cytoplasm. However, this assumption is not supported by experimental data, as the nucleo-cytosolic partitioning of miRNAs is not altered in the hst loss-of-function mutant [177,178]. A revised model, in which AGO1 is imported into the nucleus to load miRNAs and AGO1-miRNA complexes are exported to the cytoplasm by CRM1/EXPORTIN1 (EXPO1) (Fig. 2) gained traction [179]. A nuclear localization signal (NLS) and a nuclear export signal (NES) residing in the N terminal extension of AGO1 are proposed to direct the nucleo-cytosolic shuttling of AGO1 [179]. Upon treatment with Leptomycin-B, a compound that inhibits EXPO1-mediated, NES-dependent protein nuclear export, the ratio between cytoplasmic and nuclear AGO1 significantly decreased, suggesting AGO1 could be exported to the cytoplasm in an EXPO1/NES-dependent manner [179]. In addition, AGO1 with a mutated NES sequence associated with the same miRNAs as its intact counterpart, indicating that miRNAs’ loading into AGO1 could take place in the nucleus [179]. However, it is possible there are other yet unidentified NES or nuclear export pathways that transport AGO1mNES from the nucleus to the cytoplasm. Indeed, it was recently found that the TREX-2 complex core subunit THP1 interacts with the nucleoporin protein NUP1 at the nuclear envelope, together promoting the nuclear export of AGO1 or AGO1-miRISC (Fig. 2) [38]. When AGO1 was fused with the glucocorticoid receptor (GR) and transiently expressed in plants, the cytoplasmically restricted GR-AGO1 was able to associate with miR165 [180]. Fluorescence microscopy analysis and subcellular fractionation assays revealed that AGO1 is co-localized with the endoplasmic reticulum (ER) and associated with membrane-bound polysomes (MPBs) [181], [182], [183], [184]. Furthermore, cellular fractionation combined with genomic approaches shows that all miRNAs are preferably associated with MPBs instead of polysomes in general [184]. Together, these findings suggest that miRNA loading or action could take place on the ER. Further studies need to be done to determine whether miRNAs are loaded into AGO1 exclusively in the nucleus, cytoplasm, or both. And if miRNA loading occurs in the cytoplasm, does it happen on the ER and/or MBPs?
Fig 2.
miRNA-induced silencing complex (miRISC) formation for plant miRNAs. The miRNA/miRNA* duplex is loaded into AGO1, and following unwinding, the miRNA strand is selectively retained in AGO1, while the miRNA* strand is ejected. The miRISCs formed in the nucleus are exported to the cytoplasm through the nuclear pore in the CRM1/EXPO1-dependent pathway. Whether other mechanisms underlie the nucleo-cytosolic shuttling of AGO1 and/or miRISC is still unknown. The miRNA/miRNA* duplex loading may also occur in the cytoplasm. The miRNA and miRNA* strands are highlighted in red and blue, respectively. The methyl group at the 3′ end is marked with “M” in a black square. Positive and negative factors in the formation or nuclear export of AGO1 RISCs are shown.
5. Conclusion
Our knowledge of miRNA-mediated gene silencing has greatly advanced in the past two decades. Many factors involved in the regulation of miRNA biogenesis, action, and degradation have been identified, however mechanistic and cell biological details are still lacking. Most past studies have relied on molecular genetic approaches, with a dearth of biochemical and structural insights on pri-miRNA processing or miRISC formation and/or function. Structures of the dicing complex in its various forms, such as apo, pri-miRNA-bound, pre-miRNA-bound, and together with various interacting factors, will elucidate mechanisms such as substrate recognition, processing site choice, and regulation by co-factors. Structures of AGO1, AGO1-miRNA, and AGO1-miRNA-target will reveal the mechanisms of miRNA binding, target recognition, and slicing. Another area that calls for future investigations is the cell biology of miRNA biogenesis. Pieces of evidence exist for two locations of pri-miRNA processing in the nucleus: MIR gene loci [96] and D-bodies [80,90], and these locations do not overlap [96]. How is miRNA biogenesis partitioned between these locations? What RNAs and proteins constitute D-bodies? The subcellular locations of miRISC formation also deserve further investigation. Does miRNA loading into AGO1 occur in both the cytoplasm and the nucleus? If so, are certain miRNAs selectively loaded into AGO1 in one place or another? Are cytoplasmically-assembled miRISCs different from nucleus-formed miRISCs in composition or function? Future studies with innovative imaging, biochemical, and genomic tools will be needed to propel the plant miRNA biogenesis field forward.
Declaration of competing interest
The authors declare that they have no conflicts of interest in this work.
Acknowledgments
Research in the Chen laboratory on small RNAs and RNA modifications is supported by grants from National Institutes of Health (R01GM061146 and R01GM129373).
Biographies

Ye Xu received a doctorate from the University of California Riverside in 2022. Her research interests focus on understanding the molecular mechanisms underlying small non-coding RNA biogenesis, and small RNA-mediated gene and genome regulation.

Xuemei Chen received her B.S. degree from Peking University and a doctorate from Cornell University. After postdoctoral training at California Institute of Technology, she started her assistant professor position in 1999 at the Waksman Institute at Rutgers University and was promoted to associate professor in 2005. She moved to the University of California, Riverside in 2005 as an associate professor and was promoted to full professor in 2009 and distinguished professor in 2013. She was an HHMI-GBMF investigator from 2012 to 2018. She is a member of AAAS and the US National Academy of Sciences.
References
- 1.Lee R., Feinbaum R., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Wightman B., Ha L., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 3.Reinhart B., FJ S., M B., AE P., et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 4.Pasquinelli A.E., Reinhart B.J., Slack F., et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 5.Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 6.Lagos-Quintana M., Rauhut R., Lendeckel W., et al. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 7.Lau N.C., Lim L.P., Weinstein E.G., et al. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 8.Park W., Li J., Song R., et al. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12(17):1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinhart B.J., Weinstein E.G., Rhoades M.W., et al. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Llave C., Kasschau K.D., Rector M.A., et al. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14(7):1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeong D.H., Park S., Zhai J., et al. Massive analysis of rice small RNAs: mechanistic implications of regulated MicroRNAs and variants for differential target RNA cleavage. Plant Cell. 2011;23(12):4185–4207. doi: 10.1105/tpc.111.089045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfeffer S., Zavolan M., Grasser F.A., et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–737. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 13.Baumberger N., Baulcombe D.C. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. U. S. A. 2005;102(33):11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi Y., Denli A.M., Hannon G.J. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell. 2005;19(3):421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 15.Mi S., Cai T., Hu Y., et al. Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5’ terminal nucleotide. Cell. 2008;133(1):116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y., Teng C., Xia R., et al. PhasiRNAs in plants: their biogenesis, genic sources, and roles in stress responses, development, and reproduction. Plant Cell. 2020;32(10):3059–3080. doi: 10.1105/tpc.20.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nozawa M., Miura S., Nei M. Origins and evolution of microRNA genes in plant species. Genome Biol. Evol. 2012;4(3):230–239. doi: 10.1093/gbe/evs002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Z., Allen E., Fahlgren N., et al. Expression of arabidopsis MIRNA genes. Plant Physiol. 2005;138(4):2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurihara Y., Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. U. S. A. 2004;101(34):12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reinhart B.J., Weinstein E.G., Rhoades M.W., et al. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu B., Yang Z., Li J., et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307(5711):932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han M.H., Goud S., Song L., Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. U. S. A. 2004;101(4):1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vazquez F., Gasciolli V., Crété P., et al. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 2004;14(4):346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 24.Lobbes D., Rallapalli G., Schmidt D.D., et al. SERRATE: a new player on the plant microRNA scene. EMBO Rep. 2006;7(10):1052–1058. doi: 10.1038/sj.embor.7400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang L., Liu Z., Lu F., et al. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006;47(6):841–850. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- 26.Jones-Rhoades M.W., Bartel D.P. Computational identification of plant MicroRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14(6):787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 27.Zhang B.H., Pan X.P., Wang Q.L., et al. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 2005;15(5):336–360. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]
- 28.Yan K., Liu P., Wu C.A., et al. Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol. Cell. 2012;48(4):521–531. doi: 10.1016/j.molcel.2012.08.032. [DOI] [PubMed] [Google Scholar]
- 29.Knop K., Stepien A., Barciszewska-Pacak M., et al. Active 5’ splice sites regulate the biogenesis efficiency of Arabidopsis microRNAs derived from intron-containing genes. Nucleic. Acids. Res. 2017;45(5):2757–2775. doi: 10.1093/nar/gkw895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Megraw M., Baev V., Rusinov V., et al. MicroRNA promoter element discovery in Arabidopsis. RNA. 2006;12(9):1612–1619. doi: 10.1261/rna.130506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hajheidari M., Farrona S., Huettel B., et al. CDKF;1 and CDKD protein kinases regulate phosphorylation of serine residues in the C-terminal domain of arabidopsis RNA polymerase II. Plant Cell. 2012;24(4):1626–1642. doi: 10.1105/tpc.112.096834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim Y.J., Zheng B., Yu Y., et al. The role of mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 2011;30(5):814–822. doi: 10.1038/emboj.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang X., Cui Y., Li Y., et al. Transcription and processing of primary microRNAs are coupled by Elongator complex in Arabidopsis. Nat. Plants. 2015;1(6):1–9. doi: 10.1038/nplants.2015.75. [DOI] [PubMed] [Google Scholar]
- 34.Wang J., Chen S., Jiang N., et al. Spliceosome disassembly factors ILP1 and NTR1 promote miRNA biogenesis in Arabidopsis thaliana. Nucleic. Acids. Res. 2019;47(15):7886–7900. doi: 10.1093/nar/gkz526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L., Song X., Gu L., et al. NOT2 proteins promote polymerase II-dependent transcription and interact with multiple microRNA biogenesis factors in Arabidopsis. Plant Cell. 2013;25(2):715–727. doi: 10.1105/tpc.112.105882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang S., Xie M., Ren G., et al. CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proc. Natl. Acad. Sci. U. S. A. 2013;110(43):17588–17593. doi: 10.1073/pnas.1310644110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S., Quan L., Li S., et al. The protein phosphatase4 complex promotes transcription and processing of primary microRNAs in Arabidopsis. Plant Cell. 2019;31(2):486–501. doi: 10.1105/tpc.18.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang B., You C., Zhang Y., et al. Linking key steps of microRNA biogenesis by TREX-2 and the nuclear pore complex in Arabidopsis. Nat. Plants. 2020;6(8):957–969. doi: 10.1038/s41477-020-0726-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li S., Xu R., Li A., et al. SMA1, a homolog of the splicing factor Prp28, has a multifaceted role in miRNA biogenesis in Arabidopsis. Nucleic. Acids. Res. 2018;46(17):9148–9159. doi: 10.1093/nar/gky591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li S., Liua K., Zhang S., et al. STV1, a ribosomal protein, binds primary microRNA transcripts to promote their interaction with the processing complex in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2017;114(6):1424–1429. doi: 10.1073/pnas.1613069114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang C., Cai Q., Wang F., et al. Arabidopsis RBV is a conserved WD40 repeat protein that promotes microRNA biogenesis and ARGONAUTE1 loading. Nat. Commun. 2022;13(1):1–14. doi: 10.1038/s41467-022-28872-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang F., Perry S.E. Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 2013;161(3):1251–1264. doi: 10.1104/pp.112.212282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian R., Wang F., Zheng Q., et al. Direct and indirect targets of the arabidopsis seed transcription factor abscisic acid insensitive3. Plant J. 2020;103(5):1679–1694. doi: 10.1111/tpj.14854. [DOI] [PubMed] [Google Scholar]
- 44.Wu G., Park M.Y., Conway S.R., et al. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138(4):750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J.W., Czech B. Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138(4):738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 46.Yamaguchi A., Wu M.F., Yang L., et al. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of leafy, fruitfull, and apetala1. Dev. Cell. 2009;17(2):268–278. doi: 10.1016/j.devcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yumul R.E., Kim Y.J., Liu X., et al. Powerdress and diversified expression of the MIR172 gene family bolster the floral stem cell network. PLos Genet. 2013;9(1) doi: 10.1371/journal.pgen.1003218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yant L., Mathieu J., Dinh T.T., et al. Orchestration of the floral transition and floral development in arabidopsis by the bifunctional transcription factor apetala2. Plant Cell. 2010;22(7):2156–2170. doi: 10.1105/tpc.110.075606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grigorova B., Mara C., Hollender C., et al. Leunig and seuss co-repressors regulate miR172 expression in Arabidopsis flowers. Development. 2011;138(12):2451–2456. doi: 10.1242/dev.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303(5666):2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merelo P., Ram H., Caggiano M.P., et al. Regulation of MIR165/166 by class II and class III homeodomain leucine zipper proteins establishes leaf polarity. Proc. Natl. Acad. Sci. U. S. A. 2016;113(42):11973–11978. doi: 10.1073/pnas.1516110113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emery J.F., Floyd S.K., Alvarez J., et al. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003;13(20):1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 53.Kim W., Benhamed M., Servet C., et al. Histone acetyltransferase GCN5 interferes with the miRNA pathway in Arabidopsis. Cell Res. 2009;19(7):899–909. doi: 10.1038/cr.2009.59. [DOI] [PubMed] [Google Scholar]
- 54.Xu M., Hu T., Smith M.R., et al. Epigenetic regulation of vegetative phase change in Arabidopsis. Plant Cell. 2016;28(1):28–41. doi: 10.1105/tpc.15.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z., Ma Z., Castillo-González C., et al. SWI2/SNF2 ATPase CHR2 remodels pri-miRNAs via Serrate to impede miRNA production. Nature. 2018;557(7706):516–521. doi: 10.1038/s41586-018-0135-x. [DOI] [PubMed] [Google Scholar]
- 56.Choi K., Kim J., Müller S.Y., et al. Regulation of microRNA-mediated developmental changes by the SWR1 chromatin remodeling complex. Plant Physiol. 2016;171(2):1128–1143. doi: 10.1104/pp.16.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bajczyk M., Lange H., Bielewicz D., et al. SERRATE interacts with the nuclear exosome targeting (NEXT) complex to degrade primary miRNA precursors in Arabidopsis. Nucleic. Acids. Res. 2020;48(12):6839–6854. doi: 10.1093/nar/gkaa373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao S., Wang J., Jiang N., et al. Hyponastic Leaves 1 protects pri-miRNAs from nuclear exosome attack. Proc. Natl. Acad. Sci. U. S. A. 2020;117(29):17429–17437. doi: 10.1073/pnas.2007203117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu B., Bi L., Zheng B., et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. U. S. A. 2008;105(29):10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang S., Liu Y., Yu B. PRL1, an RNA-binding protein, positively regulates the accumulation of miRNAs and siRNAs in Arabidopsis. PLos Genet. 2014;10(12) doi: 10.1371/journal.pgen.1004841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li S., Liu K., Zhou B., et al. MAC3A and MAC3B, two core subunits of the MOS4-associated complex, positively influence miRNA biogenesis. Plant Cell. 2018;30(2):481–494. doi: 10.1105/tpc.17.00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li S., Li M., Liu K., et al. MAC5, an RNA-binding protein, protects pri-miRNAs from SERRATE-dependent exoribonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2020;117(38):23982–23990. doi: 10.1073/pnas.2008283117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hématy K., Bellec Y., Podicheti R., et al. The zinc-finger protein SOP1 is required for a subset of the nuclear exosome functions in Arabidopsis. PLos Genet. 2016;12(2):1–22. doi: 10.1371/journal.pgen.1005817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fan L., Gao B., Xu Y., et al. Arabidopsis AAR2, a conserved splicing factor in eukaryotes, acts in microRNA biogenesis. Proc. Natl. Acad. Sci. U. S. A. 2022;119(41) doi: 10.1073/pnas.2208415119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song L., Axtell M.J., Fedoroff N.V. RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Biol. 2010;20(1):37–41. doi: 10.1016/j.cub.2009.10.076. [DOI] [PubMed] [Google Scholar]
- 66.Bologna N.G., Mateos J.L., Bresso E.G., et al. A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 2009;28(23):3646–3656. doi: 10.1038/emboj.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Addo-Quaye C., Snyder J.A., Park Y.B., et al. Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA. 2009;15(12):2112–2121. doi: 10.1261/rna.1774909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rajagopalan R., Vaucheret H., Trejo J., et al. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20(24):3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu L., Zhou H., Zhang Q., et al. DNA methylation mediated by a microRNA pathway. Mol. Cell. 2010;38(3):465–475. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 70.Narjala A., Nair A., Tirumalai V., et al. A conserved sequence signature is essential for robust plant miRNA biogenesis. Nucleic. Acids. Res. 2020;48(6):3103–3118. doi: 10.1093/nar/gkaa077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu H., Zhou Y., Castillo-González C., et al. Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis Dicer-like1. Nat. Struct. Mol. Biol. 2013;20(9):1106–1115. doi: 10.1038/nsmb.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moro B., Chorostecki U., Arikit S., et al. Efficiency and precision of microRNA biogenesis modes in plants. Nucleic. Acids. Res. 2018;46(20):10709–10723. doi: 10.1093/nar/gky853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jia F., Rock C.D. MIR846 and MIR842 comprise a cistronic MIRNA pair that is regulated by abscisic acid by alternative splicing in roots of Arabidopsis. Plant Mol. Biol. 2013;81(4–5):447–460. doi: 10.1007/s11103-013-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bielewicz D., Kalak M., Kalyna M., et al. Introns of plant pri-miRNAs enhance miRNA biogenesis. EMBO Rep. 2013;14(7):622–628. doi: 10.1038/embor.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schwab R., Speth C., Laubinger S., et al. Enhanced microRNA accumulation through stemloop-adjacent introns. EMBO Rep. 2013;14(7):615–621. doi: 10.1038/embor.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Iki T., Cléry A., Bologna N.G., et al. Structural flexibility enables alternative maturation, ARGONAUTE sorting and activities of miR168, a global gene silencing regulator in plants. Mol. Plant. 2018;11(8):1008–1023. doi: 10.1016/j.molp.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 77.Zhang W., Gao S., Zhou X., et al. Multiple distinct small RNAs originate from the same microRNA precursors. Genome Biol. 2010;11(8):R81. doi: 10.1186/gb-2010-11-8-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song J., Wang X., Song B., et al. Prevalent cytidylation and uridylation of precursor miRNAs in Arabidopsis. Nat. Plants. 2019;5(December):1260–1272. doi: 10.1038/s41477-019-0562-1. [DOI] [PubMed] [Google Scholar]
- 79.Bhat S.S., Bielewicz D., Gulanicz T., et al. mRNA adenosine methylase (MTA) deposits m6A on pri-miRNAs to modulate miRNA biogenesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 2020;117(35):21785–21795. doi: 10.1073/pnas.2003733117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fang Y., Spector D.L. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 2007;17(9):818–823. doi: 10.1016/j.cub.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song L., Han M.H., Lesicka J., et al. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc. Natl. Acad. Sci. U S A. 2007;104(13):5437–5442. doi: 10.1073/pnas.0701061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang S.W., Chen H.Y., Yang J., et al. Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure. 2010;18(5):594–605. doi: 10.1016/j.str.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dong Z., Han M.H., Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl. Acad. Sci. U. S. A. 2008;105(29):9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kurihara Y., Takashi Y., Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12(2):206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang X., Ren W., Zhao Q., et al. Homodimerization of HYL1 ensures the correct selection of cleavage sites in primary miRNA. Nucleic. Acids. Res. 2014;42(19):12224–12236. doi: 10.1093/nar/gku907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Iwata Y., Takahashi M., Fedoroff N.V., et al. Dissecting the interactions of Serrate with RNA and dicer-like 1 in Arabidopsis microRNA precursor processing. Nucleic. Acids. Res. 2013;41(19):9129–9140. doi: 10.1093/nar/gkt667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jia T., Zhang B., You C., et al. The Arabidopsis MOS4-associated complex promotes microRNA biogenesis and precursor messenger RNA splicing. Plant Cell. 2017;29(10):2626–2643. doi: 10.1105/tpc.17.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu X., Shi Y., Li J., et al. A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis. Cell Res. 2013;23:645–657. doi: 10.1038/cr.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Qiao Y., Shi J., Zhai Y., et al. Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc. Natl. Acad. Sci. U. S. A. 2015;112(18):5850–5855. doi: 10.1073/pnas.1421475112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xie D., Chen M., Niu J., et al. Phase separation of SERRATE drives dicing body assembly and promotes miRNA processing in Arabidopsis. Nat. Cell Biol. 2021;23(1):32–39. doi: 10.1038/s41556-020-00606-5. [DOI] [PubMed] [Google Scholar]
- 91.Li Q., Liu N., Liu Q., et al. DEAD-box helicases modulate dicing body formation in Arabidopsis. Sci. Adv. 2021;7(18):1–18. doi: 10.1126/sciadv.abc6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ren G., Xie M., Dou Y., et al. Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2012;109(31):12817–12821. doi: 10.1073/pnas.1204915109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Francisco-Mangilet A.G., Karlsson P., Kim M.H., et al. THO2, a core member of the THO/TREX complex, is required for microRNA production in Arabidopsis. Plant J. 2015;82(6):1018–1029. doi: 10.1111/tpj.12874. [DOI] [PubMed] [Google Scholar]
- 94.Morlando M., Ballarino M., Gromak N., et al. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008;15(9):902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cambiagno D.A., Giudicatti A.J., Arce A.L., et al. HASTY modulates miRNA biogenesis by linking pri-miRNA transcription and processing. Mol. Plant. 2021;14(3):426–439. doi: 10.1016/j.molp.2020.12.019. [DOI] [PubMed] [Google Scholar]
- 96.Gonzalo L., Tossolini I., Gulanicz T., et al. R-loops at microRNA encoding loci promote co-transcriptional processing of pri-miRNAs in plants. Nat. Plants. 2022;8(4):402–418. doi: 10.1038/s41477-022-01125-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Speth C., Maria Willing E, Rausch S. RACK1 scaffold proteins influence miRNA abundance in Arabidopsis. Plant J. 2013;76:433–445. doi: 10.1111/tpj.12308. [DOI] [PubMed] [Google Scholar]
- 98.Hou X.L., Chen W.Q., Hou Y., et al. DEAD-BOX RNA HELICASE 27 regulates microRNA biogenesis, zygote division, and stem cell homeostasis. Plant Cell. 2021;33(1):66–84. doi: 10.1093/plcell/koaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Szarzynska B., Sobkowiak L., Pant B.D., et al. Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic. Acids. Res. 2009;37(9):3083–3093. doi: 10.1093/nar/gkp189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Laubinger S., Sachsenberg T., Zeller G., et al. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 2008;105(25) doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Raczynska K.D., Stepien A., Kierzkowski D., et al. The SERRATE protein is involved in alternative splicing in Arabidopsis thaliana. nucl. 2014;42(2):1224–1244. doi: 10.1093/nar/gkt894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim S., Yi Yang J, Xu J., et al. Two cap-binding proteins CBP20 and CBP80 are involved in processing primary microRNAs. Plant Cell Physiol. 2008;49(11):1634–1644. doi: 10.1093/pcp/pcn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen T., Cui P., Xiong L. The RNA-binding protein HOS5 and serine/arginine-rich proteins RS40 and RS41 participate in miRNA biogenesis in Arabidopsis. Nucleic. Acids. Res. 2015;43(17):8283–8298. doi: 10.1093/nar/gkv751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ben C.S., Liu R., Chinnusamy V., et al. STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic. Acids. Res. 2013;41(3):1984–1997. doi: 10.1093/nar/gks1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Köster T., Meyer K., Weinholdt C., et al. Regulation of pri-miRNA processing by the hnRNP-like protein AtGRP7 in Arabidopsis. Nucleic. Acids. Res. 2014;42(15):9925–9936. doi: 10.1093/nar/gku716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhan X., Wang B., Li H., et al. Arabidopsis proline-rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proc. Natl. Acad. Sci. U. S. A. 2012;109(44):18198–18203. doi: 10.1073/pnas.1216199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Monaghan J., Xu F., Gao M., et al. Two Prp19-Like U-Box proteins in the MOS4-associated complex play redundant roles in plant innate immunity. PLoS Pathog. 2009;5(7) doi: 10.1371/journal.ppat.1000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li Z., Wang S., Cheng J., et al. Intron lariat RNA inhibits microRNA biogenesis by sequestering the dicing complex in Arabidopsis. PLos Genet. 2016;12(11):1–25. doi: 10.1371/journal.pgen.1006422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fang X., Shi Y., Lu X., et al. CMA33/XCT regulates small RNA production through modulating the transcription of Dicer-Like genes in Arabidopsis. Mol. Plant. 2015;8(8):1227–1236. doi: 10.1016/j.molp.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 110.Huo H., Wei S., Bradford K.J. DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc. Natl. Acad. Sci. U. S. A. 2016;1(26):2199–2206. doi: 10.1073/pnas.1600558113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dolata J., Bajczyk M., Bielewicz D., et al. Salt stress reveals a new role for ARGONAUTE1 in miRNA biogenesis at the transcriptional and posttranscriptional levels. Plant Physiol. 2016;172(1):297–312. doi: 10.1104/pp.16.00830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xie Z., Kasschau K.D., Carrington J.C. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 113.Niu D., Lii Y.E., Chellappan P., et al. miRNA863-3p sequentially targets negative immune regulator ARLPKs and positive regulator SERRATE upon bacterial infection. Nat. Commun. 2016;7 doi: 10.1038/ncomms11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang Z., Guo X., Ge C., et al. KETCH1 imports HYL1 to nucleus for miRNA biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2017 doi: 10.1073/pnas.1619755114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pouch-Pélissier M.N., Pélissier T., Elmayan T., et al. SINE RNA induces severe developmental defects in Arabidopsis thaliana and interacts with HYL1 (DRB1), a key member of the DCL1 complex. PLos Genet. 2008;4(6) doi: 10.1371/journal.pgen.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li Y., Sun D., Ma Z., et al. Degradation of SERRATE via ubiquitin-independent 20S proteasome to survey RNA metabolism. Nat. Plants. 2020;6(August):970–982. doi: 10.1038/s41477-020-0721-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Machida S., Yuan Y.A. Crystal structure of Arabidopsis thaliana dawdle forkhead-associated domain reveals a conserved phospho-threonine recognition cleft for Dicer-like 1 binding. Mol. Plant. 2013;6(4):1290–1300. doi: 10.1093/mp/sst007. [DOI] [PubMed] [Google Scholar]
- 118.Zhang S., Dou Y., Li S., et al. DAWDLE interacts with DICER-LIKE proteins to mediate small RNA biogenesis. Plant Physiol. 2018;177(3):1142–1151. doi: 10.1104/pp.18.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Raghuram B., Sheikh A.H., Rustagi Y., Sinha A.K. MicroRNA biogenesis factor DRB1 is a phosphorylation target of mitogen activated protein kinase MPK3 in both rice and Arabidopsis. FEBS J. 2015;282:521–536. doi: 10.1111/febs.13159. [DOI] [PubMed] [Google Scholar]
- 120.Yan J., Wang P., Wang B., et al. The SnRK2 kinases modulate miRNA accumulation in Arabidopsis. PLos Genet. 2017;13(4):1–21. doi: 10.1371/journal.pgen.1006753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Manavella P.A., Hagmann J., Ott F., et al. Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell. 2012;151(4):859–870. doi: 10.1016/j.cell.2012.09.039. [DOI] [PubMed] [Google Scholar]
- 122.Karlsson P., Danger M., Seymour D.K., et al. KH domain protein RCF3 is a tissue-biased regulator of the plant miRNA biogenesis cofactor HYL1. Proc. Natl. Acad. Sci. U. S. A. 2015;112(45):14096–14101. doi: 10.1073/pnas.1512865112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Su C., Li Z., Cheng J., et al. The protein phosphatase 4 and SMEK1 complex dephosphorylates HYL1 to promote miRNA biogenesis by antagonizing the MAPK cascade in Arabidopsis. Dev. Cell. 2017;41(5):527–539.e5. doi: 10.1016/j.devcel.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 124.Wang L., Yan X., Li Y., et al. PRP4KA phosphorylates SERRATE for degradation via 20S proteasome to fine-tune miRNA production in Arabidopsis. Sci. Adv. 2022;8(12) doi: 10.1126/sciadv.abm8435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xie Y., Liu Y., Wang H., et al. Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nat. Commun. 2017;8(348) doi: 10.1038/s41467-017-00404-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang H., He H., Wang X., et al. Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 2011;65(3):346–358. doi: 10.1111/j.1365-313X.2010.04426.x. [DOI] [PubMed] [Google Scholar]
- 127.Tsai H.L., Li Y.H., Hsieh W.P. Hua enhancer1 is involved in posttranscriptional regulation of positive and negative regulators in Arabidopsis photomorphogenesis. Plant Cell. 2014;26(7):2858–2872. doi: 10.1105/tpc.114.126722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jagadeeswaran G., Li Y.F., Sunkar R. Redox signaling mediates the expression of a sulfate-deprivation-inducible microRNA395 in Arabidopsis. Plant J. 2014;77(1):85–96. doi: 10.1111/tpj.12364. [DOI] [PubMed] [Google Scholar]
- 129.Kawashima C.G., Yoshimoto N., Maruyama-Nakashita A., et al. Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J. 2009;57(2):313–321. doi: 10.1111/j.1365-313X.2008.03690.x. [DOI] [PubMed] [Google Scholar]
- 130.Hsieh L.C., Lin S.I., Shih A.C.C., et al. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151(4):2120–2132. doi: 10.1104/pp.109.147280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fujii H., Chiou T.J., Lin S.I., et al. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 2005;15(22):2038–2043. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 132.Beauclair L., Yu A., Bouche N. MicroRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis. Plant J. 2010;62:454–462. doi: 10.1111/j.1365-313X.2010.04162.x. [DOI] [PubMed] [Google Scholar]
- 133.Yamasaki H., Abdel-Ghany S.E., Cohu C.M., et al. Regulation of copper homeostasis by micro-RNA in Arabidopsis. J. Biol. Chem. 2007;282(22):16369–16378. doi: 10.1074/jbc.M700138200. [DOI] [PubMed] [Google Scholar]
- 134.Guan Q., Lu X., Zeng H., et al. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 2013;74(5):840–851. doi: 10.1111/tpj.12169. [DOI] [PubMed] [Google Scholar]
- 135.Fang X., Zhao G., Zhang S., et al. Chloroplast-to-nucleus signaling regulates microRNA biogenesis in Arabidopsis. Dev. Cell. 2019;48(3):371–382.e4. doi: 10.1016/j.devcel.2018.11.046. [DOI] [PubMed] [Google Scholar]
- 136.Baev V., Milev I., Naydenov M., et al. Insight into small RNA abundance and expression in high- and low-temperature stress response using deep sequencing in Arabidopsis. Plant Physiol. Biochem. 2014;84:105–114. doi: 10.1016/j.plaphy.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 137.Ha L.B., Kapoor A., Zhu J., Kang Z.J. STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell. 2006;18(July):1736–1749. doi: 10.1105/tpc.106.042184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fornara F., Panigrahi K.C.S., Gissot L., et al. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell. 2009;17:75–86. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 139.Sun Z., Guo T., Liu Y., et al. The roles of Arabidopsis CDF2 in transcriptional and posttranscriptional regulation of primary microRNAs. PLos Genet. 2015;11(10):1–19. doi: 10.1371/journal.pgen.1005598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cai Q., Liang C., Wang S., et al. The disease resistance protein SNC1 represses the biogenesis of microRNAs and phased siRNAs. Nat. Commun. 2018;9(1):1–14. doi: 10.1038/s41467-018-07516-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cho S.K., Ben Chaabane S, Shah P., et al. COP1 E3 ligase protects HYL1 to retain microRNA biogenesis. Nat. Commun. 2014:1–10. doi: 10.1038/ncomms6867. [DOI] [PubMed] [Google Scholar]
- 142.Jung H.J., Choi S.W., Boo K.H., et al. HYL1-CLEAVAGE SUBTILASE 1 (HCS1) suppresses miRNA biogenesis in response to light-to- dark transition. Proc. Natl. Acad. Sci. U. S. A. 2022;119:6. doi: 10.1073/pnas.2116757119. e2116757119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Choi S.W., Ryu M.Y., Viczián A., et al. Light triggers the miRNA-biogenetic inconsistency for de-etiolated seedling survivability in Arabidopsis thaliana. Mol. Plant. 2020;13(3):431–445. doi: 10.1016/j.molp.2019.10.011. [DOI] [PubMed] [Google Scholar]
- 144.Park S.J., Choi S.W., Kim G.M., et al. Light-stabilized FHA2 suppresses miRNA biogenesis through interactions with DCL1 and HYL1. Mol. Plant. 2021;14(4):647–663. doi: 10.1016/j.molp.2021.01.020. [DOI] [PubMed] [Google Scholar]
- 145.Achkar N.P., Cho S.K., Poulsen C., et al. A quick HYL1-dependent reactivation of microRNA production is required for a proper developmental response after extended periods of light deprivation. Dev. Cell. 2018;46(2):236–247.e6. doi: 10.1016/j.devcel.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 146.Sun Z., Li M., Zhou Y., et al. Coordinated regulation of Arabidopsis microRNA biogenesis and red light signaling through Dicer-like 1 and phytochrome-interacting factor 4. PLos Genet. 2018;14(3):1–21. doi: 10.1371/journal.pgen.1007247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jiang J., Wang B., Shen Y., et al. The Arabidopsis RNA binding protein with K homology motifs, SHINY1, interacts with the C-terminal domain Phosphatase-like 1 (CPL1) to repress stress-inducible gene expression. PLos Genet. 2013;9(7) doi: 10.1371/journal.pgen.1003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yang Z., Ebright Y.W., Yu B., et al. HEN1 recognizes 21-24nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic. Acids. Res. 2006;34(2):667–675. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li J., Yang Z., Yu B., et al. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 2005;15(16):1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Baranauske S., Mickute M., Plotnikova A., et al. Functional mapping of the plant small RNA methyltransferase: HEN1 physically interacts with HYL1 and DICER-LIKE 1 proteins. Nucleic. Acids. Res. 2015;43(5):2802–2812. doi: 10.1093/nar/gkv102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Eamens A.L., Smith N.A., Curtin S.J., et al. The arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA. 2009;15(12):2219–2235. doi: 10.1261/rna.1646909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tomari Y., Matranga C., Haley B., et al. A protein sensor for siRNA asymmetry. Science. 2004;306(5700):1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 153.Zhang X., Niu D., Carbonell A., et al. ARGONAUTE PIWI domain and microRNA duplex structure regulate small RNA sorting in Arabidopsis. Nat. Commun. 2014;5:5468. doi: 10.1038/ncomms6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Endo Y., Iwakawa H.O., Tomari Y. Arabidopsis ARGONAUTE7 selects miR390 through multiple checkpoints during RISC assembly. EMBO Rep. 2013;14(7):652–658. doi: 10.1038/embor.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Montgomery T.A., Howell M.D., Cuperus J.T., et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133(1):128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 156.Zhu H., Hu F., Wang R., et al. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell. 2011;145(2):242–256. doi: 10.1016/j.cell.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhou Y., Honda M., Zhu H., et al. Spatiotemporal sequestration of miR165/166 by arabidopsis argonaute10 promotes shoot apical meristem maintenance. Cell Rep. 2015;10(11):1819–1827. doi: 10.1016/j.celrep.2015.02.047. [DOI] [PubMed] [Google Scholar]
- 158.Zhang X., Zhao H., Gao S., et al. Arabidopsis Argonaute 2 regulates innate immunity via miRNA393*-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol. Cell. 2011;42(3):356–366. doi: 10.1016/j.molcel.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kozomara A., Griffiths-Jones S. MiRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic. Acids. Res. 2014;42(D1):68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Iki T., Yoshikawa M., Nishikiori M., et al. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol. Cell. 2010;39(2):282–291. doi: 10.1016/j.molcel.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 161.Iwasaki S., Kobayashi M., Yoda M., et al. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell. 2010;39(2):292–299. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 162.Arribas-Hernández L., Marchais A., Poulsen C., et al. The slicer activity of ARGONAUTE1 is required specifically for the phasing, not production, of trans-acting short interfering RNAs in Arabidopsis. Plant Cell. 2016;28(7):1563–1580. doi: 10.1105/tpc.16.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Carbonell A., Fahlgren N., Garcia-Ruiz H., et al. Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell. 2012;24(9):3613–3629. doi: 10.1105/tpc.112.099945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Iki T., Yoshikawa M., Meshi T., et al. Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J. 2012;31(2):267–278. doi: 10.1038/emboj.2011.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang W., Ye R., Xin Y., et al. An importin beta protein negatively regulates microRNA activity in Arabidopsis. Plant Cell. 2011;23(October):3565–3576. doi: 10.1105/tpc.111.091058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cui Y., Fang X., Qi Y. TRANSPORTIN1 promotes the association of microRNA with ARGONAUTE1 in Arabidopsis. Plant Cell. 2016;28(October):2576–2585. doi: 10.1105/tpc.16.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Vaucheret H., Mallory A.C., Bartel D.P. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell. 2006;22(1):129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Earley K., Smith M.R., Weber R., et al. An endogenous F-box protein regulates ARGONAUTE1 in Arabidopsis thaliana. Silence. 2010;1(1):1–10. doi: 10.1186/1758-907X-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Baumberger N., Tsai C.H., Lie M., Havecker E., Baulcombe D.C.C. The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 2007;17(18):1609–1614. doi: 10.1016/j.cub.2007.08.039. [DOI] [PubMed] [Google Scholar]
- 170.Fusaro A.F., Correa R.L., Nakasugi K., et al. The Enamovirus P0 protein is a silencing suppressor which inhibits local and systemic RNA silencing through AGO1 degradation. Virology. 2012;426(2):178–187. doi: 10.1016/j.virol.2012.01.026. [DOI] [PubMed] [Google Scholar]
- 171.Derrien B., Baumberger N., Schepetilnikov M., et al. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. U. S. A. 2012;109(39):15942–15946. doi: 10.1073/pnas.1209487109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chiu M.H., Chen I.H., Baulcombe D.C., et al. The silencing suppressor P25 of Potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol. Plant Pathol. 2010;11(5):641–649. doi: 10.1111/j.1364-3703.2010.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ré D.A., Cambiagno D.A., Arce A.L., et al. CURLY LEAF regulates microRNA activity by controlling ARGONAUTE 1 degradation in plants. Mol. Plant. 2020;13(1):72–87. doi: 10.1016/j.molp.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 174.Lund E., Güttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 175.Bohnsack M.T., Czaplinski K., Görlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10(2):185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yi R., Qin Y., Macara I.G., et al. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mee Yeon P., Wu G., Gonzalez-Sulser A., et al. Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl Acad. Sci. 2005;102(10):3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhu J., Li Y., Lin J., et al. CRD1, an Xpo1 domain protein, regulates miRNA accumulation and crown root development in rice. Plant J. 2019;100(2):328–342. doi: 10.1111/tpj.14445. [DOI] [PubMed] [Google Scholar]
- 179.Bologna N.G., Iselin R., Abriata L.A., et al. Nucleo-cytosolic shuttling of ARGONAUTE1 prompts a revised model of the plant microRNA pathway. Mol. Cell. 2018;69(4):709–719.e5. doi: 10.1016/j.molcel.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 180.Fan L., Zhang C., Gao B., et al. Microtubules promote the non-cell autonomous action of microRNAs by inhibiting their cytoplasmic loading onto ARGONAUTE1 in Arabidopsis. Dev. Cell. 2022;57(8):995–1008. doi: 10.1016/j.devcel.2022.03.015. .e1-e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Li S., Liu L., Zhuang X., et al. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell. 2013;153(3):562–574. doi: 10.1016/j.cell.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Brodersen P., Sakvarelidze-Achard L., Schaller H., et al. Isoprenoid biosynthesis is required for miRNA function and affects membrane association of ARGONAUTE 1 in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2012;109(5):1778–1783. doi: 10.1073/pnas.1112500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wang T., Zheng Y., Tang Q., et al. Brassinosteroids inhibit miRNA-mediated translational repression by decreasing AGO1 on the endoplasmic. JIPB. 2021;63(8):1475–1490. doi: 10.1111/jipb.13139. [DOI] [PubMed] [Google Scholar]
- 184.Li S., Le B., Ma X., et al. Biogenesis of phased siRNAS on membrane-bound polysomes in Arabidopsis. Elife. 2016 Dec 12;5:e22750. doi: 10.7554/eLife.22750. [DOI] [PMC free article] [PubMed] [Google Scholar]